Abstract
We identify and characterize 14 small heat-shock protein (sHSP) genes from the diamondback moth (DBM), Plutella xylostella (L.), a destructive pest. Phylogenetic analyses indicate that, except for sHSP18.8 and sHSP19.22, the other 12 DBM sHSPs belong to five known insect sHSP groups. Developmental expression analysis revealed that most sHSPs peaked in the pupal and adult stages. The transcripts of sHSPs display tissue specificity with two exhibiting constitutive expression in four tested tissues. Expression of sHSP18.8 in fourth instar larvae is not induced by the tested abiotic stressors, and unless sHSP21.8 is not sensitive to thermal stress, 12 sHSPs are significantly up-regulated. The messenger RNA (mRNA) levels of all sHSPs are reduced under oxidative stress. Food deprivation leads to significant down-regulation of three sHSPs. The majority of sHSPs show expression variation to various heavy metals, whereas mRNA abundances of sHSP22.1 and sHSP 28.9 are reduced by four heavy metals. The responses of sHSPs to indoxacarb and cantharidin are varied. Beta-cypermethrin and chlorfenapyr exposure results in an increase of 13 sHSP transcripts and a reduction of 12 sHSP transcripts, respectively. These results show that different sHSPs might play distinct roles in the development and regulation of physiological activities, as well as in response to various abiotic stresses of DBM.
Keywords: Small heat-shock proteins, Expression profiles, Abiotic stress responses, Plutella xylostella (L.)
Introduction
Small heat-shock proteins are probably the most diverse in structure and function among the various superfamilies of stress proteins (Franck et al. 2004). Ranging in size from ∼12 to 42 kDa, the entire sequence of small heat-shock proteins (sHSPs) shows a low degree of similarity. Compared to other types of HSPs, a unique feature of the structure of sHSPs is a conserved α-crystallin domain (ACD) of ∼90 amino acids flanked by an N-terminal arm of divergent sequence and variable length and a C-terminal extension (Kriehuber et al. 2010; Basha et al. 2012). The ACD is a conserved β-sheet sandwich facilitating several subunits of sHSP to form a larger oligomer (van Montfort et al. 2001, 2002). sHSPs play their chaperone-like roles via binding to denatured proteins and prevent irreversible protein aggregation under stress conditions, such as extreme temperature, UV irradiation, oxidation, heavy metals, and chemical intoxication (Haslbeck et al. 2005; Basha et al. 2012). In addition, sHSPs have been suggested to be involved in diverse physiological processes, such as apoptosis and autophagy, actin and intermediate filament dynamics, organization of the cytoskeleton, and membrane fluidity (Haslbeck 2002; Quinlan 2002; Tsvetkova et al. 2002; Sun and MacRae 2005).
Small heat-shock proteins are abundant and ubiquitously expressed in almost all organisms (Waters and Rioflorido 2007; Aevermann and Waters 2008; Waters et al. 2008). Genome sequence data continue to expand our understanding of the heterogeneity of sHSPs (Waters et al. 2008; Poulain et al. 2010; Kriehuber et al. 2010). Ten sHSPs (HSPB1-B10) have been identified and characterized in the human genome (Kappé et al. 2003). In plants, 19 sHSP genes, 36 sHSP genes, and 23 sHSP genes have been identified from Arabidopsis thaliana, Populus trichocarpa, and Oryza sativa genomes, respectively (Waters et al. 2008). Identification of 17 sHSPs from five diverse algal genomes has been reported (Waters and Rioflorido 2007). Eighteen sHSPs were found in the Caenorhabditis elegans genome and 20 sHSPs in the Caenorhabditis briggsae genome (Aevermann and Waters 2008). Insects are one of the most successful organisms, having evolved a strong ability to adapt to various habitats. Sixteen sHSPs have been identified in Bombyx mori, 11 in Drosophila melanogaster, 10 in Apis mellifera, and 7 in Anopheles gambiae (Li et al. 2009).
Previous studies have suggested that insect sHSPs play protective roles in response to abiotic and biotic stresses (Zhao and Jones 2012); they may also be involved in physiological processes related to developmental events (Rinehart et al. 2007; Gkouvitsas et al. 2008; Shen et al. 2011; Lu et al. 2014). However, studies of sHSPs in insects are not as extensive and exhaustive as in other organisms. To explore the diversity of structure and function of sHSPs in a worldwide destructive pest, the diamondback moth (DBM) Plutella xylostella (L.), 14 sHSPs were identified from the recently developed genomic and transcriptomic database for DBM (KONAGAbase) (Jouraku et al. 2013). In this study, we analyzed the temporal and spatial expression profiles of the 14 sHSP genes. We also monitored their responsiveness to thermal stress, oxidative stress, starvation, heavy metals, and pesticides.
Materials and methods
Insects and chemicals
An insecticide-susceptible strain of P. xylostella, maintained in the laboratory for >5 years without exposure to insecticides, was reared on pakchoi cabbage at 25 ± 2 °C, 50 ± 5 % relative humidity with a photoperiod of 16L:8D. Moths were supplied with a 5 % honey solution as nutrient and permitted to oviposition on moist gauze sterilized with a 1 % sodium hypochlorite solution gauze.
Indoxacarb, beta-cypermethrin, and chlorfenapyr were purchased from Jingchun Co. Ltd., Shanghai, China. Cantharidin was purchased from Alfa Aesar Chemical Co. Ltd (Haverhill, MA, USA). All other chemicals were of research grade or better and were obtained from commercial sources.
Identification and analysis of DBM sHSP genes
The putative DBM sHSP genes were obtained by keyword, “sHSP” or “small heat shock protein,” via searching the putative gene set (version 2) derived from the transcriptome of DBM deposited in the KONAGAbase (http://dbm.dna.affrc.go.jp/px/) (Jouraku et al. 2013). The open reading frame (ORF) of sHSP genes were deduced using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Each ORF was further searched using BLASTX against the non-redundant database at the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm its identity with other insect sHSP genes. The molecular weight of the deduced amino acid sequence of each full-length sHSP gene was predicted using the ExPASy Compute pI/Mw tool (http://web.expasy.org/compute_pi/). The alignment of deduced amino acid sequences was performed using the online tool, Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/), and sequence similarity was calculated according to the observed divergence. Secondary structure prediction was carried out with the PHD software accessed on the NPS@Web server (http://npsa-pbil.ibcp.fr). A phylogenetic tree was constructed based on the amino acid sequences by MEGA 5.0 using the neighbor-joining method with a bootstrap test of 1,000 replicates.
Sample preparation
For stage-specific expression analyses, the eggs, first to fourth instar larvae, pupae, and adults, were collected and stored at −80 °C until use. For tissue-specific expression analyses, four tissues including the head, gut, epidermis, and hemolymph from the fourth larvae were dissected on ice. These were then snap frozen and stored at −80 °C until use. Each sample was replicated three times.
Newly molted fourth instar larvae were selected for abiotic stress treatments. For temperature treatments, fourth instar larvae were exposed to 4 and 42 °C for 2 h, and then recovered at 25 °C for 1 h. For the oxidative stress treatment, adults were fed with 10 % H202 in a 5 % honey solution. For the starvation treatment, fourth instar larvae were deprived of pakchoi cabbage leaves for 24 h. The leaf-dipping method (Trisyono and Whalon 1997) was employed in the heavy metal and pesticide treatments. For the heavy metal treatments, fourth instar larvae were exposed to 10 mM Cu2+, Ni2+, Mn2+, and Pb2+ (prepared in 1 g L−1 Triton X-100 solution) for 24 h, respectively. For pesticide treatments, fourth instar larvae were exposed to 1.0 mg L−1 indoxacarb, 50.0 mg L−1 for beta-cypermethrin, 2.0 mg L−1 chlorfenapyr, and 10.0 mg L−1 cantharidin (LC50 values, prepared in 1 g L−1 Triton X-100 solution) for 24 h, respectively. Each treatment or control sample contained 16 larvae or adults with three independent replications. All samples were snap frozen and stored at −80 °C until use.
