Abstract
Rationale
The senescent cardiac phenotype is accompanied by changes in mitochondrial function and biogenesis causing impairment in energy provision. The relationship between myocardial senescence and Pim kinases deserves attention since Pim-1 kinase is cardioprotective, in part, by preservation of mitochondrial integrity. Study of the pathological effects resulting from genetic deletion of all Pim kinase family members could provide important insight regarding cardiac mitochondrial biology and the aging phenotype.
Objective
Demonstrate myocardial senescence is promoted by loss of Pim leading to premature aging and aberrant mitochondrial function.
Methods and Results
Cardiac myocyte senescence was evident at three months of age in Pim Triple KnockOut (PTKO) mice, where all three isoforms of Pim kinase family members are genetically deleted. Cellular hypertrophic remodeling and fetal gene program activation was followed by heart failure at six months in PTKO mice. Metabolic dysfunction is an underlying cause of cardiac senescence and instigates a decline in cardiac function. Altered mitochondrial morphology is evident consequential to Pim deletion together with decreased ATP levels and increased phosphorylated AMPK, exposing an energy deficiency in PTKO mice. Expression of the genes encoding master regulators of mitochondrial biogenesis, PPARγ coactivator-1 (PGC-1) α and β were diminished in PTKO hearts, as were downstream targets included in mitochondrial energy transduction, including fatty acid oxidation. Reversal of the dysregulated metabolic phenotype was observed by overexpressing c-Myc, a downstream target of Pim kinases.
Conclusion
Pim kinases prevent premature cardiac aging and maintain a healthy pool of functional mitochondria leading to efficient cellular energetics.
Keywords: Pim kinases, aging, metabolism, c-Myc, PGC-1α, hypertrophy
INTRODUCTION
Cardiac senescence and the aging phenotype are characterized by a multitude of changes on the cellular and organ level that occur over the lifetime of an organism. In contrast to rapid expansion of nascent cardiomyocytes taking place in postnatal development, de novo generation of myocytes in response to aging is very limited 1-3. The onset of ventricular hypertrophy at the cellular and organ level is a hallmark of cardiac aging, which compensates for losses in cellular density and concomitant diminution of functional hemodynamic output. The consequence of pathological cardiac hypertrophy is eventual alterations in mitochondrial metabolism and energy homeostasis promoting glucose utilization over fatty acid oxidation, exacerbating disease etiology 4-6. Preservation of mitochondrial integrity and function antagonizes aging, as myocardial senescence is associated, in part, with decreased mitochondrial content and altered metabolic function 7-9.
Transcriptional coregulators PPARγ coactivator-1 (PGC-1) α and β serve as critical regulators of mitochondrial biogenesis and cellular ATP producing pathways 5,10-12. PGC-1α and PGC-1β coactivate downstream transcription factors involved in mitochondrial biogenesis such as ERRα, NRF-1, and TFAM. PGC-1α is enriched and highly inducible in the heart. However, ablation of PGC-1α leads to compensatory upregulation of PGC-1β 4,5. PGC-1 coactivators regulate the mitochondrial fatty acid oxidation (FAO) pathway, which serves as the primary supply for bioenergetic fuel in the healthy adult heart. Heart failure and hypertrophy prompt reprogramming of fuel utilization to rely predominantly on glucose metabolism similar to the fetal heart 6. Downregulation of PGC-1 signaling and the cognate downstream target PPARα contributes to the fuel shift toward fetal metabolism in the hypertrophied heart presenting as metabolic dysfunction 4,13. Furthermore, transgenic mice with single knockdown of PGC-1α or PGC-1β demonstrate age-dependent contractile dysfunction and impaired mitochondrial function, whereas mice lacking PGC-1α and PGC-1β die shortly after birth from heart failure 4,5,14.
Pim-1, a conserved serine/threonine protein kinase, exerts multiple protective effects upon mitochondria and has recently been implicated in affecting metabolism through PGC-1α 15-17. Additionally, Pim-1 stabilizes and phosphorylates c-Myc, a known regulator of mitochondrial metabolism and biogenesis 17,18. Pim-1 also impacts upon mitochondrial dynamics through phosphorylation and cytosolic sequestration of Drp1 19. The Pim gene family consists of Pim-1, -2, and -3; three different genes transcribed from alternative start sites. All three Pim family members are constitutively active, exhibit similar substrate preferences, and differ primarily in tissue expression 20,21. Pim-1, the predominant isoform in the heart, can be genetically deleted in mice prompting compensatory upregulation of Pim-2 and -3 22,23.
Pim-1 is highly expressed in postnatal hearts but diminishes precipitously during postnatal development 22. Recently, our group documented the remarkable ability of Pim-1 overexpression to “rejuvenate” aged human cardiac stem cells by decreasing senescent markers, promoting proliferation, and survival 24. Taken together, these studies implicate Pim kinases in the maintenance and preservation of a “youthful” cellular phenotype. Since Pim-1 promotes mitochondrial integrity and antagonizes acquisition of an aging phenotype, the relationship between Pim kinase, mitochondrial biogenesis, cardiac senescence, and metabolism resulting from loss of Pim kinase activity are investigated in this report.
METHODS
Mice and Echocardiography
Echocardiography was performed under mild isoflurane sedation (.5 -1.5%) using a Vevo 770 High resolution system with wildtype FVB and PTKO mice 25. Cardiac function was analyzed in the parasternal long axis view tracking the endocardium with the supplied analysis software to obtain left ventricle inner diameter during diastole (LVIDd), left ventricle anterior wall thickness during diastole (LVAWd), ejection fraction (EF), and heart rate (HR). We used the PTKO mouse model described by Mikkers et al25. Briefly, Pim1 knockout mice were generated initially by homologus recombination using the targeting vector pGTP810 in FVB mice. Using Pim1 knockout mice PTKO mice were generated by deleting exons 1,2,and 3 for Pim2 with a PGK-Php cassette. Pim3 exons 3,4,5 and 6 were deleted using a promoterless IRES-βGeo cassette 25.
ATP assay
Lysates for ATP assay and citrate synthase assay were obtained from NTG and PTKO. Briefly, mouse hearts were extracted after cervical dislocation and immediately snap frozen in liquid nitrogen. Hearts were then homogenized in isolation buffer: 70mM Sucrose, 190mM Mannitol, 20mM HEPES solution, .2mM EDTA solution. Homegenized lysates were then employed in an ATP assay (Invitrogen) according to the manufacturer protocol.
Cell culture and viral transduction
Immortalized murine embryonic fibroblast from wildtype (iWT MEFs) and PTKO (iPTKO MEFs) were provided by Dr. Andrew S Kraft (Medical University of South Carolina, Charleston) and were cultured as previously described 15. Briefly, immortalized MEFs were maintained in DMEM (Invitrogen 11965-092) containing 10% fetal bovine serum and 1% penicillin-streptomycin-glutamine (PSG). Adenovirus EGFP was created and expanded as previously described 22,26. The PGC-1α adenovirus is identical to that used in Lehman et al. 2000 JCI 27. Adenovirus c-Myc was purchased from Vector Biolabs (Philadelphia, PA Catalog# 1285) and expanded. For adenoviral transduction iWT MEFs and iPTKO MEFs were infected in 2% fetal bovine serum DMEM for two hours followed by two wash steps of phosphate buffered saline and incubation in 10% DMEM and collected 24 hours later for analysis. A 50 multiplicity of infection was used for adenovirus EGFP, c-Myc, and 200 for PGC-1α.
NRCMs were isolated and maintained as previously published 19. Briefly, isolated NRCMs were maintained in .5% M199. siRNA transfections were performed as previously published using HiPerfect transfection reagent (Qiagen) 28,29. siRNA for Pim 1, 2, and 3 were obtained from Sigma and used at a concentration of 50nM.
