SUMMARY
The gut microbiome and daily feeding/fasting cycle influence host metabolism and contributes to obesity and metabolic diseases. However, fundamental characteristics of this relationship between the feeding/fasting cycle and the gut microbiome is unknown. Our studies show that the gut microbiome is highly dynamic, exhibiting daily cyclical fluctuations in composition. Diet-induced obesity dampens the daily feeding/fasting rhythm and diminishes many of these cyclical fluctuations. Time restricted feeding (TRF), in which feeding is consolidated to the nocturnal phase, partially restores these cyclical fluctuations. Furthermore, TRF, which protects against obesity and metabolic diseases, affects bacteria shown to influence host metabolism. Cyclical changes in the gut microbiome from feeding/fasting rhythms contribute to the diversity of gut microflora and likely represent a mechanism by which the gut microbiome affects host metabolism. Thus, feeding pattern and time of harvest, in addition to diet, are important parameters when assessing the microbiome’s contribution to host metabolism.
INTRODUCTION
The gut microbiome plays an important role in host metabolic homeostasis (Tremaroli and Backhed, 2012). However, the mechanistic basis for this metabolic effect is not well understood. Transcriptional activity in intestinal epithelial cells is highly dynamic, characterized by cyclical gene expression that is responsive to feeding and the host’s central circadian clock (Scheving, 2000). Whether the gut microbiome exhibits similar fluctuations has not yet been investigated. Characterizing this feature of the gut microbiome is necessary to better understand the relationship it may have with other drivers of host metabolism.
Obese humans and mice have gut microbiomes that are different from their lean controls (Turnbaugh et al., 2008; Turnbaugh et al., 2009a; Turnbaugh et al., 2006). In particular, obesity is associated with a reduction in bacteria from the Bacteroidetes phylum and an increase in bacteria from the Firmicutes phylum. Metagenomic analysis of the obese microflora shows that it is enriched for genes associated with lipid and carbohydrate metabolism (Turnbaugh et al., 2009a). Although it initially appeared that obesogenic microbiota (i.e., Firmicutes) contribute to obesity by harvesting more energy from the diet, more recent studies have challenged this notion. A high-fat diet can increase Firmicutes in the gut microbiome without altering host metabolism, suggesting that many observed shifts in the microflora result from dietary changes and may not have metabolic consequences (Hildebrandt et al., 2009; Murphy et al., 2010). Human studies investigating the ratio of Firmicutes and Bacteroidetes have yielded inconsistent results (Duncan et al., 2008; Ley et al., 2005; Schwiertz et al., 2010). More comprehensive analyses of the gut microbiome suggest that changes in the gut microbiome at the sub-phylum level (and involving a limited number of species) could account for metabolic changes observed between different cohorts (Cotillard et al., 2013; Le Chatelier et al., 2013; Zhang et al., 2009).
Previous studies in murine models have shown that age (Murphy et al., 2010), host genetics (Henao-Mejia et al., 2012), and diet can affect the composition of the gut microbiome (Hildebrandt et al., 2009; Murphy et al., 2010). Long-term ecological studies of the gut microbiome have revealed longitudinal stability (Faith et al., 2013; Lozupone et al., 2012; Yatsunenko et al., 2012). This has led to the hypothesis that early gut colonizers, likely acquired from parents, play a vital role in determining the composition of the host microbiota and the physiological and metabolic fate of the host (Faith et al., 2013). However, longitudinal consistencies in gut-microbiome composition are strongly associated with long-term dietary patterns (Wu et al., 2011a). A change in diet can shift the composition of the gut microbiome rapidly, often within 24 hours, in both humans and mice (David et al., 2013; Turnbaugh et al., 2009b; Wu et al., 2011a). Finally, there is an intimate relationship between the gut microbiome and host intestinal epithelial cell (IEC) circadian regulators. This is exemplified by microbiome perturbations in phase-shifted mice (Voigt et al., 2014), and IEC circadian gene dysregulation with antibiotic-induced microbiome depletion (Mukherji et al., 2013). This suggests a far more dynamic environment than previously thought.
Since the relationship between the gut microbiome and metabolism is unclear, and dietary changes lead to rapid shifts in its composition, we sought to determine whether the gut microbiome is affected by cyclical fluctuations in feeding. Natural feeding and fasting cycles result in a fluid gut milieu in terms of nutrients, pH, and secondary metabolites, but it is unclear whether the gut microbiome is similarly dynamic, and if so, whether this dynamism plays a role in host metabolism.
RESULTS
Mice Fed Normal Chow Ad Libitum Exhibit Cyclical Fluctuations in Phyla within the Gut Microbiome
We fed 12-week-old male wild-type C57BL/6 mice a normal chow or a high-fat diet (HFD) for 8 weeks. These mice were allowed ad libitum access to food or were subjected to time-restricted feeding (TRF; Figure 1, Figure S1). To assess the stability of the gut microbiome within a 24-hour period, mice were sacrificed every 4 hours and metagenomic DNA was extracted from the cecal contents. We then sequenced 16S rRNA and created microbial community profiles by clustering 16S rRNA sequences into operational taxonomic units (OTU; ≥ 97% sequence match) and used the Ribosomal Database Project classifier (with a threshold ≥ 80% bootstrap value) to assign sequences to taxonomic groups. Across mice in all conditions, we identified 298 OTUs (Table S1–3).
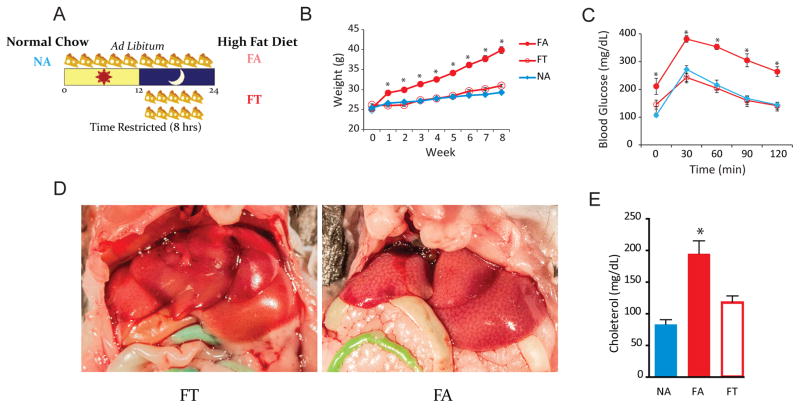
Figure 1. Study Design and Metabolic Studies of Mice in Each Condition.
(A) Study design. NA mice had ad libitum access to normal chow. FA mice had ad libitum access to HFD. FT mice had 8-hour access (ZT 13–21) to HFD. Results from our previous study were replicated (Hatori et al., 2012).
(B) Line plot showing the average weekly weight (g ± SEM) of mice in different conditions (n = 24 per condition). FA mice gained weight, whereas FT mice, despite being on a HFD were indistinguishable from NA controls. *p < 0.05 (compared to NA).
(C) Intraperitoneal glucose tolerance tests (mean ± SEM) show that TRF was protective against diabetes (n = 6 per condition). *p < 0.05 (compared to NA).
(D) On gross inspection, FA livers had steatosis, whereas FT livers did not.
(E) Serum quantification of cholesterol (n = 6 per group). Measurements (mean ± SEM) were performed twice. *p < 0.05 (compared to NA and FT).
