Abstract
Heat shock protein (Hsp) genes are stress-related genes that activate the host immune system during infection. Hsp genes expression in fish, studied during bacterial infections, is mostly confined to Hsp70 and Hsp90. The present study is an expression analysis of seven Hsp genes: Apg2, Hsp90, Hsp70, glucose-regulated protein 78 (Grp78), heat shock cognate 70 (Hsc70), Grp75, and Hsp30 during Aeromonas hydrophila infection in rohu, Labeo rohita. Forty-eight rohu juveniles were challenged with 3 × 107 cfu bacteria per 20 g of body weight intraperitoneally. The expression of these genes was studied in infected liver, anterior kidney, and spleen tissues of rohu at different time periods: 0, 1, 3, 6, 12, 24, 48, 72 h, 7, and 15 days post-infection by qPCR. The Hsp gene modulation was greater in liver as compared to spleen and kidney tissues. Induced expression of Apg2, Hsp90, Grp78, Grp75, and Hsc70 was noticed during peak periods (3 to 24 h post-challenge) of bacterial infectivity. Hsp70 was found to be down-regulated during the process of infection. In contrast to the other six genes, Hsp30 showed a variable expression pattern in all three tissues. Grp78 was found to be the most highly (1,587-fold) expressed gene in liver at 12 h post-challenge. Further, molecular characterization of Grp78 revealed it to be a highly conserved Hsp gene, essential not only during infection but also during early developmental stages of rohu, and its expression was noticed in all organs studied except in gill tissues, which indicated its potential immune regulatory role during infection process.
Keywords: Heat shock proteins, Bip or Grp78, Aeromonas hydrophila, Labeo rohita
Introduction
Heat shock protein (Hsp) genes are highly conserved and are constitutively expressed in all tissues in regular protein homeostasis and stress response (Feder and Hofmann 1999). They function as chaperones to refold aberrantly folded proteins. However, under some physiological conditions, there is a notable elevation of these genes (Sung and MacRae 2011). Stress disrupts the regular conformation of proteins demanding the aid of Hsps to regularize the effect. In fish, stress could be due to any minor change in the environment. Fish are exposed to many kinds of stressors such as microbial infection, toxic exposure, traumatic damage, radiation, or nutritional deficiency. Therefore, the modulation of Hsp genes is more apparent in fish and can be studied as molecular biomarkers of stress (Kayhan and Duman 2010).
Microbial infection is the most devastating stressor that can modulate the regular physiological activity of fish (Sung and MacRae 2011). Hsp’s activity is not limited to the post-infection stress response but can modulate the immune system as well (Roberts et al. 2010). Infection regulates stress gene expression both in the host and in parasites (Henderson et al. 2006). Molecular chaperones are probably the only protein involved in infections that have dual roles as pathogen-associated molecular patterns (PAMPs) and pattern recognition receptors (PRRs). Certain bacterial proteins were found to bind CD14 and TLR2/4 on monocytes and vascular endothelial cells of the host (Henderson 2003). Again, chaperones act as cell surface receptors for lipopolysaccharides (LPS) (Triantafilou et al. 2001). Interestingly, a host can recognize its own molecular chaperones and those of the invading bacteria. Eukaryotic Hsp genes are categorized into different families based on their function, sequence homology, and molecular weight such as HSP100, HSP90, HSP70, HSP60, HSP40, and several smaller HSP groups (Roberts et al. 2010). Hsp30 (HSP30 gene) was one among the small HSP family that acts as a molecular chaperone during complex environmental exposure (Wang et al. 2007). However, its immune-related function is as yet unknown. Apg2 (HSPA4 gene) belongs to the HSP100 family, which is mainly involved in neuronal development; its immune function is yet to be explored. The roles of the HSP90 and HSP70 families in the immune response are noteworthy (Henderson et al. 2006). The Hsp90 gene of the HSP90 family facilitates the movement of bacterial toxins from the endosomal compartment into the cytosol; it is involved in the induction of IL-8 synthesis by cells exposed to bacteria and participates in the folding and export of the Toll-like receptors (TLRs) and some integrins. The Hsp genes of the HSP70 family that are crucial during disease stress response are Hsp70 (HSP70 gene), mortalin or glucose-regulated protein 75 or Grp75 (HSPA9 gene), Hsp73 or heat shock cognate 70 or Hsc70 (HSPA8 gene), binding immunoglobulin protein (Bip) or Grp78 (HSPA5 gene). Hsp70 is released to extracellular compartments, serving as an endogenous danger signal and influencing antigen presenting cells for major histocompatibility complex (MHC)-dependent antigen processing. It can also regulate PAMP-induced TLR signaling. Mortalin or GRP75 was found to be involved in antigen processing (Wadhwa et al. 2002). Hsp73 or Hsc70 participates in apoptosis and possesses a potential role in protection against reactive oxygen species (Dastoor and Dreyer 2000). BiP or Grp78 acts as a high-affinity signaling receptor for β2-macroglobulin. Grp78 expressed on the cell surface is involved in cell signaling, viral entry, and antigen presentation. Grp78 is a highly conserved protein, which participates in protein folding, assembly and trafficking, and regulates endoplasmic reticulum (ER) homeostasis (Quinones et al. 2008).
Rohu, Labeo rohita, is one of the most important carp species cultured in India and its production is valuable to farmers. However, the fish often develop hemorrhagic septicemia caused by the Gram-negative bacterium, Aeromonas hydrophila, which is a part of the environment flora and becomes infectious during stress (van der Marel et al. 2008). Therefore, expression of Hsp genes during A. hydrophila served a dual response to infection and stress. The modulation of Hsp70 was investigated in rohu during temperature change (Das et al. 2006) and starvation (Yengkokpam et al. 2008). However, no study has yet revealed the modulation of this or any other genes of this family during bacterial infection in rohu. The current study deals with the expression of an array of heat shock protein genes such as Hsp90, Hsp70, Hsp30, mortalin or Grp75, Grp78 or Bip, Hsc70 or Hsp73, and Apg2 during A. hydrophila infection in three important tissues of rohu at different time periods in addition to the molecular characterization of the Grp78 gene.
Material and methods
Fish
Rohu juveniles were collected from nursery ponds of the Central Institute of Freshwater Aquaculture, Kausalyaganga, Bhubaneswar, Odisha, India. The fingerlings were kept in 700-L ferro-cement tanks for acclimatization prior to the experiment. The eggs before and after hatching, and hatchlings for developmental study were collected from a carp-breeding hatchery during breeding of Jayanti rohu (Nayak et al. 2011).
Virulent bacterial strain
A virulent strain of A. hydrophila (Ah 15) was an isolate from a field outbreak that occurred at a culture farm at Puri, Odisha, India (Mohanty et al. 2008).
Challenge study and sample collection
A total of 48 rohu (weighing 80–100 g) juveniles were challenged intraperitoneally with live A. hydrophila at a LD50 dose of 3 × 107 cfu/100 μl/20 g of body weight (Sahoo et al. 2008). The LD50 bacterial dose was calculated prior to the experiments with few numbers of juveniles of the same stock (Reed and Muench 1938). Tissue samples from liver, anterior kidney, and spleen were collected from infected fish at different time periods: 1, 3, 6, 12, 24, 48, 72 h, 7, and 15 days post-infection in triplicate after euthanasia with a heavy dose of MS222. Prior to the infectious challenge, tissue samples were also collected from three non-infected fish as a control. The tissue samples were kept in RNAlater (Sigma, USA) and stored at −20 °C until RNA extraction.
