Abstract
The balance between bone formation by osteoblasts and destruction of mineralized bone matrix by osteoclasts is important for bone homeostasis. The increase of osteoclast differentiation by RANKL induces bone diseases such as osteoporosis. Recent studies have shown that insulin is one of main factors mediating the cross-talk between bone remodeling and energy metabolism. However, the systemic examination of insulin-induced differential gene expression profiles in osteoclasts has not been extensively studied. Here, we investigated the global effects of insulin on osteoclast precursors at the level of gene transcription by microarray analysis. The number of genes that were up-regulated by ≥ 1.5 fold after insulin treatment for 6 h, 12 h, or 24 h was 76, 73, and 39; and 96, 83, and 54 genes were down-regulated, respectively. The genes were classified by 20 biological processes or 24 molecular functions and the number of genes involved in ‘development processes’ and ‘cell proliferation and differentiation’ was 25 and 18, respectively, including Inhba, Socs, Plk3, Tnfsf4, and Plk1. The microarray results of these genes were verified by real-time RT-PCR analysis. We also compared the effects of insulin and RANKL on the expression of these genes. Most genes had a very similar pattern of expressions in insulin- and RANKL-treated cells. Interestingly, Tnfsf4 and Inhba genes were affected by insulin but not by RANKL. Taken together, these results suggest a potential role for insulin in osteoclast biology, thus contributing to the understanding of the pathogenesis and development of therapeutics for numerous bone and metabolic diseases.
Keywords: insulin, microarray, osteoclasts
INTRODUCTION
Bone homeostasis is mediated by the balanced regulation of bone-forming osteoblasts and bone-resorbing osteoclasts (Boyle et al., 2003). Osteoclasts are derived from hematopoietic progenitors of the monocyte/macrophage lineage and the binding of receptor activator of nuclear factor-κB (NF-κB) ligand (RANKL) to its receptor, RANK, on osteoclast precursors induces osteoclast differentiation (Leibbrandt and Penninger, 2009; Teitelbaum, 2007). The elevation of bone resorption by osteoclasts causes a variety of bone diseases such as osteoporosis, Paget’s disease, rheumatoid arthritis, and periodontitis (Boyle et al., 2003; Harada and Rodan, 2003; Karsenty and Wagner, 2002; Teitelbaum, 2000).
Differentiation of osteoclasts is accompanied by changes in the expression levels of various genes (Boyle et al., 2003; Karsenty and Wagner, 2002; Teitelbaum, 2000). Several techniques including differential display-PCR (Gori et al., 2001; Han et al., 2010), subtractive hybridization (Kimura et al., 2008; Petersen et al., 2000), and cDNA microarrays (Kim et al., 2010; Wang et al., 2013) have been employed to investigate differentially-expressed genes during differentiation of osteoblasts or osteoclasts. The development of cDNA microarrays allowed the screening of quantitative differences of thousands of genes. While most studies have focused on either osteoblasts or molecular change in response to RANKL (Kim et al., 2010; Wang et al., 2013), total analyses of gene expression in response to insulin stimulation in osteoclasts have little been studied.
Insulin is an important regulator not only of glucose metabolism by increasing cellular glucose uptake but also of bone metabolism. Insulin receptors are expressed in many cell types including osteoblasts, chondrocytes, and osteoclasts and the activation of insulin receptor by insulin stimulation induces its downstream mediators, thus regulating cellular metabolism, growth, survival, and differentiation (Ferron et al., 2010; Fulzele et al., 2010; Lee and Lee, 2014; Thomas et al., 1998; Yang et al., 2010). Recently, we have shown that insulin promoted osteoclast proliferation through up-regulation of cyclinD1 and Bcl2A1 (Lee and Lee, 2014). Studies using osteoblast-specific insulin receptor-deficient mice have demonstrated that insulin receptor signaling in osteoblasts affects not only osteoblast proliferation but also energy metabolism (Ferron et al., 2010; Fulzele et al., 2010). Moreover, patients with diabetes mellitus exhibit altered bone metabolism with resultant changes in bone mineral density (BMD) (Kemink et al., 2000; Thrailkill et al., 2005). Thus, insulin is one of main factors mediating the crosstalk between bone remodeling and energy metabolism.
