Summary
Oncogenic mutations in PIK3CA, the gene encoding the catalytic subunit of phosphoinositide 3-kinase (PI 3-K), occur with high frequency in breast cancer. The protein kinase Akt is considered to be the primary effector of PIK3CA, although mechanisms by which PI 3-K mediates Akt-independent tumorigenic signals remain obscure. We show that serum and glucocorticoid-regulated kinase 3 (SGK3) is amplified in breast cancer and activated downstream of PIK3CA in a manner dependent on the phosphoinositide phosphatase INPP4B. Expression of INPP4B leads to enhanced SGK3 activation and suppression of Akt phosphorylation. Activation of SGK3 downstream of PIK3CA and INPP4B is required for 3D proliferation, invasive migration and tumorigenesis in vivo. We further show that SGK3 targets the metastasis suppressor NDRG1 for degradation by Fbw7. We propose a model in which breast cancers harboring oncogenic PIK3CA activates SGK3 signaling while suppressing Akt, indicative of oncogenic functions for both INPP4B and SGK3 in these tumors.
Introduction
Somatic mutations, amplifications and other genetic lesions in genes that encode proteins in the phosphoinositide 3-kinase (PI 3-K) pathway play a critical role in breast cancer etiology and progression by regulating phenotypes such as cell proliferation, survival and metastasis. The importance of PI 3-K signaling is highlighted by identification of activating oncogenic mutations of PIK3CA, the gene that encodes the p110α catalytic subunit of class I PI 3-K. Oncogenic PIK3CA mutations are frequent in breast cancers, particularly in estrogen receptor positive disease where approximately 40% of cases harbor one of the two most frequent mutations, H1047R and E545K (Cancer Genome Atlas, 2012; Engelman et al., 2006; Lee et al., 2005; Samuels et al., 2004). Class I PI 3-K activate signaling cascades by generating the phosphoinositides PtdIns-3,4-P2 and PtdIns-3,4,5-P3 (Manning and Cantley, 2007). Arguably the most studied and best understood effector of PI 3-K is the serine/threonine protein kinase Akt/ protein kinase B (PKB). Activation of Akt is initiated though interaction of the pleckstrin homology (PH) domain with either PtdIns-3,4-P2 or PtdIns-3,4,5-P3 (Chin and Toker, 2009; Franke et al., 1997; Woodgett, 2005). This is followed by phosphorylation of Akt by the phosphoinositide-dependent kinase-1 (PDK-1) and mammalian target of rapamycin complex 2 (mTORC2), locking the enzyme in the catalytically competent conformation (Mora et al., 2004; Sarbassov et al., 2005).
Signal termination of PI 3-K and Akt signaling is mediated by the Phosphatase and Tensin homolog (PTEN), a tumor suppressor protein that dephosphorylates PtdIns-3,4,5-P3 converting it back to PtdIns-4,5-P2 (Li et al., 1997; Maehama and Dixon, 1998). Loss of heterozygosity (LOH), inactivating mutations or deletions in PTEN are frequent in many cancers, and lead to excessive PtdIns-3,4,5-P3 accumulation and hyperactivation of downstream effectors, including Akt (Engelman et al., 2006). An alternative mechanism of negative regulation of the Akt pathway is through the SH2 domain-containing inositol phosphatase (SHIP) family of proteins that dephosphorylate PtdIns-3,4,5-P3 and generate PtdIns-3,4-P2, (Choi et al., 2002; Scheid et al., 2002). In turn, PtdIns-3,4-P2 signaling is terminated by dephosphorylation, mediated by the inositol polyphosphate-4-phosphatases type I and II (INPP4A and INPP4B), resulting in PtdIns-3-P generation (Gewinner et al., 2009; Norris et al., 1997; Norris and Majerus, 1994). INPP4A and INPP4B both function as suppressors of Akt activity (Ivetac et al., 2009), however, INPP4A expression is primarily restricted to the brain while INPP4B is expressed in most tissues, including breast (Fedele et al., 2010).
Despite numerous studies pointing to Akt as a primary transducer of the PI 3-K signal, PIK3CA mutant tumors have strikingly low levels of phosphorylated (hence activated) Akt, indicating that other PtdIns-3,4-P2 and PtdIns-3,4,5-P3 effectors link PI 3-K to tumorigenesis (Stemke-Hale et al., 2008; Vasudevan et al., 2009). Such effectors include the Tec family kinases Btk and Itk (Luo et al., 2003; Miao et al., 2010). Moreover, GTPase activating proteins for Rho family GTPases also transduce PI 3-K signaling, such as GRP1 (Lai et al., 2013). A more recent study showed that PIK3CA-mediated breast cancer cell growth and survival is dependent on the Serum and Glucocorticoid-regulated Kinase 3 (SGK3), in cases where Akt was dispensable (Vasudevan et al., 2009). SGK3 is an AGC protein kinase family member along with two other isoforms, SGK1 and SGK2. SGK isoforms share ~55 % sequence identity with the Akt1-3 catalytic domains (Firestone et al., 2003; Kobayashi et al., 1999; Tessier and Woodgett, 2006b). SGK and Akt isoforms also phosphorylate the same consensus substrate motif, RXRXXS/T, and thus possess a large number of shared substrates (Murray et al., 2004). SGK isoforms are activated by the same upstream kinases as Akt, PDK-1 at the activation loop residue and TORC2 at the hydrophobic motif (Garcia-Martinez and Alessi, 2008; Kobayashi and Cohen, 1999; Kobayashi et al., 1999; Liu et al., 2000). As with Akt, SGK phosphorylation at these residues is necessary for catalytic activity. However, unlike Akt, SGKs have unique regulatory regions. In the case of SGK3, this includes an amino-terminal Phox Homology (PX) domain (Xu et al., 2001). The SGK3 PX domain binds to PtdIns-3-P, thereby localizing a pool of the kinase to endosomal membranes (Tessier and Woodgett, 2006a; Xu et al., 2001).
Despite the nomenclature, SGK3 expression is not regulated by glucocorticoids, instead estrogen receptor (ER) signaling has been shown to induce SGK3 transcription (Wang et al., 2011a; Xu et al., 2012). Interestingly, INPP4B is also an ER-induced gene (Fedele et al., 2010). Luminal breast cancers are defined by their expression of estrogen and progesterone receptors, distinguishing them from HER2 and basal-like (triple-negative) subtypes (Fedele et al., 2010; Sorlie et al., 2001). INPP4B inactivation by LOH is a frequent event in basal-like cancers, and its loss leads to Akt hyperactivation (Cancer Genome Atlas, 2012; Fedele et al., 2010; Gewinner et al., 2009). Conversely, INPP4B has been proposed to be a novel biomarker for luminal-type breast cancers, which also harbor frequent PIK3CA oncogenic mutations.
The mechanisms linking PIK3CA to SGK3 signaling and downstream phenotypes have not been defined. Here we show that INPP4B mediates PIK3CA-dependent SGK3 activation in breast cancer cells. We also show that SGK3 regulates N-Myc downstream regulated 1 (NDRG1) leading to ubiquitination and degradation mediated by the E3 ligase F-box and WD repeat domain-containing 7 (FBW7). Finally, we show that SGK3 functions as a PI 3-K effector in the control of oncogenic signals promoting cell growth and migration of breast cancer.
Results
SGK3 is amplified and hyperactivated in breast cancer
A recent study demonstrated that SGK3 is required for the survival of certain breast cancer cell lines with oncogenic PIK3CA mutations. These same cells showed minimal Akt activity and furthermore, Akt was dispensable for survival (Vasudevan et al., 2009). The Akt PH domain binds the PI 3-K lipids PtdIns-3,4-P2 and PtdIns-3,4,5-P3, however the SGK3 regulatory region lacks a functional PH domain. Instead, SGK3 regulation is in part mediated by the PX domain that primarily binds PtdIns-3-P (Tessier and Woodgett, 2006a). Since PtdIns-3-P is not a product of class I PI 3-kinases, the mechanism by which SGK3 functions as an effector of PIK3CA remains undefined.
