Abstract
Complex genetic mutations are common in brain cancer, making gene therapy an attractive approach to repair or modulate altered genes and cellular pathways. Non-viral gene vectors can offer DNA delivery without the risk of immunogenicity and/or insertional mutagenesis that are common with viral vectors. We developed pH-responsive DNA nanoparticles, CH12K18PEG5k, by inserting a poly-L-histidine segment between PEG and poly-L-lysine to engineer a triblock copolymer. CH12K18PEG5k DNA nanoparticles trafficked through clathrin-dependent endocytosis (CME) as the primary pathway in mouse glioblastoma (GBM) cells, and protected plasmid DNA from both DNase-mediated and acidic lysosomal degradation. CH12K18PEG5k DNA nanoparticles effectively silenced a tumor-specific transgene (firefly luciferase) following direct injection into mouse intracranial GBM. Toxicity and histological analysis showed no evidence of acute or delayed inflammatory responses. These results demonstrate the utility of using this DNA nanoparticle-based technology for delivering genes to tumor cells as a possible therapeutic approach for patients with brain cancer.
Keywords: Brain Cancer, DNA Nanoparticles, GBM, Gene Therapy, shRNA
Introduction
Glioblastoma (GBM) is the most common form of malignant primary brain cancer in adults [1–4]. Due to limitations associated with current therapies, as well as the presence of chemo- and radio-resistant glioma stem cell (GSC) populations that appear to play a major role in initiating disease recurrence, the median survival for patients with GBM is less than 20 months [5–9]. With the growing knowledge of the complex genetic alterations and subtypes in GBM, modulation of defective and malfunctioning genes, cellular pathways, and the tumor microenvironment is increasingly viewed as a promising approach [10, 11]. In addition, the discovery of RNA interference (RNAi) and expanding uses of inhibitory double stranded oligonucleotides (e.g. short-hairpin ribonucleic acid, shRNA) has opened new avenues for therapeutic gene modification in diseases like GBM.
The majority of current gene therapy-based clinical trials have used viruses to deliver therapeutic DNA [12–15]. While some viruses can provide safe gene transfer in humans, Phase III clinical trials have failed to show therapeutic efficacy due in part to significant host immune responses, limited therapeutic distribution, and rapid clearance [11]. Non-viral gene vectors have emerged as an alternative to viral strategies. Benefits of non-viral vectors include reduced immunogenicity, ease of manufacturing, lack of risk of vector replication and insertion, and ability to accommodate larger plasmid DNA compared to commonly tested viruses such as adeno-associated virus. These advantages may translate to potential safety advantages for non-viral compared to viral vectors [11].
The clinical development of non-viral vectors has been hindered in part by relatively low gene transfer efficiencies compared to viral vectors. This may be due to the inability to overcome various biological barriers [16, 17]. A particularly challenging barrier involves endo-lysosomal trafficking within cells, where therapeutic DNA is often degraded in the acidic and enzyme-rich late endosomes and lysosomes before reaching the nucleus [18–20]. A common strategy to overcome this barrier is to incorporate functional groups with acid-base buffering capacity between pH 5.1–7.4, which presumably mediates escape from lower pH endo-lysosomal degradation [21]. For example, due to the presence of secondary and tertiary amines, polyetheneimine (PEI)- or poly(amidoamine) (PAMAM)-based gene carriers have been shown to facilitate endosome escape through a ‘proton sponge’ mechanism [22]. However, their clinical use is limited by toxicity induced by their highly cationic surface charge [23–25].
Highly compacted DNA nanoparticles, composed of 30-mer lysine conjugated to polyethylene glycol (PEG) via a single cysteine moiety (CK30PEG), represent a promising non-viral technology that has demonstrated remarkable effectiveness in delivering genes to the brain, eyes and lungs with minimal toxicity and immunogenicity [26–29]. In a Phase I/IIa clinical trial, DNA nanoparticles administered to the nares of Cystic Fibrosis (CF) patients were well tolerated, non-toxic and showed partial cystic fibrosis transmembrane regulator (CFTR) chloride ion channel reconstitution. We recently reported the development of pH-buffering DNA nanoparticles, CH12K18PEG5k, by inserting a poly-L-histidine segment between PEG and poly-L-lysine to engineer a triblock copolymer, which showed enhanced gene transfer efficiency in the mouse lungs [30]. The triblock copolymer design of CH12K18PEG5k DNA nanoparticles ensured that a neutral surface charge is maintained by the PEG chains and that poly-L-histidine is localized in an intermediate layer, where it is available to buffer pH without interfering with DNA compaction.
In this study, we engineered similar highly compacted CH12K18PEG5k DNA nanoparticles to test in brain tumor gene delivery studies. We hypothesized that CH12K18PEG5k DNA nanoparticles would mediate efficient gene transfer to brain tumor cells in vivo based on our previous results in the lung. In addition, we sought to test the cellular uptake and trafficking mechanisms in brain tumor cells as well as to evaluate preliminary brain toxicity.
Materials and Methods
Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and used as received, unless stated otherwise. Thiol reactive 5 kDa MW PEG, Methoxy-PEG-maleimide was purchased from Rapp Polymere (Tübingen, Germany). Lab-Tek glass-bottom tissue culture plates were from ThermoFisher Scientific (Rochester, NY). Alexa Fluor 555-labeled Cholera Toxin subunit B (CTB), tetramethylrhodamine-labeled Transferrin (TR-Transferrin), Alexa Fluor 488 sulfodichlorophenol (5-SDP) ester, Hoechst 34580, LysoTracker Red, and Organelle Lights Lysosome-RFP were purchased from Invitrogen (Carlsbad, CA). Cy5 Label-IT nucleic acid labeling kits were purchased from Mirus Bio (Madison, WI). Luciferase assay reagents were obtained from Promega (Madison, WI). Protein concentration assays were performed using the BCA Protein Assay Kit from Pierce (Rockford, IL).
Preparation of condensing peptides
The peptide, CH12K18, was synthesized by Fmoc-mediated solid-phase peptide synthesis using an automated peptide synthesizer (Symphony Quartet, Protein Technologies, Tucson, AZ). CH12K18 was purified by HPLC (Shimadzu Scientific Instruments, Columbia, MD) and molecular weights were confirmed by MALDI-TOF mass spectrometry (Voyager DE-STR, Applied Biosystems, Foster City, CA). A triblock co-polymer of poly(L-histidine)-b-poly(L-lysine)-b-PEG, CH12K18PEG5k, was prepared as previously described [30]. Briefly, 5 kDa PEG was conjugated to CH12K18 via the reaction between the sulfhydryl group of the cysteine residue and maleimide group of PEG. Completion of the reaction was verified using a 4,4’-dithiodipyridine release assay to detect unreacted sulfhydryls. For fluorescence imaging studies, CH12K18PEG5k polymer was fluorescently-labeled by conjugating the amine reactive probe Alexa Fluor 488 sulfodichlorophenol (5-SDP) ester (AF488) to the epsilon amines of lysine following the manufacturer’s protocol (reaction stoichiometry: 1 AF488 per CH12K18PEG5k). Unreacted AF488 was removed by fractionating the reaction mixture on a Sephadex G25 column (GE Healthcare, Piscataway, NJ) pre-equilibrated with 50 mM ammonium acetate (AA).
Plasmid preparation
The plasmid, pRNAT-H1.3/Hygro/siFluc (GenScript, Piscataway, NJ), encoding a GFP reporter gene, siRNA construct for luciferase, and ampicillin resistance was used. This plasmid has a coral GFP marker (cGFP) under CMV promoter control, which can be used to track the transfection efficiency; the plasmid also contains the siRNA construct for firefly luciferase, which uses a CMV-enhanced H1 promoter for siRNA expression. The plasmid were propagated in Escherichia coli DH5α under appropriate antibiotic selection, and collected and purified using EndoFree Plasmid Giga kits (Qiagen Inc., Valencia, CA) per manufacturer’s protocol. Endotoxin levels were < 5 EU/mg plasmid. For flow cytometry analysis of cellular uptake, the plasmid DNA was labeled with Cy5 (Label IT® Nucleic Acid Labeling Kit, Mirus, San Francisco, CA) according to the manufacturer’s protocol.
