Abstract
Arthropods typically show two types of segmentation: the embryonic parasegments and the adult segments that lie out of register with each other. Such a dual nature of body segmentation has not been described from Onychophora, one of the closest arthropod relatives. Hence, it is unclear whether onychophorans have segments, parasegments, or both, and which of these features was present in the last common ancestor of Onychophora and Arthropoda. To address this issue, we analysed the expression patterns of the “segment polarity genes” engrailed, cubitus interruptus, wingless and hedgehog in embryos of the onychophoran Euperipatoides rowelli. Our data revealed that these genes are expressed in repeated sets with a specific anterior-to-posterior order along the body in embryos of E. rowelli. In contrast to arthropods, the expression occurs after the segmental boundaries have formed. Moreover, the initial segmental furrow retains its position within the engrailed domain throughout development, whereas no new furrow is formed posterior to this domain. This suggests that no re-segmentation of the embryo occurs in E. rowelli. Irrespective of whether or not there is a morphological or genetic manifestation of parasegments in Onychophora, our data clearly show that parasegments, even if present, cannot be regarded as the initial metameric units of the onychophoran embryo, because the expression of key genes that define the parasegmental boundaries in arthropods occurs after the segmental boundaries have formed. This is in contrast to arthropods, in which parasegments rather than segments are the initial metameric units of the embryo. Our data further revealed that the expression patterns of “segment polarity genes” correspond to organogenesis rather than segment formation. This is in line with the concept of segmentation as a result of concerted evolution of individual periodic structures rather than with the interpretation of ‘segments’ as holistic units.
Introduction
Arthropods, including spiders, centipedes, crustaceans, insects and allies, are the most diverse and abundant animals on Earth [1], [2]. The evolutionary success of these animals might be attributed to a modular, segmented body design [3], [4]. During embryonic development, segment formation is governed by the hierarchical expression of the so-called “segmentation genes”, which in addition to playing other, pleiotropic roles in the embryo provide positional information for segmental patterning [3], [5]. However, segmentation in arthropods is more than just a simple repetition of metameric units along the body, as arthropods in fact show two types of segmentation: the embryonic parasegments, and the adult segments. The parasegments are the initial metameric units of the embryo, but due to a re-segmentation are replaced by the definitive segments that lie out of register with parasegments [6], [7].
The definitive segments comprise typical metameric units of adult arthropods, which are seen at least in those body parts that have retained the ancestral, homonomous architecture, including the trunk of centipedes and woodlice, the metasoma of scorpions, and the abdomen of insects. In these body regions, the segments are demarcated by the anterior and posterior borders of sclerites and contain additional segmental structures, such as tracheal openings, gills, ostia of the heart, and limbs with associated muscles [3], . In other body parts, the segments have fused to distinct tagmata and, thus, are no longer recognisable as individual units, for instance in the head and thorax of insects or the prosoma and opisthosoma of spiders [3], [4], [11], [12].
In contrast to the adult segments, the embryonic parasegments occur early in development and are not retained in adults. They are regarded as true metameric compartments [13], [14] and their boundaries are generated by an interaction of the canonical Hedgehog and Wnt/Wingless signalling pathways and the transcription factor Engrailed, which are expressed in cell rows flanking the parasegmental boundary [3], [7], . The expression of wingless occurs anterior to this boundary, while hedgehog and engrailed are expressed posterior to it.
At least in some chelicerate [16], crustacean [19] and insect [14], [20], [21] embryos, the parasegmental boundary is evidenced by a transverse groove between the wingless and engrailed domains. This groove, however, disappears during the re-segmentation of the embryo, after which a new (segmental) boundary arises posterior to the hedgehog and engrailed domains. This boundary corresponds to the border between adjacent sclerites in adult arthropods [5], [9]. Due to the re-segmentation of the embryo, segments and parasegments show an entirely different spatial relationship to the expression patterns of the four commonly studied “segment polarity genes”; while engrailed and hedgehog are expressed anteriorly and wingless and cubitus interruptus posteriorly in each parasegment, the opposite occurs in the definitive segments [16], .
Based on the similarities in the expression patterns of these genes, the embryonic parasegments of arthropods have been homologised with the segments of annelids [24], [25]. For example, despite apparent deviations in some species [26]–[28] the anterior-to-posterior sequence of expression of homologs of these genes is basically the same in annelids and arthropods [24]. Therefore, it has been assumed that the last common ancestor of protostomes was segmented and that the metameric exoskeleton of arthropods has evolved out of phase with this ancestral segmentation; however, the ancestral segmentation pattern is still retained in the arthropod embryo [24]. According to this scenario, one would expect that parasegments (or their vestiges) also occur as initial metameric units in embryos of one of the closest arthropod relatives, the Onychophora (velvet worms) [24], [29].
The onychophoran body exhibits a mixture of segmental and non-segmental features [30],[31]. While various structures, including limbs, crural papillae, ventral and preventral organs, cellular strands associated with midgut, nephridia, and embryonic somites ( = coelomic cavities), clearly show a metameric arrangement, no segmental organisation is evident in the cuticle or longitudinal musculature (Figure 1A–L; [30], ). Therefore, in contrast to arthropods, there are no clear segmental boundaries in adult onychophorans [8], . Consequently, the only segmental structures that might be homologous in Onychophora and Arthropoda are the limbs, the motor neurons supplying these limbs, and the nephridia and their derivatives [36]–[38].
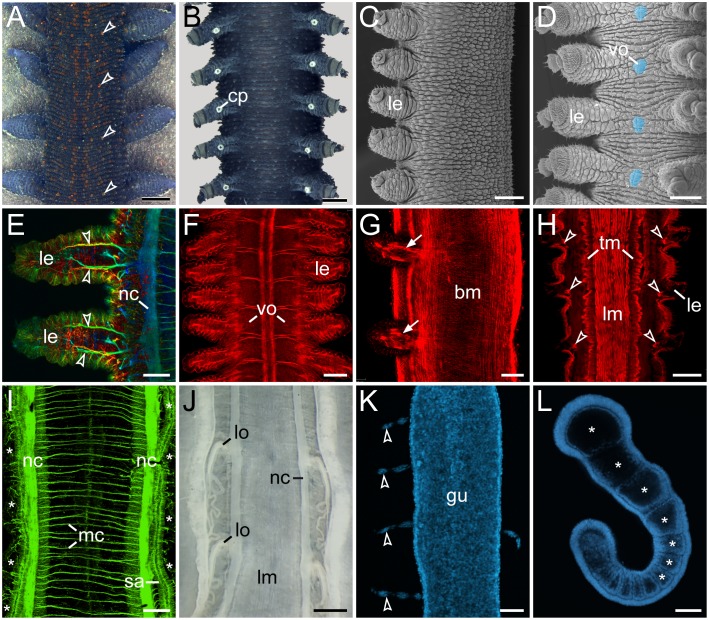
Figure 1. Segmental and non-segmental features in Onychophora.
(A, B, J) Light micrographs. (C, D) Scanning electron micrographs. (E–I, K, L) Confocal micrographs. (A) Trunk of a specimen of Euperipatoides rowelli in dorsal view. Note the segmentally repeated, terracotta-coloured papillae. (B) Trunk of a male E. rowelli in ventral view illustrating segmentally arranged crural papillae. (C) Trunk of a juvenile of Metaperipatus inae in lateral view. Note that segmentation is not evident in the integument. (D) Trunk of a juvenile of M. inae in ventral view. Segmentally arranged ventral/preventral organs between each leg pair are highlighted artificially in light-blue. (E) Legs of an embryo of Principapillatus hitoyensis in dorsal view. Anti-acetylated alpha-tubulin immunolabelling. Note the segmental arrangement of the anterior and posterior leg nerves (arrowheads). (F) Segmentally repeated anlagen of the ventral and preventral organs in an embryo of Epiperipatus biolleyi in ventral view. Phalloidin-rhodamine labelling of f-actin. (G) Segmental limb muscles (arrows) in an embryo of E. rowelli in lateral view. Phalloidin-rhodamine labelling. (H) Segmental arrangement of intrinsic leg muscles (arrowheads) in E. rowelli. Horizontal Vibratome section, phalloidin-rhodamine labelling. Note that the longitudinal and transverse musculature does not show any segmentation. (I) Ventral body wall of an embryo of P. hitoyensis in dorsal view. Anti-acetylated alpha-tubulin immunolabelling. Asterisks indicate the position of legs. Note that the median commissures are not arranged in a segmental fashion. (J) Enlarged modified nephridia ( = labyrinth organs) in the fourth and fifth leg-bearing segments. Note their segmental arrangement. Note also the lack of segmental ganglia associated with nerve cords. (K) Dissected midgut of a fully developed embryo of P. hitoyensis with segmentally arranged cellular strands (cf. [47]). DNA staining with Hoechst. (L) Embryo of P. hitoyensis in lateral view showing the segmental somites (coelomic cavities marked by asterisks). Subset of a confocal z-series showing an optical sagittal section. Abbreviations: bm, musculature of the body wall; cp, crural papilla; gu, midgut; le, leg; lm, longitudinal musculature; lo, labyrinth organ; mc, median commissures; nc, nerve cord; sa, salivary gland; tm, transverse musculature; vo, ventral/preventral organs. Scale bars: 1 mm (A), 500 µm (B), 300 µm (C–D), 50 µm (E), 100 µm (F, G, I, K, L), 200 µm (H), 1 mm (J).
Irrespective of whether or not they are homologous, various segmental structures might use a similar genetic scaffold in onychophorans and arthropods for metameric positioning along the body. Gene expression studies on embryos of the onychophoran Euperipatoides kanangrensis [29], [39] indeed revealed that the genes engrailed, cubitus interruptus, hedgehog and wingless are expressed in the same anterior-to-posterior order as in arthropods. However, based on the data available, it is impossible to determine whether or not onychophorans have true parasegmental boundaries. On the one hand, the expression of engrailed and wingless is graded and there is no precise cellular boundary between their domains at least within the trunk [29], which speaks against the existence of a parasegmental boundary in Onychophora. On the other hand, the engrailed domain extends beyond the segmental furrow [29], which would thus correspond neither to the segmental nor to the parasegmental boundary. Thus, the existence of the parasegmental boundary in the onychophoran embryo remains ambiguous.
To determine whether or not the transverse furrows retain their position within the engrailed domain throughout development, or whether a new segmental furrow arises posterior to the engrailed domain, we analysed the spatiotemporal relationship of transverse furrows and other segmental structures with respect to the engrailed domains in embryos of the onychophoran Euperipatoides rowelli. Our study covers more developmental stages than analysed before and provides a more complete picture of the anatomical changes throughout development. To further clarify whether segments or parasegments are the initial metameric units, we analysed the expression patterns of three additional genes, including cubitus interruptus, wingless and hedgehog, that are known to be involved in the developmental control of segment polarity in embryos of Drosophila melanogaster [7], [40], [41]. Furthermore, we examined in detail the spatial relationship of the expression patterns of these four genes to the individual metameric structures, such as limb buds and the anlagen of the ventral and preventral organs [30], [42], to determine whether the segmented body organisation of Onychophora is compatible with the interpretation of segments as holistic units [43]–[45] or rather with the concept of segmentation as a result of concerted evolution of individual periodic structures [8], [10], [46].
Materials and Methods
Specimen collection
Specimens of Euperipatoides rowelli Reid, 1996 (Onychophora, Peripatopsidae) were collected from rotted logs in the Tallaganda State Forest (New South Wales, Australia) in October 2010 and 2011 and maintained in the laboratory as described previously [47]. The necessary permits for the collection of onychophorans were obtained from the Forestry Commission of New South Wales, Australia (Special Purposes Permit for Research no. XX51212). We have chosen E. rowelli because this species is highly abundant and can be collected easily outside national parks [48]–[50]. Moreover, E. rowelli has become the most studied onychophoran species to date [51], the biology, anatomy, development, phylogeny and population genetics of which have been analysed extensively [30], [31], [37], [38], [42], [47]–[50], . Currently, the genome of E. rowelli is being sequenced (http://www.hgsc.bcm.tmc.edu/content/i5k-velvet-worm), which will provide additional resources for working with this “model” onychophoran species.
