Abstract
This review illustrates the value of quantitative information including concentrations, kinetic constants and equilibrium constants in modeling and simulating complex biological processes. Although much has been learned about some biological systems without these parameter values, they greatly strengthen mechanistic accounts of dynamical systems. The analysis of muscle contraction is a classic example of the value of combining an inventory of the molecules, atomic structures of the molecules, kinetic constants for the reactions, reconstitutions with purified proteins and theoretical modeling to account for the contraction of whole muscles. A similar strategy is now being used to understand the mechanism of cytokinesis using fission yeast as a favorable model system.
Introduction
Understanding the operation of complex biological systems will remain a challenge for the foreseeable future, so theoretical approaches ranging from abstract to molecularly explicit models are growing in importance in biology (1). At the abstract end of the spectrum genetic (2) and protein (3) interaction maps provide insights about the architecture of systems as well as strong constraints for models (1,4). For example, genetic analysis provided enough information to model the budding yeast cell cycle with little information about the numbers of molecules or their reaction rates (5).
My purpose is to explain the value of chemical-kinetic models and simulations to probe the dynamical aspects of biological systems. Such models depend on quantitative measurements of the concentrations of the molecules in cells, rate and equilibrium constants for the reactions, and responses of the system to perturbations. Fortunately, technology is available to obtain this mechanistic information about molecules (6) and whole systems in live cells (7,8).
I will use muscle contraction as the classic example of this approach and cytokinesis, the separation of daughter cells by a cleavage furrow, as work in process. In both systems mechanistic studies of the constituent molecules and quantitative measurements in cells have contributed to understanding dynamical aspects of the biology.
Research on Muscle Contraction Set the Standard for System Analysis in Biology
Along with action potentials and neuromuscular synapses, skeletal muscle contraction was the first biological system to be understood at a level that satisfies biophysicists (9). The pioneering scientists who deciphered muscle contraction employed a strategy that stands as a paradigm for research in systems biology. What was their strategy and what did they accomplish?
Six approaches produced the core information required to formulate and test quantitative models for muscle contraction: i), an inventory of the component molecules; ii), atomic structures of the molecules and their organization in the cellular machine; iii), kinetic and thermodynamic parameters of the chemical reactions and interactions of the molecules; iv), reconstitution of the physical and chemical reactions from purified proteins including single molecule experiments; v), characterization of the kinetics and thermodynamics of contraction of whole muscles; and vi), theoretical modeling and simulations.
-
(i)
The inventory of molecules started in the 1930s with the discovery of myosin, continued in the 1940s with the separation of actin from myosin and their reconstitution into contractile threads. Starting in the 1950s through the 1980s biochemists and molecular biologists discovered the other major proteins: tropomyosin, troponin, alpha-actinin, capping protein, tropomodulin, titin, and nebulin.
-
(ii)
In the 1960s x-ray fiber diffraction and electron microscopy revealed the arrangement of actin subunits in thin filaments, myosin molecules in thick filaments, and their sliding mechanism. Atomic structures were hard-won, taking until 1990 for the atomic structure of actin (10) and 1993 for the atomic structure of a myosin motor domain (11).
-
(iii)
Transient state kinetics in the 1970s and 1980s explained the chemical mechanism of the myosin-actin ATPase cycle (12) and established concepts that explain how other motor proteins convert the energy released by ATP hydrolysis into force and motion. Presteady-state kinetics also elucidated the mechanism of actin polymerization (13). Studies like these illustrate that no protein has a simple mechanism of action, so models may miss important features of a system if the components are treated like blobs in a wiring diagram.
-
(iv)
Albert Szent-Gyorgyi and colleagues reconstituted purified actin and myosin into threads that contract when supplied with ATP (14). Contemporary reconstitution experiments use sensitive fluorescence microscopy assays to track the movements of single myosin molecules walking on actin filaments (15) or deflecting a bead in an optical trap to measure the force produced (16).
-
(v)
For the better part of a century physiologists have collected quantitative data on muscle contraction, with much of the work done on a favorable model system, the frog. This cold-blooded amphibian offers no genetics and large-scale biochemistry is limited. However, frog muscles are ideal for physiology (17) and x-ray fiber diffraction experiments (18), because they can be removed from the animal and kept alive and contracting for hours in a cold saline solution. Starting in the 1960s experiments measured the response of whole muscles or single muscle cells to rapid changes in length or force and revealed how myosin cross-bridges respond on a millisecond timescale (17). Caged molecules and proteins with fluorescent tags were introduced into permeabilized muscle cells and used to measure conformational changes in myosin cross-bridges (19). Time resolved x-ray fiber diffraction revealed how both the thin filament and myosin heads respond when muscle is activated (20). Asynchronous and transient interactions of individual myosins with actin filaments allow filaments to slide during contraction.
-
(vi)
Strong theoretical work since the 1950s synthesized structural and biochemical discoveries into quantitative hypotheses to explain physiological phenomena (17). This approach has provided an important reality check supporting and stimulating other lines of work.
