Abstract
Ectodomain shedding mediated by tumor necrosis factor–α (TNF-α)–converting enzyme [TACE; also known as ADAM17 (a disintegrin and metalloproteinase 17)] provides an important switch in regulating cell proliferation, inflammation, and cancer progression. TACE-mediated ectodomain cleavage is activated by signaling of the mitogen-activated protein kinases (MAPKs) p38 and ERK (extracellular signal– regulated kinase). Here, we found that under basal conditions, TACE was predominantly present as dimers at the cell surface, which required its cytoplasmic domain and enabled efficient association with tissue inhibitor of metalloproteinase-3 (TIMP3) and silencing of TACE activity. Upon activation of the ERK or p38 MAPK pathway, the balance shifted from TACE dimers to monomers, and this shift was associated with increased cell surface presentation of TACE and decreased TIMP3 association, which relieved the inhibition of TACE by TIMP3 and increased TACE-mediated proteolysis of transforming growth factor–α. Thus, cell signaling altered the dimer-monomer equilibrium and inhibitor association to promote activation of TACE-mediated ectodomain shedding, a regulatory mechanism that may extend to other ADAM proteases.
INTRODUCTION
Membrane-associated metalloproteinases mediate ectodomain shedding, a posttranslational process that results in the proteolytic release of ectodomains of various transmembrane proteins, including cytokines, growth factors, receptors, and adhesion molecules. By defining the activities of its substrates, ectodomain shedding regulates cell proliferation, migration and invasion (1–4), and, consequently, inflammation and cancer progression (2, 4, 5).
The role of ectodomain shedding is well illustrated in the regulation of epidermal growth factor (EGF) receptor (EGFR) signaling by transmembrane forms of transforming growth factor–α (TGF-α) ligands (6–10). In addition to the abundance of receptor and ligands, the cell response is defined by the release of TGF-α ligands from transmembrane forms. Increased shedding of TGF-α ligands enhances EGFR activation, resulting in epithelial cell proliferation and carcinoma progression. Conversely, ectodomain shedding of the type I TGF-β receptors (TβRIs) decreases the growth inhibition by autocrine TGF-β signaling in carcinoma cells (11).
The ADAM (a disintegrin and metalloproteinase) class of structurally related, membrane-associated proteases control ectodomain shedding of various substrates (2, 4, 5). They consist sequentially of a prodomain, a catalytic domain, a disintegrin domain, a cysteine-rich region, a transmembrane domain, and a cytoplasmic tail and are related to matrix metalloproteinases (MMPs). Among the ADAM proteases, tumor necrosis factor–α (TNF-α)– converting enzyme (TACE), also known as ADAM17, mediates shedding of TGF-α, amphiregulin, and heparin-binding EGF-like growth factor (HB-EGF) and thus regulates the access of the EGFR to soluble ligands (6–8, 10, 12, 13). Increased TACE abundance in carcinomas, such as in breast carcinomas (4, 14), is thought to aid in cancer progression through increased EGFR stimulation.
TACE also mediates shedding of inflammatory cytokines and cytokine receptors and thus plays roles in inflammation (4, 5). TACE abundance is increased in the inflammatory response (15, 16), and increased TACE activity resulting from lack of tissue inhibitor of metalloproteinase-3 (TIMP3), an inhibitor of MMPs and TACE (17), leads to vascular and liver inflammation (18, 19).
Despite intensive study, the molecular regulation of activation of TACE, or any ADAM protease, remains poorly understood. Activation of TACE is thought to require removal of its prodomain by furin proteases (20), although this has been disputed (21, 22). TACE is activated upon activation of the ERK (extracellular signal–regulated kinase) or p38 MAPK (mitogen-activated protein kinase) pathway by extracellular cues, such as growth factors, inflammatory mediators, and stress (3, 4). Activation of the ERK MAPK pathway results in increased phosphorylation at two sites and decreased phosphorylation at another site in the cytoplasmic domain of TACE (23–25), and activated p38 MAPK directly phosphorylates TACE on Thr735 (10). How the changes in phosphorylation lead to TACE activation is unclear. Whereas ectodomain shedding is thought to occur at the cell surface, the bulk of TACE resides inside the cell (20, 21, 25). Therefore, TACE activation may involve rapid transport of TACE to the cell surface, which occurs in response to ERK or p38 MAPK signaling (10, 25, 26). Additionally, activation of TACE in response to phorbol 12-myristate 13-acetate (PMA) results in a rapid conformational change, exposing the catalytic site of TACE (22), which could involve disulfide bond isomerization (27). Finally, TIMP3 interacts with the ectodomain of TACE and inhibits its activity (17, 28). The increased TACE activity in the absence of TIMP3 suggests a role for TIMP3 in TACE activation (18, 19, 29), which, however, has been questioned recently (22).
Homo- or heterodimerization is a recurrent theme among cell surface receptors, and receptor activation often requires ligand-induced dimerization. There is no information on whether ADAM proteases form dimers or oligomers. MMP14 (MT1-MMP) and MMP9 form homodimers at the cell surface, although the functional importance of their dimerization is not well understood (30, 31).
We provide evidence that the activity of TACE is regulated through dimerization, which requires its cytoplasmic domain. In the absence of MAPK stimulation, TACE is presented as dimers at the cell surface, allowing TIMP3 to efficiently interact with and inhibit TACE. Activation of ERK or p38 MAPK signaling, which leads to TACE activation, results in increased monomer presentation and release of TIMP3 from TACE. These results shed light on a new aspect of the regulation of the activities of TACE and other ADAM proteases, such as ADAM10, and, together with previous observations, allow for a model of how TACE activity is induced in response to activation of ERK or p38 MAPKs.
RESULTS
Regulation of TACE-mediated ectodomain shedding by TIMP3
Because TIMP3 inhibits MMPs and TACE (17, 32), we evaluated whether it plays a role in TACE activation by p38 or ERK MAPKs. We examined the role of endogenous TIMP3 in the activity of TACE in T4-2 breast carcinoma cells (8, 10, 11) by scoring the ectodomain shedding of two endogenous substrates, the cell surface TβRI (11) and transmembrane TGF-α (10). T4-2 cells were transfected with small interfering RNA (siRNA) directed against TIMP3, resulting in a >85% reduction of TIMP3 mRNA or protein (fig. S1A).
Activation of ERK MAPK by PMA resulted in a rapid decrease of cell surface abundance of TβRI, as reported (11), and activation of p38 MAPK by anisomycin also led to cleavage of cell surface TβRI through TACE (fig. S1B). Transfection of TIMP3 siRNA increased ectodomain shedding, reducing the cell surface abundance of TβRI in unstimulated cells to the amounts seen in response to TACE activation. Treatment with PMA or anisomycin further reduced the cell surface abundance of TβRI, albeit moderately (fig. S1B). Conversely, addition of recombinant TIMP3 inhibited PMA-induced shedding of endogenous TβRI in a dose-dependent manner (fig. S1C). We also examined the role of TIMP3 in the release of TGF-α into the medium (fig. S1D). Both PMA and anisomycin activated the release of TGF-α. Transfection of TIMP3 siRNA enhanced basal TGF-α release, suggesting increased TACE activity. As with the shedding of TβRI, the TACE-mediated TGF-α shedding was only slightly increased by PMA or anisomycin. This could be explained by incomplete siRNA-mediated knockdown of TIMP3 and decreased availability of transmembrane TGF-α because of increased basal TACE activity.
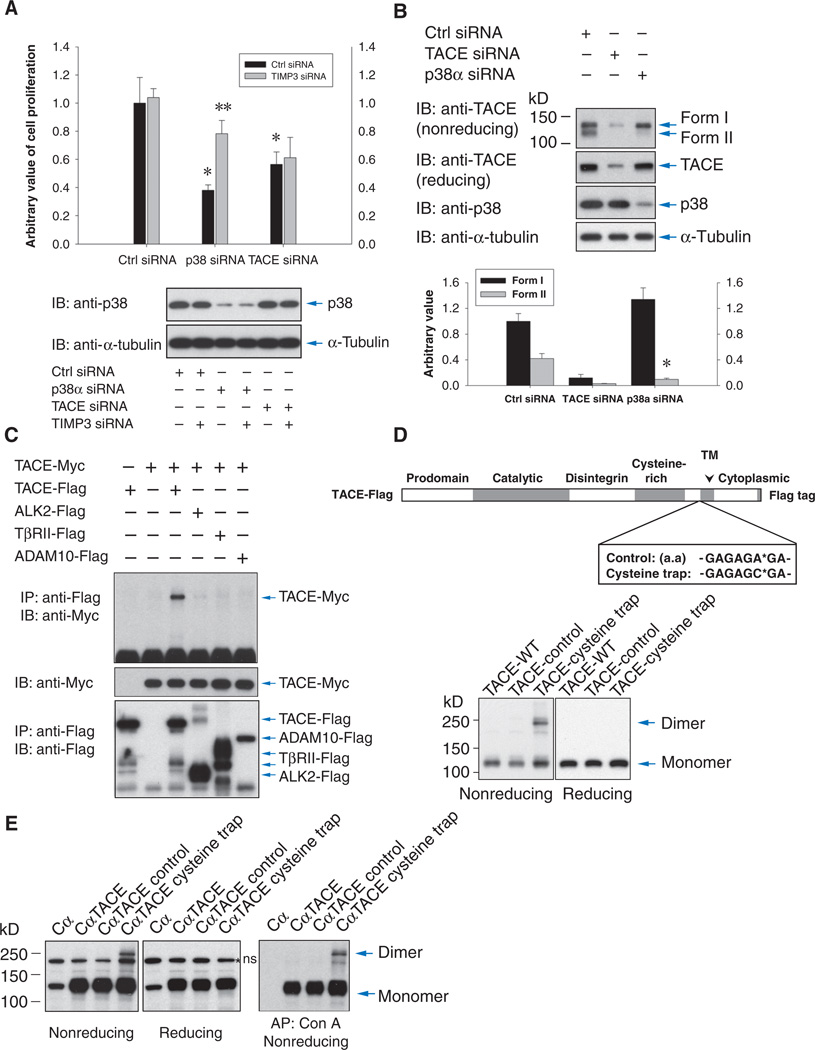
We previously showed that p38 MAPK signaling enhances T4-2 cell proliferation in part through TACE by inducing release of TGF-α and amphiregulin, thereby resulting in enhanced EGFR signaling (10). As reported, transfection of siRNAs directed against p38α MAPK resulted in decreased TACE activity (10) and inhibited cell proliferation (Fig. 1A). Concomitant transfection of TIMP3 siRNA rescued the decrease in cell proliferation, reflecting the increase in TACE activity and resulting release of EGFR ligands. As expected, transfection of TIMP3 siRNA did not rescue the decrease in proliferation in cells transfected with TACE siRNA (Fig. 1A). These results not only show that TIMP3 inhibits TACE but also implicate TIMP3 as a mediator of TACE activation in response to MAPK activation.
Fig. 1.
