Abstract
As part of a worldwide survey of the indoor mycobiota, dust was collected from nine countries. Analyses of dust samples included the culture-dependent dilution-to-extinction method and the culture-independent 454-pyrosequencing. Of the 7 904 isolates, 2 717 isolates were identified as belonging to Aspergillus, Penicillium and Talaromyces. The aim of this study was to identify isolates to species level and describe the new species found. Secondly, we wanted to create a reliable reference sequence database to be used for next-generation sequencing projects. Isolates represented 59 Aspergillus species, including eight undescribed species, 49 Penicillium species of which seven were undescribed and 18 Talaromyces species including three described here as new. In total, 568 ITS barcodes were generated, and 391 β-tubulin and 507 calmodulin sequences, which serve as alternative identification markers.
Key words: Environmental metagenomics, Indoor moulds, Eurotiales, Trichocomaceae
Introduction
Indoor environments provide humans with a protective habitat in which they spend up to 90 % of their time (Höppe & Martinac 1998). These environments reportedly have unique microbial communities, which have adapted to the specific carbon, temperature and humidity constraints of these environments (Flannigan et al. 2011). Indoor environments, especially in first world countries, are well regulated with regards to temperature and are generally dry. Carbon sources available to fungi include damaged building materials (Flannigan et al. 2011), textiles, various food products and dust (Samson et al. 2010, Flannigan et al. 2011). When actively growing on these substrates, fungi often release high concentrations of spores and fungal fragments into the air that could affect humans as pathogens (Li et al. 1998, de Hoog 2000, Garber 2001), allergens (Horner et al. 1995, Terr 2009, Aimanianda et al. 2009, Green et al. 2009, Jaakkola et al. 2010, Karvala et al. 2011), food spoilers (Pitt & Hocking 2009, Samson et al. 2010) or cause structural damage to building materials (Kauserud et al. 2007, Schmidt 2007, Chunduri 2014). Combined with the number of immuno-compromised individuals rising worldwide, many common fungi are being reported as causing infections (Vartivarian et al. 1993, Latgé 1999, Lin et al. 2001, Lyratzopoulos et al. 2002). This has lead to increased concern in the western world concerning indoor fungal communities, with much focus on Stachybotrys (commonly referred to as the “black toxic-mould”) and sick building syndrome (Mahmoudi & Gershwin 2000, Wilson & Straus 2002, Terr 2009, Straus 2009, Adams et al. 2013a). Samson et al. (2010) and Flannigan et al. (2011), based on years of experience in isolating and identifying fungi from food and indoor environments, published a list of 100 species that are commonly isolated from indoor environments in the western world. Next-generation sequencing, however, has changed the way we perceive these communities, because it allows us to detect the “unculturable” fungi (Amend et al. 2010). Culture-independent techniques rely on curated sequences (mostly from the ITS region) for correct identifications. The number of reliable databases is currently limited (Kõljalg et al. 2005, 2013). This can be attributed to the large volume of alpha taxonomy necessary to create reliable identification databases (Peterson 2012). It is crucial to get these in place, because obtaining an accurate name unlocks a large amount of associated information.
Aspergillus, Penicillium and Talaromyces (Eurotiomycetes) are considered among the commonest genera found indoors (Pitt & Hocking 2009, Samson et al. 2010, Amend et al. 2010). These species are often associated with specific food items (Frisvad & Samson 2004) and colonies can produce millions of conidia. This profuse sporulation may account for the ease with which some species are isolated. Very common indoor Aspergillus species include A. calidoustus, A. flavus (common aflatoxin producers; Codner et al. 1963, Schroeder & Boller 1973, Pildain et al. 2008, Ezekiel et al. 2014), A. fumigatus (human pathogen and most common causative agent of aspergillosis and mycetoma; Latgé 1999, de Hoog 2000, Grosjean & Weber 2007), A. penicillioides, A. restrictus, A. sydowii (a common cause of human mycoses; Takahata et al. 2008, Samson et al. 2010), A. versicolor and A. westerdijkiae (common ochratoxin A producer; Frisvad et al. 2004a). Many Penicillium species are associated with biodeterioration of specific foods and are thus commonly reported from indoor surveys. Examples include P. expansum, commonly associated with apple rot and patulin production (Frisvad et al. 2004b), while P. digitatum and P. italicum cause rots of citrus (Frisvad & Samson 2004). Other species considered very common in indoors include P. brevicompactum (produces mycophenolic acid; Frisvad et al. 2004b), P. chrysogenum (penicillin producer; Houbraken et al. 2012), P. citrinum, P. commune, P. glabrum, P. olsonii, P. oxalicum and P. rubens (penicillin producer; Houbraken et al. 2012), while common Talaromyces species include T. funiculosus, T. rugulosus (produces rugulosin; Samson et al. 2010) and T. wortmanii (produces rugulosin and wortmanin; Brian et al. 1957, Samson et al. 2010). The two species, A. versicolor and P. chrysogenum were very common in buildings with water damage and have been suggested as indicators for sick building syndrome (Andersen et al. 2011).
As of 1 January 2013, single name nomenclature for fungi was enforced in the International Code of Nomenclature for algae, fungi, and plants (ICN) (McNeill & Turland 2011, McNeill et al. 2012). The abandonment of dual nomenclature resulted in significant changes in the taxonomy and nomenclature of Aspergillus, Penicillium and Talaromyces. Based on a four gene phylogeny, Houbraken & Samson (2011) showed that species formerly classified in Penicillium subgenus Biverticillium are resolved in a monophyletic clade with the former teleomorph genus Talaromyces, while the remaining Penicillium species are associated with the younger teleomorph genus name Eupenicillium. As such, Samson et al. (2011) transferred the accepted species of Penicillium subgenus Biverticillium into Talaromyces and Houbraken & Samson (2011) transferred Eupenicillium species into Penicillium. This was well received by the general community working on these fungi (Houbraken et al. 2011a,b, Visagie & Jacobs 2012, Visagie et al. 2012, Yilmaz et al. 2012, Manoch et al. 2013, Visagie et al. 2013, Peterson & Jurjevic 2013, Sang et al. 2013, Fujii et al. 2013, Frisvad et al. 2013, Devi et al. 2014, Dufossé et al. 2014, Kanse et al. 2014).
The single name solution for Aspergillus and its large number of younger associated teleomorphic genera (such as Emericella, Neosartorya, Eurotium etc.) is controversial, although phylogenetic data seems to suggest that Aspergillus and its associated teleomorphic genera collectively represent a monophyletic clade based on Bayesian analysis of a four gene (Houbraken & Samson 2011) and 25 gene (Houbraken et al. 2014a) dataset. The International Commission of Penicillium and Aspergillus (ICPA) voted on this nomenclatural issue on 14 April 2012 and chose to retain a broad but monophyletic concept of Aspergillus, rather than splitting the genus into smaller clades correlating with the teleomorphic names.
To achieve stability in names, the opportunity exists to have them protected in the ICN. With this in mind, lists for Aspergillus, Penicillium and Talaromyces were published in Visagie et al. (2014a), Samson et al. (2014) and Yilmaz et al. (2014), with updates to these lists available on ICPA's website (http://www.aspergilluspenicillium.org). In addition to providing accepted species lists, it also provides information such as culture collection numbers for living ex-type material and GenBank accession numbers for sequences linked to these ex-types. This is considered an important step towards enabling correct species identifications in these large genera; there are currently 331 species in Aspergillus, 319 in Penicillium and 85 in Talaromyces. The ITS barcodes from this list are incorporated in the RefSeq data set intended to enhance reliable fungal identifications using the GenBank database (Schoch et al. 2014).
This paper is focused on the identification of Aspergillus, Penicillium and Talaromyces species isolated from house dust collected from nine countries and the creation of a DNA barcode database for these species. We describe 18 new species, including eight in Aspergillus, seven in Penicillium, and three in Talaromyces. This work contributes to the Alfred P. Sloan research network on the Microbiology of the Built Environment, by providing authoritative taxonomic and molecular data to be used for metagenomic studies, thereby helping bridge the gap between culture-dependent and -independent detection techniques. This is the second of a series of reports on the taxonomy of fungi isolated using dilution to extinction in the survey, the first being the brief description of a new species of Rasamsonia (Tanney & Seifert 2013). The taxonomy of the other fungi isolated, reporting a taxonomically broad and diverse range of fungi, will be the subject of future publications.
Materials and methods
Isolations and identifications
Settled dust was collected in April of 2009 using sterilised Duststream® collectors (Indoor Biotechnologies) attached to vacuum cleaners. Buildings from nine countries, including Australia, Indonesia, Mexico, Micronesia, New Zealand, South Africa, Thailand, United Kingdom and Uruguay, were included in the survey. Samples from Canada and the United States were not included in this taxonomic study. Isolations were made using a dilution-to-extinction (d2e) method modified from Collado et al. (2007), using microplates composed of capped 1.5 mL micro tubes instead of 48-well microplates (Seifert et al. unpubl.). Malt extract agar (MEA, 20 g malt extract, 15 g agar, 1 000 mL dH2O) and 20 % sucrose MEA (20SMEA) with chloramphenicol were used as isolation media and 1.0 mL of medium was dispensed into each micro tube using a multichannel pipette. House dust was suspended in a carboxymethylcellulose solution and diluted stepwise up to 1:64; the dilution yielding the maximum number of single-species colonies in the isolation was selected for subsequent study. For preliminary screening, cultures that appeared to represent Penicillium, Aspergillus, or Talaromyces were plated onto MEA in 6 cm Petri dishes and purified as necessary. All selected cultures from each country were sorted into putative species groups based on colony characters on MEA, and then up to five strains per culture-group per country were selected for more detailed study.
For our analyses of the prevalence of particular species in specific countries (Table 1), samples collected from different sites were considered to represent one sample.
Table 1.
Aspergillus, Penicillium and Talaromyces species distribution in house dust from around the world.