By the end of the 20th century this research culminated in a satisfying, quantitative account of the kinetics and thermodynamics of force generation in muscle at the molecular level (9). All six strategies were essential for this synthesis. Fascinating details are still under investigation, but the level of understanding is already higher than virtually any other process in cellular and molecular biology. As this compelling story developed, strategies employed to study muscle contraction consistently broke new ground for all of cellular and molecular biology, in part because many of the leaders in the field were trained as physicists. Their examples inspired the next generation of scientists, who asked questions about contractility in nonmuscle cells.
Cytokinesis is an Ancient Contractile Process Powered by Myosin and Actin Filaments
Cellular contractility has been a feature of eukaryotic cells for the past billion years, since the myosin-II gene arose in the common ancestor of amoebas, fungi, and animals. Before that time, primordial eukaryotes may have relied on membrane fusion to separate the daughter cells at the end of mitosis. A large clade of species including algae, plants, ciliates, and many interesting parasites still depend on fusion of vesicles derived from the Golgi apparatus to create new plasma membrane between the separated nuclei, and all eukaryotes still use similar membrane fusion proteins to separate the plasma membranes of daughter cells (21). Since the invention of myosin-II, amoeboid, fungal, and animal cells have used a tension-generating contractile ring to form a cleavage furrow in the plasma membrane during cytokinesis (22). Later multicellular animals adapted the cytokinesis proteins for contraction of specialized muscle cells, making muscle contraction the model for a wide range of biological contractile systems.
Work on cytokinesis has progressed to the point where formal models and computer simulations have tested whether ideas derived from molecular studies can explain the behavior of the whole system operating under normal conditions and when perturbed experimentally. Successful models of most of the physical events during cytokinesis are now available for one model organism, fission yeast, and evidence is accumulating about how signaling pathways control the timing of events and specify the position of the cleavage furrow.
Choice of Experimental Organisms to Study Cytokinesis
The common ancestor of amoebas, fungi, and animals seems to have had the basic cytokinesis genes, which have been conserved in the diverging species over the past billion years (22). However, over this time evolution has tested mutations of cytokinesis genes, producing fascinating variations in divergent organisms that may distract the investigator from the general principles.
Given the ancient origins of the system, one should pick an experimentally tractable organism to discover general principles about the mechanism of cytokinesis. Ideally, the organism offers efficient classical and molecular genetics to find and modify relevant genes, sufficient material to purify proteins for biochemical and biophysical analysis, and quantitative microscopy to study the dynamics of the system in wild-type and mutant cells. Community resources such as mutant collections and sophisticated experimental methods are desirable assets. Work has concentrated on a few, phylogenetically dispersed, experimental organisms including an amoeba, two species of yeast, echinoderm embryos, two invertebrate species, and cultured cells from flies and vertebrates (Table 1). Each organism has advantages for studying cytokinesis but also some limitations. All are good for microscopy, although only the two yeasts are widely used for counting molecules in live cells.
Table 1.
Model eukaryotic organisms for studying cytokinesis
| Organism | Classical genetics | Molecular genetics | Biochemistry |
|---|---|---|---|
| Slime mold Dictyostelium discoideum | None, haploid, few conditional alleles | Good | Excellent |
| Budding yeast Saccharomyces cerevisiae | Excellent haploid and diploid genetics, numerous conditional alleles, complete collection of viable knockouts | Best available, complete collection of GFP-tagged genes, excellent standards for quantitative microscopy | Excellent |
| Fission yeast Schizosaccharomyces pombe | Excellent haploid and diploid genetics, numerous conditional alleles, complete collection of viable knockouts | Excellent, many strains with GFP-tagged cytokinesis proteins, excellent standards for quantitative microscopy | Good |
| Echinoderm embryos | None | Limited | Good |
| Nematode worm C. elegans embryos | Excellent diploid genetics; few conditional alleles | Excellent RNAi including a whole genome screen assayed for defects in mitosis and cytokinesis; genome editing is making it possible to tag proteins in genome | Minimal |
| Fruit fly Drosophila melanogaster and cultured cells | Excellent diploid genetics; few conditional alleles | Excellent RNAi including screens in cultured cells assayed for defects in mitosis and cytokinesis; genome editing is making it possible to tag proteins in genome | Minimal |
| Vertebrate cells in culture | Limited diploid genetics; few conditional alleles | RNAi; genome editing making it possible to tag proteins in genome | Good |
Dictyostelium discoideum is a haploid amoeboid organism with excellent biochemistry, microscopy, and molecular genetics, but no classical genetics and few conditional mutations. Both types of yeast offer excellent haploid and diploid genetics, large collections of conditional mutations, and unparalleled molecular genetics. Strain collections are available with each nonessential gene deleted, and virtually every budding yeast protein has been tagged with green fluorescent protein (GFP) in the genome. Both yeasts are biochemically accessible. Budding yeast have one biological disadvantage—the division site is chosen early in the cell cycle rather than during mitosis like most other organisms. Another possible disadvantage of yeast is >1 billion years of divergence from animal cells. Echinoderm embryos are classic organisms to study cytokinesis, but the relevant biochemistry is not well developed and no genetics or molecular genetics are available. Their large size is advantageous for micromanipulation but presents some problems for microscopy. Decades of classical genetics identified many cytokinesis genes in the diploid invertebrates Drosophila melanogaster and Caenorhabditis elegans. Broad RNAi screens of fly cell lines and nematode embryos identified additional genes involved with mitosis and cytokinesis, but conditional mutations are rare and mechanistic biochemistry is challenging. Animal tissue culture cells are favorable for microscopy. The relevant proteins can be isolated from animal organs, but genetics are complicated by diploid genomes. RNAi has been used to look for mitosis and cytokinesis proteins and genome editing methods have recently made it possible to tag proteins in the genome.