Real-time quantitative PCR
Total RNA was extracted using Trizol Plus (TaKaRa, Dalian, China) following the manual instructions. Complementary DNAs (cDNAs) were synthesized from 1.0 μg RNA using PrimScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) and stored at −20 °C. Primer3 (http://www.simgene.com/Primer3) was employed to generate all primers (Table 1). Real-time reactions were conducted on a thermal cycler (iQ 5, Bio-Rad, Philadelphia, PA, USA) in a 20-μL total reaction volume containing 10 μL of 2xUltra SYBR Mixture (CWBIO, Beijing, China), 0.8 μL each of gene-specific primers and the cDNAs templates, and 7.6 μL of double distilled water. Thermal cycling conditions were 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min, then followed by a dissociation analysis to check the homogeneity of the PCR product. The reaction was repeated three times for each gene. Each replicate was performed with an independent RNA sample preparation and consisted of three technical replicates. Samples were normalized using the actin gene (accession: JN410820) Ct values. Basal expression levels were represented as folds over the expression levels of actin. Fold inductions were calculated with the 2−∆∆Ct method (Livak and Schmittgen 2001) between treatment and control samples for each biological replicate.
Table 1.
Sequences of qPCR primers
| Gene | Primer name | Sequence (5′–3′) |
|---|---|---|
| sHSP18.8 | qF | GTCATTTCTGCCGCTTCTTC |
| qR | AAACCCCTTGGCTGTTCTTT | |
| sHSP19.22 | qF | CCGCTGAAGTACATGAAGCA |
| qR | CCCACTGTCTTCACCTGGAT | |
| sHSP19.23 | qF | GTCTCTTCTGCCGCTGCTAT |
| qR | TTTATGTTGGAGCCGAGGTC | |
| sHSP19.5 | qF | ACGAGCACGGGTTTATATCG |
| qR | ACAGCACCCCATCTGAAGAC | |
| sHSP20.06 | qF | GCACGAAGAGAAGAAGGACG |
| qR | TTCTGGGCAGACTTTTCGTT | |
| sHSP20.09 | qF | GATGTCGGCGGACTACTACC |
| qR | TGCTCGTCCTTCTTCTCCTC | |
| sHSP20.1 | qF | GACTACGAGATCGAGCGTCC |
| qR | TCCTGCTTCTCCTCGTGTTT | |
| sHSP21.6 | qF | CTGGGACAGCCTCAACTCTC |
| qR | TGTACTCCCGGTAGACGGAC | |
| sHSP21.8 | qF | AGGAGAAGCAAGACGAGCAC |
| qR | GACCGGTTTTCGTAATTGGA | |
| sHSP21.9 | qF | CGTCTTCAGACCTTGGGAAG |
| qR | CTTCGGATAAACTGCCTGGA | |
| sHSP22.1 | qF | GGATGACCATGGCTACGTCT |
| qR | CATTGGCCTGATCTTCCACT | |
| sHSP23.4 | qF | GGATGACCATGGCTACGTCT |
| qR | CATTGGCCTGATCTTCCACT | |
| sHSP27.5 | qF | AAGGACGAGCTGAAGGTCAA |
| qR | AGGGGACAACTGACAACCAG | |
| sHSP28.9 | qF | CATCCACGGAGAAGAAGGAA |
| qR | GGGCGTAATCTAGCTCGTTG |
Statistical analysis
All data were presented as mean ± SD (standard deviation). Significant differences between treatment groups and the control group were analyzed by using Student’s t test; p < 0.05 was considered statistically significant. One-way ANOVA was used for multiple comparisons.
Results
Identification and characterization of DBM sHSPs
By searching the KONAGAbase database, 14 sHSP genes containing the full-length ORF were isolated and their deduced amino acid sequences showed high identities with other insect sHSPs. The sequence similarities were over 50 % among 6 sHSPs: sHSP23.4, sHSP19.5, sHSP20.06, sHSP20.09, sHSP19.23, and sHSP20.01. Moreover, sHSP21.8, sHSP21.9, and sHSP22.1 showed 40–48.5 % identities to each other and to those 6 sHSPs. However, the other 5 sHSPs displayed low similarities to all sHSPs (Table 2). Higher similarity means a closer evolutionary relationship. The low sequence similarities among DBM sHSPs suggested that they play diverse functional roles in physiological activities. Secondary structure analysis revealed that their deduced amino acid sequences comprise a typical α-crystallin domain which consists of eight β-strands. A characteristic C-terminal “I/V-x-I/V” motif exists in these DBM sHSPs (Fig. 1).
Table 2.
Percentage identities of amino acid residues among 14 DBM sHSPs
| sHSP27.5 | sHSP28.9 | sHSP21.6 | sHSP18.8 | sHSP19.22 | sHSP21.8 | sHSP21.9 | sHSP22.1 | sHSP23.4 | sHSP19.5 | sHSP20.06 | sHSP20.09 | sHSP19.23 | sHSP20.1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sHSP27.5 | – | 25.56 | 18.63 | 18.40 | 16.35 | 20.35 | 22.81 | 24.43 | 24.26 | 24.2 | 24.54 | 23.17 | 25.81 | 26.09 |
| sHSP28.9 | – | 20.78 | 24.84 | 25.16 | 27.27 | 26.59 | 25.97 | 25.27 | 30.25 | 26.95 | 26.79 | 29.94 | 30.06 | |
| sHSP18.8 | – | 27.35 | 27.21 | 30.27 | 29.25 | 34.9 | 32.39 | 31.03 | 28.19 | 28.19 | 30.41 | 33.33 | ||
| sHSP21.6 | – | 27.35 | 38.46 | 30.60 | 32.89 | 31.43 | 32.58 | 35.77 | 34.06 | 37.80 | 35.34 | |||
| sHSP19.22 | – | 32.64 | 29.94 | 35.04 | 28.95 | 32.06 | 35.51 | 36.23 | 32.85 | 32.37 | ||||
| sHSP21.8 | – | 45.09 | 42.08 | 40.00 | 44.91 | 47.09 | 46.82 | 48.50 | 48.26 | |||||
| sHSP21.9 | – | 47.88 | 43.75 | 47.80 | 50.61 | 50.61 | 50.63 | 51.2 | ||||||
| sHSP22.1 | – | 45.56 | 53.89 | 57.56 | 56.07 | 56.36 | 58.82 | |||||||
| sHSP23.4 | – | 50.00 | 55.37 | 55.62 | 59.41 | 57.71 | ||||||||
| sHSP19.5 | – | 59.88 | 59.88 | 54.02 | 60.95 | |||||||||
| sHSP20.06 | – | 95.00 | 73.96 | 76.00 | ||||||||||
| sHSP20.09 | – | 73.37 | 74.86 | |||||||||||
| sHSP19.23 | – | 78.49 | ||||||||||||
| sHSP20.1 | – |
Fig. 1.