Formulation of DNA nanoparticles
Compacted DNA nanoparticles were manufactured by the drop-wise addition of 9 volumes of plasmid DNA solution (0.222 mg/ml in water) to 1 volume of copolymer solution at a rate of 1 ml/min while gently mixing at room temperature (Vortex Mixer, Fisher Scientific, Rochester, NY). The final N:P ratio of positively charged amines (N) contributed by poly-L-histidine and poly-L-lysine to negatively charged DNA phosphates (P) was 2:1. For fluorescence imaging, Cy5-labeled DNA was used to assemble fluorescently-labeled DNA nanoparticles. After incubating at room temperature for 30 minutes, aggregates were removed by syringe filtration (0.2 µm). To remove free polymers and exchange water for physiologic saline, DNA nanoparticles were washed two times with 10 volumes of 0.9% NaCl and re-concentrated to 0.2 mg/ml using Amicon® Ultra Centrifugal Filters (100,000 MWCO, Millipore Corp., Billerica, MA). For in vivo experiments, DNA nanoparticles were concentrated to 4 mg/ml of DNA concentration and administered on the same day. The final DNA concentration was determined via absorbance at 260 nm using a NanoDrop ND-2000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). For long-term stability, DNA nanoparticles were stored at 4°C.
Physicochemical characterization
Assembled DNA nanoparticles were imaged by transmission electron microscopy (TEM, Hitachi H7600, Japan) to determine their morphology and size, as described previously [31, 32]. Hydrodynamic diameter and ζ-potential (surface charge) were determined by dynamic light scattering and laser Doppler anemometry, respectively, using a Nanosizer ZS90 (Malvern Instruments, Southborough, MA). Samples were diluted to 0.1 mg/ml in 10 mM NaCl pH 7.1 (viscosity 0.8894 cP and Refractive index 1.33 @ 25°C). Size measurements were performed at 25°C at a scattering angle of 90°. The zeta-potential values were calculated using the Smoluchowski equation.
DNase stability assay
The ability of compacted DNA nanoparticles to protect cargo DNA against DNase digestion was assayed as follows. Ten microliters of compacted DNA nanoparticles or free plasmid DNA (0.5 µg of DNA) were incubated with 1µl recombinant DNase I (0.5 U; RNase free; Roche Diagnostics GmbH, Mannheim, Germany) at 37°C for varying durations. After the treatment, DNase was inactivated by addition of 0.5 M EDTA followed by 15 min incubation at room temperature. To release DNA for gel electrophoresis, DNA nanoparticles were incubated with 1 µl of 0.25% Typsin at 37 °C for 10 min. Gel electrophoresis was carried out (25 min at 120 V) on a 0.8 % agarose gel containing 50 µg/ml ethidium bromide.
Cell culture
Mouse glioblastoma (GL261) cells that constitutively express the luciferase enzyme were obtained from Dr. Hua Yu (City of Hope, CA). GL261 cells were cultured at 37°C and 5% CO2 in DMEM (Invitrogen Corp., Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Invitrogen Corp.) and 1% penicillin/streptomycin (Invitrogen Corp.). In select experiments, GL261 cells were pre-transfected with Organelle Lights Lysosome-RFP kits according to the manufacturer’s protocol, which contains a gene sequence that encodes for Lamp1 (lysosome marker). For live-cell microscopy, cells were seeded between 2.0 to 2.5×103 cells per plate onto Lab-Tek glass-bottom culture plates and incubated overnight at 37°C. After overnight incubation, culture medium was replaced with fresh media before nanoparticles were added.
Confocal microscopy
LSM 510 Meta confocal microscope (Carl Zeiss Inc., Thornwood, NY) was used to capture images. For multi-color microscopy, samples were excited with 405, 488, 543 and 633nm laser lines, and images were captured by multi-tracking to avoid bleed-through between fluorophores. All samples were maintained at 37°C and 5% CO2 and observed under a 63X Plan-Apo/1.4 NA oil-immersion lens.
Co-localization with endocytic markers
In order to identify the endocytic pathways involved for CH12K18PEG5k DNA nanoparticles, co-localization studies with endocytic markers were performed in live GL261 cells. In select experiments, cells were treated with TR-Transferrin (10 µg/ml) for 1 hr or Alexa Fluor 555-CTB (3 µg/ml) for 30 min. In another experiment, cells were treated with LysoTracker (100 nM) for 30 min. Next, nanoparticles were added and then imaged under confocal microscope. The cells were subsequently washed with 1X PBS and incubated in Opti-MEM for confocal imaging. Image acquisition was limited to 200 ms per frame or less to maintain accurate co-localization information and then analyzed with MetaMorph software (Universal Imaging Corp., Downingtown, PA).
Cellular uptake and Drug inhibition
GL261 cells were seeded onto 6-well plates at an initial density of 3.0×105 cells per well and grown to 75 % confluence overnight. After 24 h, cells were treated with chlorpromazine (10 µg/ml), genistein (200 µM), amiloride (10 µM), or methyl-β-cyclodextrin (10 mM) with lovastatin (1 µg/ml) in media for 1 h at 37°C. Subsequently, 5 µg of Cy5-labeled DNA nanoparticles were added and incubated for another 2 h, after which cells were washed with 1X PBS and extracellular fluorescence was quenched with 0.4% (w/v) trypan blue. Cells were then washed with 1X PBS and harvested with 0.05% Trypsin/EDTA and resuspended in 1X PBS. Mean fluorescence intensity was analyzed using FACSCalibur flow cytometer (Becton Dickinson, Franklin Lake, NJ). Data from 10,000 events were gated using forward and side scatter parameters to exclude dying cells and debris.
In vitro gene transfer
The in vitro gene transfer efficiency of CH12K18PEG5k DNA nanoparticles was evaluated using the reporter plasmid, pRNAT-H1.3/Hygro/siFluc (GenScript, Piscataway, NJ). This plasmid has a coral GFP reporter (cGFP) under CMV promoter control, which can be used to track the transfection efficiency; the plasmid also contains the siRNA construct for firefly luciferase, which uses a CMV-enhanced H1 promoter for siRNA expression. GL261 cells were seeded onto 12-well plates at an initial density of 5.0×104 cells per well and incubated overnight at 37°C. After 24 h, cells were incubated with DNA nanoparticles (2 µg DNA/well) in media for 6 hrs at 37°C. Subsequently, nanoparticles and culture medium were replaced with fresh media. After additional 48 h incubation at 37°C, GFP positive cells were sorted and collected by FACS, after which the GFP positive cell populations were imaged under confocal microscope. Subsequently, 0.25 ml of 1X Reporter Lysis Buffer was added to each wells and then subjected to a freeze-and-thaw cycle. Cells were detached and collected using a cell scraper, and supernatants were obtained by centrifugation. Luciferase activity in the supernatants was analyzed using a standard luciferase assay kit (Promega, Madison, WI) and a 20/20n luminometer (Turner Biosystems, Sunnyvale, CA). Data are shown as relative light unit (RLU)/mg protein.