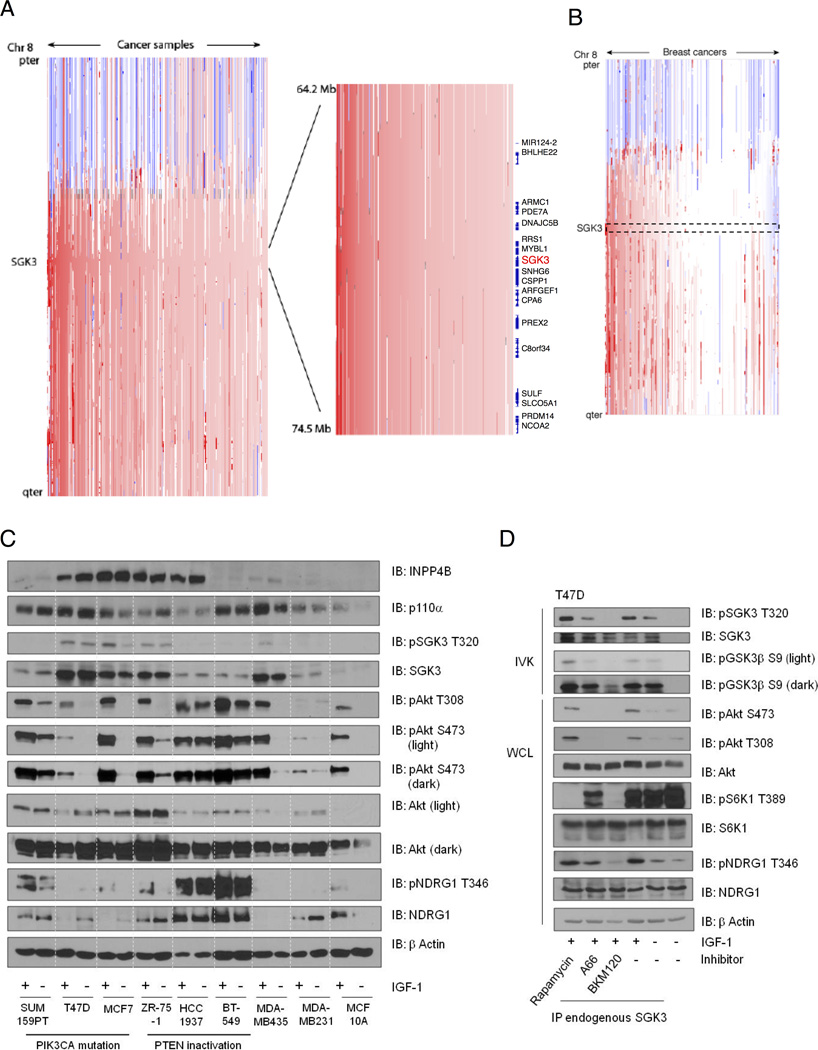
Somatic activating mutations in the SGK3 gene have not been identified with any appreciable frequency. We examined whether amplifications or deletions of SGK3 exist in human cancers and cancer cell lines in a published database of 3131 cancers (Beroukhim et al., 2010). Amplifications were present in 30% of tumors. In 4.8% of cases, these were focal events encompassing less than half of chromosome 8q, a rate significantly above the genome-wide average (q=0.00168; Figure 1A). Among the 243 breast cancers in the study, 54% exhibited amplifications of SGK3, including 12% with focal alterations (q=0.186), and indeed SGK3 is one of 64 genes within the “peak region” where these amplifications most overlap (Figure 1B). Chromosome 8q also includes MYC, which is the most commonly amplified gene among these cancers. However, the SGK3 peak was significant even after discounting amplifications which encompassed MYC.
Figure 1. SGK3 is amplified in breast cancer.
(A) Copy-number profiles across chromosome 8 (y-axis) for 3,131 cancer samples (x-axis). A magnified view of the region including the SGK3 gene locus is shown in the inset. Analysis was performed from a data set obtained from a study of 3,131 cancers of over 25 subtypes. Samples are sorted by frequency of amplification at the SGK3 locus.
(B) Copy-number profiles from 243 breast cancers selected from the data set described in (A).
(C) Cell lysates derived from the indicated breast epithelial and cancer cell lines, serum-starved (−) or stimulated with IGF-1 for 20 min (+) were immunoblotted (IB) with the indicated antibodies.
(D) T47D cells were serum-starved for 24 hr, then pretreated with the invocated inhibitors, rapamycin (100nM), A66 (1µM) or BKM120 (1µM) for 20 min, then stimulated with IGF-1 for a further 20 min. SGK3 was immunoprecipitated with anti-SGK3 and subjected to an in vitro kinase assay (IVK) using GSK-3β peptide substrate. Peptide substrate phosphorylation was detected using anti- GSK-3β pS9-antibody. SGK3 phosphorylation was detected using anti-SGK3 pT320. Total SGK3 was evaluated as control. Whole cell lysates were immunoblotted with the indicated antibodies. All results are representative of at least 3 independent experiments. See also Figures S1 and S5.
We next analyzed SGK3 protein levels in a panel of breast cancer cell lines informed by the Tumorscape analysis to have amplified SGK3, along with PIK3CA and PTEN mutation status. Cells were serum-starved and stimulated with insulin-like growth factor-1 (IGF-1) to activate PI 3-K and Akt signaling (Figure 1C). We used an antibody against the SGK3 activation loop site T320, targeted by PDK-1, as a surrogate for SGK3 activation. Concomitantly, we evaluated Akt phosphorylation at the activation loop residue (T308), also targeted by PDK-1, and hydrophobic motif (S473), phosphorylated by the TORC2 complex. An antibody that recognizes the endogenous SGK3 hydrophobic motif site at S486 is not available. We also used total and phospho-NDRG1 antibodies as surrogates for the activity of SGK3, noting that NDRG1 is a pan-SGK substrate (Murray et al., 2004; Najafov et al., 2011). We find that in cells that harbor PI 3-K pathway mutations, such as oncogenic PIK3CA or PTEN inactivation, SGK3 is basally phosphorylated and further stimulated by IGF-1 (ZR-75-1, MCF7, T47D, Figure 1C). Interestingly, in cells that display basal SGK3 phosphorylation, the total levels of NDRG1 protein are low or undetectable. Since NDRG1 is a direct SGK substrate, this correlation suggests that SGK3 activation and signaling may modulate NDRG1 degradation.
Not all breast cancer cell lines that harbor PIK3CA mutations or PTEN inactivation display elevated SGK3 phosphorylation (e.g., SUM159PT, BT-549) (Figure 1C). We therefore considered additional PI 3-K pathway alterations that would account for SGK3 phosphorylation. INPP4B is 4’ phosphoinositide phosphatase that exclusively dephosphorylates the PI 3-K lipid PtdIns-3,4-P2, converting it to PtdIns-3-P. Loss of INPP4B by LOH is frequently observed in basal-like cancers and leads to elevated PtdIns-3,4-P2 levels and in turn, Akt hyperactivation (Fedele et al., 2010; Gewinner et al., 2009). However, INPP4B loss is not observed in estrogen receptor-positive breast cancers, where PIK3CA mutations occur with the highest frequency (Cancer Genome Atlas, 2012). In our comparative analysis, we find that elevated SGK3 phosphorylation correlates directly with both elevated INPP4B expression and PIK3CA/PTEN mutation status (Figure 1C).
To provide cause-and-effect evidence for SGK3 as a PI 3-K effector, we used pathway inhibitors in cells with PIK3CA mutation and high INPP4B, as well as robust SGK3 phosphorylation. Since phosphorylation-state antibodies such as pT320 (for SGK3) or pT308 and pS473 (for Akt) do not directly measure protein kinase activity, we developed a kinase assay to measure SGK3 activity. Endogenous SGK3 was immunoprecipitated and used in in vitro kinase assays with a peptide substrate derived from GSK-3β that is a defined optimal Akt and SGK substrate, since they share a similar consensus phosphorylation motif (Murray et al., 2004). Using this assay, we show that SGK3 activity is stimulated by IGF-1, concomitant with increased T320 phosphorylation and increased NDRG1 phosphorylation (Figure 1D). SGK3 activation is attenuated by both BKM-120 and A66, pan class I and p110α specific inhibitors, respectively (Buonamici et al., 2010; Jamieson et al., 2011). Conversely, inhibition of TORC1 using rapamycin does not block SGK3 activation, consistent with the notion that SGK isoforms are TORC2 targets (Garcia-Martinez and Alessi, 2008). The increase in pSGK3 T320 phosphorylation in cells treated with rapamycin implies that SGK3 and Akt isoforms share overlapping substrates that modulate feedback inhibition of the TORC1 complex, thereby resulting in enhanced SGK3 activation (Figure 1D). Collectively, these data demonstrate that SGK3 is amplified in breast cancer and its activity is dependent on oncogenic PI 3-K signaling.
