Abstract
This study was to investigate the mechanism and role of Kif4A in doxorubicin-induced apoptosis in breast cancer. Using two human breast cancer cell lines MCF-7 (with wild-type p53) and MDA-MB-231 (with mutant p53), we quantitated the expression levels of kinesin super-family protein 4A (Kif4A) and poly (ADP-ribose) Polymerase-1 (PARP-1) by Western blot after doxorubicin treatment and examined the apoptosis by flow cytometry after treatment with doxorubicin and PARP-1 inhibitor, 3-Aminobenzamide (3-ABA). Our results showed that doxorubicin treatment could induce the apoptosis of MCF-7 and MDA-MB-231 cells, the down-regulation of Kif4A and upregulation of poly(ADP-ribose) (PAR). The activity of PARP-1 or PARP-1 activation was significantly elevated by doxorubicin treatment in dose- and time-dependent manners (P < 0.05), while doxorubicin treatment only slightly elevated the level of cleaved fragments of PARP-1 (P > 0.05). We further demonstrated that overexpression of Kif4A could reduce the level of PAR and significantly increase apoptosis. The effect of doxorubicin on apoptosis was more profound in MCF-7 cells compared with MDA-MB-231 cells (P < 0.05). Taken together, our results suggest that the novel role of Kif4A in doxorubicin-induced apoptosis in breast cancer cells is achieved by inhibiting the activity of PARP-1.
Keywords: doxorubicin, Kif4A, MCF-7, MDA-MB-231, PARP-1
INTRODUCTION
Chromokinesins are microtubule-motor molecules with chromatin-binding activity and play important roles in mitotic and meiotic regulation (Wandke et al., 2012). The kinesin super-family protein 4A (Kif4A) is a unique chromokinesin that localizes in the nucleus during the inter-phase of cell cycle (Hu et al., 2011). Kif4A protein consists of a motor N-terminal domain, a coiled-coil neck region in the middle, and a C-terminal tail domain. The motor domain has ATP hydrolysis activity for configuration changes, which makes Kif4A glide on the microtubule orbit. The neck region contains nuclear localization signals (NLSs) that promote Kif4A to translocate to cell nucleus and bind to the damaged DNA. The tail domain contains regular cysteine residues, which not only facilitates cargo transportation to cell membrane but also binds to associated proteins to regulate DNA damage repair, chromosome condensation and segregation (Hu et al., 2011; Wu et al., 2008). It has been reported that Kif4A regulates middle-spindle formation by interacting with the protein regulator of cytokinesis 1 (PRC1) (Shrestha et al., 2012). Kif4A may also promote neuronal survival by suppressing PARP-1’s enzymatic activity that is important in DNA damage repair (Borysov et al., 2011). Therefore, Kif4A is a candidate molecule that may play critical roles in DNA damage response and/or DNA repair pathways (Wu et al., 2008). However, the role of Kif4A in doxorubicin-induced apoptosis in breast cancer remains unknown.
Poly (ADP-ribose) Polymerase-1 (PARP-1) catalyzes the attachment of poly-ADP- ribose (PAR) units to target proteins, an important post-translational modification in eukaryotic cells (Lee Kraus and Hottiger, 2013). The molecular weight of PARP-1 is 113 KDa, and it consists of six domains, including two homologous zinc finger domains, Zn1 and Zn2 domains on the N-terminus. Zn1 and Zn2 contain NLSs and the KRK-X (11)-KKKSKK sequences, rendering the DNA-binding capacity of PARP-1. The highly basic 60-residue peptides in PARP-1 between BRCT domain and WGR domain contain ADP-ribose polymer attachment sites, which facilitates the DNA-dependent PAR synthesis activity of the catalytic domain (CAT). The Zn3 domain of PARP-1 is a unique zinc finger domain, distinctive in structure and function from Zn1 and Zn2. Biochemical studies indicated that the zinc ribbon motif of the Zn3 domain contains key residues for the efficient assembly of PARP-1 domains (Huambachano et al., 2011; Langelier et al., 2011). PARP-1 may serve as a co-stimulatory factor for NF-κB and hypoxia-inducible factor-1 (HIF1) in gene transcription regulation (Martinez-Romero et al., 2012; Stanisavljevic et al., 2011). PARP-1 can interact with condensin I and X-ray repair cross-complementing (XRCC). The expression levels of PARP-1, condensin I, and the base excision repair (BER) factor XRCC1 are increased dramatically upon single-strand break damage (SSB), which enhances PAR expression and the activity of PARP-1, and promotes the repair of damaged DNA (Campalans et al., 2013). The mechanism of PARP-1 in doxorubicin-induced apoptosis and/or DNA repair also remains unknown.