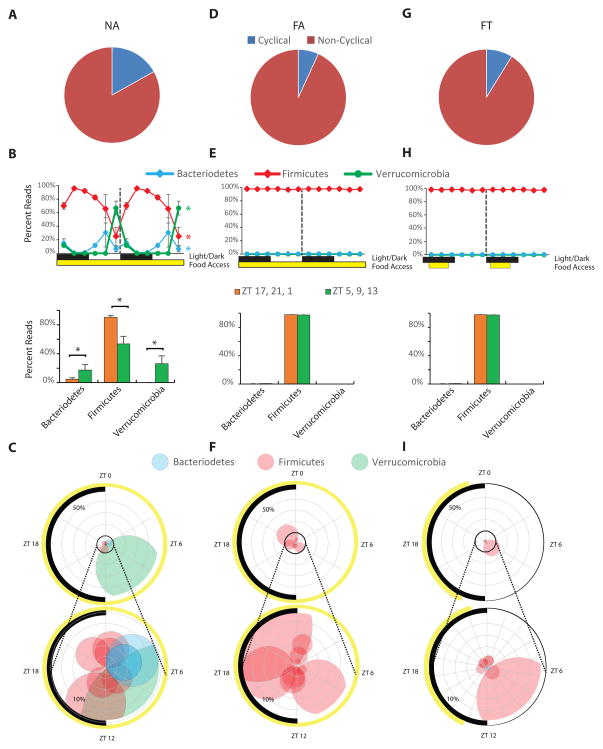
Mice with ad libitum access to normal chow (NA mice; Figure 1A) have a cyclical feeding pattern, eating most of their diet during their nighttime active phase and less during their daytime inactive phase (Hatori et al., 2012; Kohsaka et al., 2007; Liu et al., 2014). In order to determine whether a particular phylogenetic group was cyclical, we used JTK analysis, a non-parametric test that detects cycling elements (Hughes et al., 2010). In order for an OTU to be considered cyclical, its adjusted, permutation-based p-value (ADJ.P) and Benjamini-Hochberg q-values (BH.Q) both had to be less than 0.05. In NA mice, 17% of detected OTUs were cyclical (Figure 2A). Cyclical changes in the gut microbiome were apparent for all major phyla (ADJ.Ps are 2.0 × 10−6 for Firmicutes, 0.019 for Bacteroidetes, 2.9 × 10−6 for Verrucomicrobia; Figure 2B, C). The proportion of Firmicutes species peaked during nocturnal feeding and bottomed out during daytime fasting with a peak-to-trough ratio of 3:1 in NA mice. Bacteroidetes and Verrucomicrobia species peaked during daytime fasting and bottomed out during nocturnal feeding (Figure 2B, C). At any point in time, 20–83% of the reads belonged to OTUs that cycled. For NA animals, OTU peaks were not restricted to a particular time of day. They were distributed across all the time points but with a gradual rise after feeding (Figure S2A). A majority of OTUs that cycled in NA mice (80%) were underrepresented in HFD microbiomes (p < 2.0 × 10−4; Figure S2B, S2C).
Figure 2. Diurnal Rhythms of Gut Microbiome Phyla in Mice from Different Feeding Conditions.
(A) Pie chart showing the percentage of cycling and non-cycling OTUs (across all conditions) in NA mice (n = 18).
(B) Upper double-plot line graph – where the second cycle is a duplicate of the first cycle following the dashed line – shows the average percent read (± SEM) of the three most predominant phyla at each time point (n = 3 per time point). Black and white boxes indicate light off and light on, respectively. The yellow box shows when mice had access to food. Colored asterisks at the end of lines in line graph show which phyla were cycling based on JTK analysis (that is ADJ.P < 0.05 and BH.Q. < 0.05). Since it takes > 1 hour for a food bolus to reach the cecum (Padmanabhan et al., 2013), lower bar graphs show the average percent reads (± SEM, n = 9) for the dark/active feeding phase (ZT 17, 21, and 1), and the light/inactive fasting phase (ZT 5, 9, and 13). *p < 0.05.
(C) The top ten OTUs (based on percent reads) are depicted in a polar plot. The radian indicates the phase of the OTU’s peak, the distance from center is the average percent read across all time points, and the radius of each point indicates the amplitude of cycling. The colors of the circles indicate the phylum of the OTU: Firmicutes (pink), Bacteroidetes (blue), and Verrucomicrobia (green). The black arc on the left side of the plot indicates the light/dark cycle. The yellow arc depicts access to food. The bottom polar plot shows a magnified view of the inner ring (10%) of the top polar plot.
These descriptions also apply to panels for FA mice (D, E, F) and FT mice (G, H, I).
HFD Dampens the Cyclical Fluctuation of Phyla within the Gut Microbiome
Mice that have ad libitum access to HFD (FA mice; Figure 1A) spread their caloric intake, feeding during the dark/active phase and the light/inactive phase (Hatori et al., 2012; Kohsaka et al., 2007; Liu et al., 2014). They were obese (Figure 1B), had dysfunctional glucose homeostasis (Figure 1C), gross steatosis (Figure 1D), and hypercholesterolemia (Figure 1E). FA mice had fewer OTUs on 16S rRNA sequencing compared to NA mice (Figure S2D, see also Figure 5C), and fewer of these OTUs were cyclical (Figure 2D, Figure S2E, F). Interestingly, phylum-level cyclical changes observed in NA mice were dampened and did not approach significance in FA mice (ADJ.Ps are 0.03 – but BH.Q was 0.08 – for Firmicutes, 0.24 for Bacteroidetes, 1 for Verrucomicrobia, Figure 2E). The peak-to-trough ratio of Firmicutes species in FA mice was 1:1. Unlike NA mice, the maximum percentage of reads that belonged to cycling OTUs at any time point was 30%, and Firmicutes species were dominant at every time point (Figure 2F).
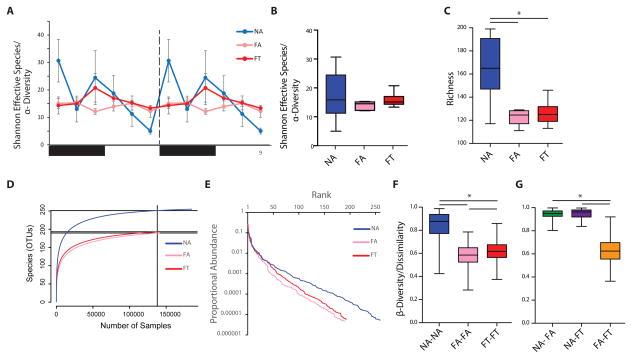
Figure 5. Diversity Analysis of Gut Microbiomes at Different Time Points.
(A) Shannon effective species (α-diversity or intra-sample diversity; ± SEM, n = 3 per time point) for each condition and time point.
(B) Box and whisker plot of Shannon effective species (α-diversity) and (C) richness averaged across all time points (n = 18). (D) Rarefaction and (E) rank-abundance curves of average reads (n= 18 per condition). Dissimilary (β-diversity) of (F) samples within the same condition (except those from the same time point), and (G) samples from different conditions. In all box plots, whiskers show minimum and maximum, the box is the 25–75th percentile, and the line is the median. *p < 0.05.
In a previous study (Hatori et al., 2012) we showed that using TRF to impose a natural feeding rhythm on mice fed a HFD (FT mice; Figure 1A) protects against diet-induced obesity and other metabolic disorders associated with FA mice. We replicated this finding in the current study (Figure 1B–E). TRF provides an ideal backdrop for studying intestinal microflora, because FT mice consume: 1) the same nutritional quality as FA mice, and 2) the same caloric quantity as NA mice (Figure S1A, B) (Hatori et al., 2012). In FT mice, therefore, microbiome differences that have metabolic consequences (e.g., obesity protective or obesogenic) are not obscured by alterations in nutritional/dietary intake. To better understand the relationship between cyclical fluctuations in the gut microbiome and diet, feeding phase, and metabolism, we expanded the study to include the FT condition.
FT mice had fewer OTUs compared to NA mice (Figure S2D, see also Figure 5C). Like FA mice, FT mice had fewer cycling OTUs than NA mice (Figure 2G, Figure S2E, S2G), and phylum-level cyclical changes were diminished (ADJ.Ps are 1 for Firmicutes, 0.06 for Bacteroidetes, 1 for Verrucomicrobia; Figure 2H). Unlike NA and FA mice, peaks in cycling OTUs in FT mice were related to the feeding schedule, occurring several hours after food was given or at the end of the fasting period (Figure S2G). Unlike NA mice and similar to FA mice, the maximum percentage of reads that belonged to cycling OTUs at any time point was 27%, and each time point was dominated by Firmicutes species (Figure 2I).
