Abstract
Patients with mammographically dense breast tissue have a greatly increased risk of developing breast cancer. Dense breast tissue contains more stromal collagen, which contributes to increased matrix stiffness and alters normal cellular responses. Stromal collagen within and surrounding mammary tumors is frequently aligned and reoriented perpendicular to the tumor boundary. We have shown that aligned collagen predicts poor outcome in breast cancer patients, and postulate this is because it facilitates invasion by providing tracks on which cells migrate out of the tumor. However, the mechanisms by which alignment may promote migration are not understood. Here, we investigated the contribution of matrix stiffness and alignment to cell migration speed and persistence. Mechanical measurements of the stiffness of collagen matrices with varying density and alignment were compared with the results of a 3D microchannel alignment assay to quantify cell migration. We further interpreted the experimental results using a computational model of cell migration. We find that collagen alignment confers an increase in stiffness, but does not increase the speed of migrating cells. Instead, alignment enhances the efficiency of migration by increasing directional persistence and restricting protrusions along aligned fibers, resulting in a greater distance traveled. These results suggest that matrix topography, rather than stiffness, is the dominant feature by which an aligned matrix can enhance invasion through 3D collagen matrices.
Introduction
Increased mammographic density is associated with a 4- to 6-fold increased risk of breast cancer (1–3), making mammographic density one of the greatest independent risk factors for breast cancer (1,3,4). This increase in density is correlated with a significantly increased deposition of extracellular matrix (ECM) proteins, most notably collagen I (5–7), which is in part responsible for the overall increase in stiffness in mammary tumors (8,9). Matrix stiffness has been shown to promote a malignant phenotype in tumor cells (8,10–12), enhance migration and invasion (13–16), and alter cell signaling, leading to increased proliferation (10,17–19). Although it is clear that matrix stiffness plays a profound role in tumor progression, we do not yet fully understand the mechanisms by which cells respond to changes in matrix stiffness.
In addition to the amount of collagen, the orientation of collagen fibers appears to play a critical role in tumor progression. Our laboratory previously characterized changes in the alignment and orientation of collagen fibers, and identified tumor-associated collagen signatures (TACS), which manifest in predictable ways during tumor progression. In particular, deposition of aligned collagen that is oriented perpendicular to the tumor boundary (termed TACS-3) creates highways on which tumor cells are observed to migrate in vivo (20), and correlates with increased invasion and metastasis in mouse models (21). More recently, we showed that TACS-3 alignment is an independent prognostic signature that correlates strongly with poor patient survival (22). These initial findings strongly indicate that matrix stiffness resulting from increased collagen deposition and matrix alignment contributes to mammary tumor progression.
Although the cellular players and mechanism for alignment generation in vivo remain elusive, in vitro studies have shown that epithelial cells and fibroblasts are capable of using Rho- and Rho kinase (ROCK)-mediated actin-myosin contractility to orient collagen fibers (23–26). Additionally, fibroblasts can deposit matrices containing aligned fibronectin or collagen in vitro (27,28). Recently, Yang et al. (28) showed that this ability of fibroblasts to produce aligned matrices is associated with expression of the cell-surface proteoglycan syndecan-1.
The high correlation of collagen alignment with breast tumor progression suggests that the mechanisms by which alignment facilitates cell migration need to be evaluated more closely. Studies of cells cultured in matrices with aligned fibers have revealed that cells polarize and orient with respect to the alignment (29–31), and that alignment is associated with increased migration and directionality (23,28,32). The underlying mechanisms for these responses to alignment, however, remain unclear. One possibility is that alignment organizes cell adhesions along fibers, resulting in more efficient migration from coordinated traction forces. It has been demonstrated that parallel-oriented fibers may also afford cells less spatial impedance and thereby enhance migration (33). Additionally, it has been suggested that alignment incurs changes in matrix stiffness, and that enhanced migration along fibers may be due to durotactic guidance. However, the effects of increasing alignment on tensile modulus have been largely assumed, without being well documented and quantified. Here, we created novel (to our knowledge) aligned matrices to parse out the contributions of 3D matrix alignment and stiffness to the migration of breast cancer cells. We find that alignment facilitates persistence without affecting migration speed, and that contact guidance via aligned fibers, rather than durotaxis due to stiffness, is the likely mechanism by which migration is enhanced. Moreover, we present a model to describe trends in cell migration induced by matrix physical and mechanical properties. Validating the model’s prediction, we found that an aligned matrix limits the number of stabilized protrusions and likely serves as mechanism that leads to more persistent migration.
Results
The aim of this study was to characterize the effects of collagen fiber alignment and matrix stiffness on the 3D cell migration of invasive ductal breast carcinoma cells. To study the mechanical properties of aligned collagen and how cells respond to changing matrix mechanics, we generated 3D collagen gels containing aligned fibers by using a device to impart mechanical strain. Similar to the method described by Vader et al. (34), we designed and used a 3D printer to create a strain device that uses a micrometer-driven arm and pin assembly to stretch one end of a collagen gel (Fig. 1 A). The pins contacted two pieces of polypropylene mesh embedded in the gel and had a width of 1 cm to produce a wide aligned region. The device was designed to fit the stage of a multiphoton microscope, and second harmonic generation (SHG) images of a 2 mg/ml gel were collected with increasing strain applied (Fig. 1 B). We used CurveAlign software (http://www.loci.wisc.edu) analysis, which employs a curvelet-based algorithm to measure fiber orientation and assigns a coefficient of alignment from zero (no alignment of one fiber relative to another) to one (all fibers in perfect alignment). The coefficient of alignment increased with strain and appeared to approach a maximum alignment at 30% strain (Fig. 1 C).
Figure 1.
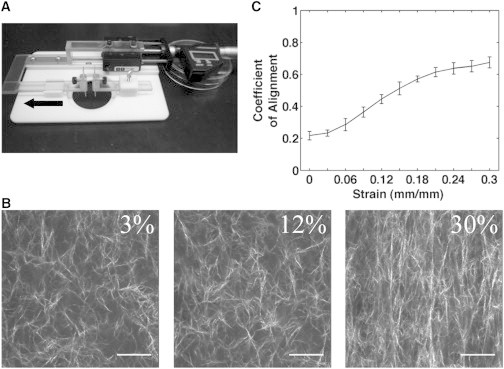
Collagen gels are aligned by mechanical strain. (A) A strain device designed and assembled for use with a multiphoton microscope is fitted with a micrometer to precisely control the amount of strain on a collagen gel placed in the center of the stage. Two pins contact the sample and the left pin/arm assembly is driven by a micrometer. The arrow indicates the direction of strain. (B) SHG images reveal collagen fiber alignment with increasing strain; scale bar, 50 μm. (C) CurveAlign software analysis of coefficient of alignment from SHG images (n = 3 gels).
To determine whether aligned collagen had a greater elastic modulus than gels with randomly oriented fibers, we cast 2 mg/ml collagen gels in a stainless-steel, dogbone-shaped mold that was machined to the dimensions specified in Roeder et al. (35). We then tested the tensile moduli of the gels using an Instron MicroTester 5548. (For an image of a representative collagen gel in the grips of the Instron, see Fig. S1 B in the Supporting Material.) Samples were either prestrained using the strain device shown in Fig. 1 or left unstrained. All samples were imaged via SHG to observe the fiber alignment and then subjected to tensile testing at a rate of 1 mm/min until failure occurred. Sample strain was computed by measuring the relative displacement of glass beads deposited on the surface of the gel. All prestrained samples demonstrated an increased coefficient of alignment due to an increase in the number of fibers oriented near 0° relative to the axis of the dogbone (71% of fibers for prestrained samples compared with 43% for unstrained samples; Fig. 2, A, left and right).