Mitochondrial respiration assay and telomere measurements
Mitochondrial isolation procedures were followed as previously published 30. Telomeres were measured in NTG and PTKO hearts using quantitative fluorescence in-situ hybridization (Q-FISH) or multiplex quantitative PCR method as previously described 23,31. Further details are in supplemental information.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0 (Graphpad Software). Statistical analysis was performed using Student's t-test. Echocardiography time course analysis was assessed by two-way ANOVA with Bonferroni's post hoc test. A p value of less than 0.05 was considered statistically significant. Error bars represent SEM. Significance indicators are *P < 0.05, **P < 0.01, and ***P < 0.001.
RESULTS
Cardiac hypertrophy followed by premature cardiac failure is evident in PTKO mice
The expression of Pim kinases are differentially regulated in the myocardium over lifespan. Pim1 expression is downregulated by 53% starting at 100 days after birth and remains 50% decreased at 324 days compared to postnatal day 3 during physiological cardiac aging (p<.01 Online Fig I A). Pim2 expression does not change with cardiac aging (Online Fig I A). Pim3 expression is upregulated at postnatal day 14 by 2.2 fold and increases at 100 days and 324 days after birth by 2.6 and 3.9 fold, respectively compared to postnatal Day 3 (p<.01 Online Fig I A). During myocardial infarction Pim1 expression increases in the infarct area at Day 1 by 2.59 fold and gradually diminishes over time by 40% within the infarct region (Online Fig I B). Pim2 expression increases in the remote region at Day 1 by 2.62 fold and also diminishes over time (Online Fig I C). Pim3 expression starts to decrease at 1 day following infarction in both the remote and infarct region and by two weeks is decreased by 78% in infarct and remote region (Online Fig I D). Increased Pim expression early during myocardial infarction is consistent with a prosurvival role for Pim genes.
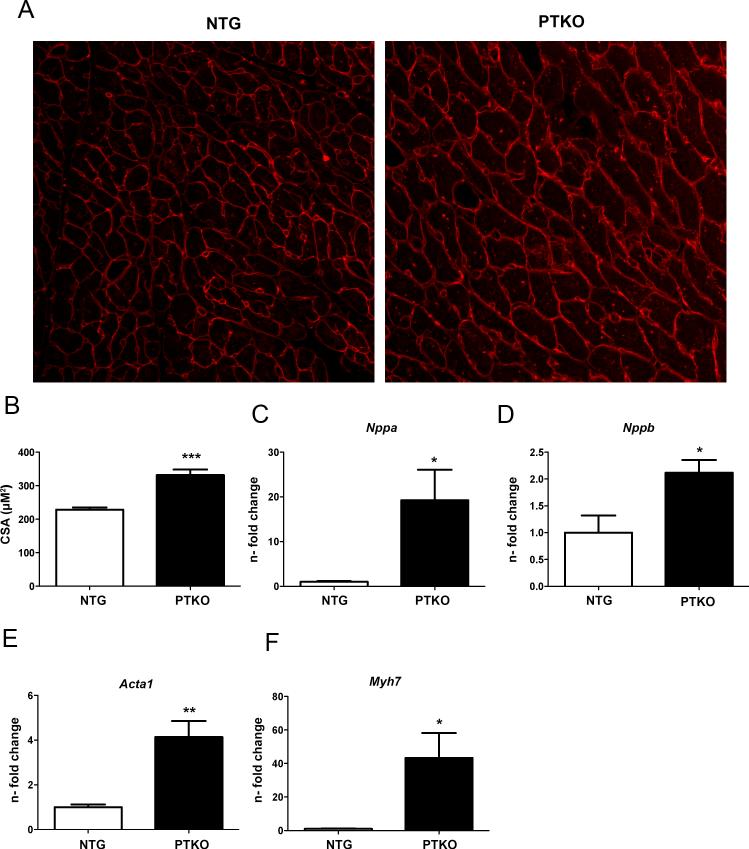
Pathological hypertrophic remodeling during cardiac aging is well documented 32,33, as is the ability of Pim-1 to antagonize myocardial hypertrophy in response to pressure overload 26. Expression of Pim1, 2,and 3 kinases decrease by 62%, 81%, and 76% at 2 weeks, respectively in response to transaortic constriction (Online Fig I E). Consistent with hypertrophic remodeling, PGC-1α expression is diminished with TAC by 59%, 72%, 88%, and 77% at 1 day, 4 day, 7 day, and 2 weeks (Online Fig I F). Therefore, blunting of cardiac hypertrophy previously reported following Pim1 overexpression 26,34 was compared to the effect of Pim kinase deletion in PTKO mice. Cell size area was increased by 1.4 fold in PTKO mice at one month (p<.001 Figure 1A & B). Increases were significant for Nppa (atrial natriuretic peptide; 19 fold (p<.05), Nppb (brain natriuretic peptide; 2 fold (p<.05), Acta1 (α-skeletal actin; 4 fold (p<.01), and Myh7 (β-myosin heavy chain; 43 fold (p<.05) following Pim deletion consistent with reactivation of the fetal gene program (Figure 1C-F). Phosphorylated ribosomal S6 and phosphorylated 4EBP1 were diminished by 53% and 52% respectively, indicating decreased mTOR activity (p<.001 and p<.05) Online Fig II A&B). However, consistent with hypertrophic remodeling phosphorylated Akt was increased by 1.4 fold and RCAN1.4 expression was upregulated by 4.4 fold in PTKO mice, a downstream readout for NFAT transcriptional activation in pathological hypertrophy (p<.001 and p<.05 Online Fig II C&D).
Figure 1. Characteristics of cardiac hypertrophy in PTKO mice.
A) Wheat germ agglutinin staining in cardiac sections demonstrating cell surface area of Non-Transgenic (NTG) and Pim Triple Knockout (PTKO) mice at 1 month. B) Quantification of cell size area (CSA) in heart tissue sections at one month (n=4 NTG and PTKO mice). C-F) mRNA levels of fetal cardiac gene markers: Nppa (atrial natriuretic peptide), Nppb (brain natriuretic peptide), αSKA (alpha skeletal actin), and β-MHC (beta myosin heavy chain) respectively. *, **, *** is significant compared to NTG mice. *P < 0.05; **P < 0.01; ***P < 0.001.
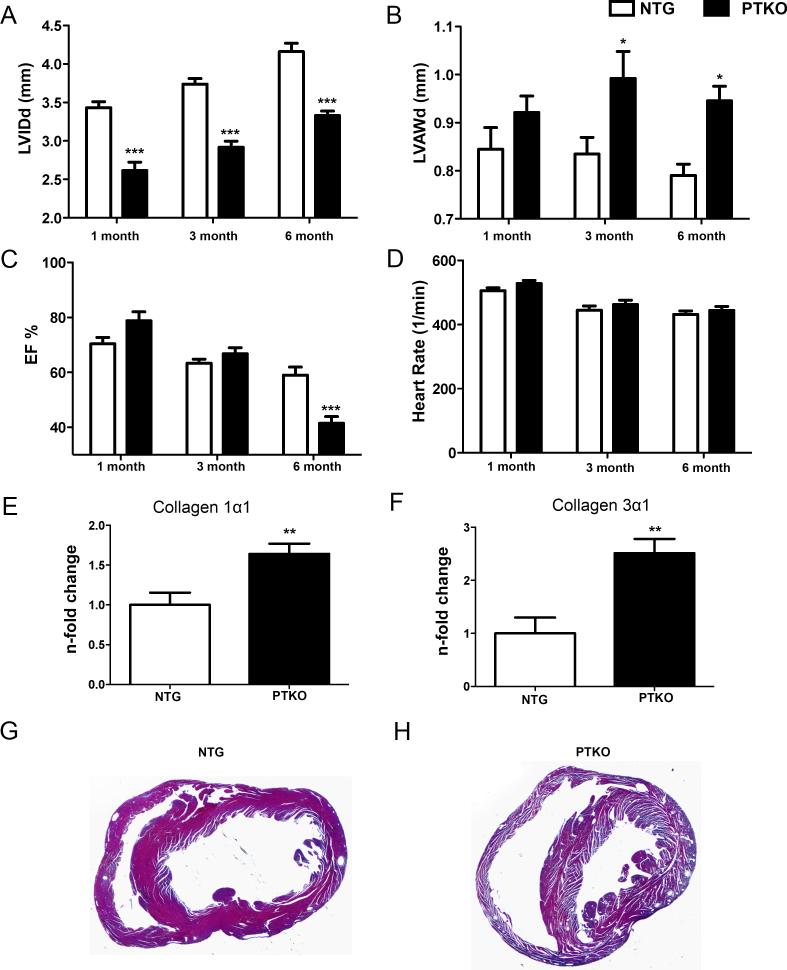
Body weight and heart weight were decreased by 22% (p<0.001) and 27% in PTKO mice, respectively (p<0.001; Online Fig III A & B). However, heart weight to body weight ratio was unchanged between PTKO and wild-type mice (Online Fig III C). Smaller left ventricle interior diameter during diastole was revealed by longitudinal echocardiography from one to six months of age in PTKO mice (p<0.001 Figure 2A). Furthermore, left ventricle anterior wall thickness during diastole demonstrated a strong trend toward increased wall thickness at one month and became significant at three months of age, showing an increase of 20% which persisted at six months (p<0.05 Figure 2B). A 30% reduction in ejection fraction was evident at six months in PTKO mice, indicating premature cardiac failure (p<0.001 Figure 2C). Heart rate was maintained at consistent levels to ensure comparability between individual measurements (Figure 2D). Consistent with cardiac hypertrophy and heart failure, markers of fibrosis collagen 1α1 and collagen 3α1 were upregulated 1.6 fold and 2.5 fold, respectively (p<.01 Figure 2 E&F). Furthermore, enlargement of the left ventricle in PTKO mice was evident by Massons trichrome staining (Figure 2 G&H). Collectively, these findings demonstrate hypertrophic remodeling in PTKO mice and progression toward failure at six months of age.
