Abstract
Background:
Campylobacter is one of the leading bacterial species causing foodborne illnesses in humans. Antimicrobial agents have been extensively used for treatment of Campylobacter infections; but in the recent years, both animal and human isolates of this bacterium have shown resistance to several antibiotics such as tetracycline.
Objectives:
The aim of this study was to investigate the presence of genetic determinants of tetracycline resistance in Campylobacter spp. recovered from poultry carcasses in Shiraz, Iran.
Materials and Methods:
Eighty-three thermophilic Campylobacter spp. Isolates were first identified based on multiplex polymerase chain reaction (PCR) and then screened for presence of tetracycline resistance genes (tet (A), tet (B), tet (O) and te (S)) by PCR.
Results:
The overall prevalence of Campylobacter jejuni and C. coli among the examined isolates was 51.8% and 48.2%, respectively. Tetracycline resistance genes of tet (B) and tet (S) were not seen among these Campylobacter spp. Isolates, whereas the most common tet gene identified was tet (O), found in 83.1% (69/83) of all the isolates. The tet (O) gene sequence comparison between C. jejuni and C. coli showed 100% similarity and these sequences (JX853721and JX853722) were also identical to the homologous sequences of other strains of Campylobacter spp. existing in the GenBank databases. In addition, tet (A) was found in 18% (15/83) of Campylobacter spp. isolates. To our knowledge, this represents the first report of tet (A) in Campylobacter spp. There was 100% homology between the sequences of tet (A) from this study (JX891463 and JX891464) and the tet (A) sequences mentioned for other bacteria in the GenBank databases.
Conclusions:
The high prevalence of tet (O) resistance gene along with new detection of tet (A) resistance gene in Campylobacter spp. isolated from poultry carcasses revealed an extensive tetracycline resistance among Campylobacter isolates from poultry in Iran. It emphasized the need for cautious use of tetracycline in poultry production to decrease the extension of tetracycline-resistant Campylobacter spp.
Keywords: Campylobacter, Tetracycline Resistance
1. Background
Campylobacter spp. has now emerged as one of the leading bacterial species, causing foodborne illnesses in humans around the world. Poultry products, especially poultry meat, are major sources of Campylobacter infection in humans (1). Campylobacter infections produce little or no clinical diseases in poultry (2), but poultry carcasses have frequently been contaminated in processing plants due to the high prevalence of Campylobacter in the intestinal tract of market-age poultry and can be transferred to human via poultry carcasses (3). For many years, Campylobacter was considered susceptible to various antimicrobial agents; but in the recent years, both animal and human isolates of this bacterium have shown resistance to several antibiotics such as fluoroquinolones and tetracycline (4). Tetracycline is a broad-spectrum antibiotic with inhibitory activity against Gram-positive and Gram-negative bacteria as well as some other atypical and noninfectious microorganisms via inhibition of protein synthesis in these harmful agents (5).
In recent years, tetracycline resistance has emerged among many pathogenic and nonpathogenic species of bacteria. This resistance is due to different tetracycline resistance (tet) genes (6). The efflux pump and ribosomal protection genes are the two most important mechanisms of tetracycline resistance in various genera of bacteria and acquisition of new tetracycline resistance genes is mostly associated with mobile components such as plasmids or transposons, which are often conjugative elements (6). These pathways of resistance genes acquisition are along with the selection pressure hypotheses, saying that tetracycline resistance genes can be exchanged by different bacteria in different ecosystems and also between human and animals isolates of microorganisms (6). Tetracycline resistance in Campylobacter spp. is primarily mediated by a ribosomal protection protein (tetO), which is transferred as plasmid-encoded gene (7), or in the chromosome where it is not self-mobile (5). Another ribosomal protection protein (tetS) has a same characters and can be transferred as plasmid-encoded gene or be in the chromosome (5). The efflux genes, tet (A) and tet (B), code for an approximately 46-kDa membrane-bound efflux protein for membrane-associated proteins that export tetracycline from the cell (5).
2. Objectives
In recent years, polymerase chain reaction (PCR) assay has increasingly been applied for detection and identification of tetracycline resistance genes, but there was no report about the distribution of tetracycline resistance genes among Campylobacter isolates in Iran. Accordingly, in this study, we tried to determine the prevalence of tet (O) gene in C. jejuni and C. coli, isolated from broiler flocks in Shiraz, southern Iran. Along with tet (O), the prevalence of two common efflux protein genes (tetA and tetB) as well as a ribosomal protection protein (tetS) was investigated in these isolates. In our knowledge, this is one of the first evidences for presence of tetracycline resistance genes other than tet (O) in Campylobacter spp. isolates.
