Abstract
In this work, we demonstrate the use of a single DNA strand and G-quadruplex-specific dye NMM as a label-free switch for the construction of series of basic logic gates (YES, NOT, OR, INHIBIT, NOR, AND). The simple GT-rich sequence could be used to interact with several molecules (K+, thrombin, Hg2+, and Pb2+) to form different structures that can be distinguished by the label-free dye NMM. Our study showed that a single G-qudruplex DNA strand can function as multiple types of one-input and two-input logic gates with different combinations of input molecules.
DNA is an information coding polymer. So it is an ideal candidate for information storage1 and molecular computation2. After the DNA computation was proposed by Adleman in 19943, numerous efforts have been made to construct DNA logic gates4,5,6,7 and computation circuits8,9,10,11,12 through a bottom-up strategy13. Recently, the study of G-quadruplex-specific dyes with light-up fluorescence14,15 provided a simple, label-free and cost-effective signal-producing approach to construct DNA-based logic gates12. Our goal here is to show that this approach can be extended by using a single G-quadruplex DNA strand from creating a single type of logic gate with two inputs to multiple types of logic gates with different combinations of several input molecules.
G-quadruplexes are unique structures formed by G-rich nucleic acid sequences and based on stacked arrays of G-quartets connected by Hoogsteen-type base pairing. These structures are further stabilized and recognized by the presence of monovalent cations (especially K+)16,17 and some biological molecules (such as thrombin)18. N-methyl mesoporphyrin IX (NMM) is an unsymmetrical anionic porphyrin characterized by a pronounced structural selectivity for G-quadruplexes but not for duplexes, triplexes, or single-stranded forms. The fluorescence of NMM is very weak, but exhibits a dramatic enhancement upon binding to G-quadruplex DNA19. This behavior is the same as the other quadruplex-specific dyes20. This allows us to utilize NMM as a label-free fluorescent probe to indicate the formation of G-quadruplex structure and monitor its conformational change. For implementing the complicated computation circuits and networks, label-free and more modular computation units are desirable. The formation of G-quadruplex could be not only triggered by many metal ions, some small molecules and proteins, but also regulated by changes of DNA secondary structure through dsDNA hybridization and aptamer binding21,22,23,24,25,26, which could bring various design strategies and indicate a great potential in label-free DNA computation. As a powerful technical tool in DNA computation, the employment of G-quadruplex as the logic component might promote the development of DNA computation in a more economical, versatile and modular way.
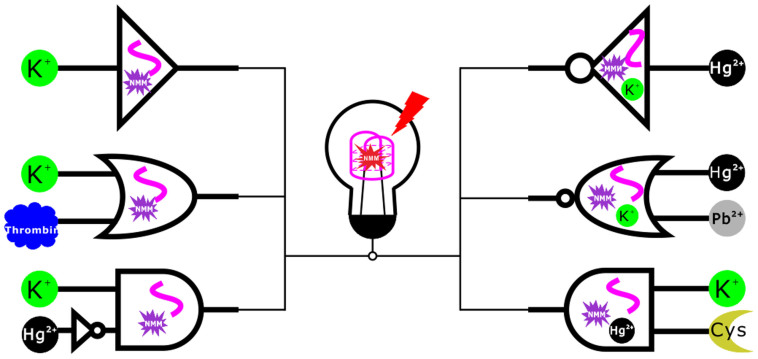
Herein, a thymine-rich and guanine-rich DNA oligomer, which could transform to G-quadruplex in the presence of several molecules, was employed to construct a series of basic logic gates with label-free fluorescent output. Fig. 1 shows multiple types of label-free fluorescent logic gates based on this single sequence, which could perform YES, NOT, OR, INHIBIT, NOR and AND logic operations under the action of different inputs.
Figure 1. Schematic representation of multiple label-free fluorescent logic gates based on a single G-quadruplex DNA.
