Abstract
Computationally supported development of small molecule inhibitors has successfully been applied to protein tyrosine phosphatases in the past, revealing a number of cell-active compounds. Similar approaches have also been used to screen for small molecule inhibitors for the cancer-related phosphatases of regenerating liver (PRL) family. Still, selective and cell-active compounds are of limited availability. Since especially PRL-3 remains an attractive drug target due to its clear role in cancer metastasis, such compounds are highly demanded. In this study, we investigated various virtual screening approaches for their applicability to identify novel small molecule entities for PRL-3 as target. Biochemical evaluation of purchasable compounds revealed ligand-based approaches as well suited for this target, compared to docking-based techniques that did not perform well in this context. The best hit of this study, a 2-cyano-2-ene-ester and hence a novel chemotype targeting the PRLs, was further optimized by a structure–activity-relationship (SAR) study, leading to a low micromolar PRL inhibitor with acceptable selectivity over other protein tyrosine phosphatases. The compound is active in cells, as shown by its ability to specifically revert PRL-3 induced cell migration, and exhibits similar effects on PRL-1 and PRL-2. It is furthermore suitable for fluorescence microscopy applications, and it is commercially available. These features make it the only purchasable, cell-active and acceptably selective PRL inhibitor to date that can be used in various cellular applications.
Keywords: Dual specificity phosphatases, Virtual screening methods, Phosphatases of regenerating liver, Enzyme inhibitors, Thienopyridone, 2-Cyano-2-ene-esters
Abbreviations: DIFMUP, 6,8-difluoro-4-methylumbelliferyl phosphate; DSP, dual specificity phosphatase; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; PRL, phosphatases of regenerating liver; PTP, protein tyrosine phosphatase; PTP1B, protein tyrosine phosphatase 1B; ROCS, rapid overlay of chemical structures; SAR, structure–activity relationship; TCPTP, T-cell protein tyrosine phosphatase; UFSRAT, ultrafast shape recognition with atom types; UPLC-MS, ultra performance liquid chromatography–mass spectrometry; USR, ultrafast shape recognition; VHR, vaccinia H1-related phosphatase; VS, virtual screening
Graphical abstract

Highlights
-
•
Computational ligand- and docking-based approaches were tested for PRL-3 as a target.
-
•
Ligand-based screening was proven a feasible approach for PRL-3 inhibitor discovery.
-
•
A low micromolar, non-competitive inhibitor with novel chemotype for PRLs was discovered.
-
•
The inhibitor efficiently blocks PRL induced cell migration.
-
•
The inhibitor is non-cytotoxic, commercially available and suitable for fluorescence microscopy applications.
1. Introduction
Cellular signaling processes are tightly regulated through phosphorylation and dephosphorylation of proteins and second messenger molecules by the action of kinases and phosphatases, respectively, thereby maintaining the correct function of cellular life. A perturbation in these highly controlled regulation processes can lead to the formation and progression of various diseases [1]. It is therefore critical to identify small organic molecules that inhibit unwanted hyperfunction of disease-promoting kinases and phosphatases; on the one hand to diminish disease progression through drug development, on the other hand to gain a better understanding of the biological mechanisms behind these processes [1].
The phosphatases of regenerating liver (PRL)-1, PRL-2 and PRL-3 are of particular interest as drug targets, because these putative oncogenes are strongly involved in cancer formation and progression [2]. They were shown to influence angiogenesis and metastasis [3], [4], each of them representing hallmarks of cancer [5], [6]. PRL-1, PRL-2 and PRL-3 are highly overexpressed in various types of cancer [3]. PRL-3 is especially found in liver metastasis rather than the primary tumors [7], making it an important drug target as cancer metastasis is the major cause for poor prognosis in cancer due to the impossibility of surgery at this late state of disease [8], [9].
Few normal functions of the PRL phosphatases are known: PRL-1 is an immediate-early response gene in regenerating liver and involved in cell differentiation, and the latter also appears to be the case for PRL-3. Additionally, all three PRLs have been suggested to regulate cell cycle progression [3]. The PRL phosphatases are members of the cysteine-based dual-specificity group of phosphatases (DSPs), which belongs to the protein tyrosine phosphatase (PTP) superfamily. Members of the DSPs not only are able to dephosphorylate phosphotyrosines like the classical PTPs but also to hydrolyze phosphomonoesters of serines, threonines, and non-protein substrates [10]. The sequence identities of PRLs compared to other phosphatases are low, but very high among the members of the PRL family. The PRLs are small (21 kDa), single domain proteins. Uncommon compared to other DSPs, their active site is both shallow and hydrophobic, allowing for the dephosphorylation of phosphotyrosines, phosphoserines and phosphothreonines as well as phosphoinositides (in the case of PRL-3) [3], [8], [11].
The development of inhibitors of the PRLs, and DSPs in general, is difficult. It is therefore not surprising that, despite their attractive features as drug targets, a drug targeting a DSP has yet to be approved. The challenge in developing phosphatase inhibitors as drug leads lays in often-found drawbacks of limited selectivity and poor pharmacokinetics [1]. This is due to the conserved active site geometry of many DSPs, and their preference to bind negatively charged entities [12]. In particular, finding selective active site inhibitors for one of the members of the PRL-family of phosphatases is a delicate task, since although their active site architecture is quite unique among phosphatases, they all share the shallow and hydrophobic binding pocket, making it difficult to find small molecules being able to only bind to one of the three PRL members [3]. So far, the developed small molecule PRL-inhibitors are either not selective when compared to other phosphatases, they do not show selectivity amongst the PRL-family, or they simply were tested for only one of the PRLs [13], [14], [15], [16], [17], [18], [19]. Selectivity within the PRL family may not be necessary for a drug lead as they are all putative oncogenes, but it is desirable to study their co-expression in cancers and potential redundancy in their roles as these aspects still need to be investigated in detail. The most potent cell-active inhibitor identified to date is thienopyridone, with a reported IC50 of 173 nM for PRL-1, 277 nM for PRL-2 and 128 nM for PRL-3 [17]. This compound, however, is not commercially available for applications in PRL biology research. Thienopyridone was found through high-throughput screening. Other experimental approaches to discover inhibitors included manual testing of single compounds based on their known cellular effect [19].
Virtual screening is an attractive alternative to search for PRL inhibitors. When a known ligand of the target is available, molecular shape similarity [20] is often used to find molecules with similar shape, but different chemical structure, to an active template (scaffold hopping). USR is a very fast shape similarity technique that has been successfully applied to the prospective identification of new inhibitors of a range of molecular and cell targets [21], [22], [23], [24], [25], [26]. A recent pharmacophoric extension of USR termed UFSRAT [27] has also identified novel cell-active inhibitors of a type II diabetes target in prospective virtual screens [28]. Other popular ligand-based techniques include ROCS [29] for shape similarity and MACCS fingerprints [30] for chemical structure similarity. On the other hand, when a known structural model of the target is available, docking techniques can also be used for structure-based virtual screening. Here we use AutoDock Vina [31], which is one of the most accurate docking techniques. A molecular docking approach using a PRL-3 homology model was already applied previously leading to a rhodanine-based micromolar inhibitor of PRL-3. The selectivity of this compound was, however, not examined [13].
In this study, we aimed at elucidating which prospective virtual screening protocols are more suitable for this target while searching for new cell-active PRL inhibitors. Due to its role in cancer metastasis, we focused our computational screens on PRL-3.
Here we applied ligand-based USR, ROCS and MACCS fingerprints to search the same chemical library for similar molecules to thienopyridone. In addition, molecules with a similar shape to thienopyridone were docked into two structural models of PRL-3 using Vina and the resulting scores were used to re-rank these hits. Furthermore, a webserver implementing UFSRAT searches over a second molecular library (see Section 4 for further details) was also used to identify molecules with similar shape and pharmacophoric properties as thienopyridone.
