Abstract
Background:
Mycoplasma synoviae is an important avian pathogen which can cause both respiratory disease and synovial joint inflammation (synovitis) in poultry. Mycoplasmas spp. may cause the respiratory system infection in ostriches with symptoms such as inflammation of the nose, trachea and also damages of lungs.
Objectives:
The current study aimed to use the M. synoviae specific Polymerase Chain Reaction (PCR) and microbiological methods in order to isolate and identify M. synoviae from suspected ostriches in Kerman Province, Iran, and compare the two methods (microbiological and PCR) employed to confirm Mycoplasmal contamination of ostrich lungs.
Materials and Methods:
Fifty three samples of different parts of lung and trachea were immediately collected after slaughtering the ostriches in Kerman Province six months. Samples were cultured in the same conditions in pleuropneumonia-like organism (PPLO) broth and to isolate and identify M. synoviae, PCR and microbiological methods were conducted. The identified isolates were confirmed by specific amplification of 16S rRNA gene (163 and 207 base pair).
Results:
In the current study, 25 and 17 out of 53 ostrich samples were identified as Mycoplasma-positive in the PCR and microbiological methods, respectively; and 13 out of 25 the mentioned Mycoplasma-positive samples were also confirmed by PCR method.
Conclusions:
The current study showed that PCR method is time consuming, effective, and efficient method to detect M. synoviae infection in ostriches. PCR method could be recommended as an alternative for culturing; M. synoviae was isolated from ostriches for first time in Kerman Province, Iran.
Keywords: Mycoplasma synoviae, PCR, Microbiological methods
1. Background
Mycoplasmas are very small prokaryotes, devoid of cell walls and bounded only by a plasma membrane. Mycoplasma synoviae is one of the most important pathogenic chickens Mycoplasmas. M. synoviae infection occurs as a subclinical upper respiratory infection. It may cause air sac lesions when combined with Newcastle Disease (ND), infectious bronchitis (IB), or both. Less frequently, M. synoviae becomes systemic and results in infectious synovitis, acute to chronic infectious disease of chickens and turkeys, involving primarily the synovial membranes of joints and tendon sheaths, and bursitis. Lameness and respiratory disorder caused by this infection results in decreased growth rate and loss of egg production. As the vertical transmission plays a major role in the spread of M. synoviae in chickens, the most effective control method is to monitor the flocks regularly and also eliminate the positive breeder flocks. Reliable and rapid diagnoses are needed to prevent infection dissemination (1). Mycoplasmas are consisted of wall-less prokaryotes which are the smallest fastidious bacteria and unusually have small genomes (2).
M. synoviae belongs to the Mycoplasmataceae family, and is a facultative anaerobic bacterium (3). Other important Mycoplasmas, namely M. mycoides, with large colonies and M. capricolum, are claimed to cause similar syndromes (4, 5). The disease occurs mainly in the Mediterranean region, but is also reported in many other areas of the world (6) including Iran. The Polymerase Chain Reaction (PCR) with Mycoplasma 16S ribosomal RNA has been applied to detect a variety of Mycoplasma species (6, 7). Computer alignment studies of these rRNA sequences which have revealed the existence of regions with highly conserved sequences and zones which display sequence variability at the genus and species levels, allow the selection of genus and species specific primers for PCR (8). The rRNA is naturally present in high copy numbers (up to 10’000 molecules per cell) (9). The PCR method has been applied to detect a variety of Mycoplasma species that cause other Mycoplasma diseases in some research works (6, 7, 10, 11).
M. synoviae has a broader host range than the other Mycoplasmas. Chickens, turkeys, and guinea fowl are the natural hosts. Several other species have been naturally infected, and others have been infected by artificial inoculation. Mycoplasmas may be transmitted horizontally, through infectious aerosols coughed and sneezed by infected birds and through contaminated feed, water, personal contact and communication with animals, mainly birds. Transmission occurs vertically from parents to their offspring, through contamination of laid eggs (transovarian transmission). Mycoplasma is commonly transmitted within species and/or between closely related species, that is, they are host-specific, with rare exceptions (12, 13).
