Abstract
MicroRNAs are small regulatory RNAs that post-transcriptionally control gene expression. Reduced expression of DICER, the enzyme involved in microRNA processing, is frequently observed in cancer and is associated with poor clinical outcome in various malignancies. Yet the underlying mechanisms are not well understood. Here, we identify tumor hypoxia as a regulator of DICER expression in large cohorts of breast cancer patients. We show that DICER expression is suppressed by hypoxia through an epigenetic mechanism that involves inhibition of oxygen-dependent H3K27me3 demethylases KDM6A/B and results in silencing of the DICER promoter. Subsequently, reduced miRNA processing leads to derepression of the miR-200 target ZEB1, stimulates the epithelial to mesenchymal transition and ultimately results in the acquisition of stem cell phenotypes in human mammary epithelial cells. Our study uncovers a previously unknown relationship between oxygen-sensitive epigenetic regulators, miRNA biogenesis and tumor stem cell phenotypes that may underlie poor outcome in breast cancer.
Cancer mortality is largely attributable to distant metastasis. The mechanisms underlying the metastatic process are complex and are part of a series of events that ultimately result in the formation of macroscopic metastasis in distant organs from cells with tumor initiating or ‘stem cell’ properties1. Acquisition of stem cell and metastatic traits that enable this process, and the conditions in tumors that stimulate it, are poorly understood. However, many recent studies indicate that some tumor cells are able to transition from an epithelial to mesenchymal phenotype through a process similar to that which occurs in development (EMT). Acquisition of the mesenchymal phenotype is associated with both increased tumor initiating properties and the ability to form metastases in experimental models. However, in tumors this process requires some degree of plasticity, as formation of a tumor at a secondary metastatic site requires transition back to the epithelial cell state (mesenchymal to epithelial transition).
miRNAs are small regulatory RNAs that play an important role in normal development and in disease by regulating the expression of a vast number of target mRNAs2. miRNA biogenesis begins with transcription of long primary miRNAs (pri-miRNA) containing one or more hairpin structures that are processed by the nuclear endonuclease DROSHA, generating a 70-nucleotide stem loop known as the precursor miRNA (pre-miRNA). The pre-miRNA is exported to the cytoplasm by XPO5, and cleaved by DICER in a complex with TRBP2 to generate a ~22-nucleotide mature miRNA duplex. One strand is loaded into the RNA-induced silencing complex (RISC), which controls gene expression through sequence-specific interactions with target mRNAs causing their degradation or translational repression3. Several members of this miRNA biogenesis pathway have been identified as haplo-insufficient tumor suppressors, including DICER itself, XPO5, and TRBP24, 5, 6, 7, 8, 9, 10. Using a variety of mouse models, these studies indicate that partial suppression of microRNA biogenesis is sufficient to accelerate tumor development. Loss of one DICER allele in mouse models, results in a reduction in overall levels of mature miRNA and increased lung and soft tissue sarcomas4, 5. These studies extend earlier clinical findings demonstrating that miRNA levels are frequently reduced in tumors6. It is not clear how a reduction in miRNA biogenesis promotes cancer, and whether loss of one or more specific miRNAs underlies this effect. However, miRNA has been hypothesized to confer ‘robustness’ to biological processes including stabilizing differentiated cell states 11. In patients, low levels of DICER in breast, ovarian, and other cancers are associated with aggressive, invasive disease, distant recurrence, and poor overall survival12, 13, 14. In several model systems, DICER repression has also been shown to stimulate metastasis7, 10.
In addition to monoallelic loss in cancer5, several mechanisms have been described as potential regulators of DICER including the transcription factors MITF15 and Tap6310, and miR-103/1077. DICER expression has also been reported to be inhibited by hypoxia through an unknown mechanism16. Hypoxia is a common feature of tumors strongly associated with poor prognosis in multiple sites including breast cancer17, 18, 19. Clinical studies show a strong association between hypoxia and distant metastasis or relapse19, 20, 21, 22, 23, 24. Laboratory data support a direct role for hypoxia in driving metastasis, including in vivo studies with cell line derived25, 26, and more recently, patient-derived xenografts grown in the orthotopic site. Hypoxia has been suggested to promote stemness in both normal tissues and tumors27, 28, 29, 30, 31, 32. However, the mechanisms driving this aggressive phenotype are poorly understood.
In this study we have identified a new mechanism linking hypoxia, reduced miRNA biogenesis and acquisition of phenotypes associated with poor outcome. We show that tumor hypoxia is associated with reduced DICER expression in large cohorts of breast cancer patients and identify an epigenetic mechanism that suppresses DICER transcription through inhibition of oxygen-dependent H3K27me3 demethylases KDM6A/B. In breast cancer, reduced expression of DICER leads to a selective decrease in processing of the miR-200 family and consequently to derepression of ZEB1 and activation of the EMT and associated stem cell phenotypes.
Results
Reduced DICER expression in hypoxic human breast cancers
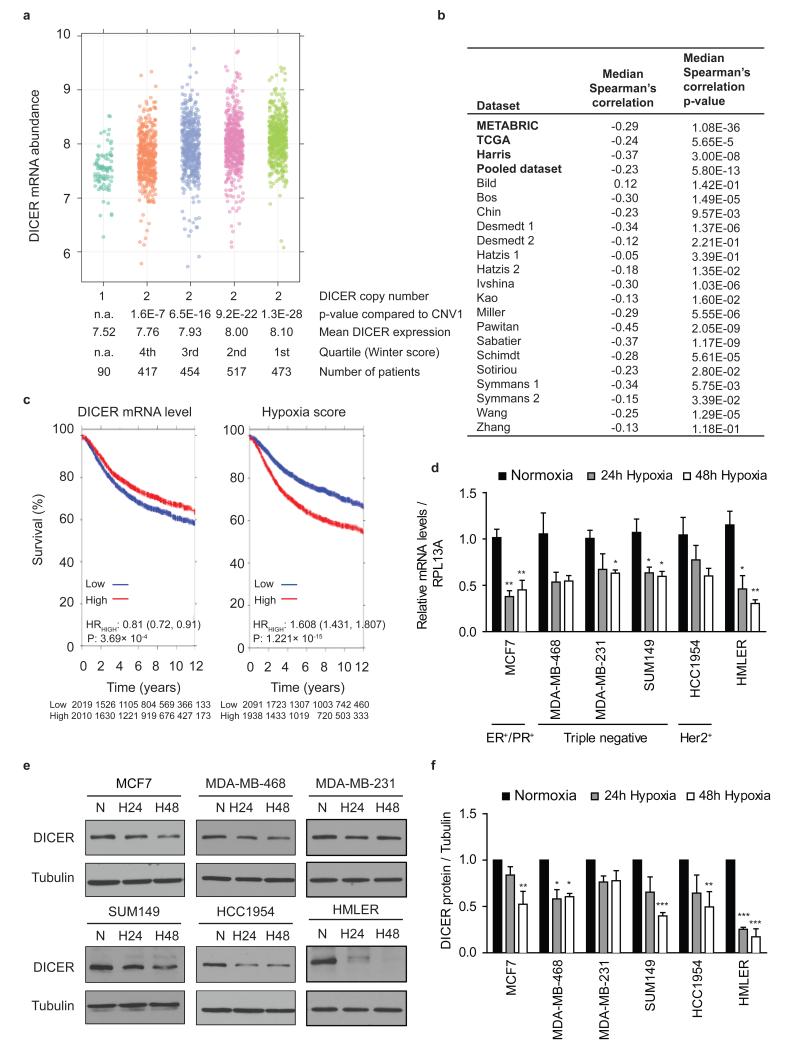
Both experimental and clinical data have demonstrated a strong correlation between hypoxia and more aggressive disease, including phenotypes recently linked to DICER suppression, such as stemness33 and metastasis18, 25. We therefore examined the association between DICER expression, DICER copy number and hypoxia in breast cancer. We stratified breast cancer patients from two datasets (METABRIC34 and TCGA35) having normal DICER copy number by the amount of hypoxia as determined using the validated Winter hypoxia signature36 (Fig. 1a and Supplementary Fig. 1a). The median RNA expression of 99 hypoxia associated genes in the Winter signature is an independent prognostic factor in head and neck squamous cell carcinoma (HNSCC) and breast cancer series. In both datasets, patients with the largest hypoxic fraction exhibited the lowest DICER mRNA expression (Fig. 1a and Supplementary Fig. 1a). A significant inverse correlation between hypoxia and DICER expression was found for the TCGA, METABRIC, Harris, and 14 out of 18 smaller breast cancer gene expression studies (Fig. 1b and Supplementary Table 1). A pooled dataset, consisting of 19 studies with long-term clinical follow-up, also demonstrated a highly significant (p= 5.80 × 10−13) inverse correlation between DICER and hypoxia. Interestingly, in both the pooled and METABRIC datasets, low levels of DICER and high levels of hypoxia were associated with poor outcome (Fig. 1c and Supplementary Figs. 2 and 3). Notably, the reduction in DICER expression in the most hypoxic quartile was reduced to levels similar to that in ∼ 5% of tumors that had monoallelic loss of DICER. These data suggest that hypoxia is a key contributor to DICER expression, and responsible for reduced DICER levels in significantly more patients than genetic loss. We confirmed that hypoxia suppresses DICER at both the mRNA and protein level in a panel of breast cancer (MCF7, MDA-MB-468, MDA-MB-231, SUM149, HCC1954), normal (MCF10A) or transformed (HMLER) mammary epithelial cell lines after exposure to oxygen levels commonly found in human tumors (<0.02 to 1.0% O2) (Fig. 1d-f and Supplementary Fig. 4). Hypoxic suppression varied from 26 to 74% at the mRNA level and 16 to 84% at the protein level over a period of 24 to 48 hours of hypoxia. DICER repression in hypoxia was not associated with any particular breast cancer subtype in either the cell lines or in the breast cancer clinical datasets (Supplementary Figs. 2, 3 and Supplementary Table 1).
