Abstract
Despite the conventional distinction between them, promoters and enhancers share many features in mammals, including divergent transcription and similar modes of transcription factor binding. Here, we examine the architecture of transcription initiation through comprehensive mapping of transcription start sites (TSSs) in human lymphoblastoid B-cell (GM12878) and chronic myelogenous leukemic (K562) tier 1, ENCODE cell lines. Using a nuclear run-on protocol called GRO-cap, which captures TSSs for both stable and unstable transcripts, we conduct detailed comparisons of thousands of promoters and enhancers in human cells. These analyses reveal a common architecture of initiation, including tightly spaced (110 bp) divergent initiation, similar frequencies of core-promoter sequence elements, highly positioned flanking nucleosomes, and two modes of transcription factor binding. Post-initiation transcript stability provides a more fundamental distinction between promoters and enhancers than patterns of histone modifications, transcription factors or co-activators. These results support a unified model of transcription initiation at promoters and enhancers.
Regulation of RNA transcription is a critical process for directing cell fates during organismal development and is necessary to maintain homeostasis throughout the lifespan of all organisms. Promoters and enhancers are major control hubs for gene regulation that integrate information from a multitude of signaling pathways through binding of signal-responsive activators and repressors. Therefore, accurately mapping and characterizing these regulatory regions is essential for defining how cell-specific transcriptomes are generated and maintained.
In mammalian cells, transcription initiation at promoters of annotated genes is accompanied by upstream antisense transcription initiation1–3. These divergent transcription start sites are tightly-spaced (<250 base pairs), and are presumed to arise from separate core promoters. The transcript representing the gene is typically stable and thus detected by standard RNA-sequencing techniques. However, the upstream, antisense RNA (uaRNA) is typically short and more difficult to detect due to a poly-A site dependent termination mechanism that rapidly targets the transcript for degradation by the exosome4, 5. Occasionally, the uaRNA appears to be replaced with another mRNA6, a lincRNA, or a tRNA gene7 to produce a pair of stable transcripts. Nearly 80% of active mammalian promoters display a bidirectional arrangement of initiation, and thus, this back-to-back arrangement of initiation has emerged as a general feature of promoters2.
Transcription initiation also occurs at enhancers. While such transcription was originally identified at several canonical enhancers, more recent high-throughput sequencing methods have demonstrated enhancer transcription to be widespread8–11. Production of enhancer RNAs (eRNAs) is also bidirectional and is associated with chromatin modifications or cofactors that are suggestive of enhancer activity (H3K4me1, p300, H3K27ac)12–14. The widespread existence of eRNAs and uaRNAs raises several important questions regarding how these RNAs are produced and whether they are functional. For example, is initiation of eRNAs governed by the same rules as promoters? RNA Polymerase II (Pol II) can operate with lower stringency when encountering naked DNA15, thus it is possible that Pol II initiates at enhancers by virtue of the open chromatin environment and high local concentration of Pol II, rather than as part of a bona fide pre-initation complex. Additionally, some studies suggest that eRNAs are important for activation of target genes16, 17, whereas others suggest that eRNA production can be dispensable for constructing a functional enhancer9. Furthermore, the process of transcription itself may be functional through modification of the chromatin architecture or the creation of negative supercoils that enhance transcription factor binding18.
Although divergent transcription at promoters and enhancers remains incompletely understood, it is nevertheless a characteristic signature that can be exploited in the identification of active regulatory elements 9, 19, 20. The signature of divergent transcription is particularly evident when transcriptional activity is assayed using the Global Nuclear Run-On sequencing (GRO-seq) method, owing to its high sensitivity for all transcriptionally-engaged RNA polymerase regardless of subsequent transcript turnover rates2, 9, 19. In addition, a variation of the GRO-seq method that enriches for 5′-7meGTP-capped RNAs, can greatly increase the sensitivity and specificity for detecting transcription initiation21, 22 (see Methods). In this article, we apply this GRO-cap method to human cells and show that it efficiently and precisely maps TSSs of coding and non-coding RNAs regardless of the resulting stability of the transcript. Thus, GRO-cap provides a more complete picture of genome-wide initiation than CAGE, which mainly detects TSSs from stable RNAs20, 23. Using our comprehensive, GRO-cap-based annotations of TSSs, we then report a detailed analysis of transcription initiation sites that sheds new light on the architecture of both promoters and enhancers across the human genome.
RESULTS
Identification of TSSs in Human Cells using GRO-cap
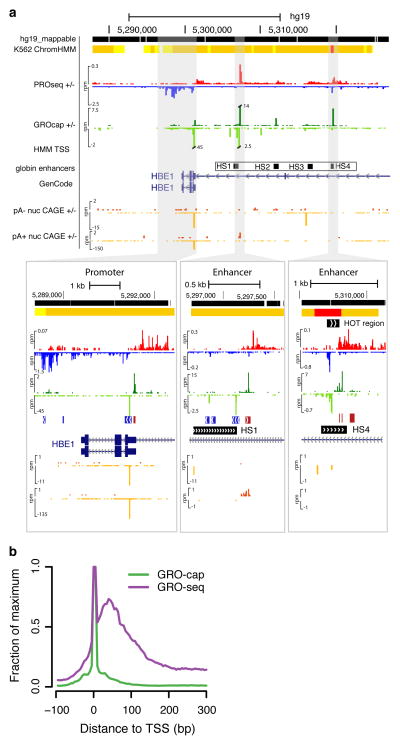
We prepared GRO-cap and GRO-seq libraries from human lymphoblastoid B-cell (GM12878) and chronic myelogenous leukemic (K562) cell lines, and PRO-seq (high resolution GRO-seq22) data from K562 cells (Supplemental Table 1). These are both “Tier 1” cell lines in the ENCODE project, allowing us to take advantage of abundant publicly available functional genomic data24. The GRO-cap assay efficiently captures TSS information from nascent transcripts, evidenced by a dramatic enrichment of GRO-cap signal at gene promoters and enhancers (Fig. 1a, b, Supplementary Fig. 1a). Figure 1a shows a specific example of the classic globin locus where divergent transcription is seen from active regions including the epsilon globin gene and the upstream hypersensitive sites (HS) that mark enhancers25 (see below).
Figure 1. GRO-cap identifies TSSs in promoters and enhancers.
(a) A UCSC genome browser54 shot of the globin locus near the LCR using K562 cell line data sets generated or used in this study. The locus contains a portion of the beta-globin locus, including the globin epsilon gene and LCR enhancers. The insets are zoomed in views of the shaded regions that show the divergent GRO-cap (+ strand: dark green, − strand: light green) signal at the epsilon-globin promoter (left) and two enhancers associated with the hypersensitive site (HS) 1 (center) and HS4 (right). The locations of the HS sites are taken from probe locations in Ashe et al. 55. ChromHMM regions track is shown on top, with predicted promoters indicated in red and enhancers in orange. Note that CAGE signal (+ strand: dark orange, - strand: light orange) is at background levels in the enhancer region. (b) GRO-cap dramatically enriches the signal for initiation sites when compared with GRO-seq. Composite GRO-seq and GRO-cap reads from the cell line plotted relative to all GENCODE TSSs.
To comprehensively identify TSS candidate sites using our data, we developed a hidden Markov model (HMM) that contrasts GRO-cap with data from control experiments, in which the critical CAP-removing enzyme, tobacco acid pyrophosphatase (TAP), is omitted (Supplemental Fig. 2a,b, Methods). This HMM identified a total of ~120K putative TSSs in each cell line, within the range previously reported (80 to 150 K)8, 10, 26, 27. The predicted TSS regions are narrow (mean 57 bp, with 95% under 140 bp), but account for 69% of all GRO-cap TAP+ reads and include both sharp and more dispersed TSSs (Supplemental Fig. 2c,d; Methods). Ninety-three percent of these TSSs are contained within enhancer or promoter regions predicted from patterns of histone modifications (ChromHMM regions) in the same cell types28. However, our mapping is more stringent and localized, identifying ~4 fold fewer regions at a ~3-fold higher resolution per site than combined ChromHMM promoter and enhancer predictions (Supplemental Fig. 2e,f).