TIMP3 regulates TACE-mediated ectodomain shedding, and TACE forms dimers. (A) Effects of transfecting control, p38α MAPK, or TACE siRNAs with TIMP3 siRNA on T4-2 cell proliferation as assessed by BrdU incorporation. Lower panel shows p38 abundance. n = 3 experiments. *P < 0.01, compared with control siRNA; **P < 0.01, compared with p38α MAPK siRNA by Student’s t tests. (B) Immunoblotting for TACE in cells transfected with TACE, p38α, or control siRNAs. Samples were subjected to SDS-PAGE without or with prior reduction with DTT. Band intensities for forms I and II were quantified by density scanning. n = 4 blots. *P < 0.001, compared with form II of control siRNA by Student’s t tests. (C) Anti-Myc immunoblotting of anti-Flag immunoprecipitates reveals association of Myc-tagged TACE with Flag-tagged TACE, but not with ALK2, TβRII, or ADAM10 in HepG2 cells. (D and E) The amino acid sequence G-A-G-A-G-A-G-A (Control) or G-A-G-A-G-C-G-A (Cysteine trap) was inserted into TACE after His692 (D, upper panel). Lysates of Cα cells transiently (D, lower panel) or stably (E) expressing control, cysteine trap, or wild-type TACE were analyzed by nonreducing SDS-PAGE and anti-TACE (D) or anti-Flag (E) immunoblotting to reveal TACE dimers, which were not detected after disulfide reduction before SDS-PAGE [(D) lower right panel and (E) middle panels]. The right panel of (E) shows anti-Flag immunoblotting of concanavalin A affinity purification to enrich glycosylated proteins including TACE. ns, nonspecific band.
TACE dimer formation
Phosphorylation of TACE on Thr735 in the cytoplasmic tail by p38α MAPK enhances cell surface presentation and stability and activity of TACE (10), and phosphorylation of TACE on Ser819 in the cytoplasmic tail occurs in response to ERK MAPK pathway activation (24). Because either pathway can induce increased TACE activity, changes in phosphorylation of the cytoplasmic tail may induce a change in conformation of the extracellular domain of TACE. Without disulfide reduction, endogenous TACE was resolved through SDS–polyacrylamide gel electrophoresis (SDS-PAGE) as two forms (Fig. 1B). The intensity of the faster migrating band (form II) was lower when TACE activation was decreased by transfection of p38α MAPK siRNA (10), suggesting that this may represent an active conformation of TACE. This observation supports the notion that TACE activation might result in a conformational change (22) and led us to study the underlying molecular mechanisms that lead to TACE-mediated ectodomain shedding.
To understand this change in presentation, we investigated whether dimerization might play a role in TACE activation. ADAM proteases are not known to dimerize and do not contain a hemopexin-like domain that mediates the dimerization of some MMPs (30, 31). We coexpressed TACE with a differentially tagged TACE or control transmembrane proteins. Coimmunoprecipitation analysis revealed association of Myc-tagged TACE with Flag-tagged TACE, but not with activin receptor–like kinase 2 (ALK2) or TβRII, two transmembrane TGF-β family receptors (11), or ADAM10 (Fig. 1C), suggesting specific homodimerization of TACE.
We next used cysteine trap analysis, a strategy that stabilizes dimer formation by introducing cysteine residues that form an extracellular disulfide bond (33). A short sequence containing a cysteine is introduced into the extracellular domain adjacent to the transmembrane domain of a cell surface protein, resulting in homodimer stabilization through intermolecular disulfide bonds. These homodimers are detected by immunoblotting after gel electrophoresis under nonreducing conditions. This approach has revealed the dimerization of several membrane-associated proteins (34–38). We introduced two different peptides at the site of the codon for His692, which is the extracellular amino acid residue proximal to the transmembrane domain of TACE. The peptide Gly-Ala-Gly-Ala-Gly-Cys-Gly-Ala incorporates a cysteine (Cys), whereas the peptide Gly-Ala-Gly-Ala-Gly-Ala-Gly-Ala does not introduce a cysteine and serves as negative control (Fig. 1D). Chinese hamster ovary (CHO) cells expressing either TACE mutant or wild-type TACE were lysed in the presence of iodoacetamide to prevent disulfide formation in cell lysates, and TACE was visualized by immunoblotting. TACE homodimers were apparent in cells transfected with the cysteine trap mutant TACE but not in cells expressing the control TACE mutant or wild-type TACE (Fig. 1D, left panel). As expected, the dimers disappeared when the samples were reduced before electrophoresis (Fig. 1D, right panel), confirming dimerization through disulfide bonds. Using Cα cells (CHO cells that express TGF-α) (39), we generated stable cells expressing cysteine trap TACE or control TACE at an abundance comparable to that of endogenous TACE in carcinoma cells. In Cα cells expressing the cysteine trap mutant, a TACE dimer band was observed by anti-Flag immunoblotting (Fig. 1E). No dimers were detected in Cα cells expressing control or wild-type TACE. These results are consistent with the coimmunoprecipitation analyses and demonstrate the ability of TACE to form dimers.
Because TACE can dimerize, we examined whether other ADAM proteases can also dimerize. We coexpressed Myc-tagged ADAM10 with Flag-tagged ADAM10 or TβRII or TACE as controls. Similarly to TACE, immunoprecipitation of Flag-tagged ADAM10 resulted in coprecipitation of Myc-tagged ADAM10 (fig. S2A). These data suggest that ADAM10 can also dimerize and that dimerization may be an inherent property of ADAM metalloproteinases.
Dimerization of endogenous TACE at the cell surface
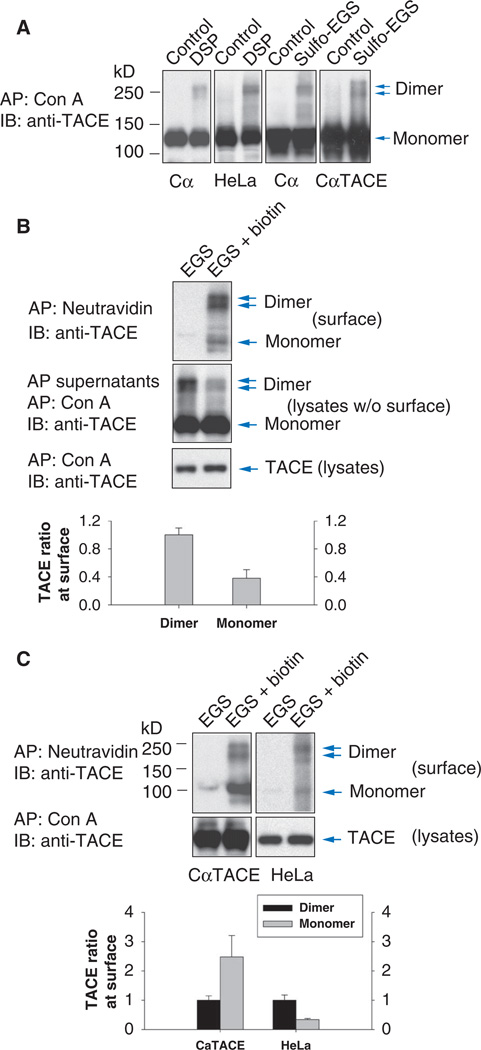
We next examined whether endogenous TACE can be detected as dimers. The bulk of TACE proteins resides inside the cells in a form containing the prodomain (proTACE), whereas a small portion of the TACE pool is present at the cell surface with the prodomain removed (20, 21, 25). Taking into account this subcellular localization, we treated Cα, CαTACE cells (Cα cells that additionally express TACE), or HeLa cells with a chemical cross-linker, the cell-permeable DSP [dithiobis(succinimidylpropionate)] or cell-impermeable sulfo-EGS [ethylene glycolbis(succinimidylsuccinate)], before lysis. Without cross-linking, adsorption of glycosylated proteins to concanavalin A Sepharose followed by anti-TACE immunoblotting revealed a pool of TACE monomers with an apparent size of 130 kD, compatible with the size of proTACE (Fig. 2A). However, after cross-linking with DSP or sulfo-EGS, TACE bands that were the size of TACE homodimers were detected. Stabilization of TACE dimers with DSP was reversed upon disulfide reduction, which is characteristic of DSP-mediated cross-linking (fig. S3A). The abundance of TACE dimers was similar in DSP-or sulfo-EGS–treated Cα and HeLa cells (Fig. 2A), suggesting that TACE dimers resided largely at the cell surface.
Fig. 2.
TACE forms dimers at the cell surface. (A) Cα cells and HeLa cells were treated or not with membrane-permeable DSP or -impermeable sulfo-EGS before cell lysis and concanavalin A affinity precipitation. Endogenous and transfected TACE were visualized by anti-TACE immunoblotting. TACE monomers and dimers were present in cells treated with DSP or sulfo-EGS. (B) Cα cells were treated with sulfo-EGS and EZ-link sulfo-NHS–LC-biotin. Biotinylated cell surface proteins adsorbed to NeutrAvidin beads were immunoblotted with anti-TACE antibody (upper panel). The remaining cell lysate proteins were subjected to concanavalin A affinity purification and anti-TACE immunoblotting (middle panel). Total endogenous TACE in Cα cells was shown by immunoblotting of concanavalin A affinity-purified proteins (lower panel). (C) TACE monomers and dimers at the cell surface in CαTACE or HeLa cells were detected as in (B). Relative density quantifications of cell surface TACE dimer and monomer, visualized after EGS cross-linking, are shown as lower panels in (B) and (C). n = 3 blots.
We performed a similar cross-linking experiment to detect whether ADAM10 dimerizes. ADAM10 is present in Cα cells, and glycosylated proteins isolated from lysed Cα cells by adsorption to concanavalin A Sepharose were immunoblotted with ADAM10 antibody (fig. S2B). Without previous cross-linking, most of the immunoreactive proteins had a size of 95 kD, corresponding to the size of ADAM10 (40). When cells were treated with sulfo-EGS before lysis, additional immunoreactive bands of 190 to 220 kD were detected, which we assumed were ADAM10 dimers. This result suggests that similarly to TACE, endogenous ADAM10 could form dimers at the cell surface.
The detection of TACE dimers by use of the cysteine trap approach, or by cross-linking with sulfo-EGS, suggested that TACE dimers are found at the cell surface. The similar abundance of TACE dimers detected with DSP or sulfo-EGS cross-linking and the similar ratios of TACE dimers to monomers suggest that these dimers may be primarily at the cell surface and that only a fraction of TACE dimerizes. To specifically address the dimerization of cell surface TACE, we incubated Cα cells with the membrane-permeable cross-linker EGS and EZ-link sulfo-NHS–LC-biotin to detect cross-linked cell surface TACE proteins that were biotinylated. Under these conditions, TACE inside the cells is subjected to cross-linking but cannot be biotinylated. Because both reagents use the same active groups [the N-hydroxysuccinimide (NHS) ester] to target primary amines in Lys residues and at the unblocked N terminus, the combined use of these two chemicals presumably reduced the cross-linking and biotinylation efficiencies. Cell surface biotinylated proteins were isolated with NeutrAvidin beads, and the supernatants, after removal of the cell surface biotinylated proteins, were adsorbed to concanavalin A beads to enrich the glycosylated proteins, including TACE. Most of the cell surface TACE presented as dimers (Fig. 2B, upper panel), and most of the TACE dimers were at the cell surface (Fig. 2B, compare dimers in the upper to the middle panel). Considering the limited efficiencies of combined cross-linking and cell surface biotinylation, these data suggest that the TACE dimers are primarily, if not exclusively, at the cell surface. Similarly to endogenous TACE in Cα cells, endogenous TACE in HeLa cells showed extensive dimerization at the cell surface, whereas the higher amount of TACE in Cα cells expressing transfected TACE resulted in a greater amount of TACE monomer (Fig. 2C).
Using a similar strategy, we detected cell surface biotinylated dimers of endogenous ADAM10 in Cα cells (fig. S2C). On the basis of their apparent molecular weights, both proADAM10 and mature ADAM10 dimers were formed, consistent with the reported presence of both ADAM10 forms at the cell surface (40). As with TACE (Fig. 2B), most ADAM10 dimers were at the cell surface (fig. S2C, compare upper and lower panels), but, compared with endogenous TACE, a relatively higher amount of ADAM10 monomers was present at the cell surface.