Figure 2.
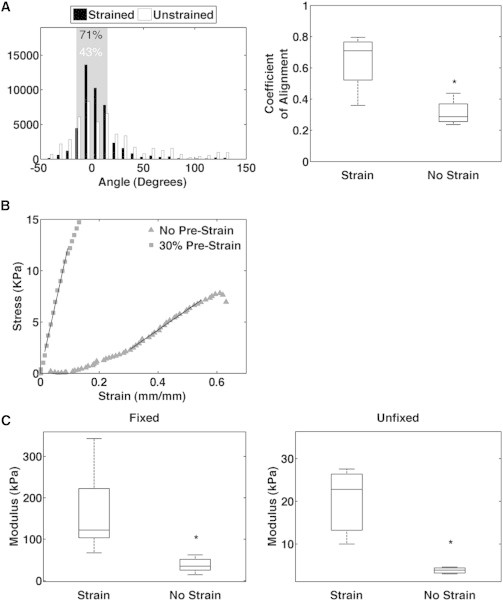
Aligned collagen gels are stiffer than unaligned gels. (A) Left: histograms of fiber angles generated from CurveAlign software of unstrained and 30% prestrained gels. Percentages represent the fraction of fiber angles ±15° from 0° relative to the axis of alignment (shaded region). (A) Right: coefficient of alignment of unstrained and 30% prestrained gels (n = 4 gels, p < 0.05). Note that even unstrained samples demonstrate fibers oriented at 0°, which is due to a stronger signal in this orientation because of laser polarization (36,37); however, all strained samples exceeded this background level. (B) Representative stress-strain curves of an unstrained gel and 30% prestrained gel fixed with PFA. (C) Tensile modulus of unstrained and 30% prestrained gels fixed with PFA (n = 13 gels, p < 0.0001) or unfixed (n = 3 gels, p < 0.05).
Stress-strain curves for unstrained samples displayed classic strain-stiffening behavior with the presence of a toe region, indicative of fiber recruitment, before the linear regime. In contrast, prestrained gels lacked a toe region, consistent with the prealignment of fibers, and the linear regime exhibited a steeper slope (Fig. 2 B). The slope of this linear regime represents the elastic modulus, and prestrained gels had greater elastic moduli than unstrained gels regardless of whether the gels were left unfixed (Fig. 2 C, right) or fixed using 4% paraformaldehyde (PFA) for 10 min immediately after prestraining (Fig. 2 C, left). Thus, prealigning the collagen matrix alters the elastic behavior and results in a significant increase in the stiffness of the matrix.
Cells are known to use durotaxis to migrate toward increased stiffness (13). Since we previously showed that cells preferentially migrate along, rather than across, aligned fibers (23), we tested whether the elastic modulus is greater along the axis of aligned fibers than across the axis of alignment. We prestrained large (2 mg/ml) collagen gels using a horizontal weight-based loading system that clamped two sides of a gel suspended in a PBS bath. One clamp remained stationary while the other was drawn with the use of weights to stretch the gel, and the gel was subsequently fixed with 4% PFA. A 3D-printed cutter was then used to cut dogbone-shaped specimens from the larger gel, with dimensions identical to those of the aforementioned dogbone mold. Specimens were cut such that the gauge region of the dogbone was either parallel or perpendicular to the direction of the applied strain (Fig. S1 A). Dogbones cut parallel to the direction of strain had aligned fibers oriented ∼0° relative to the axis of alignment, whereas those cut perpendicular to the strain direction had fibers oriented ∼90° (Fig. S1 D, left). The parallel and perpendicular cut gels had similar high coefficients of alignment (Fig. S1 D, right). However, the moduli were greatest in the parallel strained gels (Fig. S1 C), indicating that collagen is stiffest along the axis of alignment. Interestingly, the moduli for perpendicular strained and unstrained gels were similar, which served as a control for other effects of prestraining the collagen gel that were not related to orientation of the fibers.
To determine whether this increased stiffness along the axis of alignment leads cells to preferentially migrate along aligned collagen fibers, we conducted a set of experiments to assess MDA-MB-231 cell migration and matrix stiffness in aligned and random collagen gels with increasing collagen concentration. Initially, we determined the coefficient of alignment and elastic modulus for 1–4 mg/ml prestrained and unstrained collagen gels. These experiments showed that the coefficient of alignment was higher for all prestrained gels regardless of collagen concentration (Fig. 3 A). The elastic modulus increased with increasing collagen concentration for both prestrained and unstrained samples, with all prestrained specimens having a significantly greater modulus than their unstrained counterparts (Fig. 3 B). Note that the modulus of the 4 mg/ml unstrained specimens was not significantly different from that of the 1 mg/ml prestrained aligned gels, which serves below as a useful comparison for cell migration in matrices of different topography but identical stiffness.
Figure 3.
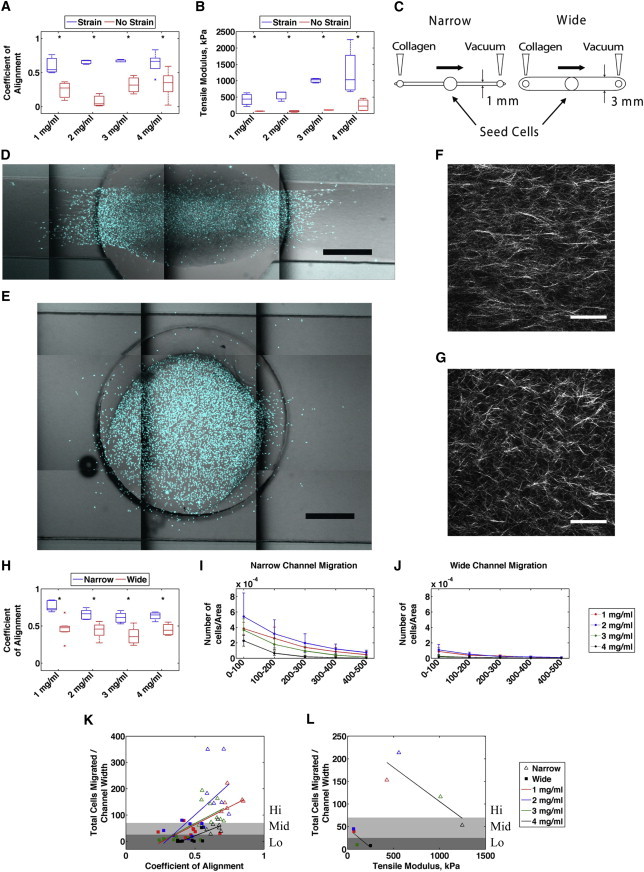
MDA-MB-231 cell migration is positively correlated with collagen alignment. (A) Coefficient of alignment of 1, 2, 3, and 4 mg/ml unstrained and 30% prestrained gels quantified using CurveAlign software (n = 3 gels, p < 0.05). (B) Tensile modulus of 1, 2, 3, and 4 mg/ml unstrained and 30% prestrained gels fixed with PFA (n = 3, p < 0.05). (C) Schematic depicting the microchannel migration assay. Flow of collagen through narrow channels results in alignment. (D and E) Representative confocal montages of narrow (D) and wide (E) channels, with cell nuclei stained with DAPI. Scale bar, 500 μm. (F and G) Representative SHG images of collagen in narrow (F) and wide (G) channels. Scale bar, 50 μm. (H) CurveAlign software analysis of coefficient of alignment in 1, 2, 3, and 4 mg/ml narrow and wide microchannels (p < 0.01). (I and J) Quantification of cell density outside of the channel port in concentric rings (100 μm wide) in narrow (I) and wide (J) channels (n = 6–9 channels). (K and L) Total cells that migrated normalized to the channel width is positively correlated with the coefficient of alignment (K) and inversely correlated with stiffness (L). Shaded regions represent statistical significance in migration and are designated as low (Lo), medium (Mid), and high (Hi) migration.
