Abstract
Context:
The gut microbiota may influence the risk of breast cancer through effects on endogenous estrogens.
Objective:
The objective of the study was to investigate whether urinary estrogens and estrogen metabolites are associated with the diversity and composition of the fecal microbiome.
Design and Setting:
This was a cross-sectional study among women enrolled in Kaiser Permanente of Colorado.
Participants:
A total of 60 women drawn from a random sample of healthy postmenopausal women (aged 55–69 y), without current or recent use of antibiotics or hormone therapy and no history of cancer or gastrointestinal disease participated in the study.
Outcome Measures and Methods:
Creatinine-standardized urinary estrogens (estrone and estradiol) and 13 hydroxylated estrogen metabolites were measured in spot urines by liquid chromatography-tandem mass spectrometry. The fecal microbiome was assessed using pyrosequencing of 16S rRNA amplicons. General linear models were used to test for associations of diversity and composition of the fecal microbiome with parent estrogen (estrone + estradiol), total estrogens, and estrogen metabolites and the ratio of estrogen metabolites to parent estrogen, which has been predictive of postmenopausal breast cancer risk in previous studies.
Results:
The ratio of metabolites to parents was directly associated with whole-tree phylogenetic diversity (R = 0.35, P = .01). Relative abundances of the order Clostridiales (R = 0.32, P = .02) and the genus Bacteroides (R = −0.30, P = .03) were also correlated with the ratio of metabolites to parents. Associations were independent of age, body mass index, and study design factors.
Conclusions:
Our data suggest that women with a more diverse gut microbiome exhibit an elevated urinary ratio of hydroxylated estrogen metabolites to parent estrogen. Further research is warranted to confirm and relate these findings to clinical disease.
Estrogens are recognized as a causal factor in the etiology of breast cancer. Interindividual variation in the metabolism of estrogens may also explain some of the differences among women in breast cancer risk (1, 2).
Metabolism of estrogens occurs both in liver and in target tissues including breast. The parent estrogens (estrone and estradiol) may be irreversibly hydroxylated at the C-2, C-4, or C-16 positions of the steroid ring, resulting in a wide array of estrogen metabolites that vary in bioavailability (3) and estrogenicity (4). The parent estrogens and their metabolites (jointly referred to as EM) can be further modified via conjugation with sulfate or glucuronide moieties, which modifies their disposition and half-life (5). Conjugated EMs are excreted in urine or bile; those excreted in bile ultimately pass into the distal gut, in which some are deconjugated by resident microbes. Once liberated, these hormones can be reabsorbed through the mucosa and enter the portal vein for enterohepatic recycling (3). EM profiles in women may be influenced by factors that influence their production, metabolism, excretion, deconjugation, or reabsorption.
In postmenopausal women, elevated circulating estrogen levels in prediagnostic sera have been consistently associated with increased breast cancer risk (6). In three recent prospective studies, several parameters derived from serum EM profiles were also consistently associated with the risk of postmenopausal breast cancer. In each of these studies, investigators observed that breast cancer risk was increased with circulating concentrations of the parent estrogens and was reduced with increasing ratios of 2- and 4-pathway estrogen metabolites to parent estrogens. Greater metabolism via the 2-hydroxylation pathway compared with the 16-hydroxylation pathway was also associated with reduced breast cancer risk in each of these studies (7–9).
The determinants of EM profiles in postmenopausal women are only partially understood. After menopause, estrogens synthesis occurs primarily in adipose tissue through aromatization of androgens, and indeed, adiposity is associated with higher circulating estrogens in postmenopausal women (10). In postmenopausal women, enterohepatic recycling of EMs may be an important determinant of their half-lives and therefore their levels in circulation (11, 12).
Therefore, we and others have hypothesized that systemic levels of EMs may be modulated, in part, by the metabolic and other activities of gut microbiota (13, 14). In a previous study that included 25 men and seven postmenopausal women, we observed that measures of fecal microbiome richness and α-diversity were directly associated with levels of urinary EMs and inversely associated with levels of fecal EMs (15). Here we present a study of the fecal microbiome in association with urinary EM profiles in a sample of 60 postmenopausal women.
Materials and Methods
Study population
The study protocol was developed by collaborators at the National Cancer Institute (NCI), Kaiser Permanente Colorado (KPCO), the Institute for Genome Sciences at the University of Maryland School of Medicine, and RTI International. The study protocol and all study materials were approved by the Institutional Review Boards at KPCO, NCI, and RTI International.
Eligible participants were sampled from the population of current female KPCO members, aged 55–69 years, who had received a normal screening mammogram within the previous 6–8 weeks (16). The virtual data warehouse (VDW) at KPCO was used to identify eligible women. The VDW is a standardized database of inpatient and outpatient diagnoses, procedures, laboratory results, and medications derived from the electronic medical record (17). Using the VDW, we excluded women with conditions or prescribed medications that could strongly impact either the normal gut microbial population or systemic hormone levels including the following: any history of prior cancer (other than nonmelanoma skin cancer), inflammatory bowel disease or diverticulitis, gastric banding or bypass surgery; other gastric or intestinal surgery (such as appendicitis) within the previous 6 months; progesterone or estrogen hormone prescription within the previous 12 months; and antibiotic prescription within the previous 6 months. From this pool of eligible women, we randomly sampled 300 women who were contacted for the study via mail and telephone contact. To manage recruitment, women were contacted in three waves of 100 women.
Data and specimen collection
After providing informed consent, participants were mailed study materials that included questionnaires (18), a specimen collection kit with illustrated instructions for collecting a urine sample and four aliquots of a fecal sample, and materials to ship the specimens overnight to the biorepository. Urine was collected in a screw-top container, without preservative. Fecal specimens were collected in four 20-mL screw top Sarstedt tubes (19), including two preloaded with 5 mL RNAlater (QIAGEN) and two with 5 mL sterile PBS. After specimen collection, the urine container and fecal tubes were secured for shipping, and two frozen gel packs were added to the shipping box. Specimens were shipped by overnight courier to the NCI biorepository, where they were stored at −80°C until use. The temperature of the specimens was recorded upon receipt.