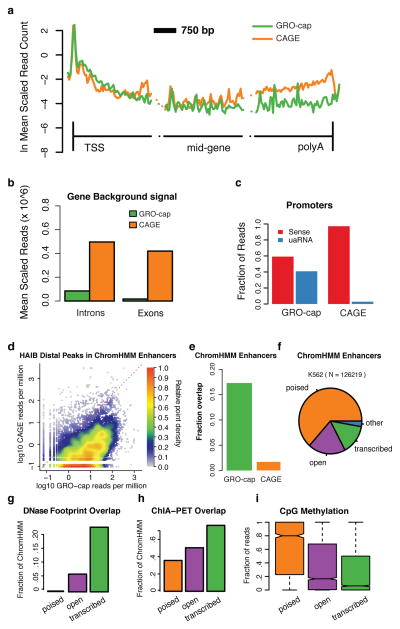
In comparison to CAGE, GRO-cap shows a similar composite profile when aligned to annotated gene TSSs (Fig. 2a). However, fewer reads map to introns and internal exons, indicating that GRO-cap has reduced background compared to CAGE (Fig. 2a,b). The decreased background for GRO-cap results in part from differences in the methodologies (cap-trapping29 versus the oligo-capping method30) used to capture capped transcripts. GRO-cap has the additional strength that it is highly sensitive to rare or rapidly degraded noncoding RNAs (ncRNAs), because it captures nascent RNAs as they are being made and before events that determine stability occur4, 21. This also eliminates background from post-transcriptionally capped RNAs31. In contrast, CAGE libraries are often dominated by highly abundant and stable RNAs (e.g. mRNAs), resulting in decreased sensitivity towards unstable RNAs5, 23, 31, such as uaRNAs at protein-coding promoters (Fig 1a, 2c). The high sensitivity and low background of GRO-cap also contribute to an increased coverage of enhancer regions predicted from histone modification patterns28 (Fig. 1a, Fig. 2d,e, Supplementary, Fig. 3a–c). As expected, GRO-cap signal correlates better with polymerase levels measured by PRO-seq in promoter-proximal regions than in the gene body (Supplemental Fig. 3d), suggesting that the signal originates primarily from nascent RNAs associated with polymerases that are paused proximal to promoters. Although this means that GRO-cap data cannot be used on its own as a measure of either initiation rates or levels of transcription elongation, GRO-cap does map comprehensively TSS locations regardless of the eventual stability of the RNA (see below).
Figure 2. Comparison of GRO-cap with CAGE.
(a) GRO-cap and CAGE profiles at protein-coding genes. Genes are broken into three 3 Kb regions covering region around the TSS, the middle of the gene, and near 3′-cleavage/poly-A site. The vertical lines represent the TSS and 3′-cleavage site. (b) Average read density in interior introns and exons (excluding the first and last of each) as a measure of GRO-cap and CAGE background signals. (c) GRO-cap and CAGE relative fraction of reads aligned to sense and divergent (uaRNA) directions at protein-coding genes (counted within underlying ChromHMM region). (d) Density scatterplot showing the signal intensity (reads per million) for GRO-cap vs. CAGE surrounding distal transcription factor ChIP-seq peaks from the Hudson Alpha Institute for Biotechnology (HAIB). (e) Fraction of ChromHMM regions containing a detectable GRO-cap (green) or CAGE (orange) TSS. (f) Comparing enhancer regions based on chromatin marks (ChromHMM Enhancers, Ernst. et al. 28) with DNAse HS (OpenChrommatin consortium) and GRO-cap, reveals three main classes of enhancer regions, poised (no DNAse HS peak nor GRO-cap TSS; orange, n = 1624), open (DNAse HS peak, but no GRO-cap TSS; purple; n= 3740) and transcribed (DNAse HS peak and GRO-cap TSS; green), and a negligible ‘other’ (no DNAse HS peak but with GRO-cap TSS; blue; n = 4703). (g–i) These three classes represent a progression in terms of functional activity, as measured by (g) an increase in detectable transcription factor footprints (Wellington footprints on DNAse HS,), (h) chromatin links (ChIA-PET overlap,) and (i) a significant reduction in CpG methylation between each transition The center line of the boxplot represents the median, the boxes encompass the interquartile range, and the whiskers extend to the minimum and maximum.
We then characterized putative enhancers captured by GRO-cap by contrasting our TSSs that are not at annotated genes with ChromHMM enhancers and open chromatin (DNase hypersensitive (DHS)) regions. This three-way comparison subdivides ChromHMM enhancers into three main classes: closed (ChromHMM only); open (ChromHMM and DNAse HS); and transcribed (ChromHMM, DNAse HS and GRO-cap TSS) (Fig. 2f). The transcribed subset, our main focus in this study, is enriched for positive regulatory activity, namely increased transcription factor binding (Wellington footprints32; Fig. 2g), distal chromatin interactions (ChIA-pet33; Fig. 2h), and reduced CpG methylation34 (Fig. 2i). In addition, the various histone modifications differ in expected patterns among poised, open, and transcribed enhancers (Supplemental Fig. 4). These results suggest that our approach identifies, with high-resolution, a subset of the sites identified by other methods, which appears to be enriched for active roles in transcriptional regulation.
“Stable” and “Unstable” RNAs at Transcription Start Sites
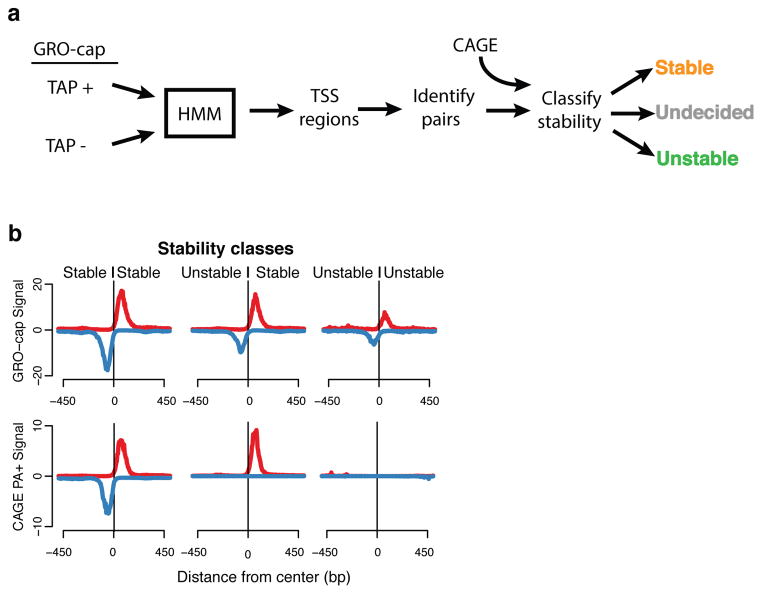
GRO-seq identified divergent transcription at promoters and enhancers2, 9, and GRO-cap has the sensitivity to detect and precisely map divergent transcription in over 90% of TSS regions (Supplemental Fig. 5d) In order to simplify downstream analyses that compare various characteristics of initiation at promoters and enhancers, we created a set of “divergent TSS pairs” that was filtered against cases of partially overlapping initiation pairs (Methods). The resulting set is composed of 22,443 TSS pairs from GM12878 and 24,894 pairs from K562 cells (38% and 39% of all TSSs respectively). As both cell lines show similar results, we will refer to GM12878 data unless otherwise stated. We then classified high-confidence GRO-cap-based TSSs into those giving rise to “stable” transcripts (captured by CAGE and GRO-cap) and those that produce “unstable” transcripts (captured only by GRO-cap) (Fig. 3a, Supplemental Fig. 5, Methods). The distinction between stable and unstable transcripts is also apparent from other RNA-based assays. For instance, stable TSSs have strong RNA-seq profiles (Supplemental Fig. 6), whereas unstable TSSs have very weak or nonexistent RNA-seq profiles. These patterns hold for both the poly-A-plus and poly-A-minus versions of CAGE and RNA-seq, indicating this difference is not simply due to differential poly-adenylation.
