Abstract
Background:
Clostridium difficile (C. difficile) is a frequent cause of nosocomial infections. During last few years, the mortality rate of C. difficile infection (CDI) increased in healthcare facilities. This organism has become a major public health concern in developed nations. Because of the increasing incidence of acquired-CDI (CA-CDI) and notable genetic overlap between C. difficile isolates from animals and humans, meat has defined as one of the probable transmission route of C. diffiicle to humans.
Objectives:
This study was performed to determine the prevalence of toxigenic C. difficile in beef and mutton meats consumed as human food in Isfahan, central part of Iran. Furthermore the polymerase chain reaction (PCR)-ribotyping employed to compare the genetic pattern of positive isolates in meat with clinical ones.
Materials and Methods:
A total of 200 raw meat samples (81 beef and 119 mutton) were purchased from meat packaging plants. The samples were anaerobically cultured in C. difficile moxalactam norfloxacin (CDMN) broth and plated on selective enrichment medium. The suspicious colonies were recultured on blood agar anaerobically. All C. difficile isolates identified by morphological and biochemical testing were screened by PCR for the presence of genes encoding the triose phosphate isomerase (tpi), toxin A (tcdA), toxin B (tcdB) and binary toxin (cdtB). The genomes of extracted isolates were analyzed by 16S-23S rRNA-based PCR ribotyping.
Results:
The overall prevalence of C. difficile with two toxigenic genes including tcdA and tcdB was estimated at 4.0%. C. difficile was detected in 2.8%, 2.1%, 3.6% and 6.2% of chopped beef, ground beef, chopped mutton and ground mutton, respectively. The cdtB gene was not found in positive isolates. Eight different ribotypes were found in isolated strains that were not identical with those belonging to patients with CDI.
Conclusions:
The results of PCR-ribotyping indicate that no relationship exists between clinical and meat isolates. We therefore conclude that other sources than meat may function as a vector for CA-CDI.
Keywords: Clostridium difficile, Meat, PCR, Iran
1. Background
Clostridium difficile is a Gram-positive, spore-forming and obligate anaerobic bacterium (1). CDI was considered a cause for symptoms of infectious diarrhea in patients after hospitalization and antibiotic treatment back in the 1970's (2). The CDI associate with two virulence factors including toxin A (tcdA) and B (tcdB) (3). Some strains of this organism have the ability to produce another toxin called binary toxin (cdt) with a yet unclear role for CDI (4). C. difficile has become an important health concern due to its high mortality, particularly in the hospitalized individuals in developed nations (5). The epidemiology of CDI in humans have been changed as a consequence of the emergence and dissemination of new strains of C. difficile called ribotype 027 and 078. The infection occurred in young individuals and other patients that were at low risk. In fact, the major risk factors for CA-CDI were not related with advanced age, antibiotic therapy or duration of hospitalization (6).
The source and transmission of C. difficile to humans altered from healthcare facilities to the outside of these places. The researchers stated that the bacterium can be transmitted via food based on the recognition of CA-CDI, detection of C. difficile in food animals as well as on the genetic similarities observed between C. difficile isolates from food animals, food and humans (7). Some previous reports pointed out that meat can very likely be a vector of C. difficile to human (8-10). In Iran, C. difficile strains of ribotype 078 was identified as the most common isolate in diarrheic hospitalized patients tested in Isfahan region. The existence of this ribotype referred to the probable incidence of C. difficile in food and food animals in the community of Isfahan (11). However, the situation of C. difficile in raw meat for human consumption is poorly understood in Iran.
2. Objectives
The aim of this surveillance study was to assess the prevalence of toxigenic C. difficile in beef and mutton meats demanded from meat packaging plants by PCR in Isfahan, Central part of Iran. In addition, PCR-ribotyping of toxigenic isolates was investigated to determine whether CA-CDI might be transmitted from consumption of meat.