Tissue specificity and ontogeny study
Tissues from liver, anterior and posterior kidneys, spleen, intestine, gill, heart, stomach, brain, eye, skin, and muscle were collected from three naive rohu juveniles. Milt, eggs before and after fertilization, and hatchlings were collected at different time periods (0, 1, 3, 6, 9, 12, 18, 24, 48 h, 3, 4, 7, and 15 days post-fertilization) during the breeding season from three pairs of broods. All the samples collected were kept in RNAlater and stored at −20 °C until RNA extraction.
RNA extraction and cDNA synthesis
Total RNA was extracted using TRI reagent (Sigma, USA). A total of 1 μg of RNA was treated with DNase I (Fermentas, Canada). First strand complementary DNA (cDNA) was synthesized using M-MLV reverse transcriptase (Genei, India) as per the manufacturer’s instructions.
Quantitative PCR (qPCR)
Four sets of primers were designed (Table 1) using the contig sequences of Apg2, Hsp73, Grp78, and Grp75 genes obtained through messenger RNA (mRNA)-seq of rohu (Robinson et al. 2012) using Primer Premier Version 5 (Lalitha 2000). Primer sequences of Hsp90, Hsp70, and Hsp30 were obtained from our previous study (Das et al. 2014). β-actin was used as the reference gene (Robinson et al. 2012). Quantitative PCR (qPCR) was performed using FastStart Essential DNA Green Master (Roche, Germany) in Light Cycler 96 (Roche, Germany). Briefly, 1 μl of cDNA synthesized was used as a template in a total reaction mixture of 10 μl containing 5 μl of 2X Light cycler SYBR green I mix, 0.5 μl of primer pairs (5 pmole), and 3 μl of H2O provided in the kit. The qPCR program consisted of pre-denaturation at 95 °C for 10 min and 45 cycles of amplification at 95 °C for 10 s, annealing temperature Ta for respective genes (Table 1) for 10 s, and 72 °C for 20 s. All reactions were performed simultaneously for each gene with β-actin, in the same plate in triplicate. qPCR specificity was verified by melt curve analysis at a temperature of 95 °C for 10 s, 65 °C for 1 min, and 95 °C for 1 min. No-template controls were included in each run.
Table 1.
List of primers used. The qPCR primers were named according to the gene name
| Sl. no. | Gene | Sequence (5′→3′) | Ta (°C) | Product length (bp) | Reference/source |
|---|---|---|---|---|---|
| 1 | HSPA90 | Forward- ACCAAAGCCGACCTCATC | 54 | 121 | Das et al. in manuscript |
| Reverse- AGAAACCCACGCCAAACT | |||||
| 2 | HSPA70 | Forward- CTACTCGGACAATCAGCC | 54 | 105 | Das et al. in manuscript |
| Reverse- GGAATGCCAATCAACTCA | |||||
| 3 | HSPA30 | Forward- TGACGCTGGACACTAAAGAC | 58 | 163 | Das et al. in manuscript |
| Reverse- CTTGAGGCAGATCAAACACTC | |||||
| 4 | HSPA9 | Forward- GCTGGTCGGAGGAATGA | 53 | 87 | Contig 87912 |
| Reverse- GGGTTCACGGACTTACTGG | |||||
| 5 | HSPA8 | Forward- TCCCAAGGTCCAGGTT | 51 | 114 | Contig 110387 |
| Reverse- GTCTTTCCCAGGTAGGC | |||||
| 6 | HSPA5 | Forward- GCGGTTTCCCTGGTCATT | 53 | 172 | Contig 54182 |
| Reverse- GCGATTGCTTTGCCTGTT | |||||
| 7 | HSPA4 | Forward- CCCATTCAAGAGCGATAC | 51 | 167 | Contig 54154 |
| Reverse- TTCACCATTCTGTCCACC | |||||
| 8 | β-actin | Forward- TTGGCAATGAGAGGTTCAGGT | All above | 153 | Robinson et al. 2012 |
| Reverse- TTGGCATACAGGTCCTTACGG | |||||
| 9 | GRP78 | Forward- ATGCGATTGCTTTGCCTGTT | 60 | 1,010 | Forward: Contig 54182 Reverse: Contig 52647 |
| Reverse-ATGGTGGAGCGGAACAAG |
Tissue-specific expression of Grp78 gene was analyzed using qPCR as described above using the gene-specific primers. Similarly, its expression during early developmental stages was also analyzed using qPCR.
Relative quantification
The quantification cycle (Cq) values were calculated using Light Cycler 96 SW 1.1 and the data were exported. N-fold differential expression was calculated using the comparative Cq method (Livak and Schmittgen 2001) by calculating the average of each Cq for the triplicate samples. The Cq value of the gene for each cDNA was subtracted from its respective Cq value of β-actin to get the ∆Cq value. Since the samples for each time period were taken in triplicate, an average of the ∆Cq values was obtained. Further, the ΔΔCq was calculated by subtracting the ∆Cq of the samples from the ∆Cq value of the calibrator. Fold difference was calculated as 2−ΔΔCq. Mean fold difference was calculated and represented as ±standard error. The ‘0’ hour control group was taken as calibrator in all the infected samples. The least expressing intestine tissue and 7-day-old larvae/hatchlings values were considered as calibrators for tissue-specific and ontogeny study of Grp78 gene, respectively.
Statistical analysis
The difference between the mean values of one gene in an organ was analyzed using one-way ANOVA followed by Duncan’s multiple range tests, with values p < 0.05 as significantly different. All values of N-fold differential expression were plotted in a graph.
