Abstract
Metastatic melanoma remains a mostly incurable disease. Although newly approved targeted therapies are efficacious in a subset of patients, resistance and relapse rapidly ensue. Alternative therapeutic strategies to manipulate epigenetic regulators and disrupt the transcriptional program that maintains tumor cell identity are emerging. Bromodomain and extraterminal domain (BET) proteins are epigenome readers known to exert key roles at the interface between chromatin remodeling and transcriptional regulation. Here, we report that BRD4, a BET family member, is significantly upregulated in primary and metastatic melanoma tissues compared with melanocytes and nevi. Treatment with BET inhibitors impaired melanoma cell proliferation in vitro and tumor growth and metastatic behavior in vivo, effects that were mostly recapitulated by individual silencing of BRD4. RNA sequencing of BET inhibitor–treated cells followed by Gene Ontology analysis showed a striking impact on transcriptional programs controlling cell growth, proliferation, cell-cycle regulation, and differentiation. In particular, we found that, rapidly after BET displacement, key cell-cycle genes (SKP2, ERK1, and c-MYC) were downregulated concomitantly with the accumulation of cyclin-dependent kinase (CDK) inhibitors (p21 and p27), followed by cell-cycle arrest. Importantly, BET inhibitor efficacy was not influenced by BRAF or NRAS mutational status, opening the possibility of using these small-molecule compounds to treat patients for whom no effective targeted therapy exists. Collectively, our study reveals a critical role for BRD4 in melanoma tumor maintenance and renders it a legitimate and novel target for epigenetic therapy directed against the core transcriptional program of melanoma.
Introduction
Despite recent advances in treatment, metastatic melanoma remains a virtually incurable disease. Intrinsic and de novo resistance to chemo- or targeted therapies in melanoma has been attributed to the underlying molecular complexity that supports functional redundancy among survival pathways. To date, extensive research efforts have been dedicated to identify genetic mutations characteristic of these tumors, with notable success (1-3). In contrast, despite their relevance, epigenetic defects that participate in melanoma pathogenesis remain understudied. Thus, defining the contribution of epigenetic dysregulation in melanoma would broaden our understanding of its underlying biology and etiology. Recent work from our laboratories and others has revealed a role for rare histones (i.e., macroH2A; ref. 4), histone methyltransferases (i.e., SETDB1; ref. 5), and loss of DNA 5-hydroxymethylation on cytosine (5-hmC; ref. 6) in the pathogenesis of melanoma. In addition to highlighting the importance of epigenetic regulation, these studies point to potential alternative or complementary therapeutic approaches to the inhibition of certain signaling pathways [e.g., extracellular signal–regulated kinase (ERK) and phosphoinositide 3-kinase].
As a highly conserved class of epigenome readers, the bromodomain (BrD)-containing proteins have been shown to exert key roles at the interface between chromatin remodeling and transcriptional regulation. A left-handed four-helix bundle characterizes the three-dimensional structure of the BrD, which consists of a hydrophobic cleft between two conserved loops that interact with acetylated lysine residues (7). In humans, there are estimated to be 61 BrDs encoded in 46 proteins (8), including chromatin regulators of the SWI/SNF superfamily of DNA helicases (9), histone acetyltransferases (HAT; refs. 10-12), as well as the BrD and extraterminal domain (BET) family of transcriptional regulators. The BET family consists of BRD2, BRD3, BRD4, and the testis-specific member BRDT (13), which share a common domain architecture. BET proteins bind to acetylated lysine residues in histones, recruit chromatin-modifying enzymes to target promoters, and function as coactivators or corepressors in a context-dependent manner (14). Recent studies have revealed important roles for BET proteins in development, inflammation, and certain types of cancer (reviewed in ref. 14). For example, high BRD2 levels have been found in a subset of human leukemia, and BRD2 overexpression in the lymphoid lineage triggers the development of B-cell lymphoma (15, 16), suggesting a prooncogenic function for this protein. In addition, BRD4-NUT or BRD3-NUT fusions in certain squamous cell carcinomas result in a prooncogenic phenotype (17, 18). In contrast, BRD4 is lost in breast cancer and may serve as a tumor suppressor in that context (19).
Recently, specific small-molecule chemical compounds have been developed to block the acetyl-lysine binding of BET proteins. The availability of these highly cell-permeable and potent inhibitors allows investigating mechanistically the roles of BET proteins in a variety of biological systems (20, 21). In particular, BET inhibitors have demonstrable efficacy in blocking tumor progression in some cancer models including acute lymphoblastic leukemia, mixed lineage leukemia, and lung adenocarcinoma (22-24). However, a role for BET proteins has yet to be described in melanoma.
In this study, we assessed the effect of pharmacologically inhibiting the BET family of proteins in melanoma cells in vitro and in vivo. We found that members of this family are amplified and/or overexpressed in a substantial subset of melanoma specimens and cell lines, suggesting a prooncogenic function for BET proteins in these tumors. In particular, our findings reveal a new role of BRD4 in melanoma tumor maintenance by supporting cellular proliferation and controlling the expression of key cell-cycle and survival regulators. Collectively, our results define a previously unappreciated role for epigenetic readers in melanoma maintenance and support a paradigm shift in therapeutic intervention against this disease.
Materials and Methods
Analysis of mRNA melanoma datasets
Gene expression data of 20 metastatic melanoma cell lines (GSE22301; ref. 25) were used to analyze the expression of BrD-containing genes. Gene expression data of human samples (GEOD3189; ref. 26) was used to determine the expression of BRD2 and BRD4 in nevi and melanoma samples.
Analysis of BRD2 and BRD4 genomic locus
Affymetrix SNP6.0 Array data for melanoma cell lines (GSE22305; ref. 25) were used to analyze the gene copy number of BRD2 and BRD4. Software used was GeneSpring (Agilent Technologies).
Immunohistochemistry
Sections were deparaffinized in xylene, rehydrated through graded alcohols (3 changes grade 100% ethanol, 3 changes grade 95% ethanol), and rinsed in distilled water. Heat-induced epitope retrieval was carried out in a 1,200-W microwave oven at 100% power in 10 mmol/L citrate buffer pH 6.0 for 20 minutes. Primary antibody incubation and detection were carried out at 37°C on a NEXes instrument (Ventana Medical Systems) using Ventana’s reagent buffer and detection kits. Endogenous peroxidase activity was blocked with hydrogen peroxide. Appropriate secondary antibodies conjugated with streptavidin–horseradish peroxidase were used. The complex was visualized with 3,3′-diaminobenzidine and enhanced with copper sulfate or with Naphthol-AS-MX phosphatase and Fast Red complex; nuclei were counterstained with hematoxylin, dehydrated, and mounted with permanent media. Immunoreactivity of BRD2 and BRD4 was scored by intensity (0–4) and percentage of positive cells (0–4). Relative expression was obtained by multiplying intensity by percentage scored by an attending pathologist (F. Darvishian).
