Abstract
For bacterial model organisms like Escherichia coli and Bacillus subtilis genetic tools to experimentally manipulate the activity of individual genes existed for decades. But for genetically less tractable yet medically important bacteria such as M. tuberculosis such tools have rarely been available. More recently several groups developed genetic switches that function efficiently in M. tuberculosis and other mycobacteria. Together these systems utilize six different transcription factors, eight different regulated promoters, and three different regulatory principles. Here we describe their design features, review their main applications, and discuss advantages and disadvantages of regulating transcription, translation, or protein stability for controlling gene activities in bacteria.
Introduction
Genetic elements that enable specific and quantitative control over the activity of individual genes are irreplaceable components of the modern genetic toolbox. They facilitate not only the purification of proteins for biochemical, structural, or immunological studies but can also be applied to improve our understanding of in vivo gene functions. Until recently only one such tool was available for use in mycobacteria and its applicability in slow growing mycobacteria was limited. But during the last decade no less than a dozen new systems have been developed. Here we review the design, components, and regulatory mechanisms of the different systems and discuss their main applications.
Genetic switches for controlling gene expression in mycobacteria
The acetamidase system (Figure 1a)
Figure 1. Regulatory systems for mycobacteria.
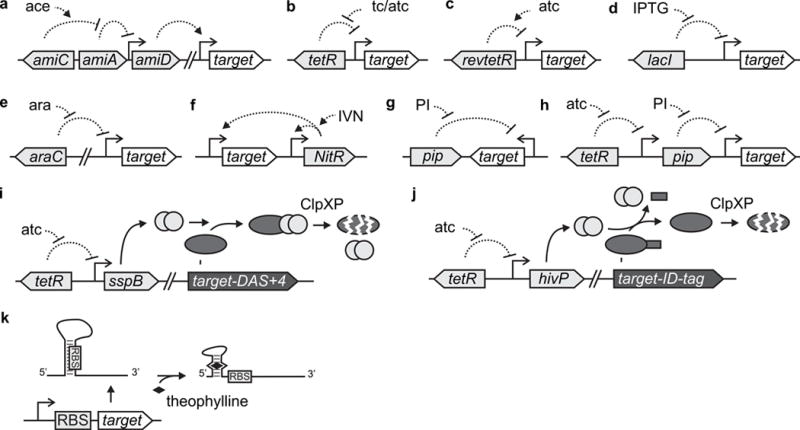
The transcriptional regulatory systems are shown in (a) to (h), the two controlled proteolysis systems in (i) and (j), and the theophylline riboswitch in (k). Dotted lines ending in a perpendicular line indicate negative regulatory interactions; dotted lines ending in an arrow represent positive regulatory interactions. Ace, acetamide; tc/atc, tetracycline / anhydrotetracycline; IPTG, isopropyl β-D-1-thiogalactopyranoside; ara, arabinose; IVN, isovaleronitrile.
During growth with short aliphatic amides (e.g. acetamide) as the primary carbon source M. smegmatis induces expression of the acetamidase encoded by amiE (1–3). The regulatory elements of this gene were utilized to generate the first inducible expression system for mycobacteria (4). The system proved valuable for the production of mycobacterial antigens (4, 5) and enabled the first silencing studies of essential genes (e.g. whmD and dnaA) in M. smegmatis (6, 7). But genetic instability limited the use of this system in M. tuberculosis (8) and its complexity – regulation of amiE involves three regulators (AmiC, AmiD, and AmiA) (9, 10) – prevented its optimization. While the acetamidase system has been largely replaced by other tools, especially in M. tuberculosis, a derivative, which incorporated the T7 RNA polymerase (RNAP), remains one of the best tools available to achieve high-level overexpression of a protein in M. smegmatis (11).
TetON and TetOFF (Figure 1b and 1c)
Tetracycline (tc) resistance of many bacteria is caused by efflux pumps whose expression is – due to the fitness defect the pumps cause in the absence of drug pressure – tightly regulated. This regulation is mediated by a single repressor protein, the tc repressor (TetR), which specifically binds two operators (tetO1 and tetO2) in the promoter that drives transcription of the efflux pump (12). In the complex with TetR the tet promoter (Ptet) is masked from access by RNAP and initiation of transcription is inhibited. When tc enters the bacterial cell it binds to TetR and induces transcription of the efflux pump before the drug can inhibit the ribosome. This sensitivity towards low drug concentrations is due to the remarkable affinity of TetR to tetracyclines, which is up to 105-fold higher than the ribosome’s affinity to tetracyclines (13).
In 2005, three groups independently reported TetR-controlled expression systems for mycobacteria (14–16). The systems shared the same basic design but differed in the origin of their regulatory components: The TetRs were derived either from the Corynebacterium glutamicum resistance determinant TetZ (14, 17) or the E. coli transposon Tn10 (15, 16); the regulated promoters were either also from TetZ (14, 17), derived from the B. subtilis xyl promoter (16), or constructed by inserting tetOs into a mycobacterial promoter (15). All three systems can be induced with low concentrations of tetracyclines in a dose-dependent manner with the preferred inducer either being tc (for the TetZ-derived systems) or anhydrotetracycline (atc) (for the two systems that utilize the Tn10 TetR). Because tc/atc has to be added to induce expression we refer to these systems as “TetON” systems.
The Tn10 TetR has been the subject of many mechanistic analyses. In a screening strain that proved particularly useful, TetR controlled expression of β–galactosidase while the lac repressor (LacI) and transcription of galK, encoding galactokinase, were repressed by LacI. This allowed to identify amino acids required for binding of TetR to tetO (mutations in these amino acids led to β–galactosidase positive and galactokinase negative colonies without atc) (18) or for induction of TetR by tetracyclines (19). A mutagenesis originally performed for the latter purpose also identified the first TetR that only bound tetO in complex with tc. Such reverse TetRs were later adapted for use in mycobacteria to construct a “TetOFF” switch in which transcription of the target gene is turned off by the addition of atc (20, 21). Optimization of these repressors for use in mycobacteria included adapting the guanine-cytosine (GC) content of the encoding genes to that of mycobacteria, which increased TetR expression and also led to an improved TetON system (21). TetON and TetOFF have been used by several groups to analyze gene functions in M. smegmatis and M. tuberculosis (Table 1). They also provide the basis for some of the other regulatory expression systems developed more recently (22–24) and a tunable coexpression system to analyze protein-protein interactions (25).
Table 1.
Regulated expression systems for mycobacteria and examples of their applications.