Using a biochemical screen we determined prospective hit rates of the different methods for PRL-3, which suggest that ligand-based screens are more suitable for PRL-3 hit identification than structure-based screens. In particular, USR led to the discovery of a novel inhibitor chemotype for PRL-3, a 2-cyano-2-ene-ester. A following structure–activity-relationship (SAR) study based on this compound revealed a low micromolar, noncompetitive inhibitor of the PRLs with acceptable selectivity over other PTPs. The compound is non-cytotoxic, active inside cells as indicated by its selective reversion of the enhanced cell migration phenotype induced by the PRLs, and commercially available. In contrast to thienopyridone that showed to block cell proliferation independently of PRL-3, our inhibitor can be applied to long-term cellular studies as it does not show an effect on cell proliferation. Furthermore, fluorescence microscopy reveals its localization to the plasma membrane, which can greatly reduce possible off-target effects due to its co-localization with PRL-3. These properties make it currently the only molecule for a broad use in PRL research.
2. Results and discussion
2.1. In silico screens and their biochemical evaluation
Ligand-based virtual screening requires at least one molecule with affinity for the target of interest. Here we used thienopyridone as a search template in all ligand-based runs because this molecule is the most potent PRL-3 inhibitor to date [17] and has a rigid chemical structure, the latter meaning that its lowest energy conformer must be similar to its bioactive conformation. USR was run to search a database with 641,485,760 conformers generated from 3,472,461 lead-like molecules (see the Experimental section for further details), which was completed in just 57 s using a single computing core. ROCS required 128 h in screening the same conformers using the same computer. Unlike USR and ROCS, MACCS fingerprints is a 2D technique and thus operates directly on the chemical structures and therefore it does not require conformational expansion. Thus, searching the 3,472,461 chemical structures, encoded as the same number of MACCS fingerprints, for those with similar structure to thienopyridone took 24 s. At a second stage, we also used the UFSRAT webserver with thienopyridone as search template, which returned the top 200 molecules in a couple of seconds.
In parallel, we explored the use of structure-based virtual screens, although we were aware of the challenges that PRL-3 presents to this approach: the active site is extremely flexible, solvent-exposed and there are no X-ray structures available. We considered two structural models of PRL-3, a NMR structure [32] and the PRL-3 homology model of the sulfate-bound PRL-1 crystal structure [11]. The apo NMR structure presents the active site in an open conformation. The general acid loop containing the aspartic acid residue is oriented away from the active site pocket, suggesting a strong conformational rearrangement occurring upon substrate binding [32]. This is further supported by a comparison of the PRL-3 apo NMR structure with crystal structures (apo and sulfate-bound) of PRL-1, where the apo and ligand-bound structures of PRL-1 adopt very similar conformations, whereas the apo structure of PRL-3 shows significant differences, adopting a much wider and flatter active site entry pocket [3]. Therefore, we thought that these structural differences could be the basis for achieving selectivity against other PRLs. For the first docking run with Vina, we assumed that thienopyridone was a competitive inhibitor and thus we only docked those molecules with similar shape to thienopyridone. The latter was done because similarly shaped molecules tend to be complementary in shape to the same binding pockets and thus should obtain higher hit rates than docking randomly selected molecules. These docking runs were performed against both the closed conformation (homology model) and the open conformation (1v3a model) of PRL-3. Thus, we called these runs docking1-hmod and docking1-1v3a, respectively. The second docking run (docking2-1v3a) was carried on all the molecules, but only against the open conformation, as this led to marginally better results than the closed conformation in the first docking run.
The screens USR, ROCS, MACCS, docking1-hmod and docking1-1v3a were carried out first. Selected compounds from these screens were purchased and tested for inhibition against PRL-3 in an in vitro phosphatase activity assay (see the Experimental section). Once the results were available, we carried out the UFSRAT and docking2-1v3a screens to investigate the impact of adding ligand-based pharmacophoric constraints and docking molecules with any shape, respectively. Inhibition was first analyzed at a single compound concentration. Active compounds in this screen were further evaluated for their respective IC50 values, and the hit rates for the computational screens were based on these. Table 1 shows the hit rates of these studies showing that overall, ligand-based similarity screening approaches worked well on PRL-3 as enzyme target and the used ligand thienopyridone.
Table 1.
Summary of results for the applied virtual screening methods. The various molecular similarity methods using the PRL-3 inhibitor thienopyridone [17] as a search template are listed (the first row refers to all ligand-based methods as a whole). Dockings were performed with the PRL-3 docking models docking1-1v3a, docking1-hmod and docking2-1v3a, as explained in the text. For the purpose of this study, we regarded as a hit a tested compound with an IC50 value of about 100 μM or less against PRL-3 using DiFMUP as substrate. The IC50 values of these five hits were 31, 48, 58, 102 and 108 μM (four additional inhibitors with potencies less than 200 μM were found but not further considered as their IC50 value was deemed too high to be promising for potential optimization). Hit rates are provided to quantify the performance of each method on PRL-3.
| In silico method | Hit rate (%) | # hits within tested compounds | # tested compounds |
|---|---|---|---|
| Similarity screens (total) | 12.2 | 5 | 41 |
| USR | 15.4 | 2 | 13 |
| MACCS | 28.6 | 2 | 7 |
| UFSRAT | 8.3 | 1 | 12 |
| ROCS | 0 | 0 | 9 |
| Docking1-1v3a | 0 | 0 | 9 |
| Docking1-hmod | 0 | 0 | 10 |
| Docking2-1v3a | 0 | 0 | 27 |
The MACCS screen obtained the highest hit rate, but as expected these hits had similar chemical structures to thienopyridone. USR obtained the second highest hit rate, with one of the two hits exhibiting a very different chemical scaffold than thienopyridone. ROCS used the same molecular database and query molecule than MACCS and USR, but found no hits. UFSRAT, which used the same query molecule but different screening database than the other three ligand-based methods, obtained the third highest hit rate.
On the other hand, docking USR hits to re-rank them according to the Vina score did not provide any hits regardless of the used structural model. Later, we established that thienopyridone is not competitive with DiFMUP as screening substrate (see Section 2.3), which means that molecules similarly shaped to this query molecule are not expected to be complementary in shape to any conformation of the active site. This suggests that the reason why structure-based screens did not find any hits is that we are docking against two conformations of the active site that are probably quite different from its unknown bioactive conformation. Overall, we observed that USR, UFSRAT and MACCS were useful for this target, which is remarkable given the low number of tested compounds required, thereby providing hits for a challenging target in a fast and cheap manner. Thus, in our hand ligand-based techniques seem much more suitable than structure-based techniques for PRL-3 hit identification.
Out of the tested compounds from the in vitro evaluation of our in silico approaches, compound 1, a 2-cyano-2-ene-ester, performed the best (Fig. 1). Compared to thienopyridone itself, compound 1 has quite different structural features, although being a top hit of a shape similarity screen. Interestingly, compound 2 was the only hit found in this screen that is structurally similar to thienopyridone, the original ligand of our in silico approaches. The rhodanine derivative [13] was furthermore chosen as additional positive control for the following biochemical studies (Fig. 1).
Fig. 1.
Structures of compounds 1 and 2 from the USR shape similarity screen, as well as of the two controls thienopyridone [17] and a rhodanine derivative (nr. 1 from Park et al. [13]).