Cell lines
SK-MEL-29, SK-MEL-187, and SK-MEL-147 melanoma cell lines were kindly provided by Dr. Alan Houghton (Memorial Sloan-Kettering Cancer Center, New York, NY), and Hermes cells by Dr. Dorothy Bennett (University of London, London, UK); HEK293T, A375, SK-MEL-2, SK-MEL-5, and SK-MEL-28 cells were acquired from American Type Culture Collection. Human melanocytes (adult and neonatal) were purchased from Lonza and Yale University (New Haven, CT). Melanocytes, Hermes, and SK-MEL were cultured as previously described (27).
Proliferation assays
Cells were seeded at 2 × 103 cells per well on a 96-well plate (n = 6/condition). The day after (day 0), cells were treated with dimethyl sulfoxide (DMSO) or 10 μmol/L BET inhibitor (MS436/MS417). In the IC50 experiments, cells were treated with DMSO or increasing concentrations of MS436 or MS417 in the 2.5 to 20 μmol/L range. At the indicated time points, cells were fixed in glutaraldehyde 0.1% solution and stored in PBS at 4°C. At the end of the experiment, cells were stained with 0.5% crystal violet. Crystals were dissolved with 15% acetic acid and optical density was read at 590 nm.
Cell-cycle analysis
Cells were seeded at 1.5 × 105 cells per 6-cm dish (n = 3/condition) in the presence of vehicle (DMSO), 10 μmol/L MS436, or 10 μmol/L MS417. The media was changed every 24 hours and supplemented with vehicle or BET bromodomain inhibitor. After 72 hours, cells were fixed and permeabilized with cold ethanol 70%. Cells were washed with PBS and resuspended in propidium iodide buffer (PI 500 ng/mL, RNAse A 10 μg/mL). Cell-cycle profiles were obtained with FlowJo cytometry analysis software.
Quantitative real-time PCR
Total RNA was extracted using RNeasy Qiagen extraction kit according to the manufacturer’s instructions. One microgram of RNA was then subjected to DNase treatment and retro-transcription. Real-time PCR of BRD2, BRD3, BRD4, CDKN1B (p27), CDKN1A (p21), MYC, ERK1, and SKP2 was conducted using SYBR green fluorescence (Applied Biosystems). GAPDH was used as an internal standard. Relative quantification of gene expression was conducted with the 2−ΔΔCt method (28).
Western blot analysis
Cells were harvested with radioimmunoprecipitation assay (RIPA) buffer 1× (Thermo) supplemented with protease and phosphatase inhibitor cocktail (Roche). Cell lysates (25–30 μg of protein) were resolved in 4% to 20% Tris–glycine SDS-PAGE gels (Invitrogen) and transferred to nitrocellulose membranes. Membranes were blocked for 1 hour with 5% nonfat milk or 5% bovine serum albumin, and probed with primary antibodies overnight at 4°C [phospho-ERK1/2 (p-ERK1/2; Cell Signaling Technology; 4370), ERK1/2 (Cell Signaling Technology; 9102), c-MYC (Cell Signaling Technology; 9402), p21 (Calbiochem; OP64), SKP2 (Invitrogen; 32-3300), p27 (Cell Signaling Technology; 3686), HSP90 (Cell Signaling Technology; 4874S)]. Membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 hour before developing with enhanced chemiluminescence (ECL) plus Western blotting detection kit (GEHealth care).
Oligonucleotide transfection
siRNA SMARTpool of siBRD2, BRD3, and BRD4 were purchased from Dharmacon. Of note, 50 nmol/L of the corresponding siRNA was transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Transfection efficiency was monitored using 50 nmol/L BLOCK-iT Fluorescent Oligo (Invitrogen).
Plasmids and lentivirus production
pGIPZ-shBRD4-#1 and pGIPZ-shBRD4-#2 were purchased from Open Biosystems. Lentiviruses were propagated using previously described methods (27) and melanoma cells were transduced with viral supernatant supplemented with polybrene (2 μg/mL).
Colony formation assay
Cells were seeded at 250 cells per well in 6-cm well plates (n = 3). After 7 to 10 days of treatment, cells were stained with crystal violet, photographed, and scored.
Mouse xenograft
A375 cells were injected (1.5 × 106/mouse) in the flank of NOD/Scid/IL2γR−/− mice (Cat# 05557; NOG; n = 20). Once tumors were palpable, mice were randomized in two groups and treated daily intraperitoneally with vehicle (5% DMSO + 10% 2-hydroxypropyl-β-cyclodextrin) or with 50 mg/kg MS417.
For the short hairpin RNA (shRNA) experiments, A375 cells were infected with NSC or shBRD4 lentivirus (pGIPZ-shBRD4-#1) for 7 days and then injected (1.5 × 106/mouse) in the flank of NOG mice (n =12). Tumor volume was measured every 2 to 3 days during 2 weeks. Tumor weight was analyzed when the tumors were excised at the end of the experiment. Lungs and liver were removed and examined under a fluorescence-equipped dissecting scope. GFP-positive lung surface lesions were photographed and counted on each lobe of every specimen. Afterward, tissues were fixed in 10% formalin, paraffin-embedded, and 5 μm sections were hematoxylin and eosin (H&E)–stained.
Statistical analysis
Unless otherwise indicated, mean values ± SEM are representative of one of three independent experiments. Statistical significance was determined by unpaired t test (GraphPad Prism Software). Of note, *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Results
BRD2 and BRD4 are overexpressed in melanoma
Data mining of our gene expression profile of 22 human melanoma cell lines (25) revealed altered levels of several BrD-containing genes compared with normal or immortal melanocytes, including GCN5L2, PBRM1, BRD4, and BRD2 (Fig. 1A). Analysis of a gene expression profile of human melanoma tissue samples (26) confirmed that mRNA levels of BRD2 and BRD4 are consistently higher in melanomas (n = 44) compared with nevi (n = 18; P < 0.001 for both; Fig. 1B). However, GCN5L2 and PBRM1 levels were not found upregulated in the same set of human melanoma samples (Supplementary Fig. S1A and S1B). We further analyzed BRD2 and BRD4 protein expression by immunostaining in a melanoma tissue microarray containing nevi (n =7), primary (n = 30), and metastatic melanoma (n = 30). Higher BRD4 expression was detected in most primary and metastatic melanoma relative to nevi (P < 0.01 and P < 0.001; Fig. 1C and D). Some metastatic and primary melanomas also showed increased BRD2 expression, suggesting that this BET family member might also be altered in a fraction of patients with melanoma (Fig. 1E and F). The increased levels of these two BET family members in primary and metastatic melanoma suggest a potential role for BET proteins in promoting melanoma tumorigenesis.
Figure 1.