Species from the Firmicutes, Bacteroidetes, and Verrucomicrobia comprised ≥ 98% of the reads in all three conditions. Analyses of other phyla revealed cyclical activity for Actinobacteria species only in the FA condition (ADJ.Ps are 0.09 for NA, 6.5 × 10−4 for FA, 0.60 for FT; Figure S2H). In contrast, Proteobacteria species did not cycle, comprising less than 0.1% of reads in most mice (ADJ.Ps are 1 for NA, FA, and FT; Figure S2I).
Sub-Phylum Analysis Reveals Dynamic Gut Microbiomes in All Conditions
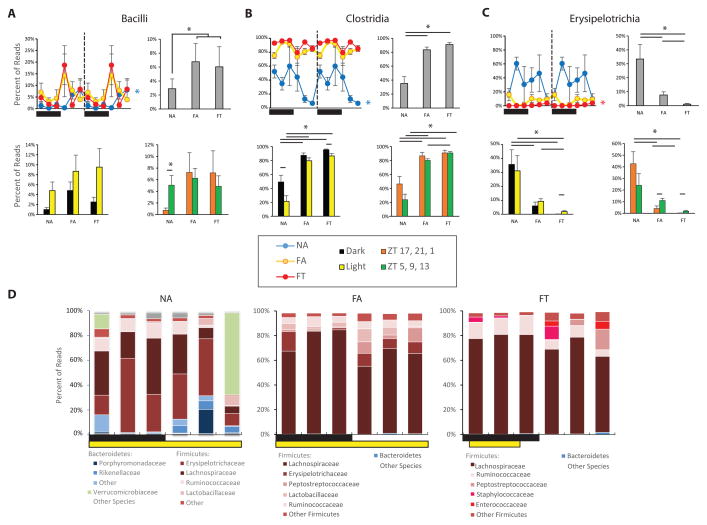
To further characterize differences between mice in three conditions, we performed sub-phylum analyses of the gut microbiome, focusing on classes, families, and genera. These analyses revealed further differences between NA mice and those in the HFD conditions. In NA mice Bacilli species were cyclical (ADJ.P = 5.0 × 10−3) with a peak that occurred after feeding (peak-to-trough 7:1; Figure 3A). On average across all time points, these species were significantly more prevalent in the gut microbiome of mice in the HFD conditions than in the NA mice (6.4% ± 1.4% vs. 2.9% ± 1.4%, respectively; p < 0.05). Unlike the NA condition, Bacilli were not cyclical in the two HFD conditions (ADJ.P = 1 for both FA and FT).
Figure 3. Subphylum Analysis of the Gut Microbiome in the Three Conditions.
Diurnal activity of several Firmicutes classes including (A) Bacilli, (B) Clostridia, and (C) Erysipelotrichia. Line graphs (left) show a double-plot of percent reads (± SEM, n = 3 per time point) for a particular class from all three conditions. Conditions are color-coded (see legend). Colored asterisks at the end of lines in line graph show which conditions were cycling based on JTK analysis. Bar graphs (top right in each panel) show percent total reads (± SEM, n = 18) for a particular class in each condition averaged over all time points. Histogram (bottom left of each panel) show percent reads (± SEM, n = 9) that are expressed when light off/light on. Histogram (bottom right of each panel) show percent reads (± SEM, n = 9) that are expressed when food from nighttime feeding has reached the cecum (ZT 17, 21, 1) and during relative fasting (ZT 5, 9, 13; see Fig 2 and Padmanabhan et al., 2013). * p < 0.05.
(D) Stacked bar graphs showing average percent reads of each family that comprised > 5% of total reads for each condition.
In addition, NA mice had a lower percentage of Clostridia species compared to the FA and FT conditions (35.2% ± 10.0%, 83.4% ± 4.0%, 90.9% ± 3.0%, respectively; Figure 3B). Clostridia as a phylogenetic class was cyclical in the NA condition, with a peak-to-trough of 9:1, but not in either of the HFD conditions (ADJ.P is 3.3 × 10−5 for NA, 0.45 for FA, 0.03 – but BH.Q was 0.08 – for FT). In both the NA and FT condition, Clostridia were more prevalent in the dark/active phase than in the light/inactive phase (for NA: 49.1% ± 9.6% vs. 21.3% ± 8.3%, p < 0.05; for FT: 95.4% ± 1.0% vs. 86.4% ± 3.6%, p < 0.05; Figure 3B).
Firmicutes species within the gut microbiome of NA mice belonged predominantly to the class Erysipelotrichia, which was far less common in the gut microbiome of mice fed the HFD, especially the FT condition (NA: 33.4% ± 10.4%; FA 7.6% ± 2.3%; FT: 1.1% ± 0.5%; one way ANOVA p < 0.0003; Figure 3C). This phylogenetic class was only cyclical in the FT condition (ADJ.P = 2.0 × 10−6) where it was twelve times more prevalent in the light/inactive phase than in the dark/active phase (1.95% ± 0.63% vs. 0.17% ± 0.05%, p < 0.05; Figure 3C).
In contrast to the analysis at the phylum level, sub-phylum analysis revealed that the microbiome of FT mice was distinct from that of FA mice. Although species in the Firmicutes phyla (and the Bacilli and Clostridia classes) dominated the gut microbiomes of both HFD cohorts, the families and genera were quite dissimilar, with differences in percent reads and cyclical activity (Figure 3D, Figure S3, Figure 4A–D). Many microfloral differences between FA and FT mice involved families and genera previously hypothesized to play roles in metabolism. Four of these will be discussed in greater detail.
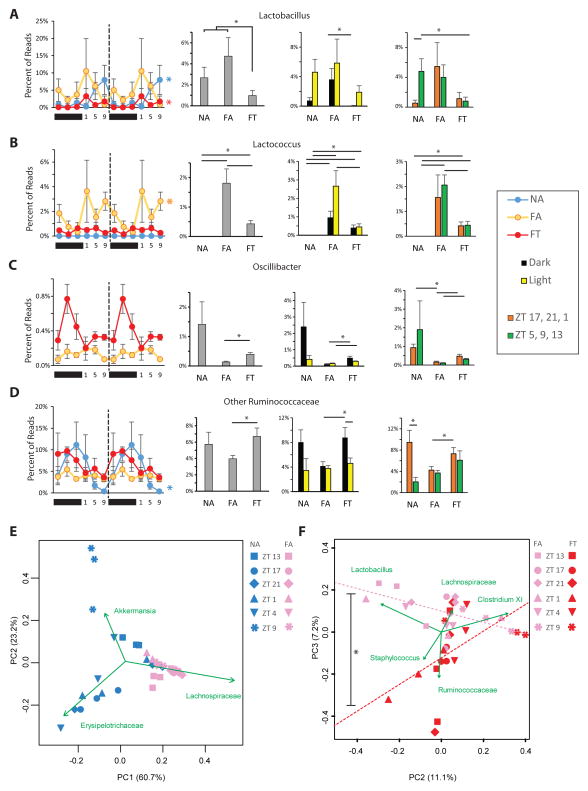
Figure 4. TRF Restores Cyclical Fluctuation in Genera Thought to be Involved in Metabolism.
Diurnal activity of (A) genus Lactobacillus, (B) genus Lactococcus, (C) genus Oscillibacter, and (D) other genera in the Ruminococcaceae family. For each, there is a double-plot of percent reads (± SEM, n = 3 per time point) for the three conditions. Colored asterisks at the end of lines in line graph show which conditions were cycling based on JTK analysis. For (C) the NA condition is excluded from the line plot but can be seen in Figure S4C. This is followed by a histogram of percent total reads (± SEM, n = 18) that this genus comprises in each condition, a histogram of percent reads (± SEM, n = 9) that are expressed when light off/light on and a histogram of percent reads (± SEM, n = 9) that are expressed when food from nighttime feeding has reached the cecum (ZT 17, 21, 1) and during relative fasting (ZT 5, 9, 13; see Fig 2 and Padmanabhan et al., 2013).
