Summary
Host genetics and the gut microbiome can both influence metabolic phenotypes. However, whether host genetic variation shapes the gut microbiome and interacts with it to affect host phenotype is unclear. Here, we compared microbiotas across > 1,000 fecal samples obtained from the TwinsUK population, including 416 twin-pairs. We identified many microbial taxa whose abundances were influenced by host genetics. The most heritable taxon, the family Christensenellaceae, formed a cooccurrence network with other heritable Bacteria and with methanogenic Archaea. Furthermore, Christensenellaceae and its partners were enriched in individuals with low body mass index (BMI). An obese-associated microbiome was amended with Christensenella minuta, a cultured member of the Christensenellaceae, and transplanted to germfree mice. C. minuta amendment reduced weight gain and altered the microbiome of recipient mice. Our findings indicate that host genetics influence the composition of the human gut microbiome and can do so in ways that impact host metabolism.
Introduction
The human gut microbiome has been linked to metabolic disease and obesity (Karlsson et al., 2013; Le Chatelier et al., 2013; Ley et al., 2005; Qin et al., 2012; Turnbaugh et al., 2009). Variation in host genetics can also underlie susceptibility to metabolic disease (Frayling et al., 2007; Frazer et al., 2009; Herbert et al., 2006; Yang et al., 2012). Despite these shared effects, the relationship between host genetic variation and the diversity of gut microbiomes is largely unknown.
The gut microbiome is environmentally acquired from birth (Costello et al., 2012; Walter and Ley, 2011), therefore it may function as an environmental factor that interacts with host genetics to shape phenotype, as well as a genetically determined attribute that is shaped by, and interacts with, the host (Bevins and Salzman, 2011; Spor et al., 2011; Tims et al., 2011). Because the microbiome can be modified for therapeutic applications (Borody and Khoruts, 2012; Hamilton et al., 2013; Khoruts et al., 2010; van Nood et al., 2013), it constitutes an attractive target for manipulation. Once the interactions between host genetics and the microbiome are understood, its manipulation could be optimized for a given host genome to reduce disease risk.
Although gut microbiomes can differ markedly in diversity across adults (Consortium, 2012; Qin et al., 2010), family members are often observed to have more similar microbiotas than unrelated individuals (Lee et al., 2011; Tims et al., 2013; Turnbaugh et al., 2009; Yatsunenko et al., 2012). Familial similarities are usually attributed to shared environmental influences, such as dietary preference, a powerful shaper of microbiome composition (Cotillard et al., 2013; David et al., 2013; Wu et al., 2011). Yet related individuals share a larger degree of genetic identity, raising the possibility that shared genetic composition underlies familial microbiome similarities.
Support for a host genetic effect on the microbiome comes mostly from studies taking a targeted approach. For instance, the concordance rate for carriage of the methanogen Methanobrevibacter smithii is higher for monozygotic (MZ) than dizygotic (DZ) twin pairs (Hansen et al., 2011), and studies comparing microbiotas between human subjects differing at specific genetic loci have shown gene-microbiota interactions (Frank et al., 2011; Khachatryan et al., 2008; Rausch et al., 2011; Rehman et al., 2011; Wacklin et al., 2011). A more general approach to this question has linked genetic loci with abundances of gut bacteria in mice (Benson et al., 2010; McKnite et al., 2012), but in humans, a general approach (e.g., using twins) has failed to reveal significant genotype effects on microbiome diversity (Turnbaugh et al., 2009; Yatsunenko et al., 2012). Thus, heritable components of the human gut microbiome remain to be identified using an unbiased approach.
Here, we assessed the heritability of the gut microbiome with a well-powered twin study. Comparisons between MZ and DZ twin pairs allowed us to assess the impact of genotype and early shared environment on their gut microbiota. Our study addressed the following questions: Which specific taxa within the gut microbiome are heritable, and to what extent? Which predicted metagenomic functions are heritable? How do heritable microbes relate to host BMI? Finally, we use fecal transplants into germfree mice to assess the phenotype effects of the most heritable taxon.
Results
Twin dataset
We obtained 1,081 fecal samples from 977 individuals: 171 MZ and 245 DZ twin pairs, 2 from twin pairs with unknown zygosity, and 143 samples from just one twin within a twinship (i.e., unrelated). In addition, we collected longitudinal samples from 98 of these individuals (see Supplemental Information). Most subjects were female, ranging in age from 23 to 86 years (average age: 60.6 ± 0.3 years). The average BMI of the subjects was 26.25 (± 0.16) with the following distribution: 433 subjects had a low to normal BMI (<25), 322 had an overweight BMI (25-30), 183 were obese (>30) and 39 individuals in which the current BMI status was unknown. We generated 78,938,079 quality-filtered sequences that mapped to the Bacteria and Archaea in the Greengenes database (average sequences per sample: 73,023 ± 889).
Microbiome composition and richness
We sorted sequences into 9,646 operational taxonomic units (OTUs, ≥97% ID). Of these OTUs, 768 were present in at least 50% of the samples. Taxonomic classification revealed a fairly typical Western diversity profile: the dominant bacterial phyla were Firmicutes (53.9% of total sequences), Bacteroidetes (35.3%), Proteobacteria (4.5%), with Verrucomicrobia, Actinobacteria, and Tenericutes each comprising 2% of the sequences, and a tail of rare bacterial phyla that together accounted for the remaining 1% of the sequences.
The most widely shared methanogen was M. smithii (64% of people, using nonrarefied data), followed by vadinCA11, a member of the Thermoplasmata with no cultured representatives (~6%), Methanospheara stadtmanae (~4%), and Methanomassiliicoccus (~4%, a member of the Thermoplasmata). Forty-six of the 61 samples in which we detected vadinCA11 also contained M. smithii, indicating that the two most dominant archaeal taxa are not mutually exclusive. Faith's PD was positively correlated with the relative abundance of the family Methanobacteriaceae (rho = 0.42 rarefied, 0.37 for transformed counts, P < 1 x 10−11), which corroborates previous reports of higher richness associating with methanogens.