Figure 3. TSS identification and classification.
(a) TSS regions were identified with a hidden Markov model (HMM) from GRO-cap reads and control (GM12878: 117,613; K562: 128,471), and combined into pairs of divergent TSSs which where then classified according to the presence of CAGE signal. (b) Composite profiles of GRO-cap and CAGE aligned to the center of GRO-cap TSS pairs after classifying pairs based on the stability of the transcript produced. Profiles are stable::stable (left), unstable::stable (center), unstable::unstable (right). Y-axes are the median read counts in 5 bp windows.
We analyzed three classes of divergent TSS pairs: Stable:Stable (SS), Unstable: Stable (US) and Unstable: Unstable (UU) pairs (Fig. 3a,b, Supplemental Fig. 5a,b). Each of these classes covers a wide range of directional transcription preferences, suggesting that directionality of initiation is not directly linked to RNA stability (Supplemental Fig. 5d,e, Methods). The stability of individual TSSs and, by extension, the classes of TSS pairs generally correspond to distinct transcript annotation types (Supplemental Fig. 5c) and histone marks (Supplemental Fig. 7). In particular, SS and US classes are enriched in chromatin signatures associated mainly with promoter regions (H3K4me3) and active transcription elongation (H3K79me2, H3K36me3), and correspond to various stable transcripts such as protein-coding genes and long intergenic non-coding RNAs (lincRNAs) (Supplemental Fig. 5c). On the other hand, UU pairs have enhancer-like chromatin features such as high H3k4me1 and low or ill-defined transcription elongation marks. Thus, our TSS pair classes generally correspond with the expected transcript annotation types, yet by using transcript stability as the basis for our analysis, we are able to reduce TSSs to three fundamental classes in a data-driven and annotation-independent fashion.
Transcriptional Level Explains Differences in Histone Modifications
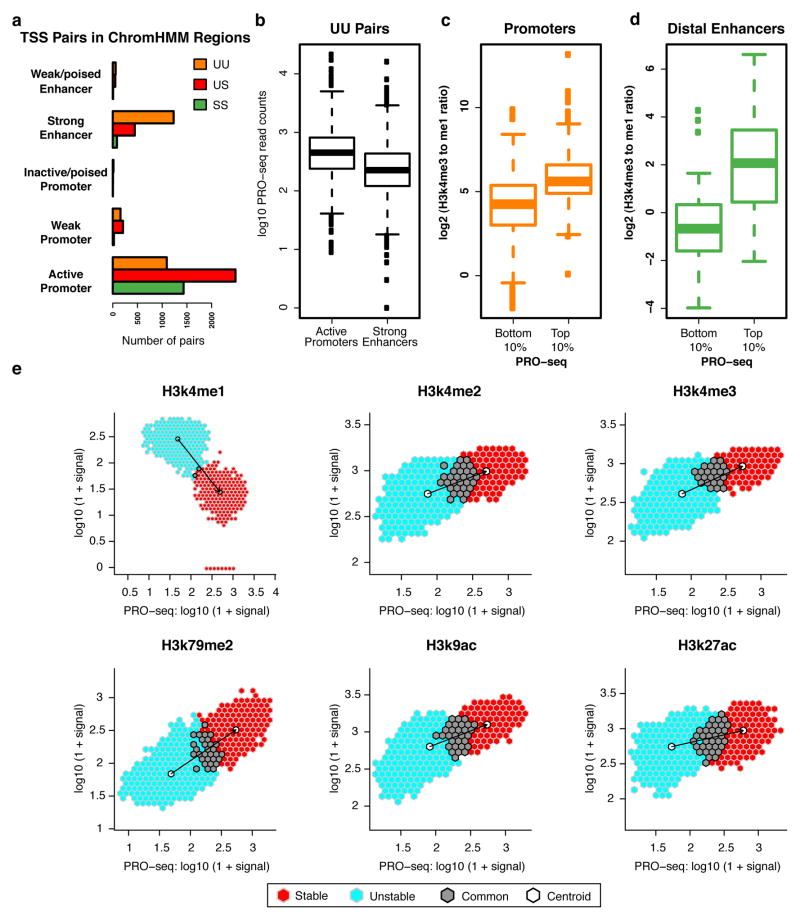
Although the ChromHMM distinction between promoters and enhancers is generally consistent with our TSS classes, with SS and US pairs mainly found at active promoters and UU pairs mainly found at enhancers (Fig. 4a), a substantial fraction of UU pairs are classified by ChromHMM as active promoter regions. This observation is unexpected given that active gene promoters should produce a stable transcript in at least one direction. Inspection of the UU pairs classified as active promoters revealed that they have stronger PRO-seq signals than UU pairs classified as enhancers (Fig. 4b). Thus, it is possible that these UU pairs are actually enhancers that are misclassified as promoters due to the presence of high levels of transcription-related histone marks (i.e. H3K4me3). A striking example occurs at the beta-globin locus, where the upstream HS4 transcribed enhancer is erroneously characterized as a promoter by ChromHMM, whereas the promoter is erroneously predicted to be an enhancer (Fig. 1a).
Figure 4. Histone marks at enhancers and promoters scale with Pol II intensity.
(a) Number of TSS pairs from each stability class mapping to different regulatory regions as designated by ChromHMM. (b) UU pairs mapping to active promoter regions (n = 1478) have a higher PRO-seq signal than those mapping to strong enhancer regions (n =3171), where active promoters and strong enhancers are defined by ChromHMM. (c-d) Ratio of mono- to tri-methylation of H3k4 at top and bottom deciles of PRO-seq signal in both (c) promoter (n = 247, 248; top and bottom deciles, respectively) and (d) enhancer TSS regions (n = 91 and 97; top and bottom deciles, respectively). (e) PRO-seq signal versus indicated histone modifications at TSS regions. Signal is further split between TSSs classified as unstable (light blue), stable (red), and points that overlap between the two (grey). Centroid for each subset in white.
In order to closely investigate the relationship between transcription level and histone marks at promoters and enhancers, we defined a set of stable TSSs from US pairs proximal to annotated protein-coding genes (putative promoters) and contrasted them with TSSs identified from UU pairs in transcription factor ChIP-seq peaks that are distal from genes (putative enhancers). Although these promoters are generally more highly transcribed than the enhancers (see Discussion), the H3k4me3/H3k4me1 ratio at both the promoters and enhancers scales with the corresponding level of Pol II (Fig. 4c, d). Expanding this analysis to all GRO-cap-identified TSSs in our TSS pairs (including both promoters and enhancers), we observed that transcription-associated histone modifications are directly related to the transcription level and this relationship is maintained independently of transcript stability (Fig. 4e). That is, as the level of transcriptionally-engaged Pol II increases at TSS pairs, so does H3K4me3 and other transcription-associated histone modifications.
