SUMMARY
Chemical damage to RNA affects its functional properties and hence may pose a significant hurdle to the translational apparatus; however, the effects of damaged mRNA on the speed and accuracy of the decoding process and their interplay with quality control processes are not known. Here, we systematically explore the effects of oxidative damage on the decoding process using a well-defined bacterial in vitro translation system. We find that the oxidative lesion 8-oxoguanosine reduces the rate of peptide-bond formation by more than three orders of magnitude independent of its position within the codon. Interestingly, 8-oxoguanosine had little effect on the fidelity of the selection process suggesting that the modification stalls the translational machinery. Consistent with these findings, 8-oxoguanosine-mRNAs were observed to accumulate and associate with polyribosomes in yeast strains in which no-go decay is compromised. Our data provide compelling evidence that mRNA-surveillance mechanisms have evolved to cope with damaged mRNA.
Keywords: ribosome, damaged mRNA, 8-oxoguanosine, no-go decay, quality control, translation
INTRODUCTION
The accurate translation of the cellular messenger RNAs is an integral feature of the ribosome and the translation factors. On intact mRNAs, the selection of aminoacylated tRNA substrates by the ribosome has evolved to utilize thermodynamic and kinetic differences between cognate and noncognate interactions (Zaher and Green, 2009a). On aberrant mRNAs, such as truncated ones and those either containing premature or lacking stop codons, a number of ribosome-based-quality-control processes ensure that these RNAs are not translated and instead are targeted for degradation (Graille and Seraphin, 2012; Kervestin and Jacobson, 2012; Shoemaker and Green, 2012). In addition to defects in its sequence, mRNA is susceptible to chemically-induced defects. Nucleic acids are constantly under assault from both endogenous and exogenous agents that can affect their chemical properties and hence their function (Wurtmann and Wolin, 2009). These assaults include reactive oxygen species (ROS), ultraviolet light and alkylating agents. In contrast to DNA, damaged RNA has received little attention presumably due to its transient nature (Li et al., 2006), even though damaged RNA is known to accumulate and has been linked to a number of neurodegenerative diseases (Nunomura et al., 1999; Shan et al., 2007; Shan and Lin, 2006; Shan et al., 2003; Tanaka et al., 2007). Indeed, in human fibroblast cells the median half-life of mRNAs is ~ 10 hours (Yang et al., 2003) indicating that damaged mRNAs may persist and are likely to pose a challenge to the cell if not corrected. Furthermore, a number of mRNA-surveillance mechanisms exist to handle aberrant mRNAs suggesting that high turnover of RNA alone is not a sufficient means to cope with damaged RNAs.
ROS are present under normal conditions as byproducts from metabolic reactions (Finkel and Holbrook, 2000) and their levels increase significantly under stress conditions. The reaction of ROS with nucleic acids results in a myriad of modifications (Barciszewski et al., 1999). Among these lesions, modification to the guanine base resulting in the 8-oxo-7,8-dihydro-guanine (8-oxoGua) is the quintessential archetype of oxidation adducts due to its ability to pair with adenosine during DNA replication (Figure 1A). In agreement with this altered base-pairing property, RNA damage appears to affect decoding, as crudely oxidized mRNA has been linked to reduced translational efficiency and accuracy (Shan et al., 2007; Shan et al., 2003; Tanaka et al., 2007). To date, mechanistic understanding of this inefficiency (i.e. whether it is due to miscoding, stalling or degradation of the damaged mRNA) is lacking. This is mainly due to the difficulty of targeting a specific transcript at an exact location for oxidation.
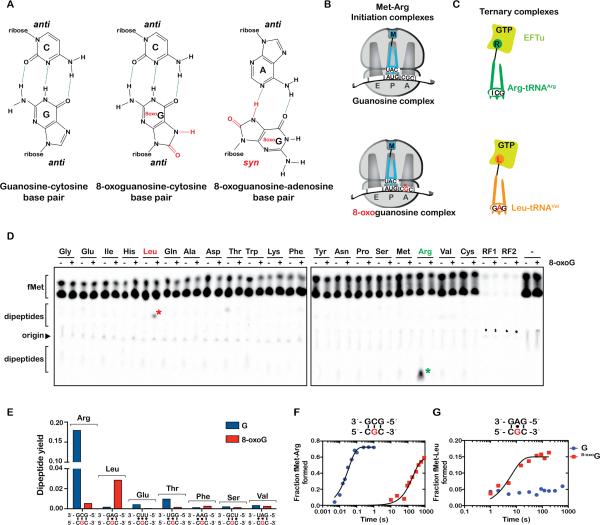
Figure 1. 8-oxoG is detrimental to the decoding process.
(A) Chemical structures of G:C, 8-oxoG:C and 8oxoG:A base pairs. 8-oxoG adopts a syn conformation allowing it to base pair with adenosine. (B) Schematic representation of guanosine and 8-oxoguanosine initiation complexes encoding for the dipeptide Met-Arg. Both complexes carry the initiator fMet-tRNAfMet in the P site; the G complex displays a CGC codon in the A site, whereas the 8-oxoG complex displays a C8-oxoGC codon in the A site. (C) Schematic representation of the cognate Arg-tRNAArg and near-cognate Leu-tRNALeu (G:A mismatch at the second position) ternary complexes. (D) Phosphorimager scan of an electrophoretic TLC showing the reactivities of the initiation complexes with the indicated ternary complexes and release factors. Green asterisk represents the cognate dipeptide, whereas the red one represents the near-cognate dipeptide resulting from a G:A mismatch. (E) Quantification of the dipeptide yield in (D); the predicted codon-anticodon interaction is shown below the x-axis and the corresponding dipeptide is shown above the bars. (F, G) Time-courses of peptide-bond formation between the initiation complexes and the cognate Arg-tRNAArg and near-cognate Leu-tRNALeu ternary complexes, respectively. See also Figure S1, S2.