Figure 2. Premature cardiac failure at six months with Pim deletion.
A) Left ventricle inner diameter during diastole (LVIDd) in millimeters. B) Left ventricle anterior wall thickness during diastole (LVAWd) in millimeters. C) Percent ejection fraction. D) Average heart rate during echocardiography measurements. E-F) Gene expression analysis of collagen 1α1 and collagen 3α1 in cardiac tissue. G and H) Low magnification image of NTG and PTKO hearts with trichrome staining. n=8 NTG and n=9 PTKO. *, **, *** is significant compared to NTG mice at each individual time point. *P < 0.05; **P < 0.01; ***P < 0.001.
Markers of senescence are elevated with loss of Pim kinases
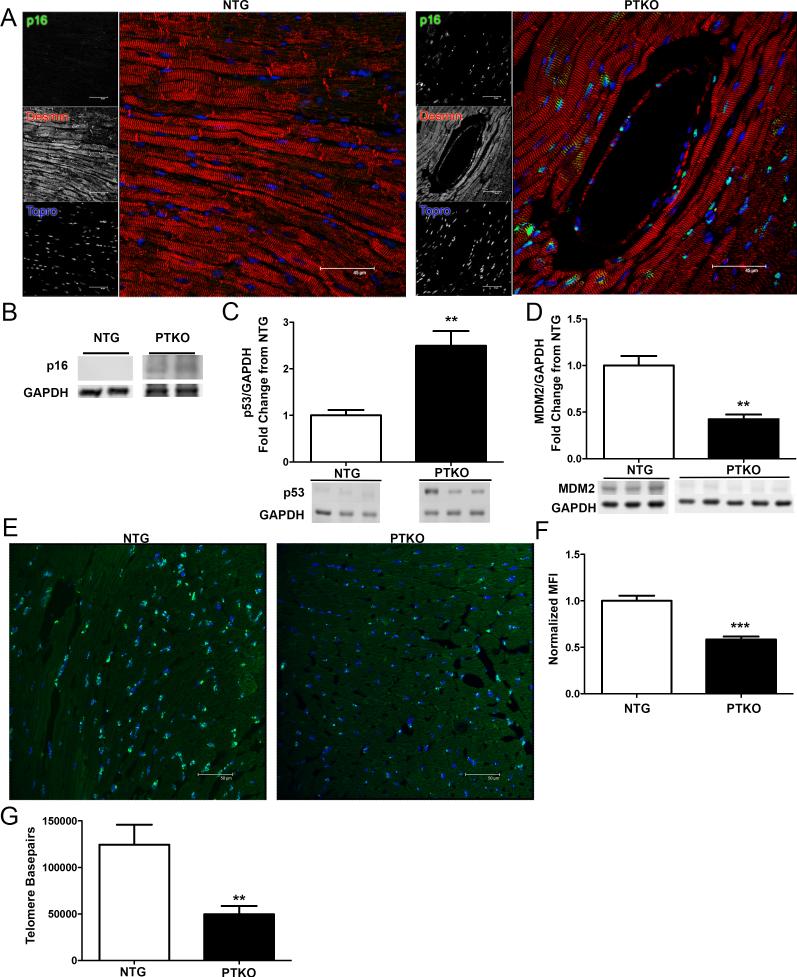
Senescent markers characterization was performed to determine the aging phenotype in PTKO mice. p16 expression was evident throughout the myocardium of PTKO mice at three months, indicating irreversible cellular senescence (Figure 3A). Increased p16 expression was confirmed by immunoblot in PTKO mice relative to non-transgenic controls (Figure 3B). p16 upregulation was validated by a concurrent reduction in p16 transcriptional repressors Id1 and Id2 by 60% and 64% in PTKO cardiac lysates respectively (p<0.05, p<.01 Online Fig IV A & B). Moreover, increased Ets-1 level (p16 transcriptional activator, 2.4 fold) also supports p16 induction (p<.01; Online Fig IV C). p53 showed a 2.5-fold increase in PTKO mice at three months (p<0.01 Figure 3C) consistent with reduction of MDM2 (p53 ubiquitin ligase MDM2; 57% lower (p<0.01; Figure 3D). Telomere mean fluorescence intensity was diminished by 41.5% in PTKO as measured by quantitative fluorescence in-situ hybridization (Q-FISH) (p<.001; Figure 3E & F). Additionally, telomere length was decreased by 40% in PTKO mice by qPCR analysis (p<.01; Figure 3G). The collective phenotypic profile observed is consistent with expectations for premature cardiac senescence and predisposition toward heart failure in PTKO mice.
Figure 3. Cellular senescence in PTKO mice.
A) Confocal images of p16 staining in NTG and PTKO mouse hearts. B-D) Protein analysis of p16, p53, and MDM2 respectively in total cardiac lysates NTG and n=3 PTKO mice at 3 months. E & F) Representative Q-FISH images and quantification normalized to mean fluorescence intensity (MFI) in NTG (n=3) and PTKO (n=3) mice at 3 months. G) q-PCR measurements of telomere lengths in NTG (n=6) and PTKO (n=6) at one month. *, **, *** is significant compared to NTG mice. *P < 0.05; **P < 0.01; ***P < 0.001.
Knockdown of Pim 1, 2, and 3 in neonatal rat cardiomyocytes (NRCMs) stimulated an increase in p53 by 1.6 fold (p<.05), whereas p16 expression was unchanged (Online Fig V A-D). Furthermore, NRCMs treated with physiological (insulin) or pathological (phenylephrine) growth stimulus promoted an increase in p53 with loss of Pim 1, 2, and 3 by 2 fold (p<.001) and 1.9 fold (p<.01) respectively (Online Fig V A&B). p16 gene expression was unchanged compared to respective scrambled controls during stress (Online Fig V C&D). Upregulation of Nppa by 1.4 fold (p<.05), Nppb by 1.4 fold (p<.05), and Myh7 by 2.5 fold was also noticed in NRCMs knocked down for Pim 1, 2, and 3, whereas alpha skeletal actin displayed a strong increasing trend (Online Fig V E). Validation of siRNA efficiency of resulted in 80%, 80%, and 67% decrease in gene expression of Pim1, 2, and 3 respectively (p<.001 Online Fig V F,G, and H). These data further endorse the premature cardiac aging phenotype of PTKO mice.