Alignment of the deduced amino acid sequences of 14 DBM sHSPs. The amino acids with over 50 % identity are shaded in gray. Sequences above black stick are regions of α-crystallin domain. Eight β-strands within the α-crystallin domain are indicated with black arrows above. The C-terminal characteristic motif, “I/V-x-I/V,” is shown in rectangle
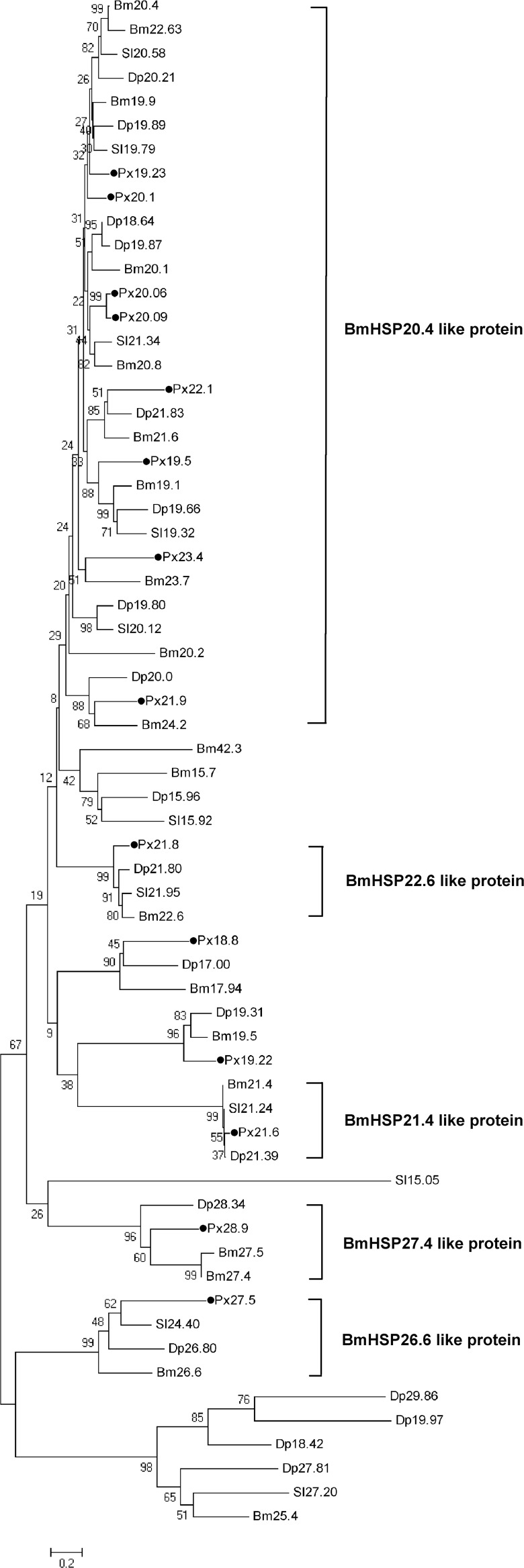
In order to analyze the relationships of DBM sHSPs to other insect sHSPs, 49 lepidopteron sHSPs, including 19 Danaus plexippus sHSPs, 10 Spodoptera litura sHSPs, and 20 B. mori sHSPs, were collected from GenBank (Table 3). The phylogenetic tree shows that 14 DBM sHSPs form at least five groups. Eight DBM sHSPs (sHSP19.23, sHSP19.5, sHSP20.06, sHSP20.09, sHSP20.1, sHSP21.9, sHSP22.1, and sHSP23.4) with high sequence similarities to each other were grouped into BmHSP20.4-like protein. Four DBM sHSPs, sHSP21.8, sHSP21.6, sHSP28.9, and sHSP27.5, belong to the group of BmHSP22.6-like protein, BmHSP21.4-like protein, BmHSP27.4-like protein, and BmHSP26.6-like protein, respectively. However, DBM sHSP18.8 and sHSP19.22 could not be assigned to any of the five known groups (Fig. 2). The sequence of sHSP19.5 (BAE48744) was available on GenBank. The sequences of the other 13 DBM sHSPs were deposited into GenBank with the accession numbers as follows: sHSP18.8 (KJ461915), sHSP19.22 (KJ461923), sHSP19.23 (KJ461913), sHSP20.06 (KJ461916), sHSP20.09 (KJ461920), sHSP20.1 (KJ461917), sHSP21.6 (KJ461922), sHSP21.8 (KJ461924), sHSP21.9 (KJ461919), sHSP22.1 (KJ461921), sHSP23.4 (KJ461918), sHSP27.5 (KJ461925), and sHSP28.9 (KJ461914).
Table 3.
The species and GenBank numbers of sHSPs sequences used for phylogenetic analysis
| Species | Gene | GenBank number |
|---|---|---|
| Plutella xylostella | sHSP19.23 | KJ461913 |
| sHSP28.9 | KJ461914 | |
| sHSP18.8 | KJ461915 | |
| sHSP20.06 | KJ461916 | |
| sHSP20.1 | KJ461917 | |
| sHSP23.4 | KJ461918 | |
| sHSP21.9 | KJ461919 | |
| sHSP20.09 | KJ461920 | |
| sHSP22.1 | KJ461921 | |
| sHSP19.5 | BAE48744 | |
| sHSP21.6 | KJ461922 | |
| sHSP19.22 | KJ461923 | |
| sHSP21.8 | KJ461924 | |
| sHSP27.5 | KJ461925 | |
| Danaus plexippus | sHSP20.21 | EHJ63989 |
| sHSP18.64 | EHJ63499 | |
| sHSP19.66 | EHJ63493 | |
| sHSP17.00 | EHJ63492 | |
| sHSP18.42 | EHJ78247 | |
| sHSP19.89 | EHJ77540 | |
| sHSP20.0 | EHJ77276 | |
| sHSP26.80 | EHJ77261 | |
| sHSP28.34 | EHJ77259 | |
| sHSP21.83 | EHJ74663 | |
| sHSP19.97 | EHJ72277 | |
| sHSP21.39 | EHJ69746 | |
| sHSP21.80 | EHJ68903 | |
| sHSP19.87 | EHJ68318 | |
| sHSP29.86 | EHJ63088 | |
| sHSP15.96 | EHJ77787 | |
| sHSP19.80 | EHJ77277 | |
| sHSP27.81 | EHJ73481 | |
| sHSP19.31 | EHJ67172 | |
| Spodoptera litura | sHSP21.95 | AFK14100 |
| sHSP19.32 | AFK14099 | |
| sHSP27.20 | AFK14101 | |
| sHSP19.79 | ADK55524 | |
| sHSP20.58 | ADK55522 | |
| sHSP20.12 | ADK55521 | |
| sHSP21.34 | ADK55520 | |
| sHSP21.24 | ADK55519 | |
| sHSP15.92 | AFK14098 | |
| sHSP15.05 | AFK14097 | |
| sHSP24.40 | ADK55523 | |
| Bombyx mori | sHSP21.4 | NP_001036985 |
| sHSP23.7 | NP_001036942 | |
| sHSP22.6 | NP_001091767 | |
| sHSP25.4 | NP_001112375 | |
| sHSP20.8 | NP_001091794 | |
| sHSP19.9 | NP_001036984 | |
| sHSP20.1 | NP_001036941 | |
| sHSP20.4 | NP_001037038 | |
| sHSP19.5 | NP_001164470 | |
| sHSP22.6 | ACM24354 | |
| sHSP27.5 | AHA85320 | |
| sHSP17.94 | XP_004923509 | |
| sHSP19.1 | XP_004923510 | |
| sHSP42.3 | XP_004922496 | |
| sHSP27.4 | NP_001276564 | |
| sHSP26.6 | NP_004923900 | |
| sHSP24.2 | NP_004923862 | |
| sHSP20.2 | NP_004923863 | |
| sHSP21.6 | NP_004933665 | |
| sHSP15.7 | NP_004926864 |
Fig. 2.