Figure 1. Impaired DICER expression in hypoxic human breast cancers.
(a) DICER mRNA abundance in breast cancer patients from METABRIC stratified by DICER CNA and hypoxic fraction as determined with the Winter hypoxia metagene. P values obtained with Wilcoxon rank sum test. (b) Inverse correlation between DICER abundance and hypoxia score. P values obtained with Spearman’s correlation. (c) Kaplan-Meier survival curves for DICER and hypoxia in pooled breast cancer dataset consisting of 19 studies. (d) RNA was extracted from indicated breast cancer cell lines exposed to 0.2% O2 for 24 or 48 hours and subjected to quantitative RTPCR (qRT-PCR) analysis of DICER with RPL13A as control (n≥3). Data represents mean ± s.e.m. P values obtained with one-way ANOVA, Bonferroni’s post hoc test. (e) Representative Western blots were performed with antibody against DICER and anti-tubulin as control. (f) densitometric analysis of Western blots in e where the intensity of the DICER bands were normalized for tubulin signal (n=3). Error bars represent s.e.m. P values obtained with one-way ANOVA, Bonferroni’s post hoc test. *P<0.05, **P<0.01, ***P<0.001
In addition to monoallelic loss in cancer, several mechanisms have been implicated in DICER regulation. These include the transcription factors MITF15 and Tap6310, which induce DICER, miR-103/107, which repress DICER7, and a Von Hippel-Lindau dependent mechanism affecting DICER protein stability16. We examined each of these and found that none could explain suppression of DICER by hypoxia in breast cancer. Reporter constructs containing 2.5kb of the DICER promoter (with or without mutations in the MITF E-Box elements) showed no regulation by hypoxia (Supplementary Fig. 5a, 5b). Similarly, no increase in miR-103/107, and no decrease in DICER transcript or protein stability were observed during hypoxia (Supplementary Fig. 5c-f). We also observed no significant difference in DICER repression in cell lines isogenic for VHL (Supplementary Fig. 5g). Importantly, MITF, Tap63, and miR-103/107 also showed weak or no correlation with DICER expression in breast cancer patients in the TCGA and METABRIC datasets (Supplementary Fig. 1b-h).
DICER is epigenetically regulated during hypoxia
The lack of regulation of the DICER reporter construct (Supplementary Fig. 5a, 5b) during hypoxia was surprising. To directly test if transcription of the endogenous DICER locus was affected we measured changes in de novo transcription during hypoxia by pulse-labeling RNA. These experiments demonstrated that DICER transcription decreased 40 to 50% during hypoxia (Fig. 2a), whereas the HIF-1 target gene carbonic anhydrase-9 (CA9) increased 20-fold (Supplementary Fig. 7a). Transcriptional suppression of DICER was not dependent on known hypoxia response pathways, including either the HIF (hypoxia inducible factor) pathway (as reported previously16) or the PERK/ATF4 arm of the unfolded protein response (Supplementary Fig. 6a-e). DICER repression occurred normally in both HIF1α knockout cells and in multiple lines where HIF1α was depleted by RNA interference (Supplementary Fig. 6a-c). Additionally, hypoxic down-regulation of DICER did not affect HIF regulation in breast cancer cell lines, which has been previously reported in other cell types16 (Supplementary Fig. 6f).
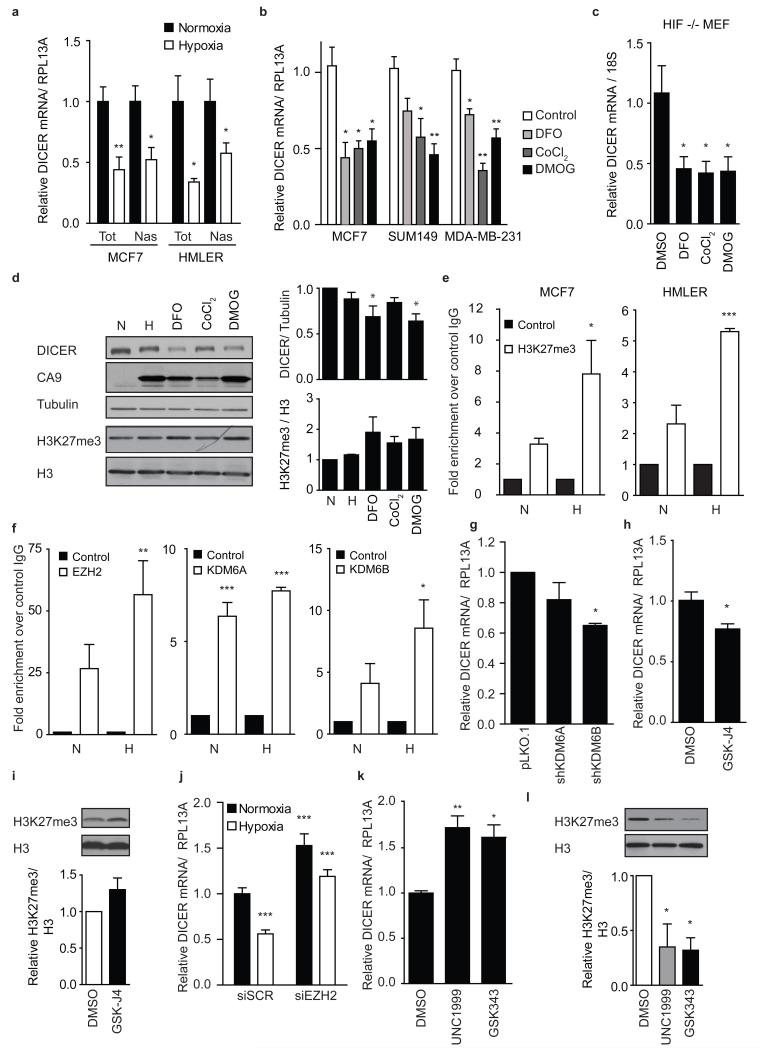
Figure 2. Epigenetic silencing of DICER promoter in response to hypoxia.
(a) Total (Tot) and nascent (Nas) DICER mRNA expression during hypoxia in MCF7 and HMLER cell lines was determined using qRT-PCR with RPL13A as control (n=4). (b) Indicated breast cancer cell lines (n=3) or (c) HIF1α null MEFS (n=4) were exposed to 500 μM iron chelator DFO, 250 μM antagonist CoCl2 or 500 μM DMOG for 24 hours. DICER mRNA expression was determined using qRT-PCR with RPL13A or 18S as control. (d) Western blot analysis of MCF7 cell extracts prepared after 24 hour exposure to 0.2% O2, 500 μM DFO, 250 μM CoCl2 or 500 μM DMOG with antibodies specific for DICER, CA9, and H3K27me3. Tubulin and H3 serve as respective loading controls. Representative blot (right) and densitometric analysis of Western blots from 4 independent experiments (left). (e) validation of the enrichment in H3K27me at the DICER promoter was done using ChIP analysis of H3K27me3 mark in combination with qRT-PCR analysis in MCF7 and HMLER cells during normoxia (N) or hypoxia (H). Fold-enrichment shown over control IgG (n=3). (f) enrichment of EZH2, KDM6A or KDM6B at the DICER promoter was done using ChIP analysis of each enzyme in combination with qRT-PCR analysis in MCF7 cells during normoxia (N) and hypoxia (H) (n=3). (g,h) qRT-PCR analysis of DICER expression with RPL13A as control in (g) MCF7 cells bearing shRNAs against KDM6A/B (n=3) or (h) treated with 10 μM KDM6A/B inhibitor GSK-J4 24hrs (n=3). (i) Representative blot (top) and quantification of global H3K27me3 (bottom) after GSK-J4 treatment in h. DICER mRNA expression in (j) MCF7 cells transiently transfected with siRNA directed against EZH2 during aerobic and hypoxic conditions (n=4) or (k) treated with 5 μM EZH2 inhibitor UNC1999 or GSK343 for 48hrs (n=3). (l) Representative Western blot (top) and quantification of global H3K27me3 levels (bottom) after EZH2 inhibition in k. Error bars represent s.e.m. P values obtained with Student’s t-test or one-way ANOVA, Bonferroni’s post hoc test. *P<0.05, **P<0.01, ***P<0.001
Although HIF1 was not required for DICER suppression during hypoxia, we found that agents that stabilize HIF by inhibiting the HIF prolyl-hydroxylases (EGLN1/2/3) did cause DICER suppression. DFO, CoCl2 and DMOG at concentrations sufficient to activate transcription of HIF target genes (e.g. CA9 – Supplementary Fig. 7b) all resulted in a significant reduction in DICER at the mRNA and protein level (Fig. 2b, 2d). DFO and CoCl2 stabilize HIF by chelating or competing with iron [Fe(II)] whereas DMOG does so by competitive inhibition of 2-oxogluterate, which in addition to oxygen, are required co-factors for the HIF prolyl-hydroxylases that mediate HIF stability. However, like hypoxia, treatment with DFO, CoCl2 and DMOG also caused DICER repression in HIF1α knockout cells (Fig. 2c).