Genera-based principle component analysis (PCA) of NA and FA mice (E) and of FA and FT mice (F). Green vectors show the axis where that particular genus accounted for most of the variability. In (F), dotted lines show trend line of FA and FT mice in the PCA, which are significantly different (p < 0.05 by ANOVA of two populations).
TRF Decreases Relative Amounts of Presumed Obesogenic Microflora and Increases Relative Amounts of Presumed Obesity-Protective Microflora
The relationship between several Lactobacillus species and obesity, as well as associated metabolic disorders, has been studied extensively (Joyce et al., 2014; Li et al., 2013; Million et al., 2013; Million et al., 2012; Raman et al., 2013). In particular, a decrease in Lactobacillus species protects against metabolic disorders associated with obesity, perhaps by altering bile acids in the lumen (Li et al., 2013). In both NA and FT mice, the Lactobacillus genera were cyclical whereas in FA mice they were not (ADJ.Ps are 4.2 × 10−6 for NA, 1 for FA, 1.2 × 10−5 for FT; Figure 4A). Furthermore, Lactobacillus species comprised a lower percentage of reads in FT mice compared to the combined ad libitum cohorts (0.97% ± 0.49% vs. 3.70% ± 1.01%, respectively, p < 0.05 Figure 4A). However, since the percentage reads were similar between FA and NA mice, it is likely that the temporal profile of these species, rather than the relative abundance, exerts greater influence on host metabolism. In particular FA mice had a higher amount of Lactobacillus species during the dark/active phase compared to FT mice (3.62% ± 1.49% vs. 0.06% ± 0.04%, respectively, p < 0.05 Figure 4A).
Previous studies have shown that the amount of Lactococcus species directly correlates with body fat percentage in mice consuming a high-fat/high-sugar diet (Parks et al., 2013). We have shown that FA mice have a much higher body fat percentage than NA mice, but body fat percentages are similar between FT and NA mice (Hatori et al., 2012). Consistent with previous results, the current study shows that Lactococcus species were barely detectable in the gut microbiome of NA mice at any time points and were much higher in the FA mice (NA: 0.00% ± 0.00%; FA 1.81% ± 0.69%; FT: 0.43% ± 0.15%; one way ANOVA p < 0.0001; Figure 4B). FT mice had a significantly lower percentage of Lactococcus species in their gut microbiome than FA mice did (Figure 4B). Lactococcus was only cyclical in the FA mice (ADJ.P = 4.9 × 10−3). In particular the main difference in Lactococcus species between FA and FT is the amount found during the light/inactive phase. The percent reads of Lactococcus species was significantly higher in the FA mice than in the FT ones (2.66% ± 0.84% vs. 0.45% ± 0.16%, respectively; p < 0.05), but not during the dark/active phase (0.96% ± 0.34% vs. 0.41% ± 0.15%, respectively; p = 0.16, Figure 4B).
High relative levels of Oscillibacter and other Ruminococcaceae species protect against obesity and non-alcoholic fatty liver disease (Raman et al., 2013). Here we found a significantly higher percentage of Oscillibacter species in FT gut microflora than we did in FA mice (0.40% ± 0.08% vs. 0.13% ± 0.04%, respectively; p < 0.05, Figure 4C). The relative reads of Oscillibacter species were cyclical in NA but not in FT or FA mice (ADJ.Ps are 2.1 × 10−4 for NA, 1 for FA, 0.09 for FT; Figure 4C, Figure S4A). Furthermore, Ruminococcaceae species comprised a higher percentage of reads in FT mice compared to FA mice (6.69% ± 1.03% vs. 3.96% ± 0.42%, respectively; p < 0.05, Figure 4D). Ruminococcaceae were highly cyclical in NA mice but not in FT and FA mice (ADJ.Ps are 3.3 × 10−5 for NA, 1 for FA, 0.09 for FT; Figure 4D). TRF increased the relative amounts of Ruminococcaceae species in the dark/feeding time of FT mice, which was significantly higher than the relative amounts in the FA mice during the same time points (8.79% ± 1.64% vs. 4.15% ± 0.71%, respectively; p < 0.05, Figure 4D). Relative reads for some of the other bacterial genera in NA, FA, and FT mice are shown in Figure S3.
Principle component analysis of NA mice revealed that the ZT 9 gut microbiome was quite distinct from gut microbiomes measured at any other time point (Figure 4E). Time within a 24 hour light:dark cycle is reported as Zeitgeber Time (ZT), or “time since lights on,” where ZT0 is when the light turns on/dawn, and ZT12 is when the light turns off. By definition ZT 9 is 9 hours after lights have been on (and 3 hours before the nocturnal feeding bout begins). After NA mice had commenced nighttime feeding, the composition of their microbiomes at ZT 13 shifted toward the FA microbiome. In NA mice, feeding changed the gut microbiome so dramatically that some time point clusters were more different from each other than they were from the FA gut microbiome (e.g., compare ZT 9 and ZT 17; Figure 4E). Principle component analysis of FA and FT gut microbiomes showed clusters that were not as dramatically different. Since Lachnospiraceae species accounted for >50% of the reads in both FA and FT mice (Figure S3), these species accounted for much of the observed variance in these cohorts (Figure S4E). However, concerning second and third principle components, the FA and FT conditions exhibited some degree of separation (Figure 4F). FT gut microbiome clusters were similar to FA gut microbiome clusters during fasting. During feeding, however, the FT clusters deviated from the “baseline” FA clusters. This suggests that if the gut microbiome directly affects metabolism in FT mice, this impact likely occurs during a narrow window of time.
Gut Microbial Ecology is Dynamic in the Gut Microbiome of Mice in NA and FT Condition
Dysbiosis, as a result of HFD, leads to alterations in microbiome ecology, particularly reductions in diversity (Ley et al., 2005; Turnbaugh et al., 2009a). Reduced diversity, particularly α-diversity (i.e., the types and relative amounts of species within a sample; intra-sample diversity), is thought to play a significant role in host metabolism and global increase in obesity (Lozupone et al., 2012), but it is unclear whether changes in microbial diversity result from changes in the nutritional composition of the diet, or from cyclical changes in luminal content.
We observed that the α-diversity of the gut microbiome of NA mice varied widely. There were usually higher levels of diversity during nighttime feeding and lower levels during daytime fasting (Figure 5A). However, gut microbiome α-diversity for FA and FT mice remained constant, without much temporal fluctuation (Figure 5A). Although NA and FT mice had significantly higher variance in their α-diversity compared to FA mice (Bartlett’s test for equal variance, p = 6.0 × 10−4), when all time points were averaged together there were no significant differences between the three conditions (NA: 21.70 ± 5.08, FA: 15.73 ± 0.81 FT: 18.75 ± 1.40,one way ANOVA p = 0.41, Figure 5A, 5B).
α-diversity can be affected by species richness (i.e., the number of unique species/OTUs) and the abundance of each species. Across all time points, NA mice had significantly higher species richness compared to the two HFD conditions (NA: 165.2 ± 11.6, FA: 122.7 ± 2.7 FT: 126.2 ± 4.5, p = 0.009, Figure 5C). Levels of richness for FT and FA mice were indistinguishable (Figure 5C). These results were confirmed using a rarefaction curve analysis (Figure 5D). However, although the rarefaction curves of the averaged FA and FT samples appeared similar, there was much more variability in species obtained from different time points. For example rarefaction plots of FT samples from ZT 21 and ZT 9 were remarkable different from each other. These plots were also different from plots obtained from FA mice at the same time points (Figure S5A). Rank-abundance analysis of samples obtained from FA, FT, and NA suggests that the main cause of the rise in the variability of FT α-diversity was an increase in the relative abundance of species, rather than an increase in the number of species (Figure 5E).
