Abstract
Food allergies are a growing public health concern. The rapidly increasing prevalence of allergic disease cannot be explained by genetic variation alone, suggesting a role for gene-by-environment interactions. The bacteria that colonize barrier surfaces, often referred to as the commensal microbiota, are dramatically affected by environmental factors and have a major impact on host health and homeostasis. Increasing evidence suggests that alterations in the composition of the microbiota, caused by factors such as antibiotic use and diet, are contributing to increased sensitization to dietary antigens. This review will discuss the cellular and molecular pathways activated by commensal bacteria to protect against allergic sensitization. By understanding the interplay between the environment, the microbiota, and the host, we may uncover novel therapeutic targets that will allow us to control the allergy epidemic.
Introduction
The hygiene hypothesis originally proposed that protection from allergic disease in children with older siblings could “be explained if allergic diseases were prevented by infection in early childhood, transmitted by unhygienic contact with older siblings, or acquired prenatally from a mother infected by contact with her older children.” [1]. This early evidence suggested that microbial factors regulate susceptibility to allergic disease. Societal efforts to improve sanitation and cleanliness have also been paralleled by an increase in autoimmune disease, broadening the original hypothesis beyond allergy. More recently the scope of the hygiene hypothesis has been further expanded to consider the role of commensal bacteria in the regulation of both allergic and inflammatory disease. It is now clear that trillions of bacteria colonize all of the body’s barrier sites, although the majority are located in the intestine [2]. Signals from these commensal bacteria are important for educating the immune system, beginning at birth and continuing throughout life. The composition of the microbiota is dynamic and strongly influenced by the external environment. It can be dramatically altered by diet, antibiotic use, mode of birth, formula feeding, vaccination and exposure to pathogens (reviewed in [3]). It is now hypothesized that changes in the composition of the microbiota, called dysbiosis, lead to a loss of protective bacterial signals, which can cause both allergic and inflammatory disease. This review, however, will focus on the influence of the microbiota on allergic disease, specifically IgE-mediated food allergy.
Evidence that commensal bacteria regulate sensitization to food allergens
Work from our laboratory first linked microbiota-derived signals to increased sensitization to food allergens. We showed that C3H/HeJ mice, which are unable to signal through Toll-like receptor (TLR) 4, had increased IgE production and allergic symptoms in response to sensitization with peanut (PN) plus cholera toxin (CT) when compared to TLR4-sufficient, C3H/FeJ mice [4]. TLR4 is the receptor for bacterial lipopolysaccharide, indicating that an inability to sense microbial products can lead to exacerbated allergic responses. Treating TLR4-sufficient mice with broad-spectrum antibiotics (Abx) to disrupt the microbial community structure of the gut increased the PN-specific IgE response to the levels seen in TLR4-deficient mice, suggesting that the microbiota was the source of the allergy-protective signal.
To identify these protective bacterial taxa our laboratory developed a gnotobiotic model of food allergy. Germ free (GF) mice, which are completely devoid of a microbiota, spontaneously generate high levels of IgE, and have increased symptoms of anaphylaxis after systemic sensitization [5]. This spontaneous IgE production is reversed by colonization with a diverse microbiota early in life, further linking commensal bacteria with the regulation of IgE. Using our model of intragastric sensitization with PN plus CT, we found that GF mice, like Abx-treated mice, had significantly increased PN-specific IgE and IgG responses when compared to SPF mice with a replete microbiota [6**]. We then colonized GF mice with representative members of the two major bacterial phyla of the intestine, Bacteroidetes and Firmicutes. Colonization with B. uniformis, a representative Bacteroidetes strain, was unable to reduce PN-specific responses. However colonization with a consortium of Clostridia, a class of anaerobic spore-forming Firmicutes, was sufficient to reduce the concentration of PN-specific IgE to that seen in SPF mice sensitized with PN plus CT. Restoration of a Clostridia containing microbiota also reduced PN-specific IgE responses in sensitized Abx-treated mice. These findings identified Clostridia as a component of the commensal microbiota critical for protection against food allergen sensitization. Other work has also postulated a role for the commensal microbiota in promoting allergic responses to food. Noval Rivas et al found that distinct microbial signatures can be induced after sensitization of allergy-susceptible mice with a gain of function mutation in the IL-4 receptor α chain (Il4raF709) compared to WT controls [7]. Allergic sensitivity was transmissible by transfer of the disease-associated microbiota, suggesting that commensal bacteria may also play a role in the pathogenesis of food allergy.
Which cells require microbiota-mediated signals?
