Abstract
Background:
Lactic acid bacteria, especially Lactobacillus spp., have been considered as excellent probiotic microorganisms, because of their activities in reducing the enteric diseases and maintaining healthy poultry.
Objectives:
The current study aimed to evaluate the phenotypic characteristics and the probiotic potentials of Lactobacillus spp. isolated from poultry.
Materials and Methods:
A total of 168 lactic acid bacteria (LAB) were isolated from healthy six and twenty-one-day old chickens and their feed samples. The isolated bacteria were identified by morphological, biochemical, and molecular tests including Polymerase Chain Reaction (PCR) and 16S rRNA gene sequencing. Biochemical fingerprinting with Phene Plate system (Ph-P) was done and the acid and bile resistant lactobacilli were subjected to the antibiotic susceptibility test.
Results:
Amongst all of the examined LAB, 30.3% were resistant to bile and acid. Most of the isolated LAB (57.1%) belonged to the genus Lactobacillus with Lactobacillus brevis (78.1%) as the dominant species followed by L. reuteri (16.6%), L. plantarum (3%), and L. vaginalis (2%). The remaining isolates were identified as Pediococcus spp. (42.9%). The Ph-P cluster analysis of 75 L. brevis and 16 L. reuteri strains showed high phenotypic diversity. Whilst the results of Ph-P typing from L. reuteri strains showed low phenotypic variations especially among the strains sensitive to acid and bile salts.
Conclusions:
Overall, the results showed that some of the high potential probiotic LAB species existed in Iranian poultry.
Keywords: Lactobacillus; Poultry, Probiotic
1. Background
The normal flora bacteria in intestinal tract of chicken is about 1011 per gram (1). They play an important role in the health of host animals by improving their intestinal balance and, to some extent, preventing ingested pathogens (2), which result in healthier chickens, and in turn, can improve the health of humans following their consumption. Commercial poultry production is one of the most important sources of animal protein for humans and an important economic activity in many countries (3). On the other hand, when poultry is exposed to stressful conditions, including diseases and unsanitary environmental conditions, serious economic losses may occur. Understanding the ecology of chicken gut's microflora may, therefore, help to maintain healthy chicken products and prevent the economic losses. Lactic acid bacteria (LAB) are among important normal microbial population in chicken (4).
The production of antimicrobial compounds, especially organic acids and bacteriocins, by LAB has provided these microorganisms with a competitive advantage over the other species as an excellent probiotic candidate (5). Some of the proposed modes of action of probiotics in poultry include their ability to tolerate acid, bile, and also adhere to the intestinal epithelium of the hosts, to show an antagonistic activity against pathogenic bacteria (6). Overall, the results have suggested that the use of probiotics in the poultry industry has resulted in the improvement of chicken body weight, feed conversion ratio and egg weight (7). The diversity of microorganisms present in digestive tract of chickens is very high. The most commonly identified Lactobacillus species are L. crispatus, L. reuteri, and L. salivarius (8, 9). In addition, a great deal of diversity could be found in lesser reported microorganisms including Bacillus, Enterococcus, Streptococcus, Pediococcus and Bifidobacteria (3).
2. Objectives
The current study aimed to isolate, and identify the phenotypic characteristics and analysis of the probiotic characteristics of the Lactobacillus spp. in the caeca, feces, and chicken feed obtained from poultry farms in the suburbs of Tehran, Iran.
3. Materials and Methods
3.1. Isolation of Lactobacilli
A total of 21 samples from ileum and caeca of healthy six (10) and twenty-one-day (11) old chickens, their fecal samples, and chicken feed were collected from two broiler chicken farms in the suburbs of Tehran, Iran. The chickens were fed with commercial corn-soy diet, without animal protein, and growth-promoting antibiotics. Serial decimal dilutions of the samples were prepared in phosphate-buffered saline and plated onto Man Rogosa and Sharpe (MRS) agar (Merck, Germany) and incubated at 37°C for 48 hours under microaerophilic condition. Colonies of different morphological appearance on MRS agar plates were picked and recultured to provide pure culture. For long-term storage, the bacteria were stored at -80°C in MRS broth with 20% glycerol.