| Expression System | Components | Regulatory range | Applications |
|---|---|---|---|
| A. Regulation of transcription | |||
| Acetamidase | AmiC, AmiD, AmiA, promoters Pc, P1, P1, and P3 (9, 10) | ~80-fold induction of M. leprae 35 kD protein in M. smegmatis (4) 22-fold induction of FtsZ in M. smegmatis (81) |
Ectopic expression of mycobacterial antigens (4, 5), PknA/B (82, 83), and toxin-antitoxin proteins (63) in M. smegmatis Silencing of whiB2 (whmD) (6), wag31 (82, 83), kasA (84), inhA (84), dnaA (7) in M. smegmatis |
| Acetamidase inducible T7 RNAP | Not reported. | Overexpression of putative drug targets in M. smegmatis (11) | |
| TetON | TetR and Ptet from TetZ (14) | 230-fold induction of luciferase activity in M. smegmatis (14, 17) 13-fold induction of luciferase activity in M. tuberculosis (14) 21-fold induction of luciferase activity in M. bovis BCG (14) |
Ectopic expression of Ms2173 (85) and toxin-antitoxin proteins (86–89) in M. smegmatis Silencing of ftsZ (14), wag31 (82, 83) clpC1, pknB, msmeg_2694 (90), parA (91) and glmM (92) in M. smegmatis. Silencing of clpC1 (90), ppk1 (93), ppk2 (94) and dosR (95) in M. tuberculosis. |
| Tn10 TetR; Pmyc1tetO (15) | 170-fold induction of GFP activity in M. smegmatis (14) 160-fold induction of β–galactosidase activity in M. tuberculosis (14) |
Ectopic expression of PhoP (96), DosR (97), various sigma factors (64), various transcription factors (65), I-SceI (98, 99) PzaA/PncA (100), BirA (101), Pbp1 (102), EspR (103) and toxin-antitoxin proteins (86, 104–107) in M. smegmatis, M. bovis BCG, and/or M. tuberculosis Silencing of ftsZ (14), secA1 (20, 108), pptT (109) ripA (110) pbp1 (102) ppm1 (111) carD (60) msmeg_3935 (112) clpP (71) in M. smegmatis. Silencing of icl (58), rv3671c (58), prcBA (58, 59) pptT (56, 109) espA (113) bioA (55) fba (114) esx-3 (115) carD (60) pckA (57), clpP (71), panC (79), lysA (79) dfrA (116) in M. tuberculosis. |
|
| Tn10 TetR; PxyltetO (16) | ~10-fold induction of GFP activity in M. smegmatis and M. tuberculosis (16) | Silencing of trpD (16), dprE1 (117), clpP1 (117), fadD32, glnA1 (117), glnE (117), pknL (117), regX3 (117), senX3 (117) in M. tuberculosis | |
| TetOFF | Reverse TetR; Pmyc1tetO (20, 21) | 50-fold repression of β–galactosidase activity in M. smegmatis (21) 10-fold repression of β–galactosidase activity in M. bovis BCG (21) |
Silencing of secA1 (20) in M. smegmatis. Silencing of prcBA (59) panC (79), lysA (79), icl (79) in M. tuberculosis. |
| pBAD | AraC, PBAD (28) | ~3-fold induction of β–galactosidase activity in M. smegmatis (28) | Ectopic expression of Rv1991c (28) |
| LacI | LacI, PT7Lac, T7 RNAP (30) | 40-fold induction of GFP protein in M. tuberculosis (30) | Identification of metabolically active M. tuberculosis in macrophages (30) |
| LacI, PtrclacO | 30-fold induction of β–galactosidase activity in M. smegmatis (31) | Silencing of ftsZ, gyrA, and gyrB in M. smegmatis (31). Silencing of gyrA, gyrB, inhA, embR, rpoB, rpoC, rplJ, rpsL, and ilvB in M. tuberculosis (31). |
|
| NitR | NitR, PnitA (36) | >100-fold induction of XylE activity in M. smegmatis (36) ~100-fold induction of XylE activity in M. tuberculosis (36) |
|
| PipON | Pip, Pptr (41) | 52-fold induction of β–galactosidase activity in M. smegmatis (41). 450-fold induction of β–galactosidase in M. tuberculosis (41). |
Silencing of fadD32 (41) and pknB (41) ftsK (68), glf (68), infB (68), leuA (68), metC (68), rne (68), rv0883c (68), rv1478 (68), rv2050 (68), rv2204c (68), secY (68), tuf (68) in M. tuberculosis |
| TetPipOFF | Tn10 TetR, Pip, Pmyc1tetO, Pptr (24) | ~60-fold repression of β–galactosidase in M. smegmatis and M. tuberculosis (24) | Silencing of ftsZ (24) in M. smegmatis. Silencing of fadD32 (118) in M. abscessus. Silencing of fadD32 (24), eccB5 (119), eccC5 (119) esx-3 (120) in M. tuberculosis. |
| B. Regulation of protein stability | |||
| DAS+4-tag | Tn10 TetR, Pmyc1tetO, SspB | 36-fold, 250-fold repression of GFP and luciferase activity in M. smegmatis, respectively (23) 7-fold repression of GFP activity in M. tuberculosis (23). |
Depletion of RpoB in M. smegmatis (23) |
| ID-tag | Tn10 TetR, Pmyc1tetO, HIV2 derived protease | ~80-fold repression of GFP activity in M. smegmatis (22) >30-fold depletion of Alr protein; ~5-fold depletion of RpoB protein in M. smegmatis (22) |
Depletion of Alr (22), DHFR (22), InhA (22), GyrA (22), KasA (22), RpoB (22), and FhaA (72) in M. smegmatis |
| C. Repression of translation | |||
| Theophyllin | Riboswitch (53) | 65-fold and 89-fold induction of GFP and β–galactosidase activities in M. smegmatis (53) 8-fold induction of GFP activity in M. tuberculosis (53) |
Silencing of katG in M. smegmatis (53) |
AraC and LacI (Figure 1d and 1e)
Leakiness, i.e. expression without inducer, is a limitation of many regulated expression systems. One of the most tightly regulated E. coli expression systems is the pBAD system (26). Its promoter, PBAD, is controlled by two regulators: AraC, which represses the promoter without arabinose and activates it in its presence, and the catabolite activator protein, CAP, which acts as a second activating factor (27). Activation of PBAD by CAP increases with the intracellular cAMP concentration. In E. coli, activity of PBAD without arabinose can thus be reduced by adding glucose to the growth medium because glucose decreases cAMP levels in this species. Unfortunately, PBAD does not function in M. smegmatis as it does in E. coli (28) and there is no apparent advantage that pBAD has over the other systems developed for mycobacteria. Tight regulation of PBAD in E. coli depends not only on protein-DNA interactions but also on direct protein-protein interactions of AraC and CAP with RNAP as well as low levels of cAMP. It therefore would be difficult to optimize the pBAD system for use in mycobacteria.
Other frequently used E. coli expression systems depend on promoters that are repressed by LacI and induced with IPTG (29). Two studies demonstrated the value of LacI for regulating gene expression in mycobacteria. The first applied LacI to repress a promoter recognized by the T7 RNA polymerase (30); the second inserted a lac operator (lacO) downstream of a mycobacterial promoter to impose susceptibility to repression by LacI (31). For both systems little expression was measured without IPTG, but no follow up studies or applications have been published and their value for broader studies remains to be determined.
NitR (Figure 1f)
The saprophytic actinomycete Rhodococcus rhodochrous encodes several nitrilases, which detoxify nitriles by hydrolyzing them into their carboxylic acid and ammonia (32). Under optimal conditions R. rhodochrous J1 increases nitrilases expression up to ~3,000-fold, which results in the nitrilase encoded by nitA accounting for ~35% of total soluble protein (32, 33). This drastic overexpression is achieved via a positive feedback loop controlled by NitR, a member of the AraC family of transcriptional regulators. The molecular mechanism by which NitR acts has not been investigated in detail. But NitR alone is sufficient to mediate induction of PnitA and its own promoter in other bacterial species, most likely functioning as a direct activator of transcription initiation (34, 35). In M. smegmatis NitR strongly activated transcription after addition of either ε-caprolactam or isovaleronitrile whereas in M. tuberculosis only isovaleronitrile was effective (36). The positive feedback loop that is generated by NitR’s activation of its own promoter distinguishes this system from all other expression systems available for mycobacteria and has three consequences: (i) Induction is strong, (ii) on a single-cell level the switch is either ON or OFF, and (iii) intermediate inducer concentrations create two sub-populations, one that has NitR-controlled gene expression turned fully ON and one that is still in the OFF-state. In contrast, intermediate concentrations of atc partially activate the TetON system so that the average expression level of most cells increases to levels between the OFF and fully induced states (36).