Table 2 shows IC50 values of these structures against PRL-3 and its close homologs PRL-2 and PRL-1. Compound 1 inhibited PRL-3 comparably well to the commercially available rhodanine control with IC50s of around 50 μM in both cases. This control compound was published with an IC50 of 13.3 μM against PRL-3 [13], which is lower but in the range of the value obtained under the conditions used here. Thienopyridone still remains the most potent PRL inhibitor published so far, with equal IC50s for all PRLs in the sub-micromolar range that are comparable to the published values [17]. Of note, as explained in the experimental section, thienopyridone seemed to spontaneously induce substrate hydrolysis in our DiFMUP assay when used at high concentrations, hence results obtained by this substrate–inhibitor combination should be taken with care. Nevertheless, since the original publication provided similar IC50 data obtained by a different assay setup in combination with a different substrate [17], we consider our result as conditionally correct. Compound 2 as thienopyridone-analog showed only minor activity against PRL-3. Since compound 1 was the most potent one found in this screen, it was chosen as starting point for follow-up studies.
Table 2.
IC50 values of thienopyridone, the rhodanine control and compounds 1 and 2 from in silico screens, obtained by in vitro DiFMUP dephosphorylation (see the Experimental section). ID of the ZINC database [33] and screening origin are indicated.
2.2. Structure–activity relationship study for compound 1
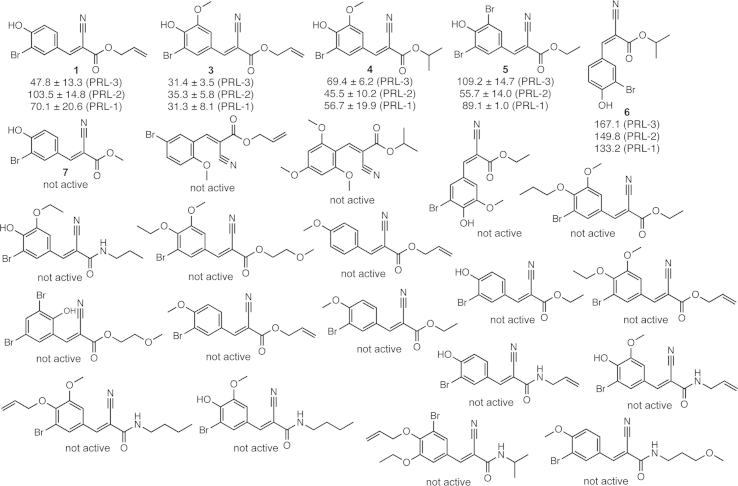
In order to optimize the inhibitory potential of compound 1 and to understand which residues are important for the activity, 23 close structural analogs were purchased and analyzed for inhibition against the PRL family members (Fig. 2, see also Supplementary Figure S1). This SAR study showed that a substitution is well tolerated in certain positions, whereas it is not at all tolerated in others. Fig. 3 highlights these findings by summarizing all changes and their outcomes in activity retention or loss. Exchanging the isomeric conformation from E to Z led to a weakened activity of the compound. Furthermore, exchanging the residues attached to the ester moiety generally diminished the inhibitory potency. Amides instead of esters were not tolerated at all, as well as any substitution in positions 4 or 6 of the aromatic ring. Addition of a methoxy-group in position 3 was well tolerated, as well as exchanging this moiety by bromine. In position 2 apparently only a hydroxyl group is accepted, because changing the features at this position led to weakening or loss of potency in all other cases. Analog 3 showed an increase in potency against all three PRL enzymes and represents the most potent analog with an IC50 of 31 μM against PRL-3, surpassing the potency of the rhodanine control toward PRL-3 in our measurements.
Fig. 2.
Structures and respective activities of all tested analogs from the SAR study of compound 1. IC50 values (μM) for inhibition of DiFMUP dephosphorylation are shown for PRL-3, PRL-2 and PRL-1, where applicable.
Fig. 3.

Result of the SAR study after analyzing 23 analogs of compound 1. Green – substitution tolerated; blue – substitution weakens potency; red – substitution leads to loss of activity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. Further in vitro evaluation of inhibition
In order to exclude the possibility of false-positive inhibition by aggregation-based mechanisms, solubility in assay buffer was ensured for all active hits by measuring and excluding light-scattering abilities in the used concentration ranges (Supplementary Table S1). Moreover, all hits and controls were shown to be reversible inhibitors by time-dependent assaying of enzyme-inhibition [34]. Compounds showed the same level of inhibition when incubated with enzyme for different time periods (data not shown). In addition, all hits and controls were tested for the possibility of being redox-cycling compounds and therefore leading to false-positive readouts due to generation of phosphatase-inhibiting hydrogen peroxide [35]. To this end, the compounds in assay buffer were treated with 1 mM catalase for 10 min prior to addition of enzyme and substrate, in order to consume any hydrogen peroxide possibly being generated. None of the compounds showed loss of activity when assayed in the presence of catalase (data not shown), and we conclude therefore that all compounds are true-positive PRL inhibitors in vitro. Since all compounds were purchased, we ensured their purity by analyzing all active compounds by UPLC-MS. According to these measurements, all active compounds were over 92% pure (Supplementary Table S2).
The type of inhibition was analyzed for analog 3 as our best hit and, additionally, for thienopyridone, since no such data was published for this compound [17]. We performed detailed kinetic analyses on analog 3 inhibiting DiFMUP hydrolysis by PRL-3. Our data showed clear noncompetitive behavior for this compound (Supplementary Figure S2). We furthermore plotted IC50 values against various used DiFMUP concentrations, showing that both compounds act noncompetitively against DiFMUP as substrate, since the obtained IC50s did not change with altered DiFMUP concentration [36] (Supplementary Figure S2B). Our results indicate that both noncompetitive inhibitors interact with the enzyme at a site different than the DiFMUP interaction site, and that this interaction is independent of binding of the substrate to the enzyme itself. Importantly, this behavior reflects the reaction of PRL-3 with DiFMUP, a non-natural phosphorylated substrate for which the exact interaction residues with the enzyme are unknown except of the catalytically active residue Cys104. Hence, we can conclude that the inhibitors interact with PRL-3-residues different from the ones used for binding DiFMUP.
2.4. Molecular docking of compound 1, analog 3 and thienopyridone
Taking into account the very flexible apo structure of PRL-3 in an open conformation with the catalytic residues Cys104 and Asp72 positioned at different loop regions pointing away from each other [32], the noncompetitive behavior of analog 3 and thienopyridone with DiFMUP as reaction substrate can be rationalized by the increased possibility of interaction sites of inhibitor and enzyme before molecular rearrangement into a closed substrate-converting state takes place.
We performed molecular docking on the open PRL-3 structure with analog 3, its parent molecule from the in silico screen (compound 1) and thienopyridone, in order to further evaluate this result. Fig. 4 shows the respective top 5 binding poses of the respective inhibitor docked to PRL-3 (pdb entry 1V3A) with the two catalytic residues indicated. Comparing the docking result of all three inhibitors, they all bind in a similar region close to the catalytic Asp72, suggesting a possible interaction with this residue. Notably, no interaction is seen with Cys104, the catalytic residue important for binding to the phosphate substrate. This is in agreement with the result of our inhibitor-type study, showing that the inhibitors bind independently of the DiFMUP substrate (which indispensably interacts with residue Cys104). In all top poses, analog 3 and compound 1 bind in a very similar manner, with the hydroxyl group pointing either towards Asp72 or alternatively downwards to a patch with a glutamine and an asparagine residue in close proximity. Since this hydroxyl group is crucial for inhibition due to complete loss of potency when substituted by other moieties, we suggest that a hydrogen bond may be formed either with Asp72, or with glutamine or asparagine when positioned downwards. Nevertheless, the remaining conclusions of the SAR study cannot be rationalized by the docking results. The same is the case for thienopyridone. In general, docking scores were remarkably low as a result of a flexible protein structure and low-molecular-weight ligands. Therefore, these docking results can only give a rough estimation of the possible binding region of the inhibitors. Without a co-crystal structure of enzyme and ligand, this remains speculative. Nevertheless, the presumable binding region of the enzyme would spare room for further modifications of the ligands by more elaborate SAR studies.
