Abstract
The endoplasmic reticulum (ER) possesses a protein quality control system that supports the efficient folding of newly synthesized glycoproteins. In this system, a series of N-linked glycan intermediates displayed on proteins serve as quality tags. The ER folding-sensor enzyme UDP-glucose:glycoprotein glucosyltransferase (UGGT) operates as the gatekeeper for ER quality control by specifically transferring monoglucose residues to incompletely folded glycoproteins, thereby allowing them to interact with lectin chaperone complexes to facilitate their folding. Despite its functional importance, no structural information is available for this key enzyme to date. To elucidate the folding-sensor mechanism in the ER, we performed a structural study of UGGT. Based on bioinformatics analyses, the folding-sensor region of UGGT was predicted to harbour three tandem thioredoxin (Trx)-like domains, which are often found in proteins involved in ER quality control. Furthermore, we determined the three-dimensional structure of the third Trx-like domain, which exhibits an extensive hydrophobic patch concealed by its flexible C-terminal helix. Our structural data suggest that this hydrophobic patch is involved in intermolecular interactions, thereby contributing to the folding-sensor mechanism of UGGT.
In eukaryotic cells, proteins destined for the secretory pathway are translocated to the endoplasmic reticulum (ER) for folding, assembly and post-translational modification, including asparagine-linked glycosylation. To guarantee that only correctly folded glycoproteins are transported to the Golgi apparatus, the ER possesses a sophisticated protein quality control system1,2,3,4,5,6,7. In this system, N-linked oligosaccharides displayed on polypeptide chains function as quality tags for the determination of glycoprotein fates, i.e. folding, transport or degradation, that are selectively recognized by certain intracellular lectins2,4,5,6.
In the ER, newly synthesized proteins are cotranslationally modified with high mannose-type tetradecasaccharide (Glc3Man9GlcNAc2), which contains three non-reducing terminal branches (designated D1, D2 and D3)8. The D1 branch is capped with the triglucosyl moiety Glc-α1,2-Glc-α1,3-Glc. Glucosidase I removes the outermost α1,2-linked glucose from the D1 branch of this triantennary glycan9,10. Subsequently, glucosidase II trims the second and third α1,3-linked glucose residues7,9,11. The monoglucosylated D1 branch, an intermediate generated during this process, exhibits a critical determinant recognized by oxidoreductase (ERp57)-associated lectins, i.e. calnexin (CNX) and/or calreticulin (CRT). UDP-glucose:glycoprotein glucosyltransferase (UGGT) catalyzes reglucosylation, thereby regenerating monoglucosylated glycoforms, which are able to revisit the chaperone complex7,12,13,14,15,16,17,18. This glucose-trimming and -tagging process is called the ‘CNX/CRT cycle’.
UGGT acts as the gatekeeper in this system because this enzyme is capable of sensing the folding states of glycoproteins as potential substrates. UGGT only transfers monoglucose residues to incompletely folded glycoproteins7,12,13,14. UGGT is a large enzyme, comprising approximately 1500 amino acid residues, which has been putatively divided into two regions: an N-terminal folding-sensor region, which accounts for approximately 80% of the enzyme and is not homologous with any known structures, and a C-terminal catalytic domain, which accounts for the remaining 20% of the enzyme and belongs to the glycosyltransferase 8 family19,20. However, no further structural information is available on this key enzyme to date. Thus, the structural basis of the working mechanism of the CNX/CRT cycle remains unclear.
In this study, to elucidate the working mechanism of UGGT, we attempted to characterize the three-dimensional (3D) structure of its N-terminal folding-sensor region. We selected Chaetomium thermophilum, a thermophilic fungus, which survives at temperatures of up to 60°C21, as the source organism for the structural study of UGGT. Our bioinformatics analyses predicted that the folding-sensor region of UGGT contains three tandem thioredoxin (Trx)-like domains. Moreover, we determined the 3D structure of a Trx domain of UGGT, thereby providing structural insights into the mechanism of substrate recognition of this folding-sensor enzyme.