Animals model
All procedures performed with mice were approved by the Johns Hopkins University Animal Care and Use Committee. Female C57BL/6 mice (age, 5–6 weeks) were purchased from Harlan Laboratories. The syngeneic cell line GL261 was transfected with firefly luciferase using lentiviral methods and used for brain tumor implantations (gift from Dr. Hua Yu, City of Hope, CA). For the implantation procedure, mice were anesthetized via intraperitoneal injection of 60 µL of ketamine hydrochloride (75 mg/kg; 100 mg/mL; ketamine HCl; Abbot Laboratories), xylazine (7.5 mg/kg; 100 mg/mL; Xyla-ject; Phoenix Pharmaceutical), and ethanol (14.25%) in a sterile 0.9% NaCl solution. Using a stereotactic frame and sterile technique, 40,000 GL261 cells were injected through a burr hole drilled 2 mm lateral to the sagittal suture and 1 mm anterior to the coronal suture at a depth of 3 mm below the dura at a rate of 1 µL/min over 3 minutes. Mice were also given the analgesic buprenorphine (Buprenex) (0.05 mg/kg, subcutaneously). Animals were observed daily for any signs of deterioration or neurological dysfunction. If the symptoms persisted and resulted in debilitation, animals were euthanized according to protocol.
Stereotactic injection of DNA nanoparticle into Brain
At day 4 after implantation of GL261 tumor cells, bioluminescent signal from the engrafted brain tumors was confirmed in each animal. Once tumor signal was confirmed, the animals were anesthetized as above and CH12K18PEG5k DNA nanoparticles or free plasmid DNA were suspended in normal saline and administered sterilely into mouse brain (n = 5 per group) through the same burr hole using a stereotactic frame. DNA nanoparticles, naked DNA, or saline control were loaded into a sterile 30-gauge Hamilton syringe needle and injected slowly: 4 µl (4 mg/ml nanoparticle/DNA solution) at a rate of 1 µl/min over 4 minutes.
In vivo Bioluminescence Imaging
Intracranial GL261 mouse tumors were imaged using a Xenogen IVIS system (Caliper Life Sciences, Hopkinton, MA). Anesthesia was induced in an induction chamber with 2.5% isoflurane in 100% oxygen at a flow rate of 1 L/min and maintained in the IVIS system with a 2.0% mixture at 0.5 L/min. The mice were injected with D-luciferin (150 mg/kg, intraperitoneally; dissolved in PBS) and returned to their home cages. Ten minutes following the D-luciferin injection, anesthesia was induced with isoflurane in an induction chamber. The animal was moved to the IVIS imaging chamber and maintained on 2 to 3% isoflurane. Photons emitted from live mice were acquired as photons/s/cm2/steradian (sr) and analyzed using LivingImage software (PerkinElmer, MA). For BLI analysis, photon emissions were integrated over 10 minutes. The same circular-shaped region of interest (ROI) templates were overlaid above the brain cortex and used for all BLI measurements.
In vivo Safety profile
To determine potential toxicity of CH12K18PEG5k DNA nanoparticles, we administered by intracranial injection a dose of 4 µl per mouse (n = 5 per group) of DNA nanoparticles (4 mg/ml) suspended in normal saline, free plasmid DNA suspended in saline, or normal saline control. The weight of tumor-bearing mice, with or without treatment, was measured daily for up to 14 days. After 14 days, mice were sacrificed, perfused with paraformaldehyde, and the brains were harvested for tissue histology. To further prepare tissues for sectioning, brains were immersed in 10% formalin for 24 h, before embedding in paraffin. Sections (5 µm) were cut from each brain and stained with hemotoxylin and eosin.
Statistical analysis
Statistical analysis of data was performed by one-way analysis of variance (ANOVA) followed by Tukey HSD or Games-Howell tests using SPSS 18.0 software (SPSS Inc., Chicago, IL). Differences were considered to be statistically significant at a level of P < 0.05.
Results
Physicochemical characterization of DNA nanoparticles
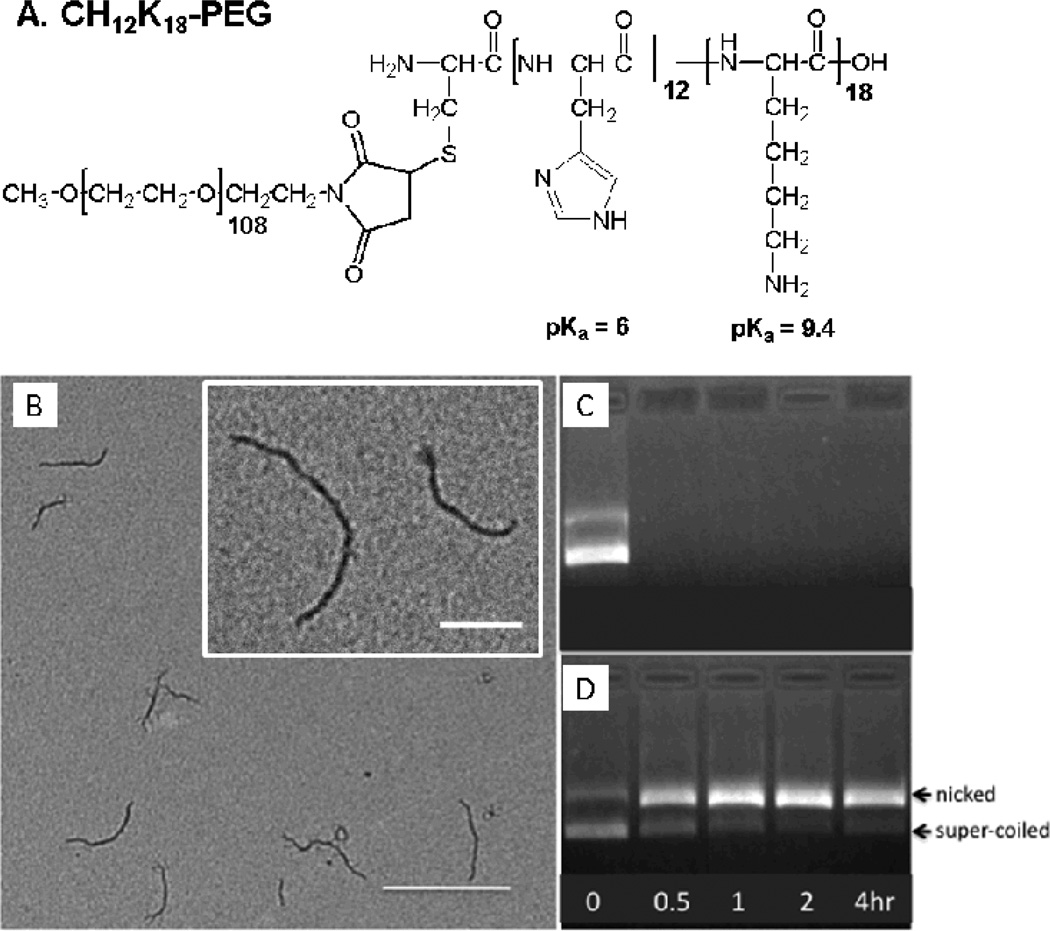
The conjugation of methoxy-PEG-maleimide to the condensing peptides, CH12K18, was confirmed by 4,4’-dithiodipyridine release assay, indicating that all free thiol groups had fully reacted to yield CH12K18PEG5k. The chemical structure of CH12K18PEG5k polymer is depicted in Figure 1A. The assembly of compacted DNA nanoparticles was confirmed by morphological examination via transmission electron microscopy (TEM), which revealed that CH12K18PEG5k DNA nanoparticles produced flexible rod-like structures (Figure 1B), in agreement with previous reports for CK30PEG DNA nanoparticles [30]. The average dimensions (length × width) for CH12K18PEG5k DNA nanoparticles were ~ 325 × 13 nm. We found that CH12K18PEG5k DNA nanoparticles had near-neutral surface charge, as indicated by ζ-potential, indicating the presence of neutrally charged PEG on the surface of nanoparticles. The particle size, polydispersity index (PDI), and ζ-potential of CH12K18PEG5k DNA nanoparticles are shown in Table 1.
