SUMMARY
Chaperones are central to the proteostasis network (PN) and safeguard the proteome from misfolding, aggregation and proteotoxicity. We categorized the human chaperome of 332 genes into network communities using function, localization, interactome, and expression datasets. During human brain aging, expression of 32% of the chaperome corresponding to ATP-dependent chaperone machines is repressed, whereas 19.5% corresponding to ATP-independent chaperones and co-chaperones are induced. These repression and induction clusters are enhanced in Alzheimer's, Huntington's, and Parkinson's brains. Functional properties of the chaperome were assessed by perturbation in C. elegans and human cell models expressing Aβ, polyglutamine and Huntingtin. Of 219 C. elegans orthologs, knockdown of sixteen enhanced both Aβ and polyQ-associated toxicity. These correspond to 28 human orthologs, of which 52% and 41% are repressed, respectively, in brain aging and disease, and 37.5% affected Huntingtin aggregation in human cells. These results identify a critical chaperome sub-network that functions in aging and disease.
INTRODUCTION
The proteomes of eukaryotic cells and tissues are represented by a collection of structurally and functionally diverse proteins that form protein-protein interaction networks to communicate within and between cells and tissues to achieve cellular healthspan and organismal lifespan (Gavin et al. 2006). Protein quality control mechanisms such as the proteostasis network (PN) protect proteome functionality and prevent accumulation of mutant, misfolded, and damaged proteins (Balch et al. 2008). Protein aggregation has profound consequences on cellular and organismal health and can cause both gain-of-function and loss-of-function (Park et al. 2013) (Yu et al. 2014).
Protein conformational diseases are widespread and include cancer, metabolic and neurodegenerative disorders (Haass and Selkoe 2007; Powers et al. 2009; Xu et al. 2011). While pathogenic pathways for neurodegenerative diseases such as Alzheimer's (AD), Huntington's (HD), and Parkinson's (PD) intersect (Ehrnhoefer et al. 2011), the clinical profiles and environmental and genetic risk factors vary substantially (Langbehn et al. 2004; Belin and Westerlund 2008; Hampel et al. 2010). For neurodegenerative diseases, the most significant and universal risk factor is aging; moreover, evidence suggests a mechanistic link between aging, aggregation-mediated proteotoxicity, and loss of proteostasis, which has been put forth as one of the nine hallmarks of aging (Cohen et al. 2006; Lopez-Otin et al. 2013). The accumulation of proteotoxic species during aging is inversely correlated with age-associated proteostasis decline (Ben-Zvi et al. 2009). Chronic expression of misfolded proteins in age-onset neurodegenerative disease leads to accumulation of misfolded species and aggregates that overwhelm proteostasis, and a basis of cellular dysfunction (Gidalevitz et al. 2006; Douglas and Dillin 2010).
A central component of the PN are molecular chaperones and co-chaperones that determine the cellular folding environment, prevent misfolding, and re-direct non-native intermediates to the native state (Hartl et al. 2011) or for clearance by the ubiquitinproteasome system (UPS) and autophagy (Schmidt and Finley 2014). The ‘chaperome’ corresponds to the ensemble of chaperones and co-chaperones that interact in a complex network of molecular folding machines to regulate proteome function (Albanese et al. 2006). An understanding of the chaperome will be instrumental to the biology of aging and how loss of proteostatic control increases the risk for protein conformational diseases. While much is known about the function of individual chaperones (Hartl et al. 2011), there is only a limited analysis of chaperome dynamics and connectivity in metazoans. We therefore compiled the human and C. elegans chaperome by a systematic literature search, as a basis for integration of human protein-protein interactions (PPIs) and aging brain expression data to achieve a chaperome interactome network. This was complemented by functional chaperome-wide RNAi screens in C. elegans models of Aβ and polyQ proteotoxicity and a human cellular model of Huntingtin aggregation.
Our study has identified chaperones clusters that exhibit striking repression and induction expression patterns in human brain aging. Repression predominates and involves all major families of cytosolic chaperones with a preponderance of ATP-dependent chaperones. We observed concordance of these dynamics with expression in brain tissues of AD, HD and PD patients. The correlation of these dynamics underlines the central role of the chaperome in aging and disease. The complement of informatics with experimentation identified a chaperome sub-network that safeguards cellular and organismal proteostasis in C. elegans models and human tissue culture cells expressing neurodegenerative disease-related misfolded proteins. This emergence of a conserved chaperome sub-network provides a resource for future studies to establish how changes in the PN affect aging and disease.
RESULTS
Composition of the Human Chaperome
We examined the expression of genes encoding molecular chaperones in human brains during normal aging and in neurodegenerative disease. For this, we compiled a list of all human chaperones and co-chaperones by combining the extensive literature on the biochemical properties of molecular chaperones together with curation and structural genomics profiling to match genes by InterPro protein domain identifiers (IPR-IDs) (Hunter et al. 2012) (Figure 1A, Table S1). This analysis identified 332 genes (Figure 1A, Tables S1, S2A, and Supplemental Experimental Procedures) that were unambiguously placed into nine chaperone gene families corresponding to HSP90, HSP70, HSP60, HSP40, Prefoldin, small HSPs (sHSPs), TPR-domain containing (Hartl and Hayer-Hartl 2002), and organellar-specific chaperones of the endoplasmic reticulum (ER) (Kleizen and Braakman 2004) and mitochondria (MITO) (Tatsuta et al. 2005). For genes with matching IPR-criteria domains, the groupings were prioritized by chaperone properties rather than localization. The organellar categorization of ER or MITO –specific represents chaperones for which the biochemical, genetic, and cell biological evidence supports both chaperone function and organellar-specific localization, and for which no IPR-domain match could be obtained.
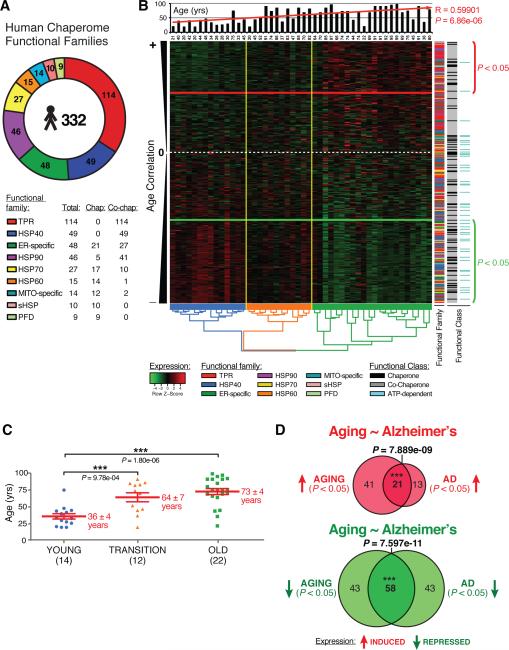
Figure 1. Differential Chaperome Responses in Human Brain Aging and Neurodegenerative Disease.
A The Human chaperome. Functional families and number of members are indicated. B. Heat map showing 318 chaperones expressed in human brain (Super Frontal Gyrus) ordered by decreasing age-correlation. The white dashed-line indicates age-expression correlation coefficient closest to zero. Genes above the red line are induced (P < 0.05), genes below the green line are repressed (P < 0.05). The histogram visualizes specimen age upon hierarchical clustering. The dendrogram visualizes hierarchical clustering of brain specimens. The y-axis color code highlights from left to right the nine chaperome families, chaperones (black), co-chaperone (grey) and ATP-dependent chaperones (turquoise). C. Three major age groups (“young” - blue, “transition” - orange, “old” - green) are visualized by dendrogram coloring in B. Values are mean age +/− SEM. *** P < 0.001, Student's t-test. D. Overlaps of chaperones induced (red) or repressed (green) in aging vs. AD. (See Figure S1, S2, S3).
