SUMMARY
Conformational fluctuations play a central role in enzymatic catalysis. However, it is not clear how the rates and the coordination of the motions affect the different catalytic steps. Here, we used NMR spectroscopy to analyze the conformational fluctuations of the catalytic subunit of the cAMP-dependent protein kinase (PKA-C), a ubiquitous enzyme involved in a myriad of cell signaling events. We found that the wild-type enzyme undergoes synchronous motions involving several structural elements located in the small lobe of the kinase, which is responsible for nucleotide binding and release. In contrast, a mutation (Y204A) located far from the active site desynchronizes the opening and closing of the active cleft without changing the enzyme’s structure, rendering it catalytically inefficient. Since the opening and closing motions govern the rate-determining product release, we conclude that optimal and coherent conformational fluctuations are necessary for efficient turnover of protein kinases.
Keywords: Protein kinase A, conformational dynamics, catalytic efficiency, synchronous motions, NMR relaxation dispersion
INTRODUCTION
Protein kinase A (PKA) is a ubiquitous phosphoryltransferase involved in a myriad of cellular events. PKA was one of the first kinases to be discovered and characterized (Krebs et al., 1959), and represents a benchmark for understanding the structure-function relationship and catalytic mechanism for the entire kinase superfamily (Taylor et al., 2012; Taylor and Kornev, 2011). Unlike metabolic enzymes optimized for fast turnover rates, PKA has evolved to orchestrate cell signaling events, mediating the transfer of external stimuli inside the cell (Taylor and Kornev, 2011). Although it targets several hundred substrates (Manning et al., 2002), this kinase is highly specific, due to a tight spatio-temporal control that ensures physiological levels of phosphorylation of its specific cellular targets (Scott and Pawson, 2009). β-adrenergic stimulation activates the PKA holoenzyme (two catalytic and two regulatory subunits, C- and R- subunits) (Zhang et al., 2012), which unleashes the active C-subunits (PKA-C), a catalytic core that PKA shares with other serine/threonine- and tyrosine-specific protein kinases. PKA-C has a bilobate structure, comprised of a small β-strand-rich lobe with a short α-helix (C-helix), which harbors the nucleotide binding pocket, and a large, mostly helical lobe containing the substrate binding site (Knighton et al., 1991). PKA-C’s catalytic cycle is a multistep process that involves nucleotide binding, substrate recognition and positioning, phosphoryl transfer and product release (ADP and phosphorylated substrate) (Lew et al., 1997). Crystal structures have captured this enzyme in the major conformational states along the reaction coordinates: apo, intermediate (nucleotide-bound), and closed (ternary complex with nucleotide and pseudo-substrate) (Johnson et al., 2001; Meharena et al., 2013). More recently the peptide substrate and product complexes have been trapped in crystals (Bastidas et al., 2013). However, these structures offers static images that do not fully describe the conformational changes and the role of motions occurring during allosteric regulation and catalysis (Huse and Kuriyan, 2002). The prominent role of conformational fluctuations for PKA-C’s catalytic cycle was inferred from the analysis of mutants aimed at disrupting the proposed allosteric network (Aimes et al., 2000; Iyer et al., 2005a; Iyer et al., 2005b; Moore et al., 2003; Yang et al., 2005; Yang et al., 2004). These studies, coupled with kinetic assays, suggest that the rate-determining step of the turnover is the ADP release following a fast phosphoryl transfer (~500 s−1), with the conformational transition from closed to open state being rate-limiting (Adams, 2001). Among these mutants, Y204A (PKA-CY204A), located in the large lobe of the enzyme and distant from the active site (Figure 1), has been shown to substantially affect both kcat/Km and kcat (Moore et al., 2003; Yang et al., 2005). Interestingly, the X-ray structure of PKA-CY204A is virtually superimposable to that of the wild-type in the presence of PKI and ATP-Mg2, with an overall rmsd of ~0.5 Å (Yang et al., 2004). From the comparison of the two X-ray structures, it emerges that Y204 couples the P+1 loop in the activation segment to both E230 and the hydrophobic core. Interestingly, Y204 does not have any direct contact with the peptide substrate, and yet this alanine substitution reduces dramatically the enzyme’s catalytic efficiency. Because the mutation induces no significant structural changes in the enzyme, let alone the active site, PKA-CY204A represents an ideal benchmark to test the relevance of the rates of motions in the PKA-C catalytic cycle.
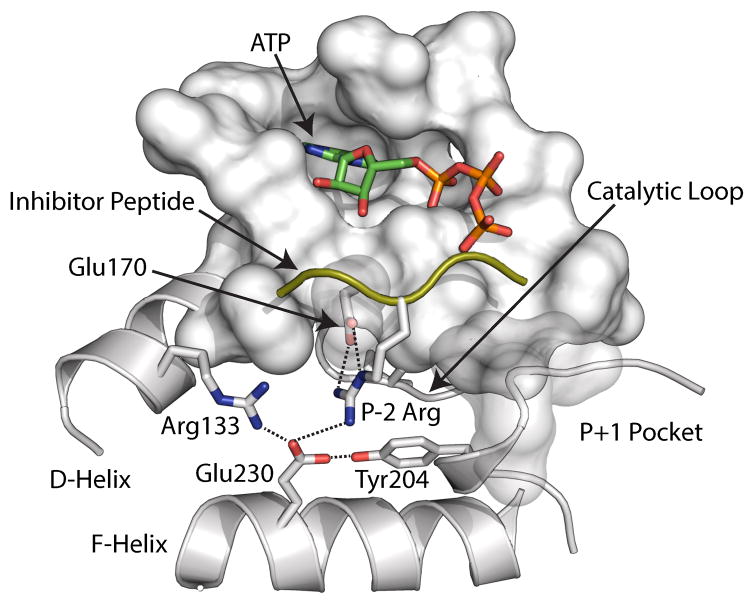
Figure 1.
Electrostatic node Y204 mapped on the crystal structure (PDB: 1ATP). The mutation is located approximately at 15 Ǻ from the active site and does not involve direct interactions with the substrate.
To analyze the effects of the Y204A mutation on the conformational dynamics of the enzyme, we carried out NMR spin relaxation measurements both in fast (ps-ns) and slow (μs-ms) timescales (Boehr et al., 2006a; Mittermaier and Kay, 2006; Palmer, 2004), focusing on the nucleotide-bound, dynamically-activated and substrate-primed forms of PKA-CWT and PKA-CY204A. We found that by disrupting the electrostatic network at the fringe of the small and large lobes, the enzyme responds with global dynamical changes; structural fluctuations in the ps-ns time-scale show regions with both enhanced and reduced mobility. Importantly, relaxation dispersion measurements for quantifying chemical exchange in the μs-ms timescale range show synchronous motions in the wild-type protein that are obliterated by the mutation. These effects are particularly evident for the Gly-rich loop and hinge region, those that mediate nucleotide binding, substrate recognition, and the rate-determining-step of the catalysis, i.e., ADP release. We conclude that synchronous (i.e., concerted) conformational fluctuations are essential for catalytic efficiency.