Fig. 4.

Molecular docking of inhibitors into the open PRL-3 structure (pdb entry 1V3A), with labeled catalytic residues Cys104 and Asp72. For each inhibitor, the top five docking poses are shown. From left to right: analog 3 – compound 1 – thienopyridone.
2.5. Selectivity of hits over other phosphatases
In order to evaluate our compounds' abilities to inhibit other phosphatases, selectivity tests were carried out by in vitro inhibition assays against the protein tyrosine phosphatases PTP1B and TCPTP as well as the unrelated dual-specificity phosphatase VHR. Compounds were assayed at a concentration roughly around their IC50 values (50 μM in all cases expect of thienopyridone, which was assayed at 0.5 μM concentration) and activities were compared between PRL-3 and the above-mentioned phosphatases. All enzymes were measured at their respective KM values towards DiFMUP (see the Experimental section). Table 3 shows a summary of these data, depicted by a selectivity index (see also Supplementary Figure S3). Compound 3 as our best analog showed higher selectivity towards PRL-3 compared to the other phosphatases, but not as good as thienopyridone. Compound 1 showed acceptable selectivity only over VHR. Notably, the rhodanine control performed not well in this context. This was not studied in the original publication [13], but given the known inherent properties of rhodanines as promiscuous binders [37], this finding is not surprising.
Table 3.
Selectivity of compounds towards PRL-3 over other phosphatases. DiFMUP dephosphorylation was measured at the respective KM of each enzyme, at a compound concentration of 50 μM (thienopyridone: 0.5 μM) and enzyme concentration of 35 nM. Selectivity index = 1 – (inhibition of PRL-3/inhibition of enzyme). The closer the value is to 1, the higher the selectivity towards PRL-3.
| Compound | Selectivity index towards PRL-3 over |
||
|---|---|---|---|
| PTP1B | TCPTP | VHR | |
| Thienopyridone | 0.93 | 0.93 | 0.93 |
| Rhodanine control | 0.34 | 0.22 | 0.54 |
| 1 | 0.38 | 0.46 | 0.64 |
| 2 | 0.25 | 0.10 | 0.25 |
| 3 | 0.65 | 0.77 | 0.85 |
| 4 | 0.08 | −0.29 | 0.24 |
| 5 | 0.48 | 0.24 | 0.44 |
2.6. Cytotoxicity and in-cell efficacy of the new inhibitors
For determination of short-term cytotoxic effects of active hits on mammalian cells, the HEK293 cell lines were incubated with compounds or DMSO vehicle for 16 h. Surviving cells were stained by MTT labeling. The results are shown in Fig. 5 for PRL-3 and empty vector control cell lines, as well as in Supplementary Figure S4 for PRL-1 and PRL-2. Only analog 5 showed a strong cytotoxic effect compared to the DMSO vehicle in all cell lines. A significant, but weak cytotoxic effect could also be seen with compound 1 at 75 μM concentration in the case of the empty vector control. Overall – with the exception of named analog 5 – all tested hits show no significant cytotoxic effects under these conditions, including thienopyridone and the rhodanine control.
Fig. 5.
Cytotoxicity of compounds with PRL-3 expressing (A) or empty vector (B) HEK293 cell lines. After 16 h incubation, surviving cells are stained with MTT and absorbance is normalized to DMSO vehicle. Results are depicted as mean ± SD, with compounds color-coded and concentrations as indicated. Two-sided t-tests were performed for p < 0.05, compared to DMSO vehicle. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
PRL-3's ability to enhance cell migration and invasion has been shown in various studies [3]. We therefore tested our newly discovered inhibitor 3 in comparison to the control compounds for reversion of PRL-3 induced cell migration by a transwell migration assay in HEK293 cell lines stably expressing PRL-3 or the empty vector control. Analog 3 was tested at three different concentrations, with 75 μM as the highest one that has been shown to be non-cytotoxic (see above). The rhodanine control was tested at 75 μM. Thienopyridone was used at 20 μM due to its higher in vitro potency [17]. As the results in Fig. 6A show, treatment of PRL-3-expressing cells with 75 μM analog 3 and the thienopyridone control significantly reverted cell migration to the basal level of the empty vector cells. Reduced migration could as well be observed in samples treated with 50 μM analog 3, but not with 12 μM. Hence, analog 3 shows concentration-dependent inhibition of PRL-3 induced cell migration. The rhodanine control showed only a negligible effect suggesting that this compound is either poorly cell-permeable, metabolized inside cells, or not selectively inhibiting PRL-3 in a cellular context, as already suggested by our in vitro selectivity study. Importantly, analog 3 as well as thienopyridone did not show a significant reduction of migration of the control cells, indicating that their action under these conditions is PRL-3-specific (Fig. 6B).
Fig. 6.
Effect of compounds on transwell cell migration. A) PRL-3 expressing (black) or empty vector control (gray) HEK293 cell lines were seeded into transwell insert chambers in the presence of compound or DMSO vehicle and allowed to migrate for 16 h (see the Experimental section). Cells that had migrated to the lower compartment were dissociated and stained with calcein-AM for fluorescence readout on a multiwell plate reader. Compound concentrations and the number of data points (N) are indicated. Results are depicted as mean ± SD, normalized to the empty vector signal. Two-sided t-tests were performed for p < 0.05. ns – not significant. B) Respective control experiment with empty vector cell line.
In the absence of clear physiological substrates as read-outs to show the ability of analog 3 to specifically inhibit PRL-3 inside cells [3], the inhibition migration of PRL expressing cells was furthermore demonstrated by microscopic means. To this end, wound healing experiments were carried out over 20 h with PRL and empty vector control cell lines by comparison of treatment with varying concentrations of compound or DMSO vehicle. Pictures of the proceeding wound closure were taken at the indicated time points, as summarized and quantified for PRL-3 in Fig. 7, and quantified for PRL-2 and PRL-1 in Supplementary Figure S5. In agreement with the results of the transwell migration experiment, a complete inhibition of PRL-3 induced cell migration to the respective basal level of the control cell line could be observed in the presence of 50 μM as well as 35 μM analog 3. A slight delay of gap closure was still visible with 25 μM analog 3. The effect was diminished with treatment of 12 μM compound concentration, as in the transwell assay. The inhibitor had no effect on migration of the empty vector control cells (Fig. 7). We conclude that analog 3 specifically inhibits migration of PRL-3 expressing cells in a concentration-dependent manner. The compound is also able to inhibit migration of PRL-2 and PRL-1 expressing cells at 50 μM concentration, however in the case of PRL-2 not as efficiently as with PRL-3 and PRL-1 (Supplementary Figure S5).
Fig. 7.
Analog 3 inhibits PRL-3 induced cell migration. Wound healing abilities of PRL-3 expressing or empty vector control HEK293 cell lines were analyzed in the presence of analog 3 or DMSO vehicle. A) Pictures of the wound gaps were taken at 0 h and 20 h time points for each condition, as indicated. a) DMSO vehicle, b) 50 μM analog 3, c) DMSO vehicle, d) 50 μM analog 3, e) 35 μM analog 3, f) 12 μM analog 3. B) Quantification of the wound healing experiment. PRL-3 data are shown in black, empty vector data in gray. Speed of migration is shown for combined data, as mean ± SD. Two-sided t-tests were performed for p < 0.05. ns – not significant.