Doxorubicin (Adriamycin, ADR) is one of the most widely used anticancer agents. It eliminates tumor cells by inhibiting DNA replication and interrupting mitosis though covalent DNA-binding (Zaremba et al., 2010). Recently, it was reported that ADR can increase the expression of PAR (an active product of PARP-1) and promote damaged DNA repair and induce chemotherapy resistance (Munoz-Gamez et al., 2005). However, the mechanisms for ADR’s roles remain unexplored. In this study, we found a novel mechanism that the increased PARP-1 activity by ADR was directly mediated through the down-regulation of Kif4A, and thus induced cell apoptosis in breast cancer cells.
MATERIAL AND METHODS
Cell culture
Breast cancer cell lines MCF-7 (with wild-type p53) and MDA-MB-231 (with mutant p53) were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences, Shanghai, China. The cells were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin, 100 U/ml streptomycin, and 2 mmol/L L-glutamine, at 37°C in a humidified 5% CO2 atmosphere.
Cell treatment
Cells were plated into 35 mm dishes (MCF-7, 3 × 105; MDA-MB-231, 5 × 105), on the next day treated with 0.05, 0.1, 0.5, 1.0, and 2.0 μmol/L ADR in 3 replicates for 24 h, followed by 12 h of recovery. Cells were lysed with 1× cell lysis buffer for protein analysis. The control cells were handled exactly same except ADR treatment. Cells treated with 1 μM ADR for 24 h and recovered for 6, 12, 24, and 48 h were collected for Western blot analysis. The HA-Kif4A plasmid was transfected into cells by using Lipofectamine2000. The 24 h after transfection, the cells were treated with 1 μM ADR for 24 h, followed by 24 h recovery. Cells transfected with HA-pcDNA3.0 plasmid but without ADR treatment were used as a control. The cells were collected to detect the expression levels of related proteins by Western blot and to measure cell apoptosis by flow cytometry (FCM). Cells were plated and treated with 2.5, 5, 10, and 20 μM PARP-1 inhibitor (3-Aminobenzamide, 3-ABA, dissolved in DMSO) for 24 h, followed by 12 h of recovery. Cells were collected to detect the expression of Kif4A and PARP-1 proteins by Western blot. Cells without 3-ABA treatment were used as a control. The cells were treated with 5 μM 3-ABA and 1 μM ADR for 24 h, followed by 12 h of recovery, then collected to measure cell apoptosis by FCM.
Western blot analysis
Cells were washed with PBS and lysed in 100 μl lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton-X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 μg/ml leupeptin) for 10 min on ice. The supernatant was collected by centrifugation and mixed with sample buffer. Proteins were resolved by SDS-PAGE (8% gel) and transferred onto poly-vinylidene difluoride (PVDF) membrane. The blot was blocked with 5% (w/v) non-fat milk in PBS containing 0.1% Tween for 1 h, washed with PBST, incubated overnight with antibodies against Kif4A (Santa Cruz Technology, USA), PARP-1 (Cell Signaling Technology, USA), PAR (SAB, USA) and α-tubulin (Sigma, USA), and incubated for 2 h with appropriate secondary antibody. Protein bands were visualized by ECL-plus (Amersham Biosciences, USA), and pictures were taken with the Chemi-Doc XPS imaging system.
Detection of cell apoptosis
The treated cells were washed twice with cold PBS, then harvested and resuspended in 1× binding buffer. The 5 μl FITC Annexin V and 5 μl PI (Propidium Iodide) were added to cell suspension and incubated for 10 min at room temperature in the dark, then 400 μl 1× binding buffer was added to each tube for FCM analysis (Becton Dickinson, USA) within 1 h.
Statistical analysis
The bands from Western blotting were quantified by Image-Pro Plus 6.0. Results were presented as mean ± standard deviation (SD). The t-test was performed using the Statistical Package for Social Science (SPSS for Windows package release 17.0). P < 0.05 was considered statistically significant. *P < 0.05; **P < 0.01.
RESULTS
ADR treatment induces Kif4A down-regulation and PARP-1 activation
Kif4A has been used to examine the status of DNA damage and DNA repair. It has been shown in many different cells that infringing DNA damage activates PARP-1. The effect of DNA damaging agent ADR on PARP-1 activity remains unknown. We hypothesized that the activation of PARP-1 induced by ADR may counteract the ADR-induced cytotxicity by promoting DNA repair, and PARP-1 activation was regulated by Kif4A. In Fig. 1, we used different concentrations of ADR to treat MCF-7 and MDA-MB-231 cells for 24 h, and found that the expression levels of Kif4A were gradually decreased in a dose-dependent manner (Figs. 1A and 1C). Moreover, ADR could induce PARP-1 activation in a dose-dependent manner (Figs. 1A and 1C). While the expression of full-length PARP-1 was not changed, smaller cleaved bands of PARP-1 were observed. Furthermore, we also treated MCF-7 and MDA-MB-231 cells with 1 μM ADR for 24 h and recovered the cells for 6, 12, 24, and 48 h. Our results showed that the expression of Kif4A was decreased gradually in a time-dependent manner, while the expression of PAR was increased gradually with longer recovery time (Figs. 2A and 2C). The expression of full-length PARP-1 was not significantly changed at all recovery time points.