Results
Bioinformatic identification of three tandem Trx-like domains in folding sensor region of UGGT
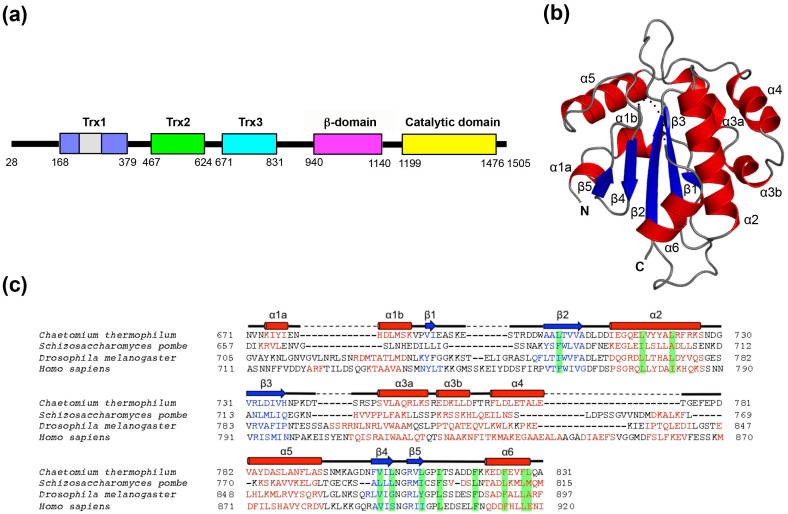
To investigate the structure of the N-terminal folding-sensor region of UGGT, we subjected its amino acid sequence (residues 28–1198) to bioinformatics analysis using the programs PSIPRED22 and DISOPRED223. The results indicate that the folding-sensor region of UGGT exhibits well-formed secondary structures: a mixed α/β region in the N-terminal part (residues 28–939) and a β-strand-rich region (termed the β-domain, residues 940–1140) around the C-terminus (Fig. 1a and Supplemental Fig. S1). Although the sequence homology of UGGT was modestly low (32.0%–34.5% identities) between the thermophilic fungus and humans (Supplemental Table S1), the secondary structure distributions appeared highly conserved across species. A remarkably disordered segment was identified at the connection between the β- and C-terminal catalytic domains (Supplemental Fig. S1). This structural feature is consistent with previously reported results of limited proteolysis20.
Figure 1.
Crystal structure of the Trx3 domain of UGGT (a) Domain structure of C. thermophilum UGGT. The Trx3 domain (residues Asn671–Ala831) was crystallized in this study. (b) Ribbon models of the Trx3 domain of C. thermophilum UGGT (Form 1). The secondary structures are highlighted (α-helix, red; β-sheet, blue) and the linker regions are shown in grey. The positions of the N- and C-termini are also indicated. Dotted line indicates disorder segment. (c) Structure-based sequence alignment of the Trx3 domains of UGGT among species (from fungi to human). The secondary structures of the Trx3 domain of C. thermophilum UGGT are indicated above the amino acid sequence. The secondary structure elements (α-helix and β-sheet) were predicted using the program PROMALS3D48 and are highlighted in red and blue, respectively. Residues involving the C-terminal α6 helix or detergent interactions are highlighted in green.
Next, we attempted to identify structural domain(s) within the N-terminal folding-sensor region using InterPro24 and Phyre225. Regarding the β-domain, no significantly homologous domains were identified. On the other hand, the folding-sensor region of UGGT was found to harbour three tandem Trx-like domains: Trx1 (residues 168–379), Trx2 (residues 467–624) and Trx3 (residues 671–831) (Fig. 1 and Supplemental Fig. S1). The arrangement of these domains is essentially identical across species, suggesting that the common structural architecture of UGGT is evolutionarily conserved. Nonetheless, the three tandem Trx-like domains share relatively low sequence identities (Trx1 versus Trx2, 22.1%; Trx1 versus Trx3, 23.3%; Trx2 versus Trx3, 16.2% in C. thermophilum), suggesting variability in their three-dimensional structures.