RESULTS
PKA-CY204A displays lower binding cooperativity
Kinetic data reported in the literature show the Y204A mutation affects both kcat/Km and kcat (Yang et al., 2004). To dissect the thermodynamics of nucleotide and substrate binding from the kinetics data, we monitored the binding of the nucleotide ATPγN and the pseudo-substrate PKI5-24 using Isothermal Titration Calorimetry (ITC) for both PKA-CWT and PKA-CY204A (Figure S1, Table S1). We found that Y204A exhibits a slightly higher affinity for ATPγN (Kd ~ 23 μM) compared to that of the wild-type (Kd ~ 58 μM), with a higher enthalpy of binding (ΔH ≈ −4.2 versus −2.7 kcal/mol). However, the affinity for PKI5-24 binding to the ATPγN-saturated enzymes is substantially different. While for PKA-CWT we observed Kd ~ 120 nM consistent with previous literature data (Herberg et al., 1999), for PKA-CY204A Kd is ~14 μM, corresponding to a loss of affinity of nearly 100 fold and a reduction of binding enthalpy of 10 kcal/mol. Since the kcat/KM and substrate affinities for the Y204A mutant is different from the wild-type enzyme, we chose to focus on the analysis of the dynamics for the nucleotide bound state.
Nucleotide-bound PKA-CWT is dynamically activated
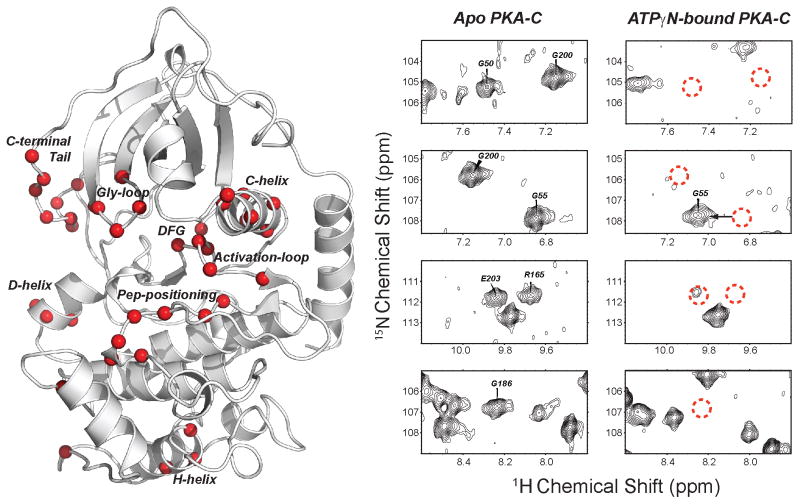
To monitor the structure and dynamics of PKA-CWT and PKA-CY204A, we collected [1H,15N]-TROSY-HSQC spectra of the apo, nucleotide-bound (ATPγN), and ternary (ATPγN and PKI) forms (Figure S2). Saturating the apo forms of both wild-type and mutant PKA-C with ATPγN resulted in significant broadening of numerous amide resonances, several of which broadened beyond detection, indicating the presence of conformational heterogeneity (Figure 2). Importantly, we detected substantial resonance broadening in the Gly-rich, DFG, and activation loops, which play major roles in the catalytic cycle (Johnson et al., 2001). Residues in the C-terminal loop flanking the Gly-rich loop and the C-helix also display notable amide resonance broadenings. Some regions with significant line broadening coincide with several loops displaying high B-factors in the crystal structure 1BKX (Figure S3).
Figure 2.
Residues broadened in PKA-CWT upon binding of ATPγN. Several broadened residues are located close to the active site (Gly-rich loop, DFG loop, C-helix, acidic residues in the C-terminal tail), others are distant (Peptide-positioning loop, activation loop, D and H helices).
Chemical shifts report only minimal structural effects for Y204A
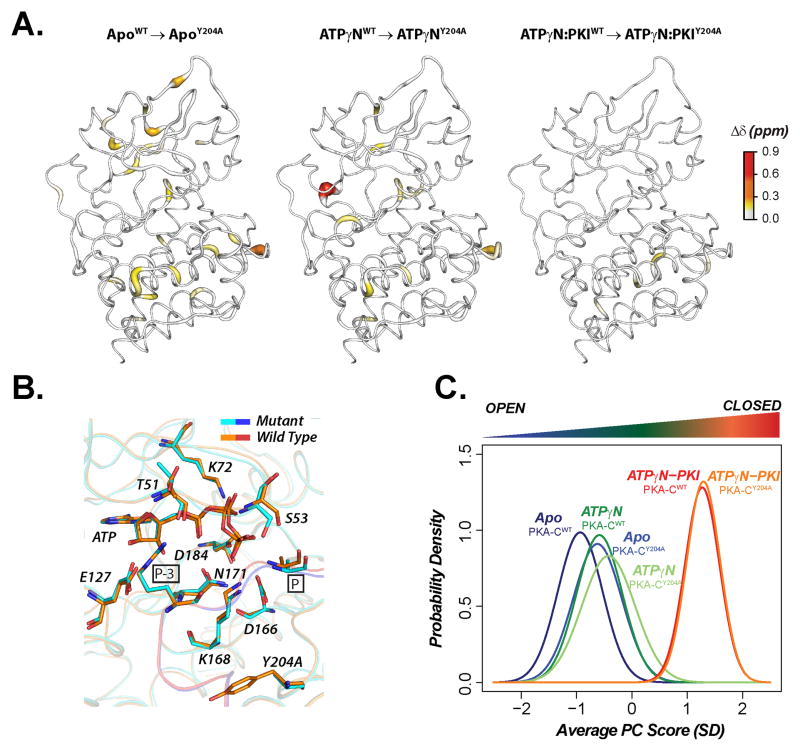
Overall, the chemical shift differences incurred by mutation at Y204A in all three states are minimal (Figure 3A), with the most prominent changes close to the mutation site, indicating that the average solution structures of PKA-CWT and PKA-CY204A are nearly identical (Figure 3B). However, tracing these subtle chemical shift changes offers useful information regarding local and global allosteric phenomena (Selvaratnam et al., 2011). Building on the chemical shift covariance analysis (Selvaratnam et al., 2011), we developed CONCISE (COordiNated ChemIcal Shifts bEhavior) that defines the probability density of each state and traces changes in the conformational equilibria upon ligand titration or mutation (Cembran et al., 2014). For both PKA-CWT and PKA-CY204A, CONCISE identified 94 residues interspersed throughout the whole structure that follow linear paths of the amide resonances upon titration with both the nucleotide and PKI5-24 (Figure S4). This indicates that the entire enzyme is involved in the open to closed transition. The probability density curves of the apo, intermediate, and closed states for the mutant are similar to those of the wild-type, with only slight differences for the apo and nucleotide-bound forms, which appear slightly more closed in the mutant. For the ternary complexes, however, the average conformations are essentially the same, with a complete overlay of the probability density curves (Figure 3C). This indicate that PKI5-24 (although with reduced binding affinity) traps for Y204A in a inhibited, dynamically quenched state (Masterson et al., 2011).
Figure 3.