Sequence analysis of Grp78 gene
A primer pair was designed (Table 1) for Grp78 from the contig sequences available from the mRNA-seq data of rohu (Robinson et al. 2012). A partial mRNA sequence of Grp78 was deduced using the above primer pair following the PCR program; pre-denaturation at 94 °C for 2 min, 45 cycles of amplification at 94 °C for 45 s, 60 °C for 45 s, and 72 °C for 1 min 30 s, and final extension at 72 °C for 10 min. The amplified fragment was run in 1.5 % low-melting agarose gel (Lonza, USA) and purified in Genei gel extraction kit (Genei, Bangalore) according to the manufacturer’s instruction. The purified fragment was cloned into a T-cloning vector using an instant cloning kit (Genei, Bangalore) and transformed into an Escherichia coli host DH5α. Positive clones were sequenced in triplicate. The final sequence of Grp78 was obtained using Clustal W multiple alignments implemented in BioEdit version 7.0.0. The partial mRNA sequence was translated into amino acid sequence and domains were identified using SMART sequence analysis tool. Amino acid alignment was carried out with sequences of the Grp78 gene of different fish species such as zebrafish, Danio rerio (AAH63946.1); grass carp, Ctenopharyngodon idella (ACJ65009.1); Mexican tetra, Astyanax mexicanus (XP_007252148.1); Japanese rice fish, Oryzias latipes (NP_001265730.1); Atlantic salmon, Salmo salar (NP_001135114.1); Japanese pufferfish, Takifugu rubripes (XP_003965205.1); olive flounder, Paralichthys olivaceus (ABG56392.1); green-spotted puffer, Tetraodon nigroviridis (CAG12424.1); Nile tilapia, Oreochromis niloticus (XP_005470418.1); flame black cichlid, Pundamilia nyererei (XP_005755726.1); southern platyfish, Xiphophorus maculatus (XP_005803813.1); zebra mbuna, Maylandia zebra (XP_004538056.1); and Burton’s mouthbrooder, Haplochromis burtoni (XP_005930177.1) along with modern humans, Homo sapiens Grp78 (NP_005338.1). A phylogenetic tree of Grp78 was constructed using the neighbor-joining method in a bootstrap test of Molecular Evolutionary Genetics Analysis 4 (MEGA 4) software by aligning the available protein sequences of several organisms in addition to the above species such as Asian swamp eel, Monopterus albus (AGO01985.1); Burmese python, Python bivittatus (XP_007420669.1); Carolina anole, Anolis carolinensis (XP_003230508.1); spotted gar, Lepisosteus oculatus (XP_006640761.1); king cobra, Ophiophagus hannah (ETE59912.1); common shrew, Sorex araneus (XP_004613383.1); American alligator, Alligator mississippiensis (XP_006271368.1); western clawed frog, Xenopus (Silurana) tropicalis (XP_002941690.1); common marmoset, Callithrix jacchus (XP_002743358.1); West Indian ocean coelacanth, Latimeria chalumnae (XP_005987377.1); black-capped squirrel monkey, Saimiri boliviensis boliviensis (XP_003940675.1); collared flycatcher, Ficedula albicollis (XP_005055797.1); painted turtle, Chrysemys picta bellii (XP_005279468.1); zebra finch, Taeniopygia guttata (XP_002192655.1); cattle, Bos taurus (DAA24281.1); degu, Octodon degus (XP_004641630.1); prairie vole, Microtus ochrogaster (XP_005346064.1); sheep, Ovis aries (XP_004005686.1); Tibetan antelope, Pantholops hodgsonii (XP_005953889.1); prairie deer mouse, Peromyscus maniculatus bairdii (XP_006988789.1); Chinese soft-shelled turtle, Pelodiscus sinensis (NP_001273821.1); lesser Egyptian jerboa, Jaculus jaculus (XP_004662863.1); brown rat, Rattus norvegicus (NP_037215.1); northern greater galago, Otolemur garnettii (XP_003785350.1); dog, Canis lupus familiaris (XP_863385.2); killer whale, Orcinus orca (XP_004269273.1); lesser hedgehog tenrec, Echinops telfairi (XP_004716507.1); red jungle fowl, Gallus gallus (NP_990822.1); Japanese quail, Coturnix japonica (BAF38391.1); star-nosed mole, Condylura cristata (XP_004677392.1); black flying fox, Pteropus alecto (XP_006918027.1); minke whale, Balaenoptera acutorostrata scammoni (XP_007194739.1); nine-banded armadillo, Dasypus novemcinctus (XP_004485131.1); thirteen-lined ground squirrel, Ictidomys tridecemlineatus (NP_001269183.1); brichardi cichlid, Neolamprologus brichardi (XP_006789208.1); common bottlenose dolphin, Tursiops truncatus (XP_004313501.1); American pika, Ochotona princeps (XP_004593628.1); Brandt's bat, Myotis brandtii (XP_005881560.1); African bush elephant, Loxodonta africana (XP_003407784.1); guinea pig, Cavia porcellus (XP_0s03470822.1); domestic goat, Capra hircus (NP_001274500.1); bactrian camel, Camelus ferus (XP_006187781.1); naked mole rat, Heterocephalus glaber (XP_004888032.1); red deer, Cervus elaphus (ACT46911.1); Japanese macaque, Macaca fuscata (BAE79724.1); wild boar, Sus scrofa (XP_001927830.3); Tasmanian devil, Sarcophilus harrisii (XP_003757187.1); and Sumatran orangutan, Pongo abelii (NP_001126927.1).
Results
Expression analysis of heat shock protein genes during A. hydrophila infection
Apg2 gene was up-regulated significantly during 3 to 24 hours post-challenge (hpc) in liver (Fig. 1a) and 3 to 48 hpc in spleen samples (Fig. 1c). However, in kidney, mild over-expression was observed at 24 hpc only (Fig. 1b). Hsp90 gene showed minute up-regulation (1.2-fold) at 6 hpc and down-regulation at 24–72 hpc in liver tissue (Fig. 2a). No significant change was observed for the remainder of the time periods in liver. In kidney, its expression was down-regulated during the initial and later time periods of challenge (Fig. 2b). The expression of Hsp90 gene was slightly up-regulated in spleen (Fig. 2c) during 3 and 24 hpc (1.5 and 1.0-folds, respectively). Hsp30 gene was up-regulated during the very first hours of infection and returned to control level, and was thereafter again up-regulated after 2 days in all three tissues examined. In liver, the initial up-regulation was up to 100- to 200-fold (174-fold in 1 hpc and 239-fold in 3 hpc) and later up to 600-fold (139-fold in 48 hpc and 595-fold in 72 hpc) (Fig. 3a). However, in kidney (Fig. 3b) and spleen (Fig. 3c), the initial up-regulation was up to 10- to 20-fold and in later time periods also remained at the same level. Hsp70 gene was down-regulated in liver (Fig. 4a) and kidney (Fig. 4b) and slightly up-regulated (1.8-fold) in spleen tissues (Fig. 4c) at 12 hpc. Mortalin gene was found to be over-expressed (10-fold) during early hours (3 to 24 hpc) of infection (Fig. 5a–c). Hsc70 gene was over-expressed during 3 to 12 hpc in liver (Fig. 6a) and 3 to 72 hpc in spleen (Fig. 6c). In kidney (Fig. 6b), Hsc70 gene expression remained almost stable during the period under study. Grp78 gene was the most highly expressed gene during A. hydrophila infection. In liver (Fig. 7a), significant up-regulation was noticed during 6 hpc (696-fold), 12 hpc (1,587-fold), and 24 hpc (334-fold). In kidney (Fig. 7b) and spleen (Fig. 7c) tissues, the over-expression was noticed during the same time periods as in the case of liver; however, the fold expression was up 10-fold only.
Fig. 1.
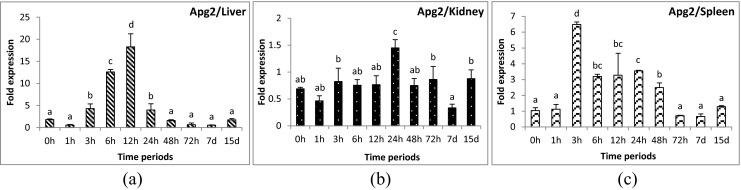
Expression analysis of Apg2 gene in liver (a), anterior kidney (b), and spleen (c) tissues of rohu, Labeo rohita, during different time periods (h, hours post-challenge, d, days post-challenge) of Aeromonas hydrophila infection. Fold expression was calculated as 2−∆∆Cq. Control group (0 h post-challenge) was taken as the calibrator
Fig. 2.
