Abstract
Epidermal keratinocytes serve as the primary barrier between the body and environmental stressors. They are subjected to numerous stress events and are likely to respond with a repertoire of heat shock proteins (HSPs). HSPA6 (HSP70B′) is described in other cell types with characteristically low to undetectable basal expression, but is highly stress induced. Despite this response in other cells, little is known about its control in keratinocytes. We examined endogenous human keratinocyte HSPA6 expression and localized some responsible transcription factor sites in a cloned HSPA6 3 kb promoter. Using promoter 5′ truncations and deletions, negative and positive regulatory regions were found throughout the 3 kb promoter. A region between −346 and −217 bp was found to be crucial to HSPA6 basal expression and stress inducibility. Site-specific mutations and DNA-binding studies show that a previously uncharacterized AP1 site contributes to the basal expression and maximal stress induction of HSPA6. Additionally, a new heat shock element (HSE) within this region was defined. While this element mediates increased transcriptional response in thermally stressed HaCaT keratinocytes, it preferentially binds a stress-inducible factor other than heat shock factor (HSF)1 or HSF2. Intriguingly, this newly characterized HSPA6 HSE competes HSF1 binding a consensus HSE and binds both HSF1 and HSF2 from other epithelial cells. Taken together, our results demonstrate that the HSPA6 promoter contains essential negative and positive promoter regions and newly identified transcription factor targets, which are key to the basal and stress-inducible expression of HSPA6. Furthermore, these results suggest that an HSF-like factor may preferentially bind this newly identified HSPA6 HSE in HaCaT cells.
Keywords: HSP70B′ promoter, HSE, AP1, HSF, Keratinocyte
Introduction
Properly controlled heat shock protein (HSP) gene expression is integral to maintaining and restoring cell homeostasis under basal and stressed conditions, respectively. Although initially known by its transcription induction from thermal stress, HSP expression is also increased in response to toxic chemical and UV light assaults (Jonak et al. 2009). Additionally, the characterization of dozens of HSP genes across multiple families established that several members are constitutively expressed, accounting for their availability in “housekeeping” chaperone function (Kampinga et al. 2009). As might be expected, within and across HSP families, there are some coding sequence similarities, common substrate targets, and shared transcriptional control by heat shock factor (HSF) (Akerfelt et al. 2010; Saibil 2013). However, these shared qualities belie the nonredundant role of several HSP identified in recent reports (Daugaard et al. 2007; Hageman et al. 2011; Heldens et al. 2012; Noonan et al. 2007a). HSPA1A (HSP70) is an important protein chaperone in unstressed conditions and is crucial to prevent stress-induced cell death. While closely related to HSPA1A, HSPA6 (HSP70B′) has similar yet distinct functions and its expression patterns (Leppa et al. 2001; Noonan et al. 2007a) vary between cell types and cell densities (Noonan et al. 2007b). In addition to expression variation between cell types, HSPA6 species expression is limited, as it has only been observed in human, swine, and goats (Banerjee et al. 2014; Dezeure et al. 1993). Unlike other widely expressed HSPs, a rodent HSPA6 homologue has not yet been discovered (Parsian et al. 2000), suggesting that it is a newly evolved HSP only found in higher mammals.
Like HSPA1A, HSPA6 expression is essential to increasing survival of cells exposed to increased temperatures or chemicals. Single or double siRNA-mediated knockdown of HSPA1A and/or HSPA6 suggests that while both HSPs are important to increasing cell survival, HSPA6 may be a secondary regulator of stress compared to HSPA1A (Noonan et al. 2007a). Decreased expression of HSPA6 did reduce the cell viability after a 42 °C heat stress or proteasome inhibitor MG-132 treatment, suggesting its importance in cell survival. HSPA6 likely forms complexes with HSPA1A and DNAJB1 (HSP40) (Chow et al. 2010; Noonan et al. 2008) to confer its protective function. Despite some HSPA6/HSPA1A overlap in facilitating cell survival, further work showed that they have distinct protein substrates. Compared to HSPA1A, HSPA6 has higher affinity for unfolded p53 but has no effect in refolding the luciferase enzyme and peroxisomal proteins (Hageman et al. 2011; Heldens et al. 2012). A better definition of HSPA6 gene expression levels and the protein factors/promoter elements contributing to it would improve our understanding as to its availability or inducibility to meet these specific protective chaperone/refolding functions.
HSPA6 production under nonstressed conditions is variable, from not detected, to low expression levels, possibly dependent on cell type (Chow et al. 2010; Gomez-Sucerquia et al. 2012) and growth condition differences (Noonan et al. 2007b). Its capacity for significant induction under stressed conditions has been well-documented, but what controls this or basal expression has mostly been elucidated using a ~287 bp minimal promoter (Rohmer et al. 2008; Wada et al. 2005; Wada et al. 2007). To enhance our understanding of HSPA6 production in other cell types and control over its basal and inducible transcription, we examined HSPA6 expression in epidermal keratinocytes and what might contribute to the control of inducible and any basal expression. Various HSPs in keratinocytes serve as a cadre of molecular chaperones and stress response proteins both for protein folding during cell differentiation and epidermal response to topical assaults. Insufficient HSP expression in keratinocyte has detrimental consequences including (i) inadequate integration of cytoskeletal and noncytoskeletal proteins to generate the skin’s barrier and (ii) failure to cope with or recover from stresses as evidenced by poor or absent wound healing (Kwon et al. 2002; Matsuda et al. 2010). In brief, we found that human keratinocytes have a significant capacity for HSPA6 induction at both the messenger RNA (mRNA) and protein levels compared to the related HSPA1A. Additionally, with computational analysis, cloning, and functional assessment of ~3 kb of the HSPA6 promoter, we found previously unidentified regions exerting negative or positive effects over basal expression as well as a novel heat shock element (HSE) upstream of that previously known (Wada et al. 2007). Constitutive and strikingly inducible HSPA6 expression in combination with complex transcriptional regulation suggests that it may be positioned to contribute significant chaperoning as well as stress-protective functions to epidermal keratinocytes.
Materials and methods
Cell culture
HaCaT (Boukamp et al. 1988), SCC13, MCF7, HeLa, HepG2, and foreskin dermal fibroblasts (FSFB) were cultured in a 3:1 Dulbecco’s modified Eagle medium (DMEM)/F12 media containing 10 % fetal bovine serum (FBS) (Thermo Scientific HyClone, Logan, UT), 100 U/ml penicillin, and 100 mg/ml streptomycin. HT29 cells were cultured in McCoy’s 5a modified media containing 10 % FBS, supplemented with 1 % nonessential amino acids, 100 U/ml penicillin, and 100 mg/ml streptomycin. Caco2 cells were cultured in MEM media containing 20 % FBS and supplemented with 1 % nonessential amino acids, 1 % pyruvate, 100 U/ml penicillin, and 100 mg/ml streptomycin. The HaCaT keratinocytes and all cell lines are derived from human tissues. All cells were grown in a 37 °C with 5 % CO2 humidified incubator. For peroxisome proliferator-activated receptor (PPAR) ligand treatment, cells were treated with 1 or 10 μM PPARα (WY14,643), β/δ (L165,043), or γ (ciglitazone) ligand 24 h after cell plating. Total RNA was collected 24 h post treatment. For heat stress experiments, cells were plated at 1.5 × 105 or 6.8 × 105 cells per well in 24- or 6-well plates, respectively. Twenty-four hours later, cells were stressed for 1 h in a 42 °C water bath (control cells were immersed in a 37 °C water bath) and returned to a 37 °C incubator for the indicated recovery time. Cells were collected for total RNA using the RNeasy kit (Qiagen, Valencia, CA) or protein using radioimmunoprecipitation assay (RIPA) lysis buffer (10 mM Tris, 150 mM NaCl, 1 % deoxycholic acid, 1 % Triton, 0.1 % sodium dodecyl sulfate (SDS)) supplemented with Halt Protease Inhibitor (Thermo Scientific, Waltham, MA). For transfection analysis, cells were plated to 70 % confluency in 24-well plates. Twenty-four hours later, cells were transfected as described below.
Real-time quantitative PCR analysis
Total RNA was reverse transcribed using the iScript reverse transcriptase (BioRad, Hercules, CA). Gene expression changes were analyzed using POWER SYBR green master mix (Life Technologies, Grand Island, NY). Real-time PCR was performed using Applied Biosystems 7500 Fast Real-Time PCR system. Data analysis was carried out on ABI 7500 software using the ΔΔCT method. All data was normalized to β-tubulin.