Results
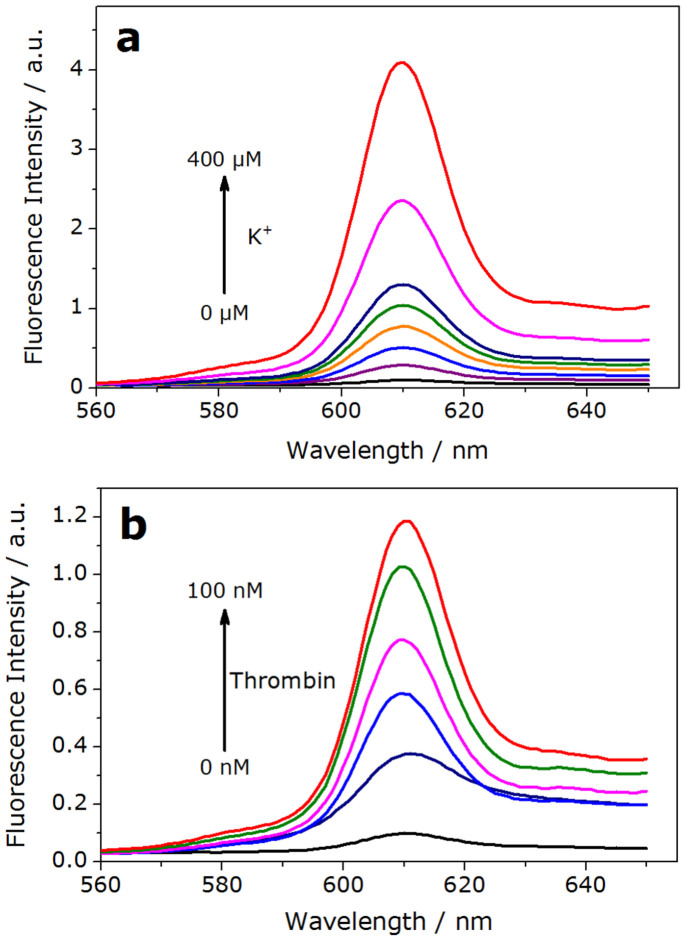
The system contained a GT-rich ssDNA (GT24) and a G-quadruplex specific fluorescent dye NMM. The fluorescence intensity of NMM was negligible when the GT24 was in “loose” status; however, in the presence of K+ or thrombin, GT24 transformed to a rigid G-quadruplex structure, which brought out a significant fluorescence increase of NMM. As shown in Fig. 2, the fluorescence intensity of NMM-GT24 complex increased gradually with the increasing concentrations of K+ and thrombin, showing a linear dependence (Fig. S1&S2). Since G-quadruplex has the characteristic peak near 290 nm in CD spectra, the formation of G-quadruplex was further validated by the circular dichroism (Fig. S3)27,28. The fluorescence intensity was normalized throughout the experiment, the background fluorescence signal (NMM-GT24 complex) at 610 nm was defined as 0.1, signals higher than 10-fold background (>1.0 a.u.) was defined as output "1", otherwise, signals lower than 0.4 a.u. as output “0”.
Figure 2. Fluorescence emission spectra of NMM-GT24 complex as a function of various concentrations of K+(a) and thrombin(b).