Fig. 1.
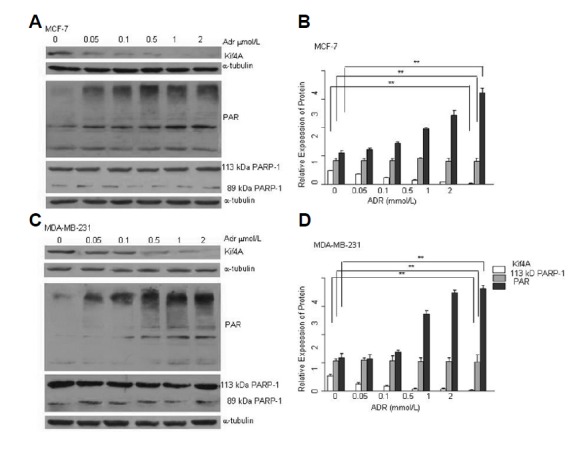
PARP-1 activation and Kif4A down-regulation after ADR treatment. Different concentrations of ADR induced PAR expression and Kif4A down-regulation in MCF-7 (A) and MDA-MB-231 (C) cell lines. After treatment with ADR (24 h) and recovery (12 h), cells were harvested and analyzed by Western blot for PAR and Kif4A expression. Relative expression levels of the three proteins were determined by Image-Pro Plus 6.0 in MCF-7 (B) and MDA-MB-231 (D) cells. **P < 0.01, compared with the control (0 μmol/L) (Kif4A) (B, D); **P < 0.01, compared with the control (PAR) (B, D); P > 0.05, compared with the control (113 KDa PARP-1) (B, D); n = 3.
Fig. 2.
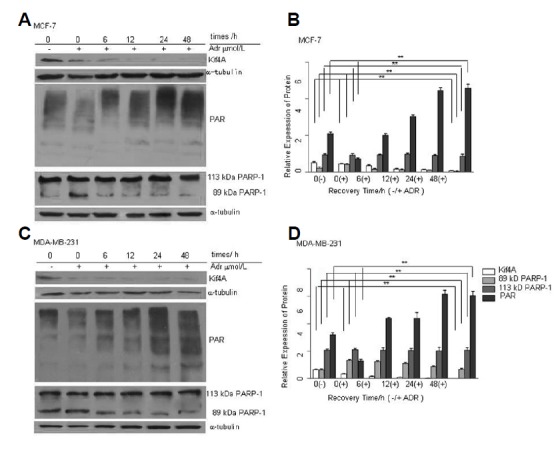
PARP-1 activation and Kif4A down-regulation after ADR treatment at different recovery time points. Time course of PAR and Kif4A expression in MCF-7 (A) and MDA-MB-231 (C) cell lines. After treatment with 1 μmol/L ADR (24 h) and different recovery time points (0, 6, 12, 24, 48 h), cells were harvested and analyzed by Western blot for PAR and Kif4A expression. Relative expression levels of the three proteins were determined by Image-Pro Plus 6.0 in MCF-7 (B) and MDA-MB-231(D) cells. **P < 0.01, compared with the 0 μmol/L doxorubicin group & the 0 recovery time group (Kif4A) (B, D); **P < 0.01 (B, D), compared with the 0 μmol/L doxorubicin group & the 0 recovery time group (PAR); **P < 0.01, compared with the 0 recovery time group (89 KDa PARP-1); P > 0.05, compared with the 0 μmol/L doxorubicin group & the 0 recovery time group (113 KDa PARP-1) (B, D); n = 3.
Kif4A expression is not affected by PARP-1 inhibitor
PARP-1 inhibitor has a potential clinical application. One of PARP-1 inhibitors, 3-ABA, can significantly decrease the expression of PAR transfer enzymes and prevent ADR-induced PARP-1 activation. To investigate whether PARP-1 inhibitor affects the expression of Kif4A, we treated MCF-7 and MDA-MB-231 cells with 2.5, 5, 10, and 20 μM 3-ABA 24 h, followed by 12 h of recovery. As shown in Fig. 3, 3-ABA inhibited the expression of PAR but did not affect the expression of Kif4A and the full-length PARP-1.
Fig. 3.