Immunoblot analysis
Whole-cell lysates were prepared in RIPA buffer, and following determination of protein concentration, 10 μg of protein was separated by SDS/PAGE, transferred to nitrocellulose membranes, rinsed with nanopure water, and treated with Qentix (Thermo Scientific Pierce). Blots were incubated in 5 % (w/v) nonfat dried milk, Tris-buffered saline (TBS), and 0.1 % Tween 20 and then subsequently probed with HSP70B′ antibody (ADI-SPA-754) at 1:1,000 dilution (Enzo Life Sciences, Farmingdale, NY) or anti-HSPA1A antibody (ADI-SPA-810) at 1:1,000 dilution (Enzo Life Sciences) (Chow et al. 2010; Noonan et al. 2007b) followed by horseradish peroxidase (HRP)-conjugated secondary goat anti-mouse antibody at 1:10,000 dilution (PerkinElmer, Branford, CT). Blots were subsequently probed for β-actin antibody (ab8227) at 1:5,000 dilution (Abcam, Cambridge, MA) followed by HRP-conjugated secondary goat anti-rabbit antibody at 1:20,000 dilution (PerkinElmer). Detection of binding was determined with enhanced chemiluminescence reagents (Thermo Scientific Pierce). Band signals were digitally captured using the Kodak image station CCF and analyzed using the Carestream molecular imaging software. The wider linear dynamic range of the software is >4 orders of magnitude and no band signals reached saturation.
Cloning and generation of wild-type and mutant HSPA6 promoter sequences
The HSPA6 promoter containing the −2,963 to +48 bp sequence (herein referred to as −3 kb-luc) was PCR amplified from human genomic DNA (cat# 636401) (BD Biosciences, San Jose, CA) using forward: 5′-GAT GGG TAC CTC ATC TTG AAT TCC CAC AAC ACA TGG-3′ and reverse: 5′-GGC TGA AGC TTA GTG AGG CTC TCC CTG CGG TTT CTC T-3′ with added KpnI and HindIII sites (underlined), respectively, for insertion into the promoterless vector pGL4.10 (Promega, Madison, WI) using the restriction sites indicated. 5′-Promoter truncations (−1,230, −647 and −70-luc) were performed by using the upstream KpnI site and native restriction enzyme sites BglII, EcoRI, and NruI, respectively. Digested sites were blunted and ligated. Internal promoter deletions were performed using the Quikchange Lightning site-directed mutagenesis kit (Agilent). Constructs ΔA, ΔB, ΔC, and ΔD were generated using the −1,230-luc; constructs ΔE, ΔF, ΔG, and ΔH were generated using the −647-luc. Fragment G site-specific mutants were generated using the Quikchange Lightning site-directed mutagenesis kit. The WHN, HSE, MZF1, C/EBP, AP1, and Zfx sites were mutated as indicated (Table 1). Sites were determined using NHR Scan (Sandelin and Wasserman 2005), Nubiscan (Podvinec et al. 2002), and MatInspector (Cartharius et al. 2005) web-based software. To generate the wild-type (WT)-G-tk-luc, AP1 mutant (mt)-G-tk-luc, and HSEmt-G-tk-luc construct, the HSPA6 G region was amplified via PCR using primers flanked by KpnI sites. A comparison HSE sequence was generated using functional HSEs derived from several HSP promoters: one site within HSPB1 (HSP27), HSPC1 (HSP90α), HSPCB (HSP90β), the downstream HSE from HSPA6 and two sites within HSPA1A (HSP70) (Dierick et al. 2007; Koizumi et al. 2013; Metz et al. 1996; Morgan et al. 1987; Shen et al. 1997; Wada et al. 2005) and visualized with WebLogo (Crooks et al. 2004). The pGL4.10 construct containing the thymidine kinase (tk-luc) minimal promoter was used to insert the HSPA6 WT, AP1mt, or HSEmt fragment G. All HSPA6 3 kb isolate, all site-directed mutants, and deletion constructs were confirmed by sequencing (University of Connecticut Biotech Center).
Table 1.
Site-directed mutagenesis primer sequences. Underlined sequences denote mutations within the binding site
| Site name | Sequence within HSPA6 | Mutated sequence |
|---|---|---|
| WHN | ACGC | ATAC |
| HSE | GGGAGGAGCTAGAACCTTCC | GGGAGGAGCTAATTCCTTCC |
| MZF1 | GCGGGGAAGGT | GCGTAGAGGGT |
| C/EBP | CTCAGGCTGCTGAAA | CTCATGCACTTGTCA |
| AP1 | TGAGTCA | TTAGTTA |
| ZFX | CTGGCCTGGCG | CTAAGATGGCG |
Plasmid transfections
HaCaT or HeLa cells were plated to 70 % confluency 24 h prior to transfections using 24-well plates. Eight hours prior to transfection, media was replaced with 0.5 ml serum-containing 3:1 DMEM/F12 media. The appropriate HSPA6 promoter pGL4.10 plasmid (200 ng) and pCMV-β galactosidase (100 ng) was transfected using Fugene6 (Promega) using 100 μl serum-free media. For PPAR experiments, cells were cotransfected with pSG5-empty vector or pSG5-PPARγ (100 ng). Twenty-four hours later, cells were treated with 10 or 100 μM ciglitazone and collected 16 h post treatment. For the thermal stress experiments, 24 h following plating, cells were stressed for 1 h in a 42 °C water bath (control cells in a 37 °C bath) and returned to a 37 °C incubator for 4 h. Cells were then collected and assayed for the luciferase activity (Promega), protein concentration (Pierce), and β-galactosidase activity (Ramirez and Aneskievich 2013).
Electrophoretic mobility shift assay
Nuclear extracts were prepared from HaCaT or HeLa cells as previously described (Encarnacao et al. 2013). Protein concentrations were determined by BCA protein assay (Pierce, Rockford, IL). Oligomers (Integrated DNA Technology, IDT, Coralville, IA) were annealed and end labeled with 32P-ATP (PerkinElmer). Electrophoretic mobility shift assay (EMSA) oligonucleotide probe sequences are shown in Table 2. Fifteen or 10 μg of nuclear extracts were incubated with radiolabeled HSE or AP1 oligomers, respectively. For AP1 EMSAs, nuclear extracts were preincubated with the appropriate antibodies or unlabeled competition oligomers 1 h or 15 min, respectively, at room temperature prior to the addition of the radiolabeled oligonucleotide probes. The mixture was incubated at room temperature for 30 min and then loaded into a 6 % polyacrylamide gel. Gels were electrophoresed at 3.5 mA/gel in 0.5× TBE buffer for 10 h at 4 °C. For HSE EMSAs, nuclear extracts were preincubated with unlabeled competition oligomers 15 min at room temperature prior to the addition of the radiolabeled oligonucleotide probes. Antibodies were added and the mixture was incubated at room temperature for 20 min, then on ice for 10 min. Samples were loaded on a 4 % polyacrylamide gel and electrophoresed at 3.5 mA/gel in 0.5× TBE buffer for 8 h at 4 °C. Anti-cJun (SC-45X), cFos (SC-52X), HSF1 (SC-9144x), and HSF2 (SC-13056X) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Gels were dried and exposed to Amersham Hyperfilm MP (GE, Buckinghamshire, UK) for at least 16 h at −80 °C with intensifying screens. Films were developed using a Kodak X-Omat 2000.
Table 2.
Oligomers used for EMSA probes. Underlined sequences denote mutations within the binding site. The predicted HSE mutation encompasses one HSE repeat, whereas the double mutant encompasses two HSE inverted repeats
| Oligomer name | Primer sequence |
|---|---|
| Predicted AP1 top | CTAGCAGCAGCCTGAGTCAGAGGCGGG |
| Predicted AP1 bottom | CTAGCCCGCCTCTGACTCAGGCTGCTG |
| AP1 consensus top | CTAGCGCTTGATGACTCAGCCGGAA |
| AP1 consensus bottom | CTAGTTCCGGCTGAGTCATCAAGCG |
| Predicted AP1 mutant top | CTAGCAGCAGCCTTAGTTAGAGGCGGG |
| Predicted AP1 mutant bottom | CTAGCCCGCCTCTAACTAAGGCTGCTG |
| Predicted HSE top | CTAGGGGAGGAGCTAGAACCTTCCCCGCA |
| Predicted HSE bottom | CTAGTGCGGGGAAGGTTCTAGCTCCTCCC |
| HSE consensus top | CTAGCGAAACCCCTGGAATATTCCCGACC |
| HSE consensus bottom | CTAGGGTCGGGAATATTCCAGGGGTTTCG |
| Predicted HSE mutant top | CTAGGGGAGGAGCTAATTCCTTCCCCGCA |
| Predicted HSE mutant bottom | CTAGTGCGGGGAAGGAATTAGCTCCTCCC |
| Predicted HSE double mutant top | CTAGGGGAGGAGCCCATTATAGTAGCGCA |
| Predicted HSE double mutant bottom | CTAGTGCGCTACTATAATGGGCTCCTCCC |
Statistical analysis
Data was analyzed using Prism Software version 5 (GraphPad) (La Jolla, CA). Student’s t test was used to compare between paired results. ANOVA with Newman Keuls or Dunnett’s post hoc was used to compare between grouped results, as specified. Statistical significance was defined as p value ≤0.05.