After characterizing the stiffness of aligned and random collagen matrices across a range of concentrations, we sought to determine the effect of changing matrix stiffness and alignment on cell migration. As the cells were not readily seeded in a reproducible way into gels that were aligned by strain, we developed a flow-based 3D microchannel assay to produce either an aligned or a random collagen matrix based on other reports using microchannels (36,37). When neutralized collagen was flowed during polymerization through narrow channels (width = 1 mm), an aligned matrix was produced. Flowing polymerizing collagen through a wider channel (width = 3 mm) produced a random matrix (Fig. 3 C). MDA-MB-231 cells were seeded in the center ports of both narrow and wide channels and allowed to migrate for 3 days, after which time the gels were fixed and stained with bisbenzimide to assess the number of cells that migrated into the channel. SHG images were collected to assess fiber alignment in narrow (Fig. 3 F) and wide (Fig. 3 G) microchannels, and confocal images of stained nuclei (Fig. 3, D and E) were analyzed in FIJI. All narrow channels had a significantly higher coefficient of alignment compared with wide channels (Fig. 3 H). Moreover, all narrow channels had a greater number of cells that migrated farther away from the port (Fig. 3 I) compared with wide channels (Fig. 3 J).
To determine whether migration correlated with matrix alignment or stiffness, total nuclei counts outside of the port boundary were normalized to the channel width and plotted against either the coefficient of alignment or the tensile modulus. Cell counts displayed a positive correlation with the coefficient of alignment for all collagen concentrations (Fig. 3 K). In contrast, cell migration displayed a negative correlation with modulus across all concentrations for both narrow and wide channels (Fig. 3 L). These results indicate that alignment improves the efficiency of cell migration, whereas increasing the matrix stiffness by increasing the collagen concentration serves to impede migration, possibly because it offers cells too many sites for cell adhesion (38,39) or because increased collagen concentration increases cell confinement by decreasing porosity (40).
To further understand how aligned collagen enhances migration, we tested the effects of alignment on migration speed and persistence in 1 and 4 mg/ml collagen. We imaged MDA-MB-231 cells seeded into narrow and wide channels every 10 min over 6 h using a confocal microscope, and tracked migrating cells using the MTrackJ plugin for FIJI. For these experiments, we only considered collagen concentrations of 1 and 4 mg/ml because these concentrations represent the extremes measured for the tensile modulus, and because the stiffness of an aligned 1 mg/ml gel was comparable to that of a random 4 mg/ml gel (Fig. 3 B). Time-lapse movies of cell migration in 1 mg/ml narrow and wide channels were used to quantify cell migration (Movie S1). Windrose plots of all of the analyzed tracks show a greater percentage of cells migrating within 10° of the direction of alignment in 1 and 4 mg/ml narrow channels (Fig. 4 A).
Figure 4.
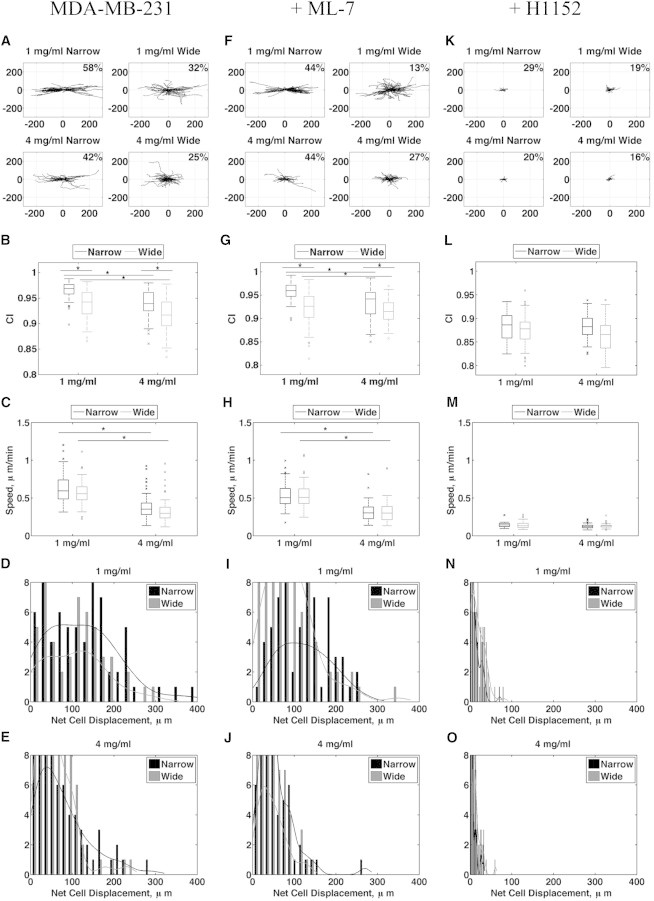
MDA-MB-231 cells migrate more slowly in stiff collagen and more directionally in aligned collagen. (A, F, and K) Windrose plots of all tracks analyzed during a 6 h confocal time lapse with a 10 min collection interval for untreated cells and cells treated with 2 μM ML-7 or 10 μM H1152 in narrow and wide channels (1 and 4 mg/ml). Percentages represent the fraction of cells with net track angles within 10° of the abscissa. (B, G, and L) Quantification of the CI for untreated and ML-7- and H1152-treated cells in narrow and wide channels (1 and 4 mg/ml, p < 0.01). (C, H, and M) Quantification of migration speed for untreated and ML-7- and H1152-treated cells in narrow and wide channels (1 and 4 mg/ml, n > 40, p < 0.01). (D, E, I, J, N, and O) Histograms of the cells’ net displacements over 6 h in 1 and 4 mg/ml for untreated and ML-7- and H1152-treated cells.
We used the chemotactic index (CI), a measure of a cell’s directional persistence that is traditionally used in reference to a chemotactic gradient, to measure the component of migration tracks in the direction of alignment, and found it to be highest in narrow channels (Fig. 4 B). Surprisingly, migration speed was unchanged between cells cultured in narrow and wide channels of the same collagen concentration (Fig. 4 C), suggesting that alignment does not affect cell speed. Moreover, cell speed was equivalently reduced at the higher collagen concentration despite the similar elastic moduli of 1 mg/ml aligned and 4 mg/ml random matrices (Fig. 3 B).