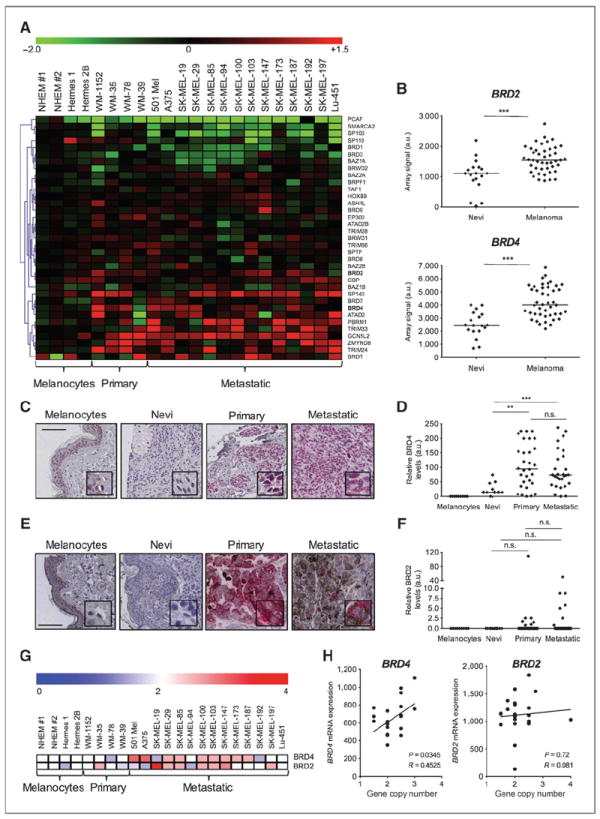
BRD2 and BRD4 are overexpressed in melanoma. A, heatmap representing the relative expression of BrD-containing proteins in melanocytes, primary melanoma cell lines and metastatic melanoma cell lines (GSE22301; ref. 25). B, microarray expression levels of BRD2 (top) and BRD4 (bottom) in nevi and melanoma human samples (GEOD3189; ref. 26). Line represents the median value. C and D, representative images of BRD4 staining of TMAs of human melanocytes, nevi, primary, and metastatic melanomas (C) followed by quantification of BRD4 signal (D). Bar, 100 μm. E and F, representative images of BRD2 staining of TMAs of human melanocytes, nevi, primary, and metastatic melanomas (E) followed by quantification of BRD2 signal (F). Bar, 100 μm. G, heatmap representing the BRD2 and BRD4 copy number in melanocytic and melanoma cell lines as extrapolated from a SNP array analysis (GSE22305; ref. 25). H, linear correlation of BRD2 (left) and BRD4 (right) mRNA expression levels with gene copy number calculated in G. n.s., not significant; a.u., arbitrary units. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
BRD2 and BRD4 loci are amplified in melanoma cell lines
Frequent chromosomal imbalances have been reported previously for the BRD4 locus (19p13) in patients with multiple myeloma (23). To investigate the mechanism(s) by which BRD4 is overexpressed in melanoma, we assessed the copy number status of the BRD4 gene in a panel of 18 melanoma cell lines using our previously reported SNP array data (ref. 25; Fig. 1G). Nine of 18 melanoma cell lines showed evidence of allele gain to various degrees, raging from 2.5 to four times more copies than human melanocytes. Moreover, we found a statistically significant correlation between increased copy number and BRD4 mRNA expression (P = 0.03; Fig. 1H), arguing that BRD4 overexpression may be explained, at least in part, by genomic amplification. Furthermore, the BRD2 locus (6p21) seemed to be amplified in 45% of melanoma cell lines but copy number did not correlate with mRNA levels, implying that other mechanism(s) might also account for BRD2 overexpression (Fig. 1G and H).
BET inhibition attenuates melanoma proliferation in vitro
A new class of diazepine-based small molecules has been shown to inhibit effectively and with high affinity the acetyllysine binding of BrD-containing proteins [i.e., JQ1 (29), iBET762 (30), and MS417 (31)]. We treated human metastatic melanoma cell lines (A375 and SK-MEL-147) with a diazobenzene BrD inhibitor MS436 (G. Zhang and M.-M. Zhou, in preparation) or with MS417, a thienotriazolodiazepine BrD inhibitor previously reported to have higher binding affinity and specificity for BET family members (31). Both compounds caused a fast occurring cytostatic effect (Fig. 2A and B) accompanied by G1 arrest (Fig. 2C and D), suggesting that specific inhibition of BET family members recapitulates the effects of general BrD inhibition in melanoma cells. Moreover, prolonged exposure to both compounds blocked colony formation and induced morphologic changes resembling a more differentiated state of melanoma cells (Fig. 2E and F). Our data suggest that inhibition of BrD proteins, and specifically of BET family members, has potent antiproliferative effects on melanoma cells, often associated with a differentiated phenotype.
Figure 2.
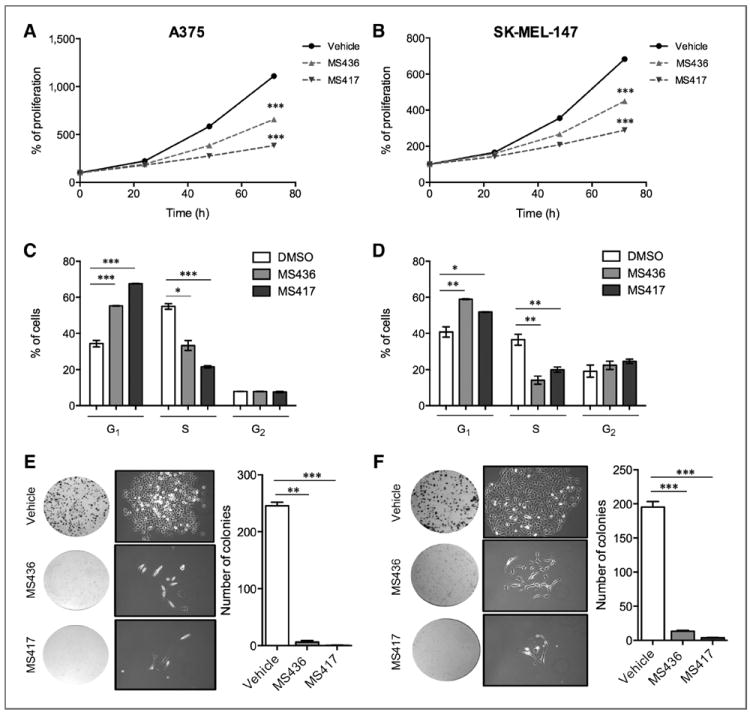
BRD or BET proteins inhibition attenuates proliferation in vitro. A and B, normalized proliferation curves of A375 (A) and SK-MEL-147 (B) treated with vehicle (DMSO), MS436 (10 μmol/L), or MS417 (10 μmol/L), measured by crystal violet staining. C and D, histograms representing the average percentage of A375 (C) or SK-MEL-147 (D) cells in G1, S, or G2–M phases after 72 hours of treatment with vehicle (DMSO), MS436 (10 μmol/L) or MS417 (10 μmol/L). E and F, macroscopic and microscopic images and quantification of colonies formed by A375 (E) or SK-MEL-147 (F) melanoma cell lines treated with vehicle (DMSO), MS436 (10 μmol/L), or MS417 (10 μmol/L). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
BET inhibition impairs melanoma tumor growth in vivo
To assess the antitumorigenic potential of BET inhibition in vivo, we tested the effects of MS417 in a xenograft mouse model. A375 metastatic melanoma cells were injected in the flanks of NOD/Scid/IL2γR−/− (NOG) mice (n = 10/group). Once tumors were palpable, we injected 50 mg/kg of MS417 daily intraperitoneally. MS417-treated mice displayed a 5-fold reduction in tumor growth and weight at the conclusion of the experiment (P < 0.001 for both; Fig. 3A–C). Mice treated with MS417 also showed a decrease in metastatic burden, with reduced number of metastasis-bearing mice and fewer lung micrometastases per lung section (Fig. 3D–F).