Results
Constitutive and inducible expression of HSPA6
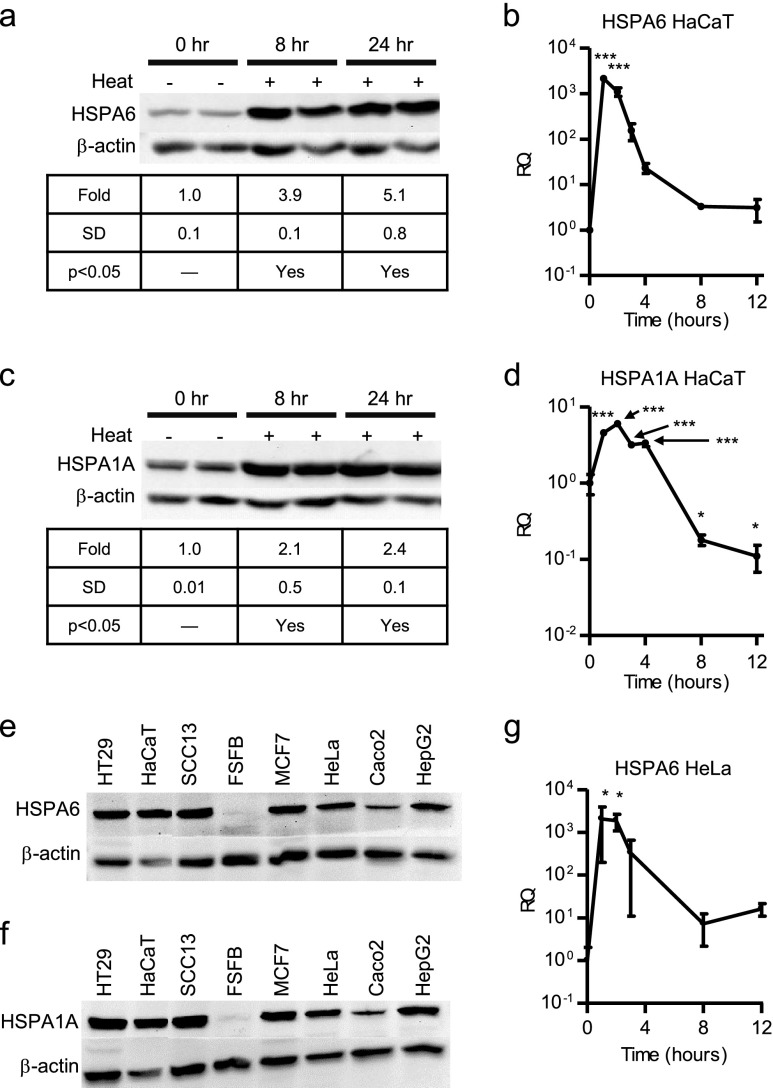
Basal and induced levels of HSPA6 were determined in HaCaT keratinocytes under standard culture and thermal stress conditions. Basal expression of HSPA6 was detected in unstressed cells with a 3.9- and 5.1-fold increase in protein levels observed at 8 and 24 h post-heat shock, respectively (Fig. 1a). HSPA6 mRNA was upregulated immediately following heat stress and then gradually decreased over 8–12 h after the stress period to levels similar to basal expression (Fig. 1b). To validate the heat shock, HSPA1A protein and mRNA expression was also examined. The induction pattern of HSPA1A protein and mRNA was similar to HSPA6, but to a lesser fold induction than HSPA6 (Fig. 1c, d). Variable levels of HSPA6 and HSPA1A were also detected in other epithelial (HT29, MCF-7, Caco2, HepG2) and epidermis-derived (SCC13) cells with relatively little detected in dermal fibroblast cells (Fig. 1e, f) under standard (nonstressed) culture conditions. HSPA6 mRNA expression was also analyzed in a nonkeratinocyte cell line, HeLa cells. The heat induction profile was similar to that observed from HaCaT keratinocytes (Fig. 1g). HSPA6 cell-type range of expression and mRNA induction in HaCaT and HeLa cell lines suggested that a combination of ubiquitous and stress-specific factors may control its gene expression.
Fig. 1.
Basal and heat-inducible expression of HSPs. Protein expression of HSPA6 (a) and HSPA1A (c) before (0 h) and post-heat shock (8 and 24 h). Samples shown from duplicate cultures. Quantitation of band intensity shown as average of duplicate cultures. mRNA expression of HSPA6 (b) and HSPA1A (d) in HaCaT cells and HSPA6 in HeLa cells (g) before (0 h) and post-heat shock (1, 2, 3, 4, 8, and 12 h). Protein expression of HSPA6 (e) and HSPA1A (f) in various unstressed cells. β-Actin used as a loading control for all Western blots. Statistically significant values are indicated using a p value <0.05 as determined using ANOVA with Dunnett’s post hoc. Points are mean + SD from one experiment, each point from replicate cultures
Determining specific transcription factor binding sites within the HSPA6 promoter
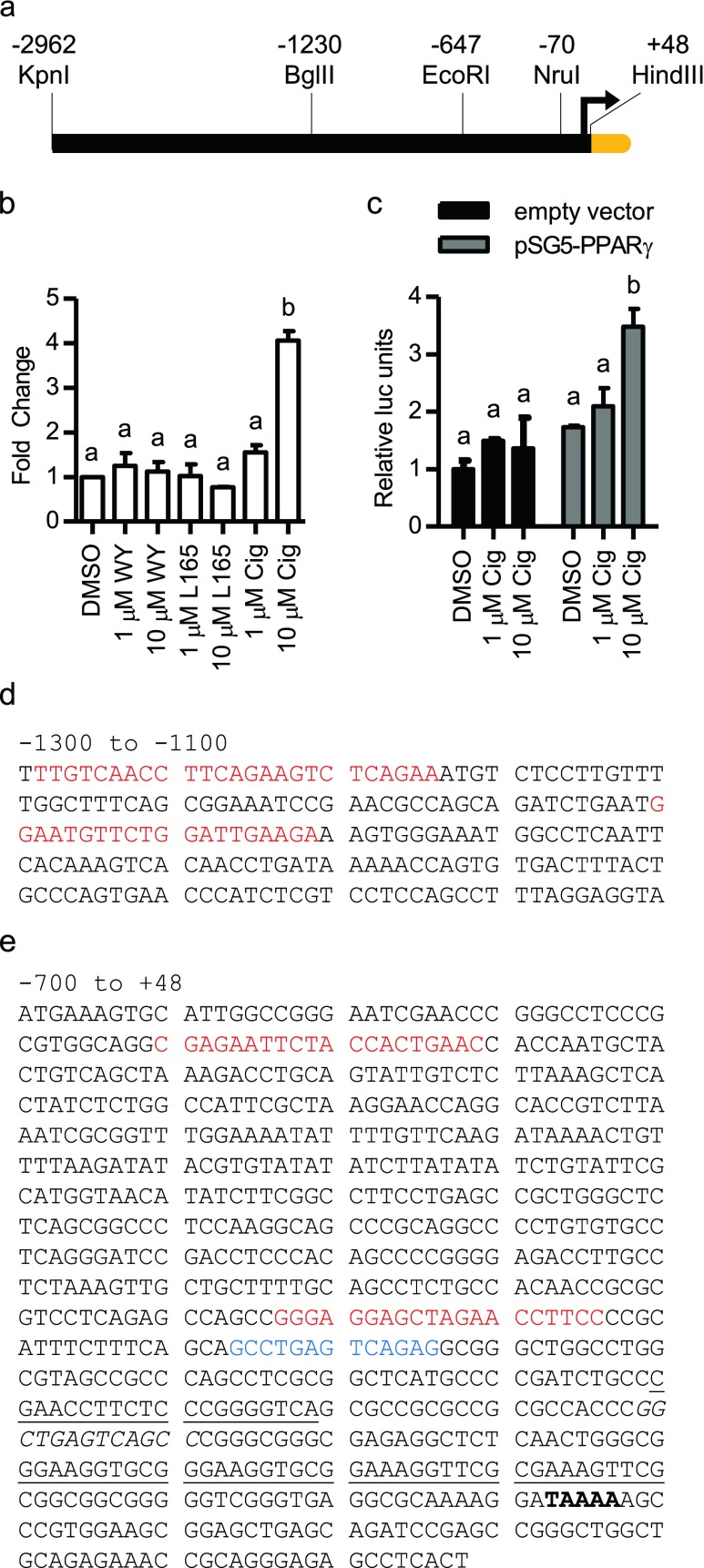
To guide physical isolation of the human HSPA6 promoter, we started with an in silico examination of sequences upstream of the referenced transcription start site (Leung et al. 1990) and 5′UTR of the NCBI mRNA reference sequence (Pruitt et al. 2005) NM_002155.3. NHR Scan (Sandelin and Wasserman 2005), Nubiscan (Podvinec et al. 2002), and MatInspector (Cartharius et al. 2005) analysis suggested high scoring hits for diverse transcription factors up through the first few thousand base pairs of DNA. Germane to our laboratory’s interest, putative PPAR and retinoic acid receptor (RAR) response elements were located between ~ −2,800 and −1,500 bp upstream of the transcriptional start site. To determine HSPA6 expression responsiveness to these and other transcription factors, we isolated and cloned its promoter region from −2,962 to +48 bp into a pGL4.10 luciferase reporter gene construct (Fig. 2a). Endogenous HSPA6 expression and HSPA6 − 2,962 kb-luc promoter activation (Fig. 2b, c) marginally increased only in the presence of a high concentration of PPARγ ligand ciglitazone or overexpressed PPARγ receptor and a high concentration of ligand, respectively. Since PPARγ has low expression in keratinocytes, we hypothesized that other transcription factor(s) may be responsible for the expression of HSPA6.
Fig. 2.