Histograms of individual cells’ net displacements demonstrate that compared with cells in wide channels, cells that migrated in narrow channels more often ended up farther away from their starting points (Fig. 4, D and E). Although the channel widths differed between narrow and wide channels, the enhanced persistence seen in narrow channels is unlikely to result from the smaller geometry. If this were true, and the dimensions of the narrow channels restricted migration to a more confined region, we would expect to see preferential migration along the edges of the matrix near the channel wall where restriction is highest. In fact, we saw cells evenly distributed across the channel width, suggesting that changing channel geometry does not impact cell migration. Taken together, these results suggest that collagen alignment affects the directional persistence, but not the speed, of migrating cells.
To deduce a potential mechanism for cells’ enhanced directional response to alignment, we asked whether Rho-mediated cell contractility plays a role in sensing alignment. We previously showed that cells require Rho and its effector, ROCK, to align collagen fibers, but once alignment is established, migration ensues independently of Rho and ROCK (23). To test whether cells require contractility to migrate in aligned collagen, we conducted time-lapse experiments similar to those shown in Fig. 4, A–E, but added to the culture media either H1152 to inhibit ROCK, or ML-7 to inhibit myosin light chain kinase (MLCK) 30 min before imaging. Inhibition of either ROCK or MLCK in MDA-MB-231 cells resulted in reduced levels of phosphorylated threonine-18 and serine-19 on MLC (data not shown), whereas ML-7 had no effect on migration speed or persistence compared with untreated cells (Fig. 4, F–J). However, H1152 dramatically inhibited cell speed across all conditions tested, which further resulted in reduced persistence (Fig. 4, K–O). These results indicate that Rho- and ROCK-mediated signaling is required for cell migration in 3D collagen matrices, but signaling from MLCK is not.
Our previous measurements of stiffness were at the bulk-material level and were aimed at assessing the relative moduli of ECM networks. However, individual cells experience the moduli of individual fibers in the network on a microscale. Therefore, to make more direct comparisons between matrix stiffness and the observed migration in microchannels, we measured the displacements of individual collagen fibers adjacent to migrating cells. Collagen was labeled with fluorescein isothiocyanate (FITC) according to Baici et al. (41), mixed with unlabeled collagen at a ratio of 1:10, and polymerized in narrow and wide microchannels. In this way, collagen fibers could be visualized by FITC immunofluorescence. FITC-collagen fibers and MDA-MB-231 cells seeded in the channels were imaged every 4 min for a period of 2 h by confocal microscopy. Time-lapse movies in 1 mg/ml narrow and wide channels were used to quantify cell-induced collagen fiber displacement (Movie S2). The fiber displacements measured in narrow channels were significantly smaller than those in wide channels for both 1 and 4 mg/ml conditions (Fig. 5 A), providing further support for the notion that aligned collagen, even at the cellular scale, is stiffer than a random matrix. Furthermore, the trends in fiber displacement in each condition correlate well with the trends in measured tensile moduli in Fig. 3 B, with the greatest fiber displacement occurring in the condition with the lowest measured bulk modulus, and vice versa. Similar fiber displacements were observed in both 1 mg/ml narrow and 4 mg/ml wide channels, which also have similar moduli (Fig. 3 B).
Figure 5.
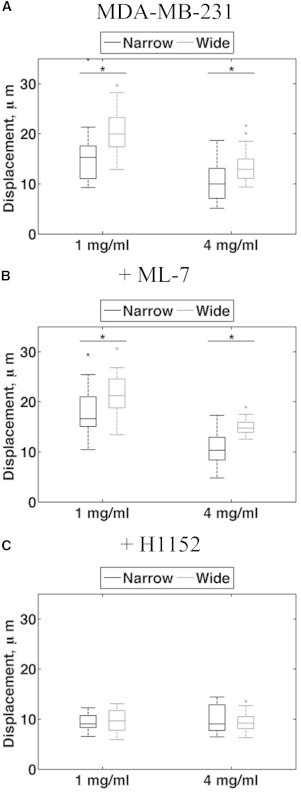
MDA-MB-231 cells displace collagen fibers in proportion to matrix stiffness. (A–C) Quantification of total collagen fiber displacement during a 2 h confocal time lapse with a 4 min collection interval for untreated and ML-7- and H1152-treated cells in narrow and wide channels (1 and 4 mg/ml, n > 15, p < 0.001).
We next determined whether the inhibition of ROCK or MLCK affects fiber displacement in addition to migration. Consistent with the results shown in Fig. 5, inhibition of MLCK with ML-7 had no effect on total fiber displacement (Fig. 5 B), whereas inhibition of ROCK with H1152 reduced fiber displacement in all conditions (Fig. 5 C). These results further suggest that ROCK, but not MLCK, is required for cells to displace the matrix and subsequently migrate.
These data suggest a complex interplay among alignment, stiffness, and ligand density in affecting cell migration. To better understand this interplay, we developed a simple mathematical model to make qualitative assessments of motility given certain matrix properties, such as alignment, stiffness, and ligand density. Our aim was to address the inherent difficulty of changing a single matrix property without affecting another one, and to shed additional light on the findings discussed herein. Our model assumed that cell speed in 3D is a bimodal function of both modulus and ligand density. This assumption was based on experimental results obtained with collagen-coated 2D polyacrylamide gels in which the modulus was altered when the ligand density was held constant or in which the ligand density was altered while the modulus was held constant (Fig. 6, A and B). This bimodal relationship is consistent with other reports of 2D cell migration (38,39,42,43). Additionally, for simplicity, we modeled cell movements in 2D, since cell displacement in 3D was measured in the x-y plane by confocal microscopy. We computed cell displacements by assigning random protrusion vectors at discrete time steps for a total of 36 time steps, corresponding to the experimental conditions under which we observed migration every 10 min for 6 h (also a total of 36 time steps). We defined coefficients to describe ligand density, stiffness, and alignment, and used these coefficients to scale the relative magnitudes of the x and y components of the protrusion vectors. A vector sum of the protrusion vectors gave the cell’s net displacement at that time point. Simulations of 100 cells were conducted in MATLAB (The MathWorks, Natick, MA) to make qualitative predictions of the trends in cell migration in 3D collagen environments within a known range of fiber alignments, ligand densities, and moduli. For further discussion about the model’s assumptions and equations, see Supplemental Materials and Methods, Fig. S2 and Table S1.
Figure 6.
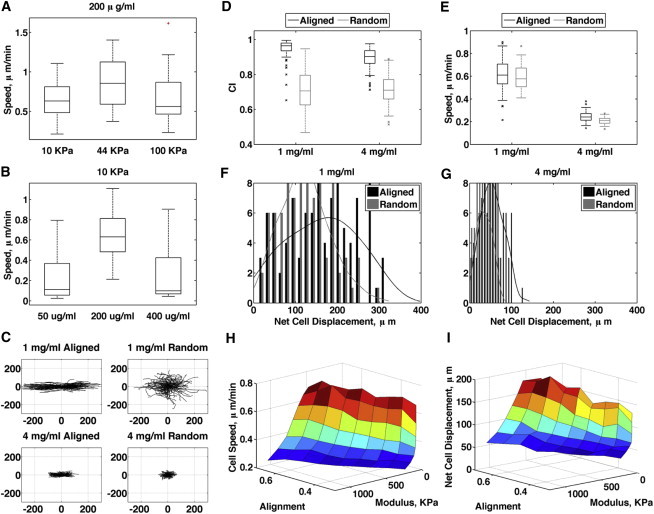
Computational model of cell migration replicates migration in 3D collagen. (A and B) 2D migration of MDA-MB-231 cells on polyacrylamide gels. (C) Windrose plots of simulated migration in 1 and 4 mg/ml aligned and random collagen from model. (D and E) Computed CI (D) and speed (E) from the model. (F and G) Histograms of computed cell displacements in 1 mg/ml (F) and 4 mg/ml (G) from the model. (H and I) Surface plot of cell speed (H) and net cell displacement (I) as a function of modulus and alignment.
