Abstract
The hereditary autosomal recessive disease ataxia telangiectasia (A-T) is caused by mutation in the DNA damage kinase ATM. ATM’s main function is to orchestrate DNA repair, thereby maintaining genomic stability. ATM activity is increased in response to several stimuli, including ionising radiation (IR) and hypotonic stress. DNMT1-associated protein 1 (DMAP1) is a member of the TIP60-p400 histone acetyl transferase (HAT) complex, which acetylates histone H4 at lysine 16 (H4K16) to affect chromatin relaxation and modulate ATM activation. Here we demonstrate that DMAP1 is required for both modes of ATM activation. Knockdown of DMAP1 impaired IR-induced ATM activation and consequently resulted in radiosensitivity and impaired the G2/M checkpoint. Moreover, DMAP1 was also required for efficient ATM signalling in response to hypotonic stress. Overexpression of DMAP1 increased IR-induced ATM substrate phosphorylation, suggesting that DMAP1 function is rate limiting for ATM signalling. DMAP1 associated with TIP60-dependent HAT activity, and depletion of DMAP1 reduced H4K16 acetylation in response to DNA damage. Treatment with histone deacetylase inhibitors rescued IR-induced ATM signalling in Dmap1-depleted cells. These results suggest that DMAP1 is a critical regulator of ATM activity and function.
Keywords: DMAP1, ATM, TIP60, double-strand break
Introduction
ATM is inactivated in the genomic instability disorder ataxia telangiectasia (A-T). A-T is characterised by neurodegeneration, cancer predisposition, premature ageing, immunodeficiency and genomic instability (1, 2). Cells from A-T patients exhibit chromosomal instability, premature senescence and severe sensitivity to agents that induce DNA double-strand breaks (DSBs) (3, 4).
ATM is a member of the phosphoinositide 3-kinase-related protein kinase family that includes ataxia telangiectasia and Rad3-related (ATR) and the catalytic subunit of DNA-dependent protein kinase (5, 6). DNA damage leads to the activation of ATM, which subsequently coordinates the cellular DNA damage response by phosphorylating numerous substrates (7). ATM functions to ensure cell survival by halting the cell cycle while the damage is repaired (8). Alternatively, in the presence of excessive damage, ATM signalling can trigger the activation of programmed cell death to maintain organismal integrity.
Following the induction of DSBs, ATM activation is likely to involve the sensing of changes in chromatin structure and consequent autoactivation of the kinase (9). This is followed by the transition of ATM from an inactive dimer to an active monomeric form and concomitant autophosphorylation of several amino acid residues, including serine 1981 (or the orthologous serine 1987 in mice). Monomeric phosphorylated ATM is then bound by Nijmegen breakage syndrome protein 1 (NBS1), a component of the MRN complex (consisting of MRE11, Rad50 and NBS1). The MRN complex recruits ATM to DSBs, where ATM phosphorylates multiple proteins at serine or threonine residues followed by glutamine (the ‘SQ/TQ’ motif) (10-13).
Chromatin modifications play an important role in ATM signalling. Specifically, acetylation of histone H4 at lysine 16 (H4K16Ac) was shown to affect chromatin relaxation and modulate ATM activation and the DNA damage response (14). The tumour suppressor TIP60 is a member of the MYST family of histone acetyltransferases. Along with its cofactor TTRAP (TRAF and TNF receptor associated protein), it is responsible for DNA damage-induced hyperacetylation of H4, which promotes the chromatin decondensation required for DNA repair (15).
DNMT1-associated protein 1 (DMAP1) was initially identified as a protein associated with the N-terminal domain of DNMT1, and was shown to function as a transcriptional corepressor by interacting with histone deacetylase 2 (16). Later, DMAP1 was biochemically identified as a component of the TIP60-p400 histone acetyltransferase complex (17, 18). However, the significance of DMAP1 for TIP60 function has not been further investigated.
Human DMAP1 and its yeast homolog Eaf2 contain a highly conserved SANT (SWI3–ADA2–NcoR–TFIIIB) domain, which mediates histone tail binding (19, 20). It was previously shown that Dmap1 knockdown in mouse embryonic fibroblasts (MEFs) led to spontaneous double-strand breaks (DSBs) (21). However, the mechanistic basis for DMAP1 function in DNA repair remains largely unknown in mammals.
Here we demonstrate that DMAP1 is required for TIP60 function in the DNA damage response. Depletion of DMAP1 results in impaired ATM signalling, corroborating the notion that histone acetylation is a fundamental and crucial requirement for ATM function.