Figure 1.
(A) Chemical Structure of CH12K18PEG5k polymer. (B) Transmission electron microscopy (TEM) of compacted DNA nanoparticles. Images reveal a minor diameter of <15 nm and major diameter of 300–350 nm. (Scale bar: 400nm; Inset scale bar: 200nm). Ability of DNA nanoparticles to protect plasmid DNA against DNase digestion: (C) plasmid alone and (D) compacted DNA nanoparticles after exposure to DNase I at 37 °C for 0 – 4 h.
Table 1.
Physicochemical properties of DNA nanoparticles.
| Formulation | pH a | PDI b | Diameter, nm c | ζ-potential, mV d |
|---|---|---|---|---|
| CH12K18-PEG5k | 7.5 | 0.26 ± 0.05 | 72 ± 0.2 | 1.8 ± 0.03 |
DNA nanoparticles were formulated at pH 7.5 and at a final N:P ratio of 2:1.
Polydispersity index (PDI), measured by dynamic light scattering, represents the relative variance in the particle size distribution. A PDI of 1 indicates a large distribution of particles size, whereas a PDI of 0 indicates a monodisperse size distribution. Data represents the average of 3 independent experiments +/− SD.
Diameter (number mean) measured by dynamic light scattering. Data represents the average of 3 independent experiments +/− SD.
Measured at pH 7.1. Data represents the average of 3 independent experiments +/− SD.
To assess the ability of DNA nanoparticles to protect the cargo DNA against DNase digestion, CH12K18PEG5k DNA nanoparticles and free plasmid DNA were incubated with DNase I for various durations before gel electrophoresis. Free plasmid DNA was completely degraded within 30 minutes of incubation with DNase I (Figure 1C). In contrast, plasmid DNA compacted with CH12K18PEG5k (DNA nanoparticles) was effectively protected from DNase I digestion for up to 4 h, although an increased ratio of nicked vs. supercoiled DNA was observed (Figure 1D).
Cellular Uptake and Intracellular trafficking of DNA nanoparticles
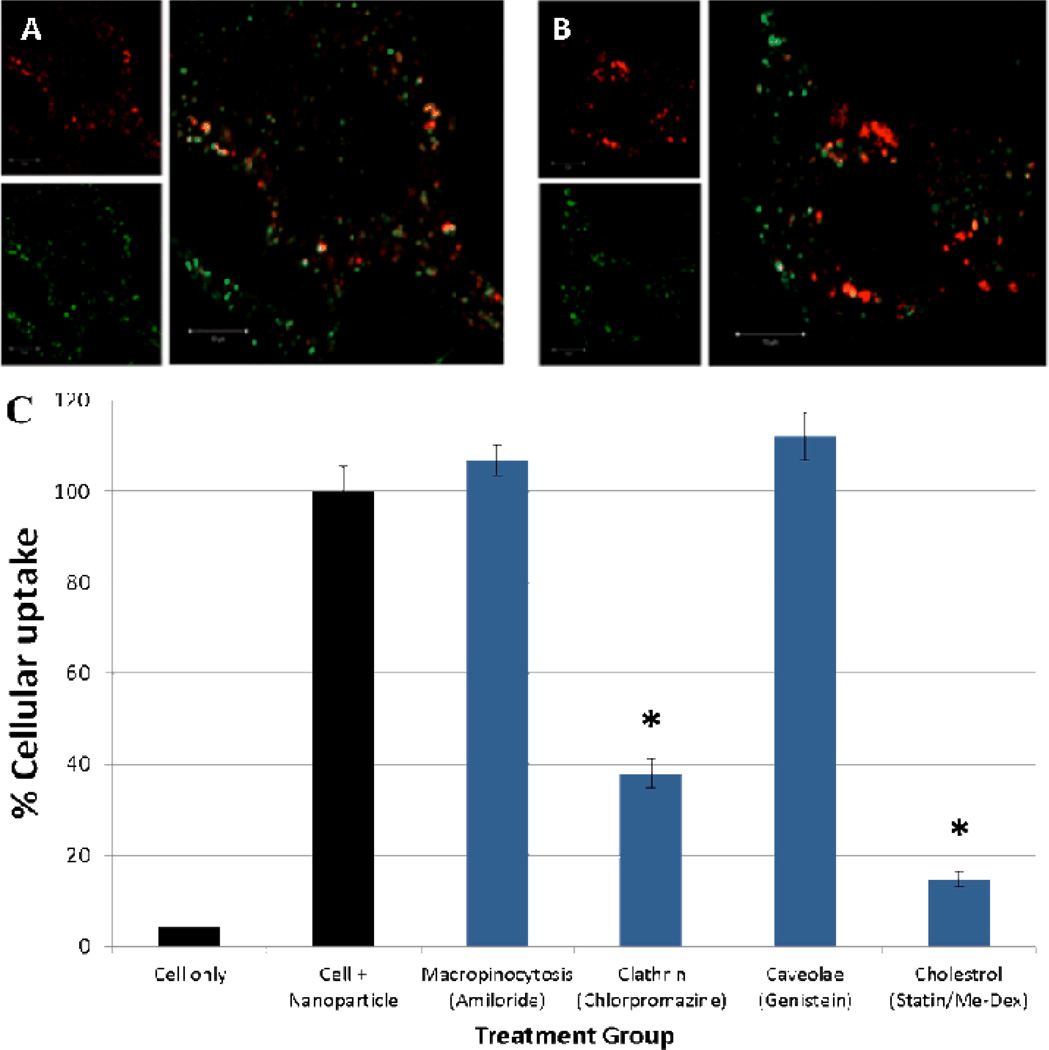
Previous studies have demonstrated that endocytic mechanisms can influence intracellular trafficking of nanoparticles [32–34]. We investigated intracellular trafficking of CH12K18PEG5k DNA nanoparticles in live GL261 murine gliobastoma cells, using confocal microscopy. We labeled CH12K18PEG5k polymer with Alexa Fluor 488, which did not affect the formation of DNA nanoparticles as confirmed by TEM and ζ-potential [35]. CH12K18PEG5k DNA nanoparticles were taken up by GL261 cells within 2 h, and they rapidly accumulated in the perinuclear region. To determine the endocytic mechanism leading to a cellular uptake, we co-incubated fluorescently-labeled CH12K18PEG5k DNA nanoparticles with various endocytic pathway markers: Transferrin (clathrin-mediated) and Cholera Toxin subunit B (CTB, caveolae-mediated), and measured their degree of co-localization using confocal microscopy. We found that a substantial fraction of CH12K18PEG5k DNA nanoparticles were co-localized with Transferrin (Figure 2A), significantly higher than with CTB (Figure 2B), suggesting they primarily enter GL261 cells via clathrin-mediated endocytosis (CME).
Figure 2.
Colocalization (yellow) of CH12K18PEG5k DNA nanoparticles (green) in GL261 mouse glioma cells after 2h with: (A) transferrin (red), a marker of clathrin-mediated endocytosis and (B) Cholera toxin B (CTB, red), a marker of caveolae-mediated uptake. CH12K18PEG5k DNA nanoparticles show minimal colocalization with CTB suggesting minimal caveolae-mediated uptake. However, substantial colocalization was observed with clathrin-mediated mechanisms (transferrin). (C) Endocytic pathway inhibition study in GL261 mouse glioma cells: Clathrin-mediated cellular uptake inhibitor, chlorpromazine, and cholesterol-dependent endocytosis inhibitors, lovastatin and methyl-β-cyclodextrin, significantly decreased nanoparticle uptake (* p <0.01).