Requirement of the cytoplasmic domain for TACE dimerization
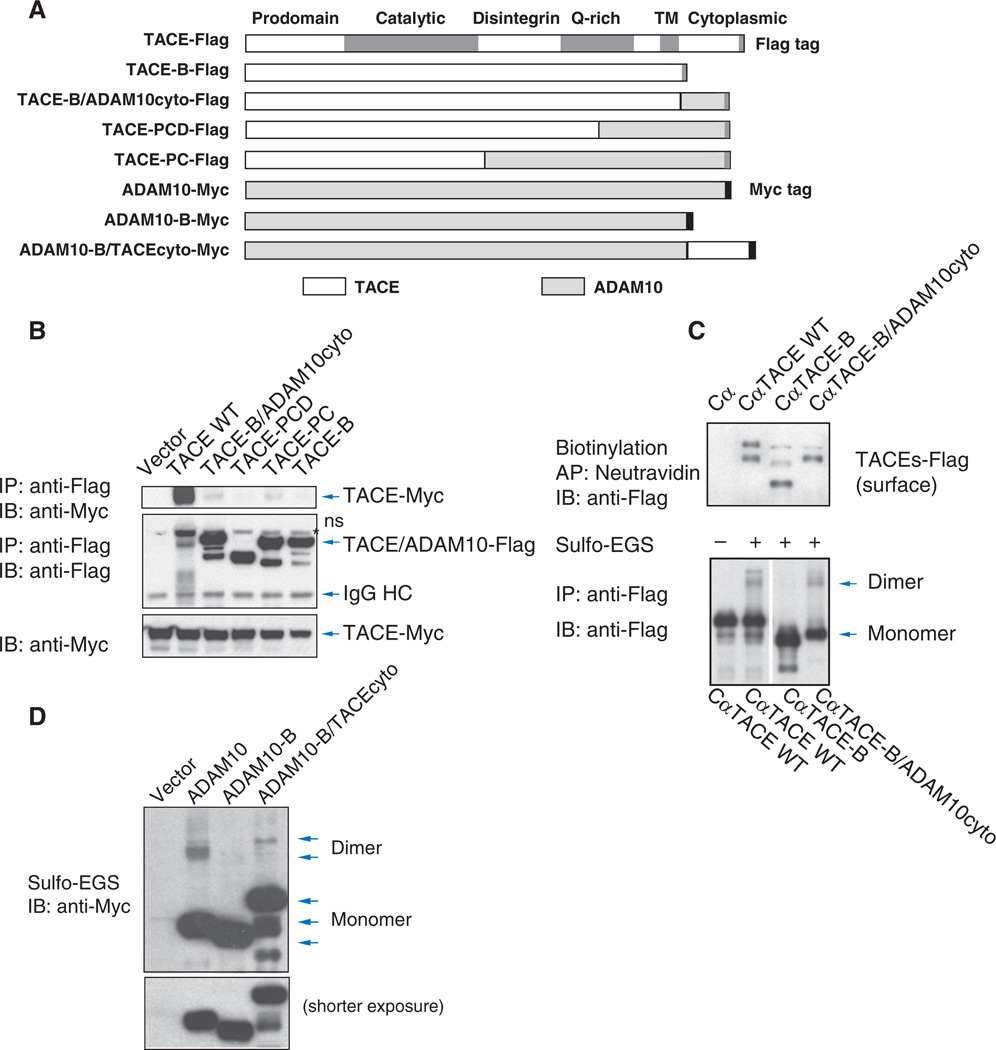
To study the role of protein domains in TACE dimerization, we generated TACE/ADAM10 chimeras by replacing the TACE sequence, starting from its C terminus, with ADAM10 domains (Fig. 3A). Individual Flag-tagged chimeras were coexpressed with Myc-tagged wild-type TACE, and their association with Myc-tagged TACE was monitored by immunoprecipitation followed by immunoblotting (Fig. 3B). Myc-tagged TACE coprecipitated only with Flag-tagged TACE (Fig. 1C) but not with the chimeras (Fig. 3B, upper panel). Additionally, removal of the cytoplasmic domain in the TACE-B mutant abolished the association with TACE, implying that the cytoplasmic domain of TACE is required for dimerization. Adding the cytoplasmic domain of ADAM10 to the truncated TACE (TACE-B/ADAM10cyto; Fig. 3A) did not allow efficient association with full-length TACE (Fig. 3B).
Fig. 3.
The cytoplasmic domain is required for TACE dimerization. (A) Schematic diagram of the TACE/ADAM10 chimeras, in which TACE domains were replaced by AD-AM10 domains. (B) Myc-tagged TACE coimmunoprecipitated with Flag-tagged TACE, but not with Flag-tagged TACE/ADAM10 chimeras, in transfected human embryonic kidney (HEK) 293 cells. ns, nonspecific band. (C) Anti-Flag immunoblotting of cell surface biotinylated Flag-tagged TACE, TACE-B, or TACE-B/ADAM10cyto chimera stably expressed in Cα cells (upper panel). The lower panel shows their monomer and dimer forms. (D) Immunoblotting for Myc-tagged ADAM10, ADAM10 with its cytoplasmic tail deleted (ADAM10-B), or ADAM10-B with a cytoplasmic domain of TACE (ADAM10-B/TACEcyto) was performed on HEK293 cells pretreated with sulfo-EGS before cell lysis. Loss of the cytoplasmic tail of ADAM10 abolished dimerization, which was partially restored by adding the TACE cytoplasmic domain.
We also compared the dimerization of stably expressed Flag-tagged TACE-B and TACE-B/ADAM10cyto to wild-type TACE. All three TACE forms were present at the cell surface at similar abundances (Fig. 3C, upper panel). Thus, the absence of the cytoplasmic domain in TACE-B did not prevent the cell surface presentation of TACE. To assess dimerization, we treated the cells with sulfo-EGS, the cell-impermeable cross-linker that cross-links only cell surface proteins. As shown by immunoblotting of the cell lysates, wild-type TACE formed dimers, but no dimers were observed for TACE-B (Fig. 3C, lower panel). These results indicate that the cytoplasmic domain of TACE is required for dimerization but not for steady-state transport to the cell surface. Additionally, the replacement of the cytoplasmic domain of TACE with that of ADAM10 resulted in dimerization at the cell surface (Fig. 3C, lower panel). Conversely, removal of the cytoplasmic domain of ADAM10 in the ADAM10-B mutant also prevented dimerization of ADAM10 (Fig. 3D), and addition of the cytoplasmic domain of TACE to the truncated ADAM10 restored dimerization, albeit not to the same extent as that of the ADAM10 cytoplasmic domain (Fig. 3D).
Together, these experiments illustrate a requirement for the cytoplasmic domain in the dimerization of TACE as well as ADAM10.
Dissociation of TACE dimers by MAPK signaling
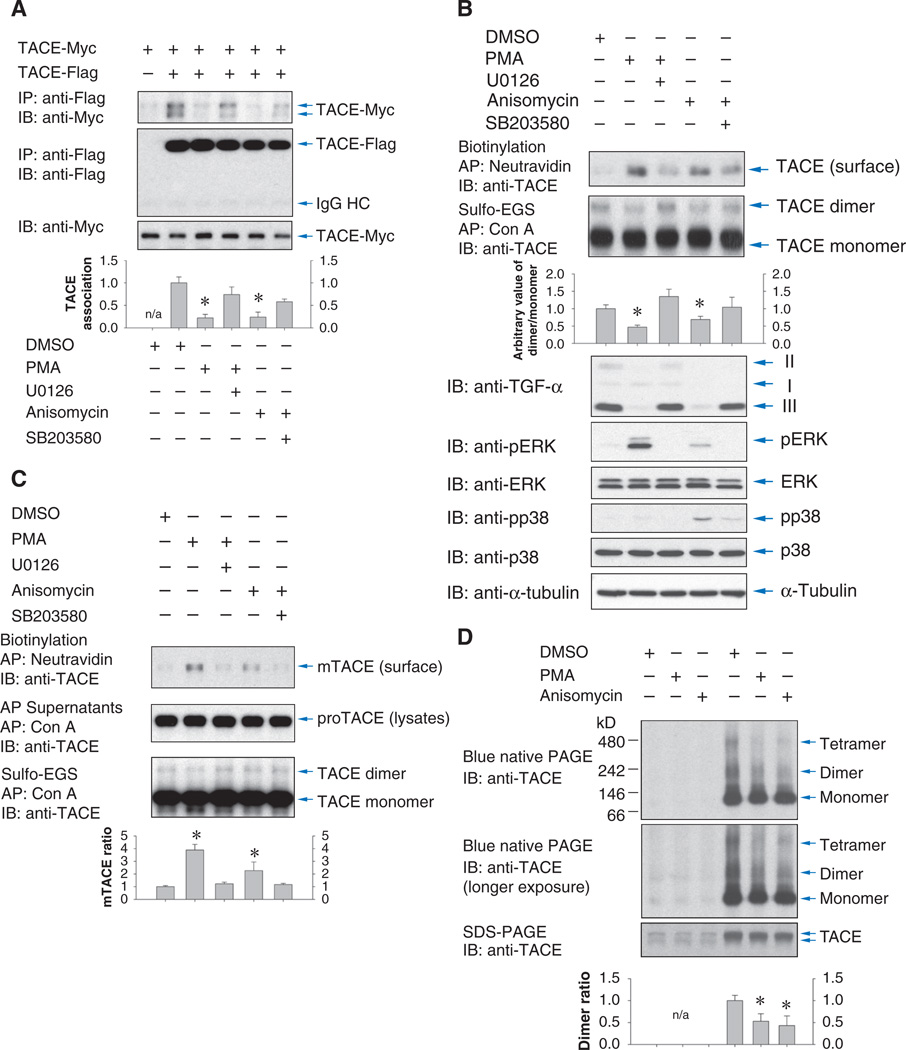
Because the cytoplasmic domain is required for dimerization of TACE, and activation of ERK or p38 MAPK induces changes in cytoplasmic domain phosphorylation, we examined the effects of these pathways on TACE dimerization. In Cα cells coexpressing differently tagged TACE forms, Flag-tagged TACE associated with Myc-tagged TACE (Fig. 4A), consistent with the dimerization in Fig. 1C. Activation of the ERK or p38 MAPK pathways with PMA or anisomycin decreased dimerization, an effect that was largely prevented when PMA treatment was combined with U0126, which inhibits the ERK MAPK pathway, or anisomycin treatment was combined with SB203580, which inhibits p38 MAPK. Similar results were observed in HepG2 cells (fig. S3B) and showed that the metalloproteinase inhibitor TAPI-1 did not affect PMA-induced dimer dissociation or TACE dimer formation in unstimulated cells (fig. S3C).
Fig. 4.