Broad diversity comparisons between MZ and DZ twin pairs
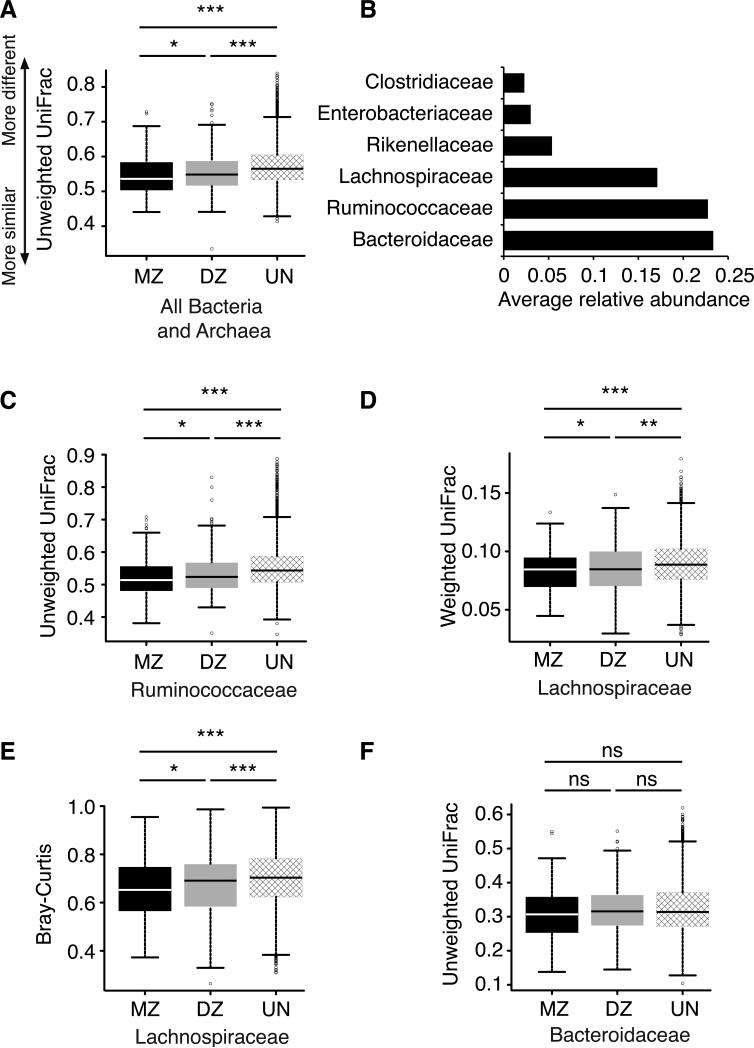
We observed that microbiotas were more similar overall within individuals (resampled) than between unrelated individuals (P < 0.001 for weighted and unweighted UniFrac and Bray-Curtis using a Student's t-test with 1,000 Monte Carlo simulations; Table S1A), and were also more similar within twin pairs compared to unrelated individuals (P < 0.009 for weighted and unweighted UniFrac and Bray-Curtis; Figure 1, S1 and Table S1). MZ twin pairs had more similar microbiotas than DZ twins for the unweighted UniFrac metric (P = 0.032), but not the weighted UniFrac and Bray-Curtis metrics (Figures 1A, S1). As greater similarities for MZ versus DZ twin pairs are seen in unweighted UniFrac but not abundance-based metrics, MZ similarities are driven by shared community membership rather than structure.
Figure 1. Microbiomes are more similar for monozygotic than dizygotic twins.
A, C-F: Box plots of beta-diversity distances between microbial communities obtained when comparing individuals within twinships for monozygotic (MZ) twin pairs and dizygotic (DZ) twin pairs, and between unrelated individuals (UN). A: the whole microbiome; C: the bacterial family Ruminococcaceae; D-E: the bacterial family Lachnospiraceae; F: the family Bacteroidaceae. The specific distance metric used in each analysis is indicated on the axes. *P<0.05, **P<0.01, ***P<0.001 for Student's t-tests with 1,000 Monte Carlo simulations. B: The average relative abundances in the whole dataset for the top six most prevalent bacterial families (unrarefied data, see Methods). Relates to Figure S1 and Table S1.
We next constrained the distance metric analyses to the three most dominant bacterial families: the Lachnospiraceae and Ruminococcaceae (Firmicutes) and Bacteroidaceae (Figure 1B). We observed greater similarities for MZ compared to DZ twins using the unweighted UniFrac metric within the Ruminococcaceae family (Figure 1C). Within the Lachnospiraceae family, significantly greater similarity for MZ compared to DZ twins emerged using the weighted UniFrac and Bray-Curtis metrics (Figures 1D, E). In contrast, when restricted to the Bacteroidaceae family, we found that MZ and DZ twins had similar pair-wise diversity using all three metrics (Figures 1F, S1B and S1E).
MZ twins have more highly correlated microbiotas
We next asked if the abundances of specific taxa were generally more highly correlated within MZ twin pairs compared to DZ twin pairs. For each twin pair we generated intraclass correlation coefficients (ICCs) for the relative abundances of OTUs. Mean ICCs were significantly greater for MZ compared to DZ twin pairs (Wilcoxon signed rank test on ICCs at the OTU level, P = 6 x 10−04; Figure 2). Since many OTUs are closely phylogenetically related, their abundances may not be independent, which may inflate levels of significance. To account for this effect, we maintained the structure of the phylogenetic tree but permuted the MZ and DZ labels in 10,000 tests to generate randomized ICCs. As an independent validation, we also applied these analyses to two previously published datasets generated originating in a population of twins from Missouri, USA: ‘Turnbaugh’ (Turnbaugh et al., 2009), which described 54 twin pairs ranging from 21-32 years of age, and ‘Yatsunenko’ (Yatsunenko et al., 2012), which included 63 twin pairs with an age range of 13-30 years of age. Mean ICCs of OTU abundances were significantly greater for MZ compared to DZ twin pairs in both of these datasets (significance by permutation: P < 0.001 and 0.047 respectively; Figure S2), corroborating our observations.