PipON and Tet/PipOFF (Figure 1g and 1h)
Pristinamycin belongs to the streptogramin group of antibiotics, which consist of at least two structurally unrelated but synergistically acting molecules. In the case of pristinamycin, these two molecules are pristinamycin I and pristinamycin II, both of which inhibit bacterial ribosomes (37). Resistance of Streptomyces pristinaespiralis to pristinamycin is due to the pristinamycin resistance gene, ptr, which encodes a multidrug efflux pump (38). The ptr promoter, Pptr, is repressed by the transcription factor Pip and can be activated with pristinamycin I, pristinamycin II, and several other antibiotics (39, 40). Pip belongs to the TetR family of transcription factors and binds to three sites in Pptr, two of which overlap with the promoters −35 and −10 hexamers (40). Pptr is a strong promoter in M. smegmatis and M. tuberculosis, can be efficiently repressed by Pip and induced with low concentrations of PI. As a consequence the PipON system has an excellent regulatory range (41).
The Pip system was also adapted to confer repression upon addition of atc. In contrast to the TetOFF system, which utilizes a reverse TetR, in Tet/PipOFF Pip is placed under the control of wt TetR so that atc increases expression of Pip. The target gene is located downstream of Pptr and thus repressed as a consequence of the increased Pip expression. When desired, PI can be used to overcome the repression caused by atc (24). A system with a similar regulatory circuit had placed TetR under the control of the acetamidase system (42).
Controlled proteolysis (Figure 1i and 1j)
Bacterial regulatory circuits often rely on posttranscriptional modifications, which include controlled degradation, to achieve rapid inactivation of a protein. In fact, posttranscriptional modification is crucial to quickly inactivate proteins with a long half live as their abundances only change slowly even after transcription and translation have stopped (43). The recognition sites of bacterial proteases include C-terminal degradation tags (44). One such tag gets added to proteins in a process called trans-translation and is encoded by the small stable RNA ssrA (45). In E. coli ssrA-tagged proteins are degraded by several proteases including ClpXP, which directly binds to the tag’s C-terminal amino acids (46). Affinity of ClpXP to the ssrA-tag is increased by the adaptor protein SspB, which binds both the tag’s N-terminus and ClpX (47, 48). Proteins containing the ssrA-derived DAS+4-tag depend on the tethering of ClpXP to the tag by SspB. As a consequence they are only degraded when SspB is expressed (Figure 1i). This SspB-dependency is due to mutations that change the tags C-terminal amino acids from Leu-Ala-Ala to Asp-Ala-Ser (hence the “DAS”) and weaken the direct interaction with ClpX and an insertion of four amino acids (hence the “+4”) that facilitates simultaneous binding of SspB and ClpX (49). Interestingly, SspB is also capable of delivering DAS+4-tagged proteins to ClpXP in bacteria that do not themselves encode an SspB homolog (50). This provided the mechanistic basis for one type of gene silencing tool that utilizes proteolysis to deplete proteins in mycobacteria (23). A second such tool was developed by placing the ssrA-tag upstream of a protecting peptide that can be removed by a site-specific protease derived from HIV-2 (labeled as hivP in Figure 1j). The resulting tag was named inducible degradation (ID) tag (22). In both systems degradation of the tagged protein is induced with atc, which turns on expression of either SspB or the HIV-2 derived protease.
The theophylline riboswitch (Figure 1k)
Riboswitches are regulatory elements, in which binding of a small molecule to an RNA aptamer results in a change in gene expression (51). They are entirely RNA-encoded and do not require any trans-factors besides the aptamer-binding ligand, which can simplify transferring functional riboswitches from one species to another (52). The riboswitch adapted for use in mycobacteria is induced by theophylline (53), a methylxanthine drug used to treat pulmonary diseases (54). In the absence of theophylline the switch forms a secondary structure that masks the ribosome binding site (RBS) and thus prevents translation. Binding of theophylline stabilizes an alternative secondary structure, which liberates the RBS and induces translation of the regulated mRNA.
Common and distinctive features of the different regulatory systems and strategies
The ideal system for manipulating gene expression would (i) be completely silent under repressing conditions, (ii) provide a large (i.e. >1,000-fold) regulatory range that can be adjusted in a dose-responsive manner with a small-molecule that has no direct effects other than controlling the targeted gene, (iii) not interfere with the target’s native regulation under inducing conditions, (iv) leave the protein sequence unchanged, and (v) allow rapid gene induction and protein depletion in growing and non-replicating bacteria in vitro and during infections of host cells and animals. Not surprisingly, such a system has yet to be developed. But the available systems approach these features to different degrees.
Regulatory range
The range of regulated expression systems can be easily assessed using reporter gene assays. It is often calculated by dividing the reporter activity under inducing conditions by that measured under maximally repressing conditions. For most systems this has been achieved using either GFP, β–galactosidase, or luciferase as reporter. A regulatory range of >100-fold was measured for several systems (i.e. two of the TetON systems, the NitR system, PipON, and SspB-mediated proteolysis) with the largest range having been reported for PipON (Table 1).
Leakiness
Identifying expression systems that permit moderate expression without inducer is straightforward and can be achieved using the same reporter gene assays used to measure their regulatory range. However, none of the reporter assays that have been used to characterize mycobacterial expression systems approach single-molecule sensitivity. Lack of detectable reporter activity under repressing conditions, which has been reported for several systems, can therefore not provide proof of complete repression. In fact, all mycobacterial expression systems most likely permit some low level of expression without inducer. Whether or not this leakiness interferes with the goals of an experiment is difficult to predict and depends on the question that is being addressed and the gene under investigation. However, when necessary, the leakiness of an expression system can be reduced by decreasing the efficiency with which the targeted mRNA is translated (55).
Dose-responsiveness
All systems besides the one regulated by NitR have either been demonstrated to be dose-responsive or are likely to be dose-responsive in the sense that intermediate concentrations of the inducer or corepressor result in intermediate expression levels within most of the bacteria. Lack of dose-responsiveness of the NitR system comes at the benefit of achieving very high expression in the induced state.
Invasiveness
Controlling a gene’s expression is not possible without changing at least its promoter, the 5′ non-coding end of its mRNA, or the 3′-end of its open reading frame. An alteration of the promoter is required to allow for transcriptional regulation, the incorporation of the riboswitch changes the mRNA’s translation initiation sequence and its 5′-end, and the gene’s 3′-end and the C-terminus of the encoded protein need to be changed to achieve controlled proteolysis. Fortunately, these modifications have little impact on the function of many genes but any one of them can prevent complementation of a particular mutant. Strategies that rely on controlling transcription and/or translation have the advantage to leave the open reading frame of the targeted gene unchanged. Controlled proteolysis on the other hand can leave a target’s native regulation of transcription and translation intact. The theophylline riboswitch can also be used in combination with a gene’s native promoter and does not require changes of the regulated protein. Riboswitches can thus provide the least invasive strategy to artificially control gene expression in bacteria.
Regulation during infections
Providing control over M. tuberculosis gene expression during infections is a key ability of expression systems designed for this pathogen. Evidence that this can be achieved during macrophage infections has been obtained for TetON/OFF, PipON, Tet/PipON and the systems controlled by LacI or NitR. However, only for TetON/OFF have experiments been reported that demonstrated efficient regulation can be achieved in animal models (55–60).
Applications
Ectopic expression
One motivation for the construction of the acetamidase system was to enable purification of Mtb or M. leprae proteins from a fast-growing mycobacterial host, which was expected to yield proteins better suited for structural and immunological studies than those expressed in E. coil (4). The need for a mycobacterial expression host is supported by the finding that >50% of all Mtb proteins can either not be efficiently produced in E. coli or accumulate as insoluble inclusion bodies (61). For these proteins M. smegmatis can be a superior expression host because its codon usage is very similar to that of pathogenic mycobacteria, which facilitates high level expression of proteins encoded by GC-rich mRNAs. Furthermore, proteins that accumulate as insoluble inclusion bodies in E. coli can – at least in some cases – be expressed as soluble proteins in M. smegmatis (61). Purification of polyhistidine-tagged recombinant proteins from M. smegmatis can be complicated by contamination with co-purified GroEL1, but this can be avoided by using an M. smegmatis strain, in which the histidine-rich C-terminus of GroEL1 has been removed (62).