Figure 3.
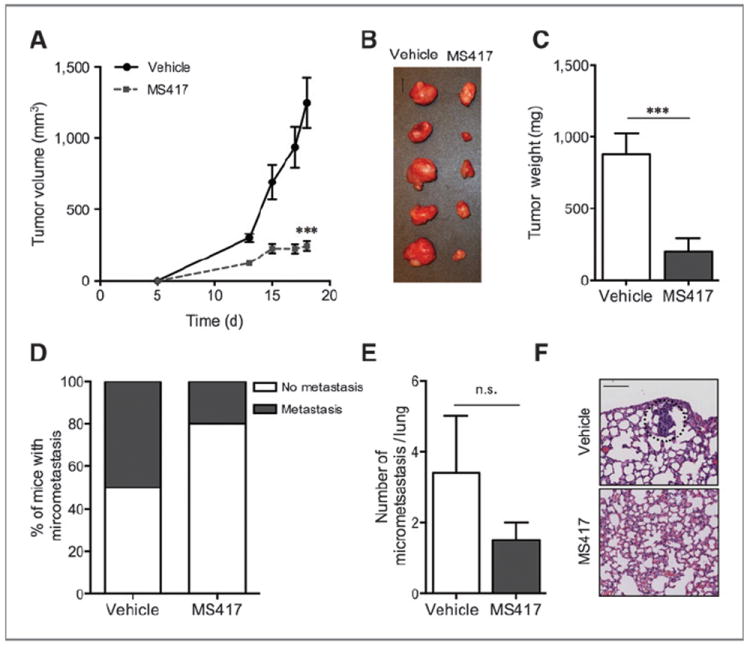
BET inhibition impairs melanoma tumor growth in vivo. A, average tumor volume of mice injected daily with either vehicle or MS417 (50 mg/kg; n = 10/treatment). B, macroscopic image of resected tumors at the conclusion of the experiment. Bar, 1 cm. C, average weight of resected tumors. D, percentage of mice bearing lung micrometastasis at the conclusion of the experiment. E, average number of micrometastasis per lung. F, representative microscopic H&E images of lungs. Metastatic focus is circled. Bar, 100 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
To determine whether the effects of BET inhibition in melanoma cell lines were reversible, we assessed the effects of drug withdrawal in vitro and in vivo. We conducted a proliferation assay in which two melanoma cell lines (A375 and SK-MEL-147) were continuously treated with MS417 (“continued” group) or switched to vehicle after 4 days of MS417 treatment (“discontinued” group). Two days after drug removal, cell morphology reverted to that of untreated cells, and cells resumed proliferation (Supplementary Fig. S2A and S2B), thus indicating that the antiproliferative effects of BET inhibition are reversible. In the xenograft model, A375 cells were injected subcutaneously in NOG mice, which were treated with MS417 or vehicle once tumors were palpable (5 days after injection). After 12 days, randomly selected mice were withdrawn from treatment (“MS417-Discontinued”; n = 9), whereas the rest remained treated (“MS417-Continued”; n = 6). Similar to the in vitro experiments, tumor growth resumed after discontinuing treatment (Supplementary Fig. S2C–S2E), highlighting melanoma cells dependence of BET proteins for proliferation.
BRD4 knockdown is sufficient to recapitulate the antitumoral effect of BET inhibitors in melanoma cells
Our comprehensive analysis of human samples and the potent effect of the BET inhibitor compounds support the relevance of BET proteins in melanoma. We depleted BRD2, BRD3, or BRD4 to further elucidate which BET family member, when inhibited, is responsible for the antiproliferative phenotype observed upon BET inhibitor treatment. Individual or combined siRNA-mediated suppression of BRD2, BRD3, or BRD4 was conducted in two independent melanoma cell lines and resulted in specific 60% to 80% mRNA reduction at 48 hours posttransfection (Fig. 4A and B). Strikingly, BRD4 knockdown caused a significant reduction in cell proliferation (Fig. 4C and D) associated with G1 arrest (Supplementary Fig. S3A and S3B), whereas BRD2 or BRD3 silencing had no or minimal effects on proliferation (Fig. 4C and D). Combinations of siBRD4 with siBRD2 or siBRD3 did not have any additive nor synergistic effect (data not shown). Similarly, stable suppression of BRD4 by shRNA lentiviral constructs consistently resulted in a stronger cytostatic effect than BRD2 or BRD3 stable silencing across a panel of four melanoma cell lines (Supplementary Fig. S3C–S3F). Furthermore, stable BRD4 silencing caused a 2- to 8-fold reduction in melanoma colony formation (A375 P < 0.001; SK-MEL-147 P < 0.01; Fig. 4E–H). Taken together, our results show that BRD4 silencing is sufficient to recapitulate the oncosuppressive effects of BET chemical inhibition on melanoma cells.
Figure 4.