Figure 2. OTU relative abundances are more highly correlated within MZ than DZ twin pairs.
At left is a phylogeny of taxa in the TwinsUK study (Greengenes tree pruned to include only OTUs shared by 50% of the TwinsUK participants) and at right are corresponding twin-pair intra class correlation coefficients (ICCs). ICCs were calculated for each OTU and the difference in correlation coefficients for MZ twin pairs versus DZ twin pairs. Bars pointing to the right indicate that the difference is positive (i.e., MZ ICCs > DZ ICCs) and bars pointing to the left indicate negative differences (DZ ICCs > MZ ICCs). The scale bar associated with the phylogeny shows substitutions/site. Relates to Figure S2.
Heritability estimates for OTUs and predicted functions
We estimated heritability using the twin-based ACE model, which partitions the total variance into three component sources: genetic effects (A), common environment (C), and unique environment (E) (Eaves et al., 1978). The largest proportion of variance in abundances of OTUs could be attributed to the twins’ unique environments (i.e., E > A; Table S2). However, for the majority of OTUs (63%), the proportion of variance attributed to genetic effects was greater than the proportion of variance attributed to common environment (A > C; Table S2).
From the ACE model we calculated 95% confidence intervals for the heritability estimates, and determined the significance of the heritability values using a permutation method to generate nominal P values (Table S2). We found a high correlation between the tail probability for inclusion of zero in the confidence interval of heritability and the P values obtained from the permutation tests (rho = 0.872, P < 10−15), indicating substantial consistency across these tests. Although heritability studies traditionally report confidence intervals and nominal P values only, we also generated FDR-corrected P values (Table S2).
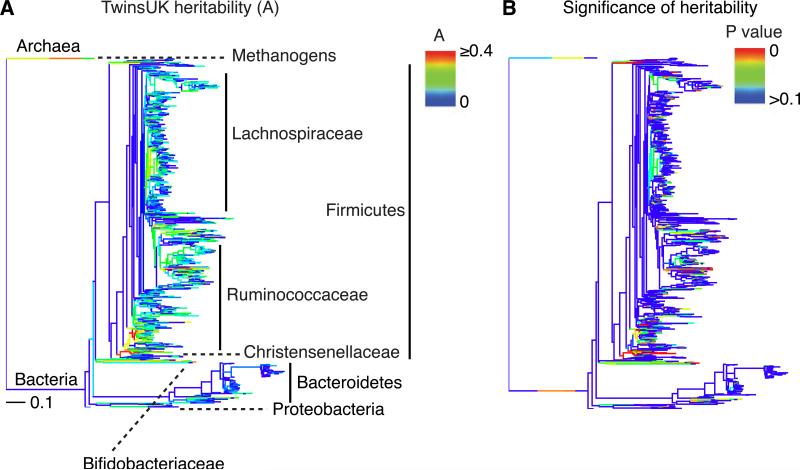
We also applied the ACE model to the abundances of sequences mapping to each node in the phylogeny. Across the three studies, the nodes of the phylogeny with the strongest heritabilities lie within the Ruminococcaceae and Lachnospiraceae families, and the Bacteroidetes are mostly environmentally determined (Figures 3 and S3). Subsets of the Archaea are also heritable in the TwinsUK and the Yatsunenko studies (the Turnbaugh study did not include data for Archaea).
Figure 3. Heritability of microbiota in the TwinsUK dataset.
A: OTU Heritability (A from ACE model) estimates mapped onto a microbial phylogeny and displayed using a rainbow gradient from blue (A = 0) to red (A ≥ 0.4). This phylogenetic tree was obtained from the Greengenes database and pruned to include only nodes for which at least 50% of the TwinsUK participants were represented. B: The significance for the heritability values shown in A was determined using a permutation test (n=1,000) and are shown on the same phylogeny as in panel A. P values range from 0 (red) to >0.1 (blue). Relates to Figure S3 and Table S2.
We characterized the longitudinal stability of each OTU by calculating the ICCs of the OTU abundance across repeat samples, which consisted of two samples collected from the same individual at different times. By permuting these repeat sample ICCs, we found that heritable OTUs (A > 0.2) were more stable (ICC > 0.6) than expected by chance (Figure S3E; P < 0.001, P value was determined as the fraction of permutations that had greater than or equal to the observed number of OTUs that are both heritable and stable).
We used PICRUSt (Langille et al., 2013) to produce predicted metagenomes from the 16S rRNA gene sequence data and applied the ACE model to estimate the heritability of predicted abundances of conserved orthologous groups (COGs). This analysis revealed 6 functions with heritabilities A > 0.2 and nominal P values < 0.05 (P values are generated by permutation testing; Supplementary Methods; Table S2). Correcting for multiple comparisons, one category, “secondary metabolites biosynthesis, transport and catabolism” (Q), passed a stringent FDR (A = 0.32, 95% CI = 0.16-0.44). We also tested alpha diversity for heritability and found that it was not heritable.
