Abstract
The Escherichia coli (E. coli) SOS response is the largest, most complex, and best characterized bacterial network induced by DNA damage. It is controlled by a complex network involving the RecA and LexA proteins. We have previously shown that the SOS response to DNA damage is inhibited by various elements involved in the expression of the E. coli toxin-antitoxin mazEF pathway. Since the mazEF module is present on the chromosomes of most E. coli strains, here we asked: Why is the SOS response found in so many E. coli strains? Is the mazEF module present but inactive in those strains? We examined three E. coli strains used for studies of the SOS response, strains AB1932, BW25113, and MG1655. We found that each of these strains is either missing or inhibiting one of several elements involved in the expression of the mazEF-mediated death pathway. Thus, the SOS response only takes place in E. coli cells in which one or more elements of the E. coli toxin-antitoxin module mazEF or its downstream pathway is not functioning.
Introduction
The enteric bacterium E. coli, like most other bacteria, carries on its chromosome the gene pair mazEF, belonging to the abundant family of toxin-antitoxin modules [1]. mazF specifies for the stable toxin MazF [2], a sequence specific endoribonuclease, which cleaves at ACA sites [3]. mazE specifies for the labile antitoxin MazE, which is degraded by the protease ClpPA [2]. E. coli mazEF is responsible for bacterial programmed cell death (PCD) under stressful conditions [4]. Under such conditions, the induced endoribonuclease MazF removes the 3′-terminal 43 nucleotides of the 16S rRNA within the ribosomes, thereby removing the anti-Shine-Dalgarno (aSD) sequence that is required for translation initiation of canonical mRNAs. Concomitantly, MazF also cleaves at ACA sites at or closely upstream from the AUG start codon of certain specific mRNAs, causing the generation of leaderless mRNAs [5]. Thus, stressful conditions lead to the generation of the alternative translation machinery [5] which is responsible for the synthesis of stress proteins, some of which are involved in cell death and the others in cell survival [6]. Therefore, mazEF can be considered as a master regulatory element, that induces downstream pathway leading to the death of most of the population, and continued survival of a small subpopulation [6]. In addition, E. coli mazEF-mediated cell death is a population phenomenon requiring the participation of NNWNN, a linear penta-peptide, which is a quorum sensing factor that we have called the Extracellular Death Factor (EDF) [7]–[8]. EDF induces the enoribonucleolytic activity of E. coli MazF [9].
Recently, using confocal microscopy and FACS analysis we showed that under condition of sever DNA damage; the triggered EDF-mazEF-mediated cell death pathway leads to the inhibition of a second cell death pathway. The latter is an Apoptotic-Like Death that we have called ALD; ALD is mediated by recA and lexA [10]. The well known, extensively studied SOS pathway (reviewed by [11]–[15]) is also a cellular response to DNA damage, and is also mediated by recA-lexA. In an uninduced cell, the lexA gene product, LexA, acts as a repressor of more than 40 genes [16]–[17], including recA and lexA, by binding to operator sequences (called SOS box) upstream to each gene or operon. Under conditions of DNA damage, regions of single-stranded DNA are generated that convert RecA to an active form that facilitate an otherwise latent capacity of LexA (and some other proteins like UmuD and the λCI repressor) to auto digest [12], [14]–[16], [18]. We have recently shown that the E. coli EDF-mazEF pathway inhibits the SOS response as it inhibits the ALD pathway (19). Since the mazEF pathway is present on the chromosomes of most E. coli strains [20], [21], we asked why is the SOS response found in so many E. coli strains? Perhaps the EDF-mazEF pathway is present but not active in those strains?