Fecal sample processing and analysis
Fecal DNA isolation, 454 Pyrosequencing of barcoded 16S rRNA genes, and filtering of reads through the Institute of Genome Sciences bioinformatics pipeline were performed as described previously by Flores et al (15). Four samples had extremely low numbers of reads, suggesting a problem with amplification steps; the extraction and amplification were repeated, but two samples were ultimately excluded from the analysis.
Operational taxonomic units (OTUs) were defined using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline as sequences with at least 97% identity and rarified (randomly sampled with replacement) to the minimum number of observed OTUs. Using the Ribosomal Data Project Bayesian classifier in QIIME, OTUs were assigned to phylum, class, order, family, and genus level taxa, and their relative abundances were calculated (20).
Measures of α-diversity were calculated for each participant, again using QIIME (20), based on samples rarefied to 2878 reads. Observed species was defined as the number of unique OTUs.
The Shannon Index has been calculated as: H′ = · lnpi where pi is the proportion of reads belonging to the ith OTU. This measure is commonly interpreted as an index of uncertainty in predicting the identity of a read drawn at random from the list of all reads, and it is understood to be most strongly influenced by relative abundances of the most prevalent phylotypes.
The Inverse Simpson has been calculated as: 2D = where pi is the proportion of reads belonging to the ith OTU. This measure is sensitive to rare OTUs and is interpreted as the reciprocal of the probability that two reads taken at random and with replacement will represent the same OTU. Phylogenetic diversity (whole tree) is a comprehensive, quantitative measure of diversity defined as the minimum total length of all the phylogenetic branches required to span the set of all sequenced OTUs from a given sample (21, 22).
Urinary estrogen assay
Stable isotope dilution liquid chromatography/tandem mass spectrometry was used to measure urinary concentrations of the parent estrogens (estrone and estradiol) and 13 estrogen metabolites including 2-hydroxylated estrogen metabolites (2-hydroxyestrone, 2-methoxyestrone, 2-hydroxyestradiol, 2-methoxyestradiol, and 2-hydroxyestrone-3-methyl ether); 4-hydroxylated estrogen metabolites (4-hydroxyestrone, 4-methoxyestrone, and 4-methoxyestradiol); and 16-hydroxylated estrogen metabolites (16α-hydroxyestrone, estriol, 17-epiestriol, 16-ketoestradiol, and 16-epiestriol). Details of the method, including sample preparation and assay conditions, have been published previously (23). For this study, we used six stable isotopically labeled standards to account for losses during sample preparation and assays: deuterated 2-hydroxyestradiol, 2-methoxyestradiol, and estriol (C/D/N Isotopes, Inc); deuterated 16-epiestriol (Medical Isotopes, Inc); and 13C-labeled estrone and estradiol (Cambridge Isotope Laboratories).
As in our study of breast cancer (7), in the present study, we considered parent estrogens (estrone plus estradiol), estrogen metabolites grouped into three major hydroxylation pathways (2-, 4-, and 16-hydroxylation); the ratio of metabolites to parent estrogen (overall and separately for each hydroxylation pathway); the ratio of 2-pathway to 16-pathway metabolites; total estrogen metabolites, and total EMs (total estrogens and estrogen metabolites). In a previous study using this assay to measure urinary concentrations of estrogens and EMs, excellent reproducibility and validity were observed, with laboratory coefficients of variation of 7% or less and intraclass correlation coefficients of 87% or greater for EM concentrations measured in samples from postmenopausal women (24).
Statistical methods
Distributions of all variables were inspected using histograms and descriptive statistics. EM levels were expressed in picomoles per milligram creatinine (which was measured in the same samples) and were grouped and summed by metabolic pathways. Concentrations also were expressed as proportions (EM percentage) of total EMs. Distributions of parent estrogens, metabolites, and metabolites to parent ratios did not deviate significantly from normality.
Associations of participant characteristics with EM and fecal microbiome measures were evaluated by using general linear models and testing either for differences across categories or for trends across categories. Participant characteristics included age in quartiles (55–58 y, 59–60 y, 61–63 y, and 64–70 y), race/ethnicity (white, black, Hispanic, unknown), and continuous body mass index (BMI). In addition, study design variables including wave of enrollment (wave 1, enrolled June-July; wave 2, enrolled August-September; wave 3, enrolled October-November) and temperature of the fecal specimen on arrival (2.0–4.9°C, 5.0–7.9°C, 8.0–12.9°C, or 13.0–22.7°C) were also included as covariates in all models. We also conducted sensitivity analyses to examine the effects of study wave and temperature of the sample when it arrived at the biorepository on associations.
Linear regression models were used to evaluate associations of each EM measure with measures of the diversity of the fecal microbiome and with taxonomic groups at phylum and family levels while adjusting for age, continuous BMI, wave of enrollment, and temperature of the fecal specimen upon arrival. Linear regression models also were used to explore the associations of EM levels with relative abundance of taxa with a median relative abundance of at least 0.1%.
Residuals from all the linear models were assessed by visual inspection and were not found to deviate substantially from normality. Values of P ≤ .05 were considered statistically significant. All tests were two sided. Analyses were conducted using SAS version 9.3 (SAS Institute).
Results
Population, microbiome metrics, and associations
A total of 62 women enrolled in the study and provided fecal and urine samples. High-quality fecal DNA 16S rRNA sequences and urinary estrogens and EM concentrations were obtained from 60 postmenopausal women. Data for two additional participating women were excluded because of low number of reads (<2000 reads per sample). Of 60 participants with complete data, 17 (27%) enrolled in wave 1, 21 (34%) in wave 2, and 24 (39%) in wave 3. As shown in Table 1, participants ranged in age from 55 to 70 years [median 64, interquartile range (IQR) 56–64] and had a median BMI of 27 (IQR 21–36), including 61.7% who were overweight or obese. The participants were predominantly white (91%) and non-Hispanic (95%), and in this way they were similar to the population from which they were drawn.
Table 1.