The family Christensenellaceae is the most highly heritable taxon
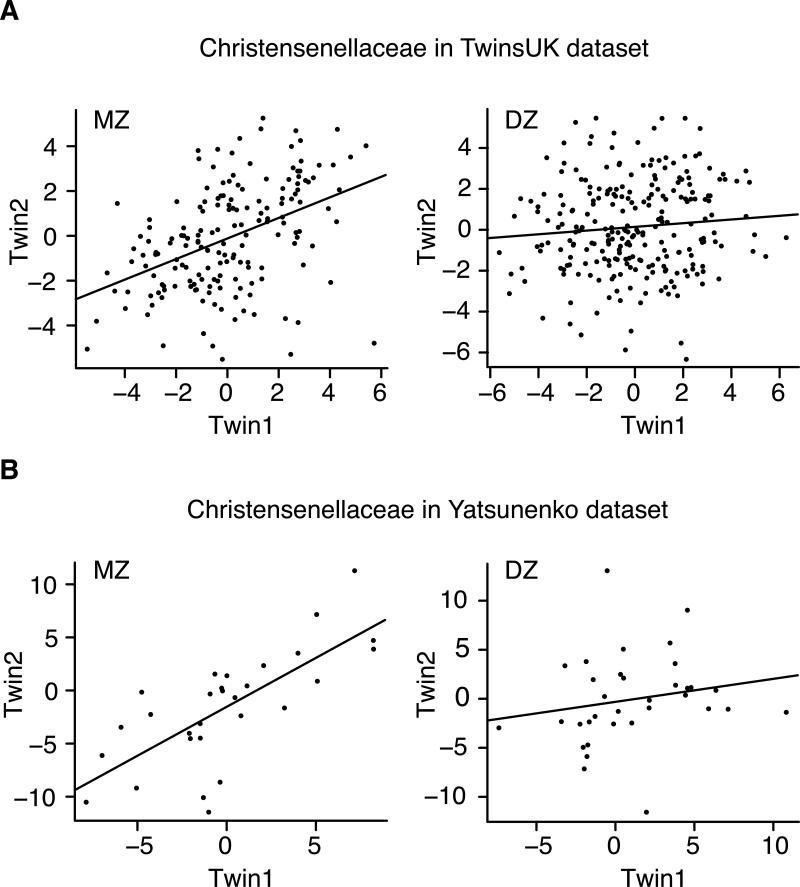
The most heritable taxon overall was the family Christensenellaceae (A = 0.39, 95% confidence interval 0.21-0.49, P = 0.001, Figure 4A and Table S2; this taxon passes a stringent FDR) of the order Clostridiales. Christensenellaceae was also highly heritable in the Yatsunenko dataset (A = 0.62, 95% confidence interval 0.38-0.77; Figure 4B and Table S2). We repeated this analysis for the taxa abundances with the effect of BMI regressed out, and results were highly correlated (Pearson correlation = 0.95, P < 1×10−15).
Figure 4. MZ twin pairs have higher correlations of Christensenellaceae than DZ twin pairs in TwinsUK and Yatsunenko datasets.
Scatter plots comparing the abundances of Christensenellaceae in the gut microbiota of MZ and DZ co-twins. Christensenellaceae abundances were transformed and adjusted to control for technical and other covariates (Residuals are plotted, see Supplemental Methods) and the data are separated by zygosity (MZ or DZ twins). A: TwinsUK dataset. B: Yatsunenko dataset.
Christensenellaceae is the hub in a co-occurrence network with other heritable taxa
We observe a module of co-occurring heritable families, and the hub (node connected to most other nodes) of this module is the family Christensenellaceae (Figures 5A and S4A). The heritable module includes the families Methanobacteriaceae (Archaea) and Dehalobacteriaceae (Firmicutes) and the orders SHA-98 (Firmicutes), RF39 (Tenericutes) and ML615J-28 (Tenericutes). The Christensenellaceae-network is anti-correlated with the Bacteroidaceae and Bifidobacteriaceae families. We validated these results by applying this method to the family-level taxonomic abundances in the Yatsunenko dataset (as this one is most technically similar to the TwinsUK dataset), where we also found the same Christensenellaceae-centered module of heritable families anti-correlated to the Bacteroidaceae/Bifidobacteriaceae module (Figure S4B).
Figure 5. Christensenellaceae is the hub of a consortium of co-occurring heritable microbes that are associated with a lean BMI.
A and B show the same network built from SparCC correlation coefficients between sequence abundances collapsed at the family level. The nodes represent families and the edges represent the correlation coefficients between families. Edges are colored blue for a positive correlation and grey for a negative correlation, and the weight of the edge reflects the strength of the correlation. Nodes are positioned using an edge-weighted force directed layout. In panel A, the nodes are colored by the heritability of the family, and in panel B, the nodes are colored by the significance of the association families and a normal vs. obese BMI. Family names are either indicated on the panel, or nodes are given a letter code. Phylum Actinobacteria: (a) Actinomycetaceae, (b) Coriobacteriaceae; Phylum Bacteroidetes: (c) Barnesiellaceae, (d) Odoribacteraceae, (e) Paraprevotellaceae, (f) Porphyromonadaceae, (g) Prevotellaceae, (h) Rikenellaceae; Phylum Firmicutes: (i) Carnobacteriaceae, (j) Clostridiaceae, (k) Erysipelotrichaceae, (l) Eubacteriaceae, (m) Lachnospiraceae, (n) Lactobacillaceae, (o) Mogibacteriaceae, (p) Peptococcaceae, (q) Peptostreptococcaceae, (r) Ruminococcaceae, (s) Streptococcaceae, (t) Tissierellaceae, (u) Turicibacteraceae, (v) Unclassified Clostridiales, (w) Veillonellaceae; Phylum Proteobacteria: (x) Alcaligenaceae, (y) Enterobacteriaceae, (z) Oxalobacteraceae, (aa) Pasteurellaceae, (ab) Unclassified RF32; Phylum Verrucomicrobia: (ac) Verrucomicrobiaceae. Relates to Figure S4.
Christensenellaceae associates with a low BMI
The family Christensenellaceae was significantly enriched in subjects with a lean BMI (< 25) compared to those with an obese BMI (> 30; Benjamini-Hochberg corrected P value < 0.05 from t-test on transformed counts; Table S2). Other members of the Christensenellaceae consortium were also enriched in lean-BMI subjects: the Dehalobacteriaceae, SHA-98, RF39, and the Methanobacteriaceae (Figure 5B). Overall, a majority (n=35) of the OTUs with highest heritability scores (A > 0.2, nominal P < 0.05) were enriched in the lean subjects. A subset of OTUs classified as Oscillospira were enriched in lean subjects, and M. smithii, though not significantly heritable, was positively associated with a lean BMI.