Results
The Extra-Cellular Death Factor (EDF) is involved in the inhibition of the SOS response
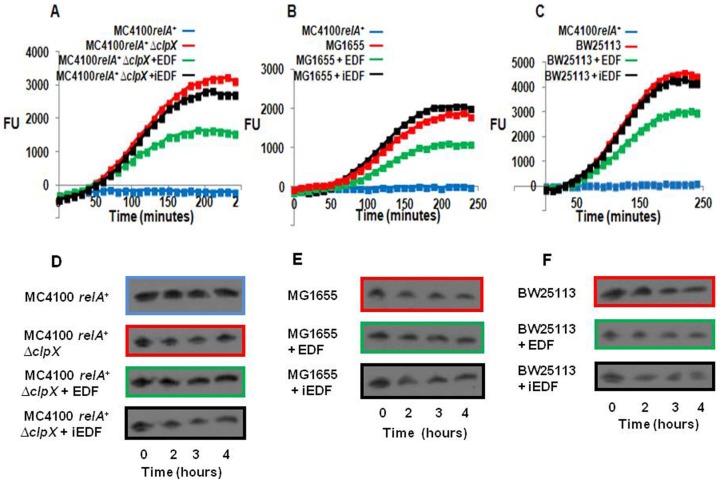
In previous studies we showed that EDF, the penta-peptide NNWNN, is involved in EDF-mazEF mediated cell death [7], and that clpX is required for the production of EDF [8]. Since, more recently we found that the action of the mazEF module prevented the SOS response [19]; here we asked if, in addition to the mazEF module, the presence of EDF is also involved in the inhibition of the SOS response. As previously [19], we also here studied the SOS response by the use of plasmid pL(lexO)-gfp [22], which bears the gene gfp under the control of the lexA operator, lexO. In this plasmid, under uninduced conditions, LexA represses gfp transcription by binding to the SOS box in the gene operator, lexO. Under DNA damage, RecA becomes activated, and acts as a co-protease stimulating the inactivation of LexA by auto-cleavage. Thus, in this system, fluorescence is a reporter for the RecA dependent SOS response. We caused DNA damage by adding nalidixic acid (NA) (10 µg/ml) to the cultures [19]. Our experiments have revealed that the SOS response was permitted not only in an E. coli MC4100relA + strain from which we deleted mazEF (MC4100relA +ΔmazEF) [19], but also when, instead of deleting mazEF, we deleted clpX (MC4100relA +ΔclpX) (Figure 1A). This effect seems to be due to the lack of EDF because: (a) the addition of EDF partially inhibits the studied SOS response (by 50%), and (b) the SOS response is not affected at all by the addition of iEDF (Figure 1A), the penta-peptide NNGNN, in which the central and crucial tryptophan has been replaced by glycine [8]. Adding iEDF to the MC4100relA +ΔclpX culture did not affect the SOS response at all (Figure 1A). Similar results were obtained when instead of studying the SOS response by the use of plasmid pL(lexO)-gfp, we studied it by following the NA-induced LexA degradation (Figure 1D). Here, LexA degradation, thus SOS response is not permitted in E. coli MC4100relA + strain carrying the mazEF module (Figure1D, first line). However, deleting clpX (MC4100relA +ΔclpX) permitted LexA degradation (Figure 1D, second line). On the other hand, the addition of EDF prevented LexA degradation (Figure 1D, third line), while LexA degradation is permitted by the addition of iEDF (Figure 1D, fourth line).
Figure 1. The inhibition of the SOS response by the mazEF pathway required the participation of EDF.
We determined the SOS response by measuring the fluorescence of the reporter plasmid pL(lexO)-gfp (A, B, C), and by LexA degradation (D, E, F). We compared E. coli strain MC4100relA + (A and D) with strains MC4100relA +ΔclpX (A and D), MG1655 (B and E), or BW25113 (C and F); the strains in A, B, and C harbored plasmid pL(lexO)-gfp. We grew the cells in M9 media supplemented with ampicillin (100 µg/ml), with shaking. When the culture reached O.D.600 0.5–0.6, we added (or not) EDF (10 ng/ml) or iEDF (100 ng/ml). These cultures were incubated without shaking at 37°C for 30 min, after which we added NA (10 µg/ml) to each sample. Immediately after adding NA, we measured fluorescence (FU) by fluorometer or LexA degradation (as described in Materials and Methods) over a period of 4 hours. The values shown are relative to those of cells that had not been treated with NA. All data are representative of three independent experiments. The colors surrounding the blots in D, E, and F correspond to the colors representing the samples in A, B, and C.
An additional support that EDF is involved in the mazEF mediated inhibition of the SOS response is derived from our studies with E. coli strain MG1655. In our previous work, we showed that E. coli strain MG1655, which carries the mazEF gene pair is defective in the production of and the response to EDF [8]. Here we found that, despite the presence of mazEF, the SOS response took place in strain MG1655 (Figure 1B). Furthermore, 240 minutes after adding EDF, we observed a 50% reduction in the SOS response; in contrast, adding iEDF did not cause any reduction in the SOS response (Figure 1B). Similar results were also obtained in E. coli strain MG1655 by studying the NA-induced LexA degradation (Figure 1E) under the SOS response condition. Also here, LexA degradation, thus the SOS response is permitted in E. coli MG1655 (Figure 1E, first line). On the other hand, the addition of EDF significantly prevented LexA degradation (Figure 1E, second line), while LexA degradation is again permitted by the addition of iEDF (Figure 1E, third line). All of these results support our hypothesis that the SOS response was permitted in the absence of EDF.
Using our fluorescence reporter system, we tested the SOS response in two additional E. coli strains. In strain AB1932 [23] the addition of EDF did not inhibit the SOS response (Figure S1). However, in E. coli strain BW25113, which has commonly been used to study the phenomena of the SOS response [23]–[24], the addition of EDF did reduce the SOS response (Figure 1C). Adding EDF to E. coli strain BW25113 led to a 30% reduction in the SOS response; again, as in the case for strains MC4100relA+ΔclpX (Figure 1A), and MG1655 (Figure 1B), adding iEDF did not lead to a reduction in the SOS response (Figure 1C). Similar results were obtained in E.coli strain BW25113 by determining the NA-induced LexA degradation. Also here, LexA degradation is permitted (Figure 1F, first line). However, its degradation is prevented by the addition of EDF (Figure 1F, second line), but not by the addition of iEDF (Figure 1F, third line). Thus, the SOS response is permitted in E.coli strains BW25113 and MG1655. This because of unknown reasons there is a deficiency of EDF in these strains (Table 1).