MAPK signaling regulates TACE dimerization. (A) Coimmunoprecipitation of Myc-tagged TACE with Flag-tagged TACE in Cα cells, treated or not with PMA or anisomycin, or the MEK inhibitor U0126 or p38 MAPK inhibitor SB203580. Cell lysates were immunoblotted for TACE. Association of Myc- and Flag-tagged TACE was quantified by density scanning (lower panel). n = 3 blots. *P < 0.01, compared with lysates from dimethyl sulfoxide (DMSO)-treated cells by Student’s t tests. IgG, immunoglobulin G. (B) Dimer formation of endogenous TACE decreased with increased cell surface presentation of TACE after activation of ERK or p38. Cα cells were treated with PMA or anisomycin and with U0126 or SB203580. Top panel: cell surface TACE detected by immunoblotting of cell surface biotinylated proteins. Second and third panels: TACE dimers and monomers detected by anti-TACE immunoblotting of concanavalin A-adsorbed proteins of cells treated with sulfo-EGS. Dimer/monomer ratios were quantified by density scanning. n = 4 experiments. *P < 0.05, compared with DMSO-treated cells by Student’s t tests. Lower panels: ectodomain shedding of TGF-α as detected by the decrease in form III of transmembrane TGF-α, and ERK or p38 activation, shown by immunoblotting for phosphorylated ERK (pERK) or phosphorylated p38 (pp38) MAPK. (C) T4-2 cells treated as in (B). Top panel: cell surface TACE detected by anti-TACE immunoblotting of NeutrAvidin-adsorbed proteins. Middle panel: proteins that did not adsorb to NeutrAvidin were adsorbed to concanavalin A beads and immunoblotted for TACE to measure TACE abundance. Lower panel: TACE dimers detected as in (B) by anti-TACE immunoblotting of concanavalin A-adsorbed proteins from cells treated with sulfo-EGS. Cell surface/total TACE ratios were quantified by density scanning. n = 3 experiments. *P < 0.05, compared with DMSO-treated cells by Student’s ttests. (D) Blue native PAGE (upper and middle panels) or SDS-PAGE (lower panel) of lysates of HEK293T cells transfected with empty vector (lanes 1 to 3) or expressing Flag-tagged TACE (lanes 4 to 6) and treated with PMA or anisomycin. TACE was visualized by anti-TACE immunoblotting, showing TACE monomers and dimers or probable tetramers under native conditions. Dimer/monomer ratios were quantified by density scanning. n = 3 experiments. *P < 0.05, compared with DMSO-treated cells by Student’s t test.
We also examined whether ERK or p38 MAPK activation affected the dimerization of endogenous TACE in Cα cells (Fig. 4B). Similarly to other cells, TGF-α is present in Cα cells as three isoforms that are derived through posttranslational processing of transmembrane TGF-α. The immature form I of transmembrane TGF-α with its nonglycosylated prosegment is intracellular, whereas form II with its glycosylated prosegment and form III lacking the prosegment are at the cell surface (41). As previously reported (10), PMA and anisomycin induced ectodomain shedding of transmembrane TGF-α, as indicated by the decrease in cell-associated transmembrane TGF-α, and these effects were inhibited by U0126 or SB203580, respectively (Fig. 4B, lower panels). Cα cells presented TACE dimers at the cell surface (Fig. 2, A to C), and treatment with PMA or anisomycin induced a rapid increase in the cell surface abundance of TACE, as indicated by cell surface biotinylation assays (Fig. 4B, top panel). These increases were inhibited by U0126 or SB203580. The substantial increase of cell surface TACE was accompanied by a decrease in TACE dimers, as detected by cross-linking by the use of sulfo-EGS, followed by adsorption to concanavalin A and immunoblotting (Fig. 4B, second panel). We conclude that TACE activation is accompanied by a rapid appearance of TACE monomers at the cell surface due to increased cell surface presentation and decreased dimerization.
The appearance of cell surface TACE monomers in response to ERK or p38 MAPK activation was also apparent in T4-2 cells (Fig. 4C). PMA and anisomycin induced the appearance of biotinylated mature TACE at the cell surface, and inhibitors of MEK1/2 (mitogen-activated or extracellular signal-regulated protein kinase kinase 1 and 2) or p38 MAPK abolished this induction (Fig. 4C, upper panel). Concomitantly, the endogenous TACE dimers at the cell surface, which were visualized after sulfo-EGS cross-linking, decreased slightly in response to PMA or anisomycin (Fig. 4C, lower panel), as with Cα cells. Finally, analysis of TACE under native conditions by blue native PAGE also revealed the presence of TACE dimers (Fig. 4D). Endogenous TACE dimers were barely detectable because of the low abundance of endogenous TACE, limited sensitivity of immunoblotting from these gels, and lack of an effective approach compatible with native PAGE to enrich for cell surface TACE. However, the TACE dimers were more prominent in cells expressing transfected TACE. PMA or anisomycin treatment decreased the abundance of TACE dimers (Fig. 4D).
Poor response of TACE mutants with stabilized or impaired dimerization to MAPK activation
Because TACE activation correlated with dimer dissociation, we attempted to address the role of this reversible dissociation in the activation response. We examined whether the cysteine trap mutant, which forms a disulfide-bonded dimer at the cell surface, and TACE-B, which lacks the cytoplasmic domain required for dimerization, could be activated in response to PMA or anisomycin.
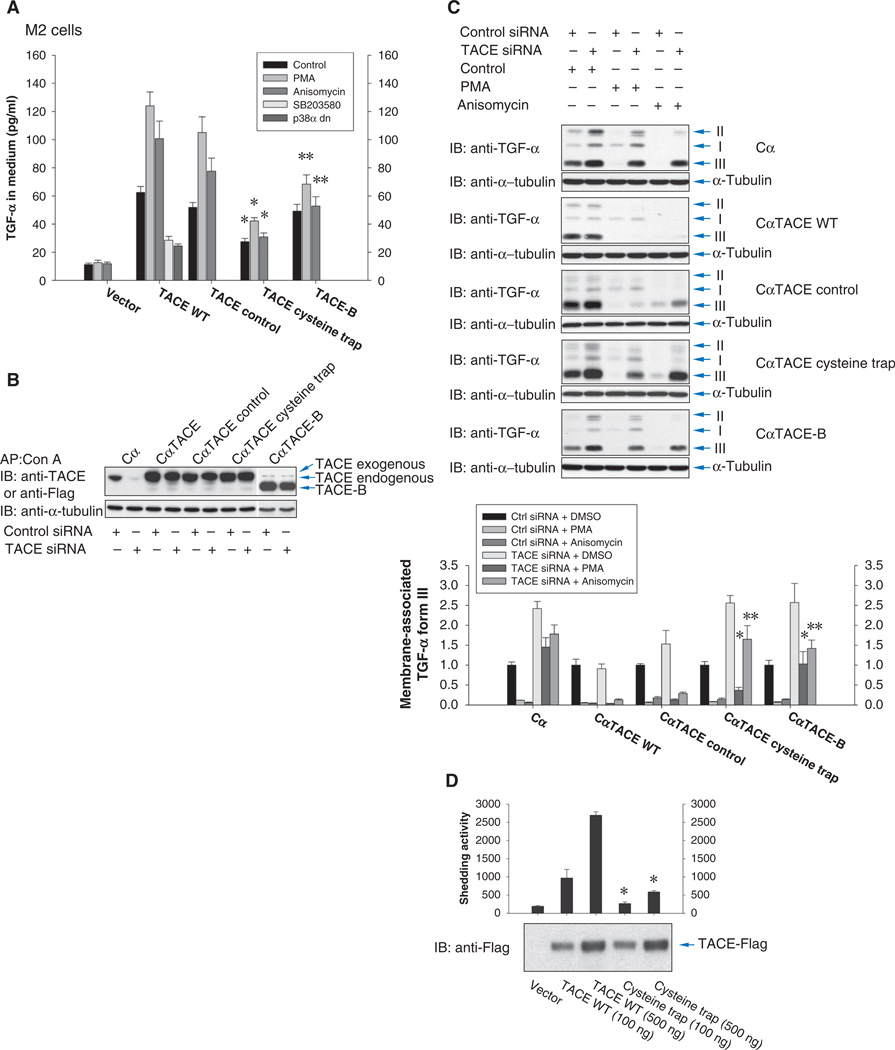
Wild-type TACE, cysteine trap TACE, or the control mutant without the cysteine residue, or TACE-B, was coexpressed at a low amount with TGF-α in M2 cells (CHO cells containing inactive TACE as a result of a C600Y mutation) (42). Whereas TACE-B does not form dimers (Fig. 3C), the abundance of TACE dimers (as detected after sulfo-EGS cross-linking) was similar in cells expressing wild-type, cysteine trap, and control TACE (fig. S3D), consistent with the similar abundances of the different forms of TACE (fig. S3E). Little TGF-α was released when TACE was not expressed, and no increase was observed upon ERK or p38 MAPK activation (Fig. 5A). Expression of wild-type TACE enhanced the release of TGF-α, consistent with a basal shedding activity of TACE (10). TGF-α release was increased in response to PMA or anisomycin, and the basal shedding was decreased when p38 MAPK activity was inhibited with SB203580 or a dominant-negative p38α MAPK (Fig. 5A), as reported (10). Similarly to wild-type TACE, the activity of the control mutant TACE was enhanced in response to PMA or anisomycin. However, the cysteine trap mutant TACE showed lower basal shedding, comparable with that of wild-type TACE after inhibition of p38 MAPK, and its activity was marginally enhanced in response to anisomycin. Although TACE-B was not activated in response to anisomycin, it showed higher basal activity. Both the cysteine trap mutant TACE and TACE-B showed a comparatively low extent of activation in response to PMA. These data suggest that dimer dissociation, which is prevented in the cysteine trap TACE mutant, has an important role in increasing TACE activity in response to ERK or p38 MAPK signaling.
Fig. 5.
TACE with stabilized or impaired dimerization responds poorly to MAPK activation. (A) Quantification of ectodomain shedding of TGF-α by M2 cells expressing TGF-α and a low amount of wild-type or mutant TACE and treated with PMA or anisomycin, or SB203580, or expressing a dominant-negative p38α MAPK mutant. n = 3 experiments. *P < 0.01, **P < 0.05, compared with wild-type TACE by Student’s t tests. (B and C) Cα cells and Cα cells expressing human wild-type TACE, cysteine trap TACE, or control TACE, or TACE-B, were transfected with control or CHO TACE siRNA. (B) Anti-TACE immunoblot. siRNA-mediated silencing of endogenous TACE was visualized only in Cα cells not expressing human TACE. In the last two lanes, TACE-B was visualized by anti-Flag immunoblotting. A weak band similar in size to TACE is nonspecific. (C) Ectodomain shedding of TGF-α induced by PMA or anisomycin was evaluated by immunoblotting for transmembrane TGF-α (forms II and III) whose abundance decreased with increased TACE activity. Relative amounts of TGF-α form III quantified by density scanning (lower panel). n = 3 blots. *P < 0.05, **P < 0.001, compared with cells expressing wild-type TACE by Student’s t tests. (D) In vitro proteolytic activity of wild-type or cysteine trap TACE immunoprecipitated from Cα cells. TACE abundance was shown by anti-Flag immunoblotting. n = 3 assays. *P < 0.001, compared with wild-type TACE by Student’s t tests.
We also generated Cα cells stably expressing wild-type, cysteine trap, or control mutant TACE, or TACE-B, and transfected these cells with siRNA directed against endogenous TACE in CHO cells. The siRNA did not interfere with expression of the transfected human TACE constructs (Fig. 5B). Ectodomain shedding of TGF-α was evaluated by examining the decrease in cell surface transmembrane TGF-α (forms II and III) (Fig. 5C). In control Cα cells, silencing of endogenous TACE decreased the basal shedding of TGF-α (Fig. 5C, top panel), allowing us to evaluate the activity of the introduced TACE mutants. Expression of wild-type or control mutant TACE enhanced basal shedding (Fig. 5C, second and third panels down), consistent with increased TACE abundance (Fig. 5B). When endogenous TACE was silenced, TGF-α shedding was increased because of activation of transfected wild-type or control mutant TACE by PMA or anisomycin (compare lane 2 to lane 4 or 6). In contrast, TGF-α shedding was only marginally increased by PMA or anisomycin in cells expressing cysteine trap TACE or TACE-B (Fig. 5C, lower two panels). It is unclear whether the slightly increased shedding in cells expressing cysteine trap TACE or TACE-B resulted from incomplete silencing of endogenous TACE or residual activity of these two mutants. Additionally, in the absence of stimulation, silencing of endogenous TACE increased the abundance of forms II and III of TGF-α only in control Cα cells or Cα cells expressing cysteine trap TACE or TACE-B, illustrating the impaired shedding by these mutants (Fig. 5C, compare lanes 1 and 2 of each panel). Our results correlate the dynamic and reversible dimerization of TACE with the increased shedding activity in response to ERK or p38 MAPK signaling.