From the simulations, we generated windrose plots of all cell simulations in 1 and 4 mg/ml aligned and random matrices, and found agreement in the distribution of tracks compared with experimental results (Fig. 6 C). Simulations showing the trends in cell speed, CI, and net displacement (Fig. 6, D–G) also correspond with the data in Fig. 4. To show trends in migration over a broader range of untested matrix conditions, we varied stiffness, ligand density, and alignment in the model, and found that migration speed was unaffected by changes in alignment (Fig. 6 H), consistent with the observed data. Increasing alignment, however, increased the net distance cells traveled (Fig. 6 I), suggesting that an increase in persistence is the main factor that determines enhanced migration due to aligned collagen in invasive breast tumors. The model best fits the data when the number of protrusions per cell is decreased as alignment increases (Fig. S2 E), which leads to the prediction that alignment enhances persistence by limiting the number of protrusions.
To better understand the mechanism of enhanced persistence in aligned collagen, and to validate our model assumption that the number of protrusions changes with the organization of the matrix, we conducted time-lapse experiments to observe protrusion dynamics. To visualize actin-rich cellular protrusions, we generated stable MDA-MB-231 cells expressing Lifeact-mRFP, which was previously used to label filamentous actin in live cells (44). Confocal imaging showed that cells in aligned collagen elongated in the direction of alignment and with fewer peripheral protrusions compared with cells in a random matrix (Fig. 7 A). Time-lapse imaging also showed that elongated cells with prominent protrusions in the direction of alignment were maintained throughout the duration of the experiment, whereas cells in randomly organized collagen had less stable protrusions (Movie S3). In addition, we also observed in some cells the formation of membrane blebs similar to those previously associated with lobopodial migration 1D adhesive surfaces (45). We wrote and used custom MATLAB code to quantify the number and length of protrusions per cell at each time point (for further description, see Supplemental Materials and Methods). We found that cells in aligned collagen had fewer protrusions overall (Fig. 7 B), which is consistent with the prediction of the model. Moreover, as the model scaled the length of protrusions by the magnitude of the protrusion vector, our finding that the lengths of individual protrusions relative to the cell centroid were greater compared with those of cells cultured in randomly organized collagen (Fig. 7 C) is also consistent with the predictions of the model. These data indicate that aligned collagen restricts protrusions to the direction of alignment, potentially stabilizing protrusions in that direction and allowing cells to maintain greater persistence in aligned collagen.
Figure 7.
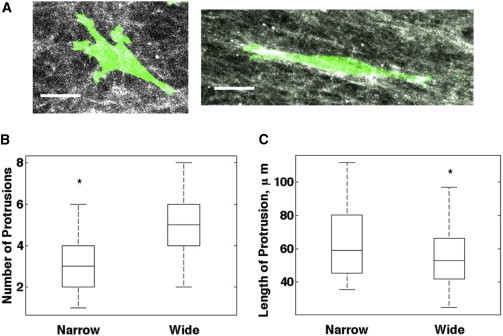
Cells in aligned collagen have fewer and longer protrusions. (A) Confocal z-compressed images of MDA-MB-231 cells expressing Lifeact-mRFP in 1 mg/ml wide (left) and narrow (right) channels. Grayscale, FITC-labeled collagen; green, Lifeact-mRFP. Scale bar: 20 μm. (B and C) Quantification of the number of protrusions per cell (B) and length of protrusion (C) relative to centroid (n = 8 cells, p < 0.0001).
Discussion
Mammographic breast density and the accompanying stiffness are associated with increased breast cancer incidence (1,4,8), but do not correlate with distal relapse (46). In contrast, collagen alignment is associated with enhanced metastasis in animal models and poor prognosis in breast cancer patients (20,22). Despite the observation that migrating carcinoma cells track along collagen fibers in vitro and in vivo (20,47–49), the mechanism by which cells recognize and migrate along fibers is poorly understood. In this work, we sought to understand the effects of matrix stiffness and alignment on migrating epithelial cells within a 3D collagen matrix. We employed a coordinated analysis of both cell migration and tensile moduli of aligned and randomly organized matrices of varying stiffness. Consistent with our previous observations (23), we find that matrix alignment is a strong promoter of cell migration and results in a greater net distance traveled by cells. Surprisingly, matrix alignment does not enhance migration speed; rather, alignment enhances migrational persistence. Cells in aligned collagen have fewer and longer protrusions that are oriented with respect to aligned fibers, suggesting that limiting protrusions is the mechanism by which alignment serves to increase persistence.
Although it is well known that collagen exhibits strain-stiffening behavior, it is less understood how migrating cells respond to matrices of different fiber orientations and stiffness. Therefore, we characterized the mechanical effects of prestraining a collagen matrix to induce fiber alignment. Our results show that collagen prealigned by strain is stiffer than a random collagen matrix of the same density and that the stiffness is greatest along the axis of alignment. Xu et al. (50) previously conducted biaxial tensile testing of collagen prealigned by flow of magnetic beads and showed that after fixation, gels aligned parallel to the direction of alignment were stiffer than gels aligned orthogonally. Here, we obtained similar findings utilizing uniaxial tensile testing and a simpler, more robust approach to induce prealignment by strain rather than flow of magnetic beads embedded in the matrix. Moreover, we found that the increased stiffness of aligned gels is preserved even in unfixed collagen gels.
Our results further suggest that alignment of collagen fibers potentially strengthens and stabilizes interactions between neighboring fibers, presumably causing the observed increase in modulus. The act of stretching a collagen matrix not only results in increased fiber proximity but can also increase subfibrillar orientation and packing (51). These stretch-induced modifications may allow for enhanced molecular interactions between adjacent fibrils (52), thus increasing the overall matrix stiffness. Similarly, we expect that the flow-induced fiber alignment in our microchannels potentially enhances interactions between adjacent fibers such that alignment, rather than the method by which the alignment is induced, is the key feature.
To study cell migration in aligned matrices, we developed an assay that makes use of the observation made by Lee et al. (36) and Sung et al. (37) that collagen alignment can be controlled by flow through a sufficiently narrow microchannel. To our knowledge, this assay represents a novel approach for studying the effects of 3D collagen alignment on cell migration. Here, we used channels 1 mm and 3 mm wide to produce highly aligned collagen matrices and a randomly organized matrix, respectively, into which we seeded cells and quantified total cell migration, speed, and persistence. We found that more cells migrated farther into the aligned collagen, but migration speed was unchanged relative to migration in the randomly organized collagen matrix.