Table 1. The identified elements related to the EDF-mazEF pathway that permitted the SOS response in the herein studied E. coli strains.
| Strains | The elements studied | |
| EDF | λ lysogen | |
| MG1655 | [−] | [−] |
| BW25113 | [−] | [−] |
| MC4100relA +λ | [+] | [+] |
| AB1932λ | [+] | [+] |
Brackets indicate the element that was missing [−] or present [+] that permitted the SOS response in each of these strains.
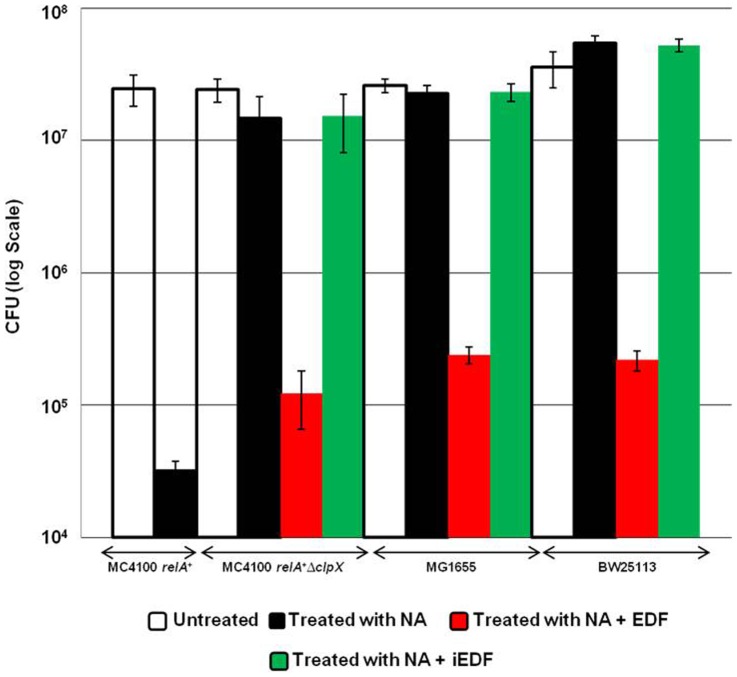
Our findings suggesting that the SOS response is permitted in E.coli strains due to the lack of EDF is also manifested by testing the viability of these strains under conditions of DNA damage (Figure 2). In E.coli strain MC4100relA +, in which the SOS responds is prevented (Figure 1A and 1D), cell viability is reduced by about 3 folds under such condition (Figure 2). In contrast, in the ΔclpX derivative of MC4100relA+ which is defective in the generation of EDF (Figure 1A and 1D), cell viability is not affected under the SOS conditions (Figure 2). However, by applying EDF cell survival is reduced by about 2 folds, and this reduction in viability does not occur by applying iEDF (Figure 2). A similar manifestation on the effect of DNA damage on cell viability was also observed in E.coli strains MG1655 and BW25113. These strains which are permitting the SOS response due to their lack of EDF (Figures 1B, 1E and Figure 1C, 1F, respectively), also permit cell survival under the SOS conditions (Figure 2). However, cell viability was reduced by about two folds when EDF was applied to these stains, and this reduction in viability does not occur by applying iEDF (Figure 2).
Figure 2. Cell viability is affected under DNA damage in E.coli strains and under conditions in which the SOS response is impaired.
We studied the effect of cell viability at low concentrations of Nalidixic acid (10 µg/ml). E. coli strains WT MC4100relA + or MC4100relA +ΔclpX or MG1655 or BW25113 were grown to OD600 0.5. We divided each culture into aliquots to which we added no NA (untreated) or added NA 10 µg/ml (treated with NA) or added NA with EDF (treated with NA + EDF) or added NA with iEDF (treated with NA + iEDF) as described in the Legend to Figure 1. We carried out viability assays on LB plates as we have described previously (19).
