A fundamental goal of nanotechnology is to integrate top-down solid state technology with bottom-up assembled nanomaterials to create functional devices that overlap in size at the nanometer scale and begin to rival the complexities of, or even directly interface with, living systems.[1-2] A variety of functional nanomaterials have been developed,[3-4] and self assembly processes are available for integrating nanomaterials of a single composition into functional devices and heirarchically structured shapes.[5-6] However, heterogeneous integration methods that are capable of reliably integrating different nanostructured materials into functional devices are needed, especially for biotechnology applications such as drug screening and biosensing.[7-9]
Here we propose the use of multimaterial intaglio printing as a solution to the problem of materials integration. Intaglio printing is a technique that was developed hundreds of years ago by the printmaking industry, but then largely forgotten except by artisans. Intaglio involves transfer of ink from the recesses of a stamp rather than from the protrusions, the latter being known as relief printing.[10] The vast majority of microcontact printing is currently relief printing, which has, for example been used to generate multilayered patterns of biological membranes using a hydrogel stamp.[11]
Intaglio printing differs from relief printing in that it makes use of the topographical dimensions of the stamp in order to control the topography of the printed features. We recently demonstrated that lipid multilayer gratings can be fabricated by printing from an elastomeric diffraction grating stamp allowing submicron lateral dimensions and control of vertical dimensions between 1-100 nm.[12] When carried out with microscopic stamp dimensions, the intaglio process has the potential to combine the material integration capabilities of pin spotting,[13] with the topographical control of nanoimprint lithography,[14] and the scalability of microcontact printing.[15] Here we show that printing from a microstructured intaglio stamp is suitable for heterogeneous integration of lipid multilayer micro- and nanostructures in a scalable manner compatible with established microarray technology, which we here refer to as nanointaglio.
Microarrays integrating multiple biomaterials onto surfaces have been successfully developed in biotechnology for screening purposes. DNA microarrays, for example, have been thoroughly developed to allow massively parallel nucleic acid hybridization experiments to be carried out on a single chip.[16-17] Similarly, protein microarrays,[18] polysaccharide arrays,[19] lipid arrays,[20] synthetic polymer arrays[21-22] and small molecule microarrays[23] have been proposed for a variety of biomolecular screening applications. Robotic pin spotting has been the primary method used to integrate different materials onto the same surface for many of these applications.[13] Several innovative solutions have been proposed to allow printing of multiple materials with micro to nanoscale feature sizes, including dip-pen nanolithography, polymer pen lithography, and soft lithography.[24]
Lipids are a promising material for nanotechnology due to their innate biocompatibity[25] and tendency to self-organize in aqueous solutions.[26] We have previously shown that dip-pen nanolithography can be used to fabricate lipid multilayer structures on surfaces,[27] which permit unique applications based on their multilayer thickness. For example, by encapsulating drugs in surface supported lipid multilayer nanostructures we were able to demonstrate their suitability for delivery of lipophilic drugs to cells in a microarray format at dosages comparable to solution delivery, offering the potential to miniaturize high-throughput screening.[28] Another promising application is in label free sensor arrays, where lipid multilayer gratings respond to analytes by a shape change and corresponding change in optical properties.[29]
The utility of lipid structures for both high-throughput screening and nanosensor array technologies has so far been limited by the number of different structured lipid materials that can be uniformly integrated onto a single surface.[28-29] Here we overcome this limit by using scalable pin spotting to integrate multiple different inks onto a single intaglio stamp, as schematically illustrated in Figure 1. In this process, robotic pin spotting is used to array lipids, dissolved in ethanol or as liposomal emulsions in water, onto an ink palette, which is used to ink a micro-structured intaglio stamp that is used in turn for printing ink from the recesses of the stamp. In this way, different liposomal concentrations were screened for optimum ink deposition and subsequent patterning (Figure S1). Figure 2a-d shows some initial prints from a stamp inked with the cationic lipid 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP) doped with 1 mol% 1,2-dioleoyl-sn-glycero-3- phosphoethanolamine-N-lissamine rhodamine B sulfonyl (Rhodamine-PE) to allow fluorescence characterization.