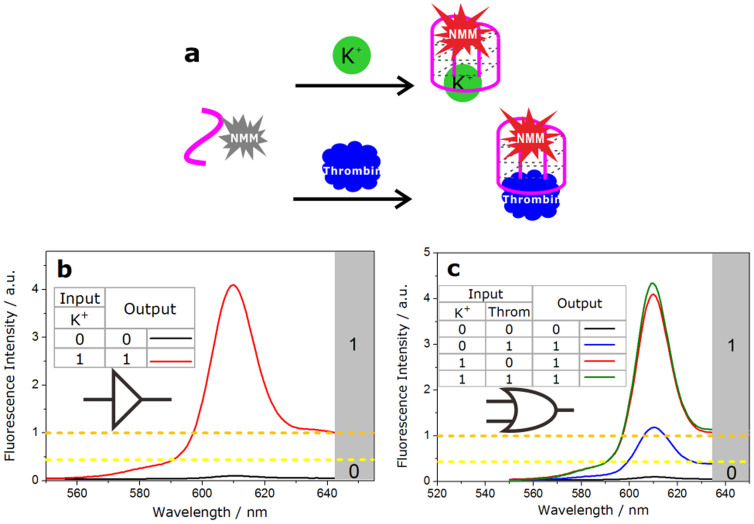
Based on aforementioned fact, we first designed YES and OR logic gates which employed NMM and GT24 as components and regarded K+ and thrombin as inputs (Fig. 3.a). For inputs, the presence of K+ or thrombin was defined as the “1” state. The presence of K+ contributed to the transformation of GT24 from a relaxed state to a taut state, turning on the fluorescence of NMM from "Off" state (>1.0 a.u., output = 1), which was defined as the simplest YES logic gate (Fig. 3.b). In the OR logic operation, the presence of both inputs (1/1) or the presence of either input (0/1, 1/0) could cause a fluorescence enhancement in 610 nm (>1.0 a.u., output = 1); while in the absence of both inputs (0/0), the system gave a low fluorescence signal due to the loose state of GT24 (<0.4 a.u., output = 0) (Fig. 3.c).
Figure 3. YES and OR logic gates.
(a), Scheme of the allosteric G-quadruplex induced by K+ (10 mM) and thrombin (100 nM). (b), Fluorescence spectra of the NMM-GT24 complex in the presence or absence of input and the truth table of YES logic gate. (c), Fluorescence spectra of the NMM-GT24 complex in all combinations of two inputs and the truth table of OR logic gate.
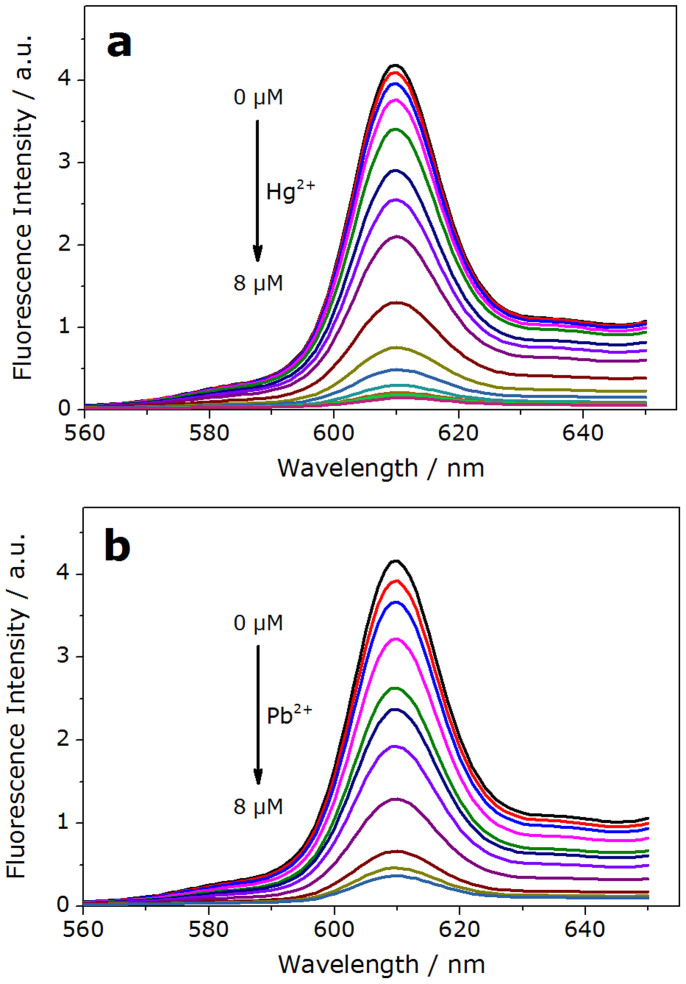
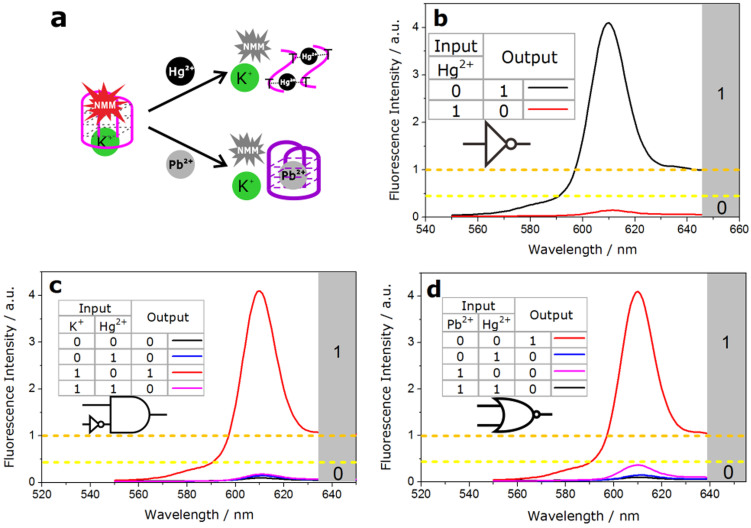
In the presence of Hg2+, GT24 was transformed from the K+-activated G-quadruplex into a hairpin structure, due to the formation of a stable T–Hg–T mismatch29,30,31,32. Consequently, as shown in Fig. 4.a & S4.a, with the increasing concentration of Hg2+, the fluorescence of NMM was restored to the low bulk value due to the release of NMM from GT24. The disappearing peak in 290 nm in circular dichroism further validated the deformation of G-quadruplex structure mediated by Hg2+ (Fig. S5). A