INPP4B regulates SGK3 activation
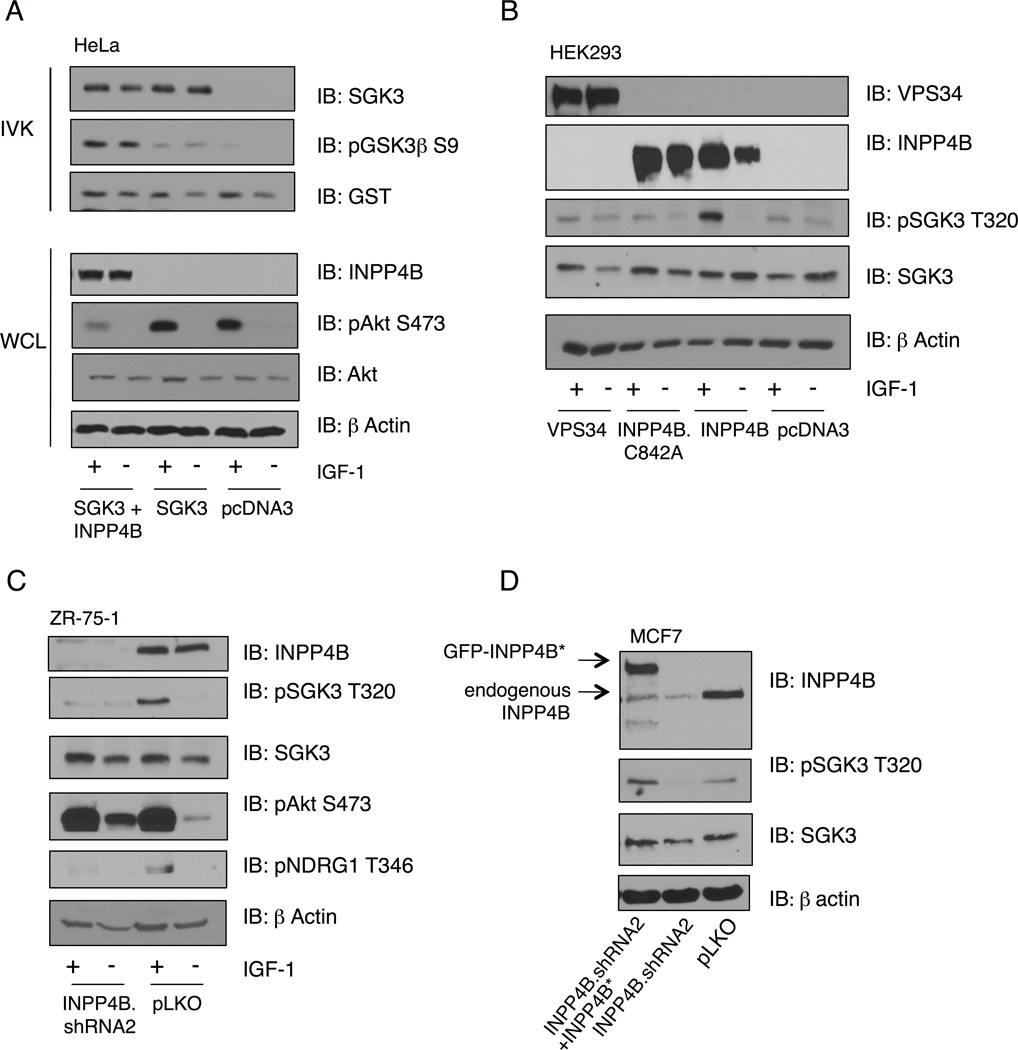
PtdIns-3-P directly binds to the SGK3 PX domain (Tessier and Woodgett, 2006a). Since high INPP4B levels correlate with the ability of IGF-1 to stimulate SGK3 in PIK3CA mutant cells (Figure 1C), and INPP4B generates PtdIns-3-P, we reasoned that INPP4B levels are rate-limiting for SGK3 activation downstream of PI 3-K. SGK3 activity measured by in vitro kinase assay is significantly enhanced in cells co-expressing INPP4B, whereas Akt phosphorylation is attenuated, as expected (Figure 2A). Moreover, endogenous SGK3 phosphorylation is also stimulated by expression of wild-type INPP4B, but not by a catalytically-inactive phosphatase mutant (Figure 2B). A requirement for INPP4B in PIK3CA-mediated SGK3 activation is further substantiated by minimal increases SGK3 phosphorylation at T320 in response to IGF-1 stimulation in MDA-MB-435 cells and the lack of effect of PI 3-K inhibitors on SGK3 phosphorylation (Figure S1), since this line expresses very low levels of INPP4B (Figure 1C). Although the primary route of PtdIns-3-P synthesis is through the class III PI 3-K Vps34, expression of an active Vps34 allele does not promote SGK3 phosphorylation (Figure 2B). Moreover, VPS34 null MEFs (fl/fl) display the same level of IGF-1-stimulated SGK3 and NDRG1 phosphorylation as control cells (Figure S2A).
Figure 2. INPP4B positively regulates SGK3 activity.
(A) HeLa cells were transfected with SGK3-GST alone or with INPP4B and serum-starved for 24 hr, prior to stimulation with IGF-1 for 20 min. SGK3 was isolated from cell lysates with glutathione beads, and incubated with GST-GSK3β peptide in an in vitro kinase assays (IVK). Peptide substrate phosphorylation was detected using anti-GSK-3β pS9-antibody. Total SGK3 was evaluated as control. Whole cell lysates were immunoblotted with the indicated antibodies.
(B) HEK293T cells were transfected with wild-type INPP4B, catalytically-inactive INPP4B.C842A or Vps34, then serum-starved for 24 hr and stimulated with IGF-1 for 20 min. Whole cell lysates were immunoblotted with the indicated antibodies.
(C) ZR-75-1 cells were infected with shRNA lentivirus targeting INPP4B, or control pLKO, serum-starved and stimulated with IGF-1 for 20 min. Cell lysates were immunoblotted with the indicated antibodies.
(D) MCF7 cells were infected with shRNA lentivirus targeting INPP4B, followed by retroviral infection with INPP4B cDNA rescue virus. Cell lysates were immunoblotted with the indicated antibodies. All results are representative of at least 3 independent experiments. See also Figure S2.
Silencing INPP4B using lentiviral shRNA completely attenuates SGK3 phosphorylation in response to IGF-1, with a concomitant decrease in phospho-NDRG1 (Figure 2C). Similarly, reduced SGK3 phosphorylation induced by INPP4B silencing is rescued by expression of an shRNA-resistant INPP4B allele (Figure 2D). Inhibition of both exogenous and endogenous SGK3 protein kinase activity is also observed in cells transduced with INPP4B shRNA (Figures S2B and S2C). Consistent with the notion that INPP4B is an estrogen-induced gene, T47D, MCF7, and ZR-75-1 cells grown in charcoal-stripped media show significantly reduced total INPP4B levels (Figure S2D). Moreover, expression of constitutively active myristoylated-Akt does not affect either total SGK3 or pT320 SGK3, indicating that signaling through Akt does not affect SGK3 signaling as part of feedback regulation (Figure S2E). We conclude that INPP4B is required for the ability of PI 3-K to activate SGK3, and that the phosphatase activity of INPP4B is essential. Moreover, we propose that Vps34 is dispensable for PIK3CA-mediated SGK3 activation.