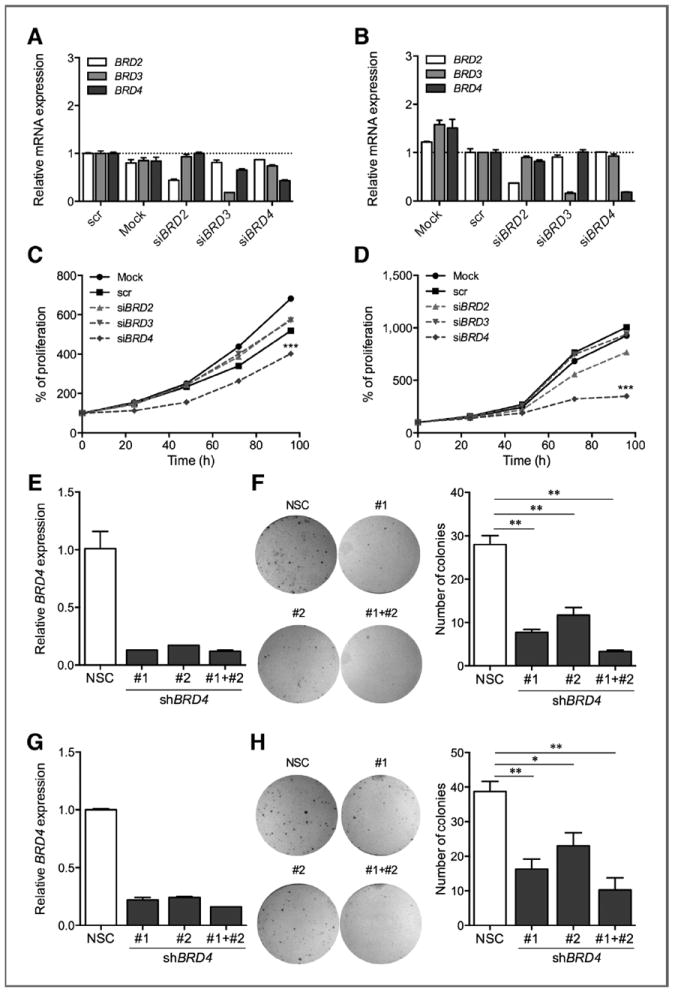
BRD4 knockdown is sufficient to recapitulate the antitumoral effects of BET inhibition in melanoma cells. A and B, mRNA levels of BET family genes in A375 (A) or SK-MEL-147 (B) melanoma cell lines treated with siRNA oligos (50 nmol/L). C and D, normalized proliferation curves of A375 (C) and SK-MEL-147 (D) treated with Mock, scr control, or siRNA oligos against BRD2, BRD3, and BRD4 (50 nmol/L), measured by crystal violet staining. E–H, relative BRD4 expression in A375 (E) or SK-MEL-147 (G) melanoma cells stably transduced with shRNA vectors against BRD4. Macroscopic images and quantification of clonal colonies formed by A375 (E and F) or SK-MEL-147 (G and H) melanoma cell lines transduced with shRNA vectors against BRD4. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
BRD4 is essential for melanoma tumor growth in vivo
Our in vitro data suggest that BRD4 could be a relevant therapeutic target in melanoma. To test the effects of BRD4 knockdown in vivo, A375 metastatic melanoma cells transduced with control (NSC; A375-NSC) or shBRD4 (A375-shBRD4) lentiviral constructs were injected in the flanks of NOG mice (n = 6/group). Tumors were first palpable at the same time in both experimental groups, suggesting that BRD4 knockdown does not affect tumor-initiation capacity. However, A375-shBRD4 injected mice showed a 3.3-fold reduction in tumor growth (P < 0.01) and a 3.2-fold reduction in tumor weight at termination (P < 0.05) compared with their A375-NSC counterparts (Fig. 5A–C). Histologic analysis of xenograft tumors revealed a strong correlation between BRD4 and Ki67 expression in both NSC and shBRD4 cohorts, suggesting that BRD4 provides a proliferative advantage (Fig. 5D). BRD4 silencing also resulted in a decrease of metastatic burden in the lungs (Fig. 5E and F). In sum, our data support BRD4 as a key contributor to melanoma proliferation and tumor maintenance and further strengthens BET inhibition as a plausible therapeutic treatment against melanoma.
Figure 5.
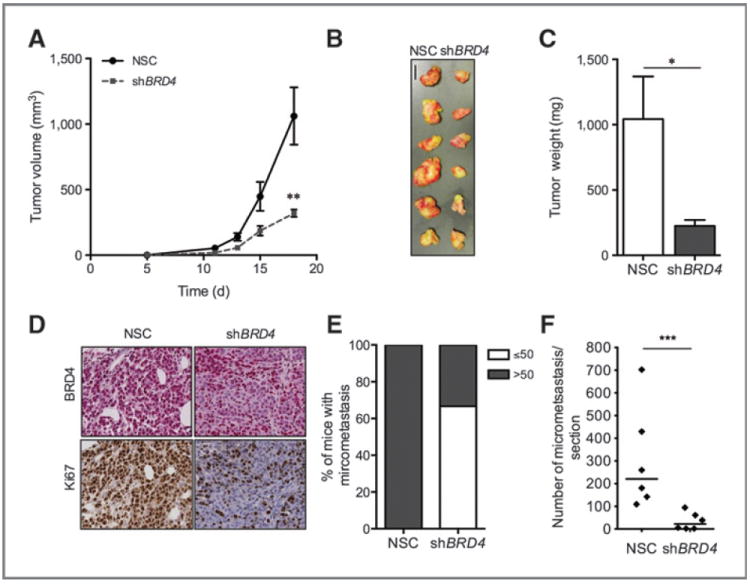
BRD4 is essential for melanoma tumor maintenance in vivo. A, tumor volume of mice injected with either NSC- or shBRD4-transduced A375 cells (n = 6/group), measured for 18 days. B, macroscopic image of resected tumors at the conclusion of the experiment. Bar, 1 cm. C, average weight of resected tumors. D, representative microscopic images of tumor histologic sections stained for either BRD4 (top) or Ki67 (bottom) in NSC- or shBRD4 A375 tumors. E, percentage of mice containing ≤50 or >50 lung micrometastasis at the conclusion of the experiment. F, average number of lung metastasis per lung. NSC, nonsilencing control. Line in F represents the median value. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
BET inhibition impacts transcriptional programs that control cell proliferation and differentiation
To investigate the cellular processes affected by BET inhibition, we analyzed the transcriptome of three melanoma cell lines (A375, SK-MEL-147, and SK-MEL-5) treated with the BrD inhibitor or vehicle for 48 hours using RNA sequencing (Illumina). Gene Ontology analysis (Ingenuity) of common differentially expressed genes showed an enrichment in biologic processes important for tumor development and progression such as cell proliferation, vasculature development, and cell differentiation (Fig. 6A).
Figure 6.