Of the 332 genes that comprise the human chaperome, 88 are functionally classified as chaperones and 244 are co-chaperones. Among the 88 chaperones are 50 ATP-dependent chaperone genes and 38 ATP-independent chaperones. The ATP-dependent chaperones are comprised of the 5 Hsp90s, 17 Hsp70s, 14 HSP60s, 6 ER-specific and 8 MITO-specific Hsp100/AAA+ ATPases, respectively. In the HSP70 family are the 17 holding and folding ATP-dependent HSP70s and 10 ATP-independent co-chaperones, corresponding to the nucleotide exchange factors (NEFs) BAG1-6, the GrpE NEFs GRPE1-2, SIL1 (Hsp110/BAP) and HSPBP1 (Mayer 2013). Likewise, the HSP90 family is comprised of 46 members including the 5 ATP-dependent Hsp90 chaperones (Pearl and Prodromou 2006) and 41 ATP-independent co-chaperones that regulate HSP90 including the 2 AHA1 cochaperones (AHSA1-2), CDC37, CDC37L1, p23, and 36 immunophilins sub-classified into 18 cyclophilins, and 18 FKBPs. The 49 HSP40s correspond to a separate family of ATP-independent co-chaperones that function as holding chaperones and HSP70 co-chaperones. Taken together, the HSP90, HSP70 and HSP60 chaperone systems correspond to 26.5% of the chaperome, with the HSP40s contributing another 15% to the chaperome.
Among the 38 ATP-independent chaperone genes are 10 sHSPs, 9 Prefoldins, 4 MITO-specific and 15 ER-specific chaperones. Another 19% of the chaperome are represented by the 48 ER-specific and 14 MITO-specific chaperones (Figure 1A, Table S2A) including 26 ER-specific oxidoreductases and 2 MITO-specific PDI-type thioredoxins. Notably, the largest chaperome subclass are the 114 TPR domain-containing genes, representing 34% of the chaperome. These include STIP1 (HOP) that functions as an HSP70 and HSP90 co-chaperone (Prodromou et al. 1999; Song and Masison 2005), and contains three TPR-classifier IPR-domains (IPR001440, IPR013026, IPR019734). Although many of these newly identified members of the TPR-domain family have not been yet shown to function as HSP70 and HSP90 co-chaperones, we opted for a comprehensive and inclusive approach. Some of these novel members have been recently validated in C. elegans using biochemical assays (Haslbeck et al. 2013). This organization into nine functional families lends itself to a systems-level evaluation of chaperome functionality in aging and disease.
Differential Chaperome Dynamics in Human Brain Aging
We examined the dynamics of the human chaperome using gene expression data from the Superior Frontal Gyrus (SFG) of 48 brains from neuropathologically and neurologically normal control individuals of 20 to 99 years (Berchtold et al. 2008; Loerch et al. 2008). Analysis of the expression patterns of the human chaperome (Figure 1B) revealed a profile that correlated with aging (P = 6.86e-06) and clustered into two groups with a mean age of 36 +/− 4 years and 73 +/− 4 years (Figure 1B, C). These age groups are highly pronounced upon hierarchical clustering, but are discernable even when ordered by chronological age (Figure S1A).
Aging correlation analysis of the human chaperome expressed in the SFG identified 101 chaperone genes (31.8%) that are repressed (corrage < 0 and P < 0.05) and 62 (19.5%) genes are induced during aging (corrage > 0 and P < 0.05) (Figure 1B, Table S3). Chaperome age-expression revealed enrichment of certain functional families in induction and repression clusters (Figure 1B, Figure S3A-D). TPR proteins tend to be induced, while HSP40s are repressed (Figure 1B). The significance of the repression and induction clusters is further supported by analysis of an independent dataset from frontal lobes of individuals of 24 to 40 years and 70 to 94 years (Loerch et al. 2008) (Figure S1D). Clustering identified isochronal aging repression and induction clusters with a significant boundary between the two age groups (Figure 1B, C, Figure S1A, B). Analysis of both datasets revealed that the chaperome genes repressed and induced in both datasets of normal brain aging overlapped significantly (P < 2.2e-16 and P = 1.7e-12, respectively) (Figure S1E, F). We observed similar age clusters with significant boundaries in Entorhinal Cortex (ENC) (Figure S1C), confirming chaperone expression dynamics in brain aging in different datasets.
Concordant Dynamics of Chaperome Expression in Brain Aging and Neurodegenerative Diseases
Since aging represents a risk factor for neurodegenerative disease we assessed the impact of AD, HD, and PD on chaperome dynamics by examining expression in patient brain samples (Hodges et al. 2006; Moran et al. 2006; Liang et al. 2008) (Table S3C-E). When analyzing brain expression datasets from AD patients, we identified 101 significantly repressed genes and 34 induced genes compared to age-matched controls. Both in aging and AD brains, chaperome repression is significantly enriched compared to overall gene repression in the genome (Figure S2A, B). We then asked whether the chaperome genes differentially regulated in AD overlapped with the aging-regulated chaperome genes, and identified 21 genes that overlapped between the 62 genes induced in aging and 34 genes induced in AD (P = 7.89e-09), and 58 genes that overlapped between 101 aging-repressed and 101 AD-repressed chaperone genes (P = 7.6e-11) (Figure 1D). Among the genes that are repressed in both aging and AD, the HSP70-HSP40 system corresponds to 36% of the 58 genes (Table S3D). Chaperome genes consistently repressed in aging and AD include members of all nine functional chaperome families, revealing that alterations in chaperone expression is not selective to specific gene families. The 21 genes induced in both aging and AD extend across all functional families except HSP60 genes. Similarly, we also examined whether the chaperome genes regulated in aging overlapped with those dynamically regulated in HD and PD. Among the 245 chaperome genes detected in all datasets, 36 genes overlapped between genes repressed in aging and HD (P = 3.96e-07) and 24 genes between aging and PD (P = 0.01062) (Figure S2C, E). Chaperome genes induced in aging also significantly overlapped with genes induced in HD (P = 2.94e-10) and PD (P = 0.00059) (Figure S2D, F) (Table S3).
We partitioned the chaperome age-expression distribution into the nine families and observed reproducible expression patterns in four distinct brain tissues (Figure S3A-D). Ranked by decreasing median aging-correlation, the induction of sHSPs and TPR genes consistently ranked high, and the HSP60s, HSP40s, and HSP70s were consistently repressed. Among repressed genes, the HSP40s exhibited significant change (P = 0.04875) with 62% of 48 HSP40 genes repressed in aging (P < 0.05), and 51% repressed in AD. Of these, 41% are repressed in both aging (SFG) and AD (P = 0.0009). Among the genes that are induced in brain aging and disease are sHSPs and TPR-containing chaperone genes (Figure S3A-D).
The distribution of ATP-dependent chaperones in aging and AD was shown to be enriched 10-fold in the repression cluster of aging SFG (P = 4.8e-06) and 11-fold in the Prefrontal Cortex (PFC) (P = 1.7e-07) (Figure S3E) whereas ATP-independent chaperones exhibited equivalent levels of repression and induction. This analysis of the chaperome in human brain aging also reveals concordant exacerbation of responses in neurodegenerative disease. These concordant chaperome changes provide evidence for significant changes in the proteostasis network in aging and neurodegenerative disease.