Results
DMAP1 is required for ATM signalling and function in response to IR
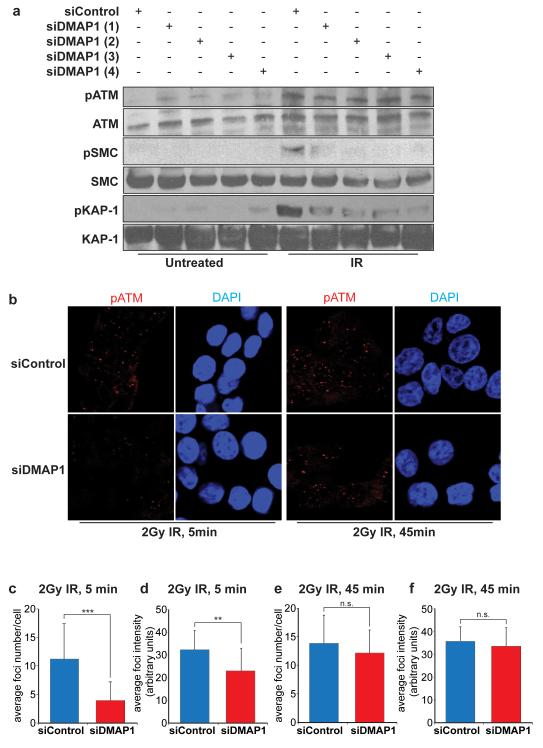
In order to understand the physiological role of DMAP1, we depleted DMAP1 function using 4 independent siRNA hairpins. DMAP1 mRNA levels were significantly reduced by each individual siRNA (Supplementary Figure 1a) and loss of DMAP1 did not adversely affect the cell cycle (Supplementary Figures 1b and c). In response to IR, DMAP1 depletion led to a substantial decrease in phosphorylation of the ATM substrates pS824-Kap1 and pS957-SMC1 (Figure 1a). These results suggest that DMAP1 is required for ATM signalling induced by DSBs.
Figure 1. DMAP1 is required for ATM signalling after IR.
(a) HCT-116 cells were transfected with control siRNA or one of four different siRNAs against DMAP1. 48 hours later cells were treated with 5 Gy IR, harvested after one hour and whole cell lysates separated and immunostained for pATM, ATM, pSMC1, SMC1, pKAP1 and KAP1. (b) Cells were treated with a pool of DMAP1 siRNAs, and after 48 hours treated with 2 Gy IR, fixed at the indicated time points and immunofluorescently stained for pATM. The mean number of foci per cell was quantified in (c, e). The pATM foci intensity was measured for at least 250 foci per experiment (d, f). The p-value of three independent experiments was calculated using the student’s t-test (***p<0.001, **p<0.01, n.s. not significant).
In order to study the role of DMAP1 in ATM signalling more closely, we investigated irradiation-induced foci (IRIF) of DNA damage signalling components. Knockdown of DMAP1 reduced IRIF of active ATM phosphorylated at serine 1981 (pATM) 5 minutes after IR treatment. Both the number of pATM foci per cell (Figures 1b and c) and the average intensity of these foci (Figures 1b and d) were significantly reduced in DMAP1-depleted cells. A similar reduction was observed for irradiation-induced 53BP1 foci in DMAP1-depleted compared with siRNA control cells (Supplementary Figure 2). At a later time point (45 minutes), no significant differences were observed, perhaps due to the actions of residual DMAP1 protein (Figures 1e and f, and Supplementary Figure 2).