Crystal structure of the third Trx-like domain of UGGT
Based on the bioinformatic prediction that folding-sensor region of UGGT possesses three tandem Trx-like domains, we performed bacterial expression, purification and crystallization of a series of Trx domains. First, we expressed each of the three Trx domains. Although we were able to express the Trx3 domain as a soluble protein, the Trx1 and Trx2 domains formed inclusion bodies in Escherichia coli cells. Therefore, we made tandem constructs for their expression. Consequently, we were able to express Trx1-Trx2, Trx2-Trx3 and Trx1-Trx2-Trx3 proteins in their soluble form. Of these constructs, we successfully crystallized the Trx3 domain with the optimization of its N- and C-terminal sequences (residues 671–831), based on the identification of proteolytically stable fragments. However, despite extensive trials, we were unable to obtain crystals of the tandem constructs Trx1-Trx2, Trx2-Trx3 or Trx1-Trx2-Trx3.
We determined two forms of the crystal structure of Trx3 domain at 3.4 and 1.7 Å resolutions. The final model of Form 1, refined to a resolution of 3.40 Å, had an Rwork of 23.5% and Rfree of 29.2% (Table 1). The crystal belonged to space group I23 with six molecules per asymmetric unit. The structures of molecules A–F were highly similar to each other with an RMSD value of 0.11–0.37 Å for superimposed Cα atoms 94–155. Molecule A in the crystal structure, which had the lowest average B value (Table 1), was used for the comparative analysis and will be primarily described hereafter. On the other hand, Form 2 of the Trx3 domain of UGGT cocrystallized with a detergent ANAPOE C12E8 belonged to space group C2221 and diffracted up to 1.70-Å resolution. In the crystal structure, one molecule was contained per asymmetric unit. The final model of Form 2 had an Rwork of 20.1% and Rfree of 24.6% (Table 1).
Table 1. Data collection and refinement statistics for UGGT-Trx3 domain.
| Form 1 | Form 2 | |
|---|---|---|
| Crystallographic data | ||
| Space group | I23 | C2221 |
| Unit cell a/b/c (Å) | 196.4/196.4/196.4 | 46.2/93.6/81.9 |
| α/β/γ (°) | 90.0/90.0/90.0 | 90.0/90.0/90.0 |
| Data processing statistics | ||
| Beam line | NSRRC 13B1 | PF-AR NW12A |
| Wavelength (Å) | 0.97888 | 0.97921 |
| Resolution (Å) | 50–3.40 (3.52–3.40) | 50–1.70 (1.73–1.70) |
| Total/unique reflections | 778,614/17,411 | 134,741/20,126 |
| Completeness (%) | 100.0 (100.0) | 98.5 (98.9) |
| Rmerge (%) | 12.7 (67.7) | 8.2 (36.6) |
| I/σ (I) | 34.1 (6.7) | 47.9 (7.2) |
| Refinement statistics | ||
| Resolution (Å) | 20.0–3.40 | 20.0–1.70 |
| Rwork/Rfree (%) | 23.5/29.2 | 20.1/24.6 |
| R.m.s. deviations from ideal | ||
| Bond lengths (Å) | 0.010 | 0.011 |
| Bond angles (°) | 1.28 | 1.47 |
| Ramachandran plot (%) | ||
| Favored | 96.5 | 98.3 |
| Allowed | 3.5 | 1.7 |
| Number of atoms | ||
| Protein atoms (A/B/C/D/E/F) | 1239/1246/1127/1231/738/871 | 1166 |
| Water molecules | - | 120 |
| Detergent molecule | - | 37 |
| Average B-values (Å2) | ||
| Protein atoms (A/B/C/D/E/F) | 79.7/80.6/92.6/95.2/135.1/139.8 | 23.8 |
| Water molecules | - | 30.1 |
| Detergent molecule | - | 64.9 |