Since hypoxia, DFO, CoCl2 and DMOG all influenced DICER expression in a HIF1α independent manner, we hypothesized that DICER was regulated through inhibition of alternative iron, oxygen, and 2-oxogluterate dependent enzymes, such as the Jumonji-domain (JMJD) containing hydroxylases KDM6A and KDM6B37 that regulate epigenetic silencing through removal of repressive histone 3 lysine 27 trimethylation (H3K27me3) marks38. Indeed, inhibition of all three required cofactors of the KDM6A/B enzymes by hypoxia, DFO, CoCl2 and DMOG resulted in an increase in total H3K27me3 in multiple cell types (Fig. 2d and Supplementary Fig. 7c). More importantly, H3K27me3 chromatin immunoprecipitation followed by sequencing (ChIP-seq) demonstrated that hypoxic exposure for as short as 8 hours led to an increase in repressive H3K27me3 marks in the DICER promoter region (Supplementary Fig. 7d). Enrichment in H3K27me3 at the DICER promoter during hypoxia was confirmed using conventional ChIP-qPCR in MCF7 (8-fold enrichment vs. hypoxic IgG control) and HMLER (5-fold enrichment vs. hypoxic IgG control) cell lines (Fig. 2e). In both cell lines this translated into an approximate doubling of H3K27me3 as compared to levels under normoxia. Furthermore, ChIP-qPCR analysis using specific antibodies against the H3K27 methyltransferase EZH2 (writer) and the oxygen-dependent H3K27me3 demethylases KDM6A and KDM6B (erasers) revealed significant enrichment over IgG controls at the DICER promoter for each respective enzyme with no significant difference in enrichment between normoxic and hypoxic conditions (Fig. 2f). Consistent with a role for epigenetic regulation of DICER, knockdown of KDM6A or KDM6B resulted in reduced DICER expression (Fig. 2g and Supplementary Fig. 7e-g). Similarly, inhibition of KDM6A/B with the inhibitor GSK-J4 at concentrations that increased overall levels of H3K27me3 by 1.3 fold, caused a 23% and 50% decrease in DICER expression in MCF7 and HMLER cells respectively (Fig. 2h,i and Supplementary Fig. 7h, 7i) without affecting HIF activity (Supplementary Fig. 7h). Conversely, knockdown of EZH2 resulted in a significant increase in DICER expression and was able to largely prevent DICER repression during hypoxia (Fig. 2j and Supplementary Fig. 7j). Similarly, inhibition of EZH2 using UNC1999 and GSK343 at levels that caused decreases in global H3K27me3 levels by 50-70% (Fig. 2l and Supplementary Fig. 7k) also increased DICER to levels comparable with EZH2 knockdown (Fig. 2k and Supplementary Fig. 7l). Together, these data indicate that basal expression of DICER is regulated by dynamic and opposing activities of KDM6A/B and EZH2, which are both constitutively present at the DICER locus, and that suppression of KDM6A/B activity under hypoxia is sufficient to increase H3K27me3 in an EZH2 dependent manner and suppress DICER transcription.
Hypoxia causes a miRNA processing defect
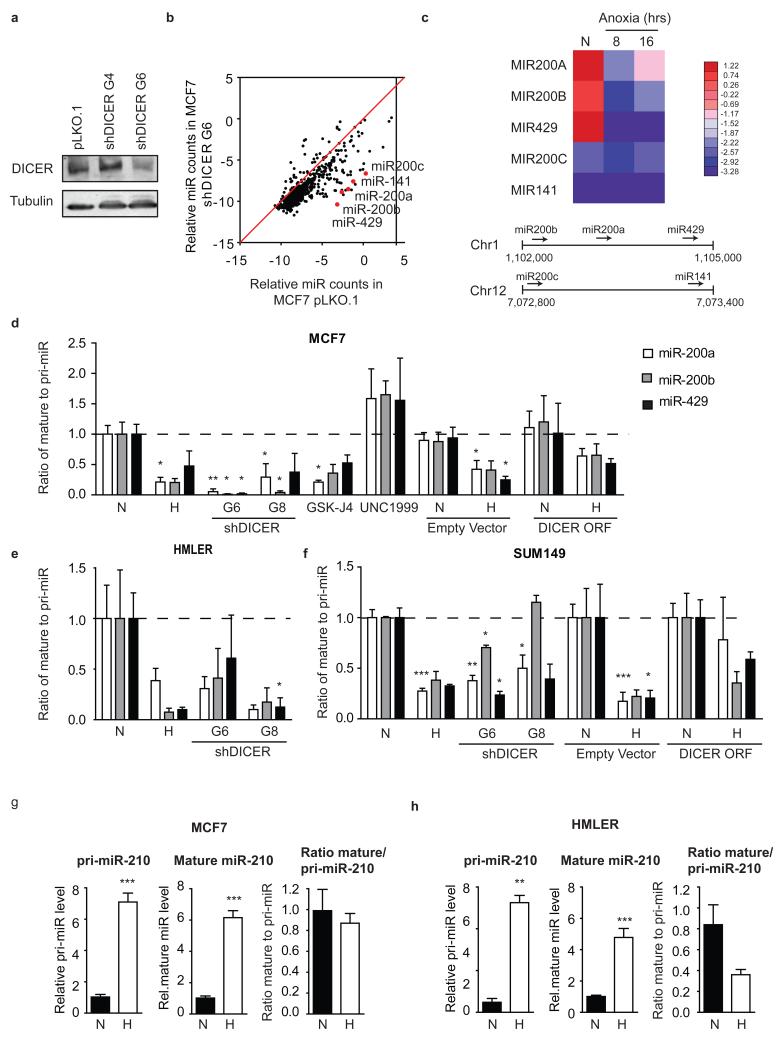
To examine the consequences of DICER suppression, we created MCF7 and HMLER breast cancer cell lines with stable knockdown of DICER and assessed changes in levels of mature miRNA. Suppression of DICER to ~30% resulted in a widespread reduction in mature miRNA for the majority of miRNA species (Fig. 3a, 3b and Supplementary Fig. 8a). Hypoxia also led to an analogous impairment in the processing of exogenously introduced shRNA (Supplementary Fig. 8b). siRNA and shRNA targeting HIF1α showed similar efficiency under aerobic conditions, but under hypoxia HIF1α shRNA, which requires DICER processing, was significantly less efficient. Under hypoxia, HIF1α siRNA reduced the HIF target gene CA9 by ~50%, whereas HIF1α shRNA failed to prevent CA9 induction.
Figure 3. Impaired miRNA processing in hypoxic cells.
(a) MCF7 cells were transduced with lentiviral shRNA constructs directed at DICER. Cell extracts from MCF7 cells bearing empty vector pLKO.1, non-functional shDICER_G4 or functional shDICER_G6 were subjected to Western blot analysis using a DICER specific antibody. Tubulin served as loading control. (b) total RNA was extracted from MCF7 pLKO.1 or shDICER_G6. miRNA levels were determined by Nanostring technology (n=3). (c) MCF7 cells exposed to hypoxia for 16 hours were used for total RNA isolation and subsequently subjected to deep-sequence analysis of miRNA content. Heat map of the miR-200 family in MCF7 during hypoxia is shown. Bottom panel shows schematic representation of the genomic organization of the miR-200 family. (d) The ratio of mature miRNA to pri-miRNA for miR-200a, b and 429 in MCF7 cells after hypoxia, DICER knockdown (shRNA G6 and G8), KDM6 inhibitor GSK-J4 (10μM), EZH2 inhibitor UNC1999 (5μM) and DICER overexpression. Mature and pri-miRNA levels were determined by qRT-PCR. Similar experiments were performed in (e) HMLER and (f) SUM149 cells. (g,h) Processing of hypoxia inducible miR-210 was assessed as described in d for (g) MCF7 and (h) HMLER cells. Expression levels of mature and pri-miRNAs for the miR-210 family was determined by qRTPCR. N indicates normoxia, H indicates hypoxia. The data d-h represents mean ratios (n=3) ± s.e.m. P values obtained with Student’s t-test. *P<0.05, **P<0.01, ***P<0.001
Sensitivity to DICER knockdown varied amongst different miRNA and the 5 members of the miR-200 family (miR-200a, miR-200b, miR-429, miR-200c, and miR-141) were amongst the most strongly repressed in both lines (Fig. 3b and Supplementary Fig. 8a). Similarly, the miR-200a/b/429 cluster was amongst the most strongly repressed miRNAs in response to hypoxia (Fig. 3c). We confirmed that both hypoxia and DICER knockdown led to a substantial defect in the processing of miR-200 family precursors (pri-miRNAs) into mature miRNAs. The ratio of mature to pri-miRNA, an indicator of DICER activity, dropped substantially in response to both DICER knockdown and hypoxia in MCF7, HMLER, and SUM149 cells (Fig. 3d-f and Supplementary Fig. 8c). Furthermore, the reduction in DICER caused by inhibition of KDM6A/B using GSK-J4 also resulted in a decrease in miR-200a/b/429 processing similar to that during hypoxia or DICER knockdown (Fig. 3d). Conversely, inhibition of EZH2 using UNC1999, which increased DICER expression, increased miR-200a/b/429 processing (Fig. 3d). Importantly, transient overexpression of DICER during hypoxia increased miR-200a/b/429 processing to near basal levels, demonstrating that these effects on miRNA processing under hypoxia are due to its effects on DICER (Fig. 3d, 3f and Supplementary Fig. 8d).