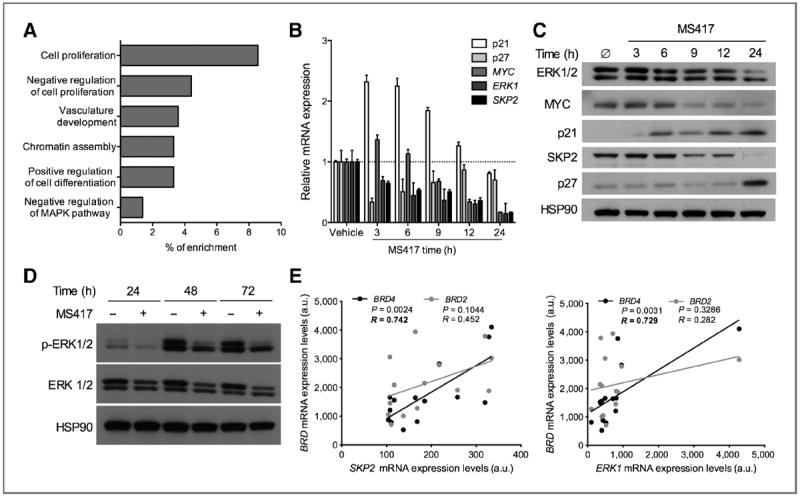
BET inhibition impacts transcriptional programs that control cell proliferation and differentiation. A, top categories of mRNA species significantly enriched (P < 0.005 for all categories) in three melanoma cell lines (SK-MEL-5, SK-MEL-147, and A375) treated with MS436 (10 μmol/L) when compared with vehicle (DMSO)–treated cells. B, relative mRNA expression of candidate downstream effectors following vehicle/MS417 treatment (10 μmol/L) over a 24-hour time course. C, protein levels of candidate downstream effectors following vehicle/MS417 treatment (10 μmol/L) over a time course of 24 hours. D, protein levels p-ERK1/2 following vehicle/MS417 treatment (10 μmol/L) for 24, 48, and 72 hours. E, a correlation between BRD2 or BRD4 mRNA levels with either SKP2 (left) or ERK1 (right) mRNA levels (EGEOD7553; ref. 32). a.u., arbitrary units.
We validated some of those pathway alterations at earlier time points, assuming that immediate gene expression changes might reflect potential direct BET targets. We found an overall reduction in total ERK1 mRNA and protein levels and reduced ERK1 phosphorylation, in response to BET inhibitor treatment (Fig. 6B–D), suggesting that ERK1 may be transcriptionally controlled by BETs. In addition, suppression of C-MYC (Fig. 6B and C), a well-established BRD4 target (14), occurred by 9 hours post-BET inhibitor treatment both at mRNA and protein levels. In contrast, the protein levels of the cyclin-dependent kinase (CDK) inhibitor p21 were rapidly upregulated, followed by those of p27, likely explaining the reduced cell proliferation and G1 accumulation observed upon BET inhibitor treatment of melanoma cells (Fig. 6C). Changes in mRNA levels of ERK1, MYC, and p21 were evident in this short time course, suggesting that protein changes of these factors could be mostly explained by transcriptional modulation. In contrast, p27 protein accumulation occurred at later time points, indicating that posttranscriptional mechanisms may account for its upregulation. In fact, the p27 ubiquitin ligase SKP2 was quickly downregulated in response to BET inhibitor treatment, likely leading to a reduced rate of p27 proteasomal degradation (Fig. 6B and C). Interestingly, SKP2 and ERK1 mRNA levels directly correlated with those of BRD4 in a panel of melanoma tissues (refs. 25, 32; R = 0.7; P = 0.02; Fig. 6E), suggesting that these two factors may be direct BRD4 targets. However, SKP2 overexpression alone was unable to rescue the antiproliferative effects of compound treatment (Supplementary Fig. S4A and S4B). Similarly, individual MYC overexpression and p21 knockdown were unable to neutralize MS417 effects (Supplementary Fig. S4C–S4F). Combinations of p21 knockdown with SKP2 (Supplementary Fig. S4G and S4H) or MYC overexpression (Supplementary Fig. S4I and S4J) were also unable to overcome MS417 cytostatic effects (Supplementary Fig. S4F), suggesting that BET inhibition leads to nonredundant, simultaneous regulation of multiple cell-cycle effectors, resulting in attenuated proliferation and increased differentiation of melanoma cells.
BRAF and NRAS mutation status does not influence melanoma cells sensitivity to BET inhibition
Because melanomas are molecularly diverse tumors (33), we decided to investigate which subset(s) of patients may benefit the most from BET inhibition. The antiproliferative capacity of BET inhibition was assessed in a panel of well-characterized melanoma cell lines representing: BRAF-mutant (A375, SK-MEL-5, SK-MEL-28, and SK-MEL-29), NRAS-mutant (SK-MEL-2 and SK-MEL-147), and BRAF/NRAS wild-type (SK-MEL-187) cell lines. In general, all cell lines tested exhibited comparable sensitivity to each drug regardless of their respective genetic background, with a notable cytostatic effect (Fig. 7 and Table 1). These data suggest that BET inhibitor response does not depend on the BRAF or NRAS mutational status of melanoma cells.
Figure 7.
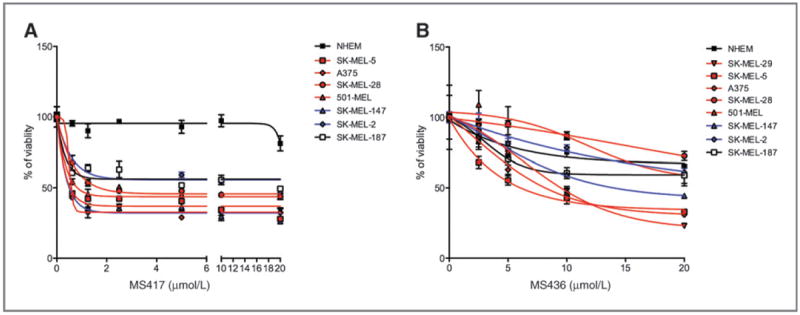
BRAF and NRAS mutation status does not influence sensitivity to BET inhibitors. A and B, sensitivity of primary melanocytes or melanoma cell lines to increasing concentrations of MS417 (A) or MS436 (B) determined by percentage of viability after 72 hours.
Table 1.
Estimated IC50 summary table of a panel of melanoma cell lines
| Cell Line | BRAF | NRAS | MS436 | MS417 |
|---|---|---|---|---|
| IC50 (μmol/L) | IC50 (μmol/L) | |||
| SK-MEL-29 | Mutant | Wild-type | 9.87 | Not determined |
| SK-MEL-5 | Mutant | Wild-type | 7.01 | 0.368 |
| A375 | Mutant | Wild-type | 9.28 | 0.057 |
| SK-MEL-28 | Mutant | Wild-type | 62.44 | 4.193 |
| 501 Mel | Mutant | Wild-type | 12.56 | 1.68 |
| SK-MEL-147 | Wild-type | Mutant | 14.69 | 0.144 |
| SK-MEL-2 | Wild-type | Mutant | 26.41 | 17.03 |
| SK-MEL-187 | Wild-type | Wild-type | 24.35 | 12.75 |
| NHEM | Wild-type | Wild-type | 104.23 | 887.29 |
Discussion
The profound effects of epigenetic modifications on cancer initiation and progression are becoming evident and are currently the subject of intensive investigation. In melanoma, epigenetic changes have been recently shown to contribute to tumor progression. The histone variant macroH2A, generally associated with condensed chromatin and fine-tuning of developmental gene expression programs, was shown to suppress melanoma progression. Its knockdown in melanoma cells resulted in increased proliferation and migration in vitro and tumor growth and metastatic potential in vivo (4). Another recent finding is the gradual loss of 5-hmC during melanoma progression. 5-hmC is a prevalent DNA modification and an intermediate of DNA demethylation, and is thought to affect gene regulation. Its loss significantly correlated with poor survival of patients with melanoma (6). Downregulation of SMARCB1 (INI1 and SNF5), a component of the SWI/SNF complex, allows bypassing BRAFV600E-induced senescence or apoptosis and promotes melanocyte transformation (34). In addition, melanoma samples display lower levels of SNF5 than nevi, correlating with poor prognosis and resistance to chemotherapy (35).