As expected from the bioinformatics analysis, the crystal structure displayed a typical Trx-like fold, i.e. a five-stranded β-sheet with a β1–β3–β2–β4–β5 arrangement surrounded by six α-helices (Fig. 1b and 1c). In the crystal structure, a part of β5–α6 loop (residues 816–818) was disordered. The C-terminal α6-containing segment showed a higher crystallographic B-factor (87.7 Å2) than the average value (79.7 Å2; Table 1). Comparison of the structure of the Trx3 domain of UGGT with known protein structures using the DALI server revealed that the protein disulfide bond isomerase (DsbA/C) homologue, Salmonella enterica ScsC26, was the most structurally similar protein (Z-score = 9.4; RMSD = 2.9 Å; identify = 18.5%; PDB code: 4GXZ). As representative of the DsbA/C structure, the well-characterized crystal structure of E. coli DsbC (PDB code: 1EEJ)27 is also shown in Supplemental Figure 2. The overall fold of Trx3 domain of UGGT was essentially identical to that of ScsC except for their variable α helical segments between 3 and 4 (α3 and α4 in UGGT-Trx3 and α3–α5 in ScsC) (Supplemental Fig. S2b). DsbC also share very similar fold with the UGGT Trx3 domain except for the N-terminal α1 helix, which directly follows the dimerization domain in DsbC, and variable α3/α4 helices (Supplemental Fig. S2c). Compared with the crystal structure of the E. coli thioredoxin trxA28 (PDB code: 2TRX; Supplemental Fig. S2d), which exhibits typical Trx fold, three contiguous helical insertions, α3, α4 and α5, were identified between β3 and β4, as observed in DsbC27. Furthermore, an N-terminal segment containing α1 and β1 regions of the Trx3 domain of UGGT was significantly different from that of E. coli trxA28 in terms of topological arrangement. In the folds shared by the Trx3 domain of UGGT, ScsC and DsbC, α1 precedes β1, which makes anti-parallel β-strands with β3 (Supplemental Fig. S2a–c). In contrast, α1 was inserted between β1 and β2, both of which were parallel with respect to β3 (Supplemental Fig. S2d). In addition, our homology modeling suggest that the Trx1 and Trx2 domains exhibit typical Trx-like folds similar to the Trx3 domain and its structural homologs, except for the N-terminal and variable α helical segments between 3 and 4 and an insertion loop (residues 226–293) in Trx1 (Supplemental Fig. S3).
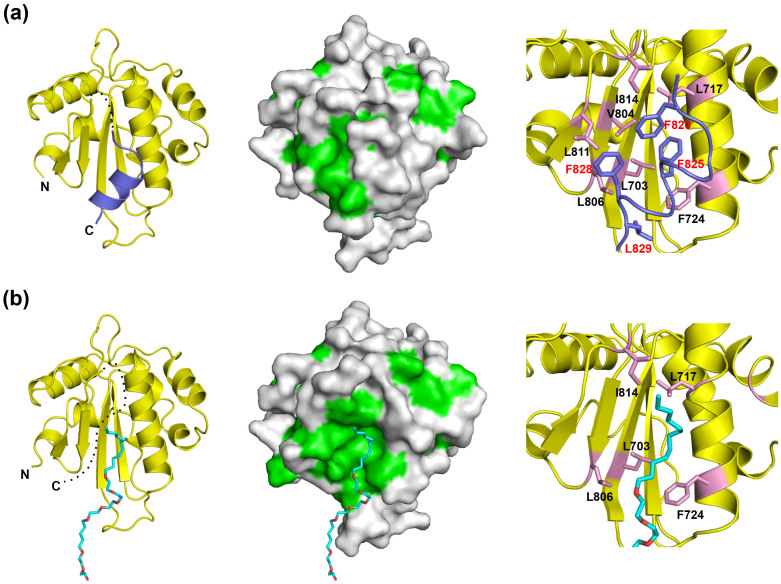
The C-terminal α6 helix, which is followed by a putatively flexible linker region in UGGT, was completely disordered in the crystal structure of Form 2, suggesting the instability of this helix (Fig. 2b, left). Because of the absence of the α6 helix, an extensive hydrophobic patch was exposed on the surface of the Trx3 domain (Fig. 2b, centre). The detergent ANAPOE C12E8 was accommodated on this exposed hydrophobic patch. The α6 helix was stabilized mainly through its hydrophobic surface, containing Phe820, Phe825, Phe828 and Leu829, which made contact with the hydrophobic patch, including Leu703 (β2), Leu717, Phe724 (α2), Val804, Leu806 (β4), Leu811 (β5) and Ile814 (β5–α6 loop) (Fig. 2a, right). Most of these hydrophobic residues were involved in the interaction with the detergent in Form 2. Thus, the C-terminal α6 helix and detergent molecule occupy the common hydrophobic surface of the Trx3 domain. These hydrophobic residues are highly conserved among species (Fig. 1 and Supplemental Fig. S1).