Characteristics of Study Participants and Summary Measures for α-Diversity of the Fecal Microbiome (n = 60 Postmenopausal Women)
| n | Mean | SD | Percentiles |
|||
|---|---|---|---|---|---|---|
| 10th | 50th | 90th | ||||
| Age, y | 60 | 60.3 | 3.2 | 56.5 | 60.0 | 64.0 |
| Height, in. | 60 | 64.0 | 2.6 | 61.0 | 64.0 | 67.8 |
| Weight, lb | 60 | 159.5 | 31.6 | 124 | 154 | 207 |
| BMI | 60 | 27.3 | 5.4 | 21.4 | 26.5 | 35.9 |
| Age at first birth among parous, y | 38 | 25.2 | 6.7 | 16 | 25.5 | 34 |
| Age at menarche, y | 54 | 12.7 | 1.4 | 11 | 13 | 14 |
| Age at menopause, y | 53 | 48.9 | 7.4 | 42 | 51 | 55 |
| Years since menopause | 53 | 11.5 | 8.7 | 5 | 10 | 21 |
| Observed speciesa | 60 | 359 | 59 | 269 | 367 | 431 |
| Shannon Indexb | 60 | 6.57 | 0.47 | 5.9 | 6.72 | 7.1 |
| Inverse Simpsonc | 60 | 38.1 | 14.1 | 19.9 | 37.6 | 58.4 |
| Phylogenetic diversity (whole tree)d | 60 | 20.1 | 3.9 | 15.2 | 20.2 | 25.2 |
| Bacteroidetes (relative abundance) | 60 | 35.6% | 9.7% | 26.4% | 34.2% | 50.4% |
| Firmicutes (relative abundance) | 60 | 63.5% | 9.8% | 48.4% | 64.9% | 73.1% |
| Other phyla (relative abundance) | 60 | 0.9% | 0.8% | 0.2% | 0.6% | 2.4% |
| Specimen temperature upon arrival, °C | 60 | 9.3 | 5.4 | 3.7 | 7.8 | 18.1 |
Observed species was defined as the number of unique OTUs.
The Shannon Index has been calculated as follows: where pi is the proportion of reads belonging to the ith OTU.
The Inverse Simpson has been calculated as follows: where pi is the proportion of reads belonging to the ith OTU.
Phylogenetic diversity (whole tree) is a quantitative measure of diversity defined as the minimum total length of all the phylogenetic branches required to span the set of all sequenced OTUs from a given sample
In the 60 specimens included in the analyses, pyrosequencing of the V1-V2 region of 16S rRNA genes generated 315 167 quality-filtered and denoised reads, with a median of 4454 reads per participant (range 2878–33 192) for analysis. These were assigned to a median of 367 (IQR 269–431) OTUs per participant and to 196 different taxa. As expected, most of these taxa were classified as belonging to the phylum Firmicutes (median relative abundance 64.9%, IQR 48.4–73.1%) or Bacteroidetes (median 34.2%, IQR 26.4–50.4%).
We observed differences in measures of fecal microbiome diversity and composition at the phylum level by wave of enrollment, temperature of the fecal specimen upon arrival, and household income (Supplemental Table 1). Samples from participants in the third wave of enrollment had higher numbers of observed taxa when compared with the previous waves (P = .05). The abundance of taxa in minor phyla (ie, neither Firmicutes nor Bacteroidetes) was greater in samples from the third wave in comparison with previous waves (P = .01). The abundance of taxa in minor phyla also was higher in specimens that arrived at higher temperature (P for trend = .02) and in women in the lowest quartile of income compared with their counterparts with higher incomes (P = .02). We did not observe statistically significant differences by age or BMI.
EM metrics and population associations
Urinary EM profiles of the study participants are shown in Table 2. Creatinine-adjusted concentrations of total EMs varied 4-fold across the interdecile range, as did concentrations of estrone, estradiol, and total estrogen metabolites. On average, the parent estrogens (estrone plus estradiol) represented 32% of total EMs, whereas 2-, 4-, and 16-hydroxylated metabolites represented, respectively, 29%, 3%, and 35% of the total EMs.
Table 2.
Urinary Concentrations of Estrogens and EMs (in Picomoles per Milligram of Creatinine) for Study Participants (n = 60 Postmenopausal Women)
| Mean | SD | Percentiles |
|||
|---|---|---|---|---|---|
| 10th | 50th | 90th | |||
| Total estrogens and metabolites | 28.10 | 17.80 | 13.08 | 22.01 | 52.87 |
| Parent estrogens | 9.41 | 8.04 | 4.00 | 6.99 | 15.83 |
| Estrone | 7.50 | 6.52 | 3.07 | 5.38 | 12.99 |
| Estradiol | 1.92 | 2.61 | 0.56 | 1.28 | 3.28 |
| Total estrogen metabolites | 18.69 | 10.92 | 9.07 | 14.97 | 36.36 |
| 2-Hydroxylation pathway | 8.08 | 4.83 | 3.91 | 6.49 | 15.50 |
| 2-Hydroxyestrone | 4.97 | 2.91 | 2.28 | 4.07 | 9.59 |
| 2-Hydroxyestradiol | 1.15 | 0.70 | 0.55 | 0.88 | 2.15 |
| 2-Methoxyestrone | 1.10 | 0.89 | 0.53 | 0.85 | 2.31 |
| 2-Methoxyestradiol | 0.47 | 0.30 | 0.19 | 0.40 | 0.83 |
| 2-Hydroxyestrone-3-methyl ether | 0.39 | 0.27 | 0.17 | 0.32 | 0.59 |
| 16-Hydroxylation pathway | 9.74 | 5.63 | 4.68 | 7.91 | 18.71 |
| Estriol | 5.64 | 3.31 | 2.70 | 4.41 | 10.07 |
| 16α-Hydroxyestrone | 1.44 | 0.84 | 0.66 | 1.17 | 2.83 |
| 16-Epiestriol | 0.68 | 0.43 | 0.28 | 0.54 | 1.24 |
| 17-Epiestriol | 0.45 | 0.31 | 0.18 | 0.33 | 0.78 |
| 16-Ketoestradiol | 1.54 | 0.92 | 0.76 | 1.25 | 3.28 |
| 4-Hydroxylation pathway | 0.87 | 0.52 | 0.41 | 0.68 | 1.67 |
| 4-Hydroxyestrone | 0.66 | 0.39 | 0.30 | 0.54 | 1.28 |
| 4-Methoxyestrone | 0.14 | 0.12 | 0.06 | 0.11 | 0.25 |
| 4-Methoxyestradiol | 0.07 | 0.04 | 0.03 | 0.05 | 0.12 |
| Metabolic ratios | |||||
| Ratio of metabolites to parent estrogens | 2.20 | 0.51 | 1.73 | 2.21 | 2.80 |
| Ratio of 2-pathway to parent estrogens | 0.95 | 0.24 | 0.71 | 0.95 | 1.22 |
| Ratio of 4 pathway to parent estrogens | 0.10 | 0.03 | 0.08 | 0.10 | 0.13 |
| Ratio of 16-pathway to parent estrogens | 1.14 | 0.26 | 0.89 | 1.16 | 1.45 |
| Ratio of 2-pathway to 16-pathway metabolites | 0.84 | 0.09 | 0.73 | 0.83 | 0.93 |
Parameters in italics represent calculated measures.