Finally, we also examined whether the change in dimerization state upon ERK or p38 MAPK resulted in increased proteolytic activity of immunoprecipitated TACE in vitro. Tethered dimerization caused by cysteine trapping substantially decreased the proteolytic activity of TACE (Fig. 5D). Additionally, activation of ERK or p38 MAPK signaling enhanced the in vitro proteolytic activity of immunoprecipitated TACE (fig. S3F), as previously reported for ERK MAPK signaling (11). In these assays, the immunoprecipitated TACE combined both the large pool of intracellular TACE and the small fraction of TACE that resides at the cell surface and is subject to regulation. These data correlate decreased dimerization with increased TACE activity.
Increased association of TIMP3 with the TACE dimer
Because TIMP3 regulates TACE activity (Fig. 1A and fig. S1, A to D) and because the TACE dimer dissociates in response to ERK or p38 MAPK activation (Fig. 4), concomitant with increased TACE activity (Fig. 5), we examined whether TACE dimerization is linked to its capacity to associate with TIMP3.
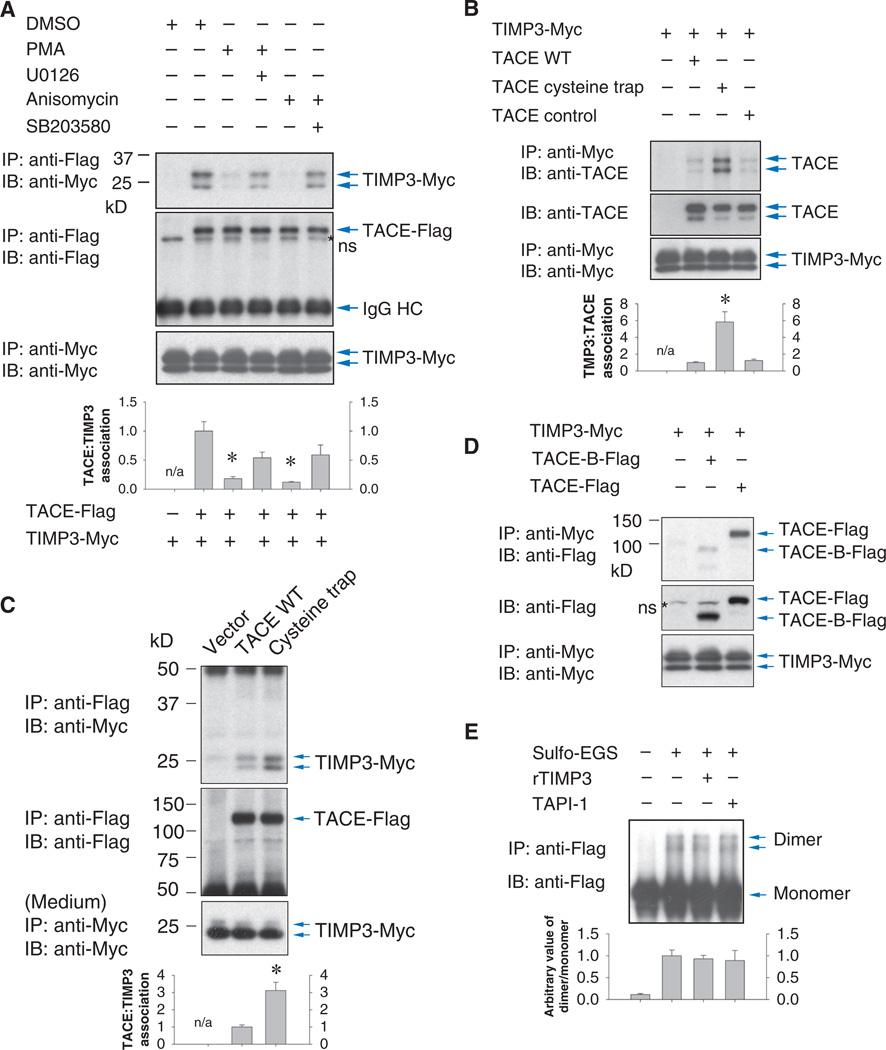
Coimmunoprecipitation analysis of cells coexpressing TIMP3 and TACE showed that the two glycosylated forms of TIMP3 associated with TACE (Fig. 6A). Both PMA and anisomycin induced dissociation of these complexes, which was inhibited by the MEK1/2 inhibitor U0126 or the p38 MAPK inhibitor SB203580, respectively (Fig. 6A). Together with the previous observations, these results suggest that dissociation of TIMP3 from TACE facilitates activation of TACE in response to MAPK signaling.
Fig. 6.
TIMP3 interacts preferentially with the TACE dimer. (A) Association of TIMP3-Myc with TACE-Flag in Cα cells decreased in response to activation of ERK or p38 by PMA or anisomycin. Cells were treated with or without the MEK inhibitor U0126 or p38 MAPK inhibitor SB203580. The association of immunoprecipitated TIMP3 with TACE was quantified by density scanning (lower panel). n = 3 blots. *P < 0.01, compared with DMSO-treated cells by Student’s t tests. ns, nonspecific band. (B) Association of Myc-tagged TIMP3 with Flag-tagged wild-type, cysteine trap, or control TACE in Cα cells, assessed by coimmunoprecipitation. (C) Conditioned medium from HEK293T cells expressing Myc-tagged TIMP3 was incubated with Cα cells expressing Flag-tagged TACE as in (B). Association of Myc-tagged TIMP3 with TACE was assessed by anti-Flag immunoprecipitation and anti-Myc immunoblotting. (B and C) Lower panels show TACE and TIMP3 in cell lysates. Cα or HEK293T cells show variability in the ratio between two forms of TIMP3. TIMP3 association with TACE was quantified by density scanning. n = 3 blots, and Student’s t tests were performed. *P < 0.01, compared with wild-type TACE. (D) More Myc-tagged TIMP3 associated with Flag-tagged wild-type TACE than with TACE-B in Cα cells as assessed by anti-Myc immunoprecipitation and anti-TACE immunoblotting. Abundance of TACE in the cell lysates or of TIMP3 in the conditioned medium was shown by immunoblotting. (E) TIMP3 does not induce TACE dimer formation. Cα cells expressing TACE were treated with TIMP3 or TAPI-1, then sulfo-EGS, before cell lysis, anti-Flag immunoprecipitation, and anti-Flag immunoblotting. Dimer/monomer ratios were quantified by density scanning (lower panel), n = 3 blots, showing no significant changes.
Supporting this notion, more TIMP3 associated with the constitutively dimerized cysteine trap TACE than with the control TACE mutant, which associated with TIMP3 to the same extent as did wild-type TACE (Fig. 6, B and C). Similar results were obtained in anti-Flag coimmunoprecipitation analyses to precipitate Flag-tagged TACEs (fig. S3G). The TACE-B/ADAM10cyto chimera, in which the TACE cytoplasmic domain was replaced by that of ADAM10, thus conferring partial restoration of dimeri-zation (Fig. 3D), showed a higher association for TIMP3 than wild-type TACE (fig. S3G), whereas the absence of the cytoplasmic domain in TACE-B resulted in impaired TIMP3 association (Fig. 6D). These data suggest that TIMP3 preferentially interacts with TACE dimers. Although the dimerization of cysteine trap TACE is constitutive, wild-type TACE and the control mutant TACE would be expected to show a dynamic balance between the monomeric and the dimeric state, with dimerization favored at the cell surface. The stronger association of TIMP3 with the TACE-B/ADAM10cyto chimera likely reflects a quantitatively different dynamic balance imposed by the cytoplasmic domain of ADAM10. Finally, although the TACE dimer had a greater capacity to associate with TIMP3, TIMP3 did not induce TACE dimerization (Fig. 6E). These data suggest that dimerization may be a prerequisite for efficient TIMP3 binding to TACE and that reversal of TACE dimerization may be the basis for the dissociation of TIMP3 in response to ERK or p38 MAPK pathway activation.
DISCUSSION
Ectodomain shedding provides an important mode of regulating processes involved in cell proliferation, inflammation, and cancer progression. How ectodomain shedding is activated at the molecular level is therefore of great interest. We provide new insights into the mechanism of activation of TACE, an ADAM protease that mediates shedding in response to growth, inflammatory, and stress stimuli. We show that dimerization and TIMP3 association are key regulatory events in the control of TACE activity. These mechanisms may be of general relevance in the control of ADAM proteases.
Role of TIMP3 in MAPK-activated TACE activation
TIMP3, an inhibitor of MMPs and TACE, inhibits ectodomain shedding in vivo. Mice lacking TIMP3 show increased TACE activity and soluble TNF-α abundance, leading to inflammation (18, 19, 43). Consistent with these observations, we show that TIMP3 silencing enhances TACE-mediated ectodomain shedding and that TIMP3 regulates TACE activity in response to ERK or p38 MAPK signaling. Thus, TIMP3 silencing compromised TACE-mediated shedding induced by ERK or p38 MAPK signaling, lowering the availability of cleavage substrate at the cell surface through increased basal shedding. TIMP3 associated with TACE, and this association was relieved in response to either pathway. Furthermore, TIMP3 related epistatically to p38 MAPK in the control of TACE activation because growth inhibition resulting from p38± MAPK silencing and consequent inhibition of TACE-mediated release of TGF-α ligands (10) was rescued by additionally silencing TIMP3. Because of the high abundance of TACE and TIMP3 in heart, kidney, lung, liver, and brain (18) and because of the changes in TIMP3 abundance in angiogenesis (44), inflammation (45), and cancer metastasis (46, 47), the regulation of TIMP3 abundance may control TACE-mediated ectodomain shedding.
PMA was recently shown to induce a rapid conformational change in TACE that exposes its catalytic site (22), possibly through disulfide bond isomerization (27). In agreement with these findings, we found that decreased TACE activity, which resulted from silencing p38α MAPK, was accompanied by a change in electrophoretic mobility that likely reflects a conformational change. This conformational change likely explains the decreased association of TIMP3 with TACE in response to ERK or p38 MAPK activation.
Consistent with several lines of evidence from cell systems and in vivo studies (18, 19, 43), but in contrast with those of Le Gall et al., who concluded that TIMP3 may not be involved in TACE activation (22), we propose that TIMP3 regulates endogenous TACE activation and that the increased TACE activity in response to ERK or p38 MAPK involves release of the inhibition by TIMP3. The basis for the discrepancy with the results of Le Gall et al. (22) is unclear but may relate to the minimal difference in TIMP3 abundance between Timp3+/− and Timp3−/− fibroblasts, which may not be adequate to regulate shedding of overexpressed substrate in their assay system. Together, our results combined with all previous reports suggest that both a conformational change and TIMP3 release are normally required for TACE activation and raise the possibility that even in the absence of TIMP3, a conformational change is still required to expose the catalytic center of TACE.
We additionally correlated TACE dimerization with increased TIMP3 binding, suggesting that TACE dimerization helps define the conformation of TACE and enhances association with TIMP3. Considering the 1:1 stoichiometry for the TIMP3 interaction with TACE (28), it is tempting to speculate that two TIMP3 proteins may interact with the TACE dimer. The structural basis of the regulation of TIMP3 association with TACE requires further study. The association of TIMP3 with TACE may be further defined by the presence of other MMPs to which TIMP3 can bind and possible effects of MAPK signaling on the abundance and endocytosis of TACE or TIMP3 (or both) (45, 48).