3.2. Identification by Biochemical Methods
Isolated lactic acid bacteria were identified by Gram-staining, catalase reaction test and sugar fermentation pattern, using different carbohydrates including salisin, arabinose, sucrose, maltose, mannose, mannitol, raffinose, rhamnose, lactose, galactose, xylose, melezitose, ribose, gluconate, and also glucose and sterile water were used as positive and negative controls.
3.3. Identification by Molecular Biology Methods
Total DNA was extracted with peqGOLD Bacterial DNA Kit (peQlab, Erlangen, Germany) according to the manufacturer's instructions. Molecular identification was carried out by Polymerase Chain Reaction (PCR) using the previously designed Lactobacillus 16S rRNA specific primers for-lac (5´-TGGAAACAGGTGCTAATACCG-3´) and Rev-lac (5´-CCATTGTGGAAGATTCCC-3´) genes (11). The PCR was performed under the following conditions: 94°C for 5 minutes; 30 cycles of 94°C for 30 seconds, 57°C for 30 seconds and 72°C for 30 seconds, and the final extension at 72°C for 7 minutes. Species-specific PCR method was used for L. brevis (10). The amplification conditions were two minutes at 94°C for initial denaturation followed by 25 cycles consisting of one minute at 94°C for denaturation, one minute at 40°C for annealing, followed by one minute at 72°C for elongation. The final extension at 72°C was prolonged to 10 minutes. Finally species-specific multiplex PCR was performed for L. acidophilus, L. casei group, L. delbrueckii, L. gasseri, L. plantarum, L. rhamnosus and L. reuteri under the following conditions: one cycle at 94°C for five minutes, 40 cycles of 94°C for 30 seconds, 51°C for 40 seconds and 72°C for 30 seconds were performed, respectively. Final extension was carried out at 72°C for seven minutes (12). The strains approved by PCR amplification for Lactobacillus 16S rRNA with different sugar fermentation patterns were subjected to sequencing. The data of sequences were compared with the sequences deposited in the GenBank database (National Center for Biotechnology Information) using the BLAST algorithm.
3.4. Phene Plate System
All of lactobacilli isolates were subjected to a biochemical fingerprinting with Phene Plate system according to the manufacturer’s instructions (PhPlate Micro-plate Techniques AB, Stockholm, Sweden) modified for typing lactobacilli (PhP-LB). Phene Plate system is based on the evaluation of the kinetics of biochemical reactions by reading test results at several occasions. The microplates contain four sets of dehydrated reagents (23 different sugar including arabinose, xylose, galactose, maltose, cellobiose, trehalose, palatinose, sucrose, lactose, melibiose, mannose, melezitose, inosine, mannitol, arbutin, sorbitol, gallactose, sorbose, rhamnose, tagatose, amigdalin, gluconate, and salicin), specifically selected for Lactobacillus genus. The plate was incubated at 37°C and the color of each well was measured after 24, 48, and 72 hours. Scanned images were analyzed by PhPWIN software package (AB, Stockholm, Sweden).
3.5. pH and Bile Tolerance
All of the Lactobacillus strains were tested for their ability to tolerate low pH and bile salt. Bacteria were grown overnight in MRS broth (1 mL), centrifuged at 6000 g, and re-suspended in phosphate buffer saline (PBS). Then, 50 µL of suspension was transferred to 5mL of MRS broth containing 0.4% bile salts (Merck, Germany) and incubated at 37°C for six hours. To examine the survival rate of different Lactobacillus strains under acidic conditions, 50µL of each bacterial suspension prepared in PBS was transferred to 5mL of phosphate-buffered saline at pH 3 and incubated at 37°C for six hours. The suspensions were centrifuged at 6000 rpm for 10 minutes and washed with sterile saline solution. Bacteria were suspended in physiological solution and a series of tenfold dilution (10-2 to 10-10) was prepared. One hundred microliter of each dilution was plated onto MRS agar (Merck, Germany) and incubated anaerobically at 37°C for 24 to 48 hours; then, the Survival rate of the Lactobacillus strains was measured by counting the cells.