Christensenellaceae is associated with health in published datasets
Because the names Christensenella and Christensenellaceae were only recently assigned to the bacterial phylogeny, we assessed the abundances of sequences assigned to these taxa in previously published studies. This analysis revealed that members of the Christensenellaceae were enriched in fecal samples of healthy versus pediatric and young adult IBD patients (P < 0.05) (Papa et al., 2012). Christensenellaceae were at greater abundance in lean-BMI compared to obese-BMI twins in the Turnbaugh dataset but the difference was not quite significant (‘time-point 2’ samples, P = 0.07). In a case study of the development of an infant's gut microbiome (Koenig et al., 2011), Christensenellaceae was present at 8.6% in the mother's stool at the time of birth, and at 20% in the infant's meconium. We also noted that Christensenellaceae is enriched in omnivorous compared to herbivorous and carnivorous mammals (Muegge et al., 2011). However, we did not find a relationship between Christensenellaceae and diet information in human studies (Wu et al., 2011; Martinez et al., 2010; Koren et al., 2012).
Christensenellaceae is associated with reduced weight gain in germfree mice inoculated with lean and obese human fecal samples
Methanogens co-occurred with Christensenellaceae in this study and have been linked to low BMI in previous studies. Because of this previous association with a low-BMI, we wanted to ensure that methanogens were present in the Christensenellaceae consortium in an initial experiment exploring its effect on weight phenotypes. Therefore, we selected 21 donors for fecal transfer to germfree mice based on BMI status (low or high) and presence or absence of the methanogen-Christensenellaceae consortium. Donors fell into one of four categories: lean with detectable methanogens (L+), lean without methanogens (L-), obese with methanogens (O+), or obese without methanogens (O-). The abundance of Christensenellaceae positively correlated with the abundance of methanogens in donor stool (rho=0.72, P=0.0002), indicating that methanogen abundance was a good proxy for the methanogen-Christensenellaceae consortium.
A 16S rRNA analysis of the fecal microbiomes before and after transfer to germfree mice showed that although members of the Christensenellaceae were present throughout the experiment in recipient mice (Figure 6A), M. smithii was undetectable in the mouse fecal or cecal samples (the first sampling was at 20hrs post-inoculation). At 20 hrs post-inoculation, the microbiota had shifted dramatically in diversity from the inoculation, but by Day 5 had shifted back partially and remained fairly stable through Day 21 (Figures 6B, 6C, S5A, and S5B). Abundances of Christensenella were correlated with PC3 (abundances rarefied at 55,000 sequences per sample vs. unweighted UniFrac; Spearman rho = 0.59, P < 2.2 x 10−16), and PC3 captured the differences between the 4 donor groups (Figure 6D). We observed a trend for Christensenella abundances as highest in the L+ group and lowest in the O- group (Figure 6A), which mirrored the weight differences between those groups: the percent change in body weights of the recipient mice was significantly lower in the L+ group compared to the O- group (Day 12, P < 0.05, t-test; Figure 6E and 6F). Cecal levels of propionate and butyrate were significantly elevated in mice receiving methanogen-positive compared to methanogen-negative microbiomes controlling for the effect of donor BMI (two-way ANOVA, P < 0.05 for both SCFAs, Figures S5C-E). Stool energy content was significantly higher for the methanogen-positive microbiomes at Day 12, when the percent changes in weight were greatest (two-way ANOVA, P = 0.004, no effect of BMI or interaction; Figure S5F). In a replicated experiment, using 21 new donors, the same weight differences were observed (a significantly lower mean weight gain for the L+ compared to the O- mouse recipients at Day 10 post-inoculation; one-way t-test, P = 0.047; Figure S5G).
Figure 6. Fecal transplants from obese and lean UK Twins to germfree mice reveal levels of Christenenallaceae post-transfer mirror delayed weight gain.
A: Median relative abundances for OTUs classified as the genus Christensenella in the four donor treatment groups over time in the recipient mouse microbiotas. B: Principal coordinates analysis of unweighted UniFrac distances for (i) the inoculum prior to transplantation, (ii) fecal samples at 4 time points, and (iii) cecal samples at Day 21 post-transplant; see panel legend for color key. The amount of variance described by the first two PCs is shown on the axes. C: Richness (Faith's PD) for the microbiomes of the transplant mice plotted against time (days post inoculation, with Day 0 = inoculation day). D: The mean values ± S.E.M. for PC3 derived for the same analysis as shown in panel B are plotted against time (Day 0 = inoculation day) for the four treatment groups. The amount of variance explained by PC3 is in parentheses. E: Percent weight change since inoculation for germfree mouse recipients of 21 donor stools that were obtained from lean or obese donors with or without detectable M. smithii, which was used as a marker for the Christensenellaceae consortium. Means for each treatment group are plotted ± S. E. M. F: Box plots for percent weight changes for the 4 groups at Day 12 post-transplant, when maximal weight differences were observed. Letters next to boxes indicate significant differences if letters are different (p < 0.05). For all panels, Dark blue = L+, lean donor with methanogens; Light blue = L-, lean donor lacking methanogens; Dark orange = O+, obese donor with methanogens; Light orange = O-, obese donor without methanogens. We repeated this experiment with a set of 21 new mice and unique human donors and recovered the same effect. Relates to Figure S5.
Christensenella minuta added to donor stool reduces adiposity gains in recipient mice
Based on the observation that Christensenella levels in the previous experiment were similar to the weight gain patterns, we performed experiments in which a donor stool lacking detectable Christensenella was amended with C. minuta and weight gain of recipient mice was monitored. One obese human donor was selected from the 21 donors from the first transplant experiment based on its lack of detectable OTUs assigned to the genus Christensenella. At Day 21 post-gavage, mice receiving the C. minuta treatment weighed significantly less than those that received unamended stool (nested ANOVA, P < 0.05, Figure 7A). Adiposity was significantly lower for mice receiving the C. minuta treatment (nested ANOVA, P = 9.4 x 10−5, Figure 7B). Energy content for stool collected at Day 21 was not different between treatments (data not shown).