Role of dimerization in TACE activation
By cross-linking, coimmunoprecipitation, blue native PAGE, and the cys-teine trap approach, we show that TACE forms dimers at the cell surface, where ectodomain shedding occurs and TACE is inhibited by TIMP3. Dimerization at the cell surface may be inherent to ADAM proteases because both TACE and ADAM10 can form homodimers. MMP14 and MMP9 can also form homodimers (30, 31), but this depends on their hemopexin-like domain (49, 50) that is absent in ADAM proteases. We also observed TACE tetramers in native PAGE (Fig. 4D), raising the possibility that high-order TACE complexes might form at the cell surface and that these could be involved in regulating the activation and substrate recognition of TACE.
We present evidence that dimerization regulates TACE-mediated shedding and that dimerization of TACE (or ADAM10) requires its cytoplasmic domain. A role of the cytoplasmic domain in TACE dimerization, together with its role in the rapid increase in the regulated cell surface presentation of TACE (10, 25, 26), may account for, or contribute to, the rapid increase in TACE activity in response to ERK or p38 MAPK signaling. Accordingly, cytoplasmically truncated TACE did not dimerize and responded poorly to ERK or p38 MAPK signaling. Conversely, disulfide-tethered TACE dimers showed impaired responsiveness to p38 MAPK, illustrating the role of reversible dimerization in TACE activity. However, cytoplasmically truncated TACE was transported to the cell surface under steady-state conditions and is enzymatically active (51), suggesting that the cytoplasmic domain does not regulate these functions. Consistent with our findings, deletion of the cytoplasmic domain impairs TACE-mediated shedding in response to PMA (10, 13, 23, 25, 26, 52, 53). However, the residual activity, which appears to depend on the amount of truncated TACE and the TACE/substrate ratio, may have led to the conclusion that the cytoplasmic domain is not essential for TACE activity (22). Expression of truncated TACE at a high abundance may make some cytoplasmic domain functions less apparent.
Because dimerization often leads to receptor activation, it may seem counterintuitive that dimer dissociation is at the basis of TACE activation. However, several examples illustrate dissociation of dimers or protein interactions in response to signaling. Some G protein (heterotrimeric guanosine triphosphate–binding protein)–coupled receptors, such as the parathyroid hormone receptor, form dimers that dissociate upon ligand binding and then activate G protein signaling (54). Dimers of the small heat shock protein HSP27 dissociate upon activation by heat shock (55), and binding of acidic phospholipids to the adenosine triphosphatase SecA results in dimer dissociation and activation (56). Finally, stress in the endoplasmic reticulum (ER) induces dissociation of the chaperone BiP (binding immunoglobulin protein) from the transmembrane kinases IRE1 (inositol-requiring enzyme 1) or PERK [PKR-like ER–localized eIF2α (eu-karyotic initiation factor 2α) kinase], thereby activating the unfolded protein response (57). In the case of TACE, dimer dissociation results in the release of TIMP3 and may contribute to a conformational change that exposes the catalytic site of TACE to allow cleavage.
Understanding the mechanism of TACE activation in response to MAPK signaling
Although TACE activation was shown more than a decade ago, the mechanism through which cell signaling leads to TACE activation has largely remained elusive. Our current and previously reported findings help to define a model of TACE activation (Fig. 7). After removal of its prodomain, TACE activity is enhanced in response to signals that activate the ERK or p38 MAPK pathways. This stimulation is rapid and does not require protein synthesis (10, 39), indicating a posttranslational event. Indeed, activation of the ERK or p38 MAPK pathway induces rapid changes in the phosphorylation of the TACE cytoplasmic domain (10, 23, 24). Although p38 MAPK has been shown to directly phosphorylate TACE on Thr735, it remains unclear whether ERK directly phosphorylates TACE or whether other effectors mediate the changes in phosphorylation. After these phosphorylation events, activation of shedding correlates with a rapid increase in mature TACE at the cell surface, resulting from mobilization of TACE from intracellular stores to the cell surface. Replacing Thr735 with alanine, which prevents phosphorylation by p38, results in degradation of TACE at the cell surface (10). Whether autocatalysis controls the cell surface presentation of TACE, as has been suggested for some ADAMs or MMPs (2, 3), remains to be determined, although inhibition of TACE activity does not affect its degradation. TACE interacts with iR-hom2, a proteolytically inactive transmembrane protein belonging to the rhomboid family, and this association controls TACE maturation and trafficking to the cell surface (58, 59). These observations illustrate the importance of cell surface presentation of TACE in the regulation of its shedding activity and raise the possibility that the interaction between TACE and iRhom2 may be a target of MAPK signaling.
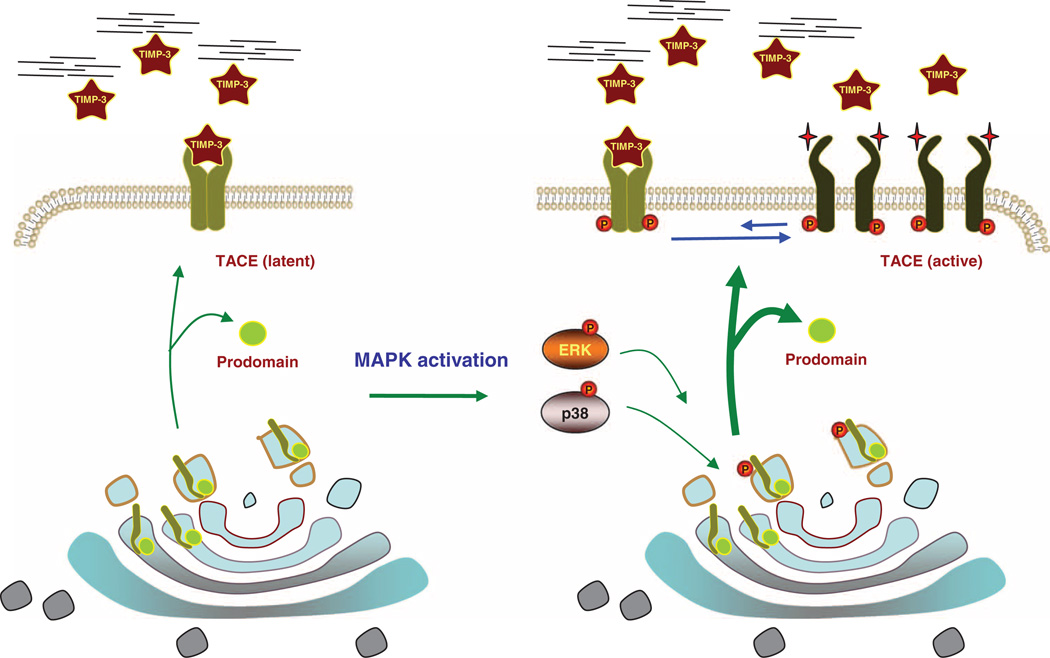
Fig. 7.
Model of TACE activation by MAPK signaling. Without MAPK signaling, cell surface TACE is primarily present as dimer and readily associates with TIMP3, possibly binding to the TACE dimer in a 2:2 ratio. TACE is therefore only partially active and its activity depends on the availability of TIMP3 (left). Upon p38 or ERK MAPK pathway activation, the cytoplasmic domain of TACE is phosphorylated, resulting in rapidly increased cell surface presentation (wider arrows), higher abundance of TACE monomers, and lower abundance of dimers. The lack of dimerization results in lower association of TACE with TIMP3 and the activation of TACE (right). The activity of TACE thus results from the dynamic balance between TACE dimers and monomers at the cell surface and the resulting differences in TIMP3 binding efficiency.
Concomitant with the rapid increase in cell surface presentation, TACE shows regulated dimerization. TACE is predominantly present at the cell surface as a dimer, whereas the bulk of TACE proteins appears to reside intracellularly in a monomeric form. The TACE dimer associates with extracellular TIMP3, which keeps TACE inactive. ERK or p38 MAPK signaling changes the dynamic balance between TACE dimers and monomers and their conformations, presumably as a result of changes in phosphorylation of its cytoplasmic domain, which is required for TACE dimerization. This change in monomer-dimer equilibrium, fueled by the rapid increase of cell surface presentation of TACE and decreased TACE dimerization, then translates into a predominance of TACE monomer at the cell surface, a conformational change in TACE, and decreased association for TIMP3, resulting in TACE activation and higher shedding activity (Fig. 7). Because ADAM10 also dimerizes, we surmise that the regulation of the dimer-monomer equilibrium in response to signaling may define the activation of other ADAM proteases and the ectodomain shedding of their substrates. However, this mode of regulation may differ for individual ADAMs because ADAM10 has a higher relative amount of monomers at the cell surface in unstimulated cells than TACE.
MATERIALS AND METHODS
Expression plasmids, reagents, and antibodies
Expression plasmids encoding TGF-α or Flag- or Myc-tagged human TACE, TβRII, and ALK2 have been previously described (10, 11). Plasmids encoding Flag- or Myc-tagged ADAM10, TACE-B, ADAM10-B, TACE/ ADAM10, or ADAM10/TACE chimeras were made with polymerase chain reaction (PCR)–based methods from pRK5-TACE-Flag and human ADAM10 complementary DNA (cDNA), which were isolated by reverse transcription–PCR (RT-PCR) from HeLa cells. A plasmid encoding mouse TIMP3 was provided by Z. Werb, and the human TIMP3 cDNA was generated by RT-PCR and inserted into pRK5 with a C-terminal Myc tag. All coding sequences were verified by DNA sequencing.
The phorbol ester PMA, anisomycin, SB203580, U0126, TAPI-1, and human TIMP3 were from Calbiochem. Iodoacetamide, concanavalin A Sepharose 4B, and anti-Flag (M2) antibody–conjugated agarose were from Sigma. Protein A Sepharose was from Amersham. The cross-linkers DSP, EGS, and sulfo-EGS, and EZ-link sulfo-NHS–LC-biotin and NeutrAvidin beads were from Pierce.
The monoclonal anti–TGF-α (ab-1) and rabbit anti-ADAM10 (ab-1) antibodies were from Calbiochem, rabbit anti-TACE was from QED Bioscience, and rabbit anti-TβRI was from Santa Cruz Biotechnology. Anti–α-tubulin, anti-Flag (M2), rabbit anti-Flag, and anti-Myc (9E10) antibodies were from Sigma.
Cell culture and transfections
CHO cells and derivative Cα (39) and M2 cells (42) were cultured in F-12 medium with 10% fetal bovine serum (FBS), and HeLa, HepG2, and 293T cells were maintained in Dulbecco’s modified Eagle’s medium with 10% FBS. T4-2 cells were provided by M. Bissell (Lawrence Berkeley National Laboratory) and cultured as described (8). CHO, Cα, M2, and 293T cells were transfected with FuGENE 6 (Roche). Cα cells were stably transfected with plasmids for TACE, cysteine trap TACE, control mutant TACE, TACE-B, or TACE-B/ADAM10cyto along with pRK7-Hyg to enable selection with hygromycin B (600 µg/ml; Roche). Colonies were obtained by serial dilution culture and analyzed by anti-Flag immunoblotting.