Figure 1.
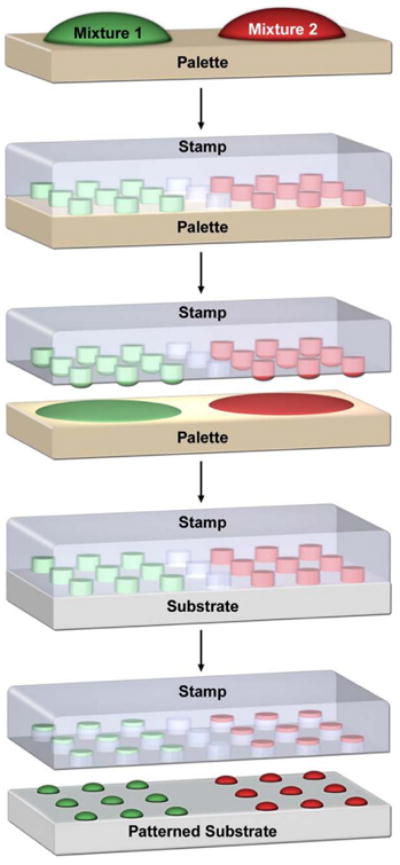
Schematic illustration showing the nanointaglio process. First, different lipid mixtures are arrayed onto an ink palette, next the stamp is brought into contact with the ink palette to effectively ink the intaglio stamp. Finally, the stamp is brought into contact with a surface, depositing three dimensional lipid multilayer structures with controlled topography.
Figure 2.
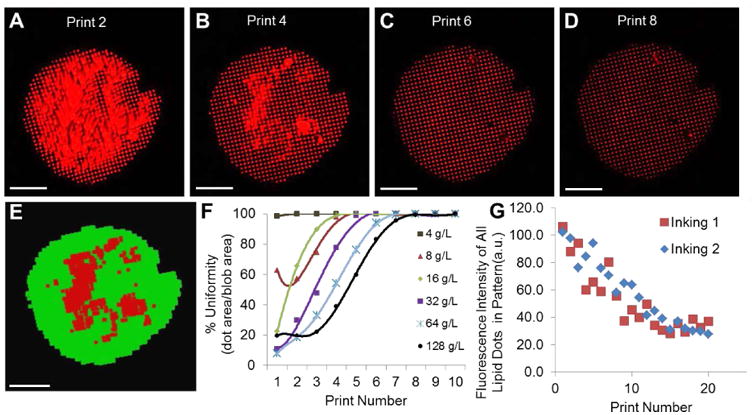
Uniformity characterization of lipid multilayer structures fabricated by nanointaglio. a-d) Fluorescence image of DOTAP dot patterns from microarrraying and multilayer stamping, representing print numbers 2, 4, 6 and 8, respectively from a liposomal concentration of 32 g/L. Bars = 100 μm. e) Automated quantification of spot uniformity based on recognition of uniform (green) and non-uniform regions (red). Bar = 100 μm. f) Spot uniformity as a function of print number for different ink concentrations; uniform prints obtained after 4-8 prints. e) Dot stamps were inked twice (shown in the graph as inking 1 and 2, respectively) from the same ink-palette. As the ink was depleted from the stamp, the intensities of the individual dots within the lipid pattern decreased linearly in intensity (height) with increasing print number for a constant force (45 Newtons) applied to the stamp. When this constant force was applied, about 20 prints were created before depletion. The similarity in the intensities between the two inkings from the same ink palette indicate the palette is capable of acting as an adequate reservoir for two successive print sequences.