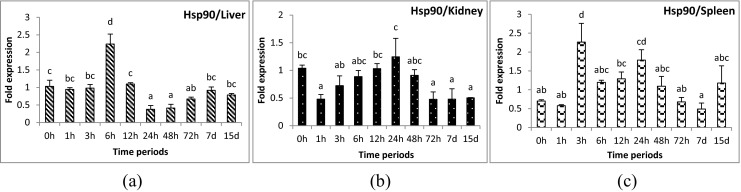
Expression analysis of heat shock protein 90 (HSP90) gene in liver (a), anterior kidney (b), and spleen (c) tissues of rohu, Labeo rohita, during different time periods (h, hours post-challenge, d, days post-challenge) of Aeromonas hydrophila infection. Fold expression was calculated as 2−∆∆Cq. Control group (0 h post-challenge) was taken as the calibrator
Fig. 3.
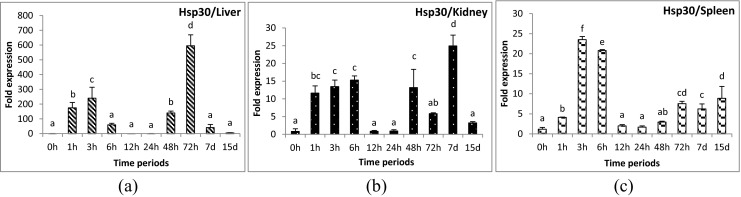
Expression analysis of heat shock protein 30 (HSP30) gene in liver (a), anterior kidney (b), and spleen (c) tissues of rohu, Labeo rohita, during different time periods (h, hours post-challenge, d, days post-challenge) of Aeromonas hydrophila infection. Fold expression was calculated as 2−∆∆Cq. Control group (0 h post-challenge) was taken as the calibrator
Fig. 4.
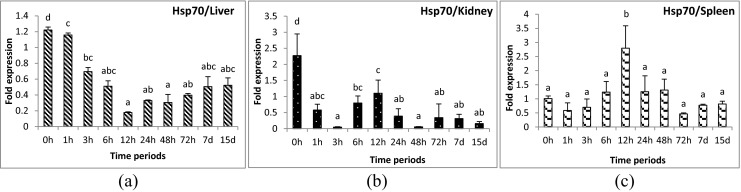
Expression analysis of heat shock protein 70 (HSP70) gene in liver (a), anterior kidney (b), and spleen (c) tissues of rohu, Labeo rohita, during different time periods (h, hours post-challenge, d, days post-challenge) of Aeromonas hydrophila infection. Fold expression was calculated as 2−∆∆Cq. Control group (0 h post-challenge) was taken as the calibrator
Fig. 5.
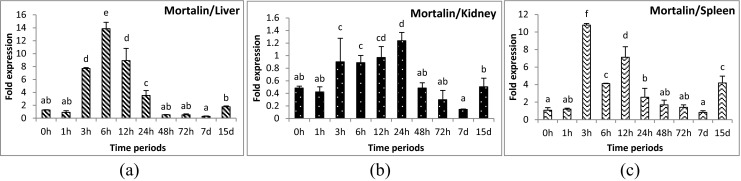
Expression analysis of glucose-regulated protein 75 (mortalin) gene in liver (a), anterior kidney (b), and spleen (c) tissues of rohu, Labeo rohita, during different time periods (h, hours post-challenge, d, days post-challenge) of Aeromonas hydrophila infection. Fold expression was calculated as 2−∆∆Cq. Control group (0 h post-challenge) was taken as the calibrator
Fig. 6.
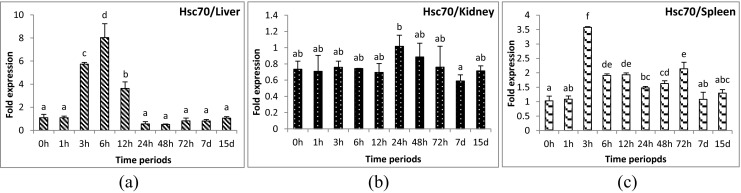
Expression analysis of heat shock cognate 70 (Hsc70) gene in liver (a), anterior kidney (b), and spleen (c) tissues of rohu, Labeo rohita, during different time periods (h, hours post-challenge, d, days post-challenge) of Aeromonas hydrophila infection. Fold expression was calculated as 2−∆∆Cq. Control group (0 h post-challenge) was taken as the calibrator
Fig. 7.
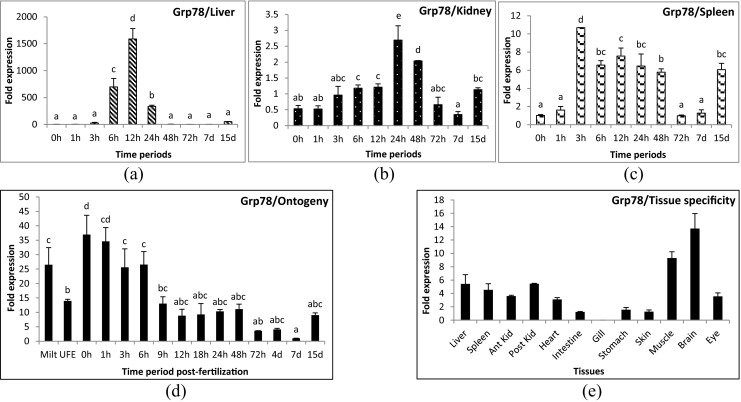
Expression analysis of glucose-regulated protein 78 (Grp78) gene in liver (a), anterior kidney (b), and spleen (c) tissues of rohu, Labeo rohita, during different time periods (h, hours post-challenge, d, days post-challenge) of Aeromonas hydrophila infection, early developmental stages (d) and in different tissues (e). Fold expression was calculated as 2−∆∆Cq. Control group (0 h post-challenge) was taken as the calibrator for infected groups. Least expressing 7-day post-fertilization and intestine samples were taken as the calibrators for ontogeny and tissue specificity study, respectively
Ontogeny, tissue specificity, and sequence analysis of Grp78 gene
Grp78 gene was expressed both in milt and unfertilized eggs prior to fertilization (Fig. 7d). After fertilization, a 20- to 30-fold rise in expression was noticed in the first few hours. Subsequently, a decrease in expression was marked at 9 h post-fertilization (13-fold onwards) and was the lowest at 7 days post-fertilization (1-fold). The expression increased to a 9-fold rise on 15 dpf. Grp78 gene expression was observed in all the tissues examined viz., liver, spleen, anterior and posterior kidneys, eye (3- to 5-fold), heart, intestine, stomach, skin (1-fold), muscle (9-fold), and brain (13-fold) except in gill tissue (Fig. 7e).