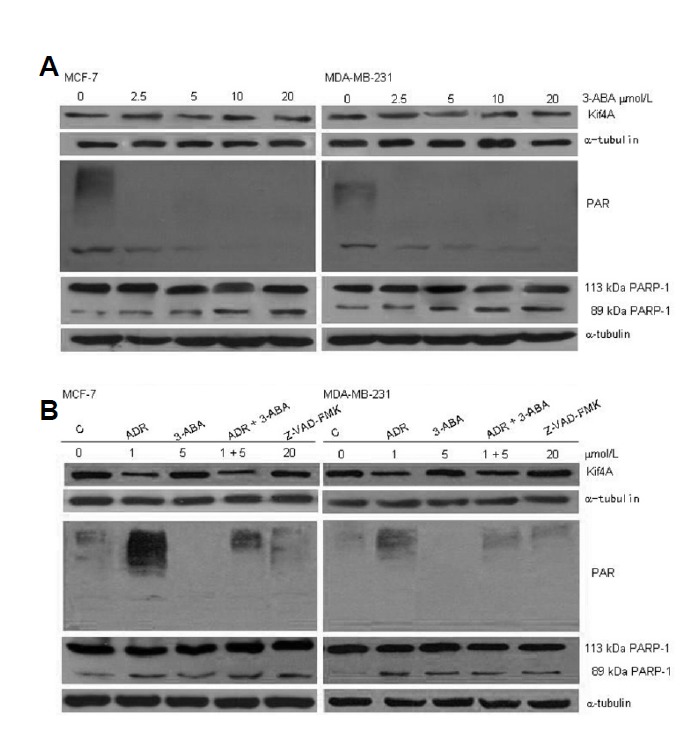
PARP-1 inhibitor 3-ABA treatment inhibits PARP-1 activation. MCF-7 and MDA-MB-231 cells were treated with 2.5, 5, 10, and 20 μmol/L 3-ABA for 24 h and recovered for 12 h. Cells were harvested and analyzed for the expression of Kif4A and PARP-1 by western blot, and α-tubulin was used as the loading control. Our results showed that 3-ABA treatment didn’t affect the expression of Kif4A and PARP-1.
Kif4A inhibits PARP-1 activation
It has been reported that Kif4A promotes neuronal survival through suppressing PARP-1 activity (Midorikawa et al., 2006). However, the relationship between Kif4A and PARP-1 in cancer has not been investigated. In this study, we transfected HA-Kif4A into MCF-7 cells using Lipofectamine® 2000. After 24 h, the cells were treated with 1 μM ADR for 24 h, followed by 12 h of recovery. We also verified the role of Kif4A in p53-deficient breast cancer cell line, MDA-MB-231. PARP-1 activation and PAR expression were significantly inhibited by Kif4A overexpression, as compared with those in the control and HA-pcDNA groups (Figs. 4A and 4C). The inhibition of PARP-1 activity in Kif4A-overexpressing cells was partially reversed by ADR treatment (Figs. 4A and 4C). However, Kif4A overexpression did not affect the expression of full-length PARP-1, suggesting that Kif4A can inhibit ADR-induced PARP-1 activation.
Fig. 4.
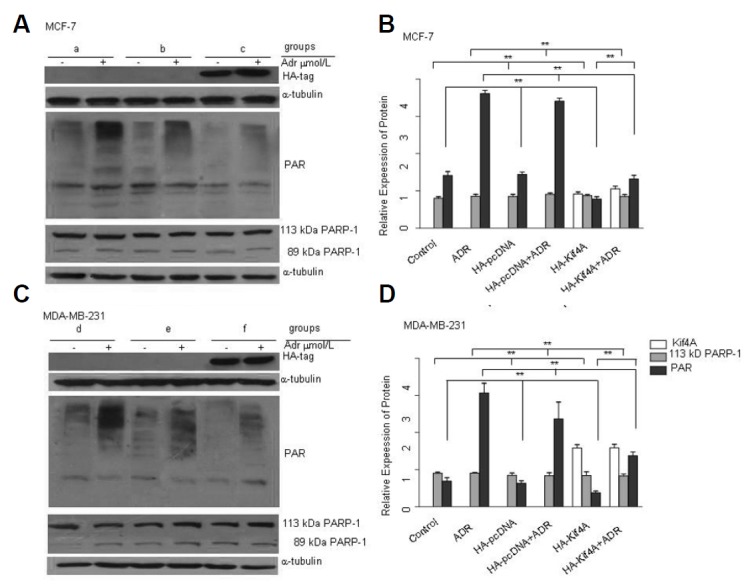
Kif4A inhibits PARP-1 activation. Overexpression of Kif4A significantly inhibited PAR formation but not PARP-1 expression in MCF-7 (A) and MDA-MB-231 (C) cells. Relative expression levels of the proteins were determined by Image-Pro Plus 6.0 in MCF-7 (B) and MDA-MB-231 (D) cells. **P < 0.01, compared with the control group & HA-pcDNA group (PAR) (B, D); **P < 0.01, compared with the ADR group & the HA-pcDNA + ADR group (PAR) (B, D); n = 3. (a: MCF-7, b: MCF-7+HA-pcDNA, c:MCF-7+HA-Kif4A, d: MDA-MB-231, e: MDA-MB-231+HA-pcDNA, f: MDA-MB-231 +HA-Kif4A)
Co-treatment of MCF-7 and MDA-MB-231 cells with doxorubicin and 3-ABA potentiates cell apoptosis
We further investigated whether the changes of Kif4A expression and PARP-1 activation induced by ADR might be directly related to the cytotoxicity. MCF-7 and MDA-MB-231 cells were treated with 5 μM 3-ABA and 1 μM ADR for 24 h, and recovered for 12 h. Cell apoptosis was examined by FCM. As shown in Fig. 5, either 3-ABA or ADR alone significantly increased cell apoptosis in MCF-7 cells to 5.94 ± 0.27% and 6.90 ± 0.17% [Figs. 5A(b) and 5A(c)], respectively, compared with the control group 3.85 ± 0.26% [Fig. 5A(a)] (P < 0.05). The co-treatment of 3-ABA and ADR led to further increase in apoptosis rate at 10.24 ± 0.81% [Fig. 5A(d)], which was significantly higher than that in 3-ABA or ADR treatment alone group (P < 0.05). In MDA-MB-231 cells, either 3-ABA or ADR alone was able to significantly increase the rates of cell apoptosis to 6.30 ± 0.48% and 7.83 ± 0.58% [Fig. 5A(f) and 5A(g)], respectively, compared with the control group 3.14 ± 0.67% [Fig. 5A(e)] (P < 0.05); the combined treatment of both drugs further increased the rate of apoptosis to 11.53 ± 0.45% [Fig. 5A(h)], compared with that of 3-ABA or ADR treatment alone (P < 0.05). Moreover, we observed that the degree of cell apoptosis was greater in MDA-MB-231 parental cells than that in MCF-7 cells [Figs. 5(d) and 5(h)].