3.6. Antibiotic Susceptibility Testing
The agar diffusion method was used to determine the antibiotic susceptibility patterns of isolated lactobacilli on MRS agar plate, instead of Muller Hinton base Medium, bases on Bauer et al. protocol (13). Fourteen antibiotics from different classes, including penicillin G (10 µg), clindamycin (2 µg), erythromycin (15 µg), tetracycline (30 µg), amoxicillin (25 µg), rifampin (5 µg), ciprofloxacin (5 µg), nitrofurantoin (300µg), amikacin (30µg), chloramphenicol (30µg), oxacillin (1 µg), augmentin (amoxicillin and clavulanate) (30 µg), tobramycin (10 µg) and streptomycin (10µg) (MAST Diagnostics, U.K) were tested in the current study. The discs were placed on the agar plates, and incubated at 37°C for 48 hours. The diameters of inhibition zones were measured and the results were expressed as sensitive and resistant according to CLSI standards.
4. Results
4.1. Isolation of Lactobacillus Species
In the current study, 168 lactic acid bacteria were isolated from rectal swabs, fecal and feed samples, out of which, 89 (53%) were isolated from 21-day-old chickens with 22, 63 and 4 isolates, and 79 (47%) isolates were collected from 6-day-old chickens with 6, 67 and 6 isolates from stools, rectal swabs and feeds, respectively.
4.2. Identification of the Isolates
Amplification with primers for 16S rRNA gene and sequencing of 247 base pair amplicon showed that 96 isolates (57.1%) belonged to Lactobacillus, and the remaining isolates were identified as Pediococcus spp. The PCR results showed the presence of 75 (78.1%), two (2%), three (3%), and 16 strains (16.6%) as L. brevis, L. vaginalis, L. plantarum and L. reuteri (Figure 1), respectively. Sequencing confirmed the gene amplification results.
Figure 1. Agarose Gel Electrophoreses of Polymerase Chain Reaction PCR Products.
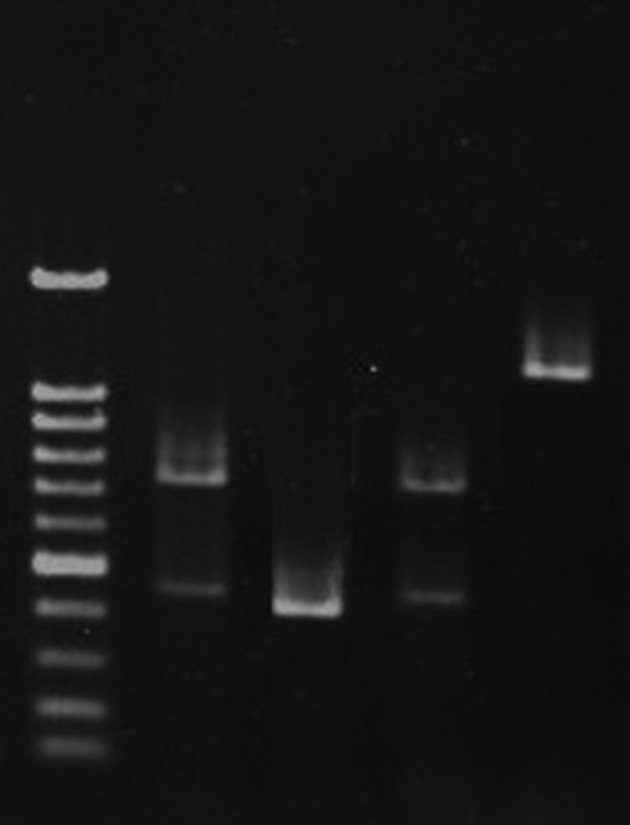
Lane M, 100 bp-DNA ladder; Lane one, L. rhamnosus; Lane two, L. plantarum; Lane three, L. rhamnosus; Lane four, L. reuteri.