Fission yeast Schizosaccharomyces pombe is currently the best model organism to seek basic mechanistic insights about cytokinesis (the frog muscle for the field), so I will compare our understanding of cytokinesis in this yeast (Fig. 1, C and D), with other organisms. One billion years of divergence from our common ancestor may be viewed as either an advantage (helping to identify shared basic principles) or a disadvantage (differences in some aspects of cytokinesis). The genetic inventory of cytokinesis proteins in fission yeast is the most complete of any organism (22), deletion and conditional alleles are freely available for most cytokinesis genes, facile molecular genetics allow an investigator to manipulate the system at will (on a timescale of days), methods are available to make quantitative measurements of protein numbers in live cells on a timescale of seconds (7,8) and an interactive community of scientists has established a general understanding of the whole process of cytokinesis (22). One difference from animal cells is that the mitotic spindle is inside the nucleus, therefore spindle microtubules cannot interact directly with the cell cortex. Another difference is a high internal turgor pressure of ∼1 MPa resisted by an elastic cell wall. Invaginating the plasma membrane against this pressure requires forces much higher than produced by any known contractile ring, so cleavage depends on extra force from the growth of cell wall material, called the septum, into the cleavage furrow (23). Septum formation and contractile ring constriction are mutually interdependent (24,25,26).
Figure 1.
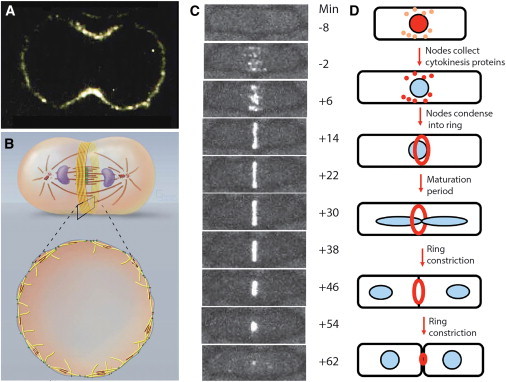
(A) Confocal fluorescence micrograph of a central section of a HeLa cell stained with Cy3.18-labeled antibodies to myosin-IIA (from Pamela Maupin, Johns Hopkins Medical School). (B) Drawings of a dividing animal cell with yellow contractile ring actin filaments and red myosin-II filaments (from Graham Johnson, University of California San Francisco). (C) Time series of fluorescence micrographs of a cell fission yeast cell expressing myosin-II regulatory light chain fused to mGFP to show formation of a contractile ring from a broad band of nodes followed by constriction. Times relative to spindle pole body separation are in minutes (from Rajesh Arasada, Yale University). (D) Drawings of the fission yeast cytokinesis time line. To see this figure in color, go online.
The Inventory of Eukaryotic Cytokinesis Proteins iS Extensive
Almost four decades of classical forward genetics revealed >150 genes that participate in cytokinesis in fission yeast (reviewed by (22)). RNAi screens in C. elegans embryos (27), (28) and cultured cell lines from flies (29,30) showed that most key cytokinesis genes have been conserved (31). These genes encode ancient contractile ring proteins including actin, myosins, formin, profilin, cofilin, Aip1, anillin, IQGAP, tropomyosin, cross-linking proteins (alpha-actinin, fimbrin), coronin, and F-BAR proteins. Other genes required for cytokinesis encode components of signaling pathways, including conserved cell cycle kinases (cyclin-dependent, aurora and polo kinases) and their regulators, as well as the septation initiation network (SIN), also known as the mitosis exit network in budding yeast and other cells. Rho family GTPases regulate contractile ring assembly in animal cells, but participate in later aspects of cytokinesis in fission yeast. Fungi have specialized cytokinesis genes for proteins that synthesize the septum, the cell wall in the cleavage furrow. Quantitative microscopy has been used to count the numbers of these proteins in whole yeast cells and document the precise times of their appearance in contractile rings and precursor structures called nodes (7).
Mechanisms of Cytokinesis Proteins
Crystal structures or useful homology models based in crystal structures are available for most of the core proteins. Important exceptions are anillin and parts of IQGAP. The mechanisms of most contractile ring proteins have been characterized in some model organism (myosins, actin, formins, profilin, cofilin, Aip1, and coronin), but additional work is required to understand the mechanisms of the more divergent proteins such as anillin and IQGAP.