Isolates from each country were placed into morpho-groups based on their characters on Czapek Yeast Autolysate agar (CYA) and Malt Extract agar (MEA), with Dichloran 18 % Glycerol agar (DG18) added for Aspergillus. From here, 5–10 strains from each morpho-group were selected for sequencing. The β-tubulin gene (BenA) and internal transcribed spacer regions of the nrDNA operon (ITS) were sequenced for Penicillium and Talaromyces, with calmodulin (CaM) and ITS sequenced for Aspergillus. Phylogenetic comparisons of newly generated sequences with a reliable reference sequence database were used for making identifications. This reference database was compiled based on the accepted species list described in the introduction. This list provides GenBank accession numbers to ITS, BenA and CaM sequences of all ex-type strains for accepted species in Aspergillus, Penicillium and Talaromyces. In the case of new species, CaM was added for Penicillium and Talaromyces, while BenA was added for new Aspergillus species.
Isolates were deposited into the working collection of the Applied and Industrial Mycology department (DTO) housed at the CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands. Strains representing new species were deposited into the public collection of the CBS-KNAW (CBS).
DNA extraction, sequencing and phylogenetic analysis
DNA was extracted from 10-day-old strains grown on MEA (some Aspergillus species on DG18) using the Ultraclean™ Microbial DNA isolation Kit (MoBio, Solano Beach, USA), with DNA preps stored at −20 °C. PCR reactions were prepared as described in Houbraken & Samson (2011). Amplification of ITS was done using the primer set ITS1 and ITS4 (White et al. 1990), primer pair Bt2a and Bt2b for BenA (Glass & Donaldson 1995) and cmd5 and cmd6 for CaM (Hong et al. 2006). A standard amplification cycle was used, which ran 35 cycles with an annealing temperature of 55 °C. Sequencing reactions were set up using the BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, CA) with the same primer sets used for amplification. Sequences were determined on an ABI PRISM 3730xl genetic analyser (Applied Biosystems, California, USA). Sequence contigs were assembled using Seqman Pro v. 9.0.4 (DNAstar Inc.) and newly generated sequences deposited into GenBank.
The phylogenies presented here were prepared using a subset of representative strains of each species. The ITS phylogenies for Aspergillus, Penicillium and Talaromyces were used to direct the allocation of sequences into the correct genera and into smaller clades (indicated by different colours in the large scale trees), to allow more robust alignments of the alternative genes BenA and CaM.
Data sets were aligned in MAFFT v. 7.058b (Katoh & Standley 2013) using the L-INS-i algorithm. When needed, manual adjustments to alignments were made in MEGA v. 5.2.2 (Tamura et al. 2011). Maximum-likelihood (ML) trees were calculated for aligned data sets using MEGA. For the multigene phylogenies presented in the Taxonomy section for new species below, data sets were concatenated in Seaview v. 4.4.1 (Gouy et al. 2010). The most suitable model was determined in MEGA based on the lowest Bayesian Information Criterion (BIC). ML analyses were run by calculating the initial tree with the Bio-Neighbour-Joining (BioNJ) option, followed by a Heuristic search with the Nearest-Neighbour-Interchange (NNI) option. Support in nodes was calculated using a bootstrap analysis with 1 000 replicates. In the phylogenies presented, thickened branches indicate bootstrap support above 80 %.
Morphology
Species were characterised using standard growth conditions (Okuda et al. 2000, Visagie et al. 2013, Visagie et al. 2014a). Strains were inoculated in three-point fashion onto CYA, MEA, Yeast Extract Sucrose agar (YES), DG18, CYA with 5 % NaCl (CYAS), Oatmeal agar (OA) and Creatine Sucrose agar (CREA). Plates were incubated in plastic boxes for 7 d in the dark at 25 °C. Additional CYA plates were incubated at 30 and 37 °C. Colour names and alphanumeric codes used in descriptions refer to Kornerup & Wanscher (1967).
Microscopic preparations were made from colonies grown on MEA, with DG18 also used for Aspergillus, after 1 to 2 wk. Lactic acid (60 %) was used as mounting fluid and excess conidia were washed away with 70 % ethanol. Characters were captured using a Zeiss SteREO Discovery.V20 dissecting microscope and Zeiss AX10 Imager.A2 compound microscope, both equipped with AxioCam MRc5 cameras using AxioVs40 v. 4.8.2.0. Microscopic measurements were done using Nikon NIS-elements D v. 4.0. Photo plates were prepared in Adobe® Photoshop® CS6 with photomicrographs cleaned up using the healing brush tool, for aesthetic reasons, without altering areas of scientific significance.
Results
Isolations and identifications
D2E dust isolations resulted in 7 904 isolates, including 1 160 Aspergillus, 1 459 Penicillium and 98 Talaromyces isolates. Isolates represented 59 Aspergillus, 49 Penicillium and 18 Talaromyces species. Of these, 18 displayed unique characters deviating from known species of these genera and are described below as new species in the taxonomy section. Species identities and their presence/absence at a country scale are provided in Table 1.
High species richness was observed in dust collected in South Africa (47 species), Thailand (44 species), Mexico (32 species), New Zealand (31 species) and Micronesia (28 species). Countries with low species richness included the United Kingdom (11 species), Australia (8 species), Indonesia (7 species) and Uruguay (5 species).
Aspergillus diversity was high in Thailand (25 species), South Africa (23 species), Mexico (23 species) and Micronesia (19 species), while no Aspergillus species were isolated from Australian house dust. Penicillium species richness was highest in New Zealand (22 species) and South Africa (16 species), with no Penicillium species isolated from Uruguay. For Talaromyces, Thailand (10 species) and South Africa (8 species) had the highest species richness, while none were isolated from Australia, Indonesia, the United Kingdom and Uruguay.
Several species were common in the house dust (Table 1), with 13 Aspergillus species isolated from more than two countries. Aspergillus sydowii occurred in dust from six countries, A. fumigatus in five and A. subramanianii, A. niger and A. versicolor in four. Six Penicillium species were isolated from more than two countries. Penicillium brevicompactum, P. citrinum and P. rubens were isolated from four and P. corylophilum, P. glabrum and P. pancosmium from three. Penicillium chrysogenum was isolated in high numbers from dust in Australia and the United Kingdom. Penicillium rubens, a close relative of P. chrysogenum and the correct name for Fleming's penicillin producer (Houbraken et al. 2011a), were also abundant in Australia, New Zealand, South Africa and the United Kingdom. Talaromyces allahabadensis and T. atroroseus occurred in three countries, with T. albobiverticillius, T. diversus and T. minioluteus occurring in two.
DNA sequences generated for identified species include 568 ITS barcodes (Aspergillus 283, Penicillium 229, Talaromyces 56). As secondary identification markers, 391 BenA (Aspergillus 126, Penicillium 203, Talaromyces 26) and 507 CaM (Aspergillus 278, Penicillium 56, Talaromyces 26) sequences were generated. All sequences were uploaded onto the Indoor Molds Database housed at the CBS-KNAW Fungal Biodiversity Center (http://www.cbs.knaw.nl/indoor/) and representative sequences for each species have been submitted to GenBank under accession numbers KJ775068–KJ775228, KJ775248–KJ775432, KJ775451–KJ775735 and KJ866960–KJ867021. Table 2 summarises GenBank numbers of strains used for multigene phylogenies in the Taxonomy section.
Table 2.
Strains used for phylogenetic analyses of new Aspergillus, Penicillium and Talaromyces species described from house dust.
| Species | Culture collection number | GenBank accession nr. |
||
|---|---|---|---|---|
| ITS | BenA | CaM | ||
| A. amoenus | NRRL 4838 | EF652480 | JN853946 | JN854035 |
| A. arenarioides | CBS 138195 = DTO 129G8 | KJ775557 | KJ775070 | KJ775256 |
| CBS 138196 = DTO 267B6 | KJ775558 | KJ775082 | KJ775347 | |
| CBS 138197 = DTO 267C7 | KJ775559 | KJ775083 | KJ775349 | |
| CBS 138198 = DTO 268E1 | KJ775560 | KJ775089 | KJ775388 | |
| CBS 138199 = DTO 268E2 | KJ775561 | KJ775090 | KJ775389 | |
| CBS 138200 = DTO 268E3 | KJ775562 | KJ775091 | KJ775390 | |
| A. arenarius | CBS 463.65 = NRRL 5012 = ATCC 16830 = IMI 055632 = IMI 055632ii = WB 4429 = WB 5012 | EU021615 | EU021674 | EU021681 |
| A. aureofulgens | CBS 653.74 = NRRL 6326 | EF669617 | EU014079 | EF669575 |
| A. austroafricanus | NRRL 233 | JQ301891 | JN853963 | JN854025 |
