SUMMARY
Vertebrate circadian rhythms are organized by the hypothalamic suprachiasmatic nucleus (SCN). Despite its physiological importance, SCN development is poorly understood. Here, we show that Lim homeodomain transcription factor 1 (Lhx1) is essential for terminal differentiation and function of the SCN. Deletion of Lhx1 in the developing SCN results in loss of SCN-enriched neuropeptides involved in synchronization and coupling to downstream oscillators, among other aspects of circadian function. Intact, albeit damped, clock gene expression rhythms persist in Lhx1-deficient SCN; however, circadian activity rhythms are highly disorganized and susceptible to surprising changes in period, phase, and consolidation following neuropeptide infusion. Our results identify a factor required for SCN terminal differentiation. In addition, our in vivo study of combinatorial SCN neuropeptide disruption uncovered synergies among SCN-enriched neuropeptides in regulating normal circadian function. These animals provide a platform for studying the central oscillator's role in physiology and cognition.
INTRODUCTION
Circadian rhythms restrict physiological functions including sleep/wake, feeding, thermoregulation, and hormone release to appropriate temporal niches. This behavior is proximally controlled by cellular circadian clocks. During the cellular clock's rising phase, Bmal1/Clock heterodimers drive transcription of clock genes including Per1/2/3 and Cry1/2, whose products inhibit their own transcription during the falling phase (reviewed in Albrecht, 2012).
Despite the ubiquity of cellular clocks in the body, the suprachiasmatic nucleus (SCN) of the anteroventral hypothalamus acts as a master regulator of light-entrained circadian rhythms, a role facilitated by a number of remarkable properties unique to its cells. In dispersed culture conditions, SCN neurons spontaneously synchronize their circadian clocks, unlike any other cell type (Ko et al., 2010). SCN slice cultures also sustain robust circadian oscillations for weeks, unlike most other neural regions (Abe et al., 2002; Tosini and Menaker, 1996). Finally, while the SCN is uniquely susceptible to direct phase-shifting by light signals from intrinsically photosensitive retinal ganglion cells (ipRGCs), it is exceptionally resistant to resetting by nonlight cues that phase-shift peripheral clocks (Buhr et al., 2010; Hattar et al., 2002). These properties rely on a robust SCN network that strengthens and coordinates the unstable oscillations of its individual cellular clocks, thereby organizing output signals that entrain the rest of the body (Albrecht, 2012; Ciarleglio et al., 2009; Romijn et al., 1997; Vasalou et al., 2009; Webb et al., 2009).
Neuropeptides prominently and selectively expressed in the SCN are heavily implicated in the regulation of the SCN network. Vasoactive intestinal peptide (Vip) is a key organizer of SCN cellular rhythms and activity; genetic disruption of Vip or its receptor, Vipr2, reduces the proportion and synchrony of circadian SCN cells (Aton et al., 2005; Maywood et al., 2006, 2011) and disrupts SCN electrical activity rhythms (Brown et al., 2007) as well as circadian- and light-entrained locomotion (Aton et al., 2005; Colwell et al., 2003; Harmar et al., 2002). Another critical component is prokineticin-2 (Prok2), a major SCN output signal (Cheng et al., 2002; Prosser et al., 2007). Gastrin-releasing peptide (Grp) and arginine vasopressin (Avp) also contribute to SCN gene expression rhythms, electrical rhythms, and output, while enkephalin (Enk) may gate SCN responsiveness to light (Albers et al., 1995; Brown et al., 2005; Harrington et al., 1993; Li et al., 2009; Maywood et al., 2006, 2011; Mihai et al., 1994; Vansteensel et al., 2005; Wideman et al., 2000). In mice, exogenous neuromedin S (Nms) alters circadian behavior but has unknown endogenous roles (Mori et al., 2005). Numerous other canonical neuropeptides are also present in SCN, and unbiased studies have identified dozens more whose characterization has only just begun (Abrahamson and Moore, 2001; Atkins et al., 2010; Lee et al., 2010).
SCN cellular heterogeneity implies complex developmental pathways guiding its differentiation and the emergence of its unique circadian properties. To date, three transcription factors involved in SCN development have been identified: Six3, Six6, and Lhx2. Deletion of any one of these factors prevents SCN specification from hypothalamic neuroepithelium (Clark et al., 2013; Roy et al., 2013; VanDunk et al., 2011). These factors are broadly expressed in developing ventral forebrain (Jean et al., 1999; Oliver et al., 1995; Xu et al., 1993), and all SCN gene expression and structural markers are undetectable after their genetic disruption (Clark et al., 2013; Roy et al., 2013; VanDunk et al., 2011), providing little insight into later stages of SCN differentiation. Therefore, nothing is yet known about the transcriptional regulatory network controlling expression of SCN-enriched neuropeptides such as Vip and Avp.
Unlike Six3, Six6, and Lhx2, hypothalamic expression of other transcription factors is restricted to the SCN during development, the earliest of which is LIM homeodomain transcription factor 1 (Lhx1) (Shimogori et al., 2010; VanDunk et al., 2011). Lhx1 guides differentiation of other neural subtypes outside the hypothalamus, including cerebellar Purkinje cells (Zhao et al., 2007) and hindbrain reticulospinal neurons (Cepeda-Nieto et al., 2005). Intriguingly, it also dictates neuropeptide fate of specific cell subpopulations during spinal cord development (Bröhl et al., 2008).
We find that Lhx1 is necessary for SCN terminal differentiation, including expression of neuropeptides profoundly important for circadian function. Despite only modest changes in variability and synchronization of cellular rhythms, circadian locomotor activity rhythms were severely disorganized and vulnerable to neuropeptidergic disturbance of consolidation, phase, and period in mice with Lhx1 selectively deleted in anterior hypothalamus. Our study identifies a transcription factor involved in SCN terminal differentiation. The Six3-Cre;Lhx1lox/lox mouse also provides a behavioral model with several emergent properties not seen in animals with deficits in a single-neuropeptide pathway, providing insight into the synergistic nature of the SCN neuro-peptide network.
RESULTS
Selective Deletion of Lhx1 in Ventral Anterior Hypothalamus
Because conventional Lhx1 null mice lack anterior neural structures (Shawlot and Behringer, 1995), we used an intersectional approach to more specifically delete Lhx1 in the developing anterior hypothalamus by crossing a conditional allele to transgenic Six3-Cre mice (Furuta et al., 2000; Kwan and Beh-ringer, 2002). As previously reported, Six3-Cre activity was more spatially restricted than native Six3 expression (Furuta et al., 2000). Six3-Cre;ROSA26::YFP mice had robust Cre activity in the ventral telencephalon and ventral anterior hypothalamus, including the SCN and subparaventricular zone (SPZ). However, Six3-Cre;ROSA26::YFP mice lacked Cre activity in more posterior regions that express Lhx1 during embryonic development, such as the intergeniculate leaflet (IGL), dorsomedial hypothalamus, and mamillary nucleus (Figures S1A and S1B). Lhx1 in situ hybridization (ISH) revealed that Six3-Cre;Lhx1lox/lox mice reliably and specifically lost Lhx1 expression in the ventral anterior hypothalamus, including the SCN and SPZ (Figures S1C–S1E and S1I). Although transient Lhx1 expression in anterior hypothalamic nucleus and anterior lateral hypothalamus were also lost at postnatal day 0 (P0) (Figure S1I), neither region has any established circa-dian function. Lhx1 was normally expressed in more posterior regions important for circadian rhythmicity and motor output (e.g., IGL, cerebellum) as well as the medial preoptic area (Figures S1F–S1H).
Lhx1 Deletion Disrupts SCN-Enriched Neuropeptide Expression
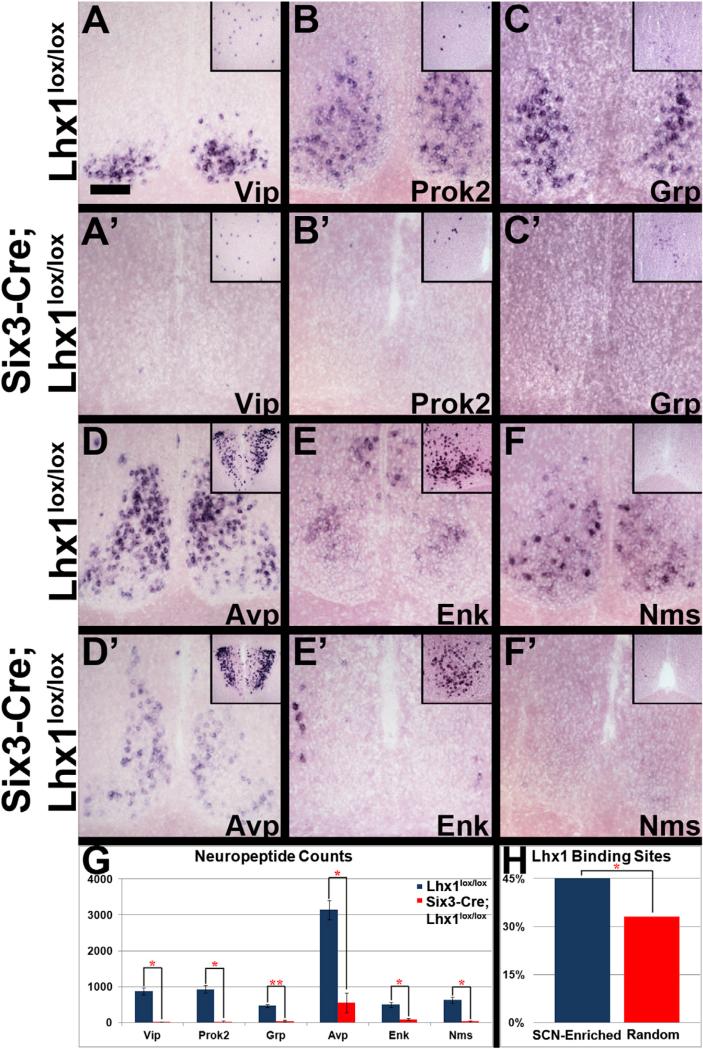
We next examined neuropeptide expression in Six3-Cre; Lhx1lox/lox and Lhx1lox/lox control SCN by ISH. Neuropeptides implicated in circadian function were downregulated (Figures 1A–1G; Table S1), including Vip, Prok2, Grp, Avp, Enk, and Nms (p < 0.05 for all except Grp, and p < 0.005 for Grp). Down-regulation was not observed in other brain regions (Figures 1A– 1F). SCN neuropeptide loss was not caused by the Cre driver alone, because Six3-Cre;Lhx1+/+ mice had normal expression of Vip, Grp, Avp, and Lhx1 (Figures S1J–S1M).