More recently ectopic expression was also used to analyze gene functions in M. smegmatis and M. tuberculosis. Many of these studies focused on type I toxin-antitoxin (TA) modules. These modules consist of two proteins that are often encoded by bicistronic operons wherein the 5′-gene encodes the antitoxin and the 3′-gene the toxin. As long as expression of the TA module continues, the toxin is bound and neutralized by its cognate antitoxin. Once expression stops, the inherently instable antitoxin is degraded leading to release and activation of the toxin. The M. tuberculosis genome encodes 88 putative TA modules, many of which are conserved within the M. tuberculosis complex yet absent from other mycobacteria (63). For many of these putative toxins, inducible overexpression was used to confirm that they are indeed functional toxins capable of arresting growth of M. smegmatis and/or M. tuberculosis (Table 1). This growth arrest generally does not occur upon simultaneous overexpression of the cognate antitoxin, i.e. the antitoxin encoded within the same TA module, but is not relieved by overexpression of other antitoxins (63). Another informative application has been to combine ectopic overexpression of DNA binding proteins witch chromatin immunoprecipitation (ChIP). This was first demonstrated in experiments that defined the in vivo binding sites of SigA and several alternative sigma factors (64). Recently, this approach has been extended to define the binding sites of many DNA binding proteins in M. tuberculosis (65).
Gene silencing
Controlled gene silencing allows studying a gene’s in vivo function under a variety of conditions even if the gene is required for growth. One conceptionally attractive strategy to conditionally inactivate a gene is to destabilize and prevent translation of its mRNA with an antisense RNA. This strategy was first applied to reduce expression of AhpC in M. bovis (66) and has since been used to inactivate several genes in M. smegmatis and M. tuberculosis (Table 1). One study in particular reported striking phenotypes for antisense-mediated gene silencing in several M. tuberculosis conditional knockdown (cKD) mutants (31). Attempts to silence different essential genes in M. smegmatis or M. tuberculosis with antisense RNAs of varying lengths in our own unpublished work have unfortunately all failed. The reasons for this failure are unclear to us and might be technical in nature. However, it is noteworthy that several research groups resorted to gene silencing approaches that are more complicated and time consuming than antisense mediated gene inactivation. Antisense mediated gene silencing thus likely failed frequently, which suggests either that expression of only a few genes is susceptible to antisense inhibition or that some of the factors important for the functionality of an antisense RNA remain to be identified.
An alternative to expressing antisense RNAs is to exchange the targeted gene’s promoter so that its transcription can be regulated directly. Promoter exchange can be achieved in situ, i.e. in the native chromosomal location, either by integrating a suicide plasmid immediately upstream of the targeted gene (15), by selecting for a double-crossover event that deletes the native promoter and replaces it with a regulated promoter (59), or by transposon insertion (67, 68). These strategies have been applied in many cases and most cKD mutants of M. tuberculosis or M. smegmatis published to date employed direct transcriptional repression (Table 1). Obtaining phenotypically well-regulated conditional knockdown mutants can, however, be challenging, especially for genes that only need to be expressed at a low level to be functional. In M. tuberculosis, bioA represents such a gene, whose mRNA is of low abundance during logarithmic growth (69, 70). It encodes the biotin biosynthetic enzyme 7,8-diaminopelargonic acid synthase, which is dispensable with extracellular biotin but essential for growth when biotin cannot be scavenged from the environment. The first BioA TetON mutant constructed with the Tn10-derived TetON system overexpressed BioA protein ~10-fold compared to wt M. tuberculosis (55). Removal of inducer decreased BioA expression by ~100-fold yet only mildly reduced growth. In its original form the Tn10-derived TetON system contains a strong Ptet located upstream of a strong translation initiation site. Strength of the promoter and the translational initiation site were likely both responsible for overexpression of BioA. It was unclear if decreasing promoter strength would sufficiently reduce bioA transcription without inducer; but weaker translational initiation sites were expected to decrease both BioA overexpression with inducer and leaky expression without inducer. Accordingly, cKD mutants containing a weak translational initiation signal upstream of the bioA open reading frame reproduced the phenotype of a bioA deletion and only grew with inducer when growth depended on biotin synthesis (55). In our hands, this strategy of minimizing the phenotypic consequences of transcriptional leakiness with weak translation initiation signals has been successful for several other targets (unpublished data) and is generally useful to improve the efficiency of transcriptional gene silencing.
Another elegant use of direct transcriptional silencing is its combination with transposon mutagenesis. This depends on a transposon carrying a regulated promoter at one end in the outward-facing direction and allows identifying well-regulated mutants based on their growth phenotypes (68).
cKD mutants that utilize transcriptional repression can be constructed by in situ promoter exchange, and similarly cKD mutants that utilize controlled proteolysis can be generated by modifying a gene’s 3′-end within its native location in the genome. This strategy has so far only been applied to the construction of cKD mutants in M. smegmatis, but shown good success in this species (22, 23, 71, 72). Nevertheless, for some targets depletion by controlled proteolysis was insufficient to produce the expected phenotypic consequences. For example, inactivation by controlled proteolysis of dihydrofolate reductase (DHFR) or alanine racemase (Alr), which are both essential for growth, depleted these enzymes by more than 97% but only modestly decreased growth of M. smegmatis (22).
Controlling gene expression during infections
Mutations that attenuate M. tuberculosis can cause growth in vivo (giv), severe giv (sgiv), and persistence (per) phenotypes in mice (73). Giv mutants replicate substantially less than wt, and sgiv mutants do not grow at all in mice whereas per mutants replicate normally but fail to persist. Genes required for growth and persistence, i.e. genes whose inactivation causes sgiv and per phenotypes can only be identified by conditional inactivation. The mycobacterial Tet systems helped demonstrate that the three sgiv genes bioA, pckA, encoding phosphenolpyruvate carboxy kinase and icl, which encodes isocitrate lyase, are required by M. tuberculosis not only to grow in mice and establish an infection, but also to persist during the chronic phase of the infection (55, 57, 58). A cKD mutant of the in vitro essential CarD revealed that M. tuberculosis depends on this transcriptional regulator for replication and persistence in mice (60). Similarly, 4′-Phosphopantetheinyl transferase PptT was shown to be required for the replication and survival of M. tuberculosis during the acute and chronic phases of infection in mice and helped validate these enzymes as a potential new drug target (56). The appearance of revertants, which are unresponsive to TetR mediated transcriptional control, can complicate the analysis of essential genes in vitro and in vivo (reference (60) and our unpublished observations). A careful analysis of the bacterial population expressing the regulated gene under investigation is therefore necessary for conclusive data interpretation.
Target-based whole cell screens
The application of regulated expression systems that can impact drug development most directly is their use in target-based whole cell screens. Such screens employ mutants in which expression of the target protein has been decreased to the extent that it limits the growth rate, which increases sensitivity towards small molecule inhibitors of that protein. This principle was initially established with Staphylococcus aureus strains, which were engineered to express growth-limiting amounts of FabF and showed an increased susceptibility to FabF-inhibitors but not to other antibiotics (74). Whole cell screens against this FabF-underexpressor identified platencin and platensimycin, the founding members of a new class of fatty acid biosynthesis inhibitors with broad-spectrum activity against Gram-positive bacteria (75–78). M. tuberculosis mutants expressing lower than wt levels of PanC, LysA, Icl1, or LepB have recently been constructed and also show target-specific changes in their susceptibility to different small molecule inhibitors (79, 80). Whole cell screens with these strains promise to identify new inhibitors of pantothenate synthase, diaminopimelate decarboxylase, isocitrate lyase, and the type I signal peptidase, respectively.