SGK3 promotes proliferation in 3D and anchorage independence of growth
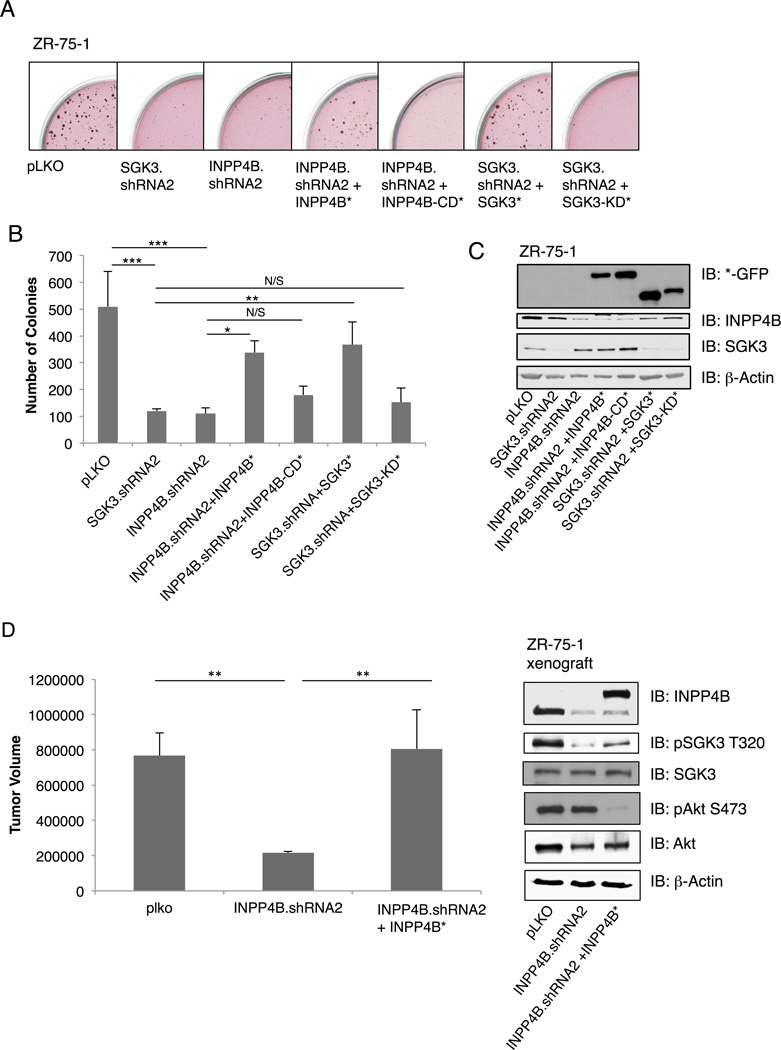
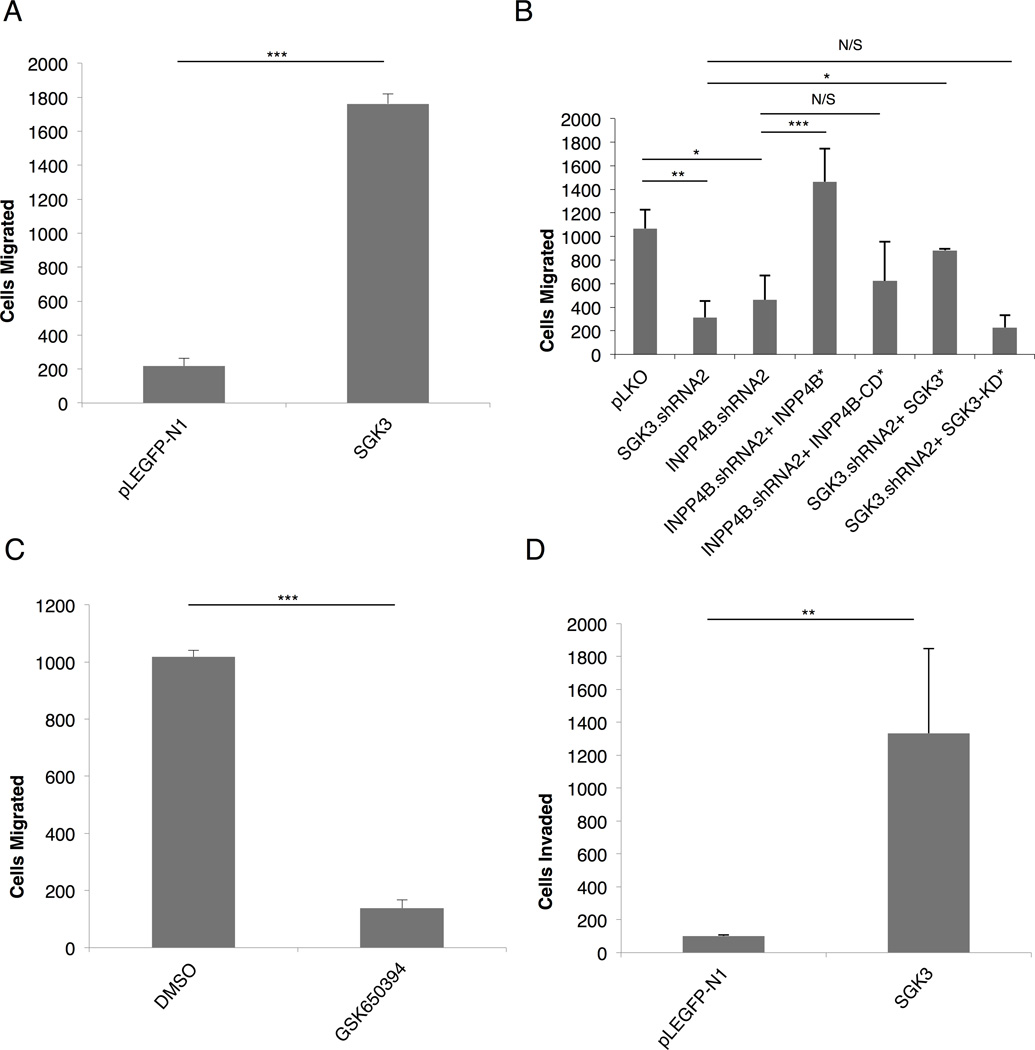
To explore the functional significance of SGK3 downstream of PIK3CA, we investigated the ability of SGK3 and INPP4B to drive anchorage independent growth, since PIK3CA has been shown to promote this phenotype (Isakoff et al., 2005). ZR-75-1 cells (PTEN missense, high SGK3 and high INPP4B, Figure 1C) were transduced with vector control, SGK3 or INPP4B shRNA, combined with cDNA rescue alleles of wild-type SGK3 or INPP4B or respective catalytically-inactive mutants, and colonies allowed to form in soft agar (Figure 3A and 3C). Whereas control cells form robust colonies in soft agar, silencing either SGK3 or INPP4B significantly inhibits the number of colonies growing in an anchorage independent manner (Figure 3B). Moreover, introduction of shRNA-resistant SGK3 and INPP4B cDNA partially rescues the defect in anchorage independent growth, whereas the catalytically-inactive mutants do not (Figure 3). Representative images of each condition are shown in Figure 3A. Similar findings are observed in MCF7 cells (PIK3CA E545K, high SGK3, and high INPP4B, Figure 1C) whereby SGK3 and INPP4B shRNA attenuate colony formation, and this is rescued by the corresponding wild-type cDNAs (Figures S3A, S3B, and S3C). To further evaluate the requirement of INPP4B in mediating SGK3 signaling to phenotypes associated with malignancy in breast cancer cells, we analyzed ZR-75-1 xenograft tumor growth in the mammary fat pad of nude mice using control, INPP4B shRNA and INPP4B cDNA rescue. INPP4B silencing significantly attenuates xenograft tumor growth, with concomitant reduction of SGK3 phosphorylation at pT320, and both tumor growth and SGK3 phosphorylation are rescued using wild-type INPP4B cDNA resistant to silencing (Figure 3D).
Figure 3. SGK3 promotes anchorage- independent growth.
(A) ZR-75-1 cells infected with pLKO vector control, INPP4B or SGK3 shRNA lentiviral vectors and grown in agar/growth media for 28 days. Additional conditions include retroviral infection of INPP4B (INPP4B*) and SGK3 (SGK*) cDNA rescue virus or INPP4B-CD* and SGK3-KD* (catalytically-inactive mutants).
(B) Number of colonies in soft agar obtained in (A) was quantitated using MatLab software.
(C) Immunoblot analysis of whole cell lysates (WCL) from (A) and (B) with indicated antibodies. All results are representative of at least 3 independent experiments.
(D) ZR-75-1cells infected with control pLKO, INPP4B.shRNA2, or shRNA-resistant INPP4B cDNA rescue were injected into the mammary fat pad of nude mice. Tumor volume was measure by calipering at 7 weeks post-injection. Error bars represent standard deviation (SD). See also Figures S3 and S5.
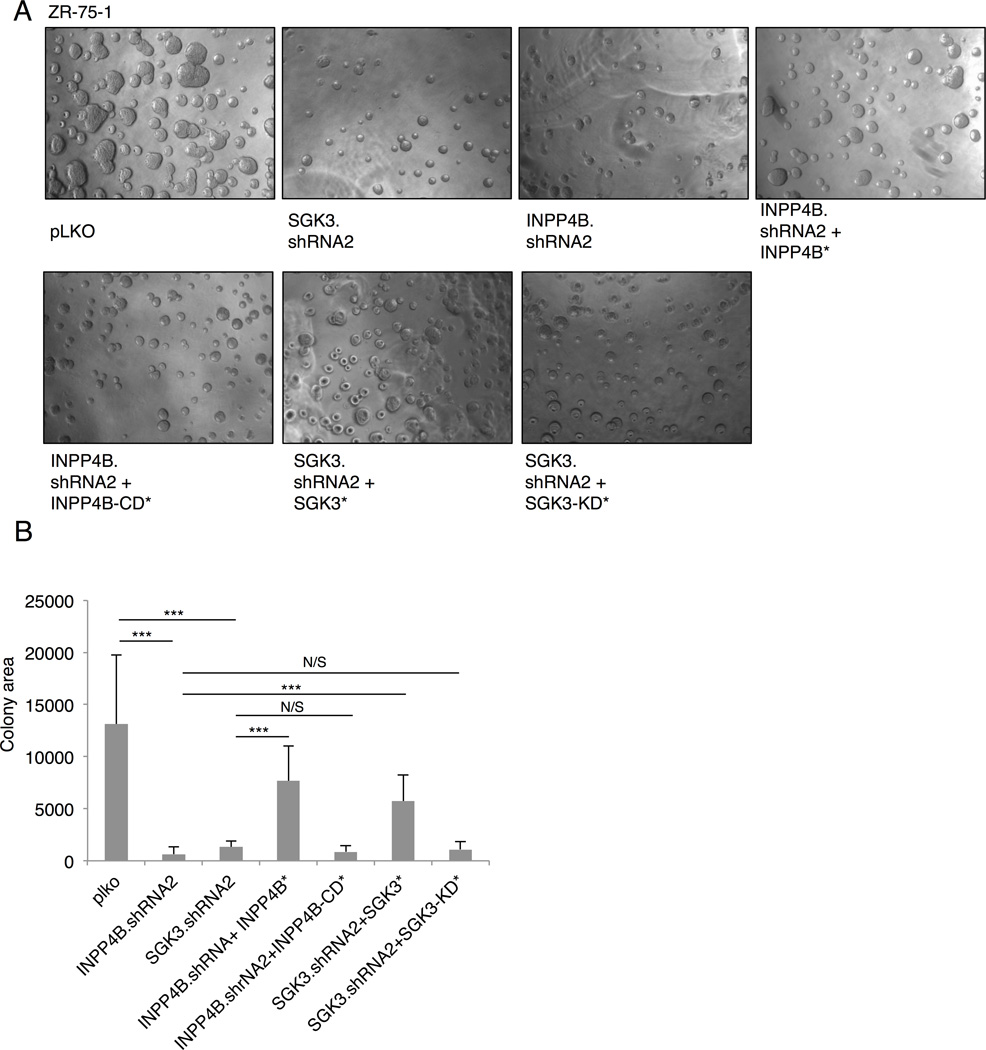
Growth of cancer cells in 3 dimensional (3D) culture further corroborates phenotypes that control tumor growth in vivo (Debnath and Brugge, 2005). 3D Matrigel growth of ZR-75-1 breast cancer cells (PTEN missense, high SGK3, high INPP4B) is significantly attenuated upon silencing of either SGK3 or INPP4B (Figures 4A, 4B, and 3C). A partial rescue of 3D growth is observed upon expression of shRNA-resistant wild type SGK3 and INPP4B, but not catalytically-inactive SGK3 or INPP4B mutants. Moreover, inhibition of 3D Matrigel growth is also observed in MCF7 cells (PIK3CA E545K, high SGK3, and high INPP4B, Figure 1C) with 2 distinct SGK3 or INPP4B shRNA sequences (Figure S3D).
Figure 4. SGK3 regulates 3D growth of breast cancer cells.