Fig. 5.
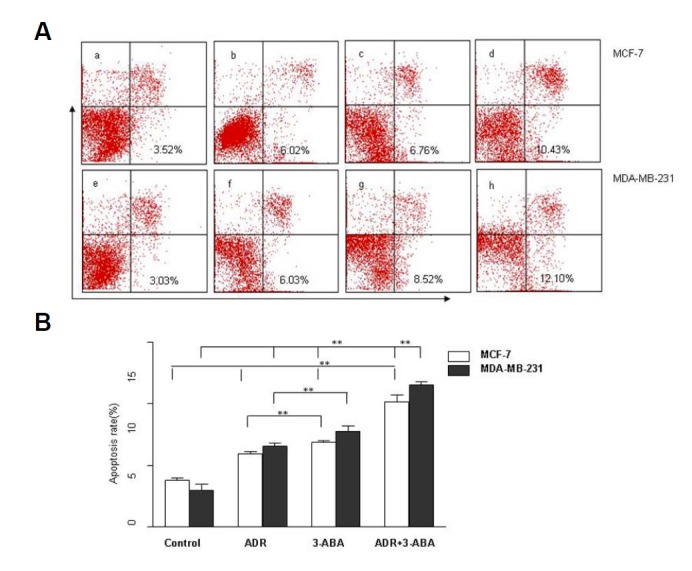
Effect of PARP-1 inhibition on cell apoptosis after ADR treatment. Cell apoptosis in MCF-7 and MDA-MB-231 cell lines was measured by FCM after co-treatment with 1 μM ADR and 5 μM 3-ABA for 24 h and recovery for 12 h. (A) The PARP-1 inhibitor 3-ABA significantly increased cell apoptosis after ADR treatment [A (d, h)]. **P < 0.01, compared with the 3-ABA group [A (b, f)] & the ADR group (MCF-7) [A (c, g)]; **P < 0.01, compared with the control group (MCF-7) [A (a, e)] **P < 0.01, compared with the ADR+3-ABA MCF-7 cell lines group [A (d)]; n = 3. (a: control group MCF-7, b: ADR-treated MCF-7, c:3-ABA-treated MCF-7, d: ADR and 3-ABA co-treated MCF-7, e: control group MDA-MB-231, f: ADR-treated MDA-MB-231, g:3-ABA-treated MDA-MB-231, h: ADR and 3-ABA co-treated MDA-MB-231)
Kif4A overexpression sensitizes breast cancer cells to apoptosis after ADR treatment
As shown in Fig. 6, Kif4A overexpression significantly increased cell apoptosis in MCF-7 cells (6.80 ± 0.23%) [Fig. 6A(e)], compared with that in the control group (4.10 ± 0.58%) [Fig. 6A(a)] and HA-pcDNA group (4.48 ± 0.42%) [Fig. 6A(c)] (P < 0.05). Moreover, Kif4A overexpression plus ADR treatment further increased cell apoptosis rate in MCF-7 cells to 9.23 ± 0.70% [Fig. 6A(f)], compared with the HA-Kif4A group (6.80 ± 0.23%) [Fig. 6A(e)] and HA-pcDNA + ADR group (7.19 ± 0.75%) [Fig. 56A(d)] (P < 0.05). In MDA-MB-231 cells, overexpression of Kif4A also significantly increased cell apoptosis (8.36 ± 0.79%) [Fig. 6A(k)], compared with the control group (3.85 ± 0.66%) [Fig. 6A(g)] and the HA-pcDNA group (4.44 ± 0.36%) [Fig. 6A(i)] (P < 0.05). Similarly, Kif4A overexpression plus ADR treatment further increased the apoptosis rate to 11.40 ± 0.68% [Fig. 6A(l)], which was significantly higher than that in the HA-Kif4A group (8.36 ± 0.79%) [Fig. 6A(k)] and HA-pcDNA+ADR group (7.10 ± 0.61%) [Fig. 6A(j)] (P < 0.05).