Many individual reactions have been reconstituted from purified proteins (actin filament nucleation and elongation by formins and profilin, actin filament gliding by myosins, actin filament severing by cofilin). Actin filaments can be reconstituted with myosin and cross-linkers into bundles or stiff networks (32), but complete contractile rings have not been reconstituted from purified cytokinesis proteins. The closest model is formation of a ring on a microfabricated surface from actin filaments nucleated by Arp2/3 complex that constricts in the presence of myosin-VI (33). Neither of these proteins participates in cytokinesis, but this work establishes that reconstitution of contractile rings should be feasible with cytokinesis proteins. Another promising approach has been to characterize constriction of fully formed contractile rings liberated from permeabilized fission yeast protoplasts (live cells removed from their protective cell wall) (34).
Mutations in yeast cells or depletion from animal cells has implicated each of these proteins in specific steps in cytokinesis. Some of these proteins are essential for viability (actin, myosin, certain formins, profilin, cofilin), but yeast cells have a remarkable capacity to maneuver around the absence or loss of function of other proteins (individual myosin isoforms, anillin, cofilin, IQGAP) and complete division.
Physiology and Modeling of Contractile Ring Assembly
In simulations of simple, conceptual models, activation of myosin in a band around the equator of a spherical (35) or cylindrical (36) network of cross-linked actin filaments can form a contractile cleavage furrow, although relatively little is known about contractile ring assembly in most cells. In animal cells some preformed actin filaments are gathered into the contractile ring from the adjacent cortex (37) and other filaments form by de novo polymerization from formin mDia2 (38). Complicated, apparently redundant signals from the poles and midzone of the mitotic spindle position the contractile ring midway between the poles of the spindle (21). The best characterized component of this system is the centralspindlin complex consisting of kinesin 6 MKlP1, Rho family GTPase-activating protein (GAP) CyK4 and MgcRacGAP (39). This complex moves from the center of the mitotic spindle to the cortex, where it activates Rho to position the contractile ring, although many mechanistic details are still being investigated. Budding yeast use a different strategy—a scaffold of septins forms early around the neck between the mother cell and the bud. The septin ring incorporates myosin-II and formins assemble actin filaments de novo for the contractile ring (21).
Contractile ring assembly is understood in the most detail in fission yeast, where events in the cytokinesis pathway occur with precise timing around the entire cell cycle (40). The contractile ring assembles from protein structures called cytokinesis nodes that associate with the inside of the plasma membrane (40,41). Two types of precursor nodes form during interphase and combine by a search and capture mechanism to form cytokinesis nodes located around the equator (Fig. 2) (42). One type node is incorporated into the contractile ring. SIN activity disperses the other class of node proteins during mitosis, so they are only present in nodes during interphase. Signals from the poles of the cell restrict cytokinesis nodes to the middle of the cell (43,44), although the details are still being investigated. This appears to differ from the positioning mechanism in animal cells, where signals from the mitotic spindle and chromosomes locate the contractile ring.
Figure 2.
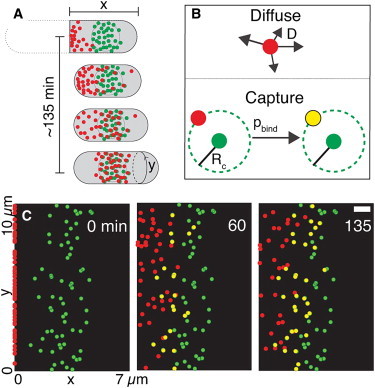
Model of cytokinesis node assembly in fission yeast by a diffuse and capture mechanism. (A) Sketches of interphase cells starting with a cell recently separated from its sister cell on the left. Type I nodes (green) form around the equator. Type II nodes emerge from the contractile ring at the end of cytokinesis and merge with type I nodes to form cytokinesis nodes. (B) Model with type II nodes diffusing on the inside of the plasma membrane and being captured by stationary type I nodes. (C) Three snapshots from a simulation of the model with 2D view of the inside of the plasma membrane. Red type II nodes start at the new pole of this daughter cell (left side) and green type I nodes start in an array around the center of the cell. Type II nodes diffuse until they are captured by an encounter with a type I node to create a yellow cytokinesis node. Times are in seconds (modified from Akamatsu et al. (42)). To see this figure in color, go online.
As the cell approaches mitosis, cytokinesis nodes accumulate myosin and other contractile ring proteins but are stationary until the arrival of formin Cdc12p (40,45). Cdc12p nucleates and elongates actin filaments in random directions (Fig. 3). Myosins in nearby nodes capture and pull on these filaments to move nodes together (45). Nodes have a two-dimensional (2D) diffusion coefficient of ∼20 nm2/s in the plane of the membrane, so a force of 3 pN is required to move them at the observed velocity of ∼30 nm/s (45). Nodes stop and start moving ∼3 times per minute because of breaks in the filaments connecting nodes (45,46).
Figure 3.