Figure 1. Lhx1 Effects on SCN Neuropeptides.
(A–F’) ISH of SCN from adult Lhx1lox/lox control (A–F) and Six3-Cre;Lhx1lox/lox mutant (A’–F’) mice for Vip (A, A’), Prok2 (B, B’), Grp (C, C’), Avp (D, D’), Enk (E, E’), and Nms (F, F’). Insets show the neocortex (A and C), preoptic area (B, E, and F), and paraventricular nucleus (D). Scale bar represents 100 μm.
(G) Mutants had fewer neuropeptide ISH-labeled SCN cells than controls (n = 3, paired two-tailed t tests, *p < 0.05, **p < 0.01; graph depicts mean ± SEM).
(H) MOPAT bioinformatics analysis showed more putative Lhx1-binding sites in enhancers of an SCN-enriched gene pool compared to a randomly selected pool (pool shown in Table S2; method described in Hu et al., 2008; binomial distribution, *p < 0.05).
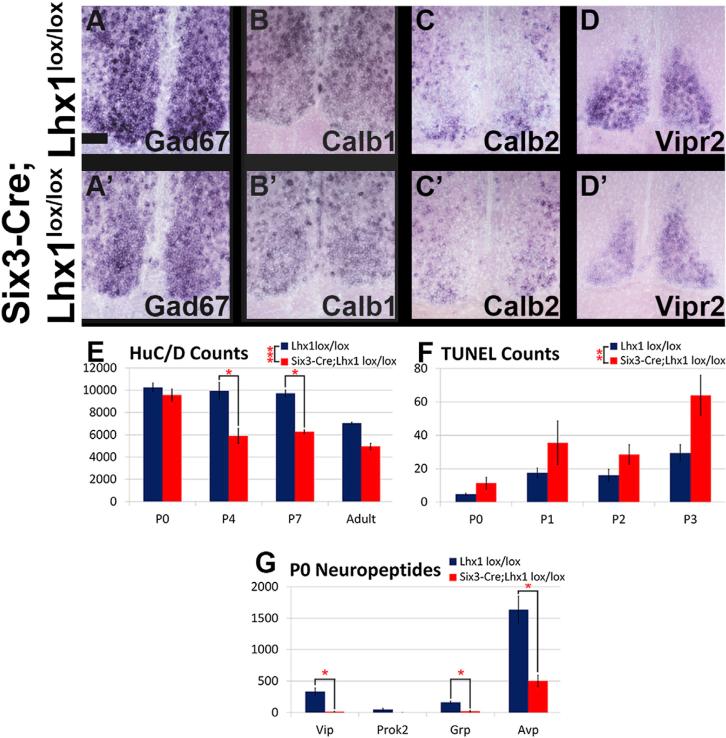
Importantly, other SCN markers were largely unaffected by Lhx1 deletion (Figures 2 and S2). GABAergic markers such as Gad67, Calb1, and Calb2 were retained (Figures 2A–2C), as were SCN markers first expressed prior to induction of circadian neuropeptides during development, such as Vipr2, Id4, Arx, Dlx2, Nr1d1, and Rorb (Figures 2D and S2A–S2E). Neuropep-tides broadly expressed in the anterior hypothalamus were also mostly preserved in Six3-Cre;Lhx1lox/lox SCN, including Sst, Agt, Nts, Cck, Gal, and Scg2 (Figures S2F–S2K). Expression of Pcsk1n, required for proteolytic processing of many neuro-peptide precursors, is also preserved (Figure S2L). Importantly, most markers normally restricted to specific SCN subdomains showed similar compartmentalization in mutants. For instance, Calb2 localized to ventral core, Rorb localized to shell, and Vipr2 expression remained weak in the center of the SCN (Figures 2C and 2D and S2E).
Figure 2. SCN Regional Identity Is Pre-served in Six3-Cre;Lhx1lox/lox Mice, but Cell Death Is Elevated in Neonatal SCN.
(A–D) ISH of SCN from Lhx1lox/lox control (A–D) and Six3-Cre;Lhx1lox/lox adult mutant (A0–D0) mice for Gad67 (A, A’), Calb1 (B, B’), Calb2 (C, C’), and Vipr2 (D, D’).
(E) Mutants had fewer SCN neurons than controls at P4 and P7 (n = 3, 3, 3, 3, ANOVA and Tukey post hoc tests, *p < 0.05, ***p < 0.0005; graph depicts mean ± SEM). Mutant SCN neuron number in adults was significantly lower than controls when initially analyzed on its own by t test (p < 0.05) but fell just short of significance in the context of the time course (p = 0.07).
(F) Mutants had increased SCN cell death in the aggregate period between P0 and P3 compared to controls, but the age(s) within this range with elevated cell death could not be pinned down (n = 3, 4, 4, 3, ANOVA with post hoc Tukey tests, **p < 0.005; graph depicts mean ± SEM).
(G) Mutants had fewer Vip, Grp, and Avp, but not Prok2, expressing SCN cells than controls at P0 (n = 3, 3, 3, 3, paired two-tailed t tests, *p < 0.05; graph depicts mean ± SEM).
Lhx1 Deletion Causes Neuron Loss and Cell Death in Developing SCN
Pan-SCN ISH markers showed that Six3-Cre;Lhx1lox/lox SCN was smaller than controls; therefore, we performed immunohistochemistry (IHC) for the pan-neuronal marker HuC/D and found that SCN neuron number was significantly reduced in Six3-Cre;Lhx1lox/lox adults (p < 0.05). To determine when neuron loss occurred in Six3-Cre;Lhx1lox/lox SCN, we conducted HuC/D IHC at multiple developmental time points (Figures 2E and S3M). There were main effects of both genotype and age (p < 0.0005) and significant interaction (p < 0.05). SCN neuron number was normal at P0 in Six3-Cre;Lhx1lox/lox mice (p = 0.95), but significantly fewer SCN neurons were present by P4 in Six3-Cre;Lhx1lox/lox SCN compared to controls (p < 0.0005) (Figures 2E and S2M). No further loss of neurons was observed in Six3-Cre;Lhx1lox/lox SCN after P4, although neuron number continued to decrease gradually in control SCN into adulthood (p < 0.05); this dropped the previously noted genotype difference in adult neuron number slightly below significance (p = 0.07) in the context of the time course.
To examine whether decreased neuron number between P0 and P4 in Lhx1-deficient SCN resulted from increased developmental apoptosis, we conducted TUNEL experiments in P0–P3 SCN. A main effect of genotype (p < 0.005) indicated an aggre-gate increase in TUNEL-positive cells across the time course in Six3-Cre; Lhx1lox/lox SCN, although no individual time points were significant (Figures 2F and S2N). There was also a main effect of age (p < 0.0005) but no interaction (p = 0.30).
Reduction in Neuropeptide Gene Expression Is Not Caused by Selective Neuron Loss in Lhx1-Deficient SCN
To examine whether neuron loss could explain SCN neuropeptide depletion in our mutants, we measured neuropeptide expression by ISH at P0, when Six3-Cre;Lhx1lox/lox SCN neuron number is normal (Figures 2G and S2O–S2R). Vip, Avp, and Grp were already profoundly downregulated in mutant SCN, indicating that loss of neuropeptide gene expression precedes neuron loss (all p < 0.05; Figure 2G). Prok2 expression was very low and variable in controls, making comparison with mutants uninterpretable (Figure 2G). We did not detect Enk or Nms in control or mutant SCN by ISH at P0 (data not shown).
We then conducted in silico MOPAT analysis to determine whether Lhx1 might directly regulate expression of SCN-enriched neuropeptides. A greater proportion of evolutionarily conserved predicted Lhx1 consensus binding sites were present within proximal regulatory regions of SCN-enriched genes relative to randomly selected genes (Figure 1H). SCN-enriched genes bearing putative Lhx1 binding sites included the three neuropep-tide genes with the greatest percent reduction in Six3-Cre; Lhx1lox/lox mice (Vip, Prok2, and Nms) as well as Enk, which was not included in our initial analysis (Figures 1G and 1H; Table S2).