| A. baeticus | NRRL 62501 = CCF 4226 = CMFISB 2153 | HE615086 | HE615092 | HE615117 |
| A. brevijanus | CBS 111.46 = NRRL 1935 = ATCC 16828 = CBS 119.45 = IMI 016066ii = IMI 16066 = NCTC 6971 = QM 7417 = WB 1935 | EF669582 | EU014078 | EF669540 |
| A. brunneus | CBS 112.26 = CBS 524.65 = NRRL 131 = NRRL 134 = ATCC 1021 = IFO 5862 = IMI 211378 = QM 7406 = Thom 4481 = Thom 5633.4 = WB 131 | EF652060 | EF651907 | EF651998 |
| NRRL 133 | EF652061 | EF651908 | EF651999 | |
| A. campestris | CBS 348.81 = NRRL 13001 = ATCC 44563 = IMI 259099 | EF669577 | EU014091 | EF669535 |
| A. candidus | CBS 566.65 = NRRL 303 = ATCC 1002 = IMI 16264 = IMI 91889 = LSHBA c .27 = NCTC 595 = QM 1995 = Thom 106 = WB 303 | EF669592 | EU014089 | EF669550 |
| NRRL 4646 | EF669605 | EU014090 | EF669563 | |
| A. capensis | CBS 138188 = DTO 179E6 | KJ775550 | KJ775072 | KJ775279 |
| A. compatibilis | CBS 488.65 = NRRL 5096 = ATCC 16847 = IMI 139277 = QM 8916 = WB 5096 | EF652499 | EF652323 | EF652411 |
| A. creber | NRRL 58592 | JQ301889 | JN853980 | JN854043 |
| A. cvjetkovicii | NRRL 227 | EF652440 | EF652264 | EF652352 |
| A. flavipes | NRRL 302 = ATCC 24487 = IMI 171885 = QM 9566 = Thom 4640.474 = WB 302 | EF669591 | EU014085 | EF669549 |
| A. fructus | NRRL 239 | EF652449 | EF652273 | EF652361 |
| A. fruticans | CBS 486.65 = NRRL 4903 = ATCC 16823 = IMI 139279 = O-1077 = QM 8033 = WB 4903 | EF652483 | EF652307 | EF652395 |
| A. glaucus | CBS 516.65 = NRRL 116 = ATCC 16469 = IMI 211383 = LCP 64.1859 = Thom 5629.C = WB 116 | EF652052 | EF651887 | EF651989 |
| NRRL 120 | EF652054 | EF651889 | EF651991 | |
| NRRL 121 | EF652055 | EF651890 | EF651992 | |
| A. griseoaurantiacus | CBS 138189 = DTO 245F5 | KJ775551 | KJ775079 | KJ775319 |
| CBS 138190 = DTO 267D2 | KJ775552 | KJ775084 | KJ775352 | |
| CBS 138191 = DTO 267D8 | KJ775553 | KJ775086 | KJ775357 | |
| A. iizukae | CBS 541.69 = NRRL 3750 = IMI 141552 = QM 9325 | EF669597 | EU014086 | EF669555 |
| NRRL 35046 | EF669596 | EU014087 | EF669554 | |
| A. janus | CBS 118.45 = NRRL 1787 = IMI 16065 = NCTC 6970 | EF669578 | EU014076 | EF669536 |
| A. jensenii | NRRL 58600 | JQ301892 | JN854007 | JN854046 |
| A. micronesiensis | CBS 138182 = DTO 245D7 | KJ775546 | KJ775078 | KJ775318 |
| CBS 138183 = DTO 267D5 | KJ775548 | KJ775085 | KJ775355 | |
| CBS 138186 = DTO 267H5 | KJ775549 | KJ775088 | KJ775372 | |
| NRRL 295 | EF669588 | EU014081 | EF669546 | |
| NRRL 4263 | EF669600 | EU014083 | EF669558 | |
| NRRL 4578 | EF669602 | EU014082 | EF669560 | |
| A. niveoglaucus | CBS 101750 | HE615135 | HE801331 | HE801323 |
| CBS 114.27 = CBS 517.65 = NRRL 127 = ATCC 10075 = IMI 32050 = LSHBA 16 = NRRL 129 = NRRL 130 = QM 1977 = Thom 5612.A16 = Thom 5633. = Thom 5633.7 = Thom 7053.2 = WB 127 = WB 130 | EF652058 | EF651905 | EF651993 | |
| NRRL 128 | EF652059 | EF651906 | EF651994 | |
| NRRL 136 | EF652062 | EF651909 | EF651995 | |
| NRRL 137 | EF652063 | EF651910 | EF651996 | |
| A. porphyreostipitatus | CBS 138202 = DTO 132D1 | KJ775563 | KJ775071 | KJ775260 |
| CBS 138203 = DTO 266D9 | KJ775564 | KJ775080 | KJ775338 | |
| A. proliferans | CBS 121.45 = NRRL 1908 = IMI 016105ii = IMI 016105iii = IMI 16105 = LSHB BB.82 = MUCL 15625 = NCTC 6546 = QM 7462 = UC 4303 = WB 1908 | EF652064 | EF651891 | EF651988 |
| NRRL 114 | EF652051 | EF651886 | EF651987 | |
| NRRL 71 | EF652053 | EF651888 | EF651990 | |
| A. protuberus | CBS 602.74 = NRRL 3505 = ATCC 18990 = QM 9804 | EF652460 | EF652284 | EF652372 |
| A. pseudoglaucus | CBS 123.28 = NRRL 40 = ATCC 10066 = IMI 016122 = IMI 016122ii = LSHBA 19 = MUCL 15624 = QM 7463 = WB 40 | EF652050 | EF651917 | EF652007 |
| A. pseudoustus | CBS 123904 = NRRL 5856 = IBT 28161 | FJ531147 | FJ531168 | FJ531129 |
| A. puniceus | CBS 495.65 = NRRL 5077 = ATCC 16800 = IMI 126692 = QM 9812 = WB 5077 | EF652498 | EF652322 | EF652410 |
| NRRL 1852 | EF652425 | EF652249 | EF652337 | |
| NRRL 4688 | EF652469 | EF652293 | EF652381 | |
| A. puulaauensis | NRRL 35641 | JQ301893 | JN853979 | JN854034 |
| A. ruber | CBS 530.65 = NRRL 52 = ATCC 16441 = IMI 211380 = QM 1973 = Thom 5599B = WB 52 | EF652066 | EF651920 | EF652009 |
| A. saccharolyticus | CBS 127449 = IBT 28509 | HM853552 | HM853553 | HM853554 |
| A. sloanii | CBS 138176 = DTO 244I8 | KJ775539 | KJ775073 | KJ775308 |
| CBS 138177 = DTO 245A1 | KJ775540 | KJ775074 | KJ775309 | |
| CBS 138231 = DTO 245A6 | KJ775541 | KJ775075 | KJ775311 | |
| CBS 138178 = DTO 245A8 | KJ775542 | KJ775076 | KJ775313 | |
| CBS 138179 = DTO 245A9 | KJ775543 | KJ775077 | KJ775314 | |
| A. subalbidus | CBS 567.65 | KJ866983 | EU076295 | EF669551 |
| CBS 138192 = DTO 129E3 | KJ775554 | KJ775068 | KJ775249 | |
| CBS 138193 = DTO 129F9 | KJ775555 | KJ775069 | KJ775250 | |
| CBS 138194 = DTO 266I9 | KJ775556 | KJ775081 | KJ775251 | |
| NRRL 4809 | EF669609 | EU014092 | EF669567 | |
| A. subversicolor | NRRL 58999 | JQ301894 | JN853970 | JN854010 |
| A. sydowii | CBS 593.65 = NRRL 250 = IMI 211384 = NRRL 254 | EF652450 | EF652274 | EF652362 |
| A. tabacinus | CBS 122718 = NRRL 4791 = IFO 4098 = QM 9766 = WB 4791 | EF652478 | EF652302 | EF652390 |
| A. taichungensis | DTO 266G2 | KJ775572 | KJ866980 | KJ775252 |
| DTO 270C9 | KJ775573 | KJ866981 | KJ775253 | |
| IBT 19404 | EU076301 | EU076297 | EU076310 | |
| A. tanneri | NRRL 62426 = NIH 1005 | JN853798 | JN896582 | JN896583 |
| A. templicola | CBS 138180 = DTO 267H4 | KJ775544 | KJ775087 | KJ775371 |
| CBS 138181 = DTO 270C6 | KJ775545 | KJ775092 | KJ775394 | |
| A. tennesseensis | NRRL 13150 | JQ301895 | JN853976 | JN854017 |
| A. tonophilus | CBS 405.65 = NRRL 5124 = ATCC 16440 = ATCC 36504 = IMI 108299 = QM 8599 = WB 5124 | EF652081 | EF651919 | EF652000 |
| A. tritici | CBS 266.81 | EU076302 | EU076293 | EU076305 |
| NRRL 313 | EF669594 | EU014093 | EF669552 | |
| A. ustus | NRRL 4991 | EF652492 | EF652316 | EF652404 |
| CBS 261.67 = NRRL 275 = ATCC 1041 = ATCC 16818 = IMI 211805 = QM 7477 = WB 275 | EF652455 | EF652279 | EF652367 | |
| A. venenatus | NRRL 13147 | JQ301896 | JN854003 | JN854014 |
| A. versicolor | CBS 583.65 = NRRL 238 = ATCC 9577 = IFO 33027 = IMI 229970 = JCM 10258 = QM 7478 = Thom 5519.57 = WB 238 | EF652442 | EF652266 | EF652354 |
| A. xerophilus | CBS 938.73 = NRRL 6131 | EF652085 | EF651923 | EF651983 |
| P. alfredii | CBS 138224 = DTO 269A4 | KJ775684 | KJ775177 | KJ775411 |
| P. atramentosum | CBS 109588 = DTO 249C3 | n.a. | KJ866976 | KJ866986 |
| CBS 109601 = DTO 249C4 | n.a. | KJ866977 | KJ866987 | |
| CBS 109611 = IBT 10565 | n.a. | KJ866972 | KJ866988 | |
| CBS 109612 = IBT 14762 | n.a. | KJ866973 | KJ866989 | |
| CBS 109613 = DTO 250G3 | n.a. | KJ866978 | KJ866990 | |
| CBS 194.88 = IBT 21504 | n.a. | KJ866974 | KJ866999 | |
| CBS 291.48 = ATCC 10104 = FRR 795 = IBT 6616 = IFO 8137 = IMI 039752 = IMI 039752ii = LSHBP 1 = MUCL 29071 = MUCL 29126 = NRRL 795 = QM 7483 | n.a. | AY674402 | FJ530964 | |
| CBS 490.84 = IBT 11800 | n.a. | KJ866975 | KJ867017 | |
| DTO 178G2 | n.a. | KJ775095 | KJ867019 | |
| P. atrovenetum | CBS 241.56 = ATCC 13352 = FRR 2571 = IFO 8138 = IMI 061837 = LSHBSm683 = QM 6963 | n.a. | JX140944 | KJ867004 |
| CBS 243.56 | n.a. | KJ866971 | KJ867005 | |
| P. brefeldianum | CBS 235.81 = NRRL 710 = FRR 710 = IFO 31731 = IMI 216896 = LCP 89.2573 = LCP 89.2578 = MUCL 38762 = QM 1872 = Thom 5296 | AF033435 | GU981623 | EU021683 |
| P. canescens | CBS 300.48 = ATCC 10419 = DSM1215 = FRR 910 = IMI 028260 = MUCL 29169 = NCTC 6607 = NRRL 910 = QM 7550 = VKMF-1148 | n.a. | JX140946 | KJ867009 |
| NRRL 35656 | n.a. | DQ658166 | DQ658167 | |
| P. chermesinum | CBS 231.81 = NRRL 2048 = FRR 2048 = IFO 31745 = IMI 191730 | AY742693 | KJ834441 | AY741728 |
| P. cinnamopurpureum | CBS 429.65 = CBS 847.68 = NRRL 162 = ATCC 18489 = CSIR 936 = FAT 362 = IAM 7016 = IFO 6032 = NHL 6359 = QM 7888 | EF626950 | EF626948 | EF626949 |
| P. coralligerum | CBS 114.69 | n.a. | KJ866970 | KJ866991 |
| CBS 123.65 = ATCC 16968 = FRR 3465 = IFO 9578 = IHEM 4511 = IMI 099159 = LCP 58.1674 = NRRL 3465 | n.a. | KJ834444 | KJ866994 | |
| P. crystallinum | CBS 479.65 = NRRL 5082 = ATCC 16833 = IMI 139270 | n.a. | EF669682 | FJ530973 |
| P. dunedinense | CBS 138218 = DTO 244G1 | n.a. | KJ775171 | KJ775405 |
| P. echinulatum | NRRL 917 | n.a. | KJ866964 | KJ867021 |
| P. ellipsoideosporum | CBS 112493 = AS 3.5688 | JX012224 | JQ965104 | AY678559 |
| P. granatense | CBS 166.81 | n.a. | KJ866967 | KJ866998 |
| P. guizhouanum | AS 3.5215 | KJ890410 | KJ890408 | KJ890406 |
| P. idahoense | CBS 341.68 = NRRL 5274 = ATCC 22055 = FRR 881 = IMI 148393 | KC411747 | EF626953 | EF626954 |
| P. incoloratum | CBS 101753 = AS 3.4672 | KJ834508 | KJ834457 | KJ866984 |
| DTO 129G5 | KJ775689 | KJ775182 | KJ775415 | |
| DTO 129I1 | KJ775690 | KJ775183 | KJ775416 | |