Model of contractile ring assembly in fission yeast. (A) Sketches of a cell with (red) cytokinesis nodes arranged in (blue) a broad band around the equator (left) and after condensation of the nodes into a contractile ring (right). (B–F) Elements of the model of contractile ring assembly. (B) Green actin filaments grow in random directions at rate Vpol from a red node. (C) An adjacent node captures actin filaments passing within capture radius rc. (D) Myosins in each node pull on an actin filament from an adjacent node with force F causing displacement at rate v. (E) Actin filaments connecting nodes break with time constant τbreak. (F) Nodes initiate new actin filaments with time constant τturn. (G) Snapshots of a stochastic simulation of the model condensing nodes into a ring with a few small gaps in 600 s. The broad band shown in (A) is opened up top to bottom showing the inside of the plasma membrane (modified from Vavylonis et al. (45)). To see this figure in color, go online.
Stochastic simulations of simple 2D models account for condensation of nodes into a contractile ring (45,47,48). Using parameters measured in live cells (node numbers, protein molecules per node, actin polymerization rates, forces and filament breaking rates) the simulations align nodes into a ring in 10 min, as observed in cells. Variation of the values of these parameters correctly predicted the consequences of reducing the rate of actin filament severing by mutating cofilin (46). Those experiments and the simulations show that breaking connections in the network serves as an error correcting mechanism to avoid the aggregation of nodes into large clumps. Including features such as a realistic three-dimensional cylindrical shape and cross-linking between actin filaments generates assembly intermediates similar to those in cells (49). The success of these simulations depended on knowledge of the numbers of molecules and their biochemical mechanisms and on constraints provided by quantitative measurements of the dynamic events in live cells.
Physiology and Modeling of Contractile Ring Constriction
Contractile rings invaginate the plasma membrane into a cleavage furrow by constricting at a rate related to the circumference, >300 nm/s in large echinoderm (50) and nematode embryos (51) but only ∼2 nm/s in small fission yeast. Actin and associated proteins leave contractile rings in proportion to the circumference in animal cells (52) and fission yeast (7). Myosin is retained and concentrated during constriction in the contractile rings of Dictyostelium (53) and fission yeast (7) but not budding yeast (54) or nematode embryos (51).
Force is the key physical parameter of constricting rings. Micromanipulation of echinoderm embryos showed that isometrically contracting rings produce forces of 10–15 nN (55), less than striated muscle per cross-sectional area. The cell wall precludes direct measurement of contractile ring tension in intact yeast cells, but the cell wall may be removed with digestive enzymes to form protoplasts providing that a solute in the medium balances the high turgor pressure. Protoplasts assemble contractile rings with about twice the circumference of a normal ring (34,56,57). As they constrict at ∼4 nm/s these rings slide to one pole of the protoplast rather than cleaving the cell in two. The tension in these rings was estimated to be ∼390 pN from the surface tension of the plasma membrane and the geometry of the tiny furrows formed by these sliding rings (57). This force per cross-sectional area is greater than contractile rings in echinoderm embryos. However, growth of the septum into the cleavage furrow formed by invaginating the plasma membrane must also contribute to overcoming the turgor pressure and can even finish cleavage in the absence of the contractile ring (23).
One goal for modeling the contractile ring is to account for the tension produced as well as the rate and extent of constriction. Although this has not yet been achieved, the following paragraphs explain a variety of approaches that are producing encouraging progress. One limitation is that the actual organization and anchoring of the filaments and motors in contractile rings is not yet known.
Contractile ring models range from purely conceptual to molecularly explicit (58). For example, Carvalho et al. (51) proposed without simulations a conceptual model with contractile units in series (like striated muscle sarcomeres) to explain why the constriction rate is proportional to the initial circumference. Numerical simulations of continuum mechanical models without molecular details can reproduce the time course of shape changes in sea urchin eggs (59), furrow formation in Dictyostelium (60), and the sliding of constricting rings in fission yeast protoplasts (34). Simulations of models with arrays of unanchored actin filaments and myosin motors established how filament density and polarity, motor density, motor polarity (unipolar and bipolar), and boundary conditions (linear array versus ring) determine the sliding behavior of the filaments including expansion or contraction of the bundle (61–64). For example, bipolar motors in a population of mixed polarity filaments slide the filaments into two groups of uniform polarity, hardly what happens in cytokinesis. This work emphasizes the importance of knowing the densities and polarities of the filaments and motors, because the predictable dynamics differ (62). Including actin filament cross-linkers along with myosin motors adds drag, but coupling cross-linking to actin filament depolymerization and severing may contribute to constriction through a ratchet mechanism (65) consistent with biological experiments (54). None of these models considered how ring molecules are anchored to the plasma membrane.