3. Materials and Methods
3.1. Samples
Samples were obtained from the Microbiology Department of Shiraz Veterinary School. These isolates were collected from 100 broiler flocks at the slaughterhouses of Shiraz county during August to September 2009 (8).
3.2. DNA Extraction
Eighty-three culture media containing Campylobacter spp. isolates were taken out from a 20°C refrigerator. The melted samples were cultured in an enriched broth media containing tryptic soy broth (TSB) (30 g/L), dextrose (2.5 g/L), sodium thioglycolate (0.5 g/L), rifampicin (10 mg/L), trimethoprim (10 mg/L), vancomycin (10 mg/L), ceftriaxone (10 mg/L), amphotericin-B (10 mg/L), incubated in a microaerophilic atmosphere (Anaerocult C, Merck) and at 37°C for hours, followed by incubation at 42°C for 44 hours (8). Unfortunately, due to the sensitive nature of this bacterium in frozen conditions (9), we could not retrieve the live bacteria. The DNA extraction was carried out directly on the culture media, using phenol-chloroform extraction (10).
3.3. Multiplex Polymerase Chain Reaction for Identification of Campylobacter Species
Multiplex PCR was used for simultaneous detection of C. jejuni and C. coli, with amplification of the mapA gene (11) and the ceuE gene, respectively (12). The primer sequence data and the annealing temperature of the three primers sets used for gene amplification are shown in Table 1.
Table 1. Primer Sequences, Annealing Temperatures, and Product Sizes of the Primers Used For Genus- and Species-Specific Gene Amplifications.
| Gene | Primer Name | Primer Sequence (5׳-3׳) | Annealing Temperature, °C | Product Size, bp |
|---|---|---|---|---|
| mapA | MapAF | CTATTTTATTTTTGAGTGCTTGTG | 52°C | 589 |
| MapAR | GCTTTATTTGCCATTTGTTTTATTA | |||
| ceuE | ColF | AATTGAAAATTGCTCCAACTATG | 52°C | 482 |
| ColR | TGATTTTATTATTTGTAGCAGCG |
Amplification reactions were performed in a 25-µL mixture, containing 1 U AmpliTaq polymerase (CinnaGen, Iran), 2 µL DNA template, 1 µL dNTPs (50 µM), 2.5 µL 10x PCR buffer (75 mM Tris-HCl, pH: 9.0, 2 mM MgCl2, 50 mMKCl, 20 mMNH42SO4 (CinnaGen, Iran)) and 1 µL (25pmoL) of each primer (CinnaGen, Iran) (Table 1). The final reaction mixture volume was adjusted to 25 µL, using distilled deionized water. The amplification reactions were carried out using a DNA thermocycler (MJ mini, BioRad, USA) with the following program: initial denaturation at 94°C for five minutes, followed by 35 cycles of denaturation at 94°C for one minute, annealing (with temperatures shown in Table 1) for one minute, and extension at 72°C for one minute, and a final extension step at 72°C for 10 minutes.
The PCR products were determined by electrophoresis of 7 μL of each reaction product in a 1.5% (w/v) agarose gel in Tris-borate-ethylenediamine tetraacetic acid (EDTA) electrophoresis buffer and visualized under the UV light. The positive controls for C. jejuni (ATCC 33291) and C. coli (RTCC 2541) were provided by Mast International Co. (USA) and Razi Vaccine and Serum Research Institute (Tehran, Iran), respectively.
3.4. Tetracycline Resistance Gene Polymerase Chain Reaction
Four tetracycline resistance genes (tet (O), tet (A), tet (B) and tet (S)) were investigated among the Campylobacter isolates, using PCR. Genes, primers and annealing temperatures are given in Table 2. The PCR reaction (25 μL) was performed in 10 Mm Tris-HCl, pH = 8.3-8.8, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs, 25 pmoL of forward and reverse primers (CinnaGen Inc., Tehran, Iran) for each tet gene stated in Table 2, and 1 U Taq DNA polymerase (Fermentas; Glen Burnie, Maryland), using 2 μL of the extracted DNA as template. The final volume of the reaction mixture was adjusted to 25 µL, using distilled deionized water. The similar amplification reactions and visual evaluation performed for Campylobacter species detection were used at this level. Only a different annealing temperature (56°C) was used for detection of tetracycline resistance genes. The positive controls for tetracycline resistance genes were obtained from the Aquatic Animal Health Unit, School of Veterinary Medicine, Shiraz University, Iran. Aeromonas sobria strain CW4 (JN806155), Lactococcus garvieae strain Ira.1s (JN998084), and Pseudomonas putida strain Fars 110 (JN937120) were used as positive controls for tet (M), tet (S), and tet (A), respectively.