| P. infrapurpureum | CBS 138219 = DTO 235F6 | KJ775679 | KJ775172 | KJ775406 |
| CBS 138220 = DTO 235G2 | KJ775680 | KJ775173 | KJ775407 | |
| CBS 138221 = DTO 235G5 | KJ775681 | KJ775174 | KJ775408 | |
| CBS 138222 = DTO 235G6 | KJ775682 | KJ775175 | KJ775409 | |
| CBS 138223 = DTO 235H5 | KJ775683 | KJ775176 | KJ775410 | |
| P. jamesonlandense | CBS 102888 = DAOM 234087 = IBT 21984 = IBT 24411 | DQ267912 | DQ309448 | KJ866985 |
| P. janczewskii | CBS 221.28 = FRR 919 = IMI 191499 = NRRL 919 | n.a. | KJ834460 | KJ867001 |
| CBS 279.47 | n.a. | KJ866968 | KJ867008 | |
| CBS 413.68 | n.a. | KJ866969 | KJ867014 | |
| CBS 414.68 | n.a. | KJ866960 | KJ867015 | |
| CBS 458.69 | n.a. | KJ866961 | KJ867016 | |
| P. janthinellum | CBS 340.48 = ATCC 10455 = IMI 040238 = NRRL 2016 = QM 6865 | GU981585 | GU981625 | KF296401 |
| P. javanicum | CBS 341.48 = ATCC 9099 = CSIR 831 = FRR 707 = IFO 31735 = IMI 039733 = MUCL 29099 = NRRL 707 = QM 1876 | GU981613 | GU981657 | KF296387 |
| P. jensenii | CBS 216.28 | n.a. | KJ866963 | KJ867000 |
| CBS 327.59 = ATCC 18317 = FRR 909 = IFO 5764 = IMI 039768 = LCP 89.1389 = NRRL 909 = QM 7587 | n.a. | JX140954 | AY443490 | |
| P. jianxiense | AS 3.6521 | KJ890411 | KJ890409 | KJ890407 |
| P. kojigenum | CBS 345.61 = ATCC 18227 = CCRC 31515 = FRR 3442 = IFO 9581 = IMI 086562 = LSHBBB394 = MUCL 2457 = NRRL 3442 = QM 7957 | AF033489 | KJ834463 | KJ867011 |
| P. lanosum | CBS 106.11 = ATCC 10458 = FRR 2009 = IFO 5851 = IFO 6099 = IMI 040224 = LSHBP 86 = MUCL 29232 = NRRL 2009 = QM 7591 | DQ304540 | DQ285627 | FJ530974 |
| P. lenticrescens | CBS 138215 = DTO 129A8 | KJ775675 | KJ775168 | KJ775404 |
| P. magnielliptisporum | CBS 138225 = DTO 128H8 | n.a. | KJ775179 | KJ775413 |
| CBS 138226 = DTO 128I1 | n.a. | KJ775180 | KJ775414 | |
| P. malacaense | CBS 160.81 = NRRL 35754 = ATCC 42241 = IJFM 7093 = IMI 253801 = VKMF-2197 | EU427300 | EU427268 | KJ866997 |
| P. malodoratum | CBS 490.65 = NRRL 5083 = IMI 172289 = ATCC 16834 | n.a. | EF669681 | FJ530972 |
| P. mexicanum | CBS 138227 = DTO 270F1 | n.a. | KJ775178 | KJ775412 |
| P. nigricans | CBS 354.48 | n.a. | KJ866965 | KJ867012 |
| P. nigricans var. sulphureum | CBS 744.70 | n.a. | KJ866966 | KJ867018 |
| P. nodulum | CBS 227.89 | KC411703 | KJ834475 | KJ867003 |
| P. novae-zeelandiae | CBS 137.41 = ATCC 10473 = IFO 31748 = IMI 040584ii = NRRL 2128 = QM 1934 = VKMF-2886 | n.a. | KJ834477 | KJ866996 |
| P. oxalicum | CBS 219.30 = ATCC 1126 = FRR 787 = IMI 192332 = MUCL 29047 = NRRL 787 = QM 7606 | AF033438 | KF296462 | KF296367 |
| P. paradoxum | NRRL 2162 = ATCC 16918 = IMI 061446 | n.a. | EF669683 | EF669692 |
| P. parvulum | CBS 132825 = NRRL 35504 | EF422845 | EF506218 | EF506225 |
| P. penarojense | CBS 113178 = IBT 23262 | GU981570 | GU981646 | KF296381 |
| P. piscarium | CBS 362.48 = ATCC 10482 = FRR 1075 = IFO 8111 = IMI 040032 = NRRL 1075 = VKMF-1823 | GU981600 | GU981668 | KF296379 |
| P. radiatolobatum | CBS 340.79 | n.a. | KJ866962 | KJ867010 |
| P. raistrickii | CBS 261.33 = ATCC 10490 = FRR 1044 = IFO 6104 = IMI 040221 = LSHBB100 = NRRL 1044 = NRRL 2039 = QM 1936 = VKMF-337 | AY373927 | KJ834485 | KJ867006 |
| P. ribeum | CBS 127809 = DAOM 234091 = IBT 16537 = IBT 24431 | DQ267916 | DQ285625 | KJ866995 |
| P. sajarovii | CBS 277.83 = CECT 2751 = IMI 259992 | KC411724 | KJ834489 | KJ867007 |
| P. scabrosum | CBS 683.89 = FRR 2950 = IBT 3736 = IMI 285533 = DAOM 214786 | DQ267906 | DQ285610 | FJ530987 |
| P. shennangjianum | CBS 228.89 | KC411705 | KJ834491 | AY678561 |
| P. simile | CBS 129191 = ATCC MYA-4591 | FJ376592 | FJ376595 | GQ979710 |
| P. singorense | CBS 138211 = DTO 129H7 | KJ775671 | KJ775164 | KJ775400 |
| CBS 138212 = DTO 129H8 | KJ775672 | KJ775165 | KJ775401 | |
| CBS 138213 = DTO 131I8 | KJ775673 | KJ775166 | KJ775402 | |
| CBS 138214 = DTO 133C6 | KJ775674 | KJ775167 | KJ775403 | |
| P. skrjabinii | CBS 439.75 = NRRL 13055 = FRR 1945 = IMI 196528 = VKMF-1940 | GU981576 | GU981626 | KF296370 |
| P. soppii | CBS 226.28 = ATCC 10496 = FRR 2023 = IFO 7766 = IMI 040217 = MUCL 29233 = NRRL 2023 = QM 1964 = IBT 18220 | AF033488 | DQ285616 | KJ867002 |
| P. swiecickii | CBS 119391 = FRR 918 = IBT 27865 = IMI 191500 = NRRL 918 | AF033490 | KJ834494 | KJ866993 |
| P. vanderhammenii | CBS 126216 = IBT 23203 | GU981574 | GU981647 | KF296382 |
| P. virgatum | CBS 114838 = BBA 65745 | AJ748692 | KJ834500 | KJ866992 |
| P. wotroi | CBS 118171 = IBT 23253 | GU981591 | GU981637 | KF296369 |
| P. yarmokense | CBS 410.69 = FRR 520 = IMI 140346 = VKMF-1076 | n.a. | KJ834502 | KJ867013 |
| P. zonatum | CBS 992.72 = ATCC 24353 | GU981581 | GU981651 | KF296380 |
| T. aculeatus | CBS 289.48 = ATCC 10409 = IMI 040588 = NRRL 2129 = NRRL A-1474 | JN899378 | KF741929 | KF741975 |
| T. allahabadensis | CBS 453.93 = ATCC 15067 = CBS 304.63 | JN899345 | KF984614 | n.a. |
| T. angelicus | KACC 46611 | KF183638 | KF183640 | KJ885259 |
| T. apiculatus | CBS 312.59 = ATCC 18315 = FRR 635 = IMI 068239 | JN899375 | KF741916 | KF741950 |
| T. atricola | CBS 255.31 = NRRL 1052 = FRR 1052 = Thom 4640.439 = ATCC 52257 | KF984859 | KF984566 | n.a. |
| T. brunneus | CBS 227.60 = ATCC 18229 = FRR 646 = IFO 6438 = IHEM 3907 = IMI 078259 = MUCL 31318 | JN899365 | KJ865722 | n.a. |
| T. cnidii | DTO 269H8 | KJ775724 | KJ775217 | KJ775426 |
| DTO 269I2 | KJ775725 | KJ775218 | KJ775427 | |
| DTO 269I6 | KJ775727 | KJ775220 | KJ775429 | |
| DTO 270A4 | KJ775729 | KJ775222 | KJ775430 | |
| DTO 270A8 | KJ775730 | KJ775223 | KJ775431 | |
| DTO 270B7 | KJ775731 | KJ775224 | KJ775432 | |
| KACC 46617 | KF183639 | KF183641 | KJ885266 | |
| T. flavovirens | CBS 102801 = IBT 27044 | JN899392 | JX091376 | KF741933 |
| T. islandicus | CBS 338.48 = ATCC 10127 = IMI 040042 = MUCL 31324 = NRRL 1036 | JN899318 | KF984655 | n.a. |
| T. liani (P. liani) | CBS 225.66 = ATCC 18325 = ATCC 18331 = IMI 098480 = NRRL 3380 = VKM F-301 | JN899395 | JX091380 | KJ885257 |
| T. loliensis | CBS 643.80 = ATCC 52252 = FRR 1798 = IMI 216901 = MUCL 31325 | JN899379 | KF984658 | n.a. |
| T. oumae-annae | CBS 138207 = DTO 180B4 | KJ775710 | KJ775203 | KJ775421 |
| CBS 138208 = DTO 269E8 | KJ775720 | KJ775213 | KJ775425 | |
| T. piceus | CBS 361.48 = ATCC 10519 = IMI 040038 = NRRL 1051 | JN899370 | KF984668 | n.a. |
| T. pinophilus | CBS 631.66 = ATCC 36839 = CECT 2809 = DSM 1944 = IAM 7013 = IMI 114933 | JN899382 | JX091381 | KF741964 |
| T. radicus | CBS 100489 = FRR 4718 | JN899324 | KF984599 | n.a. |
| T. rotundus | CBS 369.48 = ATCC 10493 = IMI 040589 = NRRL 2107 | JN899353 | KJ865730 | n.a. |
| T. rugulous | CBS 371.48 = ATCC 10128 = IMI 040041 = MUCL 31201 = NRRL 1045 | JN899374 | KF984575 | n.a. |
| T. sayulitensis | CBS 138204 = DTO 245H1 | KJ775713 | KJ775206 | KJ775422 |
| CBS 138205 = DTO 245H2 | KJ775714 | KJ775207 | KJ775423 | |
| CBS 138206 = DTO 245H3 | KJ775715 | KJ775208 | KJ775424 | |
| T. scorteus | CBS 340.34 = NRRL 1129 = FRR 1129 | KF984892 | KF984565 | n.a. |
| T. siamensis | CBS 475.88 = IMI 323204 | JN899385 | JX091379 | KF741960 |
| DTO 269I3 | KJ775726 | KJ775219 | KJ775428 | |
| T. tardifaciens | CBS 250.94 | JN899361 | KC202954 | n.a. |
| T. tratensis | CBS 133146 = KUFC 3383 | JX898040 | KF984559 | n.a. |
| DTO 270F5 | KF984889 | KF984557 | n.a. | |
| T. verruculosus | CBS 388.48 = ATCC 10513 = DSM 2263 = IMI 040039 = NRRL 1050 | JN899367 | KF741928 | KF741944 |
| DTO 129H4 | KJ775698 | KJ775191 | KJ775419 | |
| DTO 129H5 | KJ775699 | KJ775192 | KJ775420 | |
| T. viridulus | CBS 252.87 = FRR 1863 = IMI 288716 | JN899314 | JX091385 | KF741943 |
| T. wortmanii | CBS 391.48 = ATCC 10517 = IMI 040047 = NRRL 1017 | JN899352 | KF984648 | n.a. |
| T. yelensis | CBS 138209 = DTO 268E5 | KJ775717 | KJ775210 | n.a. |
| CBS 138210 = DTO 268E7 | KJ775719 | KJ775212 | n.a. | |
Phylogenetic analysis
Aspergillus phylogeny
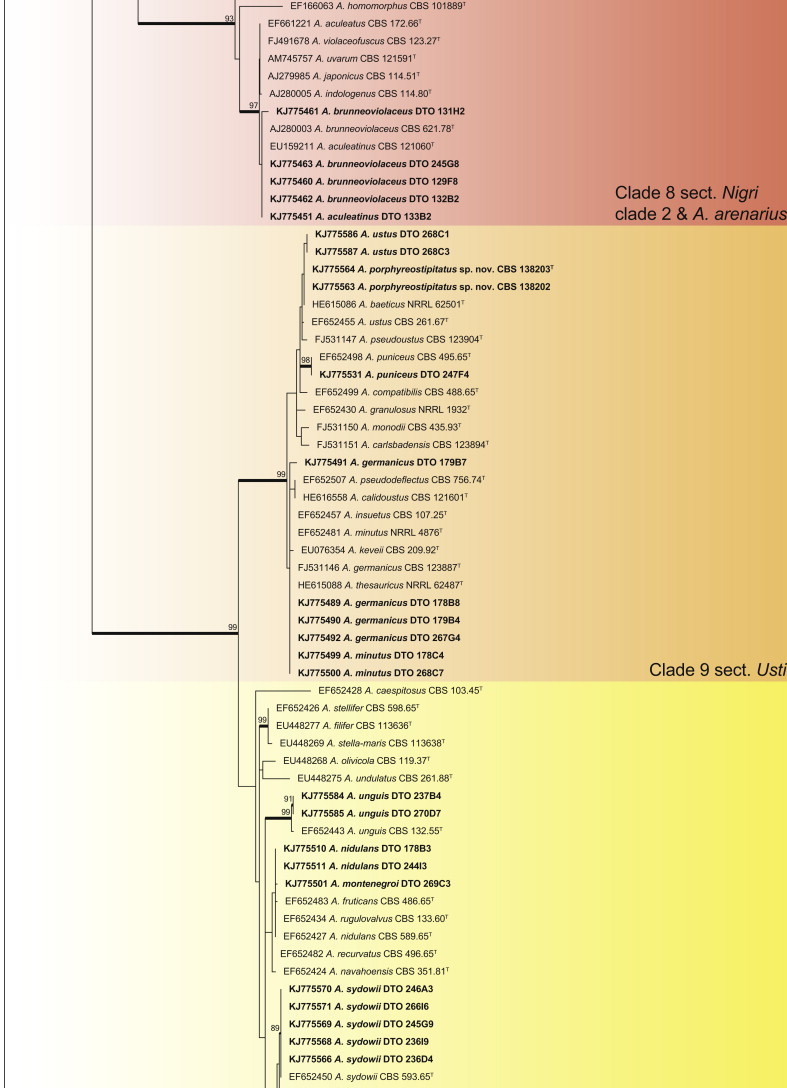
An ITS phylogeny (Fig. 1) was used to place Aspergillus isolates into their respective sections. The aligned data set included 347 strains and was 622 bp long, and the analysis employed the General Time Reversible (GTR) model with Gamma distribution (+G), with a certain fraction of sites that are evolutionary invariable (+I) selected. The analysis distributed the 59 Aspergillus species into 10 clades. The black Aspergillus species of section Nigri were resolved in two clades (clades 5 & 8), the first containing species closely related to A. nigri and the other relatives of A. aculeatus. The section is considered monophyletic following the three gene phylogeny of Peterson (2008) and four gene phylogeny of Houbraken & Samson (2011). For accurate identification, CaM phylogenies were prepared for each of the 10 clades and are presented in Figs 2–11.
Fig. 1.
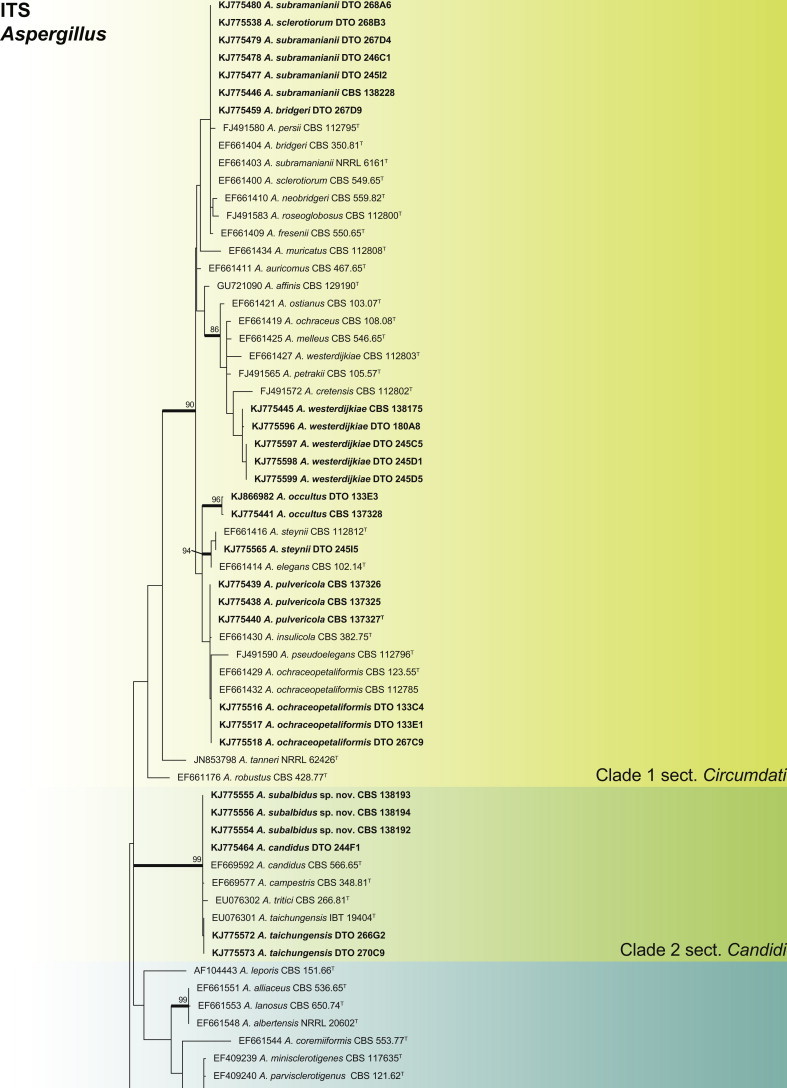