Oxidized RNA is known to accumulate in vivo, for which 8-oxoG is present at ~1 in 105 residues in total RNA (the levels could potentially be higher in the mRNA pool) under normal conditions and increases as much as 10-fold under oxidative stress (Hofer et al., 2005; Shen et al., 2000) highlighting the potential risk to cellular homeostasis. Additionally, oxidized RNA appears to turn over at a faster rate (Hofer et al., 2005), suggesting that it is selectively degraded, however the exact details of such a mechanism are currently unknown. It is conceivable that factors that recognize lesions on RNA exist and target it for degradation, but a more appealing model would involve ribosomal action. What is attractive about this model is the existence of a number of ribosome-based-mRNA-surveillance mechanisms (Shoemaker and Green, 2012). Second, as the ribosome is the only machine that sees all mRNAs, it is not difficult to imagine how a damaged-RNA-quality-control mechanism can be built into the ribosome. The signal for any degradation process, however, would have to initiate within the decoding center of the ribosome as it selects an incoming aminoacyl-tRNA (aa-tRNA) against the oxidized A-site codon. With this in mind, here we examine the effect of 8-oxoG on decoding using a high-resolution approach, providing a first look at the ribosomal response to oxidative damage.
RESULTS
8-oxoG is detrimental to the decoding process
To explore the effects of RNA damage on the decoding process, we took advantage of our high-resolution-reconstituted bacterial system (Zaher and Green, 2009b), which can monitor the efficiency of incorporation of every single amino acid (Figure S1). We produced ribosomal initiation complexes carrying the initiator tRNA f-[35S]-Met-tRNAfMet in the P site combined with a variety of intact or site-specifically damaged codons in the A site. We started our studies with a pair of complexes displaying either the intact CGC or C8-oxoGC codon in the A site (Figure 1B). Under normal conditions, this codon is decoded by Arg-tRNAArgICG with canonical Watson-Crick base pairs at the three positions. We reasoned that the oxidized complex, if 8-oxoG base pairs with A, should be decoded by Leu-tRNALeuGAG (Figure 1C). The complexes were reacted with different aa-tRNAs isoacceptors as well as the two release factors (RF1 and RF2) for 5 seconds, which should be sufficient to reach the endpoint of a normal peptidyl-transfer reaction (Figure 1D). The production of dipeptides or RF-mediated hydrolysis was visualized using an electrophoretic TLC system (Youngman et al., 2004). At first glance, the survey appears to agree well with our prediction. The intact complex efficiently reacts with Arg-tRNAArg and not with Leu-tRNALeu; the oxidized complex reacts poorly with Arg-tRNAArg but slightly better with Leu-tRNALeu, relative to the native one. Furthermore, the data suggests that the adduct affects the decoding process in a more complicated fashion than we initially anticipated; there are a set of aatRNAs that decode the intact codon but appear not to decode the modified codon (Figure 1D). Nonetheless, the reactivity of the damaged complex with the near-cognate aa-tRNA appears to proceed with an overall poor efficiency relative to normal peptide-bond formation (Figure 1E) indicating that the oxidized base may have a substantial effect on the speed of the ribosome.
Having established the crude reactivity of the complex harboring the damaged mRNA, we next determined the effects of the adduct on the rate of peptide-bond formation with cognate and near-cognate aa-tRNAs. A comparison of rates for the reaction between the CGC and C8-oxoGC complexes and the cognate Arg-tRNAArg showed that the complex displaying the modified nucleotides reacted much slower than the intact one; the apparent rate was determined to be more than 1,000-fold slower (Figure 1F). These findings suggest that 8-oxoG is likely to stall the ribosome, unless the rate with the near-cognate ternary complexes is greatly accelerated. In agreement with our endpoint analysis (Figure 1D), the apparent rate for the 8-oxoG complex with the near-cognate Leu-tRNALeu ternary complex (where G mispairs with A in the second position), albeit faster than the immeasurably slow rate for the intact complex, was very slow (0.03 s−1) – likely too slow to support efficient protein synthesis in vivo (Figure 1G). This was unexpected as our initial prediction was that 8-oxoG would lead to incorporation of the near-cognate aa-tRNA and produce some level of miscoding.