SCN Clock Gene Expression Rhythms Are Preserved after Lhx1 Deletion
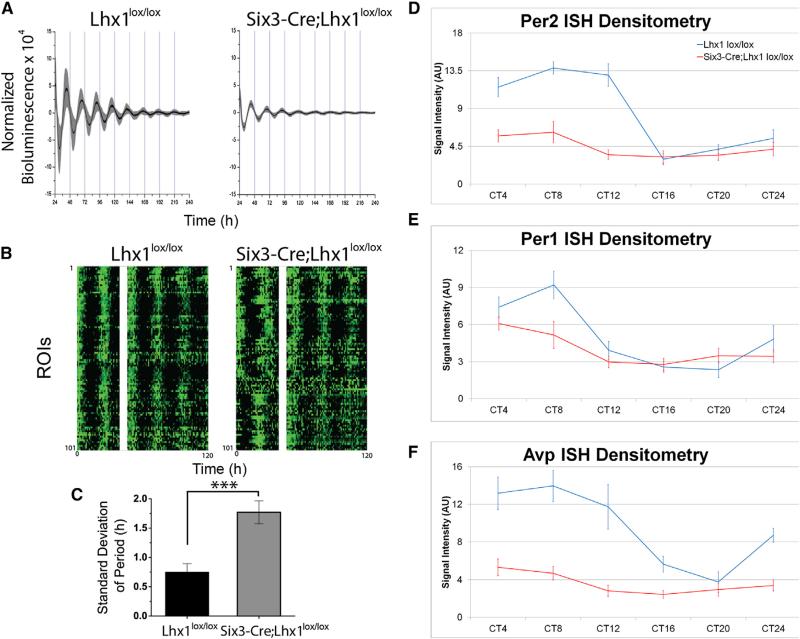
Despite profoundly decreased neuropeptide expression, bioluminescence recordings from Six3-Cre;Lhx1lox/lox;Per2luc/+ SCN cultures revealed clock gene rhythms with similar circadian period compared to controls from two independently tested groups of mice. Mean bioluminescence was similar in the first cohort but lower in the experimental group in the second cohort (Mann-Whitney U test, p < 0.01). Lhx1-deficient SCN had reduced peak-to-trough Per2 rhythm amplitude and more rapid damping in both cohorts (p < 0.05, Student's t test; Figure 3A). Higher-resolution charge-coupled device (CCD) camera data revealed that this was attributable to reduced circadian synchrony (Figure 3B) and increased circadian period variability within each Six3-Cre;Lhx1lox/lox SCN relative to controls (p < 0.001, Student's t test; Figure 3C).
Figure 3. Lhx1 Effects on SCN Clock Gene Rhythms.
(A) Adult Lhx1lox/lox (left) and Six3-Cre;Lhx1lox/lox (right) mouse SCN explants showed near-24 hr cycling of Per2::Luc bioluminescence over 10 days of recording, with moderately damped amplitude in the mutant explants. Traces show mean (black line) and SD (gray) of bioluminescence collected at 1 min intervals from 8 Lhx1lox/lox (left) and 9 Six3-Cre; Lhx1lox/lox (right) explants.
(B) Raster plots demonstrate variations in synchrony of circadian rhythms across regions of interest (ROIs) from one representative control (left) and one Six3Cre;Lhx1lox/lox SCN explant (right). White bars cover the region where data were lost while recording (40–46 hr).
(C) Loss of Lhx1 caused greater variance of circadian period in Six3-Cre;Lhx1lox/lox SCN explants (n = 5) compared to control littermates (n = 8).
(D–F) Circadian variation in expression levels of SCN clock genes Per2 (D), Per2 (E), Bmal1 (C), and Avp (F). ISH labeling intensity was present in adult Lhx1lox/lox control (D–F) and Six3-Cre;Lhx1lox/lox mutant (D’–F’) mice, with genotype and interaction effects for some genes (n = 3, ANOVA).
We confirmed a similar pattern in vivo via a 24 hr ISH time course of the core clock genes Per1 and Per2 and the clock-controlled gene Avp. There were main effects of circadian time for all three genes tested in Six3-Cre;Lhx1lox/lox SCN and controls in constant darkness (DD) (Per1 p < 0.001; Per2 p < 0.0001; Avp p < 0.0001) (Figures 3D–3F and S3A–S3C). A main effect of genotype and an interaction were observed for Per2 (p < 0.0001, p < 0.01) and Avp (both p < 0.0001), but not Per1 (p = 0.09, p = 0.27) (Figures 3D–3F and S3A–S3C). Vip mRNA was also examined but was virtually absent at all circadian time points (data not shown). Thus, both core clock and clock-controlled gene expression is still temporally regulated over circadian time, albeit with reduced amplitude for some genes, in the SCNs of intact Six3-Cre;Lhx1lox/lox mice.
Disrupted Circadian Activity Rhythms in Lhx1-Deficient Mice
Because lower-amplitude oscillations more readily phase shift to out-of-phase light exposure (Pulivarthy et al., 2007), we reasoned that Six3-Cre;Lhx1lox/lox animals would more readily entrain and reset to light cues than controls. Instead, activity of Six3-Cre;Lhx1lox/lox mice entrained less robustly than controls under a 12:12 light:dark cycle (LD) (Figure 4A), with a small advanced phase angle of nocturnal wheel running into the light phase and a reduced percentage of overall activity confined to the dark phase (Figures 4B and 4C). In many ways, these LD phenotypes seem similar to those seen in Vip−/− mice, leading us to initially hypothesize that Vip loss alone underpinned the behavioral deficits of Six3-Cre;Lhx1lox/lox mutants.
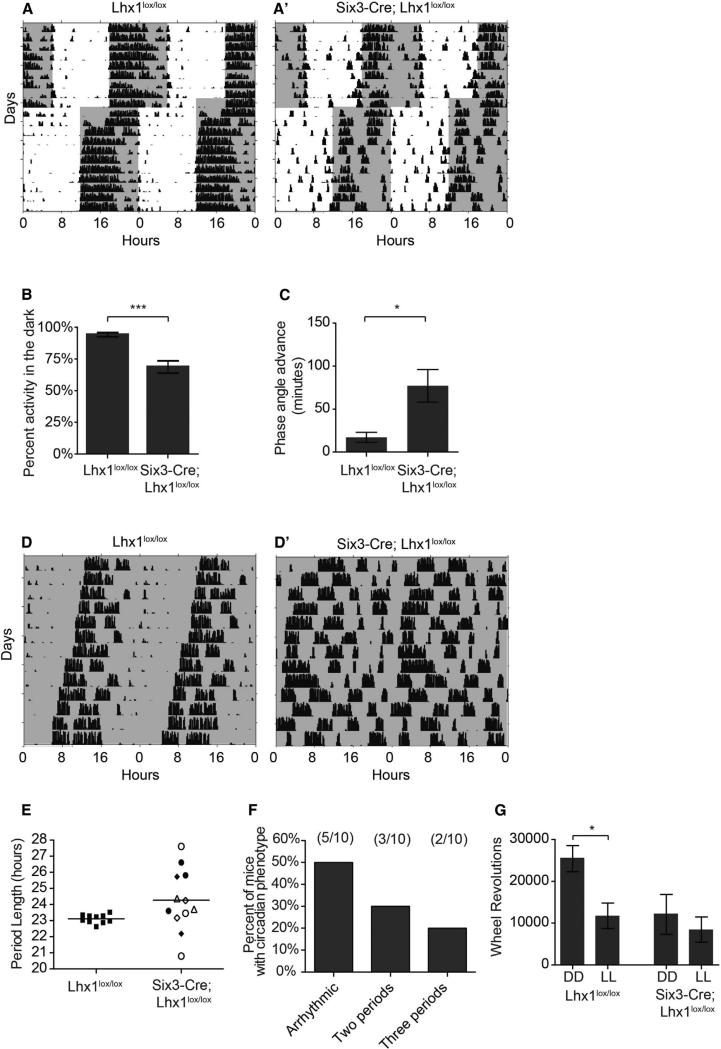
Figure 4. Loss of Lhx1 in the SCN Influences Photoentrainment and Leads to Loss of Free-Running Circadian Rhythms.
(A and A’) Representative actograms of wheel-running activity of Lhx1lox/lox (A) and Six3-Cre; Lhx1lox/lox (A’) housed in a 12 hr:12 hr light:dark cycle and subject to a 6 hr advance in the light/dark cycle. Gray shading represents lights off.
(B) Six3-Cre; Lhx1lox/lox mice show a decreased percentage of activity confined to the dark portion of the light/dark cycle (n = 10,13; p = 0.0002).
(C) Six3-Cre; Lhx1lox/lox mice show an advanced phase angle in 12:12 LD (n = 10, 11; p = 0.02). Data represent mean ± SEM. ***p < 0.001, *p < 0.05.
(D and D’) Representative actograms of wheel-running activity of Lhx1lox/lox (D) and Six3-Cre; Lhx1lox/lox (D’) mice housed in constant darkness. Six3-Cre; Lhx1lox/lox mice showed either arrhythmicitiy or split rhythms under DD.