Aspergillus phylogeny of the ITS gene region showing the placement of representative strains isolated from house dust in bold. The coloured blocks indicate the different clades referred to in the text. The tree was rooted to Talaromyces flavus.
Fig. 2.
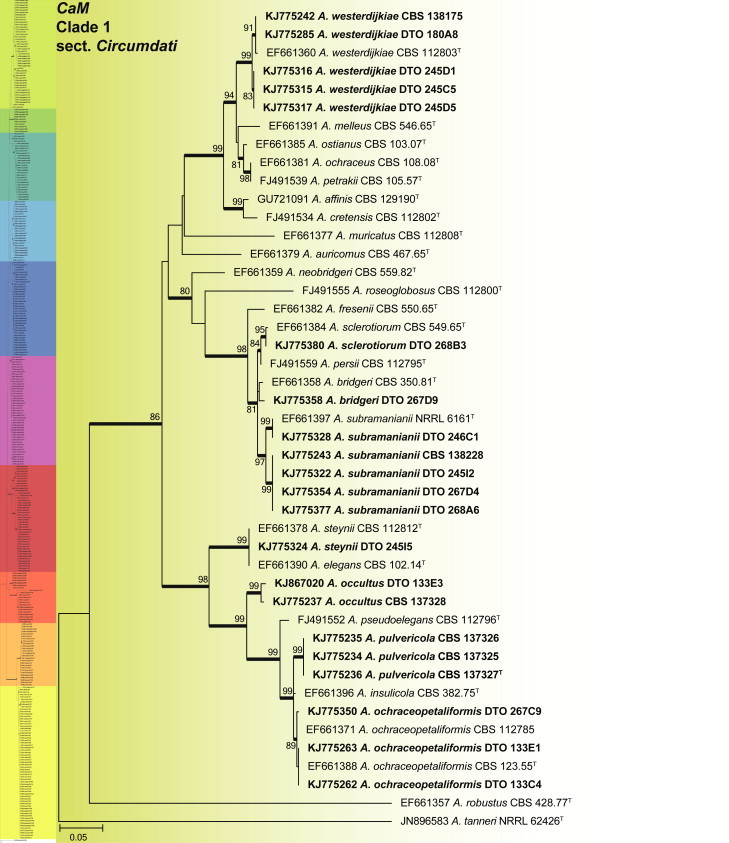
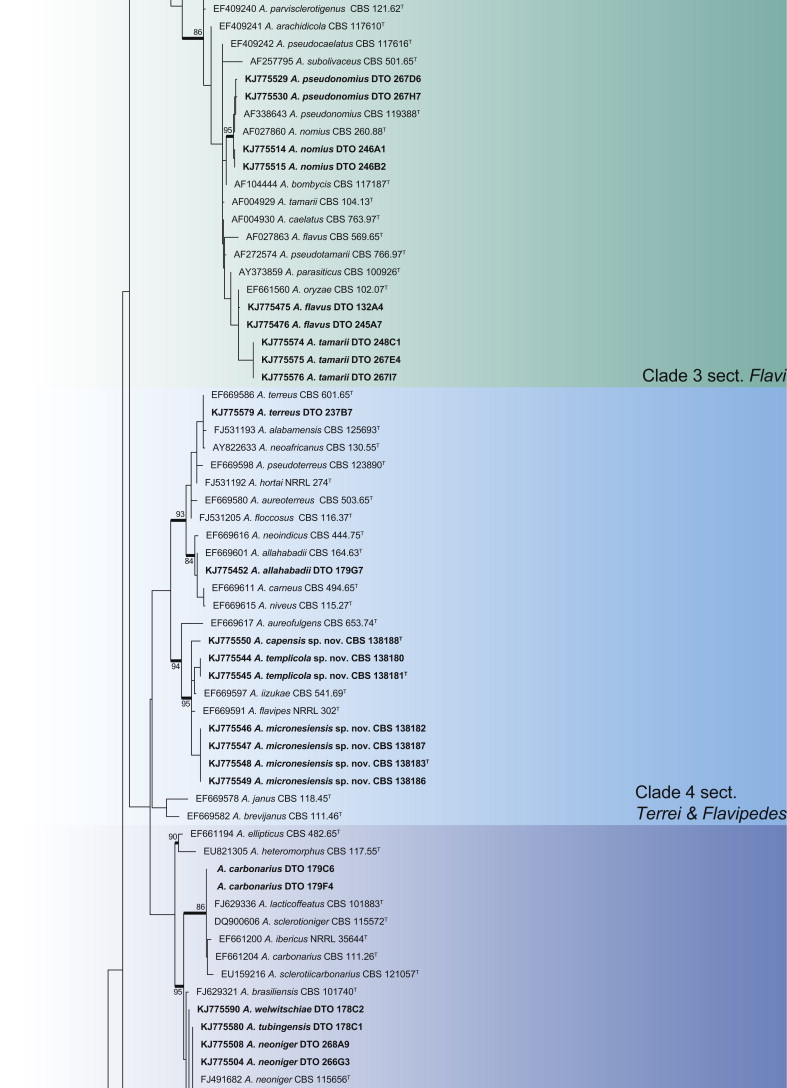
CaM phylogeny of Aspergillus section Circumdati, showing identities of species isolated from house dust in bold.
Fig. 3.