Chemical shift differences between PKA-CWT and PKA-CY204A for Apo, ATPγN-bound and ternary (ATPγN and PKI5-24-bound). (A) 1H,15N combined chemical shift differences mapped on the corresponding X-ray structures (B) Superposition of the active sites in the crystal structures of PKA-CWT (orange/red) and PKA-CY204A (cyan/blue). C. CONCISE analysis of amide resonances.
Fast timescale dynamics are redistributed throughout Y204A
In PKA-CWT, nucleotide binding drives the enzyme conformation toward a state committed to catalysis (i.e., dynamically committed state) (Masterson et al., 2010; Masterson et al., 2011). The adenine ring binds in a deep cleft located underneath the Gly-rich loop, completing the C-spine, engaging both lobes, and promoting backbone fluctuations in the ps-ns time scale (Masterson et al., 2010; Masterson et al., 2011). The total entropy changes dramatically upon binding the substrate peptide or PKI5-24. At the same time, the conformational fluctuations of the ternary complexes with the substrate and inhibitor are radically different, with the substrate binding re-distributing the motions throughout the enzyme and the pseudo-substrate quenching the conformational dynamics both in the slow and fast time scale. Based on these data, we hypothesized that the motions in the intermediate (nucleotide-bound) state of the kinase may be responsible for substrate binding cooperativity (Masterson et al., 2011). To test this hypothesis, we probed the conformational dynamics for the PKA-CY204A mutant in complex with ATPγN over a range of timescales (Summarized in Table 1). Specifically, we measured the 15N relaxation parameters R1 (longitudinal relaxation rate constant), R2 (longitudinal relaxation rate constant), and 1H-15N heteronuclear steady-state NOE (HX-NOE) (Palmer, 2004). From the R1 and R2 relaxation rates, we estimated an overall rotational correlation time of approximately 24 and 23 ns for the wild-type and Y204A mutant, respectively, values that are indicative of a monomeric enzyme. The average NOE values for wild-type PKA-C and the mutant Y204A were found to be 0.79 ± 0.13 and 0.81 ± 0.12, respectively, indicating that on average the overall fluctuations in the fast time scale are very similar in both enzymes. However, we identified local differences in the HX-NOEs between the two proteins (Figure S5). Overall, no clear patterns emerged from the data, suggesting that the conformational freedom gained by disrupting the electrostatic node impinged on Y204 is redistributed across the entire enzyme (Figure S5). This reorganization of fast timescale dynamics may contribute to the drastically reduced binding affinity for PKI5-24 as measured by ITC.
TABLE 1.
Summary of 15N relaxation and ITC data for PKA-CWT and PKA-CY204A.
Slow conformational dynamics are no longer synchronized in Y204A
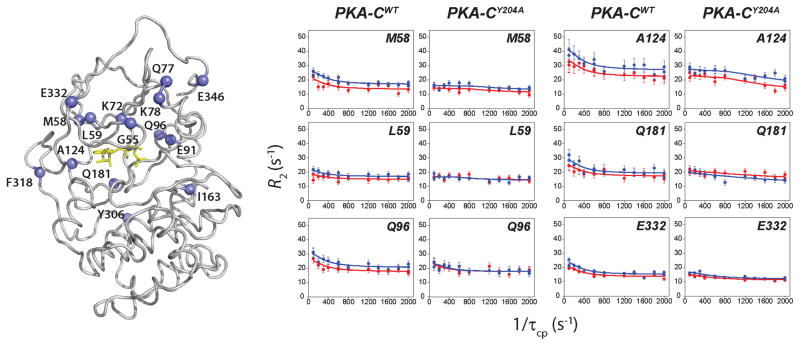
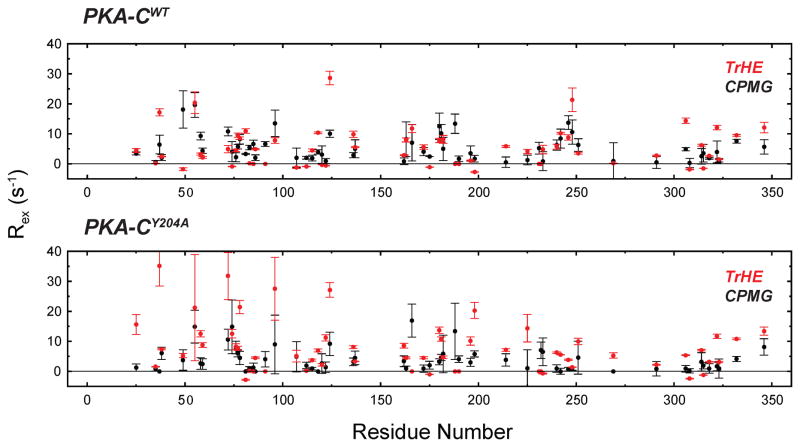
According to our proposed model (Masterson et al., 2010), the nucleotide-bound enzyme still undergoes opening and closing transitions, although with a smaller amplitude. We hypothesize that these motions are necessary for organizing the active site for substrate binding and phosphoryl transfer to occur, but also for product release. To test this hypothesis, we measured the rates of conformational exchange for those residues manifesting fast/intermediate exchange in both the wild-type and the mutant. To quantify the rates of these motions, we measured the exchange contribution to the total transverse relaxation rate constant (Rex) with two different NMR techniques: 15N Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion measurements (Wang et al., 2001) and TROSY-Hahn echo (Wang et al., 2003). The rates obtained from the CPMG relaxation dispersion experiments depend on the equilibrium populations (pclosed and popen), the chemical shift difference between the two interconverting species (Δω), and the kinetics of exchange (kex=kf+kr, where the f and r subscripts indicate the forward and reverse rates) (Palmer et al., 2001). From the individual fitting of the dispersion curves, we clustered fifteen residues manifesting similar exchange rates (Figure 4, Tables S2 and S3). These residues are distributed mainly in the small lobe, comprising G55 in the Gly-rich loop, M58 and L59 in β-sheet 2, L77 and K78 in the B-helix, E91 and Q96 in the C-helix, A124 and F318 in the hinge region between the two lobes, and E332 and E446 located in the C-terminal that wraps around the enzyme. Also included from the large lobe are I163, in the catalytic loop, and Y306. The clustering of the relaxation rates for these residues distributed among several regions indicates that in the wild-type in the nucleotide-bound state the small lobe undergoes concerted motions between the open and closed conformations present. Fitting the dispersion profiles using the full Richard-Carver equation (see Supplementary Information) resulted in populations of 94 and 6% for the closed and open states, respectively, with an exchange constant of ~1020±150 s−1 (kf ~ 66 and kr ~ 953 s−1). While the broadened resonances for both the wild-type and mutated enzymes are identical, the dispersion curves for the fast/intermediate exchange residues are quite different (Figure 5). Remarkably, the individual fitting carried out on the resonances did not result in similar kex values, and the cluster of residues identified for the wild-type as responsible for the synchronous motions is no longer present (Table S4). As a result, a global fit is not possible for multiple residues, and only sparse pairs of residues in the small lobe can be coupled. Therefore, mutation of Y204A induces a dramatic change in the dynamics of PKA-C on the μs-ms timescale in which a large region of the protein involved in active site motions is no longer synchronized. Moreover, several residues of the mutant enzyme show faster exchange dynamics as detected from the individual fits and uncorrelated motions in the small lobe (Tables S2–S4), suggesting that there is a shift in the motional regime. To support the latter, we used the 15N TROSY Hahn-echo (TrHE) experiment (Wang et al., 2003) for both PKA-CWT and PKA-CY204A (Figure S6). The window of observation of the chemical exchange for CPMG and TrHE is different, with the TrHE more sensitive to faster conformational dynamics (Wang and Palmer, 2003; Wang et al., 2003). For the wild-type, we found a good agreement between the Rex values measured by CPMG experiments and the corresponding values measured using the TrHE experiments (Figure 6). In sharp contrast, Rex values measured with these two techniques are markedly different for the mutant, as the Rex values measured with the TrHE are systematically higher than the corresponding values measured by CPMG experiments. Overall, these data suggest that the mutation increases the rates of motions throughout the enzyme with the most prominent effects localized in the small lobe (β2- and β3-sheet as well as B- and C-helix) and between the activation and positioning loop. Therefore, the disruption of the electrostatic node shifts the range of the conformational dynamics to a faster timescale that affects both substrate binding and product release.