Mitochondrial morphological and functional aberrations occur with loss of Pim
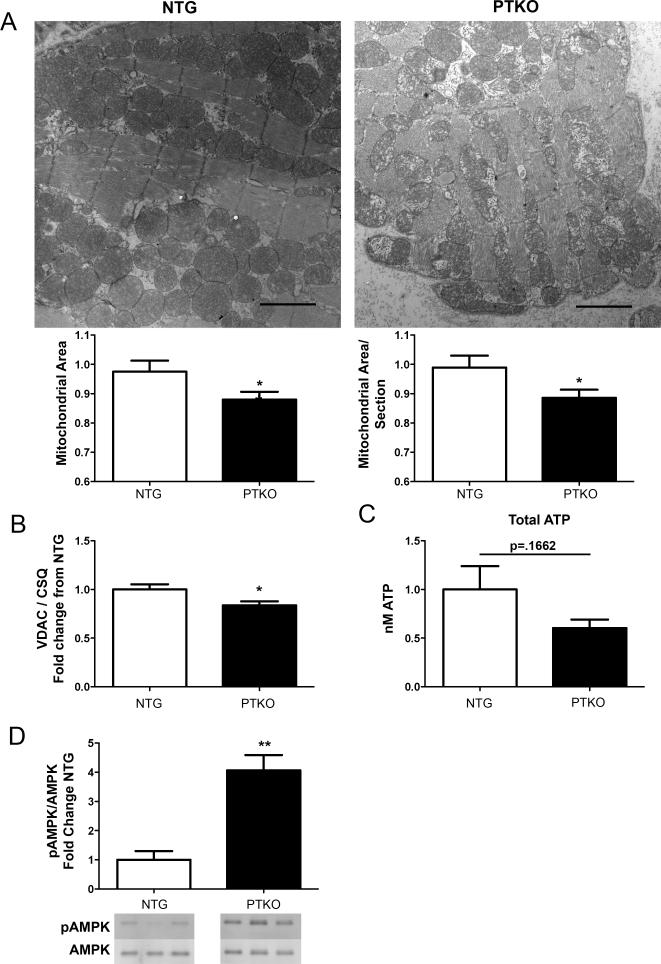
Pim1 protects mitochondrial integrity and preserves mitochondrial morphology 16,19. Mitochondrial vacuoles and disrupted cristae structures were evident by electron microscopy in hearts of PTKO mice at one month after birth (Figure 4A). Additionally, mitochondrial area and mitochondrial area per section were decreased by 11% and 10% in PTKO mouse hearts, respectively (p<0.05; Figure 4A). Validating these results, voltage-dependent anion channel (VDAC) expression was reduced by 16% in cardiac lysates of PTKO mice indicating a reduction in mitochondrial mass (p<0.05; Figure 4B).
Figure 4. Mitochondrial morphological and functional aberrations with loss of Pim.
A) Electron microscopy scans of NTG (n=3) and PTKO (n=3) hearts at one month and quantification of mitochondrial area and mitochondrial area per section. B) Protein analysis of VDAC in NTG (n=4) and PTKO (n=4) mouse hearts. C) Citrate synthase activity in cardiac lysates of NTG (n=6) and PTKO (n=6) mice. D) Total ATP levels in NTG (n=13) and PTKO (n=10) heart lysates. E) Immunoblot for pAMPK and total AMPK and analysis of pAMPK : AMPK ratio in cardiac samples of NTG (n=6) and PTKO (n=6) mice. * and ** is significant compared to NTG mice. *P < 0.05 and **P < 0.01.
Loss of mitochondrial function is consistent with a reduction in energy reserve as revealed by analysis of ATP levels (Figure 4C). Corroborating these results, an increase in the ratio of phosphorylated AMPK to total AMPK by 4 fold was noted in PTKO hearts by immunoblot (p<0.01; Figure 4D). Collectively, these results reveal alterations in mitochondrial morphology and functional capacity leading to energy deficiency resulting from loss of Pim kinases.
Metabolic regulators are decreased with deletion of Pim kinases
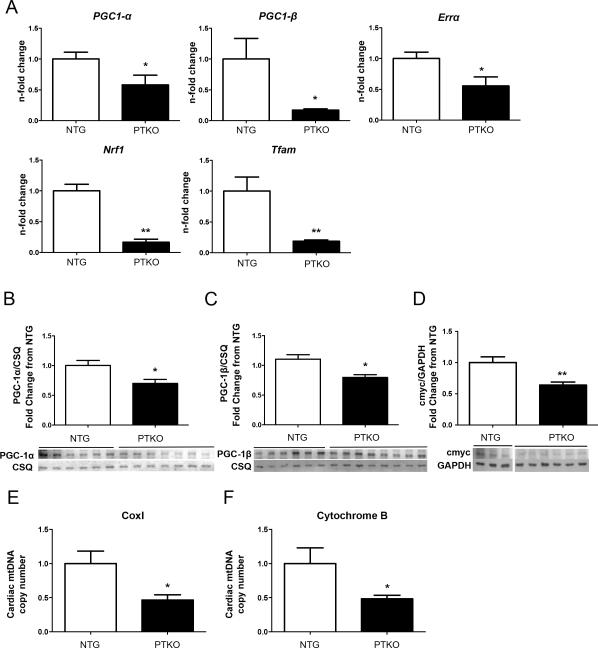
Pim kinase deletion impairs expression of PGC-1α 15, with loss of PGC-1 coactivators promoting alteration of mitochondrial morphology 4. Therefore, the impact of Pim kinase deletion upon expression of PGC-1 coactivators and downstream metabolic regulators was assessed. PGC-1α and PGC-1β expression were decreased by 42% and 82%, respectively, in PTKO cardiac lysates at one month (p<0.05; Figure 5A). Furthermore, downstream transcriptional targets of PGC-1 coactivators including Errα (estrogen-related receptor alpha), Nrf1 (nuclear respiratory factor 1), and Tfam (mitochondrial transcription factor A) were also decreased respectively by 44% (p<0.05), 83% (p<0.01), and 81% (p<0.01) in PTKO mice (Figure 5A). Peroxisome proliferative activated receptor gamma coactivator-related protein (PPRC1) was also diminished by 73% with Pim loss, evidently revealing lack of compensatory upregulation for related proteins (p<0.05; Online Fig VI A). Furthermore, decrease in PGC-1α (30%) and PGC-1β (20%) were confirmed at the protein level by immunoblot, corroborating gene expression analysis of PGC-1 coactivators (p<0.05; Figure 5B & C).
Figure 5. PPAR coactivators and downstream targets are decreased with Pim deletion.
A) mRNA levels of PGC-1α, PGC-1β, Errα, NRF1, and TFAM in 1 month NTG (n=6) and PTKO (n=6) hearts. B-D) Protein analysis of PGC-1α, PGC-1β, and c-Myc respectively in cardiac samples; NTG (n=6) and PTKO (n=7) hearts. E-F) Mitochondrial DNA analysis of Cox1 and Cytochrome B normalized to β-globin; NTG (n=4) and PTKO (n=4). * and ** is significant compared to NTG mice. *P < 0.05 and **P < 0.01.
Mitochondrial biogenesis is also dependent upon c-Myc expression that is a downstream target of Pim kinase 17,18. c-Myc protein levels were diminished by 36% in PTKO mice (p<0.01; Figure 5D). The impact of decreased regulators of mitochondrial biogenesis is apparent by measurement of mitochondrial DNA content as a correlate for mitochondrial number that was reduced by 50% in PTKO mice, consistent with decreased expression of PPARγ coactivators with Pim deletion (p<0.05; Figure 5E & F).