CaM phylogeny of Aspergillus section Candidi, showing identities of species isolated from house dust in bold.
Fig. 4.

CaM phylogeny of Aspergillus section Flavi showing identities of species isolated from house dust in bold.
Fig. 5.

CaM phylogeny of Aspergillus sections Terrei and Flavipedes, showing identities of species isolated from house dust in bold.
Fig. 6.
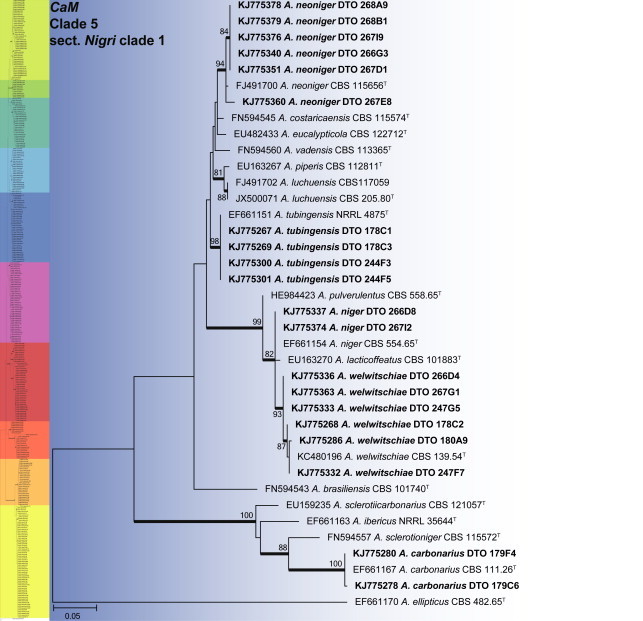
CaM phylogeny of Aspergillus section Nigri clade 1, showing identities of species isolated from house dust in bold.
Fig. 7.

CaM phylogeny of Aspergillus sections Fumigati, Clavati and Cervini, showing identities of species isolated from house dust in bold.
Fig. 8.

CaM phylogeny of Aspergillus sections Restricti, Aspergillus and Eurotium, showing identities of species isolated from house dust in bold.
Fig. 9.