3. Materials and Methods
3.1. Sampling, Preparation and C. difficile Identification
A total of 200 samples including beef and mutton were randomly purchased from meat packaging plants during an 8-month period. The examined samples were consisted of 35 (17.5%) chopped and 46 (23%) ground beef, 55 (27.5%) chopped and 64 (32%) ground mutton. The samples were transferred to the Infectious Disease and Tropical Medicine Research Center in Isfahan University of Medical Sciences in portable insulated cold boxes and analyzed in the same day of collection. Around 5 grams of each sample was cultured in 25 mL of Clostridium difficile moxalactam norfloxacin (CDMN) (Oxoid SR0048) and fortified with C. difficile selective supplement (Oxoid, SR0173) including 500 mg cysteine hydrochloride, 12 mg norfloxacin and 35 mg moxalactam per liter. The samples were incubated anaerobically for 7 days at 37°C. Subsequently, 2 mL of enriched culture were added to 2 mL of 98% ethanol (Merck, Germany) and kept for 2 h at room temperature. The tubes were centrifuged for 10 min at 10000*g. The sediment was streaked onto CDMN agar and then incubated under anaerobic conditions at 37°C for 24-48 hours. All C. difficile isolates were confirmed by morphology and L-proline- aminopeptidase test (prodisk, hardy diagnostics, Santa Maria, USA). Positive strains were recultured on blood agar and incubated at 37°C for 36 h in anaerobic conditions (8, 12).
3.2. DNA Extraction
A full loop of C. difficile grown in blood agar was suspended in 100 µL of sterile distilled water, boiled at 95°C for 3 minutes and then centrifuged at 3000*g for 15 minutes. The supernatant was collected for use as a template for amplification reaction in a thermocycler PCR (T-cY, Netherlands) (13).
3.3. Multiplex Polymerase Chain Reaction for Detection of tpi, tcdA and tcdB
The volumes of the reactions for the detection of genes encoding tpi, tcdA, tcdB of extracted DNA were based on the method described previously (13). The multiplex PCR procedure was carried out under the following conditions: initial denaturation at 95°C for 3 minutes, denaturation at 95ºC for 30 seconds, annealing for 30 seconds at temperature decrease from 65 to 55°C in 11 cycles and extension at 72°C for 30 seconds. The PCR products were then assayed by electrophoresis on 1.5% agarose gel stained with ethidium bromide (13). All PCR materials were purchased from SinaClon BioScience Company, Iran.
3.4. Identification of cdtB by Polymerase Chain Reaction
All conditions including volumes and temperatures of the reactions for detection of cdtB were performed from the method described by Stubbs et al. (14).
3.5. Polymerase Chain Reaction -Ribotyping of C. difficile isolates
PCR-ribotyping reaction was performed in a total volume of 100 µL containing 200 µM of each dNTPs mix, 1.5 mM MgCl2, 2.5 U of Taq DNA polymerase, 50 µl of each primer, 10 mM Tris-HCl (pH 8.8), 50 mM KCl and 10 µL of DNA extract. Amplification programmed for 30 cycles consisting of 95°C for 6 minutes in initial denaturation, 92°C for 60 seconds in denaturation, 55°C for 60 seconds in annealing, 72°C for 6 minutes in extension steps. Amplicon product was loaded on 1.5% agarose gel for 6 hours at 80 V. Scanning by UV light was done after staining with ethidium bromide (15). The patterns of ribotype were visually compared with over 3000 isolates from humans and animals which gathered in the international collections of the University of Guelph, Canada. For ribotype pattern included in the collection, international name were choosen. Otherwise an internal nomenclature selected for unavailable ribotype patterns in the collections (11).
3.6. Bacteria
The strain of C. difficile ribotype 027 for microbiological analysis kindly obtained from the database of the University of Guelph, Canada.