4.3. Phplate
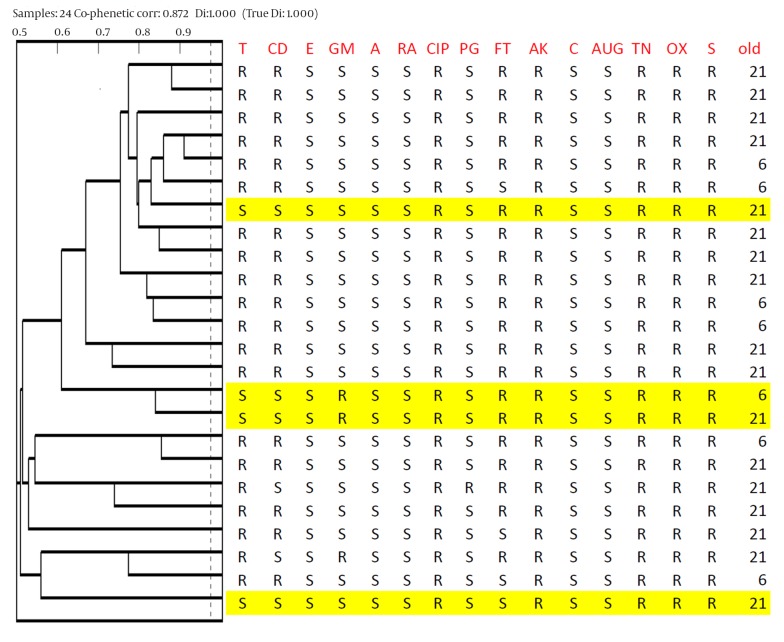
Seventy five L. brevis and sixteen L. reuteri strains were differentiated into primary groups based on carbohydrate fermentation assays for Lactobacillus strains typing (PhP-LB). Data were obtained after three continuous readings of change color by bacteria on microtiter plates, including 23 different sugars, and analyzed with PhPWIN software. Data regarding the biochemical reactions of all isolates were compared and clustered. All Lactobacillus strains were run in the same intra-assay and identify level ID level was set at 0.975. Strains with similarity more than 0.975 were defined as the same PhP type. The cluster analysis of 75 L. brevis strains showed that 42 (56%) isolates were single types (STs) and the remaining 33 isolates belonged to six common types (CTs). The most frequent CTs were CT1 and CT3 which included eight isolates, five isolates from twenty-one-day-old and three isolates from six-day-old chickens, and 13 isolates, five isolates from twenty-one-day-old and eight isolates from six-day-old chickens, respectively. The other CTs, CT2, CT4, CT5 and CT6 included four, four, two and two isolates, respectively. The results of PhP test for 24 bile and acid resistant L. brevis strains are shown in Figure 2.
Figure 2. Dendrogram derived from UPGMA clustering of the Phene Plate Typing Data of 24 bile and acid resistant Lactobacillus brevis isolates from chicken.
PhP typing divided the 16 L. reuteri isolates into one CT (CT1) and six STs. The cluster analysis of 16 L. reuteri strains showed high phenotypic similarity. For example, CT1 included 10 isolates collected from six-day-old chickens.
4.4. pH and Bile Tolerance
All 96 Lactobacillus isolates were tested for resistance to acid and bile salts. Out of which, 31 isolates were identified as resistant to low pH and bile salts. Only 24 L. brevis (32%) and two L. reuteri (12.5%) strains showed resistance to acid and bile salts. Two L. vaginali and three L. plantarum strains were resistant to acid and bile salts.
4.5. Antibiotic Susceptibility Testing
Antibiotic susceptibility tests showed that all strains were sensitive to Augmentin, amoxicillin, erythromycin, chloramphenicol, and rifampin. The isolates showed different degrees of resistance to the other antibiotics as follow: tetracycline 20 (64.5%), ciprofloxacin 31 (100%), clindamycin 21 (67.7%), penicillinG1 (3.2 %), nitrofurantoin 22 (70.9%), amikacin 31 (100%), oxacillin 31 (100%), tobramycin 31 (100%), and streptomycin 31 (100%) (Table 1)
Table 1. Antibiotic Susceptibility of Lactobacillus Strains Isolated From Chickensa.