Conclusions and future perspectives
When the first edition of this book was published the only regulated expression system available was the acetamide system. Since then a dozen new regulatory systems have been developed that together utilize six different transcription factors (TetR, revTetR, AraC, LacI, NitR, and Pip) and eight different regulated promoters (PBAD, PT7Lac, PtrclacO, PnitA, Pptr, and three different Ptet promoters). They were applied not only to facilitate purification of correctly folded proteins but also to study mycobacterial gene functions within their native hosts either by ectopic expression or conditional inactivation. By now several mycobacterial expression systems function so efficiently that their use in most applications is straightforward. However, the isolation of phenotypically well-regulated cKD mutants remains challenging, irrespectively of the regulatory system one chooses for mutant construction. Reducing expression with antisense RNAs has been successful for some genes, but failed to silence at least as many. This is unfortunate, because antisense-based gene silencing does not require manipulation of the host chromosome by homologous recombination. It would thus become the most straightforward approach to generate cKD mutants if its success rate could be improved.
Direct transcriptional silencing was often but not always successful. Due to the inherent leakiness of most regulated promoters direct transcriptional silencing is most inefficient for genes whose products are only needed in low amounts. The opposite is likely true for controlled proteolysis because highly expressed proteins will burden the host’s proteolytic machinery more than proteins expressed at a lower level. That transcriptional silencing and controlled proteolysis can both fail to produce phenotypically well-regulate cKD is essentially a consequence of their limited dynamic range, which spans only 2 orders of magnitude. In contrast, M. tuberculosis gene expression, as measured by RNA sequencing, spans at least 4 to 5 orders of magnitude (69, 70). One of the main remaining challenges in the development of regulated expression systems for mycobacteria is thus to expand their dynamic range. In ongoing work we observed that this can be achieved by combining transcriptional repression with controlled proteolysis. This strategy of combining existing regulatory systems that differ in their mechanism of regulation could be further extended. For example, it should be possible to combine the theophylline riboswitch with any of the transcriptional regulation systems to reduce their effective leakiness yet still allow high level expression when necessary.
Acknowledgments
We are grateful for the support we have received from the National Institutes of Health, Bill & Melinda Gates Foundation, Wellcome Trust, Heiser Program for Research in Leprosy and Tuberculosis, Ellison Medical Foundation and Cornell University.
References
- 1.Draper P. Aliphatic Acylamide Amidohydrolase of Mycobacterium Smegmatis – Its Inducible Nature and Relation to Acyl-Transfer to Hydroxylamine. J Gen Microbiol. 1967;46:111. doi: 10.1099/00221287-46-1-111. [DOI] [PubMed] [Google Scholar]
- 2.Mahenthiralingam E, Draper P, Davis EO, Colston MJ. Cloning and Sequencing of the Gene Which Encodes the Highly Inducible Acetamidase of Mycobacterium-Smegmatis. J Gen Microbiol. 1993;139:575–583. doi: 10.1099/00221287-139-3-575. [DOI] [PubMed] [Google Scholar]
- 3.Parish T, Mahenthiralingam E, Draper P, Davis EO, Colston MJ. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiol-Uk. 1997;143:2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 4.Triccas JA, Parish T, Britton WJ, Gicquel B. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. Fems Microbiol Lett. 1998;167:151–156. doi: 10.1111/j.1574-6968.1998.tb13221.x. [DOI] [PubMed] [Google Scholar]
- 5.Daugelat S, Kowall J, Mattow J, Bumann D, Winter R, Hurwitz R, Kaufmann SH. The RD1 proteins of Mycobacterium tuberculosis: expression in Mycobacterium smegmatis and biochemical characterization. Microbes and infection / Institut Pasteur. 2003;5:1082–1095. doi: 10.1016/s1286-4579(03)00205-3. [DOI] [PubMed] [Google Scholar]
- 6.Gomez JE, Bishai WR. whmD is an essential mycobacterial gene required for proper septation and cell division. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8554–8559. doi: 10.1073/pnas.140225297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greendyke R, Rajagopalan M, Parish T, Madiraju MV. Conditional expression of Mycobacterium smegmatis dnaA, an essential DNA replication gene. Microbiology. 2002;148:3887–3900. doi: 10.1099/00221287-148-12-3887. [DOI] [PubMed] [Google Scholar]
- 8.Brown AC, Parish T. Instability of the acetamide-inducible expression vector pJAM2 in Mycobacterium tuberculosis. Plasmid. 2006;55:81–86. doi: 10.1016/j.plasmid.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Parish T, Turner J, Stoker NG. amiA is a negative regulator of acetamidase expression in Mycobacterium smegmatis. BMC microbiology. 2001;1:19. doi: 10.1186/1471-2180-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts G, Muttucumaru DG, Parish T. Control of the acetamidase gene of Mycobacterium smegmatis by multiple regulators. Fems Microbiol Lett. 2003;221:131–136. doi: 10.1016/S0378-1097(03)00177-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Jain P, Gulten G, Liu Z, Feng YC, Ganesula K, Motiwala AS, Ioerger TR, Alland D, Vilcheze C, Jacobs WR, Sacchettini JC. Mycobacterium tuberculosis Dihydrofolate Reductase Is Not a Target Relevant to the Antitubercular Activity of Isoniazid. Antimicrob Agents Ch. 2010;54:3776–3782. doi: 10.1128/AAC.00453-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillen W, Berens C. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annual review of microbiology. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 13.Lederer T, Kintrup M, Takahashi M, Sum PE, Ellestad GA, Hillen W. Tetracycline analogs affecting binding to Tn10-Encoded Tet repressor trigger the same mechanism of induction. Biochemistry. 1996;35:7439–7446. doi: 10.1021/bi952683e. [DOI] [PubMed] [Google Scholar]
- 14.Blokpoel MC, Murphy HN, O’Toole R, Wiles S, Runn ES, Stewart GR, Young DB, Robertson BD. Tetracycline-inducible gene regulation in mycobacteria. Nucleic acids research. 2005;33:e22. doi: 10.1093/nar/gni023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic acids research. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll P, Muttucumaru DG, Parish T. Use of a tetracycline-inducible system for conditional expression in Mycobacterium tuberculosis and Mycobacterium smegmatis. Applied and environmental microbiology. 2005;71:3077–3084. doi: 10.1128/AEM.71.6.3077-3084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams KJ, Joyce G, Robertson BD. Improved mycobacterial tetracycline inducible vectors. Plasmid. 2010;64:69–73. doi: 10.1016/j.plasmid.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wissmann A, Wray LV, Jr, Somaggio U, Baumeister R, Geissendorfer M, Hillen W. Selection for Tn10 tet repressor binding to tet operator in Escherichia coli: isolation of temperature-sensitive mutants and combinatorial mutagenesis in the DNA binding motif. Genetics. 1991;128:225–232. doi: 10.1093/genetics/128.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecht B, Muller G, Hillen W. Noninducible Tet repressor mutations map from the operator binding motif to the C terminus. Journal of bacteriology. 1993;175:1206–1210. doi: 10.1128/jb.175.4.1206-1210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo XV, Monteleone M, Klotzsche M, Kamionka A, Hillen W, Braunstein M, Ehrt S, Schnappinger D. Silencing Mycobacterium smegmatis by using tetracycline repressors. Journal of bacteriology. 2007;189:4614–4623. doi: 10.1128/JB.