Despite the defect in miRNA processing caused by DICER repression, some mature miRNAs increased during hypoxia, including the widely reported hypoxia inducible miR-210. However, in this case the increase is due entirely to a transcriptional effect (Supplementary Fig. 8e, 8f and Fig. 3g, 3h). The processing of miR-210 mediated by DICER is reduced under hypoxia but this effect is smaller than the overall transcriptional increase resulting in increased mature levels of the miRNA.
Hypoxia stimulates EMT and CSC associated properties
The miR-200 family is implicated in regulation of the ZEB1 and ZEB2 transcription factors, which repress E-cadherin and stimulate the EMT39. DICER knockdown has previously been implicated in promoting the EMT and metastasis through the miR-200 family7, and miR-200b repression and ZEB1 induction during hypoxia has been reported40, 41. We hypothesized that DICER suppression during hypoxia may similarly regulate the EMT and perhaps underlie the known association of tumor hypoxia with metastasis, stemness and aggressive disease. Indeed, DICER repression in response to hypoxia or knockdown resulted in enhanced expression of ZEB1, loss of epithelial marker expression (E-cadherin) and increased expression of mesenchymal markers (N-cadherin and Vimentin) (Fig. 4a-c). The transcription factor TWIST, previously implicated in hypoxia-induced EMT and metastasis42 remained unchanged, as did the transcription factor SNAIL (Supplementary Fig. 9a, 9b). The hypoxia and DICER knockdown induced EMT is mechanistically distinct from the classical EMT inducer TGFβ1, which was associated with increased expression of TWIST and SNAIL, but caused no change in DICER expression or miRNA biogenesis (Supplementary Fig. 9c-e).
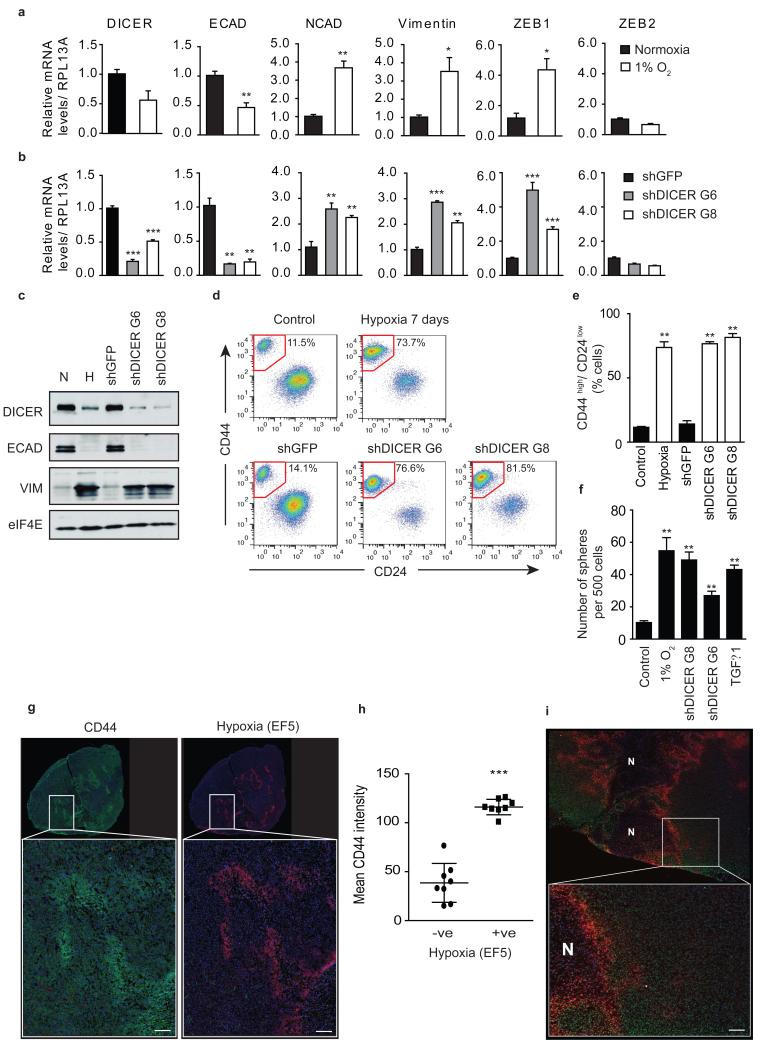
Figure 4. DICER repression promotes cancer stem cell associated phenotypes by reduced miR-200 expression during hypoxia.
(a) HMLER cells were grown for 7 days under 1% O2. RNA was extracted and used to determine DICER mRNA levels by qRT-PCR analysis. Expression of epithelial and mesenchymal associated genes was assessed simultaneously. RPL13A served as control (n=3). (b) Similar analysis as in a was performed on HMLER cells transduced with 2 independent shRNA constructs targeting DICER (n=3). (c) HMLER protein extracts were subjected to Western blot analysis of DICER, E-cadherin, Vimentin and eIF4E as loading control. (d) Representative FACS analysis of HMLER cells using antibodies specific for CD44 and CD24. (e) Quantification of the percentage of cells with CD44highCD24low for 3 independent experiments. (f) Sphere formation assay, number spheres formed per 500 cells plotted (n=3). (g) Representative image of whole tumor (top) and 25X region (bottom) of CD44 (green), EF5 (red) and DAPI (blue) staining in HMLER orthotopic xenograft. (h) Mean CD44 intensity in hypoxic (EF5 negative) versus non-hypoxic (EF5 positive) tumor regions of HMLER xenografts (n=8 mice). (i) Representative image of whole tumor (top) and 25X region (bottom) of H3K27me3 (green), EF5 (red) and DAPI (blue) staining in HMLER orthotopic xenograft. N indicates regions of tumor necrosis. Scale bar, 100 μm. Error bars represent s.e.m. P values obtained with Student’s t-test or one-way ANOVA, Bonferroni’s post hoc test. *P<0.05, **P<0.01, ***P<0.001
In breast cancer, the EMT has been linked to acquisition of stem cell phenotypes, including expression of cell surface antigens associated with human breast stem cells (CD44high CD24low), increased mammosphere formation, and increased tumor initiation capacity43, 44, 45. In HMLER cells, hypoxic exposure led to an increase in the frequency of CD44high CD24low cells from 11.5% to 73.7% (Fig. 4d, 4e), comparable to levels observed following exposure to TGFβ1 (Supplementary Fig. 9e) or reported following forced exposure of SNAIL or TWIST43. A similar increase in the fraction of CD44highCD24low cells occurred following DICER knockdown alone (76.6% and 81.5% vs. 14.1%) (Fig. 4d, 4e). Both hypoxic exposure and DICER knockdown resulted in a greater than 5-fold increase in mammosphere formation similar to that observed following TGFβ1 exposure (Fig. 4f). Importantly, hypoxia is similarly able to influence H3K27me3, DICER, EMT, and stem cell phenotypes in vivo. We established HMLER xenografts and examined the spatial relationship between hypoxia, H3K27me3, and CD44 using multi-fluorescence immunohistochemistry. As shown in Fig. 4g and 4h, tumor hypoxia in vivo (as assessed by EF5) is strongly associated with increased expression of the stem cell marker CD44 (Fig. 4g, 4h). Hypoxic tumor areas also show increased overall levels of H3K27me3, with discernable gradients in expression away from hypoxic areas (Fig. 4i). Furthermore, we assessed DICER, ZEB1, and E-cadherin expression in vivo using a panel of breast cancer xenografts and found that DICER and E-cadherin were inversely correlated with the endogenous hypoxia marker CA9 whereas ZEB1 showed a positive correlation with CA9 (Supplementary Fig. 9f).