As control experiment, similar wound healing experiments were carried out with varying concentrations of thienopyridone. The inhibitor was reported to fully inhibit HUVEC cell migration at a concentration of about 15 μM [17]. Here, thienopyridone fully inhibited migration of PRL-3 expressing HEK293 cells at a concentration of about 20 μM (Supplementary Figure S6). This is in good agreement with the published result and close to the effective concentration of analog 3 (35 μM). When comparing the effective cellular concentrations of thienopyridone and analog 3 taking into consideration their IC50 values in vitro (0.2 versus 31 μM for thienopyridone and analog 3, respectively), it becomes clear that analog 3 shows similar potencies in cells as in the in vitro experiments whereas thienopyridone does not. This could indicate that thienopyridone might be degraded, does not enter cells efficiently or has other targets and effects inside cells.
In order to rule out any cell-proliferative side effects of PRL-3 overexpressing cells that could influence time needed for the gap closure, we conducted a proliferation experiment on both PRL-3 and control HEK293 cell lines. Both cell lines show similar proliferation profiles (Supplementary Figure S7), confirming the unbiased basis for the comparative migration studies. Furthermore, we analyzed the effect of our tested inhibitors on cell proliferation of PRL-3 overexpressing or empty vector control HEK293 cells. Compounds that interfere with the cell proliferation machinery could potentially also influence the cell's migration abilities. In order to exclude such indirect effects, we conducted cell proliferation experiments on PRL-3 and control cell lines in the absence or presence of analog 3 or thienopyridone. Fig. 8 summarizes these results, showing that analog 3 had no significant influence on HEK293 cell proliferation, whether or not PRL-3 is overexpressed. Thienopyridone, on the other hand, severely influenced cell proliferation potentially by blocking cell cycle progression, which we reason because we did not observe a short-term (within one cell cycle) cytotoxicity of the compound (Fig. 5). Importantly, this anti-proliferative effect is entirely independent of PRL-3, since the compound had the same effect on the empty vector control cells (Fig. 8). Thus, thienopyridone showed strong cellular effects that cannot be attributed to its activity toward PRL-3. We cannot confidently conclude if this effect plays a causative role in the observed inhibition of cell migration. However, this result strongly suggests that biological experiments carried out with thienopyridone should be interpreted with caution.
Fig. 8.
Proliferation experiment of PRL-3 expressing (A) and empty vector control (B) HEK293 cell lines with analog 3 and thienopyridone. Respective cells were seeded in equal numbers into a 96-well plate on day 1 and incubated with the respective compound or DMSO vehicle, as explained in the Experimental Section. Live cells were stained with MTT and analyzed on a plate reader every indicated day.
Finally, we tested the inhibitors' suitability for cellular applications in fluorescence microscopy by monitoring mKate-PRL-3 and GFP constructs on a confocal microscope in the presence and absence of inhibitor. We observed that the presence of analog 3 did not influence the fluorescence readouts of mKate-PRL-3 and GFP (Fig. 9a and b), indicating that the compound is suitable for such imaging applications. Thienopyridone, on the other hand, showed a strong auto-fluorescence in both observed GFP and mKate channels (Fig. 9c and d). The compound is therefore not useful for microscopy experiments, at least when using green and red fluorescent proteins. Interestingly, analog 3 showed auto-fluorescence when excited at 405 nm and viewed at about 450 nm that does not interfere with green or red channels. Surprisingly, the compound localized to the plasma membrane and to a lesser extent to inner membrane compartments (Fig. 9a and b), apparently co-localizing with its target PRLs. This is the case both in cells overexpressing (mKate-PRL-3 construct) and lacking (GFP construct) PRL-3, indicating that the inhibitor's localization is a structure-inherent feature. In order to test this further, we investigated the localization of analog 7, which is equally fluorescent when excited at 405 nm. The analog was inactive in vitro and did not affect wound healing abilities of PRL-3 expressing cells (Supplementary Figure S8) Like analog 3, this compound localized to the plasma membrane and inner membrane compartments, although showing a slightly bigger proportion of vesicular localization (Fig. 9e and f). This suggests that the structure rather than the activity determines the compounds' localization. The co-localization of analog 3 with PRL-3 at the plasma membrane could explain the inhibitor's equally strong cellular potency compared to its in vitro activity. We consider this localization advantageous for any cellular studies targeting PRL proteins, since co-localization of both enzyme and inhibitor should greatly reduce any off-target effects.
Fig. 9.
Confocal fluorescence microscopy of compounds with mKate-PRL-3 or GFP transfected HEK293 cells, as indicated, after 30 min incubation. The distribution did not change after 1.5 h (measured with analog 3 only, data not shown). For thienopyridone and mKate-PRL-3 (d), a PRL-3 high-expressing example was chosen for presentation in order to ensure a visible difference to the thineopyridone signal. Compounds were visualized by exciting at 405 nm. GFP was excited at 488 nm, mKate at 561 nm. Scale bar = 10 μm.
3. Conclusion
In the computational part of this study, we applied a number of virtual screening techniques in order to elucidate which is the most suitable for applying it on a larger scale for PRL-3 as target. This only required testing a total of 87 compounds, which is far cheaper and quicker than screening a much larger library of randomly selected compounds. Importantly, most of these hits contain new PRL-3-active scaffolds with the focused compound of this study being a 2-cyano-2-ene-ester, produced by the USR screening method. As the most suitable virtual screening methods for this target have obtained hit rates in the range of 16–22%, we expect that exclusively using these on much larger molecular databases using several diverse PRL-3 inhibitors as templates will lead to many more additional inhibitors. The latter is necessary to build accurate QSAR models for each member of the PRL family as a way to ultimately search for selective and potent PRL-3 inhibitors.
Testing a further 23 purchasable analogs of our preliminary hit has led to a low micromolar, noncompetitive PRL inhibitor that is quite selective over other PTPs compared to a published rhodanine inhibitor with similar potency [13], but not as good as the potent PRL inhibitor thienopyridone [17]. Rhodanines are compounds that are considered PAINS – so-called frequent hitters with promiscuous binding properties [37] – and should be used with caution. Of note, the rhodanine we used as control inhibitor in our study was not sufficiently active inside cells to modify the studied phenotypes, even at high concentration.
We showed that our developed inhibitor (analog 3) is non-cytotoxic and able to efficiently inhibit the PRLs inside cells. It reverts PRL-3-induced cell migration back to basal levels in a concentration-dependent manner, and it is furthermore able to inhibit PRL-1 and PRL-2 activity inside cells. The compound exerts full cellular activity against PRL-3 at a concentration of around 35 μM, which is similar to its in vitro potency of 31 μM, and similar to the in-cell active concentration of thienopyridone. Furthermore, analog 3 unexpectedly showed localization to the plasma membrane, independently of PRL-3 overexpression. As PRL-3 localizes to the plasma membrane as well, this feature potentially minimizes off-target effects that could occur in other cellular compartments and likely represents the reason for the inhibitor's comparable potency in cells and in vitro.
An interesting finding of our study is that thienopyridone entirely blocks proliferation of HEK293 cells without being short-term cytotoxic. This is a strong side effect of the compound that is clearly independent of the expression of PRL-3. Consequently, this behavior could influence the cells' migration abilities and may lead to false positive inhibition in wound healing experiments, and studies of molecular mechanisms of PRLs with thienopyridone should be viewed with caution. In the original publication, the authors demonstrate that thienopyridone induces anoikis in cancer cells, a type of apoptosis initiated by cleavage of p130Cas, and that there is a functional link between the PRLs and p130Cas. They could not observe p130Cas cleavage after PRL knockdown, and a clear link between the observed phenotypes after thienopyridone treatment and knockdown of the PRLs could not be made [17]. Overall, while thienopyridone clearly is a highly potent inhibitor of PRL activity, this suggests that thienopyridone has also an influence on the cells' apoptotic and proliferative machinery that does not involve PRL activity. By contrast, our compound does not affect cell proliferation and survival and therefore constitutes the only PRL inhibitor developed to date that is active inside cells with no obvious long-term side effects. Nevertheless, thienopyridone as a strong PRL inhibitor was the starting point of our study, showing the value of potent known inhibitors for computational ligand-based studies.