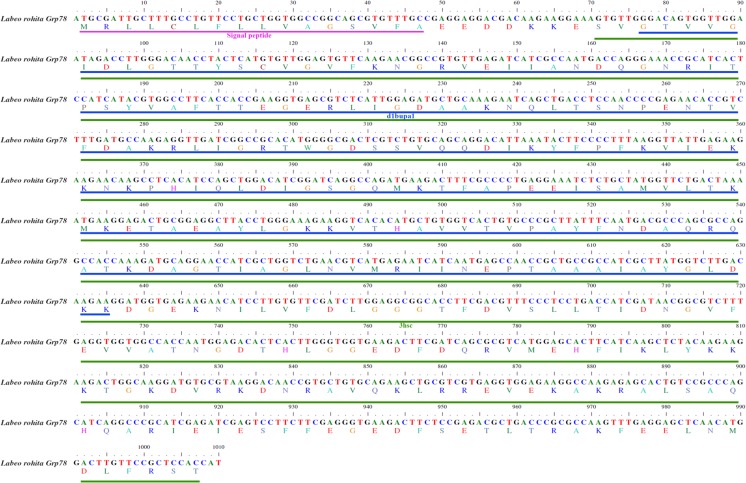
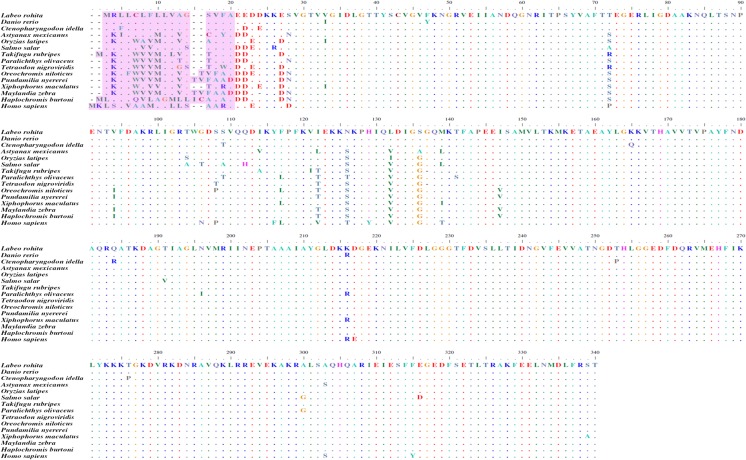
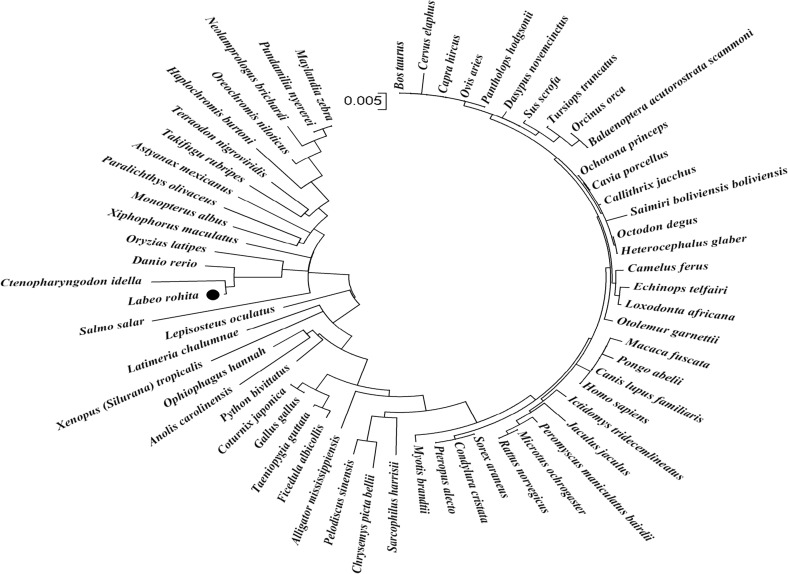
A partial mRNA sequence of 1,010 bp rohu Grp78 (Fig. 8) was deduced and submitted to GenBank (Accession no. KJ755846). The translated protein comprised 336 amino acids with a signal peptide of 16 amino acids. Two domains identified from the partial protein sequence were 3hsc (24–336 amino acids) from Protein Data Base (PDB) and d1bupa1 (26–212 amino acids) from Structural Classifications of Proteins (SCOP). The amino acid alignment (Fig. 9) results revealed that the Grp78 amino acid sequence is highly conserved not only among teleosts but also with higher vertebrates as well. Rohu Grp78 sequence showed maximum similarity with zebrafish and grass carp. Sequences of Japanese rice fish and southern platyfish shared more similarity than other similar sequences such as Mexican tetra, Japanese pufferfish, olive flounder, green-spotted puffer, Nile tilapia, flame black cichlid, zebra mbuna, and Burton’s mouthbrooder. Atlantic salmon Grp78 amino acid sequence was the most divergent of them all. However, all the above sequences differ slightly from human Grp78 sequence. Maximum dissimilarity was seen in the signal peptide region which comprised 16–18 amino acids. A phylogenetic analysis of Grp78 (Fig. 10) showed two distinct clades, one having all the teleosts and the other with higher vertebrates. Among the teleosts, Atlantic salmon formed a different group in contrast to the other teleosts which formed a single group. Rohu shared a place with grass carp, zebrafish, and Japanese rice fish. Southern platyfish was in a separate group and the rest of the fish species were in a single group.
Fig. 8.
Partial mRNA sequence of Grp78 gene from rohu, Labeo rohita (GenBank accession no. KJ755846), showing two domains such as 3hsc (green) and d1bupa1 (blue) with signal peptide (pink)
Fig. 9.
Amino acid alignment of L. rohita Grp78 (KJ755846) with D. rerio (AAH63946.1), C. idella (ACJ65009.1), A. mexicanus (XP_007252148.1), O. latipes (NP_001265730.1), S. salar (NP_001135114.1), T. rubripes (XP_003965205.1), P. olivaceus (ABG56392.1), T. nigroviridis (CAG12424.1), O. niloticus (XP_005470418.1), P. nyererei (XP_005755726.1), X. maculatus (XP_005803813.1), M. zebra (XP_004538056.1), H. burtoni (XP_005930177.1), and H. sapiens (NP_005338.1) where periods denote identical residues and letter e’s denote blanks/gaps. The colored regions indicated the signal peptide sequence
Fig. 10.
Phylogenetic analysis of Grp78 gene constructed using neighbor-joining method in bootstrap test based on a Clustal W multiple sequence alignment of amino acid sequences. Scale bar indicates the degree of divergence represented by a given length of branch. The black circle symbol indicated rohu Grp78
Discussion
Heat shock protein genes influence fish physiology in various ways including development and aging, stress physiology and endocrinology, immunology, environmental physiology, stress tolerance, and acclimatization (Basu et al. 2003). Hsp gene response can vary according to tissue, distinct HSP families, and stressors. The sensitivity of Hsp gene expression can vary according to species, developmental stage, and season (Kayhan and Duman 2010). Hsp gene expression during bacterial infection was previously studied in fish. However, the studies were limited mostly to Hsp90 and Hsp70 (Sung and MacRae 2011). The present communication investigates an array of Hsp genes to establish their possible role in immunity in a comparative mode, and the results obtained with regard to Grp78 gene expression in liver tissue illustrated the major roles played by these molecules during bacterial pathogenesis.