(A) ZR-75-1 cells infected with control pLKO, SGK3 shRNA, or INPP4B shRNA, shRNA rescue, and cDNA catalytic mutants and grown for 8 days in 3D Matrigel/growth media. Representative images spheroids at 4× magnification are shown.
(B) Quantitation of 2D spheroid size from (A) was performed using ImageJ software. Error bars represent SD. *P < 0.05; **P < 0.01; ***P < 0.001. Results are representative of at least 3 independent experiments. See also Figures S3 and S5.
SGK3 promotes breast cancer cell invasive migration
Signaling through PIK3CA also modulates breast cancer cell migration and invasion, pre-requisite phenotypes for metastatic dissemination of tumor cells (Pang et al., 2009). We therefore examined if SGK3 is required for breast cancer cell invasive migration. Using in vitro Transwell migration assays, we first evaluated SUM159PT cells that express low levels of endogenous SGK3 (Figure 1C). Ectopic expression of SGK3 robustly enhances SUM159PT cell migration (Figure 5A). Conversely, SGK3 or INPP4B shRNA both significantly attenuate ZR-75-1 and MCF7 cell migration (Figure 5B, Figure S4A). A catalytic small molecule pan-SGK inhibitor, GSK650394 (Sherk et al., 2008) also significantly blocks MCF7, ZR-75-1, and T47D cell migration (Figure 5C, Figures S4B–C). Finally, ectopic expression of SGK3 also promotes invasive migration through Matrigel (Figure 5D). Therefore, SGK3 protein kinase activity promotes migration of breast cancer cells that display elevated levels of INPP4B. Notably, silencing of SGK3 or INPP4B in MDA-MB-435 cells does not lead to significant alterations in anchorage independent growth, 3D colony formation, or cell migration (Figure S5), and since this cell line does not robustly express INPP4B, this further corroborates the requirement for INPP4B activity in mediating PI 3-K-dependent SGK3 activation.
Figure 5. SGK3 promotes breast cancer cell invasive migration.
(A) SUM159PT breast cancer cells were infected with SGK3 or pLEGFP followed by 2 hr migration through Transwell assays. Relative migration is measured along the Y-axis as the ratio of migrated cells in the test condition versus the control.
(B) ZR-75-1 breast cancer cells infected with SGK3 or INPP4B shRNA lentiviral vectors or control pLKO for 48 hr followed by Transwell migration assays.
(C) MCF7 cells were inhibited with SGK inhibitor, GSK650394 (10uM), or control DMSO for 20 min prior to Transwell migration.
(D) HS578T breast cancer cells were infected with SGK3 of control pLEGFP-N1 followed by Transwell Matrigel invasion assays. Error bars represent standard deviation. See also Figures S4 and S5.
SGK3 mediates NDRG1 proteolytic processing
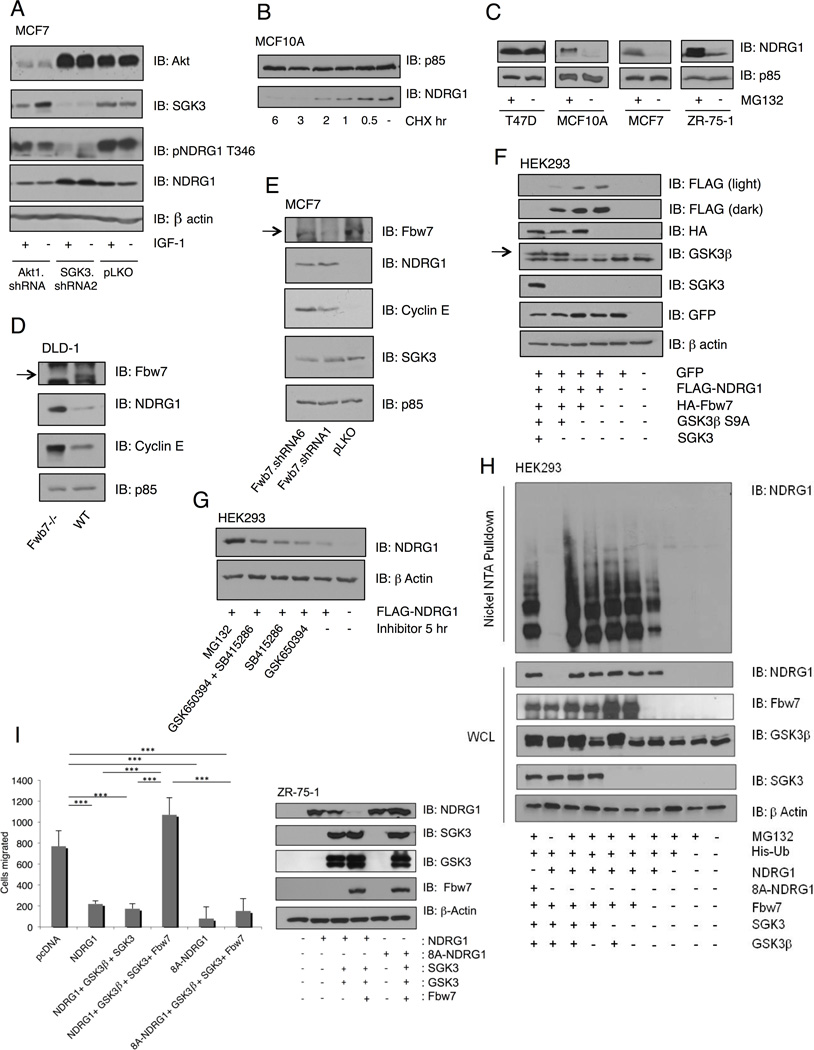
Finally, since NDRG1 is an established metastasis suppressor and is also an SGK substrate (Kachhap et al., 2007; Murray et al., 2004; Najafov et al., 2011), we evaluated the ability of SGK3 to regulate NDRG1 stability. We initially noted a correlation of NDRG1 protein levels and SGK3 phosphorylation (Figure 1C). Consistent with this, silencing SGK3, but not Akt1, results in a decrease in phosphorylated NDRG1 (pT346), with a concomitant increase in total NDRG1 (Figure 6A). A cycloheximide chase experiment reveals that the half-life of NDRG1 in serum-containing media is approximately 1 hour (Figure 6B). We also examined NDRG1 protein stability in cells treated with a proteasome inhibitor, and find that NDRG1 levels are dramatically enhanced upon inhibition of the 26S proteasome with MG132 (Figure 6C). Curiously, in T47D cells, NDRG1 levels are not affected by MG132. T47D cells are PIK3CA H1047R mutant, have high SGK3 and INPP4B, and moreover are FBXW7 null (Mao et al., 2008). Fbw7 is an F-box E3 ubiquitin ligase and substrate recognition domain of the SKP1-cullin-F-box (SCF) ubiquitin ligase complex. A cycloheximide chase in T47D cells shows no significant changes in NDRG1 total protein levels, as predicted (Figure S6A). Moreover, expression of Fbw7 in T47D cells leads to a decrease in NDRG1 total protein levels (Figure S6B). We also made use of wild-type and Fbw7 null DLD-1 colon carcinoma cells (Rajagopalan et al., 2004) and show that Fbw7 deletion leads to a dramatic increase in NDRG1 protein levels, compared to wild-type cells, whereas NDRG1 message levels remain unchanged (Figure 6D and Figure S6C). Moreover, silencing of Fbw7 using two distinct shRNA sequences leads to significantly increased NDRG1 protein levels compared to control cells (Figure 6E). Increased NDRG1 levels upon Fbw7 silencing are also observed in a distinct breast cancer cell line, BT-20 (Figure S6D). Importantly, the fold increase in NDRG1 is comparable to that observed with Cyclin E, a known Fbw7 substrate (Figure 6E).
Figure 6. NDRG1 is an FBW7 substrate.
(A) MCF7 cells infected with SGK3 shRNA, Akt1 shRNA or control pLKO, serum-starved then stimulated with IGF-1 for 20 min. Cell lysates were immunoblotted with the indicated antibodies.
(B) MCF10A cells were treated with cycloheximide or control DMSO for the indicated times and cell lysates immunoblotted with anti-NDRG1 and anti-p85.
(C) T47D, MCF10A, MCF7, and ZR-75-1 cells were treated with MG132 for 16 hr and immunoblotted with anti-NDRG1 and anti-p85.
(D) Cell lysates of wild-type (Fbw7 WT) or DLD-1 colon cancer cells null for Fbw7 (Fbw7 −/−) were immunoblotted with the indicated antibodies.
(E) MCF7 cells were infected with two distinct Fbw7 shRNA’s or control pLKO, and lysates immunoblotted with the indicated antibodies.
(F) HEK293T cells were transfected with control GFP vector, alone or with the indicated combinations of NDRG1-FLAG, HA-Fbw7, HA-GSK3β.S9A and SGK3-GST. NDRG1 stability was evaluated by immunoblotting whole cell lysates with anti-FLAG, and indicated antibodies.
(G) HEK293T cells were transfected for 48 hr with NDRG1 -FLAG prior to 5 hr treatment with GSK650394, SB415286, GSK650394 and SB415286, or MG132 in order to examine NDRG1 protein stability.
(H) A His-Ubiquitin assay was performed after transfection of HEK293T cells with FLAG-NDRG1, GST-Fbw7, HA-GSK3β.S9A, SGK3-GFP, and His-Ub, followed by treatment with MG132 for 12 hr. Nickel NTA beads were used to isolate His-ubiquitin complexes, followed by immunoblotting with the indicated antibodies.