Analysis of the HSPA6 promoter. a Diagram of the HSPA6 promoter from −2,962 to +48 bp. Restriction enzyme sites shown and used for generating promoter truncation constructs. Yellow region indicates start of the luciferase reporter gene. b Endogenous HSPA6 mRNA expression following treatment with PPARα (WY14,643), β/δ (L165,041), or γ (ciglitazone) ligand. c Promoter activation of the −2962-luc construct due to treatment with ciglitazone and/or increasing amounts of PPARγ protein. Statistically significant values are indicated using a p value <0.05 as determined using ANOVA with Newman Keuls post hoc. Bars are mean + SD from one experiment, each bar from triplicate cultures. HSPA6 promoter sequence between −1,300 and −1,100 bp (d) and between −700 and +48 bp (e). Red sequences denote putative HSE. Blue sequences denote putative AP1 sites. Nucleotides in boldface denote predicted TATA box. Underlined sections denote previously identified HSEs. Italicized sections denote previously identified AP1 site
Searching the promoter for other predicted sites, further in silico promoter analysis was performed. In addition to searching for factors that contribute to HSPA6’s basal expression, we searched for HSEs that may contribute to the stress inducibility of HSPA6. Our analysis found previously recognized sites such as two HSEs at −181 to −161 bp and −100 to −60 bp, an AP1 site at −139 to −132 bp, and a predicted TATA box within the characterized minimal promoter (Wada et al. 2007; Leung et al. 1990). Importantly, previously unrecognized potential transcription factor binding sites (such as AP1, C/EBP, and upstream HSEs) (Fig. 2d, e) were identified suggesting additional control possibilities for basal and inducible HSPA6 promoter activity.
HSPA6 promoter contains negative and positive basal regulatory regions
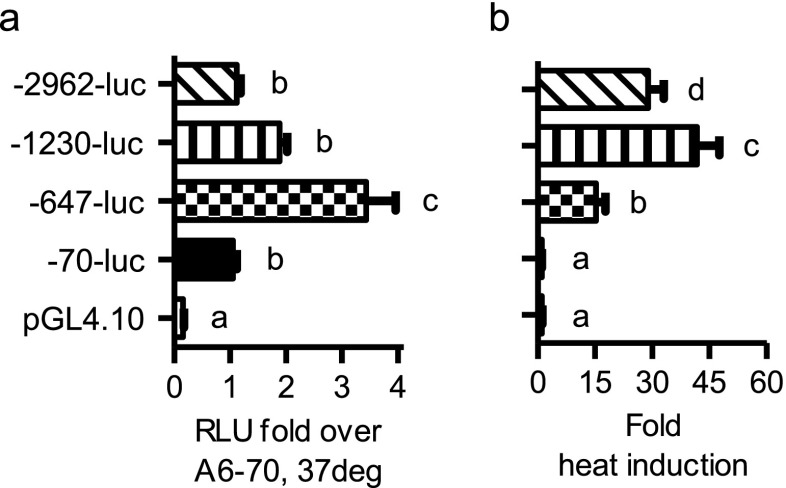
To narrow down the responsive regions involved in HSPA6 regulation, we generated a series of 5′ truncation constructs using naturally occurring restriction enzyme sites (Fig. 2a). Transfection results from the truncated HSPA6 promoter constructs from unstressed cells showed the −647-luc had increased luciferase activity compared to the “minimal” −70-luc. Interestingly, extending the 5′ promoter to −1,230 bp further led to a reduction of luciferase activity back to the minimal promoter levels (Fig. 3a). These results suggest that the regulation of unstressed HSPA6 includes both negative (between −1,230 and −648 bp) and positive (between −647 and −70 bp) elements. Heat-induced expression (Fig. 3b) was observed with promoter regions inclusive of sequence upstream of the previously described −181 and −100 bp HSEs. Notably, however, the fold induction of the −1,230 and −3 kb was significantly greater than the fold induction of the −647 bp reporter suggesting other proactive elements involved in maximal stress response.
Fig. 3.
Determining the transcriptionally regulated regions within the HSPA6 promoter. a Promoter activation at basal, 37 °C conditions. The graph is normalized to the −70-luc. b Promoter induction due to a 1 h, 42 °C heat stress. Graph shown as fold inducibility compared to 37 °C condition. Statistically significant values are indicated by different letters using a p value <0.05 as determined using ANOVA with Newman Keuls post hoc. Bars are mean + SEM from three experimental repeats, each bar from triplicate cultures
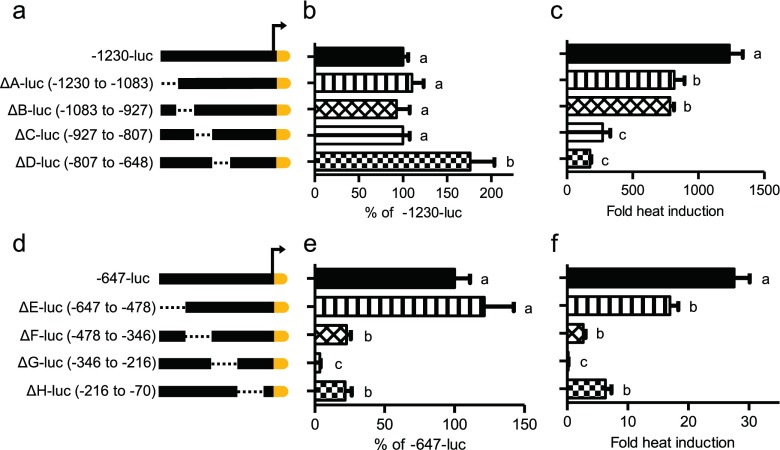
To further guide our search for HSPA6 promoter regions contributing to its overall basal activity and stress response, we generated several constructs deleting ~150 bp lengths within the −1,230-luc or −647-luc constructs. Deletions within the −1,230-luc, ΔA-, ΔB-, ΔC-, and ΔD-luc (Fig. 4a) showed that removal of fragment D (−806 to −648) increased the promoter activity 76 %, suggesting possible negative regulatory element(s) within this region (Fig. 4b). The stress responsiveness of ΔA-, ΔB-, ΔC-, and ΔD-luc were also compromised as a decrease in fold induction was observed with these constructs compared to full-length −1,230-luc (Fig. 4c). Internal promoter deletions within the −647-luc, ΔE-, ΔF-, ΔG-, and ΔH-luc (Fig. 4d) suggested that fragments F, G, and H each contribute to the activation of the HSPA6 promoter. Removal of fragments F or H reduced the promoter activity to ~35 % compared to the full-length −647-luc. Deletion of fragment G (−346 to −217 bp) (Fig. 4e, f) led to the complete loss of the HSPA6 promoter activity. While deletion of each individual fragment reduced the heat inducibility of the promoter, the induction of ΔG-luc was significantly reduced back to basal levels of −647-luc. Because removal of fragment G lost all promoter activation, we focused on determining the possible site(s) within this region that are crucial for the activation of the HSPA6 promoter.
Fig. 4.