2. Objectives
The current study aimed to isolate and identify M. synoviae from suspected Iranian ostrich samples by PCR and microbiological methods in Kerman Province, and the obtained results to confirm Mycoplasmal contamination of ostrich lung.
3. Materials and Methods
3.1. Mycoplasmas, Isolation and Enrichment
Fifty three suspected samples were collected by multiple random sampling of different parts of ostrich lungs within six months, stored at 4°C and transferred to the Reference Mycoplasma Laboratory in Razi Vaccine and Serum Research Institute, Karaj, Iran for examination. Overall, 53 samples were placed into the PPLO broth as transport medium on ice packs and were transferred to laboratory within 12 hours (14).
3.2. Bacteriology
For primary isolation, the specimens were diluted and filtered by 0.45μL syringe filter (Nunk, Denmark) in fresh PPLO broth and inoculated onto PPLO agar medium (BBL, Becton Dickinson and company Cockeyville, Sparks, MD, USA). The media was supplemented with 12% of equal volumes of inactivated horse or swine sera. Thallium acetate (1:4000) and penicillin (Roche company, Germany) (1000 IU/mL) as fungal and bacterial inhibitors, nicotinamide adenine dinucleotide (NAD) (Roche company, Germany) (1:10000 w/v) as a necessary requirement of M. synoviae, and cysteine hydrochloride (1:10000 w/v) (Roche company, Germany) as a reducing agent for NAD were also added to the media. Inoculated broth and agar media were incubated under microaerophilic condition (7% CO2) at 37ºC with 98% relative humidity and checked for color change of broth and typical Mycoplasma colonies on agar. As soon as the phenol red indicator changed to yellow, the subculture onto the fresh broth and agar were carried out (7, 15). Several passages until 21 to 28 days were subcultured. Identification of the M. synoviae samples was carried out by serological methods, after appearing the specific colonies of Mycoplasma.
3.3. Agglutination Test
Suspension of the suspected colony was made in drops of saline on slide. The slide was discarded if auto-agglutination occurred or the suspension was rough in saline. A drop (approximately 50 μL) of M. synoviae specific antiserum (Material No. 10100761) (SPAFAS Canada) was added and mixed to one drop (approximately 50 μL) of smooth suspension. If agglutination was observed within one or two minutes, the isolate was considered as M. synoviae (14).
3.4. DNA Extraction
DNA was extracted from the enriched samples, according to Kojima et al. protocol (7). As per European Pharmacopoeia 2005, negative and positive controls were PPLO broth media and a standard strain of M. synoviae (MS-NCTC 10124-05), respectively. About 500 µL of the samples were placed in a 1 × 5 µL Eppendorf tube (Roche Company, Germany) and micro centrifuged at 13000 rpm for 15 minutes. Then, 100 µL of lyses buffer was added to 100 µL of precipitate, which the tube was placed in a 56°C bath for four hours. Then, 200 µL saturated phenol was added and the tube was centrifuged at 13000 rpm for 20 minutes.
The supernatant was transferred to another tube and an equal volume of mixed phenol/chloroform (1:1) was added. After centrifuging at 13000 rpm for 20 minutes, the supernatant was transferred to another tube and an equal volume of pure chloroform was added, and then, centrifuged at 13000 rpm for five minutes. The supernatant was transferred to a new tube, mixed with 1/10 volume of 3 M acetate sodium and was precipitated in -20°C with two-fold cool and pure ethanol for 20 minutes. Next, the tube was centrifuged at 13000 rpm for 15 minutes, then 200 µL of 70% ethanol was added and the tube was centrifuged at 13000 rpm for five minutes, and finally the DNA was dried and re-suspended at 4°C to be used for PCR (4, 7).