NOT gate performs logical negation on its input. In other words, if the input is present, then the output will be false (“0”). Similarly, the absence of input results in a true output(“1”). The logic of an INHIBIT gate is that the output is “1” only when one particular input is present when the second input is not present. Established on these properties, this GT24 sequence with K+ could be used to construct NOT logic gate by using the addition of Hg2+ as the input. And the INHIBIT logic gate was constructed with GT24 by using the addition of K+/Hg2+ as the inputs (Fig. 5). In the NOT gate, the output is “1” when the input (Hg2+) was “0” (Fig. 5.b). In the INHIBIT gate, the output was “1” only upon the addition of K+ (input = 1/0), while it was “0” for all other combination of inputs (0/0, 0/1, 1/1) (Fig. 5.c).
Figure 4.
(a), Fluorescent spectra of the NMM-GT24 complex with K+ (10 mM) upon the addition of Hg2+ (0, 0.05, 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0 μM). (b), Fluorescent spectra of the NMM-GT24 complex with K+ (10 mM) upon the addition of Pb2+ (0, 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 2.0, 4.0, 6.0, 8.0 μM).
Figure 5. NOT, INHIBIT and NOR logic gates.
(a), Graphic illustration of the NMM-GT24 complex with K+ (10 mM) as a switch by employing Hg2+ (10 μM) and Pb2+ (10 μM) as inputs. (b), Fluorescence spectra of the NMM-GT24 complex with K+ in the presence or absence of Hg2+ and the truth table of NOT logic gate. (c), Fluorescence spectra of the NMM-GT24 complex in the presence of all input combinations of K+ and Hg2+ and the truth table of INHIBIT logic gate. (d), Fluorescence changes for NMM-GT24 complex with K+ in the presence of all input combinations of Pb2+ and Hg2+ and the truth table of NOR logic gate.