Estrogen measures were not statistically significantly associated with any of the variables selected a priori as potential confounders, including age, race, Hispanic ethnicity, education level, BMI, study wave, and specimen temperature upon receipt by the biorepository (data not shown). Total EMs tended to increase across the following categories of BMI: underweight/normal, overweight, and obese (P for trend = .11).
EM and microbiome associations
In Table 3 we present multivariable-adjusted estrogen measures by quintiled measures of diversity of the fecal microbiome. Total EMs tended to increase over quintiles of diversity measures, but these associations were not statistically significant [P for trend = .23, P for trend = .19, P for trend = .24, and P for trend = .12, for observed species, Shannon index, Inverse Simpson, and phylogenetic diversity (whole tree), respectively]. The ratio of metabolites to parent estrogen increased statistically significantly across quintiles of whole-tree phylogenetic diversity (P for trend = .004) but not for observed species (P for trend = 0.22), Shannon index (P for trend = .24), or Inverse Simpson (P for trend = .77). On a continuous scale, all three pathway-specific metabolites to parent ratios were directly correlated with phylogenetic diversity (Table 4), as shown in Figure 1. The ratio of 2- to 16-hydroxylation pathway metabolites also showed a statistically significant increasing trend across quintiles of phylogenetic diversity (P for trend = .05, Table 3), but not for the other α-diversity measures (P ≥ .79). Relative abundance of taxa in the phyla Firmicutes, Bacteroidetes, and others were not associated with estrogen measures, with one exception: the metabolites to parent ratio was positively correlated with the relative abundance of other phyla (P for trend = .04).
Table 3.
Adjusted Meansa (95% Confidence Limits) for Urinary Concentrations of Estrogens and Estrogen Metabolites (Jointly Referred to as EM, in Picomoles per Milligram of Creatinine) by Quintiles of Measures of Diversity and Composition of the Fecal Microbiome in 60 Postmenopausal Women
| Quintile | n | Total EM |
Ratio of Metabolites to Parent Estrogens |
Ratio of 2-Pathway Metabolites to Parent Estrogens |
Ratio of 2- to 16-Pathway Metabolites |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adjusted Mean | 95% CL | P Valueb,c | Adjusted Mean | 95% CL | P Valueb,c | Adjusted Mean | 95% CL | P Valueb,c | Adjusted Mean | 95% CL | P Valueb,c | ||||||
| Observed speciesd | |||||||||||||||||
| <310 | 12 | 21.5 | 10.3 | 32.8 | .23 | 2.00 | 1.67 | 2.33 | .22 | 0.83 | 0.68 | 0.99 | .19 | 0.80 | 0.75 | 0.85 | .79 |
| 311–355 | 12 | 30.7 | 18.9 | 42.5 | .25 | 2.15 | 1.81 | 2.49 | .09 | 0.95 | 0.79 | 1.11 | .08 | 0.85 | 0.80 | 0.91 | .81 |
| 356–378 | 12 | 30.7 | 19.0 | 42.5 | 2.34 | 2.00 | 2.69 | 1.03 | 0.87 | 1.19 | 0.86 | 0.80 | 0.91 | ||||
| 379–407 | 12 | 24.5 | 12.9 | 36.0 | 2.17 | 1.84 | 2.51 | 0.96 | 0.80 | 1.11 | 0.86 | 0.81 | 0.91 | ||||
| >408 | 12 | 35.8 | 24.0 | 47.6 | 2.32 | 1.97 | 2.66 | 0.99 | 0.83 | 1.15 | 0.81 | 0.76 | 0.86 | ||||
| Shannon Indexe | |||||||||||||||||
| <6.21 | 12 | 29.2 | 18.5 | 39.8 | .19 | 1.92 | 1.61 | 2.24 | .24 | 0.81 | 0.67 | 0.96 | .23 | 0.81 | 0.76 | 0.86 | .88 |
| 6.21–6.53 | 12 | 19.2 | 8.2 | 30.2 | .19 | 2.39 | 2.07 | 2.71 | .11 | 1.05 | 0.90 | 1.20 | .10 | 0.86 | 0.81 | 0.91 | .80 |
| 6.53–6.79 | 12 | 24.2 | 13.7 | 34.6 | 2.13 | 1.83 | 2.44 | 0.93 | 0.78 | 1.07 | 0.83 | 0.78 | 0.88 | ||||
| 6.79–6.97 | 12 | 36.9 | 26.0 | 47.8 | 2.24 | 1.92 | 2.55 | 0.98 | 0.83 | 1.13 | 0.85 | 0.80 | 0.90 | ||||
| >6.98 | 12 | 31.2 | 20.5 | 42.0 | 2.30 | 1.99 | 2.62 | 0.99 | 0.85 | 1.14 | 0.82 | 0.77 | 0.87 | ||||
| Inverse Simpsonf | |||||||||||||||||