Cross-linking and cell surface protein biotinylation
Cells were treated with U0126 (10 µM) or SB203580 (10 µM) 30 min before and during PMA (25 nM) or anisomycin (1 µM) treatment. Cells were washed three times with ice-cold phosphate-buffered saline (PBS) and then incubated with 1 mM cross-linker DSP, EGS, or sulfo-EGS and/or 1 mM EZ-link sulfo-NHS–LC-biotin in PBS at 4°C for 60 min. Cross-linking or biotinylation reactions were stopped with 100 mM glyCine in PBS, and cells were lysed in Myc lysis buffer (11). After a 2-hour incubation with concanavalin A Sepharose (Sigma), NeutrAvidin (Pierce), or anti-Flag M2 beads (Sigma), adsorbed proteins were washed three times with magnesium lysis buffer (MLB), separated by 4 to 20% SDS-PAGE, and immunoblotted with anti-TACE, anti-ADAM10, or anti-Flag antibodies.
Blue native PAGE and immunoblotting
293T cells were transfected with 0.1 µg of plasmid encoding C-terminal Flag-tagged TACE or control vector. Thirty-six hours after transfection, cells were treated with PMA (25 nM) or anisomycin (1 µM) for 30 min and lysed in NativePAGE sample buffer with 1% DDM (n-dodecyl-D-maltoside; Invitrogen). After addition of 0.25% Coomassie G-250, cell lysates were separated by blue native PAGE with the NativePAGE Bis-Tris Gel System (Invitrogen). After electrophoresis, the gel was soaked in 0.1% SDS buffer, transferred to polyvinylidene difluoride membrane, fixed by 8% acetic acid, air-dried, soaked in methanol to remove Coomassie dye, and subjected to immunoblotting with anti-TACE antibody and chemiluminescence.
Cysteine trap analysis
Cysteine trap analysis of dimerization of membrane-associated proteins was done as previously described (33). To generate cysteine trap or control mutant TACE, we introduced the DNA sequences GGTGCAGGTGCTG-GTTGCGGTGCT or GGTGCAGGTGCTGGTGCAGGTGCT into the pRK5-TACE-Flag after nucleotide 2172 of the human TACE cDNA sequence (ref: NM_003183.4), thus inserting the peptide Gly-Ala-Gly-Ala-Gly-Cys-Gly-Ala or Gly-Ala-Gly-Ala-Gly-Ala-Gly-Ala, respectively, before His672. Cα cells were then transfected with 0.2 µg of plasmid encoding TACE, cysteine trap TACE, or control mutant TACE with FuGENE 6 (Roche) and lysed 48 hours later with MLB containing 10 mM iodoAcetamide to stabilize disulfide bonds. Cell lysates were subjected to 4 to 20% gradient SDS-PAGE, with or without reduction with dithiothreitol (DTT), and anti-TACE immunoblotting. Cα cells stably expressing TACE, TACE-MC, or TACE-Con were similarly analyzed but without previous disulfide reduction.
Coimmunoprecipitations and Western blotting
Cα, HepG2, or 293T cells were transfected with 0.2 µg of plasmid encoding C-terminal Myc- or Flag-tagged TACE, TACE-B, ADAM10, TACE-B/ ADAM10cyto, and/or TIMP3. Forty-eight hours after transfection, cells were treated with anisomycin, PMA, and/or U0126 for the indicated times and lysed with MLB. Lysates were subjected to immunoprecipitation with anti-Flag (M2)–conjugated Sepharose, anti-Flag rabbit polyclonal antibody, or anti-Myc (9E10) antibody with protein A Sepharose. After four washes with MLB, adsorbed proteins were analyzed by 4 to 20% gradient SDS-PAGE (Invitrogen) and anti-Myc or anti-Flag immunoblotting. Cell lysates were also analyzed by SDS-PAGE and immunoblotting to control protein abundance.
RNA interference
siRNA (QIAGEN) to silence endogenous TACE in CHO or T4-2 cells targeted the Chinese hamster TACE mRNA sequence 5′-GAGGAUUUAAAG-GUUAUGGAA-3′ (nucleotides 900 to 920; ref: AY313173Z) or human TACE mRNA sequence 5′-AAGAAACAGAGUGCUAAUUUA-3′ (Hs_ ADAM17_8) (nucleotides 2642 to 2662; ref: NM_003183.4), respectively. siRNA to silence TIMP3 or p38α MAPK in T4-2 cells targeted the human mRNA sequence 5′-CACGCUGGUCUACACCAUCAA-3′ or 5′-GUGGCCUUAAGCUGGAGGUCA-3′ (Hs_TIMP3_2 and Hs_TIMP3_5, nucleotides 1360 to 1380 and 515 to 535; ref: NM000362) or 5′-CUCAGU-GAUACGUACAGCCAA-3′ (Hs_MAPK14_7, nucleotides 1933 to 1953; ref: NM_001315). siRNA was transfected with Lipofectamine RNAiMAX (Invitrogen) for 48 hours before further assay, and reverse transfection was used to reach optimal efficiency.
Ectodomain shedding assay
Twenty-four hours after transfection with TIMP3, p38α MAPK, or control siRNA, T4-2 cells were starved overnight, washed with fresh medium, and treated with PMA (25 nM) or anisomycin (1 µM) for 60 min. Cells were then surface-biotinylated and lysed, and proteins adsorbed to NeutrAvidin beads were analyzed by 4 to 20% gradient SDS-PAGE and anti-TβRI (Santa Cruz Biotechnology) immunoblotting to visualize cell surface TβRI. Shedding of transmembrane TGF-α in T4-2 cells, Cα cells, or transfected M2 cells and soluble TGF-α release were detected as previously described (10). For quantif ication of soluble TGF-α, media were collected, cooled, and cleared at 5000 rpm for 10 min to remove unattached cells and debris, and TGF-α was quantified with Quantikine Immunoassay kit (R&D Systems).
TACE proteolytic activity assay
TACE activity was measured with the InnoZyme TACE Activity Kit (Calbiochem). Cα transfected to express wild-type or cysteine trap TACE or HepG2 cells transfected with control or TACE siRNA were treated for 30 min with PMA (25 nM), anisomycin (1 µM), U0126 (10 µM), or SB203580 (10 µM). Cells were lysed with MLB; a portion of the cell ly-sates was subjected to the TACE activity assay, and another portion was subjected to anti-Flag immunoblotting to assess TACE abundance. TACE in cell lysates was visualized by anti-TACE monoclonal antibody and its activity was measured with an internally quenched fluorescent substrate. Cleavage of the amide bond of the substrate released the fluorophore, resulting in increased fluorescence, which directly relates to the activity of TACE.
Cell proliferation assay
Cell proliferation was quantified by 5-bromo-2′-deoxyuridine (BrdU) incorporation. T4-2 cells were transfected with siRNA to TACE, p38α MAPK, TIMP3, or control siRNA and were seeded 24 hours later in a 48-well plate. After 72 hours, cells were incubated with BrdU for 3 hours, and incorporated BrdU was measured with the BrdU Cell Proliferation Assay kit (Calbiochem).
Statistics
Quantitative data are presented as means ± SEM from at least three independent experiments. Data presented as either fold change or as percentage were log-transformed before statistical analysis. When appropriate, statistical differences between each group were analyzed with unpaired Student’s t test by SigmaPlot 10.0. Differences were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
We are grateful to L. Wu for suggestions, Z. Werb for mouse TIMP3 plasmid, and M. J. Bissell for T4-2 cells. Funding: This research was sponsored by grants RO1-CA63101 and PO1-HL60231 to R.D. J.L. was supported by a postdoctoral fellowship from the American Heart Association.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/5/222/ra34/DC1
Fig. S1. TIMP3 regulates TACE-mediated shedding of endogenous substrates.
Fig. S2. ADAM10 forms dimers at the cell surface.
Fig. S3. TACE
Author contributions: P.X. and R.D. conceived the study and experimental design and wrote the manuscript. P.X. carried out most experiments. J.L. and M.S.-Y. contributed with several experiments and helped with data analyses and through discussions.
Competing interests:
The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Blobel CP. ADAMs: Key components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 2.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: Multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 3.Huovila AP, Turner AJ, Pelto-Huikko M, Kärkkäinen I, Ortiz RM. Shedding light on ADAM metalloproteinases. Trends Biochem. Sci. 2005;30:413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Murphy G. The ADAMs: Signalling scissors in the tumour microenvironment. Nat. Rev. Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 5.Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin. Cell Dev. Biol. 2009;20:164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Borrell-Pages M, Rojo F, Albanell J, Baselga J, Arribas J. TACE is required for the activation of the EGFR by TGF-α in tumors. EMBO J. 2003;22:1114–1124. doi: 10.1093/emboj/cdg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, Lo Y, Baribaud F, Mikami I, Reguart N, Yang G, Li Y, Yao W, Vaddi K, Gazdar AF, Friedman SM, Jablons DM, Newton RC, Fridman JS, Minna JD, Scherle PA. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenny PA, Bissell MJ. Targeting TACE-dependent EGFR ligand shedding in breast cancer. J. Clin. Invest. 2007;117:337–345. doi: 10.1172/JCI29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalaris A, Adam N, Sina C, Rosenstiel P, Lehmann-Koch J, Schirmacher P, Hartmann D, Cichy J, Gavrilova O, Schreiber S, Jostock T, Matthews V, Häsler R, Becker C, Neurath MF, Reiss K, Saftig P, Scheller J, Rose-John S. Critical role of the disintegrin metalloprotease ADAM17 for intestinal inflammation and regeneration in mice. J. Exp. Med. 2010;207:1617–1624. doi: 10.1084/jem.20092366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu P, Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol. Cell. 2010;37:551–566. doi: 10.1016/j.molcel.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Xu P, Lamouille S, Xu J, Derynck R. TACE-mediated ectodomain shedding of the type I TGF-β receptor downregulates TGF-β signaling. Mol. Cell. 2009;35:26–36. doi: 10.1016/j.molcel.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 2004;164:769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horiuchi K, Le Gall S, Schulte M, Yamaguchi T, Reiss K, Murphy G, Toyama Y, Hartmann D, Saftig P, Blobel CP. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol. Biol. Cell. 2007;18:176–188. doi: 10.1091/mbc.E06-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98:621–628. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel IR, Attur MG, Patel RN, Stuchin SA, Abagyan RA, Abramson SB, Amin AR. TNF-α convertase enzyme from human arthritis-affected cartilage: Isolation of cDNA by differential display, expression of the active enzyme, and regulation of TNF-α. J. Immunol. 1998;160:4570–4579. [PubMed] [Google Scholar]
- 16.Colón AL, Menchén LA, Hurtado O, De Cristóbal J, Lizasoain I, Leza JC, Lorenzo P, Moro MA. Implication of TNF-α convertase (TACE/ADAM17) in inducible nitric oxide synthase expression and inflammation in an experimental model of colitis. Cytokine. 2001;16:220–226. doi: 10.1006/cyto.2001.0969. [DOI] [PubMed] [Google Scholar]
- 17.Amour A, Slocombe PM, Webster A, Butler M, Knight CG, Smith BJ, Stephens PE, Shelley C, Hutton M, Knäuper V, Docherty AJ, Murphy G. TNF-α converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 1998;435:39–44. doi: 10.1016/s0014-5793(98)01031-x. [DOI] [PubMed] [Google Scholar]
- 18.Mohammed FF, Smookler DS, Taylor SE, Fingleton B, Kassiri Z, Sanchez OH, English JL, Matrisian LM, Au B, Yeh WC, Khokha R. Abnormal TNF activity in Timp3−/− mice leads to chronic hepatic inflammation and failure of liver regeneration. Nat. Genet. 2004;36:969–977. doi: 10.1038/ng1413. [DOI] [PubMed] [Google Scholar]