Exactly which cells in the intestine receive microbial signals to regulate sensitization to food allergens remains an outstanding question. The intestinal lumen is a complex site, filled with hundreds of bacterial species as well as diverse food antigens. The gut associated lymphoid tissue must be able to differentiate between these innocuous dietary or microbial antigens and potentially harmful pathogens. The intestinal epithelium forms the physical barrier between the immune system and the contents of the intestinal lumen (both dietary and microbial) and is also the first site of host-microbe interaction. As part of the epithelial barrier, intestinal epithelial cells (IEC) regulate the passage of solutes and antigen from the lumen into the lamina propria (LP) via tight junctions (TJ) [8]. Changes in the cytokine milieu can alter the expression and function of different TJ proteins to regulate the antigens that cross into the LP. IEC express TLRs [9–15] and produce cytokines in response to microbial stimulation (reviewed in [16]). Other evidence suggests that specialized epithelial cells like M cells can shuttle antigen to dendritic cells (DC) and other antigen presenting cells (APC) [17,18]. Goblet cells produce mucus, which forms a thick matrix that promotes spatial segregation and prevents direct contact between most bacteria and the epithelium. Recently, it has been shown that goblet cells can also pass luminal antigen to LP DC [19*–20]. Paneth cells produce antimicrobial peptides that can regulate the composition of the microbiota [21–23]. Extensive interaction between the epithelium and commensal bacteria is therefore critical for the maintenance of mucosal homeostasis.
Our study supports a role for IECs in Clostridia-mediated protection against food allergen sensitization. Because Clostridia reside close to the intestinal epithelium [24,25] we hypothesized that Clostridia would induce a unique set of genes in IEC that would not be induced by non-protective bacteria like B. uniformis. Microarray analysis of IEC from GF, B. uniformis, and Clostridia-colonized mice showed that Clostridia colonization upregulated expression of Reg3b, the gene that encodes the antimicrobial peptide REGIIIβ [6**]. Expression of the REG family of antimicrobial peptides depends on both MyD88-signaling and IL-22/IL-22 receptor signaling [22]; we found that the expression of IL-22 was increased in the colonic LP of Clostridia colonized mice [6**]. IL-22 is a barrier protective cytokine that is produced by hematopoietic cells but acts on stromal cells [26]. In the intestine, IL-22 can stimulate epithelial proliferation, increase antimicrobial peptide production, and promote mucus secretion by goblet cells [27,28]. The role of this suite of responses in protection against intestinal pathogens and in promoting tissue repair is well documented [27,28]. We hypothesized that the barrier protective effects of IL-22 would also reduce epithelial permeability to food antigens, thereby reducing the concentration of PN antigen in the blood and limiting the opportunity for sensitization. This proved to be the case; mice colonized with Clostridia had reduced concentrations of two immunodominant PN allergens, Ara h 6 and Ara h 2, in their serum compared to GF or Abx-treated mice. Treating Clostridia-colonized mice with neutralizing antibody to IL-22 abrogated this effect and also increased PN-specific Ig and T cell responses to sensitization with PN plus CT. Thus Clostridia-induced IL-22 is a novel innate mechanism by which the microbiota can regulate the permeability of the epithelial barrier and contribute to protection against food allergen sensitization.
There are also several subsets of DC that are uniquely adapted to their niche in the LP that may also respond to commensal bacteria and mediate host-microbiota cross-talk [29]. DC are professional APC and express diverse pattern recognition receptors and cytokines to receive signals from and respond to microbial stimulation. Some of these specialized DC populations are essential for establishing and maintaining tolerance to food. In particular CD103+ DC can traffic and carry antigen from the LP to the mesenteric lymph node (MLN), where they interact with naïve T cells [30,31]. Under healthy conditions, they are particularly good at driving these naïve T cells toward the Foxp3+ regulatory T cell (Treg) lineage. They produce TGF-β and express aldehyde dehydrogenase (ALDH) enzymes that can convert dietary vitamin A to retinoic acid (RA) which, together, promote Foxp3 expression [32–34]. These Foxp3+ Treg then migrate back to the LP to suppress subsequent responses to food. When this process is disrupted, food allergies can result. New evidence suggests that intestinal eosinophils can release eosinophil peroxidase (EPO) to activate these same CD103+ DCs but skew them to stimulate food antigen-specific Th2 responses in the MLN instead of inducing Tregs [35]. CD103+ DC can also respond to microbial signals, such as flagellin, and express proinflammatory cytokines, such as IL-23 [36]. IL-23 induces IL-22 production by other cells (including innate lymphoid cells, ILC, see below), so these DC also indirectly impact IEC function. Their ability to integrate cues from both innate immune cells and the microbiota suggests that CD103+ DC play a central role in the induction of tolerance to dietary antigen and the prevention of food allergen sensitization.