| <24.4 | 12 | 31.1 | 20.7 | 41.6 | .24 | 1.98 | 1.67 | 2.29 | .77 | 0.84 | 0.69 | 0.98 | .68 | 0.81 | 0.76 | 0.86 | .89 |
| 24.5–34.7 | 12 | 15.8 | 4.6 | 27.0 | .22 | 2.37 | 2.04 | 2.71 | .66 | 1.05 | 0.89 | 1.21 | .59 | 0.87 | 0.82 | 0.93 | .88 |
| 34.8–41.4 | 12 | 25.0 | 14.5 | 35.6 | 2.35 | 2.03 | 2.66 | 1.00 | 0.86 | 1.15 | 0.81 | 0.77 | 0.86 | ||||
| 41.5–52.1 | 12 | 35.8 | 25.3 | 46.4 | 2.07 | 1.75 | 2.38 | 0.90 | 0.76 | 1.05 | 0.84 | 0.79 | 0.89 | ||||
| >52.2 | 12 | 31.1 | 20.3 | 41.8 | 2.19 | 1.87 | 2.51 | 0.96 | 0.81 | 1.11 | 0.84 | 0.79 | 0.89 | ||||
| Phylogenetic diversity (whole tree)g | |||||||||||||||||
| <16.5 | 12 | 22.7 | 11.5 | 33.9 | .12 | 1.92 | 1.61 | 2.22 | .004 | 0.81 | 0.67 | 0.95 | .002 | 0.81 | 0.76 | 0.86 | .045 |
| 16.6–19.2 | 12 | 28.4 | 17.2 | 39.6 | .14 | 1.94 | 1.64 | 2.24 | .01 | 0.82 | 0.68 | 0.96 | .003 | 0.80 | 0.75 | 0.84 | .08 |
| 19.3–20.7 | 12 | 24.6 | 13.1 | 36.2 | 2.26 | 1.95 | 2.57 | 1.00 | 0.86 | 1.15 | 0.86 | 0.82 | 0.91 | ||||
| 20.8–23.6 | 12 | 29.3 | 18.2 | 40.4 | 2.39 | 2.09 | 2.69 | 1.04 | 0.90 | 1.18 | 0.84 | 0.79 | 0.88 | ||||
| >23.7 | 12 | 36.3 | 24.7 | 47.9 | 2.41 | 2.09 | 2.72 | 1.06 | 0.91 | 1.20 | 0.87 | 0.82 | 0.92 | ||||
| Bacteroidetes (relative abundance) | |||||||||||||||||
| <27.7% | 12 | 27.3 | 15.5 | 39.0 | .85 | 1.85 | 1.55 | 2.16 | .59 | 0.77 | 0.63 | 0.91 | .57 | 0.78 | 0.74 | 0.83 | .56 |
| 27.7–31.9% | 12 | 29.2 | 17.0 | 41.3 | .56 | 2.40 | 2.08 | 2.71 | .69 | 1.05 | 0.91 | 1.20 | .67 | 0.86 | 0.81 | 0.90 | .70 |
| 32.0–37.9% | 13 | 27.9 | 16.9 | 38.9 | 2.32 | 2.04 | 2.61 | 1.04 | 0.91 | 1.17 | 0.89 | 0.84 | 0.93 | ||||
| 38.0–40.9% | 12 | 25.3 | 13.2 | 37.4 | 2.34 | 2.03 | 2.66 | 0.98 | 0.84 | 1.13 | 0.79 | 0.74 | 0.83 | ||||
| >41.0 | 11 | 31.1 | 19.1 | 43.1 | 1.98 | 1.67 | 2.30 | 0.86 | 0.72 | 1.01 | 0.85 | 0.80 | 0.89 | ||||
| Firmicutes (relative abundance) | |||||||||||||||||
| <57.2% | 11 | 30.8 | 18.8 | 42.8 | .98 | 2.00 | 1.69 | 2.32 | .52 | 0.87 | 0.72 | 1.02 | .51 | 0.85 | 0.80 | 0.89 | .50 |
| 57.2–62.0% | 12 | 25.9 | 13.7 | 38.0 | .50 | 2.33 | 2.00 | 2.65 | .63 | 0.98 | 0.83 | 1.13 | .62 | 0.79 | 0.74 | 0.83 | .72 |
| 62.1–67.4% | 13 | 28.2 | 17.4 | 39.0 | 2.37 | 2.09 | 2.66 | 1.07 | 0.94 | 1.20 | 0.90 | 0.86 | 0.94 | ||||
| 67.5–71.3% | 12 | 25.8 | 14.0 | 37.6 | 2.30 | 1.99 | 2.61 | 1.00 | 0.85 | 1.14 | 0.83 | 0.79 | 0.88 | ||||
| >71.4% | 12 | 30.2 | 18.2 | 42.2 | 1.86 | 1.54 | 2.18 | 0.78 | 0.63 | 0.93 | 0.79 | 0.74 | 0.83 | ||||
| Other phyla (relative abundance) | |||||||||||||||||
| <0.39% | 12 | 21.7 | 10.6 | 32.8 | .15 | 2.02 | 1.70 | 2.34 | .12 | 0.86 | 0.71 | 1.00 | .12 | 0.80 | 0.76 | 0.85 | .67 |
| 0.40–0.48% | 11 | 29.3 | 17.9 | 40.7 | .15 | 2.11 | 1.79 | 2.44 | .29 | 0.92 | 0.76 | 1.07 | .42 | 0.83 | 0.78 | 0.87 | .66 |
| 0.49–0.61% | 13 | 25.5 | 14.3 | 36.7 | 2.13 | 1.81 | 2.46 | 0.95 | 0.80 | 1.10 | 0.88 | 0.83 | 0.92 | ||||
| 0.62–0.72% | 13 | 32.3 | 20.7 | 43.8 | 2.40 | 2.07 | 2.73 | 1.07 | 0.91 | 1.22 | 0.87 | 0.82 | 0.92 | ||||
| >0.72% | 11 | 32.9 | 21.6 | 44.3 | 2.29 | 1.96 | 2.61 | 0.96 | 0.81 | 1.11 | 0.80 | 0.75 | 0.85 | ||||
Adjusted for age, BMI, wave, and temperature of the fecal sample upon arrival.
P for trend across categories of diversity or relative abundance.
P for the continuous measure of diversity or relative abundance.
Observed species was defined as the number of unique OTUs.
The Shannon Index has been calculated as follows: where pi is the proportion of reads belonging to the ith OTU.
The Inverse Simpson has been calculated as follows: where pi is the proportion of reads belonging to the ith OTU.
Phylogenetic diversity whole tree is a quantitative measure of diversity defined as the minimum total length of all the phylogenetic branches required to span the set of all sequenced OTUs from a given sample.