Phylogenetic tree of sHSPs from P. xylostella (Px), D. plexippus (Dp), S. litura (Sl), and B. mori (Bm). Percentage bootstrap values (1,000 replicates) are indicated on the nodes of the tree. The DBM sHSPs are labeled with black dots
Stage- and tissue-specific expression profile of DBM sHSPs
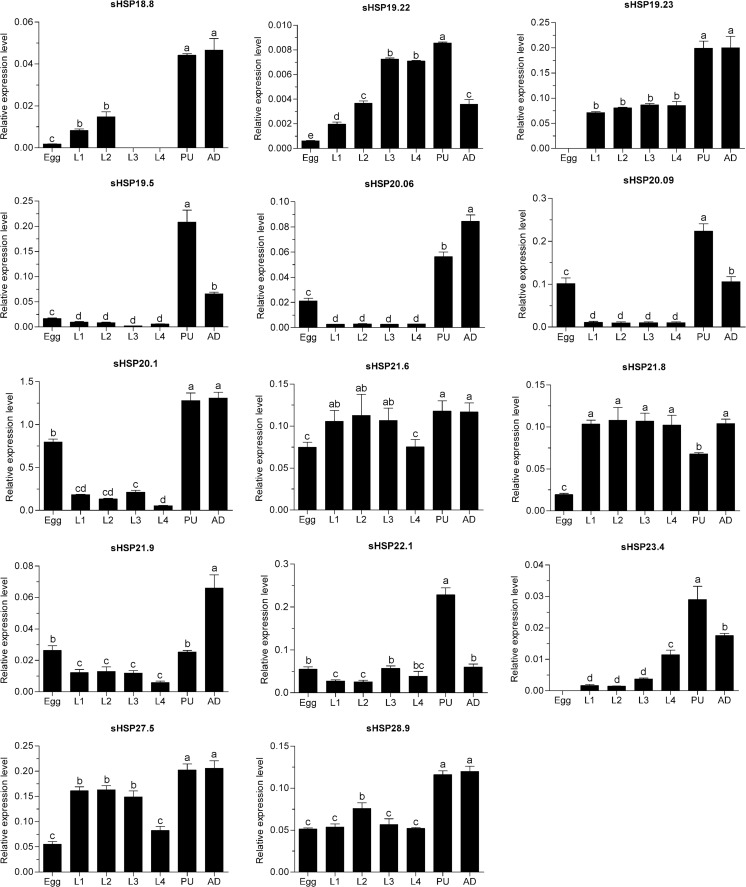
Stage-specific expression patterns of the DBM sHSPs were determined in the egg, larval, pupal, and adult stages by quantitative PCR (qPCR) reactions. All sHSPs were variously expressed throughout these developmental stages. sHSP19.23 and sHSP23.4 were not expressed in the egg stage, while sHSP18.8 expression was undetectable in the third and fourth larval stages. Most of the sHSPs were overexpressed in the pupal and adult stages. The messenger RNA (mRNA) levels of sHSP19.5, sHSP20.09, sHSP22.1, and sHSP23.4 in the pupal stage were significantly higher than that in other stages, while sHSP21.9 was more highly expressed in the adult stage. We also found that the expression levels of most sHSPs in the egg stage were relatively low. Moreover, the majority of the sHSPs showed similar expression levels throughout larval stages. sHSP19.23, sHSP19.5, sHSP20.06, sHSP20.09, sHSP20.1, sHSP21.9, and sHSP22.1 were expressed at lower levels in larval stages than that in other stages (Fig. 3).
Fig. 3.
Relative mRNA expression levels of the DBM sHSPs in different developmental stages. Expression levels were assessed using actin gene for normalization. Different letters on the tops of the column indicate significance in the different expression levels by ANOVA. L1 the first instar larvae, L2 the second instar larvae, L3 the third instar larvae, L4 the fourth instar larvae, PU pupae, AD adult
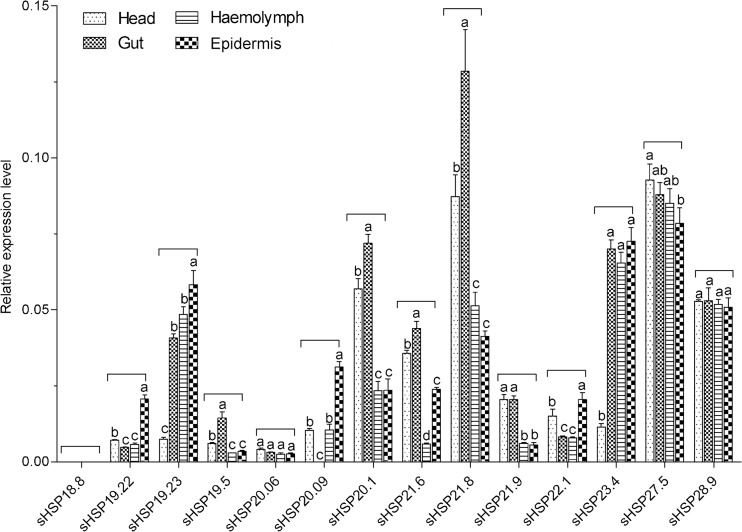
Tissue-specific expression profiles of DBM sHSPs were investigated in the head, gut, hemolymph, and epidermis of the fourth instar larvae. Expression of sHSP18.8 was not monitored in tested tissues, which is consistent with the finding that it was undetectable in fourth larval stages. sHSP20.09 was not expressed in the gut. Two sHSPs, sHSP20.06 and sHSP28.9, were uniformly expressed in four tested tissues. Only sHSP27.5 was found to have a high expression in the head. The mRNA levels of sHSP19.5, sHSP20.1, sHSP21.6, and sHSP21.8 in the gut were higher than those in other tissues. In addition, most sHSPs were moderately expressed in hemolymph. In the epidermis, sHSP19.22, sHSP19.23, sHSP20.09, sHSP22.1, and sHSP23.4 were highly expressed (Fig. 4).
Fig. 4.