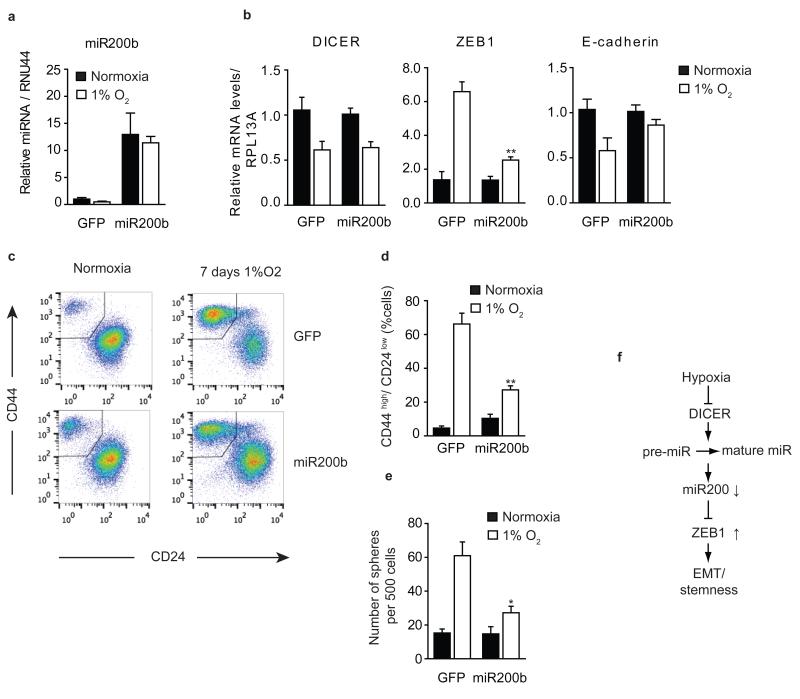
Finally, we tested directly if hypoxia and DICER knockdown induced EMT and acquisition of stem cell phenotypes is dependent on loss of mature miR-200 levels. DICER overexpression during hypoxia, which rescued the defect in miR-200 processing (Fig. 3d), also prevented increased expression of ZEB1 (Supplementary Fig. 9g). Furthermore, overexpression of miR-200b alone, prevented increased expression of ZEB1 and loss of E-cadherin during hypoxia, without affecting DICER repression and the defect in miRNA biogenesis (Fig. 5a, 5b). miR-200b overexpression during hypoxia also prevented the increase in the CD44highCD24low population and mammosphere formation (Fig. 5c-e). Together, these demonstrate that hypoxic suppression of DICER causes an EMT driven acquisition of stem cell properties in breast cancer through a reduction in the biogenesis of mature miR-200 family members (Fig. 5f).
Figure 5. Hypoxia stimulates cancer stem cell associated phenotypes in a miR-200b dependent manner.
HMLER cells overexpressing miR-200b or GFP (n=3) grown for 7 days under 1% O2 or control conditions. (a,b) qRT-PCR analysis was performed to measure (a) mature miR200b levels, (b) DICER, ZEB1 and E-cadherin mRNA level where RPL13A served as control (n=3). (c) Representative FACS analysis using antibodies specific for CD44 and CD24. (d) Quantification of the percentage of cells with CD44highCD24low for 3 independent experiments. (e) Sphere formation assay, number spheres formed per 500 cells plotted (n=3). (f) Model of hypoxia mediated EMT and stemness. Error bars represent s.e.m. P values obtained with Student’s t-test. *P<0.05, **P<0.01, ***P<0.001
Discussion
Our study demonstrates that hypoxia in the tumor microenvironment is a contributor to DICER expression and repression of miRNA biogenesis. In large independent cohorts of breast cancer patients, the association between hypoxia and DICER is stronger than any previously reported regulator of DICER, and the number of patients affected by this mechanism exceeds those harboring monoallelic loss. This analysis also indicates that DICER suppression may underlie part of the known association of hypoxia with metastasis and poor outcome, although hypoxia remains a better discriminator than DICER in breast cancer patients. This is perhaps not surprising given that hypoxia influences other important biological processes including angiogenesis, altered metabolism, and chromosomal instability. The changes in histone methylation and DICER expression are relatively small (~2-fold) in response to hypoxia or inhibition of EZH2 and KDM6A/B. Previous studies unambiguously identified DICER as a haploinsufficient tumor suppressor and an important driver of tumorigenesis under conditions of less than 50% repression4, 5. Our study is consistent with these earlier reports, and demonstrates that relatively small changes in DICER expression during hypoxia underlie functional changes in miRNA biogenesis, EMT, and properties associated with stemness. Interestingly, Rupaimoole et al. have demonstrated that hypoxia causes additional suppression of miRNA biogenesis through silencing of DROSHA (in print in the same issue), and that the combined defect in miRNA biogenesis resulting from DICER and DROSHA suppression leads to increased metastasis in ovarian cancer. The coordinated suppression of these two key enzymes required for miRNA biogenesis as well as hypoxia dependent suppression of AGO246 suggests a particularly important and broad role for oxygen in the regulation of miRNA levels. The consequences of such regulation are likely to be different in specific tissues and cancer types, depending upon the expression of different miRNAs. In breast cancer, we show that selective sensitivity of the miR-200 family to loss of DICER plays a dominant role in regulation of EMT and cancer stem cell phenotypes, which may underlie the known association of hypoxia with aggressive disease in breast and other cancer types12, 13, 14, 17, 18.
Previous studies have reported multiple mechanisms of DICER regulation, which can occur at the level of transcription through MITF15 and Tap6310, mRNA stability through miR-103/107 as well as at the protein level in a VHL dependent manner16. Our study reveals that DICER transcription is also regulated dynamically by acquisition or loss of the repressive H3K27me3 polycomb mark that is typically associated with gene silencing38. This mark is also found on promoters of so-called ‘poised’ genes expressed at low levels in embryonic stem cells where it is often associated with co-occurrence of the activating H3K4 mark47. Several previous studies have indicated that the balance between writing (EZH2) and erasing (KDM6A/B) H3K27me3 marks within the genome can play an important role in cancer48, 49. This is supported by the finding that EZH2 is frequently overexpressed or activated in cancer50, 51, 52, and associated with increased metastasis53. Conversely, the H3K27me3 demethylase KDM6A is frequently mutated in human cancers54, 55 and KDM6B expression is decreased in subsets of human cancers56, 57. Our study identifies DICER as an important integrator of EZH2 methyltransferase and KDM6A/B demethylase activity. We showed that EZH2 and KDM6A/B are present at the DICER locus, and that inhibition of these enzymes can cause acute changes in the levels of H3K27me3, DICER expression, and DICER activity. The hydroxylase activity of KDM6A/B requires molecular oxygen, 2-oxogluterate, and Fe(II) in order to demethylate H3K27me3. Correspondingly we found that hypoxia, or depletion/competition of the other cofactors with DFO, CoCl2 or DMOG all suppress DICER. To our knowledge, this is the first such example of a gene whose expression is dynamically regulated at the epigenetic level by the availability of metabolic enzymatic co-factors. Since the basal levels of H3K27me3 are influenced by both EZH2 and KDM6A/B activities, we expect that additional metabolic cofactors that regulate their activity including methyl donation (EZH2) or 2-oxogluterate (KDM6A/B) will similarly influence DICER H3K27me3 levels and its expression.
In contrast to genetic alteration of EZH2 and/or KDM6A/B, hypoxia driven changes in the epigenetic state of DICER enables transient changes in cell phenotype in response to the local tumor microenvironment. In breast cancer, our findings demonstrate that suppression of DICER during hypoxia is sufficient to reduce miR-200 levels and enable cells to undergo an EMT and acquire stem cell properties. The fact that this occurs at the epigenetic level in an EZH2 dependent manner is consistent with a recent report that implicates EZH2 in controlling the EMT through epigenetic reprogramming58, but provides a potential mechanism for cells to retain plasticity and restore DICER and miR-200 levels when oxygen becomes available. Cancer cells need to undergo the reverse process, mesenchymal to epithelial transition, to ensure successful colonization and metastatic outgrowth59, 60. We speculate that the clinical importance of hypoxia in driving metastasis and poor outcome is linked to this ability to transiently influence cell phenotype via DICER expression and miRNA biogenesis.
Finally, our results provide new potential therapeutic opportunities for targeting hypoxia and its influence on poor outcome in cancer. Hypoxia has been demonstrated to influence patient outcome both through its ability to promote metastatic growth of disseminated stem cells, and for regrowth of tumors following treatment. Hypoxic cells are intrinsically resistant to radiation and other forms of chemotherapy, and small numbers of these cells can ‘re-seed’ the tumor, enabling regrowth following therapy. Hypoxic tumor cells also arise as a consequence of treatment with anti-angiogenic agents, and can contribute to tumor regrowth post therapy61. Consequently, the ability of hypoxic cells to stimulate tumor initiation – either in naïve metastases or following therapy of primary tumors, is considered to play a large role in treatment outcome. It may be possible to interfere with the epigenetic regulation of DICER during hypoxia and/or its influence on the expression of key miRNAs that promote these adverse clinical effects. Recent potent, orally available inhibitors of EZH2 have been reported62, 63, and our results suggest that treatment with these agents induce DICER expression. Alternatively, a siRNA and/ or miRNA strategy that does not rely on Drosha/Dicer processing and is not compromised within the tumor microenvironment could be applied. Effective delivery of siRNA and/or miRNA in vivo has improved remarkably and pre-clinical studies have demonstrated the potential of miR-200b delivery in breast, ovarian, and other orthotopic models64.