Table 4.
Pearson Correlations (R, P Value) Between Measures of Diversity of the Fecal Microbiome and Measures of Estrogen Metabolism in 60 Postmenopausal Women
| Total EM |
Parent Estrogens |
Total Estrogen Metabolites |
Ratio of All Metabolites to Parent Estrogens |
Ratio of 2-Pathway Metabolites to Parent Estrogens |
Ratio of 4-Pathway Metabolites to Parent Estrogens |
Ratio of 16-Pathway Metabolites to Parent Estrogens |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | P Value | R | P Value | R | P Value | R | P Value | R | P Value | R | P Value | R | P Value | |
| Observed species | 0.14 | 0.29 | 0.06 | 0.66 | 0.18 | 0.16 | 0.21 | 0.11 | 0.21 | 0.11 | 0.16 | 0.21 | 0.20 | 0.13 |
| Shannon Index | 0.17 | 0.18 | 0.07 | 0.60 | 0.23 | 0.07 | 0.19 | 0.14 | 0.18 | 0.16 | 0.14 | 0.28 | 0.20 | 0.13 |
| Inverse Simpson | 0.18 | 0.17 | 0.09 | 0.51 | 0.23 | 0.07 | 0.06 | 0.65 | 0.05 | 0.72 | 0.02 | 0.90 | 0.07 | 0.59 |
| Phylogenetic diversity (whole tree) | 0.22 | 0.09 | 0.10 | 0.46 | 0.29 | 0.02 | 0.31 | 0.02 | 0.33 | 0.01 | 0.28 | 0.03 | 0.28 | 0.03 |
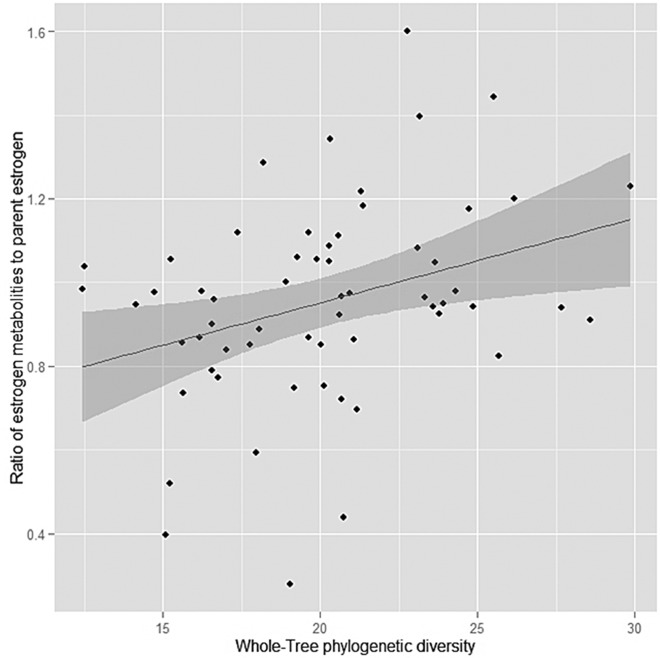
Figure 1.
Linear association (with 95% confidence interval) of whole-tree phylogenetic diversity with the ratio of estrogen metabolites to parent estrogen. From a linear regression model adjusted for age, BMI, wave, and temperature of the specimens upon arrival at the biorepository.
Several statistically significant correlations were observed between particular taxa and estrogen measures (Supplemental Table 2). The ratio of all estrogen metabolites to parent estrogen was directly associated with the order Clostridiales (r = 0.32, P = .02) and, among the Clostridiales, with the family Ruminococcaceae (r = 0.37, P = .005). The total EM level was significantly associated with the genus Ruminococcus (r = 0.28, P = .04). The genus Bacteroides was inversely associated with the ratio of estrogen metabolites to parent estrogens (r = −0.30, P = .03).
In sensitivity analyses that excluded women with specimens that may have been compromised by elevated temperatures during shipping [samples with temperature at receipt >13°C (n = 14)], results were consistent with presented findings (data not shown). The ratio of the metabolites to parent estrogen was observed to increase significantly across quintiles of phylogenetic diversity as seen before (P = .02). No statistically significant interaction between sample temperature and phylogenetic diversity was observed.
Discussion
The gut microbiota affects host metabolism, physiology, and pathophysiology in ways that remain largely obscure. We and others have hypothesized that one important action of the gut microbiota is the modulation of systemic estrogens (13–15).
In the present study, we observed that the composition and diversity of the intestinal microbiota were associated with patterns of estrogen metabolism that are predictive of the risk of breast cancer in postmenopausal women (7–9). Specifically, high fecal microbial diversity, as measured by whole-tree phylogenetic diversity, was statistically significantly associated with a high ratio of metabolites to parent estrogen; these measures of microbial diversity had similar correlations with analogous ratios for each metabolic pathway (2-, 4-, and 16-hydroxylation). Because higher ratios of 2- and 4-hydroxylated metabolites to parent estrogens have been associated with a reduced risk of postmenopausal breast cancer, our data suggest that postmenopausal breast cancer risk may be reduced for women who have high intestinal microbial diversity.
The ratio of estrogen metabolites to parent estrogen was also associated with relative abundances of taxa at the phylum, class, and genus levels. Specifically, relative abundances of a number of taxa in the class Clostridia, including the order Clostridiales and the family Ruminococcaceae, were directly associated with the ratio of metabolites to parent estrogens, whereas the genus Bacteroides was inversely associated with this ratio. These findings are consistent with our previous study of 25 men and seven postmenopausal women, in which total urinary EM was correlated directly with fecal microbial alpha diversity and abundance of four Clostridia taxa (15).
Systemic EM profiles may be influenced by the composition and diversity of the gut microbiota through a number of plausible mechanisms. Our a priori hypothesis was that the gut microbial population influences systemic EM profiles by deconjugating and thus liberating the EMs secreted in bile. Enterohepatic circulation of a 16-hydroxylation metabolite, estriol, has been clearly demonstrated (11), and both urinary and fecal levels of estriol and parent estrogens are altered by the administration of antibiotics (25). We observed a weak trend, suggesting a direct association between total EM and diversity; the association appeared to be stronger for estrogen metabolites than for parent estrogens, suggesting that the microbial population may differentially affect the components of total EM. Bacterial β-glucuronidase activity can be pharmacologically blocked (26), but an effect on estrogens has not yet been demonstrated.