Figure 4.
Conformational motions in the slow timescale probed by 15N CPMG relaxation dispersion. Left: residues showing concerted motions in the wild-type enzyme are mapped on the structure. Right: 15N CPMG dispersion curves for residues of PKA-CWT and PKA-CY204A reveal a change in the nature of the dynamics upon Y204A mutation.
Figure 5.
Comparison of the Rex measured by CPMG relaxation dispersion and TrHE. Only the residues with both CPMG and TrHE values are compared. All of the TrHE data are reported in Figure S6.
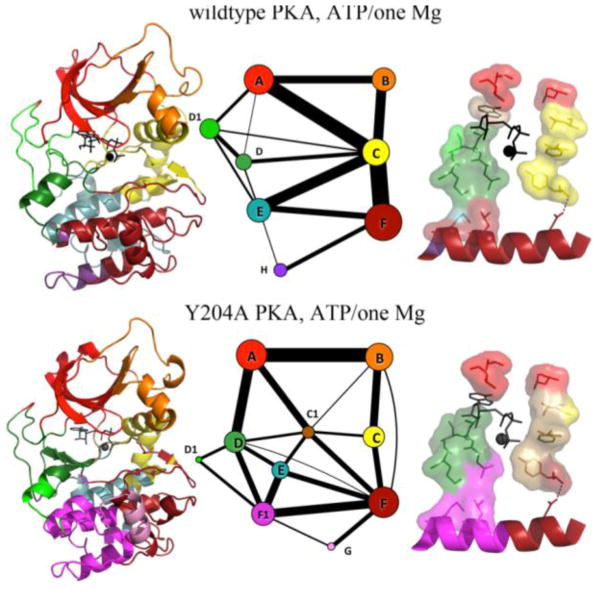
Figure 6.
Molecular dynamics simulations (5–6 μs) community analysis of PKA-CWT and PKA-CY204A. Left: The community map is colored on the structure. Middle: A graph representation of the community map where the thickness of the lines connecting the communities indicates the presence of correlation of the motions. Right: The C-spine and R-spine anchored to the F-helix. Y204A mutation disrupts semi-rigid communities and causes the dihedral populations to shift in a number of areas related to activation, ATP-binding, and substrate binding. Definition of the communities: ComA: ATP binding and substrate coordination; ComB: αC-helix adjusting; ComC: regulatory and assembly the R spine; ComD: ATP binding and catalytic site; ComE: C-spine supporting; ComF: Activation and substrate binding. ComF1 and ComG: substrate binding.
Molecular dynamics simulations highlight loss of correlated motions in Y204A
To interpret the motions highlighted by the nuclear spin relaxation measurements and visualize how the Y204A mutation alters inter-residue contacts and correlated motions at a global level, we performed multiple-microsecond timescale molecular dynamics simulations for both the wild-type (5.7 μs) and Y204A (5.1 μs) kinases. The MD trajectories were analyzed using the community analysis (Kappel et al., 2012; Newman, 2004; Sethi et al., 2009), which identifies structurally-contiguous regions exhibiting correlated motions. The community analysis views motions in the protein as a dynamical network and highlights correlated motions in proteins as semi-rigid, semi-flexible units of three-dimensional sub-structures that are grouped together and connected to other units by physical contacts and correlated motions. Community analysis of PKAWT clusters the kinase residues in eight major communities (Figure 6). A prominent feature of the wild-type enzyme is the regulatory ComC, which acts as a hub and is strongly connected to the rest of the community (thick lines in Figure 6). In the mutated enzyme, these correlations are completely rearranged, with ComC splitting into two different sub-communities (ComC and ComC1), losing the role of central communication hub. As a result, the αC-β4 linker, DFG-motif, and several R-spine residues pull away from ComC that contains the αC-helix, a critical domain for catalysis. The loss of correlations within and between the communities parallels the effects of the mutation on the slow time scale motion as probed by NMR and is consistent with the loss of concerted motion in Y204A. Overall the changes in the distribution of motions and the reduced couplings within and between communities throughout the entire enzyme upon mutation support the conclusions reached by the NMR analysis and identify subtle but important changes that reduce the binding affinity and catalytic efficiency for the enzyme
DISCUSSION
There is a passionate debate about the role of conformational fluctuations in enzymatic catalysis (Garcia-Viloca et al., 2004; Hammes-Schiffer, 2002; Hammes-Schiffer and Benkovic, 2006; Pisliakov et al., 2009; Schwartz and Schramm, 2009). Recent studies have inferred that motions in the NMR-detectable timescale might affect the chemical step of catalysis (Bhabha et al., 2011; Boehr et al., 2006b; Whittier et al., 2013). On the other hand, theoretical calculations seem to dismiss this argument (Pisliakov et al., 2009). Nonetheless, enzyme motions are implicated in several steps of catalysis, including allosteric activation (Cui and Karplus, 2008; Lipchock and Loria, 2010; Popovych et al., 2006; Tzeng and Kalodimos, 2009; Tzeng and Kalodimos, 2013), active site optimization (Boehr et al., 2006b; Henzler-Wildman et al., 2007; Mauldin et al., 2009), co-factors or substrate binding, as well as product release (Masterson et al., 2010). An intriguing hypothesis is that enzyme structure and conformational dynamics may have co-evolved (Bhabha et al., 2013; Chao et al., 2012; Jackson et al., 2009; Wolf-Watz et al., 2004). Indeed, the importance of conformational flexibility in protein kinases is emerging in several different instances. In particular, motions in the intermediate-exchange timescale (μs-to-ms) have been detected by NMR in several different kinases, e.g., p38a mitogen-activated protein kinase (Honndorf et al., 2008), Abelson kinase (Skora et al., 2013), the CheA histidine kinase (Wang et al., 2014), and extracellular signal-regulated kinase 2 (ERK2) (Xiao et al., 2014). In past years, we have used nuclear spin relaxation measurements to trace the conformational changes as well as structural fluctuations of kinase A, mimicking major states along the reaction coordinates (Masterson et al., 2010; Masterson et al., 2011). We found that the apo state is malleable and water accessible, but does not display conformational fluctuations that are synchronous to the enzymatic turnover. Nucleotide binding shifts the population of the enzyme toward an intermediate state, providing the phosphoryl group for the reaction and connecting the small lobe to the large lobe via the hydrophobic C-spine. Upon substrate binding, the conformational fluctuations of the enzyme are redistributed, allowing the enzyme to adopt a dynamically competent state (Masterson et al., 2010). In sharp contrast, binding of a pseudo substrate peptide inhibitor such as PKI obliterates the conformational fluctuations, trapping the enzyme in a dynamically quenched state (Masterson et al., 2011).