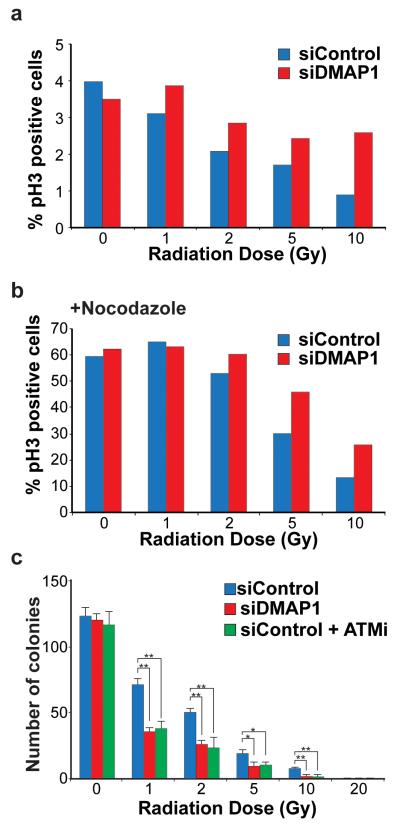
ATM function is required for cell cycle checkpoints, including the G2/M checkpoint in response to IR (22). We monitored the tendency of cells to escape the G2/M checkpoint after IR treatment and enter mitosis, indicated by immunofluorescent detection of phosphorylated histone H3 by fluorescence-activated cell sorting. The experiment was performed with and without the mitotic inhibitor nocodazole, to arrest cells that had passed through the G2/M checkpoint in mitosis. A549 cells treated with DMAP1 siRNA arrested in mitosis to the same extent as control cells in the absence of IR treatment (Figures 2a and b). Increasing doses of IR from 1-10 Gy decreased entry into mitosis, but this tendency was reduced in DMAP1 siRNA-treated cells. DMAP1 depletion consistently resulted in a higher percentage of cells escaping arrest (Figures 2a and b) indicating that the ATM-regulated G2/M checkpoint requires DMAP1 function. Representative flow cytometry plots for all IR doses are shown in Supplementary Figure 3. To test the importance of DMAP1 in the radiation response further, we investigated clonogenic survival after IR. DMAP1 depletion reduced the numbers of colonies compared with controls to a similar extent as treatment with a pharmacological ATM inhibitor (Figure 2c). It is worth noting that DMAP1 depletion was performed by transient transfection, so DMAP1 protein levels are reduced at the time of IR treatment but presumably return to normal at later stages of the experiment (which lasts for 7 days). Thus the apparent difference in clonogenic survival is likely to be a direct reflection of DMAP1 function in the early cellular response to IR.
Figure 2. DMAP1-deficient cells are radiosensitive.
A549 cells were transfected with DMAP1 or control siRNA, and after 48 hours were irradiated at the doses indicated. Cells were immediately incubated in the presence of either (a) DMSO or (b) nocodazole and after 18 hours fixed and analysed for phospho-serine 10-Histone 3 (pH3) by FACS analysis. In (c) A549 cells were transfected with siDMAP1 or control siRNA. After 48 hours, DMSO or ATM inhibitor (ATMi) was added, and one hour later cells were treated with the indicated radiation doses and counted. 1000 cells were plated per condition, and the number of colonies formed was counted seven days later. At least three independent experiments were performed and p values calculated using the student’s t-test, (*p<0.05; **p<0.01).
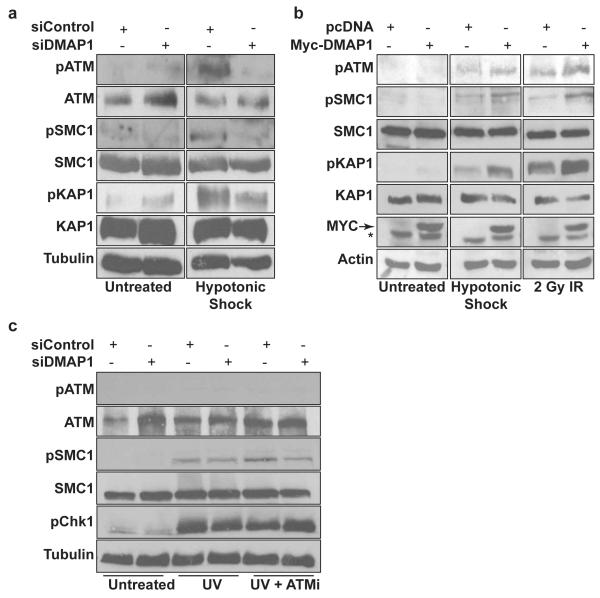
To investigate the role of DMAP1 in non-canonical ATM signalling, cultured cells were subjected to hypotonic stress. DMAP1 depletion also reduced ATM substrate phosphorylation in these conditions. S957-SMC1 phosphorylation and S824-Kap1 phosphorylation were reduced (Figure 3a). Conversely, overexpression of DMAP1 increased ATM substrate phosphorylation in response to both hypotonic shock and IR treatment. Basal ATM activity did not change (Figure 3b). We also investigated the role of DMAP1 in response to stimuli that activate ATR signalling. DMAP1 depletion did not alter pS957-SMC1 and pS317-Chk1 phosphorylation in response to ultraviolet radiation (Figure 3c). This is likely due to the minor role of ATM in pS957-SMC1 and pS317-Chk1 phosphorylation after ultraviolet radiation, as pharmacological ATM inhibition only led to a modest decrease in phosphorylation (Figure 3c). Thus, DMAP1 function is dispensable for ATR signalling, but required for ATM pathway activation in response to both canonical and non-canonical ATM stimuli.