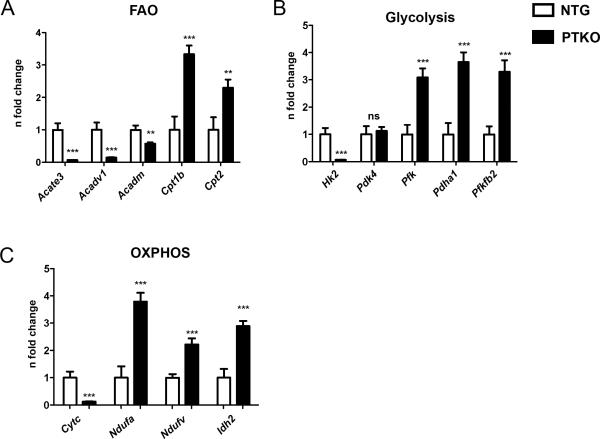
Decreased expression of PGC-1 coactivators alters the expression profile of cardiac metabolic genes 5,6,9, so representative genes involved in fatty acid oxidation, glycolysis, and oxidative phosphorylation were assessed. Fatty acid oxidation related enzymes located in the inner mitochondrial membrane were decreased including Acate3 (acyl coenzyme A thioesterase 3; 14.9 fold (p<0.001), Acadv1 (aceyl coenzyme A long chain; 6.9 fold (p<.001)), and Acadm (FAO-acetyl CoenzymeA dehydrogenase; 1.7 fold (p<.01)) (Figure 6A). In contrast, transporters of fatty acids such as Cpt1b (carnatine palmitoyl transferase) and Cpt2 (carnatine palmitoyl transferase 2) were increased by 3.3 fold (p<0.001) and 2.3 fold (p<.01), respectively, consistent with previous literature demonstrating upregulation of fatty acid transporters with prolonged AMPK activation (Figure 6A) 35. The rate-limiting step of glycolysis involves Hk2 (hexokinase 2) that was reduced by 15.3 fold (p<.001 Figure 6B) in PTKO hearts. Conversely, downstream targets involved in the glycolytic pathway: Pfk (phosphofructokinase), Pdha1 (pyruvate dehydrogenase E1 alpha 1), Pfkfb2 (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2) were highly upregulated by 3 fold (p<.001), 3.6 fold (p<.001), and 3.3 fold (p<.001) (Figure 6B). Furthermore, pyruvate respiration was increased whereas succinate respiration was unaltered (p<0.05; Online Fig VI B & C) indicating a shift away form fatty acid oxidation and toward glucose metabolism similar to PGC-1α/β double knockouts 4. Genes involved in lipid sterol biosynthesis were also dramatically reduced in cardiac tissue of PTKO mice indicating disruption in the PPAR signaling circuit (p<0.05; Online Fig VI D). mRNA analysis for genes involved in oxidative phosphorylation, NDUFA (NADH dehydrogenase 1 alpha subcomplex subunit 9), NDUFV (NADH dehydrogenase ubiquinone flavoprotein 1), and Idh2 (isocitrate dehydrogenase 2), revealed upregulation of 3.8 fold (p<.001), 2.2 fold (p<.001), and 2.9 fold (p<.001) respectively (Figure 6C). However, Cytc (cytochrome C) a target of PGC-1α was decreased by 8 fold (p<.001 Figure 6C). To assess global changes a microarray analysis of PPAR signaling genes involved in fatty acid oxidation, transport, and lipid sterol biosynthesis was performed, confirming altered expression of genes involved in the PPAR signaling pathway (Online Fig VII). 62 PPAR-related signaling genes were detected in cardiac tissue, 32 showed a downregulation, 21 genes remained unchanged, and 8 genes were induced. Three out of the four genes included in the targeted screen (Acadm, PGC-1α, Cpt1b, and Cpt2) demonstrated similar results to our targeted approach. Acadm and PGC-1α were downregulated, whereas Cpt1b was upregulated. Cpt2 was upregulated in our screen and was unchanged in the microarray, this discrepancy might be due to differences in primer pairs as Cpt2 is predicted to have 5 alternatively spliced transcripts (NCBI: AceView). Taken together, these results demonstrate disruption of metabolic processes with Pim ablation mimicking the phenotype of PGC-1α/β double knockouts 4.
Figure 6. Alterations in metabolic gene expression profile ensue with loss of Pim kinases.
mRNA levels of A) Fatty acid oxidation genes (Enzymes: Acate3, acyl coenzyme A thioesterase 3, mitochondrial; Acadv1, aceyl coenzyme A long chain; Acadm, FAO-acetyl CoenzymeA dehydrogenase. Transporters: Cpt1b, carnatine palmitoyl transferase; Cpt2, carnatine palmitoyl transferase 2. B) Glycolysis genes (Hk2, hexokinase 2; Pdk4, pyruvate dehydrogenase kinase 4; Pfk, phosphofructokinase; PDHA1, pyruvate dehydrogenase E1 alpha 1; PFKFB2, (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2). C) Oxidative Phosphorylation (OXPHOS) (Cytc, cytochrome c; Ndufa, NADH dehydrogenase 1 alpha subcomplex subunit 1; Ndufv, NADH dehydrogenase ubiquinone flavoprotein 1); Idh2, isocitrate dehydrogenase 2. n=6 NTG and n=6 PTKO. *, **, *** is significant compared to NTG mice. *P < 0.05; **P < 0.01; ***P < 0.001.
c-Myc rescues expression of metabolic regulators
The metabolic phenotype of PTKO mice correlates with loss of c-Myc expression secondary to Pim kinase ablation, so the participation of c-Myc was assessed by a rescue experiment involving overexpression of PGC-1α, c-Myc, and Pim1 using immortalized murine embryonic fibroblast from wildtype (iWT MEFs) and PTKO (iPTKO MEFs) mice. Metabolic deficiencies in the iPTKO MEFs were confirmed to be comparable to those of PTKO cardiac lysates. Expression of c-Myc and PGC-1α were diminished by 82% and 55% (respectively) in iPTKO MEFs (p<0.05; Online Fig VIII A) similar to PTKO mouse hearts. Phosphorylated AMPK: Total AMPK ratio was increased in iPTKO MEFs by 2.3 fold consistent with decreased cellular energy stores (p<0.05; Online Fig VIII B). Similarly, alterations of metabolic regulators and reduction in fatty acid oxidation enzymes were comparable between iPTKO MEFs and PTKO heart lysates (Online Fig VIII C). Since the iPTKO MEFs possess analogous metabolic derangements to PTKO heart samples, molecular restorations were attempted by overexpression of either c-Myc, PGC-1α, and Pim1. Overexpression of EGPF, Pim1-EGFP, PGC-1α-EGFP, and c-Myc-EGFP adenovirus was confirmed in iPTKO MEFs (Online Fig VIII D).