Adiposity is positively correlated with circulating levels of parent estrogens and inversely associated with the SHBG in postmenopausal women (27). Increasing adiposity has also been associated with reduced 2-hydroxylation of estrogens; this association has been seen in studies of overweight and obese women (28), whereas increased 2-hydroxylation has been found in lean women, including those with anorexia nervosa (29) or who participate in frequent, strenuous exercise (30, 31). These observations are supported by a recent finding that a factor secreted by adipocytes could inhibit 2-hydroxylation of estradiol by MCF-7 cells (32).
Recent literature has also demonstrated associations between BMI and variations in the composition and function of the gut microbiota (33, 34). We did not observe statistically significant associations between BMI and measures of diversity in our data, perhaps in part because of the relatively narrow range of BMI in this population. We controlled for BMI in our analyses, but there may still be residual confounding because BMI is an imperfect measure of adiposity.
Adiposity and inflammation have both been hypothesized to increase estrogen production. In particular, disruption of the gut microbiota may alter estrogen reabsorption and also contribute to obesity-related phenotypes by reduced integrity of the gut mucosa, allowing bacterial translocation into circulation and, as a consequence, chronic low-grade systemic inflammation (35). Chronic exposure to lipopolysaccharide and other toxins shed by bacteria induces inflammation by activation of toll-like receptors in adipocytes, in turn releasing proinflammatory cytokines (35). In vitro exposure of breast epithelial cells to the proinflammatory cytokine TNF-α leads to increased total EM, 2- and 4-pathway metabolites, and the ratio of estradiol to estrone (36). Functional effects on estrogen-metabolizing enzymes induced by TNF-α include an increased expression of aromatase (CYP19), CYP1A1, and CYP1B1; reduced expression of catechol O-methyltransferase and nicotinamide adenine dinucleotide phosphate-quinone oxidoreductase 1; and modulated expression of 17β-hydroxysteroid dehydrogenase type I (36, 37). Other in vivo studies have demonstrated that inflammation can drive estrogen production in the breast through the up-regulation of aromatase (38). These observations suggest that systemic inflammation would result in increased EM and in EM profiles with higher ratios of metabolites to parents. Clinical studies have found that regular use of aspirin, which can reduce systemic inflammation, is associated with lower circulating estrogen levels among postmenopausal women (39, 40).
Determinants of estrogen and EM profiles in postmenopausal women are largely unknown but may be important for a number of health outcomes in this population. Our study is noteworthy for its finding that, in a random sample of healthy, postmenopausal women, microbial diversity in the distal gut was positively associated with the ratio of hydroxylated estrogen metabolites to parent estrogens even after adjustment for age, BMI, and study design variables. Because the levels and metabolism of endogenous estrogens play important roles in the etiology of breast cancer (7–9), these observations support the hypothesis that breast cancer risk in postmenopausal women may be affected by differences in systemic estrogens attributable to differences in the intestinal microbiota.
The major limitations of our study must be noted, especially its small size and cross-sectional design, which preclude detection of weaker associations and temporal relationships. In addition, because we did not adjust for multiple comparisons, the associations that we found with particular taxa must be considered exploratory. Our assessment of the microbiome relied merely on 16S rRNA profiles, without more extensive whole-genome sequencing or functional assays. Nonetheless, if our findings are independently replicated, they provide an early step toward the ultimate goal of understanding how the gut microbiota may affect estrogen homeostasis and its impact on human health.
Acknowledgments
We thank Doug Fadrosh (Institute for Genome Sciences, University of Maryland Baltimore, Maryland) for managing the extraction and microbiome profiling of the fecal specimens; Jeffrey Basilio and Kathleen Spaid (National Institutes of Health Clinical Center, Bethesda, Maryland) for measuring creatinine in the urine specimens; and especially the study participants.
This research was supported by the Intramural Research Program of the NCI, National Institutes of Health, but this support did not influence the data analysis, manuscript preparation, or decision to submit for publication.
This work was supported in whole or in part by federal funds from the National Cancer Institute Intramural Research Program under Contract HHSN261200800001E.
Disclosure Summary: The authors have nothing to declare.
Footnotes
- BMI
- body mass index
- EM
- estrogens and their metabolites
- KPCO
- Kaiser Permanente Colorado
- NCI
- National Cancer Institute
- OTU
- operational taxonomic unit
- QIIME
- Quantitative Insights Into Microbial Ecology
- VDW
- virtual data warehouse.