Differences in β-diversity (i.e., the measure of dissimilarity between two samples; inter-sample diversity) were also measured. We first analyzed the difference between the gut microbiome of mice subjected to the same condition, but at different time points (i.e., within condition, Figure 5F) The within condition β-diversity was significantly different between all three conditions (NAvNA: 0.845 ± 0.010, FAvFA: 0.581 ± 0.008, FTvFT 0.625 ± 0.009, one way ANOVA p < 0.0001). This indicates that the NA gut microbiome undergoes dramatic temporal fluctuations compared to the HFD conditions (Figure 5F, Figure S5B). Furthermore, the gut microbiome of FT mice also experience significant fluctuations when compared to that of the FA mice. Dissimilarity between NA gut microbiomes and those of the FA and FT condition (i.e., outside condition) was nearly as high as that measured for the NA within condition dissimilarity (NAvFA: 0.946 ± 0.002, NAvFT: 0.949 ± 0.002, FAvFT 0.632 ± 0.006, one way ANOVA p < 0.0001, Figure 5G, Figure S5B).
Changes in the Gut Microbiome Is Accompanied with Changes in Stool Metabolites
It is unclear how cyclical (or TRF-induced) variation in the gut microbiome affects host metabolism. To better understand the specific effects of the gut microbiome, we analyzed stool metabolites that are altered by gut microbes (not by host enzymes) from dark/feeding and light/fasting cages. Pooled stool was collected from fresh nighttime and daytime cages from each condition. Samples were analyzed for specific metabolites.
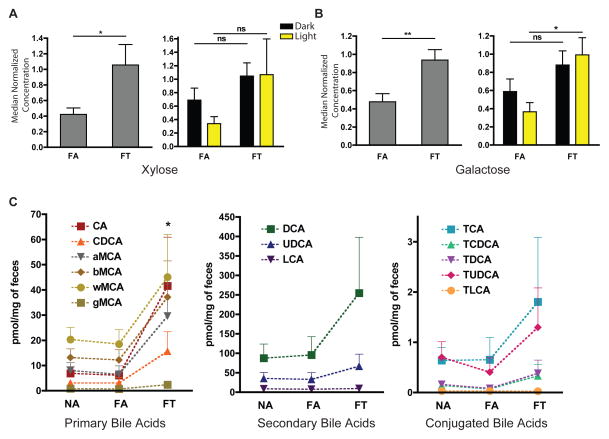
We first investigated stool metabolites that would suggest that FA and FT mice extract different amounts of energy from chow. Hemicellulose, which is a component of the plant cell wall, is a non-absorbable complex sugar present in both normal and HFD murine chow. It is normally broken down into more absorbable sugars, xylose and galactose, with the aid of cellulase enzymes from gut bacteria. FT mice excreted significantly more xylose but there was no difference in the stool collected in the light and dark periods (Figure 6A). FT also excreted more galactose, with a significantly higher amount excreted in the stool collected during the light period (Figure 6B). Thus, FT mice excreted rather than absorbed these calories. NA mice excreted much higher amounts of xylose and galactose than FA and FT mice (Figure S6A). However, it is difficult to perform direct comparisons between mice fed normal or HFD chow, since the diets contain different amounts of hemicellulose.
Figure 6. Metabolites Processed by Gut Microbes are Differentially Excreted in the Feces of Mice in Different Conditions.
Average relative quantification of (A) xylose and (B) galactose (± SEM) in the feces of mice fed a HFD (n = 4 per condition, from separate cages). Histogram on left shows average across all samples collected (n=8). Histogram on right shows differences between samples collected from dark and light (n = 4). See Figure S6A for NA results.
(C) Average absolute quantification (± SEM) of primary, secondary, and tauro-conjugated bile acids within feces. Dashed line connects similar bile acids to allow easy comparison across conditions. CA: cholate, CDCA: chenodeoxycholate, MCA: Muricholate (a: alpha-, b: beta-, g: gamma-, w: omega-), DCA: deoxycholate, UDCA: ursodeoxycholate, LCA: lithocholate, T-: Tauro-. *p < 0.05, **p < 0.01.
Another way in which the gut microbiome may affect host metabolism is through altering luminal bile acid signaling (Matsubara et al., 2013; Sayin et al., 2013). Fecal/stool bile acids are structurally complex, and this complexity is driven by microbial species within the intestinal tract (Hagey and Krasowski, 2013). We therefore assessed stool bile acids from mice in the three conditions. More primary bile acids were found in FT mice compared to NA and FA mice (Figure 6C, Figure S6B). Furthermore, there tended to be more secondary and tauro-conjugated bile acids in FT mice compared to NA and FA mice, although these trends were not significant (Figure 6C; Figure S6B). Hence, alterations in the gut microbiome of FT mice led to increased excretion of bile acids, and likely increased concentrations of bile acids within the gut lumen. High levels of bile acids in FT mice could affect metabolism through luminal bile acid signaling.
DISCUSSION
Previous studies have shown long-term stability of the gut microbiome, although variation driven by age, diet, and the environment has also been reported (Faith et al., 2013; Lozupone et al., 2012). This long-term stability has been tempered with recent evidence that the gut microbiome rapidly changes when the diet is altered (David et al., 2013; Turnbaugh et al., 2009b). Here we have shown that the gut microbiome exhibited daily cyclical variation in a variety of dietary and feeding pattern conditions, with the assumption that the changes in the gut microbiome are similar to those in host gene expression to make 24 hours a sufficient amount of time to assess circadian changes. Of note, during the review of this manuscript, another group also report diurnal changes in the mammalian gut microbiome (Thaiss et al., 2014).
NA mice showed cyclical fluctuation in the composition of the gut microbiome. Firmicutes species were most abundant with feeding during the dark/active phase and reached their low point with fasting during the light/inactive phase. Conversely, Bacteroidetes and Verrucomicrobia species rose during fasting and fell during feeding. These cyclical fluctuations in specific members of the gut microbiome were accompanied by changes in the diversity of the microflora environment. The α-diversity of the gut microbiome fluctuated with the time of day and was highly variable, rising with feeding and falling with fasting. The wide fluctuations between time points was confirmed with significantly higher β-diversity between NA samples taken at different time points.
Mice given ad libitum access to a HFD (a model of diet-induced obesity) lose diurnal feeding (Hatori et al., 2012; Kohsaka et al., 2007). As a consequence, their gut microbiome contained half as many cycling OTUs, and cycling OTUs comprised less of the overall gut microbiome. Firmicutes species, particularly of the Clostridia order, dominated the gut microbiome of FA mice, and overall species numbers were reduced. In these mice α-diversity did not fluctuate; rather it stayed constantly low. Likewise, within-condition β-diversity of the gut microbiome remained low.
We originally hypothesized that TRF in FT mice would make the gut microbiome highly dynamic, similar to the gut microbiome of NA mice. However, this was not the case. Superficially, FT and FA gut microbiomes were very similar, highlighting the important role of diet in forming the gut microbial environment. At the subphylum level, however, there were key differences between FT and FA gut microbiomes. In addition, FT mice had a higher β-diversity, but not α-diversity compared to FA mice. These differences in microbial ecology reinforce the fact that the gut microbiome of FA and FT mice are quite different in subtle ways.
Temporal changes in the diversity are particularly important in understanding the contribution of the gut microbiome to obesity and metabolic disease (Ley et al., 2005; Turnbaugh et al., 2009a). Many have observed a decrease in α-diversity in diet-induced obesity/FA mice compared to control/NA mice and, hence, have hypothesized that increasing it can be protective against obesity (Lozupone et al., 2012). However, our data shows that α-diversity can vary widely throughout the day in NA mice and that, when all time points are taken into account, there is no significant differences between any conditions that we have tested. Changes in β-diversity, however, are significant, further showing that fluctuations in the gut microbiome are important for host metabolism, not necessarily richness of species which is affected by diet.
A key finding of the paper is that the feeding pattern affected the composition of the luminal microflora, even when animals were fed the same diet. Analysis of the gut microbiome of FA and FT mice revealed differences when analyzed at the subphylum level. Many of the OTUs that regained cycling or were significantly different in the FT gut microbiome (compared to the FA one) belong to genera that have been hypothesized to play a role in host metabolism. Lactobacillus species, which are thought to be obesogenic (Joyce et al., 2014; Li et al., 2013; Million et al., 2013; Million et al., 2012; Parks et al., 2013; Raman et al., 2013), were cyclical in the gut microbiome of NA and FT mice, but not in FA mice. Compared to FA mice, FT mice had lower levels of these species. Lactococcus are also thought to be obesogenic (Million et al., 2012). FT mice had significant reduction in Lactococcus species compared to FA mice, especially during the light phase, suggesting that fasting may play a crucial role in keeping these species at check. In addition, protective species, such as those belonging to the Ruminococcacea family, including the genus Ocillibacter (Raman et al., 2013), were elevated in the gut microbiome of NA and FT mice but not in FA mice. These results suggest that the benefits of TRF could be due, at least in part, to an alteration in the gut microbiome.