(E) Quantification of free-running circadian period lengths in DD. Period lengths for Six3-Cre; Lhx1lox/lox are those found in mice with split rhythms. The multiple period lengths measured within a single animal are indicated by the same symbol in the mutant graph. (F) Distribution of circadian phenotypes observed in Six3-Cre;Lhx1lox/lox mice.
(G) Lhx1lox/lox activity is decreased in constant light as compared to constant darkness. This difference was not observed in Six3-Cre; Lhx1lox/lox mice (n = 9–11 per group; two-way ANOVA plight = 0.02, pgenotype = 0.02, Bonferroni posttest Lhx1lox/lox DD versus LL, p < 0.05). Data represent mean ± SEM. *p < 0.05.
However, this proved incomplete when we examined the animals in DD. Despite moderate damping and desynchrony phenotypes in Lhx1-deficient SCN similar to or milder than previous descriptions of Vip−/− and Vipr2−/− SCN rhythms (Aton et al., 2005; Brown et al., 2007; Maywood et al., 2011), free-running activity rhythms of Lhx1-deficient mice were profoundly disorganized (Figure 4D). A total of 50% of Six3-Cre;Lhx1lox/lox mice lacked measurable rhythms within 3 days of entry into DD, while the remainder showed two or even three distinct split rhythms, sometimes of dramatically different periods (Figure 4F). In contrast, 13% of Vip−/− mice on a similar mixed background became arrhythmic within 3 days of entry into DD; this increased only to 26% of Vip−/− mice over the course of several weeks in DD (Colwell et al., 2003). A subsequent study of Vip−/− mice backcrossed to C57BL/6 found that 64% of these animals had badly disrupted rhythms, but most were multirhythmic, not arrhythmic (Aton et al., 2005).
When a behavioral rhythm is measurable, Six3-Cre;Lhx1lox/lox free-running rhythms often lacked a clearly dominant rhythm and were not of significantly different average period than controls, although individual variability in period length was markedly greater (Figure 4E). This is in contrast to Vip−/− mice, which regardless of genetic background consistently have a clear dominant rhythm with a significantly shorter period than wild-type mice (Aton et al., 2005; Colwell et al., 2003).
Advanced phase angle and increased activity during the LD light phase also suggested Six3-Cre;Lhx1lox/lox mice have defects in light-dependent activity suppression. Although such a decrease was supported by the observation that overall activity in constant light (LL) and DD was similar, we noticed a substantial decrease in activity in mutants under DD relative to controls (Figure 4G), precluding a conclusion about masking from these data alone. To more rigorously measure masking, we first examined acute activity suppression of wheel-running activity by a 3 hr light pulse from zeitgeber time (ZT) 14–17 and found no significant difference in masking in Lhx1lox/lox and Six3-Cre;Lhx1lox/lox mice (Figure S4G). We then examined masking under an ultradian light cycle (3.5 hr light: 3.5 hr dark). Mice do not entrain to this cycle; instead, light pulses suppress wheel-running activity, permitting separation of light-masking effects from photoentrainment. The percentage of activity during the dark portion of the ultradian cycle in Lhx1lox/lox and Six3-Cre;Lhx1lox/lox mice was similar (Figure S4H). Interestingly, masking does not require the SCN (Redlin and Mrosovsky, 1999), indicating that specifically SCN-mediated circadian behaviors are impaired in this mouse line. This again contrasts with Vip−/− mice, which show substantially elevated activity in response to a dark pulse (Colwell et al., 2003).
Taken together, these data show that Six3-Cre;Lhx1lox/lox mice have disrupted circadian behavior including abnormal photoentrainment and disrupted free running rhythms, with the latter seeming substantially more severe than in Vip−/− mice. However, light-dependent masking of locomotor activity is normal in our mice.
Intact IpRGC Innervation of SCN After Lhx1 Deletion
The abnormal photoentrainment but intact masking of Six3-Cre;Lhx1lox/lox mice then led us to investigate whether Lhx1 loss altered retinal innervation of SCN. We utilized the Opn4tau-LacZ mouse to selectively label ipRGCs by X-gal staining (Güler et al., 2008; Hattar et al., 2002). Six3-Cre;Lhx1lox/lox; Opn4taulacZ/+ mutants and Lhx1lox/lox;Opn4taulacZ/+ controls had similar gross innervation of SCN, olivary pretectal nucleus, and lateral geniculate nucleus (Figures S4A and S4B). This was corroborated by labeling all RGCs using intravitreal cholera toxin tract tracing in Six3-Cre;Lhx1lox/lox mice and controls (Figures S4C and S4D). We then measured pupillary light reflex and visual acuity (optokinetic response) to assess the function of ipRGCs and RGCs, respectively. We found that ipRGCs are functional in Six3-Cre;Lhx1lox/lox mice, because pupillary light reflex was comparable to controls under both dim and bright light (Figure S4E). Six3-Cre;Lhx1lox/lox mice also show an optokinetic response, but we observed a small but significant reduction in visual acuity (Figure S4F). This is likely due to defects in horizontal cell lamination known to occur after Lhx1 deletion in the developing retina (Poché et al., 2007), which is targeted by our Cre driver (Furuta et al., 2000). Lhx1 does not play a specific role in the development of any other retinal cell type. Taken together, these results suggest that deficits in locomotor activity of Six3-Cre;Lhx1lox/lox mice were not a result of abnormal retinal input to the SCN.
Behavioral Rhythms of Six3-Cre;Lhx1lox/lox Mice Are Hypersensitive to Pulses of the SCN Synchronizing Neuropeptide Grp and Output Neuropeptide Prok2
Given the low-amplitude but persistent molecular and cellular rhythms in Lhx1-deficient SCN, we hypothesized that an in vivo pulse of Grp, known to synchronize disorganized SCN cellular oscillators in culture (Brown et al., 2005; Maywood et al., 2011), should consolidate our mutants’ behavioral rhythms. To allow approximate calculation of circadian time (CT) for the injections, we selected Six3-Cre;Lhx1lox/lox mice exhibiting the mildest subset of splitting phenotypes, including one exceptional mouse that usually exhibited essentially wild-type behavior, for our cannulation studies (Figures 5A, 5B, and S5A–S5C). When ISH was subsequently performed on the SCNs of the cannulated cohort, higher numbers of cells appeared to be both Lhx1 positive and neuropeptide positive than in previously examined Six3-Cre;Lhx1lox/lox mice (Figure S5D). This suggested that lower Cre efficiency was responsible for the better-preserved rhythms of these animals.
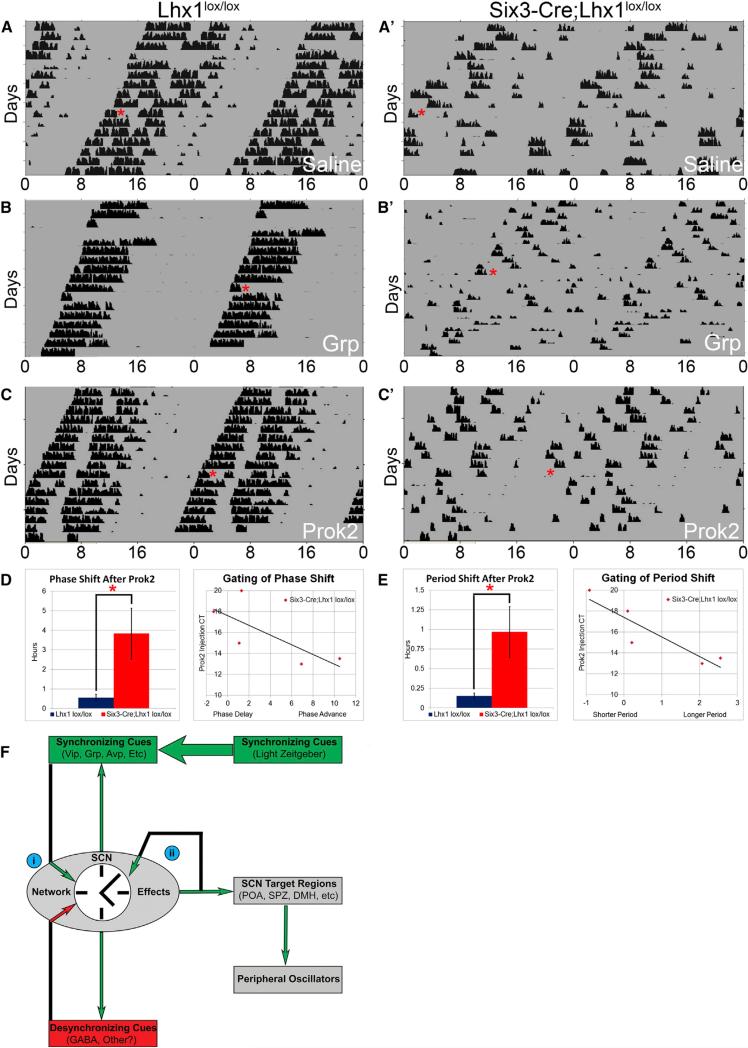
Figure 5. The Lhx1-Deficient Circadian System Is Hypersensitive to Cannulation of the SCN Output Molecule Prok2.