One defining feature of mammalian promoters is a higher CpG nucleotide content than enhancers, which is thought to contribute to transcription-independent deposition of H3K4me3. For instance, the CpG-binding protein, Cfp1, has been implicated in deposition of H3K4me3 through its recruitment of Setd135. However, the DNA binding domain of Cfp1 is dispensable for targeting H3K4me3 to active genes, suggesting the relationship between CpG content and H3K4me3 may be indirect. Furthermore, there is a clear disconnect between CpG content and histone modifications (H3K4me3 and others) at promoters and enhancers (Supplemental Fig. 8), suggesting that H3K4me3 level at these enhancers is not tied directly to CpG content. Thus, the difference in histone modifications at promoters and enhancers is not specific to the type of regulatory element, but rather, this difference appears to be more fundamentally associated with the level of transcription.
Architecture of Initiation at Promoters and Enhancers
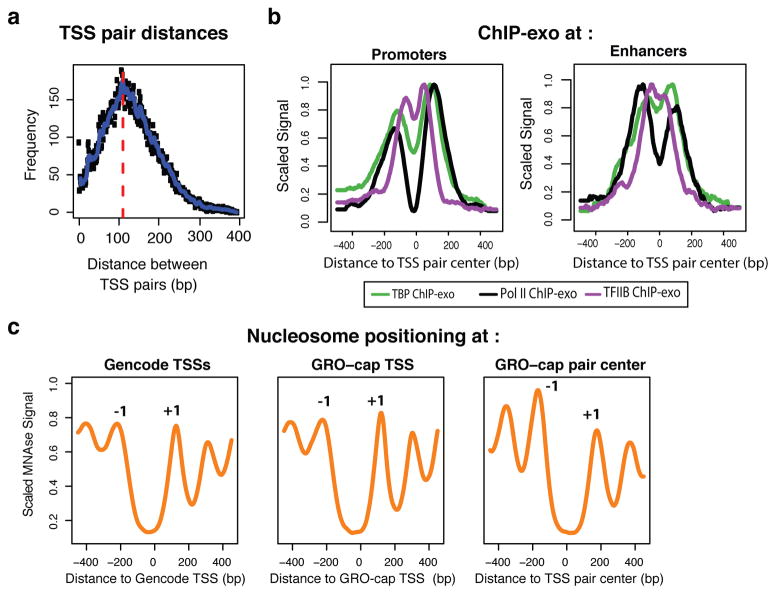
To identify features of initiation regions that might distinguish promoters from enhancers, we closely examined the architecture of TSS regions. Using our high-confidence TSS pairs, we show that divergent initiation occurs, on average, 110bp apart (Fig. 5a) with relatively small variations between TSS pair classes (Supplemental Fig. 9). While divergent initiation is less common in C. elegans, our estimates of the distance between divergent pairs in that species is nearly identical21. Despite the narrow distance, a high-resolution ChIP-exo26 localization of two general transcription factors (GTFs) that bind core promoters (TBP and TFIIB) reveals an independent transcription initiation complex forms in each direction at divergent TSS pairs at promoters and enhancers (Fig. 5b).
Figure 5. Architecture of TSS pairs.
(a) Divergent TSSs are tightly packed, with an estimated 110 bp inter-TSS distance, as estimated from the overall distribution of opposing strand read distances. (b) ChIP-exo profile26 for Pol II (black), TBP (green) and TFIIB (purple), centered on TSS pairs and split between promoter (top) and enhancer (bottom) regions (ChromHMM). (c) Mnase-seq profiles at protein-coding promoters, aligned either by GENCODE annotations (left; also positive for GRO-cap signal), GRO-cap TSS at GENCODE promoters (center), or to GRO-cap TSS pair centers (right). Peaks corresponding to -1 and +1 nucleosomes are indicated.
Transcription initiation is often closely followed by promoter-proximal pausing. ChIP-exo data has revealed that the majority of Pol II at promoters is downstream of TBP and TFIIB and likely to be in a paused state26. Thus, we hypothesized that there might be some interplay between the strength and location of pausing and divergent TSS distances. Although we observe distinct pause modes (proximal-focused and distal-dispersed, as previously found in Drosophila22), we find no effect of these modes on divergent initiation distances (Supplemental Fig 10a–c), or peak locations of TFIIB (Supplemental Fig. 10d). Together with the similar divergent TSS distance results from C. elegans (where pausing is rare), this observation suggests that pausing location does not feed back and influence the locations of divergent TSSs.
Although we find symmetric initiation and GTF binding at divergent promoter TSSs, nucleosome positioning is thought to be asymmetric at promoters. Typically, with respect to GENCODE TSSs, there is a well-positioned downstream nucleosome (+1 nucleosome), whereas the upstream nucleosome (–1 nucleosome) has more variable positioning36 (Fig. 5c, top). In contrast, nucleosomes are reported to be strongly positioned at both sides of transcription factor-bound enhancers37 (Supplemental Fig. 11). However, when we align to the center of our TSS pairs, we clearly see that both nucleosomes flanking the protein-coding US and SS TSSs are well-positioned (Fig. 5c, bottom), with similar profiles to those at enhancers. Thus, the symmetric architecture of initiation regions applies universally to promoters and enhancers.
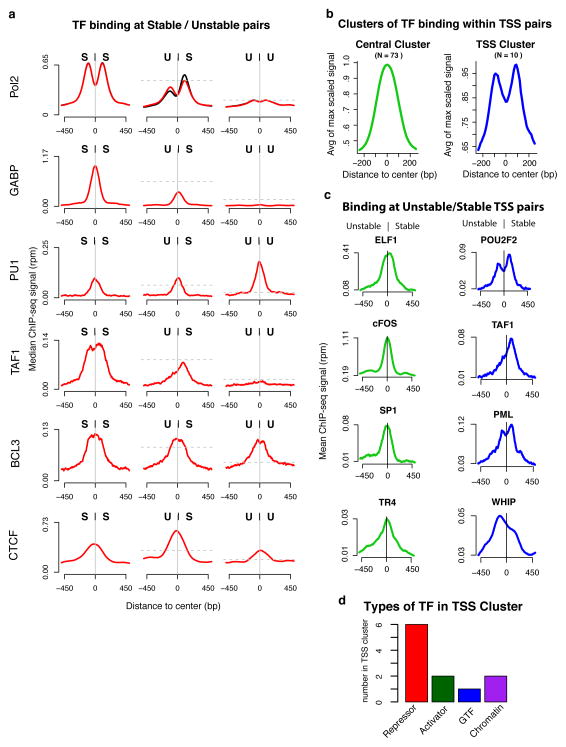
The observed symmetries of nucleosome positioning and core promoter factors raise the question of how sequence-specific transcription factors bind within this context. Using transcription factor ChIP-seq data from ENCODE, we observed four main preferences for pair classes by transcription factors (Fig. 6a, Supplemental Fig. 12): factors that bind preferentially at SS pairs (e.g., GABP); factors that bind preferentially at UU pairs (e.g., PU1); factors that bind indiscriminately at all pair classes (e.g., BCL3) and factors with a preference for US pairs (e.g., CTCF). In addition, we observed two clusters of transcription factors by relative position of binding sites within divergent TSS pairs (Fig. 6b,c): central binding factors (e.g., SP1) and TSS-proximal binding factors (e.g., PML). We are limited by the ChIP-seq sets available, but given the datasets used (N = 84), most factors fall into the central binding cluster (binding profile peaks in the center between divergent TSSs; N = 73) versus the TSS binding cluster (binding profile peaks over TSS position; N = 10) (Supplemental Table 2). Interestingly, the TSS-proximal binding cluster includes both GTFs such as TAF1 and transcriptional repressors such as NRSF and Pml (Fig. 6d), suggesting a potential involvement in transcript stability determination or preferential targeting of these factors to stable transcripts. These results provide a clear relationship between transcription factor binding and TSS structure and suggest that central-binding transcription factors and the symmetrical structure of initiating regions may be mechanistically linked.