Hsp90 gene seemed to be involved in bacterial immunity, and an increased expression was also found during viral infections in fish (Chen et al. 2010; Lee et al. 1996). During bacterial infections, the role of this gene was uncertain and may be specific to particular environments. In the present experiment, Hsp90 gene was up-regulated during 6 hpc in liver, and 3 and 24 hpc in spleen tissue of rohu during A. hydrophila infection. Down-regulation was noticed during 24–72 hpc in liver, 72 h to 15 days post challenge (dpc) in kidney, and 7 dpc in spleen tissues of A. hydrophila infected rohu. This type of fluctuation in expression patterns was also noticed in previous studies. For example, increased expression of the gene was found at 3, 12, and 24 h, no change after 24 h and down-regulation at 48 h post-infection in gills of black tiger shrimp Penaeus monodon when challenged with Vibrio harveyi (Rungrassamee et al. 2010). Similar patterns were noticed when the expression of Hsp90 gene was induced after 1 h of Vibrio splendidus exposure in hemocytes of the soft-shell clam Mya arenaria and was down-regulated thereafter (Araya et al. 2010). The initial up-regulation may be due to bacterial multiplication and toxicity, since Hsp90 gene is required for the movement of bacterial toxins from the endosomal compartments into the cytosol (Haug et al. 2004). However, the down-regulation in the later stage indicated a confined role for this gene during the initial periods of infection possibly due to a reduction in bacterial load in the survivors.
Hsp70 gene was the most widely studied heat shock protein in fish during infections. An increased expression of Hsp70 gene in liver and kidney tissues of Coho salmon, Oncorhynchus kisutch during Renibacterium salmoninarum infection up to 63 days post-exposure (Forsyth et al. 1997) was the first report to explain the influence of heat shock protein genes in fish in response to pathogens. Hsp70 gene was also over-expressed in rainbow trout after acute Vibrio anguillarum challenge (Ackerman and Iwama 2001). Induced expression of Hsp70 gene was noticed in sea bream liver tissue at 36 h post-infection with Vibrio alginolyticus (Deane et al. 2004). The induction was not limited to fish, but higher expression was also reported in infected crabs, clams, and shrimps as well during Gram-negative bacterial exposure (Cui et al. 2010; Rungrassamee et al. 2010; Yue et al. 2011). However, in the present study, Hsp70 gene was down-regulated in liver and kidney tissues. In spleen tissue, the up-regulation at 12 hpc to A. hydrophila was not noteworthy in terms of fold change. The transcript studied in the present experiment was the third isoform of Hsp70. Previous studies of the same isoform showed down-regulation in the resistant line compared to the susceptible line of rohu to A. hydrophila infection (Das et al. in manuscript). There were six Hsp70 gene isoforms identified in eukaryotes with functional specificity (Kabani and Martineau 2008). Thus, variable function Hsp70 gene isoforms need to be investigated further to conclude which isoform is being activated during infection in fishes.
Mortalins or Grp75 gene is a member of the HSP70 family. Induced expression of mortalin gene was observed during 3 to 24 hpc in rohu, down-regulated at 7 dpc, and subsequently increased at 15 dpc in all the three tissues examined. Although little has been studied on mortalin in fish, it was found to be involved in antigen processing in higher vertebrates (Wadhwa et al. 2002). The increased expression at initial time periods may be due to its possible involvement in the processing of bacterial antigens that were infecting the cells. However, the down-regulation at 7 dpc seems to be correlated with presence of the bacterial load at the specific site. The rise in gene expression at 15 dpc may be a protective mechanism for shedding of membrane vesicles loaded with complement and membrane attack complex (MAC) to protect the cells from complement-mediated lysis (Pilzer and Fishelson 2005).
Phagocytes are major innate immune molecules which show respiratory or oxidative burst activity by releasing bactericidal reactive oxygen species (ROS) upon stimulated by PAMPs through receptors or by the uptake of pathogens (Magnadottir 2010). However, excessive production of these ROS may lead to oxidative stress in fish. Over-expression of Hsc70 protects cells from oxidative stress (Dastoor and Dreyer 2000; Gebhardt et al. 1999). Up-regulation of Hsc70 gene in A. hydrophila-infected liver and spleen tissues of rohu during 3 to 12 and 3 to 72 hpc, respectively, signifies the protective mechanism of the host cell from infection-related oxidative stress. Previous reports suggested an increased level of superoxide production in Puntius sarana during A. hydrophila infection (Das et al. 2011). However, kidney tissue was found to lack this mechanism as no major change in expression was noticed.
Induced expression of Apg2 gene during 3–24 hpc in liver and 3–48 hpc in spleen tissues indicated the participation of this gene in immune function. However, this is the first report of induced expression of this gene in any teleosts during bacterial infection. Hsp30 gene also showed a specific expression pattern in all three tissues examined in contrast to the other genes studied. An initial increase in expression occurred at 6 hpc, then again sudden decrease in expression was observed during 12–24 hpc, and again induction in expression level was noticed after 48 hpc. The increase in expression level might be due to the need for chaperone activities during bacterial infection. It has been reported that this bacterium causes high mortality during 12–24 hpc; possibly, the infected fish might experience a high load of bacteria and bacteria-laden damaged tissues or organs in the system during this period. The up- or down-regulation of these Hsp genes might be correlated with the level of bacterial persistence in the host and the degree of tissue damage caused therein. A previous study also reported the reduction of mortality in challenged rohu within 48 hpc. Hence, a further rise in expression after 48 hpc in all tissues confirms its potential role in chaperone activities during the tissue-healing process after a toxic insult.
The expression pattern of Grp78 gene in teleosts during any bacterial infection was first studied in olive flounder infected with Streptococcus parauberis in which the gene was up-regulated at 6 hpc in kidney tissue (Cha et al. 2013). Induced expression of Grp78 in liver tissue of rohu was remarkably high during 6 to 24 hpc compared to any other genes examined. An increase in expression level was also noticed during 3–48 hpc in kidney and spleen tissues. Its increased expression during bacterial infection indicated that the gene is an important innate immune molecule in fish that needs further characterization and in-depth study. Grp78 or Bip is a cell surface protein which participates in cell signaling, viral entry, and antigen presentation (Quinones et al. 2008). It has an unidentified role in bacterial infection in mammals (Henderson et al. 2006). It was reported as an infection-specific gene expressed during the controlled expression of antimicrobial peptides in Caenorhabditis elegans in Drechmeria coniospora infection (Couillault et al. 2012). Increased expression of Grp78 gene was also noticed during viral infections. Up-regulation of this gene was studied in poly I:C-induced liver and kidney tissues of grass carp (Chuxin et al. 2009). Interaction of host Grp78 or Bip protein with viral capsid was also evident in grouper, Epinephelus spp. (Lu et al. 2012). However, a functional characterization of Grp78 protein during bacterial infection in fish is needed to further identify its mechanism of action.