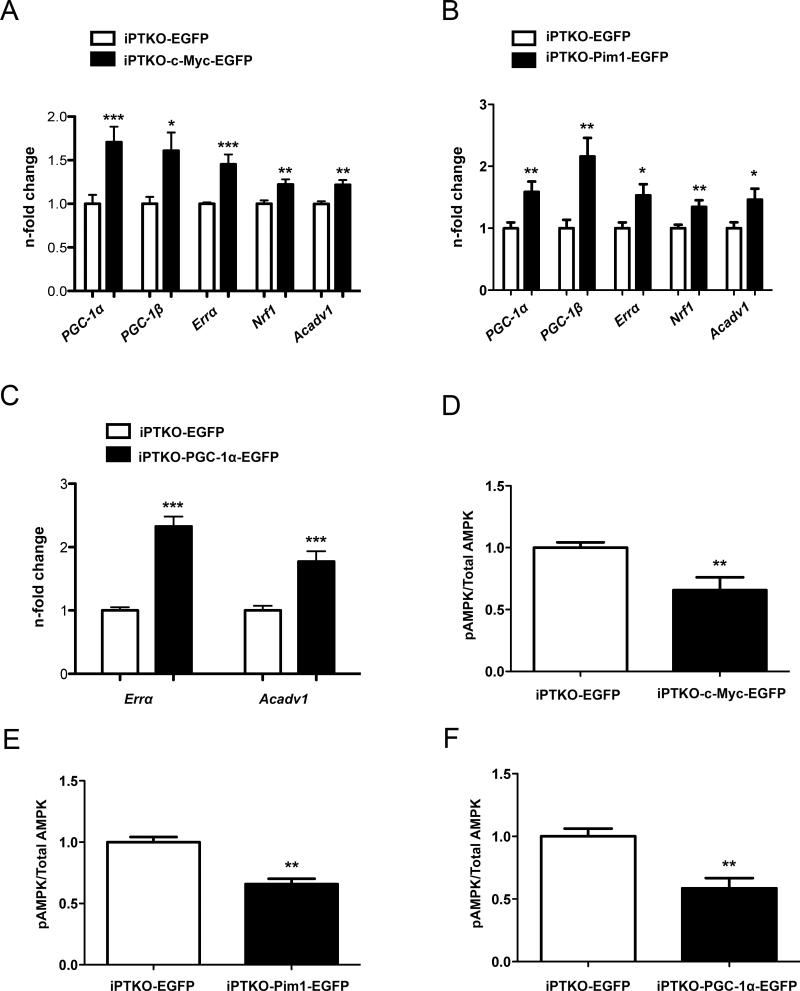
c-Myc overexpression in iPTKO MEFs increased expression of metabolic regulators PGC-1α (1.7 fold; p<0.05), PGC-1β (1.6 fold; p<0.05), Errα (1.4 fold; p<0.05), Nrf1 (1.2 fold; p<0.05), and Avadv1 (1.2 fold; p<0.05) (Figure 7A). Pim1 overexpression increased expression of metabolic regulators PGC-1α (1.6 fold; p<0.05), PGC-1β (2.2 fold; p<0.05), Errα (1.5 fold; p<0.05), Nrf1 (1.3 fold; p<0.05), and Avadv1 (1.5 fold; p<0.05) (Figure 7B). PGC-1α overexpression in iPTKO MEFs restored gene expression of the PGC-1α downstream targets: Errα and Acadv1 by 2.3 fold and 1.8 fold respectively (p<0.001; Figure 7C). Consequential to increased metabolic regulator expression, the ratio of phosphorylated AMPK to total AMPK decreased by 34% (p<.01) with c-Myc, 35% with Pim1, and 41% (p<.01) with PGC-1α overexpression, indicating improvement in cellular energy stores following interventions to overexpress c-Myc, Pim1, or PGC-1α (Figure 7 D,E, and F). In summary, the phenotype resulting from loss of Pim kinases can be rescued with overexpression of c-Myc, Pim1, or PGC-1α.
Figure 7. Rescue of metabolic genes with c-Myc, Pim1, and PGC-1α overexpression.
Overexpression of c-Myc, Pim1, and PGC-1α adenovirus respectively in iPTKO MEFs followed by: A-C) qPCR of PPAR coactivators and downstream targets. D-F) Quantification of pAMPK : Total AMPK ratio. n=3 individual experiments and *, **, *** is significant compared to iPTKO MEFs infected with EGFP adenovirus mice. *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
Cardiac aging is characterized by pathological hypertrophic remodeling that is a primary indicator of and confirmed risk factor for heart failure 33. Pathological hypertrophy and progression toward heart failure were evident at six months in PTKO mice (Figure 1 and 2) consistent with previous published studies demonstrating antagonism of myocardial hypertrophy by Pim1 26. Increased AKT phosphorylation and RCAN1.4 expression, a readout for activated NFAT, was evident with loss of Pim, which serve as molecular underpinnings for hypertrophic signaling (Online Fig II) and remodeling. Additionally, fetal gene reprogramming typical for hypertrophic stimulation is blunted by Pim1 overexpression 26. Promotion of hypertrophic effects at the molecular, cellular, and organ level in PTKO hearts relatively early in life (Figure 1) and enhanced senescence marker expression (Figure 3) is consistent with Pim kinase deletion manifesting with features of an accelerated aging phenotype.
Pathological hypertrophic remodeling causes an energetic fuel shift from fatty acid oxidation to glucose metabolism 6. Cellular metabolic changes are potent regulators of the aging phenotype and PTKO mice demonstrate an up-regulation of glycolytic enzymes and diminished fatty acid oxidation enzymes, exhibiting metabolic alterations (Figure 6). Mitochondrial metabolism and energetics converge upon the PPAR signaling circuit and the influence of Pim kinases upon PGC-1α is established 5,15. Now for the first time to our knowledge, the connection between downregulation of PGC-1 α and β and downstream targets impacted by Pim kinase deletion has been revealed in the heart by our results (Figure5). Furthermore, PGC-1α is important and necessary to maintain a pool of healthy mitochondria, as PGC-1α regulates mitochondrial biogenesis 27. Disruption in mitochondrial biogenesis occurs during aging causing accumulation of unhealthy mitochondria and potential increases in toxic metabolic byproducts that potentially accelerate cellular aging 36. Diminished PPAR signaling occurs with aging in human and murine animal models, supporting our contention that disrupted metabolism and diminished mitochondrial biogenesis through loss of PGC-1α in PTKO mice contributes to premature aging 37,38.
Downstream of Pim kinases is c-Myc, a potent regulator of mitochondrial biogenesis and PGC-1β 18,39,40. Diminished expression of c-Myc was evident in PTKO hearts and iPTKO MEFs (Figure 5D and Online Fig VIII A). Acute overexpression of c-Myc in iPTKO MEFs promoted upregulation of PGC-1α and PGC-1β in addition to downstream regulators of mitochondrial biogenesis. c-Myc activation during pathological cardiac injury is shown to promote glucose metabolism by down regulating PPARγ coactivators 18. In comparison, the observed loss of c-Myc still prompted enhanced glycolytic enzyme expression in PTKO mice, suggesting alternative mechanism(s) for promotion of glucose metabolism in PTKO mice. The loss of Pim kinase consequently promotes diminished expression of c-Myc along with PPARγ coactivators presenting as cellular metabolic alterations consistent with acquisition of an aging phenotype.
Mitochondria play a central role in energy metabolism, with mitochondrial morphology intimately tied to mitochondrial function 36. Pim kinases protect mitochondrial integrity by up-regulating anti-apoptotic Bcl-2 family members (Bcl-2 and Bcl-XL) and phosphorylating BAD on serine 112 16,34. Furthermore, Pim1 regulates mitochondrial morphological dynamics by regulating Drp1 localization and phosphorylation 19. Imbalance in mitochondrial fission sensitizes mitochondria to cell death in addition to promoting formation of smaller mitochondria as seen in PTKO hearts (Figure 4A). Pim deletion also prompted altered mitochondrial morphology and disarray of myofibrils (Figure 4A). Mitochondrial morphology alterations can increase apoptotic sensitivity to cell death and propensity to permeabilize in response to stress. Disruption of mitochondrial dynamics occurring with Pim loss promotes instability in mitochondrial DNA 19,41 and reduction in mitochondrial DNA content precedes pathological hypertrophy 42. Consistent with these assertions, PTKO mice display diminished levels of mitochondrial DNA content prior to heart failure (Figure 5E & F) that can serve as an additional predisposing factor for premature aging at the mitochondrial level with loss of Pim.
Telomere attrition disrupts mitochondrial function though a molecular mechanism involving p53 mediated cellular senescence leading to a p53 dependent repression of PGC-1α 43. Telomere length decrease in PTKO mice (Figure 3E-G) is a plausible inductive mechanism for upregulation of p53 activity and subsequent blunting of PGC-1α. Conversely, increased telomere length resulting from Pim1 kinase overexpression is associated with decreased expression of senescent markers including p53 and increased cell cycling in human cardiac progenitor cells 24. Comparable beneficial effects of telomere length maintenance are also observed following Pim1 kinase overexpresssion in murine cardiac progenitor cells 23. Furthermore, PTKO mice at 4 weeks of age exhibit an accelerated aging phenotype evident by telomere attrition of 40% by qPCR and FISH (Figure 3E-G). This dramatic decrease would be comparable to the relative age of 30 months in a normal mouse3. Increased cellular turnover during physiological aging leads to a reduction in telomere length with each cell division44-46. Although Pim1 deletion blunts cellular proliferation that may help slow telomeric shortening, any modest beneficial effect is apparently overwhelmed by depression of TERT activity that compromises preservation of telomere length 23, leading to telomeric shortening in PTKO mice consistent with accelerated physiological aging. Therefore, the impact of Pim kinase upon telomere biology highlights an important potential mechanism affecting mitochondrial function and causing cellular senescence that is the subject of ongoing investigation.