The increase in the total number of migrating cells in the aligned matrix can be accounted for by the marked increase in the persistence of cells in aligned collagen. Furthermore, our results demonstrate that the effects of collagen alignment on persistence are not due to the increased stiffness of an aligned matrix, or durotaxis, because cells migrated faster and were more persistent in 1 mg/ml aligned collagen than in 4 mg/ml random matrix, even though the two conditions had nearly the same stiffness. In contrast, we found that when increased matrix stiffness was generated by increased collagen concentration, which also increased the ligand density and reduced the porosity of the matrix, cell speed was reduced. This result is in contrast to the well-documented durotactic response of cells cultured on 2D substrates (14) and may reflect the greater inherent complexity of 3D matrices. Importantly, the mechanical properties extrapolated from measurements of fiber displacement correlate well with the measured bulk tensile moduli of aligned and random gels, suggesting that the differences in the mechanical properties of aligned and random collagen gels at the microscale correlate to the macroscale. Moreover, this finding demonstrates that increased stiffness along the axis of alignment is observed regardless of the mechanism of alignment (flow or strain).
We further show that migration is dependent on Rho/ROCK-generated contractility and independent of MLCK. Previously, we demonstrated that cells first require a contractile mechanism involving Rho and ROCK to generate aligned collagen, and that the requirement for Rho/ROCK ceases for migration in a prealigned collagen matrix (23). One possible explanation for this discrepancy is the difference in the duration of inhibitor treatment between the two studies (8 h in this study compared with every 24 h for a period of 3 days in the previous study), suggesting that compensation for Rho inhibition may occur over longer treatment time frames. As in our previous study, we again find that fiber displacement is also dependent on ROCK, not MLCK, which agrees with other reports of matrix deformation by contractile forces (53–55). These results provide additional support for the idea proposed by Grinnell et al. (56) and Miron-Mendoza et al. (57) that cell migration and matrix deformation are both consequences of the same Rho- and ROCK-driven contractile mechanism and could occur in concert. Matrix stiffness likely determines the extent of fiber deformation as well as the amount of traction a migrating cell can generate by utilizing Rho-mediated contractility.
From our results, we infer that the collagen fibers that are most constrained in the matrix move the least when pulled and exhibit the greatest stiffness. This finding is also supported by the observation of Lopez-Garcia et al. (58) that NMuMG cells produce larger strains in matrices of lower stiffness. These results lead to the assumption that stiffer conditions may provide the most traction and thus allow cells to use their contractile forces to achieve the fastest migration. Consistent with this idea, previous studies showed that stiffness increases the amount of force adhesions generate, and force-generating active Rho is upregulated in stiff matrices (10,53,59). These findings on their own would predict that increased stiffness in 3D would lead to enhanced migration speeds. However, increasing stiffness in 3D matrices is also typically accompanied by increased ligand density and decreased matrix porosity, both of which are known to significantly impact migration speed (39,40). For this reason, it has often been challenging to develop techniques that allow one to separate stiffness from other factors in a 3D environment to assess changes in migratory behavior. Recently, however, Harley et al. (60) were able to alter the individual strut stiffness of a collagen-GAG scaffold, and identified a bimodal relationship between stiffness and cell migration speed similar to that seen on 2D substrates. As new methods for engineering matrices with precisely controlled physical and mechanical properties become available, we will be able to study additional relationships involving cell speed and persistence.
To address the multifactorial responses to the topographical and mechanical properties of the matrix, we developed a mathematical model that incorporates matrix ligand density, stiffness, and alignment. Our model allows us to better understand the trends over the range of experimental conditions tested, where it is difficult to deduce effects incurred by isolating a single 3D matrix parameter. Others have proposed a model to describe the effect of fibroblast migration speed on the alignment of fibrin and collagen in dermal wound healing (61). Our model considers protrusion generation as a stochastic process that leads to adhesion formation with ECM fibers, whereas other groups have used models to suggest that in vivo migration in discontinuous environments may not require adhesions (62). Our model predicts that the number of protrusions diminish with increased alignment and increase as ligand density increases. We show experimentally that cells in aligned collagen indeed have fewer protrusions, which is likely responsible for this observed increase in persistence. To better replicate cell persistence, the model incorporates a probability for a protrusion vector to maintain its orientation across multiple time steps that increases as a function of matrix alignment. Although we are unsure of the exact cellular mechanism of persistence, the predictions of our model would be consistent with the hypothesis that persistence reflects the presentation of ligands along an axis allowing cells to form and stabilize adhesions in a given direction, enabling more efficient migration. This notion is consistent with the observation that integrin adhesions are localized along collagen fibers in 3D (63) and on 1D collagen fibers, where cell migration along 1D fibers recapitulates migration in 3D matrices (64). Moreover, we expect that persistence also reflects the loss of competing ligand binding choices in other directions. We predict that as ligand concentration increases, there will be more opportunities for adhesion-stabilized protrusions to occur outside the direction of alignment, consistent with the observation that persistence decreases as ligand concentration increases.
A limitation of our model is that, for simplicity, it assumes all matrix parameters are held constant over the simulated time frame, when in reality cells are constantly modifying their environment via Rho/ROCK contractility, which causes local changes in alignment, stiffness, and ligand density. Moreover, there are additional effects of proteolytic modification of collagen that we have not taken into account (63). Furthermore, the model input parameters require tested modulus values for given alignments and ligand densities, which can also change with different matrix components and the extent of cross-linking. Once we are able to measure the stiffness of a broader range of matrix compositions and more reliably establish the relationships among matrix composition, alignment, and stiffness, we can revise the model parameters. Despite these limitations, however, we show that our model can accurately replicate trends in migration speed and persistence in 1 mg/ml and 4 mg/ml aligned and random collagen matrices. Furthermore, the model is consistent with the observation that migration speed across a range of matrix conditions is unaffected by alignment, but alignment profoundly increases the net distance cells travel. This finding is significant in the context of breast tumor progression, where the presence of TACS-3 aligned fibers is correlated with a dramatic increase in metastases and poor long-term survival (21,22). Our results suggest that highly aligned regions in the tumor microenvironment can provide cells that have a high migratory capacity with the most reliable and robust escape route, and provide additional mechanistic understanding of how the aligned collagen characterized by TACS-3 relates to its role as a potential biomarker for breast cancer.
Acknowledgments
The authors thank Drs. Kyung Sung and David Beebe for assistance with microfluidic channels, Dr. Maddy Parsons for generously supplying the Lifeact-mRFP construct, and Curtis Rueden for assistance with the FIJI software and analysis.
This work was funded by NIH grants U01CA143069 to A.W., Y.J., and P.J.K., and R01 CA142833 and R01 CA114462 to P.J.K. and supported by the University of Wisconsin-Madison Graduate School for M.R.S. and W.C.C.
Supporting Material
Six-hour confocal time-lapse images of MDA-MB-231 cells migrating in wide (top panels) and narrow (bottom panels) microchannels. DIC images (left panels) and fluorescence images of FITC-labeled collagen fibers (right panels) were collected at 10 min intervals. Scale bar, 500 μm.
Two-hour confocal time-lapse images of MDA-MB-231 cells displacing collagen fibers in wide (top panels) and narrow (bottom panels) microchannels. DIC images (left panels) and fluorescence images of FITC-labeled collagen fibers (right panels) were collected at 4-min intervals. Scale bar, 200 μm.