In traditional intaglio, excess ink is wiped from the stamp, leaving only the ink in the recesses for transfer. For the nanointaglio process, we removed excess ink by sacrificial printing to minimize cross contamination for multi-material integration, as shown in Figure 2a-d. To determine how many prints were required to remove excess ink, several prints were made of fluorescently labeled lipid inks, and the uniformity of the patterns was characterized by fluorescence microscopy (Figure 2a-d). For this purpose, we define the “uniformity” of an intaglio print as the percentage of area covered by the desired pattern (e.g. 5 μm dots, stamp charactization is shown in Figure S2), relative to the total patterned area. Software was written to quantify the uniformity from fluorescence microscopy data based on this criterion (Figure 2e) as a function of the lipid ink concentration used for robotic pin spotting onto the palette (Figure 2f). We found that the number of sacrificial prints needed to obtain uniform prints depends on the concentration of the lipids in the ink used for robotic pin spotting onto the palette (Figure 2f). Liposomal ink concentrations below 2 g/L failed to provide adequate transfer from the ink palette to the structured stamp (Figure S1). For ink concentrations greater than 4 g/L, the number of prints required to reach >95% uniformity ranged from 4-7 depending on ink concentration. Importantly, the variation in dot height between different concentrations at the same print number was found to be below 10%, except for those structures made from 4 g/L, which suffered from ink depletion (Figure S3). This indicates that uniform nanointaglio patterns can be generated from a wide range of ink concentrations. We have previously shown that the fluorescence intensities of fluorescent lipid multilayer structures can be used as an indicator of multilayer thickness[30], and have therefore used their intensities as indicators of how lipid multilayer heights change with print number, as ink depletes from the stamp. One inking from the PDMS palette allows a series of prints to be created. We found that for dot arrays, the multilayer height decreases with print number for up to about 20 prints (Figure 2g). We have previously found that lipid diffraction gratings can be printed at least 6 times without a significant decrease in the heights of the resulting structures,[12] suggesting that the number of prints that can be made from a single stamp depends on the stamp geometry, i.e. pattern and depth.
The aspect ratio (i.e. the ratio of multilayer height to lateral width at half-height) of the lipid multilayer structures is a critical dimension for both cell culture and biosensor applications.[28-29] Higher aspect ratios (defined as height/width)are desirable, and for lipids the maximum aspect ratios achieved are about 0.1 (height/width).[12,29] We found that, in the nanointaglio process, the lipid multilayer height and corresponding aspect ratio can be controlled by stamp geometry and by print number. Figure 3a shows an atomic force microscopy (AFM) image of a dot array with heights 423.4 nm ± 44.7 nm and widths at half-height 4.18 μm ± 0.29 μm. This produces an aspect ratio of 0.102 ± 0.0170 (height/width). Figure 3b shows an AFM image of DOPC gratings with heights of 30.1 nm ± 4.80 nm with line width at half-height 285 nm ±18.0 nm. The grating apect ratio (height/w) is 0.105 ± 0.0131. The lipid pattern aspect ratio is ultimately determined by the dimensions of the stamp, with height limitations based upon the intaglio grooves. The results suggest that the minimum line width is 10 times wider than the multilayer height.
Figure 3.
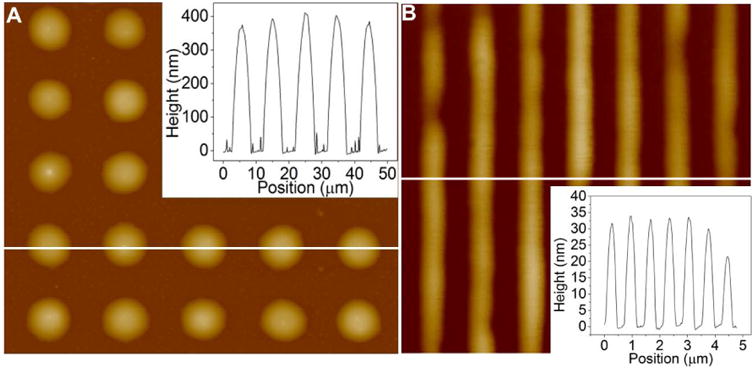
Aspect ratio of 0.1 as a critical dimension in nanointaglio. a) AFM topographical profile of micron scale lipid dots imaged on glass. The inset shows a section analysis of dots with width at half height of 4.18 μm ± 0.29 μm, heights of 423.4 nm ± 44.7 nm and aspect ratios of 0.102 ± 0.0170 (height/width). b) AFM topographical profile of of DOPC diffraction gratings imaged on spin coated PMMA on glass. Grating height analysis of the above cross section shows grating line features with heights of 30.1 nm ± 4.80 nm, widths at half height of 285 nm ±18.0 nm and aspect ratios of 0.105 ± 0.0131 (line height/line width). The results suggest that line widths are limited to 10 times the multilayer height.