Methods
Data set analyses
Retrieval and processing of breast cancer datasets
Preprocessing of raw mRNA abundance datasets was performed in R statistical environment (v2.14.1). Raw affymetrix-based data (PMIDs: 16273092, 17545524, 16141321, 16280042, 16478745, 18498629, 17157792, 19421193, 20098429, 20064235, 20490655, 20697068, 18821012, 18593943, 15721472, 21501481, 17079448, 21422418 and 21558518) were normalized using RMA (robust multi-array average) algorithm65 (R package: affy v1.32.1). ProbeSet annotation to Entrez Gene IDs was performed using custom CDFs66 (R packages: hgu133ahsentrezgcdf v15.0.0, hgu133bhsentrezgcdf v15.0.0, hgu133plus2hsentrezgcdf v15.0.0, hthgu133ahsentrezgcdf v15.0.0, hgu95av2hsentrezgcdf v15.0.0 for affymetrix-based breast cancer datasets). The METABRIC breast cancer dataset was preprocessed, summarized and quantile-normalized from the raw expression files generated by Illumina BeadStudio (R packages: beadarray v2.4.2 and illuminaHuman v3.db_1.12.2). Raw data files were downloaded from European genome-phenome archive (EGA) (Study ID: EGAS00000000083). Data files of one sample were not available at the time of this analysis, and were therefore excluded. All datasets were normalized independently. Preprocessed segmented genome copy number aberration (CNA) data from the METABRIC cohort was downloaded from EGA. Preprocessed mRNA abundance data from The Cancer Genome Atlas (TCGA) were downloaded from cBioPortal for Cancer Genomics of Memorial Sloan-Kettering Cancer Center (MSKCC) http://www.cbioportal.org/public-portal/ on Nov. 6, 2012. Level 4 processed copy number aberration (CNA) data from TCGA were downloaded from GISTIC2 from the Broad Institute at http://gdac.broadinstitute.org/runs/analyses__2012_09_13/reports/cancer/BRCA/copynumber/gistic2/nozzle.html Discrete amplification and deletion calls for each sample based on the obtained CNA data were tabulated. For both mRNA abundance and CNA data, patient clinical information were obtained online from the original publication35.
Calculation of Winter signature scores
For each gene in the Winter signature, each patient in a given cohort was assigned an initial score of either +1 or −1 as follows: A patient was assigned +1 if her expression of that gene exceeded the median expression of that gene in that complete cohort. Otherwise, the patient was assigned −1 for that gene. For each patient in a given cohort, the +1’s and −1’s obtained as described above over all genes in the Winter signature were summed, and the resulting sum was the Winter signature score for that patient in that cohort.
mRNA abundance correlation analysis of Dicer with MITF or TP63
For each of the Metabric (Training and Validation datasets combined) and TCGA breast cancer mRNA abundance data sets, a scatter plot was generated of the Dicer mRNA abundance against that of MITF. Pearson’s product-moment correlation coefficient of the mRNA abundance of the two genes was computed and its p-value, based on the two-sided t-test for the Pearson’s product-moment correlation, was computed. This analysis was repeated for the correlation of Dicer mRNA abundance and TP63 mRNA abundance.
Comparison of Dicer mRNA abundance and Winter signature
For each of the Metabric (Training and Validation datasets combined) and TCGA breast cancer data sets, only patients with no Dicer copy-number variation and those with monoallelic Dicer loss were retained. These retained patients were then divided into five groups as follows: Those with monoallelic Dicer loss formed one group, and those with no Dicer copy-number variation were ordered by their Winter signature scores and divided into four quarters, with the first quarter comprising patients with the lowest Winter signature scores while the fourth represents those with highest Winter signature scores. A strip plot was generated to visualize the differences in Dicer mRNA abundance levels among these five groups. The Dicer mRNA abundance of patients in each of the four quarters of the group with no Dicer copy-number variation was compared against the group with monoallelic Dicer loss, by computing the p-value based on the Wilcoxon rank sum test.
Comparison of miR-103/107 expression and Winter signature
Analyses were carried out in R statistical environment (version 3.0.1) (http://www.r-project.org/). All tests were two-sided and considered statistical significant at the 0.05 level. Clinical and miRNA expression were downloaded from the Cancer Genome Atlas Project (TCGA) available through the associated files of the paper: Comprehensive molecular portraits of human breast tumors, Nature, September 27, 2012. https://tcga-data.nci.nih.gov/docs/publications/brca_2012/. We also downloaded Level 3 (RNASeqV2) genes profiles for Breast Invasive Carcinoma from TCGA Data Portal: https://tcga-data.nci.nih.gov/tcga/. A hypoxia score was assigned to each TCGA sample based on the Winter hypoxia metagene signature and following the previously reported methodology67. The Shapiro-Wilk test was applied to verify if the data follows a normal distribution. Accordingly, an analysis of variance test with Tukey post-hoc test was applied to assess the relationship between miR-103 and Winter hypoxia score, whereas a non-parametric test Kruskal–Wallis test with Nemenyi post-hoc test was applied to assess the relationship between miR-107 expression and Winter hypoxia score.
Cell culture and treatment
The following cell lines were obtained from ATCC and grown according to provided subculturing instructions: MCF7, MDA-MB-468, MDA-MB-231, T47D, U373, HCT116, Hela, ME180 and SiHa. HCC1954 and SUM149 cell lines were provided as a gift from Dr. Benjamin Neel (Princess Margaret Cancer Centre). HCC1954 cells were grown in RPMI medium supplemented with 10% FBS and SUM149 cells were grown in Ham’s F12 medium supplemented with 5% FBS, insulin (5 μg/ml), hydrocortisone (1 μg/ml), and 10mM Hepes (pH 7.4). HMLER cells were a gift from Dr. Robert Weinberg (MIT) and were grown as previously described68. MCF10A cells were provided by Dr. Senthil Muthuswamy (Princess Margaret Cancer Centre) and were grown in DMEM/F12 (Gibco BRL) supplemented with 5% donor horse serum, 20 ng/ml EGF, 10 μg/ml insulin, 1 ng/ml cholera toxin, 100 μg/ml hydrocortisone, 50 μg/ml penicillin and 50 μg/ml streptomycin. WT and HIF1α−/− MEFs, RCC4 and RCC4+pVHL were grown in DMEM medium supplemented with 10% FBS. All cell lines were routinely tested to confirm the absence of Mycoplasma. For hypoxic exposure cells were transferred into a HypOxygen H35 workstation. The atmosphere in the chamber consisted of 5% H2, 5% CO2, the desired % O2, and residual N2. For protein stability experiments, MCF7 cells were exposed to 100 μg/ml cycloheximide (Sigma) for 0, 2, 4, or 8 hours of normoxia or 0.2% O2 hypoxia. For exposure to stress inducing agents, indicated cell lines were grown for 24 hrs in 250 μM cobalt chloride (CoCl2), 500 μM deferoxamine (DFO) or 500 μM dimethyloxalylglycine (DMOG). For inhibition of KDM6A/B activity, MCF7 and HMLER cells were grown for 24 hrs in GSK-J4 at indicated concentrations. For inhibition of EZH2 activity, MCF7 and HMLER cells were grown for 48-72 hrs in UNC1999 or GSK343 at indicated concentrations. For exposure to TGFβ1, HMLER cells were cultured in standard growth medium with the addition of 5% FBS and treated with 2.5 ng/ml TGFβ1 for 12 days.
RNA extraction and quantitative RT-PCR
RNA was isolated using TRI reagent (Sigma) and samples were reverse transcribed using qScript cDNA SuperMix (Quantas). Quantitative real-time PCR was performed on an Eppendorf Realplex2 mastercycler using SYBR green (Quantas). Specific primers used are listed in Supplementary Table 2. For determination of DICER mRNA half life cells were treated with 5 μg/ml actinomycin D (Sigma) for the indicated times. De novo RNA synthesis was measured using the Click-iT Nascent RNA Capture Kit (Molecular Probes). Briefly, MCF7 and HMLER cells were pulse labeled with 0.2mM 5-ethynyl Uridine for 1 hour during aerobic conditions or 24 hrs hypoxia (0.2% O2) after which RNA was isolated as described above. Nascent RNA was captured using magnetic streptavidin beads, reverse transcribed and analyzed by quantitative real-time PCR. For determination of pri-miRNA and mature miRNA levels the following assays from Applied Biosystems were used: pri-miR200a (Hs03303376_pri), pri-miR200b (Hs03303027_pri), pri-miR429 (Hs03303727_pri), pri-miR200c (Hs03303157_pri), pri-miR141 (Hs03303157_pri), pri-miR210 (Hs03302948_pri), hsa-miR200a (000502), hsa-miR200b (002251), hsa-miR429 (001024), hsa-miR200c (002300), miR141 (002145), hsa-miR103 (000439), hsa-miR107 (000443), hsa-miR210 (000512), RNU44 (001094), RNU48 (001006).