Figure 2. An extensive hydrophobic patch of the Trx3 domain is concealed by a flexible C-terminal helix.
The crystal structures of the Trx3 domain in Forms 1 and 2 are indicated in (a) and (b), respectively. The ribbon and surface models are shown in the left and centre. Dotted lines indicate disordered segments. In the surface model (centre), the hydrophobic residues are shown in green. Close-up views of the C-terminal helix or detergent-interacting regions are represented on the right. Residues involved in these interactions are highlighted in the pink stick model. In Form 1 (a), the C-terminal α6 helix is highlighted in slate. In Form 2 (b), the detergent ANAPOE C12E8 is shown as a stick model.
Discussion
In this study, we proposed that the folding-sensor region of UGGT contains three tandem Trx-like domains and, solved the first 3D structure of a structural domain, i.e. the third Trx-like domain, of this functional region (Fig. 1 and Supplemental Fig. S1). Trx-like domains are common to members of the protein disulfide isomerase (PDI) family, which are responsible for assisting protein folding in the ER29. Most PDI family members are multidomain proteins containing both redox-active and -inactive Trx-like domains in different arrangement29,30. For example, PDI (PDIA1) as a representative member of PDI family possesses four tandem Trx-like domains (designated a, b, b′ and a′), of which a and a′ domains have a CXXC catalytic motif, whereas b and b′ domains do not31,32. None of the Trx-like domains of UGGT possess the CXXC catalytic motif, indicating that this enzyme is not directly involved in thiol/disulfide exchange reactions. In this context, the cis-Pro loop adjacent to the CXXC motif, a hallmark of redox-active Trx-fold proteins29 and involved in substrate recognition in DsbA32, is not present in the Trx3 domain of UGGT. Noncatalytic Trx-like domains are often involved in substrate recognition33,34,35, co-factor interaction36 and functional intradomain interactions34. UGGT forms a stable complex with Sep15, a 15-kDa selenocystein-containing oxidoreductase37 which possesses one redox-active Trx-like domain and enhances the glucosyltransferase activity of UGGT38. It is plausible that Sep15 serves as a structural extension of UGGT with a complementary function.
Growing evidence implies that UGGT exhibits glucosyltransferase activity only against incompletely folded glycoproteins, suggesting that the folding-sensor region has exposed the hydrophobic patch as a principal substrate-binding site7,12,13,14. The Trx3 domain possesses an extensive hydrophobic patch, which is covered by the flexible C-terminal helix and can participate in interactions with hydrophobic molecules (Fig. 2). The hydrophobic residues involved in these intramolecular and intermolecular interactions are conserved across species (Supplemental Fig. S1). Thus, our crystallographic study provides an atomic view of the potential substrate-binding site of UGGT. In addition, our homology modeling data suggested that Trx1 and Trx2 domains also exhibit larger hydrophobic patches located at the opposite site as compared with that of the Trx3 domain, suggesting the possibility of their involvement in substrate recognition (Supplemental Fig. S4). Concomitantly, this may be the cause of inclusion body formation of the isolated Trx1 and Trx2 domains. In general, molecular chaperones undergo conformational transitions coupled with the shielding and exposure of their hydrophobic patches as substrate-binding sites35,39. Although we cannot exclude the possibility that the hydrophobic patch of the Trx3 domain is covered by other domain(s) in intact UGGT, the flexible properties of the C-terminal helix of Trx3 may contribute to regulatory mechanisms underlying the folding-sensing function of this domain.
In summary, our bioinformatic analyses predicted that the folding-sensor region of UGGT harbours three tandem Trx-like domains. Moreover, we provided snapshots of the 3D structure of the third Trx-like domain, in which a putative substrate-binding hydrophobic patch is intramolecularly masked or involved in an intermolecular interaction, offering a key breakthrough toward understanding of the functional mechanisms of this ER folding-sensor enzyme.