The SOS response is permitted in E. coli strains carrying prophage lambda
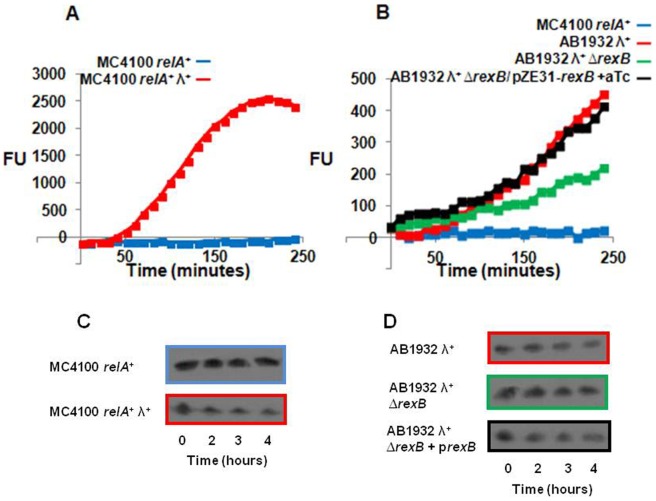
One of the few genes expressed by phage λ in its lysogenic state is λrexB [25]–[27]. In previous work, we showed that its product, λRexB, inhibits the degradation of the antitoxic labile compound, MazE, thereby preventing mazF mediated death pathway [28]. Therefore, we anticipated that, in contrast to E. coli strain MC4100relA + in which the SOS response is prevented (Figure 1A and 1D), in the presence of a λ prophage the SOS response would be permitted in this strain. As we expected, the presence of the λ prophage overcame the inhibitory effect of mazEF on the SOS response (Figure 3A). Similar results were obtained in E. coli strain MC4100relA + when instead of studying the SOS response by the use of plasmid pL(lexO)-gfp, we studied it by following LexA degradation (Figure 3C). Here, the NA-induced LexA degradation, thus the SOS response, is permitted in an E. coli strain carrying prophage λ, MC4100relA +λ (Figure 3C, second line), and not in the absence of the prophage from the chromosome of this bacterial strain (Figure 3C, first line). Also, in E.coli strain AB1932, in which the SOS response has been observed [23], and which has been reported to bear a λ prophage on its chromosome [23], we observed the SOS response both by the use of plasmid pL(lexO)-gfp (Figure 3B) and by following LexA degradation (Figure 3D, first line). Furthermore, deleting rexB from its λ prophage reduced the SOS response by 50%, while introducing a plasmid bearing λrexB and inducing it permitted the SOS response (Figure 3B). In addition, deleting rexB from its λ prophage stabilized LexA degradation (Figure 3D, second line), while introducing a plasmid bearing λrexB and inducing it permitted LexA degradation (Figure 3D, third line), Thus, our results provide an explanation for the SOS response in strain AB1932λ (Table 1).
Figure 3. λ Lysogens overcome the inhibitory effect of mazEF on the SOS response.
We determined the SOS response by measuring the fluorescence of the reporter plasmid pL(lexO)-gfp (A, B), and by LexA degradation (C, D). We used E. coli MC4100relA + as a control strain, and two experimental parent strains lysogenized by phage λ: MC4100relA + λ + (A, C), and AB1932λ +, AB1932λ +ΔrexB, AB1932λ +ΔrexB/pZA31-rexB (B, D). The strains in A and B harbored plasmid pL(lexO)-gfp. Cells were grown as described in the Legend to Figure 1, except that the ampicillin concentration was (25 µg/ml), and chloramphenicol (6.25 µg/ml) was added to cells harboring plasmid pZE31-rexB. At O.D.600 0.5–0.6, the strains harboring plasmid pZE31-rexB were induced by the addition of aTc (0.5 µg/ml), and incubated without shaking at 37°C for 30 min after which we added NA (10 µg/ml). We measured fluorescence (FU) (A and B) by fluorometer or LexA degradation (C and D) over a period of 4 hours. The values shown are relative to those of cells not treated by NA. All data are representative of three independent experiments. The colors surrounding the gels in C and D correspond to the colors representing the samples in A and B.
Our findings suggesting that the SOS response is permitted in an E.coli strain carrying prophage lambda was further supported by testing the viability of strain AB1932λ under conditions of DNA damage (Figure 4A). In this strain, in which we have shown that the SOS responds is permitted due to the presence of λrexB gene (Figures 3B and 3D), We found that also cell viability is reduced by about two folds under conditions of DNA damage (Figure 4A). In contrast, in the ΔrexB derivative of strain AB1932λ in which the SOS response is severely affected (Figures 3B and 3D); cell viability is not affected under the SOS conditions (Figure 4A). However, introducing a plasmid bearing λrexB and inducing it, lead to the reduction of cell viability by about two orders (Figure 4A).
Figure 4. λrexB is involved in the transfer from lysogenic to lytic stage.