Although these studies have advanced our understanding of the role of motions on enzymatic turnover, they did not clearly demonstrate the importance of the rates and the correlations of the conformational fluctuations with respect to the structural changes. The X-ray structures of the different states (Taylor et al., 2005) as well as molecular dynamics simulations (Masterson et al., 2011) suggest that motion is a driving force in the catalytic cycle (Lu et al., 2005). Fluctuations in kinase A involve a mechanical hinge-bending motion of the two lobes, featuring an asymmetric twist that brings the glycine-rich loop close to the triphosphate moiety, thus causing the complete closure of the active cleft over the substrate (Li et al., 2002). The kinetics of exchange for these large amplitude motions occurring during the turnover were estimated at a kex of ~200 s−1 (Masterson et al., 2010), a value remarkably similar to that of ERK2 (Xiao et al., 2014). However, the importance of the faster rates of motions that we measured in the nucleotide bound form (i.e., kex ~ 1020 s−1) has not been fully appreciated. The Y204A mutant offers a unique opportunity to test the role of conformational dynamics as the structures of the PKA-CWT and PKA-CY204A are virtually superimposable (Yang et al., 2004). Yet, the mutation causes a 400-fold decrease of the catalytic efficiency (Moore et al., 2003) and approximately a 100-fold reduction in the affinity for the pseudo-substrate, two phenomena essentially inexplicable from the structural data alone. Thermocalorimetric studies and deuterium exchange mass spectrometry data showed an overall reduction of the PKA-CY204A stability, inferring an increase of conformational dynamics (Yang et al., 2005). Indeed, our data show that the key to understanding the anomalous behavior of the mutant is the nucleotide binding event. In contrast to ERK2 (Xiao et al., 2014), where the nucleotide binding seems to have a marginal role in modulating the conformational fluctuations, our data emphasize the central role of the nucleotide in orchestrating phosphoryl transfer in PKA-C. Binding of the nucleotide first links the two hydrophobic spines so that all of the catalytic machinery is integrated. In addition, the nucleotide provides the phosphoryl group necessary for chemistry to take place; however it also pre-organizes the substrate binding site, enhancing its binding affinity, shifting the range of the dynamics in a regime that is competent for catalysis. Importantly, the changes in the rates of the opening and closing motions detected for the nucleotide-bound form of PKA-CY204A by the relaxation dispersion measurements are probably responsible for both the decreased affinity for the substrate and the decrease in kcat, affecting product release. In the wild-type enzyme, the opening and closing motions of the nucleotide-bound form are synchronous for several residues in the small lobe and in the hinge regions. For the Y204A mutant, these conformational fluctuations are no longer synchronous, reducing the enzyme’s catalytic efficiency. These dysfunctional conformational fluctuations of PKA-CY204A are too fast to be captured by the CPMG experiments, but are detectable by TrHE experiments. Since the chemical step of the catalysis is too fast to be determined by NMR experiments, a logical explanation for the reduced catalytic turnover in PKA-CY204A is that these dysfunctional dynamics reduce the probability of the enzyme reaching a conformation competent for phosphoryl transfer (Daily et al., 2010; Mauldin et al., 2009). While our previous studies have shown that absence of motion renders the kinase inactive, relaxation dispersion data show that the presence of asynchronous motions affects the opening and closing of the enzyme, reducing the enzyme catalytic turnover.
Using a mutation of PKA-C that removes a crucial electrostatic node far from the active site, we demonstrated the relevance of both exchange rates and coordination of the conformational fluctuations for the kinase A catalytic turnover. When the fluctuations of the small lobes are synchronous, the enzyme functions with proper turnover rates. Upon removal of the electrostatic node, the rates of opening and closing are no longer synchronous and the enzyme becomes sluggish. Since different kinases display a wide range of catalytic efficiencies while maintaining the same fold, modulation of the conformational fluctuations across the entire kinome are probably responsible for tuning the signaling efficiency of these enzymes.
EXPERIMENTAL PROCEDURES
ITC
Both PKA-CWT and PKA-CY204A were dialyzed into 20 mM MOPS, 90 mM KCl, 10 mM DTT, 10 mM MgCl2, 1 mM NaN3 at pH 6.5. PKA-C concentrations for ITC measurements were between 11.8 – 32 μM. ATPγN was added to reach a final concentration of 2 mM. All ITC measurements were performed with a Microcal VP-ITC instrument at 27°C. Approximately 1.7 mL of PKA-C was used for each experiment and 280 μL of 2–4 mM of ATPγN or 140–210μM of PKI5-24 in the titrant syringe. All experiments were performed in triplicate. The heat of dilution of the ligand to the buffer was taken into account by measuring the heat of dilution of the ligand to the buffer and was subtracted from the experiment accordingly. The binding curves were analyzed with the NanoAnalyze software (TA Instruments) assuming 1:1 stoichiometry and using the Wiseman Isotherm:
where the change of the total complex, d[MX] with respect to the change of the ligand concentration, d[Xtot] is dependent on r, the ratio of the Kd with respect to the total protein concentration, and Rm, the ratio between the total ligand and total protein concentration. The free energy of binding was determined using the following:
where R is the universal gas constant and T is the temperature at measurement (300K). The entropic contribution to binding was calculated using the following:
NMR Spectroscopy
Uniformly 15N-labeded perdeuterated wild-type PKA-C and its Y204A mutant were overexpressed and purified as described elsewhere (Masterson et al., 2008). The NMR samples contained 90 mM KCl, 20 mM KH2PO4, 10 mM MEGA-8 (Octanoyl-N-methylglucamide), 5% glycerol, 12 mM ATPγN, 10 mM MgCl2, 20 mM DTT, 1 mM NaN3 in a mixed solvent of 5% D2O in water, pH 6.5. The protein concentrations for PKA-CWT and PKA-CY204A were ~400 μM, as determined from A280 measurements. Experiments were performed at 27 °C on Varian Inova spectrometers operating at 1H frequency of 800 MHz and 900 MHz. The [1H,15N] TROSY-HSQC spectra of PKA-CWT and PKA-CY204A in the apo, nucleotide bound (ATPγN), and inhibitor bound (ATPγN and PKI) forms were re-referenced to remove any shifting artifacts. To improve our statistics we carried out the CONCISE analysis using a total of six states (Cembran et al., 2014). The 15N shifts were then scaled by 0.154 to balance the weight of perturbations along the nitrogen and proton dimensions, and principal component analysis (PCA) was applied independently to each residue to identify the direction of largest variance across the six states defined above. The ratio between the standard deviations along PC1 and PC2 constitutes a measure of how well the different states follow a linear pattern for a given residue. Residues characterized by poor linearity (σ-PC1/σ-PC2) < 3.0) were discarded from our analysis, together with residues with small perturbations (range of chemical shift along PC1 < 0.05 ppm) and thus more heavily affected by experimental error. These thresholds reduced the number of residues available for all states from 155 to 94 (Figure S4). Nuclear spin (15N) relaxation experiments performed with TROSY detected T1, T1ρ and 15N[1H]-NOE, TROSY-Hahn-echo, and TROSY-CPMG experiments for the PKA-CWT and PKA-CY204A samples were carried out using 50×1600 points and spectral widths of 2403×10504 Hz in the indirect and direct dimensions. The 1H and 15N carrier frequencies were set on water resonance and 120.5 ppm, respectively.