CX3CR1+ macrophages may also contribute to protection against food allergen sensitization. CX3CR1+ cells have been shown to extend processes into the intestinal lumen to acquire antigen [37,38] and a new study reports that this antigen is then passed to the CD103+ DC, which carry it to the MLN [39*]. They can also produce IL-10 that expands Foxp3+ Tregs in the LP [39*,40]. In the absence of CX3CR1+ cells, there is a reduction in food antigen-specific Tregs and increased delayed-type hypersensitivity responses in sensitized mice, indicating that transfer of food antigen to DC by these cells is critical to establish tolerance [40]. CX3CR1+ cells were originally described as resident in the LP [41]. They can, however, migrate out of the intestine under conditions of dysbiosis, so they are also sensitive to changes in the microbiota [42]. If their localization is altered, it may limit antigen capture and prevent the establishment of tolerance, increasing the chances for food allergen sensitization.
Final cell types that may be important for receiving microbial signals to prevent allergic sensitization are innate lymphoid cells (ILC), particularly RORγt+ Group 3 ILC. Group 2 ILC are often associated with allergy because they produce Th2 cytokines and can exacerbate disease [43]. Group 3 ILC, on the other hand, were first identified as important for protection against bacterial infection [44]. Recent data suggests they are more versatile than that, though, and contribute to the maintenance of homeostasis. Group 3 ILC can present antigen on MHC class II and can induce anergy in CD4 T cells to prevent aberrant responses to the microbiota [45]. They can also be stimulated to promote IL-22 by DC-produced cytokines such as IL-23 and IL-6 [46]. Interestingly, RA can also promote IL-22 production by ILC [47*]. In our model, the IL-22 required to prevent allergen uptake into the blood is produced primarily by these RORγt+ ILC [6**]. Treating Clostridia-colonized Rag−/− mice with anti-CD90 to deplete ILC significantly reduces the expression of IL-22 and also abrogates the barrier protective effect of Clostridia. Serum Ara h 6 and Ara h 2 concentrations are higher after ILC depletion, even when Clostridia are present. Another study demonstrated that IL-1β produced by commensal bacteria-activated CX3CR1+ macrophages could also stimulate ILC3 to produce GM-CSF [48**]. GM-CSF then drives the production of IL-10 and RA by macrophages and DC, which promote the conversion of naïve T cells into Foxp3+ Treg. Altering the commensal bacteria may disrupt this macrophage-ILC-DC axis, contributing to the development of food allergic responses by limiting the production of anti-inflammatory cytokines and reducing bacterially induced Treg conversion.
What signals are provided by the microbiota to protect against the generation of food allergy?
In addition to not knowing which cell type (or types) interact with the microbiota, the signals that are required from the bacteria to prevent allergic disease are also not well understood. TLR-mediated signaling, aryl hydrocarbon receptor (AhR)-mediated signaling, and diet-derived compounds may all be involved in the interaction between bacteria and the host. There is evidence that each of these classes of signals has the potential to modulate the host immune response and to protect against allergy. It is also possible, however, that it is a balance or combination of multiple signals that is required to prevent sensitization to food. If the composition of the microbiota is disrupted, this balance may be lost.
A large body of work supports a role for TLR-mediated signals in regulating host homeostasis and interactions with the microbiota (reviewed in [49,50]). Indeed, the first clue that the microbiota may be involved in allergic sensitization to food came from the exacerbated disease phenotype in TLR4-deficient mice [4]. TLRs are expressed on IEC and DC and an essential role for signaling in these cell types is well established. In the absence of individual TLRs or the downstream adaptor molecule MyD88, there is increased inflammation, indicating these signals can be protective in the intestine [51]. TLRs may also be expressed in certain subsets of ILC and can control the expression of effector cytokines [52]. A lack of TLR-mediated signals has also been linked to deficiencies in regulatory cell populations, leading to the expansion of effector cells that may drive or exacerbate disease. For example, loss of TLR2 signaling reduces Treg proportions in the colon [53] while DC-specific deletion of TRAF6, a downstream molecule in the TLR signaling pathway, promotes Th2 cell differentiation in the LP in response to stimulation by commensals and impaired Treg conversion in response to fed antigen [54**]. Furthermore, stimulation of DC by TLR ligands can trigger expression of a variety of cytokines, both pro- and anti-inflammatory [55]. In the TRAF6 model, DC-derived IL-2 was critical to maintain tolerance through Tregs. TLR signaling in DC can also drive the upregulation of IL-23, IL-6, and IL-1β, which can, in turn, drive the expression of IL-22 in other cell types. Given the wide variety of TLR ligands expressed by the diverse members of the microbiota, it is clear that these signals can have a profound effect on the nature of an immune response, including the response to food.