Figure 6. Modes of transcription factor binding at TSS pairs.
(a) Representative ChIP-seq profiles of different modes of transcription factor binding at different TSS pair stability classes. Signals are subject to paired subsampling to correct for Pol II signal dependency (top plot, Methods). The y axes are the median read density in 5bp windows. The horizontal dashed lines represent the expected peak signal level if the signal followed the scaling of Pol II relative to the SS panel. (b) ENCODE transcription factor ChIP-seq profiles, anchored on TSS pairs, cluster into two distinct groups, central binders (green) and TSS binders (blue). (c) Examples of the two positional modes of binding at US (Unstable, Stable) pairs. (d) Classification of factors within the TSS binding cluster. The total number of factors in d are greater than the number of TSS binding factors because factors can be part of more than one functional group (see Supplemental table 2).
Sequence Predictors of Transcript Stability
Because DNA sequence is known to influence initiation, productive transcription, RNA processing and stability, we also examined the sequence composition near our TSS pairs. In general, we find that sequence conservation and nucleotide frequency are indicative of transcript stability (Supplemental Fig. 13a–c). In particular, SS TSSs are associated with increased C and G nucleotides and increased CpG di-nucleotides within and around pairs. In contrast, UU TSS pairs are depleted for C, G, and CpG. US TSS pairs display a combination of these two patterns. Despite these biases, we see similar frequencies of core promoter elements (TATA and Inr) in the expected positions at all classes of TSS pairs (Supplemental Fig. 14a,b). This observation is consistent with ChIP-exo detection of GTFs at all classes of TSS pairs (Supplemental Fig. 14c), indicating that other mechanisms might be dictating the production of stable versus unstable transcripts. Indeed, recent work has shown that sequences that direct the binding and activity of poly-A dependent termination machinery or the U1 splicing complex work antagonistically to direct unstable or stable transcription, respectively, at protein-coding genes4, 5. In this model, 5′-splice sites (SS5) that bind U1 can suppress poly-A site (PAS)-dependent termination, thus promoting productive elongation of protein-coding mRNAs.
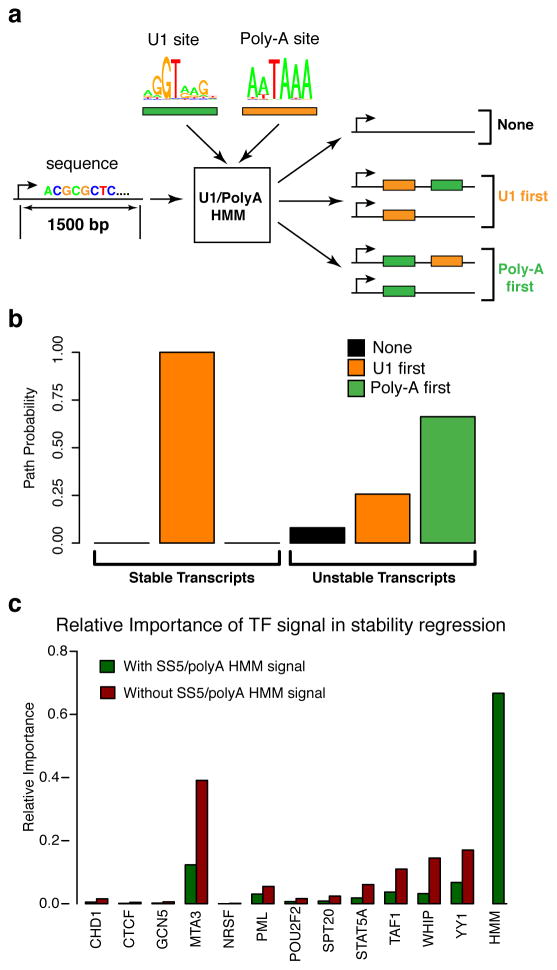
To determine if there is a direct relationship between our transcript stability classes and the premature PAS-dependent termination, we scanned the regions downstream of TSSs for matches to the poly-A and SS5 motifs and observed that our stable and unstable TSS classes follow a pattern consistent with these reports (Supplemental Fig. 15a,b). That is, the SS5 motif is enriched downstream of stable transcripts but depleted at unstable transcripts, and vice-versa for the PAS motif. We devised an HMM that incorporates SS5 and PAS motif models and used it to compare the likelihoods of SS5 binding sites before and after a poly-A site (Fig. 7a, Supplemental Fig. 15c). Our results indicate that SS5 binding sites strongly tend to precede the PAS on stable transcripts but not on unstable transcripts (Fig. 7b). In the case of single exon genes (N = 105), both SS5 and PAS sites are less frequent, but PAS sites are more depleted than SS5 sites (Supplemental Fig. 15d,e). These results are consistent with previous observations for protein-coding genes, and importantly, they demonstrate that these sequence predictors of elongation hold for all TSSs, including those at enhancers. Furthermore, our HMM can be used to predict transcript stability to high accuracy (63%), suggesting that these motifs and their spatial relationship are strong determinants in this process.
Figure 7. Determinants of RNA stability for both promoters and enhancers.
(a) Diagram of transcript U1/poly-A classification. Each transcript (first 1.5kbp) is processed through an HMM to determine relative order and occurrence of SS5 and PAS elements. (b) Estimated path probabilities of alternative element occurrences (neither SS5 nor PAS: black, SS5 first: orange, PAS first: green) obtained by applying the EM algorithm to each transcript subset (stable and unstable TSS stability classes). (c) Relative importance of various transcript factors in a logistic regression of the stability classes, with (green) and without (red) including the U1/poly-A HMM derived signal (posterior path probability of being in unstable class).
Finally, we used logistic regression to assess the relevance of transcription factors in the TSS-binding cluster to transcription stability. transcription factors, by themselves, explain only a small fraction of the variance in stability (R2 = 0.05). Furthermore, when the signal from the poly-A/U1 HMM is also considered, their relative importance drops considerably (Fig. 7c). These observations suggest that most of the information about stability comes from the presence or absence of early poly-A sites and U1 splicing signals, but they do not rule out the possibility that some of these transcription factors may be components of the splicing pathway or contribute to feedback between splicing and transcription levels.