Figure 7. Addition of Christensenella minuta to donor stool leads to reduced weight and adiposity gains in recipient mice.
A: Box plot of percent weight change for germfree mouse recipients of a single donor stool only (lacking detectable Christensenella in unrarefied 16S rRNA data) or the donor stool amended with live C. minuta. B: Box plots showing percent body fat for mice in each group at Day 21 N = 12 mice per treatment. C, D: Principal coordinates analysis of unweighted UniFrac distances for (i) the inoculum prior to transplantation, (ii) fecal samples at 5 time points post-transplant; see panel legend for color key. The amount of variance described by the first two PCs is shown on the axes. The same data projection is shown in panels C and D; sample symbols are colored by time point (C) and by treatment (D). E: Relationship between PCs from the PCoA analysis and levels of Oscillospira at Day 21 (rho = −0.71, P = P < 0.001). Symbols are colored by treatment. Relates to Figure S6.
Analysis of the microbial community by 16S rRNA gene sequencing showed an impact on the overall community diversity that persisted over time (Figure 7C, D). After an initial acclimation (20 h), the communities within recipient mice began to separate by treatment regardless of the effects of time and co-caging (Figures 7C, D and S6). At 5 days post-inoculation, the relative abundance of C. minuta was similar to that observed in the previous transplant experiment and persisted throughout the duration of the study. We identified two genera that discriminated the two treatments at Day 21: Oscillospira and a genus within the Rikenellaceae were enriched in the C. minuta treatment (misclassification error rate of 0.06). Oscillospira abundances were significantly correlated with PC2 in the unweighted UniFrac analysis of the communities (rho = −0.71, P = 0.0009; Figure 7E), which is the PC that separates the C. minuta-amended and unamended microbiotas.
Discussion
Our results represent the first strong evidence that the abundances of specific members of the gut microbiota are influenced in part by the genetic makeup of the host. Earlier studies using fingerprinting approaches also reported host genetic effects (Stewart et al., 2005; Zoetendal et al., 2001), but without sequence data it is not possible to know if the taxa shown here to be heritable were also driving those patterns. The Turnbaugh et al. and Yatsunenko et al. studies, which are quite similar in experimental approach, reported a lack of host genetic effect on the gut microbiome, most likely because both studies were underpowered. Nevertheless, re-analysis of the data from both studies validated our observation that the abundances of taxa are more highly correlated within MZ than DZ twin pairs. Thus, host genetic interactions with specific taxa are likely widespread across human populations, with profound implications for human biology.
The most highly heritable taxon in our dataset was the family Christensenellaceae, which was also the hub of a co-occurrence network that includes other taxa with high heritability. A notable component of this network was the archaeal family Methanobacteriaceae. Similarly, Hansen et al. had previously identified members of the Christensenellaceae (reported as relatives of Catabacter) as co-occurring with M. smithii (Hansen et al., 2010). These co-occurrence patterns could derive from different scenarios: for instance, multiple taxa may be heritable and co-occur while each taxon is affected by host genetics independently, or alternatively one (or a few) taxa may be heritable and other taxa correlate with host genetics due to their co-occurrence with these key heritable taxa. Further experimental research will be required to elucidate if the co-occurring heritable taxa interact in syntrophic partnerships or simply respond similarly to host-influenced environmental cues in the gut.
Our results suggest that environmental factors mostly shape the Bacteroidetes community, since most were not heritable. These results are consistent with those of a recent study of Finnish MZ twins, in which levels of Bacteroides spp. were more similar between twins when their diets were similar (Simoes et al., 2013). Members of the Bacteroidetes have been shown to respond to diet interventions (Wu et al., 2011; David et al. 2013)
Importantly, the family Christensenellaceae is heritable in the Yatsunenko dataset and its network is also present. This validation did not involve a directed search using the taxa identified in this study but was made by applying the ACE model independently. In the TwinsUK as well as the Missouri twins datasets, the majority of OTUs with the highest heritability estimates fell within the Ruminococcaceae and Lachnospiraceae families. The Missouri and TwinsUK studies differed somewhat in the levels and structure of heritability, which may relate to study size (Kuczynski et al., 2010), participant age (Claesson et al., 2011), population (Yatsunenko et al., 2012), and/or diet (Wu et al., 2011), all of which have been shown to affect microbiome structure.
The high heritability of the Christensenellaceae raises questions about the nature of interactions between the host and members of this family, but to date there is little published work with which to infer their roles. Christensenella minuta is Gram-negative, non-spore forming, non-motile, and produces SCFAs (Morotomi et al., 2012). A review of publicly available data suggests it is present from birth and associates with a healthy state but not with diet. Thus, although diet is a heritable trait in the same population (Menni et al., 2013; Teucher et al., 2007), it does not appear to be driving the heritability of the Christensenellaceae. Obesity is also strongly heritable in the TwinsUK population, raising the question of whether the heritabilities we saw for gut microbes were driven by BMI. To test this, we reran the heritability calculations using residuals after regressing out the effect of BMI and found that results of the two analyses were highly correlated. This suggests that the effect of host genetics on Christensenellaceae abundance is independent of an effect of BMI.
Our transplantation experiments showed a moderating effect of methanogen-presence in the human donor on weight gain of recipient mice, although strikingly, M. smithii did not persist in mice. In contrast, Christensenellaceae levels in mice mirrored their weight gain. Transfer to germfree mice of microbiomes from obese and lean donors generally results in greater adiposity gains for obese compared to lean donors (Ridaura et al., 2013; Turnbaugh et al., 2008; Vijay-Kumar et al., 2010). These studies have not reported the methanogen or Christensenellaceae status of the donors, so whether these microbes affect the host phenotype is unknown. M. smithii has been associated with a lean phenotype in multiple studies (Million et al., 2013; Million et al., 2012; Schwiertz et al., 2010; Armougom et al., 2009; Le Chatelier, 2013), raising the possibility that methanogens are key components of the consortium for regulating host phenotype. The results of our methanogen-Christensenellaceae transfer revealed that although methanogens may be a marker for a low BMI in humans, they are not required to promote a lean phenotype in the germfree mouse experimental model. This result suggests that methanogens may be functionally replaced by another consortium member in the mouse, while the Christensenellaceae are not.