Stochastic simulations of a model based on molecular details produces the tension measured in fission yeast and explains how rapid turnover of the components contributes to constriction (57). This ∼2D model (Fig. 4) places the numbers of molecules counted in cells in a band 200 nm wide corresponding to the inside of the plasma membrane. Formins polymerize flexible actin filaments that turn over on a timescale tens of seconds by dissociation of formins from the membrane and severing of the filaments. Clusters of 20 dimeric myosin molecules bind and dissociate on the same timescale. Drag on the anchors of formins and myosins was estimated from direct observations and modeling. Myosins pull on the actin filaments producing forces resisted by drag on the anchors and transient alpha-actinin cross-links between the filaments. In stochastic simulations the molecules self-assemble into a bundle of actin filaments; actin filaments grow from formin anchors in random directions and those close to the 200 nm band are captured by myosin and aligned into tension-producing units with antiparallel actin filaments overlapping myosin clusters. Anchoring of the formins and myosins allows the self-assembled, disorganized bundle of filaments to produce tension. At steady state the ring operates near its maximum isometric tension of ∼350 pN as observed in protoplasts. Continuous turnover and reassembly of new contractile units are required to maintain the geometrical relations between the filaments that produce tension. The rings determine the tension and the resistance determines the constriction rate, therefore rings detached from the plasma membrane constrict rapidly (66). The septum-synthesizing enzyme Bgs1p is part of the anchoring mechanism (67), and growth of the septum may provide centripetal force to overcome turgor pressure (23) but its growth may also limit the rate of constriction. The success of this model did not depend on additional features such as filament buckling (64), actin depolymerization (54), or cross-linkers that track ends (65).
Figure 4.

Model of contractile ring constriction in fission yeast. (A) Molecular components and their reactions. (B) Forces in the system. (C) Snapshot from a simulation of the model at steady state with myosin clusters in orange, formins in blue, and actin filaments in gray. Horizontal lines show the edges of the 200 nm wide zone of the membrane that binds myosins and formins (modified from Stachowiak et al. (57)). To see this figure in color, go online.
Physiology and Modeling of Contractile Ring Disassembly
More than 40 years ago Schroeder (52) discovered that constricting contractile rings in animal cells maintain a constant density of actin filaments. This behavior has been verified in C. elegans and fission yeast, but very little has been learned about the mechanism, in large part because formins exclude tagged actin molecules from the contractile ring (7,68,51). At least three mechanisms could control the actin filament content of the ring. The most likely mechanism in fission yeast is a decline in formin Cdc12p numbers during constriction of the ring (7). The total Cdc12p in the ring peaks at the onset of constriction and then declines in proportion to the circumference. This could be due to either slower binding or faster dissociation of Cdc12p, but the mechanism has not been investigated. Less likely explanations are changes in the rates of actin polymerization from formins or actin filament severing. In fact, the severing activity of cofilin is much more important for contractile ring assembly than constriction or disassembly (46).
Lessons Learned
Comparing the models of cytokinesis illustrates my main point—abstract models can provide an overview of what is possible, while models based on quantitative information at both the molecular and cellular levels provide additional insights into the dynamics of the system. Even the Stachowiak model (57) will be improved with better information about the numbers, lengths, and organization of the actin filaments, organization of myosin clusters, physical attachments of formins and myosin clusters to the plasma membrane, regulation of myosin and formin activity, and turnover of each component in cells.
Models also provide the physical context for investigating how cells regulate cytokinesis. Virtually all cytokinesis proteins are present throughout the cell cycle (7), therefore the chemistry of the cell must change at each transition in the cytokinesis pathway. For example, SIN activity disperses type 1 nodes and is required for contractile ring constriction and septum formation, but other changes allow interphase nodes to collect each contractile ring protein at a specific time and turn on actin polymerization to begin the constriction and disassembly of the ring.
Readers will be interested in the perspective of Ditlev et al. (69) on the value of parameter values in models of actin filament assembly during cellular motility and endocytosis.
Acknowledgments
I am grateful to the peer reviewers who improved the manuscript with thoughtful criticisms and suggestions.