3.7. Oligonucleotide Primers
The primers for the amplification of the tpi housekeeping, tcdA, tcdB and cdtB genes provided by Metabion International AG, Martinsried, Germany has listed in Table 1. The primer sequences of PCR-ribotyping were 5'-GTGCGGCTGGATCACCTCCT-3' and 5'-CCCTGCACCCTTAATAACTTGACC-3' for 16s and 23s, respectively (15).
Table 1. The Primers Used for Identification of Toxigenic C. difficle in This Study .
| Gene | Sequence (5'→3') | Product Size, bp | Reference |
|---|---|---|---|
| tpi | 230 | (13) | |
| Forward | AAAGAAGCTACTAAGGGTACAAA | ||
| Reverse | CATAATATTGGGTCTATTCCTAC | ||
| tcdA | 369 | (13) | |
| Forward | AGATTCCTATATTTACATGACAATAT | ||
| Reverse | GTATCAGGCATAAAGTAATATACTTT | ||
| tcdB | 160 | (13) | |
| Forward | GGAAAAGAGAATGGTTTTATTAA | ||
| Reverse | ATCTTTAGTTATAACTTTGACATCTTT | ||
| CdtB | 510 | (14) | |
| Forward | CTTAATGCAAGTAAATACTGAG | ||
| Reverse | AACGGATCTCTTGCTTCAGTC |
4. Results
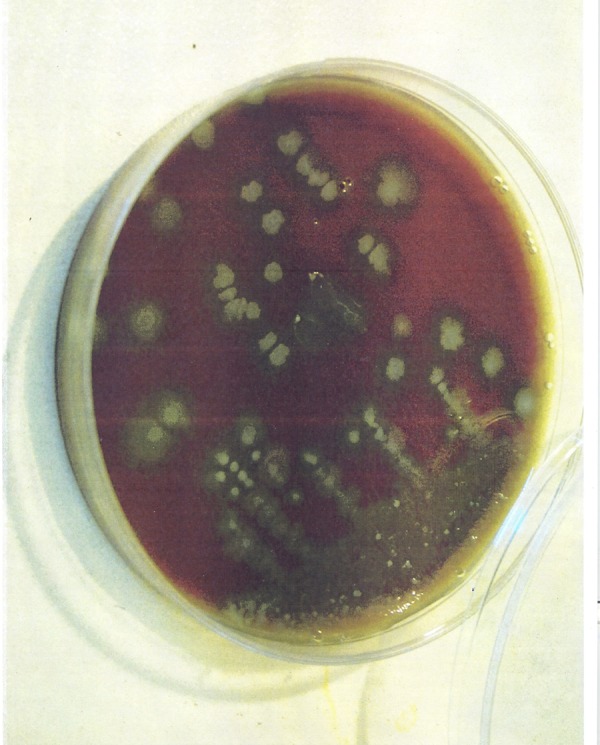
The morphology of C. difficile strains subcultured on CDMN agar is presented in Figure 1.
Figure 1. Irregular and Grey Colony with Opaque Appearance of C. difficile.