Pb2+ competitively binds to the K+-stabilized G-quadruplex to form more compact DNA fold33,34. As judged by the M−O6 bond length, O6−O6 diagonal distance and inter-tetramer separation, structure analysis indicated that the divalent Pb2+ templates a smaller G-quadruplex than K+ does35. As a result, the Pb2+-stabilized G-quadruplex prohibits the formation of the complex of G-quadruplex with NMM36, which resulted in fluorescence decrease (Fig. 4.b). This enabled us to design a NOR gate, in which Hg2+ and Pb2+ serve as inputs, in the initial state, the enhanced fluorescence signal was observed since NMM bound with the K+-activated G-quadruplex. In the presence of either or both inputs (0/1, 1/0, 1/1), the signal of fluorescence decreased significantly (<0.4 a.u., output = 0), which was in accord with the proper execution of a NOR logic gate (Fig. 5.d).
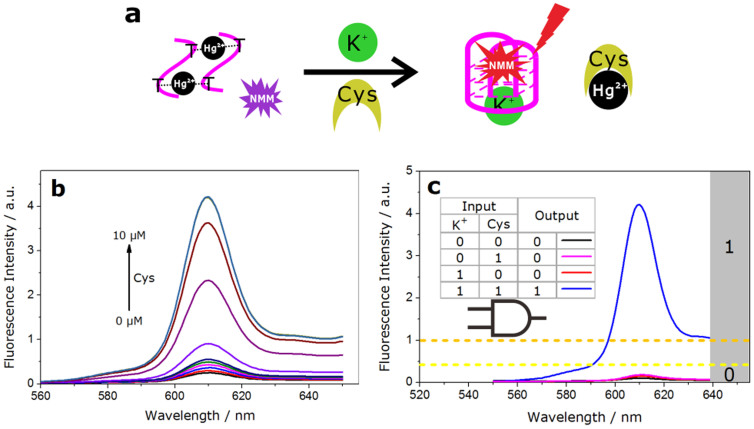
Cysteine is a strong Hg2+ binder that can withdraw Hg2+ ions from the T−Hg−T complex and sequester Hg2+ ions in the form of stable Hg-Cys complexes37. So we constructed an AND logic gate with Hg2+ and GT24 as components and employed K+ and cysteine as inputs. As shown in Fig. 6.a, initially the sequence held the structure in the T-Hg-T state, then the cooperative binding of cysteine with Hg2+ turned it back into a quadruplex structure with the assistant of monovalent cation K+, therefore the signal intensity of NMM was enhanced. Fig. 6b. shows that the enhanced fluorescence intensity depended on the increasing concentration of Cys. Hence, like in a typical AND operation, the output was “1” (enhanced FI>1.0 a.u.) only when both K+ and cysteine were added into the system (input = 1/1), and it was “0” when only one of them or neither was added (input = 0/0, 0/1, 1/0; Fig. 6.c).
Figure 6. AND logic gate.
(a), Graphic illustration of AND logic gate switched by K+ and cysteine as two inputs. (b), Fluorescence titration of the NMM-GT24 complex (with 10 mM K+ and 8 μM Hg2+ in solution) in the presence of various concentrations of cysteine (0, 0.2, 0.4, 0.6, 0.8, 1.0, 2.0, 4.0, 6.0, 8.0, 10 μM). (c), Fluorescence spectra in the presence of all input combinations and the truth table of AND logic gate.
Discussion
The light-up fluorescence from the interaction between NMM and G-quadruplex provided a label-free approach to indicate the formation of G-quadruplex structure and monitor its conformational change. G-quadruplex-forming sequence is a simple and cost-effective signal-producing logic component to construct DNA-based logic gates. The GT-rich sequence GT24 in this study could interact with several molecules (potassium, thrombin, mercury, lead and cysteine), which could act as a versatile recognition unit in multiple types of logic gates with different combinations of several input molecules as showed. This might be regarded as particular case since the single GT24 DNA in this study contains only G-rich quadruplex-forming segments and T-rich Hg2+-binding segments, but this technical report using a single strand could give us a clue that we might embed multiple aptamer sequences in one DNA strand to undertake programmable computing tasks. There have been reports on artificial aptazyme-based riboswitch by inserting more than one RNA aptamer sequence, which bind specifically to their target ligands, into the 5′UTR of mRNA, making a cell to be the arithmetic unit38. Aptamers could be modulated by interactions between aptamers and their targets including various ions, molecules, even cells39,40,41, which brings more input elements in DNA computation and indicates the applications of intelligent diagnosis or drug delivery in biomedicine42,43,44. The introduction and integrating of aptamer sequences in DNA computation would make DNA computation in a more versatile and practical way.