To confirm our microscopy observations with endocytic markers, we measured the cellular uptake of CH12K18PEG5k DNA nanoparticles in GL261 cells treated with different endocytic pathway inhibitors using flow cytometry. DNA plasmid was fluorescently-labeled with Cy5, which did not affect the formation of DNA nanoparticles as confirmed by TEM and ζ-potential [30]. The uptake of CH12K18PEG5k DNA nanoparticles was significantly reduced (P < 0.01) following treatment of cells with chlorpromazine, which blocks CME by causing clathrin to accumulate in late endosomes (Figure 2C). In contrast, the uptake of CH12K18PEG5k DNA nanoparticles was not affected by genistein or amiloride, inhibitors of caveolae-mediated uptake and macropinocytosis, respectively (Figure 2C). The cellular uptake of CH12K18PEG5k DNA nanoparticles was significantly reduced (P < 0.01) by cholesterol depletion, using methyl-β-cyclodextrin (extracts cholesterol from plasma membrane) and lovastatin (inhibitor of de novo cholesterol synthesis), in good agreement with the cholesterol-dependent nature of CME (Figure 2C). Together, these findings suggest that CH12K18PEG5k DNA nanoparticles entered GL261 cells primarily via CME.
Endo-lysosomal trafficking of DNA nanoparticles
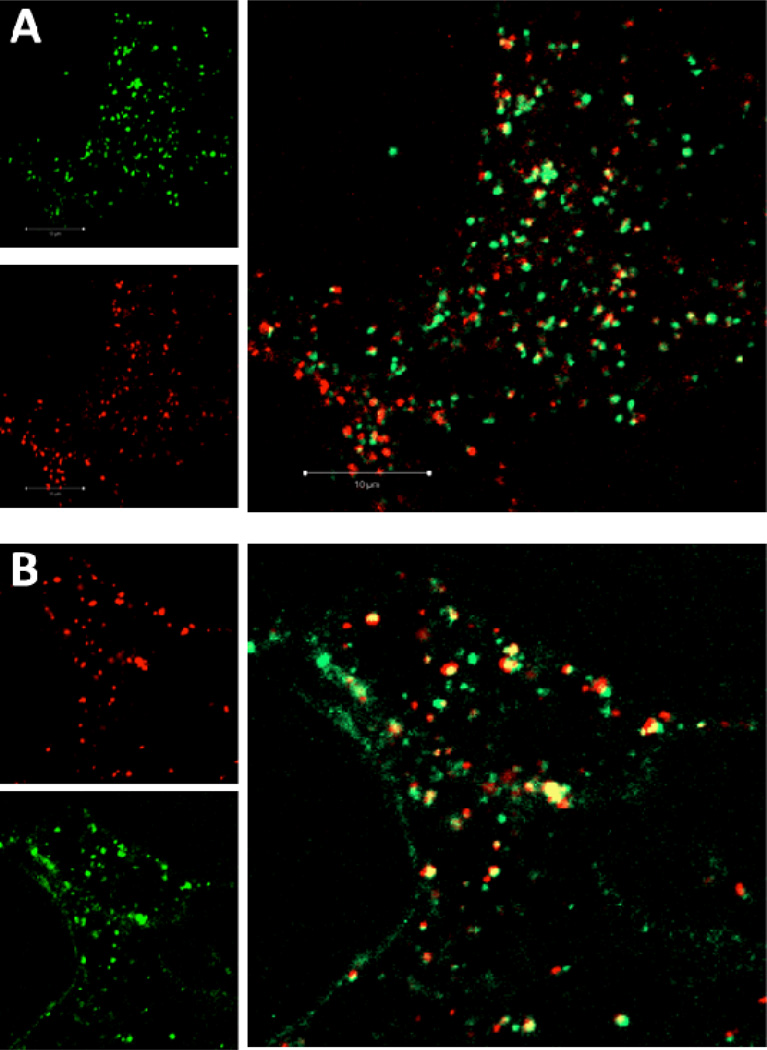
To determine whether CH12K18PEG5k DNA nanoparticles are trafficked via the endo-lysosomal pathway, we performed co-localization studies using Lysotracker-Red and Lamp-1-RFP, well-characterized markers for late endosomes and lysosomes, respectively. Interestingly, CH12K18PEG5k DNA nanoparticles showed low co-localization with LysoTracker, an acid sensitive probe that labels late endosomes (Figure 3A). We found that CH12K18PEG5k DNA nanoparticles were highly co-localized with Lamp-1-RFP, after 2 h (Figure 3B). This indicates that CH12K18PEG5k DNA nanoparticles traffic through CME into the late endosomal stage while effectively buffering the pH, thereby blocking this degradative mechanism.
Figure 3.
Endolysosomal trafficking of CH12K18PEG5k DNA nanoparticles in mouse glioma cells at 2 h. (A) DNA nanoparticles (green) showed little colocalization (yellow) with low pH endolysosomes (pH sensitive LysoTracker, red), (B) but showed substantial colocalization with the late endolysomal marker (Lamp1, red).
In vitro gene transfer in mouse glioblastoma cells
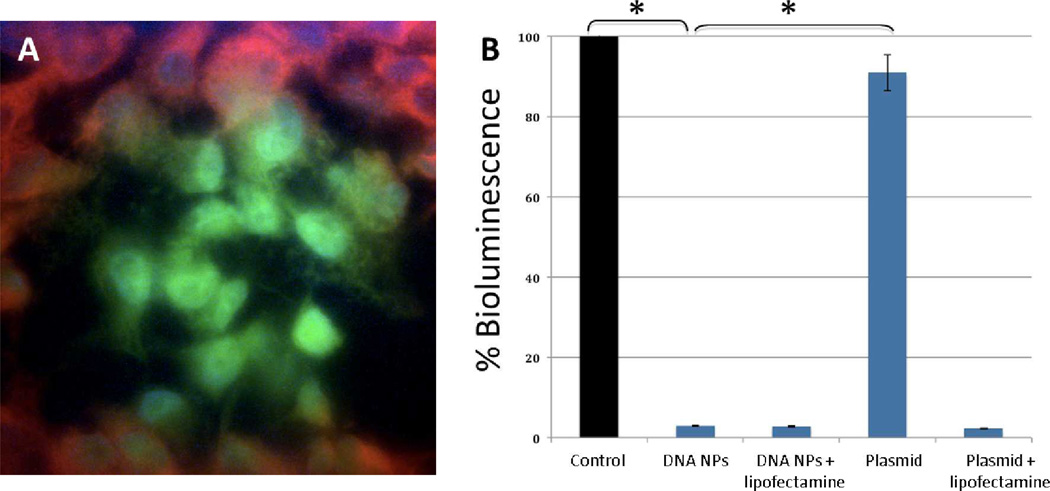
The in vitro gene transfer efficiency of CH12K18PEG5k DNA nanoparticles was evaluated in GL261 cells using the reporter plasmid, pRNAT-H1.3/Hygro/siFluc. This plasmid contains the coral GFP reporter (cGFP) and the siRNA construct for firefly luciferase. The gene transfer in GL261 cells was confirmed by imaging GFP positive cells using fluorescent confocal microscopy (Figure 4A). The FACS sorted GFP positive cell populations were then assayed for knockdown of luciferase (Figure 4B). The gene knockdown efficiency of CH12K18PEG5k DNA nanoparticles was comparable to that obtained using a commercially available transfection agent, lipofectamine 2000 (Figure 4B). In comparison, free plasmid DNA did not show appreciable gene knockdown of luciferase activity, similar to cell control (P < 0.01).
Figure 4.
(A) In vitro gene transfer of CH12K18PEG5k DNA nanoparticles in mouse glioma cells constiuitively expressing the luciferase enzyme. pRNAT plasmid contains GFP and shLuc sequences, Red = Luciferase, Green = GFP, Blue = Dapi (Nuclei). (B) Quantification of in vitro gene knockdown of CH12K18PEG5k DNA nanoparticles (DNA NPs): GFP-positive (transfected) cells were sorted out using FACS and 105 cells from each group were then analyzed using quantitative bioluminescence assay (* p < 0.01).