Figure 3. DMAP1 is required for non-canonical ATM signalling.
(a) HCT-116 cells were transfected with a pool of siRNAs against DMAP1 or control siRNA, then 48 hours later were treated with 135mOsm NaCl for one hour. (b) HCT-116 cells were transfected with Myc-DMAP1, and 48 hours later treated with 2 Gy IR or 135mOsm NaCl, then harvested one hour later. (c) Cells were pre-treated for one hour with ATM inhibitor (ATMi) or DMSO, and treated with 50 J m−2 UV. Whole cell lysates were immunoblotted for pATM, ATM, pSMC1, SMC1, pKAP1, KAP1, phospho-p53, p53, pChk1, Myc, actin or tubulin. * denotes a non-specific band.
DMAP1 interacts with TIP60 and is required for histone H4 acetylation
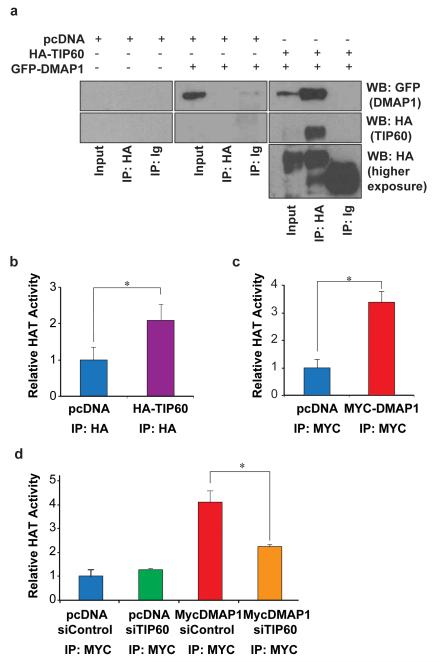
DMAP1 has been shown to be a stable subunit of the human NuA4 complex (17, 18), but the relevance of DMAP1 function for TIP60 activity has not been analysed. Co-overexpression of HA-TIP60 with GFP-tagged DMAP1 followed by immunoprecipitation (IP) confirmed the biochemical interaction between the proteins (Figure 4a), in agreement with the previous studies that used IP/mass spectrometry approaches.
Figure 4. DMAP1 interacts with the histone acetyltransferase TIP60.
a) 293A cells were transfected with the indicated combinations of pcDNA, HA-TIP60 and GFP-DMAP1. After 48 hours cells were harvested, and lysates immunoprecipitated using either an HA antibody or Ig control. Inputs and immunoprecipitates were immunoblotted for HA and GFP. Two different exposures of the HA immunoblot are shown. For (b), (c) and (d) cells were transfected with the indicated combinations of pcDNA, Myc-DMAP1, HA-TIP60, siControl and siTIP60. After 48 hours lysates were immunoprecipitated with either HA antibody, Myc antibody or Ig control. Washed immunoprecipitates were incubated with a colourimetric histone substrate, and HAT activity determined by spectrophotometry. Machine background was subtracted, and experimental values were normalised to a reaction containing no immunoprecipitate. The p-value of three independent experiments was calculated using the student’s t-test (*p<0.05).
We next investigated the role of DMAP1 in regulating TIP60 histone acetyltransferase (HAT) activity. Overexpressed tagged TIP60 was immunoprecipitated, washed IPs were incubated with a colourimetric histone substrate, and HAT activity determined by spectrophotometry. As expected, TIP60 IPs contained significant HAT activity (Figure 4b). Similarly, DMAP1 IPs were also associated with HAT activity (Figure 4c). To investigate the relative contribution of TIP60 to DMAP1-associated HAT activity, TIP60 mRNA was depleted by transfection of siRNAs (Supplementary Figure 4a). Knockdown of TIP60 did not alter the cell cycle distribution significantly (Supplementary Figures 4b and c), but reduced DMAP1-associated HAT activity (Figure 4d). Thus TIP60 accounts for a significant proportion of the HAT activity connected with DMAP1.