Grp78 binds to hydrophobic patches on nascent polypeptides within the endoplasmic reticulum (ER) during signaling of the unfolded protein response (Quinones et al. 2008). Increased expression of this gene during early developmental stages may control the protein function especially in gametes, since a high level of expression was seen in milt and unfertilized eggs. Moreover, increased expression of Grp78 gene after fertilization also indicated its major role as a molecular chaperone during developmental days in fish. Developmental regulation of many Hsps genes such as Hsp90α, Hsp90β, and Hsp47 was observed in zebrafish previously (Basu et al. 2002). Constitutive expression of Grp78 gene in several important tissues emphasized its importance in regulating ER homeostasis in fish. It participates in the unfolded protein response (UPR) during accumulation of mis-folded proteins, Ca2+ depletion, or oxidative stress (Lee 2001). The two domains identified from the partial protein sequence, 3hsc and d1bupa1, were characteristic features of 70 kd heat shock protein cognate. The 3hsc is a hydrolase acting on acid anhydrides. The d1bupa1, a member of actin-like ATPase domain superfamily, is an alpha and beta protein containing ribonuclease H-like motif. The gene is highly conserved throughout the evolutionary hierarchy from yeast to mammals. Amino acid alignment of Grp78 of different fish species showed diversity in the signal peptide region. The signal sequences are sufficiently degenerative that there is little evolutionary pressure on them. As long as certain general features such as hydrophobicity are maintained, there will be no change in function. Therefore, these diversities may be due to diversity in substrates for which the signal is being optimized (Kim et al. 2002). Further translocation studies are needed to study the effect of these variations in signal peptide sequences on Grp78 targets. The phylogenetic tree of Grp78 was as per the vertebrate evolution. All the carps were grouped in one cluster like the cichlids, amphibians, reptiles, birds, mammals, and rodents. Teleosts were in a separate clade from higher vertebrates in the phylogenetic tree. The presence of West Indian Ocean coelacanth Grp78 sequence in the higher vertebrates’ clade was expected since it is more closely related to reptiles and mammals than fishes (Amemiya et al. 2013). However, the greater similarity of the spotted gar Grp78 sequence with higher vertebrates than with other fishes signified the conserved nature of this gene throughout the vertebrate evolution since spotted gar is one of the most primitive fish (Carman 2002). The diversity in other fish species may be an environmental adaptation of this gene to deal better with the environmental stress that is more evident in aquatic organisms than higher vertebrates.
The present study thus depicted the involvement of major heat shock proteins during bacterial exposure in fish. In addition to the earlier studies that are mostly confined to Hsp90 and Hsp70 genes in fish during infection, the present study included other members of HSP70 family such as Grp78, Grp75, and Hsc70 and indicated their potential role in the immune process during bacterial infection in fish. Further, the modulation of heat shock proteins was more evident in liver tissue followed by spleen and kidney tissues of infected fish. Grp78 gene was found to be a conserved heat shock protein that plays an important role not only during infection but also during early developmental stages and maintains the regular physiological processes of fish.
Acknowledgments
The authors are grateful to the Director, Central Institute of Freshwater Aquaculture, Bhubaneswar, India for providing the necessary facilities for conducting experiments. The authors are thankful to the Fish Genetics and Biotechnology Division, CIFA for providing fish for conducting experiments. The Council of Scientific and Industrial Research (CSIR), Government of India is duly acknowledged for financial support to the first author in terms of research fellowship grant (CSIR Sanction no. 9/694(0006)/2013-EMR-1).
References
- Ackerman PA, Iwama GK. Physiological and cellular stress responses of juvenile rainbow trout to vibriosis. J Aquat Anim Health. 2001;13:173–180. doi: 10.1577/1548-8667(2001)013<0173:PACSRO>2.0.CO;2. [DOI] [Google Scholar]
- Amemiya CT, Alfo¨ldi J, Lee AP, Fan S, Philippe H, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya MT, Markham F, Mateo DR, McKenna P, Johnson GR, et al. Identification and expression of immune-related genes in hemocytes of soft-shell clams, Mya arenaria, challenged with Vibrio splendidus. Fish Shellfish Immunol. 2010;29:557–564. doi: 10.1016/j.fsi.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Basu N, Todgham AE, Ackerman PA, Bibeau MR, Nakano K, Schulte PM, Iwama GK. Heat shock protein genes and their functional significance in fish. Gene. 2002;295:173–183. doi: 10.1016/S0378-1119(02)00687-X. [DOI] [PubMed] [Google Scholar]
- Basu N, Kennedy CJ, Iwama GK. The effects of stress on the association between Hsp70 and the glucocorticoid receptor in rainbow trout. Comp Biochem Physiol. 2003;134:655–663. doi: 10.1016/S1095-6433(02)00372-0. [DOI] [PubMed] [Google Scholar]
- Carman SM. Special animal abstract for Lepisosteus oculatus (spotted gar) Lansing: Michigan Natural Features Inventory; 2002. p. 3. [Google Scholar]
- Cha IS, Kwon J, Park SB, Jang HB, Nho SW, Kim YK, Hikima J, Aoki T, Jung TS. Heat shock protein profiles on the protein and gene expression levels in olive flounder kidney infected with Streptococcus parauberis. Fish Shellfish Immunol. 2013;34:1455–1462. doi: 10.1016/j.fsi.2013.03.355. [DOI] [PubMed] [Google Scholar]
- Chen YM, Kuo CE, Wang TY, Shie PS, Wang WC, Huang SL, et al. Cloning of an orange-spotted grouper Epinephelus coioides heat shock protein 90AB (HSP90AB) and characterization of its expression in response to nodavirus. Fish Shellfish Immunol. 2010;28:895–904. doi: 10.1016/j.fsi.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Chuxin WU, Yi L, Meisheng M, Likun W, Chengyu H. Up-regulation of grass carp GRP78 gene expression under heat shock and poly I:C stress. Chin J Appl Environ Biol. 2009;15:814–818. [Google Scholar]
- Couillault C, Fourquet P, Pophillat M, Ewbank JJ. A UPR independent infection-specific role for a BiP/GRP78 protein in the control of antimicrobial peptide expression in C. elegans epidermis. Virulence. 2012;3:299–308. doi: 10.4161/viru.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Liu Y, Luan W, Li Q, Wu D, et al. Molecular cloning and characterization of a heat shock protein 70 gene in swimming crab (Portunustri tuberculatus) Fish Shellfish Immunol. 2010;28:56–64. doi: 10.1016/j.fsi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Das T, Pal AK, Chakraborty SK, Manush SM, Chatterjee N, Apte SK. Metabolic elasticity and induction of heat shock protein 70 in Labeo rohita acclimated to three temperatures. Asian Austral J Anim Sci. 2006;19:1033–1039. doi: 10.5713/ajas.2006.1033. [DOI] [Google Scholar]
- Das A, Sahoo PK, Mohanty BR, Jena JK. Pathophysiology of experimental Aeromonas hydrophila infection in Puntius sarana: early changes in blood and aspects of the innate immune-related gene expression in survivors. Vet Immunol Immunopathol. 2011;142:207–218. doi: 10.1016/j.vetimm.2011.05.017. [DOI] [PubMed] [Google Scholar]