Forty-minute confocal time-lapse images of MDA-MB-231 cells expressing Lifeact-mRFP to visualize protrusions in narrow (top) and wide (bottom) microchannels. Scale bar, 20 μm.
Supporting Citations
References (65–74) appear in the Supporting Material.
References
- 1.Boyd N.F., Lockwood G.A., Yaffe M.J. Mammographic densities and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 1998;7:1133–1144. [PubMed] [Google Scholar]
- 2.Boyd N.F., Martin L.J., Yaffe M.J. Mammographic densities as a marker of human breast cancer risk and their use in chemoprevention. Curr. Oncol. Rep. 2001;3:314–321. doi: 10.1007/s11912-001-0083-7. [DOI] [PubMed] [Google Scholar]
- 3.McCormack V.A., dos Santos Silva I. Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 2006;15:1159–1169. doi: 10.1158/1055-9965.EPI-06-0034. [DOI] [PubMed] [Google Scholar]
- 4.Boyd N.F., Dite G.S., Hopper J.L. Heritability of mammographic density, a risk factor for breast cancer. N. Engl. J. Med. 2002;347:886–894. doi: 10.1056/NEJMoa013390. [DOI] [PubMed] [Google Scholar]
- 5.Alowami S., Troup S., Watson P.H. Mammographic density is related to stroma and stromal proteoglycan expression. Breast Cancer Res. 2003;5:R129–R135. doi: 10.1186/bcr622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T., Sun L., Boyd N. The association of measured breast tissue characteristics with mammographic density and other risk factors for breast cancer. Cancer Epidemiol. Biomarkers Prev. 2005;14:343–349. doi: 10.1158/1055-9965.EPI-04-0490. [DOI] [PubMed] [Google Scholar]
- 7.Guo Y.P., Martin L.J., Boyd N.F. Growth factors and stromal matrix proteins associated with mammographic densities. Cancer Epidemiol. Biomarkers Prev. 2001;10:243–248. [PubMed] [Google Scholar]
- 8.Paszek M.J., Zahir N., Weaver V.M. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Lopez J.I., Kang I., Weaver V.M. In situ force mapping of mammary gland transformation. Integr. Biol. (Camb.) 2011;3:910–921. doi: 10.1039/c1ib00043h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Provenzano P.P., Inman D.R., Keely P.J. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tilghman R.W., Cowan C.R., Parsons J.T. Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS ONE. 2010;5:e12905. doi: 10.1371/journal.pone.0012905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander N.R., Branch K.M., Weaver A.M. Extracellular matrix rigidity promotes invadopodia activity. Curr. Biol. 2008;18:1295–1299. doi: 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo C.M., Wang H.B., Wang Y.L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelham R.J., Jr., Wang Yl. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menon S., Beningo K.A. Cancer cell invasion is enhanced by applied mechanical stimulation. PLoS ONE. 2011;6:e17277. doi: 10.1371/journal.pone.0017277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostic A., Lynch C.D., Sheetz M.P. Differential matrix rigidity response in breast cancer cell lines correlates with the tissue tropism. PLoS ONE. 2009;4:e6361. doi: 10.1371/journal.pone.0006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wozniak M.A., Modzelewska K., Keely P.J. Focal adhesion regulation of cell behavior. Biochim. Biophys. Acta. 2004;1692:103–119. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Klein E.A., Yin L., Assoian R.K. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr. Biol. 2009;19:1511–1518. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulrich T.A., de Juan Pardo E.M., Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Provenzano P.P., Eliceiri K.W., Keely P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Provenzano P.P., Inman D.R., Keely P.J. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conklin M.W., Eickhoff J.C., Keely P.J. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Provenzano P.P., Inman D.R., Keely P.J. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys. J. 2008;95:5374–5384. doi: 10.1529/biophysj.108.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim A., Lakshman N., Petroll W.M. Quantitative assessment of local collagen matrix remodeling in 3-D culture: the role of Rho kinase. Exp. Cell Res. 2006;312:3683–3692. doi: 10.1016/j.yexcr.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brownfield D.G., Venugopalan G., Bissell M.J. Patterned collagen fibers orient branching mammary epithelium through distinct signaling modules. Curr. Biol. 2013;23:703–709. doi: 10.1016/j.cub.2013.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sander E.A., Barocas V.H., Tranquillo R.T. Initial fiber alignment pattern alters extracellular matrix synthesis in fibroblast-populated fibrin gel cruciforms and correlates with predicted tension. Ann. Biomed. Eng. 2011;39:714–729. doi: 10.1007/s10439-010-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J.H., Jia F., Woo S.L. Cell orientation determines the alignment of cell-produced collagenous matrix. J. Biomech. 2003;36:97–102. doi: 10.1016/s0021-9290(02)00233-6. [DOI] [PubMed] [Google Scholar]
- 28.Yang N., Mosher R., Friedl A. Syndecan-1 in breast cancer stroma fibroblasts regulates extracellular matrix fiber organization and carcinoma cell motility. Am. J. Pathol. 2011;178:325–335. doi: 10.1016/j.ajpath.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruschwitz R., Friedrichs J., Engelmann K. Alignment and cell-matrix interactions of human corneal endothelial cells on nanostructured collagen type I matrices. Invest. Ophthalmol. Vis. Sci. 2010;51:6303–6310. doi: 10.1167/iovs.10-5368. [DOI] [PubMed] [Google Scholar]
- 30.Vernon R.B., Gooden M.D., Wight T.N. Microgrooved fibrillar collagen membranes as scaffolds for cell support and alignment. Biomaterials. 2005;26:3131–3140. doi: 10.1016/j.biomaterials.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Kim D.H., Provenzano P.P., Levchenko A. Matrix nanotopography as a regulator of cell function. J. Cell Biol. 2012;197:351–360. doi: 10.1083/jcb.201108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai E.S., Huang N.F., Fuller G.G. Aligned nanofibrillar collagen regulates endothelial organization and migration. Regen. Med. 2012;7:649–661. doi: 10.2217/rme.12.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedl P., Wolf K. Tube travel: the role of proteases in individual and collective cancer cell invasion. Cancer Res. 2008;68:7247–7249. doi: 10.1158/0008-5472.CAN-08-0784. [DOI] [PubMed] [Google Scholar]
- 34.Vader D., Kabla A., Mahadevan L. Strain-induced alignment in collagen gels. PLoS ONE. 2009;4:e5902. doi: 10.1371/journal.pone.0005902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roeder B.A., Kokini K., Voytik-Harbin S.L. Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J. Biomech. Eng. 2002;124:214–222. doi: 10.1115/1.1449904. [DOI] [PubMed] [Google Scholar]
- 36.Lee P., Lin R., Lee L.P. Microfluidic alignment of collagen fibers for in vitro cell culture. Biomed. Microdevices. 2006;8:35–41. doi: 10.1007/s10544-006-6380-z. [DOI] [PubMed] [Google Scholar]
- 37.Sung K.E., Su G., Beebe D.J. Control of 3-dimensional collagen matrix polymerization for reproducible human mammary fibroblast cell culture in microfluidic devices. Biomaterials. 2009;30:4833–4841. doi: 10.1016/j.biomaterials.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palecek S.P., Loftus J.C., Horwitz A.F. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 39.Zaman M.H., Trapani L.M., Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolf K., Te Lindert M., Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baici A., Cohen G., Böni A. A handy assay for collagenase using reconstituted fluorescein-labeled collagen fibrils. Anal. Biochem. 1980;108:230–232. doi: 10.1016/0003-2697(80)90574-6. [DOI] [PubMed] [Google Scholar]