Localization of the repressible and inducible regions within the HSPA6 promoter. Internal deletions between −1,230 and −648 (a) and between −647 and −70 (d) denoted as the dotted line. Promoter activation at basal, 37 °C conditions. Graphs are shown as percent of −1,230-luc (b) and −647-luc (e). Promoter induction due to a 1 h, 42 °C heat stress. Graphs are shown as fold inducibility compared to 37 °C condition (c, f). Statistically significant values are indicated by different letters using a p value <0.05 as determined using ANOVA with Newman Keuls post hoc. Bars are mean + SEM from three experimental repeats, each bar from triplicate cultures
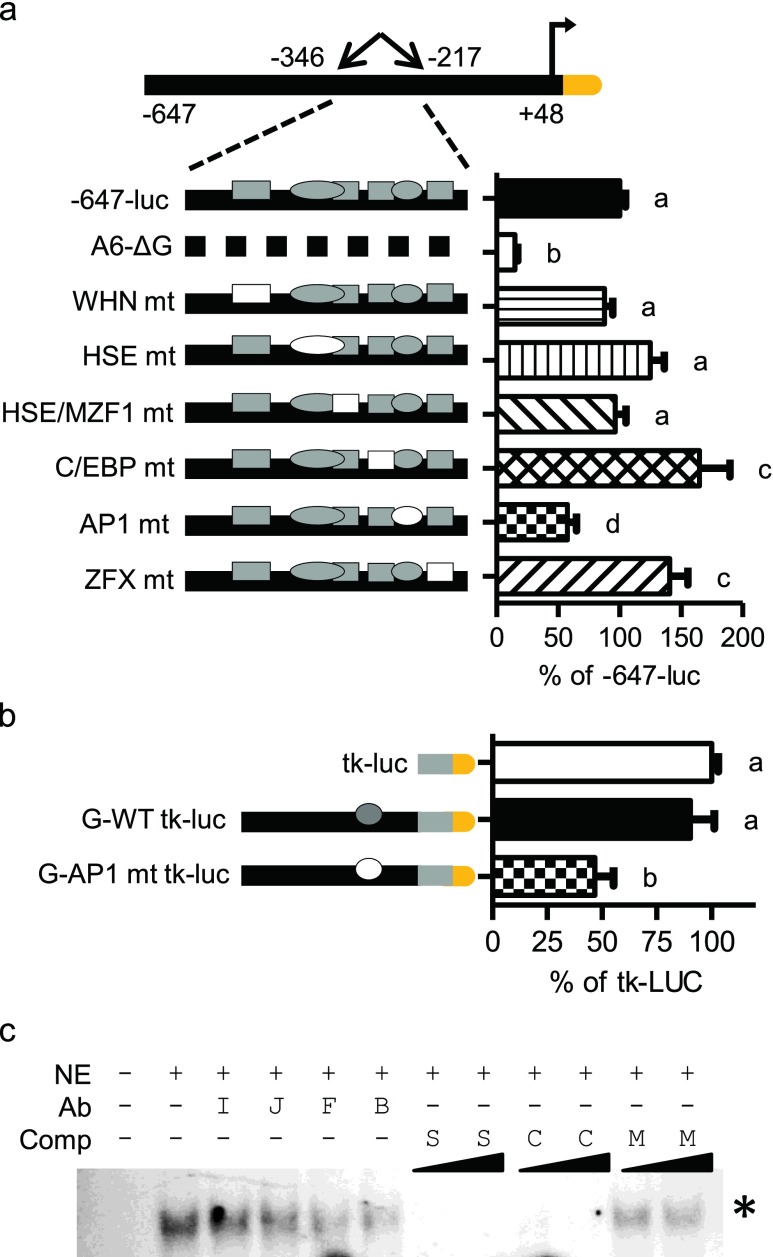
A novel AP1 site within −346 to −217 bp contributes to the activation of the HSPA6 promoter
A transcription factor binding site search within fragment G (−346 to −217 bp) determined the six top-scoring predicted elements: WHN, HSE, MZF1, C/EBP, AP1, and ZFX (Fig. 5a). We generated site-specific mutants within the −647-luc constructs to determine which binding site may contribute to the unstressed (37 °C) expression of HSPA6. Of these candidate sites, WHN, HSE, and MZF1 had no significant effect on the basal HSPA6 promoter activation, whereas C/EBP- and ZFX-specific mutants resulted in an ~50 % increase in the −647-luc promoter activity. Only mutation of AP1 resulted in activity loss. This local length of the HSPA6 promoter seems to share the trait of positive and negative control we saw upstream with −1,230 and −3 kb lengths. Thus, the −346 to −217 bp region is likely crucial for proper positive and negative promoter regulation within the HSPA6 native promoter.
Fig. 5.
Characterization of the −244 bp AP1 site. a Site-specific mutations within the −647-luc construct. Filled shapes indicate wild-type elements. Specific site mutants are shown as empty shapes. Graph shown as percent of −647-luc. b AP1-specific mutant within the fragment G-tk-luc construct. Graph shown as percent of tk-luc. Statistically significant values are indicated by different letters using a p value <0.05 as determined using ANOVA with Newman Keuls post hoc. Bars are mean + SEM from three experimental repeats, each bar from triplicate cultures. c EMSA of AP1. Antibodies include nonspecific IgG (I), anti-cJun (J), anti-cFos (F), or both anti-cJun and anti-cFos (B). Nonradiolabeled competitor oligomers include self HSPA6 AP1 site (S), consensus oligomer (C), or mutated AP1 (M) at 5- or 50-fold excess. Asterisk denotes disrupted binding species
To independently assess regulation conferred by region G, we fused the −346 to −217 bp region to the minimal tk promoter (Fig. 5b). This G-tk-luc construct did not significantly change the luciferase activity compared to the tk-luc construct. When the AP1 site was mutated, a ~54 % reduction was observed compared to wild-type fragment G-tk-luc (G-WT tk-luc). These results suggest that fragment G activation potential is promoter context dependent, contributing to the native HSPA6 promoter but not a heterologous (tk) promoter. These results are consistent with the predicted −244 bp AP1 site contributing to the promoter activation of HSPA6.
To test for physical association of AP1 to the predicted binding site, EMSAs were performed. Using the predicted HSPA6 − 244 bp AP1 sites as the 32P-labeled oligomers, our EMSA results show an AP1-specific band competed by an unlabeled HSPA6 AP1 site and consensus AP1 site, but not by the mutated AP1 site. Addition of AP1 subunit (cJun, cFos, or combined)-specific antibody disrupted the band species in the cFos and the combined cJun/cFos lanes (Fig. 5c). Based on reporter construct and the protein-DNA interaction experiments, HSPA6 − 244 to −237 is a newly identified, functional AP1 site contributing to transcriptional activity of the HSPA6 promoter in unstressed conditions.
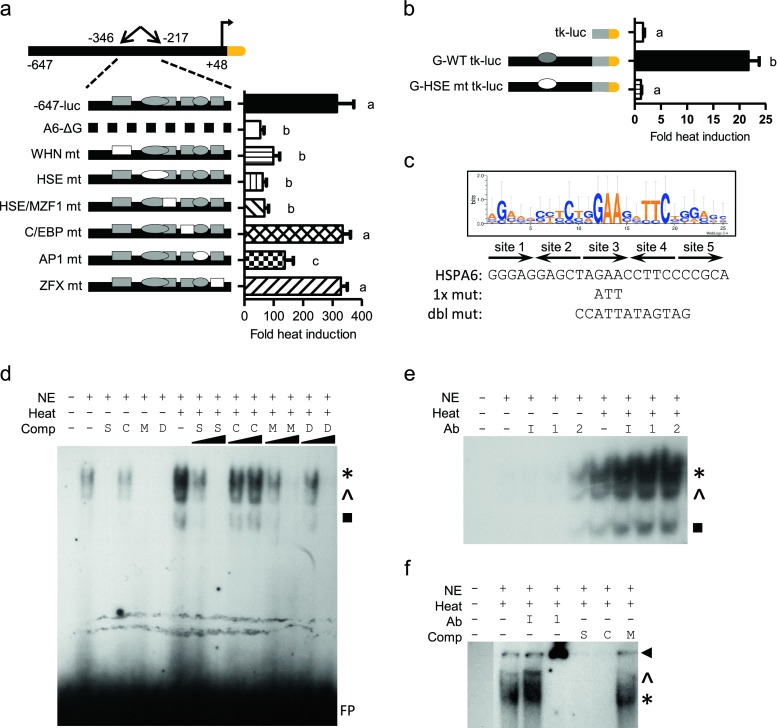
A novel heat shock element within −346 to −217 bp contributes to the activation and thermal induction of the HSPA6 promoter
In addition to promoter elements that might contribute to control of basal activity, our transcription factor search predicted a previously unreported heat shock element within region G. To test its contribution, or how other sites might affect overall stress response, we tested six site-specific mutants (Fig. 6a) under basal versus stressed conditions to determine fold induction. Mutations within the WHN, HSE, HSE/MZF1, and AP1 sites significantly reduced the stress inducibility of −647-luc, whereas C/EBP- and ZFX-specific mutations had no significant effect on stress inducibility (Fig. 6a). Taken together, these separate binding sites each contribute to the heat inducibility of the promoter and likely have an additive effect in order to gain maximal stress inducibility.
Fig. 6.
Characterization of the −284 bp HSE in HaCaT cells. Site-specific mutations within the −647-luc construct (a) or fragment G-tk-luc construct (b). Filled shapes indicate wild-type elements. Specific site mutants are shown as empty shapes. Graphs shown as fold heat induction. Statistically significant values are indicated by different letters using a p value <0.05 as determined using ANOVA with Newman Keuls post hoc. Bars are mean + SEM from three experimental repeats, each bar from triplicate cultures. c Sequence alignment of functional 5× inverted HSE repeats from several human HSP promoters shown as a WebLogo graphical representation. Letter height reflects occurrence of the nucleotide. Below graphic are the predicted HSPA6 − 284 bp HSE, the single and double mutant sequences. Arrows indicate single HSE repeat. EMSA of HSPA6 HSE incubated with unlabeled competitor oligomers (d) or with antibodies (e). EMSA of consensus HSE (f). Nonradiolabeled competitor oligomers include self HSPA6 HSE site (S), consensus HSE (C), mutated HSE (M), or double mutated HSE (D) at 10- or 100-fold excess. Antibodies include nonspecific IgG (I), anti-HSF1 (1), or anti-HSF2 (2). Asterisks denote major binding species, while caret and black box denote minor binding species. FP denotes free probe. Arrowheads denote supershift
To address the heat stress inducibility of the fragment G HSE, we fused the −346 to −217 bp region to the tk-luc construct. This specific construct is advantageous as it tests the fragment G candidate HSE independent of the previously described HSEs. The G-WT-tk-luc was heat induced ~20-fold compared to the tk-luc. In contrast, the G-HSEmt tk-luc abolished all stress induction (Fig. 6b). These results show that the −284 bp HSE contributes to the increased expression of HSPA6 due to stress induction.