Once the parameters for uniform patterning were established, we were able to integrate multiple materials onto the same surface, as shown in Figure 4a-d. Three different nanointaglio inks were prepared by doping DOTAP with the fluorophores Marina Blue – DHPE (2 mol%), carboxyfluorescein-PE (2 mol%), and rhodamine-PE (1 mol%), microarrayed onto an ink palette (Figure S4) and stamped onto a glass substrate. Figure 4b-d shows fluorescence micrographs of the different lipid dot patterns, demonstrating pattern uniformity. The coefficients of variation between the average intensities of each dot in Figure 4b-d was well below 10% (Figure S5).
Figure 4.
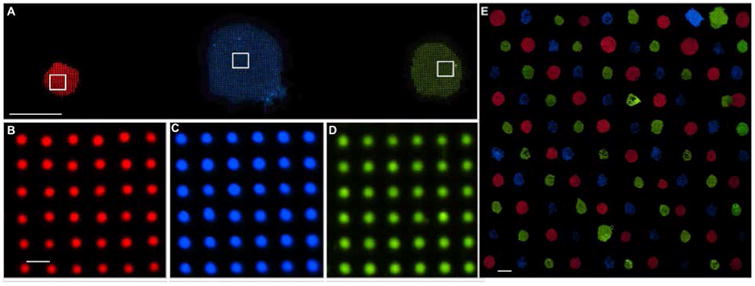
Materials integration. a) Fluorescence image of lipid dot patterns from different inks (1 Mol % Rhodamine-DOPE, 2 Mol % Marina Blue-DHPE, and Carboxyfluorescein-DOPE, respectively) printed onto a glass slide, demonstrating material integration. Bar = 200 μm. b-d) 40× fluorescent images of Rhodamine-PE, Marina Blue-DHPE, and Carboxyfluorescein-PE doped patterns shown in panel (a); Bar = 10 μm. e) 10×10 dot pattern array of DOTAP doped with 1 Mol % Rhodamine-DOPE, Marina Blue-DHPE, and Carboxyfluorescein-DOPE for red, blue and green fluorescence, respectively. This demonstrates the scalability of nanointaglio for printing different lipids onto the same surface without significant cross contamination. Shown is 100 lipid patterns printed over and area of ∼15 mm2. Scale bar = 200 μm.
A promising application of lipid multilayer micro and nanostructures is as biosensors. We have previously shown that fluid lipid multilayer diffraction gratings fabricated by dip-pen nanolithography can respond to analytes by changing shape and their corresponding optical properties.[29] However, the quantitative response of these gratings is highly dependant on their nanometer scale height, and so far we have lacked a method for integrating a sufficient number of lipid multilayer nanostructures of different composition with high enough uniformity to carry out multiplexed sensing using arrays of different lipids. Nanointaglio provides a scalable solution to this materials integration problem, in which lipid patterns can be placed at a minimum of 400 microns apart, enabling four lipid patterns to be printing in an area of roughly 0.2 mm2 (Figure 4e). Theoretically, this enables a 16 pin tool to print 1600 lipid patterns 400 micron apart over an area of 2.31 cm2.