| Antibiotic Resistance | ||||
|---|---|---|---|---|
| Antibiotic | Lactobacillusbrevis | Lactobacillus plantarum | Lactobacillus vaginalis | Lactobacillus reuteri |
| Tetracycline | 20 (83.3) | 0 (0) | 0 (0) | 0 (0) |
| Clindamycin | 18 (75) | 3 (100) | 0 (0) | 0 (0) |
| Erythromycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Amoxicillin | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Rifampin | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Ciprofloxacin | 24 (100) | 3 (100) | 2 (100) | 2 (100) |
| Penicillin G | 1 (4.1) | 0 (0) | 0 (0) | 0 (0) |
| Nitrofurantoin | 21 (87) | 0 (0) | 1 (50) | 0 (0) |
| Amikacin | 24 (100) | 3 (100) | 2 (100) | 2 (100) |
| Chloramphenicol | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Augmentin | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Tobramycin | 24 (100) | 3 (100) | 2 (100) | 2 (100) |
| Oxacillin | 24 (100) | 3 (100) | 2 (100) | 2 (100) |
| Streptomycin | 24 (100) | 3 (100) | 2 (100) | 2 (100) |
a Data are presented as No. (%).
5. Discussion
In broiler nutrition, Lactobacillus, Streptococcus, Pediococcus, Bacillus, Bifidobacterium, Enterococcus, Aspergillus, Candida, and Saccharomyces were the probiotic species (7), which play significant role in animal health. In the current study, the search for LAB with probiotic properties was conducted with rectal swaps, fecal, and the feed samples collected from a poultry farm, near Tehran, Iran. Phenotypic characteristics and specific PCR were used to identify 168 samples. Only 96 (57.1%) isolates were confirmed by 16s DNA sequencing as Lactobacillus species. The results showed that the primers used in the current study were probably not specific enough to detect all Lactobacillus species. The reason for the primers unspecificity is under investigation in this laboratory. The remaining isolates (42.9%) were identified as Pediococcus spp. , considered as common normal flora with excellent probiotic activities (3, 14, 15). In the current study, 75 of the isolated Lactobacillus spp. (78.1%) were L. brevis , considered as one of the most frequently isolated species from dairy products and animals (16, 17). The present study further indicated that L. brevis was the dominant species in two separate broiler farms at distinctive locations and in different age groups. The other Lactobacillus species isolated in the current study were L. reuteri (16.6%), and L. plantarum (3%). Lactobacillus plantarum and L.reuteri were also considered as good candidates for probiotic in humans and animals (18, 19). An investigation by Majidzade Heravi et al. (18) showed that L. crispatus, L. salivarius, and L. reuteri were the most frequently species isolated from gastrointestinal tract of broiler chickens in Mashhad, Iran. Taheri et al. (20) also found that L. crispatus showed potential probiotic characteristics compared to the other strains isolated from the gastrointestinal tract of chickens. The difference between the present study results and the reports by other investigators about different isolated Lactobacillus species could be due to the fact that in the current study, the obtained isolates were from broilers with no antibiotics feeding, diet composition, age of the chicken, breed, geographic locations and the section of the intestinal tract such as small intestine, ileum, and cecum. All these factors were found to affect the composition of the intestinal normal bacterial content in particular Lactobacillus species (3, 7, 21).
In the current study, 30.3% of the rectal isolates showed resistance to acid and bile , which was significantly higher than the reports by Belkacem et al. (22) and Jin et al. (23) who showed that 17% and 16% of their isolates were resistant to acid and bile, respectively. In addition, PhPlate analysis showed high diversity among the isolated L. brevis strains. This was consistent with the reports published by the other investigators who showed a great deal of diversity, using other phenotypic techniques, within Lactobacillus species in the poultry raised under commercial conditions (24). One of the safety aspects of the probiotics which should be considered is their ability to transfer or obtain antibiotic resistance genes. Non-pathogenic enteric bacteria may transfer their antibiotic resistance genes to pathogenic bacteria in gut microbiota. It is, therefore, important to select the probiotic strains without promiscuous antibiotic resistance elements. In the current study, antibiotic susceptibility tests showed that all of the strains were sensitive to common antibiotics such as amoxicillin, erythromycin, chloramphenicol, Augmentin, and rifampin.