Dissection and fixation of embryos
For cytochemical and gene expression studies, females of E. rowelli were anaesthetised in chloroform vapour and the reproductive tracts dissected and transferred into dishes containing a physiological saline [71]. After dissecting the embryos from the uteri, the embryonic membranes were removed manually using two forceps. The embryos were then fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS; 0.1 mol/L, pH 7.4) and staged according to Walker and Tait [72] with the following modification. We classified stage V embryos in a more restrictive way by using the following features: (i) cerebral grooves ( = anlagen of the hypocerebral organs) appear as longitudinal slits in the middle of each cephalic lobe, and (ii) the anlagen of the last (15th) pair of walking legs have formed. After staging, the embryos were either processed further for cytochemical experiments or dehydrated in a graded methanol series and stored at −20°C for subsequent gene expression experiments.
Cytochemistry
For cytochemical studies, the embryos fixed in 4% paraformaldehyde were rinsed several times in PBS and incubated overnight at room temperature in a solution containing the f-actin marker phalloidin-rhodamine (Invitrogen, Carlsbad, CA) as described previously [30]. After repeated rinses in PBS, the embryos were counterstained with the DNA-selective fluorescent dye Bisbenzimide (H33258; Sigma-Aldrich, St. Louis, MO, USA; 1 µg/mL in PBS) and rinsed again in PBS. The embryos were then mounted between two coverslips in Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA) and analysed with the confocal laser-scanning microscope Zeiss LSM 510 META (Carl Zeiss MicroImaging GmbH, Jena, Germany).
Identification and amplification of gene fragments
Library preparation and assembly of the embryonic transcriptomes from E. rowelli were performed as described previously [58]. Local tBLASTn searches [73] were conducted using transcriptome libraries from different embryonic stages [58]. Previously published sequences from other onychophoran and arthropod species were used as queries [29], [74], [75]. RNA was isolated from pooled embryos of different developmental stages using TRIzol Reagent (Invitrogen) and RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany) according to the manufacturers’ protocols. First-strand synthesis was performed using random hexamer primers and Superscript III polymerase (Invitrogen). Second-strand synthesis was carried out with DNA Pol I polymerase (Invitrogen). The obtained cDNA was purified using NucleoSpin Extract II-Kit (Macherey-Nagel, Düren, Germany) following the manufacturer's protocol. Fragments of engrailed, cubitus interruptus, wingless and hedgehog were amplified using specific primers (Table 1). The corresponding sequences were made available under the GenBank accession numbers KF218600–KF218603.
Table 1. Specific primers used for PCR.
| Gene | Fragmentlength (in bases) | Direction | Primer sequence |
| engrailed | 668 | forward | CTGAACTGGGTCGATCTGAATATCTCG |
| reverse | CGTATATTTGCCTCGCTTACAAG | ||
| cubitus interruptus | 602 | forward | TCCTCGCCCGTTCTGCCACT |
| reverse | TCCAGGCAGTTCACGCCGTT | ||
| wingless | 1019 | forward | TCCGTGCCGCGTACTCTACCT |
| reverse | CCTCACTTTTATAACCTCTACCACA | ||
| hedgehog | 607 | forward | TCACAGGGGCAAAAGCCCAGT |
| reverse | CGATTGGCGGTTGAGGCTGGT |
Sequence alignment and phylogenetic analyses
The identified sequences of E. rowelli homologs of engrailed (Er-en), cubitus interruptus (Er-ci), wingless (Er-wg) and hedgehog (Er-hh) were compared to the sequences available from the NCBI database using BLAST searches. The corresponding amino acid sequences from E. rowelli were analysed together with those from several other metazoan species, including the closely related onychophoran species E. kanangrensis (see Table S1). Sequence alignments (481, 200, 572 and 429 amino acid positions for engrailed, cubitus interruptus, hedgehog and wingless, respectively) were generated with the online version of MAFFT [76] using the FFT-NS-i strategy (see Figure S1). The appropriate models for protein evolution (LG+G+F for engrailed DAYHOFF+I+G for cubitus interruptus, and LG+I+G for hedgehog and wingless) were selected using ProtTest 3.2 [77] according to the Akaike Information Criterion (AIC) [78]. Maximum Likelihood analyses were performed using Pthread-based version of RAxML v7.2.8 [79]. Nodal support was calculated using 100 bootstrap replicates. Phylogenetic trees were visualised with iTol [80] and edited with Adobe (San Jose, CA, USA) Illustrator CS5.1.
Molecular cloning, probe preparation and whole-mount in situ hybridization
Gene fragments were cloned into the pGEM-T Vector System I (Promega Corporation, Madison, WI, USA). Digoxigenin- and biotin-labelled RNA probes were prepared using DIG RNA Labeling Kit SP6/T7 and Biotin RNA Labeling Mix (Roche, Mannheim, Germany). Whole-mount in situ hybridization was performed as described previously [22], [81] with the following modifications. The embryos stored in 100% methanol were rehydrated in a graded methanol series (2×100%, 75%, 50% and 25% in PBST [PBS+0.1% Tween-20], 7 min each). Pre-hybridization (six hours) and hybridization steps (three days) were carried out at 60°C. 100–500 ng of the probes were diluted in 500 µl hybridization solution (50% formamide, 5xSSC, 50 µg/mL heparin, 50 µg/mL yeast tRNA, 5% Dextran sulphate, 0.1% Tween-20). Post-hybridization washes included several rinses in hybridization buffer at 60°C, followed by several rinses in a washing solution (2xSSC+0.1% Tween-20) at 60°C and in PBST at room temperature. The embryos were then incubated for 3 hours in a blocking solution (10% normal goat serum in PBST) at room temperature, followed by an incubation with anti-digoxigenin alkaline phosphatase-conjugated antibody (Roche), diluted 1∶1000 in blocking solution for two days at 4°C. After several washes with PBST at room temperature, NBT/BCIP staining solution (Roth, Karlsruhe, Germany) was added. The reaction was stopped after the desired staining was achieved by several washes with PBST. Double whole-mount in situ hybridization was carried out as described by Schinko et al. [82] with the following modifications. After detection of the first colour, the embryos were incubated in inactivation buffer (50% formamide, 5xSSC, 0.1% Tween-20, 10% sodium dodecyl sulphate) at 60°C in a heating block. Embryos were then washed in blocking solution for two hours and the antibody for the second staining was added at a dilution of 1∶100 in blocking solution. The second colour reaction was stopped by several washes with PBST and the embryos were then re-fixed in 4% paraformaldehyde and stored at 4°C. For nuclear staining, the DNA-selective fluorescent dye SYBR Green (Invitrogen) was applied according to the manufacturer’s protocol.
Microscopy and image processing
The embryos were analysed under a stereomicroscope (Leica WILD M10 with a WILD MDG 17 Stand; Leica Microsystems, Wetzlar, Germany) and a transmitted-light microscope (Leica Leitz DMR; Leica Microsystems) equipped with a colour digital camera (PCO AG SensiCam, Kelheim, Germany). Several micrographs were taken from each embryo at different focal planes and merged to a single image using the Auto-Blend Layers function in Adobe Photoshop CS5.1. Brightness and contrast were adjusted using Photoshop CS5.1. Final panels and diagrams were designed with Illustrator CS5.1 and exported to Tagged Image File Format files. Confocal laser-scanning microscopy and image processing were performed as described previously [30].
Results
Developmental origin and fate of transverse segmental furrows in embryos of E. rowelli
The initial segmental structures appearing in the embryo of E. rowelli are the paired mesodermal somites ( = coelomic cavities; Figure 2A; see also Figure S2). Due to an anterior-to-posterior progression in development (i.e., the anterior segments are further advanced than the posterior ones), the somites arise sequentially in the antennal, jaw and slime papilla segments, followed by the trunk segments (Figure 2A). As soon as the mesodermal somites have formed, transverse furrows appear at regular intervals in the overlying ectoderm (Figure 2A–D; see Figure S2). Although the formation of these furrows lags behind that of somites, their position along the body corresponds exactly to the border between each adjacent somite (see Figure S2).
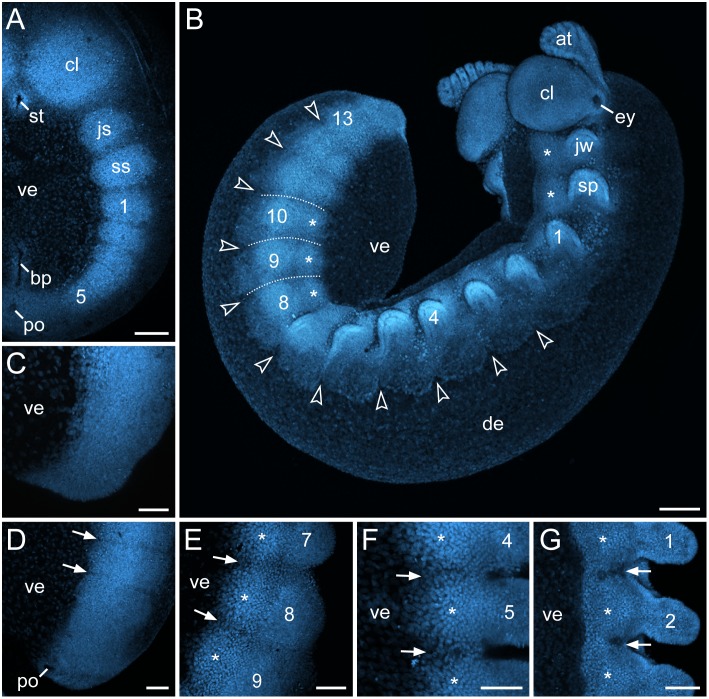
Figure 2. Developmental origin and fate of transverse segmental furrows in embryos of E. rowelli at successive developmental stages.
Confocal micrographs of embryos labelled with Bisbenzimide. Leg-bearing segments and corresponding limbs are numbered. Asterisks indicate the anlagen of the ventral and preventral organs. (A) Stage I embryo in ventral view. Note distinct borders between the developing segments. (B) Late stage III embryo in ventro-lateral view. Arrowheads point to the dorsal indentations in the ectoderm. Dotted lines demarcate the transverse furrows. (C–G) Chronological sequence of the formation of the segmental furrows. Embryos in lateral (C–E) and ventral view (F, G). Arrows point to the segmental furrows. (C) Posterior end of a stage II embryo. Note that furrows are not visible yet. (D) Posterior end of a stage III embryo. (E) Midbody of an early stage IV embryo. Note an additional longitudinal furrow separating the anlagen of the ventral/preventral organs and the developing limbs. (F) Anterior body region of a stage IV embryo. (G) Anterior body region of a stage V embryo. Abbreviations: at, developing antenna; bp, blastoporus; cl, cephalic lobe; de, dorsal extra-embryonic tissue; ey, eye; js, jaw segment; jw, jaw; st, stomodaeum; sp, slime papilla; ss, slime papilla segment; po, proctodaeum; ve, ventral extra-embryonic tissue. Scale bars: 200 µm (A, B), 100 µm (C–G).
During further development, paired segmental limb buds (numbered in Figure 2B, E–G) arise in an anterior-to-posterior progression from lateral portions of the germ band. Simultaneously, the median portions of the germ band give rise to segmental thickenings (asterisks in Figure 2B, E–G) that are the anlagen of the ventral and preventral organs (see ref. [42] for details on the developmental fate of these thickenings). Shortly after the limb buds and the anlagen of the ventral and preventral organs have formed, the lateral ectodermal portions of the germ band extend dorsally in a regular, undulating fashion. This regular growth gives rise to conspicuous segmental indentations in the ectoderm (arrowheads in Figure 2B), which correspond in position to the transverse furrows that separate the segmental anlagen of limbs and the ventral/preventral organs along the body (dotted lines in Figure 2B and arrows in Figure 2E–G). These indentations show that there is a defined structure in the ectoderm and that the furrows are not simple undulations caused by the bulging, underlying mesodermal somites.
Following the anterior-to-posterior progression in development, the transverse segmental furrows and the dorsal indentations of the ectoderm become less prominent at the anterior end in stage IV embryos and are hardly detectable at stage V (Figure 2B, F, G). Neither the transverse furrows nor the segmental indentations persist beyond this developmental stage in E. rowelli (cf. [30], [42], [56], [62]).