TLRs are not the only receptors that may be involved in host/microbiota interaction, however. Recent work has shown an important role for AhR in regulating and responding to the microbiota. Although originally described as a receptor for xenobiotics [56], it is now known that AhR can have a profound effect on the immune system. AhR is a transcription factor found in a variety of cells, including T cells, DC, and ILC [57–59]. AhR is therefore expressed in several of the major cell types found in the gut that may interact with commensals. Endogenous ligands for the AhR are now known, including products of tryptophan metabolism, some of which are bacterially derived [60]. AhR can also be stimulated by compounds found in cruciferous vegetables, so dietary signals may be important for the activation of this pathway as well [61].
One of the most notable effects of AhR is regulation of IL-22 expression. In the absence of AhR, little IL-22 is produced and the ability to fight intestinal pathogens is reduced [58*]. Loss of AhR signaling also changes the balance of Th cell subsets in the intestine, which subsequently alters the composition of the microbiota [62]. Based on our work and proposed model, the loss of Clostridia-induced IL-22 by Group 3 ILC could have consequences for intestinal permeability to food antigens as well. In the setting of low IL-22 levels as a result of diminished AhR signaling (due to lack of stimulation by bacterial or dietary components), more antigen may cross the epithelial barrier, leading to increased allergic sensitization. In support of this idea, administration of AhR ligands can suppress sensitization induced by PN plus CT [63]. Other work has revealed that AhR activation reduces effector T cell number [64] and expands and activates the CD103+ DC population to promote tolerance to orally administered antigen [65]. Together, these models suggest that AhR may be an important signaling pathway in each of the cellular compartments discussed above that are thought to control the establishment and maintenance of tolerance to food.
A final source of signals that are involved in regulating the interaction between the host and the microbiota are diet-derived compounds. One of the major contributions of the microbiota to host health is the metabolism of dietary components that the host cannot digest itself. Different diets, however, support different microbial populations based on the substrates provided for bacterial metabolism. An important example of this is dietary fiber. The microbiota metabolizes dietary fiber into short chain fatty acids (SCFA) such as acetate, butyrate, and propionate, which the host can then use in a variety of ways. A study comparing the microbiota of different human populations showed that a diet high in plant fiber promotes the growth of SCFA-producing bacteria to a greater extent than a high fat Western style diet [66]. SCFAs are an energy source for colonocytes [67,68] but can also be actively transported into cells via monocarboxylate transporters [69], act as histone deacetylase (HDAC) inhibitors [70], and signal through G-protein coupled receptors (GPCRs) [71–73*]. Through these different pathways, SCFAs can act on almost all cells of the immune system to alter their function [68]. Feeding a high fiber diet [74,75] or administering acetate, propionate, or butyrate orally [76] can all increase the proportion of Foxp3+ Tregs in the colonic lamina propria of GF or antibiotic-treated mice. Induction of Tregs has been linked to both signaling through GPCRs [76] and HDAC inhibition [74,75]. SCFA also circulate in the blood to reach sites far from the intestine to protect against airway hyperreactivity via GPCR signaling [77]. Other work from our group has shown that treating cow’s milk allergic infants with a probiotic-supplemented formula leads to an expansion of butyrate-producing Clostridiales and increased fecal butyrate levels, which correlates with accelerated acquisition of tolerance to cow’s milk [78,79].
Further evidence that SCFA, particularly butyrate, may be involved in establishing tolerance to food or protecting against allergic sensitization comes from a recent report by Singh et al, which links the production of butyrate to the production of another diet-derived compound, RA [73*]. Butyrate can signal through GPR109a, a GPCR and the receptor for niacin as well as butyrate. In GPR109a knock out (Niacr1−/−) mice, the proportion of Foxp3+ Tregs in the colonic lamina propria is reduced. When the cause of this deficit was examined, the authors found that treatment of GPR109a-sufficient DCs with butyrate improved their ability to promote conversion of naïve T cells into Foxp3+ Tregs. Butyrate-treated DCs also had increased expression of ALDH1 (Aldh1a1), an enzyme involved in metabolizing RA. As mentioned previously, RA is produced from Vitamin A and is critical for the development of food-specific Foxp3+ Tregs. When DCs from Niacr1−/− mice were treated with butyrate, however, there was no increase in expression of Aldh1a1, demonstrating that signaling through this receptor is necessary for expression of ALDH1. Antibiotic treatment reversed this phenotype, confirming that the microbiota is required for this process. The increase in Aldh1a1 expression suggests that butyrate induces Tregs by promoting the production of RA by DCs. Perhaps this butyrate-mediated pathway plays a role in the induction of tolerance to food antigens and prevents food allergen sensitization.