Other species of gut bacteria have been hypothesized to play a role in host metabolism. These include Bifidobacterium (Million et al., 2012) and Akkermansia (Everard et al., 2013), both of which are highly cyclical species. Bifidobacterium species did not seem to be affected by the different dietary conditions (once data were averaged over all time points). Akkermansia seemed to thrive most in the fasting lumen of NA mice. Our findings suggest that changes in these genera may not be sufficient to improve metabolism since they were not different between FA and FT conditions.
Mechanism by which the gut microbiome affects host metabolism is not well understood. Modification and fluctuation of the gut microbiome does not indicate that the host metabolism has changed, since this association may be correlative. However, our analysis of stool metabolites revealed how differences in the gut microbiome between FA and FT mice could explain their distinct phenotypes. For example, analysis of sugars revealed high levels of xylose and galactose in the stool of FT mice. This suggests that these sugars more readily absorbed by the guy of FA mice than FT mice.
The analysis of stool bile acid also yielded interesting results, especially given that Lactobacillus species were underrepresented in the gut microbiome of FT mice. Some Lactobacillus species produce bile acid hydrolase, which can conjugate bile acids that are antagonists of the main ileal bile acid receptor, the farnesoid X receptor (FXR) (Li et al., 2013). Enterohepatic bile acid signaling mediated by FXR most directly affects the activity of cyotchorme p450 7A1 (Cyp7a1, also known as cholesterol 7 alpha-hydroxylase), which is the main enzyme for de novo bile acid synthesis from cholesterol (Calkin and Tontonoz, 2012; Gilardi et al., 2007; Matsubara et al., 2013). This enzyme exhibits high levels of activity in FT mice compared to FA mice (Hatori et al., 2012), which corresponds with decreased serum cholesterol in FT mice (Hatori et al., 2012). In the current study we also observed low levels of serum cholesterol in FT mice (Figure 1E). In addition, bile acid signaling through FXR, which is highly circadian, can modulate glucose homeostasis and lipid/triglyceride metabolism (Calkin and Tontonoz, 2012; Gilardi et al., 2007; Matsubara et al., 2013; Zarrinpar and Loomba, 2012; Zhang et al., 2011). Hence, TRF may have beneficial effects by restoring the cyclical fluctuation of gut microflora, which in turn would alter bile acids signaling to affect metabolism. However, additional experiments using selective FXR agonists/antagonists or FXR-related knockout mice will be needed to prove this relationship.
The gut microbiome is essential for normal gut homeostasis, not only for its immunological and metabolic roles, but also in host circadian gene expression (Mukherji et al., 2013). Antibiotic-induced microbiota depletion eliminates gut circadian gene expression and perturbs intestinal epithelial cell homeostasis. It is unlikely that a static gut microbiome could drive cyclical gene expression of the gut. Cyclical variation in the gut microbiome may also account for large variation in human microbiome results observed within and between studies (Duncan et al., 2008; Schwiertz et al., 2010). The realization that the gut microbiome has fine-scale cyclical variation in many species indicates that the time of day should be controlled for when characterizing the gut microbiome. A better record of animal diet, feeding patterns, and time of specimen collection is necessary to interpret studies of the gut microbiome, and to understand host-microbiome relationship.
EXPERIMENTAL PROCEDURES
Animals
All animal experiments were carried out in accordance with the guidelines of the Animal Care and Use Committee of the Salk Institute. Eight-week old male C57BL/6J mice (Jackson Laboratory) were group housed (three mice per cage) under a 12-hour light:12-hour dark schedule and ad libitum access to normal chow for 2 weeks to adapt to the housing conditions before being randomly assigned to one of the feeding conditions (see below).
Feeding Schedule and Diets
NA mice were fed a normal chow diet (LabDiet 5001, 29% protein, 13% fat, 58% carbohydrates) with unrestricted (ad libitum) access. FA and FT mice received HFD (LabDiet 58Y1; 18% protein, 61% fat, 21% carbohydrates) with either ad libitum access (FA mice), or during an 8 hour window between ZT 13 and ZT 21 (time restricted feeding - FT mice; Figure 1A). Access to food was controlled by transferring mice daily between cages with food and water to cages that had only water. Animal weights and food intake were measured weekly. Animals were maintained on assigned diets and feeding paradigms for 8 weeks.
Metabolic Measurements
Glucose tolerance tests (6 mice per group) were performed 7 weeks after initiation of the feeding paradigm. Animals were fasted for 16 hours on paper bedding before the test. Blood glucose levels were measured using One Touch Ultra glucometer prior to injection of glucose (1 g/kg intraperitonally) and every 30 minutes after injection. Serum from fasting animals (6 per condition) at 8 weeks was used to measure ALT, cholesterol, and triglycerides using Thermo Scientific Infinity Reagents according to manufacturer’s instructions.
Metagenomic DNA Extraction and 16S rRNA Sequencing
Three mice (from different cages) from each feeding condition were sacrificed every 4 hours over a 24-hour period, and each individual’s cecum was flash frozen. Individual ceca were then resuspended in PBS and digested with RNAse A and proteinase K at 55 °C for 1 hour before lysis with a bead beater. DNA from the lysate was extracted using Phenol/Chloroform/Isoamyl alcohol, precipitated, and washed with ethanol. Resulting DNA was resuspended in TE. 16S rRNA gene sequence tags, corresponding to the hypervariable V1–V3 region, were generated using the 454 pyrosequencing platform. These sequences were used to generate microbial community profiles using OTU-based classification.
Sequence Analysis
We used UPARSE (Edgar, 2013) to process the raw data (including the QC step) and to compute OTUs. We computed 97% identity OTUs and, using representative OTU sequences, classified them using Ribosomal Database Project classifiers. A threshold of ≥ 80% bootstrap value was used to assign a sequence to a taxonomic group. Since so many identified species were in the Firmicutes phylum, another analysis searching for the closest neighbor to the OTU sequence (the match had to be at least 97% identity) was performed using the CD-HIT program (Wu et al., 2011b). We used quantitative PCR of selected taxa to confirm results.
Determining Cyclical Fluctuations
For each OTU, the percent of total reads was calculated for each mouse and then averaged per time point per condition (3 mice). The data from 6 time points were analyzed by JTK-Cycler to detect cyclical variation (Hughes et al., 2010). Significance was determined if the permutation-based p-values (i.e. adjusted p-value) and the Benjamini–Hochberg q-values (BH.Q) were both less than 0.05. This strict criteria was used to reduce false positive rates. ANOVA and t-tests were used in parallel to detect overall changes in the microbiome or transcripts between feeding paradigms.
Diversity Analysis
All diversity analyses were performed using the vegan package (Oksanen et al., 2013) in R for Windows (The Comprehensive R Archive Network). For α-diversity, a Shannon-index was initially calculated, which was then converted to a Shannon effective species (Jost, 2006) for easier comparisons between different populations. ANOVA was used to compare the three groups. For β-diversity, the Jaccard dissimilarity index was used to measure dissimilarity between gut microbiomes of samples within each condition (but different time points) and between each condition (Koleff et al., 2003). Rarefaction and rank-abundance plots were measured by taking diversity of average reads for each condition, unless otherwise stated.