In summary, we have successfully demonstrated that the single GT-rich DNA sequence can be used as label-free fluorescent switch by binding with G-quadruplex-specific dye NMM for constructing multiple types of DNA logic gates. Our study suggests a strategy that one could insert different aptamer domains into a single oligonucleotide to undertake more complex computing functions.
Methods
Materials
Oligonucleotide (GT24: 5′-GGGTTTTGGGTTTTGGGTTTTGGG-3′) was purchased from Invitrogen Biotechnology Co., Ltd. (Shanghai, China). N-methyl mesoporphyrin IX (NMM) was bought from J&K Scientific Ltd. (Beijing, China) and other chemicals were of analytical grade purity and purchased from Sigma-Aldrich (St. Louis, U.S.A.). All measurements were performed in 20 mM Tris-HCl, 10 mM MgCl2 and pH 7.5. The concentration of GT24 and NMM was 0.5 μM throughout the experiments. Computations for YES, OR logic gates were completed in 10 min, and for NOT, INHIBIT, NOR each input combination was incubated for 0.5 h, in the AND logic operation the fluorescence intensity was recorded after 1.5 h.
Instruments
Ultrapure water from MILLI-Q Synthesis (Millipore Int., Molsheim, France) source was used throughout the experiments. The concentration of NMM was measured on a UV-2550 spectrophotometer (SHIMADZU, Japan). Fluorescence spectra data were collected with an F-4600 fluorescence spectrophotometer (Hitachi Co. Ltd., Japan) equipped with a Xenon lamp excitation source. Slit widths for the excitation (the excited wavelength 399 nm) and emission were set at 10 and 10 nm respectively. CD spectra were measured with a Chirascan dichrograph (Applied Photophysics Ltd) in 1 cm path length quartz. The scan speed was 50 nm min−1 and bandwidth was 1 nm.
Author Contributions
Y.G. designed the experiments, performed the majority of experiments, analyzed results and wrote the paper. L.Z., L.X. and X.Z. performed experiments and analysed results. J.H. supervised the project and analysed results. R.P. supervised the project, analysed results and wrote the paper.
Supplementary Material
supplementary information
Acknowledgments
This work is supported by the National Natural Science Foundation of China (21275156, 41273093, 21175101) and the State Key Laboratory of Electroanalytical Chemistry.