Table 2. Genes, Primers, and Annealing Temperatures Used for Detection of Tetracycline Resistance Genes.
| Target Gene | Sequence (5'-3') | Temperature,°C | Amplicon Size, bp | Reference |
|---|---|---|---|---|
| Tet (O) | F: AACTTAGGCATTCTGGCTCAC | 56°C | 515 | (28) |
| R: TCCCACTGTTCCATATCGTCA | ||||
| Tet (A) | F: GTGAAACCCAACATACCCC | 56°C | 888 | (29) |
| R: GAAGGCAAGCAGGATGTAG | ||||
| Tet (B) | F: CCTTATCATGCCAGTCTTGC | 56°C | 774 | (29) |
| R: ACTGCCGTTTTTTCGCC | ||||
| Tet (S) | CATAGACAAGCCGTTGACC | 56°C | 667 | (28) |
3.5. Sequencing
The PCR products generated from different tet (O) and tet (A) genes in C. coli and C. jejuni were purified and sequenced (ABI 3730 capillary DNA analyzer; Applied Biosystems, Foster City, CA, USA) to assess the diversity of the genes and they were compared to the GenBank databases using the BLAST program maintained by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). In addition, multiple sequence alignments were made by the ClustalW method, using MEGA4 software (13).
4. Results
The multiplex PCR results showed that 51.8% (43/83) and 48.2% (40/83) of the 83 thermophilic Campylobacter spp. isolates were C. jejuni and C. coli, respectively. In the screening of tetracycline resistance genes, 83.1% of Campylobacter spp. isolates (69/83) were positive for tet (O) (Figure 1). tet (O) was seen in 92.5% of C. coli isolates (37/40) and 74.4% of C. jejuni isolates (32/43). tet (A) was found in 18% of Campylobacter spp. isolates (15/83), including 7 (16.3%) C. jejuni and 8 (20%) C. coli (Figure 2). Nine (11%) Campylobacter spp. isolates were positive for both tet (O) and tet (A). tet (B) and tet (S) resistance genes were not seen in these Campylobacter spp. isolates.
Figure 1. Polymerase Chain Reaction Amplification of the tet (O) Gene.
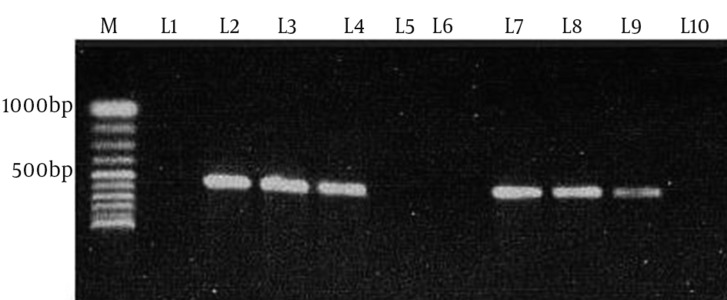
M, 100bp DNA ladder (Vivantis, Malaysia); L1, negative control (distilled water); L2, tet (O) positive control (JX853721); L3, L4, L7, L8 and L9, tet (O) positive samples; L5, L6 and L10, tet (O) negative samples.
Figure 2. Polymerase Chain Reaction Amplification of the tet (A) Gene.
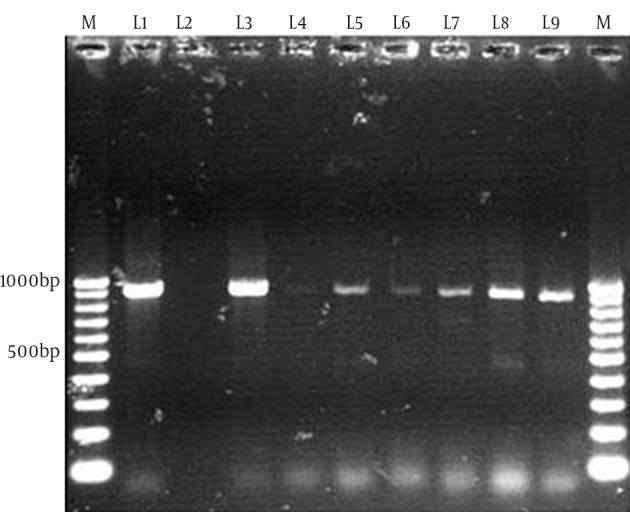
M, 100bp DNA ladder (Vivantis, Malaysia); L1: tet (A) positive control (JX891463); L2, tet (A) negative control (distilled water); L3-L9, tet (A) positive samples.