The effect on decoding is insensitive to the position of the lesion
The first two positions of the codon and anticodon minihelix require strict Watson-Crick base pairing, whereas the third position allows some wobble base pairing. Additionally, the decoding center of the ribosome utilizes different interactions to recognize the correct geometry for the different positions (Ogle et al., 2001). As a result, we reasoned that the position of the modification within the codon might have differential effects on tRNA selection by the ribosome. In particular, we expected the decoding process to be normal in the presence of damage at the third position of the codon. To address this hypothesis, we generated three new sets of complexes , placing the 8-oxoG at either the first, second or third position of the codon (see Figure S2). In each case, the 8-oxoG complex reacted much less efficiently with the cognate aa-tRNA than its intact counterpart. The complexes with 8-oxoG in the first or second position instead reacted more efficiently with the predicted (if 8-oxoG pairs with A, or G at the wobble position) near cognate aa-tRNA, but again with an overall poor yield. Lastly the third set of complexes, carrying the 8-oxoG at the wobble position, reacted poorly with its cognate aa-tRNA, in direct contrast to our prediction that the third position is insensitive to damage. Furthermore, the complex reactivity with near cognate aa-tRNAs displayed a distinct profile relative to its intact counterpart, highlighting the detrimental effect of the adduct on the decoding process even at the third position of the codon (Figure S2). Taken together, the data reveals an unprecedented and comprehensive overview of the potential havoc caused by oxidative damage to mRNA on the translation machinery.
We next tested if modification of other positions would elicit a less profound effect on the rate of peptide-bond formation. We measured the rate of peptide-bond formation for the same set of complexes described earlier with their respective cognate and near cognate aa-tRNAs. Cognate aa-tRNA incorporation was three to four orders of magnitude slower for 8-oxoG codons than undamaged controls (Figure 2A). Conversely, 8-oxoG codons incorporated near-cognate aa-tRNAs nearly tenfold faster than intact codons, although the observed rates (0.002-0.03 s−1) remained dramatically slower than that measured for normal peptide-bond formation (20-40 s−1) (Figure 2A). These observations collectively argue that 8-oxoG likely stalls the elongation phase of translation regardless of its location within the anticodon. Furthermore the drastic inhibitory effect of 8-oxoG on the decoding process was confirmed under competitive conditions. In particular, in contrast to the native GGC complex, which produced the expected full-length peptide when incubated in the presence of the full complement of aa-tRNAs, elongation factors and release factors (PURE system, NEB), the oxidized G8-oxoGC complex failed to produce any detectable peptide products (Figure 2B).
Figure 2. 8-oxoG inhibits peptide-bond formation.
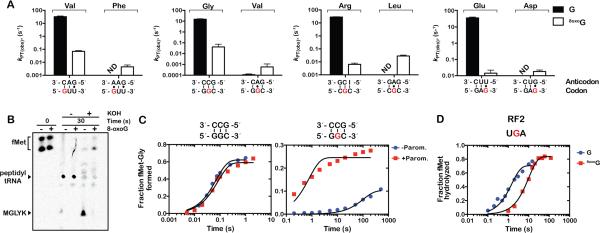
(A) Observed rates for peptidyl transfer (kPT) measured on native (GUU, GGC, CGC, GAG) and 8-oxoG complexes (8-oxoGUU, G8-oxoGC, C8-oxoGC, GA8-oxoG) with cognate - left two bars in each graph - and near-cognate (A:G mismatches at the 8-oxoG position) – right two bars in each graph tRNAs. The codon-anticodon interaction is shown below the x-axis and the corresponding dipeptide is shown above the bars. Clear bars represent rates observed with native complexes; black ones represent those observed with 8-oxoG complexes. Reactions were carried out at least in duplicate ± SEM. (B) Phosphorimager scan of an electrophoretic TLC of reactions between native complex (GGC) or 8-oxoG complex (G8-oxoGC), which encode MGLYK peptide, and the full complement of aa-tRNAs, elongation and release factors (PURE system, NEB). Reactions were allowed to proceed for 30 seconds before addition of 100mM KOH. (C) Time-courses of peptide-bond formation between the native GGC complex and Gly-tRNAGly (left panel) or oxidized G8oxoGC complex and Gly-tRNAGly (right panel) in the absence (blue circles) and presence of paromomycin (red squares). (D) Time-courses of RF2-mediated release on native UGA (blue circles) and U8-oxoGA (red squares) initiation complexes.
To provide mechanistic insight into the deleterious effects of oxidized mRNA on the decoding process, we set out to explore if the effects of 8-oxoG on peptidyl transfer result from inhibition of conformational changes known to be important for the decoding process (Ogle et al., 2002). In particular, we added the aminoglycoside paromomycin to our reaction, which binds the decoding center and induces a conformation in the 30S subunit similar to that observed when cognate tRNA is bound; in doing so, it allows the ribosome to accept near-cognate tRNAs as if they were cognate ones (Carter et al., 2000). As expected (Pape et al., 2000), the addition of paromomycin to a reaction between the GGC complex and its cognate Gly-tRNAGly ternary complex had no effect on the observed rate of peptide-bond formation (Figure 2C left panel). In contrast, addition of the antibiotic to the oxidized G8oxoGC complex accelerated the rate of peptide-bond formation more than 100-fold (Figure 2C right panel). These observations strongly argue that 8-oxoG prevents the small subunit from adopting the active conformation required for proper tRNA selection.