Identification of homologs of engrailed, cubitus interruptus, wingless and hedgehog in E. rowelli and phylogenetic analyses
Irrespective of the assembly filters used (F15, F25, and F30; see ref. [58] for details on methodology), we identified contigs of only one homolog of each gene (Er-en, Er-ci, Er-hh and Er-wg) in our transcriptomic data. However, since the complete genome sequence is unavailable for Onychophora, we cannot rule out the possibility that there might be additional copies of these genes in the genome, although these might not be expressed during development. To determine whether the identified homologs from the onychophoran E. rowelli are orthologous to the corresponding sequences from arthropods, we carried out phylogenetic analyses (see Table S1 and Figure S3). In the resulting cladograms, the identified homologs Er-en, Er-hh, Er-ci and Er-wg form sister groups to the corresponding sequences from E. kanangrensis (see Figure S3). These results confirm that the identified sequences of E. rowelli are indeed orthologs rather than paralogs of the corresponding arthropod genes.
Expression of engrailed during development in E. rowelli
During embryogenesis of E. rowelli, engrailed is expressed in segmentally repeated stripes along the germ band, whereas no expression is seen in the ventral and dorsal extra-embryonic tissue (Figures 3A–F, 4A–D). As the embryo grows, engrailed stripes are added posteriorly and increase in size towards the anterior end, following the anterior-to-posterior progression along the body (Figure 3A–F). Notably, the stripes occur after the segmental furrows have formed and the signal is graded, as it does not show clear expression boundaries (cf. Figure 3C, D; see ref. [29] for similar data obtained from the closely related species E. kanangrensis). In contrast to other segments, the engrailed domain is weaker and situated more dorsally in the antennal segment (Figure 3A, B). Shortly after the antenna has formed, the shape of this domain transforms from a stripe to a spot-shaped domain, which follows the elongation of the developing antenna (arrowheads in Figure 3E, F). This results in an elongated domain at the antennal basis, which extends further posteriorly and is still seen above the eye anlage later in development (Figures 3F, 4D).
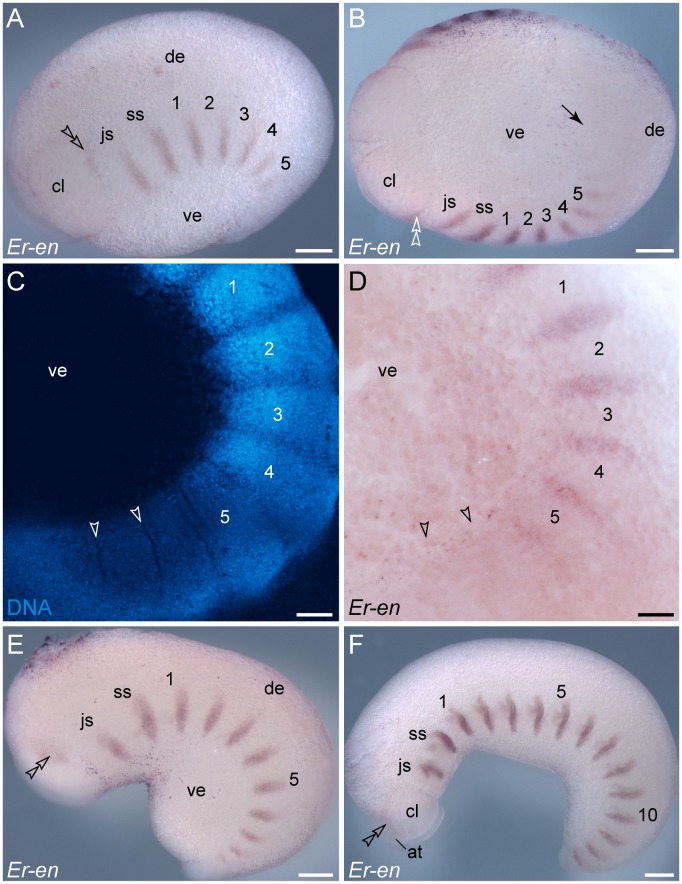
Figure 3. Expression of engrailed in embryos of the onychophoran E. rowelli at subsequent developmental stages.
Anterior is left in A, B, E, F and up in C, D. Leg-bearing segments and corresponding limbs are numbered. Note the repeated stripes along the body and the lack of expression in the dorsal and ventral extra-embryonic tissue. Double-arrowheads in A, B, E and F indicate the dorsally located domain of engrailed in the cephalic lobe. Arrowheads in C and D point to the transverse ectodermal furrows, which lack engrailed expression. (A) Early stage II embryo in lateral view. (B) The same embryo as in A in ventral view. Note the lack of expression in the posterior region and around the proctodaeum (black arrow). (C) Confocal micrograph of the posterior end of the same embryo as in A labelled with the DNA marker SYBR Green. (D) Light micrograph of the same embryo as in C. (E) Early stage III embryo in lateral view with eleven engrailed stripes along the body and a spot-shaped domain in the developing antenna (arrowhead). (F) Late stage III embryo in lateral view with 15 engrailed stripes along the body (13 within the trunk) and an additional domain in the developing antenna (arrowhead). Abbreviations: at, developing antenna; cl, cephalic lobe; de, dorsal extra-embryonic tissue; js, jaw segment; ss, slime papilla segment; ve, ventral extra-embryonic tissue. Scale bars: 250 µm (A, B, E, F), 100 µm (C, D).
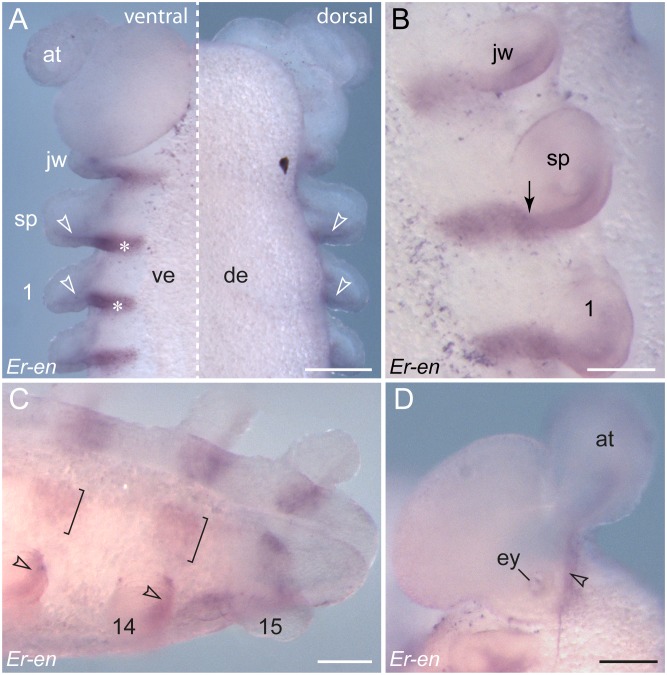
Figure 4. Details of engrailed expression in embryos of E. rowelli.
Leg-bearing segments and corresponding limbs are numbered. (A) Ventral (left) and dorsal perspective (right) of the anterior end of a stage IV embryo. Note the expression in the ventral ectoderm (asterisks) and in the ectoderm and mesoderm of each developing limb (arrowheads). (B) Details of the anlagen of a jaw, a slime papilla and the first leg in a stage IV embryo. Arrow points to the transition between a wide ventral stripe and a narrow domain within the limb, which become separate during further development. (C) Ventro-lateral view of the posterior end of a stage V embryo. Note that the domains within the limb buds (arrowheads) and those on the ventral body surface (square brackets) have become separate. The latter correspond to the anlagen of the ventral and preventral organs [30], [42], [56]. (D) Lateral view of a developing antenna in a stage IV embryo. Abbreviations: at, developing antenna; de, dorsal extra-embryonic tissue; ey, developing eye; jw, embryonic jaw; sp, embryonic slime papilla; ve, ventral extra-embryonic tissue. Scale bars: 250 µm (A), 125 µm (B–D).
The expression of the remaining engrailed stripes in the embryo precedes the formation of limb buds (Figure 3A). Notably, each engrailed stripe extends beyond the segmental furrow in the early embryo (Figure 5A). This pattern persists throughout development and no migration or shift of the initial furrow, or establishment of a new segmental furrow is evident (Figure 5A–E). When the limb buds arise, each engrailed stripe follows the curvature of the corresponding limb bud, in which it is located posteriorly in both ectoderm and mesoderm (Figures 3F, 4A–C, 5C–E). While no engrailed expression occurs dorsally, each engrailed stripe continues from the limb bud to the ventral ectoderm (Figure 4A, B). After all fifteen leg-bearing segments have formed, the initial stripes of engrailed are subdivided in two separate domains: a lateral domain associated with the limb bud, and a median domain corresponding to the segmental anlage of the ventral and preventral organs (Figure 4C).
Figure 5. Spatial relationship between segmental furrows and repeated engrailed domains in embryos of E. rowelli.
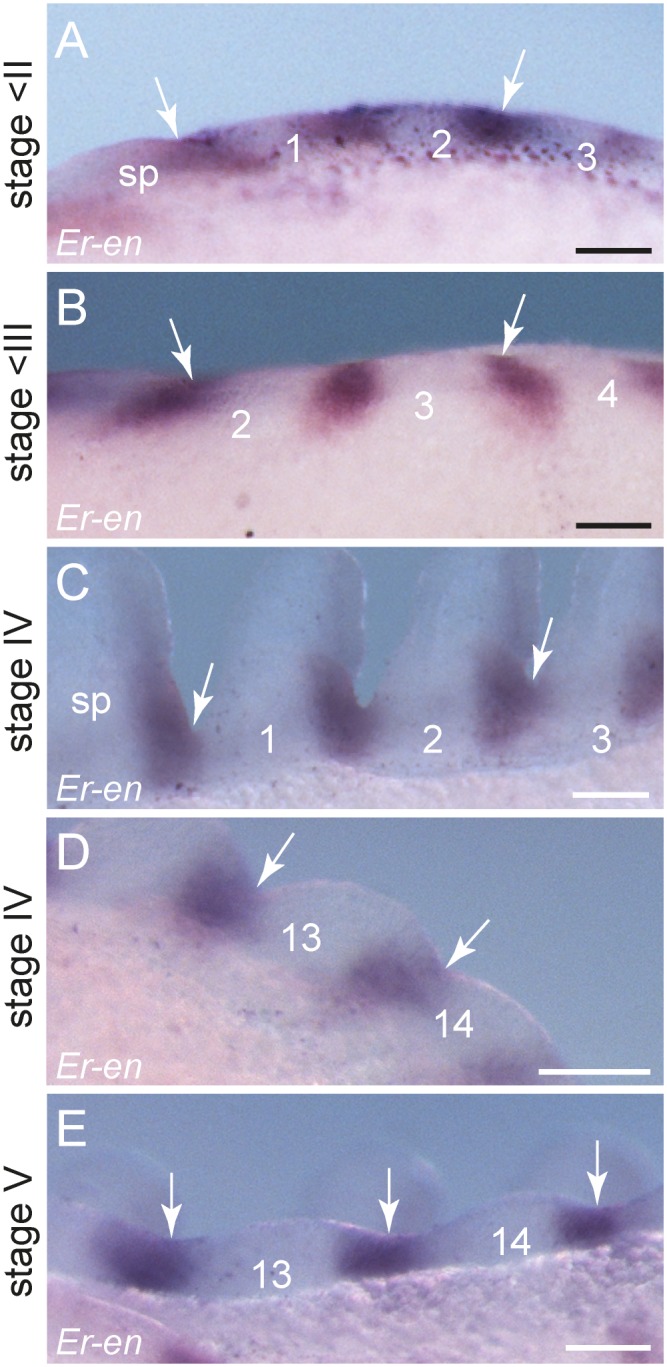
Anterior is left in all images. Leg-bearing segments and corresponding limbs are numbered. Arrows indicate the position of segmental furrows in embryos of E. rowelli at subsequent developmental stages in ventral (A, C, D), dorsal (B), and ventro-lateral perspectives (E). Abbreviation: sp, anlage of the slime papilla/slime papilla segment. Scale bars: 100 µm (A, B, D, E,), 125 µm (C).