Four amplified PCR products (tetO and tetA from both C. jejuni and C. coli sources) were sequenced and aligned with the other related tet gene sequences obtained from GenBank. The tet (O) genes from C. jejuni (JX853722) and C. coli (JX853721) detected in this study showed 100% homology with the sequences of tet (O) gene, previously reported for Campylobacter spp. The sequences of tet (A) genes for C. coli and C. jejuni have been deposited in the GenBank database under the accession numbers JX891463 and JX891464, respectively. tet (A) sequence was not mentioned previously for Campylobacter spp.; thus, the tet (A) sequences from this study was compared with the tet (A) gene of other bacteria instead of Campylobacter. In common with tet (O), there was 100% homology between the present Iranian tet (A) sequence and other tet (A) sequences mentioned in GenBank database.
5. Discussion
There is a significant concern about the increasing antibiotic resistance in Campylobacter spp. isolated from both humans and animals. In developing countries like Iran, most of the antimicrobial agents in human pharmacopeia are also used in poultry industry (14). Between these antimicrobial agents, tetracycline family is the most commonly used antibiotic in domestic animals farming, including poultry industry, because of its low cost, efficacy, and lack of side effects (5). Tetracycline has been extremely used in poultry industry of Iran for many years and mass application of this antibiotic for grow promotions and treatment aims has led to a high tetracycline resistance in different bacteria isolated from different poultry samples in Iran (14-17). Campylobacter spp. is one of the most important bacteria separated from poultry carcasses in Iran poultry slaughterhouses, with high resistance to different antimicrobial agents. Resistance to tetracycline has been one of the most common findings among antimicrobial susceptibility tests of Campylobacter isolates from poultry carcasses and meats in Iran (15-17). Moreover, there were some reports from Iran about the increasing tetracycline resistance in human Campylobacter isolates (18).
Due to the fastidious and slowly growing nature of Campylobacter spp., detection and species differentiation of this organism by biochemical tests is not reliable and leads to controversial results. These difficulties have led to introducing various DNA-based PCR methods for genus and species detection of Campylobacter spp. Among these methods, multiplex PCR proved to be accurate and simple to perform (19). In our study, multiplex PCR results showed that of 83 thermophilic Campylobacter spp. isolates, 51.8% were C. jejuni and 48.2% were C. coli. Among tetracycline resistance determinants investigated in these isolates, tet (O) was detected in 83.1% of Campylobacter spp. isolates. The high prevalence of tet (O) genes in these isolates represents high tetracycline resistance in Campylobacter spp. isolates obtained from poultry carcasses in Iran, because the tet (O) gene is the most commonly reported determinant conferring resistance to tetracycline in the Campylobacter genus, and in many previous study, this gene was detected in all tetracycline-resistant Campylobacter spp. isolates (20-23).
Previous studies via antimicrobial susceptibility testing in Iran have reported the prevalence rates of tetracycline resistance of Campylobacter spp.to be 70.6% and 78.3% in chicken carcasses of Shahrekord and Ahvaz, respectively (15, 16). High rates of tetracycline resistance in these studies might be due to this fact that tetracycline has been commonly used in the poultry industry of Iran as a therapeutic or prophylactic agent (16). Therapeutic and sub therapeutic uses of tetracycline for a long period may evolutionally generate tetracycline-resistant Campylobacter species and lead to widespread distribution of tetracycline-resistant Campylobacter in animal reservoirs (2).
Most of C. coli isolates harbored the tet (O) resistance gene, so that the presence of tet (O) gene in C. coli (92.5%) was higher than C. jejuni (74.4%). C. coli has demonstrated faster resistance increasing ability to larger number of antimicrobial agents, compared with C. jejuni (24). There was no evidence of tet (B) or tet (S) existence, but tet (A) was found in 18% of the Campylobacter spp. isolates. This is the first evidence for tet (A) presence in Campylobacter spp. isolates. Tetracycline resistance in Campylobacter spp. is primarily mediated by a ribosomal protection protein, tet (O); however, in recent years, with the finding of a novel transferable antibiotic resistance gene, tet (24), in C. fetus, this monopoly has been questioned (25). Between different tetracycline resistance genes, tet (A) has some special features, predisposing its spread to new genera. Tet (A) has a broad host range and is carried by various environmental genera (26). Furthermore, recent studies have demonstrated that tet (A) gene can be located on mobile elements such as plasmids and can be horizontally transferred among bacterial strains (27). Falsafi et al., (14) reported high prevalence (50%) of tet (A) resistance gene among Escherichia coli isolates from the Iranian poultry farms samples. Poultry E. coli strains with this high prevalence of tet (A) resistance gene can be a reservoir for this antimicrobial resistance gene and may play a role in distribution of this resistance gene to other bacteria such as Campylobacter spp. in poultry industry.