Peptide release is only marginally affected in the presence of 8-oxoG
Two of the three stop codons contain a guanosine: UAG and UGA. In E. coli these are recognized by release factor 1 (RF1) and release factor 2 (RF2), respectively. Stop-codon recognition by RFs is inherently different than sense-codon recognition by aa-tRNA due to the intrinsic differences in the polymer (protein vs RNA) carrying out the recognition (Zaher and Green, 2009a). Unlike aa-tRNA selection, which requires many residues of the decoding center to adopt a distinct conformation from the ground state, release-factor selection involves fewer and dissimilar conformational changes within the decoding center (Korostelev et al., 2008; Laurberg et al., 2008; Weixlbaumer et al., 2008). As a result we reasoned that the effects of 8-oxoG on sense-codon recognition could be different than those determined for RF-recognition on stop codons. Since we expected 8-oxoG to have the greatest effect at the second position, we programmed ribosomes with f-[35S]-Met-tRNAfMet in the P site and either a UGA or U8-oxoGA in the A site and assessed their reactivity with RF2. In direct contrast to peptide bond formation, the presence of 8-oxoG only marginally affected the rate of peptide release (0.7 s−1 and 0.1 s−1 for control and 8-oxoG respectively) (Figure 2D). Together, these results suggest that 8-oxoG prevents conformational changes associated with RNA-RNA interactions (i.e. codon-anticodon), but not protein-RNA interactions (i.e. RF-stop codon) in the decoding center.
8-oxoG inhibits protein synthesis in cell extracts
So far, our characterization of the effects of 8-oxoG on the decoding process has focused on initiation complexes in a bacterial reconstituted system and the effects of adducts on authentic elongation during protein synthesis were not discerned. Nevertheless, a number of studies have looked at the consequences of oxidative RNA damage on its function and all hinted at a profound correlation between oxidized mRNA and reduced translational efficiency and accuracy (Shan et al., 2007; Shan et al., 2003; Tanaka et al., 2007). However, in all of these studies the identities of the adducts formed from the crude hydrogen peroxide treatment and their relative abundance was not quantified. Furthermore, the effect of the treatment on the yield of full-length protein products was only marginal at best (Tanaka et al., 2007), presumably due to the inefficiency of the hydrogen peroxide treatment.
To improve upon this and provide a better understanding of the effects of the adduct on translation, we generated synthetic reporter mRNAs containing the lesion at a specific location within the coding sequence. We used in vitro transcription to synthesize a ~300 nt transcript and ligated this to an 8-oxoG-containing synthetic RNA oligonucleotide (Figure S3). We note that the final construct has the modification on the first position of a GAC codon. We also constructed three control reporters (one with the equivalent native codon, a second with a stop codon and a third shorter transcript with no stop codon) (Figure 3A and Figure S3). Using these reporters, we could assess how detrimental the 8-oxoG adduct is to translational speed and whether our reconstituted system recapitulates what happens during bona fide translation in bacterial and eukaryotic extracts. We initially analyzed the translation of these reporters in an S30 bacterial extract. In agreement with the reconstituted system, the 8-oxoG transcript produced a protein product of a size similar to that of the stop transcript (Figure 3B). A priori, one would expect the incubation of 8-oxoG and no-stop transcripts in the S30 extract to result in the accumulation of peptidyl-tRNA as these transcripts are expected to stall the ribosome, unless a ribosomal rescue system is operational in these extracts. Interestingly, we observe no peptidyl-tRNA accumulation with either reporter, even though the products were resolved on bis-Tris gels (Figure 3B), which are known to maintain the integrity of peptidyl-tRNA.
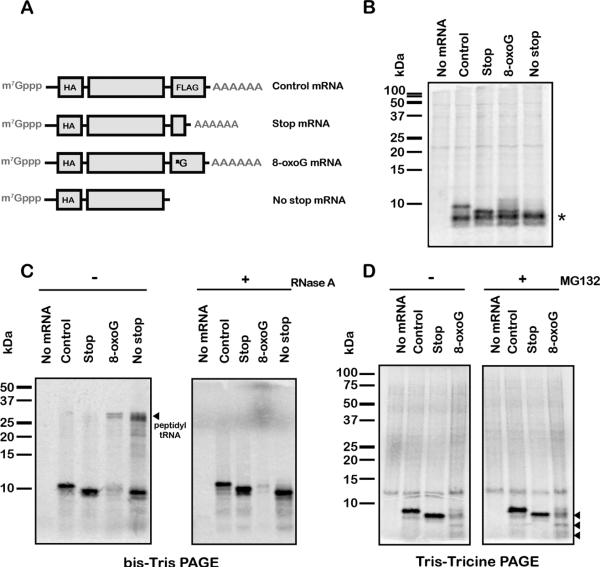
Figure 3. 8-oxoG stalls translation in cell extracts.
(A) Schematic of the mRNA reporters used in eukaryotic extracts. The full-length mRNA encodes a peptide that has a molecular weight of ~9 kDa, whereas the stop mRNA, which has a stop codon at the position of the 8-oxoG codon in the 8-oxoG mRNA, encodes a peptide that has a molecular weight of ~8.4 kDa. Reporters used in S30 reactions were identical minus the cap and polyadenylation. (B) Autoradiograph of a bis-Tris gel of translation assays using bacterial S30 extracts. The 8-oxoG transcript yields a peptide of a size similar to the stop reporter. * indicates non-specific band. (C) Autoradiograph of bis-Tris and Tris-Tricine gels of in vitro translation assays in wheat germ extracts. Proteins were labeled by the addition of [35S]-Methionine to the reactions. The 8-oxoG mRNA and no-stop mRNA both accumulate peptidyl-tRNA, visible on the bis-Tris gel, which disappear upon the addition of RNase A. Reactions separated on Tris-Tricine were incubated with and without MG132. The 8-oxoG mRNA produces truncated protein products (marked by arrows); the largest of these has a size similar to that observed in the presence of stop mRNA. See also Figure S3.