Concluding remarks
Although it is clear from mouse models and human studies that the microbiota influences the development of allergic disease, exactly how this occurs remains incompletely understood. It is most likely that a variety of signals from the microbiota must be integrated by the various cell subsets in the intestine to fine-tune the balance between activation and tolerance. When this balance is disrupted, sensitization can be induced. TLR, AhR, and dietary signals may be received by IEC, DC, macrophages, and ILC to fortify the epithelial barrier, promote Foxp3+ Treg induction, and guard against the generation of food-specific Th2 effector populations (Fig. 1). Other effector populations like basophils and natural killer T cells can be regulated by the microbiota as well [80,81], further reinforcing the tolerogenic environment. An increased understanding of how all these pathways intersect will inform the development of targeted strategies to promote tolerance to dietary antigen. Ultimately, this knowledge will help us treat and prevent food allergen sensitization to effectively halt this epidemic.
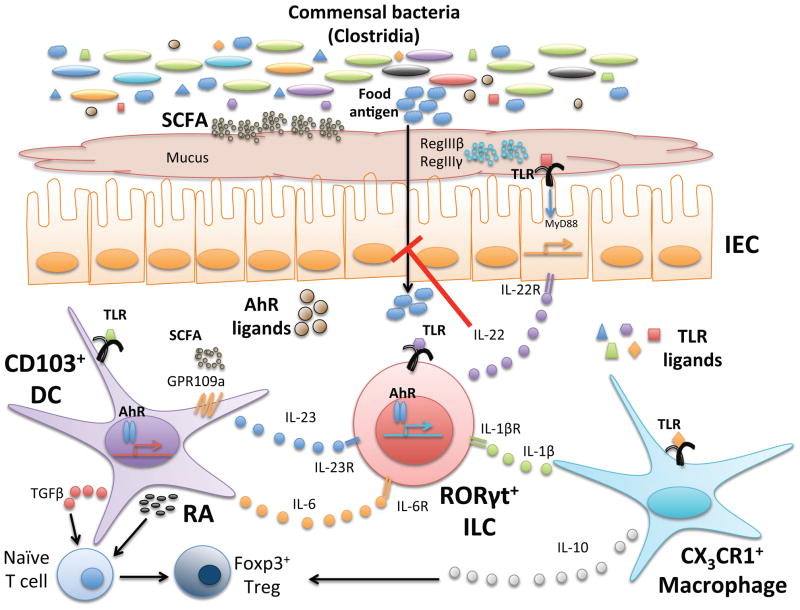
Figure 1. Cellular and molecular pathways through which commensal bacteria regulate allergic responses to food.
Commensal bacteria and their products, particularly TLR ligands, AhR ligands, and short chain fatty acids (SCFA) can all influence intestinal homeostasis. TLR ligands act on IEC, CD103+ DC, CX3CR1+ macrophages, and RORγt+ ILC to promote cytokine secretion. TGF-β and dietary RA produced by activated DC and IL-10 from macrophages induce conversion of naïve T cells to Foxp3+ Treg and expand this regulatory population. SCFAs produced by bacterial fermentation of dietary fiber act on DC via GPCRs to further promote RA production and reinforce the tolerogenic environment. AhR ligands, derived from the diet or produced during bacterial metabolism, can also act on DC and ILC. IL-22 produced by ILC in response to cytokine stimulation (IL-23, IL6, or IL-1β) by DC or macrophages or by AhR stimulation, can act on the epithelium to promote barrier integrity by inducing expression of antimicrobial peptides RegIIIβ and RegIIIγ, increasing epithelial proliferation, and promoting mucus secretion. Together, this network of signals maintains homeostasis in the host and prevents responses to food. When these signals are altered or lost due to changes in the microbiota (dysbiosis), food allergies may develop.
Highlights.
The commensal microbiota regulates sensitization to food antigens.
Many cell types interact with commensal bacteria including IEC, DC, and ILC.
Signaling by TLR or AhR ligands and dietary products may prevent sensitization.
Bacterially induced cytokines like IL-22 are important to reinforce the epithelial barrier.