Methods
Protein expression and purification
C. thermophilum var. thermophilum La Touche (DSM 1495) was obtained from DSMZ, Braunschweig, Germany. Total RNA was isolated using TRIzol® reagent (Life Technologies). The cDNA was synthesized using SuperScript® III Reverse Transcriptase (Life Technologies) with oligo d(T) primers according to the manufacturer's instructions. Full-length UGGT cDNA was cloned by PCR using a C. thermophilum genomic DNA database21. Recombinant UGGT proteins were expressed as glutathione S-transferase (GST)-fused proteins. The Trx1 (residues 168–379), Trx2 (residues 467–624), Trx3 (residues 671–831), Trx1-Trx2 (residues 168–624), Trx2-Trx3 (residues 467–831) and Trx1-Trx2-Trx3 (residues 168–831) domains were amplified by PCR and subcloned into the BamHI and XbaI sites of a modified pCold-GST vector (Takara Bio Inc.)40, in which the factor Xa site was replaced with the tobacco etch virus (TEV) protease recognition site. Recombinant proteins were expressed in E. coli BL21 Star™ cells (Life Technologies) according to the manufacturer's protocols (Takara Bio Inc.). GST-fused proteins were purified using glutathione-Sepharose™ columns (GE Healthcare). Subsequently, the GST tag was removed by adding TEV protease to the resin for 12 h at 277 K, leaving two additional residues Gly-Ser at the N-terminus. The resultant proteins were further purified by size-exclusion chromatography (Superdex-200; GE Healthcare) using a buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.1 mM EDTA. The selenomethione (SeMet)-labelled Trx3 domain was expressed in E. coli B834 (DE3) using M9 minimal medium with SeMet. Expression and purification were performed following the same protocol as that for the native protein. Purified proteins were dialyzed against a buffer containing 10 mM Tris-HCl (pH 7.5) and 100 mM NaCl. The integrity of the protein samples was validated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) analysis using an AXIMA-CFR™ spectrometer (Shimadzu) and N-terminal Edman sequencing with a Procise 494HT protein sequenator (ABI/Life Technologies).
Protein crystallization, X-ray data collection and structure determination
The crystals of the Trx3 domain of UGGT (Form 1, 10 mg/ml) were grown in a buffer containing 60% Tacsimate (pH 7.0) for 2 weeks at 289 K. The crystals of the Trx3 domain of UGGT (Form 2) were obtained by equilibrating a solution of 8 mg/ml protein with 1.2 mM ANAPOE C12E8 (polyoxyethylene[8]dodecyl ether · 3,6,9,12,15,18,21,24-octaoxahexatriacontan-1-ol) mixed with an equal volume of precipitant solution containing 23% PEG3350, 0.1 M Tris-HCl (pH 7.0) and 0.2 M ammonium acetate for 6 days at 289 K. The crystals were transferred into the reservoir solution and flash-cooled in liquid nitrogen. Data sets for Forms 1 and 2 were collected using synchrotron radiation at 13B1 of the National Synchrotron Radiation Research Center (Hsinchu, Taiwan) and AR-NW12A of the Photon Factory (Tsukuba, Japan), respectively. All diffraction data were processed using HKL200041. Crystal parameters are summarized in Table 1.
The 1.70 Å-resolution crystal structure of the Trx3 domain of UGGT (Form 2) was solved using the SAD method. The initial phase was determined using the SHELX C/D/E program42. The initial model was automatically built using ARP/wARP43. Further manual model building into the electron density maps and refinement were performed using COOT44 and REFMAC545, respectively. The 3.40 Å-resolution structure of the Trx3 domain of UGGT (Form 1) was solved by molecular replacement using the program Phaser46 with the crystal structure of Form 2 as a search model. The stereochemical quality of the final model was assessed by RAMPAGE47. The final refinement statistics are summarized in Table 1. Graphic figures were prepared using PyMOL (http://www.pymol.org/). Homology modeling of the Trx1 and Trx2 domains were performed using Phyre225 with Neisseria gonorrhoeae DsbC-like protein (PDB code: 3GV1) and Neisseria meningitidis DsbA1 (PDB code: 3DVW) as templates, respectively.
Author Contributions
T.S. and K.K. conceived and designed the experiments; T.Z. and T.S. performed the bioinformatics analyses and crystallographic experiments; all authors wrote and reviewed the manuscript.
Additional Information
Accession codes The coordinates and structural factors of the crystal structures of the Trx3 domain of C. thermophilum UGGT (Forms 1 and 2) have been deposited in the Protein Data Bank under the accession numbers 3WZT and 3WZS, respectively.