Western blot analysis
Cells were washed twice with cold PBS and scraped in 50 mM Tris HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS supplemented with protease and phosphatase inhibitors (Roche). After centrifugation at 10,000 g supernatants were boiled in Laemmli buffer for 10 min and proteins were resolved by SDS-PAGE. Proteins were subsequently transferred onto PVDF membrane and blocked for 1 hour in TBS containing 0.05% Tween 20 (TBS-T) supplemented with 5% skim milk powder. Membranes were probed overnight at 4°C with antibodies directed against: DICER (1:200, H-212; Santa Cruz Biotechnology), CA9 (1:1000, M75; gift from Dr. Silvia Pastorekova), H3K27me3 (1:20000, 07-449; Upstate), H3 (1:5000, D1H2; Cell Signaling Technology), E-cadherin (1:1000, Cell Signaling Technology), Vimentin (1:500, clone V9; Sigma), β-tubulin (1:20000, Abcam), β-actin (1:20000, Sigma) or eIF4E (1:1000, BD Transduction Laboratories). Bound antibodies were visualized using HRP-linked secondary antibodies (GE Healthcare) and ECL luminescence (Pierce).
Luciferase reporter assay
A 2.5 kb fragment from the 5′ flanking region of the DICER1 gene was previously described15. MCF7 cells were transiently co-transfected with the DICER or CA9 promoter69 constructs and pcDNALacZ using Lipofectamine (Invitrogen). Transfected cells were subcultured 16 hrs post-transfection, exposed to hypoxia and finally harvested 48 hrs after transfection. Luciferase and β-galactosidase activity was measured using a commercial kit (Applied Biosystems) and measured on the Fluorstar Optima plate reader (BMG Labtech).
Plasmids and viral infections
Knockdown was achieved using lentiviral shRNA constructs directed against DICER: TRCN0000004386, TRCN000000439; HIF-1α: TRCN0000003810; KDM6A: TRCN0000107760; KDM6B: TRCN0000236677; GFP as control: TRCN0000072181. DICER ORF was cloned into pLenti CMV DEST using Gateway LR Clonase II (Invitrogen). PrimiR200b was cloned into pLJM1 as Age1/EcoR1 fragment. Lentiviral particles were generated by co-transfection of 293T cells with packaging plasmids pCMVdR8.74psPAX2 and pMD2.G together with shRNA vector pLKO.1 using Lipofectamine 2000. Virus supernatant was harvested 48 and 72 hrs post-transfection. Cell lines were transduced with lentiviral supernatant in the presence of 8 μg/ml polybrene. Infected cells were selected for 48 hrs in 2 μg/ml puromycin containing media or 7 days in 5 μg/ml blasticidin containing media. Validated short interfering RNA (siRNA) duplexes directed against EZH251 AAGACTCTGAATGCAGTTGCT and HIF1α CUGAUGACCAGCAACUUGA were ordered from Sigma. Stealth RNAi negative control was ordered from Invitrogen (12935-300). For siRNA experiments cells were transfected 72 hours before analysis with 2 nM siRNA duplex using lipofectamine (Invitrogen).
Chromatin immunoprecipitation (ChIP)
MCF7 and HMLER cells were fixed in 1% formaldehyde. Cross-linking was allowed to proceed for 10 min at room temperature and stopped by addition of glycine at a final concentration of 0.125 M, followed by an additional incubation for 5 min. Fixed cells were washed twice with PBS and harvested in SDS Buffer (50 mM Tris at pH 8.1, 0.5% SDS, 100 mM NaCl, 5 mM EDTA), supplemented with protease inhibitors (Aprotinin, Antipain and Leupeptin all at 5μg/mL and 1mM PMSF). Cells were pelleted by centrifugation, and suspended in IP Buffer (100 mM Tris at pH 8.6, 100 mM NaCl , 0.3% SDS, 1.7% Triton X-100, and 5 mM EDTA), containing protease inhibitors. Cells were disrupted by sonication, yielding genomic DNA fragments with a bulk size of 200-500 bp. For each immunoprecipitation, 1 mL of lysate was precleared by addition of 35 μL of blocked protein A beads (50% slurry protein A-Sepharose, Amersham; 0.5 mg/mL fatty acidfree BSA, Sigma; and 0.2 mg/mL herring sperm DNA in TE), followed by clarification by centrifugation. 10μl aliquots of precleared suspension were reserved as input DNA and kept at 4°C. Samples were immunoprecipitated overnight at 4°C using 1 μg of antibodies for either HA as a negative control (sc-805; Santa Cruz), H3K27me3 (07-449; Upstate), EZH2 (AC22, Millipore), KDM6A (ab36938, Abcam) or KDM6B (ab85392, Abcam). Immune complexes were recovered by adding 40 μL of blocked protein A beads and incubated for 4 hrs at 4°C. Beads were washed three times in 1 mL of Mixed Micelle Buffer (20 mM Tris at pH 8.1, 150 mM NaCl, 5 mM EDTA, 5% w/v sucrose, 1% Triton X-100, and 0.2% SDS), twice in 1mL of Buffer 500 (50 mM HEPES at pH 7.5, 0.1% w/v deoxycholic acid, 1% Triton X-100, and 1 mM EDTA), twice in 1mL of LiCl Detergent Wash Buffer (10 mM Tris at pH 8.0, 0.5% deoxycholic acid, 0.5% NP-40, 250 mM LiCl, and 1 mM EDTA), and once in 1mL of TE. Immuno-complexes were eluted from beads in 250μL elution buffer (1% SDS; and 0.1M NaHCO3) for 2 hrs at 65°C with continuous shaking at 1000 rpm, and after centrifugation supernatants were collected. 250μL elution buffer was added to input DNA samples and these were processed in parallel with eluted samples. Crosslinks were reversed overnight at 65°C followed by a 2h digestion with RNAseA at 37°C and 2h proteinase K (0.2μg/μL) at 55°C. DNA fragments were recovered using QIAquick PCR purification columns, according to manufacturers’ instructions. Samples were eluted in 75 μL EB buffer and then further 1/5 diluted in TE buffer. The immunoprecipitated DNA was quantified by real-time qPCR using SYBR® Green I (Applied Biosystems) and following forward and reverse primers: DICER: FCGGTGGGCGTTAAATAAGTG and R-CCCCCATACTGAGATGCTGT. After PCR melting curves were acquired to ensure that a single product was amplified in the reaction.
Global miRNA profiling
Total RNA was isolated from cells bearing shDICER G6 and pLKO.1 as control or cells exposed to hypoxia and samples were subjected to Nanostring analysis using the Nanostring Human v2 miRNA code set at the UHN Microarray Centre. miRNAs with counts equal or lower than the negative controls were discarded from the analysis. miRNA counts were normalized to housekeeping gene RPLP0 and averaged for 3 independent experiments.
Fluorescence-activated cell sorting
FACS analysis was performed on a BD FACS Calibur (Becton Dickinson) using PE-conjugated anti-CD24 antibody (clone ML5) and APC-conjugated anti-CD44 (clone G44-26) (BD bioscience).
Mammosphere culture
Mammosphere culture was performed as previously described70 with slight modifications. The mammospheres were cultured for 7-10 days in MammoCult media (StemCell Technologies) supplemented with 4 μg/ml heparin (Sigma) and 0.5 μg/ml hydrocortisone (Sigma). For sphere formation assays, mammospheres were dissociated to single cells with trypsin and 500 dissociated cells were plated in a 96-well plate and cultured for 10 days. Mammospheres with diameter >75 μm were counted.
In vivo models
All animal experiments were performed under protocols approved by the Ontario Cancer Institute’s Animal Care Committee, according the regulations of the Canadian Council on Animal Care. Female NOD-SCID mice at 6-8 weeks old were used to inject HMLER cells into the inguinal mammary fat pad (1 × 106 cells in a 1:1 mixture of BD Matrigel and media) following anesthetization with isoflurane. One week prior to the injection of cells, a 60-day release pellet containing 2 mg 17β-estradiol and 20 mg progesterone (Innovative Research of America) was implanted subcutaneously into each mouse. At end point, mice were sacrificed, tumors harvested, and OCT embedded for immunohistochemical staining and analysis.
Immunohistochemical staining and image analysis
The expression of CD44 and EF5 in orthotopic xenografts was investigated as follows: Flash frozen tissue samples were embedded in OCT and stored at −80C until sectioned. Sections were thawed at room temperature prior to fixation in 2% paraformaldehyde for 20 mins. After washing sections in PBS, sections were permeabilized in PBS containing 0.5% Triton X-100 for 15 minutes, wash 3×5 mins in PBS-T and then incubated in a primary antibody cocktail of mouse anti-human CD44 (1:75, BD Pharmingen) and rat anti-mouse CD31 (1:300, BD Pharmingen) or anti-human H3K27me3 (1:500, C36B11; Cell Signalling) overnight at room temperature. Sections were subsequently washed 3×5 mins in PBS-T followed by incubation in a secondary antibody cocktail of goat anti-mouse Alexafluor 488 (1:200, Life Technologies) and goat anti-rat Alexafluor 555 (1:200, Life technologies) or goat anti-mouse Alexafluor 488 (1:200, Life Technologies) for 1 hour at room temperature. After washing in PBS-T, sections were incubated in EF5-Cy5 (1:50, provided by Dr. Cameron Koch) for 3 hours at room temperature. After washing, sections were finally incubated in a working solution of DAPI for 5 minutes, washed, dried and imaged on a laser scanning microscope (Huron Technologies). Images were analyzed using Definiens Tissue Studio software, which allows for semi-automatic histology image analysis. Briefly, the software was trained to identify viable tumor areas, necrotic areas, tumor stroma, and empty areas within the scanned section. A threshold was determined by mean +2SD intensity in the EF5 and CD44 channels. The average CD44 staining intensity within the tumor area was measured in both EF5 negative and positive areas. EF5-positive area above a background threshold was obtained from the average intensity of all tumor sections.