(I) ZR-75-1 cells transfected for 24 hr with pcDNA3, NDRG1, 8A–NDRG1, Fbw7, GSK-3β, and SGK3 followed by an 18 hr Transwell migration assay. Quantification of migrated cells and representative immunoblots of whole cell lysates (WCL) with indicated antibodies are shown. Error bars represent standard deviation. See also Figure S6.
Studies have shown NDRG1 phosphorylation by SGK3 at T346/T356/T366 primes for phosphorylation by GSK-3β at S342/S352/S462 (Murray et al., 2004). GSK-3β phosphorylation is a common priming event for Fbw7 substrate binding (Wang et al., 2011b). To assess whether NDRG1 stability is dependent on the concerted action of SGK3, GSK-3β and Fbw7, we co-expressed each component in combination and measured NDRG1 stability. Maximal NDRG1 degradation is only observed when both SGK3 and GSK-3β are simultaneously co-expressed with Fbw7 (Figure 6F). Treatment of cells with SGK inhibitor, GSK650394, or GSK3 inhibitor, SB415286, conversely causes an increase in NDRG1 levels, while dual treatment with both GSK650394 and SB415286 increases NDRG1 stability even further (Figure 6G). Finally, we used an established cell-based ubiquitination assay to evaluate NDRG1 ubiquitination. NDRG1 ubiquitination is increased when Fbw7 is co-expressed, and ubiquitination is maximal when SGK3 and GSK-3β are co-expressed (Figure 6H). In the absence of MG132, NDRG1 ubiquitination is completely eliminated. Furthermore, NDRG1 ubiquitination is significantly attenuated in a mutant that lacks SGK3 and GSK-3β consensus phosphorylation sites (Figure 6H; FLAG-8A-NDRG1). Therefore, NDRG1 stability is regulated by SGK3 activity and subsequent Fbw7-mediated degradation. To address Fbw7 binding to NDRG1 a GST-binding assay was performed. NDRG1. T346A has reduced ability to bind Fbw7-GST and the 8A-NDRG1 eliminates binding to Fbw7, demonstrating SGK3 and GSK-3β phosphorylation is necessary for Fbw7 binding (Figure S6E). In order to determine the phenotypic significance of SGK3 regulation of NDRG1 we investigated the ability of NDRG1 to mediate migration. In ZR-75-1 cells the expression of NDRG1 dramatically inhibits Transwell cellular migration, while co-expression with SGK3, GSK-3β, and Fbw7 causes a dramatic increase in migration and decrease in NDRG1 protein levels, rescuing the NDRG1 suppression of motility (Figure 6I). Expression of the 8A-NDRG1 mutant phenocopies wild-type NDRG1-mediated inhibition of cell migration, however this is not rescued by co-expression of SGK3, GSK-3β and Fbw7, to the extent that is seen with wild type NDRG1 (Figure 6I). We argue that NDRG1 degradation provides a significant contribution to SGK3 downstream signaling including cell migration.
Discussion
The frequency of PI 3-K pathway mutations in many human solid tumors, including breast cancer, has led to the development of numerous small molecule inhibitors currently in phase I and II clinical trials (Baselga, 2011). These include inhibitors to PIK3CA, such as the pan class I PI 3-K inhibitor BKM-120, as well as p110α-specific inhibitors such as BYL-719 and A66. Various Akt catalytic inhibitors are also undergoing pre-clinical validation for many tumor types (Cleary and Shapiro, 2010; Courtney et al., 2010). Arguably, most studies that have evaluated the mechanism by which PI 3-kinases transduce signals to phenotypes associated with malignancy have focused on Akt as the effector molecule. However, other PI 3-K targets have been shown to signal downstream of PtdIns-3,4,5-P3, for example the Tec family kinases such as Btk in hematological malignancies (Davids and Brown, 2012). A more recent study pointed to SGK3 as a critical effector of oncogenic PIK3CA-mutant breast cancer cells in which Akt is dispensable (Vasudevan et al., 2009). This finding provides insight into an additional target for the development of new drugs for combination therapy in PI 3-K pathway mutant tumors.
Since SGK3 does not possess a PH domain in the regulatory region that would bind PtdIns-3,4,5-P3, the mechanism by which PIK3CA can activate SGK3 has remained obscure (Tessier and Woodgett, 2006b). We reasoned that such a mechanism would have to take into account the lipid-binding property of the SGK3 PX domain, which binds PtdIns-3-P (Tessier and Woodgett, 2006a). Although PI 3-K has been shown to activate SGK isoforms through PDK-1 and TORC2 (Garcia-Martinez and Alessi, 2008), it has also been shown that a PX domain lipid-binding mutant of SGK3 is catalytically inactive (Tessier and Woodgett, 2006a). This same PX domain mutant is not phosphorylated at the corresponding Thr320 and Ser486 sites, indicating PtdIns-3-P binding as a pre-requisite for phosphorylation and activation. Since class I PI 3-kinases do not synthesize PtdIns-3-P directly, an intermediate step(s) is required for SGK3 activity. Our data show that INPP4B is both necessary and sufficient to transduce the PIK3CA signal to SGK3 activation. INPP4B is a PtdIns-3,4-P2 phosphatase, such that inactivation of INPP4B by LOH in human tumors leads to Akt hyperactivation (Fedele et al., 2010; Gewinner et al., 2009). Conversely, one would predict that in tumors with normal or elevated expression of INPP4B, the synthesis of PtdIns-3-P would promote the activation of an alternative effector(s). We propose that SGK3 is one such effector, whose activation by PIK3CA is mediated by INPP4B in breast cancer cells. This model is supported by several lines of evidence. First, breast cancer cell lines with high INPP4B levels show elevated SGK3 phosphorylation and activity, whereas lines with low or undetectable INPP4B do not (Figure 1C). Expression of catalytically active, but not inactive INPP4B leads to activation of SGK3 with a concomitant inhibition of IGF-1-stimulated Akt phosphorylation (Figure 2A). Moreover, silencing INPP4B with shRNA attenuates SGK3 activation, and also leads to inhibition of anchorage-independent growth and proliferation in 3D (Figure 2D, Figure 3 and Figure 4). In a previous study, silencing INPP4B with shRNA was shown to result in increased anchorage-independent growth, also in MCF7 cells (Fedele et al., 2010). The reason for the discrepancies between the two studies is unclear, but could be due to differences in shRNA sequences used. In our studies, two distinct shRNA sequences were used, and introduction of INPP4B cDNA rescued the phenotype. Regardless, our data support a model in which INPP4B functions as both an oncogene and a tumor suppressor. A tumor suppressor function is evident in basal-like tumors which show a high frequency of INPP4B LOH, and a concomitant increase in Akt activity (Fedele et al., 2010). Conversely, an oncogenic role for INPP4B is supported by breast cancer cell lines derived from luminal tumors that show elevated SGK3 activation and high INPP4B expression.
INPP4B dephosphorylates PtdIns-3,4-P2 to generate PtdIns-3-P, and our results show that INPP4B catalytic activity is required for SGK3 activation. Since the SGK3 PX domain binds PtdIns-3-P, this would argue that SGK3 activation would occur at sites of PtdIns-3-P accumulation. PtdIns-3-P is also generated by class I and II PI 3-kinases. Specifically, a significant pool of PtdIns-3-P is generated by the class III enzyme VPS34 at endosomal membranes (Backer, 2008). Previous studies have shown that SGK3 is localized at the endosome in a manner that depends on an intact PX domain (Tessier and Woodgett, 2006a). Since SGK3 phosphorylation and subsequent activation are dependent on a functional PX domain, this would argue that SGK3 activation occurs primarily at the endosome. However, in our studies we find that PIK3CA-mediated activation of SGK3 does not require VPS34 (Figure S2A). The products of PIK3CA, PtdIns-3,4,5-P3 and PtdIns-3,4-P2 are synthesized primarily at the plasma membrane, indicating that the pool of PtdIns-3-P generated by INPP4B would also reside at the plasma membrane. In turn, a plasma membrane pool of PtdIns-3-P would recruit SGK3 leading to its activation. Although we cannot formally rule out that the PtdIns-3-P generated by INPP4B recruits SGK3 at the endosome, this is unlikely because studies have demonstrated that the endosomal recruitment of SGK3 is PI 3-K-independent (Tessier and Woodgett, 2006a). Tools for detecting endogenous SGK3 localization, such as activation-state antibodies, are not of sufficient sensitivity for use in immunofluorescence or immunohistochemistry, and to date our results aimed at localizing SGK3 in response PI 3-K and INPP4B signaling have been inconclusive. Future studies using more specific reagents will be required to address the relevant localization of SGK3 downstream of PIK3CA.
Oncogenic PIK3CA mutations are particularly prevalent in ER-positive breast cancers (Cancer Genome Atlas, 2012). Interestingly, INPP4B has been identified as a potential biomarker for luminal-type breast cancers (Fedele et al., 2010). Moreover, both INPP4B and SGK3 are transcriptionally regulated by estrogen (Harvell et al., 2006; Wang et al., 2011a; Xu et al., 2012). This suggests that the PIK3CA/INPP4B/SGK3 signaling axis is particularly important at promoting tumorigenesis in hormone receptor-positive cancers. It is also interesting to note that the original characterization of SGK3 as a required target for PIK3CA-mediated growth and survival was conducted in MCF7 cells, which express estrogen receptor (Vasudevan et al., 2009). Whether ER expression is necessary or rate-limiting for the PIK3CA/INPP4B/SGK3 pathway remains to be determined, since other mechanisms contribute to the transcriptional induction of both INPP4B and SGK3.