Pim kinases promote proliferation by regulating cell cycle proteins such as p21, p27, and cdc25A through phosphorylation 47-49. Furthermore, Pim phosphorylation and stabilization of c-Myc enhances cell cycle entry 17, leading to the plausible idea that Pim kinases antagonize the aging phenotype and favor ongoing mitotic activity by blunting the impact of cell cycle inhibitors. Indeed, downregulation of cell cycle inhibitors p16 and p53 leading to increased proliferation occurs in aged and senescent human cardiac progenitor cells overexpressing Pim1 kinase 24. Conversely, in the PTKO mice the increased expression of p16 and p53 support a role for the Pim kinase family in preserving phenotypic characteristics of cellular “youthfulness” (Figure 3 and Online Fig V).
The cardioprotective activity of Pim1 through the preservation of mitochondrial structure and morphology is well established primarily based upon the overexpression of Pim1 16,19. However, the converse of this experimental approach implies that the loss of Pim kinase would promote deterioration of mitochondrial structure and function. Such diminution of Pim kinase occurs naturally with organismal aging, but ours is the first assessment of germline deletion for all three Pim kinase family members upon myocardial biology. Degenerative changes in mitochondria such as altered structural integrity, shifts in metabolic fuel selectivity, and decreased ATP production are hallmarks of cellular aging and senescence 50. Clearly, select essential features of the aging and senescent phenotype are recapitulated with accelerated timing in the PTKO line. Now a link can be placed to connect Pim kinases to aging and metabolic changes through the PPAR signaling circuit. Important to note as a limitation of this study is the PTKO mouse models are global knockouts and not cardiac-specific. Therefore, in vivo findings although consistent with expectations remain correlative in nature, so these findings were buttressed by mechanistic molecular signaling in vitro studies with iPTKO MEFs. At present, the circumstantial evidence points toward alterations in mitochondrial biogenesis and metabolism coincident with premature cardiac aging of PTKO hearts. Looking forward to an interventional strategy, prevention of metabolic derangements is important to antagonize the cardiac aging phenotype. So too, delineating underlying causes of aging are important to develop therapeutic interventions to treat aging myopathy in the elderly population. The PTKO mouse model could serve in this capacity as an excellent model system to study cardiac aging and senescence. Collectively, this study gives credence to the possibility of utilizing Pim1 as a therapeutic agent for treatment of and intervention in the pathogenesis of cardiac aging.
Supplementary Material
Novelty and Significance.
What Is Known?
Mitochondrial dysfunction is associated with aging.
Overexpression of Pim1 kinase decreases the expression of senescent markers in human cardiac progenitor cells (hCPCs).
Pim1 kinase protects mitochondrial integrity and loss of Pim1 kinase promotes mitochondrial fission.
What New Information Does This Article Contribute?
Loss of Pim kinases causes premature cardiac aging.
Mitochondrial and metabolic alterations are evident with loss of Pim and converges upon PPARγ coactivator-1 (PGC-1) α signaling.
Overexpression of Pim1 rescued metabolic alterations in Pim Triple Knockout (PTKO) immortalized murine embryonic fibroblast (iPTKO MEFs).
Myocardial aging leads to deterioration of cardiac function promoting onset of heart failure in part by altered metabolism. Preventing deterioration of mitochondrial function is known to blunt cardiac aging and avert heart failure. Pim kinases promote cell proliferation, survival, and preserve mitochondrial integrity in the cardiac context. Therefore, Pim kinase-mediated protection of mitochondrial function was hypothesized to antagonize phenotypic consequences of aging associated with metabolic derangement. Cardiac metabolism depends upon mitochondrial function that is regulated by PGC-1α signaling to stimulate fatty acid oxidation (FAO). Heart failure prompts decreased FAO and a concomitant increase in glycolytic metabolism that devolves into declining energy reserves. Loss of Pim kinases caused premature heart failure and was coincident with decreased PGC-1 α signaling, altered cardiac metabolism, and decreased ATP levels. Conversely, overexpression of Pim1, the major cardiac isoform, in iPTKO MEFs rescued metabolic gene expression and increased energy reserves. Therefore, taken together with ability of Pim1 overexpression to blunt the expression of senescence markers in hCPCs and maintain mitochondrial integrity, our findings reveal novel beneficial properties imparted by Pim-1, lending further support for activation of Pim-1 as a therapeutic modality to inhibit cardiac aging and failure by preserving mitochondrial function and healthy metabolic activity.
ACKNOWLEDGEMENTS
We thank all members of the M.A.S laboratory for helpful discussion and comments.
SOURCES OF FUNDING
M.A. Sussman is supported by National Institutes of Health grants R01HL067245, R37HL091102, R01HL105759, R01 HL113656, R01 HL117163, and R01 HL113647. D.P. Kelly is supported by RO1-HL058493 and RO1-HL101189. S. Din is supported by American Heart Association (AHA) pre doctoral award 12PRE12060248 and the Rees Stealy Foundation. M.H. Konstandin and M. Volkers are supported by Deutsche Forschungsgemeinschaft Grants KO 3900/1-1 and MV 1659 1/1 respectively.
Nonstandard Abbreviations and Acronyms
- Acta1
α-skeletal actin
- AMPK
AMP-activated protein kinase
- CSA
Cell Size Area
- %EF
Percent ejection fraction
- Err-α
Estrogen-related receptor alpha
- iPTKO MEFs
Immortalized Pim Triple KnockOut murine embryonic fibroblast
- iWT MEFs
Immortalized wild-type murine embryonic fibroblast
- LVIDd
Left ventricle inner diameter diastole
- LVAWd
Left ventricle anterior wall diastole
- Myh7
β-myosin heavy chain
- Nppa
Atrial natriuretic peptide
- Nppb
Brain natriuretic peptide
- NRF1
Nuclear respiratory factor 1
- NTG
Non-transgenic wild type
- PGC-1α
PPAR gamma coactivator 1 alpha
- PGC-1β
PPAR gamma coactivator 1 beta
- PTKO
Pim Triple KnockOut
- TFAM
Mitochondrial transcription factor A
Footnotes
DISCLOSURES
None
The other authors report no conflicts.