- 19.Federici M, Hribal ML, Menghini R, Kanno H, Marchetti V, Porzio O, Sunnarborg SW, Rizza S, Serino M, Cunsolo V, Lauro D, Mauriello A, Smookler DS, Sbraccia P, Sesti G, Lee DC, Khokha R, Accili D, Lauro R. Timp3 deficiency in insulin receptor-haploinsufficient mice promotes diabetes and vascular inflammation via increased TNF-α. J. Clin. Invest. 2005;115:3494–3505. doi: 10.1172/JCI26052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlöndorff J, Becherer JD, Blobel CP. Intracellular maturation and localization of the tumour necrosis factor a convertase (TACE) Biochem. J. 2000;347(Pt. 1):131–138. [PMC free article] [PubMed] [Google Scholar]
- 21.Peiretti F, Canault M, Deprez-Beauclair P, Berthet V, Bonardo B, Juhan-Vague I, Nalbone G. Intracellular maturation and transport of tumor necrosis factor a converting enzyme. Exp. Cell Res. 2003;285:278–285. doi: 10.1016/s0014-4827(03)00052-1. [DOI] [PubMed] [Google Scholar]
- 22.Le Gall SM, Maretzky T, Issuree PD, Niu XD, Reiss K, Saftig P, Khokha R, Lundell D, Blobel CP. ADAM17 is regulated by a rapid and reversible mechanism that controls access to its catalytic site. J. Cell Sci. 2010;123:3913–3922. doi: 10.1242/jcs.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Díaz-Rodríguez E, Montero JC, Esparís-Ogando A, Yuste L, Pandiella A. Extracellular signal-regulated kinase phosphorylates tumor necrosis factor a-converting enzyme at threonine 735: A potential role in regulated shedding. Mol. Biol. Cell. 2002;13:2031–2044. doi: 10.1091/mbc.01-11-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan H, Turck CW, Derynck R. Characterization of growth factor-induced serine phosphorylation of tumor necrosis factor-α converting enzyme and of an alternatively translated polypeptide. J. Biol. Chem. 2003;278:18617–18627. doi: 10.1074/jbc.M300331200. [DOI] [PubMed] [Google Scholar]
- 25.Soond SM, Everson B, Riches DW, Murphy G. ERK-mediated phosphorylation of Thr735 in TNFα-converting enzyme and its potential role in TACE protein trafficking. J. Cell Sci. 2005;118:2371–2380. doi: 10.1242/jcs.02357. [DOI] [PubMed] [Google Scholar]
- 26.Killock DJ, Ivetić A. The cytoplasmic domains of TNFα-converting enzyme (TACE/ ADAM17) and L-selectin are regulated differently by p38 MAPK and PKC to promote ectodomain shedding. Biochem. J. 2010;428:293–304. doi: 10.1042/BJ20091611. [DOI] [PubMed] [Google Scholar]
- 27.Willems SH, Tape CJ, Stanley PL, Taylor NA, Mills IG, Neal DE, McCafferty J, Murphy G. Thiol isomerases negatively regulate the cellular shedding activity of ADAM17. Biochem. J. 2010;428:439–450. doi: 10.1042/BJ20100179. [DOI] [PubMed] [Google Scholar]
- 28.Wisniewska M, Goettig P, Maskos K, Belouski E, Winters D, Hecht R, Black R, Bode W. Structural determinants of the ADAM inhibition by TIMP-3: Crystal structure of the TACE-N-TIMP-3 complex. J. Mol. Biol. 2008;381:1307–1319. doi: 10.1016/j.jmb.2008.06.088. [DOI] [PubMed] [Google Scholar]
- 29.Guinea-Viniegra J, Zenz R, Scheuch H, Hnisz D, Holcmann M, Bakiri L, Schonthaler HB, Sibilia M, Wagner EF. TNFα shedding and epidermal inflammation are controlled by Jun proteins. Genes Dev. 2009;23:2663–2674. doi: 10.1101/gad.543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itoh Y, Takamura A, Ito N, Maru Y, Sato H, Suenaga N, Aoki T, Seiki M. Homo-philic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J. 2001;20:4782–4793. doi: 10.1093/emboj/20.17.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson MW, Bernardo MM, Pietila M, Gervasi DC, Toth M, Kotra LP, Massova I, Mobashery S, Fridman R. Characterization of the monomeric and dimeric forms of latent and active matrix metalloproteinase-9. Differential rates for activation by stromelysin 1. J. Biol. Chem. 2000;275:2661–2668. doi: 10.1074/jbc.275.4.2661. [DOI] [PubMed] [Google Scholar]
- 32.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): An ancient family with structural and functional diversity. Biochim. Biophys. Acta. 2010;1803:55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorokin A, Lemmon MA, Ullrich A, Schlessinger J. Stabilization of an active dimeric form of the epidermal growth factor receptor by introduction of an inter-receptor disulfide bond. J. Biol. Chem. 1994;269:9752–9759. [PubMed] [Google Scholar]
- 34.Burke CL, Stern DF. Activation of Neu (ErbB-2) mediated by disulfide bond-induced dimerization reveals a receptor tyrosine kinase dimer interface. Mol. Cell. Biol. 1998;18:5371–5379. doi: 10.1128/mcb.18.9.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galvin BD, Hart KC, Meyer AN, Webster MK, Donoghue DJ. Constitutive receptor activation by Crouzon syndrome mutations in fibroblast growth factor receptor (FGFR) 2 and FGFR2/Neu chimeras. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7894–7899. doi: 10.1073/pnas.93.15.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang G, den Hertog J, Su J, Noel J, Sap J, Hunter T. Dimerization inhibits the activity of receptor-like protein-tyrosine phosphatase-a. Nature. 1999;401:606–610. doi: 10.1038/44170. [DOI] [PubMed] [Google Scholar]
- 37.Majeti R, Xu Z, Parslow TG, Olson JL, Daikh DI, Killeen N, Weiss A. An inactivating point mutation in the inhibitory wedge of CD45 causes lymphoproliferation and autoimmunity. Cell. 2000;103:1059–1070. doi: 10.1016/s0092-8674(00)00209-9. [DOI] [PubMed] [Google Scholar]
- 38.Siegel PM, Muller WJ. Mutations affecting conserved cysteine residues within the extracellular domain of Neu promote receptor dimerization and activation. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8878–8883. doi: 10.1073/pnas.93.17.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan H, Derynck R. Ectodomain shedding of TGF-α and other transmembrane proteins is induced by receptor tyrosine kinase activation and MAP kinase signaling cascades. EMBO J. 1999;18:6962–6972. doi: 10.1093/emboj/18.24.6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Constitutive and regulated a-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bringman TS, Lindquist PB, Derynck R. Different transforming growth factor-α species are derived from a glycosylated and palmitoylated transmembrane precursor. Cell. 1987;48:429–440. doi: 10.1016/0092-8674(87)90194-2. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Fan H. Loss of ectodomain shedding due to mutations in the metalloprotease and cysteine-rich/disintegrin domains of the tumor necrosis factor-α converting enzyme (TACE) J. Biol. Chem. 2004;279:27365–27375. doi: 10.1074/jbc.M401690200. [DOI] [PubMed] [Google Scholar]
- 43.Murthy A, Defamie V, Smookler DS, Di Grappa MA, Horiuchi K, Federici M, Sibilia M, Blobel CP, Khokha R. Ectodomain shedding of EGFR ligands and TNFR1 dictates hepatocyte apoptosis during fulminant hepatitis in mice. J. Clin. Invest. 2010;120:2731–2744. doi: 10.1172/JCI42686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saunders WB, Bohnsack BL, Faske JB, Anthis NJ, Bayless KJ, Hirschi KK, Davis GE. Coregulation of vascular tube stabilization by endothelial cell TIMP-2 and pericyte TIMP-3. J. Cell Biol. 2006;175:179–191. doi: 10.1083/jcb.200603176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louis E, Ribbens C, Godon A, Franchimont D, De Groote D, Hardy N, Boniver J, Belaiche J, Malaise M. Increased production of matrix metalloproteinase-3 and tissue inhibitor of metalloproteinase-1 by inflamed mucosa in inflammatory bowel disease. Clin. Exp. Immunol. 2000;120:241–246. doi: 10.1046/j.1365-2249.2000.01227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyazaki T, Kato H, Nakajima M, Faried A, Takita J, Sohda M, Fukai Y, Yamaguchi S, Masuda N, Manda R, Fukuchi M, Ojima H, Tsukada K, Kuwano H. An immuno-histochemical study of TIMP-3 expression in oesophageal squamous cell carcinoma. Br. J. Cancer. 2004;91:1556–1560. doi: 10.1038/sj.bjc.6602185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mylona E, Magkou C, Giannopoulou I, Agrogiannis G, Markaki S, Keramopoulos A, Nakopoulou L. Expression of tissue inhibitor of matrix metalloproteinases (TIMP)-3 protein in invasive breast carcinoma: Relation to tumor phenotype and clinical outcome. Breast Cancer Res. 2006;8:R57. doi: 10.1186/bcr1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sampieri CL, Nuttall RK, Young DA, Goldspink D, Clark IM, Edwards DR. Activation of p38 and JNK MAPK pathways abrogates requirement for new protein synthesis for phorbol ester mediated induction of select MMP and TIMP genes. Matrix Biol. 2008;27:128–138. doi: 10.1016/j.matbio.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Osenkowski P, Toth M, Fridman R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP) J. Cell. Physiol. 2004;200:2–10. doi: 10.1002/jcp.20064. [DOI] [PubMed] [Google Scholar]
- 50.Itoh Y, Ito N, Nagase H, Evans RD, Bird SA, Seiki M. Cell surface collagenolysis requires homodimerization of the membrane-bound collagenase MT1-MMP. Mol. Biol. Cell. 2006;17:5390–5399. doi: 10.1091/mbc.E06-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy P, Slack JL, Davis R, Cerretti DP, Kozlosky CJ, Blanton RA, Shows D, Peschon JJ, Black RA. Functional analysis of the domain structure of tumor necrosis factor-α converting enzyme. J. Biol. Chem. 2000;275:14608–14614. doi: 10.1074/jbc.275.19.14608. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Q, Thomas SM, Lui VW, Xi S, Siegfried JM, Fan H, Smithgall TE, Mills GB, Grandis JR. Phosphorylation of TNF-α converting enzyme by gastrin-releasing peptide induces amphiregulin release and EGF receptor activation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6901–6906. doi: 10.1073/pnas.0509719103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng Y, Schlondorff J, Blobel CP. Evidence for regulation of the tumor necrosis factor α-convertase (TACE) by protein-tyrosine phosphatase PTPH1. J. Biol. Chem. 2002;277:42463–42470. doi: 10.1074/jbc.M207459200. [DOI] [PubMed] [Google Scholar]
- 54.Pioszak AA, Harikumar KG, Parker NR, Miller LJ, Xu HE. Dimeric arrangement of the parathyroid hormone receptor and a structural mechanism for ligand-induced dissociation. J. Biol. Chem. 2010;285:12435–12444. doi: 10.1074/jbc.M109.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Almeida-Souza L, Goethals S, de Winter V, Dierick I, Gallardo R, Van Durme J, Irobi J, Gettemans J, Rousseau F, Schymkowitz J, Timmerman V, Janssens S. Increased monomerization of mutant HSPB1 leads to protein hyperactivity in Charcot-Marie-Tooth neuropathy. J. Biol. Chem. 2010;285:12778–12786. doi: 10.1074/jbc.M109.082644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Or E, Navon A, Rapoport T. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21:4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 58.McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, Lang KS, Häussinger D, Wakeham A, Itie-Youten A, Khokha R, Ohashi PS, Blobel CP, Mak TW. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335:229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.