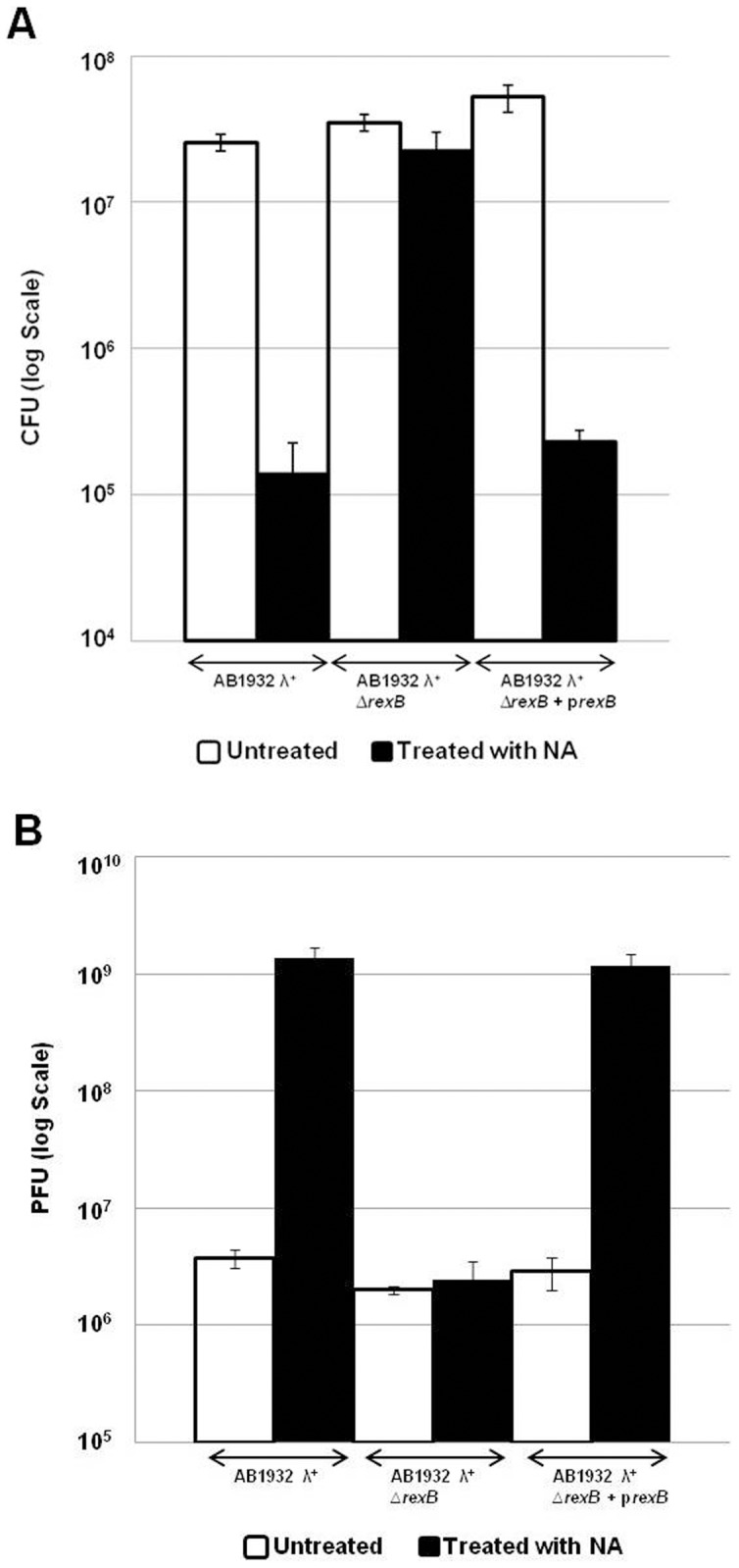
(A) Cell viability (CFU/ml) and (B) λ plaque forming units (PFU/ml). The E. coli strains AB1932λ + or AB1932λ +ΔrexB or AB1932λ +ΔrexB/pZA31-rexB were grown as described in the Legend to Figure 2. We carried out viability assays on LB plates as we have described previously (19) and the PFU were determined as described in Materials and Methods.
Furthermore, as expected, the observed reduction in cell viability by applying DNA damaging conditions to E.coli strain AB1932 λ carrying the rexB gene (Figure 4A) is due to the transfer of the phage from its lysogenic to its lytic stage (Figure 4B). This is here shown by the determination of the plaque forming ability of the phage which we found to be 109 PFU/ml when the λ lysogens carried rexB or when λ rexB was carried on a plasmid, and 106 PFU/ml in the absence of λrexB (Figure 4B). Such a result is an outcome of the process of the induction of prophage λ which is a classical manifestation of the SOS response (see under Discussion).
Discussion
The bacterial response to DNA damage was first described by Radman in the 70′s, who called it the “SOS response” [13]. To date, it is the largest, most complex, and best understood bacterial DNA damage-inducible network to be characterized [11]–[15], [18]. The expression of genes in the SOS regulatory network is controlled by a complex circuit involving the RecA and LexA proteins [15]. In response to an SOS-inducing treatment or conditions, regions of single-stranded DNA are generated. The binding of RecA to these regions of single-stranded DNA in the presence of nucleotide triphosphate generates a nucleotide filament, and as a result converts RecA to an active form that facilitates an otherwise latent capacity of LexA (and some other proteins like UmuD and the λ repressor CI) to undergo self-cleavage [12], [14]–[16], [18].
We have previously shown that the EDF-mazEF mediated death pathway inhibits the SOS response (19). This inhibition was caused by the expression of mazF itself, as well as the genes yfbU, slyD, yfiD, clpP and ygcR (19) specifying for proteins acting in the death pathway downstream to mazEF [6]. Moreover, as we have shown here the extra-cellular death factor EDF, the peptide NNWNN, is also involved in the inhibition of the SOS response (Figure 1). These results suggested that E. coli strains commonly used in studies of the SOS response may be defective in the expression of the EDF-mazEF pathway. Here we studied the following E. coli strains used in SOS response studies: AB1932 [23], BW25113 [24], [29] and MG1655 [16], [23]. We found that although these strains are carrying the mazEF module in their chromosome (Figure S2), they were missing or inhibiting other elements of the EDF-mazEF mediated death pathway (summarized in Table 1). Strains BW25113 and MG1655 are defective in the production of EDF (Figure 1 and Table 1). The SOS response was also studied in E.coli strain MC4100relA1 [22] which is defective in the production of the starvation signaling molecule ppGpp [30]. We have previously shown that this strain indeed permits the SOS response [19], probably because ppGpp triggers the mazEF pathway [2].