3.5. Performing Polymerase Chain Reaction
According to the protocol of Kojima et al., two forward and reverse Mycoplasma specific primers (7), which amplify a 163 base pair region of 16S rRNA gene, were used. M. synoviae specific primers (MS-1 and MS-2) had been already designed by Lauerman et al. (5) who amplified a 207bp region of lipoprotein gene. The primers sequences with their corresponding genes are shown in Table 1.
Table 1. Nucleotide sequences and Primers Used for Identification of Mycoplasma synoviae.
Amplifications were carried out in a total volume of 25 μL containing 17.5 μL DNA, 0.1 µL of each primer, 0.5 µL dNTP mix (10 mM) (Cinna Gen Inc.), 4 µL Mgcl2 (25 mM) (Cinna Gen Inc.), 2.5 µL PCR buffer (10X) (Cinna Gen Inc.) and 0.25 µL Tag DNA polymerase (5 units/µL) (Cina Gen Inc.). The reaction mixture was thermocycled (Eppendorf, Germany) for 34 cycles, beginning with an initial denaturation step of minimum 95°C. The temperature and time profile of each cycle was as follows: 94°C for one minute (Denaturation), 55ºC for one minute (Annealing) and 72°C for one minute (Extension), while PCRs were finished with a final extension step at 72°C for five minutes. PCR products were stored at 4°C. PCRs were carried out using two programmable thermocycler apparatuses (Primus and Master gradient). Positive (MS-H vaccine strain) and negative (PPLO broth media) controls were included in all tests (4, 7, 16).
3.6. Amplicon Electrophoresis
An 8 μL aliquot of each PCR products was mixed with 2 μL loading buffer (6X). The PCR products and 100 bp DNA ladder were separated by electrophoresis (Apelex, France) in a 1% agarose gel (Agarose MP, Roche) with 0.5 μL/mL ethidium bromide (100 volts for one hour) following illustration under UV transilluminator and then photographed (Visi-Doc-It system, UVP, UK) (7).
3.7. Specificity and Sensitivity of the PCR Test
The sensitivity of the PCR procedure was carried out by testing the DNA extractions of serial log dilutions of bacterial suspension of MS. The specificity of the PCR was confirmed by testing the extracted DNA of M. gallisepticum, Salmonella entritidis, and water (14).
4. Results
4.1. Bacteriology
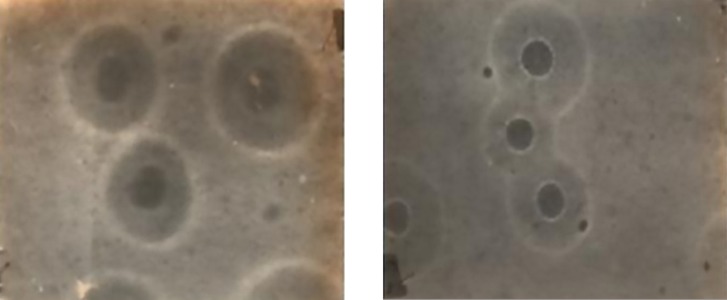
In the current study, 17 out of 53 ostrich samples were found Mycoplasma-positive. Figure 1 shows the colonies of MS, after confirmation by PCR.
Figure 1. Colonies of Mycoplasma synoviae on the PPLO Agar (X 40).
4.2. Polymerase Chain Reaction
The growth of Mycoplasmas in PPLO broth media was demonstrated by changes in color or turbidity due to biochemical activity and metabolism of the bacteria. Fifty-three suspected samples were evaluated by PCR test. Mycoplasma-positive isolates were detected in 25 out of 53 ostrich samples in the PCR method, and also all isolations showed a specific amplicon at 163bp to the Mycoplasma genus (Figure 2). All Mycoplasma-positive samples were analyzed for M. synoviae by PCR method; out of 25 Mycoplasma-positive samples, 52% were identified as M. synoviae (Figure 3). Thirteen out of 25 Mycoplasma-positive samples were also positive for M. synoviae species. On the other side, 17 out of 53 ostrich samples were identified positive in the microbiological method. The results of the PCR and microbiological tests are presented in Table 2.