Here, we investigated the genes that were differentially-expressed by insulin on osteoclast precursors using cDNA microarray. The identification of these genes provided the first systematic information on the response of the osteoclast precursors to insulin at the level of gene transcription. It could be useful in identifying biochemical markers for the diagnosis of numerous bone and metabolic diseases, or as potential targets for the development of therapeutic agents.
MATERIALS AND METHODS
Isolation of bone marrow precursors and cell culture
Isolation of bone marrow precursors was performed as described previously (Choi et al., 2013). Briefly, bone marrow cells were flushed out from the femur of 4–6-week-old C57BL/6 mice with a sterile 21-gauge syringe and incubated in alpha-MEM media containing 10% FBS and 10 ng/ml M-CSF (R&D Systems). After 24 h, non-adherent cells were harvested and cultured in the presence of M-CSF (20 ng/ml) for 3 days. After washing out the non-adherent cells, adherent cells were used as bone marrow-derived monocytes/macrophages (BMMs). Culture media was changed every 2 days.
RNA preparation
Total RNA was extracted using Trizol (Invitrogen Life Technologies, USA) and then purified using RNeasy columns (Qiagen, USA) according to the manufacturer’s protocol. For quality control, the RNA purity and integrity were evaluated by denaturing gel electrophoresis, measuring the absorbance at 260 and 280 nm, and analyzed on Agilent 2100 Bioanalyzer (Agilent Technologies, USA).
Microarray experiments
Total RNA was amplified and purified using an Ambion Illumina RNA amplification kit (Ambion, USA) to yield biotinylated cRNA according to the manufacturer’s instructions. Briefly, 550 ng of total RNA were reverse- transcribed to cDNA using a T7 oligo (dT) primer. Second- strand cDNA was synthesized, transcribed in vitro, and labeled with biotin-NTP. After purification, the cRNA was quantified using a ND-1000 spectrophotometer (NanoDrop, USA). A total of 750 ng of labeled cRNA samples were hybridized to each mouse-8 expression bead array for 16–18 h at 58°C, according to the manufacturer’s instructions (Illumina, Inc., USA). Detection of an array signal was conducted using Amersham fluorolink streptavidin-Cy3 (GE Healthcare Bio-Sciences, UK) following the bead array manual. Arrays were scanned with an Illumina bead array reader confocal scanner according to the manufacturer’s instructions. The quality of hybridization and overall chip performance were monitored by visual inspection of both internal quality control checks and the raw scanned data. Raw data were extracted by the software provided by the manufacturer [Illumina GenomeStudio v2009.2 (Gene Expression Module v1.5.4)]. Probe signal value was transformed by logarithm and normalized by the quantile method. The comparative analysis between the test and control groups was carried out using fold-change, LPE test adjusted FDR p value. False discovery rate (FDR) was controlled by adjusting the p value using the Benjamini-Hochberg algorithm. Go-ontology analysis for the list of significant probes was performed using PANTHER (http://www.pantherdb.org/panther/ontologies.jsp), using text files containing Gene ID list and the accession number of illumina probe ID.
cDNA synthesis and real-time PCR
Total RNA isolated from cells treated with RANKL (Peprotech) or insulin (Sigma) was used as a template for cDNA synthesis. The reverse transcription of total RNA to cDNA was performed with Superscript III reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. Real-time PCRs were performed with the Brilliant UltraFast SYBR Green QPCR Master Mix (Agilent Technologies) and specific primers for target genes and HPRT (for endogenous control) from QIAGEN in triplicates on an MX3000 instrument (Agilent Technologies). HPRT was used for normalization of all quantitation.
Statistical analysis
Data were expressed as mean ± S.D. from at least 3 independent experiments. Statistical differences were analyzed by student’s t-test. P < 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
Analysis of microarray expression data
We isolated BMMs from mice femur and then treated the cells with 10 nM of insulin for 6 h, 12 h or 24 h in the presence of MCSF, to explore genes regulated by insulin in osteoclast precursors. Stimulation with insulin for 6 h resulted in significant up-regulation of 76 genes and down-regulation of 96 genes; and for 12 h, up-regulation of 73 genes and down-regulation of 83 genes. Insulin treatment for 24 h caused up-regulation of 39 genes and down-regulation of 54 genes compared to the non-treated cells (Fig. 1). These data suggested that stimulation with insulin induced a change of immediate-early genes rather than late genes in osteoclast precursors. Furthermore, the results indicated that insulin signaling in osteoclasts may be associated with cellular and molecular events of proliferation or differentiation in osteoclasts.