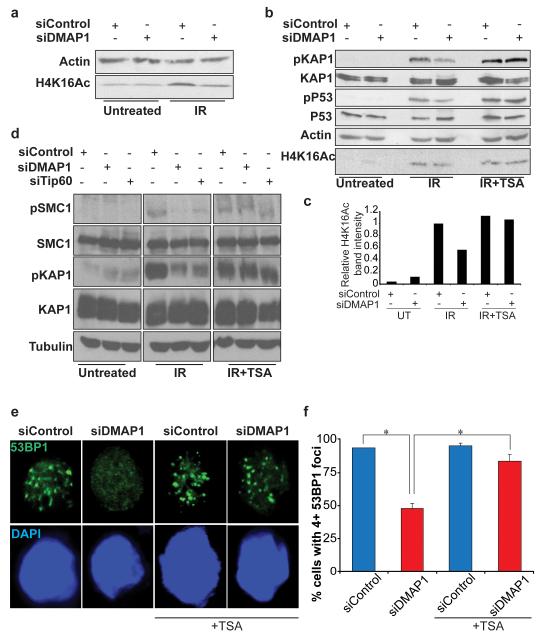
H4K16 is mainly acetylated by the histone acetyltransferases MOF and TIP60 (13, 23, 24) and H4K16Ac has recently been shown to be required for efficient ATM signalling (14). We thus tested the hypothesis that DMAP1 might control ATM signalling by affecting the levels of H4K16Ac. Irradiation resulted in an increase in H4K16Ac levels, which were reduced upon depletion of DMAP1 (Figure 5a). Trichostatin A (TSA) is an organic compound that selectively inhibits the class I and II mammalian histone deacetylase families of enzymes, thereby increasing substrate acetylation levels. TSA treatment rescued the decrease in H4K16Ac levels in DMAP1-depleted conditions (Figures 5b and c). Importantly, TSA treatment also ameliorated the defects in IR-induced Smc1, Kap1 and p53 phosphorylation caused by DMAP1 depletion (Figures 5b and d). Depletion of TIP60 had a similar effect on IR-induced ATM substrate phosphorylation as DMAP1 depletion, consistent with the idea that they work together (Figure 5d). TSA treatment also rescued impaired 53BP1 IRIF formation in DMAP1-depleted cells (Figures 5e and f). Thus, DMAP1 appears to be required for ATM signalling in response to DSBs by regulating the extent of acetylation of histone H4 at lysine 16, via TIP60.
Figure 5. DMAP1 contributes to H4K16 acetylation after DNA damage.
(a) HCT-116 cells were transfected with control or DMAP1 siRNA and after 48 hours were harvested untreated or one hour after 5 Gy IR. Whole cell lysates were probed for actin and H4K16Ac. (b) After 48 hours HCT-116 cells transfected with control or DMAP1 siRNA were pre-treated with TSA or DMSO for six hours, then irradiated at 5 Gy and harvested after an hour. Whole cell lysates were probed with the indicated antibodies. (c) The intensity of the H4K16Ac bands was measured, and normalised to actin loading control intensity. (d) Cells transfected with the indicated siRNAs were pre-treated with TSA or DMSO for six hours, then treated with 5 Gy IR and harvested after one hour. Whole cell lysates were immunoblotted for pSMC1, SMC1, pKAP1, KAP1 and tubulin. (e) HCT-116 were cultured in the presence of either DMSO or TSA for six hours, treated with 2 Gy IR then fixed after five minutes. Cells were fluorescently immunostained for 53BP1. Representative images are shown in (e) and a quantification in (f); p values were calculated using the student’s t-test, (*p<0.05).
Discussion
In this study, we identify a role of DMAP1 in ATM signalling. DMAP1 is important for histone acetylation, which in turn regulates chromatin structure and the ATM-dependent DNA damage response.
It has been known that ATM activation in response to DNA damage is associated with chromatin changes (9, 25). Moreover, previously it was shown that H4K16 acetylation is an important histone modification for ATM signalling (14). H4K16 acetylation is dramatically downregulated with the loss of DMAP1. Thus, it is likely that DMAP1 promotes ATM activation by affecting chromatin modifications, including H4K16Ac. However, we cannot rule out that DMAP1, and the TIP60 acetyltransferase, also affect ATM signalling by regulating the acetylation of non-histone substrates. Most importantly, it has been shown that acetylation of ATM itself may regulate its activation (26, 27). However, we have not been able to detect ATM acetylation reliably (data not shown).
It is an open question how DMAP1 regulates TIP60 activity. Although DMAP1 was found to be associated with the NuA4/TIP60 complex, its biochemical function within the complex is not known. The SANT domain of mammalian DMAP1, which is also present in the yeast homolog Eaf2, interacts with histone tails (19, 20). It is therefore tempting to speculate that DMAP1 might impart some positional information on TIP60, to determine the induction and precise location of H4K16 acetylation.