The majority of SGK3 substrates identified to date have also been shown to be phosphorylated by Akt, and thus likely share overlapping phenotypes during tumor progression (Tessier and Woodgett, 2006b). However, NDRG1 is the only known specific SGK substrate, although the functional consequence of SGK-mediated phosphorylation of NDRG1 is not known. We explored NDRG1 as a target of SGK3 in breast cancer cells, since NDRG1 is a known metastasis suppressor and its expression inversely correlates with breast cancer grade (Kovacevic and Richardson, 2006). Our results show that SGK3 is required for invasive migration, a pre-requisite for metastatic dissemination (Figure 5). We also find that NDRG1 protein levels are inversely correlated with SGK3 activity, suggesting that SGK3 might control NDRG1 stability (Figure 1C). It has been shown that SGK3 phosphorylation of NDRG1 primes for subsequent phosphorylation by GSK-3β (Murray et al., 2004). Our studies provide additional insight into NDRG1 regulation, as we show that SGK3 and GSK-3β control the interaction of NDRG1 with the substrate recognition domain of the E3 ligase SCF complex, Fbw7 (Figure 6). Fbw7 targets NDRG1 for degradation by the 26S-proteasome. In turn, loss of NDRG1 expression is known to reduce the recycling of E-cadherin to the plasma membrane, increase cell migration and eventual metastatic dissemination (Kachhap et al., 2007). Consistent with this, we find that silencing SGK3 inhibits cell migration, an effect that is phenocopied by expression of NDRG1 (Figure 5 and Figure 6). In this model, PIK3CA-mutant tumor cells with elevated SGK3 activity would maintain low NDRG1 levels, resulting in enhanced invasive migration.
In summary, we have identified a new mechanism by which PIK3CA transduces tumorigenic phenotypes in breast cancer, specifically though SGK3 activation. INPP4B catalytic activity is required for the ability of breast cancer cells with oncogenic PIK3CA to activate SGK3. This provides an alternative mechanism for the PI 3-K signaling axis to drive cancer progression under conditions in which Akt is inactive. These findings advocate for the development of small molecule inhibitors targeting SGK3, for PIK3CA-addicted tumors resistant to Akt inhibition.
Experimental Procedures
Cell culture and transfection
BT-20, DLD-1, HEK293T, HCC1937, HeLa, MDA-MB-435, MDA-MB-231, NIH/3T3 and MCF7 cells were cultured in DMEM (Cellgro) supplemented with 10% Fetal Bovine Serum (FBS; Gibco). ZR-75-1 cells were grown in RPMI 1640 supplemented with 10% FBS. MCF10A cells were maintained in DMEM/Ham’s F12 medium with 5% equine serum (Cellgro), 10 µg/ml insulin, 500 ng/ml hydrocortisone (Sigma-Aldrich), 20 ng/ml epidermal growth factor (EGF) (R&D systems) and 100 ng/ml cholera toxin (List Biological Labs). T47D and BT-549 were cultured in RPMI 1640 supplemented with 10% FBS and 10 µg/ml insulin. SUM159PT were grown in HAM’s F12 with 5% FBS and 5 µg/ml insulin and 1 µg/ml hydrocortisone. Hs578T were grown in 10% FBS/DMEM and 0.01 mg/ml bovine insulin. VPS34 fl/fl MEFs were provided by Wei-Xing Zong (Stony Brook University) and cultured as described (Jaber et al., 2012). Transient transfections were performed using X-tremeGENE HP(Roche).
Growth factors and inhibitors
Cells were stimulated for 20 min with 100 ng/ml recombinant human IGF-1 (R&D systems). MG132 (Cayman Chemicals) was used at 10 µM for 16 hr. Cycloheximide was used at 10 µg/ml. Inhibitors were as follows: BKM120, 1µM; A66, 1µM; rapamycin, 100nM; GSK650394, 10µM; and SB415286, 25 µM. Charcoal stripping was used to deplete cell growth media of estrogen. Cells were grown in 10% charcoal-stripped FBS/ phenol red-free media for 5 days prior to lysate collection.
Antibodies
Anti-Akt (#4685), anti-phospho-Akt S473 (#4060), anti-phospho-Akt T308 (#9275), anti-Cyclin E (#4129), anti-GSK3β (#9315), anti-phospho-GSK3β (#9336), anti-NDRG1 (# 5196), anti-phospho-NDRG1 (#3217), anti-p110α (#4249), anti-S6K (#2705), anti-pS6K T389 (#9205), anti-SGK3 (#8156), anti-Vps34 (#4263) and anti-β-actin (#4970) were purchased from Cell Signaling Technologies. Anti-FLAG M2 (#F3165) was from Sigma-Aldrich. Anti-SGK3 pT320 (# S1010-85W8) was from US Biologicals. Anti-GFP (# sc-9996), anti-GST (# sc-459) and anti-INPP4B (#sc-12318) were from Santa Cruz. Anti-Fbw7 (#A301–720A) was from Bethyl Laboratories. Anti-SGK3 (#LS-C132061) for immunoprecipitation was from LifeSpan BioSciences. Anti-HA was generated and purified from the 12CA5 hybridoma. Anti-p85, generated in house, has been described. Horseradish peroxidase-conjugated anti-goat was from Millipore. Horseradish peroxidase-conjugated anti-rabbit and anti-mouse immunoglobulin G antibodies were from Chemicon.
In vitro protein kinase assays
SGK3 was isolated from cell lysate with glutathione beads and incubated with 75ng GST-GSK3β substrate peptide and 250 µM cold ATP in kinase assay buffer (Cell Signaling Technologies) for 35 min at 30°C.
Transwell migration and invasion assays
Transwell assays were carried out as previously described (Chin and Toker, 2010). Briefly, NIH 3T3 conditioned media was added to the lower chamber of 8 µm pore Transwells (Corning). For invasion assays, wells were coated with 10 mg Matrigel (BD Biosciences). Cells were added to the upper chamber and migration was allowed to proceed for 2–24 hr at 37°C, depending on the cell line.
Immunoblotting
Immunoblotting was carried out as previously described (Chin and Toker, 2010).
Quantitative RT-PCR analysis
RNA was extracted using the Qiagen RNeasy. Reverse transcription reaction was carried out with ABI Taqman Reverse Transcriptional Reagent. Quantitative RT-PCR was performed using SYBR Green PCR master mix in an ABI Prism 7500 sequence detector (Applied Biosystems, Foster City, CA) with primers described in the supplemental methods.
Copy number analysis
The algorithm GISTIC (Genomic Identification of Significant Targets in Cancer) was used to analyze chromosome 8 gene copy numbers of the Tumorscape database. Tumorscape data analysis was performed as described (Beroukhim et al., 2010). SGK3 GISTIC profile was generated using 3131 cell lines and tumors. GISTIC analysis for breast cancer was performed using 243 breast samples and a limit of Q-value < 0.250 for focal peak amplification. Significance values were determined using the FDR (false discovery rate) test.
3D Culture Assay
3D cultures were performed as described (Debnath et al., 2003). Chamber slides were coated with growth factor-reduced Matrigel (BD Biosciences). Subsequently, 3000 cells were seeded to each chamber slide in assay media (10% FBS/RPMI, 2 µg/ml puromycin, and 2% Matrigel). Media was replaced 4 days after seeding. ImageJ was used to quantify 2D surface area on Day 8(NIH).
Soft Agar Colony Formation Assay
Soft agar assays were performed by coating 6 cm3 tissue culture plates with 5 mL of 0.8% Noble agar / growth media (10% FBS/DMEM and 2 µg/ml puromycin) and allowed to solidify at 20°C. 5×104 cells were plated in 1 mL top layer 0.4% Noble agar/ growth media. Growth media was added every 4 days and cells were counted and measured 28 days after seeding. Quantitation was performed using MatLab software (MathWorks).
Tumor xenografts
6–8 week-old female nude mice were purchased from Taconic and maintained in a pathogen-free environment. All procedures were carried out under the approval of the Institutional Animal Care and Use Committee at the Beth Israel Deaconess Medical Center and comply with the federal guidelines for maintenance and care of laboratory animals. 3 mice for each experimental condition were injected in the mammary fat pad with 1.5 × 106 ZR-75-1 cells in media with 50% Matrigel. Tumor volume was measured by: volume=0.52 × L × W^2.
Ubiquitination Assay
Cell based in vivo ubiquitination assays was performed as described (Chan et al., 2012). 293T cells were transfected with indicated plasmids for 48 hr followed by lysis in denatured buffer (6M guanidine-HCl, 0.1M Na2HPO4/NaH2PO4, 10 mM imidazole). The cell lysates were incubated with nickel beads for 3 hr, washed, and subjected to immunoblotting analysis.
Supplementary Material
Acknowledgements
We thank Lewis Cantley, John Blenis, Wade Harper and members of the Toker and Wei laboratories for productive discussions; Wade Harper, John Blenis, Lewis Cantley, Donald P. McDonnell, Sandra Marmiroli, Junying Yuan, Wei-Xing Zong, and Sushant Kachhap for providing reagents. This study was supported in part by grants from the Department of Defense Breast Cancer Research Program (J.A.G., BC093630), the National Institutes of Health (A. T., CA177910; R.B., U54CA143798; W.W., GM089763 and GM094777), and Men's Collaborative for Women's Cancers (R.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Supplementary material can be found online within the Pubmed electronic article.