We next asked whether the inhibitory effects of 8-oxoG on translation were evident in eukaryotic extracts. For efficient translation in eukaryotes, the transcripts were capped using vaccinia capping enzyme and polyadenylated (except for the no stop reporter) using E. coli polyA polymerase. Consistent with our bacterial studies, 8-oxoG prevented the translation of full-length protein. The 8-oxoG reporter, when incubated in wheat germ extracts or rabbit reticulocyte lysates, failed to produce the full-length protein product and instead generated a product of similar length to that of the stop codon reporter (Figure 3C,D and Figure S3). However, in contrast to the bacterial extract results, we could detect accumulation of peptidyl tRNA in the presence of the 8-oxoG and no-stop transcripts suggesting that the ribosomal rescue system in the wheat germ extract is not efficient at clearing a stalled ribosome on the oxidized RNA. Additionally, the yield of the truncated protein products is much less than those produced in the presence of the control and stop mRNA, suggesting that 8-oxoG indeed inhibits the progress of translation. To gain an improved resolution of the products being produced from these reactions, we utilized Tris-Tricine gels to resolve the peptide products. With this gel system, we noted the appearance of several smaller protein bands in the presence of 8-oxoG mRNA (Figure 3D), likely resulting from ribosomes stacked behind the ribosome that stalls on the adduct. Furthermore, we observed no improvement in the yield of the truncated protein products upon addition of the proteasome inhibitor MG132 (Figure 3D) suggesting that the diminished yield of the truncated product is likely due to slowed translation and not to turnover of the protein products. We also note that incubation of labeled RNA in extracts did not reveal significant degradation of the RNA during the course of the reaction (Figure S3). These observations strongly suggest that, similar to what we observe in the bacterial reconstituted system, 8-oxoG halts translational elongation in eukaryotes.
8-oxoG mRNAs accumulate in vivo in the absence of no-go decay
Three mRNA-quality control processes are known to exist in eukaryotes, nonsense-mediated decay, no-go decay and non-stop decay. In no-go decay (NGD), transcripts containing secondary structures such as hairpins or stretches of rare codons are cleaved just upstream of the ribosome (Doma and Parker, 2006; Tsuboi et al., 2012). In yeast, the process involves the proteins Dom34, Hbs1 and Rli1 that together are responsible for recognizing and subsequent recycling of the stalled ribosome (Pisareva et al., 2011; Shoemaker et al., 2010; Shoemaker and Green, 2011). The process results in an endonucleolytic cleavage of the mRNA; the 3’-end piece is then degraded by the 5’-3’ exonuclease Xrn1 and the 5’-end piece is degraded by the exosome (Tsuboi et al., 2012). The exact role of these various quality control processes in recognizing damaged RNA (e.g. oxidized RNA) has not been studied. Intriguingly, however, quality control processes have been linked to damaged RNA where enzymatic depurination at specific sites within a transcript was found to stall the ribosome and is likely to elicit no-go decay (Gandhi et al., 2008).
Our observation that the 8-oxoG modification stalls protein synthesis suggests that 8-oxoG-containing mRNAs might be subject to degradation through no-go decay. To address this possibility, we quantified 8-oxoG levels in total and polyA-enriched RNA from different yeast strains using a competitive ELISA protocol (Yin et al., 1995) that specifically recognizes the oxidized base (Figure S4). Interestingly, the levels of the adduct in wild-type cells under normal conditions were approximately 5-fold higher in polyA-enriched RNA than in total RNA (~0.03 ng 8-oxoG/μg total RNA versus ~0.15 8-oxoG/μg polyA-enriched RNA), suggesting that mRNAs are more susceptible to oxidative stress, perhaps due to their naked nature and reduced secondary structure relative to rRNA or tRNA. Furthermore, we observe little to no change in the level of 8-oxoG adducts in total RNA in the absence of quality control or mRNA-decay pathways. In contrast, the levels of 8-oxoG in polyA-enriched RNA increase significantly by almost 2-fold (1.85-fold ±0.17, p=0.028) in the absence of Dom34 (Figure 4A), suggesting that 8-oxoG-containing mRNA is subject to NGD and is stabilized in the absence of Dom34. Additionally, Xrn1-deficient cells show a significant accumulation of 8-oxoG (2.18-fold ±0.25, p=0.011), whereas abrogating exosome function had no effect (Figure S4). We next sought to confirm that the changes in 8-oxoG levels are not the result of strain to strain variation by complementing the dom34Δ and xrn1Δ strains and measuring the levels of 8-oxoG in these rescued strains (dom34Δ and xrn1Δ comp., respectively). These strains were complemented by integrating the native genes at the his3 locus of chromosome XV (under the control of the endogenous promoter and carrying the endogenous UTR), resulting in transcript levels that were similar to wild-type (Figure S4). Consistent with our proposal, 8-oxoG levels were restored to wild-type upon reintroduction of Dom34 and Xrn1 (Figure 4A). It is worth noting that our measurements of 8-oxoG levels are likely an underrepresentation, since our polyA-enriched samples still contain some rRNA (Figure S4). Furthermore, the twofold increase in the steady-state levels of 8-oxoG in the absence of Dom34 and Xrn1 is consistent with the effects of these factors on the steady-state levels of typical NGD reporters (Tsuboi et al., 2012).