Using the functionally defined HSPA6 − 284 bp HSE as wild-type EMSA probe, single site mutations within HSE site 3 were chosen as used by Koizumi et al. (2013) and a double site mutation was guided by the WebLogo sequence to avoid any unintentional HSE formation (Fig. 6c). A band pattern bound the −284 bp HSE in unstressed HaCaT keratinocyte nuclear extracts and increased in intensity with heat shock. The binding species could be competed with unlabeled wild-type, single mutant, and double mutant HSPA6 HSE site, but not with unlabeled consensus HSE oligomers (Fig. 6d). Additionally, HSF1- or HSF2-specific antibodies did not affect the band pattern (Fig. 6e). Using 32P-labeled consensus HSE oligomers as EMSA probe, our unlabeled HSPA6 HSE was able to compete the HSF1 specific band, suggesting that the −284 bp HSE can compete for HSF1 under these circumstances (Fig. 6f).
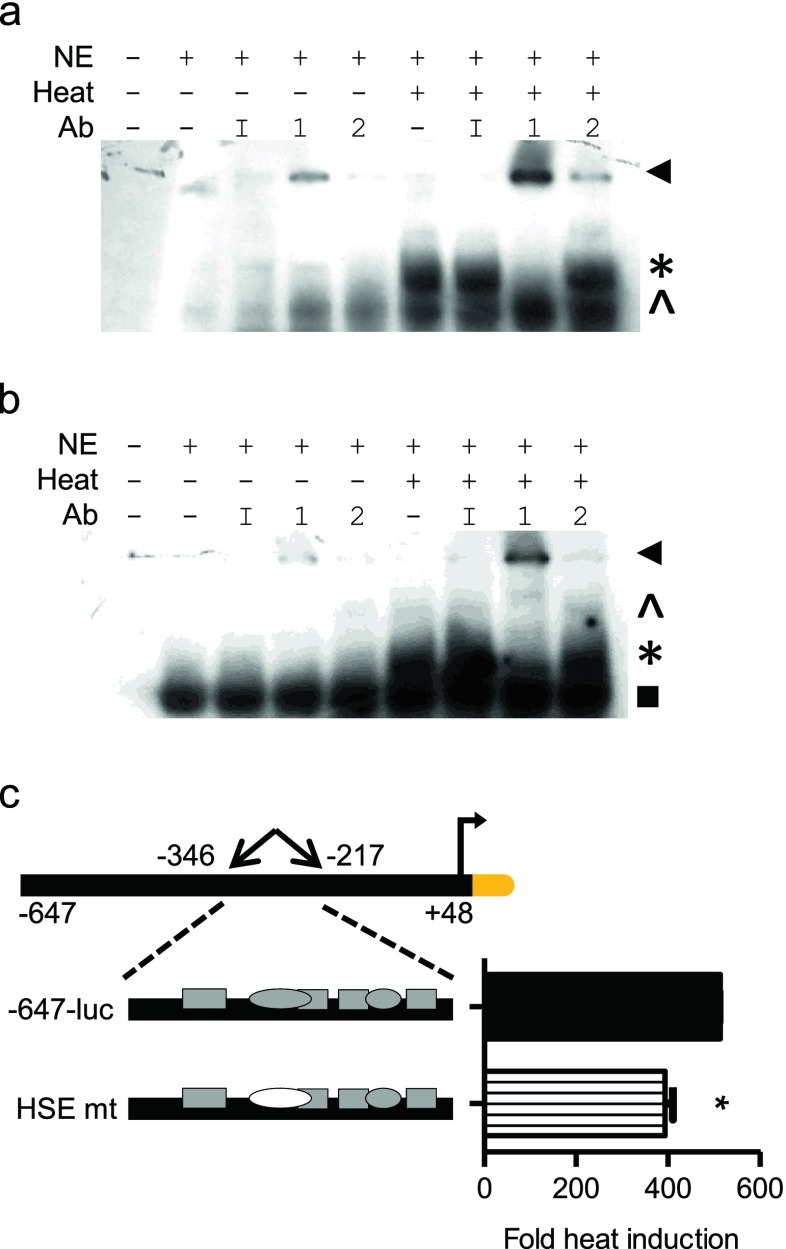
In an attempt to resolve these disparate results (HSE −284 transcriptionally active in mediating thermal stress response but not performing as a classic HSF-binding HSE in vitro), we compared the HSPA6 − 284 bp HSE binding with that of a consensus HSE (derived from the HSPA1A promoter) (Morgan et al. 1987) with an alternate nuclear protein source, HeLa cells. Using HeLa nuclear extracts, the HSPA6 − 284 bp HSE generated a binding complex recognized by HSF1 antibody from unstressed and stressed cells, while a HSF2 complex was recognized from stressed cells (Fig. 7a). As a control, we confirmed that the same extracts were generating a HSF1-containing complex on the A1A consensus HSE (Fig. 7b). These transcription activation and EMSA results suggest that a heat stress-associated transcription factor, other than HSF1 or HSF2, binds the −284 bp HSE in HaCaT cells to activate the HSPA6 promoter. Transfection of the −647-luc wild-type or with the HSE mutant into HeLa cells significantly reduced the stress induction of the −647 bp HSPA6 promoter (Fig. 7c). Together, the functionality of the −284 bp HSE in HeLa cells indicates that HSF1 and HSF2 can bind this site suggesting potential differential occupation across cell types.
Fig. 7.
Characterization of the −284 bp HSE in HeLa cells. EMSA of HSPA6 HSE (a) or consensus HSE (b) with HeLa nuclear extracts. Antibodies include nonspecific IgG (I), anti-HSF1 (1), or anti-HSF2 (2). Asterisks denote major binding species. Carets denote minor binding species. Arrowheads denote supershift. c Site-specific HSE mutation within the −647-luc construct. Graph shown as fold heat induction. The asterisk indicates a statistically significant difference from the parental −647-luc construct (p value <0.05) as determined using Student’s t test. Bars are mean + SD from one experiment, each bar from triplicate cultures
Discussion
Although discovered over 20 years ago (Leung et al. 1990), HSPA6’s role as a chaperone and stress-responding protein has only been recently studied (Chow et al. 2010; Khalouei et al. 2013; Leppa et al. 2001; Noonan et al. 2007a). Relative to other members of the HSP70 family, little is known about the transcriptional regulation of HSPA6 in stressed and unstressed cells (Rohmer et al. 2008; Wada et al. 2007). Undetectable basal and highly stress-inducible expression of HSPA6 was noted in varying cell types with a wide range of expression (Chow et al. 2010; Kolker et al. 2012; Noonan et al. 2007b). Consistent with a recent report (Gomez-Sucerquia et al. 2012), our studies demonstrate detectable but variable levels of constitutive HSPA6 protein in all human cell types examined. Basal expression of HSPA6 could be dependent on different cell growth conditions, such as cell density (Noonan et al. 2007b) or cell culture media (Wu and Morimoto 1985; Zachova et al. 2009), and may address the HSPA6 detection here but not in prior analysis of HaCaT keratinocytes (Ravagnan et al. 2013). Upon thermal stress, HaCaT keratinocytes responded with induction of both HSPA1A and A6, although the latter to many more fold at the mRNA level. Given its detection in nonstressed conditions (Gomez-Sucerquia et al. 2012 and our report), its capacity for significant fold induction (Chow et al. 2010; Qiao et al. 2012), and its likely contribution to post-stress cell survival (Noonan et al. 2007b), we sought to better define the control of its basal and stress-inducible expression in keratinocytes, a cell type with wide dependence on chaperone function and likely to encounter diverse stress conditions. Nevertheless, even with this better-defined control of its expression, any HSPA6 contribution to chaperoning or stress recovery would be restricted to species carrying the gene such as human, goat, and swine (Banerjee et al. 2014; Dezeure et al. 1993, and Parsian et al. 2000). We found novel elements contributing to its basal expression and importantly an upstream HSE likely contributing to its maximal induction during a stress response.
An in silico promoter analysis indicated several candidate regulatory elements throughout the first several thousand base pairs of the promoter guiding our cloning of a 3 kb region (relative to the transcriptional start site −2,962 to +48 bp). Prior to our work, Wada and colleagues examined the contribution of an AP1 site and two HSEs within the proximal region (−287 to +110 bp) of the HSPA6 promoter to develop a reporter construct sensitive to cadmium chloride exposure (Wada et al. 2005; Wada et al. 2007). Taking advantage of these longer promoter regions, we found that a region further upstream (−807 to −648 bp, here named fragment D) confers a negative regulation and contributes to heat stress inducibility on HSPA6. The presence of repressive regions is consistent not only in the transfection results but may also explain the reduced endogenous expression in several cell types, such as dermal fibroblasts and Caco2 cells. Despite this region’s overall limiting effect on promoter activity, we established a novel HSE in this region (−807 to −648 bp) that contributes to the HSPA6 maximal stress inducibility. Furthermore, we observed that a region (−346 to −216 bp, fragment G) in our extended promoter, directly upstream of previously identified AP1 (−139 bp) and HSEs (−181 and −100 bp) sites, is crucial for the basal and heat inducibility of HSPA6 likely by its provision of an additional AP1 site and another HSE. To test the activating potential of this region, fragment G was fused to a (tk) promoter where it could confer stress responsiveness to this heterologous promoter but not raise its basal activity. Interestingly, when the AP1 site was mutated in this fragment, basal activity was reduced suggesting that fragment G was a combination of both positive (like AP1) and negative elements (as yet to be identified) and that loss of AP1 favored the remaining negative control from this region. Nevertheless, given that under basal, unstressed conditions, this fragment G region can confer promoter activation in the context of the HSPA6 promoter, we sought to determine possible factors contributing to this region’s basal and stress inducibility.