Chaperome Network Community Dynamics in Brain Aging and Disease
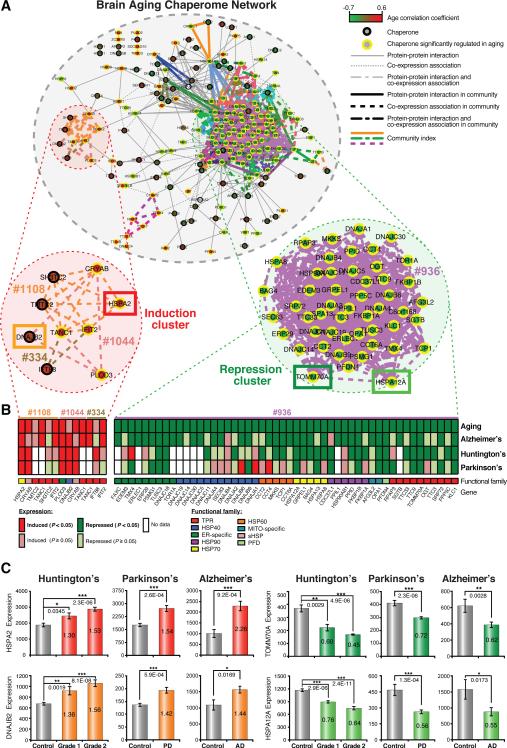
The repression and induction of chaperones in brain aging likely affects the cellular balance of chaperone machines and functionality of the PN, leading us to consider how these components of the chaperome are physically and functionally associated. We visualized the systems-level connectivity of the chaperome by integrating physical protein-protein interactions (PPI edges) from public databases together with co-expression pairs in aging brains (COX edges) (Tables S4, S5) into a PPI-COX interactome (Figure 2A). The COX edges were identified using transcriptome data from Superior Frontal Gyrus (SFG) tissue given its highly significant and pronounced aging dynamics. Among all 50,403 chaperome pairs, we found 1,193 significant COX edges (corrage > 0.8 and P < 0.05), of which fifteen of 191 unique PPIs are also COX edges. Applying the link-community clustering algorithm (Ahn et al. 2010) to the PPI-COX network, we identified 40 link communities, of which 34 are repression communities compared to only 6 induction communities (Figure 2A, Table S6).
Figure 2. Chaperome Network Community Dynamics in Brain Aging and Age-Onset Neurodegenerative Diseases.
A. Integrated human chaperome network based on physical protein-protein interaction (PPI) and co-expression (COX) edges and link communities of chaperones concordantly induced or repressed during brain aging. PPIs (solid edges). COX > 0.8 (dashed edges). Color-scale indicates positive to negative correlation between age and gene expression. Link communities highlighted by edge color, yellow node borders indicate significant age expression correlation (P < 0.05). B. Chaperones and co-chaperones selected from induction and repression communities and their expression in brains from AD, HD and PD patients. (*P < 0.05, ** P < 0.01, *** P < 0.001, Student's t test). C. Heat maps visualize induction or repression in aging, AD, HD and PD at node resolution for communities highlighted in A. Community numbers and gene names are indicated. Color code micrographic visualizes functional family. Significantly induced and repressed genes (P < 0.05) shown in dark red and green, respectively and non-significantly induced and repressed genes with P ≥ 0.05 are shown in light red and light green. (See Figure S2, S3).
This network community clustering analysis also revealed that the majority of genes residing in induction or repression link-communities, showed concordant patterns in aging and neurodegenerative diseases, respectively (Figure 2A, B), leading to the hypothesis that these aging expression changes can affect the cellular balance of chaperone machines and functionality of the proteostasis network (PN). For example, concordantly aggravated expression patterns for the aging-induced genes HSPA2 (HSP70) and DNAJB2 (HSP40) and the aging-repressed HSPA12A (HSP70) and TOMM70A (TPR) were observed in brain biopsies from AD, HD and PD patients (Figure 2C). In terms of gene expression dynamics in aging and disease these exemplary genes are representative for induction and repression cluster dynamics. Included in the repression cluster are members across eight of nine chaperone gene families with predominance of ATP-dependent chaperones and HSP40 cochaperones, while induction communities are enriched for TPR-domain co-chaperones and sHSPs. These observations provide support that prominent changes in chaperone expression levels could accelerate AD, HD or PD disease pathology characterized by elevated toxic gain-of-function aggregation. Chaperome repression communities dominate the network landscape in human brain aging and disease, linking known chaperones and co-chaperones with less well characterized chaperones, possibly revealing novel interactions important for the aging and disease PN.
Chaperome Function in C. elegans Models of Protein Aggregation
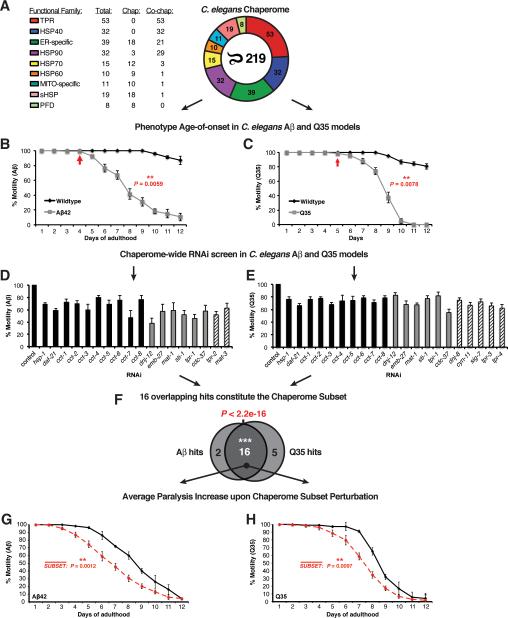
To complement the bioinformatics analysis of the human chaperome with in vivo functional data, we experimentally validated the functional consequences of repressed chaperome gene expression and function during aging and disease using two established C. elegans models expressing the cytotoxic aggregation-prone proteins, Aβ (Aβ42) and polyQ (Q35), implicated in AD and polyglutamine (HD) diseases, respectively (Link 1995; Satyal et al. 2000; Morley et al. 2002). In both models, aggregation and toxicity, measured by decreased motility, increases in an age-dependent manner and can be suppressed by lifespan-enhancing pathways such as the insulin-like signalling pathway (daf-2, age-1) and the heat shock response (hsf-1) (Morley et al. 2002; Cohen et al. 2006). We assembled and curated the C. elegans chaperome by orthology mapping and manual curation (Harris et al. 2010; Sayers et al. 2012) (see Supplemental Methods), and identified 219 C. elegans chaperone and co-chaperone genes corresponding to the same nine functional gene families (Table S2B). Comparison of C. elegans and human chaperomes revealed similar proportions of most functional families, with notably a reduction in TPR domain co-chaperones to 24% of the C. elegans chaperome from 34% in the human chaperome, and increased number of sHSPs to 19 genes in C. elegans from 10 sHSP genes in humans (Figure 3A).