Figure 2. Specificity of the PCR Assay Using the Specific Primers.
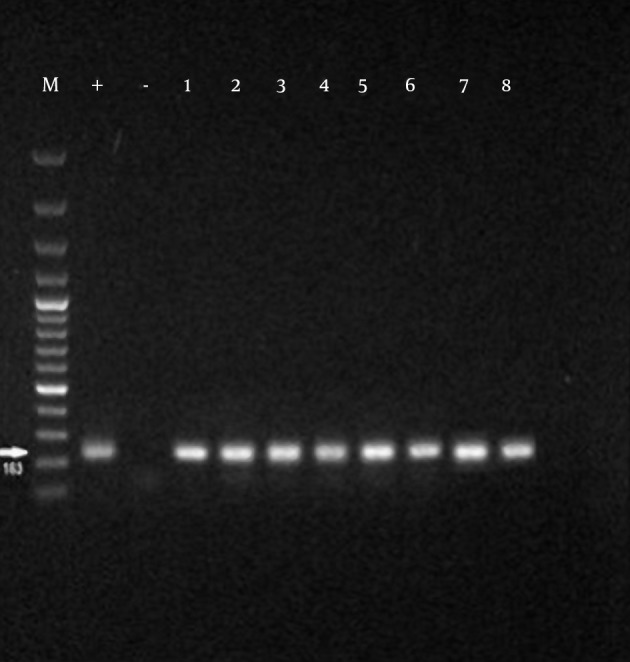
M, marker 100 bp; PC (+), positive control [M. synoviae (MS-NCTC 10124-05)]; NC (-), negative control; 1 to 8 suspected samples;163bp bands were formed in positive samples.
Figure 3. Specificity of the PCR Assay Using the Primers.
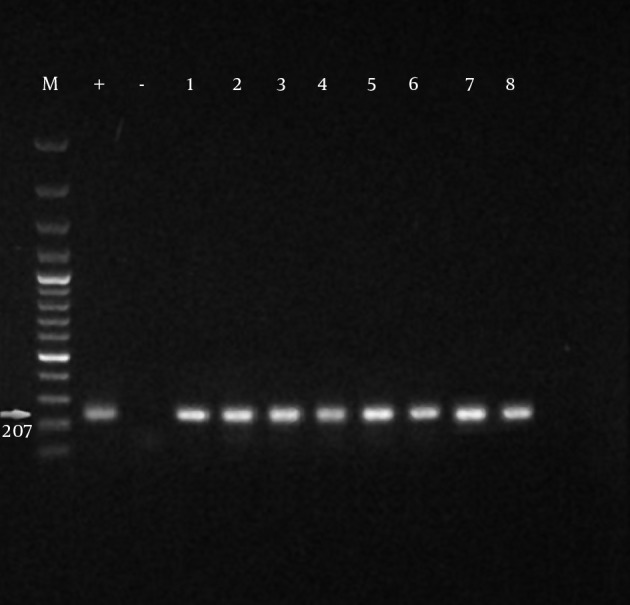
MS-1 and MS-2; M: marker 100 bp; PC (+): positive control (M. synoviae (MS-NCTC 10124-05)); NC (-): negative control. Positive Mycoplasma sp. samples; 207 bp bands were formed in positive species.
Table 2. Distribution of Samples for Mycoplasma-PCR, M. Synoviae-PCR and Microbiological Results.