(A–C) Representative actograms of wheel-running activity of Lhx1lox/lox (A–C) and Six3-Cre;Lhx1lox/lox (A’–C’) mice before and after an i.c.v. injection (red asterisk) of saline (A), Grp (B), or Prok2 (C) in DD.
(D and E) Mutants had higher-magnitude phase shifts (D) and period shifts (E) in response to Prok2 than controls (left) (n = 8, two-tailed heteroscedastic t tests, *p < 0.05; bar graphs depict mean ± SEM). Both phase and period shifts in mutants were gated by injection CT (right) (n = 5, linear regression, scatterplots depict CT versus phase or period shift of individual injections).
(F) A model of SCN neuropeptide dynamics. Synchronizing factors are shown in green, desynchronizing factors are shown in red, and factors without clear implications for synchrony are shown in black. The following insights gained from our study are highlighted: (i) synchronizing and desynchronizing effects of SCN signals like Grp can be contingent on interpretation by the broader network, and (ii) core neuropeptides may buffer circadian period length, stabilizing it in the face of feedback signaling from SCN outputs like Prok2 that would otherwise alter it.
We first determined that an intracerebroventricular (i.c.v.) saline injection produced no effect regardless of genotype (Figure 5A). Control mice exhibited phase shifts after Grp cannulation (–0.55 hr ± 0.26, p < 0.05, one-tailed paired Student's t test), consistent with previous work (Piggins et al., 1995) (Figures 5B and S5A). Acute inhibition of locomotion was also often observed (Figure S5A). Surprisingly, visual inspection of acto-grams from Six3-Cre;Lhx1lox/lox mice after i.c.v. Grp injection showed considerable deconsolidation of activity rhythms (Figures 5B’ and S5A’). To quantify this, we compared the number of animals of both genotypes with measurable circadian phase before, but not following, Grp administration. We found a significant deleterious effect of Grp on mutant activity rhythms (post-injection phase measurable after 7/7 control injections versus 1/5 mutant injections, Fisher's exact test, p = 0.01).
Given the dramatic deconsolidating effect of Grp, we hypothesized that Lhx1-deficient SCN might be sensitive to pulses of SCN output neuropeptides that do not normally produce phase changes in wild-type animals. We tested this by i.c.v. injection of the SCN output neuropeptide Prok2. We found that i.c.v. Prok2 cannulation acutely inhibited locomotion without affecting either phase or period in control mice, as previously observed in rat (Figures 5C and S5B; Cheng et al., 2002). Remarkably, Prok2 significantly altered not just phase but also period in Six3-Cre;Lhx1lox/lox mice (p < 0.05; Figures 5C–5E and S5B’). Period shift after a single injection often lasted for many cycles (Figure S5B’). When rhythms were robust enough to calculate a more accurate post hoc circadian time of the injection (5/8 injections), there was clear circadian gating of both period shift (r2 = 0.82) and phase shift (r2 = 0.56), with the largest shifts occurring closest to the target injection time of CT14 (Figures 5D and 5E).
A prediction from these studies is that Prok2 will produce larger shifts in phase and period in mutant mice with the greatest reduction of SCN neuropeptides. The number of neuropeptidergic cells in Six3-Cre;Lhx1lox/lox SCN correlated well with the magnitude of phase shifts of mutant mice after Prok2 injection (r2 = 0.92 [Vip], r2 = 0.88 [Grp], r2 = 0.86 [Avp]) and, to some extent, also with their period shifts (r2 = 0.76 [Vip], r2 = 0.43 [Grp], r2 = 0.38 [Avp]). While fewer Vip and Grp cells correlated with larger phase and period shifts, Avp cell number had the opposite relationship (Figures S5D and S5E). In sum, our cannulation studies highlighted the importance of the SCN neuropep-tide network for buffering and transforming the circadian effects of its own signaling molecules.
DISCUSSION
Lhx1 Controls Terminal Differentiation of SCN Neurons
In this study, we found that Lhx1 deletion dramatically downregulates expression of several SCN-enriched neuropeptides crucial for circadian function (Figure 1; Table S1). However, the expression of pan-neuronal, GABAergic interneuron and other genes broadly expressed in anterior hypothalamus is largely intact in Six3-Cre;Lhx1lox/lox SCN, as are SCN-specific genes first detectable prior to embryonic day 16.5 (E16.5) (Figures 2A–2D and S2A–S2L). This suggests that transcriptional networks regulating regional identity, neuropeptidergic and GABAergic cell fate are separate developmental programs in the SCN. This contrasts with the only other neural system where Lhx1 is known to dictate neuropeptidergic fate; in spinal cord inhibitory interneurons, Lhx1 both drives Npy and Enk expression in conjunction with Lhx5 (Bröhl et al., 2008) and helps maintain GABAergic fate (Pillai et al., 2007). Lhx5 is not expressed in developing mammalian SCN, which may lead to Lhx1-dependent, SCN-specific expression of neuropeptides different from those in spinal interneurons (Bröhl et al., 2008).
While Lhx1 deletion also causes a comparatively moderate loss of neurons by P4 (Figures 2E and S2M), the region's gross anatomy remains essentially intact, with most surviving domain-specific markers retaining normal compartmentalization (Figures 2A–2D and S2A–S2L). Because SCN neurogenesis is mostly complete by E16 and TUNEL revealed elevated cell death in Lhx1-deficient SCN between P0 and P3 (Figures 2F and S2N), increased neuronal apoptosis likely decreased neuron number. Apoptosis also increased between P0 and P3 in both control and mutant SCN. This is in line with the timing of peak cell death in hamster SCN, which immediately precedes innervation by the retinohypothalamic tract (Müller and Torrealba, 1998). Neuron loss does not cause SCN neuropeptide downregulation, because many neuropeptidergic populations lost in adult Lhx1-deficient SCN (Vip, Avp, Grp, and possibly Prok2) are already reduced in Lhx1-deficient SCN at P0, prior to neuron loss (Figures 2G and S2O–S2R). Taken together, these data indicate that increased apoptosis and neuron loss are likely secondary to defects in SCN terminal differentiation in Six3-Cre;Lhx1lox/lox SCN.
Given that Lhx1 is first detectable at E11.5 when SCN neurogenesis begins, we initially hypothesized that Six3-Cre; Lhx1lox/lox mice would have an early SCN specification defect, akin to neuronal progenitor-specific deletion of Six3 (VanDunk et al., 2011) or constitutive deletion of Six6 or Lhx2 (Clark et al., 2013; Roy et al., 2013), rather than a terminal differentiation defect. Preservation of early-onset markers in Lhx1 mutants suggests that any role of Lhx1 in early SCN differentiation is redundant with other factors up to E16.5. One candidate is Lhx8, a close Lhx1 homolog coexpressed with Lhx1 in developing SCN from E11.5 until E16.5, when Lhx8 is downregulated (Shimogori et al., 2010). Future analysis of Lhx1/Lhx8 double mutants should resolve whether the latter is partially compensating for Lhx1 deletion.
More broadly, Lhx1's central role in terminal differentiation of multiple neuropeptidergic cell types in SCN presents an opportunity to dissect the transcriptional network guiding SCN cell fate determination. Leveraging this starting point, future investigations will be able to isolate both upstream regulators of Lhx1 expression and downstream factors that further subdivide SCN neuropeptidergic populations. Combined with identification of factors involved in initial SCN specification, such as Six3, Six6, and Lhx2 (Clark et al., 2013; Roy et al., 2013; VanDunk et al., 2011), this line of study could eventually allow stem cell lineages to be differentiated into specific SCN cell types, with profound implications for treatment of circadian-related sleep disturbances, mood disorders, and other pathologies.
Lhx1 Is Required for Normal SCN Cellular and Behavioral Rhythmicity
Despite loss of expression of multiple neuropeptides involved in synchronization of SCN oscillators in Six3-Cre;Lhx1lox/lox mice, rhythms of Per2::luciferase expression in SCN slice cultures showed normal average period and only moderately damped amplitude (Figure 3A). CCD imaging showed this was due to reduced cellular synchrony and increased period variability in Lhx1-deficient SCN (Figure 3B-C). Furthermore, our ISH time course showed that clock and clock-controlled gene expression is still temporally regulated in vivo after Lhx1 deletion (Figures 3D–3F and S3A–S3C). These deficits are similar to what has been observed shortly after culturing Vipr2−/− SCN slices (Brown et al., 2007).
The severe circadian deficits in Six3-Cre;Lhx1lox/lox DD activity rhythms were unexpected given the moderate effects on SCN oscillators in these animals (Figures 3 and 4). Although some mutants had milder splitting phenotypes (Figures 4F, 5, S5A, and S5B), most had multiple rhythms with distinct periods or had no measurable rhythms at all (Figures 4D and 4F). Typically, behavioral rhythms are more, not less, well organized than those of cellular clocks (Aton et al., 2005; Colwell et al., 2003; Harmar et al., 2002), making our observations unusual. Severe behavioral disorganization, normal average circadian period, a lack of altered activity rhythm masking by darkness, and the disconnect between robustness of cellular and behavioral rhythms all suggest that the phenotype of Lhx1 deletion in SCN is not simply a product of Vip loss (Aton et al., 2005; Colwell et al., 2003).