Relative mRNA expression levels of sHSPs in different tissues of DBM fourth instar larvae. Expression levels were assessed using the actin gene for normalization. Different letters on the tops of the column indicate significance in the different expression levels by ANOVA
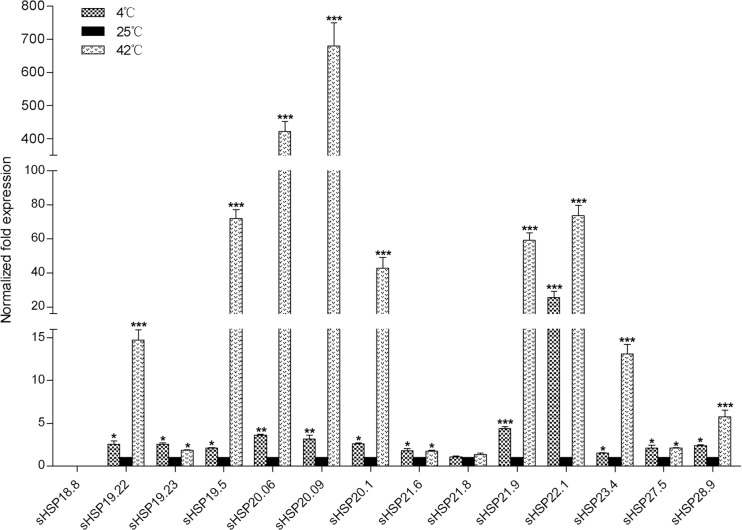
Effects of thermal stress on DBM sHSPs expression
Expression of sHSP18.8 was not detected either in cold- or in heat-shock showing that sHSP21.8 did not respond to either heat or cold stress. Other sHSPs were all up-regulated. After cold-shock treatment at 4 °C for 2 h, the expression level of sHSP22.1 increased most intensively by 25.5-fold. The mRNA levels of the other 12 sHSPs showed moderate increases of 1.5- to 4.7-fold. After heat-shock treatment at 42 °C for 2 h, the remaining 12 sHSPs showed quite different degrees of up-regulation. sHSP20.09 displayed a most remarkable increase of 680.3-fold. The expression of sHSP20.06 was also intensively increased by 421.5-fold. The expression levels of six sHSPs, sHSP19.22, sHSP19.5, sHSP20.1, sHSP21.9, sHSP22.1, and sHSP23.4, were highly increased by 14.7- to 73.5-fold. The other four sHSPs transcripts were overexpressed by 1.85- to 5.7-fold (Fig. 5).
Fig. 5.
Normalized mRNA expression levels of sHSPs in DBM fourth instar larvae in response to the cold and thermal stresses. The expression levels of the 25 °C treatment were set to 1. *P < 0.05; **P < 0.01; and ***P < 0.001 on the tops of the column indicate significance in the different expression levels
Effects of oxidative and starvation stress on DBM sHSPs expression
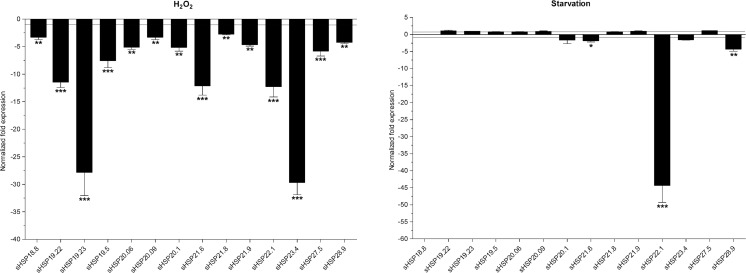
To investigate the responses of DBM sHSPs expression to oxidative stress, adults were exposed to H2O2 for 24 h. Transcript abundances of all sHSPs were significantly decreased, and mRNA expression levels of five sHSPs were down-regulated by over 10-fold, with sHSP19.22 at 11.5-fold, sHSP19.23 at 27.8-fold, sHSP21.6 at 12.2-fold, sHSP22.1 at 12.3-fold, and sHSP23.4 at 29.7-fold. The decreases in other sHSPs expression were between 2.7- and 5.8-fold (Fig. 6).
Fig. 6.
Normalized mRNA expression levels of the DBM sHSPs in response to the starvation and oxidative stresses. The expression levels of control samples were set to 1. *P < 0.05; **P < 0.01; and ***P < 0.001 on the tops of the column indicate significance in the different expression levels
To determine whether DBM sHSP expression changed in response to food deprivation, fourth larvae were starved for 24 h. Expression levels of four sHSPs, sHSP20.1, sHSP21.6, sHSP22.1, and sHSP28.9, were significantly down-regulated by 1.7-, 1.8-, 4.3-, and 44.3-fold, respectively. Other sHSPs seemed not to respond to the starvation treatment (Fig. 6).
Effect of heavy metals on DBM sHSPs expression
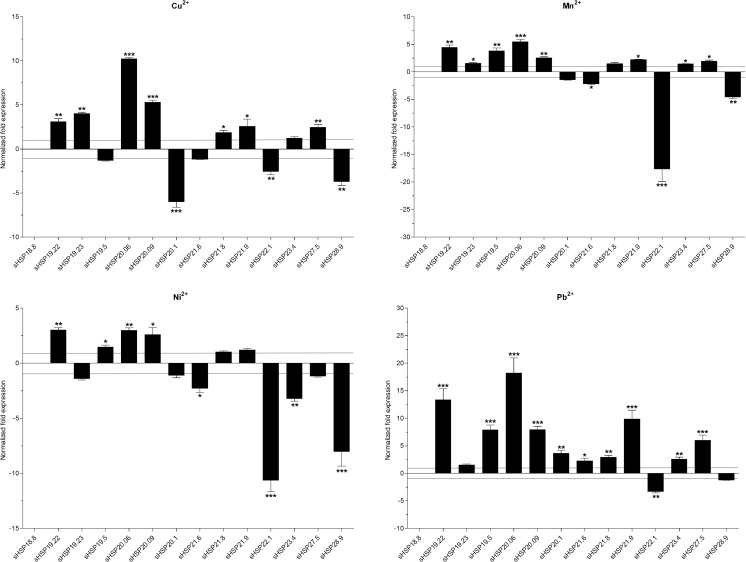
To examine the responses of sHSPs to heavy metals, fourth instar larvae were exposed to Cu2+, Mn2+, Ni2+, and Pb2+ for 24 h. Expression of sHSP18.8 was not detected in all treatments. The response of the other 13 sHSPs to individual heavy metal varied. Cu2+ exposure resulted in a significant reduction in expression levels of sHSP20.1 (6.1-fold), sHSP22.1 (2.6-fold), and sHSP28.9 (3.7-fold). However, expression levels of seven sHSPs were significantly increased with sHSP19.22 at 3.1-fold, sHSP19.23 at 4.1-fold, sHSP20.06 at 10.2-fold, sHSP20.09 at 5.3-fold, sHSP21.8 at 1.9-fold, sHSP21.9 at 2.6-fold, and sHSP27.5 at 2.5-fold. For the Mn2+ treatment, expression levels of four sHSPs, sHSP20.1, sHSP21.6, sHSP22.1, and sHSP28.9, were significantly decreased by 1.4-, 2.2-, 17.6-, and 4.5-fold, respectively. Other sHSP expressions were significantly up-regulated by 1.4- to 5.5-fold. Following Ni2+ treatment, three sHSPs, sHSP20.1, sHSP21.8, and sHSP21.9, were not induced, while expression levels of four sHSPs, sHSP19.22, sHSP19.5, sHSP20.06, and sHSP20.09, were significantly increased by 1.5- to 2.9-fold. The expression levels of other sHSPs were down-regulated by 1.4- to 10.0-fold. Exposure to Pb2+ led to significant decreases in the expression level of sHSP22.1 by 3.2-fold. Expression levels of sHSP19.23 and sHSP28.9 were not affected. The mRNA levels of other sHSPs were significantly up-regulated. The change for two sHSPs were over 10-fold, sHSP19.22 at 13.4-fold and sHSP20.06 at 18.3-fold, respectively (Fig. 7).
Fig. 7.