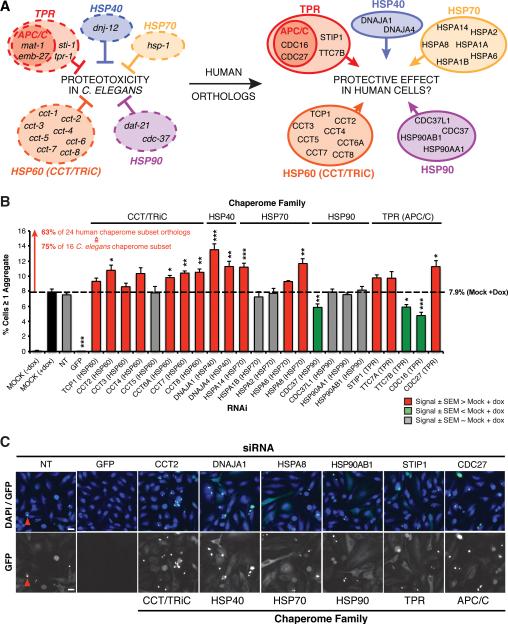
Figure 3. Functional Chaperome Perturbation Analyses in C.elegans Models of Protein Misfolding.
A. The C. elegans Chaperome with its functional families and numbers of members per family are shown. B. Paralysis (% motility) for wild type and Aβ42 expressing C. elegans from day 1 to day 12 of adulthood. Arrow indicates paralysis age-of-onset. C. Motility defects for wild type and Q35-expressing C. elegans from day 1 to day 12. Arrow indicates age-of-onset. D. RNAi paralysis phenotypes on day 4 of adulthood (% motility) for C. elegans expressing Aβ42 (Mean ± SEM, n=3 and n ≥ 25 animals/trial). E. RNAi motility defects on day 2 of adulthood (% motility) for C. elegans expressing Q35 (Mean ± SEM, n=3 and n ≥ 25 animals/trial). F. Venn diagram indicating significant overlap of 16 hits from both screens (P < 2.2e-16, Fisher's exact test). G. Average paralysis (% motility) for RNAi of all 16 chaperome subset genes in Aβ42 expressing C. elegans throughout adulthood (days 1 to 12). Data points are means of corresponding data points in each RNAi experiment, each based on n ≥ 3 independent experiments and n ≥ 25 animals/trial. ** P < 0.01, Student's t-test. H. Average paralysis (% motility) for chaperome subset RNAi in Q35 expressing C. elegans throughout adulthood (days 1 to 12). Data points as in G. (See Figure S4).
We first examined the functional requirements of all 219 C. elegans chaperome genes using RNAi knockdown in animals expressing Aβ42 by monitoring early-onset paralysis on day 4 of adulthood (Figure 3B). Based on our methodology of screening, assessing age-dependent aggregation and toxicity phenotypes (measured by decreased motility), genes when knocked down that caused motility defects in both wild type and disease models were eliminated; in other words, genes that gave the same magnitude of phenotypic change in wild type animals were not further considered. This functional screen identified 18 genes (Figure 3D), corresponding to 10 ATP-dependent chaperones (HSC70 hsp-1, HSP90 daf-21, and eight subunits of the CCT/TRiC chaperonin complex), the co-chaperones, HSP40 dnj-12, cdc-37, and the TPR-domain protein,STI1 that upon knockdown significantly enhanced Aβ42 proteotoxicity (Figure 3D). Included among these modifiers were four TPR-domain proteins, the anaphase promoting complex (APC/C) subunits mat-1, mat-3 and emb-27, and the uncharacterized ORFs, Y39A3CR.3 (tpr-1) and Y57G7A.10 (tpr-2) conserved in human as TTC7A/TTC7B and EMC2, with a phenotype on the Aβ42 animals that were not previously implicated in proteostasis.
To test whether the chaperone genes that regulate Aβ42 proteotoxicity have more general effects, we performed a subsequent RNAi screen using a C. elegans model for expression of aggregation-prone polyQ (Q35), associated with HD pathogenesis. This model of chronic Q35 protein expression and aggregation exhibits an early onset age-dependent decline in muscle function (Figure 3C) (Morley et al. 2002). Knockdown of 21 of the 219 C. elegans chaperome genes significantly enhanced polyQ-dependent proteotoxicity (Figure 3E). These correspond to the same ATP-dependent chaperones (daf-21, hsp-1 and the eight CCT/TRiC complex subunits) that overlapped with the Aβ42 screen, and 11 co-chaperones of which six genes were identified in both screens, including the TPR-domain APC/C subunits mat-1 and emb-27 and the TPR protein, Y39A3CR.3 (tpr-1) (Figure 3D, E). These results reveal an overlapping common subset of 16 chaperome genes including the newly identified members of the APC/C (P < 2.2e-16, Fisher's exact test) (Figure 3D, E, F). Additionally, these screens also identified seven chaperones with more selective phenotypes on either Aβ42 or Q35 expressing animals.
To investigate whether the common chaperome subset also affects age-related proteotoxicity, we assessed the functionality of C. elegans muscle throughout adulthood (days 1 to 12) following RNAi knockdown of all 16 subset genes in both models. All 16 genes were highly protective in the Aβ42 (P = 0.0012) and Q35 models (P = 0.0097) and showed significant reduction in motility upon RNAi suggesting that repression of the chaperome subset renders aged organisms more susceptible to proteotoxicity. (Figure 3G, H, S4).
A Chaperome Subset Safeguards Proteostasis in Ageing C. elegans
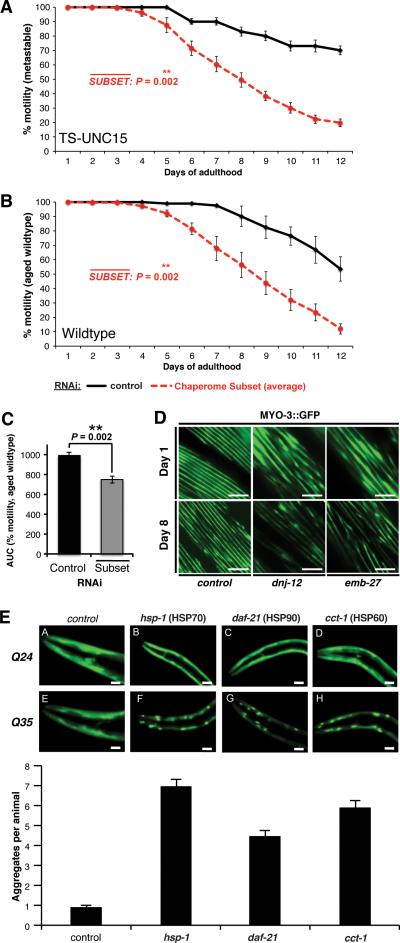
Our network analysis of chaperome repression demonstrated concordant changes in brain aging and neurodegenerative disease (Figure 1D, Figure S2). This led us to speculate whether expression of these chaperones also change during aging, thus affecting the susceptibility of Aβ42 and Q35 expressing animals to proteotoxic insult (Figure 3G, H). To address this, we monitored the age-dependent changes in motility of C. elegans that express a metastable temperature-sensitive mutation in paramyosin (UNC-15TS) that misfolds in early adulthood at the permissive temperature (Gidalevitz et al. 2006). At the permissive temperature, knockdown of the chaperome subset (Figure 4A, S4C) substantially exacerbated the loss of muscle cell function during aging of UNC-15TS animals, providing direct functional evidence for a protective role in aging.
Figure 4. A Chaperome Subset Safeguards C. elegans Proteostasis against Aging-Related Proteotoxicity.