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klotzsche M, Ehrt S, Schnappinger D. Improved tetracycline repressors for gene silencing in mycobacteria. Nucleic acids research. 2009;37:1778–1788. doi: 10.1093/nar/gkp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei JR, Krishnamoorthy V, Murphy K, Kim JH, Schnappinger D, Alber T, Sassetti CM, Rhee KY, Rubin EJ. Depletion of antibiotic targets has widely varying effects on growth. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4176–4181. doi: 10.1073/pnas.1018301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JH, Wei JR, Wallach JB, Robbins RS, Rubin EJ, Schnappinger D. Protein inactivation in mycobacteria by controlled proteolysis and its application to deplete the beta subunit of RNA polymerase. Nucleic acids research. 2011;39:2210–2220. doi: 10.1093/nar/gkq1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boldrin F, Casonato S, Dainese E, Sala C, Dhar N, Palu G, Riccardi G, Cole ST, Manganelli R. Development of a repressible mycobacterial promoter system based on two transcriptional repressors. Nucleic acids research. 2010;38:e134. doi: 10.1093/nar/gkq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang Y, Mead D, Dhodda V, Brumm P, Fox BG. One-plasmid tunable coexpression for mycobacterial protein-protein interaction studies. Protein science : a publication of the Protein Society. 2009;18:2316–2325. doi: 10.1002/pro.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. Journal of bacteriology. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schleif R. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS microbiology reviews. 2010;34:779–796. doi: 10.1111/j.1574-6976.2010.00226.x. [DOI] [PubMed] [Google Scholar]
- 28.Carroll P, Brown AC, Hartridge AR, Parish T. Expression of Mycobacterium tuberculosis Rv1991c using an arabinose-inducible promoter demonstrates its role as a toxin. Fems Microbiol Lett. 2007;274:73–82. doi: 10.1111/j.1574-6968.2007.00842.x. [DOI] [PubMed] [Google Scholar]
- 29.Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Applied microbiology and biotechnology. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 30.Lee BY, Clemens DL, Horwitz MA. The metabolic activity of Mycobacterium tuberculosis, assessed by use of a novel inducible GFP expression system, correlates with its capacity to inhibit phagosomal maturation and acidification in human macrophages. Molecular microbiology. 2008;68:1047–1060. doi: 10.1111/j.1365-2958.2008.06214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaur P, Agarwal S, Datta S. Delineating bacteriostatic and bactericidal targets in mycobacteria using IPTG inducible antisense expression. PloS one. 2009;4:e5923. doi: 10.1371/journal.pone.0005923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi M, Shimizu S. Versatile Nitrilases – Nitrile-Hydrolyzing Enzymes. Fems Microbiol Lett. 1994;120:217–223. [Google Scholar]
- 33.Nagasawa T, Kobayashi M, Yamada H. Optimum Culture Conditions for the Production of Benzonitrilase by Rhodococcus-Rhodochrous J1. Arch Microbiol. 1988;150:89–94. [Google Scholar]
- 34.Komeda H, Hori Y, Kobayashi M, Shimizu S. Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10572–10577. doi: 10.1073/pnas.93.20.10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herai S, Hashimoto Y, Higashibata H, Maseda H, Ikeda H, Omura S, Kobayashi M. Hyper-inducible expression system for streptomycetes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14031–14035. doi: 10.1073/pnas.0406058101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandey AK, Raman S, Proff R, Joshi S, Kang CM, Rubin EJ, Husson RN, Sassetti CM. Nitrile-inducible gene expression in mycobacteria. Tuberculosis. 2009;89:12–16. doi: 10.1016/j.tube.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukhtar TA, Wright GD. Streptogramins, oxazolidinones, and other inhibitors of bacterial protein synthesis. Chemical reviews. 2005;105:529–542. doi: 10.1021/cr030110z. [DOI] [PubMed] [Google Scholar]
- 38.Blanc V, Salah-Bey K, Folcher M, Thompson CJ. Molecular characterization and transcriptional analysis of a multidrug resistance gene cloned from the pristinamycin-producing organism, Streptomyces pristinaespiralis. Molecular microbiology. 1995;17:989–999. doi: 10.1111/j.1365-2958.1995.mmi_17050989.x. [DOI] [PubMed] [Google Scholar]
- 39.Salah-Bey K, Blanc V, Thompson CJ. Stress-activated expression of a Streptomyces pristinaespiralis multidrug resistance gene (ptr) in various Streptomyces spp. and Escherichia coli. Molecular microbiology. 1995;17:1001–1012. doi: 10.1111/j.1365-2958.1995.mmi_17051001.x. [DOI] [PubMed] [Google Scholar]
- 40.Folcher M, Morris RP, Dale G, Salah-Bey-Hocini K, Viollier PH, Thompson CJ. A transcriptional regulator of a pristinamycin resistance gene in Streptomyces coelicolor. The Journal of biological chemistry. 2001;276:1479–1485. doi: 10.1074/jbc.M007690200. [DOI] [PubMed] [Google Scholar]
- 41.Forti F, Crosta A, Ghisotti D. Pristinamycin-inducible gene regulation in mycobacteria. Journal of biotechnology. 2009;140:270–277. doi: 10.1016/j.jbiotec.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Hernandez-Abanto SM, Woolwine SC, Jain SK, Bishai WR. Tetracycline-inducible gene expression in mycobacteria within an animal host using modified Streptomyces tcp830 regulatory elements. Arch Microbiol. 2006;186:459–464. doi: 10.1007/s00203-006-0160-2. [DOI] [PubMed] [Google Scholar]
- 43.Gur E, Biran D, Ron EZ. Regulated proteolysis in Gram-negative bacteria--how and when? Nature reviews. Microbiology. 2011;9:839–848. doi: 10.1038/nrmicro2669. [DOI] [PubMed] [Google Scholar]
- 44.Gottesman S. Proteolysis in bacterial regulatory circuits. Annual review of cell and developmental biology. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 45.Keiler KC. Biology of trans-translation. Annual review of microbiology. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- 46.Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 48.Lessner FH, Venters BJ, Keiler KC. Proteolytic adaptor for transfer-messenger RNA-tagged proteins from alpha-proteobacteria. Journal of bacteriology. 2007;189:272–275. doi: 10.1128/JB.01387-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Molecular cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 50.Griffith KL, Grossman AD. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Molecular microbiology. 2008;70:1012–1025. doi: 10.1111/j.1365-2958.2008.06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends in biochemical sciences. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Topp S, Reynoso CM, Seeliger JC, Goldlust IS, Desai SK, Murat D, Shen A, Puri AW, Komeili A, Bertozzi CR, Scott JR, Gallivan JP. Synthetic riboswitches that induce gene expression in diverse bacterial species. Applied and environmental microbiology. 2010;76:7881–7884. doi: 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seeliger JC, Topp S, Sogi KM, Previti ML, Gallivan JP, Bertozzi CR. A riboswitch-based inducible gene expression system for mycobacteria. PloS one. 2012;7:e29266. doi: 10.1371/journal.pone.0029266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barnes PJ. Theophylline: new perspectives for an old drug. American journal of respiratory and critical care medicine. 2003;167:813–818. doi: 10.1164/rccm.200210-1142PP. [DOI] [PubMed] [Google Scholar]