Normalized mRNA expression levels of the DBM sHSPs in response to the heavy metals. The expression levels of control samples were set to 1. *P < 0.05; **P < 0.01; and ***P < 0.001 on the tops of the column indicate significance in the different expression levels
Effects of pesticides on DBM sHSP expression
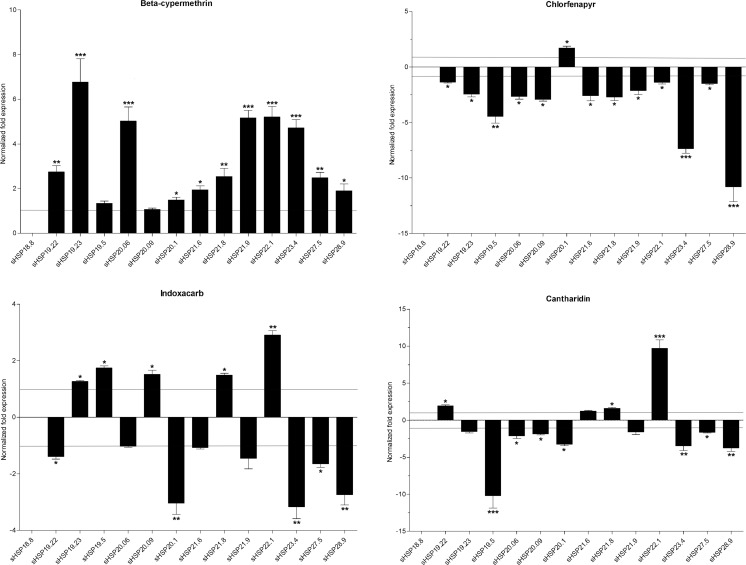
To monitor the responses of sHSPs to pesticides, fourth instar larvae were exposed to beta-cypermethrin, chlorfenapyr, indoxacarb, and cantharidin for 24 h. Expression of sHSP18.8 was not detected in all treatments. After beta-cypermethrin exposure, expression levels of all sHSPs, except for sHSP20.09, were significantly increased by 1.3- to 6.8-fold. The sHSP19.23 showed the highest increase. Chlorfenapyr treatment caused the down-regulation of almost all sHSPs expression levels by 1.3- to 10.8-fold, with the biggest change occurring in sHSP28.9 expression levels. However, the up-regulation of sHSP20.1 expression level was determined to be 1.7-fold. Five sHSPs were up-regulated by 1.3- to 2.9-fold in their expression levels following indoxacarb exposure, while six sHSPs were observed to be down-regulated by 1.4- to 3.2-fold. Expression levels of most sHSPs were slightly affected by cantharidin. There was down-regulation in sHSP19.5 by 9.7-fold and up-regulation in sHSP22.1 by 10.2-fold (Fig. 8).
Fig. 8.
Normalized mRNA expression levels of the DBM sHSPs in response to pesticides. The expression levels of control samples were set to 1. *P < 0.05; **P < 0.01; and ***P < 0.001 on the tops of the column indicate significance in the different expression levels
Discussion
Abundantly expressed sHSPs are important modulators of insect survival. Previous research has suggested that insect sHSPs participate in normal development and diapause (Hayward et al. 2005; Gkouvitsas et al. 2008; Kokolakis et al. 2009). In addition, the expression of sHSPs has been reported to be induced and modulated in response to abiotic stresses such as heat and cold shock (Concha et al. 2012; Lu et al. 2014), ultraviolet radiation (Nguyen et al. 2009; Sang et al. 2012), heavy metals (Sonoda et al. 2007; Wang et al. 2012), chemical pesticides (Sonoda and Tsumuki 2007), etc.
In this work, we identified and characterized 14 sHSPs from a destructive pest, the diamondback moth (DBM), P. xylostella. The amino acid sequences of these DBM sHSPs shared considerable sequence similarities with sHSPs from other insects. The phylogenetic tree analysis placed 12 DBM sHSPs into five clusters with a corresponding B. mori ortholog (Fig. 2). However, sHSP18.8 and sHSP19.22 could not be grouped into these five clusters suggesting that they might have undergone a different evolutionary pathway. These two sHSPs also show low sequence identities to other sHSPs. Moreover, of 14 DBM sHSPs, 8 had over 40 % similarity and belonged to the largest BmHSP20.4-like protein cluster. The same situation is also present in other insects (Li et al. 2009; Shen et al. 2011).
sHSPs are the most diverse in structure and function among the various stress protein families (Franck et al. 2004). A defining feature of small heat-shock proteins is the conserved α-crystallin domain toward the carboxyl terminus (Kriehuber et al. 2010), which existed in all 14 DBM sHSPs, and every DBM sHSP had a characteristic motif “I/V-x-I/V” following the α-crystallin domain. The core α-crystallin domain of sHSPs is a platform for oligomer assembly, and the “I/V-x-I/V” motif is also believed to play a key role in this process (Basha et al. 2012, 2013). Sequence alignment of DBM sHSPs shows that their N-terminal is highly variable. This region has been suggested as determining substrate specificity and chaperone activity (Basha et al. 2006).
sHSPs play important roles in insect development. They participate in the regulation of development in insects such as Chilo suppressalis (Lu et al. 2014), Lucilia cuprina (Concha et al. 2012), S. litura (Shen et al. 2011), D. melanogaster (Takahashi et al. 2010), and Liriomyza sativa (Huang et al. 2009). The developmental expression patterns of different sHSPs among these insects are quite varied. Two sHSP23 genes in Ceratitis capitata were highly expressed in the larval stage (Kokolakis et al. 2009). Highest expression levels of sHSP19.8, sHSP21.4, and sHSP21.5 in C. suppressalis (Lu et al. 2014); sHSP19.7, sHSP20, and sHSP20.7 in S. litura (Shen et al. 2011); and sHSP19.7 and sHSP19.8 in Cydia pomonella were observed in adults (Garczynski et al. 2011). Up-regulation of sHSP20.8 in S. litura (Shen et al. 2011), and sHSP19.5, sHSP20.8, and sHSP21.7 in L. sativa (Huang et al. 2009) have been reported in the pupal stage. In this study, we found that the expression profiles of most DBM sHSPs exhibited a common trend: the relative levels of sHSPs were low in the egg and larval stages and reached a peak in the pupal or adult stages. It is noticeable that sHSP18.8 was not expressed in the third and fourth instar larvae. Moreover, transcripts of sHSP19.23 and sHSP23.4 were not detectable in the egg stage. This might be the first finding that insect sHSPs are not expressed in certain developmental stages, implying that they may not be involved in any physiological activities in these stages. The exact underlying mechanism needs to be further investigated. In general, the various expression patterns of DBM sHSPs also indicate that they could have evolved specific roles in development.
The transcripts of insect sHSPs display tissue specificity. In this work, only two DBM sHSPs, sHSP20.06 and sHSP28.9, exhibited constitutive expression patterns in the four tested tissues suggesting their fundamental roles in in vivo activities. In B. mori, sHSP20.4 was found to be selectively expressed in the midgut (Saravanakumar et al. 2008). Four DBM sHSPs, sHSP19.5, sHSP20.1, sHSP21.6, and sHSP21.8, were highly expressed in the gut. The transcript of Apis cerana cerana HSP27.6 was scarce in the head (Liu et al. 2012); however, high expression of sHSP27.5 was observed in the head suggesting its potential role in nervous activity. In S. litura, six sHSPs showed very low mRNA levels in the epidermis (Shen et al. 2011), whereas five DBM sHSPs were significantly overexpressed in the epidermis. The role of small heat-shock proteins in the tissues is not well defined. One explanation is that sHSP genes expressed in specific tissues may play an important role in maintaining normal organism functioning and may also protect the protein’s ability to function in tissues under stress conditions (Gu et al. 2012).