In vivo gene transfer and Bioluminescence Imaging
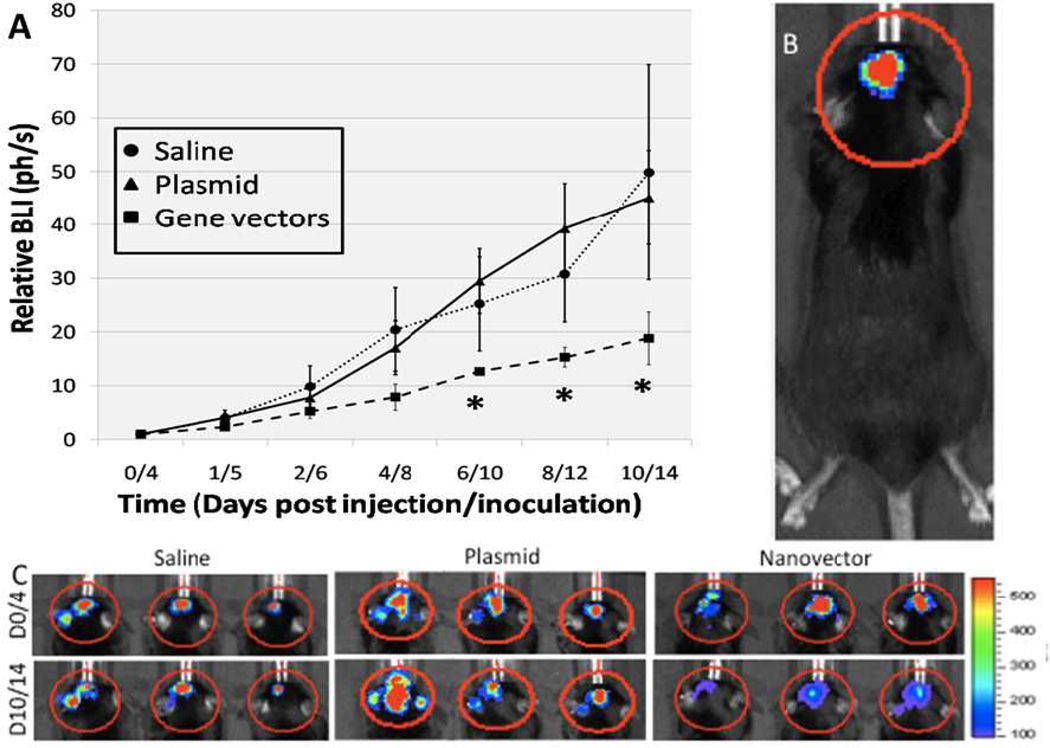
We tested the ability of CH12K18PEG5k DNA nanoparticles to knockdown luciferase in an intracranial GL261 mouse glioma model. Luciferase-expressing intracranial GL261 tumors were imaged via bioluminescence with a Xenogen IVIS live animal imaging system. Luciferase-expressing tumors were evident in the brain 4 days after tumor implantation (Figure 5B). The luciferase activity of intracranial GL261 tumors was followed via BLI for up to 14 days. At 4 days after tumor implantation, 4 µg of CH12K18PEG5k DNA nanoparticles carrying pRNAT-H1.3/Hygro/siFluc plasmid or free plasmid DNA were administered into the location of the original tumor cells (n = 5 per group) using a stereotactic frame. Control mice were treated with normal saline. At 2 days post-treatment, there was no significant difference in the luciferase activity of intracranial GL261 tumors between CH12K18PEG5k DNA nanoparticles, naked DNA, or saline control treated mice groups. However, by 6 days post-treatment, there was a statistically significant decrease in the cumulative BLI signals for the group that received CH12K18PEG5k DNA nanoparticles, compared to the groups that received free plasmid DNA alone or normal saline control (Figure 5A). Luciferase knockdown persisted for up to 14 days after inoculation of tumor cells, as evident by the reduced intensity in BLI shown in Figure 5C.
Figure 5.
(A) Cumulative bioluminescenc imaging (BLI) of luminescent intracranial tumors following normal saline, plasmid alone, and compacted CH12K18PEG5k DNA nanoparticles injection (n=5 per group). By day 6/10 (6 days post nanoparticle injection/10 days post tumor innoculation) there was a stastically signifcant decrease in the BLI signal in the DNA nanoparticles treatment group. (B) Representative BLI signal from an intracranial GL261 glioma. (C) BLI of animals in the saline, plasmid, and CH12K18PEG5k DNA nanoparticles groups on days 0/4 and 10/14. Note reduced BLI in tumor luminescence signal over this time course in the DNA nanoparticles treatment group (* p < 0.05).
In vivo safety profile
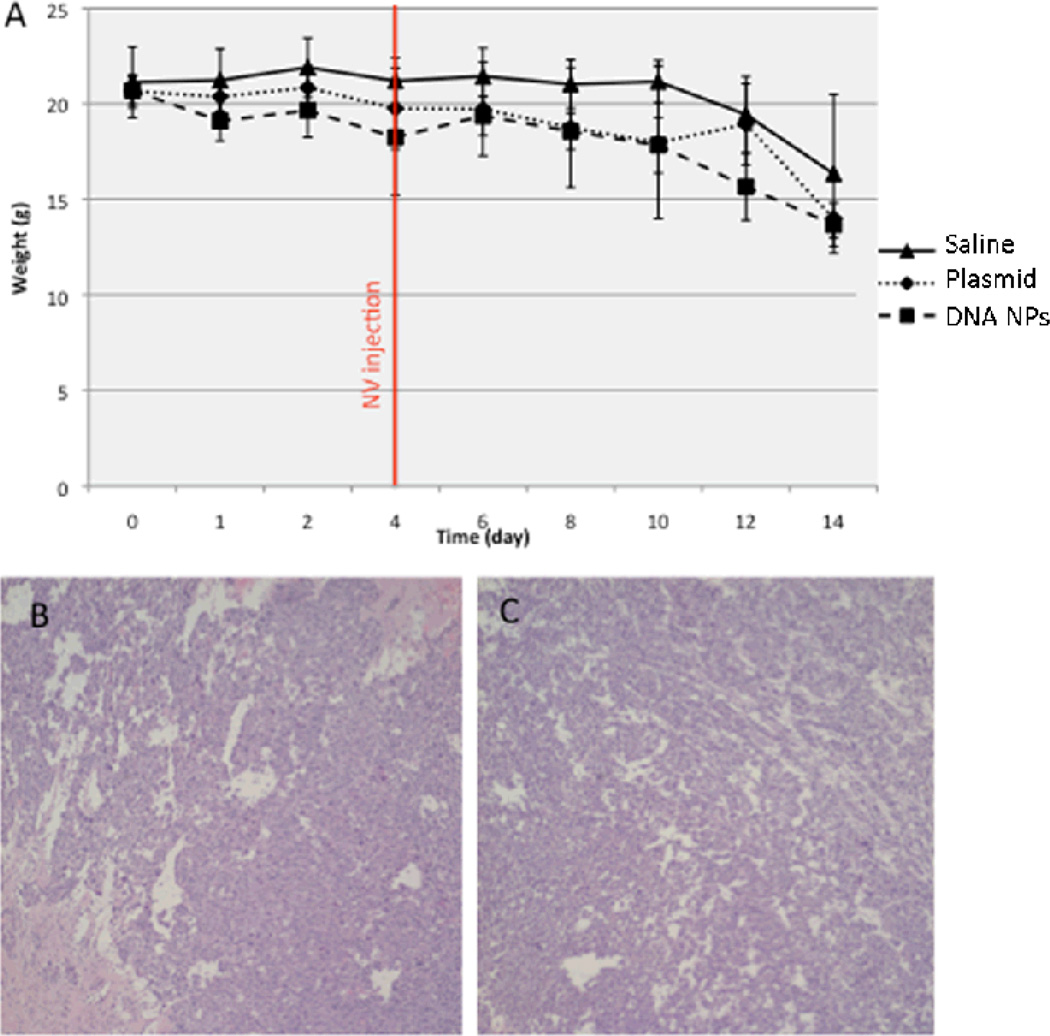
Despite their wide use as gene carrier platform, several groups have reported on the toxicity of cationic polymers, such as PEI, PAMAM, and PLL [23–25]. We thus investigated whether pH-buffering PEG-coated CH12K18PEG5k DNA nanoparticles may cause accelerated weight loss or inflammation in the mouse brain. We administered 4 µg per mouse (n = 5 per group) of CH12K18PEG5k DNA nanoparticles, free plasmid DNA, or normal saline control into mice bearing GL261 tumors. We did not observe a significant difference in the weight of tumor-bearing mice following treatment, suggesting minimal systemic or neurotoxicity from DNA nanoparticle administration (Figure 6A). At 14 days post-treatment, mice were sacrificed and brains were harvested for tissue histology. We found that the brains of the mice treated with CH12K18PEG5k DNA nanoparticles did not show significant tissue damage or increase in immune cell infiltration (Figure 6B), compared to saline treated controls (Figure 6C). These results suggest that CH12K18PEG5k DNA nanoparticles did not trigger acute inflammation in brain tumors.