The most likely explanation rationalizing our SCN clock and behavioral data is that loss of expression of SCN neuropeptides following Lhx1 deletion leads the SCN to become not only internally disorganized but also uncoupled from downstream oscillators in the absence of external light cues. This suggests that Lhx1-regulated neuropeptide(s) and/or other signals could constitute the unidentified paracrine signals proposed to be sufficient for synchronization of peripheral oscillators by SCN (Silver et al., 1996). Prok2 is one likely component, because Prok2−/− SCN also seems to have a reduced ability to drive circadian rhythms (Li et al., 2006); however, the low percentages of Prok2−/− mice with severely disrupted rhythms (13% in mixed background and 20% after backcrossing to C57BL/6) suggest that it alone cannot account for the entirety of the Six3-Cre; Lhx1lox/lox phenotype. On the other hand, phase-advanced photoentrainment in Six3-Cre;Lhx1lox/lox mice, with an activity onset before dark, suggests that the Lhx1-deficient SCN does engage downstream oscillators when driven by light. Light entrainment may coordinate remaining SCN outputs, such as GABA (Moore and Speh, 1993), to preserve activity rhythms under light-dark conditions.
Finally, Lhx1-deficient mice did not decrease their activity in response to constant light, reflecting a decrease in overall activity (Figure 4G), despite having intact masking (Figures S4G and S4H). IpRGCs do not appear to contribute to this defect; they are functional and innervate their targets normally (Figures S4A–S4D). Both Vip−/− and Prok2−/− mice have decreased overall activity levels under DD (Colwell et al., 2003; Li et al., 2006); loss of these and other neuropeptides following Lhx1 deletion may act synergistically to drop DD activity to the same low, common baseline seen in wild-type mice in LL.
Implications for SCN Clock Function
Induction of arrhythmicity in Six3-Cre;Lhx1lox/lox mice given i.c.v. Grp (Figure 5B) suggests that whether a given neuropeptide promotes synchronization or desynchronization is not an intrinsic property of the molecule and its receptors alone. Instead, the effects of a given neuropeptide on synchrony also seem to depend on interpretation by the broader SCN neuropeptide network. It is known that large doses of Vip can transiently disrupt rhythms in normal mice, both in slice culture and in vivo, but that even very high doses of Grp fail to mimic this effect (An et al., 2013), and it has long been known that normal mice merely phase shift in response to exogenous Grp (Piggins et al., 1995). Based on the literature, one would expect excess Grp to help consolidate activity in Lhx1-deficient mice, as it does in slice cultures of Vip−/− SCN (Brown et al., 2005; Maywood et al., 2011). Instead, quite the opposite occurs, suggesting that another signaling molecule(s) present in the Vip−/− SCN underpins Grp's synchrony-promoting activity.
Similarly, after i.c.v. Prok2 injection in Six3-Cre;Lhx1lox/lox mice, dramatic changes in not just phase-shift magnitude, as might be predicted when the SCN oscillator is damped (Vitaterna et al., 2006), but also period length (Figures 5C–5E and S5C– S5E) were surprising. This neuropeptide has no effect on phase or period in wild-type animals (Cheng et al., 2002), which we reproduce in our study. The dramatic differences in the effects of Prok2 infusion in wild-type and Lhx1-deficient mice indicate a previously unappreciated Lhx1-dependent role for the SCN in buffering changes in phase and period induced by infusion of SCN-derived output neuropeptides. This provides insight into a central problem faced by the SCN: how to prevent changes in the central oscillator induced by both its own activity and afferent inputs from other brain regions, some of which use the same neurotransmitters, including Prok2 (Cheng et al., 2006; Krout et al., 2002).
We propose that period buffering is at least partially mediated by the core neuropeptides Vip and Grp. Their mRNA expression negatively correlated with the magnitude of both phase and period changes after i.c.v. Prok2 infusion (Figures S5D and S5E), consistent with extending the peptides’ well-documented ability to increase central oscillator robustness and thus control of circadian phase (Kuhlman et al., 2003; McArthur et al., 2000; Piggins et al., 1995) to stabilization of circadian period in the face of perturbation by SCN output molecules. Meanwhile, expression of Avp positively correlated with both phase-shift magnitude and change in period length (Figure S5D and S5E), consistent with its role in relaying circadian clock signals to the SCN core (Leak and Moore 2001; Schwartz et al., 2010). To what extent the in vivo circadian system is susceptible to Prok2 infusion when only core neuropeptides are lost, as in the Vip−/− mouse, presents an interesting question for future study.
In sum, our functional findings may explain why the SCN selectively expresses so many different neuropeptides, with signals from each individual neuropeptide cooperating to synergistically enhance behavioral stabilization (Figure 5F). Because the SCN expresses at least half a dozen different neuropeptides that are absent or greatly reduced in Six3-Cre;Lhx1lox/lox mice, this is a finding that is only readily obtainable using a developmental genetic approach that globally disrupts SCN neuropeptide expression and that would have been missed using conventional behavioral, pharmacological, or individual neuropeptide-mutation approaches.
Summary
Our work identifies distinct transcriptional networks selectively regulating SCN terminal differentiation, as opposed to initial SCN specification. Specifically, our study shows that Lhx1 loss selectively disrupts SCN terminal differentiation, leading to a failure to induce many neuropeptides crucial for normal circadian function (Figures 1 and 2). This discovery provides an entry point to dissect the transcriptional networks underlying development of the master clock and eventually determine how the region acquires its many unique circadian properties during early post-natal life.
Despite profound disruption of many components of the neuropeptide network, Six3-Cre;Lhx1lox/lox SCN rhythms are no more disrupted than those seen in Vip−/− SCN; however, Six3-Cre;Lhx1lox/lox mice show much more defective free-running circadian activity rhythms than anticipated from the disruption of their cellular clocks, particularly in the absence of light input (Figures 3 and 4). We believe that these phenotypes suggest a profound SCN output deficit in these animals; therefore, they will be useful for determining the contribution of multiple SCN neuropeptides in controlling coupling between the SCN and peripheral oscillators and the role of this coupling in regulating physiology and cognition.
Finally, our data extend the field's understanding of the highly interdependent nature of individual SCN neuropeptides’ functions (Figure 5F). The desynchronizing effects of exogenous Grp in our Six3-Cre;Lhx1lox/lox mice suggest that interactions among SCN neuropeptide signals can completely transform the effect of some signals on behavioral synchrony. Furthermore, our Prok2 cannulation studies identify a role for Lhx1-dependent elements of the SCN network, likely including the core neuropep-tides Vip and Grp, in maintaining robust circadian period length, as well as phase, in response to changing levels of secreted SCN output neuropeptides such as Prok2.
EXPERIMENTAL PROCEDURES
Animals and Housing
Six3-Cre;Lhx1lox/lox and control Lhx1lox/lox mice were used, with the following exceptions: Six3-Cre;Lhx1lox/lox;Opn4lacZ/+ and Lhx1lox/lox;Opn4lacZ/+ mice for X-gal staining, Six3-Cre;Lhx1lox/lox; Per2luc/+ and Lhx1lox/lox;Per2luc/+ mice for luciferase imaging, and Six3-Cre;ROSA26::YFP mice for Cre mapping. Genetically modified mice were provided by Y. Furuta (Six3-Cre), R. Behringer (Lhx1lox/lox), and Jackson Laboratories (Per2::luc). Behavioral studies used males; other experiments used both sexes. Except where otherwise noted, mice were on a 15 hr:9 hr light:dark cycle. All mice were mixed C57BL6/ Sv129 background. All controls were littermates. All procedures and care were approved by Johns Hopkins Institutional Animal Care and Use Committee, in line with National Institutes of Health guidelines for experimental animals.
Genotyping
Transnetyx conducted most genotyping using proprietary methods. Some floxed Lhx1 alleles were genotyped as described previously (Kwan and Beh-ringer, 2002), and some Cre was detected by presence of a ~300 bp band amplified using primers 5’-TTCCCGCAGAACCTGAAGAT-3’ and 5’-CCCCA GAAATGCCAGATTAC-3’.
Tissue Collection and Preparation
Brains were dissected, fresh frozen in OCT (Tissue-Tek) at the desired age, and stored at −80° C. Each experimental/control pair was collected in the same session. Clock gene time course mice were entrained to LD, moved to DD for 24 hr, and sacrificed in 4 hr increments on DD day 2. Next, 25 mm serial sections were cut on a Leica CM3050-S cryostat, thaw mounted on Superfrost Plus slides (Fisher), and stored at −80°C. Five adjacent slide sets were prepared for ISH; two were prepared for IHC and TUNEL.
In Situ Hybridization
Probes were selected and tissue processed as previously described (Shimogori et al., 2010).
Immunohistochemistry
Slides were fixed in 4% paraformaldehyde in 13 PBS, blocked in Superblock (Thermo Scientific), and then incubated overnight at 4°C in 1:200 monoclonal mouse immunoglobulin G (IgG) 2b anti-human-HuC/D (Invitrogen) or 1:500 polyclonal rabbit IgG anti-GFP (Molecular Probes) in PBS (5% horse serum, 0.16% Triton X-100 in 1X PBS). Slides were then incubated in 1:500 goat anti-mouse Alexa 568 IgG (Molecular Probes) or 1:500 donkey anti-rabbit Alexa 488 IgG (Molecular Probes), DAPI stained, and coverslipped with Gelvatol.