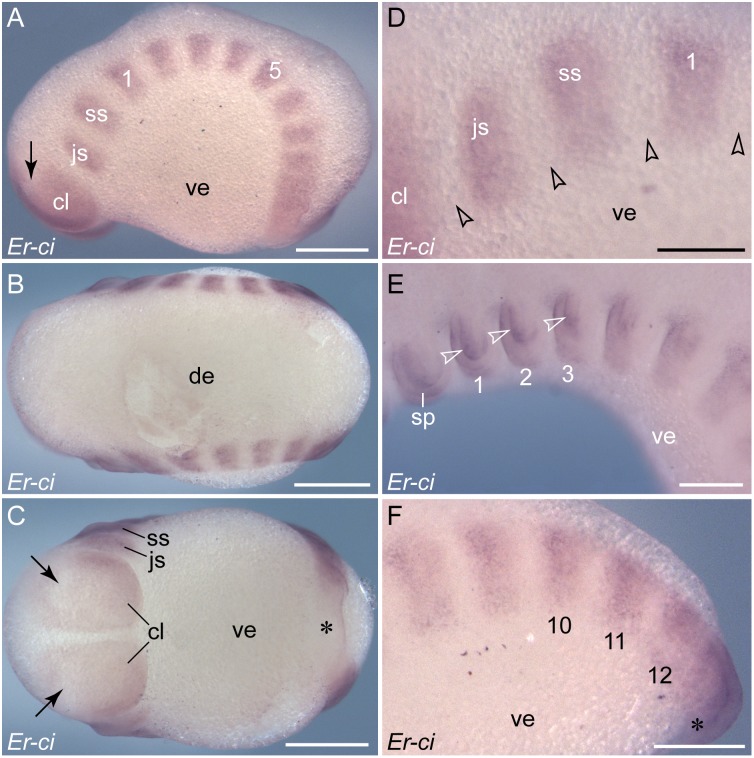
Expression of cubitus interruptus is anterior to each engrailed domain in embryos of E. rowelli
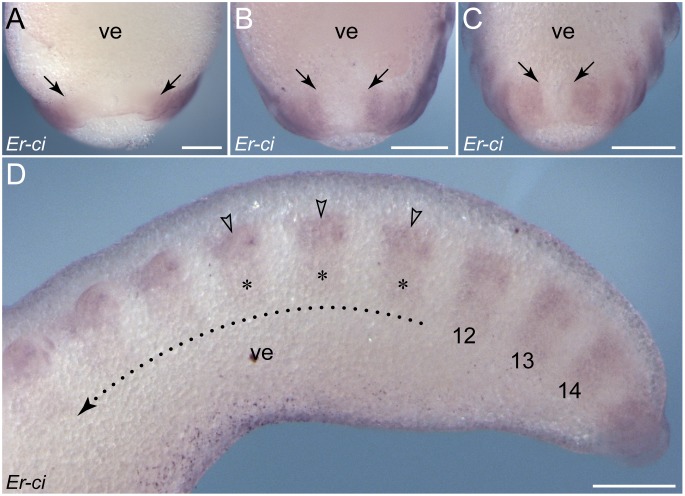
In contrast to engrailed, which is expressed in stripes, cubitus interruptus is first expressed as a continuous belt at the posterior end of the embryo, excluding the region around the proctodaeum (Figures 6A–C, F, 7A–C). We were unable to detect cubitus interruptus expression in embryos earlier than stage II and, therefore, cannot exclude that this gene might be also expressed in a continuous belt at the anterior end. The continuous belt of expression persists at the posterior end, whereas its anterior part dissociates into increasingly well-defined, segmental, rectangular domains (Figures 6A–F, 7A–D). From stage IV onwards, the continuous belt disappears and only the rectangular domains are visible along the antero-posterior body axis. Similar to the engrailed stripes, the cubitus interruptus domains do not show clear expression boundaries but rather a graded signal towards the margins of each domain (Figures 6D, F, 7D). The largest unitary expression domain occurs in the cephalic lobes of the antennal segment, in which only two transverse dorsal regions lack expression (arrows in Figure 6A, C).
Figure 6. Expression of cubitus interruptus in embryos of E. rowelli.
Anterior is left in all images. Leg-bearing segments and corresponding limbs are numbered. (A–C) Stage II embryo in lateral, dorsal and ventral views. Note the continuous belt of expression on each side of the body at the posterior end, which dissolves into nearly rectangular domains towards the anterior end. Note also the expression-free areas in the cephalic lobes (arrows) and around the proctodaeum (asterisk). (D) Detail of the anterior-most expression domains in a stage II embryo in lateral view. Arrowheads point to wide gaps between the domains. (E) Anterior portion of a stage IV embryo in lateral view showing expression in the ectoderm and mesoderm (somite walls) of the limb anlagen (arrowheads). (F) Posterior end of the same embryo as in E. Note the remnant of the continuous belt of expression near the proctodaeum (asterisk), which dissolves into solitary domains towards the anterior end. Abbreviations: cl, cephalic lobe; de, dorsal extra-embryonic tissue; js, jaw segment; sp, embryonic slime papilla; ss, slime papilla segment; ve, ventral extra-embryonic tissue. Scale bars: 500 µm (A–C), 250 µm (D–F).
Figure 7. Details of cubitus interruptus expression in embryos of E. rowelli.
(A–C) Posterior ends of embryos at developmental stages II, III and IV in ventral view. Arrows indicate the posterior-most regions of expression, which subsequently move towards the proctodaeum. (D) Posterior end of a stage IV embryo in lateral view. Leg-bearing segments and corresponding limbs are numbered. Dotted line with an arrow indicates the direction of decreasing expression in the ventral ectoderm (asterisks) but increasing expression in the limb anlagen (arrowheads) towards the anterior end. Abbreviation: ve, ventral extra-embryonic tissue. Scale bars: 250 µm (A–D).
When the limbs arise, the expression of cubitus interruptus persists in the anterior region of each limb bud, where it is expressed in both ectoderm and mesoderm (Figure 6E). At advanced developmental stages, the continuous belt of cubitus interruptus expression is no longer evident, as it disintegrates completely into separate segmental domains (Figures 6F, 7D). Similar to engrailed stripes (cf. Figure 4C), each cubitus interruptus domain consists of a lateral and a median portion, each of which has a different fate further in development, while the lateral portion (including the limb bud; arrowheads in Figure 7D) persists until late in development, the median portion (including the anlage of ventral/preventral organs; asterisks in Figure 7D) disappears earlier, following the anterior-to-posterior progression.
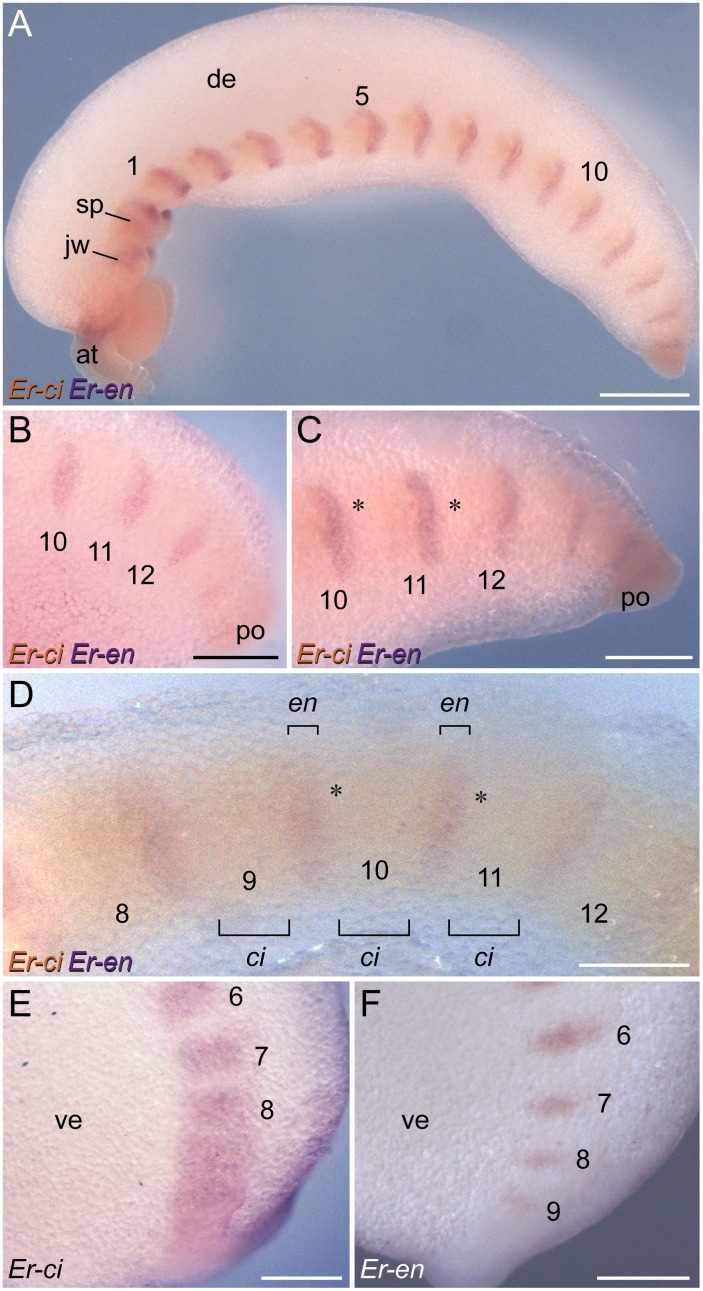
To analyse the spatial relationship between the expression patterns of cubitus interruptus and engrailed, we conducted additional single and double in situ hybridization experiments (Figure 8A–F). Due to a persisting continuous belt of cubitus interruptus expression at the posterior end of the embryo (cf. Figure 6A), the domains of engrailed and cubitus interruptus overlap initially in this body region (Figure 8B, E, F). After the subdivision of this belt into separate, segmentally repeated domains, gaps occur between the cubitus interruptus and engrailed domains (Figure 8C, D). This is in line with the results of our single in situ hybridization experiments, which revealed gaps between adjacent cubitus interruptus domains that are wider than each engrailed stripe (cf. Figures 3A, 6A).
Figure 8. Localisation of engrailed and cubitus interruptus expression in embryos of E. rowelli.
Labelling for engrailed is illustrated in purple and for cubitus interruptus in orange in A–D. Embryos in lateral view. Anterior is left and dorsal is up in all images. Leg-bearing segments are numbered. (A) Overview of a stage IV embryo. (B) Posterior end of a stage III embryo. Note that no gaps are evident between the engrailed and cubitus interruptus domains. Note also that there might be a co-expression of the two genes at least in cells located within the posterior-most engrailed stripes, as there is a continuous belt of cubitus interruptus expression in this body region at that stage (cf. E and F; see also Figures 6F, 7B). (C) Posterior end of a stage IV embryo. Asterisks indicate gaps between subsequent engrailed and cubitus interruptus domains. (D) Through-light micrograph showing subsequent domains of engrailed and cubitus interruptus in a stage IV embryo. Note gaps between the engrailed and cubitus interruptus domains (asterisks). (E) Expression of cubitus interruptus at the posterior end of a stage II embryo. (F) Expression of engrailed at the posterior end of a stage II embryo. Abbreviations: at, antenna; ci, cubitus interruptus domains; de, dorsal extra-embryonic tissue; en, engrailed domains; jw, jaw; po, proctodaeum; sp, slime papilla; ve, ventral extra-embryonic tissue. Scale bars: 500 µm (A), 250 µm (B, C, E, F), 200 µm (D).
Dynamic patterns of wingless expression during development in E. rowelli
Similar to engrailed, wingless is expressed in a reiterated pattern along the germ band (Figure 9A–F). However, in contrast to engrailed, this gene is initially expressed in the early anlagen of limbs (arrowheads in Figure 9B) and each segmental domain extends subsequently in a stripe-like fashion to the ventral ectoderm (arrows in Figure 9B), following the anterior-to-posterior progression in development. Each stripe demarcates the middle of each limb basis, from which it runs further medially (Figure 9C–F). When the limb buds elongate, the spot-shaped domains become more prominent at the tip of each developing limb, including the slime papillae and the jaws (Figure 9B–D). As development proceeds, the initial unitary domain associated with each limb is subdivided into two separate domains: a spot-shaped domain at the tip of each limb, and a stripe-shaped domain in the ventral ectoderm, including the limb basis (Figure 9D–F). In addition to these domains along the body, a ubiquitous expression of wingless occurs in the cephalic lobes and the antennae and a conspicuous ring-shaped domain is seen around the proctodaeum (Figure 9A, C, E).