References
- 1. Yager JD. Endogenous estrogens as carcinogens through metabolic activation. J Natl Cancer Inst Monogr. 2000:67–73. [DOI] [PubMed] [Google Scholar]
- 2. Cavalieri E, Frenkel K, Liehr JG, Rogan E, Roy D. Estrogens as endogenous genotoxic agents—DNA adducts and mutations. J Natl Cancer Inst Monogr. 2000;75–93. [DOI] [PubMed] [Google Scholar]
- 3. Adlercreutz H, Martin F. Biliary excretion and intestinal metabolism of progesterone and estrogens in man. J Steroid Biochem. 1980;13:231–244. [DOI] [PubMed] [Google Scholar]
- 4. Zhu BT, Han GZ, Shim JY, Wen Y, Jiang XR. Quantitative structure-activity relationship of various endogenous estrogen metabolites for human estrogen receptor α and β subtypes: Insights into the structural determinants favoring a differential subtype binding. Endocrinology. 2006;147:4132–4150. [DOI] [PubMed] [Google Scholar]
- 5. Raftogianis R, Creveling C, Weinshilboum R, Weisz J. Estrogen metabolism by conjugation. J Natl Cancer Inst Monographs. 2000:113–124. [DOI] [PubMed] [Google Scholar]
- 6. Key T, Appleby P, Barnes I, Reeves G, Endogenous Hormones and Breast Cancer Collaborative Group. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J Natl Cancer Inst. 2002;94:606–616. [DOI] [PubMed] [Google Scholar]
- 7. Fuhrman BJ, Schairer C, Gail MH, et al. Estrogen metabolism and risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 2012;104:326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falk RT, Brinton LA, Dorgan JF, et al. Relationship of serum estrogens and estrogen metabolites to postmenopausal breast cancer risk: a nested case-control study. Breast Cancer Res. 2013;15:R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dallal CM, Tice JA, Buist DS, et al. Estrogen metabolism and breast cancer risk among postmenopausal women: a case-cohort study within B∼FIT. Carcinogenesis. 2014;35:346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Key TJ, Appleby PN, Reeves GK, et al. Body mass index, serum sex hormones, and breast cancer risk in postmenopausal women. J Natl Cancer Inst. 2003;95:1218–1226. [DOI] [PubMed] [Google Scholar]
- 11. Heimer GM, Englund DE. Enterohepatic recirculation of oestriol studied in cholecystectomized and non-cholecystectomized menopausal women. Upsala J Med Sci. 1984;89:107–115. [DOI] [PubMed] [Google Scholar]
- 12. Goldin BR, Adlercreutz H, Dwyer JT, Swenson L, Warram JH, Gorbach SL. Effect of diet on excretion of estrogens in pre- and postmenopausal women. Cancer Res. 1981;41:3771–3773. [PubMed] [Google Scholar]
- 13. Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10:324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rose DP. Diet, hormones, and cancer. Annu Rev Public Health. 1993;14:1–17. [DOI] [PubMed] [Google Scholar]
- 15. Flores R, Shi J, Fuhrman B, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012;10:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feigelson HS, Bischoff K, Ardini MA, et al. Feasibility of self-collection of fecal specimens by randomly sampled women for health-related studies of the gut microbiome. BMC Res Notes. 2014;7:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delate T, Bowles EJ, Pardee R, et al. Validity of eight integrated healthcare delivery organizations' administrative clinical data to capture breast cancer chemotherapy exposure. Cancer Epidemiol Biomarkers Prev. 2012;21:673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Block G, Hartman AM, Naughton D. A reduced dietary questionnaire: development and validation. Epidemiology. 1990;1:58–64. [DOI] [PubMed] [Google Scholar]
- 19. Flores R, Shi J, Gail MH, Gajer P, Ravel J, Goedert JJ. Assessment of the human faecal microbiota: II. Reproducibility and associations of 16S rRNA pyrosequences. Eur J Clin Invest. 2012;42:855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crozier RH, Dunnett LJ, Agapow PM. Phylogenetic biodiversity assessment based on systematic nomenclature. Evol Bioinformatics Online. 2005;1:11–36. [PMC free article] [PubMed] [Google Scholar]
- 22. Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 23. Xu X, Keefer LK, Ziegler RG, Veenstra TD. A liquid chromatography-mass spectrometry method for the quantitative analysis of urinary endogenous estrogen metabolites. Nat Protocols. 2007;2:1350–1355. [DOI] [PubMed] [Google Scholar]
- 24. Falk RT, Xu X, Keefer L, Veenstra TD, Ziegler RG. A liquid chromatography-mass spectrometry method for the simultaneous measurement of 15 urinary estrogens and estrogen metabolites: assay reproducibility and interindividual variability. Cancer Epidemiol Biomarkers Prev. 2008;17:3411–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adlercreutz H, Pulkkinen M, Hämäläinen E, Korpela J. Studies on the role of intestinal bacteria in metabolism of synthetic and natural steroid hormones. J Steroid Biochem. 1984;20:217–229. [DOI] [PubMed] [Google Scholar]
- 26. Wallace B, Wang H, Lane K, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rinaldi S, Key TJ, Peeters PH, et al. Anthropometric measures, endogenous sex steroids and breast cancer risk in postmenopausal women: a study within the EPIC cohort. Int J Cancer. 2006;118:2832–2839. [DOI] [PubMed] [Google Scholar]
- 28. Schneider J, Bradlow HL, Strain G, Levin J, Anderson K, Fishman J. Effects of obesity on estradiol metabolism: decreased formation of nonuterotropic metabolites. J Clin Endocrinol Metab. 1983;56:973–978. [DOI] [PubMed] [Google Scholar]
- 29. Fishman J, Boyar RM, Hellman L. Influence of body weight on estradiol metabolism in young women. J Clin Endocrinol Metab. 1975;41:989–991. [DOI] [PubMed] [Google Scholar]
- 30. Matthews CE, Fowke JH, Dai Q, et al. Physical activity, body size, and estrogen metabolism in women. Cancer Causes Control. 2004;15:473–481. [DOI] [PubMed] [Google Scholar]
- 31. Snow RC, Barbieri RL, Frisch RE. Estrogen 2-hydroxylase oxidation and menstrual function among elite oarswomen. J Clin Endocrinol Metab. 1989;69:369–376. [DOI] [PubMed] [Google Scholar]
- 32. Bradlow HL, Sepkovic DW, Telang N, Tiwari R. Adipocyte-derived factor as a modulator of oxidative estrogen metabolism: implications for obesity and estrogen-dependent breast cancer. In Vivo. 2011;25:585–588. [PubMed] [Google Scholar]
- 33. Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Creely SJ, McTernan PG, Kusminski CM, et al. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;292:E740–E747. [DOI] [PubMed] [Google Scholar]
- 36. Kamel M, Shouman S, El-Merzebany M, et al. Effect of tumour necrosis factor-α on estrogen metabolic pathways in breast cancer cells. J Cancer. 2012;3:310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salama SA, Kamel MW, Diaz-Arrastia CR, et al. Effect of tumor necrosis factor-α on estrogen metabolism and endometrial cells: potential physiological and pathological relevance. J Clin Endocrinol Metab. 2009;94:285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Subbaramaiah K, Howe LR, Bhardwaj P, et al. Obesity is associated with inflammation and elevated aromatase expression in the mouse mammary gland. Cancer Prev Res. 2011;4:329–346. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39. Hudson AG, Gierach GL, Modugno F, et al. Nonsteroidal anti-inflammatory drug use and serum total estradiol in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2008;17:680–687. [DOI] [PubMed] [Google Scholar]
- 40. Gates MA, Tworoger SS, Eliassen AH, Missmer SA, Hankinson SE. Analgesic use and sex steroid hormone concentrations in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2010;19:1033–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]