Stool Metabolite Measurements
Fresh stool was collected from the nighttime feeding and daytime fasting cages and flash frozen. The stool was then powderized in liquid nitrogen before further analysis. Relative quantification of xylose and galactose was performed using Metabolon, Inc. (Evans et al., 2009). Bile acids quantification was performed by the metabolomics core services at the University of Michigan. Bile acids were extracted from feces (50 mg) using a two-step solvent extraction. Supernatants were combined, dried, and re-suspended for LC-MS separation by RPLC. Results were quantified using standard curves of authentic standards (Griffiths and Sjovall, 2010). For both analyses, data were averaged from four samples obtained from different light/dark cages from each condition.
Statistical Analysis
Unless otherwise stated, all statistical analyses among the three groups were nonparametric, one-way ANOVAs (Kruskal-Wallis test) with Dunns post test. All comparisons between two groups were two-tailed nonparametric t-tests (Mann Whitney U Test).
Supplementary Material
Figure S1, related to Figure 1: Food consumption and serum biomarker results. (A) Cumulative food consumption (kcal/mouse ± SEM, n=24) and (B) individual calories consumed (kcal/mouse/week ± SEM, n=24). As in previous experiments (see ref 11), there is no difference between FT and NA mice. Serum quantification (± SEM) of (C) alanine transaminase (ALT) activity and (D) triglycerides in overnight fasted mice (n=6 per groups; measures were repeated twice independently).
Figure S2, related to Figure 2: Gut microbiome dynamics. (A) Heatmap of cycling OTUs organized by peak from NA mice. Corresponding heatmap of the OTUs from other conditions are shown to illustrate that phases and cycling changed with diet and TRF. Bar graph at the bottom of column shows the time point at which the OTUs peaked. Black and white boxes show light on/light off timing. Yellow box shows when mice had access to food. (B) Log2 plot of percent reads of OTUs that are found in all three feeding conditions. The x-axis is log2 of the average (across all time points, n=18) of percent reads of an OTU in NA divided by its percent reads in FA. When positive, this would indicate that a particular OTU is more common in NA than in FA, and when negative, that it is less common in NA compared to FA. Likewise, the y-axis is the log2 of the average (across all time points, n=18) of percent reads of an OTU in FT divided by its percent reads in FA. When positive this would indicate that a particular OTU is more common in FT than in FA, and when negative, that a particular OTU is less common in FT compared to FA. This panel shows OTUs that are non-cycling and those that are cycling in NA. OTUs that cycled in NA were much more common in the NA gut microbiome than they were in FA gut microbiome (p <0.0005, Chi-square test). (C) Is a similar log2 plot as (B) but shows all OTUs that were common to all three feeding conditions. (D) Venn diagrams showing the number of total OTUs that were shared between the three feeding conditions and (E) the number of cycling OTUs that were shared between the three feeding conditions. Heatmap of cycling OTUs organized by peak from (F) FA mice and (G) FT mice. Corresponding heatmap of the OTUs from other conditions are shown to illustrate that phases and cycling changed with diet and TRF. Bar graph at the bottom of column shows the time point at which the OTUs peaked. Black and white boxes show light on/light off timing. Yellow box shows when mice had access to food. Diurnal activity of the phylum Actinobacteria (H) and Proteobacteria (I). First column: double plot of percent read for a particular genus from all three conditions (n=3/condition/time point). Second column: double plot of percent reads for the genus from the two HFD conditions (n=3/condition/time point). Third column: histogram of percent total reads that this genus comprises in each condition (n=18/condition). Fourth column: histogram showing percent reads that are expressed when light off/light on (n=9). Fifth column: histogram showing percent reads that are expressed when food from nighttime feeding has reached the cecum (ZT 17, 21, 1) and during relative fasting (ZT 5, 9, 13; n=9, see Figure 1 and ref 20). * p <0.05.
Figure S3, related to Figure 3: Subphylum analysis of the gut microbiome. Diurnal activity of genera identified in our study. First column: double plot of percent read (± SEM, n=3 per time point) for a particular genus from all three conditions. Second column: double plot of percent reads (± SEM, n=3 per time point) for the genus from the two HFD conditions. Third column: histogram of percent total reads (± SEM, n=18) that this genus comprises in each condition. Fourth column: histogram showing percent reads (± SEM, n=9) that are expressed when light off/light on. Fifth column: histogram showing percent reads (± SEM, n=9) that are expressed when food from nighttime feeding has reached the cecum (ZT 17, 21, 1) and during relative fasting (ZT 5, 9, 13; see Figure 1 and ref 20). * p < 0.05.
Figure S4, related to Figure 4: TRF restores cyclical fluctuation in genera thought to be involved in metabolism. (A) Percent reads (± SEM, n=3 per time point) of a double plot of genus Oscillibacter from all three conditions. (B) Genera based principle component analysis (PCA) of FA and FT mice. Green vectors show the axis where that particular genus accounted for most of the variability.
Figure S5, related to Figure 5: Diversity analysis. (A) Rarefaction curve of average reads from specific time points in FA and FT (number in box is ZT time point) (n=3/condition). The rarefaction plots of FT mice at ZT 9 and 21 were remarkably different, whereas those from FA mice at ZT 9 and 21 were quite similar. (B) Triangle plots illustrating β-diversity (see Koleff et al., 2003 and Fig 3F, 3G). Briefly, points that are close to apex (a′) share many OTUs. However points that are near the base (b′ or c′) have many dissimilarities. These plots show that NAvFA and NAvFT have the most dissimilarities and are nearly indistinguishable. NAvNA have much more dissimilarity than the FAvFA and FTvFT.
Figure S6, related to Figure 6: Metabolites that are processed by gut microbes are differentially excreted in the feces of mice in different conditions. (A) Average relative quantification of xylose and galactose (± SEM) in the feces (n = 4 samples from each condition, from separate cages) which includes results from all three conditions. Absolute quantification in pmol/mg of feces of the primary biles acids MCA, CA and DCA, the secondary bile acids DCA, UDCA and LCA and their respective tauro (T-) and glyco (G-) conjugates. CA: Cholate, CDCA: Chenodeoxycholate, MCA: Muricholate (a: alpha-, b: beta-, g: gamma-, w:omega-), DCA: Deoxycholate, UDCA: ursodeoxycholate, LCA: lithocholate, T-: Tauro-, G-:Glyco.
Acknowledgments
Raw data for our analysis (Table S1–3) is located in the Supplementary Materials as an excel file. We would like to thank Russel Van Gelder and Lakshmi Akileswaran for performing confirmatory studies. AZ received support from NIH T32 DK07202, AASLD Liver Scholar Award, and the American Gastroenterological Association Research Foundation Elsevier Pilot Research Award. AC received salary support from an American Diabetes Association Mentor-Based Postdoctoral Fellowship (7-12-MN-64). SP received support from DK091618. The Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002, Glenn Foundation, and NIH center grants P50 GM085764, P30 CA014195, P30 EY019005, KL2 TR00099, and R24 DK080506 supported various aspects of the study. Metabolomic Core Services were supported by NIH DK097153 to the University of Michigan.