DISCUSSION
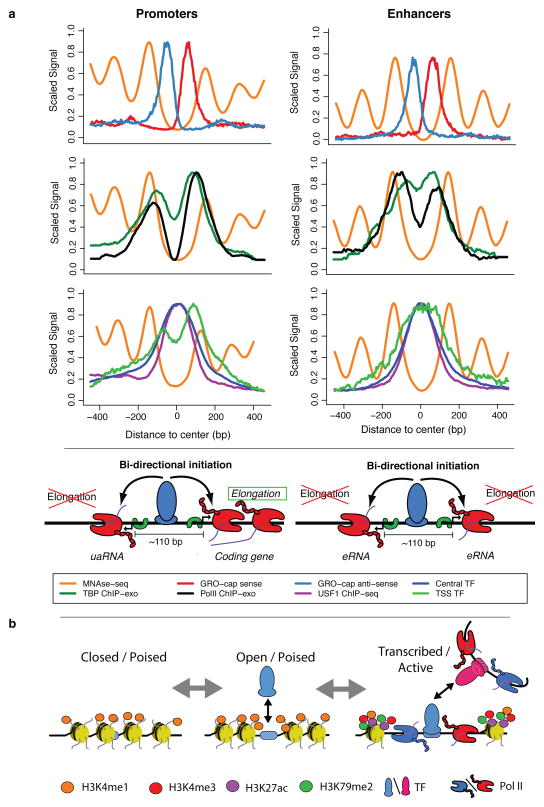
Several studies have documented divergent transcription at promoters and enhancers2, 3, 8, 9, 38, however, the nature and organization of initiation sites, their underlying DNA elements, and their relationships with transcription factor binding and nucleosome positions have yet to be reconciled. In this article, we show that assaying nascent RNAs dramatically increases sensitivity for enhancer detection compared with methods that map accumulated RNAs. By contrasting our GRO-cap data with CAGE data, we are able to classify TSS pairs based on the stability of the resulting transcripts. Unstable transcripts are those that are likely targeted for immediate degradation by the exosome, and thus are unable (or less likely) to be discovered in assays that detect accumulated RNAs, such as CAGE. By contrast, stable transcripts are detectable in both nascent and accumulated RNA pools. These classifications allow us to work directly from genome-wide functional genomic assays without reliance on genomic annotations. By analyzing these annotation-free TSSs together with DNA sequences and functional genomic data, we are able to catalog the precise nature of the structure and chromatin content at initiation sites. We find that the divergent TSS pairs at both promoters and active enhancers: 1) have similar frequencies of canonical core promoter elements, 2) have distinct transcription complexes at each member of a pair, 3) are separated by 110bp on average, 4) are bound by central transcription activators, 5) are flanked on both sides by positioned nucleosomes, and 6) have histone modifications typically associated with transcription initiation, present in proportion to the amount of transcription. These results suggest a unified model for the mechanisms that govern transcription initiation at both enhancers and promoters (Fig. 8a).
Figure 8. Summary of transcription initiation at regulatory regions.
(a) Our analysis of TSSs reveals a common structure across all initiation regions, including promoters and enhancers. In both cases, (first row) a tightly packed (110 bp \apart) divergent TSS pair (+ strand: red, − strand: blue) surrounded by well-positioned nucleosomes (orange), with independent pre-initiation complexes (separate TBP (green) and Pol II ChIP-exo peaks (black), second row) and sharing two distinct transcription factor cluster binding modes (central: green, over TSS: blue; third row). We propose that central, activator transcription factor binding (USF1 example: purple), in conjunction with core promoter elements, determines the positioning of the divergent initiation sites. Finally, DNA sequence properties (not depicted here), possibly in cooperation with other factors, determine the resulting transcript type (stable/elongating: protein coding, unstable/terminating: uaRNA, eRNA, etc.). (b) A model depicting possible progression of enhancer states from chromatin marked but largely inaccessible regions (left), followed by more open regions through transcription factor binding (center) and finally, active transcription, which brings with it the associated chromatin marks (in particular, H3K79me2 and H3K27ac and increased methylation levels of H3K4; right).
We show that divergent initiation occurs within a window of 90–120 bp, which is a surprisingly narrow interval considering that a PIC makes contacts up to 50bp upstream and downstream from the TSS39. The close proximity of divergent initiation events and the evidence for bound transcription factors between them makes it difficult to imagine that multiple independent polymerase complexes and transcription activators simultaneously occupy the same promoter. One possible alternative is that one polymerase initiates first and then pauses downstream, allowing enough space for a second polymerase to initiate upstream and in the opposite direction. Consistent with this hypothesis, high-resolution ChIP-exo data suggests that the majority of Pol II on chromatin of human cells (K562) is paused approximately 50bp downstream of the initiation site26. We also show that these independent and divergent transcription complexes have similar frequencies of well-known core promoter elements in the underlying DNA. This result is fundamentally important as it suggests that recruited Pol II is not randomly initiating at open DNA regions associated with enhancers and divergent promoters15. Rather, the normal cohort of general transcription factors is positioned to facilitate initiation at these sites.
We also find evidence for positional modes for transcription factor binding in divergent TSS regions. Most factors bind between the two divergent TSSs (central binders), suggesting that they play a role in activation and are likely a major determinant or result of the overall architecture of initiation sites. On the other hand, the TSS-proximal transcription factors are primarily enriched for repressors, suggesting that certain repressors can act by preventing access of the transcription machinery to critical parts of the core promoter. The apparent tight spacing and organization of binding suggests that few factors simultaneously bind at any given initiation region. This observation is in agreement with evidence for a small number of identifiable sequence motifs even when numerous factors are found in narrow regions by ChIP-seq40. Coinciding signals may reflect indirect binding of transcription factors or binding events that occur in a subset of cells within a population. Finally, the close relationship between transcription factor binding and initiation in our model provides a possible explanation for why protein-coding genes typically have multiple associated mRNAs with small differences in TSS location. These alternative TSSs likely result from the presence of multiple neighboring binding sites for transcription factors that compete as anchors for initiation. As a result, depending on cell type and condition, different transcription factor binding events lead to small shifts of the initiation site.
Promoter regions are generally assumed to be quite broad, with promoter-associated transcription factor binding sites spanning a multi-kilobase region near the TSS, but our results suggest that initiation regions are primarily defined by a relatively narrow 100-to-200 bp window. Part of this discrepancy can be attributed to poor or incomplete annotation of genes, but it may also indicate that multiple independent initiation regions often act as neighboring enhancers. Although we have focused here on non-overlapping TSS pairs to simplify our analyses, we expect that overlapping TSS pairs will represent an aggregate of the local transcription factor occupancies. In the future, it will be interesting to further investigate transcription factor occupancy at these more complex regions with the help of higher-resolution assays, such as ChIP-exo45.
Previous work suggests that enhancer chromatin undergoes a progression from a closed state to an open state required for transcription factor binding14, 41–43. Our analyses of DNase-hypersensitivity and GRO-cap data at enhancers generally support the existence of, and potential progression through, at least three enhancer states: closed, open, and transcriptionally active (Figure 8b). Comparisons of these states with other functional genomics data suggest that the transcribed enhancers are the most active, whereas the closed and open classes represent a poised state. We envision that it is equally plausible to progress in either direction between states, thus, the poised states could represent enhancers that have yet to be activated, or dormant enhancers that are vestiges of past activity44. Interestingly, the poised enhancers resemble a form of pre-activated promoters recently observed during developmental transitions45, providing yet another similarity between regulation at promoters and enhancers. Although we see less evidence for transcription factor binding at open and untranscribed enhancers, these regions could arise through binding of a small number of ‘pioneering’ transcription factors. Also, some poised enhancers could be open simply because they have relatively poor affinity for nucleosomes due to underlying sequences. Alternatively, permissive chromatin could arise concomitantly with transcription factor binding and transcription14. In either case, the transition from the open or poised states to transcriptionally active state is clearly related to binding of central, activating transcription factors (Fig. 8b). It will require further work to determine whether or not all functionally active enhancers (influencing the activity of target transcripts) generate local transcription.
It is generally thought that distinct mechanisms selectively mark histones at enhancers and promoters. In particular, enhancers are typically identified as having high levels of H3K4me1 relative to H3K4me312, 13. However, we observe a strong positive correlation between absolute levels of transcription and the H3K4me3/H3K4me1 ratio at active enhancers, suggesting that differences in H3K4 methylation patterns at enhancers and promoters may simply reflect differences in transcription levels. Consistent with this observation, H3K4me3 has been detected at some active enhancers11, 46, and can be deposited in a transcription-dependent manner11, 47. Why, then, are enhancers generally observed to have less transcription initiation and hence less H3K4me3 than promoters? One possible explanation comes from observations of feedback mechanisms whereby elongation of transcription positively contributes to subsequent rounds of initiation. A related possibility, consistent with our observation of splicing-dependent difference in transcript stability at promoters and enhancers, would be feedback from the splicing machinery. Indeed, the presence of a U1 splice site can positively influence recruitment of GTFs to promoters48. In addition, the GTF, TAF15, has been shown to interact with the U1 snRNP providing another link between the splicing and initiation complex. Therefore, splicing-dependent elongation of transcription not only distinguishes promoters from enhancers, but may also help explain different intensities of transcription initiation and hence, histone modifications at these regions.