Overexpression of insect sHSPs has long been implicated in responsiveness to thermal stress (Gehring and Wehner 1995). In our present work, 12 DBM sHSPs were significantly up-regulated by both heat and cold treatments which is consistent with most recent reports on sHSPs in other insects (Sakano et al. 2006; Huang et al. 2009; Colinet et al. 2010; Shen et al. 2011 ; Concha et al. 2012; Lu et al. 2014). Our results also revealed that heat shock induces the expression of most sHSPs more intensively than cold shock. It has been suggested that cross-resistance may not be present between heat and cold adaptations in insects (Huang and Kang 2007). Moreover, sHSP21.8 was not sensitive to thermal stress. The same finding was also observed for sHSP20 and sHSP21.4 in S. litura (Shen et al. 2011). It appears that the thermal adaptation of DBM and other insects is modulated by comprehensively regulating the expression of different sHSPs.
To date, reports on the response of insect sHSPs to other abiotic stresses, such as starvation, oxidative stress, heavy metals, and chemical pesticides, are not as extensive as on their response to thermal stress. We investigated the responses of DBM sHSPs to the above four abiotic stresses. We found that three DBM sHSPs were significantly down-regulated after food deprivation for 24 h, which corresponds with the expression of sHSP20 found in Pteromalus puparum (Wang et al. 2012). In A. cerana cerana, sHSP27.6 expression was significantly induced by H2O2 (Liu et al. 2012); however, we found mRNA levels of DBM sHSPs were all reduced. sHSPs in different insects may play different roles in response to oxidative stress.
sHSPs are capable of sensing the cellular stress caused by various environmental pollutants, such as heavy metals and the harmful chemicals, used in pesticides. Exposure to Cd2+ significantly increased sHSP27 mRNA levels in Chironomus riparius (Martínez-Paz et al. 2013). The expression of sHSP20 in P. puparum increased at a low concentration of Cu2+ and Cd2+ but decreased at high concentrations of Cu2+ and Cd2+ (Wang et al. 2012). It has been reported that sHSP19.7 and sHSP20.7 in cultured cells of Mamestra brassicae could be induced by exposure to Cu2+, Cd2+, and Pb2+ (Sonoda et al. 2007). Our results reveal that transcript abundance for three sHSPs, sHSP19.22, sHSP20.06, and sHSP20.09, were all increased, while mRNA levels of sHSP22.1 and sHSP 28.9 were all reduced by the four heavy metals tested. Other sHSPs showed variable expression patterns to different heavy metals. In addition, Pb2+ treatment increased transcription of most sHSPs. Heavy metals are capable of denaturing proteins. The overexpression of sHSPs might contribute to prevent protein denaturation or degradation. Previous research found that expression levels of sHSP19.7 and sHSP20.7 in cultured cells of M. brassicae were significantly up-regulated in response to high concentrations of chlorfenapyr but remained unchanged at low concentrations (Sonoda and Tsumuki 2007). The LC50 value amount of chlorfenapyr used in our study decreased the expression of 12 DBM sHSPs, yet, only increased the expression of sHSP20.1 by 1.7-fold. In contrast, an increase in mRNA abundance for all DBM sHSPs occurred after beta-cypermethrin exposure. The responses of sHSPs to indoxacarb and cantharidin were irregular. Our results suggest that sHSPs play roles in the physiological processes that are affected by pesticides. In addition, expression of DBM sHSPs might be induced in a heavy metal- or pesticide-specific manner indicating their potential use as a biomarker.
In conclusion, 14 sHSPs were identified from DBM and characterized which showed different transcriptional expression profiles in various tissues and at different developmental stages, as well as in response to various abiotic stresses. The findings provide valuable insights for further investigation of the functions of sHSPs superfamily in insects and help us understand the adaptability of insects to hostile environments.
Acknowledgments
We sincerely thank Prof. John Richard Schrock (Emporia State University, USA) for revising the manuscript. This research is supported by the Special Fund for the Public Interest (Agriculture) (200903052) by The Ministry of Science and Technology and The Ministry of Agriculture of China and the ‘13115’ Sci-Tech Innovation Project of Shaanxi Province (2007ZDKG-14).
References
- Aevermann BD, Waters ER. A comparative genomic analysis of the small heat shock proteins in Caenorhabditis elegans and briggsae. Genetica. 2008;133:307–319. doi: 10.1007/s10709-007-9215-9. [DOI] [PubMed] [Google Scholar]
- Basha E, Friedrich KL, Vierling E. The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J Biol Chem. 2006;281:39943–39952. doi: 10.1074/jbc.M607677200. [DOI] [PubMed] [Google Scholar]
- Basha E, O’Neill H, Vierling E. Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–117. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha E, Jones C, Blackwell AE, Cheng G, Waters ER, Samsel KA, Siddique M, Pett V, Wysocki V, Vierling E. An unusual dimeric small heat shock protein provides insight into the mechanism of this class of chaperones. J Mol Biol. 2013;425:1683–1696. doi: 10.1016/j.jmb.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colinet H, Lee SF, Hoffmann A. Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster. FEBS J. 2010;277:174–185. doi: 10.1111/j.1742-4658.2009.07470.x. [DOI] [PubMed] [Google Scholar]
- Concha C, Edman RM, Belikoff EJ, Schiemann AH, Carey B, Scott MJ. Organization and expression of the Australian sheep blowfly (Lucilia cuprina) hsp23, hsp24, hsp70 and hsp83 genes. Insect Mol Biol. 2012;21:169–180. doi: 10.1111/j.1365-2583.2011.01123.x. [DOI] [PubMed] [Google Scholar]
- Franck E, Madsen O, van Rheede T, Ricard G, Huynen MA, de Jong WW. Evolutionary diversity of vertebrate small heat shock proteins. J Mol Evol. 2004;59:792–805. doi: 10.1007/s00239-004-0013-z. [DOI] [PubMed] [Google Scholar]
- Garczynski SF, Unruh TR, Guédot C, Neven LG. Characterization of three transcripts encoding small heat shock proteins expressed in the codling moth, Cydia pomonella (Lepidoptera: Tortricidae) Insect Sci. 2011;18:473–483. doi: 10.1111/j.1744-7917.2010.01401.x. [DOI] [Google Scholar]
- Gehring WJ, Wehner R. Heat shock protein synthesis and thermotolerance in Cataglyphis, an ant from the Sahara desert. Proc Natl Acad Sci U S A. 1995;92:2994–2998. doi: 10.1073/pnas.92.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkouvitsas T, Kontogiannatos D, Kourti A. Differential expression of two small Hsps during diapause in the corn stalk borer Sesamia nonagrioides (Lef.) J Insect Physiol. 2008;54:1503–1510. doi: 10.1016/j.jinsphys.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Gu J, Huang LX, Shen Y, Huang LH, Feng QL. Hsp70 and small Hsps are the major heat shock protein members involved in midgut metamorphosis in the common cutworm, Spodoptera litura. Insect Mol Biol. 2012;5:535–543. doi: 10.1111/j.1365-2583.2012.01158.x. [DOI] [PubMed] [Google Scholar]