Finally, among the E. coli strains commonly used in studies of the SOS response, the most interesting strain studied here is AB1932. Our results indicate that this strain is defective in the mazEF-mediated pathway because of being a λ lysogen (Figure 3B, 3D and Table 1), that carries the rexB gene of the phage [23]. Moreover, strain MC4100relA+ in which the SOS response is inhibited (Figure 3A and the first line of 3C), enabled the SOS response when lysogenised with phage λ carrying rexB (Figure 3A and second line of 3B). We have previously reported that λRexB, the product of the λrexB gene, prevents the degradation of the antitoxin MazE [28]. Thus, it is expected that in the presence of λRexB, MazF activity is prevented and therefore the SOS response is permitted. Indeed, here we found that, in strain AB1932λ (Figure 3B and 3D), simply deleting the λrexB gene significantly reduced the SOS response. Moreover, we found that λrexB is involved in the transfer from the lysogenic to the lytic stage of the phage (Figure 4B). This result is a self evident outcome of the process of λ phage induction which is a classical manifestation of the SOS response. Under conditions of DNA damage, RecA is converted to an active form that facilitates an otherwise latent capacity of λCI repressor to auto digest [18]. Thereby λ phage can be transferred from its lysogenic to its lytic stage. Thus, the product of λrexB gene that prevents the degradation of the antitoxin MazE [28], and thereby prevents MazF activity, permits in turn the SOS response (Figure 3), Therefore, λRexB is a crucial factor that enables the transfer of phage λ from its lysogenic to its lytic stage.
Furthermore, our herein described study, showing that the SOS response was discovered and is permitted due to the use of E. coli strains deficient in the expression of the EDF-mazEF mediated pathway, is a striking example for the reason of different experimental results obtained by the use of different bacterial and animal strains during research of a specific biological phenomena, and thereby belongs to the history of genetic studies. Furthermore, our herein results on, the SOS response to DNA damage in E. coli, reflects the complexity of the interplay between cellular networks, and as such reflects the importance of personalized medicine in general, and specifically in the use of antibiotics due to the expected diversity of individual microbiota.
Materials and Methods
Bacterial strains and plasmids
We used the following E. coli strains: MC4100relA + (WT) [28] and its derivatives MC4100relA +ΔmazEF [28], MC4100relA +ΔclpX [8. We also used strain MG1655 that we have found previously to be defective in EDF production [8]. For testing E. coli strains that are commonly used for SOS studies, we used the following strains (obtained from the E. coli Stock Center, CGSC, at Yale University in New Haven, Connecticut, USA): AB1932 (λ+, F−, argE3 or argH1, metA28, lacY1 or lacZ4, thi-1, xyl-5 or xyl-7, galK2, tsx-6) {23], and BW25113 Δ(araDaraB)567,ΔlacZ4787(::rrn3),rph1,Δ(rhaDrhaB)568,hsdR514lacIqrrnBT14ΔlacZWJ16hsdR514ΔaraBADAH33ΔrhaBADLD78), [24], [29]. We constructed E. coli MC4100relA + λ+ by its lysogenization from strain MC4100relA1λ+, which was kindly provided by Dr. Ilan Rosenstein. We also constructed E. coli strain AB1932λΔrexB, by the use of Datsenko method [31]. For our fluorescence measurements, we used plasmid pL(lexO)-gfp [22], which carries a ampR and thus confers resistance to ampicillin, and which was kindly provided by Dr. Lyle A. Simmons. We constructed plasmid pZE31-rexB in which the promoter is inducible by aTc (anhydrotetracycline) [32].
Materials and Media
We grew the various E. coli strains in liquid M9 minimal medium containing 1% glucose and a mixture of 2 mg/ml of each of the essential amino-acids excluding tyrosine and cysteine. Synthetic EDF (NNWNN) and synthetic iEDF (NNGNN) were purchased from GeneScript Corp (Piscataway, NJ, USA).
Growth conditions
For the experiments in which we measured fluorescence, we diluted over-night cultures harboring plasmid pL(lexO)-gfp [22] 1∶100 in 10 ml of M9 minimal medium, with 100 µg/ml of ampicillin. We grew the cells at 37°C, with shaking (220 rpm), to O.D600 0.5–0.6. We divided each culture into 500 µl aliquots, and to each aliquot we added the appropriate antibiotics and/or EDF at the concentrations described in the figure legends. These samples were used for fluorescence assays as described below.