Acknowledgments
The authors would like to thank V. Guzzetta, P. Johnston, and S. Pawar for critical review of this manuscript. This work was supported by Food Allergy Research and Education (FARE) and by NIAID (AI106302).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strachan DP. Hay fever, hygiene, and household size. Bmj. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feehley T, Stefka AT, Cao S, Nagler CR. Microbial regulation of allergic responses to food. Semin Immunopathol. 2012;34:671–688. doi: 10.1007/s00281-012-0337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172:6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 5.Cahenzli J, Köller Y, Wyss M, Geuking Markus B, McCoy Kathy D. Intestinal Microbial Diversity during Early-Life Colonization Shapes Long-Term IgE Levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **6.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo G-Y, Cao S, Theriault BR, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. The authors demonstrate that alterations to the commensal microbiota increase PN-specific responses and identify Clostridia as a protective bacterial population. They also identify IL-22 as an important innate cytokine induced by bacteria that limits allergen uptake. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, Chehoud C, Kuczynski J, DeSantis T, Warrington J, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201–212. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 9.Otte JM, Cario E, Podolsky DK. Mechanisms of cross hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology. 2004;126:1054–1070. doi: 10.1053/j.gastro.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Abreu MT, Arnold ET, Thomas LS, Gonsky R, Zhou Y, Hu B, Arditi M. TLR-4 and MD-2 expression is regulated by immune-mediated signals in human intestinal epithelial cells. J Biol Chem. 2002;277:20431–20437. doi: 10.1074/jbc.M110333200. [DOI] [PubMed] [Google Scholar]
- 11.Bambou JC, Giraud A, Menard S, Begue B, Rakotobe S, Heyman M, Taddei F, Cerf-Bensussan N, Gaboriau-Routhiau V. In vitro and ex vivo activation of the TLR5 signaling pathway in intestinal epithelial cells by a commensal Escherichia coli strain. J Biol Chem. 2004;279:42984–42992. doi: 10.1074/jbc.M405410200. [DOI] [PubMed] [Google Scholar]
- 12.Gribar SC, Richardson WM, Sodhi CP, Hackam DJ. No longer an innocent bystander: epithelial toll-like receptor signaling in the development of mucosal inflammation. Mol Med. 2008;14:645–659. doi: 10.2119/2008-00035.Gribar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neal MD, Leaphart C, Levy R, Prince J, Billiar TR, Watkins S, Li J, Cetin S, Ford H, Schreiber A, et al. Enterocyte TLR4 mediates phagocytosis and translocation of bacteria across the intestinal barrier. J Immunol. 2006;176:3070–3079. doi: 10.4049/jimmunol.176.5.3070. [DOI] [PubMed] [Google Scholar]
- 14.Shang L, Fukata M, Thirunarayanan N, Martin AP, Arnaboldi P, Maussang D, Berin C, Unkeless JC, Mayer L, Abreu MT, et al. Toll-Like Receptor Signaling in Small Intestinal Epithelium Promotes B-Cell Recruitment and IgA Production in Lamina Propria. Gastroenterology. 2008;135:529–538. doi: 10.1053/j.gastro.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vora P, Youdim A, Thomas LS, Fukata M, Tesfay SY, Lukasek K, Michelsen KS, Wada A, Hirayama T, Arditi M, et al. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J Immunol. 2004;173:5398–5405. doi: 10.4049/jimmunol.173.9.5398. [DOI] [PubMed] [Google Scholar]
- 16.Prescott D, Lee J, Philpott DJ. An epithelial armamentarium to sense the microbiota. Sem Immunol. 2013;25:323–333. doi: 10.1016/j.smim.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Neutra MR, Mantis NJ, Frey A, Giannasca PJ. The composition and function of M cell apical membranes: implications for microbial pathogenesis. Semin Immunol. 1999;11:171–181. doi: 10.1006/smim.1999.0173. [DOI] [PubMed] [Google Scholar]
- 18.Neutra MR, Mantis NJ, Kraehenbuhl JP. Collaboration of epithelial cells with organized mucosal lymphoid tissues. Nat Immunol. 2001;2:1004–1009. doi: 10.1038/ni1101-1004. [DOI] [PubMed] [Google Scholar]
- *19.McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, Newberry RD, Miller MJ. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. Using fluorescence microscopy, the authors demonstrate that goblet cells can form passages from the lumen to the LP, allowing antigen to be passed to CD103+ DC. DCs that acquire fed antigen from goblet cells can then stimulate antigen-specific T cells to divide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoop KA, Miller MJ, Newberry RD. Transepithelial antigen delivery in the small intestine: different paths, different outcomes. Curr Opin Gastroenterol. 2013;29:112–118. doi: 10.1097/MOG.0b013e32835cf1cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes. 2011;2:99–104. doi: 10.4161/gmic.2.2.15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13:21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 27.Sanos SL, Vonarbourg C, Mortha A, Diefenbach A. Control of epithelial cell function by interleukin-22-producing RORgammat+ innate lymphoid cells. Immunology. 