- Haslbeck M. sHsps and their role in the chaperone network. Cell Mol Life Sci. 2002;59:1649–1657. doi: 10.1007/PL00012492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- Hayward SAL, Pavlides SC, Tammariello SP, Rinehart JP, Denlinger DL. Temporal expression patterns of diapause-associated genes in flesh fly pupae from the onset of diapause through post-diapause quiescence. J Insect Physiol. 2005;51:631–640. doi: 10.1016/j.jinsphys.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Huang LH, Kang L. Cloning and interspecific altered expression of heat shock protein genes in two leafminer species in response to thermal stress. Insect Mol Biol. 2007;16:491–500. doi: 10.1111/j.1365-2583.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- Huang LH, Wang CZ, Kang L. Cloning and expression of five heat shock protein genes in relation to cold hardening and development in the leafminer, Liriomyza sativa. J Insect Physiol. 2009;55:279–285. doi: 10.1016/j.jinsphys.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Jouraku A, Yamamoto K, Kuwazaki S, Urio M, Suetsugu Y, Narukawa J, Miyamoto K, Kurita K, Kanamori H, Katayose Y, Matsumoto T, Noda H. KONAGAbase: a genomic and transcriptomic database for the diamondback moth, Plutella xylostella. BMC Genomics. 2013;14:464. doi: 10.1186/1471-2164-14-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappé G, Franck E, Verschuure P, Boelens WC, Leunissen JA, de Jong WW. The human genome encodes 10 α-crystalline-related small heat shock proteins: HspB1-10. Cell Stress Chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:THGECS>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokolakis G, Kritsidima M, Tkachenko T, Mintzas AC. Two hsp23 genes in the Mediterranean fruit fly, Ceratitis capitata: structural characterization, heat shock regulation and developmental expression. Insect Mol Biol. 2009;18:171–181. doi: 10.1111/j.1365-2583.2009.00868.x. [DOI] [PubMed] [Google Scholar]
- Kriehuber T, Rattei T, Weinmaier T, Bepperling A, Haslbeck M, Buchner J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010;24:3633–3642. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- Li ZW, Li X, Yu QY, Xiang ZH, Kishino H, Zhang Z. The small heat shock protein (sHSP) genes in the silkworm, Bombyx mori, and comparative analysis with other insect sHSP genes. BMC Evol Biol. 2009;9:215. doi: 10.1186/1471-2148-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xi D, Kang M, Guo X, Xu B. Molecular cloning and characterization of Hsp27. 6: the first reported small heat shock protein from Apis cerana cerana. Cell Stress Chaperones. 2012;17:539–551. doi: 10.1007/s12192-012-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu MX, Hua J, Cui YD, Du YZ. Five small heat shock protein genes from Chilo suppressalis: characteristics of gene, genomic organization, structural analysis, and transcription profiles. Cell Stress Chaperones. 2014;19:91–104. doi: 10.1007/s12192-013-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Paz P, Morales M, Martín R, Martínez-Guitarte JL, Morcillo G. Characterization of the small heat shock protein Hsp27 gene in Chironomus riparius (Diptera) and its expression profile in response to temperature changes and xenobiotic exposures. Cell Stress Chaperones. 2013 doi: 10.1007/s12192-013-0479-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TTA, Michaud D, Cloutier C. A proteomic analysis of the aphid Macrosiphum euphorbiae under heat and radiation stress. Insect Biochem Mol Biol. 2009;39:20–30. doi: 10.1016/j.ibmb.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Poulain P, Gelly JC, Flatters D. Detection and architecture of small heat shock protein monomers. PLoS One. 2010;5:e9990. doi: 10.1371/journal.pone.0009990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan R. Cytoskeletal competence requires protein chaperones. Prog Mol Subcell Biol. 2002;28:219–234. doi: 10.1007/978-3-642-56348-5_12. [DOI] [PubMed] [Google Scholar]
- Rinehart JP, Li A, Yocum GD, Robich RM, Hayward SA, Denlinger DL. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc Natl Acad Sci U S A. 2007;104:11130–11137. doi: 10.1073/pnas.0703538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano D, Li B, Xia Q, Yamamoto K, Fujii H, Aso Y. Genes encoding small heat shock proteins of the silkworm, Bombyx mori. Biosci Biotechnol Biochem. 2006;70:2443. doi: 10.1271/bbb.60176. [DOI] [PubMed] [Google Scholar]
- Sang W, Ma WH, Qiu L, Zhu ZH, Lei CL. The involvement of heat shock protein and cytochrome P450 genes in response to UV-A exposure in the beetle Tribolium castaneum. J Insect Physiol. 2012;58:830–836. doi: 10.1016/j.jinsphys.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Saravanakumar R, Ponnuvel KM, Qadri SMH. Expression of metabolic enzyme genes and heat-shock protein genes during embryonic development in diapause and non-diapause egg of multivoltine silkworm Bombyx mori. Biologia. 2008;63:737–744. doi: 10.2478/s11756-008-0124-x. [DOI] [Google Scholar]
- Shen Y, Gu J, Huang LH, Zheng SC, Liu L, Xu WH, Feng QL, Kang L. Cloning and expression analysis of six small heat shock protein genes in the common cutworm, Spodoptera litura. J Insect Physiol. 2011;57:908–914. doi: 10.1016/j.jinsphys.2011.03.026. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Tsumuki H. Induction of heat shock protein genes by chlorfenapyr in cultured cells of the cabbage armyworm, Mamestra brassicae. Pestic Biochem Physiol. 2007;89:185–189. doi: 10.1016/j.pestbp.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Ashfaq M, Tsumuki H. A comparison of heat shock protein genes from cultured cells of the cabbage armyworm, Mamestra brassicae, in response to heavy metals. Arch Insect Biochem. 2007;65:210–222. doi: 10.1002/arch.20178. [DOI] [PubMed] [Google Scholar]
- Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi KH, Rako L, Takano-Shimizu T, Hoffmann AA, Lee SF. Effects of small Hsp genes on developmental stability and microenvironmental canalization. BMC Evol Biol. 2010;10:284. doi: 10.1186/1471-2148-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trisyono A, Whalon ME. Fitness costs of resistance to Bacillus thuringiensis in Colorado potato beetle (Coleoptera: Chrysomelidae) J Econ Entomol. 1997;90:267–271. doi: 10.1093/jee/88.1.21. [DOI] [PubMed] [Google Scholar]
- Tsvetkova NM, Horvath I, Torok Z, Wolkers WF, Balogi Z, Shigapova N, Crowe LM, Tablin F, Vierling E, Crowe JH, Vigh L. Small heat shock proteins regulate membrane lipid polymorphism. Proc Natl Acad Sci U S A. 2002;99:13504–13509. doi: 10.1073/pnas.192468399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Mol Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- van Montfort R, Slingsby C, Vierling E. Structure and function of the small heat shock protein/alpha-crystallin family of molecular chaperones. Adv Protein Chem. 2002;59:105–156. doi: 10.1016/S0065-3233(01)59004-X. [DOI] [PubMed] [Google Scholar]
- Wang H, Li K, Zhu JY, Fang Q, Ye GY. Cloning and expression pattern of heat shock protein genes from the endoparasitoid wasp, Pteromalus puparum in response to environmental stresses. Arch Insect Biochem. 2012;79:247–263. doi: 10.1002/arch.21013. [DOI] [PubMed] [Google Scholar]
- Waters ER, Rioflorido I. Evolutionary analysis of the small heat shock proteins in five complete algal genomes. J Mol Evol. 2007;65:162–174. doi: 10.1007/s00239-006-0223-7. [DOI] [PubMed] [Google Scholar]
- Waters ER, Aevermann BD, Sanders-Reed Z. Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones. 2008;13:127–142. doi: 10.1007/s12192-008-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Jones WA. Expression of heat shock protein genes in insect stress responses. Invertebr Surviv J. 2012;9:93–101. [Google Scholar]