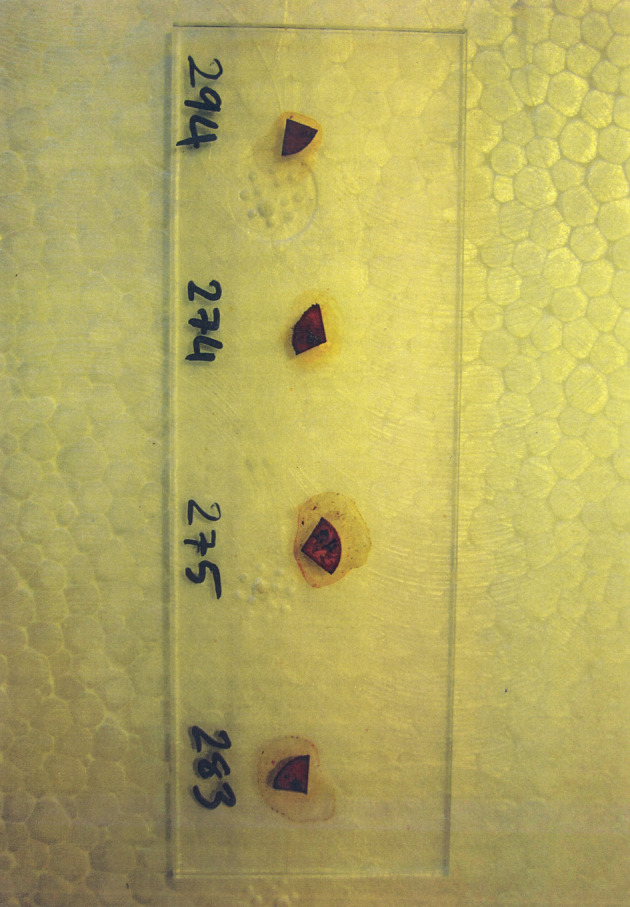
L-proline aminopeptidase test by the proline disk was used for the detection of C. difficile (Figure 2). This test is used to examine enzymatic hydrolysis of L-proline-β-naphthylamide extant in the disc based on releasing free β-naththylamine by the color changes after addition PEP (Para-dimethyl amino-cinnamaldehyle in a weak hydrochloric acid solution) reagent. The aforementioned test is approved especially useful in screening for C. difficile.
Figure 2. Proline Disk Staining of C. difficile From Meat Samples.
We isolated strain of C. difficile with tpi housekeeping gene (230 bp) and two toxigenic genes including tcdA (369 bp) and tcdB (160 bp) in 8 (4%) of the 200 samples of beef and mutton meats for human consumption as follows: one (2.8%) in the 35 chopped beef, one (2.1%) in the 46 ground beef, two (3.6%) in the 55 chopped mutton and four (6.2%) in the 64 ground mutton samples (Figure 3).
Figure 3. The Result of the Multiplex PCR for C. difficile in Agarose Gel Electrophoresis.
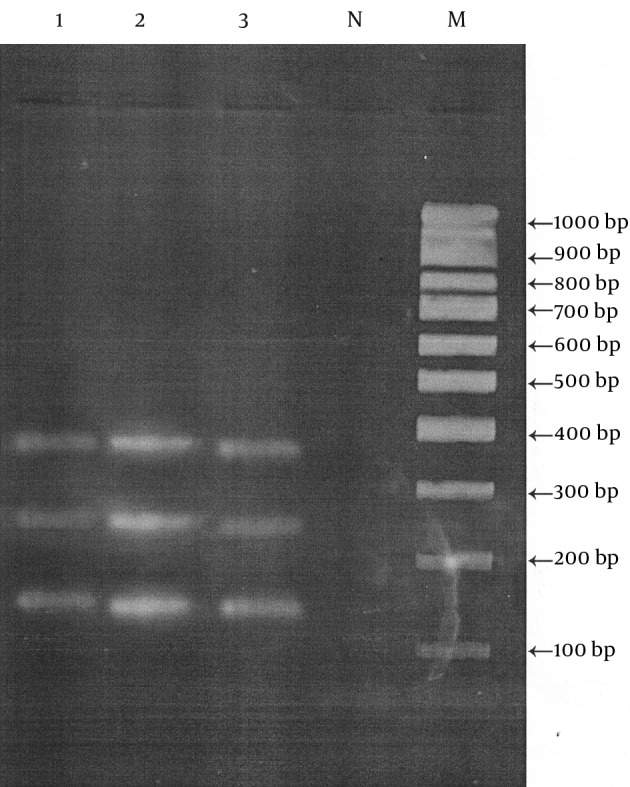
Lane M, marker 100-1000 bp; Lane N, negative control; Lane 1 and 3, tpi, tcdA and tcdB fragments of isolated C. difficile in meat samples; Lane 2, C. difficile strandard ribotypye 027.