- 42.Wu P., Hoying J.B., Lauffenburger D.A. Integrin-binding peptide in solution inhibits or enhances endothelial cell migration, predictably from cell adhesion. Ann. Biomed. Eng. 1994;22:144–152. doi: 10.1007/BF02390372. [DOI] [PubMed] [Google Scholar]
- 43.Peyton S.R., Putnam A.J. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell. Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 44.Riedl J., Crevenna A.H., Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrie R.J., Gavara N., Yamada K.M. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 2012;197:439–455. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park C.C., Rembert J., Kerlikowske K. High mammographic breast density is independent predictor of local but not distant recurrence after lumpectomy and radiotherapy for invasive breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 2009;73:75–79. doi: 10.1016/j.ijrobp.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 47.Wang W., Wyckoff J.B., Condeelis J.S. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62:6278–6288. [PubMed] [Google Scholar]
- 48.Dickinson R.B., Guido S., Tranquillo R.T. Biased cell migration of fibroblasts exhibiting contact guidance in oriented collagen gels. Ann. Biomed. Eng. 1994;22:342–356. doi: 10.1007/BF02368241. [DOI] [PubMed] [Google Scholar]
- 49.Petrie R.J., Doyle A.D., Yamada K.M. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu B., Chow M.J., Zhang Y. Experimental and modeling study of collagen scaffolds with the effects of crosslinking and fiber alignment. Int. J. Biomater. 2011;2011:172389. doi: 10.1155/2011/172389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pins G.D., Christiansen D.L., Silver F.H. Self-assembly of collagen fibers. Influence of fibrillar alignment and decorin on mechanical properties. Biophys. J. 1997;73:2164–2172. doi: 10.1016/S0006-3495(97)78247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freeman J.W., Silver F.H. The effects of prestrain and collagen fibril alignment on in vitro mineralization of self-assembled collagen fibers. Connect. Tissue Res. 2005;46:107–115. doi: 10.1080/03008200590954140. [DOI] [PubMed] [Google Scholar]
- 53.Wozniak M.A., Desai R., Keely P.J. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol. 2003;163:583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyckoff J.B., Pinner S.E., Gschmeissner S., Condeelis J.S., Sahai E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. Aug 8. 2006;16(15):1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 55.Lakshman N., Kim A., Petroll W.M. Rho plays a central role in regulating local cell-matrix mechanical interactions in 3D culture. Cell Motil. Cytoskeleton. 2007;64:434–445. doi: 10.1002/cm.20194. [DOI] [PubMed] [Google Scholar]
- 56.Grinnell F., Rocha L.B., Jiang H. Nested collagen matrices: a new model to study migration of human fibroblast populations in three dimensions. Exp. Cell Res. 2006;312:86–94. doi: 10.1016/j.yexcr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Miron-Mendoza M., Seemann J., Grinnell F. Collagen fibril flow and tissue translocation coupled to fibroblast migration in 3D collagen matrices. Mol. Biol. Cell. 2008;19:2051–2058. doi: 10.1091/mbc.E07-09-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopez-Garcia M.D., Beebe D.J., Crone W.C. Mechanical interactions of mouse mammary gland cells with collagen in a three-dimensional construct. Ann. Biomed. Eng. 2010;38:2485–2498. doi: 10.1007/s10439-010-0015-5. [DOI] [PubMed] [Google Scholar]
- 59.Bhadriraju K., Yang M., Chen C.S. Activation of ROCK by RhoA is regulated by cell adhesion, shape, and cytoskeletal tension. Exp. Cell Res. 2007;313:3616–3623. doi: 10.1016/j.yexcr.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harley B.A., Kim H.D., Gibson L.J. Microarchitecture of three-dimensional scaffolds influences cell migration behavior via junction interactions. Biophys. J. 2008;95:4013–4024. doi: 10.1529/biophysj.107.122598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dallon J.C., Sherratt J.A., Maini P.K. Modeling the effects of transforming growth factor-beta on extracellular matrix alignment in dermal wound repair. Wound Repair Regen. 2001;9:278–286. doi: 10.1046/j.1524-475x.2001.00278.x. [DOI] [PubMed] [Google Scholar]
- 62.Tozluoğlu M., Tournier A.L., Sahai E. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat. Cell Biol. 2013;15:751–762. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- 63.Wolf K., Wu Y.I., Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 64.Doyle A.D., Wang F.W., Yamada K.M. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heck J.N., Ponik S.M., Keely P.J. Microtubules regulate GEF-H1 in response to extracellular matrix stiffness. Mol. Biol. Cell. 2012;23:2583–2592. doi: 10.1091/mbc.E11-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chua Chee K. Three-dimensional rapid prototyping technologies and key development areas. Comput. Contr. Eng. J. 1994;5:200–206. [Google Scholar]
- 67.Wozniak M.A., Keely P.J. Use of three-dimensional collagen gels to study mechanotransduction in T47D breast epithelial cells. Biol. Proced. Online. 2005;7:144–161. doi: 10.1251/bpo112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez-Garcia M.D., Beebe D.J., Crone W.C. Young’s modulus of collagen at slow displacement rates. Biomed. Mater. Eng. 2010;20:361–369. doi: 10.3233/BME-2010-0649. [DOI] [PubMed] [Google Scholar]
- 69.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeung T., Georges P.C., Janmey P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 71.Aratyn-Schaus Y., Oakes P.W., Gardel M.L. Preparation of complaint matrices for quantifying cellular contraction. J. Vis. Exp. 2010;(46) doi: 10.3791/2173. pii: 2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schoen I., Pruitt B.L., Vogel V. The yin-yang of rigidity sensing: how forces and mechanical properties regulate the cellular response to materials. Annu. Rev. Mater. Res. 2013;43:589–618. [Google Scholar]
- 73.Smith A.S., Sengupta K., Sackmann E. Force-induced growth of adhesion domains is controlled by receptor mobility. Proc. Natl. Acad. Sci. USA. 2008;105:6906–6911. doi: 10.1073/pnas.0801706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huttenlocher A., Horwitz A.R. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Six-hour confocal time-lapse images of MDA-MB-231 cells migrating in wide (top panels) and narrow (bottom panels) microchannels. DIC images (left panels) and fluorescence images of FITC-labeled collagen fibers (right panels) were collected at 10 min intervals. Scale bar, 500 μm.
Two-hour confocal time-lapse images of MDA-MB-231 cells displacing collagen fibers in wide (top panels) and narrow (bottom panels) microchannels. DIC images (left panels) and fluorescence images of FITC-labeled collagen fibers (right panels) were collected at 4-min intervals. Scale bar, 200 μm.
Forty-minute confocal time-lapse images of MDA-MB-231 cells expressing Lifeact-mRFP to visualize protrusions in narrow (top) and wide (bottom) microchannels. Scale bar, 20 μm.