The results of the C. minuta spike-in experiments supported the hypothesis that members of the Christensenellaceae promote a lean host phenotype. Addition of C. minuta also remodeled the diversity of the community. Intriguingly, Oscillospira, which includes heritable OTUs in the TwinsUK dataset and is associated with a lean BMI, was enriched in the C. minuta-amended microbiomes. How C. minuta reshapes the community remains to be explored. The relatively low levels of C. minuta and its profound effects on the community and the host may indicate that it is a keystone taxon. Together these findings indicate that the Christensenellaceae are highly heritable bacteria that can directly contribute to the host phenotype with which they associate.
Conclusion
Host genetic variation drives phenotype variation, and this study solidifies the notion that our microbial phenotype is also influenced by our genetic state. We have shown that the host genetic effect varies across taxa and includes members of different phyla. The host alleles underlying the heritability of gut microbes, once identified, should allow us to understand the nature of our association with these health-associated bacteria, and eventually to exploit them to promote health.
Experimental Procedures
Human subjects and sample collection
Fecal samples were obtained from adult twin pair participants of the TwinsUK registry (Moayyeri, 2013). Most participants were women (only 20 men were included). Twins collected fecal samples at home, and the samples were refrigerated for up to 2 days prior to their annual clinical visit at King's College London, at which pointed they were stored at −80°C until processing.
Diversity and phylogenetic analyses
We amplified 16S rRNA genes (V4) from bulk DNA by PCR prior to sequencing on the Illumina MiSeq 2x250bp platform at Cornell Biotechnology Resource Center Genomics Facility. We performed quality filtering and analysis of the 16S rRNA gene sequence data with QIIME 1.7.0 (Caporaso et al., 2010).
Predicted metagenomes
PICRUSt v1.0.0 was used to predict abundances of COGs from the OTU abundances rarefied at 10,000 sequences per sample.
Heritability estimations
Heritability estimates were calculated on the OTU abundances, taxon bins, nodes throughout the bacterial phylogenetic tree, α-diversity, and PICRUSt-predicted COGs using the structural equation modeling software OpenMx (Boker et al., 2011).
Microbiota transfer experiments
Stool samples from the Twins UK cohort were selected as described in the main text and inoculated into 6-week old germ-free Swiss Webster mice via oral gavage, with one recipient mouse per fecal donor. Mice were single-housed. For the Christensenella minuta addition, three experiments were conducted: In the first experiment, one treatment group received only donor stool inoculum, whereas the other received donor stool amended with 1 × 108 C. minuta cells;for the second experiment, a heat-killed C. minuta control was added; in the third experiment, the heat-killed control was compared to live C. minuta only (no vehicle-only control). Mice were housed 4 per cage, with 3 cages per treatment. In all experiments, mice were provided with autoclaved food and water ad libitum and maintained on a 12-hr light/dark cycle. Body weight and chow consumption were monitored and fecal pellets were collected weekly. At sacrifice, adiposity was analyzed using DEXA. Body, mesenteric adipose tissue, and gonadal fat pad tissue weights were collected at this time. Gross energy content of mouse stool samples was measured by bomb calorimetry using an IKA C2000 calorimeter (Dairy One, Ithaca, NY). Wet cecal contents were weighed and resuspended in 2% (v/v) formic acid by vortexing and measured using a gas chromatograph (HP series 6890) with flame ionization detection.
Statistical analysis
Values are expressed as the mean ± SEM unless otherwise indicated. Full methods are described in the Supplemental Information
Supplementary Material
Acknowledgements
We thank Wei Zhang, Sara Di Rienzi, Lauren Harroff, Largus Angenent, Hannah de Jong, Gabe Fox, Nick Scalfone, Aymé Spor, and Beth Bell for their help. We also thank three anonymous reviewers for their helpful comments, and Mary-Claire King for her advice and encouragement. This work was funded by NIH RO1 DK093595, DP2 OD007444, The Cornell Center for Comparative Population Genomics, the Wellcome Trust, and the European Community's Seventh Framework Programme (FP7/2007-2013). The study also received support from the National Institute for Health Research (NIHR) BioResource Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas’ NHS Foundation Trust and King's College London. R.E.L. is a Fellow of the David and Lucile Packard Foundation and of the Arnold and Mabel Beckman Foundation; J.K.G. is a National Academy of Sciences predoctoral Fellow; Tim Spector is holder of an ERC Advanced Researcher Award; R.K. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: R.E.L. and A.G.C. supervised the study, and J.T.B. and T.D.S. helped design study and provided comments and discussion. J.T.B. and T.D.S. oversaw collection of samples; J.K.G., R.E.L., O.K., J.L.S., A.C.P, J.L.W. oversaw microbial data generation; J.K.G. performed the analysis with contributions from R.E.L, R.B., A.G.C., J.L.W., O.K., A.C.P, M.B., W.V.T. and R.K; J.K.G. and J.L.W. performed microbiota transfer experiments; J.K.G., J. L.W and R.E.L. prepared the manuscript, with comments from A.G.C, T.D.S, J.T.B, R.B., and R.K.
Author Information: The 16S rRNA gene sequences have been deposited in the European Nucleotide Archive (ENA), European Bioinformatics Institute, with accession numbers ERP006339 and ERP006342.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and data and can be found with this article online.