REFERENCES
- 1.Anversa P, Fitzpatrick D, Argani S, Capasso JM. Myocyte mitotic division in the aging mammalian rat heart. Circ Res. 1991;69:1159–1164. doi: 10.1161/01.res.69.4.1159. [DOI] [PubMed] [Google Scholar]
- 2.Cesselli D, Beltrami AP, D'Aurizio F, et al. Effects of age and heart failure on human cardiac stem cell function. Am J Pathol. 2011;179:349–366. doi: 10.1016/j.ajpath.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez A, Rota M, Nurzynska D, et al. Activation of cardiac progenitor cells reverses the failing heart senescent phenotype and prolongs lifespan. Circ Res. 2008;102:597–606. doi: 10.1161/CIRCRESAHA.107.165464. [DOI] [PubMed] [Google Scholar]
- 4.Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arany Z, He H, Lin J, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol. 2002;29:339–345. doi: 10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 7.Wenz T. Regulation of mitochondrial biogenesis and PGC-1alpha under cellular stress. Mitochondrion. 2013;13:134–142. doi: 10.1016/j.mito.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Finkelstein J, Gray N, Heemels MT, Marte B, Nath D. Metabolism and disease. Nature. 2012;491:347. doi: 10.1038/491347a. [DOI] [PubMed] [Google Scholar]
- 9.Aubert G, Vega RB, Kelly DP. Perturbations in the gene regulatory pathways controlling mitochondrial energy production in the failing heart. Biochim Biophys Acta. 2013;1833:840–847. doi: 10.1016/j.bbamcr.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan JG, Fong JL, Medeiros DM, Finck BN, Kelly DP. Insulin-resistant heart exhibits a mitochondrial biogenic response driven by the peroxisome proliferator-activated receptor-alpha/PGC-1alpha gene regulatory pathway. Circulation. 2007;115:909–917. doi: 10.1161/CIRCULATIONAHA.106.662296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leone TC, Kelly DP. Transcriptional control of cardiac fuel metabolism and mitochondrial function. Cold Spring Harb Symp Quant Biol. 2011;76:175–182. doi: 10.1101/sqb.2011.76.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 13.Riehle C, Wende AR, Zaha VG, et al. PGC-1beta deficiency accelerates the transition to heart failure in pressure overload hypertrophy. Circ Res. 2011;109:783–793. doi: 10.1161/CIRCRESAHA.111.243964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lelliott CJ, Medina-Gomez G, Petrovic N, et al. Ablation of PGC-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beharry Z, Mahajan S, Zemskova M, Lin YW, Tholanikunnel BG, Xia Z, Smith CD, Kraft AS. The Pim protein kinases regulate energy metabolism and cell growth. Proc Natl Acad Sci U S A. 2011;108:528–533. doi: 10.1073/pnas.1013214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borillo GA, Mason M, Quijada P, et al. Pim-1 kinase protects mitochondrial integrity in cardiomyocytes. Circ Res. 2010;106:1265–1274. doi: 10.1161/CIRCRESAHA.109.212035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Wang Z, Li X, Magnuson NS. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27:4809–4819. doi: 10.1038/onc.2008.123. [DOI] [PubMed] [Google Scholar]
- 18.Ahuja P, Zhao P, Angelis E, Ruan H, Korge P, Olson A, Wang Y, Jin ES, Jeffrey FM, Portman M, Maclellan WR. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J Clin Invest. 2010;120:1494–1505. doi: 10.1172/JCI38331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Din S, Mason M, Volkers M, Johnson B, Cottage CT, Wang Z, Joyo AY, Quijada P, Erhardt P, Magnuson NS, Konstandin MH, Sussman MA. Pim-1 preserves mitochondrial morphology by inhibiting dynamin-related protein 1 translocation. Proc Natl Acad Sci U S A. 2013;110:5969–5974. doi: 10.1073/pnas.1213294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sussman MA. Mitochondrial integrity: preservation through Akt/Pim-1 kinase signaling in the cardiomyocyte. Expert Rev Cardiovasc Ther. 2009;7:929–938. doi: 10.1586/erc.09.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005;115:2618–2624. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muraski JA, Rota M, Misao Y, et al. Pim-1 regulates cardiomyocyte survival downstream of Akt. Nat Med. 2007;13:1467–1475. doi: 10.1038/nm1671. [DOI] [PubMed] [Google Scholar]
- 23.Cottage CT, Neidig L, Sundararaman B, Din S, Joyo AY, Bailey B, Gude N, Hariharan N, Sussman MA. Increased mitotic rate coincident with transient telomere lengthening resulting from pim-1 overexpression in cardiac progenitor cells. Stem Cells. 2012;30:2512–2522. doi: 10.1002/stem.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohsin S, Khan M, Nguyen J, Alkatib M, Siddiqi S, Hariharan N, Wallach K, Monsanto M, Gude NA, Dembitsky WP, Sussman MA. Rejuvenation of Human Cardiac Progenitor Cells with Pim-1 Kinase. Circ Res. 2013;113:1169–1179. doi: 10.1161/CIRCRESAHA.113.302302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, Berns A. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24:6104–6115. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muraski JA, Fischer KM, Wu W, et al. Pim-1 kinase antagonizes aspects of myocardial hypertrophy and compensation to pathological pressure overload. Proc Natl Acad Sci U S A. 2008;105:13889–13894. doi: 10.1073/pnas.0709135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konstandin MH, Toko H, Gastelum GM, Quijada P, De La Torre A, Quintana M, Collins B, Din S, Avitabile D, Volkers M, Gude N, Fassler R, Sussman MA. Fibronectin is essential for reparative cardiac progenitor cell response after myocardial infarction. Circ Res. 2013;113:115–125. doi: 10.1161/CIRCRESAHA.113.301152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konstandin MH, Volkers M, Collins B, Quijada P, Quintana M, De La Torre A, Ormachea L, Din S, Gude N, Toko H, Sussman MA. Fibronectin contributes to pathological cardiac hypertrophy but not physiological growth. Basic Res Cardiol. 2013;108:375. doi: 10.1007/s00395-013-0375-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, Jimenez R, Petrosyan S, Murphy AN, Gustafsson AB. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turdi S, Fan X, Li J, Zhao J, Huff AF, Du M, Ren J. AMP-activated protein kinase deficiency exacerbates aging-induced myocardial contractile dysfunction. Aging Cell. 2010;9:592–606. doi: 10.1111/j.1474-9726.2010.00586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Susic D, Frohlich ED. The aging hypertensive heart: a brief update. Nat Clin Pract Cardiovasc Med. 2008;5:104–110. doi: 10.1038/ncpcardio1091. [DOI] [PubMed] [Google Scholar]
- 34.Fischer KM, Cottage CT, Konstandin MH, Volkers M, Khan M, Sussman MA. Pim-1 kinase inhibits pathological injury by promoting cardioprotective signaling. J Mol Cell Cardiol. 2011;51:554–558. doi: 10.1016/j.yjmcc.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chabowski A, Momken I, Coort SL, Calles-Escandon J, Tandon NN, Glatz JF, Luiken JJ, Bonen A. Prolonged AMPK activation increases the expression of fatty acid transporters in cardiac myocytes and perfused hearts. Mol Cell Biochem. 2006;288:201–212. doi: 10.1007/s11010-006-9140-8. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iemitsu M, Miyauchi T, Maeda S, Tanabe T, Takanashi M, Irukayama-Tomobe Y, Sakai S, Ohmori H, Matsuda M, Yamaguchi I. Aging-induced decrease in the PPAR-alpha level in hearts is improved by exercise training. Am J Physiol Heart Circ Physiol. 2002;283:H1750–1760. doi: 10.1152/ajpheart.01051.2001. [DOI] [PubMed] [Google Scholar]
- 38.Sihag S, Cresci S, Li AY, Sucharov CC, Lehman JJ. PGC-1alpha and ERRalpha target gene downregulation is a signature of the failing human heart. J Mol Cell Cardiol. 2009;46:201–212. doi: 10.1016/j.yjmcc.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 42.Karamanlidis G, Bautista-Hernandez V, Fynn-Thompson F, Del Nido P, Tian R. Impaired mitochondrial biogenesis precedes heart failure in right ventricular hypertrophy in congenital heart disease. Circ Heart Fail. 2011;4:707–713. doi: 10.1161/CIRCHEARTFAILURE.111.961474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahin E, Colla S, Liesa M, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kajstura J, Pertoldi B, Leri A, Beltrami CA, Deptala A, Darzynkiewicz Z, Anversa P. Telomere shortening is an in vivo marker of myocyte replication and aging. Am J Pathol. 2000;156:813–819. doi: 10.1016/S0002-9440(10)64949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kajstura J, Gurusamy N, Ogorek B, et al. Myocyte turnover in the aging human heart. Circ Res. 2010;107:1374–1386. doi: 10.1161/CIRCRESAHA.110.231498. [DOI] [PubMed] [Google Scholar]
- 46.Kajstura J, Rota M, Cappetta D, et al. Cardiomyogenesis in the aging and failing human heart. Circulation. 2012;126:1869–1881. doi: 10.1161/CIRCULATIONAHA.112.118380. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Mochizuki T, Kitanaka C, Noguchi K, Muramatsu T, Asai A, Kuchino Y. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J Biol Chem. 1999;274:18659–18666. doi: 10.1074/jbc.274.26.18659. [DOI] [PubMed] [Google Scholar]
- 48.Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68:5076–5085. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Wang Z, Magnuson NS. Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol Cancer Res. 2007;5:909–922. doi: 10.1158/1541-7786.MCR-06-0388. [DOI] [PubMed] [Google Scholar]
- 50.Lee HC, Wei YH. Mitochondria and aging. Adv Exp Med Biol. 2012;942:311–327. doi: 10.1007/978-94-007-2869-1_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.