Molecular Dynamics Simulations
The details of the systems simulated are described in the Supporting Information. The systems were prepared using AMBER12 (Gotz et al., 2012; Le Grand et al., 2013). Production simulations (wild-type, 5.7 μs, and Y204A, 5.1 μs) were run on a 512-node Anton supercomputer, and the trajectories were analyzed using mutual information values to capture correlated motions involving semi-rigid regions (Morcos et al., 2010).
Community Analysis
For our community analysis, we computed Cartesian mutual information values instead of dihedral mutual information values, as the former better capture correlated motions involving semi-rigid regions (Morcos et al., 2010), while the latter are more suitable for analyzing couplings between surface sites (McClendon et al., 2009; Wan et al., 2013). We calculated the Cartesian mutual information for backbone Cα atoms, as well as for an additional representative terminal atom for each residue. The matrix of mutual information among these atoms was calculated with the MutInf software package (McClendon et al., 2009; McClendon et al., 2012), splitting each trajectory into six blocks of equal size and using uniformly binned histograms with 24 bins per degree of freedom. We used 20 sets of scrambled data to estimate the distribution of mutual information under the null hypothesis of independence, and calculated the excess mutual information with the previously-described under sampling correction (McClendon et al., 2009).
Supplementary Material
Figure S1: Isothermal Titration Calorimetry (ITC) with ATPγN to the apo form and the pseudo-substrate PKI5-24 to the ATPγN-bound form of PKA-CWT and PKA-CY204A. Top: ATPγN binding to Apo PKA-CWT and the PKA-CY204A mutant. Bottom: Pseudo-substrate, PKI5-24, binding to PKA-CWT and PKA-CY204A in the presence of 2 mM ATPγN. All measurements were performed in triplicate and the error reflects the standard deviation.
Figure S2: Overlays of the [1H, 15N]-TROSY-HSQC spectra for PKA-CWT and PKA-CY204A for the three forms of the kinase: apo (top), ATPγN-bound (middle), and ATPγN and PKI5-24-bound (bottom).
Figure S3: Qualitative comparison of the B-factor from the crystal structure 1BKX (left) with the resonances undergoing broadening beyond detection upon binding the nucleotide (also shown in Figure 2). B-factors are shown in a rainbow scale from blue (1) to red (100).
Figure S4. Left: List of residues that defined linear trajectories for the 6 states analyzed with CONCISE using the thresholds reported in the Methods section. Right: The linear residues are mapped onto the kinase structure (1ATP).
Figure S5: 1H-15N HX-NOE as probe of fast conformational dynamics. Top: The HX-NOE is plotted as a function of the PKA sequence where the red colored values represent NOEs above or below one standard deviation from the average. Bottom: HX-NOE values reported on the crystal structure 1ATP of the kinase. Bulges and colors show regions with higher mobility. S10, Q12, E13, E17, T37, I46, K61, K105 are more rigid in the mutant (higher values of HX-NOEs), while K7, N67, K78 are more dynamic (lower HX-NOE values). Residues near or belonging to the linker regions and preceding the D-helix (Y117, M120 and Y122) are relatively rigid; whereas G125, located near a C-spine residue, and A188, right after the DFG-loop, are more mobile. In the peptide positioning loop, A206 becomes rigid, while E208 becomes mobile in the Y204A enzyme.
Figure S6: 15N TROSY Hahn-echo as a probe for slow conformational dynamics. Top: Rex values obtained from the TrHE experiments are plotted as a function of the PKA sequence where the red colored values represent Rex > 7 s−1 and |ΔRex| > 7 s−1. Due to the magnitude and orientation of the 15N chemical shift tensor, the Rex values for the non-exchanging residues are scattered around zero with a standard deviation of approximately 2 s−1. Bottom: Rex values are shown on the crystal structure 1ATP of the kinase with bulges and colors highlighting the regions with higher mobility.
TABLE S1, related to Table 1: Thermodynamics of ligand binding to PKA-CWT and PKA-CY204A determined by ITC. Binding of the ATP mimic, ATPγN, to Apo form of the enzyme (top) was measured as well as PKI5-24 binding of the nucleotide bound form of the enzyme (bottom).
TABLE S2, related to Table 1. 15N CPMG relaxation dispersion group fit of PKA-CWT.
TABLE S3, related to Figure 4. 15N CPMG relaxation dispersion individual fit of PKA-CWT.
TABLE S4, related to Figure 5. 15N CPMG relaxation dispersion individual fit of PKA-CY204A.
HIGHLIGHTS.
Synchronous motions are essential for kinase A catalytic efficiency
Allosteric mutation globally de-synchronizes motions
Optimal conformational fluctuations are central to kinase turnover.
Acknowledgments
This work is supported by the NIH (GM 100301).
The NMR experiments were acquired at the Minnesota NMR Center and at the University of Colorado. Many thanks to Dr. G. Armstrong for the NMR experiments carried out in Colorado, Dr. A. Kornev for assistance with visualization of the community maps, and Prof. L.E. Kay and Prof. D. Korznev for providing the CPMG-fit software. This work is supported by the NIH (GM 100301).
Footnotes
AUTHORS CONTRIBUTIONS
G.V., S.S.T. conceived, directed and analyzed all experimental research; G.V. and A.S. prepared the manuscript. L.R.M., J.K., L.R.M., and A.S. prepared the samples and performed NMR spectroscopy experiments. C.M. performed molecular dynamics simulations and analysis and determined the community maps, and A.C. performed molecular dynamics simulations. All authors discussed the results and implications and commented on the manuscript at all stages.