Figure 6.
(A) Weight curves of tumor-bearing mice (inoculated day 0), following normal saline (control), plasmid DNA alone, or CH12K18PEG5k DNA nanoparticles administration (DNA NPs) given at day 4. No significant difference was observed between the groups (n=5 per group), suggesting minimal systemic or neuro-toxicity from the DNA nanoparticles or plasmid DNA administration. Representative intracranial tumor histology (post-injection day 10) in the region of the direct injections of either (B) normal saline or (C) CH12K18PEG5k DNA nanoparticles. Note the lack of tissue damage or inflammatory infiltrate in both groups.
Discussion
We report efficient gene transfer and sustained shRNA-mediated, tumor –specific transgene silencing using highly compacted pH-responsive CH12K18PEG5k DNA nanoparticles in an intracranial mouse glioblastoma model. Negligible knockdown of luciferase expression was observed in mice that received free plasmid DNA encoding for Luciferase shRNA, suggesting that compaction was necessary for efficient uptake and gene expression in glioma cells. Recently, significant resources have been devoted to understanding the pathobiology of GBM and determine the specific cellular pathways that drive the malignant behavior of these tumors [10, 11]. DNA nanoparticles are an attractive alternative to viral vectors for this purpose because of their relatively nontoxic and nonimmunogenic properties; immunogenicity reduces gene transfer efficacy upon repeated dosing, which is generally observed with viral vectors due to the generation of neutralizing immune responses [11].
Similar to our previous finding for human bronchial epithelial (BEAS-2B) cells [30], we found that CH12K18PEG5k nanoparticles entered mouse glioma (GL261) cells by CME. The trafficking of therapeutic DNA and gene vectors in the acidic endolysosomal pathway has long been considered a critical bottleneck in non-viral gene therapy. We incorporated a pH-buffering polymer, CH12K18PEG5k, to overcome the degradative endo-lysosomal intracellular barrier. Inclusion of poly-L-histidine into CH12K18PEG5k DNA nanoparticles increased the buffering capacity, between pH 5.1–7.4, which may facilitate endosomal rupture and escape from lysosomal degradation [30]. Our results are consistent with this hypothesis, since we observed minimal co-localization between CH12K18PEG5k DNA nanoparticles and late endosomes/lysosomes labeled with the LysoTracker, suggesting that DNA nanoparticles successfully buffered pH inside the endosomes.
Furthermore, our preliminary safety studies in tumor-bearing mice were encouraging. The brains of tumor-bearing mice treated with CH12K18PEG5k DNA nanoparticles were histologically indistinguishable from those treated with normal saline at 14 days post-exposure. These results are also consistent with our previous finding, where mice treated with CH12K18PEG5k DNA nanoparticles by inhalation displayed only mild and transient inflammatory response in the lungs [30]. Conjugation of PEG to CH12K18 polymer may have mitigated/reduced inflammation induced by highly positive surface property intrinsic to the cationic polymer-based gene carriers [27, 30, 35, 36]. Advantages of PEG coating on nanoparticles for drug and gene delivery have been widely reported in the literature [37, 38]. PEG coatings can offer resistance to degradation by nucleases, provide increased colloidal stability, provide bio-inert surfaces that minimize interactions with extracellular components, and confer stealth-like properties to evade the reticuloendothelial system for long circulation in the bloodstream [35, 39–41].
The therapeutic potential of PEG-poly-lysine-based DNA nanoparticles was previously highlighted by the promising results from a Phase I/IIa study in CF patients. The level of gene transfer to human nasal mucosa mediated by DNA nanoparticles, as measured by PCR analysis of vector DNA, was comparable to the highest levels observed in an AAV intrasinus trial [36]. In addition, our DNA nanoparticles are stable for > 3 years at 4 °C, non-immunogenic (repeat dosing in the lungs of mice has been demonstrated), non-inflammatory, and have been shown to be effective in the airway, brain, and eye [27, 28, 42].
An increasing number of clinical trials with a focus on brain cancer gene delivery are currently ongoing [11, 12, 15]. This trend confirms the emerging importance and feasibility of genetic modulation as a viable therapeutic strategy for brain cancer. Our results indicate that pH-responsive CH12K18PEG5k DNA nanoparticles represent a promising platform for brain tumor gene therapy. These results support further investigation into the use of this promising polymer-based CH12K18PEG5k DNA nanoparticles with convection-enhanced delivery (CED) and other novel delivery approaches in brain tumor patients [8, 43, 44].
Conclusion
In summary, we report successful brain tumor specific transgene knockdown using pH-responsive CH12K18PEG5k DNA nanoparticles as a vehicle for gene delivery. This synthetic DNA nanoparticle platform is capable of protecting plasmid DNA against DNase digestion, pH buffering to minimize lysosomal degradation, and enhanced gene transfer both in vitro and in vivo, while eliciting negligible toxic and inflammatory responses. Our results demonstrate the utility of using this DNA nanoparticle-based technology for delivering genes to tumor cells as a possible therapeutic approach for patients with brain cancer.