CaM phylogeny of Aspergillus section Nigri clade 2 and A. arenarius, showing identities of species isolated from house dust in bold.
Fig. 10.
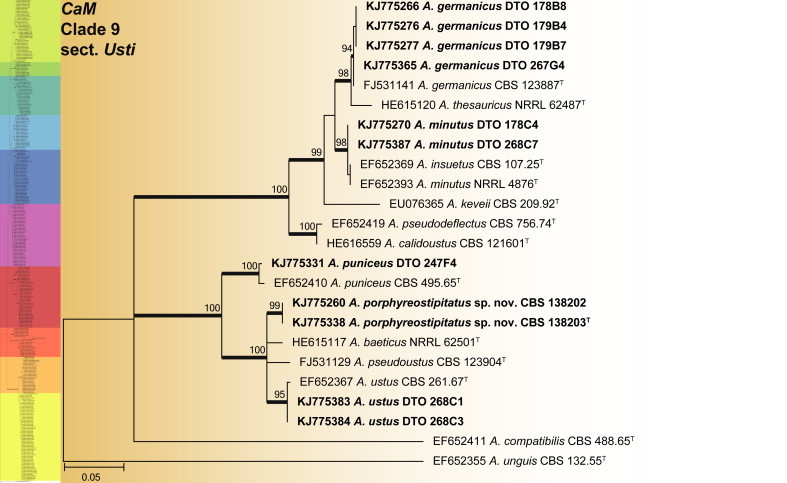
CaM phylogeny of Aspergillus section Usti, showing identities of species isolated from house dust in bold.
Fig. 11.
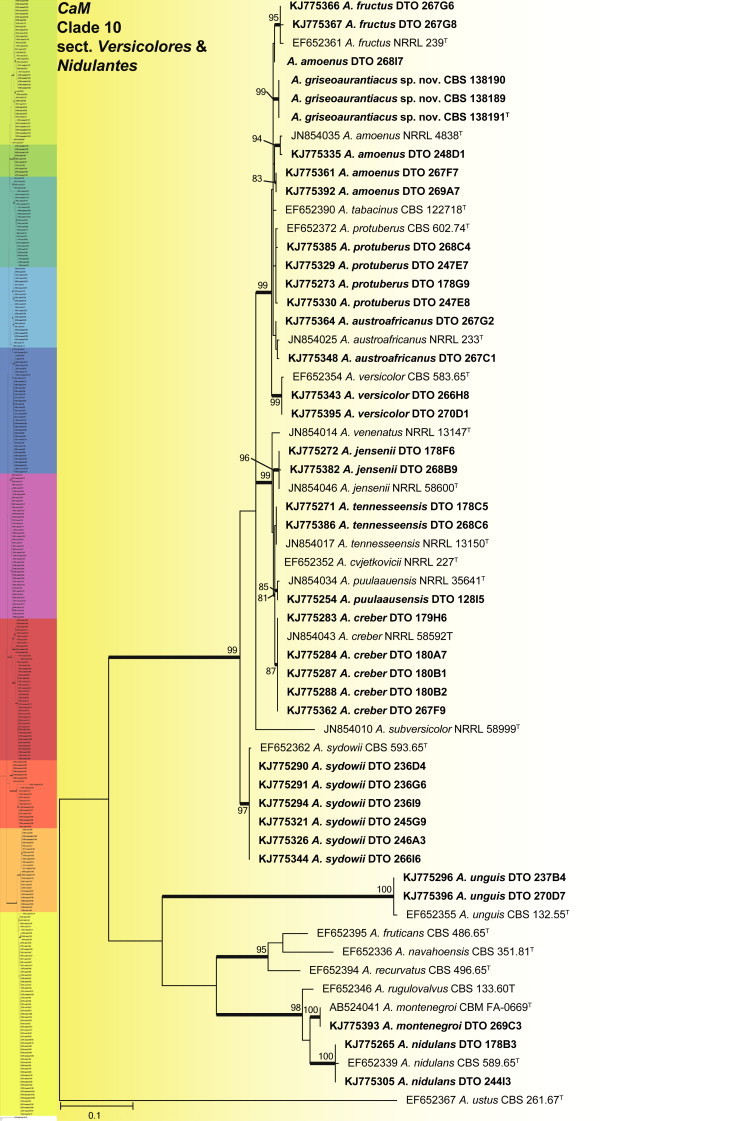
CaM phylogeny of Aspergillus sections Versicolores and Nidulantes, showing identities of species isolated from house dust in bold.
Clade 1 consisted of species classified in section Circumdati (Fig. 2). The aligned data set was 554 bp long, with Kimura 2-parameter (K2 + G + I) the most suitable model. This group of species is generally recognised by their ochre coloured conidiophore heads and a large number of species produce ochratoxins (Frisvad et al. 2004a, Visagie et al. 2014b). One of these ochratoxin producers is A. westerdijkiae, which was isolated in very high numbers from the South African house dust. A monographic treatment on the section is published in this issue of Studies in Mycology and includes descriptions for two new species, A. occultus and A. pulvericola isolated in this study. We note that A. elegans and A. steynii have identical CaM sequences (Fig. 2), even though ITS (Fig. 1a) and BenA distinguishes them.
Clade 2 represents section Candidi (Fig. 3). The aligned data set was 527 bp long, with K2 + G the most suitable model. Within the clade, we identified a new species with similar morphological features to A. candidus. Phylogenetically it is distinct and is described as A. subalbidus in the taxonomy section below. Two isolates identified as A. taichungensis had sequence variation compared to the ex-type strain, but morphologically they were all identical. As such, the sequence variation was considered insufficient to justify describing a new species. Sequences for A. candidus are highly variable based on Varga et al. (2007). A new species, A. pragensis, was recently described in the A. candidus complex (Hubka et al. 2014). However, a number of strains analysed in Varga et al. (2007) do not phylogenetically conform to the clades accepted by Hubka et al. (2014) as A. candidus and A. pregensis. As such, this clade needs more revision and we tentatively identify DTO 244F1 as A. candidus, even though it most probably represents a new species.
Aspergillus section Flavi is resolved in clade 3 (Fig. 4). The CaM alignment was 517 bp long and K2 + G selected for ML analysis. The species isolated from dust include A. flavus, A. nomius, A. pseudonomius and A. tamarii.
Clade 4 contains sections Terrei and Flavipedes (Fig. 5). The aligned data set was 571 bp long with the K2 + G model selected for ML analysis. In section Terrei, we isolated A. terreus and A. allahabadii. In section Flavipedes, the three isolated species are considered new and described as A. capensis, A. micronesiensis and A. templicola in the taxonomy section.
Clade 5, labelled section Nigri clade 1 (Fig. 6), contains the black Aspergilli closely related to A. niger. The aligned data set was 442 bp long, with the K2 + G model selected for ML analysis. Five species were identified in this clade as A. carbonarius, A. neoniger, A. niger, A. tubingensis and A. welwitschiae.
Sections Fumigati, Clavati and Cervini all occurred in clade 6 (Fig. 7). The aligned data set was 578 bp long and the K2 + G model was selected for ML analysis. Strains were identified as A. clavatus, A. hiratsukae, A. lentulus and A. fumigatus. The latter species was isolated in high numbers from five countries, namely Mexico, Micronesia, New Zealand, South Africa and Thailand.
Clade 7 resolves sections Restricti and Aspergillus in one clade (Fig. 8). The aligned data set was 610 bp long and the K2 + G + I model was used for ML analysis. Within the clade, we isolated one new species closely related to A. glaucus and A. proliferans, the latter also found in the dust samples together with A. chevalieri, A. montevidensis, A. penicillioides, A. pseudoglaucus, A. restrictus and A. ruber.
Clade 8 contains the black Aspergillus species closely related to A. aculeatus, labelled as Nigri clade 2 (Fig. 9). Even though the ITS phylogeny places A. arenarius closest to this clade, its taxonomic placement is currently uncertain and will be the focus of a future paper. The aligned data set was 470 bp long and the most suitable model was K2 + I. Two black species were identified as A. aculeatinus and A. brunneoviolaceus. A new species, closely related to A. arenarius, is described as A. arenarioides in the taxonomy section.
Clade 9 contains section Usti species (Fig. 10). The aligned data set was 510 bp long and K2 + I selected for the ML analysis. Five species were identified as A. germanicus, A. minutus, A. puniceus, A. ustus and a new species described as A. porphyreostipitatus.
Clade 10 contains sections Versicolores and Nidulantes (Fig. 11). The aligned data set was 545 bp long and K2 + G was selected as the most suitable model. Aspergillus versicolor is often isolated from indoor environments. Jurjević (2012) considered it to represent a complex and accepted nine species. We isolated and identified all of the new species they accepted, namely A. amoenus, A. austroafricanus, A. creber, A. fructus, A. jensenii, A. protuberus, A. puulaausensis, A. tennesseensis, and A. versicolor. Aspergillus sydowii was abundant and had a wide distribution in the house dust. In addition, we introduce a new species in the section as A. griseoaurantiacus. In section Nidulantes, we identified strains as A. unguis, A. montenegroi and A. nidulans.
Penicillium phylogeny
An ITS phylogeny was used to place Penicillium house dust isolates in their respective sections (Fig. 12). The aligned data set included 380 strains and was 585 bp long. The GTR + G + I model was the most suitable for the ML analysis. The phylogeny resolved the 49 house dust species, distributed among 12 clades. Clades corresponded well with the sections proposed by Houbraken & Samson (2011). To obtain more accurate identifications, BenA gene trees were analysed for each ITS clade and are presented in Figs 13–24.
Fig. 12.

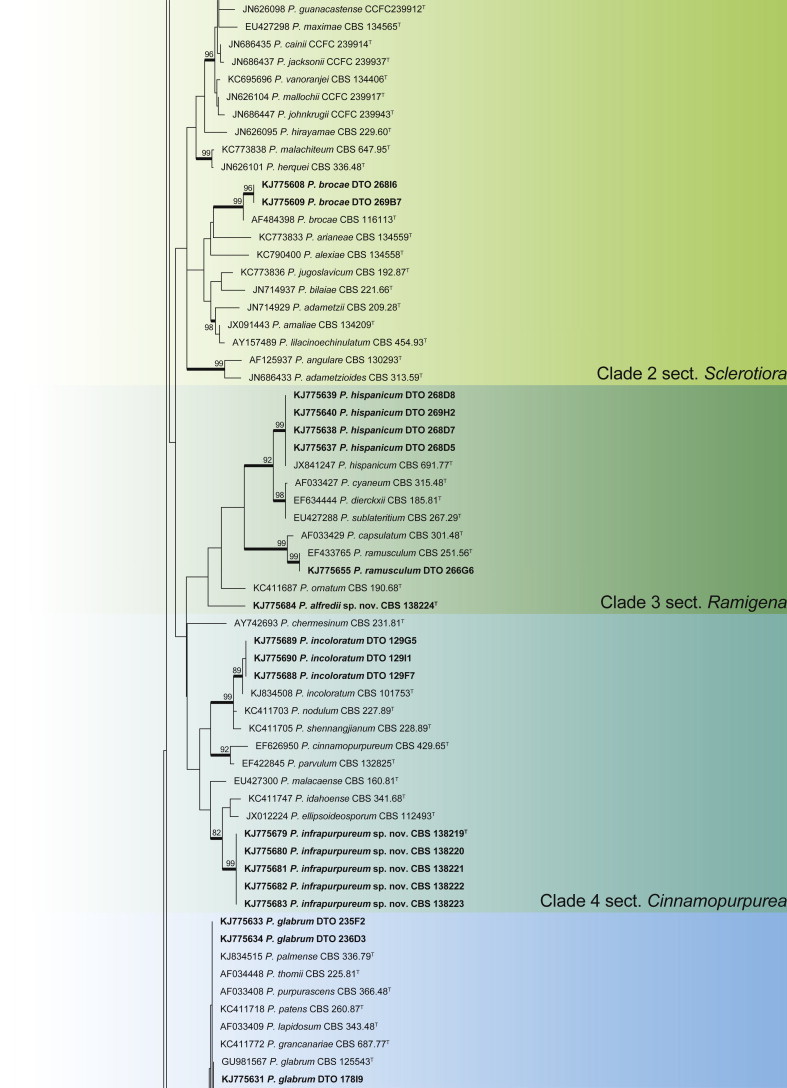
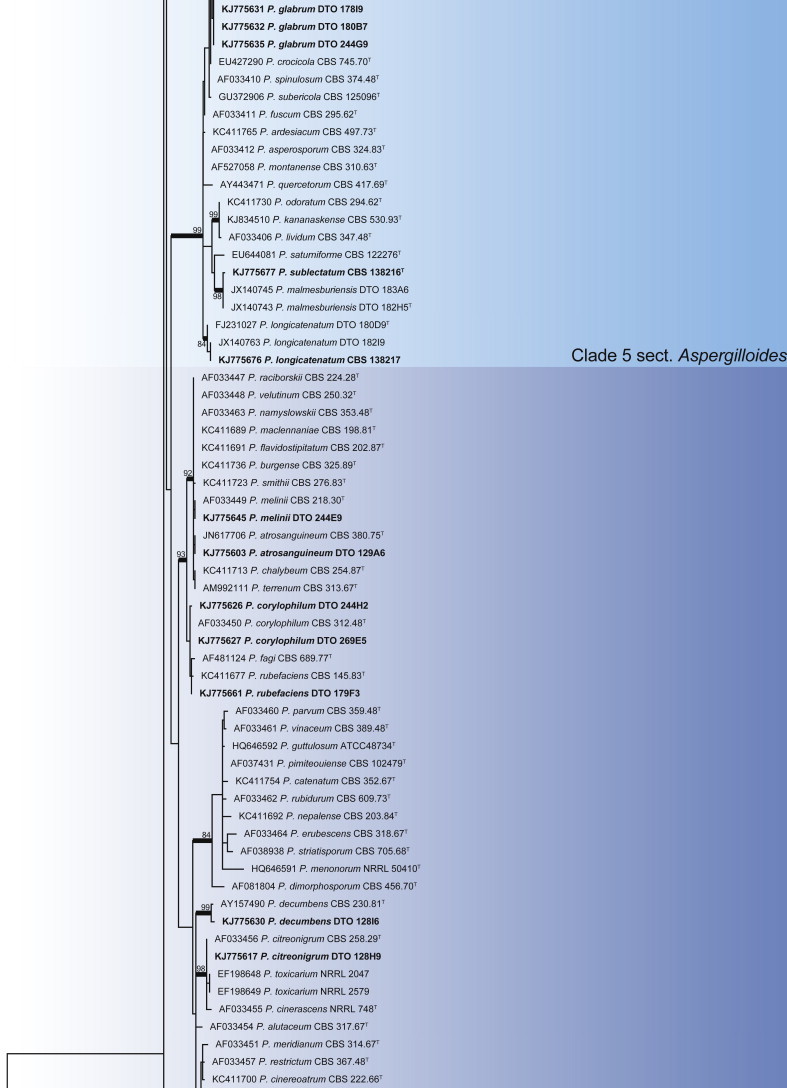