Cell viability
For the experiments on cell viability, we grew cells in 10 ml M9 minimal medium to an optical density at 600 nm (OD600) of 0.5 to 0.6. Then, we divided each culture into 500-µl aliquots to which we added the appropriate concentration of NA. We incubated each aliquot at 37°C for 4 h (or as described in the figure legends) and then washed them twice with phosphate-buffered saline (PBS) (pH 7.2). We carried out viability assays on LB plates as we have described previously (6).
Plaque assay
We grew cells in 10 ml M9 minimal medium with or without antibiotic, as described above, and treated them with appropriate concentration of NA for 4 h (or as described in the figure legends). The supernatants were collected, serially diluted and mixed with soft agar containing TG1 cells. Then, the mixtures were poured on LB agar plates. The plates were incubated at 37°C for overnight and plaque forming units (PFU) were calculated by counting plagues.
Fluorescence measurements
From the 500 µl aliquot samples of the growing E. coli cultures harboring plasmid pL(lexO)-gfp treated with EDF we placed 250 µl samples into each well of a 96 well plate. In each well, using a 485±15 nm excitation filter and a 530±15 nm emission filter, we measured the fluorescence 25 times at intervals of 10 min. The fluorophore was excited with 1000 CW lamp energy, and the fluorescence in each well was measured for 1 s (FLUOstar galaxy, BMG Labtechnologies).
Determining the LexA degradation by Western blot analysis
We grew cells in 10 ml M9 minimal medium, as described above, and treated them with appropriate concentration of NA for different periods of time as described in the figure legends. To lyse the cells, we centrifuged samples for 2 min and then suspended the cell pellets in 50 µl of Bugbuster master mix (Novagen), incubating them with vigorous shaking at room temperature for 10 min. Then, we centrifuged them at 4°C for 10 min and transferred the supernatants containing the crude extract to fresh tubes. We determined protein concentrations using the Bradford assay (Bio-Rad, Hercules, CA, USA). For Western blot analysis, we used rabbit polyclonal antibody to the LexA DNA binding region as the primary antibody (Abcam) and donkey polyclonal antibody to rabbit IgG (horseradish peroxidase [HRP]) as the secondary antibody (Abcam).
Supporting Information
In strain AB1932 the addition of EDF did not inhibit the SOS response. We compared E. coli strain MC4100relA + with strain AB1932; it harbored plasmid pL(lexO)-gfp. We grew the cells as described in the legend to Figure 1. When the culture reached O.D.600 0.5–0.6, we added (or not) EDF (10 ng/ml). These cultures were incubated without shaking at 37°C for 30 min, after which we added NA (10µg/ml) to each sample. Immediately after adding NA, we measured fluorescence (FU) by fluorometer over a period of 4 hours. The values shown are relative to those of cells that had not been treated with NA. All data are representative of three independent experiments.
(TIF)
E. coli strains commonly used for SOS studies bear the mazEF module on their chromosomes. Using two primers, (i) forward primer-GCCGAAATTTGCTCGTATCT and (ii) reverse primer-CTGAAAATTGCGGGTCTGTC, we performed PCR to detect the mazEF module in four E. coli strains: (1) MC4100relA +, (2) MC4100relA +ΔmazEF, (3) BW25113, (4) AB1932.
(TIF)
Acknowledgments
We thank F. R. Warshaw-Dadon (Jerusalem, Israel) for her critical reading of the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Israel Science Foundation (grant 66/10) and the United States Army (grant W911NF-09-1-0212). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Engelberg-Kulka H, Hazan R, Amitai S (2005) mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. of Cell Science 118:4327–4332. [DOI] [PubMed] [Google Scholar]
- 2. Aizenman E, Engelberg-Kulka H, Glaser G (1996) An Escherichia coli chromosomal “addiction module” regulated by 3′, 5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA. 93:6059–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, et al. (2003) MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell. 12:913–923. [DOI] [PubMed] [Google Scholar]
- 4. Hazan R, Sat B, Engelberg-Kulka H (2003) Escherichia coli mazEF- mediated cell death is triggered by various stressful conditions. J. of Bacteriol. 186:3663–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, et al. (2011) Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli . Cell 14:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amitai S, Kolodkin-Gal I, Hannanya-Meltabashi M, Shacher A, Engelberg-Kulka H (2009) Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genetics 5(3):e1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H (2007) A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli . Science 318:652–655. [DOI] [PubMed] [Google Scholar]
- 8. Kolodkin-Gal I, Engelberg-Kulka H (2008) The extracellular death factor: physiological and genetic factors influencing its production and response in Escherichia coli . J. of Bacteriol. 190:3169–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belitsky M, Avshalom H, Erental A, Yelin I, Kumar S, et al. (2011) The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxin MazF and ChpBK. Mol. Cell. 41:625–635. [DOI] [PubMed] [Google Scholar]