Figure 9. Expression of wingless in embryos of E. rowelli.
Anterior is up in all images. Leg-bearing segments and corresponding limbs are numbered. (A) Stage II embryo in ventral view. Note that wingless is expressed in segmentally repeated stripes in the anlagen of limbs and in areas in which the limb buds arise. Note also wingless expression in the cephalic lobes and the proctodaeum. (B) Detail of wingless expression in the same embryo as in A in lateral view. Note that the expression first appears in the limb buds (arrowheads), followed by stripe-shaped domains in the ventral ectoderm (arrows). (C) Stage III embryo in lateral view. Note a weak and ubiquitous expression of wingless in the cephalic lobes and a strong expression in the anlagen of antennae. (D) Detail of wingless expression of the same embryo as in D in lateral view. Arrows point to appearing stripes along the ventral body surface, whereas arrowheads denote the spot-shaped domains in the distal portion of each developing limb. (E) Posterior end of a stage IV embryo in ventro-lateral view. As in the stage II and III embryos, the expression first appears in the limb buds (arrowheads), followed by stripe-shaped domains in the ventral ectoderm (arrows). (F) Detail of a stage IV embryo in ventral view. Note that the stripes in the ventral ectoderm are separated by non-expressing cells from the spot-shaped domains in the distal limb portions. Abbreviations: at, developing antenna; cl, cephalic lobe; js, jaw segment; jw, embryonic jaw; lb, limb buds; po, proctodaeum; sp, embryonic slime papilla; ss, slime papilla segment; ve, ventral extra-embryonic tissue. Scale bars: 250 µm (A, C), 125 µm (B, D), 200 µm (E), 100 µm (F).
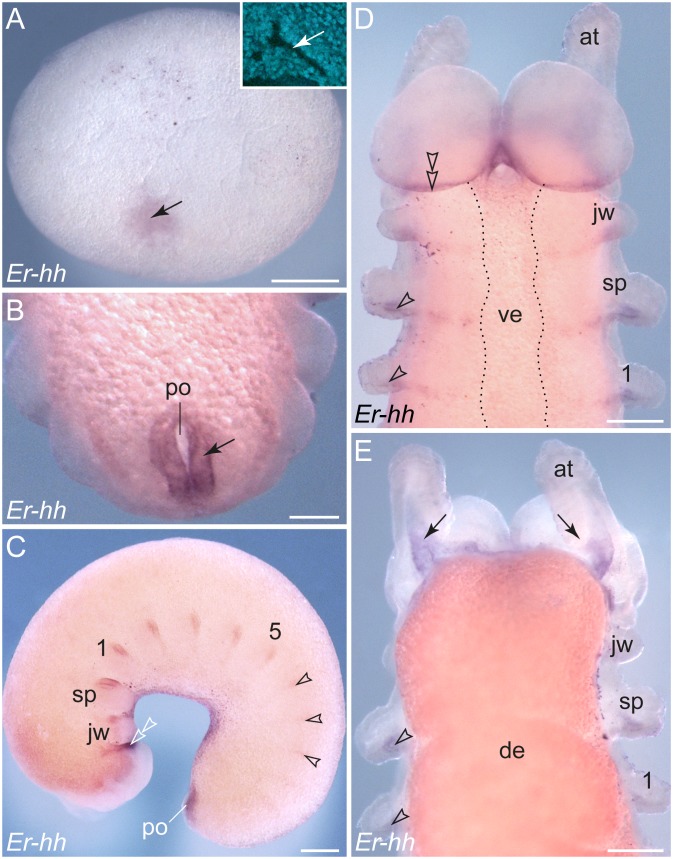
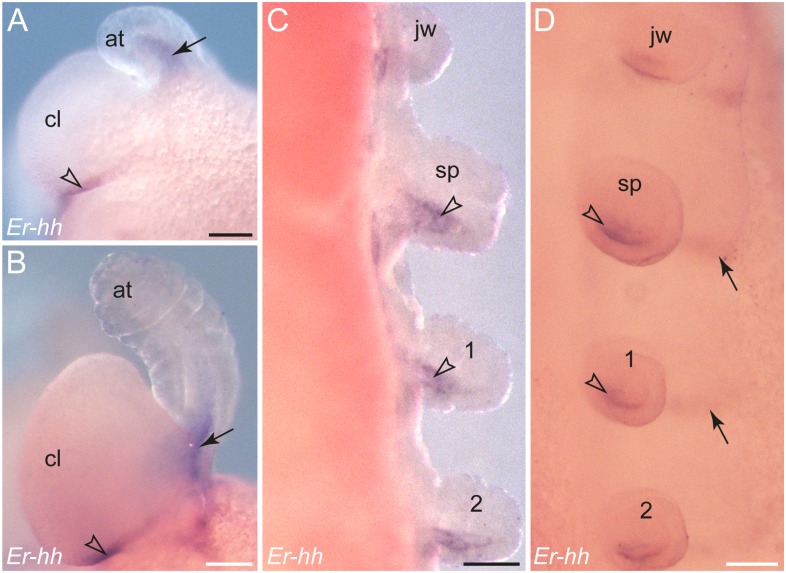
Posterior expression of hedgehog in each segment in embryos of E. rowelli
The earliest detectable expression of hedgehog occurs around the early elongating blastopore (Figure 10A). Later in development, a ring-shaped domain is still seen around the proctodaeum (Figure 10B, C), thus resembling the expression pattern of wingless in the same region (cf. Figure 9A, E). In addition to this posterior domain, stripes of hedgehog expression emerge sequentially along the embryo, following the anterior-to-posterior progression in development (Figure 10C). This pattern is similar to the expression of engrailed, but the hedgehog stripes are thinner and shorter than the engrailed stripes and are expressed later, first appearing in each developing limb bud (Figure 10C).
Figure 10. Expression of hedgehog in embryos of E. rowelli.
Leg-bearing segments and corresponding limbs are numbered. Arrowheads indicate segmentally repeated stripes in the posterior portion of each developing limb. (A) Stage I embryo. Inset in the upper right corner shows a fluorescent micrograph (DNA labelling) of the blastoporal area from the same embryo. Arrows point to the position of the blastopore, which is surrounded by hedgehog expressing cells. (B) Posterior end of a stage IV embryo. Arrow indicates the expression around the proctodaeum. (C) Stage III embryo in lateral view showing repeated stripes of expression along the body. Arrowheads point to the emerging hedgehog domains in the posterior portion of each developing limb in the posterior half of the embryo. Note an elongated domain at the posterior border of the cephalic lobes (double-arrowhead). (D) Anterior end of a stage IV embryo in ventral view. Double-arrowhead points to the elongated domain at the posterior border of the cephalic lobes. Dotted line indicates the border between the ectoderm and the ventral extra-embryonic tissue. (E) Dorsal view of the same embryo as in D. Arrows point to the expression in the mesoderm at the bases of the developing antennae. Abbreviations: at, developing antenna; de, dorsal extra-embryonic tissue; jw, embryonic jaw; po, proctodaeum; sp, embryonic slime papilla; ve, ventral extra-embryonic tissue. Scale bars: 500 µm (A), 100 µm (B), 250 µm (C), 200 µm (D, E).
Within the limb buds, hedgehog is expressed posteriorly in both ectoderm and mesoderm as a graded signal; only in the antennae is this gene expressed dorsally (Figures 10C–E, 11A–D). During further development, the hedgehog stripes extend medially in the ventral ectoderm (Figure 10D, 11D). These median domains are located posterior to the corresponding wingless domains (see Figure S4). The most prominent stripe of hedgehog expression demarcates the posterior boundary of the cephalic lobes in the antennal segment (Figures 10C, D, 11A, B, D).
Figure 11. Details of hedgehog expression in embryos of E. rowelli.
Leg-bearing segments and corresponding limbs are numbered. (A, B) Anterior ends of a stage III and a stage IV embryo in lateral view. Arrows point to the expression in the mesoderm of the developing antennae, whereas arrowheads indicate the expression at the posterior border of the cephalic lobe. (C, D) Details of limbs in a stage IV embryo in dorsal (in C) and ventro-lateral views (in D). Arrowheads indicate the expression in the mesoderm of each developing limb, although expression is also evident in the ectoderm of limb anlagen. Arrows point to stripes of expression in the ventral ectoderm. Abbreviations: at, antenna; cl, cephalic lobe; jw, jaw segment; po, proctodaeum; sp, slime papilla. Scale bars: 100 µm (A–D).
Discussion
Segments, rather than parasegments, are the initial metameric units in the onychophoran embryo
Parasegments are believed to be the initial metameric compartments of the arthropod embryo [3], [7], [13], [14], . They arise early in development and are recognised by several features, including (i) the juxtaposed pattern of wingless and engrailed/hedgehog expression (via an autoregulatory interaction of these genes, which was initially demonstrated in embryos of Drosophila melanogaster [7], [15], [83], [84]); (ii) conspicuous expression patterns of some Hox genes, obeying the parasegmental boundaries [7], [11], [16], [83], [85]; and (iii) cell lineages that at least in crustaceans are restricted to the genealogical units corresponding to parasegments [86]–[90]. Thus, if present, parasegments should be recognisable in the onychophoran embryo based on these criteria.
Our data from E. rowelli, as well as those from the closely related species E. kanangrensis [29], [39], show that engrailed, cubitus interruptus, wingless and hedgehog are all expressed in a reiterated pattern in the onychophoran embryo (Figure 12). The expression occurs in repeated sets along the body, the relative order of which corresponds to that in arthropods; cubitus interruptus and wingless are expressed anterior to the engrailed and hedgehog domains (Figure 13A–D) [16], [17], [22], [83], [91], [92]. These data are in line with the assumption of an autoregulatory interaction of these genes [7], [13], [14], although the graded expression pattern in the trunk lacks boundaries in onychophorans. Nonetheless, one could argue that a distinct boundary might still exist at the post-transcriptional level [29], which would correspond to the parasegmental boundary of arthropods (Figure 13D).
Figure 12. Diagrams of engrailed, cubitus interruptus, wingless and hedgehog expression in embryos of the onychophoran E. rowelli.
The upper row illustrates stage III embryos in lateral view, whereas the lower row shows stage IV embryos in ventral and lateral views, respectively. Note the segmentally repeated patterns of expression of all four “segment polarity genes” and their specific order within each segment (see text for further details).
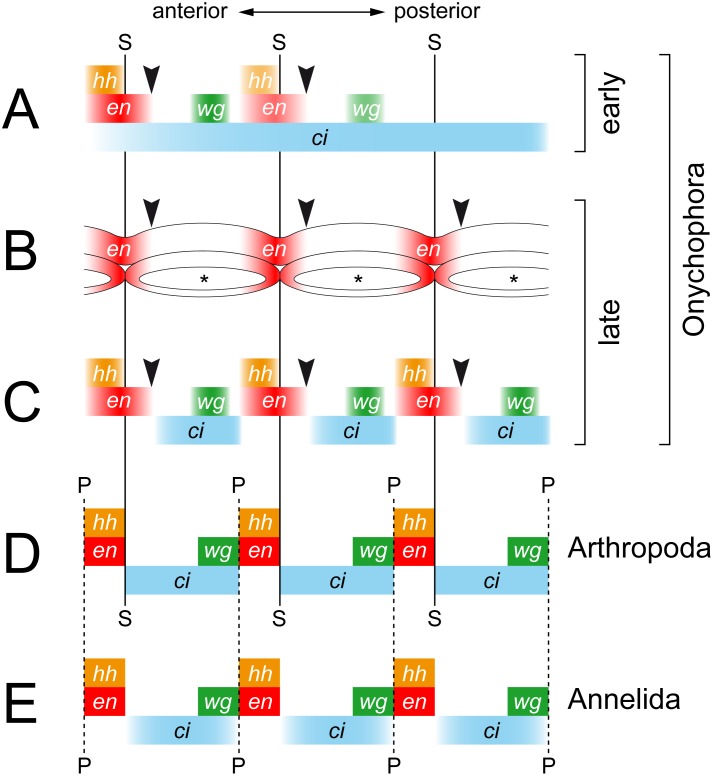
Figure 13. Diagrams comparing the expression patterns of homologs of engrailed, cubitus interruptus, wingless and hedgehog along the body in onychophorans, arthropods and annelids.