Potency and selectivity of our inhibitor within the class of PRL enzymes could potentially be improved by conducting more elaborative SAR studies. To this end, a combination of chemical and computational approaches as well as co-crystal structures would be desirable. With its physicochemical properties and a clogP value of about 2.5, our compound 3 follows Lipinski's rule of five [38]. Since optimization tends to increase the molecular weight and logP, there is scope for more comprehensive SAR studies to optimize its selectivity and potency. Interestingly, the inhibitor shares very similar structural features with Entacapone, an approved anti-Parkinson drug [39], although the latter does not inhibit PRL activity (data not shown). As molecules with similar chemical structure tend to have a similar pharmacology, our PRL-3 inhibitor and its future optimized derivatives might be as safe as Entacapone when administered in humans, which increases its therapeutic potential.
Overall, we show here that ligand-based computational approaches work well with the target PRL-3 and its strong inhibitor thienopyridone, as opposed to structure-based techniques. Biochemical screening of only a modest number of compounds led us to a micromolar inhibitor that we could further optimize by a basic SAR study. The obtained molecule shows a remarkable in-cell efficacy by specifically inhibiting PRL-3 induced cell migration in a dose-dependent manner, with similar effects on PRL-1 and PRL-2. It is acceptably selective over other tested PTPs, does not disturb cell proliferation, can successfully be applied to fluorescence microscopy and is commercially available.
4. Experimental section
4.1. Materials
6,8-Difluoro-4-methylumbelliferyl phosphate (DiFMUP) was purchased from Life Technologies. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was from Sigma. Thienopyridone was purchased from Enamine by synthesis-on-demand. Entacapone was a kind gift from Christian Klein, Heidelberg University. Tested compounds from in silico screens were from Enamine and Vitas-M. Specifically, compound 1 and analog 3 were from Enamine and the rhodanine control was from Vitas-M. All other chemicals were purchased from Sigma and Merck.
4.2. Ligand-based and structure-based virtual screening
The main screening library was downloaded from the ZINC database version 8 [33], more concretely subset 1 of all usual lead-like ligands containing 3,472,461 molecules in MDL SD format. OE omega 2.4.3 was run with default settings on this set to generate an average of 185 3D conformers per molecule, resulting in a molecular database with 641,485,760 conformers (total CPU time was 15 days). USR descriptors were pre-calculated for each of the 641 million conformers, taking a total of 48 min on a single computing core. MACCS fingerprints were also precalculated with OpenBabel 2.3.1 (4.3 h for 3,472,461 molecules). The following similarity thresholds were used: 0.95 (USR score), 0.9 (ROCS shape tanimoto score) and 0.8 (MACCS tanimoto score) leading to 693, 23 and 30 highlighted molecules respectively. UFSRAT is available as a webserver [40] and thus it screened a different multi-conformer database of commercially available molecules (19,412,000 conformers, with an average of four conformers per molecule). UFSRAT returns the top 200 molecules in a couple of seconds.
Regarding docking, two structural models of PRL-3 were used. The homology model is fully described in McParland et al. [11]. Python scripts from AutoDockTools (ADT) 1.5.4 were used to protonate the 1v3a model and database molecules at pH = 7.4 as well as converting them to pdbqt format. For each structure, the center of the binding pocket was manually selected with the ADT GUI and a cube with size 24 Å in each dimension set. Vina was run with default exhaustiveness.
Each supplier catalog only covers part of the ZINC database. We selected the two vendors with the best coverage of our hits (Enamine and Vitas-M) to purchase compounds for in vitro tests. In other words, we did not select every vendor so as to minimize shipping costs. This means that not all high-ranking compounds were tested. Furthermore, some of the ordered compounds turned out to be unavailable, which again resulted in more high-ranking compounds not being tested. However, this is not an issue as our goal was not to be exhaustive, but rather to implement a fast and very cost-effective protocol leading to new PRL-3 inhibitors.
4.3. Recombinant proteins
VHR, PTP1B and TCPTP were prepared as previously described [41]. PRL-3, as well as PRL-1 and PRL-2, were prepared as in McParland et al. [11], with the following modifications: Plasmid vector pETM-20 was used to overexpress His-TEV-tagged fusion proteins. After lysing and purifying His-tagged proteins accordingly using a FPLC Histras HP 1 mL column, His-tags were cleaved by His-TEV protease and cleared by another round of Histrap column chromatography.
4.4. In vitro phosphatase activity assay
In vitro evaluation of phosphatase and –inhibitor activity was determined using DiFMUP as fluorogenic phosphate-substrate. Recombinant PRL protein (35 nM final assaying concentration) was incubated in assay buffer (20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 10 mM DTT, 0.01% Triton-X 100, 5% DMSO) for about 20 min before proceeding, in order to ensure full reduction and therefore maximal activity.
KM values were determined by measuring reaction kinetics of substrate dilution series on a 96-well plate reader (excitation 358 nm, emission 452 nm). Initial velocities were plotted against DiFMUP concentration and the data was analyzed by Michaelis–Menten kinetics curve fitting using GraphPad Prism 6.
Enzyme inhibition was determined at a substrate concentration corresponding to the respective KM value of the individual enzyme (21 μM for PRL-3; 24 μM for PRL-1, PRL-2, PTP1B and TCPTP; 90 μM for VHR). PTP1B and TCPTP were assayed in a buffer described by Meyer et al. [40]. IC50 data were obtained by measuring inhibitor concentration series. After baseline correction (respective inhibitor in the absence of enzyme), initial velocities were plotted against inhibitor concentration (logarithmic scale) and analyzed with GraphPad Prism 6.
When measuring PRL-inhibition with thienopyridone, we observed spontaneous DiFMUP hydrolysis in the presence of high concentrations of inhibitor (and the absence of enzyme) leading to the blank-subtracted data curves showing a potentially false positive “full inhibition”. This effect could not be seen at inhibitor concentrations below 2 μM. Although measuring concentrations around the IC50 value (0.2 μM) was apparently acceptable, we consider this behavior as unusual and this could lead to a false IC50 value measured by our method. Nevertheless, in the original publication the measurement of the IC50 value for thienopyridone was carried out using a different assay setup including a different substrate, and our IC50 value matches the published ones [17]. Therefore our measured value should be accurate in spite of the unusual behavior at high inhibitor concentration.
4.5. Solubility and purity of compounds
Solubility of compounds in assay buffer (see corresponding paragraph) was measured in the same concentration range as used for IC50 determination. Light scattering was measured on a NepheloStar (BMG Labtech) in comparison to a buffer blank.
Purity of hits was validated by UPLC-MS on a Waters Acquity System with a ACQUITY UPLC BEH C18 Column and a gradient of 5–100% acetonitrile containing 0.2% formic acid. Absorbance was detected between 210 and 400 nm by a PDA detector. Peak areas were integrated from signals at 254 nm.
4.6. Cell culture and generation of stable cell lines
HEK Flp-In T-rex 293 cells (Invitrogen) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS (Invitrogen), 1% l-glutamine, 1% penicillin and 1% streptomycin in a humidified atmosphere containing 5% CO2.
Cell lines containing stably integrated human PRL-3, PRL-2, PRL-1 or the respective empty vector construct were prepared as previously described [11]. Stable cell lines were grown in selection medium (supplemented with 15 μg/mL blasticidin and 100 μg/mL hygromycin). Protein expression was induced by addition of tetracycline (1 μg/mL) 20 h prior to performing follow-up experiments.