References
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. The Biochemical journal. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Baselga J. Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. The oncologist. 2011;16(Suppl 1):12–19. doi: 10.1634/theoncologist.2011-S1-12. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, Hsiao K, Yuan J, Green J, Ospina B, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Science translational medicine. 2010;2:51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin YR, Toker A. Function of Akt/PKB signaling to cell motility, invasion and the tumor stroma in cancer. Cell Signal. 2009;21:470–476. doi: 10.1016/j.cellsig.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin YR, Toker A. The Actin-Bundling Protein Palladin Is an Akt1-Specific Substrate that Regulates Breast Cancer Cell Migration. Molecular cell. 2010;38:333–344. doi: 10.1016/j.molcel.2010.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Zhang J, Murga C, Yu H, Koller E, Monia BP, Gutkind JS, Li W. PTEN, but not SHIP and SHIP2, suppresses the PI3K/Akt pathway and induces growth inhibition and apoptosis of myeloma cells. Oncogene. 2002;21:5289–5300. doi: 10.1038/sj.onc.1205650. [DOI] [PubMed] [Google Scholar]
- Cleary JM, Shapiro GI. Development of phosphoinositide-3 kinase pathway inhibitors for advanced cancer. Current oncology reports. 2010;12:87–94. doi: 10.1007/s11912-010-0091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davids MS, Brown JR. Targeting the B cell receptor pathway in chronic lymphocytic leukemia. Leukemia & lymphoma. 2012;53:2362–2370. doi: 10.3109/10428194.2012.695781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nature reviews. Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nature reviews. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Fedele CG, Ooms LM, Ho M, Vieusseux J, O'Toole SA, Millar EK, Lopez-Knowles E, Sriratana A, Gurung R, Baglietto L, et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22231–22236. doi: 10.1073/pnas.1015245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestone GL, Giampaolo JR, O'Keeffe BA. Stimulus-dependent regulation of serum and glucocorticoid inducible protein kinase (SGK) transcription, subcellular localization and enzymatic activity. Cell Physiol Biochem. 2003;13:1–12. doi: 10.1159/000070244. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) The Biochemical journal. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, Barretina J, Lin WM, Rameh L, Salmena L, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvell DM, Richer JK, Allred DC, Sartorius CA, Horwitz KB. Estradiol regulates different genes in human breast tumor xenografts compared with the identical cells in culture. Endocrinology. 2006;147:700–713. doi: 10.1210/en.2005-0617. [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, Cantley LC, Brugge JS. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer research. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- Ivetac I, Gurung R, Hakim S, Horan KA, Sheffield DA, Binge LC, Majerus PW, Tiganis T, Mitchell CA. Regulation of PI(3)K/Akt signalling and cellular transformation by inositol polyphosphate 4-phosphatase-1. EMBO reports. 2009;10:487–493. doi: 10.1038/embor.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson S, Flanagan JU, Kolekar S, Buchanan C, Kendall JD, Lee WJ, Rewcastle GW, Denny WA, Singh R, Dickson J, et al. A drug targeting only p110alpha can block phosphoinositide 3-kinase signalling and tumour growth in certain cell types. The Biochemical journal. 2011;438:53–62. doi: 10.1042/BJ20110502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachhap SK, Faith D, Qian DZ, Shabbeer S, Galloway NL, Pili R, Denmeade SR, DeMarzo AM, Carducci MA. The N-Myc down regulated Gene1 (NDRG1) Is a Rab4a effector involved in vesicular recycling of E-cadherin. PLoS One. 2007;2:e844. doi: 10.1371/journal.pone.0000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. The Biochemical journal. 1999;339(Pt 2):319–328. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Deak M, Morrice N, Cohen P. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. The Biochemical journal. 1999;344(Pt 1):189–197. [PMC free article] [PubMed] [Google Scholar]
- Kovacevic Z, Richardson DR. The metastasis suppressor, Ndrg-1: a new ally in the fight against cancer. Carcinogenesis. 2006;27:2355–2366. doi: 10.1093/carcin/bgl146. [DOI] [PubMed] [Google Scholar]
- Lai CL, Srivastava A, Pilling C, Chase AR, Falke JJ, Voth GA. Molecular Mechanism of Membrane Binding of the GRP1 PH Domain. Journal of molecular biology. 2013;425:3073–3090. doi: 10.1016/j.jmb.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer [see comments] Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- Liu D, Yang X, Songyang Z. Identification of CISK, a new member of the SGK kinase family that promotes IL-3-dependent survival. Curr Biol. 2000;10:1233–1236. doi: 10.1016/s0960-9822(00)00733-8. [DOI] [PubMed] [Google Scholar]
- Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of biological chemistry. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, Balmain A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Skidan I, Yang J, Lugovskoy A, Reibarkh M, Long K, Brazell T, Durugkar KA, Maki J, Ramana CV, et al. Small molecule inhibition of phosphatidylinositol-3,4,5-triphosphate (PIP3) binding to pleckstrin homology domains. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20126–20131. doi: 10.1073/pnas.1004522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, Peggie M, Bain J, Bloomberg GB, Grahammer F, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. The Biochemical journal. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafov A, Sommer EM, Axten JM, Deyoung MP, Alessi DR. Characterization of GSK2334470, a novel and highly specific inhibitor of PDK1. The Biochemical journal. 2011;433:357–369. doi: 10.1042/BJ20101732. [DOI] [PubMed] [Google Scholar]
- Norris FA, Atkins RC, Majerus PW. The cDNA cloning and characterization of inositol polyphosphate 4-phosphatase type II. Evidence for conserved alternative splicing in the 4-phosphatase family. The Journal of biological chemistry. 1997;272:23859–23864. doi: 10.1074/jbc.272.38.23859. [DOI] [PubMed] [Google Scholar]
- Norris FA, Majerus PW. Hydrolysis of phosphatidylinositol 3,4-bisphosphate by inositol polyphosphate 4-phosphatase isolated by affinity elution chromatography. The Journal of biological chemistry. 1994;269:8716–8720. [PubMed] [Google Scholar]
- Pang H, Flinn R, Patsialou A, Wyckoff J, Roussos ET, Wu H, Pozzuto M, Goswami S, Condeelis JS, Bresnick AR, et al. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase. Cancer research. 2009;69:8868–8876. doi: 10.1158/0008-5472.CAN-09-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Scheid MP, Huber M, Damen JE, Hughes M, Kang V, Neilsen P, Prestwich GD, Krystal G, Duronio V. Phosphatidylinositol (3,4,5)P3 is essential but not sufficient for protein kinase B (PKB) activation; phosphatidylinositol (3,4)P2 is required for PKB phosphorylation at Ser-473: studies using cells from SH2-containing inositol-5-phosphatase knockout mice. The Journal of biological chemistry. 2002;277:9027–9035. doi: 10.1074/jbc.M106755200. [DOI] [PubMed] [Google Scholar]
- Sherk AB, Frigo DE, Schnackenberg CG, Bray JD, Laping NJ, Trizna W, Hammond M, Patterson JR, Thompson SK, Kazmin D, et al. Development of a small-molecule serum- and glucocorticoid-regulated kinase-1 antagonist and its evaluation as a prostate cancer therapeutic. Cancer research. 2008;68:7475–7483. doi: 10.1158/0008-5472.CAN-08-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer research. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier M, Woodgett JR. Role of the Phox homology domain and phosphorylation in activation of serum and glucocorticoid-regulated kinase-3. The Journal of biological chemistry. 2006a;281:23978–23989. doi: 10.1074/jbc.M604333200. [DOI] [PubMed] [Google Scholar]
- Tessier M, Woodgett JR. Serum and glucocorticoid-regulated protein kinases: variations on a theme. J Cell Biochem. 2006b;98:1391–1407. doi: 10.1002/jcb.20894. [DOI] [PubMed] [Google Scholar]
- Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, Hennessy BT, Tseng H, Pochanard P, Kim SY, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou D, Phung S, Masri S, Smith D, Chen S. SGK3 is an estrogen-inducible kinase promoting estrogen-mediated survival of breast cancer cells. Molecular endocrinology. 2011a;25:72–82. doi: 10.1210/me.2010-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fukushima H, Gao D, Inuzuka H, Wan L, Lau AW, Liu P, Wei W. The two faces of FBW7 in cancer drug resistance. Bioessays. 2011b;33:851–859. doi: 10.1002/bies.201100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Woodgett JR. Recent advances in the protein kinase B signaling pathway. Curr Opin Cell Biol. 2005;17:150–157. doi: 10.1016/j.ceb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Xu J, Liu D, Gill G, Songyang Z. Regulation of cytokine-independent survival kinase (CISK) by the Phox homology domain and phosphoinositides. J Cell Biol. 2001;154:699–705. doi: 10.1083/jcb.200105089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wan M, He Q, Bassett RL, Jr, Fu X, Chen AC, Shi F, Creighton CJ, Schiff R, Huo L, et al. SGK3 is associated with estrogen receptor expression in breast cancer. Breast cancer research and treatment. 2012;134:531–541. doi: 10.1007/s10549-012-2081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.