Within fragment G (−346 to −216 bp), six top-scoring transcription factor binding sites were predicted: WHN, HSE, HSE/MZF1, C/EBP, AP1, and ZFX. In unstressed conditions, site-specific mutants within fragment G showed that an AP1 site at −240 bp contributes to the HSPA6 promoter activation. Surprisingly, our mutation analysis discovered two presumptive repressive elements, C/EBP and ZFX (Gokhman et al. 2013; Sachdeva et al. 2012), within this region. Similar to the full HSPA6 3 kb promoter, this shorter fragment also contains both positive and negative elements which contribute to the overall basal transcription of HSPA6. Consistent with this AP1 site contributing to the basal activation of HSPA6, we found that cFos, a typical subunit of the AP1 protein dimer, but not cJun binds the AP1 site within fragment G. We expect that since the AP1 heterodimer can be made from several subunits (cJun, junB, junC, Fra-1, Fra-2, cFos, or fosB), the dominant AP1 dimer bound to the −240 AP1 binding site could be cFos and another subunit other than cJun. Results from the promoter mutation and EMSA analyses confirm that a new AP1 site within fragment G contributes to the basal and inducible expression of HSPA6. Other non-HSF proteins have also been recently shown to contribute control over basal and stress-induced HSP expression. Ataxin-3, possibly through DNA binding but more likely through its interaction with transcription factors, augments the full capacity of the HSP70 (HSPA1A) promoter to thermal and chemical stress (Reina et al. 2012).
In addition to its basal regulation, we characterized the functional elements within −346 to −216 that contribute to its stress inducibility. The predicted HSE at −284 bp (gGGAg gAGCt aGAAc cTTCc cCGCa) contains one imperfect site (site 1) and two perfect sites (sites 3 and 4). The HSE-specific mutation of site 3 prevented the stress inducibility to a similar effect of the deletion of entire fragment G, indicating that these nucleotides contribute to the only HSE in this region. Sites other than the HSE (WHN, HSE/MZF1, and AP1) also appear to contribute to the maximal heat induction of HSPA6. The HSE/MZF1 site is labeled as such due to the overlap between key nucleotides within these two sites. The mutations within MZF1 also affect HSE site 4. We believe that the decrease in stress inducibility due to the MZF1 mutation is due to the loss of a core HSE repeat. Mutating the AP1 site, which plays a role in basal activation of HSPA6, also reduced the induction compared to the full-length, wild-type −647-luc. Similar to HSPA1A, multiple HSEs (Koizumi et al. 2013) and the elements conferring basal activation (Williams and Morimoto 1990) of HSPA6 may be necessary to obtain the maximal stress inducibility. The HSE within fragment G may be working in concert with other factors to achieve the maximal stress inducibility of HSPA6. Our results show that this HSE is necessary for the stress inducibility within the −346 to −216 bp region of the HSPA6 promoter but not sufficient for the maximal stress induction. The combined effect of the HSE we characterized and the previously determined sites (Wada et al. 2005; Wada et al. 2007) contribute to the maximal stress inducibility of HSPA6, which may reflect the binding of HSF to these three HSE sites. Further, the binding of either HSF1 or HSF2 varies on the type of stressor (Mathew et al. 2001). While our report used increased temperature as a stressor, Wada et al. conducted their experiments using treatment with cadmium. The contribution of the −284 bp site to binding HSF to the upstream HSEs could be dependent on both the type of stressor and the availability of HSF proteins.
The new HSPA6 HSE we demonstrated at −284 bp provided thermal responsiveness both in the context of the HSPA6 promoter and when transferred to the heterologous tk promoter. While this sequence was able to bind HaCaT keratinocyte stress-associated nuclear protein(s) in vitro, it was surprising that these binding factors were not recognized by either HSF1 or HSF2 antibodies. Intriguingly, when this site was used as a competitor for the 32P-labeled consensus HSE, again with HaCaT keratinocyte nuclear extracts, it competed HSF1 from the consensus probe. To test the binding of our site to HSF, we performed the EMSA analysis using nuclear extracts from a different cell type, HeLa cells. Data from the HeLa EMSAs suggest that our site binds HSF1 under unstressed and stressed conditions and HSF2 primarily in stressed nuclear extracts. It is possible that a different HSF family member may bind this HSE, HSF4. This transcription factor can compete for HSF1 binding (Fujimoto et al. 2004); however, expression of HSF4 has not been tested in keratinocytes. Thus, our site may bind one of these HSFs, or possibly a different stress-inducible factor. To date, HSF4’s function and expression have mainly been characterized in eye development, playing a role in lens development (Fujimoto and Nakai 2010). Additionally, we tested the contribution of this HSE in HeLa cells. Mutating the −284 bp HSE reduced the stress inducibility of the −647-luc construct, suggesting that this HSE contributes to the maximal stress inducibility of HSPA6 in various cell types. Altogether, the results from the promoter report and DNA-binding assays show our site as a functional HSE which can bind HSF1 and HSF2, but preferentially binds another, yet characterized stress-inducible factor when presented with HaCaT keratinocyte nuclear proteins.
In the present study, we performed an analysis of HSPA6 promoter to search for core basal and inducible transcriptional elements, finding both positive and negative regulatory regions. Two factors, AP1 and HSF, contribute to the expression of HSPA6. AP1 regulates HSPA6 transcription under both unstressed and stressed conditions, whereas a HSF-like factor HSF contributes to the heat inducibility of HSPA6. In addition to characterizing these regions, our HSE EMSA results suggest that an HSF-like factor from HaCaT keratinocytes may preferentially bind the HSPA6 promoter warranting further investigation of this possibly novel factor. The role of HSPA6 in cutaneous biology and wound healing could be further investigated in species where HSPA6 is expressed, such as human cell or porcine systems, the latter of which is a well-characterized animal model for epithelial skin research (Sullivan et al. 2001).
Acknowledgments
We thank Dr. C. Giardina for helpful discussions and N. Fusenig for HaCaT keratinocytes. This work was supported by a National Institute of Arthritis and Musculoskeletal and Skin Diseases grant (AR048660 to BJA) and University of Connecticut Research Foundation grant (BJA). Partial support was provided by summer fellowships from the University of Connecticut Center for Regenerative Biology Excellence in Graduate Research Award (VPR), the Edward A. Khairallah Summer Graduate Fellowship (VPR), and the Cross-Disciplinary Fellowship in Molecular & Cellular Biology and Pharmaceutical Sciences (VPR). MS is a recipient of a University of Connecticut Life Sciences Honors Thesis Research Award and K.A. Nieforth Pharmacy Student Research Fellowship; AS is a recipient of a K.A. Nieforth Pharmacy Student Research Fellowship.