The original definition of an enhancer describes a genomic interval that stimulates transcription of another locus independently of its position and orientation relative to the transcribed locus49. Our analyses reveal that mechanisms governing chromatin content and architecture at enhancers are quite similar to those at promoters. What then is a proper description of an enhancer? 3D chromatin links bridging different initiation regions have been observed both between traditional enhancers and promoters and between pairs of promoters33. Thus, the implication is that any initiation region can function as an enhancer, through the central binding activator, irrespective of the fate or function of the local transcripts that are generated. Conversely, it is currently not clear whether some transcription factors can enhance distal transcription activity without generating local transcription.
Our observations have implications for an intriguing potential relationship between divergent transcription and the origin of new genes. It has recently been shown that asymmetries in productive transcriptional elongation favoring the sense-coding direction at gene promoters can be explained by a disproportional tendency for promoter-proximal cleavage and polyadenylation shortly after initiation in the antisense direction, which appears to be associated with an enrichment for PASs in upstream antisense regions of genes4, 5. Furthermore, PASs are depleted and U1 snRNP recognition sites (SS5s) are enriched in the sense direction, consistent with observations that the U1 snRNP complex protects pre-mRNAs from cleavage and polyadenylation50, 51. Building on these observations, Wu and Sharp recently proposed a model for the evolutionary origin of new genes whereby short, unstable upstream antisense RNAs (uaRNAs) gradually increase in length and stability as mutations eliminate PASs and create new SS5s52. In this way, uaRNAs or eRNAs could develop, in a stepwise fashion, first into noncoding RNAs and then into protein-coding mRNAs, perhaps acquiring splicing capabilities along the way (which, in turn, would further improve stability). This process could be encouraged by positive feedback with transcription-associated mutational asymmetries, which are biased toward G and T nucleotides53 and therefore would favor the formation of SS5s and the abolishment of PASs. In this article, we have shown that transcription initiation occurs in a bidirectional fashion at thousands of enhancers that have fundamentally the same architecture of initiation as traditional promoters. Thus, if uaRNAs and eRNAs do indeed sometimes develop into genes, then the genome is replete with potential new genes, many of them far from existing genes. Additional studies of nascent RNAs across cell types and species may help to shed light on these important evolutionary questions.
METHODS
Preparation of GRO-cap, PRO-seq and GRO-seq libraries
GRO-cap libraries for K562 and GM12878 cells were produced precisely as described in Kruesi et. al21. 1×107 nuclei were used for each GRO-cap library or control. GRO-seq libraries for K562 or GM12878 cells were produced as described in Wang et al56. PRO-seq libraries were produced as described previously22, using the TruSeq™ small RNA adapters (Illumina), and 5×106 nuclei.
Mapping of sequencing data
After sequencing GRO-seq and GRO-cap reads were trimmed to 30 bases, and mapped first to a single copy of the rDNA locus to remove related transcribed sequences. Reads that did not map to the rDNA were then mapped to the hg19 version of the human genome. Reads were required to be unique and have no more than two mismatches. PRO-seq reads (100 bases) were processed essentially as in Kwak et al. 22. Adapters were removed with cutadapt57, and then unique sequences 15bp or greater were then mapped to the hg19 genome were kept for further analysis.
Prediction of Transcription Start Sites
Pre-processing of GRO-cap Data
GRO-cap aligned data, normalized by total read counts, was summarized in fixed intervals of 10 bp along the reference genome, to increase the signal in low intensity initiation sites and “smooth” away minor misalignments between the TAP+ and TAP- conditions. Each 10 bp interval was assigned two values, one summarizing the TAP+ to TAP- signal differences and the other indicating the presence of a TAP+ “peak”. To summarize the TAP+ to TAP- signal difference in each interval we assigned the interval to one of three categories: 1) “no signal” (TAP+ has zero reads); 2) “enriched” (TAP+ > TAP-); or 3) “depleted” (TAP- > TAP+ > 0). To compute the binary “peak” indicator for an interval, we searched for “depleted” intervals (as per the above definition) within ten 10-bp intervals (100 bp) in either direction, and if at least two were found, we used their mean normalized read counts as an estimate of the local background level. The interval in question was then called “peaked” if its normalized read count was greater than twice the estimated local background level. We found that our final predictions were not very sensitive to the threshold for calling peaks, with a wide range of fold-enrichments producing numbers of predictions that differed by no more than 3%.
Design of the Hidden Markov Model
Previous CAGE studies have shown that TSS regions can be both “sharp” (highly peaked) and “broad”58–60. Therefore, we designed our hidden Markov model (HMM) to have a single background state (B) and two groups of alternative states, representing non-peaked (M1) and peaked (M2) TSS regions (Supplemental Fig. 2a). The M1 and M2 groups are each composed of three states, and within each group, these states share the same multinomial emission distribution for “no signal”, “enriched”, and “depleted” TAP+ read counts. In addition, the states have a conditionally independent emission distribution for the peak signal, set such that only the middle state of the M2 group permits “peaked” intervals. Because multiple peaks can occur in a single peaked TSS regions, the transitions among the states in the M2 group allow for zero or more steps between consecutive peaks (middle state). This design enforces a distinction between sharp and broad TSSs, while avoiding false positives due to highly local spikes in the data.
Parameter Estimation and Transcription Start Site Prediction
The free parameters of the model were set as follows. Most transition probabilities were set to zero or one according to the constraints of the model design (Supplemental Fig. 2a), or were assigned values reflecting a non-informative uniform prior distribution over possible state transitions (for example, the transitions out of the first and last states of the M2 group). The two exceptions to this rule were the self-transition probabilities for the background state and the middle (peak-emitting) M2 state, which were assigned high (0.99) and low (0.1) values, respectively, because we expect peaks to be sparse along the genome. The emission parameters were set approximately based on empirical observations of TSS regions. In particular, we observed that background regions are mostly devoid of reads (P(“no signal”) = 0.9; P(“enriched”) = P(“depleted”) = 0.05). By contrast, non-peaked regions (M1 group; broad TSSs) are dense in “enriched” intervals (P(“no signal”) = 0.09; P(“enriched”) = 0.9; P(“depleted”) = 0.01). Peaked regions (M2 group; peaked TSSs) have both “enriched” and “depleted” intervals, in varying proportions, but because this group is anchored by the “peaked” indicator, it is not sensitive to the exact emission probabilities as long as “no signal” is unlikely; therefore, for these states we used P(“no signal”) = 0.1; P(“enriched”) = 0.45; P(“depleted”) = 0.45.
TSS regions were obtained by running the Viterbi algorithm60, 61 on the preprocessed GRO-cap data, which finds the most likely path through the HMM given the data and the model parameters. The predicted TSS regions were then refined for further analysis as follows. First, regions of longer than 100 bp that were assigned to the M2 state group were split into constituent “peaked” subregions such that distances of at least 30 bp were maintained between them. In addition, all regions were trimmed of leading and trailing “depleted” (TAP- > TAP+) intervals. The effects of these postprocessing steps can be seen in Supplemental Fig. 2b
TSS Paired Regions
A divergent TSS pair is composed of adjacent TSS regions in opposing orientations (a minus strand TSS region followed by plus strand TSS region) within 150 bp of each other (nearest edges). This threshold was set empirically, after manual observation of initiation sites, in order to capture the observed distances between divergent TSS regions (median nearest edge distance was 40 bp). TSS pairs were further filtered by requiring a high GRO-cap signal (minimum number of reads above the 20% quantile), so that we could reliably scale the various signals of interest by expression level in downstream analysis.