In the past decade, new small-molecule drugs that modify the epigenetic landscape of tumors have been found successful in improving patients’ survival (reviewed in ref. 36). U.S. Food and Drug Administration–approved histone deacetylase (HDAC) inhibitors vorinostat and valproic acid have been shown to potentiate the cytotoxic effects of chemotherapeutic agents (i.e., doxorubicin, flavopiridol, karenitecin) in phase I/II clinical trials (37, 38). However, melanoma remains largely resistant to current epigenetic therapy. For instance, phase II trials of HDAC inhibitor pyridylmethyl-N-{4-[(2-aminophenyl)-carbamoyl]-benzyl}-carbamate (MS-274) on refractory melanomas showed no objective response (39). The lack of response to tested epigenetic treatments in patients with melanoma suggests that the relevant dysregulated epigenetic factors have not yet been identified or targeted.
For a high percentage of patients for whom no targeted therapy is currently available, discovery of new druggable targets is a crucial line of investigation. A promising potent agent used in patients with late-stage melanoma is the monoclonal antibody ipilimumab, designed against the CTLA-4 molecule, which blocks CTLA-4 signaling and stimulates T-cell activation. The use of ipilimumab improves overall survival, but only 11% of patients exhibit complete responses (40). Selective BRAF inhibitors such as PLX4032 (vemurafenib) and GSK2118436 (dabrafenib) have shown unparalleled clinical efficacy in BRAFV600E-mutant metastatic melanoma, with approximately 60% of patients showing significant tumor regression (41). However, despite the spectacular initial responses, resistance to BRAF inhibitors rapidly follows and almost without exception. This phenomenon is attributed to functional redundancy between signaling pathways in tumor cells (42, 43) or by additional mechanisms, such as a spliced variant form of mutated BRAF that enhances dimerization in vemurafenib-treated cells (44). The progression-free survival for both ipilimumab- and vemurafenib-treated patients, although significantly improved, is still very short, suggesting that targeting one dysregulated signaling molecule or pathway may be insufficient to provide durable responses. To address this unmet need, we have investigated whether and how proteins of the BET family, for which small-molecule inhibitors have recently become available, could play a significant role in melanoma maintenance and progression.
Recent studies have shown that pharmacologic inhibition of BET/acetylated histone binding causes cell growth arrest, differentiation, or apoptosis in disease models including multiple myeloma (23), Burkitt’s lymphoma (45), acute myeloid leukemia (46), mixed lineage leukemia (22), and lung adeno-carcinoma (ref. 24; reviewed in ref. 47). Evidence of efficacy and safety of BET inhibition in preclinical testing is fueling the pharmacologic development of these compounds for further clinical evaluation.
Given our observation that BRD2 and BRD4 are overexpressed in human primary and metastatic melanomas, we hypothesized that their epigenetic and/or transcriptional regulation of certain target genes may support melanoma tumor-igenicity. Using our well-characterized small-molecule inhibitors (refs. 31, 48; G. Zhang and M.-M. Zhou, in preparation), we showed that BET inhibition reduces melanoma growth and metastatic capacity in vitro and in vivo. Most melanoma cell lines treated with BET inhibitors underwent G1 arrest with notable morphologic changes resembling a more differentiated state. BET inhibition in cell lines derived from hematologic malignances was shown to impair cell-cycle progression mainly by affecting the MYC transcriptional program and, in the case of Burkitt’s lymphoma, by triggering apoptosis. However, the mechanism by which BET proteins support tumorigenicity differs between cancer types. For example, FOSL1, and not MYC, has recently been described as the main effector of BET inhibitor-induced cell-cycle arrest in lung adenocarcinoma, suggesting that the BET-regulated transcriptome is likely cell context–dependent (24). Indeed, BET family members have been shown to control specific subsets of genes in different cell types. Thus, BETs regulate key inflammatory genes in activated macrophages (30), and BRDT controls germ cell transcripts in testis (13).
In our experimental system, RNA-sequencing of BET inhibitor–treated melanoma cells revealed that these compounds impair melanoma tumorigenicity mainly by affecting the transcriptional program that controls cell-cycle progression. In particular, MYC, ERK1, and SKP2 were rapidly down-regulated after BET displacement, whereas prominent cell-cycle inhibitors such as p21 and subsequently p27 were upregulated, providing a plausible mechanistic explanation to the robust cell-cycle arrest and attenuated tumor growth observed in response to BET inhibitor treatment. Whether the effects on transcription occurring after BET displacement are the result of changes in chromatin conformation, reduced transcriptional initiation, or defective elongation remains to be elucidated.
Although both BRD2 and BRD4 seem altered in melanoma samples, only BRD4 knockdown recapitulated the cell-cycle effects of BET inhibition in four different melanoma cell lines in vitro and suppressed tumor growth and metastasis in vivo. BET proteins contain two conserved BrDs in the N-terminal region of the protein that allow them to function as epigenome readers through binding to the acetylated lysine residues in histone tails. Protein–protein interactions through other conserved functional domains in the BET proteins such as the extraterminal domain or the more variable C-terminus domain (CTD) give them the capacity to assemble chromatin regulator complexes and broadly regulate gene transcription in the context of chromatin. BRD4-specific CTD confers it a unique control of gene expression. This domain promotes the assembly of the active transcriptional machinery directed by transcription factors such as p53 (49), NF-κB (50), and STAT3 (51). Moreover, it facilitates the recruitment of the positive transcriptional elongation factor-β (p-TEFβ) complex to activate RNA-Pol II and trigger pause release and transcriptional elongation (52, 53). It is plausible that this unique function of BRD4 in promoting transcriptional elongation distinguishes it from the rest of the BET family and critically contributes to its prooncogenic functions in melanoma. Interestingly, a class of inhibitors of dihydroorotate dehydrogenase (DHODH), such leflunomide, that inhibits the transcriptional elongation of genes required for neural crest development, has also recently shown to inhibit melanoma growth (54).
Our analysis of human specimens showing significant direct correlation between mRNA levels of SKP2 and ERK1 with BRD4 support these genes as potential BRD4 targets by which it may exert its prooncogenic role in melanoma. Interestingly, MYC levels correlated with BRD2 mRNA levels (data not shown), suggesting that although most cell cycle–related effects of BET inhibition can be attributed to BRD4 displacement, it is possible that the concomitant displacement of other BET proteins may broaden the oncosuppressive effects of these small-molecule compounds.