A. Average paralysis (% motility) for chaperome subset RNAi in the unc-15(e1402) TS-strain at 15°C throughout adulthood (days 1 to 12). Data points are means of corresponding data points in each RNAi experiment, based on n ≥ 3 experiments and n ≥ 20 animals/trial. ** P < 0.01, Student's t-test. B. Average paralysis (% motility) in aged wild type animals throughout adulthood (days 1 to 12). Data points as in A. C. Early-onset paralysis (% motility) upon chaperome subset RNAi in aged wild type animals (compare B.) as area under the curve (AUC) for control aging wild type worms (n=5) and average of RNAi-treated aging wild type worms (n=3). ** P < 0.01, Student's t-test. D. Sarcopenia phenotype upon control, dnj-12 and emb-27 RNAi visualized by MYO3::GFP fluorescence at day 1 vs. day 8 of adulthood. Scale bar, 10μm E. Early-onset aggregation of polyQ (Q35) expressed in body wall muscle cells upon chaperome subset RNAi. Q25, sub-threshold polyQ. Graph shows increased aggregate count upon RNAi compared to control (Mean ± SEM, n ≥ 100 animals, n = 3, scale bar: 0.02mm). (See Figure S4).
To further characterize the chaperome subset during aging, we monitored various physiological phenotypes in wild type and RNAi treated animals and showed that this subset of the chaperome are important to prevent or delay age-dependent paralysis (Figure 4B, C, S4D). Knockdown of daf-21 (Hsp90) or hsp-1 (HSC70) led to increased paralysis in 45% and 44% of day 6 animals, respectively, and knockdown of TPR co-chaperones tpr-1 and dnj-12 resulted in 70% impairment (Figure S4D). The loss of motility in wild type animals was due to deterioration of myofilament structure (sarcopenia). We monitored the integrity of MYO-3, a heavy chain component of myofilaments by MYO-3::GFP fluorescence, and upon knockdown of dnj-12 and emb-27 observed more severe sarcopenia at day 8 compared to day 1, while structural integrity of muscle was maintained up to day 8 in controls (Figure 4D). Consistent with these observations, RNAi of cct-1, hsp-1, and daf-21 leads to premature Q35 aggregation (Figure 4E), recapitulating the misfolding of disease associated polyQ proteins, a hallmark of HD. Our evidence at cellular and organismal level suggests that the chaperome subset safeguards proteostasis during aging.
The Chaperome Subset Safeguards Proteostasis in Human Cells
To test whether the chaperome subset identified in C. elegans models of Aβ and Q35 proteotoxicity functionally extends to the human chaperome, and to test its potential contribution to HD, we used a high-content imaging assay to quantify aggregation of doxycyclin-inducible Huntingtin-exon1(Q78)-GFP (Htt-GFP) expressed in HeLa cells. The 16 subset C. elegans chaperones and co-chaperones correspond to 28 genes in the human genome, of which 24 genes are expressed in HeLa cells (Nagaraj et al. 2011) (Figure 5A). We confirmed siRNA knockdown efficiency of HTT-GFP, CCT2, DNAJA1 and HSPA8 by immunoblot analysis to show reduced levels of these chaperones (Figure S5A). An increase in the fraction of Htt-GFP expressing cells with protein aggregates was observed upon knockdown of 15 human chaperones compared to control, corresponding to 63% of the human chaperome subset, and 75% of the worm chaperome subset, respectively (Figure 5B, Table S7). These included all subunits of the CCT/TRiC complex (except CCT5), HSP40 and HSP70 family members DNAJA1 (HDJ-2), DNAJA4, HSPA8 (HSC70) and HSPA14 (Figure 5B, C), the TPR-domain APC/C subunits CDC23 and CDC27 that, upon knockdown, led to significantly elevated aggregation (Figure S5B). The overall protective effect of the chaperome subset in human cells against Htt-GFP aggregation provides additional functional evidence to support the results from the C. elegans RNAi analysis of the chaperome subset in proteotoxicity.
Figure 5. Human Chaperome Sub-Network Safeguards Proteostasis against Huntingtin aggregation.
A. 16 chaperome subset members identified by RNAi screens in Aβ42 and Q35 C. elegans models grouped by functional family. APC/C, anaphase-promoting complex/cyclosome and the corresponding 24 human chaperome subset members identified by orthology mapping. B. Percentage of HeLa cells with ≥ one Huntingtin-exon1(Q78)-GFP (HTT-GFP) aggregate and upon siRNA. Results shown for all 24 human orthologs expressed in HeLa, corresponding to the 16 C. elegans chaperome subset members (mean ± SEM, n=6). Red, grey and green bars represent increased, unchanged and decreased aggregation, respectively measured as “%Cells ≥ 1 aggregate”. * P < 0.05, ** P < 0.01, *** P < 0.001, Student's t-test. C. Representative images for non-targeting (NT) siRNA, siRNA-GFP (positive control) and siRNA against each one member of the chaperome subset functional families are shown. Red arrows exemplify HTT-GFP aggregates. Scale bar, 20μm. (See Figure S5).
The Chaperome Sub-Network is Repressed in Brain Aging and Disease
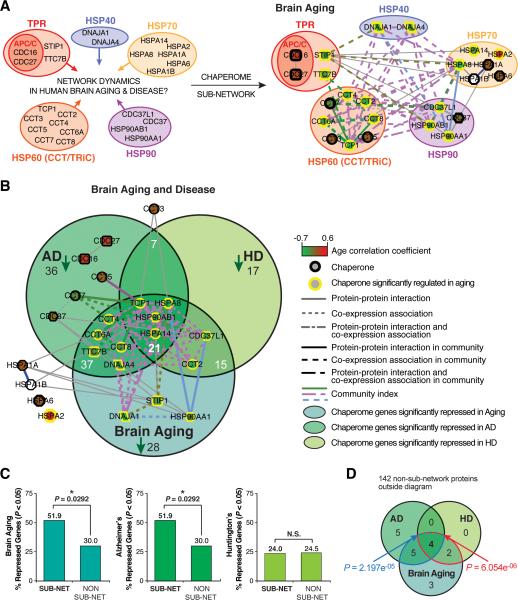
While broad chaperome repression in aging human brains pointed towards proteostasis functional decline and increased susceptibility to proteotoxic insults in neurodegenerative diseases, our functional screens in C. elegans led to the identification of a common chaperome subset. To investigate the dynamics of the corresponding chaperome subset in human brain aging and disease, we examined the connectivity of the chaperome interactome and partitioning across communities of induction and repression. We matched orthologous human chaperome subset nodes in the PPI-COX chaperome interactome to visualize their expression dynamics and interactome topology (Figure 6A). Of the 28 human nodes, 27 genes are expressed in the human brain (Berchtold et al 2008), of which the majority (20 genes) are inter-connected in a major component involving 60 edges. This human chaperome sub-network is significantly interconnected (P < 0.01) (Figure 6A, Figure S6A-C). The sub-network nodes include five chaperone families and have a higher degree of connectivity than non-sub-network nodes (P = 0.04) (Figure S6D). Overall, 52% of subnetwork nodes are repressed in aging brain compared to 30% repression of non-sub-network nodes (P = 0.0292, Fisher's exact test) (Figure 6C). Likewise 52% of sub-network nodes are significantly repressed in AD compared to 30% repression of non-sub-network nodes (P = 0.0292, Fisher's exact test), 24% in HD and 16% are repressed in both diseases (Figure 6C). The chaperome sub-network is nearly 2-fold more repressed in brain aging and AD as opposed to non-sub-network chaperome genes. We further tested the significance of the overlaps of sub-network genes repressed in all conditions, aging, AD, and HD as compared to non-sub-network genes asking whether the subset of chaperome sub-network genes repressed in both AD and HD is significantly enriched. Four genes that are significantly repressed both in AD and HD (HSP90AB1, HSPA8, HSPA14 and TCP1) are also repressed in aging (Figure 6B). We applied a Fisher exact test considering all chaperome genes significantly repressed in at least one condition, AD, HD or aging, which includes 19 chaperome sub-network and 142 non-sub-network chaperome genes. The significance of the observation that a set of 4 genes repressed in AD and HD is also repressed in aging (P < 10e-05, Fisher's exact test, Figure 6D) demonstrates the power of integrating data from more than one disease and aging. Some of the sub-network chaperome nodes that were not significantly repressed in aging are repressed in AD, including the APC/C complex TPR-domain subunits CDC16 and CDC27. These results reveal that the chaperome sub-network is significantly more interconnected in the human interactome and concordantly more repressed in brain aging. The significantly enriched fraction of aging-repressed sub-network chaperome nodes versus non-sub-network nodes, and their aggravated repression in aging and disease is in agreement with the chaperome sub-network phenotypes observed in our RNAi functional perturbation experiments.