2011;132:453–465. doi: 10.1111/j.1365-2567.2011.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- 29.Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, et al. Origin of the Lamina Propria Dendritic Cell Network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL, Berg P-L, Davidsson T, Powrie F, Johansson-Lindbom B, et al. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med. 2008;205:2139–2149. doi: 10.1084/jem.20080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Marquez G, Forster R, Agace WW. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. 2005;202:1063–1073. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall JA, Cannons JL, Grainger JR, Dos Santos LM, Hand TW, Naik S, Wohlfert EA, Chou DB, Oldenhove G, Robinson M, et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity. 2011;34 :435–447. doi: 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu DK, Jimenez-Saiz R, Verschoor CP, Walker TD, Goncharova S, Llop-Guevara A, Shen P, Gordon ME, Barra NG, Bassett JD, et al. Indigenous enteric eosinophils control DCs to initiate a primary Th2 immune response in vivo. J Exp Med. 2014;211:1657–1672. doi: 10.1084/jem.20131800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203:2841–2852. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niess JH, Adler G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J Immunol. 2010;184:2026–2037. doi: 10.4049/jimmunol.0901936. [DOI] [PubMed] [Google Scholar]
- *39.Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral Tolerance Can Be Established via Gap Junction Transfer of Fed Antigens from CX3CR1+ Macrophages to CD103+ Dendritic Cells. Immunity. 40:248–261. This paper demonstrates that CX3CR1+ cells can extend processes into the intestinal lumen to acquire orally administered antigen and can pass this antigen on to migratory DC. When this pathway is blocked, inflammatory responses to oral antigen are increased. [Google Scholar]
- 40.Hadis U, Wahl B, Schulz O, Hardtke-Wolenski M, Schippers A, Wagner N, Muller W, Sparwasser T, Forster R, Pabst O. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monticelli LA, Sonnenberg GF, Artis D. Innate lymphoid cells: critical regulators of allergic inflammation and tissue repair in the lung. Curr Opin Immunol. 2012;24:284–289. doi: 10.1016/j.coi.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanos SL, Diefenbach A. Innate lymphoid cells: from border protection to the initiation of inflammatory diseases. Immunol Cell Biol. 2013;91:215–224. doi: 10.1038/icb.2013.3. [DOI] [PubMed] [Google Scholar]
- 45.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutz S, Eidenschenk C, Ouyang W. IL-22, not simply a Th17 cytokine. Immunol Rev. 2013;252:116–132. doi: 10.1111/imr.12027. [DOI] [PubMed] [Google Scholar]
- *47.Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, Groom JR, Misiak A, Dungan LS, Sutton CE, Streubel G, et al. Retinoic acid expression associates with enhanced IL-22 production by gammadelta T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med. 2013;210:1117–1124. doi: 10.1084/jem.20121588. Most of the work on RA and tolerance focuses on the effects of CD103+ DC. This paper finds a role for RA in regulating innate responses, including improving epithelial barrier function via IL-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **48.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, Merad M. Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science. 2014;343:1477–1487. doi: 10.1126/science.1249288. This paper demonstrates that commensal bacterial stimulation of macrophages drives IL-1β production which in turn promotes ILC3 to produce GM-CSF. This GM-CSF then expands DC populations that can promote Treg induction in the colon. In the absence of this signaling axis, tolerance to fed antigens is significantly impaired. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rakoff-Nahoum S, Medzhitov R. Innate immune recognition of the indigenous microbial flora. Mucosal Immunol. 2008;1:S10–S14. doi: 10.1038/mi.2008.49. [DOI] [PubMed] [Google Scholar]
- 50.Kamdar K, Nguyen V, DePaolo RW. Toll-like receptor signaling and regulation of intestinal immunity. Virulence. 2013;4:207–212. doi: 10.4161/viru.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Crellin NK, Trifari S, Kaplan CD, Satoh-Takayama N, Di Santo JP, Spits H. Regulation of Cytokine Secretion in Human CD127+ LTi-like Innate Lymphoid Cells by Toll-like Receptor 2. Immunity. 2010;33:752–764. doi: 10.1016/j.immuni.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 53.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **54.Han D, Walsh Matthew C, Cejas Pedro J, Dang Nicholas N, Kim Youngmi F, Kim J, Charrier-Hisamuddin L, Chau L, Zhang Q, Bittinger K, et al. Dendritic Cell Expression of the Signaling Molecule TRAF6 Is Critical for Gut Microbiota-Dependent Immune Tolerance. Immunity. 2013;38:1211–1222. doi: 10.1016/j.immuni.2013.05.012. Using conditional ablation of TRAF6 in DC, the authors demonstrate that TLR signaling in DC is required to prevent spontaneous Th2 skewing in the small intestine and promote Treg generation. Administration of antibiotics reverses these effects, indicating commensal bacteria are the source of the TLR ligands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, Luche H, Fehling HJ, Hardt WD, Shakhar G, Jung S. Intestinal Lamina Propria Dendritic Cell Subsets Have Different Origin and Functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 56.Mandal P. Dioxin: a review of its environmental effects and its aryl hydrocarbon receptor biology. J Comp Physiol B. 2005;175:221–230. doi: 10.1007/s00360-005-0483-3. [DOI] [PubMed] [Google Scholar]