All isolated colonies were identified in different carcasses collected in different dates. No cdtB gene was found in positive isolates of toxigenic C. difficile. In the present study, a series of eight different ribotypes were identified that genetic overlap were not observed with those belonging to animal and patients with CDI (Table 2).
Table 2. Ribotype Data for Isolates of C. difficile Strains From Beef and Mutton Samples.
| Sample Code | Meat Type | Ribotype |
|---|---|---|
| 1 | Chopped beef | IR11 |
| 2 | Ground beef | IR12 |
| 3 | Chopped mutton | IR13 |
| 4 | Chopped mutton | IR14 |
| 5 | Ground mutton | IR15 |
| 6 | Ground mutton | IR16 |
| 7 | Ground mutton | IR17 |
| 8 | Ground mutton | IR18 |
5. Discussion
The present study evaluated the prevalence of C. difficile in beef and mutton collected from the meat packaging plants in an attempt to show how the Isfahan community may be exposed to this organism by consumption of meat. The prevalence of C. difficile considered in our study was much lower than several of the previous studies. A relatively high incidence of C. difficile contamination was observed in the United States, where 37 (42%) of 88 different retailed meat products and raw meats were found to be contaminated with this organism (9). Another study in the United States showed C. difficile prevalence in four (8%) out of 50 retailed ground meat samples (16). C. difficile has also been detected in experiments run in Canada with an incidence of 20% (12 out of 60) meat samples analyzed by Rodriguez-Palacios et al. (8). Afterwards, the aforementioned authors identified C. difficile in 13 (6.1%) out of 214 samples of ground and chopped beef meat (17). Similarly, C. difficile was isolated in 28 (12%) out of 230 meat samples submitted to analysis in another Canadian study (18). However, lower incidence rates of C. difficile have been reported in the studies led by other authors.
In a survey in Costa Rica, 200 meat samples were tested, out of which four (2%) were contaminated with C. difficile similarly to a study in the United States where 2% (2 out of 102) of the samples were contaminated (19, 20). Furthermore, the contamination rate of C. difficile was reported 2% in 13 out of 660 different meat samples tested in recent survey in Iran (10). In Sweden, France, Austria and the Netherlands, incidence rates of C. difficile contamination were 2.4%, 1.9%, 3% and 1.6% in the meat samples analyzed, respectively (6, 21-23). The results of the present investigation indicated a higher percentage of toxigenic strains of C. difficile in ground meats than in chopped ones (5% vs. 3%). This cannot be regarded as an unusual event because of further handling of minced meats. Our findings are in close agreement with the evidences of other authors that indicate the persistence of C. difficile spores which may exist in the environment and facilities of processing plants (16).
Care must be taken when interpreting available studies because the different data about the frequency of C. difficile are mainly affected by variable numbers of samples with different sampling, isolation methods and variations in regions. Nonetheless, the finding of present study would contribute to the international data of C. difficile prevalence in raw meat. In accordance to the results of the present survey concerning the ribotyping, the study led by de Boer et al., showed that around 80% of the isolated strains were not identical with the ribotypes found in patients with CDI in the Netherlands (6).
A Canadian report managed to evidence a collection of 12 ribotypes from C. difficile isolates in meat samples through PCR-ribotyping. Overall, eight (67%) of the 12 isolates hadn’t previously been identified and designated as belonging to the ribotype M31. The other identified ribotypes were 014, 077 and M26 (8). Furthermore, these authors cited the genetic diversity of C. difficile in retail raw meat in another study, including ribotypes 014, 077, M26, C, F, H, K and J (17). In contrast, some different studies showed a link between the presence of similar ribotypes in meat samples and clinical ones. For example, ribotype 012, 027, 078 identified in meat samples in Canada, Iran, USA and Europe (9, 10, 18, 20, 22). Unfortunately, no comprehensive surveys or reference laboratory data exist in Iran. It seems that establishment of an association to gather routine information about C. difficile as a causative agent of CDI in Iranian hospitals is necessary by regulatory agencies. Not having adequate information on the prevalence of C. difficile in food and food animal in Iran and the geographical aspects constitute the limitations of this study.