Vertical lines demarcate the position of segmental (solid lines) and parasegmental boundaries (hatched lines) only for taxa, in which they are identifiable morphologically. Black arrowheads indicate the position of expected, albeit non-existent, segmental boundaries in Onychophora, which is in contrast to arthropods. (A) Diagram of expression in embryos of the onychophoran Euperipatoides rowelli at an early developmental stage. Note the continuous belt of cubitus interruptus expression. (B) Diagram of expression of engrailed in the ectoderm and the mesodermal somites in the onychophoran Euperipatoides kanangrensis (based on previous data [29]). Asterisks indicate coelomic cavities. Note that the expression pattern does not correspond to the segmental boundaries and that each somite shows an anterior and a posterior expression domain. (C) Diagram of expression in embryos of the onychophoran E. rowelli at a late developmental stage. Note the graded expression patterns that do not form distinct boundaries (cf. ref. [29]). (D) Diagram of expression in the fruit fly Drosophila melanogaster, modified from [24]. (E) Diagram of expression in the annelid Platynereis dumerilii, modified from [24]. Abbreviations: ci, cubitus interruptus; en, engrailed; hh, hedgehog; P, parasegmental boundary; S, segmental boundary (in onychophorans only seen during embryogenesis); wg, wingless.
Another piece of evidence for parasegments in arthropods comes from the anterior expression borders of the posterior Hox genes, which typically correspond to the parasegmental boundaries, including Antennapedia, Ultrabithorax, abdominal-A, Abdominal-B in chelicerates, fushi tarazu, Abdominal-B in myriapods, proboscipedia in crustaceans, and Sex combs reduced, Antennapedia, abdominal-A and Abdominal-B in hexapods [7], [11], [14], [16], [83], [85], [93]. However, the expression patterns of Hox genes in the onychophoran E. kanangrensis and localisation of the Ultrabithorax and abdominal-A proteins in Acanthokara kaputensis mostly revealed segmental rather than parasegmental patterns, which is different from the situation in arthropods [3], [93]. The anterior expression borders of seven of the ten Hox genes in E. kanangrensis clearly do not lie between the wingless and engrailed/hedgehog domains but instead correspond to the segmental furrows [94], [95]. In contrast to this, the anterior expression borders of Hox3, Sex combs reduced and fushi tarazu have been interpreted as being positioned “in the middle” [95] of the corresponding segments. However, the position of the anterior expression borders of these three genes in relation to the engrailed and wingless domains still needs to be analysed to clarify whether or not these genes are expressed in a “parasegmental” pattern.
In addition to gene expression studies, cell lineage analyses revealed a correlation between the clonal and parasegmental boundaries in crustaceans [86−90], . The progeny of cells on either side of the parasegmental boundary remain separated, suggesting that parasegments are real genealogical units [6], [89], [96]. These units are reflected in the parasegmental organisation of the ventral nerve cord in various arthropods, including the fruit fly Drosophila melanogaster [6], [20], [21]. During Drosophila development, each neuromere originates from cells located between two consecutive parasegmental furrows [21] and eventually gives rise to a ganglion. Therefore, due to their out-of-register nature, each resulting ganglion is shifted anteriorly once the embryo is re-segmented [6], [7]. This shifted arrangement of ganglia is a common feature of all arthropod groups, especially in body regions that have retained the ancestral, ladder-like organisation of the ventral nerve cord with separate, metameric ganglia [6]).
In contrast to arthropods, the onychophoran nerve cord displays no such metameric ganglia (cf. Figure 1I, J; [31], [97], [98]). Unfortunately, cell lineage analyses, which would unveil the boundaries of putative genealogical units and their relationship to the nervous system, are currently unfeasible in Onychophora. However, neuronal tracing of leg nerves revealed no anterior shift in the arrangement of motor neurons in the nerve cords of the onychophoran E. rowelli [37], which contrasts with the anteriorly shifted ganglia and motor neurons in arthropods [6]. Thus, there is currently no neuroanatomical indication of parasegments in adult onychophorans.
In addition to the ganglia of arthropods, the parasegmental boundaries are also manifested morphologically as transverse grooves in the embryonic ectoderm of some chelicerate [16], crustacean [19] and insect species [14], [20], [21], although embryonic grooves are not always recognisable e.g., in decapod crustaceans [99]. During re-segmentation of the embryo, each ectodermal groove situated in a parasegmental position (anterior to the engrailed domain) is replaced by a new groove in a segmental position (posterior to the engrailed domain) (Figure 13D; [16], [19], [100], [101]). This contrasts with our findings, which instead show that the transverse furrow in E. rowelli embryos does not change its position during development, thus providing no indication for a re-segmentation of the embryo. Moreover, the furrow is not located anterior to the engrailed domain but rather corresponds to the segmental border between adjacent somites (Figure 13B). Therefore, in contrast to arthropods, neither the embryonic ectoderm nor the organisation of the adult nervous system provides evidence for a morphological manifestation of parasegments in Onychophora.
Despite the lack of morphological evidence, we cannot exclude that parasegments, as defined by the autoregulatory interaction of “segment polarity genes” [7], [14], [16], [18], might still exist in Onychophora, at least at the post-transcriptional level [29]. However, the present and previous data from the onychophoran embryo clearly show that wingless, engrailed and hedgehog are expressed after the segmental boundaries have been established [29], [39] (Figure 13A–C). Therefore, the parasegments cannot be the initial metameric units in the onychophoran embryo, because they are preceded by segments, which are recognisable by segmental furrows, dorsal indentations of the germ band, and metameric somites. This clearly contrasts with the situation in arthropods, in which parasegments rather than segments are the initial metameric units of the embryo [3], [7], [13], [14], [16], [18].
“Segment polarity genes” are not involved in segment formation in Onychophora
Our data from E. rowelli show that the timing of expression of engrailed, cubitus interruptus, wingless and hedgehog is entirely different from that in arthropods (Figure 13A–D). An initial belt of expression of cubitus interruptus occurs at the posterior end in E. rowelli, but this gene is unlikely to be involved in segment formation because this posterior belt dissociates into metameric domains only after the segmental furrows have formed. Likewise, the segmental domains of the three remaining genes occur after the establishment of the segmental furrows, suggesting that these genes play no role in segment formation, in contrast to what occurs in arthropods [17], [40], [102]. Additionally, our data confirm that each engrailed domain extends beyond the segmental furrow in the ectoderm of the onychophoran embryo [29]. The same holds true for the mesoderm, in which engrailed is expressed beyond the border of adjacent somites (Figure 13B; [29]). Thus, there is no clear spatial relationship between the formation of segmental boundaries and the expression of engrailed in Onychophora (arrowheads in Figure 13B, C).
Although the anterior-to-posterior order of expression of “segment polarity genes” might be conserved in arthropods and onychophorans [39], the lack of a spatial and temporal relationship between the expression domains and the segmental boundaries speaks against the involvement of these genes in segment formation in Onychophora. A similar lack of correlation has been demonstrated recently for most pair rule genes in the onychophoran E. kanangrensis [39], indicating that there might be an additional, early segment patterning mechanism in the onychophoran embryo. Identifying this underlying mechanism would be key to understanding the evolution of segmentation in Panarthropoda (Onychophora + Tardigrada + Arthropoda).
Concerted patterning of segmental structures during onychophoran development
Our data further revealed spatiotemporal differences in the expression patterns between the median and lateral portions of the germ band in E. rowelli embryo for all four “segment polarity genes” studied. These differences are more evident in advanced developmental stages, in which the lateral domains are associated with the developing limbs, whereas the median domains correspond to the anlagen of the ventral and preventral organs [30], [42], [56]. These findings correspond to the previously published data on engrailed and wingless expression in E. kanangrensis, where the anlagen of the ventral and preventral organs (cf. refs [30], [42]) were instead interpreted as the developing nerve cords [29].
A similar correlation between the expression patterns of “segmentation genes” and organogenesis was observed recently in E. kanangrensis, in which the “pair rule gene” odd-skipped is expressed in the segmental anlagen of nephridia [39]. These findings imply that the observed expression patterns are associated with the individual segmental structures [8], [10], [46] rather than with a segment as a holistic unit [43]–[45]. This might explain why the sets of substructures comprising a ‘segment’ [9], [45] differ between onychophorans and arthropods. Therefore, we suggest that the segmental structures and organs that do not have any homologues in arthropods (and other animals; cf. Figure 1A–L) might have evolved in the onychophoran lineage, after the segmentation of the body and the corresponding patterning mechanisms responsible for a concerted positioning of such structures were already present.
Conclusions
Based on the relative position of segmental and parasegmental boundaries and similarities in the expression patterns of the segment polarity genes (Figure 13D, E), the embryonic parasegments of arthropods have been homologised with the adult segments of annelids [24], [25]. This implies that the last common ancestor of protostomes possessed parasegments, whereas definitive segments evolved in arthropods [24], [25], [103]. According to this hypothesis, one would expect that parasegments also occur in one of the closest arthropod relatives, the Onychophora [24], [29]. However, our data suggest that despite the conserved anterior-to-posterior order of expression of the segment polarity genes, the mechanisms of segment formation might be fundamentally different in Onychophora. Although gene expression studies have provided useful insights into segment formation and body patterning in various animals, the complexity and plasticity of the mechanisms involved are still poorly understood, especially in non-model organisms, such as onychophorans. The fundamental differences in segment patterning in Onychophora revealed in this and previous studies [29], [39], [104] suggest that it might be premature to speculate on the common origin of segmentation in distantly related animal groups, such as annelids and arthropods [24], [25], [105]. We caution that the similarities at the transcriptional level might be superficial due to an independent recruitment of the same canonical signalling pathways, most of which are certainly older than the origin of segmentation [106]–[108].
Supporting Information
Sequence alignments for cubitus interruptus , engrailed , hedgehog and wingless .
(PDF)
Early development of the embryonic furrows in embryos of E. rowelli . Confocal micrographs of embryos, double-labelled with the DNA marker Bisbenzimide (A, C) and the f-actin marker phalloidin-rhodamine (B, D). The images in A and C are from the same embryos as in Figure 2C and D. (A, B) Posterior end of a stage II embryo. Note that the segmental furrows have not been formed yet. (C, D) Posterior end of a stage III embryo. Arrows point to the segmental furrows in the ectoderm (in C) and between the mesodermal somites ( = coelomic cavities, marked by asterisks in D). Abbreviations: po, proctodaeum; ve, ventral extra-embryonic tissue. Scale bars: 100 µm (A–D).
(TIF)
Cladograms based on phylogenetic analyses of engrailed , cubitus interruptus , hedgehog and wingless sequences using RAxML. Numbers at nodes are maximum likelihood bootstrap values (100 replicates). Sequences of the onychophoran E. rowelli are highlighted in bold/red. For the analyses of engrailed and hedgehog phylogenies, Platynereis dumerilii was used as an outgroup. For the analysis of cubitus interruptus phylogeny, Achaearanea tepidariorum was selected as an outgroup. For the analysis of wingless, we set up an alignment of several Wnt1 and Wnt6 sequences from [109] and used Wnt6 from different taxa as an outgroup (note that the sequences from the onychophorans E. rowelli and E. kanangrensis cluster together within the Wnt1 clade).
(TIF)
Expression of wingless and hedgehog in embryos of E. rowelli . Leg segments of stage IV embryos in ventral view. Note that the hedgehog stripes are located posterior to the corresponding wingless domains. Abbreviation: le, legs. Scale bars: 100 µm (A, B).
(TIF)
List of species and genes with corresponding accession numbers used for phylogenetic analyses.