Supplementary Material
Supplementary information
Acknowledgments
We thank Drs Masato Kawasaki, Hisayoshi Makyio (KEK, Tsukuba, Japan) and Chiang Cheng-Hung [National Synchrotron Radiation Research Center (NSRRC), Hsinchu, Taiwan] for help with X-ray data collection. We are grateful to the NSRRC 13B1 and PF-AR NW12A beamline staff for providing the data collection facilities and supports. We acknowledge Dr Hirokazu Yagi (Nagoya City University, Nagoya, Japan) and Ms Yumiko Makino (National Institute for Basic Biology, National Institutes of Natural Sciences, Okazaki, Japan) for help with MALDI-TOF/MS and N-terminal sequence analyses, respectively. This work was supported in part by the Okazaki ORION project and Grants-in-Aid for Scientific Research (Grant Numbers 24770102, 25121730 to T.S. and 25102008, 24249002 to K.K.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and by PRESTO project (Grant Number 13417569 to T.S.) from the Japan Science and Technology Agency.
References
- Ellgaard L. & Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4, 181–91 (2003). [DOI] [PubMed] [Google Scholar]
- Kato K. & Kamiya Y. Structural views of glycoprotein-fate determination in cells. Glycobiology 17, 1031–44 (2007). [DOI] [PubMed] [Google Scholar]
- Takeda Y., Totani K., Matsuo I. & Ito Y. Chemical approaches toward understanding glycan-mediated protein quality control. Curr Opin Chem Biol 13, 582–91 (2009). [DOI] [PubMed] [Google Scholar]
- Lederkremer G. Z. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol 19, 515–23 (2009). [DOI] [PubMed] [Google Scholar]
- Aebi M., Bernasconi R., Clerc S. & Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem Sci 35, 74–82 (2010). [DOI] [PubMed] [Google Scholar]
- Kamiya Y., Satoh T. & Kato K. Molecular and structural basis for N-glycan-dependent determination of glycoprotein fates in cells. Biochim Biophys Acta 1820, 1327–37 (2012). [DOI] [PubMed] [Google Scholar]
- D'Alessio C., Caramelo J. J. & Parodi A. J. UDP-GlC:glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin Cell Dev Biol 21, 491–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher D. J. & Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R (2006). [DOI] [PubMed] [Google Scholar]
- Grinna L. S. & Robbins P. W. Substrate specificities of rat liver microsomal glucosidases which process glycoproteins. J Biol Chem 255, 2255–8 (1980). [PubMed] [Google Scholar]
- Deprez P., Gautschi M. & Helenius A. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol Cell 19, 183–95 (2005). [DOI] [PubMed] [Google Scholar]
- Totani K., Ihara Y., Matsuo I. & Ito Y. Substrate specificity analysis of endoplasmic reticulum glucosidase II using synthetic high mannose-type glycans. J Biol Chem 281, 31502–8 (2006). [DOI] [PubMed] [Google Scholar]
- Taylor S. C., Ferguson A. D., Bergeron J. J. & Thomas D. Y. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat Struct Mol Biol 11, 128–34 (2004). [DOI] [PubMed] [Google Scholar]
- Caramelo J. J., Castro O. A., Alonso L. G., De Prat-Gay G. & Parodi A. J. UDP-Glc:glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates. Proc Natl Acad Sci U S A 100, 86–91 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totani K., Ihara Y., Tsujimoto T., Matsuo I. & Ito Y. The recognition motif of the glycoprotein-folding sensor enzyme UDP-Glc:glycoprotein glucosyltransferase. Biochemistry 48, 2933–40 (2009). [DOI] [PubMed] [Google Scholar]
- Schrag J. D. et al. The Structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol Cell 8, 633–44 (2001). [DOI] [PubMed] [Google Scholar]
- Caramelo J. J. & Parodi A. J. Getting in and out from calnexin/calreticulin cycles. J Biol Chem 283, 10221–5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G. et al. Structural basis of carbohydrate recognition by calreticulin. J Biol Chem 285, 38612–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouquet A. et al. X-Ray structure of the human calreticulin globular domain reveals a peptide-binding area and suggests a multi-molecular mechanism. PLoS ONE 6, e17886 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S. M. & Kaufman R. J. The noncatalytic portion of human UDP-glucose: glycoprotein glucosyltransferase I confers UDP-glucose binding and transferase function to the catalytic domain. J Biol Chem 278, 43320–8 (2003). [DOI] [PubMed] [Google Scholar]