- 55.Woong Park S, Klotzsche M, Wilson DJ, Boshoff HI, Eoh H, Manjunatha U, Blumenthal A, Rhee K, Barry CE, 3rd, Aldrich CC, Ehrt S, Schnappinger D. Evaluating the sensitivity of Mycobacterium tuberculosis to biotin deprivation using regulated gene expression. PLoS pathogens. 2011;7:e1002264. doi: 10.1371/journal.ppat.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leblanc C, Prudhomme T, Tabouret G, Ray A, Burbaud S, Cabantous S, Mourey L, Guilhot C, Chalut C. 4′-Phosphopantetheinyl transferase PptT, a new drug target required for Mycobacterium tuberculosis growth and persistence in vivo. PLoS pathogens. 2012;8:e1003097. doi: 10.1371/journal.ppat.1003097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blumenthal A, Trujillo C, Ehrt S, Schnappinger D. Simultaneous analysis of multiple Mycobacterium tuberculosis knockdown mutants in vitro and in vivo. PloS one. 2010;5:e15667. doi: 10.1371/journal.pone.0015667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nature medicine. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bashiri G, Squire CJ, Baker EN, Moreland NJ. Expression, purification and crystallization of native and selenomethionine labeled Mycobacterium tuberculosis FGD1 (Rv0407) using a Mycobacterium smegmatis expression system. Protein expression and purification. 2007;54:38–44. doi: 10.1016/j.pep.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 62.Noens EE, Williams C, Anandhakrishnan M, Poulsen C, Ehebauer MT, Wilmanns M. Improved mycobacterial protein production using a Mycobacterium smegmatis groEL1DeltaC expression strain. BMC biotechnology. 2011;11:27. doi: 10.1186/1472-6750-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS genetics. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodrigue S, Brodeur J, Jacques PE, Gervais AL, Brzezinski R, Gaudreau L. Identification of mycobacterial sigma factor binding sites by chromatin immunoprecipitation assays. Journal of bacteriology. 2007;189:1505–1513. doi: 10.1128/JB.01371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T, Mahwinney C, Kennedy AD, Allard R, Brabant W, Krueger A, Jaini S, Honda B, Yu WH, Hickey MJ, Zucker J, Garay C, Weiner B, Sisk P, Stolte C, Winkler JK, Van de Peer Y, Iazzetti P, Camacho D, Dreyfuss J, Liu Y, Dorhoi A, Mollenkopf HJ, Drogaris P, Lamontagne J, Zhou Y, Piquenot J, Park ST, Raman S, Kaufmann SH, Mohney RP, Chelsky D, Moody DB, Sherman DR, Schoolnik GK. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson T, de Lisle GW, Marcinkeviciene JA, Blanchard JS, Collins DM. Antisense RNA to ahpC, an oxidative stress defence gene involved in isoniazid resistance, indicates that AhpC of Mycobacterium bovis has virulence properties. Microbiology. 1998;144(Pt 10):2687–2695. doi: 10.1099/00221287-144-10-2687. [DOI] [PubMed] [Google Scholar]
- 67.Rubin EJ, Akerley BJ, Novik VN, Lampe DJ, Husson RN, Mekalanos JJ. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forti F, Mauri V, Deho G, Ghisotti D. Isolation of conditional expression mutants in Mycobacterium tuberculosis by transposon mutagenesis. Tuberculosis. 2011;91:569–578. doi: 10.1016/j.tube.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 69.Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, Perkins TT, Parkhill J, Dougan G, Young DB. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS pathogens. 2011;7:e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uplekar S, Rougemont J, Cole ST, Sala C. High-resolution transcriptome and genome-wide dynamics of RNA polymerase and NusA in Mycobacterium tuberculosis. Nucleic acids research. 2013;41:961–977. doi: 10.1093/nar/gks1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raju RM, Unnikrishnan M, Rubin DH, Krishnamoorthy V, Kandror O, Akopian TN, Goldberg AL, Rubin EJ. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS pathogens. 2012;8:e1002511. doi: 10.1371/journal.ppat.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, Griffin JE, Venghatakrishnan H, Zukauskas A, Wei JR, Dhiman RK, Crick DC, Rubin EJ, Sassetti CM, Alber T. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Science signaling. 2012;5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Glickman MS, Jacobs WR., Jr Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell. 2001;104:477–485. doi: 10.1016/s0092-8674(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 74.Young K, Jayasuriya H, Ondeyka JG, Herath K, Zhang CW, Kodali S, Galgoci A, Painter R, Brown-Driver V, Yamamoto R, Silver LL, Zheng YC, Ventura JI, Sigmund J, Ha S, Basilio A, Vicente F, Tormo JR, Pelaez F, Youngman P, Cully D, Barrett JF, Schmatz D, Singh SB, Wang J. Discovery of FabH/FabF inhibitors from natural products. Antimicrob Agents Ch. 2006;50:519–526. doi: 10.1128/AAC.50.2.519-526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jayasuriya H, Herath KB, Zhang C, Zink DL, Basilio A, Genilloud O, Diez MT, Vicente F, Gonzalez I, Salazar O, Pelaez F, Cummings R, Ha S, Wang J, Singh SB. Isolation and structure of platencin: a FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angew Chem Int Ed Engl. 2007;46:4684–4688. doi: 10.1002/anie.200701058. [DOI] [PubMed] [Google Scholar]
- 76.Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, Colletti SL, Herath K, Cummings R, Salazar O, Gonzalez I, Basilio A, Vicente F, Genilloud O, Pelaez F, Jayasuriya H, Young K, Cully DF, Singh SB. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7612–7616. doi: 10.1073/pnas.0700746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 78.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abrahams GL, Kumar A, Savvi S, Hung AW, Wen S, Abell C, Barry CE, 3rd, Sherman DR, Boshoff HI, Mizrahi V. Pathway-selective sensitization of Mycobacterium tuberculosis for target-based whole-cell screening. Chemistry & biology. 2012;19:844–854. doi: 10.1016/j.chembiol.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ollinger J, O’Malley T, Ahn J, Odingo J, Parish T. Inhibition of the sole type I signal peptidase of Mycobacterium tuberculosis is bactericidal under replicating and nonreplicating conditions. Journal of bacteriology. 2012;194:2614–2619. doi: 10.1128/JB.00224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dziadek J, Rutherford SA, Madiraju MV, Atkinson MA, Rajagopalan M. Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology. 2003;149:1593–1603. doi: 10.1099/mic.0.26023-0. [DOI] [PubMed] [Google Scholar]
- 82.Jani C, Eoh H, Lee JJ, Hamasha K, Sahana MB, Han JS, Nyayapathy S, Lee JY, Suh JW, Lee SH, Rehse SJ, Crick DC, Kang CM. Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC microbiology. 2010;10:327. doi: 10.1186/1471-2180-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154:725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 84.Bhatt A, Kremer L, Dai AZ, Sacchettini JC, Jacobs WR., Jr Conditional depletion of KasA, a key enzyme of mycolic acid biosynthesis, leads to mycobacterial cell lysis. Journal of bacteriology. 2005;187:7596–7606. doi: 10.1128/JB.187.22.7596-7606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rao M, Liu H, Yang M, Zhao C, He ZG. A copper-responsive global repressor regulates expression of diverse membrane-associated transporters and bacterial drug resistance in mycobacteria. The Journal of biological chemistry. 2012;287:39721–39731. doi: 10.1074/jbc.M112.383604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frampton R, Aggio RB, Villas-Boas SG, Arcus VL, Cook GM. Toxin-antitoxin systems of Mycobacterium smegmatis are essential for cell survival. The Journal of biological chemistry. 2012;287:5340–5356. doi: 10.1074/jbc.M111.286856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang F, He ZG. Characterization of an interplay between a Mycobacterium tuberculosis MazF homolog, Rv1495 and its sole DNA topoisomerase I. Nucleic acids research. 2010;38:8219–8230. doi: 10.1093/nar/gkq737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robson J, McKenzie JL, Cursons R, Cook GM, Arcus VL. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. Journal of molecular biology. 2009;390:353–367. doi: 10.1016/j.jmb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 89.Yang M, Gao C, Wang Y, Zhang H, He ZG. Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PloS one. 2010;5:e10672. doi: 10.1371/journal.pone.0010672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barik S, Sureka K, Mukherjee P, Basu J, Kundu M. RseA, the SigE specific anti-sigma factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Molecular microbiology. 2010;75:592–606. doi: 10.1111/j.1365-2958.2009.07008.x. [DOI] [PubMed] [Google Scholar]