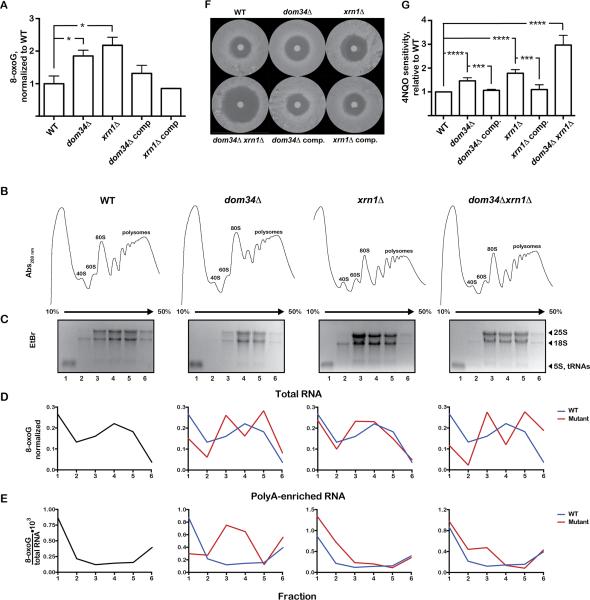
Figure 4. 8-oxoG mRNA is stabilized in yeast in the absence of no-go-decay factors and associates with polysomes in the absence of Dom34.
(A) Bar graph showing the levels of 8-oxoG, relative to WT, in polyA-enriched RNAs isolated from the indicated yeast strains. The values shown are the means of at least three measurements ±SEM. * p<0.05 (B) Polysomes profiles of the indicated strains. (C) Denaturing agarose electrophoresis of the RNA samples isolated from the different fractions in the sucrose gradient. (D, E) Distribution of 8-oxoG levels across the gradients in total RNA and polyA-enriched RNA samples, respectively. Levels of 8-oxoG in (D) were normalized to the total amount of 8-oxoG across the gradient, whereas in (E) they were divided by the amount of polyA-enriched RNA. (F) Representative images of zone of inhibition assays on the indicated yeast strains using 4-nitroquinoline-1-oxide (4NQO). (G) Quantification of 4NQO sensitivity for the indicated strains, relative to WT. Data are mean ±SEM, from results of at least three biological replicates. *** p<0.001, **** p<0.0001 See also Figure S4.
These observations are in full agreement with our current understanding of the NGD process, wherein: 1) translation arrest results in an endonucleolytic cleavage upstream of the stall sequence (in this case 8-oxoG); 2) ribosomes are recycled by Dom34:Hbs1; 3) the 3’-fragment, i.e. 8-oxoG containing, is degraded by Xrn1 whereas the 5’-fragment is degraded by the exosome (Doma and Parker, 2006; Tsuboi et al., 2012). It is worth noting that while endonucleolytic cleavage is not directly mediated by Dom34:Hbs1, it is significantly stimulated in the presence of these factors (Tsuboi et al., 2012). Two key predictions emerge from this model: 1) In the absence of Dom34, 8-oxoG-containing mRNA is more likely to associate with the ribosomes; 2) In the absence of Xrn1, 8-oxoG-containing mRNA is stabilized but would not be associated with the ribosomes. We set out to test these predictions by performing polysome analysis of the different yeast strains and quantifying the distribution of 8-oxoG among the different fractions of RNPs (Figure 4B-E). xrn1Δ strains have been shown to accumulate polyA mRNAs in the light fraction of the polysome due to stabilization of uncapped mRNA that cannot be translated (Hu et al., 2009). To correct for this, we normalized the levels of 8-oxoG to the total amount of RNA during our polysomes analysis of polyA-enriched RNA, which should adjust for variation in polyA-RNA occupancy across the sucrose gradient. As expected, for all strains we observed very similar distributions of 8-oxoG in total RNA (Figure 4D), consistent with our measurements from whole cells (Figure S4). However, in the absence of Dom34, 8-oxoG levels in polyA-enriched RNA are significantly higher in the 80S and polysome fractions (Figure 4E) demonstrating that Dom34p plays a role in clearing stalled ribosomes on damaged mRNA. Additionally, in the absence of Xrn1, 8-oxoG levels were elevated in the light fractions, containing non-ribosome-bound RNAs, and unaltered in the 80S and polysome fractions. Supporting the model that Dom34 acts upstream of Xrn1 in NGD, Xrn1/Dom34 double mutant 8-oxoG levels were similar to those of Dom34 (Figure 4E).