The nanointaglio process described here provides a method of integrating different functionally microstructured materials onto a single surface. By using robotic pin spotting to integrate multiple inks onto a palette, we are able to produce a scalable manufacturing process. Uniform structures can be produced, and importantly the lipid multilayer thickness can be controlled by the print number (Figure 2g) and stamp geometry (Figure 3a-b). As the nanointaglio fabrication process is scalable, we expect it to be widely useful for the production of high throughput drug screening microarrays and massively parallel biosensor applications.
Experimental Section
Ink Preparation
Lipids used for arraying, screening and microscopy were 1,2-dioeoyl-sn-glycero-3-phosphocholine (DOPC); 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP); 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-lissamine rhodamine B sulfonyl (DOPE-RB). These were purchased from Avanti Polar Lipids, Inc. 1, 2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (Marina Blue DHPE) was purchased from Invitrogen. Chloroform solutions of the different lipids were mixed to obtain the desired molar ratios. When making liposomal formulations, chloroform was evaporated off under a Nitrogen stream, then further dried in vacuum overnight to form a thin film of lipid on the bottom of the glass vials. After drying, water was added to the vials containing the dried lipids. Samples were then lightly vortexed for 10 seconds, then sonicated for 10 minutes. After sonication, vortexing was used as needed to ensure suspension.
Microarraying
The different lipid solutions were microarrayed from standard 384 well microtitre plates (Axygen, Inc., PMI110-07 V1., Union city, CA) using a BioRobotics pinspotter model BG600 (Comberton, Cambridge, England) onto the substrate of choice, using a 200 μm 4×4 stainless steel microspot pin tool. For integration of multiple fluorescent inks, DOTAP was doped with 1 Mol% rhodamine-PE, and 2 Mol% Marina Blue DHPE and carboxyfluorescein,-PE, respectively; the results were microarrayed in a 2×3 array pattern onto a PDMS ink pallet. Microarray pins were washed to ensure no cross contamination between inks. We found that 2 minute washes in acetone and then water, followed by 30 seconds of drying sufficed.
Intaglio Printing
Using the microarraying procedure, the DOTAP stamp was inked onto a palette, which was then left in vacuum overnight to evaporate residual water. The PDMS stamp was then inked by firm, uniform contact with the ink palette. Once inked, the PDMS stamp was pressed onto a glass substrate by hand being careful to apply uniform, firm pressure measured on a bathroom scale to be about 45 Newtons for ∼20 seconds before careful removal. The PDMS stamps can be reused after adequate sonication in isopropanol, then water for 10 minutes each followed by drying under nitrogen. Cleanliness was verifed using fluorescence and brightfield microscopy.
Substrate and Sample Preparation
For Fig. 1, glass cover slips were used as received as the patterned substrate (No. 1.5, 22×22 mm, VWR, Radnor, PA). For spin coating, a 4% solution of PMMA dissolved in chlorobenzene was spin coated onto a glass cover slip and baked in an oven at 150 degrees celcius for 20 minutes.
Characterization and Imaging Techniques
A Ti-E epifluorescence inverted microscope (Nikon Instruments, Melville, NY) fitted with a Retiga SRV (QImaging, Canada) CCD camera (1.4 MP, Peltier cooled to -45 ° C) was used for fluorescence and brightfield imaging of lipid structures. Heights and topography of the lipid prints were measured using tapping mode with a Dimension 3000 AFM (Veeco Instruments, Plainview, NY) and tapping mode AFM cantilevers (# OMCLAC160TS-W2, 7 nm nominal tip radius, 15 μm tip height, 42 N m-1 spring constant, Olympus, Center Valley, PA). The data from Figure 3 was analyzed using Nanoscope Analysis (Version 1.4, Bruker). Each image was flattened. Sections shown in panels a and b were exported to OriginPro 8.1 and plotted. Quantitative values were found for the dot patterns by sections drawn through the center of each of the five rows of dots. The height and width at half height were found from the sectional analysis data. For the gratings, four line sections were chosen at random and height and width at half height found as mentioned above. Error was found by calculating the standard deviation for each of the values. Characterization of the arrays made by nanointaglio printing of different lipid concentrations, each of the patterns was imaged using AFM. Samples of 25 lipid dots from each pattern were imaged and analyzed to determine the height of each dot. Using ImageJ downloaded from the NIH website, the average intensity was calculated for round regions around each of the lipid dots. The average height of each of the 25 lipid dots was found using AFM image height conversion. Supplemental figure 3 graphs the coefficient of variation of the average heights, such that CV = st.dev of average height/mean average height×100%, plotted with each liposomal concentration from which the surface structures were derived.