TUNEL Assay
Slides were processed with TMR Red In Situ Cell Death Kit (Roche) following the manufacturer's instructions for cryopreserved tissue.
X-Gal Stain
Slides were processed as described previously (Hattar et al., 2002).
Cholera Toxin Labeling
Mice were anesthetized with Avertin (20 mg/ml). Eyes were injected intravitreally with 2 ml cholera toxin B subunit (CTb) conjugated to Alexa Fluor 488 (Invitrogen). Three days later, mice were deeply anesthetized with 1 ml Avertin and transcardially perfused with 4% formalin. Brains were removed, postfixed 2 hr in 4% formalin, cryoprotected in 30% sucrose, and frozen in OCT (Tissue-Tek). Next, 50 mm sections were cut and free-floated in 0.1 M phosphate buffer, blocked (0.1 M phosphate buffer, 3% Triton X-100, 0.5% BSA, 1.5% rabbit serum) for 1 hr, incubated in goat anti-CTb (List Biologicals, 1:8,000) overnight at 4°C, and visualized with rabbit anti-goat Vectastain horseradish peroxidase kit (Vector Labs) using 3,3-diaminobenzidine (Sigma). Sections were mounted, dehydrated, and coverslipped with Permount and then viewed on a Zeiss Axio Imager M1 microscope.
Cell Counting
Slides were imaged by epifluorescence or bright field (Axioskop 2 Mot Plus, Zeiss Microscopy). After blinded manual counting of labeled SCN cells, total cells per hemisphere were estimated. Three brains per genotype per marker were counted. Statistical analysis was done in Excel (paired two-way t test) or Prism (two-way ANOVA with Tukey post hoc tests).
Circadian Time Course Densitometry
Slides were imaged by bright field (Axioskop 2 Mot Plus, Zeiss Microscopy) and analyzed in ImageJ. Rolling ball correction was applied to reduce background. Signal intensity from 0 to 255 arbitrary units was calculated in both SCN hemispheres and two cell-poor areas per image. Intensity of cell-poor regions was subtracted from SCN intensity, and mean and SEM d-intensity for each brain was computed in Excel. Two-way ANOVA analysis was done in Prism. Three brains per genotype per time point were used.
Real-Time Per2::Luciferase Measurement and Analysis
SCN slices were cultured and maintained at 36°C in darkness. Bioluminescence from each culture was either integrated every minute with a photomultiplier tube or for 15 s every 10 min with a CCD camera. Photomultiplier tube data were analyzed using modified published methods (Herzog et al., 2004). CCD data were stacked as previously described (Schindelin et al., 2012) and analyzed over 101 2 3 2 pixel regions of interest. See Supplemental Experimental Procedures for details.
Wheel-Running Activity
Mice were housed individually with a 4.5-inch running wheel and ad libitum food and water. Wheel running was monitored using Vitalview (Mini Mitter). Photoentrainment was assayed in a 12 hr:12 hr light:dark cycle (LD). Free-running circadian rhythms were assessed in constant darkness (DD) and constant light (LL). Light intensity in LD and LL was ~600 lux, provided by Philips Daylight deluxe fluorescent lamps. Clocklab (Actimetrics) was used to generate actograms and measure circadian periods. A 3 hr light pulse from ZT 14–17 was used to measure acute activity suppression (masking) by light. Wheel revolutions were quantified during the pulse and normalized to the previous day where mice were undisturbed in the dark. A 1-week 3.5 hr:3.5 hr light:dark ultradian light cycle was also used to measure masking. Wheel revolutions were quantified to determine total activity during light and dark phases. Data were analyzed by Student's t test.
Stereotaxic Cannulation and i.c.v. Injections
Surgeries and targeting validation were performed as described previously (Byerly et al., 2013). Briefly, unilateral cannulas were implanted in lateral cerebral ventricle of anesthetized mice. Successful cannula placement was verified in four Six3Cre;Lhx1lox/lox and four Lhx1 lox/lox mice by measuring feeding after NPY i.c.v. injection. A total of 2 μl of 114 μM Prok2, Grp, or saline vehicle was i.c.v. injected under dim red light into mice free-running in DD, at estimated CT 14. See Supplemental Experimental Procedures for details.
Optomotry
Behavior was assessed as described previously (Chen et al., 2011). Briefly, animals were adapted for 5 min to an elevated platform centered in a virtual cylinder. A sine wave grating was presented and acuity was assessed by the staircase method; stimulus tracking was recorded from above. Each mouse was tested in triplicate.
Pupillary Light Reflex
Behavior was assessed as described previously (Chen et al., 2011). Briefly, pupil size of one eye was video recorded for 15 s after 1 hr dark adaptation and then 30 s while the opposite eye was stimulated with a 470 nm light-emitting diode. Percent constriction relative to dark-adapted baseline was calculated from images captured from the video. Measurements were restricted to midday (ZT 4–8). Responses to low (22 μW cm2) and high (5.66 mW cm2) light intensities were recorded.
Motif Pair Tree Bioinformatic Analysis
The Lhx1 transcription factor binding site position weight matrix was downloaded from the Bulyk PDM Database (http://the_brain.bwh.harvard.edu/pbms/webworks2/explore.php). Proximal enhancer regions (2 kb upstream and 1kb downstream of transcription start site) were extracted from annotated reference genes, and only sequences conserved between mouse and human genomes were considered. We identified Lhx1 binding sites with motif pair tree (MOPAT) software (Hu et al., 2008), with 0.0001 as the p value cutoff defining positive hits.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Newman, J. Salvatierra, T. Thein, T. Pak, and D. Lee for blinding images for analysis; L. Jiang for technical assistance; B. Maier for Chronostar software; and J. Nathans, T. Shimogori, D. Lee, and W. Yap for comments on the manuscript. This work was supported by a Johns Hopkins Brain Science Institute grant to S.B. and S.H. and NIMH grant 63104 to E.D.H. J.L.B. was supported by a Visual Neuroscience Training Program fellowship from the Wilmer Eye Institute and a Graduate Research Fellowship from the National Science Foundation. S.B. was a W.M. Keck Distinguished Young Scholar in Medical Research.
Footnotes
AUTHOR CONTRIBUTIONS
J.L.B., T.A.L., E.A.S., E.D.H., S.H., and S.B. designed the study. J.L.B. performed most histology and analysis and assisted with cholera toxin tracing. T.A.L. performed most behavior and cholera toxin tracing and analysis and assisted with ISH time course; A.C.R. also conducted some behavior. E.A.S. and E.D.H. performed all Per2::luciferase imaging experiments and analysis. M.S.B. conducted cannulation experiments, assisted by J.L.B. and T.A.L. J.H. performed all bioinformatic analysis. H.W. conducted pilot ISH studies. J.L.B. and H.W. crossed and maintained all mice used. All authors contributed intellectually. J.L.B., T.A.L., S.H., E.A.S., E.D.H., and S.B. wrote the paper.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.03.060.