- 10. Erental A, Sharon I, Engelberg-Kulka H (2012) Two programmed cell death systems: an apoptotic-like death is inhibited by the mazEF mediated death pathway. PLoS Biology 10(3):e1001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hersh MN, Ponder RG, Hastings PJ, Rosenberg SM (2004) Adaptive mutation and amplification in Escherichia coli: two pathways of genome adaptation under stress. Res Microbiol. 155:352–359. [DOI] [PubMed] [Google Scholar]
- 12. Little JW (1991) Mechanism of specific LexA cleavage: Autodigestion and the role of RecA coprotease. Biochimie. 74:411–421. [DOI] [PubMed] [Google Scholar]
- 13. Radman M (1975) SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Molecular mechanisms for repair of DNA 5A:355–367. [DOI] [PubMed] [Google Scholar]
- 14. Sutton MD, Smith BT, Godoy VG, Walker GC (2000) The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479–497. [DOI] [PubMed] [Google Scholar]
- 15.Walker GC (1996) Escherichia coli and Salmonella- Cellular and molecular biology; 2th edition. Washington D.C: ASM press, pp. 1400–1415. [Google Scholar]
- 16. Courcella J, Khodursky A, Peter B, Brown PO, Hanawalt PC (2001) Comperative gene expression profiles following UV exposure in wild-type and SOS -deficient Escherichia coli . Genetics 158:41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fernandez DH, Tomoo O, Sayura A, David C, Jeffrey JH, et al. (2000) Identification of additional genes belonging to the LexA regulon in Escherichia coli . Mol Microbiol. 35:1560–1572. [DOI] [PubMed] [Google Scholar]
- 18. Little JW (1984) Autodigestion of lexA and phage λ repressors. Proc Natl Acad Sci USA 81:1375–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erental A, Kalderon Z, Saada A, Smith Y, Engelberg-Kulka H (2014) Apoptotic-LikeDeath, An Extreme SOS Response in Escherichia coli . Mbio 5(4):e01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mittenhuber G (1999) Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J. Mol. Microbiol. Biothechnol. 1:295–1302. [PubMed] [Google Scholar]
- 21. Pandey DP, Gerdes K (2005) Toxin-antitoxin loci are highly abundant in free living but lost from host-associated prokaryotes. Nuc. Acids Res. 33:966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davies BW, Kohanski MA, Simmons LA, Winkler JA, Collins JJ, et al. (2009) Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli . Molecular Cell 36:845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mount DW, Low KB, Edmeston JS (1972) Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet light-induced mutations. Journal of Bacteriology 112:886–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beaber JW, Hochhut B, Waldor MK (2004) SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. [DOI] [PubMed] [Google Scholar]
- 25. Landsman J, Kroger M, Hobom G (1982) The rex region of bacteriophage lambda: two genes under three-way control. Gene 20:11–24. [DOI] [PubMed] [Google Scholar]
- 26. Hayes S, Szybalski W (1973) Control of short leftward transcripts from the immunity and ori regions in induced coliphage lambda. Mol Gen Genet 126:257–290. [DOI] [PubMed] [Google Scholar]
- 27. Belfort M (1978) Anomalous behavior of bacteriophage lambda polypeptides in polyacrylamide gels: resolution, identification, and control of the lambda rex gene product. J Virol. 28:270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Engelberg-Kulka H, Reches M, Narasimhan M, Schoulaker-Schwarz R, Klemes Y, et al. (1998) rexB of bacteriophage lambda is an anti-cell death gene. Proc. Natl. Acad. Sci USA 26:15481–15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moolenaar GF, Van Rossum-Fikkert S, Van Kesteren M, Goosen N (2002) Cho, a second endonuclease involved in Escherichia coli nucleotide excision repair Proc Natl Acad Sci USA. 99:1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Metzger S, Schrelber G, Aizenman E, Cashel M, Glaser G (1989) Characterization of the relA mutation and a comparison of relA1 with new relA Null alleles in Escherichia coli . J. of Biological Chemistry 264:21146–21152. [PubMed] [Google Scholar]
- 31. Datsenko A, Wanner LB (2005) One- step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci USA 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bujard H, Lutz R (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1–I2 regulatory elements Nucleic Acids Research. 25:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In strain AB1932 the addition of EDF did not inhibit the SOS response. We compared E. coli strain MC4100relA + with strain AB1932; it harbored plasmid pL(lexO)-gfp. We grew the cells as described in the legend to Figure 1. When the culture reached O.D.600 0.5–0.6, we added (or not) EDF (10 ng/ml). These cultures were incubated without shaking at 37°C for 30 min, after which we added NA (10µg/ml) to each sample. Immediately after adding NA, we measured fluorescence (FU) by fluorometer over a period of 4 hours. The values shown are relative to those of cells that had not been treated with NA. All data are representative of three independent experiments.
(TIF)
E. coli strains commonly used for SOS studies bear the mazEF module on their chromosomes. Using two primers, (i) forward primer-GCCGAAATTTGCTCGTATCT and (ii) reverse primer-CTGAAAATTGCGGGTCTGTC, we performed PCR to detect the mazEF module in four E. coli strains: (1) MC4100relA +, (2) MC4100relA +ΔmazEF, (3) BW25113, (4) AB1932.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.