Footnotes
AUTHOR CONTRIBUTIONS
AZ and SP designed the study. AZ and SY analyzed the microbiome data. AZ and AC carried out the wet lab experiments and analyzed the metabolomic results. AZ, SY, and AC wrote the manuscript. SP analyzed the data and supervised the entire study. All authors discussed the results and commented on the manuscript.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nature reviews Molecular cell biology. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013 doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, Flint HJ. Human colonic microbiota associated with diet, obesity and weight loss. International journal of obesity. 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical chemistry. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi F, Mitro N, Godio C, Scotti E, Caruso D, Crestani M, De Fabiani E. The pharmacological exploitation of cholesterol 7alpha-hydroxylase, the key enzyme in bile acid synthesis: from binding resins to chromatin remodelling to reduce plasma cholesterol. Pharmacology & therapeutics. 2007;116:449–472. doi: 10.1016/j.pharmthera.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Griffiths WJ, Sjovall J. Bile acids: analysis in biological fluids and tissues. Journal of lipid research. 2010;51:23–41. doi: 10.1194/jlr.R001941-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagey LR, Krasowski MD. Microbial biotransformations of bile acids as detected by electrospray mass spectrometry. Advances in nutrition. 2013;4:29–35. doi: 10.3945/an.112.003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell metabolism. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724. e1711–1712. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of biological rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. Entropy and diversity. Oikos. 2006;113:363–374. [Google Scholar]
- Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CG. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell metabolism. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Koleff P, Gaston KJ, Lennon JJ. Measuring beta diversity for presence-absence data. Journal of Animal Ecology. 2003;72:367–382. [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, Fabre KM, Mitchell JB, Patterson AD, Gonzalez FJ. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nature communications. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Huang M, Wu X, Shi G, Xing L, Dong Z, Qu Z, Yan J, Yang L, Panda S, et al. PER1 phosphorylation specifies feeding rhythm in mice. Cell reports. 2014;7:1509–1520. doi: 10.1016/j.celrep.2014.04.032. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T, Li F, Gonzalez FJ. FXR signaling in the enterohepatic system. Molecular and cellular endocrinology. 2013;368:17–29. doi: 10.1016/j.mce.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million M, Angelakis E, Maraninchi M, Henry M, Giorgi R, Valero R, Vialettes B, Raoult D. Correlation between body mass index and gut concentrations of Lactobacillus reuteri, Bifidobacterium animalis, Methanobrevibacter smithii and Escherichia coli. International journal of obesity. 2013;37:1460–1466. doi: 10.1038/ijo.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. International journal of obesity. 2012;36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, Clarke SF, O’Toole PW, Quigley EM, Stanton C, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. vegan: Community Ecology Package 2013 [Google Scholar]
- Padmanabhan P, Grosse J, Asad AB, Radda GK, Golay X. Gastrointestinal transit measurements in mice with 99mTc-DTPA-labeled activated charcoal using NanoSPECT-CT. EJNMMI research. 2013;3:60. doi: 10.1186/2191-219X-3-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell metabolism. 2013;17:141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2013;11:868–875. e861–863. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell metabolism. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Scheving LA. Biological Clocks and the Digestive System. Gastroenterology. 2000;119:536–549. doi: 10.1053/gast.2000.9305. [DOI] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell host & microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009a;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science translational medicine. 2009b;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, Turek FW, Keshavarzian A. Circadian disorganization alters intestinal microbiota. PloS one. 2014;9:e97500. doi: 10.1371/journal.pone.0097500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011a;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC genomics. 2011b;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Loomba R. Review article: the emerging interplay among the gastrointestinal tract, bile acids and incretins in the pathogenesis of diabetes and non-alcoholic fatty liver disease. Alimentary pharmacology & therapeutics. 2012;36:909–921. doi: 10.1111/apt.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, et al. Human gut microbiota in obesity and after gastric bypass. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YK, Guo GL, Klaassen CD. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PloS one. 2011;6:e16683. doi: 10.1371/journal.pone.0016683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1: Food consumption and serum biomarker results. (A) Cumulative food consumption (kcal/mouse ± SEM, n=24) and (B) individual calories consumed (kcal/mouse/week ± SEM, n=24). As in previous experiments (see ref 11), there is no difference between FT and NA mice. Serum quantification (± SEM) of (C) alanine transaminase (ALT) activity and (D) triglycerides in overnight fasted mice (n=6 per groups; measures were repeated twice independently).
Figure S2, related to Figure 2: Gut microbiome dynamics. (A) Heatmap of cycling OTUs organized by peak from NA mice. Corresponding heatmap of the OTUs from other conditions are shown to illustrate that phases and cycling changed with diet and TRF. Bar graph at the bottom of column shows the time point at which the OTUs peaked. Black and white boxes show light on/light off timing. Yellow box shows when mice had access to food. (B) Log2 plot of percent reads of OTUs that are found in all three feeding conditions. The x-axis is log2 of the average (across all time points, n=18) of percent reads of an OTU in NA divided by its percent reads in FA. When positive, this would indicate that a particular OTU is more common in NA than in FA, and when negative, that it is less common in NA compared to FA. Likewise, the y-axis is the log2 of the average (across all time points, n=18) of percent reads of an OTU in FT divided by its percent reads in FA. When positive this would indicate that a particular OTU is more common in FT than in FA, and when negative, that a particular OTU is less common in FT compared to FA. This panel shows OTUs that are non-cycling and those that are cycling in NA. OTUs that cycled in NA were much more common in the NA gut microbiome than they were in FA gut microbiome (p <0.0005, Chi-square test). (C) Is a similar log2 plot as (B) but shows all OTUs that were common to all three feeding conditions. (D) Venn diagrams showing the number of total OTUs that were shared between the three feeding conditions and (E) the number of cycling OTUs that were shared between the three feeding conditions. Heatmap of cycling OTUs organized by peak from (F) FA mice and (G) FT mice. Corresponding heatmap of the OTUs from other conditions are shown to illustrate that phases and cycling changed with diet and TRF. Bar graph at the bottom of column shows the time point at which the OTUs peaked. Black and white boxes show light on/light off timing. Yellow box shows when mice had access to food. Diurnal activity of the phylum Actinobacteria (H) and Proteobacteria (I). First column: double plot of percent read for a particular genus from all three conditions (n=3/condition/time point). Second column: double plot of percent reads for the genus from the two HFD conditions (n=3/condition/time point). Third column: histogram of percent total reads that this genus comprises in each condition (n=18/condition). Fourth column: histogram showing percent reads that are expressed when light off/light on (n=9). Fifth column: histogram showing percent reads that are expressed when food from nighttime feeding has reached the cecum (ZT 17, 21, 1) and during relative fasting (ZT 5, 9, 13; n=9, see Figure 1 and ref 20). * p <0.05.
Figure S3, related to Figure 3: Subphylum analysis of the gut microbiome. Diurnal activity of genera identified in our study. First column: double plot of percent read (± SEM, n=3 per time point) for a particular genus from all three conditions. Second column: double plot of percent reads (± SEM, n=3 per time point) for the genus from the two HFD conditions. Third column: histogram of percent total reads (± SEM, n=18) that this genus comprises in each condition. Fourth column: histogram showing percent reads (± SEM, n=9) that are expressed when light off/light on. Fifth column: histogram showing percent reads (± SEM, n=9) that are expressed when food from nighttime feeding has reached the cecum (ZT 17, 21, 1) and during relative fasting (ZT 5, 9, 13; see Figure 1 and ref 20). * p < 0.05.
Figure S4, related to Figure 4: TRF restores cyclical fluctuation in genera thought to be involved in metabolism. (A) Percent reads (± SEM, n=3 per time point) of a double plot of genus Oscillibacter from all three conditions. (B) Genera based principle component analysis (PCA) of FA and FT mice. Green vectors show the axis where that particular genus accounted for most of the variability.
Figure S5, related to Figure 5: Diversity analysis. (A) Rarefaction curve of average reads from specific time points in FA and FT (number in box is ZT time point) (n=3/condition). The rarefaction plots of FT mice at ZT 9 and 21 were remarkably different, whereas those from FA mice at ZT 9 and 21 were quite similar. (B) Triangle plots illustrating β-diversity (see Koleff et al., 2003 and Fig 3F, 3G). Briefly, points that are close to apex (a′) share many OTUs. However points that are near the base (b′ or c′) have many dissimilarities. These plots show that NAvFA and NAvFT have the most dissimilarities and are nearly indistinguishable. NAvNA have much more dissimilarity than the FAvFA and FTvFT.
Figure S6, related to Figure 6: Metabolites that are processed by gut microbes are differentially excreted in the feces of mice in different conditions. (A) Average relative quantification of xylose and galactose (± SEM) in the feces (n = 4 samples from each condition, from separate cages) which includes results from all three conditions. Absolute quantification in pmol/mg of feces of the primary biles acids MCA, CA and DCA, the secondary bile acids DCA, UDCA and LCA and their respective tauro (T-) and glyco (G-) conjugates. CA: Cholate, CDCA: Chenodeoxycholate, MCA: Muricholate (a: alpha-, b: beta-, g: gamma-, w:omega-), DCA: Deoxycholate, UDCA: ursodeoxycholate, LCA: lithocholate, T-: Tauro-, G-:Glyco.