Supplemental information includes 6 figures and 4 tables and can be found with this article online at ***
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chemical reviews. 2001:101. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- Aimes RT, Hemmer W, Taylor SS. Serine-53 at the tip of the glycine-rich loop of cAMP-dependent protein kinase: Role in catalysis, P-site specificity, and interaction with inhibitors. Biochemistry. 2000;39:8325–8332. doi: 10.1021/bi992800w. [DOI] [PubMed] [Google Scholar]
- Bastidas AC, Deal MS, Steichen JM, Guo Y, Wu J, Taylor SS. Phosphoryl transfer by protein kinase A is captured in a crystal lattice. J Am Chem Soc. 2013;135:4788–4798. doi: 10.1021/ja312237q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabha G, Ekiert DC, Jennewein M, Zmasek CM, Tuttle LM, Kroon G, Dyson HJ, Godzik A, Wilson IA, Wright PE. Divergent evolution of protein conformational dynamics in dihydrofolate reductase. Nat Struct Mol Biol. 2013;20:1243–1249. doi: 10.1038/nsmb.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabha G, Lee J, Ekiert DC, Gam J, Wilson IA, Dyson HJ, Benkovic SJ, Wright PE. A Dynamic Knockout Reveals That Conformational Fluctuations Influence the Chemical Step of Enzyme Catalysis. Science. 2011;332:234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chemical Reviews. 2006a;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006b;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- Cembran A, Kim J, Gao J, Veglia G. NMR mapping of protein conformational landscapes using coordinated behavior of chemical shifts upon ligand binding. Physical chemistry chemical physics: PCCP. 2014;16:6508–6518. doi: 10.1039/c4cp00110a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao F-A, Morelli A, Haughner JCI, Churchfield L, Hagmann LN, Shi L, Masterson LR, Sarangi R, Veglia G, Seelig B. Structure and dynamics of a primordial catalytic fold generated by in vitro evolution. Nat Chem Biol. 2012 doi: 10.1038/nchembio.1138. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Science. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily MD, Phillips GN, Jr, Cui Q. Many local motions cooperate to produce the adenylate kinase conformational transition. J Mol Biol. 2010;400:618–631. doi: 10.1016/j.jmb.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Viloca M, Gao J, Karplus M, Truhlar DG. How enzymes work: Analysis by modern rate theory and computer simulations. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- Gotz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. Journal of Chemical Theory and Computation. 2012;8:1542–1555. doi: 10.1021/ct200909j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes-Schiffer S. Impact of enzyme motion on activity. Biochemistry. 2002;41:13335–13343. doi: 10.1021/bi0267137. [DOI] [PubMed] [Google Scholar]
- Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. In Annual Review of Biochemistry. 2006:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–U813. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- Herberg FW, Doyle ML, Cox S, Taylor SS. Dissection of the nucleotide and metal-phosphate binding sites in cAMP-dependent protein kinase. Biochemistry. 1999:38. doi: 10.1021/bi982672w. [DOI] [PubMed] [Google Scholar]
- Honndorf VS, Coudevylle N, Laufer S, Becker S, Griesinger C. Dynamics in the p38alpha MAP kinase-SB203580 complex observed by liquid-state NMR spectroscopy. Angewandte Chemie. 2008;47:3548–3551. doi: 10.1002/anie.200705614. [DOI] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Iyer GH, Garrod S, Woods VL, Taylor SS. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: Study of the Lys72His mutant of cAMP-dependent kinase. Journal of Molecular Biology. 2005a;351:1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Iyer GH, Moore MJ, Taylor SS. Consequences of lysine 72 mutation on the phosphorylation and activation state of cAMP-dependent kinase. Journal of Biological Chemistry. 2005b;280:8800–8807. doi: 10.1074/jbc.M407586200. [DOI] [PubMed] [Google Scholar]
- Jackson CJ, Foo JL, Tokuriki N, Afriat L, Carr PD, Kim HK, Schenk G, Tawfik DS, Ollis DL. Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21631–21636. doi: 10.1073/pnas.0907548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chemical Reviews. 2001;101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- Kappel K, Wereszczynski J, Clubb RT, McCammon JA. The binding mechanism, multiple binding modes, and allosteric regulation of Staphylococcus aureus Sortase A probed by molecular dynamics simulations. Protein science: a publication of the Protein Society. 2012;21:1858–1871. doi: 10.1002/pro.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, Sowadski JM. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- Krebs EG, Graves DJ, Fischer EH. Factors affecting the activity of muscle phosphorylase b kinase. J Biol Chem. 1959;234:2867–2873. [PubMed] [Google Scholar]
- Le Grand S, Gotz AW, Walker RC. SPFP: Speed without compromise-A mixed precision model for GPU accelerated molecular dynamics simulations. Comput Phys Commun. 2013;184:374–380. [Google Scholar]
- Lew J, Taylor SS, Adams JA. Identification of a partially rate-determining step in the catalytic mechanism of cAMP-dependent protein kinase: a transient kinetic study using stopped-flow fluorescence spectroscopy. Biochemistry. 1997:36. doi: 10.1021/bi963164u. [DOI] [PubMed] [Google Scholar]
- Li F, Gangal M, Juliano C, Gorfain E, Taylor SS, Johnson DA. Evidence for an internal entropy contribution to phosphoryl transfer: A study of domain closure, backbone flexibility, and the catalytic cycle of cAMP-dependent protein kinase. Journal of Molecular Biology. 2002;315:459–469. doi: 10.1006/jmbi.2001.5256. [DOI] [PubMed] [Google Scholar]
- Lipchock JM, Loria JP. Nanometer propagation of millisecond motions in V-type allostery. Structure. 2010;18:1596–1607. doi: 10.1016/j.str.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BZ, Wong CF, McCammon JA. Release of ADP from the catalytic subunit of protein kinase A: A molecular dynamics simulation study. Protein Science. 2005;14:159–168. doi: 10.1110/ps.04894605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci U S A. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson LR, Cheng C, Yu T, Tonelli M, Kornev A, Taylor SS, Veglia G. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6:821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson LR, Shi L, Metcalfe E, Gao J, Taylor SS, Veglia G. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc Natl Acad Sci U S A. 2011;108:6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauldin RV, Carroll MJ, Lee AL. Dynamic dysfunction in dihydrofolate reductase results from antifolate drug binding: modulation of dynamics within a structural state. Structure. 2009;17:386–394. doi: 10.1016/j.str.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meharena HS, Chang P, Keshwani MM, Oruganty K, Nene AK, Kannan N, Taylor SS, Kornev AP. Deciphering the structural basis of eukaryotic protein kinase regulation. PLoS biology. 2013;11:e1001680. doi: 10.1371/journal.pbio.1001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittermaier A, Kay LE. New tools provide new insights in NMR studies of protein dynamics. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Adams JA, Taylor SS. Structural basis for peptide binding in protein kinase A. Role of glutamic acid 203 and tyrosine 204 in the peptide-positioning loop. The Journal of biological chemistry. 2003:278. doi: 10.1074/jbc.M210807200. [DOI] [PubMed] [Google Scholar]