| Control | Mycoplasma-PCR, No. (%) | M .synoviae-PCR, No. (%) | Microbiologic, No. (%) |
|---|---|---|---|
| + | 25 (47) | 13 (52) | 17 (32) |
| - | 28 (53) | 12 (48) | 36 (68) |
| Total | 53 (100) | 25 (100) | 25 (100) |
5. Discussion
In the current study, M. synoviae species was isolated and identified from ostrich samples by PCR and microbiological methods in Kerman Province, Iran. M. synoviae infection among slaughtered ostriches was truly confirmed. Efficient M. synoviae microbiological and molecular methods are necessary for diagnosis, eradication and research goals of this important poultry pathogen. Although respiratory infections with M. synoviae are generally considered sub clinical, an increasing number of reports have documented economic losses related to respiratory infections and arthropathic strains (14, 17). The eggshell pathology and the concomitant egg production losses that result from M. synoviae infection, further highlight the economic significance of these bacteria in commercial poultry (18). In the current study, the most specific microbiological and PCR methods, without false negative, were set up and established. The M. synoviae infection in ostrich samples was confidently confirmed. Although serological screening is still widely used, it may not detect subclinical M. synoviae infections, and monitoring programs that depend solely on detecting serocon version may be inadequate and nonspecific (14).
Traditionally, M. synoviae has been diagnosed by serological screening or microbiological method followed by identification of the Mycoplasma. Serological screening is relatively inexpensive but not always sufficiently specific or sensitive, while microbiological tests can be insensitive and time consuming (17). Detection of M. synoviae by microbiological methods was not reported from Kerman Province slaughtered ostriches before the current report. The current study successfully isolated 17 M. synoviae species by microbiological method. M. synoviae culture and isolation are not easy and almost are not accurate in all the Iranian poultry laboratories. Sample collection is important because M. synoviae tend to disappear from the lesions after a few weeks. Since M. synoviae die rapidly at room temperature, chilling the specimens during transportation should be considered. To control the bacterial and fungal contamination of samples, resistant antibiotic resistance as well as filtration should be considered. Microbiological method is needed for some research projects and even for diagnosis; to prepare the PCR samples, many false negative PCR results might occur without enrichment (19); therefore, culturing should not be ignored. But culturing can be costly and time-consuming, and can also be inconclusive because of low sensitivity (20). In the current study, microbiological tests had some limitations such as: ostrich slaughter was not widely performed, suspected samples were hardly collected, and M. synoviae isolation was not so easy.
There are few works on isolation and molecular identification of Mycoplasmas. In the Fars Province of Iran, Ghaleh Golab et al. isolated and detected M. smagallicepticum in chicken farms using PCR and RFLP assays and microbiological methods (21). Hosseini et al. analyzed the M. gallisepticum isolates of different geographical areas of Iran by random amplification of polymorphic DNA (RAPD) (22). Ghaleh Golab et al. identified four M. synoviae species from Fars Province using PCR and RFLP assays. The primers in Ghaleh Golab et al. study selectively amplified a 780 bp DNA fragment within the 16S rRNA gene of M. gallisepticum, M. synoviae and M. iowae (23).
Comparing the PCR and culture methods, the current research showed that 17 and 25 out of 53 samples were positive by microbiological and PCR methods, respectively. Furthermore, the specific PCR can be used to identify the Mycoplasma sp. After obtaining positive results from the culture tests. To eradicate M. synoviae infection, it is important to identify M. synoviae rapidly and accurately and molecular methods such as PCR are helpful. Previously, M. synoviae specific PCRs were performed based on the 16S rRNA gene (5, 14) and recently they are based on haemagglutinin genes (19, 24).
The current study specific primers of Lauerman et al. were used (5). They indicated a sensitivity of 82% for their M. synoviae PCR and a specificity of 100% compared with those of microbiological and serological tests, and epizootiology, and history. To increase the PCR sensitivity, they suggested extraction of RNA and performing reverse transcription of the rRNA. These M. synoviae primers were selected from the 16S rRNA gene, and each cell is reported to have 104 ribosomes (5). However, it is suggested to apply other specific species PCR based on haemagglutinin genes. To obtain better M. synoviae PCRs results, it is also recommended to enrich the isolates, which is consisted as specimen’s incubation for 24 hours prior to detergent treatment in the DNA extraction stage. This is certainly due to an increase of the DNA starting material and the reduction of PCR inhibitors present in the original samples (19). Because of its sensitivity and specificity, PCR should circumvent the overgrowth problems encountered with culturing the field samples (20). These results strongly support the use of this PCR assay as an efficient alternative or supplement to culture and serological identification, which are labor-intensive, extremely time-consuming, and often provide confusing results. Overall, it is suggested that the PCR could be an alternative method for accurate identification of the M. synoviae infection especially in slaughter ostriches.