Most of the isolates were sensitive to penicillin G (96.8%) which was in accordance with the findings reported by Danielsen (25) and Coppola (26) et al. Natural resistance against ciprofloxacin, nitrofurantoin, and vancomycin was also observed amongst the isolated species. In accordance with the other reposts, the current study also found resistance to streptomycin, amikacin, ciprofloxacin, tobramycin, and oxacillin (26-28). Overall, the results of the current study showed no significant antibiotic resistance amongst the isolates. Diverse populations of Lactobacillus, as determined by PhPlate, found in the current study were proportional to the isolated large number of acid and bile resistant Lactobacillus species, and the proportion was higher than the reported figures by the other investigators.
Footnotes
Author’s Contribution:1- Study concept and design: Malihe Talebi, Mohammad R. Pourshafie 2- Acquisition of data: Nasrin Noohi, Mahdi Rohani 3- Analysis and interpretation of data: Malihe Talebi, Gholamhosein Ebrahimipour , Mohammad R. Pourshafie 4- Drafting of the manuscript: Nasrin Noohi ,Malihe Talebi 5- Critical revision of the manuscript for important intellectual content: Mohammad R. Pourshafie 6- Administrative, technical, and material support: Gholamhosein Ebrahimipour, Mohammad R. Pourshafie 8- Study supervision: Mohammad R. Pourshafie.
Financial Disclosure:This study was supported by a grant (No. 90007454) from the Deputy of Research, Iran National Science Foundation.
Funding/Support:This project was supported by Iran National Science Foundation.
References
- 1.Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol. 2008;74(16):4985–96. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patterson JA, Burkholder KM. Application of prebiotics and probiotics in poultry production. Poult Sci. 2003;82(4):627–31. doi: 10.1093/ps/82.4.627. [DOI] [PubMed] [Google Scholar]
- 3.Ohimain EI, Ofongo RTS. The Effect of Probiotic and Prebiotic Feed Supplementation on Chicken Health and Gut Microflora. Int J Anim Veter. 2012;4(2):135–43. [Google Scholar]
- 4.Musikasang H, Tani A, H-kittikun A, Maneerat S. Probiotic potential of lactic acid bacteria isolated from chicken gastrointestinal digestive tract. World J Microbiol Biotechnol. 2009;25(8):1337–45. doi: 10.1007/s11274-009-0020-8. [DOI] [Google Scholar]
- 5.Corcionivoschi N, Drinceanu D, Stef L, Luca I, Julean C, Mingyart O. Probiotics-identification and ways of action. Innov Rom Food Biotechnol. 2010;6:1–11. [Google Scholar]
- 6.Ghadban GS. Probiotics in broiler production. Arch Geflugelk. 2002;66(2):49–58. [Google Scholar]
- 7.Lutful Kabir SM. The role of probiotics in the poultry industry. Int J Mol Sci. 2009;10(8):3531–46. doi: 10.3390/ijms10083531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephenson DP, Moore RJ, Allison GE. Lactobacillus strain ecology and persistence within broiler chickens fed different diets: identification of persistent strains. Appl Environ Microbiol. 2010;76(19):6494–503. doi: 10.1128/AEM.01137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raja MMM, Raja A, Imran MM. Lactobacillus as a probiotic feed for chickens. Int J Poultry Sci. 2009;8:763–7. doi: 10.3923/ijps.2009.763.767. [DOI] [Google Scholar]
- 10.Guarneri T, Rossetti L, Giraffa G. Rapid identification of Lactobacillus brevis using the polymerase chain reaction. Lett Appl Microbiol. 2001;33(5):377–81. doi: 10.1046/j.1472-765x.2001.01014.x. [DOI] [PubMed] [Google Scholar]
- 11.McOrist AL, Jackson M, Bird AR. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J Microbiol Methods. 2002;50(2):131–9. doi: 10.1016/s0167-7012(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 12.Kwon HS, Yang EH, Yeon SW, Kang BH, Kim TY. Rapid identification of probiotic Lactobacillus species by multiplex PCR using species-specific primers based on the region extending from 16S rRNA through 23S rRNA. FEMS Microbiol Lett. 2004;239(2):267–75. doi: 10.1016/j.femsle.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 13.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–6. [PubMed] [Google Scholar]