Figure 6. The Human Chaperome Sub-Network is Repressed in Aging and Disease.
A. Extraction of the human orthologous chaperome sub-network from the chaperome interactome shown in Figure 2A, to highlight chaperome sub-network dynamics in human aging brain and neurodegenerative disease. Nodes, edges, shapes and edge strengths as in Figure 2A. B. Human chaperome sub-network superimposed on Venn overlaps of chaperome genes significantly repressed in aging, AD, and HD. C. Graphs show % sub-network (sub-net) vs % non-sub-network (non sub-net) chaperome nodes repressed in human aging brain (SFG) and brains from Alzheimer's (SFG) and Huntington's disease (PFC) patients (See Figure S6). SFG = Superior Frontal Gyrus, PFC = Prefrontal Cortex. D. Venn diagram of overlaps of chaperome sub-network genes significantly repressed in both AD and HD as well as in aging. P values are based on Fisher's exact test, considering only chaperones that are significantly repressed in at least one of the three conditions, aging, AD or HD. The blue overlap area and P value indicate the significance of the overlap of subnetwork genes repressed in all three conditions against the union of sub-network genes repressed in ‘aging only’ and in ‘aging and AD’, the red overlap area and P value indicate the significance of the overlap of sub-network genes repressed in all three conditions against the union of sub-network genes repressed in ‘aging only’ and in ‘aging and HD’.
Thus, our orthogonally integrated chaperome-scale approach successfully identified a small subset of chaperones and specific co-chaperones from hundreds of chaperone and co-chaperone factors, including well-established and novel regulators of proteostasis maintenance in aging and disease that are highly interconnected in a chaperome sub-network.
DISCUSSION
Molecular chaperones, being amongst the most highly conserved genes with essential functions for protein biogenesis might have been thought to be equally important for proteome maintenance. Our analysis of chaperome dynamics combines the analysis of expression in human brain and neurodegenerative disease, experimental validation using C. elegans and human cells expressing disease-associated aggregates, and protein-protein interaction network analysis, from which we have identified a chaperome sub-network affected in aging and disease. This study has identified a subset of the chaperome that is critical to maintain proteostasis in aging and upon challenge with neurodegenerative disease-associated proteins. This combined approach led to identification of a conserved subset of 16 genes in C. elegans, comprised of the ATP-dependent chaperones HSC70, HSP90, the CCT/TRiC complex, select HSP40 and TPR-domain co-chaperones that exhibit altered expression during human brain aging and are functionally required in C. elegans models to prevent proteotoxicity of neurodegenerative disease associated proteins. Of the human chaperome of 332 genes, 32% are repressed in brain aging corresponding mostly to ATP-dependent chaperone machines involved in holding intermediates and folding to the native states, and is represented by highly connected repression clusters that concordantly decline in brains of AD, PD, and HD patients. We propose that changes in expression of the sub-network may signify early events leading to age-associated proteostasis collapse with implications for the pathogenesis of neurodegenerative disease.
The composition of the chaperome sub-network highlights the central importance of specific ATP-dependent cytoplasmic chaperone machines during aging from C. elegans to human essential to achieve a healthy tissue proteostatic state that can withstand challenge by expression of cytoplasmic polyQ expansion proteins or Aβ. Some of these chaperones such as the HSP70-HSP40 machine have been previously implicated in the modulation of polyQ-expanded protein aggregation (Cummings et al. 1998; Jana et al. 2000). Likewise, the level of cytosolic chaperonin affects polyQ toxicity by actively modulating the aggregation state, a major cause of polyQ cytotoxicity in Huntington's disease (Nollen et al. 2004; Behrends et al. 2006; Kitamura et al. 2006; Tam et al. 2006). Differences are revealed that may point towards aging or disease-specific sub-network modules or contribution of only parts of the sub-network to pathways involved in molecular pathology of these diseases. Notably, Hsp90 resides at the intersection of aging, AD and HD. The lower overall number of sub-network vs. non-sub-network genes repressed in HD compared to those repressed in AD could correspond to differences in the molecular underpinnings of these diseases, while the 6 subnetwork genes significantly repressed in HD are also significantly repressed in aging, including Hsp90, Hsp70 and chaperonin (TCP1), representative of the core cytoplasmic molecular folding machineries. In addition to identification of the major ATP-dependent chaperones in our functional screens, we show that components of the anaphase-promoting complex (APC), an E3 ubiquitin ligase that targets proteins for degradation are highly effective modifiers of Aβ and polyQ proteotoxicity phenotypes. Despite its established role in the exit from mitosis it has been shown that the APC is functional in post-mitotic neurons (Gieffers et al. 1999; Kim et al. 2009; Marrocco et al. 2009) with a role in cognitive processes (Kuczera et al. 2011). Further evidence suggests that deregulated APC function are associated with neurodegeneration and cognitive decline (Almeida et al. 2005; Li et al. 2008; Maestre et al. 2008) and a screen in yeast identified the APC as modifier of polyQ toxicity (Bocharova et al. 2008). Considering existing evidence, we propose that these APC components represent novel proteostasis modifiers with a role in degradation through their E3 ubiquitin ligase function, while mechanistically it is possible that they may function as co-chaperones. Previous studies using RNAi in C. elegans have identified components acting in protein degradation as modifiers of polyQ proteotoxicity (Nollen et al. 2004).
The largest chaperome gene class are the 114 TPR domain-containing proteins corresponding to 53 orthologs in C. elegans, accommodating, in an unbiased way, the large degree of functional heterogeneity of TPR-domain proteins in chaperone-related function. Four C. elegans proteins contribute to the chaperome sub-network (sti-1, mat-1, emb-27 and tpr-1) and another four were identified to affect proteostasis in the Aβ (tpr-2 and mat-3) and polyQ (tpr-3 and tpr-4) models, of which none except sti-1 had been previously described to have a role in proteostasis. The TPR family contains the largest number of novel members that have not yet been shown to function as HSP90 and HSP70 co-chaperones, although a recent biochemical analysis in C. elegans identified 13 TPR proteins that interact with HSP70 or HSP90 (Haslbeck et al. 2013). Based on our inclusive and un-biased chaperome-scale in vivo results from two independent screens, we identified additional TPR-domain proteins to be functionally equivalent to HSP90 or HSC70, leading us to propose that these factors harbor properties that are highly linked to proteostasis network maintenance and concordantly regulated with global chaperome dynamics in aging and disease.