Image analysis
An ImageJ macro was written to producethe measurement of print uniformity for different lipid concentrations (Fig. 2g). The macro created a mask, then dilated the pixels of the pattern to define a total area. The macro output the total area in pixels and the processed image, shown in Figure. 2f in green. The macro then repeated, first eroding pixels to erase the uniform regions, then dilating the same number of times to show only non-uniform regions (panel 2f in red). The two images were then overlaid. Percent uniformity of the entire lipid pattern is defined as [1 − (non-uniform regions/total area)]× 100%.
Storage
Patterned lipid arrays may be stored in a nitrogen glovebox (Mbraun, Inc., Model Labstar (1200/780) Stratham, NH, USA) until use. They will remain stable under inert atmospheric conditions. The lipid patterns can be disrupted by exposure to humidity or the air - water interface. By dehydrating them in an anhydrous N2 atmosphere within glovebox, it is possible to reporducibly immerse them in water without disruption of the patterned interface.[29]
Supplementary Material
Supporting Figure 1. Variation of the ink concentration. 4× red fluorescence overview of lipid multilayer patterns created from microarrraying and multilayer stamping using a 5um well pattern. DOTAP concentrations of 4, 8, 16, 32, 64, and 128 g/L stamped onto a glass substrate (4th print). Lipid concentrations at 2 g/L do not microarray sufficiently to allow pattern formation by lipid multilayer stamping.
Supporting Figure 2. Characterization of a PDMS Stamp with 5μm diameter holes at a center-to-center spacing of 10 μm. a, 40× bright field image of a section of the PDMS stamp the size of the white box in a. Bar = 25 μm. b, AFM topographical image, and c sectional analysis shows that the wells are ∼2.7 μm deep, with 5 μm diameters.
Supporting Figure 3. Coefficient of variation (CV) of the average height of the lipid dots derived from the series of AFM measurements on glass as shown in panel e of Figure 2 of the concentrations 4, 8, 16, 32, 64 and 128 g/L liposomal concentrations. The CV of the average height is 10 % or less for all concentrations except 4 and 64 g/L, indicating uniform patterning of the structures sampled.
Supporting figure 4. Multicomponent patterning from microarraying. 2×3 ink pallet of DOTAP doped with 1 Mol % Rhodamine-DOPE, 2 Mol % Marina Blue-DHPE, and Carboxyfluorescein-DOPE. There was no detectable contamination of the different fluorescently tagged lipids. Scale bar = 200 μm
Supporting figure 5. Uniformity of integrated lipid dot patterns. The coefficient of variation (CV) of the three different fluorescently doped lipid dot patterns from figure panels 1 j-l is well below 10%, demonstrating uniform patterning. The CV was calculated by finding the average intensity for each dot shown in panels 1 j-l and then finding the intensities' average and standard deviation.
Acknowledgments
The authors thank Eric Lochner for help with AFM and Kevin John for the schematic illustrations in Figs. 1 and 4. SL Acknowledges support from the National Institute of Health grant R01 GM107172-01.
Footnotes
Supporting Information: Supporting Information is available online from the Wiley Online Library or from the author.
Contributor Information
Troy W. Lowry, Department of Biological Sciences and Integrative Nanoscience Institute, Florida State University, Tallahassee, Florida 32306-4370, USA; Department of Physics Florida State University Tallahassee, Florida, 32306-4350, USA
Aubrey Kusi-Appiah, Department of Biological Sciences and Integrative Nanoscience Institute, Florida State University, Tallahassee, Florida 32306-4370, USA.