TSS Stability Classification
GRO-cap-based TSSs were classified into those giving rise to “stable” transcripts (captured by CAGE and GRO-cap) and those that produce “unstable” transcripts (captured only by GRO-cap). In practice, our TSS regions were classified as unstable in the absence of CAGE reads and as stable if they contained at least 8 CAGE reads. These thresholds are conservative, and the latter is above the estimated CAGE background in introns (Supplemental Fig. 5a; grey bars). We focused on high-confidence sets of both stable and unstable transcripts by further requiring a high GRO-cap signal (minimum number of reads above the 20% quantile). Interestingly, regardless of whether lincRNAs arise from regions classified as promoters or enhancers62, GENCODE63 lincRNAs are largely stable by our classification.
Paired Subsampling
In our analysis of divergent initiation regions, we produced composite profiles for paired TSSs in a variety of ChIP-based assays. A challenge in interpreting these profiles is that the marginal distributions of transcription levels often differ significantly at members of each pair, and other signals of interest, such as ChIP-seq measures of transcription factor binding, correlate strongly with transcription level. Thus, apparent differences in the signals of interest may simply reflect differences in overall transcription level. This is especially a problem for US pairs, because unstable TSSs tend to have substantially lower transcription levels than their stable counterparts.
To improve the interpretability of these plots, we generated composite profiles by a sub-sampling method that ensures the marginal Pol II ChIP-seq distributions are the same at the left and right TSSs. Briefly, we summarize each TSS pair by four values: the Pol II ChIP-seq values and the signal of interest, both at the left and right TSS. For convenience, the Pol II ChIP-seq values are discretized into bins. We then define a shared “target” distribution for Pol II by pooling the data for the left and right TSSs. Finally, we subsample from the collection of TSS pairs (summarized by their four values) in such a way that the left and right Pol II distributions exactly match the target distribution. This subsampling step is complicated by the dependency between the left and right Pol II distributions, but this complication can be addressed by a simple algorithm that performs a depth-first search over possible combinations of samples from the original distribution, branches of which are terminated whenever the constraints on the subsample are violated. The induced marginal distributions of values for the signal of interest at the left and right TSS are then compared. In this way, differences in the profiles that simply reflect differences in Pol II (a surrogate for transcription level) are eliminated.
Splicing Signal Hidden Markov Model
To define the hidden Markov model (HMM) for splicing signals, we start with a 5′ splice site (SS5) position weight matrix (PWM) estimated from GENCODE 16 annotations of the first exon for protein-coding genes (Supplemental Fig. 15b). In addition, a PWM for poly(A) sites (PAS) was estimated from the sequences reported in (Beaudoing et al.64). Finally, a background model was estimated from the full DNA sequences, assuming independence of sites.
Our HMM combines these motif models in such a way that we can make inferences about the relative positioning of SS5 and PAS sequence motifs. In particular, the HMM permits branching from an initial background state into five alternative paths. Two of these paths visit the SS5 site before an optional PAS; two others visit the poly-A site before an optional U1 site; and a final path includes none of the two motif signals. The HMM is structured such that the transition from the initial background state is taken once and only once (Supplemental Fig. 15c).
We applied this HMM to sequences spanning the first 1.5 kb of TSSs in each class (stable and unstable). To estimate the relative likelihood of each path, we computed maximum likelihood estimates of the transition probabilities into each of the five alternative paths using the Baum-Welch algorithm65. Because the number of free parameters is the same for all paths, no model complexity penalty is needed for this comparison.
Additionally, the probability of each alternative path for each sequence can be estimated by setting the uniform transition probabilities out of the initial background state and then computing the respective the posterior probabilities. This enables the use of the HMM model as a sequence classifier (by thresholding the sum of the posterior over the sequence) and it is used as the input for the stability regression (below).
Stability Regression
The relative contribution of individual transcription factors and the splicing signal HMM towards predicting the TSS class (stable or unstable) was assessed by logistic regression. Transcription factor signals correspond to sums of ChIP-seq signal in the predicted TSS region. Relative importance of regression weights was computed according to Johnson et al.66. Because transcription factor binding patterns are often strongly correlated with transcription level, we applied the logistic regression to subsamples of stable and unstable TSSs with matching Pol II signal distributions (as described above).
Supplementary Material
Acknowledgments
We would like to thank Vivian Cheung for deep sequencing of the GM12878 GRO-seq and K562 PRO-seq libraries. We would also like to thank members of the Lis and Siepel labs for helpful comments on this work. This work was supported by grants: GM25232 to JL, and HG0070707 to AS and JL.
Footnotes
Database accession numbers
All data files are available on the Gene Expression Omnibus (GEO ref. number GSE60456).
Links to data tracks
All data are also available as tracks on the UCSC genome browser54 link: http://compgen.bscb.cornell.edu/~alm253/hubs/grocap/hub.txt Tracks for the TSS calls and stability classifications are also available in Supplementary Data Set 1.
Author contributions
LC and JL designed the experiments. LC and CW produced the data sets. AM designed and implemented software for data analysis. AM, LC, AS, JL, and CD analyzed the data and interpreted the results. LC, AM, AS, JL, and CD wrote the manuscript.
References
- 1.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 2.Core LJ, Waterfall J, Lis J. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008 doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ntini E, et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- 6.Trinklein ND, et al. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oler AJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koch F, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 12.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 13.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 16.Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- 19.Melgar MF, Collins FS, Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol. 2011;12 doi: 10.1186/gb-2011-12-11-r113. R113-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife. 2013;2:e00808. doi: 10.7554/eLife.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson R, et al. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. bioRxiv. 2014 doi: 10.1038/ncomms6336. [DOI] [PubMed] [Google Scholar]
- 24.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orkin SH. Regulation of globin gene expression in erythroid cells. Eur J Biochem. 1995;231:271–281. doi: 10.1111/j.1432-1033.1995.tb20697.x. [DOI] [PubMed] [Google Scholar]
- 26.Venters BJ, Pugh BF. Genomic organization of human transcription initiation complexes. Nature. 2013;502:53–58. doi: 10.1038/nature12535. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiraki T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100:15776–81. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maruyama K, Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 31.Affymetrix ENCODE Transcriptome Project & Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piper J, et al. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 2013;41:e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clouaire T, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaffney DJ, et al. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 39.Coulombe B, Burton ZF. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63:457–78. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foley JW, Sidow A. Transcription-factor occupancy at HOT regions quantitatively predicts RNA polymerase recruitment in five human cell lines. BMC Genomics. 2013;14:720–2164. 14–720. doi: 10.1186/1471-2164-14-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stergachis AB, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wamstad JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pekowska A, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Damgaard CK, et al. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 49.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 50.Kaida D, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu X, Sharp PA. Divergent transcription: a driving force for new gene origination? Cell. 2013;155(5):990–6. doi: 10.1016/j.cell.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green P, et al. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 54.Kent WJ, et al. The human genome browser at UCSC. 2002 doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang IX, et al. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Rep. 2014;6:906–915. doi: 10.1016/j.celrep.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–12. [Google Scholar]
- 58.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 59.Sandelin A, et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 60.Viterbi AJ. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Transactions on Information Theory. 1967;13 (2):260–269. [Google Scholar]
- 61.Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proceedings of the IEEE. 1989;77 (2):257–286. [Google Scholar]
- 62.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durbin R, Eddy SR, Krogh A, Mitchison GJ. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press; 1998. [Google Scholar]
- 66.Johnson JW. A heuristic method for estimating the relative weight of predictor variables in multiple regression. Multivariate Behavioral Research, MVB. 2000;35:1–19. doi: 10.1207/S15327906MBR3501_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.