We next hypothesized that if no-go decay is important in rescuing ribosomes stalled on damaged mRNA, then deleting dom34 and xrn1 should make yeast cells sensitive to nucleic-acids damaging agents. In particular, we were interested in determining the sensitivity of the different yeast strains to the carcinogen 4-nitroquinoline-1-oxide (4NQO), which is known to react with RNA to form the 8-oxoguanine lesion (Tada and Kohda, 1993). We utilized a zone of inhibition (halo) assay to compare the growth of the deletion strains in the presence of 4NQO (Figure 4F). Consistent with a role for no-go decay in clearing damaged mRNA, deletion of dom34 and xrn1 rendered yeast cells significantly more sensitive to 4NQO (1.5-fold ±0.05, p<0.0001 and 1.8-fold ±0.06, p<0.0001, respectively), and reintroduction of the genes restored normal growth relative to the wild-type strain (Figure 4G). Finally, the dom34Δ xrn1Δ double mutant was found to exhibit dramatic sensitivity to 4NQO compared to the single mutants (3.0-fold ±0.18, p<0.0001) (Figure 4G). These observations strongly suggest that Dom34 and Xrn1 protect the cell from the effects of RNA-damaging agents.
DISCUSSION
Here, we demonstrated that mRNA containing the oxidized nucleotide 8-oxoguanosine stalls the ribosome and that NGD may have evolved to clear these and other aberrant mRNAs. A priori we expected the adduct, based on its chemical properties, to increase miscoding by allowing G:A mismatches between the codon and anticodon. In contrast to this prediction, 8-oxoG was found to inhibit tRNA selection in a well-defined reconstituted system, with rates of peptide-bond formation that are three to four orders of magnitude slower than those measured on intact complexes. Remarkably, the effects of the modifications on the accuracy of tRNA selection appear to be minimal; the rates with near-cognate ternary complexes in the presence of 8-oxoG are slightly faster (<10-fold) relative to intact complexes. Furthermore, the position of the adduct, even at the wobble position, resulted in a similar effect on the decoding process highlighting the severity of mRNA oxidation on the protein synthesis machinery. In agreement with observations from the reconstituted systems, we find near complete inhibition of protein synthesis in the presence of 8-oxoG during authentic elongation in eukaryotic extracts. The effects of 8-oxoG on the decoding process appear to be a direct result of impaired RNA-RNA interaction between the codon and anticodon, as recognition of stop codons by protein factors was only marginally affected.
How can a small change to the nucleobase result in such drastic effects on the decoding process? One could imagine that the addition of the carbonyl oxygen to the non Watson-Crick face of the base introduces a polar group that needs to be hydrated. The introduction of this energetic penalty is likely to be inhibitory to the decoding process. On the other hand, in solution 8-oxoG exists in equilibrium between the anti and syn conformation, but on the ribosome the adduct might favor the anti conformation, which is detrimental to proper A-minor interaction in the decoding center. Regardless of the conformation of 8-oxoG in the decoding center, the modification appears to prevent the decoding center from adopting an active conformation. This is supported by our data showing that the addition of paromomycin alleviates most of the inhibitory effects observed in the presence of 8-oxoG. These observations highlight the sensitive nature of the decoding center, which is reinforced by recent reports demonstrating a uridine to pseudouridine change (a small change that does not affect the base-pairing properties) has a dramatic affect on the decoding process and increases stop-codon readthrough (Fernandez et al., 2013; Karijolich and Yu, 2011).
Our in vitro studies suggested that oxidized RNA is subject to no-go decay. No-go decay was initially characterized by Doma and Parker (Doma and Parker, 2006), who showed that mRNAs with strong secondary structure or stretches of rare codons are targeted for degradation in a process that is dependent on translation. While mRNAs with hairpins for the most part are not biologically relevant, this study established a direct relationship between translational speed and rapid degradation of the corresponding mRNA. Consistent with this, we find 8-oxoG mRNA to be stabilized in the absence of Dom34 (a key component in no-go decay), and associated with polysomes. These observations strongly suggest oxidized mRNA is a primary target of ribosome-based mRNA quality control processes. We note that no-go decay and in particular Dom34 has been implicated in a number of processes; for example Inada and colleagues have shown the factor to be important in rescuing ribosomes trapped on truncated mRNA (Tsuboi et al., 2012). Recently Guydosh and Green used ribosomal profiling to show that in the absence of the factor, ribosomes venture into the 3’-UTR (Guydosh and Green, 2014). Furthermore, the factor is critical for nonfunctional rRNA decay (NRD) (LaRiviere et al., 2006; Soudet et al., 2010) and ribosomal assembly (Strunk et al., 2012). Interestingly, all of these processes share a common feature whereby the target for the factors is a stalled ribosome with an empty A site (van den Elzen et al., 2014). It is highly likely that the list of NGD targets is long and may include other modified mRNAs such as those resulting from alkylative and UV damage.
Oxidative damage to RNA appears to be correlated with a number of age-related neurodegenerative diseases and is likely to be a contributing factor to their pathogenicity. As an example, Shan et al. showed that in Alzheimer's patients, greater than 50% of mRNA isolated from cerebral cortex were oxidized (Shan and Lin, 2006). Our results indicate that RNA damage represents a real and serious threat to the cell and RNA surveillance pathways such as NGD may have evolved to provide a mechanism to cope with these damages. RNA metabolism could be a useful target for delaying or eliminating the onset of age-related neurodegenerative diseases.