Because the effect of depleting DMAP1 was strongest a few minutes after irradiation, it appears that IRIF formation is delayed but not abolished in DMAP1-depleted cells. Although H4K16Ac levels were reduced, some acetylation was retained (Figure 5a), which could be sufficient for slower foci formation. This H4K16 acetylation could be from residual DMAP1 remaining after the knockdown, or from other histone acetyltransferases, such as MOF, partially compensating for the reduction in acetylation by the DMAP1–TIP60 complex. A previous study showing no effect of DMAP1 depletion on ATM phosphorylation 6–8 hours after irradiation (21) is consistent with our results at the later time point of 45 min, and with a delay in IRIF formation in DMAP1-depleted cells.
Downregulation of H4K16 acetylation is a hallmark of human cancers (28). Besides being considered as a biomarker of human cancers, loss of H4K16 acetylation could promote tumourigenesis by abrogation of the ATM-dependent DNA damage response. In line with this, mono-allelic loss of the human TIP60 gene occurs frequently in human lymphomas and head-and-neck and mammary carcinomas. Also in mice, TIP60 is a potent haploinsufficient tumour suppressor of Myc-induced lymphomagenesis (29). These data indicate that TSA and other HDAC inhibitors that impact on H4K16 acetylation and ATM signalling may have therapeutic potential for cancer treatment.
Materials and Methods
Cell culture and treatment
HCT-116, A549 and 293A cells were cultured in DMEM supplemented with 10% FCS. Transfections were performed with Lipofectamine-2000 (Invitrogen, 11668-019) according to the manufacturer’s protocol. In some cases cells were treated with 10μM ATM inhibitor (118500, Calbiochem, Germany) or 5μM TSA (Sigma, T8552) during the course of the experiment. IR experiments were performed using a Cs137 Gamma Irradiator at 2.1 Gy/min. Hypotonic swelling was performed using 50 mM NaCl in 1% FBS/ PBS supplemented with 0.45% glucose. Cells were harvested in Buffer A (150mM NaCl, 80mM Tris–HCl (pH 7.2), 0.2% NP-40, 10% Glycerol) supplemented with 1mM Na3VO4, 1mM β-glycerophosphate, 50mM NaF and complete protease inhibitor cocktail (Sigma, UK). For cell cycle profiling, cells were stained with Hoechst and analysed using a Becton Dickinson FACScan.
Western blotting
Western blots were performed using standard procedures. Protein samples were separated by SDS–PAGE, and subsequently transferred onto PVDF membranes. The following antibodies were used: pS1981-ATM (4526, Cell Signalling); ATM (2C1, Santa Cruz), pS824-Kap1 (Bethyl Laboratories, Montgomery, USA); Kap1 (Bethyl Laboratories Montgomery, USA); β-actin (A5060, Sigma, UK); α-tubulin (7291, Abcam, UK); pS957-SMC1 (5D11G5, Millipore, UK); SMC1 (AB3908, Millipore, UK); pS317-Chk1 (2344, Cell Signalling, UK); pS15-p53 (9284, Cell Signalling, UK); p53 (sc126, Santa Cruz) H4K16-Ac (05-1232 Millipore, UK); Myc (C3956, Sigma, UK), GFP (TA155041, Origene, USA) and HRP-conjugated goat anti-mouse/rabbit IgG (Jackson, PA, USA.). All primary antibodies were used at 1:1000 dilution and secondary antibodies at 1:5000.
Immunofluorescence
HCT-116 cells were adhered onto slides, treated as indicated and fixed with 4% PFA. Cells were permeabilised with 0.5% Triton/PBS and blocked with 10% FCS/PBS/0.1% Triton X100 before addition of primary antibody diluted 1:400 in blocking buffer: pS1981-ATM (4526, Cell Signalling, UK) or 53BP1 (sc22760, Santa Cruz, UK). Cells were washed three times with blocking buffer then incubated with secondary antibody (AlexaFluor 568 and AlexaFluor 488; Invitrogen, UK) diluted 1:400 in blocking buffer for one hour. Cells were washed with PBS and stained with DAPI before mounting in Mowiol (475904, Calbiochem, Germany). Cells were imaged using a LSM 510 (Zeiss) confocal microscope. For quantification of IF images, a minimum of 100 cells for each experimental condition were counted.