Research from the author’s laboratory reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01GM026132 and R01GM026338. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Janes K.A., Lauffenburger D.A. Models of signalling networks - what cell biologists can gain from them and give to them. J. Cell Sci. 2013;126:1913–1921. doi: 10.1242/jcs.112045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costanzo M., Baryshnikova A., Boone C. Charting the genetic interaction map of a cell. Curr. Opin. Biotechnol. 2011;22:66–74. doi: 10.1016/j.copbio.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Westermarck J., Ivaska J., Corthals G.L. Identification of protein interactions involved in cellular signaling. Mol. Cell. Proteomics. 2013;12:1752–1763. doi: 10.1074/mcp.R113.027771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis N.E., Nagarajan H., Palsson B.O. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 2012;10:291–305. doi: 10.1038/nrmicro2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen K.C., Csikasz-Nagy A., Tyson J.J. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol. Biol. Cell. 2000;11:369–391. doi: 10.1091/mbc.11.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard T.D., De La Cruz E.M. Take advantage of time in your experiments: a guide to simple, informative kinetics assays. Mol. Biol. Cell. 2013;24:1103–1110. doi: 10.1091/mbc.E13-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J.Q., Pollard T.D. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 8.Joglekar A.P., Bouck D.C., Salmon E.D. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geeves M.A., Holmes K.C. The molecular mechanism of muscle contraction. Adv. Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 10.Kabsch W., Mannherz H.G., Holmes K.C. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 11.Rayment I., Rypniewski W.R., Holden H.M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 12.Lymn R.W., Taylor E.W. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara I., Vavylonis D., Pollard T.D. Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 2007;104:8827–8832. doi: 10.1073/pnas.0702510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szent-Gyorgyi A. Studies on muscle. Acta Physiol. Scand. 1945;9:1–115. doi: 10.1111/j.1748-1716.1989.tb08599.x. [DOI] [PubMed] [Google Scholar]
- 15.Yildiz A., Forkey J.N., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 16.Finer J.T., Simmons R.M., Spudich J.A. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 17.Huxley A.F., Simmons R.M. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 18.Huxley H.E., Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J. Mol. Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 19.Corrie J.E., Brandmeier B.D., Irving M. Dynamic measurement of myosin light-chain-domain tilt and twist in muscle contraction. Nature. 1999;400:425–430. doi: 10.1038/22704. [DOI] [PubMed] [Google Scholar]
- 20.Huxley H.E., Faruqi A.R., Milch J.R. The use of synchrotron radiation in time-resolved X-ray diffraction studies of myosin layer-line reflections during muscle contraction. Nature. 1980;284:140–143. doi: 10.1038/284140a0. [DOI] [PubMed] [Google Scholar]
- 21.Oliferenko S., Chew T.G., Balasubramanian M.K. Positioning cytokinesis. Genes Dev. 2009;23:660–674. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- 22.Pollard T.D., Wu J.-Q. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Proctor S.A., Minc N., Chang F. Contributions of turgor pressure, the contractile ring, and septum assembly to forces in cytokinesis in fission yeast. Curr. Biol. 2012;22:1601–1608. doi: 10.1016/j.cub.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Wang H., Balasubramanian M.K. Drc1p/Cps1p, a 1,3-beta-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics. 1999;153:1193–1203. doi: 10.1093/genetics/153.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortés J.C., Ishiguro J., Ribas J.C. Localization of the (1,3)beta-D-glucan synthase catalytic subunit homologue Bgs1p/Cps1p from fission yeast suggests that it is involved in septation, polarized growth, mating, spore wall formation and spore germination. J. Cell Sci. 2002;115:4081–4096. doi: 10.1242/jcs.00085. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt M., Bowers B., Cabib E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 2002;115:293–302. doi: 10.1242/jcs.115.2.293. [DOI] [PubMed] [Google Scholar]
- 27.Goshima G., Vale R.D. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Somma M.P., Fasulo B., Gatti M. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell. 2002;13:2448–2460. doi: 10.1091/mbc.01-12-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gönczy P., Echeverri C., Hyman A.A. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 30.Sönnichsen B., Koski L.B., Echeverri C.J. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 31.Pollard T.D. Functional genomics of cell morphology using RNA interference: pick your style, broad or deep. J. Biol. 2003;2:25. doi: 10.1186/1475-4924-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koenderink G.H., Dogic Z., Weitz D.A. An active biopolymer network controlled by molecular motors. Proc. Natl. Acad. Sci. USA. 2009;106:15192–15197. doi: 10.1073/pnas.0903974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reymann A.C., Boujemaa-Paterski R., Blanchoin L. Actin network architecture can determine myosin motor activity. Science. 2012;336:1310–1314. doi: 10.1126/science.1221708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra M., Huang Y., Balasubramanian M. Cylindrical cellular geometry ensures fidelity of division site placement in fission yeast. J. Cell Sci. 2012;125:3850–3857. doi: 10.1242/jcs.103788. [DOI] [PubMed] [Google Scholar]
- 35.Astrom J.A., von Alfthan S., Karttunen M. Myosin motor mediated contraction is enough to produce cytokinesis in the absence of polymerisation. Soft Matter. 2010;6:5375–5381. [Google Scholar]