EXPERIMENTAL PROCEDURES
Detailed methods are provided in Supplemental Experimental Procedures
Dipeptide formation survey/rates of peptide-bond formation
For survey reactions, equal volume of purified initiation complexes (2 μM), pre-incubated at 37°C, was added to ternary complex. The mixture was incubated at 37°C for 5 seconds before the reaction was quenched by the addition of 100 mM KOH. Peptide release was carried out in a similar manner, except initiation complexes were combined with RF1 or RF2 at 10 μM and the reaction stopped using 20 mM EDTA pH 6.0. To determine rates of peptide-bond formation, equal volumes of initiation and ternary complexes were mixed on a quench-flow instrument (RQF-3 quench-flow, KinTek Corporation). For all reactions, dipeptides were resolved from unreacted fMet using cellulose TLCs electrophoresis in Pyridine-acetate buffer (pH 2.7) (Youngman et al., 2004).
To determine the observed rates of peptide-bond formation or peptide release, dipeptide yield (PT rates) or fraction of released fMet (peptide release) was plotted against time and the curve fit to a first-order rate equation using GraphPad prism software. Observed rates reported were calculated by multiplying the fit rate with the end point and then divided by the maximal end point (observed with the native complexes).
ELISA
8-oxoG was quantified using a competitive ELISA assay. Each experiment included a set of standards (8-oxoG in 0.1% BSA), enabling quantification by a standard curve (Abs = (Absmax)/(1 + ([8-OHG]/IC50)).
Zone of inhibition assay
Yeast cultures were grown to mid-log phase and plated in triplicate at 0.5 OD per plate. After drying thoroughly, 25 μg of 4-nitroquinoline-1-oxide in DMSO was spotted on a 0.5 cm filter paper disc and placed in the center of the plate. Plates were incubated at 30°C for ~48 hours before imaging. The area of inhibition was measured using ImageJ. Data were normalized to WT and analyzed using Student's t-test.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by an NIH grant (R00GM094210) and funding from the Searle Scholars Program to H.S.Z. We thank Allen Buskirk and Joseph Jez for helpful comments on the manuscript and members of the laboratory for useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
C.L.S, B.H.H. and H.S.Z. designed the experiments and wrote the manuscript. C.L.S, B.H.H., J.W.M., A.S.R. and H.S.Z. performed the experiments.
REFERENCES
- Barciszewski J, Barciszewska MZ, Siboska G, Rattan SI, Clark BF. Some unusual nucleic acid bases are products of hydroxyl radical oxidation of DNA and RNA. Mol. Biol. Rep. 1999;26:231–238. doi: 10.1023/a:1007058602594. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez IS, Ng CL, Kelley AC, Wu G, Yu YT, Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500:107–110. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Gandhi R, Manzoor M, Hudak KA. Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. J. Biol. Chem. 2008;283:32218–32228. doi: 10.1074/jbc.M803785200. [DOI] [PubMed] [Google Scholar]
- Graille M, Seraphin B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat. Rev. Mol. Cell Biol. 2012;13:727–735. doi: 10.1038/nrm3457. [DOI] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer T, Badouard C, Bajak E, Ravanat JL, Mattsson A, Cotgreave IA. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol. Chem. 2005;386:333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474:395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. USA. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere FJ, Cole SE, Ferullo DJ, Moore MJ. A late-acting quality control process for mature eukaryotic rRNAs. Mol. Cell. 2006;24:619–626. doi: 10.1016/j.molcel.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- Li Z, Wu J, Deleo CJ. RNA damage and surveillance under oxidative stress. IUBMB life. 2006;58:581–588. doi: 10.1080/15216540600946456. [DOI] [PubMed] [Google Scholar]
- Nunomura A, Perry G, Pappolla MA, Wade R, Hirai K, Chiba S, Smith MA. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer's disease. J. Neurosci. 1999;19:1959–1964. doi: 10.1523/JNEUROSCI.19-06-01959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Chang Y, Lin CL. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 2007;21:2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- Shan X, Lin CL. Quantification of oxidized RNAs in Alzheimer's disease. Neurobiol. Aging. 2006;27:657–662. doi: 10.1016/j.neurobiolaging.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Shan X, Tashiro H, Lin CL. The identification and characterization of oxidized RNAs in Alzheimer's disease. J. Neurosci. 2003;23:4913–4921. doi: 10.1523/JNEUROSCI.23-12-04913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Wu W, Hazen SL. Activated leukocytes oxidatively damage DNA, RNA, and the nucleotide pool through halide-dependent formation of hydroxyl radical. Biochemistry. 2000;39:5474–5482. doi: 10.1021/bi992809y. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. USA. 2011;108:E1392–1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2010;29:80–92. doi: 10.1038/emboj.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Kohda K. Identification of N4-(guanosin-7-yl)-4-aminoquinoline 1- oxide and its possible role in guanine C8 adduction. Nucleic acids symposium series. 1993:25–26. [PubMed] [Google Scholar]
- Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proc. Natl Acad. Sci. USA. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- van den Elzen AM, Schuller A, Green R, Seraphin B. Dom34-Hbs1 mediated dissociation of inactive 80S ribosomes promotes restart of translation after stress. EMBO J. 2014;33:265–276. doi: 10.1002/embj.201386123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtmann EJ, Wolin SL. RNA under attack: cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 2009;44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE., Jr. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin B, Whyatt RM, Perera FP, Randall MC, Cooper TB, Santella RM. Determination of 8-hydroxydeoxyguanosine by an immunoaffinity chromatography-monoclonal antibody-based ELISA. Free Radical Biol. Med. 1995;18:1023–1032. doi: 10.1016/0891-5849(95)00003-g. [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009a;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009b;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.