References
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee D, Upadhyay RC, Chaudhary UB, Kumar R, Singh S, Ashutosh GJM, Polley S, Mukherjee A, Das TK, De S. Seasonal variation in expression pattern of genes under HSP70: seasonal variation in expression pattern of genes under HSP70 family in heat- and cold-adapted goats (Capra hircus) Cell Stress Chaperones. 2014;19:401–408. doi: 10.1007/s12192-013-0469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chow AM, Mok P, Xiao D, Khalouei S, Brown IR. Heteromeric complexes of heat shock protein 70 (HSP70) family members, including Hsp70B′, in differentiated human neuronal cells. Cell Stress Chaperones. 2010;15:545–553. doi: 10.1007/s12192-009-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Dezeure F, Vaiman M, Chardon P. Characterization of a polymorphic heat shock protein 70 gene in swine outside the SLA major histocompatibility complex. Biochim Biophys Acta. 1993;1174:17–26. doi: 10.1016/0167-4781(93)90087-T. [DOI] [PubMed] [Google Scholar]
- Dierick I, Irobi J, Janssens S, Theuns J, Lemmens R, Jacobs A, Corsmit E, Hersmus N, Van Den Bosch L, Robberecht W, De Jonghe P, Van Broeckhoven C, Timmerman V. Genetic variant in the HSPB1 promoter region impairs the HSP27 stress response. Hum Mutat. 2007;28:830. doi: 10.1002/humu.9503. [DOI] [PubMed] [Google Scholar]
- Encarnacao PC, Ramirez VP, Zhang C, Aneskievich BJ. Sp sites contribute to basal and inducible expression of the human TNIP1 (TNFalpha-inducible protein 3-interacting protein 1) promoter. Biochem J. 2013;452:519–529. doi: 10.1042/BJ20121666. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Nakai A. The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 2010;277:4112–4125. doi: 10.1111/j.1742-4658.2010.07827.x. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Izu H, Seki K, Fukuda K, Nishida T, Yamada S, Kato K, Yonemura S, Inouye S, Nakai A. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 2004;23:4297–4306. doi: 10.1038/sj.emboj.7600435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhman D, Livyatan I, Sailaja BS, Melcer S, Meshorer E. Multilayered chromatin analysis reveals E2f, Smad and Zfx as transcriptional regulators of histones. Nat Struct Mol Biol. 2013;20:119–126. doi: 10.1038/nsmb.2448. [DOI] [PubMed] [Google Scholar]
- Gomez-Sucerquia LJ, Blas-Garcia A, Marti-Cabrera M, Esplugues JV, Apostolova N. Profile of stress and toxicity gene expression in human hepatic cells treated with Efavirenz. Antiviral Res. 2012;94:232–241. doi: 10.1016/j.antiviral.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Hageman J, van Waarde MA, Zylicz A, Walerych D, Kampinga HH. The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem J. 2011;435:127–142. doi: 10.1042/BJ20101247. [DOI] [PubMed] [Google Scholar]
- Heldens L, van Genesen ST, Hanssen LL, Hageman J, Kampinga HH, Lubsen NH. Protein refolding in peroxisomes is dependent upon an HSF1-regulated function. Cell Stress Chaperones. 2012;17:603–613. doi: 10.1007/s12192-012-0335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Klosner G, Trautinger F. Significance of heat shock proteins in the skin upon UV exposure. Front Biosci. 2009;14:4758–4768. doi: 10.2741/3565. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalouei S, Chow AM, Brown IR. Stress-induced localization of HSPA6 (HSP70B′) and HSPA1A (HSP70-1) proteins to centrioles in human neuronal cells. Cell Stress Chaperones. 2013;19:321–327. doi: 10.1007/s12192-013-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S, Suzuki K, Yamaguchi S. Heavy metal response of the heat shock protein 70 gene is mediated by duplicated heat shock elements and heat shock factor 1. Gene. 2013;522:184–191. doi: 10.1016/j.gene.2013.03.090. [DOI] [PubMed] [Google Scholar]
- Kolker E, Higdon R, Haynes W, Welch D, Broomall W, Lancet D, Stanberry L, Kolker N. MOPED: Model Organism Protein Expression Database. Nucleic Acids Res. 2012;40:D1093–D1099. doi: 10.1093/nar/gkr1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SB, Young C, Kim DS, Choi HO, Kim KH, Chung JH, Eun HC, Park KC, Oh CK, Seo JS. Impaired repair ability of hsp70.1 KO mouse after UVB irradiation. J Dermatol Sci. 2002;28:144–151. doi: 10.1016/S0923-1811(01)00156-6. [DOI] [PubMed] [Google Scholar]
- Leppa S, Kajanne R, Arminen L, Sistonen L. Differential induction of Hsp70-encoding genes in human hematopoietic cells. J Biol Chem. 2001;276:31713–31719. doi: 10.1074/jbc.M104375200. [DOI] [PubMed] [Google Scholar]
- Leung TK, Rajendran MY, Monfries C, Hall C, Lim L. The human heat-shock protein family. Expression of a novel heat-inducible HSP70 (HSP70B′) and isolation of its cDNA and genomic DNA. Biochem J. 1990;267:125–132. doi: 10.1042/bj2670125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Mathur SK, Jolly C, Fox SG, Kim S, Morimoto RI. Stress-specific activation and repression of heat shock factors 1 and 2. Mol Cell Biol. 2001;21:7163–7171. doi: 10.1128/MCB.21.21.7163-7171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Hoshino T, Yamashita Y, Tanaka K, Maji D, Sato K, Adachi H, Sobue G, Ihn H, Funasaka Y, Mizushima T. Prevention of UVB radiation-induced epidermal damage by expression of heat shock protein 70. J Biol Chem. 2010;285:5848–5858. doi: 10.1074/jbc.M109.063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz K, Ezernieks J, Sebald W, Duschl A. Interleukin-4 upregulates the heat shock protein Hsp90alpha and enhances transcription of a reporter gene coupled to a single heat shock element. FEBS Lett. 1996;385:25–28. doi: 10.1016/0014-5793(96)00341-9. [DOI] [PubMed] [Google Scholar]
- Morgan WD, Williams GT, Morimoto RI, Greene J, Kingston RE, Tjian R. Two transcriptional activators, CCAAT-box-binding transcription factor and heat shock transcription factor, interact with a human hsp70 gene promoter. Mol Cell Biol. 1987;7:1129–1138. doi: 10.1128/mcb.7.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan EJ, Place RF, Giardina C, Hightower LE. Hsp70B′ regulation and function. Cell Stress Chaperones. 2007;12:393–402. doi: 10.1379/CSC-278e.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan EJ, Place RF, Rasoulpour RJ, Giardina C, Hightower LE. Cell number-dependent regulation of Hsp70B' expression: evidence of an extracellular regulator. J Cell Physiol. 2007;210:201–211. doi: 10.1002/jcp.20875. [DOI] [PubMed] [Google Scholar]
- Noonan E, Giardina C, Hightower L. Hsp70B′ and Hsp72 form a complex in stressed human colon cells and each contributes to cytoprotection. Exp Cell Res. 2008;314:2468–2476. doi: 10.1016/j.yexcr.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Parsian AJ, Sheren JE, Tao TY, Goswami PC, Malyapa R, Van Rheeden R, Watson MS, Hunt CR. The human Hsp70B gene at the HSPA7 locus of chromosome 1 is transcribed but non-functional. Biochim Biophys Acta. 2000;1494:201–205. doi: 10.1016/S0167-4781(00)00203-7. [DOI] [PubMed] [Google Scholar]
- Podvinec M, Kaufmann MR, Handschin C, Meyer UA. NUBIScan, an in silico approach for prediction of nuclear receptor response elements. Mol Endocrinol. 2002;16:1269–1279. doi: 10.1210/mend.16.6.0851. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S, Lamore SD, Cabello CM, Lesson JL, Munoz-Rodriguez JL, Wondrak GT. Thiostrepton is an inducer of oxidative and proteotoxic stress that impairs viability of human melanoma cells but not primary melanocytes. Biochem Pharmacol. 2012;83:1229–1240. doi: 10.1016/j.bcp.2012.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez VP, Aneskievich BJ (2013) Transgene delivery to cultured keratinocytes via replication-deficient adenovirus vectors. Methods Mol Biol [DOI] [PubMed]
- Ravagnan G, De Filippis A, Carteni M, De Maria S, Cozza V, Petrazzuolo M, Tufano MA, Donnarumma G. Polydatin, a natural precursor of resveratrol, induces beta-defensin production and reduces inflammatory response. Inflammation. 2013;36:26–34. doi: 10.1007/s10753-012-9516-8. [DOI] [PubMed] [Google Scholar]
- Reina CP, Nabet BY, Young PD, Pittman RN. Basal and stress-induced Hsp70 are modulated by ataxin-3. Cell Stress Chaperones. 2012;17:729–742. doi: 10.1007/s12192-012-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer S, Mainka A, Knippertz I, Hesse A, Nettelbeck DM. Insulated hsp70B′ promoter: stringent heat-inducible activity in replication-deficient, but not replication-competent adenoviruses. J Gene Med. 2008;10:340–354. doi: 10.1002/jgm.1157. [DOI] [PubMed] [Google Scholar]
- Sachdeva M, Liu Q, Cao J, Lu Z, Mo YY. Negative regulation of miR-145 by C/EBP-beta through the Akt pathway in cancer cells. Nucleic Acids Res. 2012;40:6683–6692. doi: 10.1093/nar/gks324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW. Prediction of nuclear hormone receptor response elements. Mol Endocrinol. 2005;19:595–606. doi: 10.1210/me.2004-0101. [DOI] [PubMed] [Google Scholar]
- Shen Y, Liu J, Wang X, Cheng X, Wang Y, Wu N. Essential role of the first intron in the transcription of hsp90beta gene. FEBS Lett. 1997;413:92–98. doi: 10.1016/S0014-5793(97)00883-1. [DOI] [PubMed] [Google Scholar]
- Sullivan TP, Eaglstein WH, Davis SC, Mertz P. The pig as a model for human wound healing. Wound Repair Regen. 2001;9:66–76. doi: 10.1046/j.1524-475x.2001.00066.x. [DOI] [PubMed] [Google Scholar]
- Wada K, Taniguchi A, Xu L, Okano T. Rapid and highly sensitive detection of cadmium chloride induced cytotoxicity using the HSP70B′ promoter in live cells. Biotechnol Bioeng. 2005;92:410–415. doi: 10.1002/bit.20601. [DOI] [PubMed] [Google Scholar]
- Wada K, Taniguchi A, Okano T. Highly sensitive detection of cytotoxicity using a modified HSP70B′ promoter. Biotechnol Bioeng. 2007;97:871–876. doi: 10.1002/bit.21293. [DOI] [PubMed] [Google Scholar]
- Williams GT, Morimoto RI. Maximal stress-induced transcription from the human HSP70 promoter requires interactions with the basal promoter elements independent of rotational alignment. Mol Cell Biol. 1990;10:3125–3136. doi: 10.1128/mcb.10.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BJ, Morimoto RI. Transcription of the human hsp70 gene is induced by serum stimulation. Proc Natl Acad Sci U S A. 1985;82:6070–6074. doi: 10.1073/pnas.82.18.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachova K, Krupka M, Chamrad I, Belakova J, Horynova M, Weigl E, Sebela M, Raska M. Novel modification of growth medium enables efficient E. coli expression and simple purification of an endotoxin-free recombinant murine hsp70 protein. J Microbiol Biotechnol. 2009;19:727–733. [PubMed] [Google Scholar]