RT-PCR
mRNA was isolated from cells using a Qiagen RNeasy Micro Kit and cDNA was synthesised according to the manufacturer’s protocol (Roche, 11483188001). RT-PCR was performed using the following primers:
DMAP1-F 5′-ACGGAGCAATGTTCTTCCAC-3′
DMAP1-R, 5′-CAGGCACCTGCACAGTCTTA-3
TIP60-F, 5′-CAGGACAGCTCTGATGGAATAC-3′
TIP60-R, 5′-AGAGGACAGGCAATGTGGTGAG-3′
Actin-F, 5′-GGATGCAGAAGGAGATCACTG-3′;
Actin-R, 5′-CGATCCACACGGAGTACTTG-3′
Radiosensitivity Assays
For G2/M checkpoint analysis, 10μM ATM inhibitor (Calbiochem, 118500) or DMSO was added to transfected cells one hour prior to irradiation. After irradiation at the indicated dosages, cells were treated with 40nM nocodazole (M1404, Sigma, UK) or DMSO, for 18 hours. Cells were fixed in 70% ethanol, stained with pSer10-H3 antibody (06-570, Cell signalling, UK) followed by incubation with Alexa647 secondary antibody (A-31571, Invitrogen, UK). Fluorescence-activated cell sorting was performed using an LSR II Flow Cytometer (BD). Analysis was carried out using FlowJo (Tree Star) software. For post-irradiation survival analysis, transfected cells were irradiated at the indicated doses. Cells were immediately trypsinised, counted using a Vi-Cell XR cell counter (Beckman Coulter) and 1000 cells plated out per condition in 60mm dishes. Seven days later crystal violet was used to count the number of colonies formed.
Co-Immunoprecipitations
For co-immunoprecipitations, 293A cells co-expressing GFP-DMAP1 and HA-TIP60 were lysed in buffer A, sonicated and cleared by centrifugation for 10 minutes at 4°C. Lysates were immunoprecipitated overnight with either antibodies against GFP (Origene TA150041), HA (H6908, Sigma, UK) or IgG control (02-6102, Invitrogen, UK). Immunocomplexes were captured using Protein A and Protein G beads (Sigma, UK), washed four times with buffer A and resuspended in Laemmli sample buffer.
HAT Assays
For HAT assays, cell lysates were immunoprecipitated as described, using either HA (H6908, Sigma, UK) or Myc (C3956, Sigma, UK) antibodies. Immunocomplexes were resuspended in Buffer A, and HAT activity was measured with a colourimetric Histone Acetyltransferase Activity Assay Kit (65352, AbCam, UK) according to the manufacturer’s protocol.
Supplementary Material
(a) qPCR quantification of the relative levels of DMAP1 mRNA 48 hours after transfection with four different siRNAs targeted against DMAP1, normalised to the mRNA level in cells transfected with control siRNA. (b) FACS analysis of the cell cycle profile of siDMAP1 or siControl cells 48 hours after transfection. (c) Quantification of (b). Error bars represent the standard deviation of three independent experiments.
HCT-116 cells were treated with a pool of four siRNAs targeting DMAP1. After 48 hours cells were treated with 2 Gy IR, fixed at the indicated time points and immunofluorescently stained for 53BP1. The mean number of foci per cell was quantified (left panels), and 53BP1 focus intensity was measured for at least 250 foci per experiment (right panels). The p-value of three independent experiments was calculated using the student’s t-test (***p<0.001; n.s., not significant).
A549 cells transfected with DMAP1 siRNA or control siRNA were irradiated at the indicated doses of IR 48 hours after transfection, and immediately treated with nocodazole (NOC) or DMSO. Cells were analysed by FACS 18 hours later and the percentage of pS10-H3 (pH3)-positive cells quantified. FACS profiles for each IR dose and condition are shown. PI, propidium iodide.
(a) qPCR quantification of the relative levels of TIP60 mRNA in HCT-116 cells 48 hours after transfection with two different siRNAs targeted against TIP60, normalised to the mRNA level in cells transfected with control siRNA. (b) FACS analysis of the cell cycle profile of siTIP60 or siControl cells 48 hours after transfection. (c) Quantification of (b). Error bars represent the standard deviation of three independent experiments.