- 57.Frericks M, Meissner M, Esser C. Microarray analysis of the AHR system: Tissue-specific flexibility in signal and target genes. Toxicol Appl Pharmacol. 2007;220:320–332. doi: 10.1016/j.taap.2007.01.014. [DOI] [PubMed] [Google Scholar]
- *58.Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36 :92–104. doi: 10.1016/j.immuni.2011.11.011. This paper demonstrates that IL-22 production by ILC and T cells is significantly impaired in the absence of AhR signaling. It is one of the first papers to find a role for AhR in regulating intestinal homeostasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 60.Stockinger B, Meglio PD, Gialitakis M, Duarte JH. The Aryl Hydrocarbon Receptor: Multitasking in the Immune System. Ann Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 61.Bjeldanes LF, Kim JY, Grose KR, Bartholomew JC, Bradfield CA. Aromatic hydrocarbon responsiveness-receptor agonists generated from indole-3-carbinol in vitro and in vivo: comparisons with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Proc Natl Acad Sci U S A. 1991;88:9543–9547. doi: 10.1073/pnas.88.21.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 Innate Lymphoid Cells Inhibit T-Cell-Mediated Intestinal Inflammation through Aryl Hydrocarbon Receptor Signaling and Regulation of Microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schulz VJ, Smit JJ, Willemsen KJ, Fiechter D, Hassing I, Bleumink R, Boon L, van den Berg M, van Duursen MB, Pieters RH. Activation of the aryl hydrocarbon receptor suppresses sensitization in a mouse peanut allergy model. Toxicol Sci. 2011;123:491–500. doi: 10.1093/toxsci/kfr175. [DOI] [PubMed] [Google Scholar]
- 64.Schulz VJ, Smit JJ, Bol-Schoenmakers M, van Duursen MB, van den Berg M, Pieters RH. Activation of the aryl hydrocarbon receptor reduces the number of precursor and effector T cells, but preserves thymic CD4+CD25+Foxp3+ regulatory T cells. Toxicol Lett. 2012;215:100–109. doi: 10.1016/j.toxlet.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 65.Schulz VJ, van Roest M, Bol-Schoenmakers M, van Duursen MBM, van den Berg M, Pieters RHH, Smit JJ. Aryl hydrocarbon receptor activation affects the dendritic cell phenotype and function during allergic sensitization. Immunobiology. 2013;218:1055–1062. doi: 10.1016/j.imbio.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 66.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 68.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganapathy V, Thangaraju M, Gopal E, Martin P, Itagaki S, Miyauchi S, Prasad P. Sodium-coupled Monocarboxylate Transporters in Normal Tissues and in Cancer. The AAPS Journal. 2008;10:193–199. doi: 10.1208/s12248-008-9022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 71.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. The Orphan G Protein-coupled Receptors GPR41 and GPR43 Are Activated by Propionate and Other Short Chain Carboxylic Acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 72.Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, et al. GPR109A Is a G-protein–Coupled Receptor for the Bacterial Fermentation Product Butyrate and Functions as a Tumor Suppressor in Colon. Cancer Research. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *73.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, et al. Activation of gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. The authors demonstrate that butyrate signaling through GPR109a is required for induction of Tregs by LP DC. In the absence of GPR109a, there is increased intestinal inflammation and reduced RA production by DC. Altering the microbiota with antibiotics or feeding a fiber free diet to reduce butyrate production has a similar effect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 75.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 78.Berni Canani R, Nocerino R, Terrin G, Coruzzo A, Cosenza L, Leone L, Troncone R. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: A randomized trial. J Allergy Clin Immunol. 2012;129:580–582. doi: 10.1016/j.jaci.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 79.Berni Canani R, Stefka AT, Khan AA, Nocerino R, Aitoro R, Patton TJ, Calignano A, Meli R, Mattace Raso G, Simeoli R, et al. Lactobacillus rhamnosus GG supplemented formula expands butyrate producing bacteria in cow’s milk allergic infants. doi: 10.1038/ismej.2015.151. under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, Kambayashi T, Larosa DF, Renner ED, Orange JS, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]