Footnotes
Authors’ Contributions:All authors had equal role in design, work, statistical analysis and manuscript writing.
Funding/Support:This research was supported by Islamic Azad University, Kerman Science and Research Branch.
References
- 1.Seifi S, Shirzad M. Incidence and risk factors of Mycoplasma synoviae infection in broiler breeder farms of Iran. Veterinary World J. 2012;5(5):265. doi: 10.5455/vetworld.2012.265-268. [DOI] [Google Scholar]
- 2.Whitcomb RF, Gasparich GE, French FE, Tully JG, Rose DL, Carle P, et al. Spiroplasma syrphidicola sp. nov., from a Syrphid Fly (Diptera: Syrphidae). J. Syst. Evol. 1996;46(3):797–801. doi: 10.1099/00207713-46-3-797. [DOI] [PubMed] [Google Scholar]
- 3.Salisch H, Hinz KH, Graack HD, Ryll M. A comparison of a commercial PCR-based test to culture methods for detection of Mycoplasma gallisepticum and Mycoplasma synoviae in concurrently infected chickens. Avian Pathol. 1998;27(2):142–7. doi: 10.1080/03079459808419315. [DOI] [PubMed] [Google Scholar]
- 4.Kleven SH, Fletcher OJ, Davis RB. Influence of strain of Mycoplasma synoviae and route of infection on development of synovitis or airsacculitis in broilers. Avian Dis. 1975;19(1):126–35. [PubMed] [Google Scholar]
- 5.Lauerman LH, Hoerr FJ, Sharpton AR, Shah SM, van Santen VL. Development and application of a polymerase chain reaction assay for Mycoplasma synoviae. Avian Dis. 1993;37(3):829–34. [PubMed] [Google Scholar]
- 6.Zendulková D, Madanat A, Lány P, Rosenbergová K, Pospíšil Z. Detection of Mycoplasma agalactiae by Polymerase Chain Reaction in Jordanian Sheep and Goat Herds. Acta Veterinaria Brno. 2007;76(1):71–7. doi: 10.2754/avb200776010071. [DOI] [Google Scholar]
- 7.Kojima A, Takahashi T, Kijima M, Ogikubo Y, Nishimura M, Nishimura S, et al. Detection of Mycoplasma in avian live virus vaccines by polymerase chain reaction. Biologicals. 1997;25(4):365–71. doi: 10.1006/biol.1997.0108. [DOI] [PubMed] [Google Scholar]
- 8.van Kuppeveld FJ, van der Logt JT, Angulo AF, van Zoest MJ, Quint WG, Niesters HG, et al. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl Environ Microbiol. 1992;58(8):2606–15. doi: 10.1128/aem.58.8.2606-2615.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waters AP, McCuthan TF. Ribosomal RNA: nature's own polymerase-amplified target for diagnosis. Parasitol Today. 1990;6(2):56–9. doi: 10.1016/0169-4758(90)90071-b. [DOI] [PubMed] [Google Scholar]
- 10.Azevedao EO, Alcantara MDB, Nascimento ER, Tabosa IM, Barreto ML, Almeida J. Contagious agalactia by Mycoplasma agalactiae in small ruminants in Brazil Braz J Microbiol. Brazil Braz J Microbiol. 2006;37(5):576–81. [Google Scholar]
- 11.Blanchard A, Gautier M, Mayau V. Detection and identification of mycoplasmas by amplification of rDNA. FEMS Microbiol Lett. 1991;65(1):37–42. doi: 10.1016/0378-1097(91)90467-o. [DOI] [PubMed] [Google Scholar]
- 12.Nascimento ER, Pereira VLA, Nascimento MGF, Barreto ML. Avian Mycoplasmosis Update. Brazilian J Poult Sci. 2005;7(1):1–9. [Google Scholar]