Fig. 1.
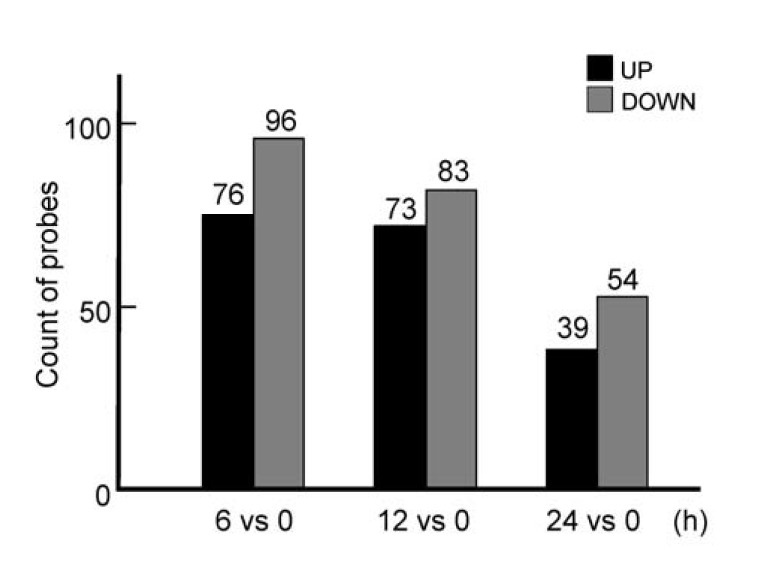
The number of genes changed by insulin treatment. The number of genes changed by ≥ 1.5 fold in cells treated with 10 nM of insulin for 6 h, 12 h or 24 h compared to un-treated controls, were determined after microarray.
Gene expression profiles in insulin-treated cells
Up- or down-regulated genes in response to insulin treatment were classified according to the Panther group analysis based on 20 biological processes or 24 molecular functions (http://www.pantherdb.org) as presented in Figs. 2A and 2B, respectively. Among the groups classified by biological processes, the biggest was ‘signal transduction’, i.e. 109 genes, in which cytokines such as interleukin-15 (IL-15) and chemokine (C-X-C motif) ligand 10 were included (data not shown). The next was ‘immunity- and defense’, i.e. 69 genes. 25 genes were found to be associated with ‘developmental processes’ and 18 genes with ‘cell proliferation and differentiation’. The number of ‘receptor’- and ‘signaling molecules’-included genes based on 24 molecular functions, were 35 and 31 respectively. This result revealed that insulin induced changes in expression of genes involved in various cellular and molecular events in osteoclasts.
Fig. 2.
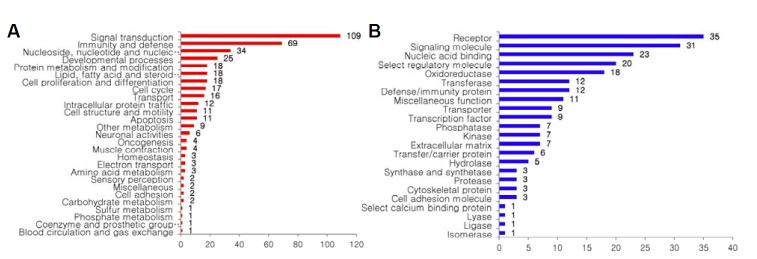
Analyses of genes changed by insulin treatment. The genes were analyzed by grouping according to biological process (A) or molecular function (B) and counted.
Most of these genes are involved in ‘housekeeping’ activities of many cell types. We focused on the ‘developmental processes’- and ‘cell proliferation and differentiation’-related genes since the identification of these genes can help to elucidating the molecular mechanism that underlies bone remodeling and energy metabolism. The lists of ‘developmental processes’- and ‘cell proliferation and differentiation’-related genes were presented in Tables 1 and 2, respectively. As shown in Table 1, Inhba, Socs, Mllt11, Clec2d, Dscr1, Tnc, Plk3, and Egr2 were up-regulated in the insulin-treated groups. Conversely, Emb, Tlr8, Tlr13, Ednrb, Vim, Itm2b, Fabp5, Plk1, Tgfbr2, Ang, and Fgd2 were down-regulated. In the ‘cell proliferation and differentiation’ group, IL1a, Cxcl10, Sphk1, Il1b, Cdkn1a, Plk3, Inhba, Ccnd1, Cd86, and Pdgfb were up-regulated whereas Tnfsf4, Ednrb, Uhrf1, and Plk1 were down-regulated (Table 2).