Prof. Jingjiao Guan, Department of Chemical and Biomedical Engineering, Florida State University, Tallahassee, Florida 32310-6046, USA
Prof. David H. Van Winkle, Department of Physics Florida State University Tallahassee, Florida, 32306-4350, USA
Dr. Michael W. Davidson, The National High Magnetic Field Laboratory, Florida State University, Tallahassee, Florida 32310-3706, USA
Prof. Steven Lenhert, Email: lenhert@bio.fsu.edu, Department of Biological Sciences and Integrative Nanoscience Institute, Florida State University, Tallahassee, Florida 32306-4370, USA.
References
- 1.Rohrer H. The nanoworld: Chances and challenges. Microelectron Eng. 1996;32:5–14. [Google Scholar]
- 2.Tian BZ, et al. Three-Dimensional, Flexible Nanoscale Field-Effect Transistors as Localized Bioprobes. Science. 2010;329:830–834. doi: 10.1126/science.1192033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia YN, Xiong YJ, Lim B, Skrabalak SE. Shape-Controlled Synthesis of Metal Nanocrystals: Simple Chemistry Meets Complex Physics? Angew Chem Int Edit. 2009;48:60–103. doi: 10.1002/anie.200802248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geim AK, Novoselov KS. The rise of graphene. Nat Mater. 2007;6:183–191. doi: 10.1038/nmat1849. [DOI] [PubMed] [Google Scholar]
- 5.Park H, et al. High-density integration of carbon nanotubes via chemical self-assembly. Nat Nanotechnol. 2012;7:787–791. doi: 10.1038/nnano.2012.189. [DOI] [PubMed] [Google Scholar]
- 6.Ke YG, Ong LL, Shih WM, Yin P. Three-Dimensional Structures Self-Assembled from DNA Bricks. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavin RK, Lugli P, Zhirnov VV. Science and Engineering Beyond Moore's Law. P Ieee. 2012;100:1720–1749. [Google Scholar]
- 8.Song YJ et al. Multiplexed volumetric bar-chart chip for point-of-care diagnostics. Nat Commun. 2012;3 doi: 10.1038/ncomms2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting Organ-Level Lung Functions on a Chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel B, Bernard A, Bietsch A, Delamarche E, Geissler M, Juncker D, Kind H, Renault JP, Rothuizen H, Schmid H, SchmidtWinkel P, Stutz R, Wolf H. Printing meets lithography: Soft approaches to high-resolution patterning. IBM Journal of Research and Development. 2001;45:870–870. [Google Scholar]
- 11.Majd S, Mayer M. Generating Arrays with High Content and Minimal Consumption of Functional Membrane Proteins. Journal of the American Chemical Society. 2008;130:16060–16064. doi: 10.1021/ja8055485. [DOI] [PubMed] [Google Scholar]
- 12.Nafday OA, Lowry TW, Lenhert S. Multifunctional Lipid Multilayer Stamping. Small. 2012;8:1021–1028. doi: 10.1002/smll.201102096. [DOI] [PubMed] [Google Scholar]
- 13.Barbulovic-Nad I, et al. Bio-microarray fabrication techniques - A review. Crit Rev Biotechnol. 2006;26:237–259. doi: 10.1080/07388550600978358. [DOI] [PubMed] [Google Scholar]
- 14.Guo LJ. Nanoimprint lithography: Methods and material requirements. Adv Mater. 2007;19:495–513. [Google Scholar]
- 15.Whitesides GM, Ostuni E, Takayama S, Jiang XY, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 16.Schena M, Shalon D, Davis RW, Brown PO. Quantitative Monitoring of Gene-Expression Patterns with a Complementary-DNA Microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 17.Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nat Rev Genet. 2006;7:200–210. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 18.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 19.Ban L, et al. Discovery of glycosyltransferases using carbohydrate arrays and mass spectrometry. Nat Chem Biol. 2012;8:769–773. doi: 10.1038/nchembio.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bally M, et al. Liposome and Lipid Bilayer Arrays Towards Biosensing Applications. Small. 2010;6:2481–2497. doi: 10.1002/smll.201000644. [DOI] [PubMed] [Google Scholar]