Fig. 6.
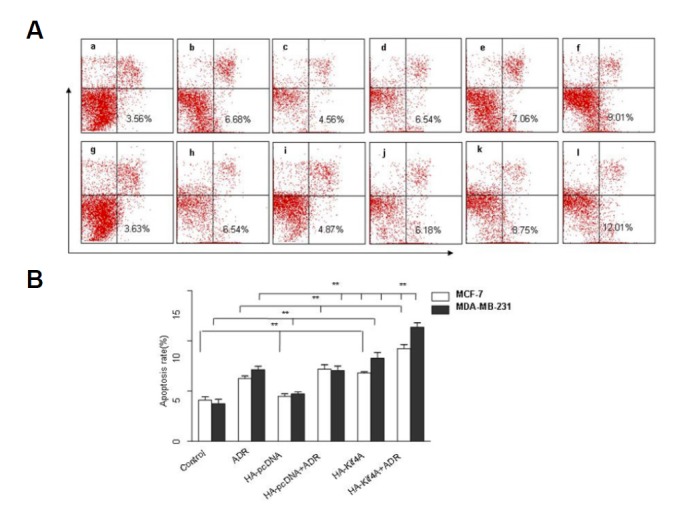
Effects of Kif4A overexpression on cell apoptosis after ADR treatment. Cell apoptosis in Kif4A-overexpressing MCF-7 and MDA-MB-231 cells was examined by FCM after treatment with 1 μmol/L ADR. (A) Kif4A over-expression inhibited PARP-1 activation and significantly increased cell apoptosis after ADR treatment [A (f, l)]. **P<0.01, compared with the control group [A (a, g)]& the HA-pcDNA group (MCF-7) [A (c, i)]; **P<0.01, compared with the ADR group [A (b, h)]& the HA-pcDNA + ADR group [A (d, j)] & the HA-Kif4A group (MCF-7) [A (e, k)]; **P<0.01, compared with the HA-Kif4A +ADR MCF-7 cells group [A (f)]; n=3. (a: control group MCF-7, b: ADR-treated MCF-7, c: HA-pcDNA-treated MCF-7, d: ADR and HA-pcDNA co-treated MCF-7, e: HA-Kif4A-treated MCF-7, f: ADR and HA-Kif4A co-treated MCF-7, g: control group MDA-MB-231, h: ADR-treated MDA-MB-231, i: HA-pcDNA-treated MDA-MB-231, j: ADR and HA-pcDNA co-treated MDA-MB-231, k: HA-Kif4A-treated MDA-MB-231, l: ADR and HA-Kif4A co-treated MDA-MB-231).
DISCUSSION
ADR is an widely used anticancer agent with broad spectrum of activities involving DNA intercalation, free-radical formation, covalent DNA binding, inhibition of the topoisomerase II, and DNA strand breakage (Hydock et al., 2012; Zaremba et al., 2010). Extensive studies have contributed to understanding ADR chemoresistance (Xu et al., 2013). Recent studies demonstrated that permeability glycoprotein (P-gp) (Sun et al., 2012; Zheng et al., 2010), breast cancer resistance protein (BCRP)(Sukowati et al., 2012), multidrug resistance-associated proteins (MRPs) (Villar et al., 2012), breast cancer associated 1 (BRCA1), and PARP-1 may all play important roles in chemotherapy resistance (Alli et al., 2011). PARP-1 may be activated by ADR for DNA repair (Farmer et al., 2005; Javle and Curtin, 2011). PARP-1 has caused much attention because PARP-1 inhibitors have been used as new therapeutic drugs for different cancers. Studies have shown that compare with ADR, PARP-1 inhibitors are more effective to BRCA1 mutant breast cancer (Ibragimova and Cairns, 2011) and/or triple-negative breast cancer (Oonk et al., 2012), suggesting a correlation between ADR resistance and PARP-1 activation. It has been demonstrated that both PARP and BRCA regulate the pathways of DNA repair. Due to the accumulation of mutations induced by error-prone DNA repair, breast cancers mostly develop in the setting of BRCA deficiency. A series of recent clinical trials have tested the application of PARP inhibition as a therapeutic strategy to target BRCA-deficient tumors (Annunziata and Bates, 2010). PARP-1 activity can also be a biomarker of DNA repair deficiency in breast cancer (Jacot et al., 2013). It has been reported that Kif4A regulates middle-spindle formation by interacting with the protein regulator of cytokinesis 1, PRC1 (Shrestha et al., 2012). Kif4A is a candidate molecule that may play critical roles in DNA damage response and/or DNA repair pathways (Wu et al., 2008). Midorikawa et al. (2006) demonstrated that Kif4A promotes neuron apoptosis by suppressing PARP-1 activity. These findings prompt us to investigate the impact of ADR on Kif4A and PARP-1 as a potential novel mechanism of resistance mechanism. Furthermore, it has been reported that PARP inhibition may represent a novel way of selectively targeting p53-deficient breast cancer cells (Munoz-Gamez et al., 2005). To further verify our hypothesis, we used two breast cancer cells MCF-7 (with wild-type p53) and MDA-MB-231 (p53-deficient) to study whether p53 play a role in PARP-1 inhibition-induced apoptosis. The results showed that PARP inhibition was more selective in p53-deficient MDA-MB-231 cells compared with wild-type p53 MCF-7 cells.