It is well accepted that the presence of C. difficile could result from different factors such as pollutants in environment, slaughtering process, poor operational and operators' hygiene. This is an issue rarely subjected to serious studies; therefore, comprehensive surveys are recommended in this regards. Overall, the results of the current study confirm the existence of C. difficile in beef and mutton meats mostly in their ground form in Iran which could be related to the wide distribution of C. difficile spores in the environment of meat packaging plants especially in meat grinder. No relationship was observed between meat isolates and clinical ones based on PCR-ribotyping results. Regarding with our finding, it suggests further investigations on other food matrices in Iran to determine the real situation of C. difficile contamination.
Acknowledgments
Sincere thanks are forwarded to all involved and especially Mrs. Parisa Shoaei, in Isfahan University of Medical Sciences and Dr. Mamnpoosh, the head of veterinary organization in Najafabad and Dr. Ehsan Kabiri for their assistance in receiving meat samples from meat packaging plants.
Footnotes
Funding/Support:This study is extracted from the PhD dissertation entitled "Molecular-Epidemiology of Clostridium difficile in different red meats (goat, camel, cow and mutton) and meat product "Hamburger" in Isfahan, Iran" and supported by Department of Food Science and Technology, College of Food Science and Technology, Tehran Science and Research Branch, Islamic Azad University, Tehran, Iran.
Authors' Contributions:The project was performed by ZE, MJ, HE, JSW and MC. DNA extraction, PCR techniques were performed by ZE. Sample collection, culture, statistical analysis and manuscript writing were performed by ZE, MJ, HE and JSW. All authors read and approve the final manuscript.
References
- 1.Borriell SP, Murray PR, Funke G. Clostridium perfringens, Clostridium difficile, and other Clostridium species. In: Borriello SP, Aktories K, editors. Topley and Wilson's microbiology and microbial infections. Vol. 2. ASM press; 2005. pp. 1098–136. [Google Scholar]
- 2.Bartlett JG, Chang TW, Gurwith M, Gorbach SL, Onderdonk AB. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N Engl J Med. 1978;298(10):531–4. doi: 10.1056/NEJM197803092981003. [DOI] [PubMed] [Google Scholar]
- 3.Kuehne SA, Cartman ST, Minton NP. Both, toxin A and toxin B, are important in Clostridium difficile infection. Gut Microbes. 2011;2(4):252–5. doi: 10.4161/gmic.2.4.16109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goncalves C, Decre D, Barbut F, Burghoffer B, Petit JC. Prevalence and characterization of a binary toxin (actin-specific ADP-ribosyltransferase) from Clostridium difficile. J Clin Microbiol. 2004;42(5):1933–9. doi: 10.1128/JCM.42.5.1933-1939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redelings MD, Sorvillo F, Mascola L. Increase in Clostridium difficile-related mortality rates, United States, 1999-2004. Emerg Infect Dis. 2007;13(9):1417–9. doi: 10.3201/eid1309.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer E, Zwartkruis-Nahuis A, Heuvelink AE, Harmanus C, Kuijper EJ. Prevalence of Clostridium difficile in retailed meat in the Netherlands. Int J Food Microbiol. 2011;144(3):561–4. doi: 10.1016/j.ijfoodmicro.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Weese JS. Clostridium difficile in food--innocent bystander or serious threat? Clin Microbiol Infect. 2010;16(1):3–10. doi: 10.1111/j.1469-0691.2009.03108.x. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Palacios A, Staempfli HR, Duffield T, Weese JS. Clostridium difficile in retail ground meat, Canada. Emerg Infect Dis. 2007;13(3):485–7. doi: 10.3201/eid1303.