Sequencing of tet (O) and tet (A) showed 100% homogeneity with other related tet (O) and tet (A) sequences in GenBank. This homogeneity is a persist finding in recent years; for example, sequence analysis was performed for tetracycline resistance (Tcr) plasmids from C. jejuni by other groups and our sequences appeared to be highly conserved, in view of the fact that recently sequenced Tcr plasmids are identical with first Tcr plasmid isolated in the late 1970s (24). Presence of efflux genes such as tet (A) has not been demonstrated in previous studies in genus Campylobacter (24), but our study showed new evidence of tet (A) presence in this genus that may have a complementary rule in the appearance of high level resistance to tetracycline in Campylobacter spp.
High prevalence of tet (O) resistance gene along with new detection of tet (A) resistance gene in Campylobacter spp. isolated from poultry carcasses, revealed an extensive tetracycline resistance among Campylobacter isolates obtained from poultry in Iran. It emphasizes the need for cautious use of tetracycline in poultry production to decrease the extension of tetracycline-resistant Campylobacter spp.
Acknowledgments
There were no acknowledgments.
Footnotes
Authors’ Contributions:For preparing this manuscript, all the authors contributed equally.
References
- 1.Humphrey T, O'Brien S, Madsen M. Campylobacters as zoonotic pathogens: a food production perspective. Int J Food Microbiol. 2007;117(3):237–57. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Luangtongkum T, Morishita TY, Ison AJ, Huang S, McDermott PF, Zhang Q. Effect of conventional and organic production practices on the prevalence and antimicrobial resistance of Campylobacter spp. in poultry. Appl Environ Microbiol. 2006;72(5):3600–7. doi: 10.1128/AEM.72.5.3600-3607.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeffrey JS, Tonooka KH, Lozanot J. Prevalence of campylobacter spp. from skin, crop, and intestine of commercial broiler chicken carcasses at processing. Poult Sci. 2001;80(9):1390–2. doi: 10.1093/ps/80.9.1390. [DOI] [PubMed] [Google Scholar]
- 4.Iovine NM. Resistance mechanisms in Campylobacter jejuni. Virulence. 2013;4(3):230–40. doi: 10.4161/viru.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232–60. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245(2):195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 7.Gibreel A, Tracz DM, Nonaka L, Ngo TM, Connell SR, Taylor DE. Incidence of antibiotic resistance in Campylobacter jejuni isolated in Alberta, Canada, from 1999 to 2002, with special reference to tet(O)-mediated tetracycline resistance. Antimicrob Agents Chemother. 2004;48(9):3442–50. doi: 10.1128/AAC.48.9.3442-3450.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ansari-Lari M, Hosseinzadeh S, Shekarforoush SS, Abdollahi M, Berizi E. Prevalence and risk factors associated with campylobacter infections in broiler flocks in Shiraz, southern Iran. Int J Food Microbiol. 2011;144(3):475–9. doi: 10.1016/j.ijfoodmicro.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Murinda SE, Nguyen LT, Oliver SP. Problems in isolation of Campylobacter jejuni from frozen-stored raw milk and bovine fecal samples: genetic confirmation using multiplex PCR. Foodborne Pathog Dis. 2004;1(3):166–71. doi: 10.1089/fpd.2004.1.166. [DOI] [PubMed] [Google Scholar]
- 10.Mayer MP. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 1995;163(1):41–6. doi: 10.1016/0378-1119(95)00389-n. [DOI] [PubMed] [Google Scholar]