In this study, we found that in two of breast cancer cell lines: (1) ADR treatment inhibited the expression Kif4A but activated PARP-1; (2) More directly, ectopic expression of Kif4A inhibited PARP-1 activity, decreased DNA-repair function, and induced cell apoptosis; (3) ADR treatment induced cell apoptosis by inhibiting Kif4A expression along with increasing the PARP-1 activity to enhance the effect on DNA-damage. We also found that the combined treatment of ADR and PARP-1 inhibitor further enlarged the rate of apoptosis compared with ADR treatment alone, suggesting a role of PARP-1 in ADR resistance. Furthermore, we showed that ADR did not affect exogenous Kif4A; ADR might mainly regulate Kif4A in the process of transcription. Our findings may help further understand the mechanisms of PARP-1-mediated ADR resistance.
Therefore, a clear understanding of the relationships among ADR, Kif4A, PARP-1, BRCA1 and BRCA2 in the future may help to reveal the mechanism of ADR resistance in some cancer cells.
Acknowledgments
This research project was supported by national natural science foundation of China (No.81171653, 30972703), natural science foundation of Jiangsu province (BK2011246, BK2011247), the project of Six batch of major talent summit (BRA2010037), society development plans, department of science and technology Changzhou (CE20135048, CJ20112020, CZ20110024, CS20102020).
REFERENCES
- Alli E., Sharma V.B., Hartman A.R., Lin P.S., McPherson L., Ford J.M. Enhanced sensitivity to cisplatin and gemcitabine in Brca1-deficient murine mammary epithelial cells. BMC Pharmacol. 2011;11:7. doi: 10.1186/1471-2210-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata C.M., Bates S.E. PARP inhibitors in BRCA1/BRCA2 germline mutation carriers with ovarian and breast cancer. F1000 Biol. Rep. 2010;2 doi: 10.3410/B2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borysov S.I., Granic A., Padmanabhan J., Walczak C.E., Potter H. Alzheimer Abeta disrupts the mitotic spindle and directly inhibits mitotic microtubule motors. Cell Cycle. 2011;10:1397–1410. doi: 10.4161/cc.10.9.15478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campalans A., Kortulewski T., Amouroux R., Menoni H., Vermeulen W., Radicella J.P. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res. 2013;41:3115–3129. doi: 10.1093/nar/gkt025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Hu C.K., Coughlin M., Field C.M., Mitchison T.J. KIF4 regulates midzone length during cytokinesis. Curr. Biol. 2011;21:815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huambachano O., Herrera F., Rancourt A., Satoh M.S. Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J. Biol. Chem. 2011;286:7149–7160. doi: 10.1074/jbc.M110.175190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hydock D.S., Lien C.Y., Jensen B.T., Parry T.L., Schneider C.M., Hayward R. Rehabilitative exercise in a rat model of doxorubicin cardiotoxicity. Exp. Biol. Med. 2012;237:1483–1492. doi: 10.1258/ebm.2012.012137. [DOI] [PubMed] [Google Scholar]
- Ibragimova I., Cairns P. Assays for hypermethylation of the BRCA1 gene promoter in tumor cells to predict sensitivity to PARP-inhibitor therapy. Methods Mol. Biol. 2011;780:277–291. doi: 10.1007/978-1-61779-270-0_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot W., Thezenas S., Senal R., Viglianti C., Laberenne A.C., Lopez-Crapez E., Bibeau F., Bleuse J.P., Romieu G., Lamy P.J. BRCA1 promoter hypermethylation, 53BP1 protein expression and PARP-1 activity as biomarkers of DNA repair deficit in breast cancer. BMC Cancer. 2013;13:523. doi: 10.1186/1471-2407-13-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javle M., Curtin N.J. The role of PARP in DNA repair and its therapeutic exploitation. Br. J. Cancer. 2011;105:1114–1122. doi: 10.1038/bjc.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier M.F., Planck J.L., Roy S., Pascal J.M. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 2011;286:10690–10701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Kraus W., Hottiger M.O. PARP-1 and gene regulation: progress and puzzles. Mol. Aspects Med. 2013;34:1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Martinez-Romero R., Canuelo A., Siles E., Oliver F.J., Martinez-Lara E. Nitric oxide modulates hypoxia-inducible factor-1 and poly(ADP-ribose) polymerase-1 cross talk in response to hypobaric hypoxia. J. Appl. Physiol. 2012;112:816–823. doi: 10.1152/japplphysiol.00898.2011. [DOI] [PubMed] [Google Scholar]