- Morcos F, Chatterjee S, McClendon CL, Brenner PR, Lopez-Rendon R, Zintsmaster J, Ercsey-Ravasz M, Sweet CR, Jacobson MP, Peng JW, et al. Modeling conformational ensembles of slow functional motions in Pin1-WW. PLoS Comput Biol. 2010;6:e1001015. doi: 10.1371/journal.pcbi.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Analysis of weighted networks. Phys Rev E. 2004:70. doi: 10.1103/PhysRevE.70.056131. [DOI] [PubMed] [Google Scholar]
- Palmer AG. NMR characterization of the dynamics of biomacromolecules. Chemical Reviews. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- Pisliakov AV, Cao J, Kamerlin SCL, Warshel A. Enzyme millisecond conformational dynamics do not catalyze the chemical step. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17359–17364. doi: 10.1073/pnas.0909150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nature Structural & Molecular Biology. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SD, Schramm VL. Enzymatic transition states and dynamic motion in barrier crossing. Nat Chem Biol. 2009;5:551–558. doi: 10.1038/nchembio.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JD, Pawson T. Cell Signaling in Space and Time: Where Proteins Come Together and When They’re Apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc Natl Acad Sci U S A. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. P Natl Acad Sci USA. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skora L, Mestan J, Fabbro D, Jahnke W, Grzesiek S. NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc Natl Acad Sci U S A. 2013;110:E4437–4445. doi: 10.1073/pnas.1314712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Ilouz R, Zhang P, Kornev AP. Assembly of allosteric macromolecular switches: lessons from PKA. Nature reviews Molecular cell biology. 2012;13:646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Kim C, Vigil D, Haste NM, Yang J, Wu J, Anand GS. Dynamics of signaling by PKA. Biochimica et biophysica acta. 2005:1754. doi: 10.1016/j.bbapap.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends in Biochemical Sciences. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–U139. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Allosteric inhibition through suppression of transient conformational states. Nat Chem Biol. 2013;9:462–465. doi: 10.1038/nchembio.1250. [DOI] [PubMed] [Google Scholar]
- Wang CY, Grey MJ, Palmer AG. CPMG sequences with enhanced sensitivity to chemical exchange. Journal of Biomolecular Nmr. 2001;21:361–366. doi: 10.1023/a:1013328206498. [DOI] [PubMed] [Google Scholar]
- Wang CY, Palmer AG. Solution NMR methods for quantitative identification of chemical exchange in N-15-labeled proteins. Magnetic Resonance in Chemistry. 2003;41:866–876. [Google Scholar]
- Wang CY, Rance M, Palmer AG. Mapping chemical exchange in proteins with MW > 50 kD. Journal of the American Chemical Society. 2003;125:8968–8969. doi: 10.1021/ja035139z. [DOI] [PubMed] [Google Scholar]
- Wang X, Vallurupalli P, Vu A, Lee K, Sun S, Bai WJ, Wu C, Zhou H, Shea JE, Kay LE, et al. The linker between the dimerization and catalytic domains of the CheA histidine kinase propagates changes in structure and dynamics that are important for enzymatic activity. Biochemistry. 2014;53:855–861. doi: 10.1021/bi4012379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittier SK, Hengge AC, Loria JP. Conformational motions regulate phosphoryl transfer in related protein tyrosine phosphatases. Science. 2013;341:899–903. doi: 10.1126/science.1241735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Watz M, Thai V, Henzler-Wildman K, Hadjipavlou G, Eisenmesser EZ, Kern D. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Lee T, Latham MP, Warner LR, Tanimoto A, Pardi A, Ahn NG. Phosphorylation releases constraints to domain motion in ERK2. Proc Natl Acad Sci U S A. 2014;111:2506–2511. doi: 10.1073/pnas.1318899111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Garrod SM, Deal MS, Anand GS, Woods VL, Taylor S. Allosteric network of cAMP-dependent protein kinase revealed by mutation of Tyr204 in the P+1 loop. Journal of Molecular Biology. 2005;346:191–201. doi: 10.1016/j.jmb.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Yang J, Ten Eyck LF, Xuong NH, Taylor SS. Crystal structure of a cAMP-dependent protein kinase mutant at 1.26A: new insights into the catalytic mechanism. Journal of molecular biology. 2004:336. doi: 10.1016/j.jmb.2003.11.044. [DOI] [PubMed] [Google Scholar]
- Zhang P, Smith-Nguyen EV, Keshwani MM, Deal MS, Kornev AP, Taylor SS. Structure and allostery of the PKA RIIbeta tetrameric holoenzyme. Science. 2012;335:712–716. doi: 10.1126/science.1213979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Isothermal Titration Calorimetry (ITC) with ATPγN to the apo form and the pseudo-substrate PKI5-24 to the ATPγN-bound form of PKA-CWT and PKA-CY204A. Top: ATPγN binding to Apo PKA-CWT and the PKA-CY204A mutant. Bottom: Pseudo-substrate, PKI5-24, binding to PKA-CWT and PKA-CY204A in the presence of 2 mM ATPγN. All measurements were performed in triplicate and the error reflects the standard deviation.
Figure S2: Overlays of the [1H, 15N]-TROSY-HSQC spectra for PKA-CWT and PKA-CY204A for the three forms of the kinase: apo (top), ATPγN-bound (middle), and ATPγN and PKI5-24-bound (bottom).
Figure S3: Qualitative comparison of the B-factor from the crystal structure 1BKX (left) with the resonances undergoing broadening beyond detection upon binding the nucleotide (also shown in Figure 2). B-factors are shown in a rainbow scale from blue (1) to red (100).
Figure S4. Left: List of residues that defined linear trajectories for the 6 states analyzed with CONCISE using the thresholds reported in the Methods section. Right: The linear residues are mapped onto the kinase structure (1ATP).
Figure S5: 1H-15N HX-NOE as probe of fast conformational dynamics. Top: The HX-NOE is plotted as a function of the PKA sequence where the red colored values represent NOEs above or below one standard deviation from the average. Bottom: HX-NOE values reported on the crystal structure 1ATP of the kinase. Bulges and colors show regions with higher mobility. S10, Q12, E13, E17, T37, I46, K61, K105 are more rigid in the mutant (higher values of HX-NOEs), while K7, N67, K78 are more dynamic (lower HX-NOE values). Residues near or belonging to the linker regions and preceding the D-helix (Y117, M120 and Y122) are relatively rigid; whereas G125, located near a C-spine residue, and A188, right after the DFG-loop, are more mobile. In the peptide positioning loop, A206 becomes rigid, while E208 becomes mobile in the Y204A enzyme.
Figure S6: 15N TROSY Hahn-echo as a probe for slow conformational dynamics. Top: Rex values obtained from the TrHE experiments are plotted as a function of the PKA sequence where the red colored values represent Rex > 7 s−1 and |ΔRex| > 7 s−1. Due to the magnitude and orientation of the 15N chemical shift tensor, the Rex values for the non-exchanging residues are scattered around zero with a standard deviation of approximately 2 s−1. Bottom: Rex values are shown on the crystal structure 1ATP of the kinase with bulges and colors highlighting the regions with higher mobility.
TABLE S1, related to Table 1: Thermodynamics of ligand binding to PKA-CWT and PKA-CY204A determined by ITC. Binding of the ATP mimic, ATPγN, to Apo form of the enzyme (top) was measured as well as PKI5-24 binding of the nucleotide bound form of the enzyme (bottom).
TABLE S2, related to Table 1. 15N CPMG relaxation dispersion group fit of PKA-CWT.
TABLE S3, related to Figure 4. 15N CPMG relaxation dispersion individual fit of PKA-CWT.
TABLE S4, related to Figure 5. 15N CPMG relaxation dispersion individual fit of PKA-CY204A.