- 11.Stucki U, Frey J, Nicolet J, Burnens AP. Identification of Campylobacter jejuni on the basis of a species-specific gene that encodes a membrane protein. J Clin Microbiol. 1995;33(4):855–9. doi: 10.1128/jcm.33.4.855-859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez I, Grant KA, Richardson PT, Park SF, Collins MD. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J Clin Microbiol. 1997;35(3):759–63. doi: 10.1128/jcm.35.3.759-763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 14.Falsafi T, Ebrahimi M, Asgarani E, Mirtorabi V. The pattern, association with multidrug-resistance and transferability of plasmid-mediated tetracycline resistance inEscherichia coli isolates from the poultry in Iran. A Microbiol. 2009;59(2):199–205. doi: 10.1007/bf03178318. [DOI] [Google Scholar]
- 15.Rahimi E, Ameri M. Antimicrobial resistance patterns of Campylobacter spp. isolated from raw chicken, turkey, quail, partridge, and ostrich meat in Iran. Food Control. 2011;22(8):1165–70. doi: 10.1016/j.foodcont.2011.01.010. [DOI] [Google Scholar]
- 16.Rahimi E, Momtaz H, Ameri M, Ghasemian-Safaei H, Ali-Kasemi M. Prevalence and antimicrobial resistance of Campylobacter species isolated from chicken carcasses during processing in Iran. Poult Sci. 2010;89(5):1015–20. doi: 10.3382/ps.2009-00090. [DOI] [PubMed] [Google Scholar]
- 17.Taremi M, Mehdi Soltan Dallal M, Gachkar L, MoezArdalan S, Zolfagharian K, Reza Zali M. Prevalence and antimicrobial resistance of Campylobacter isolated from retail raw chicken and beef meat, Tehran, Iran. Int J Food Microbiol. 2006;108(3):401–3. doi: 10.1016/j.ijfoodmicro.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 18.Hamidian M, Sanaei M, Azimi-Rad M, Tajbakhsh M, Dabiri H, Zali M. fla-typing, RAPD analysis, isolation rate and antimicrobial resistance profile of Campylobacter jejuni and Campylobacter coli of human origin collected from hospitals in Tehran, Iran. Ann Microbiol. 2010;61(2):315–21. doi: 10.1007/s13213-010-0141-1. [DOI] [Google Scholar]
- 19.Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J Clin Microbiol. 2002;40(12):4744–7. doi: 10.1128/JCM.40.12.4744-4747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazi W, Senok A, Al-Mahmeed A, Arzese A, Bindayna K, Botta G. Trends in antibiotic sensitivity pattern and molecular detection of tet(O)-mediated tetracycline resistance in campylobacter jejuni isolates from human and poultry sources. Jpn J Infect Dis. 2008;61(1):82–4. [PubMed] [Google Scholar]
- 21.Piddock LJ, Griggs D, Johnson MM, Ricci V, Elviss NC, Williams LK, et al. Persistence of Campylobacter species, strain types, antibiotic resistance and mechanisms of tetracycline resistance in poultry flocks treated with chlortetracycline. J Antimicrob Chemother. 2008;62(2):303–15. doi: 10.1093/jac/dkn190. [DOI] [PubMed] [Google Scholar]
- 22.Pratt A, Korolik V. Tetracycline resistance of Australian Campylobacter jejuni and Campylobacter coli isolates. J Antimicrob Chemother. 2005;55(4):452–60. doi: 10.1093/jac/dki040. [DOI] [PubMed] [Google Scholar]
- 23.Wardak S, Szych J, Zasada AA, Gierczynski R. Antibiotic resistance of Campylobacter jejuni and Campylobacter coli clinical isolates from Poland. Antimicrob Agents Chemother. 2007;51(3):1123–5. doi: 10.1128/AAC.01187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebreyes WA, Thakur S, Morrow WE. Campylobacter coli: prevalence and antimicrobial resistance in antimicrobial-free (ABF) swine production systems. J Antimicrob Chemother. 2005;56(4):765–8. doi: 10.1093/jac/dki305. [DOI] [PubMed] [Google Scholar]
- 25.Abril C, Brodard I, Perreten V. Two novel antibiotic resistance genes, tet(44) and ant(6)-Ib, are located within a transferable pathogenicity island in Campylobacter fetus subsp. fetus. Antimicrob Agents Chemother. 2010;54(7):3052–5. doi: 10.1128/AAC.00304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang XX, Zhang T, Fang HH. Antibiotic resistance genes in water environment. Appl Microbiol Biotechnol. 2009;82(3):397–414. doi: 10.1007/s00253-008-1829-z. [DOI] [PubMed] [Google Scholar]
- 27.Szczepanowski R, Krahn I, Linke B, Goesmann A, Puhler A, Schluter A. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology. 2004;150(Pt 11):3613–30. doi: 10.1099/mic.0.27317-0. [DOI] [PubMed] [Google Scholar]