- 36.Shlomovitz R., Gov N.S. Physical model of contractile ring initiation in dividing cells. Biophys. J. 2008;94:1155–1168. doi: 10.1529/biophysj.107.111351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao L.G., Wang Y.L. Mechanism of the formation of contractile ring in dividing cultured animal cells. I. Recruitment of preexisting actin filaments into the cleavage furrow. J. Cell Biol. 1990;110:1089–1095. doi: 10.1083/jcb.110.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe S., Ando Y., Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol. Biol. Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White E.A., Glotzer M. Centralspindlin: at the heart of cytokinesis. Cytoskeleton (Hoboken) 2012;69:882–892. doi: 10.1002/cm.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J.Q., Kuhn J.R., Pollard T.D. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 41.Wu J.Q., Sirotkin V., Pollard T.D. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akamatsu M., Berro J., Pollard T.D. Cytokinetic nodes in fission yeast arise from two distinct types of nodes that merge during interphase. J. Cell Biol. 2014;204:977–988. doi: 10.1083/jcb.201307174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin S.G., Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- 44.Moseley J.B., Mayeux A., Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 45.Vavylonis D., Wu J.-Q., Pollard T.D. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 46.Chen Q., Pollard T.D. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J. Cell Biol. 2011;195:485–498. doi: 10.1083/jcb.201103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laporte D., Ojkic N., Wu J.Q. α-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol. Biol. Cell. 2012;23:3094–3110. doi: 10.1091/mbc.E12-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ojkic N., Wu J.Q., Vavylonis D. Model of myosin node aggregation into a contractile ring: the effect of local alignment. J. Phys. Condens. Matter. 2011;23:374103. doi: 10.1088/0953-8984/23/37/374103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bidone T.C., Tang H., Vavylonis D. Dynamic network morphology and tension buildup in 3D model of cytokinetic ring assembly. Biophys J. 2014;107:2618–2628. doi: 10.1016/j.bpj.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mabuchi I. Cleavage furrow: timing of emergence of contractile ring actin filaments and establishment of the contractile ring by filament bundling in sea urchin eggs. J. Cell Sci. 1994;107:1853–1862. doi: 10.1242/jcs.107.7.1853. [DOI] [PubMed] [Google Scholar]
- 51.Carvalho A., Desai A., Oegema K. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell. 2009;137:926–937. doi: 10.1016/j.cell.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 52.Schroeder T.E. The contractile ring. II. Determining its brief existence, volumetric changes, and vital role in cleaving Arbacia eggs. J. Cell Biol. 1972;53:419–434. doi: 10.1083/jcb.53.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson D.N., Cavet G., Spudich J.A. Quantitation of the distribution and flux of myosin-II during cytokinesis. BMC Cell Biol. 2002;3:4. doi: 10.1186/1471-2121-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendes Pinto I., Rubinstein B., Li R. Actin depolymerization drives actomyosin ring contraction during budding yeast cytokinesis. Dev. Cell. 2012;22:1247–1260. doi: 10.1016/j.devcel.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rappaport R. Cell division: direct measurement of maximum tension exerted by furrow of echinoderm eggs. Science. 1967;156:1241–1243. doi: 10.1126/science.156.3779.1241. [DOI] [PubMed] [Google Scholar]
- 56.Jochová J., Rupes I., Streiblová E. F-actin contractile rings in protoplasts of the yeast Schizosaccharomyces. Cell Biol. Int. Rep. 1991;15:607–610. doi: 10.1016/0309-1651(91)90007-6. [DOI] [PubMed] [Google Scholar]
- 57.Stachowiak M.R., Laplante C., O’Shaughnessy B. Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev. Cell. 2014;29:547–561. doi: 10.1016/j.devcel.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mendes Pinto I., Rubinstein B., Li R. Force to divide: structural and mechanical requirements for actomyosin ring contraction. Biophys. J. 2013;105:547–554. doi: 10.1016/j.bpj.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He X., Dembo M. On the mechanics of the first cleavage division of the sea urchin egg. Exp. Cell Res. 1997;233:252–273. doi: 10.1006/excr.1997.3585. [DOI] [PubMed] [Google Scholar]
- 60.Zhang W., Robinson D.N. Balance of actively generated contractile and resistive forces controls cytokinesis dynamics. Proc. Natl. Acad. Sci. USA. 2005;102:7186–7191. doi: 10.1073/pnas.0502545102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biron D., Alvarez-Lacalle E., Moses E. Molecular model of the contractile ring. Phys. Rev. Lett. 2005;95:098102. doi: 10.1103/PhysRevLett.95.098102. [DOI] [PubMed] [Google Scholar]
- 62.Zemel A., Mogilner A. Motor-induced sliding of microtubule and actin bundles. Phys. Chem. Chem. Phys. 2009;11:4821–4833. doi: 10.1039/b818482h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inoue Y., Tsuda S., Adachi T. Modeling myosin-dependent rearrangement and force generation in an actomyosin network. J. Theor. Biol. 2011;281:65–73. doi: 10.1016/j.jtbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 64.Lenz M., Gardel M.L., Dinner A.R. Requirements for contractility in disordered cytoskeletal bundles. New J. Phys. 2012;14:033037. doi: 10.1088/1367-2630/14/3/033037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zumdieck A., Kruse K., Jülicher F. Stress generation and filament turnover during actin ring constriction. PLoS ONE. 2007;2:e696. doi: 10.1371/journal.pone.0000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mishra M., Kashiwazaki J., Mabuchi I. In vitro contraction of cytokinetic ring depends on myosin II but not on actin dynamics. Nat. Cell Biol. 2013;15:853–859. doi: 10.1038/ncb2781. [DOI] [PubMed] [Google Scholar]
- 67.Arasada R., Pollard T.D. Transport of β-glucan synthetase from the Golgi apparatus explains why F-BAR protein Cdc15p is required to anchor the cytokinetic contractile ring in fission yeast. Cell Reports. 2014;8:1533–1544. doi: 10.1016/j.celrep.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Q., Nag S., Pollard T.D. Formins filter modified actin subunits during processive elongation. J. Struct. Biol. 2012;177:32–39. doi: 10.1016/j.jsb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ditlev J.A., Mayer B.J., Loew L.M. There is more than one way to model an elephant. Experiment-driven modeling of the actin cytoskeleton. Biophys. J. 2013;104:520–532. doi: 10.1016/j.bpj.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]