Table 1.
List of “developmental processes”-related genes regulated by insulin treatment in primary osteoclast precursors
| Gene symbol | Description | 6 h (fold) | 12 h (fold) | 24 h (fold) |
|---|---|---|---|---|
| Emb | Embigin | −1.52 | −1.34 | −1.38 |
| Tlr8 | Toll-like receptor 8 | −1.58 | −1.39 | −1.30 |
| Tlr13 | Toll-like receptor 13 | −1.58 | −1.43 | −1.32 |
| Inhba | Inhibin beta-A | 1.78 | 1.54 | 1.36 |
| Socs2 | Suppressor of cytokine signaling 2 | 1.59 | 1.54 | 1.26 |
| Mllt11 | Mixed-lineage leukemia; translocated to,11 | 1.58 | 1.51 | 1.34 |
| Clec2d | C-type lectin domain family 2, member d | 1.43 | 1.54 | 1.39 |
| Ednrb | Endothelin receptor type B | −1.46 | −1.53 | −1.65 |
| Vim | Vimentin | −1.17 | −1.27 | −1.53 |
| Itm2b | Integral membrane protein 2B | −1.50 | −1.32 | −1.26 |
| Fabp5 | Fatty acid binding protein 5 | −1.61 | −1.44 | −1.24 |
| Dscr1 | Down syndrome critical region gene 1 | 1.97 | 2.06 | 2.11 |
| Tnc | Tenascin C | 1.56 | 1.71 | 1.71 |
| Plk1 | Polo-like kinase 1 (Drosophila) | −1.66 | −1.70 | −1.42 |
| Plk3 | Polo-like kinase 3 (Drosophila) | 1.89 | 1.82 | 1.40 |
| Tgfbr2 | Transforming growth factor, beta receptor 2 | −1.81 | −1.63 | −1.36 |
| Egr2 | Early growth response 2 | 1.74 | 1.70 | 1.22 |
| Ang | Angiogenin | −1.69 | −1.59 | −1.40 |
| Fgd2 | FYVE, RhoGEF and PH domain containing 2 | −1.80 | −1.56 | −1.37 |
Table 2.
List of “cell proliferation & differentiation”-related genes regulated by insulin treatment in primary osteoclast precursors
| Gene symbol | Description | 6 h (fold) | 12 h (fold) | 24 h (fold) |
|---|---|---|---|---|
| Tnfsf4 | Tumor necrosis factor superfamily, member 4 | −1.41 | −1.55 | −1.56 |
| Ednrb | Endothelin receptor type B | −1.46 | −1.53 | −1.65 |
| Uhrf1 | Ubiquitin-like, containing PHD and RING finger domains, 1 | −1.63 | −1.55 | −1.33 |
| Plk1 | Polo-like kinase 1 (Drosophila) | −1.66 | −1.70 | −1.42 |
| Il1a | Interleukin1 alpha | 2.93 | 2.48 | 1.72 |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | 2.29 | 1.96 | 1.71 |
| Sphk1 | Sphingosine kinase 1 | 1.99 | 1.96 | 1.73 |
| Il1b | Interleukin1 beta | 1.71 | 1.78 | 1.51 |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A | 1.65 | 1.54 | 1.62 |
| Plk3 | Polo-like kinase 3 (Drosophila) | 1.89 | 1.82 | 1.40 |
| Inhba | Inhibin beta-A | 1.78 | 1.54 | 1.36 |
| Ccnd1 | CyclinD1 | 1.60 | 1.56 | 1.41 |
| Cd86 | CD86 antigen | 1.59 | 1.63 | 1.41 |
| Pdgfb | Platelet-derived growth factor beta polypeptide | 1.48 | 1.54 | 1.39 |
Validation of microarray results and comparison with the effects of RANKL
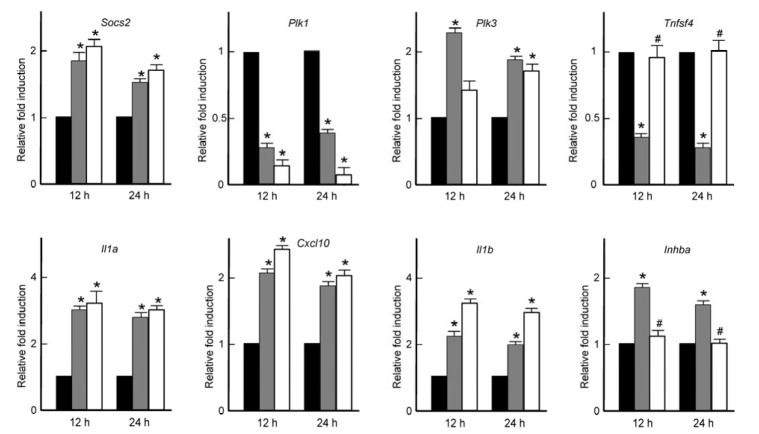
Real-time PCR was performed to verify the gene expression profiles provided by the microarray analysis. Microarray results were confirmed by real-time PCR and, overall, real-time PCR results were found to be very similar to the microarray results, as shown in Fig. 3: insulin induced mRNA expression of Socs2, Plk3, Il1a, Cxcl10, Il1b, and Inhba, while it repressed the expression of Plk1 and Tnfsf4 in osteoclast precursors. These results corroborated microarray as an efficient technique to identify a large number of differentially expressed genes.