4.7. Cytotoxicity assay
Tetracycline-induced HEK293 stable cell lines were seeded in a transparent 96-well plate with 10000 cells/well in growth medium. After 24 h incubation, compounds and DMSO controls were added to the respective wells in the indicated final concentrations and incubated for 16 h. The medium was removed, fresh medium was added to each well, MTT (5 mg/mL in PBS; used 1:10 in growth medium) was added and the cells were further incubated for 4 h at 37 °C. Finally, the medium was removed, 100 μL DMSO was added to each well, pipetted thoroughly to dissolve the produced formazan crystals, and incubated 10 min at RT. Absorbance was determined at 570 nm in a multiwell plate reader and the data was normalized to the respective DMSO vehicle. Each measurement was carried out in triplicates in at least two independent experiments.
4.8. Proliferation assay
HEK293 stable cell lines were seeded in a transparent 96-well plate with 1500 cells/well in growth medium supplemented with tetracycline and grown for the indicated time. For experiments containing inhibitors, respective compounds or DMSO vehicle were added immediately after seeding. Analog 3 was used at 50 μM, thienopyridone at 20 μM concentration. At each indicated day, the medium was removed from the respective wells and MTT was added like described above. After 4 h incubation at 37 °C, 100 μL DMSO was added into the respective wells and absorbance was read at 570 nm. The measurement was performed in triplicates.
4.9. Transwell migration assay
HEK293 stable cell lines were induced with tetracycline 20 h prior to seeding into transwell inserts. Cells were starved for 6 h and washed twice with serum-free growth medium. Cells (5–10 × 105 per well) were seeded into the upper chambers of 24-well transwell inserts (100 μL, 8 μm membrane pore size; Brand) and supplemented with the respective inhibitor or DMSO control. The lower chambers were filled with 800 μL growth medium containing 5% FBS. Migration was allowed for 16 h at 37 °C. Insert-membranes were washed once by carefully dipping into PBS. The medium was removed from the lower chamber. Inserts were returned to the respective compartment and 700 μL of Cell Dissociation Solution (amsbio) containing 1.2 μL/mL Calcein-AM (1.67 μg/μL in DMSO; Life Technologies) was added. Dissociation and staining was allowed for 1 h at 37 °C. 100 μL solution of each well was pipetted into a black 96-well plate (in triplicates per trans-well) and stained cells were analyzed on a multiwell plate reader (excitation 485 nm, emission 520 nm). The experiment was carried out repeatedly in four replicates per condition. The number of individual data points is indicated in the results section.
4.10. Wound healing assay
Wound healing experiments were carried out with μ-dish 35 mm culture inserts from Ibidi. 9 × 105 cells/mL of the respective HEK293 stable cell lines were seeded into each of the two insert wells and allowed to settle for 20 h in the presence of tetracycline. On the day of experimentation the inserts were gently removed and growth medium supplemented with 5% FBS and the respective compound or DMSO vehicle were added. Pictures of the wound gap were taken at 0 h and 20 h time points. A Zeiss Cellobserver HS microscope was used with 10× magnitude.
4.11. Confocal microscopy
HEK293 cells were transfected with 0.1 μg of mKate-PRL-3 or GFP constructs 18 h before imaging on a Leica SP2 confocal microscope equipped with a 63× oil lense. Compounds were added in imaging buffer (50 μM analog 3 and analog 7, 20 μM thienopyridone) and incubated for 30 min before imaging. The following imaging buffer was used: 20 mM HEPES, pH 7.4, 115 mM NaCl, 1.2 mM CaCl2, 1.2 mM MgCl2, 1.2 mM K2HPO4. GFP was excited with a 488 nm laser, mKate with a 561 nm laser. Compounds were visualized with a 405 nm laser.
Author contributions
B.H. designed and carried out all in vitro and cellular experiments, interpreted the data and wrote the manuscript. M.D. participated in dephosphorylation assays and preparation of stable cell lines. P.J.B. designed and performed all in silico experiments and analysis, interpreted the data and wrote the manuscript. M.K. coordinated and planned this study, designed the experiments, interpreted the data and wrote the manuscript.
Acknowledgments
M.K. is grateful for the support of the German Science Foundation (DFG) within the Emmy-Noether program (KO 4013/1-1). B.H. thanks the Austrian Academy of Sciences (ÖAW) for a DOC fellowship. P.J.B. gratefully acknowledges funding from the UK Medical Research Council (Methodology Research Fellowship G0902106) and the French Inserm. The authors thank the EMBL-Technology Development Funds for funding, Janet Thornton, EMBL-EBI, for helpful discussions, the Chemical Biology Core Facility and the Protein Expression and Purification Core Facility at EMBL for their support, G. Varsano and P. Luján Miralles for assistance with the confocal microscopy, M. Mülbaier and A. Rüger, Cellzome (GSK), for their support with the UPLC-MS, C. Klein (University of Heidelberg) for sharing Entacapone, and R. Breinbauer (TU Graz) for helpful discussions.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ejmech.2014.08.060.
Contributor Information
Pedro J. Ballester, Email: pedro.ballester@inserm.fr.
Maja Köhn, Email: koehn@embl.de.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Vintonyak V.V., Waldmann H., Rauh D. Using small molecules to target protein phosphatases. Bioorg. Med. Chem. 2011;19:2145–2155. doi: 10.1016/j.bmc.2011.02.047. [DOI] [PubMed] [Google Scholar]
- 2.Bessette D.C., Qiu D., Pallen J.C. PRL PTPs: mediators and markers of cancer progression. Cancer Metastasis Rev. 2008;27:231–252. doi: 10.1007/s10555-008-9121-3. [DOI] [PubMed] [Google Scholar]
- 3.Rios P., Li X., Köhn M. Molecular mechanisms of the PRL phosphatases. FEBS J. 2013;280:505–524. doi: 10.1111/j.1742-4658.2012.08565.x. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman M.W., Homanics G.E., Lazo J.S. Targeted deletion of the metastasis-associated phosphatase Ptp4a3 (PRL-3) suppresses murine colon cancer. PLoS One. 2013;8:e58300. doi: 10.1371/journal.pone.0058300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 7.Saha S., Bardelli A., Buckhaults P., Velculescu V.E., Rago C., StCroix B., Romans K.E., Choti M.A., Lengauer C., Kinzler K.W., Vogelstein B. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 8.Al-Aidaroos A.Q.O., Zeng Q. PRL-3 phosphatase and cancer metastasis. J. Cell. Biochem. 2010;111:1087–1098. doi: 10.1002/jcb.22913. [DOI] [PubMed] [Google Scholar]
- 9.Sharlow E.R., Wipf P., McQueeney K.E., Bakan A., Lazo J.S. Investigational inhibitors of PTP4A3 phosphatase as antineoplastic agents. Expert Opin. Investig. Drugs. 2014;23:661–673. doi: 10.1517/13543784.2014.892579. [DOI] [PubMed] [Google Scholar]
- 10.Li X., Wilmanns M., Thornton J., Köhn M. Elucidating human phosphatase-substrate networks. Sci. Signal. 2013;6:rs10. doi: 10.1126/scisignal.2003203. [DOI] [PubMed] [Google Scholar]