- 21.Mei Y, et al. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat Mater. 2010;9:768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 23.Bailey SN, Sabatini DM, Stockwell BR. Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. P Natl Acad Sci USA. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braunschweig AB, Huo FW, Mirkin CA. Molecular printing. Nat Chem. 2009;1:353–358. doi: 10.1038/nchem.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salaita K, et al. Restriction of Receptor Movement Alters Cellular Response: Physical Force Sensing by EphA2. Science. 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tayebi L, et al. Long-range interlayer alignment of intralayer domains in stacked lipid bilayers. Nat Mater. 2012;11:1074–1080. doi: 10.1038/nmat3451. [DOI] [PubMed] [Google Scholar]
- 27.Lenhert S, Sun P, Wang Y, Fuchs H, Mirkin CA. Massively parallel dip-pen nanolithography of heterogeneous supported phospholipid multilayer patterns. Small. 2007;3:71–75. doi: 10.1002/smll.200600431. [DOI] [PubMed] [Google Scholar]
- 28.Kusi-Appiah AE, Vafai N, Cranfill PJ, Davidson MW, Lenhert S. Lipid multilayer microarrays for in vitro liposomal drug delivery and screening. Biomaterials. 2012;33:4187–4194. doi: 10.1016/j.biomaterials.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 29.Lenhert S, et al. Lipid multilayer gratings. Nat Nanotechnol. 2010;5:275–279. doi: 10.1038/nnano.2010.17. [DOI] [PubMed] [Google Scholar]
- 30.Nafday OA, Lenhert S. High-throughput optical quality control of lipid multilayers fabricated by dip-pen nanolithography. Nanotechnology. 2011;22(22) doi: 10.1088/0957-4484/22/22/225301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure 1. Variation of the ink concentration. 4× red fluorescence overview of lipid multilayer patterns created from microarrraying and multilayer stamping using a 5um well pattern. DOTAP concentrations of 4, 8, 16, 32, 64, and 128 g/L stamped onto a glass substrate (4th print). Lipid concentrations at 2 g/L do not microarray sufficiently to allow pattern formation by lipid multilayer stamping.
Supporting Figure 2. Characterization of a PDMS Stamp with 5μm diameter holes at a center-to-center spacing of 10 μm. a, 40× bright field image of a section of the PDMS stamp the size of the white box in a. Bar = 25 μm. b, AFM topographical image, and c sectional analysis shows that the wells are ∼2.7 μm deep, with 5 μm diameters.
Supporting Figure 3. Coefficient of variation (CV) of the average height of the lipid dots derived from the series of AFM measurements on glass as shown in panel e of Figure 2 of the concentrations 4, 8, 16, 32, 64 and 128 g/L liposomal concentrations. The CV of the average height is 10 % or less for all concentrations except 4 and 64 g/L, indicating uniform patterning of the structures sampled.
Supporting figure 4. Multicomponent patterning from microarraying. 2×3 ink pallet of DOTAP doped with 1 Mol % Rhodamine-DOPE, 2 Mol % Marina Blue-DHPE, and Carboxyfluorescein-DOPE. There was no detectable contamination of the different fluorescently tagged lipids. Scale bar = 200 μm
Supporting figure 5. Uniformity of integrated lipid dot patterns. The coefficient of variation (CV) of the three different fluorescently doped lipid dot patterns from figure panels 1 j-l is well below 10%, demonstrating uniform patterning. The CV was calculated by finding the average intensity for each dot shown in panels 1 j-l and then finding the intensities' average and standard deviation.