Fig. 3.
Validation of microarray results and comparison with the effects of RANKL. Relative expression of each gene induced by 10 nM of insulin or 200 nM of RANKL for the indicated times by real-time PCR. *P < 0.05 vs. cells without stimulation, #P < 0.05 vs. cells treated with insulin.
Changes in the genes expression were examined after RANKL treatment for 12 h and 24 h, to study the comparative effects of insulin and RANKL in osteoclast precursors. Most of the genes, except Tnfsf4 and Inhba, showed very similar expression patterns between insulin- and RANKL-treated cells. Among these genes, Socs2 was increased in response to both insulin and RANKL in osteoclast precursors. Suppressor of cytokine signaling 2 (SOCS2) is a suppressor of the Janus kinase (JAK) and signal transducers and activators of transcription (STAT) signal transduction pathway (Krebs and Hilton, 2001; Lorentzon et al., 2005). The role of SOCS2 seems to be controversial since Socs2−/− mice displayed an increased longitudinal skeletal growth associated with a deregulated GH/IGF-I signaling (Metcalf et al., 2000) whereas it resulted in reduced trabecular and cortical volumetric BMD (Lorentzon et al., 2005). The cellular role of SOCS2 by insulin and RANKL remains to be investigated further. Polo-like kinases (PLKs) consist of a family of kinases which play critical roles during multiple stages of cell cycle progression (Smits et al., 2000; Takai et al., 2005; van Vugt et al., 2001). Whereas Plk1 has been linked to cell proliferation, several studies have observed that Plk3 expression was negatively correlated with cancer development (Dai et al., 2000; 2002; Takai et al., 2005). In the present study, we found that whereas Plk3 was up-regulated, Plk1 was down-regulated by insulin and RANKL treatment, which implied that insulin and RANKL may contribute to osteoclast differentiation by regulating proliferation through diametrically opposed control on PLK1 and PLK3.
Interestingly, Tnfsf4 and Inhba genes were differentially affected by insulin and RANKL: both were not changed by RANKL but notably decreased or increased by insulin, respectively. OX40L, a member of the TNF family, encoded by Tnfsf4 gene (Ohshima et al., 1997), played a role as a late checkpoint in diabetes development (Pakala et al., 2004). The targeting of OX40L reduced the incidence of diabetes (Pakala et al., 2004) but promoted osteoclastogenesis (Gwyer Findlay et al., 2014). Activins betaA (encoded by Inhba) is a member of the TGF-beta superfamily. Inhba-deficient mice were small and lean because of reduced IGF-1 levels, the abnormalities of growth plates, and the increased mitochondrial energy metabolism (Brown et al., 2003; Li et al., 2009). Based on these earlier observations, our results implied that OX40L and activin may be involved in osteoclast insulin receptor signaling, rather than RANKL, and thus specifically on osteoclasts or islet biology.