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Songer JG, Trinh HT, Killgore GE, Thompson AD, McDonald LC, Limbago BM. Clostridium difficile in retail meat products, USA, 2007. Emerg Infect Dis. 2009;15(5):819–21. doi: 10.3201/eid1505.081071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahimi E, Jalali M, Weese JS. Prevalence of Clostridium difficile in raw beef, cow, sheep, goat, camel and buffalo meat in Iran. BMC Public Health. 2014;14:119. doi: 10.1186/1471-2458-14-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jalali M, Khorvash F, Warriner K, Weese JS. Clostridium difficile infection in an Iranian hospital. BMC Res Notes. 2012;5:159. doi: 10.1186/1756-0500-5-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedorko DP, Williams EC. Use of cycloserine-cefoxitin-fructose agar and L-proline-aminopeptidase (PRO Discs) in the rapid identification of Clostridium difficile. J Clin Microbiol. 1997;35(5):1258–9. doi: 10.1128/jcm.35.5.1258-1259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, et al. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004;42(12):5710–4. doi: 10.1128/JCM.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stubbs S, Rupnik M, Gibert M, Brazier J, Duerden B, Popoff M. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett. 2000;186(2):307–12. doi: 10.1111/j.1574-6968.2000.tb09122.x. [DOI] [PubMed] [Google Scholar]
- 15.Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175(2):261–6. doi: 10.1111/j.1574-6968.1999.tb13629.x. [DOI] [PubMed] [Google Scholar]
- 16.Houser BA, Soehnlen MK, Wolfgang DR, Lysczek HR, Burns CM, Jayarao BM. Prevalence of Clostridium difficile toxin genes in the feces of veal calves and incidence of ground veal contamination. Foodborne Pathog Dis. 2012;9(1):32–6. doi: 10.1089/fpd.2011.0955. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Palacios A, Reid-Smith RJ, Staempfli HR, Daignault D, Janecko N, Avery BP, et al. Possible seasonality of Clostridium difficile in retail meat, Canada. Emerg Infect Dis. 2009;15(5):802–5. doi: 10.3201/eid1505.081084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weese JS, Avery BP, Rousseau J, Reid-Smith RJ. Detection and enumeration of Clostridium difficile spores in retail beef and pork. Appl Environ Microbiol. 2009;75(15):5009–11. doi: 10.1128/AEM.00480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quesada-Gomez C, Mulvey MR, Vargas P, Gamboa-Coronado Mdel M, Rodriguez C, Rodriguez-Cavillini E. Isolation of a toxigenic and clinical genotype of clostridium difficile in retail meats in Costa Rica. J Food Prot. 2013;76(2):348–51. doi: 10.4315/0362-028X.JFP-12-169. [DOI] [PubMed] [Google Scholar]
- 20.Curry SR, Marsh JW, Schlackman JL, Harrison LH. Prevalence of Clostridium difficile in uncooked ground meat products from Pittsburgh, Pennsylvania. Appl Environ Microbiol. 2012;78(12):4183–6. doi: 10.1128/AEM.00842-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Von Abercron SM, Karlsson F, Wigh GT, Wierup M, Krovacek K. Low occurrence of Clostridium difficile in retail ground meat in Sweden. J Food Prot. 2009;72(8):1732–4. doi: 10.4315/0362-028x-72.8.1732. [DOI] [PubMed] [Google Scholar]
- 22.Bouttier S, Barc MC, Felix B, Lambert S, Collignon A, Barbut F. Clostridium difficile in ground meat, France. Emerg Infect Dis. 2010;16(4):733–5. doi: 10.3201/eid1604.091138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jobstl M, Heuberger S, Indra A, Nepf R, Kofer J, Wagner M. Clostridium difficile in raw products of animal origin. Int J Food Microbiol. 2010;138(1-2):172–5. doi: 10.1016/j.ijfoodmicro.2009.12.022. [DOI] [PubMed] [Google Scholar]