Acknowledgements
Support was given from the Experimental HistoPathology Unit, Bioinformatics and Biostatistics, the Equipment Park, Biological Resources and the FACS Lab in the London Research Institute (CR-UK). We thank Dr Cremona and Dr Kanu for critical reading of the manuscript and the Mammalian Genetics Lab for input and discussions. This work was supported by an ERC grant (281661 ATMINDDR) to A.B. The London Research Institute is funded by Cancer Research UK.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Chun HH, Gatti RA. Ataxia-telangiectasia, an evolving phenotype. DNA repair. 2004;3(8-9):1187–96. doi: 10.1016/j.dnarep.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Frappart PO, McKinnon PJ. Ataxia-telangiectasia and related diseases. Neuromolecular Med. 2006;8(4):495–511. doi: 10.1385/NMM:8:4:495. [DOI] [PubMed] [Google Scholar]
- 3.Kuhne M, Riballo E, Rief N, Rothkamm K, Jeggo PA, Lobrich M. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer research. 2004;64(2):500–8. doi: 10.1158/0008-5472.can-03-2384. [DOI] [PubMed] [Google Scholar]
- 4.Shiloh Y, Tabor E, Becker Y. Colony-forming ability of ataxia-telangiectasia skin fibroblasts is an indicator of their early senescence and increased demand for growth factors. Experimental cell research. 1982;140(1):191–9. doi: 10.1016/0014-4827(82)90169-0. [DOI] [PubMed] [Google Scholar]
- 5.Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118(1):9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol. 2001;13(2):225–31. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 7.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 8.Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA repair. 2004;3(8-9):997–1007. doi: 10.1016/j.dnarep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 10.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature reviews Cancer. 2003;3(3):155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 11.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. The Journal of biological chemistry. 1999;274(53):37538–43. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 12.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. The EMBO journal. 2003;22(20):5612–21. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26(56):7741–8. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Chen Y, Lu LY, Wu Y, Paulsen MT, Ljungman M, et al. Chfr and RNF8 synergistically regulate ATM activation. Nature structural & molecular biology. 2011;18(7):761–8. doi: 10.1038/nsmb.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature cell biology. 2006;8(1):91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 16.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature genetics. 2000;25(3):269–77. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 17.Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. The Journal of biological chemistry. 2003;278(44):42733–6. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 18.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Molecular and cellular biology. 2004;24(5):1884–96. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Molecular cell. 2002;10(4):935–42. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 20.Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nature reviews Molecular cell biology. 2004;5(2):158–63. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 21.Negishi M, Chiba T, Saraya A, Miyagi S, Iwama A. Dmap1 plays an essential role in the maintenance of genome integrity through the DNA repair process. Genes to cells: devoted to molecular & cellular mechanisms. 2009;14(11):1347–57. doi: 10.1111/j.1365-2443.2009.01352.x. [DOI] [PubMed] [Google Scholar]
- 22.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nature reviews Molecular cell biology. 2008;9(10):759–69. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 23.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306(5704):2084–7. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 24.Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Molecular and cellular biology. 2010;30(14):3582–95. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, et al. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat Cell Biol. 2009;11(1):92–6. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(37):13182–7. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Molecular and cellular biology. 2007;27(24):8502–9. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37(4):391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 29.Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448(7157):1063–7. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) qPCR quantification of the relative levels of DMAP1 mRNA 48 hours after transfection with four different siRNAs targeted against DMAP1, normalised to the mRNA level in cells transfected with control siRNA. (b) FACS analysis of the cell cycle profile of siDMAP1 or siControl cells 48 hours after transfection. (c) Quantification of (b). Error bars represent the standard deviation of three independent experiments.
HCT-116 cells were treated with a pool of four siRNAs targeting DMAP1. After 48 hours cells were treated with 2 Gy IR, fixed at the indicated time points and immunofluorescently stained for 53BP1. The mean number of foci per cell was quantified (left panels), and 53BP1 focus intensity was measured for at least 250 foci per experiment (right panels). The p-value of three independent experiments was calculated using the student’s t-test (***p<0.001; n.s., not significant).
A549 cells transfected with DMAP1 siRNA or control siRNA were irradiated at the indicated doses of IR 48 hours after transfection, and immediately treated with nocodazole (NOC) or DMSO. Cells were analysed by FACS 18 hours later and the percentage of pS10-H3 (pH3)-positive cells quantified. FACS profiles for each IR dose and condition are shown. PI, propidium iodide.
(a) qPCR quantification of the relative levels of TIP60 mRNA in HCT-116 cells 48 hours after transfection with two different siRNAs targeted against TIP60, normalised to the mRNA level in cells transfected with control siRNA. (b) FACS analysis of the cell cycle profile of siTIP60 or siControl cells 48 hours after transfection. (c) Quantification of (b). Error bars represent the standard deviation of three independent experiments.