Taken together, these results implied that insulin could play not only similar roles on osteoclast precursors as RANKL, or enhance the effects of RANKL, but also have unique independent roles. Although, the direct role of insulin on osteoclast differentiation or RANKL signaling and the examination to molecular pathways involving these genes are still the subjects of further research, this study gives hints for the development of therapeutic agents for bone and metabolic diseases.
Acknowledgments
This work was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (NRF-2010-0025761) and the Soonchunhyang University Research Fund.
REFERENCES
- Boyle W.J., Simonet W.S., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Brown C.W., Li L., Houston-Hawkins D.E., Matzuk M.M. Activins are critical modulators of growth and survival. Mol. Endocrinol. 2003;17:2404–2417. doi: 10.1210/me.2003-0051. [DOI] [PubMed] [Google Scholar]
- Choi J., Choi S.Y., Lee S.Y., Lee J.Y., Kim H.S., Lee S.Y., Lee N.K. Caffeine enhances osteoclast differentiation and maturation through p38 MAP kinase/Mitf and DC-STAMP/CtsK and TRAP pathway. Cell. Signal. 2013;25:1222–1227. doi: 10.1016/j.cellsig.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Dai W., Li Y., Ouyang B., Pan H., Reissmann P., Li J., Wiest J., Stambrook P., Gluckman J.L., Noffsinger A., Bejarano P. PRK, a cell cycle gene localized to 8p21, is downregulated in head and neck cancer. Genes Chromosomes Cancer. 2000;27:332–336. doi: 10.1002/(sici)1098-2264(200003)27:3<332::aid-gcc15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Dai W., Liu T., Wang Q., Rao C.V., Reddy B.S. Down-regulation of PLK3 gene expression by types and amount of dietary fat in rat colon tumors. Int. J. Oncol. 2002;20:121–126. [PubMed] [Google Scholar]
- Ferron M., Wei J., Yoshizawa T., Del Fattore A., DePinho R.A., Teti A., Ducy P., Karsenty G. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell. 2010;142:296–308. doi: 10.1016/j.cell.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulzele K., Riddle R.C., DiGirolamo D.J., Cao X., Wan C., Chen D, Faugere M.C., Aja S., Hussain M.A., Brüning J.C., Clemens T.L. Insulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body composition. Cell. 2010;142:309–319. doi: 10.1016/j.cell.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori F., Divieti P., Demay M.B. Cloning and characterization of a novel WD-40 repeat protein that dramatically accelerates osteoblastic differentiation. J. Biol. Chem. 2001;276:46515–46522. doi: 10.1074/jbc.M105757200. [DOI] [PubMed] [Google Scholar]
- Gwyer Findlay E., Danks L., Madden J., Cavanagh M.M., McNamee K., McCann F., Snelgrove R.J., Shaw S., Feldmann M., Taylor P.C., Horwood N.J., Hussell T. OX40L blockade is therapeutic in arthritis, despite promoting osteoclastogenesis. Proc. Natl. Acad. Sci. USA. 2014;111:2289–2294. doi: 10.1073/pnas.1321071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.H., Odathurai Saminathan S., Kim S.J. Insulin stimulates gene expression of ferritin light chain in osteoblast cells. J. Cell. Biochem. 2010;111:1493–1500. doi: 10.1002/jcb.22879. [DOI] [PubMed] [Google Scholar]
- Harada S., Rodan G.A. Control of osteoblast function and regulation of bone mass. Nature. 2003;423:349–355. doi: 10.1038/nature01660. [DOI] [PubMed] [Google Scholar]
- Karsenty G., Wagner E.F. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- Kemink S.A., Hermus A.R., Swinkels L.M., Lutterman J.A., Smals A.G. Osteopenia in insulin-dependent diabetes mellitus; prevalence and aspects of pathophysiology. J. Endocrinol. Invest. 2000;23:295–303. doi: 10.1007/BF03343726. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kim K., Youn B.U., Jin H.M., Kim N. MHC class II transactivator negatively regulates RANKL-mediated osteoclast differentiation by downregulating NFATc1 and OSCAR. Cell. Signal. 2010;22:1341–1349. doi: 10.1016/j.cellsig.