Acknowledgements
This work was supported by the National Institutes of Health grants R01CA164789, 5T32CA130840, K12NS080223, and F32HL103137. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Pollack IF. Brain tumors in children. N Engl J Med. 1994;331(22):1500–1507. doi: 10.1056/NEJM199412013312207. [DOI] [PubMed] [Google Scholar]
- 3.Sanders RP, et al. High-grade astrocytoma in very young children. Pediatr Blood Cancer. 2007;49(7):888–893. doi: 10.1002/pbc.21272. [DOI] [PubMed] [Google Scholar]
- 4.Wilson CB. Current concepts in cancer: brain tumors. N Engl J Med. 1979;300(26):1469–1471. doi: 10.1056/NEJM197906283002605. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Fung LK, et al. Chemotherapeutic drugs released from polymers: distribution of 1,3-bis(2-chloroethyl)-1-nitrosourea in the rat brain. Pharm Res. 1996;13(5):671–682. doi: 10.1023/a:1016083113123. [DOI] [PubMed] [Google Scholar]
- 7.Degen JW, et al. Safety and efficacy of convection-enhanced delivery of gemcitabine or carboplatin in a malignant glioma model in rats. J Neurosurg. 2003;99(5):893–898. doi: 10.3171/jns.2003.99.5.0893. [DOI] [PubMed] [Google Scholar]
- 8.Bobo RH, et al. Convection-enhanced delivery of macromolecules in the brain. Proc Natl Acad Sci U S A. 1994;91(6):2076–2080. doi: 10.1073/pnas.91.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeAngelis LM. Brain tumors. N Engl J Med. 2001;344(2):114–123. doi: 10.1056/NEJM200101113440207. [DOI] [PubMed] [Google Scholar]
- 10.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tobias A, et al. The art of gene therapy for glioma: a review of the challenging road to the bedside. J Neurol Neurosurg Psychiatry. 2013;84(2):213–222. doi: 10.1136/jnnp-2012-302946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiocca EA, et al. A phase I trial of Ad.hIFN-beta gene therapy for glioma. Mol Ther. 2008;16(3):618–626. doi: 10.1038/sj.mt.6300396. [DOI] [PubMed] [Google Scholar]
- 13.Lang FF, et al. Phase I trial of adenovirus-mediated p53 gene therapy for recurrent glioma: biological and clinical results. J Clin Oncol. 2003;21(13):2508–2518. doi: 10.1200/JCO.2003.21.13.2508. [DOI] [PubMed] [Google Scholar]
- 14.Lesniak MS. Gene therapy for malignant glioma. Expert Rev Neurother. 2006;6(4):479–488. doi: 10.1586/14737175.6.4.479. [DOI] [PubMed] [Google Scholar]
- 15.Wakabayashi T, et al. A phase I clinical trial of interferon-beta gene therapy for high-grade glioma: novel findings from gene expression profiling and autopsy. J Gene Med. 2008;10(4):329–339. doi: 10.1002/jgm.1160. [DOI] [PubMed] [Google Scholar]
- 16.Suh J, Wirtz D, Hanes J. >Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci U S A. 2003;100(7):3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaffer DV, et al. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen DN, et al. Polymeric Materials for Gene Delivery and DNA Vaccination. Advanced Materials. 2009;21(8):847–867. doi: 10.1002/adma.200801478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pack DW, et al. Design and development of polymers for gene delivery. Nature Reviews Drug Discovery. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 20.Wong SY, Pelet JM, Putnam D. Polymer systems for gene delivery--Past, present, and future. Progress in Polymer Science. 2007;32(8–9):799–837. [Google Scholar]
- 21.Behr J-P. The Proton Sponge: a Trick to Enter Cells the Viruses Did Not Exploit. CHIMIA International Journal for Chemistry. 1997;51:34–36. [Google Scholar]
- 22.Sonawane ND, Szoka FC, Verkman AS. Chloride Accumulation and Swelling in Endosomes Enhances DNA Transfer by Polyamine-DNA Polyplexes. Journal of Biological Chemistry. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 23.Hunter AC, Moghimi SM. Cationic carriers of genetic material and cell death: a mitochondrial tale. Biochim Biophys Acta. 2010;1797(6–7):1203–1209. doi: 10.1016/j.bbabio.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 24.Malek A, Czubayko F, Aigner A. PEG grafting of polyethylenimine (PEI) exerts different effects on DNA transfection and siRNA-induced gene targeting efficacy. J Drug Target. 2008;16(2):124–139. doi: 10.1080/10611860701849058. [DOI] [PubMed] [Google Scholar]
- 25.Petersen H, et al. Polyethylenimine-graft-poly(ethylene glycol) copolymers: influence of copolymer block structure on DNA complexation and biological activities as gene delivery system. Bioconjug Chem. 2002;13(4):845–854. doi: 10.1021/bc025529v. [DOI] [PubMed] [Google Scholar]
- 26.Liu G, et al. Nanoparticles of compacted DNA transfect postmitotic cells. Journal of Biological Chemistry. 2003;278(35):32578–32586. doi: 10.1074/jbc.M305776200. [DOI] [PubMed] [Google Scholar]
- 27.Ziady AG, et al. Minimal toxicity of stabilized compacted DNA nanoparticles in the murine lung. Mol Ther. 2003;8(6):948–956. doi: 10.1016/j.ymthe.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Yurek DM, et al. Long-term transgene expression in the central nervous system using DNA nanoparticles. Mol Ther. 2009;17(4):641–650. doi: 10.1038/mt.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farjo R, et al. Efficient non-viral ocular gene transfer with compacted DNA nanoparticles. PLoS ONE. 2006;1:e38. doi: 10.1371/journal.pone.0000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boylan NJ, et al. Enhancement of airway gene transfer by DNA nanoparticles using a pH-responsive block copolymer of polyethylene glycol and poly-L-lysine. Biomaterials. 2012;33(7):2361–2371. doi: 10.1016/j.biomaterials.2011.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boylan NJ, et al. Highly compacted DNA nanoparticles with low MW PEG coatings: In vitro, ex vivo and in vivo evaluation. Journal of Controlled Release. 2011 doi: 10.1016/j.jconrel.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim AJ, et al. Non-degradative intracellular trafficking of highly compacted polymeric DNA nanoparticles. J Control Release. 2012;158(1):102–107. doi: 10.1016/j.jconrel.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim A, Hanes J. The emergence of multiple particle tracking in intracellular trafficking of nanomedicines. Biophysical Reviews. 2012;4(2):83–92. doi: 10.1007/s12551-012-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai SK, et al. Privileged delivery of polymer nanoparticles to the perinuclear region of live cells via a non-clathrin, non-degradative pathway. Biomaterials. 2007;28(18):2876–2884. doi: 10.1016/j.biomaterials.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 35.Kim AJ, et al. Use of single-site-functionalized PEG dendrons to prepare gene vectors that penetrate human mucus barriers. Angew Chem Int Ed Engl. 2013;52(14):3985–3988. doi: 10.1002/anie.201208556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konstan MW, et al. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum Gene Ther. 2004;15(12):1255–1269. doi: 10.1089/hum.2004.15.1255. [DOI] [PubMed] [Google Scholar]
- 37.DeRouchey J, et al. Decorated rods: a "bottom-up" self-assembly of monomolecular DNA complexes. J Phys Chem B. 2006;110(10):4548–4554. doi: 10.1021/jp053760a. [DOI] [PubMed] [Google Scholar]
- 38.Li SD, Huang L. Nanoparticles evading the reticuloendothelial system: role of the supported bilayer. Biochim Biophys Acta. 2009;1788(10):2259–2266. doi: 10.1016/j.bbamem.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogris M, et al. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6(4):595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 40.Nance EA, et al. A dense poly(ethylene glycol) coating improves penetration of large polymeric nanoparticles within brain tissue. Sci Transl Med. 2012;4(149) doi: 10.1126/scitranslmed.3003594. 149ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ensign LM, et al. Mucus penetrating nanoparticles: biophysical tool and method of drug and gene delivery. Adv Mater. 2012;24(28):3887–3894. doi: 10.1002/adma.201201800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yurek DM, et al. DNA nanoparticles: detection of long-term transgene activity in brain using bioluminescence imaging. Mol Imaging. 2011;10(5):327–339. doi: 10.2310/7290.2010.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruce JN, et al. Regression of recurrent malignant gliomas with convection-enhanced delivery of topotecan. Neurosurgery. 2011;69(6):1272–1279. doi: 10.1227/NEU.0b013e3182233e24. discussion 1279-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunwar S. Convection enhanced delivery of IL13-PE38QQR for treatment of recurrent malignant glioma: presentation of interim findings from ongoing phase 1 studies. Acta Neurochir Suppl. 2003;88:105–111. doi: 10.1007/978-3-7091-6090-9_16. [DOI] [PubMed] [Google Scholar]