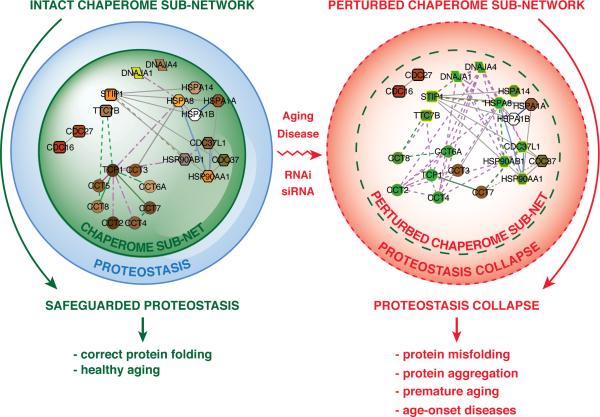
Previous efforts to characterize chaperone networks have proposed sub-networks that participate in the folding of newly synthesized proteins and those that interact with preexisting proteins that become denatured upon acute stress (Albanese et al, 2006). Our network analysis combines chaperome expression analysis in aging brains with community network clustering to organize the connectivity dynamics of the human chaperome network and provides a three-dimensional representation of the chaperome sub-network within the context of the complete chaperome interactome. The sub-network nodes are more interconnected and repressed than non-sub-network chaperome nodes, and suggests that perturbation of a sub-network node and its edges would result in a more severe perturbation of network integrity compared to the more peripheral low-degree non-sub-network nodes. Accordingly, loss of protective chaperome sub-network buffering capacity compromises proteostasis capacity, leading to an increased sensitivity to endogenous and exogenous stress and disease predisposition (Figure 7).
Figure 7. The Chaperome Sub-Network as Proteostasis Safeguard and its Collapse in Disease.
A. Chaperome sub-network action as buffer that safeguards proteostasis from proteotoxic stress, facilitating healthy development and aging. B. Chaperome sub-network perturbation entails proteostasis collapse, exposure to proteotoxicity, accelerated aging and age-onset disease. Perturbation exemplified by loss of edges and repression of gene expression (green nodes).
The Hsp90 chaperone has been proposed to have buffering capacity in its role as a capacitor of morphological variation and facilitator of evolution. Reduction of Hsp90 levels below a critical threshold exposes cryptic endogenous variants such as genetic mutations that would otherwise be corrected or suppressed by Hsp90 (Rutherford and Lindquist 1998; Queitsch et al. 2002). Local fluctuations in the expression of Hsp90 in one tissue is recognized by transcellular chaperone signalling by the compensatory expression of chaperones in distant tissues to achieve organismal proteostasis (van Oosten-Hawle et al. 2013). The chaperome subset described here unites Hsp90 and its co-chaperones with several novel and previously undescribed PN components in a highly interconnected subnetwork. We propose that the chaperome sub-network, rather than individual chaperones are important for the properties of the PN in aging and disease and suggest that this sub-network is a functional “core chaperome” within the global chaperome (Powers and Balch 2013). Based on the importance of proteostasis in health and disease, we suggest that chaperome sub-network-targeted therapeutic interventions may be beneficial for a large number of age-related protein misfolding disorders (Powers et al. 2009).
EXPERIMENTAL PROCEDURES
Chaperome gene list curation, annotation and orthology mapping
We curated the literature for chaperone and co-chaperone families covering all structural and functional categories and subcellular localizations relevant to chaperone-assisted protein folding and comprehensively annotated the human and C. elegans family members. We matched genes with domain structures referenced in UniProt prioritizing 44 bona-fide IPR-criteria domains (IPR-IDs) (Table S1) characteristic of each family to consolidate our literature-based annotation and to identify new members not previously associated with these families. We organized the genes into 9 functional families based on literature evidence on activities and the IPR-criteria domains. Chaperones with exclusive function in the ER and mitochondrial (MITO) compartments for which no IPR-domain could be matched were grouped as ‘ER-specific’ and ‘MITO-specific’. Small co-chaperone families with unambiguous functional association with a chaperone were grouped within the respective chaperone system (family). HSP40 and TPR-domain co-chaperones were organized in separate families. Human and C. elegans chaperome gene lists were matched and reconciled using orthology pairs with the NCBI HomoloGene database. The annotations were re-curated based on the WormBase (release WS234) comparative genomics tool that associates C. elegans genes with human orthologs based on curated and automated predictions by NCBI KOGS, InParanoid, TreeFam, precomputed BLAST results, Ensembl COMPARA and the orthologs matrix project (OMA). We applied bipartite mapping to identify orthology pairs and the respective species-specific chaperome subsets.
Chaperome expression correlation analyses
Expression profiles for 318 human chaperome genes were extracted from two independent transcriptome datasets of human brain biopsy tissue samples covering subjects from a variety of ages (Berchtold et al. 2008; Loerch et al. 2008). We calculated Pearson correlation between age and each chaperone's expression and evaluated significance of correlation (P < 0,05). Hierarchical clustering was performed to cluster samples by expression profile similarity. For further details see supplemental experimental procedures.
Construction of the integrated brain aging chaperome interactome
We retrieved 64,738 unique human protein-protein interactions (PPIs) from MINT, BioGRID, HPRD, and IntACT and extracted the human chaperome interactome. Amongst these we identified PPIs involving chaperome genes expressed in Superior Frontal Gyrus of aging human brains (Berchtold et al. 2008). Co-expression correlation coefficients (corrage) were calculated for all-by-all gene pairs to identify pairs with significant Pearson correlation of co-expression. Pairs with corrage ≥ 0.8 (P < 9.0e-12) were considered significantly co-expressed (COX) and combined with PPIs into an integrated PPI-COX network. Visualizations were generated in Cytoscape v2.8.1 (Smoot et al. 2011).
Chaperome network community clustering
We applied the link-community network clustering algorithm gauged at a community size cut-off of ≥ 3 nodes and ≥ 2 edges (Ahn et al. 2010) to identify communities of interconnected aging-co-regulated chaperome genes. See supplemental experimental procedures.
C. elegans strains and maintenance
C. elegans wild type Bristol strain N2, Aβ42 CL2006 (dvIs2), Q35 AM140 (rmIs132[Punc-54::q35::yfp]), the temperature sensitive (TS) mutant strain CB1402 (unc-15(e1402)) and the RW1596 (myo-3(st386);stEx30[myo-3::GFP;rol-6(su1006)] strains were maintained according to standard methods at 20°C on nematode growth media (NGM) with OP50 E. coli (Brenner 1974).
C. elegans
RNAi screens for chaperome modifiers of protein misfolding-related proteotoxicity
Chaperome-wide RNAi screens for enhancement of motility defects in C. elegans body wall muscle cells were performed using the commercial RNAi library. Missing or incorrect clones were cloned into L4440 (Kamath and Ahringer 2003). Synchronized L1 animals expressing Aβ42 or polyQ were fed bacteria expressing RNAi for each target. In case of lethality or larval arrest worms were fed at the L4 stage. Adult worms from age-synchronized populations were scored for paralysis (Link 1995) (Aβ42 screen) on day 4 or for motility defects (Silva et al. 2011) (Q35 screen) on day 2 of adulthood. RNAi candidates with 20% decrease in movement compared to control in ≥ 3 experiments constitute the final set. For aging-related proteotoxicity, synchronized adult animals fed chaperome subset RNAi were assayed daily and non-responders to prodding were scored as paralyzed. unc-15(e1402) animals were grown at 15°C and assayed daily for paralysis. To assess myofilament structure we monitored MYO-3::GFP fluorescence in animals fed bacteria expressing dsRNA against each target. Images were taken on days 1 and 8 of adulthood using a Zeiss Axiovert 200 microscope. See supplemental experimental procedures.