- 91.Nisa S, Blokpoel MC, Robertson BD, Tyndall JD, Lun S, Bishai WR, O’Toole R. Targeting the chromosome partitioning protein ParA in tuberculosis drug discovery. The Journal of antimicrobial chemotherapy. 2010;65:2347–2358. doi: 10.1093/jac/dkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kang J, Xu L, Yang S, Yu W, Liu S, Xin Y, Ma Y. Effect of Phosphoglucosamine Mutase on Biofilm Formation and Antimicrobial Susceptibilities in M. smegmatis glmM Gene Knockdown Strain. PloS one. 2013;8:e61589. doi: 10.1371/journal.pone.0061589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sureka K, Dey S, Datta P, Singh AK, Dasgupta A, Rodrigue S, Basu J, Kundu M. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Molecular microbiology. 2007;65:261–276. doi: 10.1111/j.1365-2958.2007.05814.x. [DOI] [PubMed] [Google Scholar]
- 94.Sureka K, Sanyal S, Basu J, Kundu M. Polyphosphate kinase 2: a modulator of nucleoside diphosphate kinase activity in mycobacteria. Molecular microbiology. 2009;74:1187–1197. doi: 10.1111/j.1365-2958.2009.06925.x. [DOI] [PubMed] [Google Scholar]
- 95.Rao SP, Camacho L, Huat Tan B, Boon C, Russel DG, Dick T, Pethe K. Recombinase-based reporter system and antisense technology to study gene expression and essentiality in hypoxic nonreplicating mycobacteria. Fems Microbiol Lett. 2008;284:68–75. doi: 10.1111/j.1574-6968.2008.01193.x. [DOI] [PubMed] [Google Scholar]
- 96.Goyal R, Das AK, Singh R, Singh PK, Korpole S, Sarkar D. Phosphorylation of PhoP protein plays direct regulatory role in lipid biosynthesis of Mycobacterium tuberculosis. The Journal of biological chemistry. 2011;286:45197–45208. doi: 10.1074/jbc.M111.307447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Minch K, Rustad T, Sherman DR. Mycobacterium tuberculosis growth following aerobic expression of the DosR regulon. PloS one. 2012;7:e35935. doi: 10.1371/journal.pone.0035935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sinha KM, Stephanou NC, Gao F, Glickman MS, Shuman S. Mycobacterial UvrD1 is a Ku-dependent DNA helicase that plays a role in multiple DNA repair events, including double-strand break repair. The Journal of biological chemistry. 2007;282:15114–15125. doi: 10.1074/jbc.M701167200. [DOI] [PubMed] [Google Scholar]
- 99.Stephanou NC, Gao F, Bongiorno P, Ehrt S, Schnappinger D, Shuman S, Glickman MS. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. Journal of bacteriology. 2007;189:5237–5246. doi: 10.1128/JB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baughn AD, Deng J, Vilcheze C, Riestra A, Welch JT, Jacobs WR, Jr, Zimhony O. Mutually exclusive genotypes for pyrazinamide and 5-chloropyrazinamide resistance reveal a potential resistance-proofing strategy. Antimicrob Agents Chemother. 2010;54:5323–5328. doi: 10.1128/AAC.00529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duckworth BP, Geders TW, Tiwari D, Boshoff HI, Sibbald PA, Barry CE, 3rd, Schnappinger D, Finzel BC, Aldrich CC. Bisubstrate adenylation inhibitors of biotin protein ligase from Mycobacterium tuberculosis. Chemistry & biology. 2011;18:1432–1441. doi: 10.1016/j.chembiol.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hett EC, Chao MC, Rubin EJ. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS pathogens. 2010;6:e1001020. doi: 10.1371/journal.ppat.1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature. 2008;454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Korch SB, Contreras H, Clark-Curtiss JE. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. Journal of bacteriology. 2009;191:1618–1630. doi: 10.1128/JB.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ahidjo BA, Kuhnert D, McKenzie JL, Machowski EE, Gordhan BG, Arcus V, Abrahams GL, Mizrahi V. VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PloS one. 2011;6:e21738. doi: 10.1371/journal.pone.0021738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharp JD, Cruz JW, Raman S, Inouye M, Husson RN, Woychik NA. Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. The Journal of biological chemistry. 2012;287:12835–12847. doi: 10.1074/jbc.M112.340109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Singh R, Barry CE, 3rd, Boshoff HI. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. Journal of bacteriology. 2010;192:1279–1291. doi: 10.1128/JB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rigel NW, Gibbons HS, McCann JR, McDonough JA, Kurtz S, Braunstein M. The Accessory SecA2 System of Mycobacteria Requires ATP Binding and the Canonical SecA1. The Journal of biological chemistry. 2009;284:9927–9936. doi: 10.1074/jbc.M900325200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chalut C, Botella L, de Sousa-D’Auria C, Houssin C, Guilhot C. The nonredundant roles of two 4′-phosphopantetheinyl transferases in vital processes of Mycobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8511–8516. doi: 10.1073/pnas.0511129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hett EC, Chao MC, Deng LL, Rubin EJ. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS pathogens. 2008;4:e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rana AK, Singh A, Gurcha SS, Cox LR, Bhatt A, Besra GS. Ppm1-encoded polyprenyl monophosphomannose synthase activity is essential for lipoglycan synthesis and survival in mycobacteria. PloS one. 2012;7:e48211. doi: 10.1371/journal.pone.0048211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Trauner A, Lougheed KE, Bennett MH, Hingley-Wilson SM, Williams HD. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. The Journal of biological chemistry. 2012;287:24053–24063. doi: 10.1074/jbc.M112.364851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Garces A, Atmakuri K, Chase MR, Woodworth JS, Krastins B, Rothchild AC, Ramsdell TL, Lopez MF, Behar SM, Sarracino DA, Fortune SM. EspA acts as a critical mediator of ESX1-dependent virulence in Mycobacterium tuberculosis by affecting bacterial cell wall integrity. PLoS pathogens. 2010;6:e1000957. doi: 10.1371/journal.ppat.1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de la Paz Santangelo M, Gest PM, Guerin ME, Coincon M, Pham H, Ryan G, Puckett SE, Spencer JS, Gonzalez-Juarrero M, Daher R, Lenaerts AJ, Schnappinger D, Therisod M, Ehrt S, Sygusch J, Jackson M. Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1,6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli. The Journal of biological chemistry. 2011;286:40219–40231. doi: 10.1074/jbc.M111.259440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kumar A, Zhang M, Zhu L, Liao RP, Mutai C, Hafsat S, Sherman DR, Wang MW. High-throughput screening and sensitized bacteria identify an M. tuberculosis dihydrofolate reductase inhibitor with whole cell activity. PloS one. 2012;7:e39961. doi: 10.1371/journal.pone.0039961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carroll P, Faray-Kele MC, Parish T. Identifying vulnerable pathways in Mycobacterium tuberculosis by using a knockdown approach. Applied and environmental microbiology. 2011;77:5040–5043. doi: 10.1128/AEM.02880-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cortes M, Singh AK, Reyrat JM, Gaillard JL, Nassif X, Herrmann JL. Conditional gene expression in Mycobacterium abscessus. PloS one. 2011;6:e29306. doi: 10.1371/journal.pone.0029306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Di Luca M, Bottai D, Batoni G, Orgeur M, Aulicino A, Counoupas C, Campa M, Brosch R, Esin S. The ESX-5 associated eccB-EccC locus is essential for Mycobacterium tuberculosis viability. PloS one. 2012;7:e52059. doi: 10.1371/journal.pone.0052059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Serafini A, Boldrin F, Palu G, Manganelli R. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. Journal of bacteriology. 2009;191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