- Guerin M. & Parodi A. J. The UDP-glucose:glycoprotein glucosyltransferase is organized in at least two tightly bound domains from yeast to mammals. J Biol Chem 278, 20540–6 (2003). [DOI] [PubMed] [Google Scholar]
- Amlacher S. et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–89 (2011). [DOI] [PubMed] [Google Scholar]
- Jones D. T. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292, 195–202 (1999). [DOI] [PubMed] [Google Scholar]
- Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F. & Jones D. T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337, 635–45 (2004). [DOI] [PubMed] [Google Scholar]
- Hunter S. et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40, D306–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A. & Sternberg M. J. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4, 363–71 (2009). [DOI] [PubMed] [Google Scholar]
- Shepherd M. et al. Structural and functional characterization of ScsC, a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium. Antioxid Redox Signal 19, 1494–506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A. A. et al. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat Struct Biol 7, 196–9 (2000). [DOI] [PubMed] [Google Scholar]
- Katti S. K., LeMaster D. M. & Eklund H. Crystal structure of thioredoxin from Escherichia coli at 1.68 Å resolution. J Mol Biol 212, 167–84 (1990). [DOI] [PubMed] [Google Scholar]
- Heras B., Kurz M., Shouldice S. R. & Martin J. L. The name's bond.disulfide bond. Curr Opin Struct Biol 17, 691–8 (2007). [DOI] [PubMed] [Google Scholar]
- Kozlov G., Määttänen P., Thomas D. Y. & Gehring K. A structural overview of the PDI family of proteins. FEBS J 277, 3924–36 (2010). [DOI] [PubMed] [Google Scholar]
- Edman J. C., Ellis L., Blacher R. W., Roth R. A. & Rutter W. J. Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature 317, 267–70 (1985). [DOI] [PubMed] [Google Scholar]
- Freedman R. B., Klappa P. & Ruddock L. W. Protein disulfide isomerases exploit synergy between catalytic and specific binding domains. EMBO Rep 3, 136–40 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klappa P., Ruddock L. W., Darby N. J. & Freedman R. B. The b' domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J 17, 927–35 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serve O. et al. Redox-dependent domain rearrangement of protein disulfide isomerase coupled with exposure of its substrate-binding hydrophobic surface. J Mol Biol 396, 361–74 (2010). [DOI] [PubMed] [Google Scholar]
- Serve O., Kamiya Y. & Kato K. Protein Folding: Redox-dependent chaperoning, following PDI footsteps (Walters E. C. ed) 489–500 (NOVA Science Publishers, New York, 2011). [Google Scholar]
- Russell S. J. et al. The primary substrate binding site in the b' domain of ERp57 is adapted for endoplasmic reticulum lectin association. J Biol Chem 279, 18861–9 (2004). [DOI] [PubMed] [Google Scholar]
- Korotkov K. V., Kumaraswamy E., Zhou Y., Hatfield D. L. & Gladyshev V. N. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J Biol Chem 276, 15330–6 (2001). [DOI] [PubMed] [Google Scholar]
- Takeda Y. et al. Both isoforms of human UDP-glucose:glycoprotein glucosyltransferase are enzymatically active. Glycobiology 24, 344–50 (2014). [DOI] [PubMed] [Google Scholar]
- Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol 14, 630–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K. & Kojima C. pCold-GST vector: a novel cold-shock vector containing GST tag for soluble protein production. Protein Expr Purif 62, 120–7 (2008). [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. & Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M. A short history of SHELX. Acta Crystallogr A 64, 112–22 (2008). [DOI] [PubMed] [Google Scholar]
- Langer G., Cohen S. X., Lamzin V. S. & Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc 3, 1171–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N., Vagin A. A. & Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53, 240–55 (1997). [DOI] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell S. C. et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 50, 437–50 (2003). [DOI] [PubMed] [Google Scholar]
- Pei J., Kim B. H. & Grishin N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36, 2295–300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information