Statistical analyses
Unless otherwise stated, a Student’s t-test or one-way ANOVA with Bonferroni’s post hoc test were used to test significance between populations. A significance threshold of p<0.05 was applied. Points and error bars plotted in graphs represent the mean ± s.e.m for 3 or more independent experiments..
Supplementary Material
Acknowledgements
We want to thank R. Weinberg for providing the HMLER cells. We would also like to thank C. Arrowsmith for providing the SGC inhibitors. We would like to thank Alison Casey, Milan Ganguly and Trevor Do for advice and technical assistance on the immunohistochemical staining in these and other tissue samples. This work was financially supported by the Dutch Cancer Society (KWF grant UM 2008-4068 to BW), the Ontario Ministry of Health and Long Term Care (OMOHLTC), the Terry Fox New Frontiers Research Program (PPG09-020005 to B.G.W and M.K.), the Ontario Institute for Cancer Research and Terry Fox Research Institute (Stem Cell Program to B.G.W.), the Canadian Institute for Health Research (CIHR grant 201592 to B.G.W. and M.K.) and the EU 7th framework program (METOXIA project 222741 to B.G.W. and M.K.). The views expressed do not necessarily reflect those of the OMOHLTC. This study was conducted with the support of the Ontario Institute for Cancer Research to P.C.B. through funding provided by the Government of Ontario. E.K. and C.Q.Y. were supported by fellowships from the Canadian Breast Cancer Foundation (CBCF). This study makes use of data generated by the Molecular Taxonomy of Breast Cancer International Consortium34, which was funded by Cancer Research UK and the British Columbia Cancer Agency Branch.
A.L.H. was supported by funding from Cancer Research UK (CRUK_A11359) and the Breast Cancer Research Foundation (BCRF). A.K.S. was supported by funding from the National Institute of Health (U54 151668, P50 CA083639, and UH2 TR000943). M.I. was supported by funding from the National Institute of Health (R01 CA155332-01).
Footnotes
References
- 1.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 5.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Martello G, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Melo SA, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Melo SA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Su X, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grelier G, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karube Y, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merritt WM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy C, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho JJ, et al. Functional importance of Dicer protein in the adaptive cellular response to hypoxia. The Journal of biological chemistry. 2012;287:29003–29020. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaupel P. Prognostic potential of the pre-therapeutic tumor oxygenation status. Advances in experimental medicine and biology. 2009;645:241–246. doi: 10.1007/978-0-387-85998-9_36. [DOI] [PubMed] [Google Scholar]
- 18.Fyles A, et al. Tumor hypoxia has independent predictor impact only in patients with node-negative cervix cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2002;20:680–687. doi: 10.1200/JCO.2002.20.3.680. [DOI] [PubMed] [Google Scholar]
- 19.Schindl M, et al. Overexpression of hypoxia-inducible factor 1alpha is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:1831–1837. [PubMed] [Google Scholar]
- 20.Hussain SA, et al. Hypoxia-regulated carbonic anhydrase IX expression is associated with poor survival in patients with invasive breast cancer. Br J Cancer. 2007;96:104–109. doi: 10.1038/sj.bjc.6603530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan M, Rayoo M, Takano EA, Fox SB. BRCA1 tumours correlate with a HIF-1alpha phenotype and have a poor prognosis through modulation of hydroxylase enzyme profile expression. Br J Cancer. 2009;101:1168–1174. doi: 10.1038/sj.bjc.6605287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiraga T, Kizaka-Kondoh S, Hirota K, Hiraoka M, Yoneda T. Hypoxia and hypoxia inducible factor-1 expression enhance osteolytic bone metastases of breast cancer. Cancer Res. 2007;67:4157–4163. doi: 10.1158/0008-5472.CAN-06-2355. [DOI] [PubMed] [Google Scholar]
- 23.Brennan DJ, et al. CA IX is an independent prognostic marker in premenopausal breast cancer patients with one to three positive lymph nodes and a putative marker of radiation resistance. Clin Cancer Res. 2006;12:6421–6431. doi: 10.1158/1078-0432.CCR-06-0480. [DOI] [PubMed] [Google Scholar]
- 24.Vaupel P, Briest S, Hockel M. Hypoxia in breast cancer: pathogenesis, characterization and biological/therapeutic implications. Wien Med Wochenschr. 2002;152:334–342. doi: 10.1046/j.1563-258x.2002.02032.x. [DOI] [PubMed] [Google Scholar]
- 25.Cairns RA, Hill RP. Acute hypoxia enhances spontaneous lymph node metastasis in an orthotopic murine model of human cervical carcinoma. Cancer research. 2004;64:2054–2061. doi: 10.1158/0008-5472.can-03-3196. [DOI] [PubMed] [Google Scholar]
- 26.Cairns RA, Kalliomaki T, Hill RP. Acute (cyclic) hypoxia enhances spontaneous metastasis of KHT murine tumors. Cancer Res. 2001;61:8903–8908. [PubMed] [Google Scholar]
- 27.Tamara Marie-Egyptienne D, Lohse I, Hill RP. Cancer stem cells, the epithelial to mesenchymal transition (EMT) and radioresistance: Potential role of hypoxia. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Hill RP, Marie-Egyptienne DT, Hedley DW. Cancer stem cells, hypoxia and metastasis. Semin Radiat Oncol. 2009;19:106–111. doi: 10.1016/j.semradonc.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Das B, Tsuchida R, Malkin D, Koren G, Baruchel S, Yeger H. Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cells. 2008;26:1818–1830. doi: 10.1634/stemcells.2007-0724. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaturvedi P, et al. Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirectional signaling promotes metastasis. J Clin Invest. 2013;123:189–205. doi: 10.1172/JCI64993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lock FE, et al. Targeting carbonic anhydrase IX depletes breast cancer stem cells within the hypoxic niche. Oncogene. 2012 doi: 10.1038/onc.2012.550. [DOI] [PubMed] [Google Scholar]
- 34.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winter SC, et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67:3441–3449. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- 37.Pollard PJ, et al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem J. 2008;416:387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 39.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 40.Okajima M, et al. Anoxia/reoxygenation induces epithelial-mesenchymal transition in human colon cancer cell lines. Oncology reports. 2013;29:2311–2317. doi: 10.3892/or.2013.2401. [DOI] [PubMed] [Google Scholar]
- 41.Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. The Journal of biological chemistry. 2011;286:2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang MH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 43.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimono Y, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blick T, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in human breast cancer. Journal of mammary gland biology and neoplasia. 2010;15:235–252. doi: 10.1007/s10911-010-9175-z. [DOI] [PubMed] [Google Scholar]
- 46.Shen J, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 48.Yap DB, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varambally S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. The EMBO journal. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banerjee R, et al. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene. 2011;30:4339–4349. doi: 10.1038/onc.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crea F, Paolicchi E, Marquez VE, Danesi R. Polycomb genes and cancer: time for clinical application? Crit Rev Oncol Hematol. 2012;83:184–193. doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature genetics. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agger K, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes & development. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barradas M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes & development. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiwari N, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–783. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 59.Ocana OH, et al. Metastatic colonization requires the repression of the epithelial mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 60.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehta S, et al. Assessing early therapeutic response to bevacizumab in primary breast cancer using magnetic resonance imaging and gene expression profiles. J Natl Cancer Inst Monogr. 2011;2011:71–74. doi: 10.1093/jncimonographs/lgr027. [DOI] [PubMed] [Google Scholar]
- 62.Konze KD, et al. An Orally Bioavailable Chemical Probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013 doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amatangelo MD, Garipov A, Li H, Conejo-Garcia JR, Speicher DW, Zhang R. Three-dimensional culture sensitizes epithelial ovarian cancer cells to EZH2 methyltransferase inhibition. Cell Cycle. 2013;12:2113–2119. doi: 10.4161/cc.25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 66.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic acids research. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Starmans MH, et al. The prognostic value of temporal in vitro and in vivo derived hypoxia gene-expression signatures in breast cancer. Radiother Oncol. 102:436–443. doi: 10.1016/j.radonc.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 68.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van den Beucken T, et al. Hypoxia-induced expression of carbonic anhydrase 9 is dependent on the unfolded protein response. The Journal of biological chemistry. 2009;284:24204–24212. doi: 10.1074/jbc.M109.006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & development. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.