Our promising results open the possibility to test BET inhibitors in the clinical setting especially as targeted therapies are only available for patients with tumors bearing certain mutations. Interestingly, we observed that BRAF or NRAS status does not influence the response to BET inhibitor treatment. This may be due to the pleiotropic effects of BET inhibition in genome regulation that are evident by the lack of phenotypic rescue by single overexpression of MYC and SKP2 or knockdown of p21. Independence of BRAF status suggests BET inhibition as a possible new line of treatment for patients for whom no targeted therapy currently exists. In addition, because BET inhibitor treatment led to suppression of ERK1 and reduced its activated phosphorylated state, it is possible that its use in combination with BRAF inhibitor therapy may bypass or overcome resistance by impacting simultaneously both upstream and downstream of the MAPK pathway.
In summary, our findings reveal BRD4 as a prooncogenic factor in melanoma, which functions to regulate multiple key processes such as cell-cycle progression, survival, and proliferation. Our experimental results support the possibility of using BET inhibitors efficiently in the clinic, allowing for potent regulation of the epigenetic and transcriptional machinery in control of gene expression in the disease state.
Acknowledgments
The authors thank Dr. Jenny Xiang (Cornell University Genomics Core Laboratories, Ithaca, NY) for RNA sequencing experiments. The authors also thank Dr. Dorothy Bennett (UCL) for kindly providing us with human immortal melanocytes (Hermes), Dr. Alan Houghton (MSKCC) for some of the SK-MEL cell lines, and to the New York University (NYU) Histopathology and Immunohistochemistry Cores, supported in part by grant 5P30CA016087-32 from the National Cancer Institute, for tissue processing and histologic stainings.
Grant Support
This work was funded by the Elsa U. Pardee Foundation grant and the Melanoma Research Alliance Pilot Award (E. Hernando), and research grants from the NIH (M.-M. Zhou and E. Hernando).
Footnotes
Authors’ Contributions
Conception and design: M.F. Segura, B. Fontanals-Cirera, G. Zhang, M. Ohlmeyer, M.-M. Zhou, E. Hernando
Development of methodology: M.F. Segura, P. Gonzalez-Gomez, M. Morante, F. Darvishian
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): M.F. Segura, B. Fontanals-Cirera, M.V. Guijarro, L. Jubierre, F. Darvishian, I. Osman
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M.F. Segura, B. Fontanals-Cirera, A. Gaziel-Sovran, D. Hanniford, W. Zhang, I. Osman, E. Hernando
Writing, review, and/or revision of the manuscript: M.F. Segura, B. Fontanals-Cirera, A. Gaziel-Sovran, M.V. Guijarro, D. Hanniford, G. Zhang, I. Osman, M.-M. Zhou, E. Hernando
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M.F. Segura, G. Zhang, M. Morante, M.-M. Zhou
Study supervision: M.F. Segura, M.-M. Zhou, E. Hernando
Disclosure of Potential Conflicts of Interest
M.-M. Zhou, G. Zhang, and M. Ohlmeyer have ownership interest (including patents) in patent on BrD ligands. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev. 2012;26:1131–55. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–9. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–7. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–46. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–6. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Dev. 2009;12:659–65. [PMC free article] [PubMed] [Google Scholar]
- 9.Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, et al. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–72. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 10.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–9. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 11.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–3. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 12.Haynes SR, Dollard C, Winston F, Beck S, Trowsdale J, Dawid IB. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–84. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465–77. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenwald RJ, Tumang JR, Sinha A, Currier N, Cardiff RD, Rothstein TL, et al. E mu-BRD2 transgenic mice develop B-cell lymphoma and leukemia. Blood. 2004;103:1475–84. doi: 10.1182/blood-2003-06-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denis GV, Green MR. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 1996;10:261–71. doi: 10.1101/gad.10.3.261. [DOI] [PubMed] [Google Scholar]
- 17.French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–7. [PubMed] [Google Scholar]
- 18.Reynoird N, Schwartz BE, Delvecchio M, Sadoul K, Meyers D, Mukherjee C, et al. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J. 2010;29:2943–52. doi: 10.1038/emboj.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford NP, Alsarraj J, Lukes L, Walker RC, Officewala JS, Yang HH, et al. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci U S A. 2008;105:6380–5. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–45. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 21.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–34. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 22.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A. 2012;109:19408–13. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose AE, Poliseno L, Wang J, Clark M, Pearlman A, Wang G, et al. Integrative genomics identifies molecular alterations that challenge the linear model of melanoma progression. Cancer Res. 2011;71:2561–71. doi: 10.1158/0008-5472.CAN-10-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 27.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–9. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–23. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G, Liu R, Zhong Y, Plotnikov AN, Zhang W, Zeng L, et al. Down-regulation of NF-kappaB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem. 2012;287:28840–51. doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–82. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 34.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–74. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin H, Wong RP, Martinka M, Li G. Loss of SNF5 expression correlates with poor patient survival in melanoma. Clin Cancer Res. 2009;15:6404–11. doi: 10.1158/1078-0432.CCR-09-1135. [DOI] [PubMed] [Google Scholar]
- 36.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 37.Munster PN, Marchion D, Thomas S, Egorin M, Minton S, Springett G, et al. Phase I trial of vorinostat and doxorubicin in solid tumours: histone deacetylase 2 expression as a predictive marker. Br J Cancer. 2009;101:1044–50. doi: 10.1038/sj.bjc.6605293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dickson MA, Rathkopf DE, Carvajal RD, Grant S, Roberts JD, Reid JM, et al. A phase I pharmacokinetic study of pulse-dose vorinostat with flavopiridol in solid tumors. Invest New Drugs. 2011;29:1004–12. doi: 10.1007/s10637-010-9447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hauschild A, Trefzer U, Garbe C, Kaehler KC, Ugurel S, Kiecker F, et al. Multicenter phase II trial of the histone deacetylase inhibitor pyridylmethyl-N-{4-[(2-aminophenyl)-carbamoyl]-benzyl}-carbamate in pretreated metastatic melanoma. Melanoma Res. 2008;18:274–8. doi: 10.1097/CMR.0b013e328307c248. [DOI] [PubMed] [Google Scholar]
- 40.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–57. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 48.Borah JC, Mujtaba S, Karakikes I, Zeng L, Muller M, Patel J, et al. A small molecule binding to the coactivator CREB-binding protein blocks apoptosis in cardiomyocytes. Chem Biol. 2011;18:531–41. doi: 10.1016/j.chembiol.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, et al. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–63. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 50.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–87. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou T, Ray S, Lee C, Brasier AR. The STAT3 NH2-terminal domain stabilizes enhanceosome assembly by interacting with the p300 bromodomain. J Biol Chem. 2008;283:30725–34. doi: 10.1074/jbc.M805941200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biol Rep. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–54. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–22. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]