- 13.Stipkovits LS, zatmari Z. controlling mycoplasma infection in poultry. World Poult. 2006;22(7):42–3. [Google Scholar]
- 14.Pourbakhsh SA, Shokril GR, Banani M, Elhamriia F, Ashtari A. Detection of Mycoplasma synoviae infection in broiler breeder farms of Tehran province using PCR and culture methods. Arch Razi Inst. 2010;65(2):75–81. [Google Scholar]
- 15.Kleven SH, Swayne DE, Glisson JR, Jackwood MW, Pearson JE, Reed WM. A laboratory manual for the isolation and identification of avian pathogens. 4 ed. USA: American Association of avian pathologists; 1998. [Google Scholar]
- 16.Tola S, Angioi A, Rocchigiani AM, Idini G, Manunta D, Galleri G, et al. Detection of Mycoplasma agalactiae in sheep milk samples by polymerase chain reaction. Vet Microbiol. 1997;54(1):17–22. doi: 10.1016/s0378-1135(96)01269-2. [DOI] [PubMed] [Google Scholar]
- 17.Grim T, Kleven O, Mikulica O. Nestling discrimination without recognition: a possible defence mechanism for hosts towards cuckoo parasitism? Proc Biol Sci. 2003;270 Suppl 1:S73–5. doi: 10.1098/rsbl.2003.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feberwee A, de Wit JJ, Landman WJ. Induction of eggshell apex abnormalities by Mycoplasma synoviae: field and experimental studies. Avian Pathol. 2009;38(1):77–85. doi: 10.1080/03079450802662772. [DOI] [PubMed] [Google Scholar]
- 19.Mardassi BB, Mohamed RB, Gueriri I, Boughattas S, Mlik B. Duplex PCR to differentiate between Mycoplasma synoviae and Mycoplasma gallisepticum on the basis of conserved species-specific sequences of their hemagglutinin genes. J Clin Microbiol. 2005;43(2):948–58. doi: 10.1128/JCM.43.2.948-950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewing ML, Cookson KC, Phillips RA, Turner KR, Kleven SH. Experimental infection and transmissibility of Mycoplasma synoviae with delayed serologic response in chickens. Avian Dis. 1998;42(2):230–8. [PubMed] [Google Scholar]
- 21.Ghaleh Golab Behbahan N, Asasi AR, Afsharifar AR, Pourbakhsh S. Isolation and detection of Mycoplasma gallisepticum by polymerase chain reaction and restriction fragment length polymorphism. IJVR. 2005;6:35–41. [Google Scholar]
- 22.Hosseini H, Bozorgmehrifard MH, Peighambari SM, Pourbakhsh SA, Razzazian M. Random amplified polymorphic DNA (RAPD) fingerprinting of Mycoplasma gallisepticum isolates from chickens. Arch Razi Inst. 2006;61(2):67–71. [Google Scholar]
- 23.Ghaleh Golab Behbahan N, Asasi K, Afsharifar AR, Pourbakhsh SA. Susceptibilities of Mycoplasma gallisepticum and Mycoplasma synoviea Isolates to Antimicrobial Agents in vitro. Inter J Poult Sci. 2008;7(11):1058–64. doi: 10.3923/ijps.2008.1058.1064. [DOI] [Google Scholar]
- 24.Hong Y, Garcia M, Leiting V, Bencina D, Dufour-Zavala L, Zavala G, et al. Specific detection and typing of Mycoplasma synoviae strains in poultry with PCR and DNA sequence analysis targeting the hemagglutinin encoding gene vlhA. Avian Dis. 2004;48(3):606–16. doi: 10.1637/7156-011504R. [DOI] [PubMed] [Google Scholar]