REFERENCES
- Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. J. Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Albers HE, Gillespie CF, Babagbemi TO, Huhman KL. Analysis of the phase shifting effects of gastrin releasing peptide when microinjected into the suprachiasmatic region. Neurosci. Lett. 1995;191:63–66. doi: 10.1016/0304-3940(95)11559-1. [DOI] [PubMed] [Google Scholar]
- Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74:246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- An S, Harang R, Meeker K, Granados-Fuentes D, Tsai CA, Mazuski C, Kim J, Doyle FJ, 3rd, Petzold LR, Herzog ED. A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc. Natl. Acad. Sci. USA. 2013;110:E4355–E4361. doi: 10.1073/pnas.1307088110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins N, Jr., Mitchell JW, Romanova EV, Morgan DJ, Cominski TP, Ecker JL, Pintar JE, Sweedler JV, Gillette MU. Circadian integration of glutamatergic signals by little SAAS in novel suprachiasmatic circuits. PLoS ONE. 2010;5:e12612. doi: 10.1371/journal.pone.0012612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröhl D, Strehle M, Wende H, Hori K, Bormuth I, Nave KA, Müller T, Birchmeier C. A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Dev. Biol. 2008;322:381–393. doi: 10.1016/j.ydbio.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Brown TM, Hughes AT, Piggins HD. Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J. Neurosci. 2005;25:11155–11164. doi: 10.1523/JNEUROSCI.3821-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Colwell CS, Waschek JA, Piggins HD. Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J. Neurophysiol. 2007;97:2553–2558. doi: 10.1152/jn.01206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly MS, Swanson RD, Semsarzadeh NN, McCulloh PS, Kwon K, Aja S, Moran TH, Wong GW, Blackshaw S. Identification of hypothalamic neuron-derived neurotrophic factor as a novel factor modulating appetite. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013;304:R1085–R1095. doi: 10.1152/ajpregu.00368.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda-Nieto AC, Pfaff SL, Varela-Echavarría A. Homeodomain transcription factors in the development of subsets of hindbrain reticulospinal neurons. Mol. Cell. Neurosci. 2005;28:30–41. doi: 10.1016/j.mcn.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Chen S-K, Badea TC, Hattar S. Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature. 2011;476:92–95. doi: 10.1038/nature10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou QY. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–410. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- Cheng MY, Leslie FM, Zhou QY. Expression of prokineticins and their receptors in the adult mouse brain. J. Comp. Neurol. 2006;498:796–809. doi: 10.1002/cne.21087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarleglio CM, Gamble KL, Axley JC, Strauss BR, Cohen JY, Colwell CS, McMahon DG. Population encoding by circadian clock neurons organizes circadian behavior. J. Neurosci. 2009;29:1670–1676. doi: 10.1523/JNEUROSCI.3801-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DD, Gorman MR, Hatori M, Meadows JD, Panda S, Mellon PL. Aberrant development of the suprachiasmatic nucleus and circadian rhythms in mice lacking the homeodomain protein Six6. J. Biol. Rhythms. 2013;28:15–25. doi: 10.1177/0748730412468084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;285:R939–R949. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis. 2000;26:130–132. [PubMed] [Google Scholar]
- Güler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao HW, Barnard AR, Cahill H, Badea TC, Zhao H, et al. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008;453:102–105. doi: 10.1038/nature06829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Harrington ME, Rahmani T, Lee CA. Effects of damage to SCN neurons and efferent pathways on circadian activity rhythms of hamsters. Brain Res. Bull. 1993;30:655–669. doi: 10.1016/0361-9230(93)90097-u. [DOI] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H. Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J. Biol. Rhythms. 2004;19:35–46. doi: 10.1177/0748730403260776. [DOI] [PubMed] [Google Scholar]
- Hu J, Hu H, Li X. MOPAT: a graph-based method to predict recurrent cis-regulatory modules from known motifs. Nucleic Acids Res. 2008;36:4488–4497. doi: 10.1093/nar/gkn407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean D, Bernier G, Gruss P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech. Dev. 1999;84:31–40. doi: 10.1016/s0925-4773(99)00068-4. [DOI] [PubMed] [Google Scholar]
- Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, Takahashi JS. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 2010;8:e1000513. doi: 10.1371/journal.pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krout KE, Kawano J, Mettenleiter TC, Loewy AD. CNS inputs to the suprachiasmatic nucleus of the rat. Neuroscience. 2002;110:73–92. doi: 10.1016/s0306-4522(01)00551-6. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, Silver R, Le Sauter J, Bult-Ito A, McMahon DG. Phase resetting light pulses induce Per1 and persistent spike activity in a sub-population of biological clock neurons. J. Neurosci. 2003;23:1441–1450. doi: 10.1523/JNEUROSCI.23-04-01441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Behringer RR. Conditional inactivation of Lim1 function. Genesis. 2002;32:118–120. doi: 10.1002/gene.10074. [DOI] [PubMed] [Google Scholar]
- Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J. Comp. Neurol. 2001;433:312–334. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- Lee JE, Atkins N, Jr., Hatcher NG, Zamdborg L, Gillette MU, Sweedler JV, Kelleher NL. Endogenous peptide discovery of the rat circadian clock: a focused study of the suprachiasmatic nucleus by ultrahigh performance tandem mass spectrometry. Mol. Cell. Proteomics. 2010;9:285–297. doi: 10.1074/mcp.M900362-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JD, Hu WP, Boehmer L, Cheng MY, Lee AG, Jilek A, Siegel JM, Zhou QY. Attenuated circadian rhythms in mice lacking the prokineticin 2 gene. J. Neurosci. 2006;26:11615–11623. doi: 10.1523/JNEUROSCI.3679-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JD, Burton KJ, Zhang C, Hu SB, Zhou QY. Vasopressin receptor V1a regulates circadian rhythms of locomotor activity and expression of clock-controlled genes in the suprachiasmatic nuclei. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;296:R824–R830. doi: 10.1152/ajpregu.90463.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O'Neill JS, O'Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O'Brien JA, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc. Natl. Acad. Sci. USA. 2011;108:14306–14311. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur AJ, Coogan AN, Ajpru S, Sugden D, Biello SM, Piggins HD. Gastrin-releasing peptide phase-shifts suprachiasmatic nuclei neuronal rhythms in vitro. J. Neurosci. 2000;20:5496–5502. doi: 10.1523/JNEUROSCI.20-14-05496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihai R, Juss TS, Ingram CD. Suppression of suprachiasmatic nucleus neurone activity with a vasopressin receptor antagonist: possible role for endogenous vasopressin in circadian activity cycles in vitro. Neurosci. Lett. 1994;179:95–99. doi: 10.1016/0304-3940(94)90943-1. [DOI] [PubMed] [Google Scholar]
- Moore RY, Speh JC. GABA is the principal neurotransmitter of the circadian system. Neurosci. Lett. 1993;150:112–116. doi: 10.1016/0304-3940(93)90120-a. [DOI] [PubMed] [Google Scholar]
- Mori K, Miyazato M, Ida T, Murakami N, Serino R, Ueta Y, Kojima M, Kangawa K. Identification of neuromedin S and its possible role in the mammalian circadian oscillator system. EMBO J. 2005;24:325–335. doi: 10.1038/sj.emboj.7600526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C, Torrealba F. Postnatal development of neuron number and connections in the suprachiasmatic nucleus of the hamster. Brain Res. Dev. Brain Res. 1998;110:203–213. doi: 10.1016/s0165-3806(98)00108-4. [DOI] [PubMed] [Google Scholar]
- Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- Piggins HD, Antle MC, Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J. Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai A, Mansouri A, Behringer R, Westphal H, Goulding M. Lhx1 and Lhx5 maintain the inhibitory-neurotransmitter status of interneurons in the dorsal spinal cord. Development. 2007;134:357–366. doi: 10.1242/dev.02717. [DOI] [PubMed] [Google Scholar]
- Poché RA, Kwan KM, Raven MA, Furuta Y, Reese BE, Behringer RR. Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J. Neurosci. 2007;27:14099–14107. doi: 10.1523/JNEUROSCI.4046-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser HM, Bradley A, Chesham JE, Ebling FJ, Hastings MH, Maywood ES. Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc. Natl. Acad. Sci. USA. 2007;104:648–653. doi: 10.1073/pnas.0606884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulivarthy SR, Tanaka N, Welsh DK, De Haro L, Verma IM, Panda S. Reciprocity between phase shifts and amplitude changes in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA. 2007;104:20356–20361. doi: 10.1073/pnas.0708877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redlin U, Mrosovsky N. Masking by light in hamsters with SCN lesions. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 1999;184:439–448. doi: 10.1007/s003590050343. [DOI] [PubMed] [Google Scholar]
- Romijn HJ, Sluiter AA, Pool CW, Wortel J, Buijs RM. Evidence from confocal fluorescence microscopy for a dense, reciprocal innervation between AVP-, somatostatin-, VIP/PHI-, GRP-, and VIP/PHI/GRP-immunoreactive neurons in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 1997;9:2613–2623. doi: 10.1111/j.1460-9568.1997.tb01691.x. [DOI] [PubMed] [Google Scholar]
- Roy A, de Melo J, Chaturvedi D, Thein T, Cabrera-Socorro A, Houart C, Meyer G, Blackshaw S, Tole S. LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J. Neurosci. 2013;33:6877–6884. doi: 10.1523/JNEUROSCI.4216-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MD, Congdon S, de la Iglesia HO. Phase misalignment between suprachiasmatic neuronal oscillators impairs photic behavioral phase shifts but not photic induction of gene expression. J. Neurosci. 2010;30:13150–13156. doi: 10.1523/JNEUROSCI.1853-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, et al. A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 2010;13:767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- VanDunk C, Hunter LA, Gray PA. Development, maturation, and necessity of transcription factors in the mouse suprachiasmatic nucleus. J. Neurosci. 2011;31:6457–6467. doi: 10.1523/JNEUROSCI.5385-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansteensel MJ, Magnone MC, van Oosterhout F, Baeriswyl S, Albrecht U, Albus H, Dahan A, Meijer JH. The opioid fentanyl affects light input, electrical activity and Per gene expression in the hamster suprachiasmatic nuclei. Eur. J. Neurosci. 2005;21:2958–2966. doi: 10.1111/j.1460-9568.2005.04131.x. [DOI] [PubMed] [Google Scholar]
- Vasalou C, Herzog ED, Henson MA. Small-world network models of intercellular coupling predict enhanced synchronization in the suprachiasmatic nucleus. J. Biol. Rhythms. 2009;24:243–254. doi: 10.1177/0748730409333220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Ko CH, Chang AM, Buhr ED, Fruechte EM, Schook A, Antoch MP, Turek FW, Takahashi JS. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc. Natl. Acad. Sci. USA. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AB, Angelo N, Huettner JE, Herzog ED. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc. Natl. Acad. Sci. USA. 2009;106:16493–16498. doi: 10.1073/pnas.0902768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wideman CH, Murphy HM, Nadzam GR. Vasopressin deficiency provides evidence for separate circadian oscillators of activity and temperature. Peptides. 2000;21:811–816. doi: 10.1016/s0196-9781(00)00213-8. [DOI] [PubMed] [Google Scholar]
- Xu Y, Baldassare M, Fisher P, Rathbun G, Oltz EM, Yancopoulos GD, Jessell TM, Alt FW. LH-2: a LIM/homeodomain gene expressed in developing lymphocytes and neural cells. Proc. Natl. Acad. Sci. USA. 1993;90:227–231. doi: 10.1073/pnas.90.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR, Westphal H. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc. Natl. Acad. Sci. USA. 2007;104:13182–13186. doi: 10.1073/pnas.0705464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.