Human chaperome sub-network interactome
We extracted 28 human orthologs of the 16 chaperome subset genes as described. The human chaperome sub-network was obtained as a sub-network of the PPI-COX chaperome interactome. To test significance we built 100 randomized control PPI-COX networks, keeping number of nodes, edges and node degree constant, but rewiring edges between nodes and treating PPI and COX edges separately to conform to the different nature of these interactions. Network figures were generated with Cytoscape.
siRNA-HCI for modifiers of Huntingtin aggregation
Chaperome modifiers of Huntingtin-exon1(Q78)-GFP (Htt-GFP) aggregation (% cells ≥ 1 aggregates) were identified by siRNA perturbation coupled to high-content imaging (HCI) in HeLa cells. Monoclonal doxycycline-inducible cells expressing Htt-GFP were transfected with non-targeting or quadruplex siRNA smart-pools. Cells were fixed, stained with Hoechst dye and analyzed. See supplemental experimental procedures.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of the Morimoto Laboratory, the Center for Cancer Systems Biology (CCSB), and Proteostasis Therapeutics Inc. for their support and critical reading of the manuscript. These studies were supported by Proteostasis Therapeutics, Inc, Institute Sponsored Research funds from the Dana-Farber Cancer Institute (DFCI) Strategic Initiative, grants from the National Institutes of Health (NIGMS, NIA, NIMS), the Ellison Medical Foundation, a foundation that requests to be anonymous, and the Daniel F. and Ada L. Rice Foundation to RIM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.B., C.V., J.H.S., and A.V. performed experiments or contributed new reagents. M.B., T.R., S.W., Y.Z. and K.O. performed computational analyses. M.B., C.V., H.G. and R.I.M. wrote the manuscript. D.G., M.V., H.G. and R.I.M. designed and/or advised research. M.V., H.G. and R.I.M. should be considered joint senior authors.
CONFLICT OF INTEREST
The authors do not declare competing financial interests.
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Experimental Procedures, six figures, and seven tables and can be found with this article online.
REFERENCES
- Ahn YY, Bagrow JP, et al. Link communities reveal multiscale complexity in networks. Nature. 2010;466(7307):761–764. doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- Albanese V, Yam AY, et al. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124(1):75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Almeida A, Bolanos JP, et al. Cdh1/Hct1-APC is essential for the survival of postmitotic neurons. J Neurosci. 2005;25(36):8115–8121. doi: 10.1523/JNEUROSCI.1143-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, et al. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Behrends C, Langer CA, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Molecular cell. 2006;23(6):887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Belin AC, Westerlund M. Parkinson's disease: a genetic perspective. The FEBS journal. 2008;275(7):1377–1383. doi: 10.1111/j.1742-4658.2008.06301.x. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, et al. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(35):14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(40):15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharova NA, Sokolov SS, et al. Unexpected link between anaphase promoting complex and the toxicity of expanded polyglutamines expressed in yeast. Cell Cycle. 2008;7(24):3943–3946. doi: 10.4161/cc.7.24.7398. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, et al. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313(5793):1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Mancini MA, et al. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nature genetics. 1998;19(2):148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- Douglas PM, Dillin A. Protein homeostasis and aging in neurodegeneration. The Journal of Cell Biology. 2010;190(5):719–729. doi: 10.1083/jcb.201005144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnhoefer DE, Wong BKY, et al. Convergent pathogenic pathways in Alzheimer's and Huntington's diseases: shared targets for drug development. Nat Rev Drug Discov. 2011;10(11):853–867. doi: 10.1038/nrd3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, et al. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311(5766):1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Gieffers C, Peters BH, et al. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci U S A. 1999;96(20):11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nature reviews. Molecular cell biology. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hampel H, Frank R, et al. Biomarkers for Alzheimer's disease: academic, industry and regulatory perspectives. Nat Rev Drug Discov. 2010;9(7):560–574. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- Harris TW, Antoshechkin I, et al. WormBase: a comprehensive resource for nematode research. Nucleic acids research. 2010;38(Database issue):D463–467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, et al. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295(5561):1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Haslbeck V, Eckl JM, et al. Chaperone-interacting TPR proteins in Caenorhabditis elegans. Journal of molecular biology. 2013;425(16):2922–2939. doi: 10.1016/j.jmb.2013.05.019. [DOI] [PubMed] [Google Scholar]
- Hodges A, Strand AD, et al. Regional and cellular gene expression changes in human Huntington's disease brain. Human molecular genetics. 2006;15(6):965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Hunter S, Jones P, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic acids research. 2012;40(Database issue):D306–312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana NR, Tanaka M, et al. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Human molecular genetics. 2000;9(13):2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30(4):313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kim AH, Puram SV, et al. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136(2):322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nature cell biology. 2006;8(10):1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- Kleizen B, Braakman I. Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol. 2004;16(4):343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Kuczera T, Stilling RM, et al. The anaphase promoting complex is required for memory function in mice. Learn Mem. 2011;18(1):49–57. doi: 10.1101/lm.1998411. [DOI] [PubMed] [Google Scholar]
- Langbehn DR, Brinkman RR, et al. A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clinical genetics. 2004;65(4):267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- Li M, Shin YH, et al. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat Cell Biol. 2008;10(9):1083–1089. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Reiman EM, et al. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(11):4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(20):9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerch PM, Lu T, et al. Evolution of the aging brain transcriptome and synaptic regulation. PloS one. 2008;3(10):e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, et al. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre C, Delgado-Esteban M, et al. Cdk5 phosphorylates Cdh1 and modulates cyclin B1 stability in excitotoxicity. EMBO J. 2008;27(20):2736–2745. doi: 10.1038/emboj.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco K, Thomann A, et al. The APC/C E3 ligase remains active in most post-mitotic Arabidopsis cells and is required for proper vasculature development and organization. Development. 2009;136(9):1475–1485. doi: 10.1242/dev.035535. [DOI] [PubMed] [Google Scholar]
- Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends in biochemical sciences. 2013;38(10):507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Moran LB, Duke DC, et al. Whole genome expression profiling of the medial and lateral substantia nigra in Parkinson's disease. Neurogenetics. 2006;7(1):1–11. doi: 10.1007/s10048-005-0020-2. [DOI] [PubMed] [Google Scholar]
- Morley JF, Brignull HR, et al. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Molecular systems biology. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EA, Garcia SM, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(17):6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Kukushkin Y, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154(1):134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annual review of biochemistry. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Powers ET, Balch WE. Diversity in the origins of proteostasis networks -a driver for protein function in evolution. Nature reviews. Molecular cell biology. 2013;14(4):237–248. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, et al. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Siligardi G, et al. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. The EMBO journal. 1999;18(3):754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, et al. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417(6889):618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396(6709):336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Satyal SH, Schmidt E, et al. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(11):5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, et al. Database resources of the National Center for Biotechnology Information. Nucleic acids research. 2012;40(Database issue):D13–25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochimica et biophysica acta. 2014;1843(1):13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Fox S, et al. A genetic screening strategy identifies novel regulators of the proteostasis network. PLoS genetics. 2011;7(12):e1002438. doi: 10.1371/journal.pgen.1002438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Masison DC. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1). The Journal of biological chemistry. 2005;280(40):34178–34185. doi: 10.1074/jbc.M505420200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Geller R, et al. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nature cell biology. 2006;8(10):1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T, Model K, et al. Formation of membrane-bound ring complexes by prohibitins in mitochondria. Molecular biology of the cell. 2005;16(1):248–259. doi: 10.1091/mbc.E04-09-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Reumers J, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nature chemical biology. 2011;7(5):285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- Yu A, Shibata Y, et al. Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(15):E1481–1490. doi: 10.1073/pnas.1321811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.