- 11.McParland V., Varsano G., Li X., Thornton J., Baby J., Aravind A., Meyer C., Pavic K., Rios P., Köhn M. The metastasis-promoting phosphatase PRL-3 shown activity toward phosphoinositides. Biochemistry. 2011;50:7579–7590. doi: 10.1021/bi201095z. [DOI] [PubMed] [Google Scholar]
- 12.He R., Zeng L.-F., He Y., Zhang S., Zhang Z.-Y. Small molecule tools for functional interrogation of protein tyrosine phosphatases. FEBS J. 2013;280:731–750. doi: 10.1111/j.1742-4658.2012.08718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park H., Jung S.-K., Jeong D.G., Ryu S.E., Kim S.J. Discovery of novel PRL-3 inhibitors based in the structure-based virtual screening. Bioorg. Med. Chem. Lett. 2008;18:2250–2255. doi: 10.1016/j.bmcl.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Ahn J.H., Kim S.J., Park W.S., Cho S.Y., Ha J.D., Kim S.S., Kang S.K., Jeong D.G., Jung S.-K., Lee S.-H., Kim H.M., Park S.K., Lee K.H., Lee C.W., Ryu S.E., Choi J.-K. Synthesis and biological evaluation of rhodanine derivatives as PRL-3 inhibitors. Bioorg. Med. Chem. Lett. 2006;16:2996–2999. doi: 10.1016/j.bmcl.2006.02.060. [DOI] [PubMed] [Google Scholar]
- 15.Pathak M.K., Dhawan D., Lindner D.J., Borden E.C., Farver C., Yi T. Pentamidine is an inhibitor of PRL phosphatases with anticancer activity. Mol. Cancer Ther. 2002;1:1255–1264. [PubMed] [Google Scholar]
- 16.Han Y.-M., Lee S.-K., Jeong D.G., Ryu S.E., Han D.C., Kim D.K., Kwon B.-M. Emodin inhibits migration and invasion of DLD-1 (PRL-3) cells via inhibition of PRL-3 phosphatase activity. Bioorg. Med. Chem. Lett. 2012;22:323–326. doi: 10.1016/j.bmcl.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Daouti S., Li W.-H., Qian H., Huang K.-S., Holmgren J., Levin W., Reik L., McGady D.L., Gillespie P., Perrotta A., Bian H., Reidhaar-Olson J.F., Bliss S.A., Olivier A.R., Sergi J.A., Fry D., Danho W., Ritland S., Fotouhi N., Heimbrook D., Niu H. A selective phosphatase of regenerating liver phosphatase inhibitor suppresses tumor cell anchorage-independent growth by a novel mechanism involving p130Cas cleavage. Cancer Res. 2008;68:1162–1169. doi: 10.1158/0008-5472.CAN-07-2349. [DOI] [PubMed] [Google Scholar]
- 18.Moon M.K., Han Y.-M., Lee Y.-J., Lee L.H., Yang J.H., Kwon B.-M., Kim D.K. Inhibitory activities of anthraquinones from Rubia akane on phosphatase regenerating liver-3. Arch. Pharm. Res. 2010;33:1747–1751. doi: 10.1007/s12272-010-1106-4. [DOI] [PubMed] [Google Scholar]
- 19.Choi S.-K., Oh H.-M., Lee S.-K., Jeong D.G., Ryu S.E., Son K.-H., Han D.C., Sung N.-D., Baek N.-I., Kwon B.-M. Biflavonoids inhibited phosphatase of regenerating liver-3 (PRL-3) Nat. Prod. Res. 2006;20:341–346. doi: 10.1080/14786410500463312. [DOI] [PubMed] [Google Scholar]
- 20.Ballester P.J., Brown N. Molecular shape. In: Brown N., editor; Mannhold R., Kubinyi H., Folkers G., editors. Bioisosteres in Medicinal Chemistry. Wiley-VCH; Germany: 2012. pp. 155–166. (Methods and Principles in Medicinal Chemistry). [Google Scholar]
- 21.Ballester P.J. Ultrafast shape recognition: method and applications. Future Med. Chem. 2011;3:65–78. doi: 10.4155/fmc.10.280. [DOI] [PubMed] [Google Scholar]
- 22.Li H., Huang J., Chen L., Liu X., Chen T., Zhu J., Lu W., Shen X., Li J., Hilgenfeld R., Jiang H. Identification of novel falcipain-2 inhibitors as potential antimalarial agents through structure-based virtual screening. J. Med. Chem. 2009;52:4936–4940. doi: 10.1021/jm801622x. [DOI] [PubMed] [Google Scholar]
- 23.Ballester P.J., Westwood I., Laurieri N., Sim E., Richards W.G. Prospective virtual screening with ultrafast shape recognition: the identification of novel inhibitors of arylamine N-acetyltransferases. J. R. Soc. Interface. 2010;7:335–342. doi: 10.1098/rsif.2009.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ballester P.J., Mangold M., Howard N.I., Marchese-Robinson R.L., Abell C., Blumberger J., Mitchell J.B.O. Hierarchical virtual screening for the discovery of new molecular scaffolds in antibacterial hit identification. J. R. Soc. Interface. 2012;9:3196–3207. doi: 10.1098/rsif.2012.0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teo C.Y., Rahman M.B.A., Chor A.L.T., Salleh A.B., Ballester P.J., Tejo B. Ligand-based virtual screening for the discovery of inhibitors for protein arginine deiminase type 4 (PAD4) Metabolomics. 2013;3:118. [Google Scholar]
- 26.Patil S.P., Ballester P.J., Kerezsi C. Prospective virtual screening for novel p53-MDM2 inhibitors using ultrafast shape recognition. J. Comput. Aided Mol. Des. 2014;28:89–97. doi: 10.1007/s10822-014-9732-4. [DOI] [PubMed] [Google Scholar]
- 27.Hsin K.-Y., Morgan H.P., Shave S.R., Hinton A.C., Taylor P., Walkinshaw M.D. EDULISS: a small-molecule database with data-mining and pharmacophore searching capabilities. Nucleic Acids Res. 2010;39:D1042–D1048. doi: 10.1093/nar/gkq878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adie J.E. 2010. Structure-based Drug Design of 11β-hydroxysteroid Dehydrogenase Type 1 Inhibitors.https://www.era.lib.ed.ac.uk/handle/1842/4673 (PhD thesis) [Google Scholar]
- 29.Nicholls A., Gaughey G.B., Sheridan R.P., Good A.C., Warren G., Mathieu M., Muchmore S.W., Brown S.P., Grant J.A., Haigh J.A., Nevins N., Jain A.N., Kelley B. Molecular shape and medicinal chemistry: a perspective. J. Med. Chem. 2010;53:3862–3886. doi: 10.1021/jm900818s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.http://openbabel.org/wiki/Tutorial:Fingerprints, April 15, 2014.
- 31.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozlov G., Cheng J., Ziomek E., Banville D., Gehring K., Ekiel I. Structural insights into molecular function of the metastasis-associated phosphatase PRL-3. J. Biol. Chem. 2004;279:11882–11889. doi: 10.1074/jbc.M312905200. [DOI] [PubMed] [Google Scholar]
- 33.http://zinc.docking.org/, May 5, 2014.
- 34.Arangoni A.G. Wiley; New York: 2003. Enzyme Kinetics – a Modern Approach. [Google Scholar]
- 35.Johnston P.A. Redox cycling compounds generate H2O2 in HTS buffers containing strong reducing reagents—real hits or promiscuous artifacts? Curr. Opin. Chem. Biol. 2011;15:174–182. doi: 10.1016/j.cbpa.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Copeland R.A. second ed. Wiley; New York: 2013. Evaluation of Enzyme Inhibitors in Drug Discovery. [Google Scholar]
- 37.Baell J.B., Holloway G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 38.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 39.http://www.fda.gov/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/ucm206513.htm, May 5, 2014.
- 40.http://opus.bch.ed.ac.uk/ufsrat/, April 15, 2014.
- 41.Meyer C., Hoeger B., Temmerman K., Tatarek-Nossol M., Pogenberg V., Bernhagen J., Wilmanns M., Kapurniotu A., Köhn M. Development of accessible peptidic tool compounds to study the phosphatase PTP1B in intact cells. ACS Chem. Biol. 2014;9:769–776. doi: 10.1021/cb400903u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.