References cited
- Armougom F, Henry M, Vialettes B, Raccah D, Raoult D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS ONE. 2009;4:e7125. doi: 10.1371/journal.pone.0007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basseri RJ, Basseri B, Pimentel M, Chong K, Youdim A, Low K, Hwang L, Soffer E, Chang C, Mathur R. Intestinal methane production in obese individuals is associated with a higher body mass index. Gastr Hepatol. 2012;8:22–28. [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins CL, Salzman NH. The potter's wheel: the host's role in sculpting its microbiota. Cell Mol Life Sci. 2011;68:3675–3685. doi: 10.1007/s00018-011-0830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemen JG, Venema K, van de Poll MC, Olde Damink SW, Buurman WA, Dejong CH. Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin Nutr. 2009;28:657–661. doi: 10.1016/j.clnu.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T, Spies J, Estabrook R, Kenny S, Bates T, et al. OpenMx: An open source extended structural equation modeling framework. Psychometrika. 2011;76:306–317. doi: 10.1007/s11336-010-9200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9:88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SF, Murphy EF, Nilaweera K, Ross PR, Shanahan F, O'Toole PW, Cotter PD. The gut microbiota and its relationship to diet and obesity: new insights. Gut Microb. 2012;3:186–202. doi: 10.4161/gmic.20168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, Goodrich JK, Bell JT, Spector TD, Banfield JF, et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. eLife. 2013;2:e01102. doi: 10.7554/eLife.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield TF, Merrill JK, Bagg RN. Meta-analysis of the effects of monensin in beef cattle on feed efficiency, body weight gain, and dry matter intake. J Anim Sci. 2012;90:4583–4592. doi: 10.2527/jas.2011-5018. [DOI] [PubMed] [Google Scholar]
- Eaves LJ, Last KA, Young PA, Martin NG. Model-fitting approaches to the analysis of human behaviour. Heredity. 1978;41:249–320. doi: 10.1038/hdy.1978.101. [DOI] [PubMed] [Google Scholar]
- Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, Zhu W, Sartor RB, Boedeker EC, Harpaz N, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Hamilton MJ, Weingarden AR, Unno T, Khoruts A, Sadowsky MJ. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut microbes. 2013;4:125–135. doi: 10.4161/gmic.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, McSweeney CS. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS ONE. 2008;3:e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Bäckhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J, Costello EK, Nemergut DR, Zaneveld J, Lauber CL, Knights D, Koren O, Fierer N, Kelley ST, Ley RE, et al. Direct sequencing of the human microbiome readily reveals community differences. Genome Biol. 2010;11:210. doi: 10.1186/gb-2010-11-5-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto J-M, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Lee S, Sung J, Lee J, Ko G. Comparison of the gut microbiotas of healthy adult twins living in South Korea and the United States. Appl Environ Microbiol. 2011;77:7433–7437. doi: 10.1128/AEM.05490-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2013;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J. Resistant Starches Types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS ONE. 2010;5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnite AM, Perez-Munoz ME, Lu L, Williams EG, Brewer S, Andreux PA, Bastiaansen JW, Wang X, Kachman SD, Auwerx J, et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One. 2012;7:e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menni C, Zhai G, Macgregor A, Prehn C, Romisch-Margl W, Suhre K, Adamski J, Cassidy A, Illig T, Spector TD, et al. Targeted metabolomics profiles are strongly correlated with nutritional patterns in women. Metabolomics. 2013;9:506–514. doi: 10.1007/s11306-012-0469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million M, Angelakis E, Maraninchi M, Henry M, Giorgi R, Valero R, Vialettes B, Raoult D. Correlation between body mass index and gut concentrations of Lactobacillus reuteri, Bifidobacterium animalis, Methanobrevibacter smithii and Escherichia coli. Int J Obes. 2013;37:1460–1466. doi: 10.1038/ijo.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes. 2012;36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort Profile: TwinsUK and healthy ageing twin study. Int J oEpidemiol. 2013;42:76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondot S, Kang S, Furet JP, Aguirre de Carcer D, McSweeney C, Morrison M, Marteau P, Dore J, Leclerc M. Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflamm Bowel Dis. 2011;17:185–192. doi: 10.1002/ibd.21436. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morotomi M, Nagai F, Watanabe Y. Description of Christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int J Syst Evol Microbiol. 2012;62:144–149. doi: 10.1099/ijs.0.026989-0. [DOI] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One. 2012;7:e39242. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf K, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Rausch P, Rehman A, Kunzel S, Hasler R, Ott SJ, Schreiber S, Rosenstiel P, Franke A, Baines JF. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci U S A. 2011;108:19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, Schreiber S, Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Simoes CD, Maukonen J, Kaprio J, Rissanen A, Pietilainen KH, Saarela M. Habitual dietary intake is associated with stool microbiota composition in monozygotic twins. J Nutr. 2013;143:417–423. doi: 10.3945/jn.112.166322. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Stewart JA, Chadwick VS, Murray A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol. 2005;54:1239–1242. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- Teucher B, Skinner J, Skidmore PM, Cassidy A, Fairweather-Tait SJ, Hooper L, Roe MA, Foxall R, Oyston SL, Cherkas LF, et al. Dietary patterns and heritability of food choice in a UK female twin cohort. Twin Res Hum Genet. 2007;10:734–748. doi: 10.1375/twin.10.5.734. [DOI] [PubMed] [Google Scholar]
- Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, de Vos WM, Zoetendal EG. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013;7:707–717. doi: 10.1038/ismej.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tims S, Zoetendal EG, Vos WM, Kleerebezem M. Host Genotype and the Effect on Microbial Communities. In: Nelson KE, editor. Metagenomics of the Human Body. Springer; New York: 2011. pp. 15–41. [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacklin P, Makivuokko H, Alakulppi N, Nikkila J, Tenkanen H, Rabina J, Partanen J, Aranko K, Matto J. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One. 2011;6:e20113. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Loos RJ, Powell JE, Medland SE, Speliotes EK, Chasman DI, Rose LM, Thorleifsson G, Steinthorsdottir V, Magi R, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490:267–272. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, Visser J.A.G.M.d., Vos W.M.d. The host genotype affects the bacterial community in the human gastrointestinal tract. Microbial Ecology in Health and Disease. 2001;13:129–134. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.