References
- Goldman N. et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei R., Matamoros E., Liu M., Stefanovic D. & Stojanovic M. N. Training a molecular automaton to play a game. Nat. Nanotechnol. 5, 773–777 (2010). [DOI] [PubMed] [Google Scholar]
- Adleman L. M. Molecular computation of solutions to combinatorial problems. Science 266, 1021–1024 (1994). [DOI] [PubMed] [Google Scholar]
- Lake A., Shang S. & Kolpashchikov D. M. Molecular logic gates connected via DNA four way junctions. Angew. Chem. Int. Ed. 49, 4459–4462 (2010). [DOI] [PubMed] [Google Scholar]
- Freeman R., Finder T. & Willner I. Multiplexed analysis of Hg2+ and Ag+ ions by nucleic acid functionalized CdSe/ZnS quantum dots and their use for logic gate operations. Angew. Chem. Int. Ed. 48, 7818–7821 (2009). [DOI] [PubMed] [Google Scholar]
- Li T., Zhang L., Ai J., Dong S. & Wang E. Ion-tuned DNA/Ag fluorescent nanoclusters as versatile logic device. ACS Nano 5, 6334–6338 (2011). [DOI] [PubMed] [Google Scholar]
- Li T., Ackermann D., Hall A. M. & Famulok M. Input-dependent induction of oligonucleotide structural motifs for performing molecular logic. J. Am. Chem. Soc. 134, 3508–3516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig G., Soloveichik D., Zhang D. Y. & Winfree E. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006). [DOI] [PubMed] [Google Scholar]
- Qian L. & Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011). [DOI] [PubMed] [Google Scholar]
- Rudchenko M. et al. Autonomous molecular cascades for evaluation of cell surfaces. Nat. Nanotechnol. 8, 580–586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yang Y., Yan H. & Liu Y. Three-input majority logic gate and multiple input logic circuit based on DNA strand displacement. Nano Lett. 13, 2980–2988 (2013). [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhang L., Li T., Dong S. & Wang E. Enzyme-free unlabeled DNA logic circuits based on toehold-mediated strand displacement and split G-quadruplex enhanced fluorescence. Adv. Mater. 25, 2440–2444 (2013). [DOI] [PubMed] [Google Scholar]
- Krishnan Y. & Simmel F. C. Nucleic acid based molecular devices. Angew. Chem. Int. Ed. 50, 3124–3156 (2011). [DOI] [PubMed] [Google Scholar]
- He H. Z., Chan D. S., Leung C. H. & Ma D. L. G-quadruplexes for luminescent sensing and logic gates. Nucleic Acids Res. 41, 4345–4359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Largy E., Granzhan A., Hamon F., Verga D. & Teulade-Fichou M. P. Visualizing the quadruplex: from fluorescent ligands to light-up probes. Top. Curr. Chem. 330, 111–177 (2013). [DOI] [PubMed] [Google Scholar]
- Sen D. & Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334, 364–366 (1988). [DOI] [PubMed] [Google Scholar]
- Williamson J. R., Raghuraman M. K. & Cech T. R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59, 871–880 (1989). [DOI] [PubMed] [Google Scholar]
- Macaya R. F., Schultze P., Smith F. W., Roe J. A. & Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA. 90, 3745–3749 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthanari H., Basu S., Kawano T. L. & Bolton P. H. Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res. 26, 3724–3728 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. & Sen D. A catalytic DNA for porphyrin metallation. Nat. Struct. Biol. 3, 743–747 (1996). [DOI] [PubMed] [Google Scholar]
- Kong D. M., Guo J. H., Yang W., Ma Y. E. & Shen H. X. Crystal violet-G-quadruplex complexes as fluorescent sensors for homogeneous detection of potassium ion. Biosens. Bioelectron. 25, 88–93 (2009). [DOI] [PubMed] [Google Scholar]
- Zhao C., Wu L., Ren J. & Qu X. A label-free fluorescent turn-on enzymatic amplification assay for DNA detection using ligand-responsive G-quadruplex formation. Chem. Commun. 47, 5461–5463 (2011). [DOI] [PubMed] [Google Scholar]
- Guo Y., Xu P., Hu H., Zhou X. & Hu J. A label-free biosensor for DNA detection based on ligand-responsive G-quadruplex formation. Talanta 114, 138–142 (2013). [DOI] [PubMed] [Google Scholar]