- Midorikawa R., Takei Y., Hirokawa N. KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell. 2006;125:371–383. doi: 10.1016/j.cell.2006.02.039. [DOI] [PubMed] [Google Scholar]
- Munoz-Gamez J.A., Martin-Oliva D., Aguilar-Quesada R., Canuelo A., Nunez M.I., Valenzuela M.T., Ruiz de Almodovar J.M., De Murcia G., Oliver F.J. PARP inhibition sensitizes p53-deficient breast cancer cells to doxorubicin-induced apoptosis. Biochem. J. 2005;386:119–125. doi: 10.1042/BJ20040776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oonk A.M., van Rijn C., Smits M.M., Mulder L., Laddach N., Savola S.P., Wesseling J., Rodenhuis S., Imholz A.L., Lips E.H. Clinical correlates of ‘BRCAness’ in triple-negative breast cancer of patients receiving adjuvant chemotherapy. Ann. Oncol. 2012;23:2301–2305. doi: 10.1093/annonc/mdr621. [DOI] [PubMed] [Google Scholar]
- Shrestha S., Wilmeth L.J., Eyer J., Shuster C.B. PRC1 controls spindle polarization and recruitment of cytokinetic factors during monopolar cytokinesis. Mol. Biol. Cell. 2012;23:1196–1207. doi: 10.1091/mbc.E11-12-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisavljevic J., Porta-de-la-Riva M., Batlle R., de Herreros A.G., Baulida J. The p65 subunit of NF-kappaB and PARP1 assist Snail1 in activating fibronectin transcription. J. Cell Sci. 2011;124:4161–4171. doi: 10.1242/jcs.078824. [DOI] [PubMed] [Google Scholar]
- Sukowati C.H., Rosso N., Pascut D., Anfuso B., Torre G., Francalanci P., Croce L.S., Tiribelli C. Gene and functional up-regulation of the BCRP/ABCG2 transporter in hepatocellular carcinoma. BMC Gastroenterol. 2012;12:160. doi: 10.1186/1471-230X-12-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Chen Z., Li L., Sun D., Tian Y., Pan H., Bi H., Huang M., Zeng S., Jiang H. The two enantiomers of tetrahydropalmatine are inhibitors of P-gp, but not inhibitors of MRP1 or BCRP. Xenobiotica. 2012;42:1197–1205. doi: 10.3109/00498254.2012.702247. [DOI] [PubMed] [Google Scholar]
- Villar V.H., Vogler O., Martinez-Serra J., Ramos R., Calabuig-Farinas S., Gutierrez A., Barcelo F., Martin-Broto J., Alemany R. Nilotinib counteracts P-glycoprotein-mediated multidrug resistance and synergizes the antitumoral effect of doxorubicin in soft tissue sarcomas. PLoS One. 2012;7:e37735. doi: 10.1371/journal.pone.0037735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandke C., Barisic M., Sigl R., Rauch V., Wolf F., Amaro A.C., Tan C.H., Pereira A.J., Kutay U., Maiato H., et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 2012;198:847–863. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Zhou L., Khidr L., Guo X.E., Kim W., Lee Y.M., Krasieva T., Chen P.L. A novel role of the chromokinesin Kif4A in DNA damage response. Cell Cycle. 2008;7:2013–2020. doi: 10.4161/cc.7.13.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Diao Y., Qi S., Pan X., Wang Q., Xin Y., Cao X., Ruan J., Zhao Z., Luo L., et al. Phosphorylated Hsp27 activates ATM-dependent p53 signaling and mediates the resistance of MCF-7 cells to doxorubicin-induced apoptosis. Cell. Signal. 2013;25:1176–1185. doi: 10.1016/j.cellsig.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Zaremba T., Thomas H., Cole M., Plummer E.R., Curtin N.J. Doxorubicin-induced suppression of poly(ADP-ribose) polymerase-1 (PARP-1) activity and expression and its implication for PARP inhibitors in clinical trials. Cancer Chemother. Pharmacol. 2010;66:807–812. doi: 10.1007/s00280-010-1359-0. [DOI] [PubMed] [Google Scholar]
- Zheng Z., Aojula H., Clarke D. Reduction of doxorubicin resistance in P-glycoprotein overexpressing cells by hybrid cell-penetrating and drug-binding peptide. J. Drug Target. 2010;18:477–487. doi: 10.3109/10611860903548347. [DOI] [PubMed] [Google Scholar]