- 14.Mikulski D, Jankowski J, Naczmanski J, Mikulska M, Demey V. Effects of dietary probiotic (Pediococcus acidilactici) supplementation on performance, nutrient digestibility, egg traits, egg yolk cholesterol, and fatty acid profile in laying hens. Poult Sci. 2012;91(10):2691–700. doi: 10.3382/ps.2012-02370. [DOI] [PubMed] [Google Scholar]
- 15.Mountzouris KC, Tsirtsikos P, Kalamara E, Nitsch S, Schatzmayr G, Fegeros K. Evaluation of the efficacy of a probiotic containing Lactobacillus, Bifidobacterium, Enterococcus, and Pediococcus strains in promoting broiler performance and modulating cecal microflora composition and metabolic activities. Poult Sci. 2007;86(2):309–17. doi: 10.1093/ps/86.2.309. [DOI] [PubMed] [Google Scholar]
- 16.Skelin A, Mrkonjić Fuka M, Canzek Majhenic A, Redžepovic S, Samarzija D, Bogovič Matijasic B. Phenotypic and genotypic characterization of indigenous Lactobacillus community from traditional Istrian ewe's cheese. Food Technol Biotechnol. 2012;50(3):362–70. [Google Scholar]
- 17.Ramos CL, Thorsen L, Schwan RF, Jespersen L. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 2013;36(1):22–9. doi: 10.1016/j.fm.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Majidzadeh Heravi R, Kermanshahi H, Nassiri MR, Heravi Moussavi A. Screening of lactobacilli bacteria isolated from gastrointestinal tract of broiler chickens for their use as probiotic. Afr J Microbiol Rese. 2011;5 [Google Scholar]
- 19.Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Moller PL, Michaelsen KF, Paerregaard A, et al. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65(11):4949–56. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taheri HR, Moravej H, Tabandeh F, Zaghari M, Shivazad M. Screening of lactic acid bacteria toward their selection as a source of chicken probiotic. Poult Sci. 2009;88(8):1586–93. doi: 10.3382/ps.2009-00041. [DOI] [PubMed] [Google Scholar]
- 21.Knarreborg A, Simon MA, Engberg RM, Jensen BB, Tannock GW. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl Environ Microbiol. 2002;68(12):5918–24. doi: 10.1128/AEM.68.12.5918-5924.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belkacem B, Meriem M, Mebrouk K. Probiotic Potential of Thermotolerants Lactobacilli Isolated from Chicken Gastrointestinal Digestive and Their Use as Poultry Feed. World Appl Sci J. 2009;7:951–7. [Google Scholar]
- 23.Jin LZ, Ho YW, Abdullah N, Jalaludin S. Acid and bile tolerance of Lactobacillus isolated from chicken intestine. Lett Appl Microbiol. 1998;27(3):183–5. doi: 10.1046/j.1472-765x.1998.00405.x. [DOI] [PubMed] [Google Scholar]
- 24.Stephenson DP, Moore RJ, Allison GE. Comparison and utilization of repetitive-element PCR techniques for typing Lactobacillus isolates from the chicken gastrointestinal tract. Appl Environ Microbiol. 2009;75(21):6764–76. doi: 10.1128/AEM.01150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danielsen M, Wind A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int J Food Microbiol. 2003;82(1):1–11. doi: 10.1016/s0168-1605(02)00254-4. [DOI] [PubMed] [Google Scholar]
- 26.Coppola R, Succi M, Tremonte P, Reale A, Salzano G, Sorrentino E. Antibiotic susceptibility of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. Le Lait. 2005;85(3):193–204. doi: 10.1051/lait:2005007. [DOI] [PubMed] [Google Scholar]
- 27.Ashraf R, Shah NP. Antibiotic resistance of probiotic organisms and safety of probiotic dairy products. Int Food Res J. 2011;18(3):837–53. [Google Scholar]
- 28.Zhou JS, Pillidge CJ, Gopal PK, Gill HS. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int J Food Microbiol. 2005;98(2):211–7. doi: 10.1016/j.ijfoodmicro.2004.05.011. [DOI] [PubMed] [Google Scholar]