- Guo Y. et al. Label-free logic modules and two-layer cascade based on stem-loop probes containing a g-quadruplex domain. Chem. Asian. J. 9, 2397–2401 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang Z., Sharon E., Freeman R., Liu X. & Willner I. Fluorescence detection of DNA, adenosine-5′-triphosphate (ATP), and telomerase activity by zinc(II)-protoporphyrin IX/G-quadruplex labels. Anal. Chem. 84, 4789–4797 (2012). [DOI] [PubMed] [Google Scholar]
- Li D., Shlyahovsky B., Elbaz J. & Willner I. Amplified analysis of low-molecular-weight substrates or proteins by the self-assembly of DNAzyme-aptamer conjugates. J. Am. Chem. Soc. 129, 5804–5805 (2007). [DOI] [PubMed] [Google Scholar]
- Li T., Wang E. K. & Dong S. J. Parallel G-quadruplex-specific fluorescent probe for monitoring DNA structural changes and label-free detection of potassium ion. Anal. Chem. 82, 7576–7580 (2010). [DOI] [PubMed] [Google Scholar]
- Yuminova A. V., Spiridonova V. A., Arutyunyan A. M. & Kopylov A. M. Structural study of thrombin binding DNA aptamers by the circular dichroism. Dokl. Biochem. Biophys. 442, 36–38 (2012). [DOI] [PubMed] [Google Scholar]
- Miyake Y. et al. MercuryII-mediated formation of thymine-HgII-thymine base pairs in DNA duplexes. J. Am. Chem. Soc. 128, 2172–2173 (2006). [DOI] [PubMed] [Google Scholar]
- Li T., Dong S. & Wang E. Label-free colorimetric detection of aqueous mercury ion (Hg2+) using Hg2+-modulated G-quadruplex-based DNAzymes. Anal. Chem. 81, 2144–2140 (2009). [DOI] [PubMed] [Google Scholar]
- Wang Z., Heon L. J. & Lu Y. Highly sensitive “turn on” fluorescent sensor for Hg2+ in aqueous solution based on structure-switching DNA. Chem. Commun. 45, 6005–6007 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. et al. A Hg2+-mediated label-free fluorescent sensing strategy based on G-quadruplex formation for selective detection of glutathione and cysteine. Analyst. 138, 1713–1718 (2013). [DOI] [PubMed] [Google Scholar]
- Smirnov I. & Shafer R. H. Lead is unusually effective in sequence-specific folding of DNA. J. Mol. Biol. 296, 1–5 (2000). [DOI] [PubMed] [Google Scholar]
- Li T., Wang E. & Dong S. Potassium-lead-switched G-quadruplexes: a new class of DNA logic gates. J. Am. Chem. Soc. 131, 15082–15083 (2009). [DOI] [PubMed] [Google Scholar]
- Kotch F. W., Fettinger J. C. & Davis J. T. A lead-filled G-quadruplex: insight into the G-quartet's selectivity for Pb2+ over K+. Org. Lett. 2, 3277–3280 (2000). [DOI] [PubMed] [Google Scholar]
- Wang X. Y. et al. A novel molecular logic system based on lead-induced substitution of potassium from a G-quadruplex as a fluorescent lead sensor. Anal. Methods 5, 5597–5601 (2013). [Google Scholar]
- Stricks W. & Kolthoff I. M. Reactions between mercuric mercury and cysteine and glutathione - apparent dissociation constants, heats and entropies of formation of various forms of mercuric mercapto-cysteine and mercapto-glutathione. J. Am. Chem. Soc. 75, 5673–5681 (1953). [Google Scholar]
- Win M. N. & Smolke C. D. Higher-order cellular information processing with synthetic RNA devices. Science 322, 456–460 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Donovan M. J. & Jiang J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 113, 2842–2862 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao Z. & Lu Y. Functional nucleic acid sensors. Chem. Rev. 109, 1948–1998 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Li B. & Wang E. "Fitting" makes "sensing" simple: label-free detection strategies based on nucleic acid aptamers. Acc. Chem. Res. 46, 203–213 (2013). [DOI] [PubMed] [Google Scholar]
- Pei H. et al. Reconfigurable three-dimensional DNA nanostructures for the construction of intracellular logic sensors. Angew. Chem. Int. Ed. 51, 9020–9024 (2012). [DOI] [PubMed] [Google Scholar]
- Liang H. et al. Functional DNA-containing nanomaterials: cellular applications in biosensing, imaging, and targeted therapy. Acc. Chem. Res. 47, 1891–1901 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M. et al. DNA "nano-claw": logic-based autonomous cancer targeting and therapy. J. Am. Chem. Soc. 136, 1256–1259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information