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Kimura H., Kwan K.M., Zhang Z., Deng J.M., Darnay B.G., Behringer R.R., Nakamura T., de Crombrugghe B., Akiyama H. Cthrc1 is a positive regulator of osteoblastic bone formation. PLoS One. 2008;3:e3174. doi: 10.1371/journal.pone.0003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs D.L., Hilton D.J. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001;19:378–387. doi: 10.1634/stemcells.19-5-378. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Lee N.K. Up-regulation of cyclinD1 and Bcl2A1 by insulin is involved in osteoclasts proliferation. Life Sci. 2014;114:57–61. doi: 10.1016/j.lfs.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Leibbrandt A., Penninger J.M. RANKL/RANK as key factors for osteoclast development and bone loss in arthropathies. Adv. Exp. Med. Biol. 2009;649:100–113. doi: 10.1007/978-1-4419-0298-6_7. [DOI] [PubMed] [Google Scholar]
- Li L., Shen J.J., Bournat J.C., Huang L., Chattopadhyay A., Li Z., Shaw C., Graham B.H., Brown C.W. Activin signaling: effects on body composition and mitochondrial energy metabolism. Endocrinology. 2009;150:3521–3529. doi: 10.1210/en.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzon M., Greenhalgh C.J., Mohan S., Alexander W.S., Ohlsson C. Reduced bone mineral density in SOCS-2-deficient mice. Pediatr. Res. 2005;57:223–226. doi: 10.1203/01.PDR.0000148735.21084.D3. [DOI] [PubMed] [Google Scholar]
- Metcalf D., Greenhalgh C.J., Viney E., Willson T.A., Starr R., Nicola N.A., Hilton D.J., Alexander W.S. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature. 2000;405:1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Tanaka Y., Tozawa H., Takahashi Y., Maliszewski C., Delespesse G. Expression and function of OX40 ligand on human dendritic cells. J. Immunol. 1997;159:3838–3848. [PubMed] [Google Scholar]
- Pakala S.V., Bansal-Pakala P., Halteman B.S., Croft M. Prevention of diabetes in NOD mice at a late stage by targeting OX40/OX40 ligand interactions. Eur. J. Immunol. 2004;34:3039–3046. doi: 10.1002/eji.200425141. [DOI] [PubMed] [Google Scholar]
- Petersen D.N., Tkalcevic G.T., Mansolf A.L., Rivera-Gonzalez R., Brown T.A. Identification of osteoblast/osteocyte factor 45 (OF45), a bone-specific cDNA encoding an RGD-containing protein that is highly expressed in osteoblasts and osteocytes. J. Biol. Chem. 2000;275:36172–36180. doi: 10.1074/jbc.M003622200. [DOI] [PubMed] [Google Scholar]
- Smits V.A., Klompmaker R., Arnaud L., Rijksen G., Nigg E.A., Medema R.H. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- Takai N., Hamanaka R., Yoshimatsu J., Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24:287–291. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- Teitelbaum S.L. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- Teitelbaum S.L. Osteoclasts: what do they do and how do they do it? Am. J. Pathol. 2007;170:427–435. doi: 10.2353/ajpath.2007.060834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.M., Udagawa N., Hards D.K., Quinn J.M., Moseley J.M., Findlay D.M., Best J.D. Insulin receptor expression in primary and cultured osteoclast-like cells. Bone. 1998;23:181–186. doi: 10.1016/s8756-3282(98)00095-7. [DOI] [PubMed] [Google Scholar]
- Thrailkill K.M., Lumpkin C.K., Jr., Bunn R.C., Kemp S.F., Fowlkes J.L. Is insulin an anabolic agent in bone? Dissecting the diabetic bone for clues. Am. J. Physiol. Endocrinol. Metab. 2005;289:E735–745. doi: 10.1152/ajpendo.00159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vugt M.A., Smits V.A., Klompmaker R., Medema R.H. Inhibition of Polo-like kinase-1 by DNA damage occurs in an ATM- or ATR-dependent fashion. J. Biol. Chem. 2001;276:41656–41660. doi: 10.1074/jbc.M101831200. [DOI] [PubMed] [Google Scholar]
- Wang Y., Inger M., Jiang H., Tenenbaum H., Glogauer M. CD109 plays a role in osteoclastogenesis. PLoS One. 2013;8:e61213. doi: 10.1371/journal.pone.0061213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang X., Wang W., Liu J. Insulin stimulates osteoblast proliferation and differentiation through ERK and PI3K in MG-63 cells. Cell. Biochem. Funct. 2010;28:334–341. doi: 10.1002/cbf.1668. [DOI] [PubMed] [Google Scholar]