(PDF)
Acknowledgments
We are thankful to all members of the Mayer laboratory for animal husbandry, to Sandra Treffkorn for help with dissecting the embryos, to Lars Hering for providing the transcriptomic data, to Ivo de Sena Oliveira for help with macrophotography, to Christine Martin for assistance with confocal laser-scanning microscopy, to Johannes Schinko for providing protocols for double in situ hybridization, to Michael Gerth for critical discussions and to Vladimir Gross for proofreading the manuscript and providing useful and insightful comments. We gratefully acknowledge Dave M. Rowell, Ivo de Sena Oliveira, Lars Hering, Stefan Schaffer and Theresa Quaas for collecting the specimens and to Noel N. Tait for his help with permits. The staff of Forests NSW (New South Wales, Australia) is gratefully acknowledged for providing the collecting permits.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Emmy Noether Programme of the German Research Foundation (DFG, http://www.dfg.de/index.jsp) to GM (Ma 4147/3-1). The authors acknowledge support from the German Research Foundation (DFG) and Leipzig University within the program of Open Access Publishing. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhang Z-Q (2011) Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Zootaxa 3148:1–237. [DOI] [PubMed] [Google Scholar]
- 2. Zhang Z-Q (2013) Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness (Addenda 2013). Zootaxa 3703:1–82. [DOI] [PubMed] [Google Scholar]
- 3.Carroll S, Grenier JK, Weatherbee SD (2005) From DNA to Diversity. Molecular Genetics and the Evolution of Animal Design. Malden, USA: Blackwell Publishing. 258 p. [Google Scholar]
- 4. Giribet G, Edgecombe GD (2012) Reevaluating the arthropod tree of life. Annu Rev Entomol 57:167–186. [DOI] [PubMed] [Google Scholar]
- 5. Sanson B (2001) Generating patterns from fields of cells–examples from Drosophila segmentation. EMBO Rep 2:1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deutsch JS (2004) Segments and parasegments in arthropods: a functional perspective. BioEssays 26:1117–1125. [DOI] [PubMed] [Google Scholar]
- 7. Damen WGM (2007) Evolutionary conservation and divergence of the segmentation process in arthropods. Dev Dyn 236:1379–1391. [DOI] [PubMed] [Google Scholar]
- 8. Budd GE (2001) Why are arthropods segmented? Evol Dev 3:332–342. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen C (2012) Animal Evolution: Interrelationships of the Living Phyla. Oxford: Oxford University Press. 402 p. [Google Scholar]
- 10.Fusco G, Minelli A (2013) Arthropod segmentation and tagmosis. In:A. Minelli, G. Boxshall, G. Fuscoeditors. Arthropod Biology and Evolution. Springer Berlin Heidelberg. pp. 197−221.
- 11. Hughes CL, Kaufman TC (2002) Hox genes and the evolution of the arthropod body plan. Evol Dev 4:459–499. [DOI] [PubMed] [Google Scholar]
- 12. Hughes CL, Kaufman TC (2002) Exploring the myriapod body plan: expression patterns of the ten Hox genes in a centipede. Development 129:1225–1238. [DOI] [PubMed] [Google Scholar]
- 13. Martinez-Arias A, Lawrence PA (1985) Parasegments and compartments in the Drosophila embryo. Nature 313:639–642. [DOI] [PubMed] [Google Scholar]
- 14. Lawrence PA (1988) The present status of the parasegment. Development 104:61–65.2472942 [Google Scholar]
- 15. Ingham PW, Martinez-Arias A (1992) Boundaries and fields in early embryos. Cell 68:221–235. [DOI] [PubMed] [Google Scholar]
- 16. Damen WGM (2002) Parasegmental organization of the spider embryo implies that the parasegment is an evolutionary conserved entity in arthropod embryogenesis. Development 129:1239–1250. [DOI] [PubMed] [Google Scholar]
- 17. Simonnet F, Deutsch J, Queinnec E (2004) hedgehog is a segment polarity gene in a crustacean and a chelicerate. Dev Genes Evol 214:537–545. [DOI] [PubMed] [Google Scholar]
- 18. Tautz D (2004) Segmentation. Dev Cell 7:301–312. [DOI] [PubMed] [Google Scholar]
- 19. Prpic NM (2008) Parasegmental appendage allocation in annelids and arthropods and the homology of parapodia and arthropodia. Front Zool 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence PA (1992) The Making of a Fly. Oxford: Blackwell Scientific Publications. 228 p. [Google Scholar]
- 21.Campos-Ortega JA, Hartenstein V (1997) The Embryonic Development of Drosophila melanogaster. Berlin, Heidelberg, New York: Springer. 405 p. [Google Scholar]
- 22. Janssen R, Prpic NM, Damen WGM (2004) Gene expression suggests decoupled dorsal and ventral segmentation in the millipede Glomeris marginata (Myriapoda: Diplopoda). Dev Biol 268:89–104. [DOI] [PubMed] [Google Scholar]
- 23. Farzana L, Brown SJ (2008) Hedgehog signaling pathway function conserved in Tribolium segmentation. Dev Genes Evol 218:181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dray N, Tessmar-Raible K, Le Gouar M, Vibert L, Christodoulou F, et al. (2010) Hedgehog signaling regulates segment formation in the annelid Platynereis . Science 329:339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Prud’homme B, de Rosa R, Arendt D, Julien JF, Pajaziti R, et al. (2003) Arthropod-like expression patterns of engrailed and wingless in the annelid Platynereis dumerilii suggest a role in segment formation. Curr Biol 13:1876–1881. [DOI] [PubMed] [Google Scholar]
- 26. Seaver EC, Paulson DA, Irvine SQ, Martindale MQ (2001) The spatial and temporal expression of Ch-en, the engrailed gene in the polychaete Chaetopterus, does not support a role in body axis segmentation. Dev Biol 236:195–209. [DOI] [PubMed] [Google Scholar]
- 27. Kang DM, Huang F, Li DL, Shankland M, Gaffield W, et al. (2003) A hedgehog homolog regulates gut formation in leech (Helobdella). Development 130:1645–1657. [DOI] [PubMed] [Google Scholar]
- 28. Seaver EC, Yamaguchi E, Richards GS, Meyer NP (2012) Expression of the pair-rule gene homologs runt, Pax3/7, even-skipped-1 and even-skipped-2 during larval and juvenile development of the polychaete annelid Capitella teleta does not support a role in segmentation. EvoDevo 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eriksson BJ, Tait NN, Budd GE, Akam M (2009) The involvement of engrailed and wingless during segmentation in the onychophoran Euperipatoides kanangrensis (Peripatopsidae: Onychophora) (Reid 1996). Dev Genes Evol 219:249–264. [DOI] [PubMed] [Google Scholar]
- 30. Mayer G, Whitington PM (2009) Neural development in Onychophora (velvet worms) suggests a step-wise evolution of segmentation in the nervous system of Panarthropoda. Dev Biol 335:263–275. [DOI] [PubMed] [Google Scholar]
- 31. Whitington PM, Mayer G (2011) The origins of the arthropod nervous system: insights from the Onychophora. Arthropod Struct Dev 40:193–209. [DOI] [PubMed] [Google Scholar]
- 32. Mayer G, Koch M (2005) Ultrastructure and fate of the nephridial anlagen in the antennal segment of Epiperipatus biolleyi (Onychophora, Peripatidae)–evidence for the onychophoran antennae being modified legs. Arthropod Struct Dev 34:471–480. [Google Scholar]
- 33. Mayer G (2007) Metaperipatus inae sp. nov. (Onychophora: Peripatopsidae) from Chile with a novel ovarian type and dermal insemination. Zootaxa 1440:21–37. [Google Scholar]
- 34. Oliveira IS, Franke FA, Hering L, Schaffer S, Rowell DM, et al. (2012) Unexplored character diversity in Onychophora (velvet worms): a comparative study of three peripatid species. PLoS ONE 7:e51220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oliveira IS, Schaffer S, Kvartalnov PV, Galoyan EA, Palko IV, et al. (2013) A new species of Eoperipatus (Onychophora) from Vietnam reveals novel morphological characters for the South-East Asian Peripatidae. Zool Anz 252:495–510. [Google Scholar]
- 36. Mayer G (2006) Origin and differentiation of nephridia in the Onychophora provide no support for the Articulata. Zoomorphology 125:1–12. [Google Scholar]
- 37. Mayer G, Martin C, Rüdiger J, Kauschke S, Stevenson P, et al. (2013) Selective neuronal staining in tardigrades and onychophorans provides insights into the evolution of segmental ganglia in panarthropods. BMC Evol Biol 13:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oliveira IS, Mayer G (2013) Apodemes associated with limbs support serial homology of claws and jaws in onychophora (velvet worms). J Morphol 274:1180–1190. [DOI] [PubMed] [Google Scholar]
- 39. Janssen R, Budd GE (2013) Deciphering the onychophoran ‘segmentation gene cascade’: Gene expression reveals limited involvement of pair rule gene orthologs in segmentation, but a highly conserved segment polarity gene network. Dev Biol 382:224–234. [DOI] [PubMed] [Google Scholar]
- 40. Martinez-Arias A, Baker NE, Ingham PW (1988) Role of segment polarity genes in the definition and maintenance of cell states in the Drosophila embryo. Development 103:157–170. [DOI] [PubMed] [Google Scholar]
- 41. DiNardo S, Sher E, Heemskerk-Jongens J, Kassis JA, O’Farrell PH (1988) Two-tiered regulation of spatially patterned engrailed gene expression during Drosophila embryogenesis. Nature 332:604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oliveira IS, Tait N, Strübing I, Mayer G (2013) The role of ventral and preventral organs as attachment sites for segmental limb muscles in Onychophora. Front Zool 10:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goodrich ES (1897) On the relation of the arthropod head to the annelid prostomium. Q J Microsc Sci 40:247–268. [Google Scholar]
- 44. Goodrich ES (1945) The study of nephridia and genital ducts since 1895. Q J Microsc Sci 86:113–392. [PubMed] [Google Scholar]
- 45. Scholtz G (2002) The Articulata hypothesis–or what is a segment? Org Divers Evol 2:197–215. [Google Scholar]
- 46. Hannibal R, Patel N (2013) What is a segment? EvoDevo 4:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baer A, Mayer G (2012) Comparative anatomy of slime glands in Onychophora (velvet worms). J Morphol 273:1079–1088. [DOI] [PubMed] [Google Scholar]
- 48. Barclay S, Ash JE, Rowell DM (2000) Environmental factors influencing the presence and abundance of a log-dwelling invertebrate, Euperipatoides rowelli (Onychophora: Peripatopsidae). J Zool 250:425–436. [Google Scholar]
- 49. Barclay SD, Rowell DM, Ash JE (2000) Pheromonally mediated colonization patterns in the velvet worm Euperipatoides rowelli (Onychophora). J Zool 250:437–446. [Google Scholar]
- 50. Sunnucks P, Curach NC, Young A, French J, Cameron R, et al. (2000) Reproductive biology of the onychophoran Euperipatoides rowelli . J Zool 250:447–460. [Google Scholar]
- 51. Blaxter M, Sunnucks P (2011) Velvet worms. Curr Biol 21:R238–R240. [DOI] [PubMed] [Google Scholar]
- 52. Reid AL (1996) Review of the Peripatopsidae (Onychophora) in Australia, with comments on peripatopsid relationships. Invertebr Taxon 10:663–936. [Google Scholar]
- 53. Mayer G, Tait NN (2009) Position and development of oocytes in velvet worms shed light on the evolution of the ovary in Onychophora and Arthropoda. Zool J Linn Soc 157:17–33. [Google Scholar]
- 54. Mayer G, Whitington PM (2009) Velvet worm development links myriapods with chelicerates. Proc R Soc B 276:3571–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Allwood J, Gleeson D, Mayer G, Daniels S, Beggs JR, et al. (2010) Support for vicariant origins of the New Zealand Onychophora. J Biogeogr 37:669–681. [Google Scholar]
- 56. Mayer G, Kato C, Quast B, Chisholm R, Landman K, et al. (2010) Growth patterns in Onychophora (velvet worms): lack of a localised posterior proliferation zone. BMC Evol Biol 10:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayer G, Whitington PM, Sunnucks P, Pflüger H-J (2010) A revision of brain composition in Onychophora (velvet worms) suggests that the tritocerebrum evolved in arthropods. BMC Evol Biol 10. [DOI] [PMC free article] [PubMed]
- 58. Hering L, Henze MJ, Kohler M, Kelber A, Bleidorn C, et al. (2012) Opsins in Onychophora (velvet worms) suggest a single origin and subsequent diversification of visual pigments in arthropods. Mol Biol Evol 29:3451–3458. [DOI] [PubMed] [Google Scholar]
- 59. Jeffery NW, Oliveira IS, Gregory TR, Rowell DM, Mayer G (2012) Genome size and chromosome number in velvet worms (Onychophora). Genetica 140:497–504. [DOI] [PubMed] [Google Scholar]
- 60. Ou Q, Shu D, Mayer G (2012) Cambrian lobopodians and extant onychophorans provide new insights into early cephalization in Panarthropoda. Nat Commun 3:1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mayer G, Kauschke S, Rüdiger J, Stevenson PA (2013) Neural markers reveal a one-segmented head in tardigrades (water bears). PLoS ONE 8:e59090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Treffkorn S, Mayer G (2013) Expression of the decapentaplegic ortholog in embryos of the onychophoran Euperipatoides rowelli . Gene Expr Patterns 13:384–394. [DOI] [PubMed] [Google Scholar]
- 63. Martin C, Mayer G (2014) Neuronal tracing of oral nerves in a velvet worm–Implications for the evolution of the ecdysozoan brain. Front Neuroanat 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murienne J, Daniels SR, Buckley TR, Mayer G, Giribet G (2014) A living fossil tale of Pangaean biogeography. Proc R Soc B 281. [DOI] [PMC free article] [PubMed]
- 65. Baer A, Oliveira IS, Steinhagen M, Beck-Sickinger AG, Mayer G (2014) Slime protein profiling: a non-invasive tool for species identification in Onychophora (velvet worms). J Zool Syst Evol Res 52:265–272. [Google Scholar]
- 66.Mayer G, Franke FA, Treffkorn S, Gross V, Oliveira IS (in press) Onychophora. In: A. Wanninger, editor. Evolutionary Developmental Biology of Invertebrates. Berlin: Springer. ca. 65.