- Das S, Chhottaray C, Mahapatra KD, Saha JN, Baranski M, Robinson N, Sahoo PK (2014) Analysis of immune-related ESTs and differential expression analysis of few important genes in lines of rohu (Labeo rohita) selected for resistance and susceptibility to Aeromonas hydrophila infection. Manuscript under review in Mol Biol Rep [DOI] [PubMed]
- Dastoor Z, Dreyer JL. Nuclear translocation and aggregate formation of heat shock cognate protein 70 (Hsc70) in oxidative stress and apoptosis. J Cell Sci. 2000;113:2845–2854. doi: 10.1242/jcs.113.16.2845. [DOI] [PubMed] [Google Scholar]
- Deane EE, Li J, Woo NYS. Modulated heat shock protein expression during pathogenic Vibrio alginolyticus stress of sea bream. Dis Aquat Org. 2004;62:205–215. doi: 10.3354/dao062205. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Hsps, molecular chaperons and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Forsyth RB, Candido EPM, Babich SL, Iwama GK. Stress protein expression in coho salmon with bacterial kidney disease. J Aquat Anim Health. 1997;9:18–25. doi: 10.1577/1548-8667(1997)009<0018:SPEICS>2.3.CO;2. [DOI] [Google Scholar]
- Gebhardt BR, Ries J, Caspary WF, Boehles H, Stein J. Superoxide: a major factor for stress protein induction in reoxygenation injury in the intestinal cell line Caco-2. Digestion. 1999;60:238–245. doi: 10.1159/000007664. [DOI] [PubMed] [Google Scholar]
- Haug G, Aktories K, Barth H. The host cell chaperone Hsp90 is necessary for cytotoxic action of the binary iota-like toxins. Infect Immun. 2004;72:3066–3068. doi: 10.1128/IAI.72.5.3066-3068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B. Chaperonins: chameleon proteins that influence myeloid cells. In: van Eden W, editor. Heat shock proteins and inflammation. Basle: Birkhauser Verlag; 2003. pp. 175–192. [Google Scholar]
- Henderson B, Allan E, Coates AR. Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect Immun. 2006;74:3693–3706. doi: 10.1128/IAI.01882-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, Martineau CN. Multiple Hsp70 isoforms in the eukaryotic cytosol: mere redundancy or functional specificity? Curr Genomics. 2008;9:338–248. doi: 10.2174/138920208785133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayhan FE, Duman BS. Heat shock protein genes in fish. Turk J Fish Aquat Sci. 2010;10:287–293. doi: 10.4194/trjfas.2010.0218. [DOI] [Google Scholar]
- Kim SJ, Mitra D, Salerno JR, Hegde RS. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev Cell. 2002;2:207–217. doi: 10.1016/S1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- Lalitha S. Primer premier 5. Biotech Software Internet Rep. 2000;1:270–272. doi: 10.1089/152791600459894. [DOI] [Google Scholar]
- Lee AS (2001) The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci 26:504–510 [DOI] [PubMed]
- Lee JY, Cho WJ, Do JW, Kim HJ, Park JW. Monoclonal antibodies raised against infectious haematopoietic necrosis virus (IHNV) G protein and a cellular 90 kDa protein neutralize IHNV infection in vitro. J Gen Virol. 1996;77:1731–1737. doi: 10.1099/0022-1317-77-8-1731. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2-∆∆Ct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu MW, Ngou FH, Chao YM, Lai YS, Chen NY, Lee FY, Chiou PP. Transcriptome characterization and gene expression of Epinephelus spp in endoplasmic reticulum stress-related pathway during betanodavirus infection in vitro. BMC Genomics. 2012;13:651. doi: 10.1186/1471-2164-13-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnadottir B. Immunological control of fish diseases. Mar Biotechnol. 2010;12:361–379. doi: 10.1007/s10126-010-9279-x. [DOI] [PubMed] [Google Scholar]
- Mohanty BR, Mishra J, Das S, Jena JK, Sahoo PK. An outbreak of aeromoniasis in an organized composite carp culture farm in India: experimental pathogenicity and antibiogram study. J Aqua. 2008;16:27–37. [Google Scholar]
- Nayak SP, Mohanty BR, Mishra J, Rauta PR, Das A, Eknath AE, Sahoo PK. Ontogeny and tissue-specific expression of innate immune related genes in rohu, Labeo rohita (Hamilton) Fish Shellfish Immunol. 2011;30:1197–1201. doi: 10.1016/j.fsi.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Pilzer D, Fishelson Z. Mortalin/GRP75 promotes release of membrane vesicles from immune attacked cells and protection from complement-mediated lysis. Int Immunol. 2005;17:1239–1248. doi: 10.1093/intimm/dxh300. [DOI] [PubMed] [Google Scholar]
- Quinones QJ, de Ridder GG, Pizzo SV. GRP78: a chaperone with diverse roles beyond the endoplasmic reticulum. Histol Histopathol. 2008;23:1409–1416. doi: 10.14670/HH-23.1409. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent end points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- Roberts RJ, Agius C, Saliba C, Bossier P, Sung YY. Heat shock proteins (chaperones) in fishes and shellfishes and their potential role in health and welfare: a review. J Fish Dis. 2010;33:789–801. doi: 10.1111/j.1365-2761.2010.01183.x. [DOI] [PubMed] [Google Scholar]
- Robinson N, Sahoo PK, Baranski M, Mahapatra KD, Saha JN, Das S, Mishra Y, Das P, Barman HK, Eknath AE. Expressed sequences and polymorphisms in rohu carp (Labeo rohita, Hamilton) revealed by mRNA-seq. Mar Biotechnol. 2012;14:620–633. doi: 10.1007/s10126-012-9433-8. [DOI] [PubMed] [Google Scholar]
- Rungrassamee W, Leelatanawit R, Jiravanichpaisal P, Klinbunga S, Karoonuthaisiri N. Expression and distribution of three heat shock protein genes under heat shock stress and under exposure to Vibrio harveyi in Penaeus monodon. Dev Comp Immunol. 2010;34:1082–1089. doi: 10.1016/j.dci.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Sahoo PK, Mahapatra KD, Saha JN, Barat A, Sahoo M, Mohanty BR, Gjerde B, Odegard J, Rye M, Salte R. Family association between immune parameters and resistance to Aeromonas hydrophila infection in the Indian major carp, Labeo rohita. Fish Shellfish Immunol. 2008;25:163–169. doi: 10.1016/j.fsi.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Sung YY, MacRae TH. Heat shock proteins and disease control in aquatic organisms. J Aquac Res Dev. 2011;S2:006. [Google Scholar]
- Triantafilou K, Triantafilou M, Dedrick RL. A CD14-independent LPS receptor cluster. Nat Immunol. 2001;2:338–345. doi: 10.1038/86342. [DOI] [PubMed] [Google Scholar]
- van der Marel M, Schroers V, Neuhaus H, Steinhagen D. Chemotaxis towards, adhesion to, and growth in carp gut mucus of two Aeromonas hydrophila strains with different pathogenicity for common carp, Cyprinus carpio L. J Fish Dis. 2008;31:321–330. doi: 10.1111/j.1365-2761.2008.00902.x. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Taira K, Kaul SC. An Hsp70 family chaperone, mortalin/mthsp70/PBP74/Grp75: what, when, and where? Cell Stress Chaperones. 2002;7:309–316. doi: 10.1379/1466-1268(2002)007<0309:AHFCMM>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wei Y, Li X, Cao H, Xu M, Dai J. The identification of heat shock protein genes in goldfish (Carassius auratus) and their expression in a complex environment in Gaobeidian Lake, Beijing, China. Comp Biochem Physiol. 2007;145:350–362. doi: 10.1016/j.cbpc.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Yengkokpam S, Pal AK, Sahu NP, Jain KK, Dalvi R, Misra S, Debnath D. Metabolic modulation in Labeo rohita fingerlings during starvation: Hsp70 expression and oxygen consumption. Aquaculture. 2008;285:234–237. doi: 10.1016/j.aquaculture.2008.08.034. [DOI] [Google Scholar]
- Yue X, Liu B, Sun L, Tang B. Cloning and characterization of a hsp70 gene from Asiatic hard clam Meretrix meretrix which is involved in the immune response against bacterial infection. Fish Shellfish Immunol. 2011;30:791–799. doi: 10.1016/j.fsi.2010.12.027. [DOI] [PubMed] [Google Scholar]