- 67.Mayer G (in press) Onychophora. In: A. Schmidt-Rhaesa, S. Harzsch, G. Purschke, editors. Structure and Evolution of Invertebrate Nervous Systems. Oxford Oxford University Press. ca. 20.
- 68.Murdock DJE, Gabbott SE, Mayer G, Purnell MA (in press) Decay of velvet worms (Onychophora), and bias in the fossil record of lobopodians. BMC Evol Biol. [DOI] [PMC free article] [PubMed]
- 69.Mayer G, Martin C, Oliveira IS, Franke FA, Gross V (in press) Latest anomalocaridid affinities challenged. Nature. [DOI] [PubMed]
- 70.Franke FA, Schumann I, Hering L, Mayer G (in press) Phylogenetic analysis and expression patterns of Pax genes in the onychophoran Euperipatoides rowelli reveal a novel bilaterian Pax subfamily. Evol Dev. [DOI] [PubMed]
- 71. Robson EA, Lockwood AP, Ralph R (1966) Composition of blood in Onychophora. Nature 209:533.5919597 [Google Scholar]
- 72. Walker MH, Tait NN (2004) Studies of embryonic development and the reproductive cycle in ovoviviparous Australian Onychophora (Peripatopsidae). J Zool 264:333–354. [Google Scholar]
- 73. Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Oda H, Nishimura O, Hirao Y, Tarui H, Agata K, et al. (2007) Progressive activation of Delta-Notch signaling from around the blastopore is required to set up a functional caudal lobe in the spider Achaearanea tepidariorum . Development 134:2195–2205. [DOI] [PubMed] [Google Scholar]
- 75. Kanayama M, Akiyama-Oda Y, Nishimura O, Tarui H, Agata K, et al. (2011) Travelling and splitting of a wave of hedgehog expression involved in spider-head segmentation. Nat Commun 2:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Akaike H (1974) A new look at the statistical model identification. Automatic Control, IEEE Transactions on 19:716–723. [Google Scholar]
- 79. Stamatakis A (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- 80. Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Prpic NM, Tautz D (2003) The expression of the proximodistal axis patterning genes Distal-less and dachshund in the appendages of Glomeris marginata (Myriapoda: Diplopoda) suggests a special role of these genes in patterning the head appendages. Dev Biol 260:97–112. [DOI] [PubMed] [Google Scholar]
- 82. Schinko J, Posnien N, Kittelmann S, Koniszewski N, Bucher G (2009) Single and double whole-mount in situ hybridization in red flour beetle (Tribolium) embryos. Cold Spring Harb Protoc 8:pdb.prot5258. [DOI] [PubMed] [Google Scholar]
- 83. Hughes CL, Kaufman TC (2002) Exploring myriapod segmentation: the expression patterns of even-skipped, engrailed, and wingless in a centipede. Dev Biol 247:47–61. [DOI] [PubMed] [Google Scholar]
- 84. DiNardo S, Heemskerk J, Dougan S, O’Farrell PH (1994) The making of a maggot: patterning the Drosophila embryonic epidermis. Curr Opin Genet Dev 4:529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schwager E, Schoppmeier M, Pechmann M, Damen W (2007) Duplicated Hox genes in the spider Cupiennius salei . Front Zool 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patel NH (1994) The evolution of arthropod segmentation: insights from comparisions of gene expression patterns. Development: 201–207. [PubMed]
- 87. Scholtz G (1995) Expression of the engrailed gene reveals nine putative segment-anlagen in the embryonic pleon of the freshwater crayfish Cheras destructor (Curstacea, Malacostraca, Decapoda). Biol Bull 188:157–165. [DOI] [PubMed] [Google Scholar]
- 88. Scholtz G, Dohle W (1996) Cell lineage and cell fate in crustacean embryos–a comparative approach. Int J Dev Biol 40:211–220. [PubMed] [Google Scholar]
- 89.Dohle W, Gerberding M, Hejnol A, Scholtz G (2004) Cell lineage, segment differentiation, and gene expression in crustaceans. Evolutionary Developmental Biology of Crustacea. A.A. Balkema Publishers. 95–133.
- 90. Browne WE, Price AL, Gerberding M, Patel NH (2005) Stages of embryonic development in the amphipod crustacean, Parhyale hawaiensis . Genesis 42:124–149. [DOI] [PubMed] [Google Scholar]
- 91. Nagy LM, Carroll S (1994) Conservation of wingless patterning functions in the short-germ embryos of Tribolium castaneum . Nature 367:460–463. [DOI] [PubMed] [Google Scholar]
- 92. Janssen R, Budd GE, Damen WGM, Prpic N-M (2008) Evidence for Wg-independent tergite boundary formation in the millipede Glomeris marginata . Dev Genes Evol 218:361–370. [DOI] [PubMed] [Google Scholar]
- 93. Grenier JK, Garber TL, Warren R, Whitington PM, Carroll S (1997) Evolution of the entire arthropod Hox gene set predated the origin and radiation of the onychophoran/arthropod clade. Curr Biol 7:547–553. [DOI] [PubMed] [Google Scholar]
- 94. Eriksson BJ, Tait NN, Budd GE, Janssen R, Akam M (2010) Head patterning and Hox gene expression in an onychophoran and its implications for the arthropod head problem. Dev Genes Evol 220:117–122. [DOI] [PubMed] [Google Scholar]
- 95. Janssen R, Eriksson B, Tait N, Budd G (2014) Onychophoran Hox genes and the evolution of arthropod Hox gene expression. Front Zool 11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dohle W, Scholtz G (1988) Clonal analysis of the crustacean segment–the discordance between genealogical and segmental borders. Development 104:147–160.3253055 [Google Scholar]
- 97.Mayer G, Harzsch S (2007) Immunolocalization of serotonin in Onychophora argues against segmental ganglia being an ancestral feature of arthropods. BMC Evol Biol 7. [DOI] [PMC free article] [PubMed]
- 98. Mayer G, Harzsch S (2008) Distribution of serotonin in the trunk of Metaperipatus blainvillei (Onychophora, Peripatopsidae): Implications for the evolution of the nervous system in Arthropoda. J Comp Neurol 507:1196–1208. [DOI] [PubMed] [Google Scholar]
- 99. Patel NH, Kornberg TB, Goodman CS (1989) Expression of engrailed during segmentation in grasshopper and crayfish. Development 107:201–212. [DOI] [PubMed] [Google Scholar]
- 100. Dearden PK, Donly C, Grbic M (2002) Expression of pair-rule gene homologues in a chelicerate: early patterning of the two-spotted spider mite Tetranychus urticae . Development 129:5461–5472. [DOI] [PubMed] [Google Scholar]
- 101. Piepenburg O, Vorbrüggen G, Jäckle H (2000) Drosophila segment borders result from unilateral repression of Hedgehog activity by Wingless signaling. Mol Cell 6:203–209. [PubMed] [Google Scholar]
- 102. Patel NH (1989) The role of segment polarity genes during Drosophila neurogenesis. Genes Dev 3:890–904. [DOI] [PubMed] [Google Scholar]
- 103. Saudemont A, Dray N, Hudry B, Le Gouar M, Vervoort M, et al. (2008) Complementary striped expression patterns of NK homeobox genes during segment formation in the annelid Platynereis . Dev Biol 317:430–443. [DOI] [PubMed] [Google Scholar]
- 104. Hogvall M, Schonauer A, Budd G, McGregor A, Posnien N, et al. (2014) Analysis of the Wnt gene repertoire in an onychophoran provides new insights into the evolution of segmentation. EvoDevo 5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Balavoine G, Adoutte A (2003) The segmented Urbilateria: A testable scenario. Integr Comp Biol 43:137–147. [DOI] [PubMed] [Google Scholar]
- 106. Pang K, Ryan J. Program NCS, Mullikin J, Baxevanis A, et al (2010) Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi . EvoDevo 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ryan JF, Pang K, Schnitzler CE, Nguyen A-D, Moreland RT, et al. (2013) The genome of the Ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342:1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Graham A, Butts T, Lumsden A, Kiecker C (2014) What can vertebrates tell us about segmentation? EvoDevo 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Janssen R, Le Gouar M, Pechmann M, Poulin F, Bolognesi R, et al. (2010) Conservation, loss, and redeployment of Wnt ligands in protostomes: implications for understanding the evolution of segment formation. BMC Evol Biol 10. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignments for cubitus interruptus , engrailed , hedgehog and wingless .
(PDF)
Early development of the embryonic furrows in embryos of E. rowelli . Confocal micrographs of embryos, double-labelled with the DNA marker Bisbenzimide (A, C) and the f-actin marker phalloidin-rhodamine (B, D). The images in A and C are from the same embryos as in Figure 2C and D. (A, B) Posterior end of a stage II embryo. Note that the segmental furrows have not been formed yet. (C, D) Posterior end of a stage III embryo. Arrows point to the segmental furrows in the ectoderm (in C) and between the mesodermal somites ( = coelomic cavities, marked by asterisks in D). Abbreviations: po, proctodaeum; ve, ventral extra-embryonic tissue. Scale bars: 100 µm (A–D).
(TIF)
Cladograms based on phylogenetic analyses of engrailed , cubitus interruptus , hedgehog and wingless sequences using RAxML. Numbers at nodes are maximum likelihood bootstrap values (100 replicates). Sequences of the onychophoran E. rowelli are highlighted in bold/red. For the analyses of engrailed and hedgehog phylogenies, Platynereis dumerilii was used as an outgroup. For the analysis of cubitus interruptus phylogeny, Achaearanea tepidariorum was selected as an outgroup. For the analysis of wingless, we set up an alignment of several Wnt1 and Wnt6 sequences from [109] and used Wnt6 from different taxa as an outgroup (note that the sequences from the onychophorans E. rowelli and E. kanangrensis cluster together within the Wnt1 clade).
(TIF)
Expression of wingless and hedgehog in embryos of E. rowelli . Leg segments of stage IV embryos in ventral view. Note that the hedgehog stripes are located posterior to the corresponding wingless domains. Abbreviation: le, legs. Scale bars: 100 µm (A, B).
(TIF)
List of species and genes with corresponding accession numbers used for phylogenetic analyses.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.