Penicillium phylogeny of the ITS gene region showing the placement of representative strains isolated from house dust in bold. The coloured blocks indicate the different clades referred to in the text. The tree was rooted to Talaromyces flavus.
Fig. 13.

BenA phylogeny of Penicillium section Citrina, showing identities of species isolated from house dust in bold.
Fig. 14.

BenA phylogeny of Penicillium section Sclerotiora, showing identities of species isolated from house dust in bold.
Fig. 15.
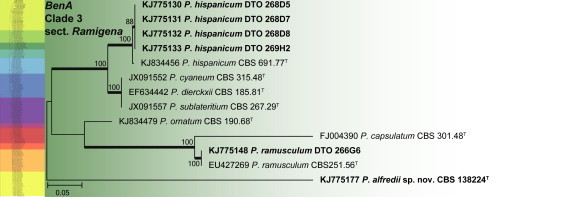
BenA phylogeny of Penicillium section Ramigena, showing identities of species isolated from house dust in bold.
Fig. 16.

BenA phylogeny of Penicillium section Cinnamopurpurea, showing identities of species isolated from house dust in bold.
Fig. 17.
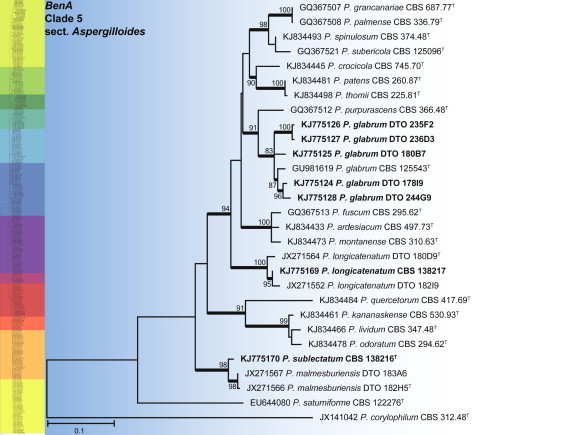
BenA phylogeny of Penicillium section Aspergilloides, showing identities of species isolated from house dust in bold.
Fig. 18.
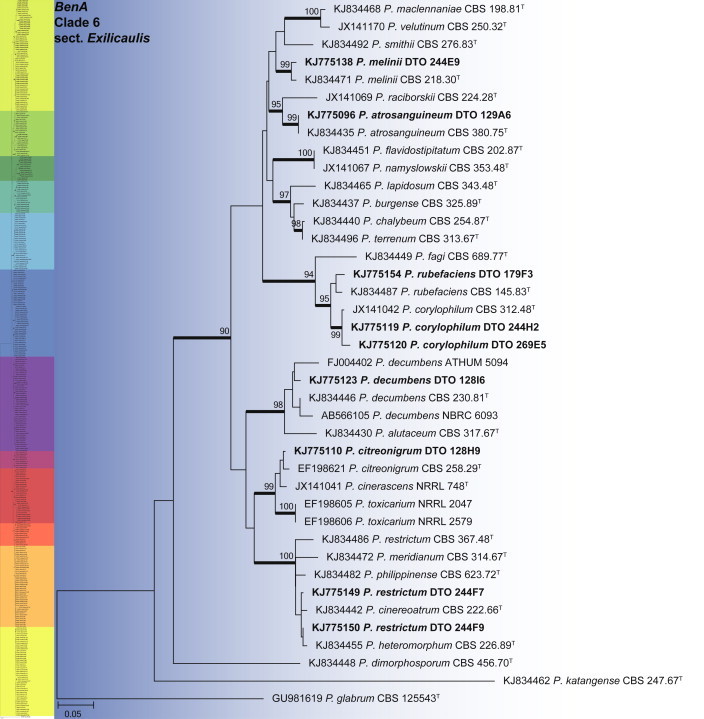
BenA phylogeny of Penicillium section Exilicaulis, showing identities of species isolated from house dust in bold.
Fig. 19.

BenA phylogeny of Penicillium section Lanata-Divaricata, showing identities of species isolated from house dust in bold.
Fig. 20.

BenA phylogeny of Penicillium section Canescentia, showing identities of species isolated from house dust in bold.
Fig. 21.

BenA phylogeny of Penicillium sections Brevicompacta & Ramosa, showing identities of species isolated from house dust in bold.
Fig. 22.
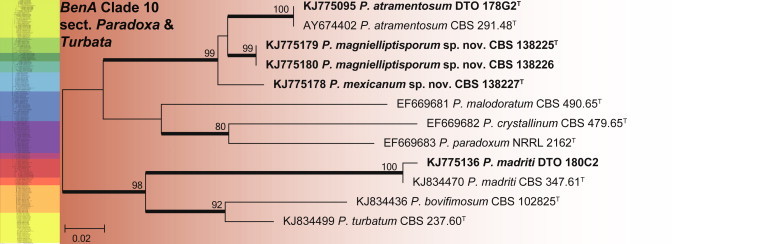
BenA phylogeny of Penicillium sections Paradoxa & Turbata, showing identities of species isolated from house dust in bold.
Fig. 23.
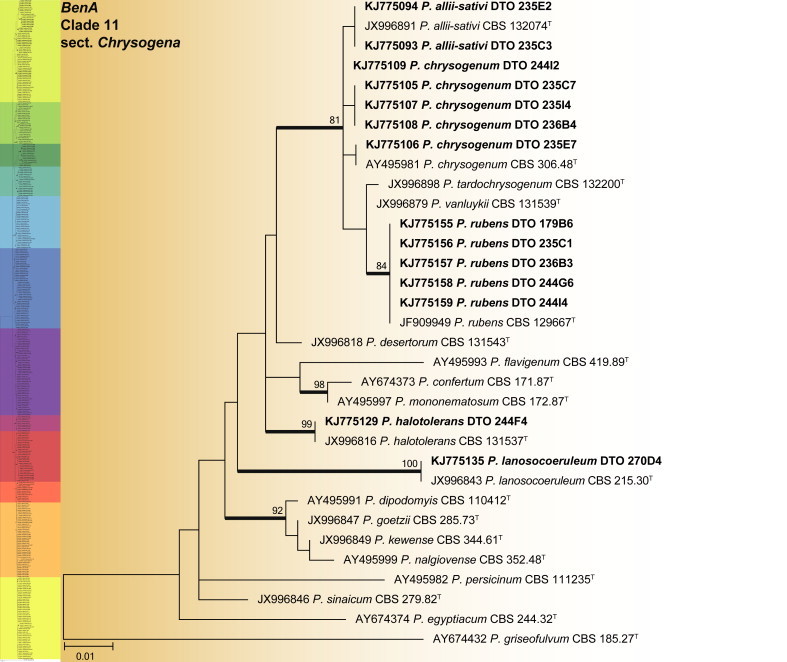
BenA phylogeny of Penicillium section Chrysogena, showing identities of species isolated from house dust in bold.
Fig. 24.

BenA phylogeny of Penicillium sections Penicillium & Fasciculata, showing identities of species isolated from house dust in bold.
Clade 1 contains section Citrina (Fig. 13), a group of species of wide distribution and isolated from a wide range of sources (Houbraken et al. 2011b). The aligned data set was 448 bp long, with the K2 + G model selected for ML analysis. Species were identified as P. citrinum, P. pancosmium, P. roseopurpureum, P. sanguifluum, P. sizovae, P. steckii and P. sumatraense. Penicillium pancosmium was abundant in samples collected from South Africa, Indonesia and Micronesia. It is also extremely common in isolations from house dust samples collected in Regina, Canada (Hirooka, Tanney & Seifert, unpubl.).
Clade 2 corresponds with the recently revised section Sclerotiora (Fig. 14) (Rivera et al. 2012, Visagie et al. 2013). The aligned data set was 374 bp long, with the K2 + G model selected for ML analysis. Penicillium brocae was the only species isolated from house dust that belongs to the section.
Clade 3 includes section Ramigena (Fig. 15). The aligned data set was 402 bp long and K2 + I was the most suitable model for ML analysis. Two species, P. hispanicum and P. ramusculum, were identified from house dust. BenA also shows that P. cyaneum, P. dierckii and P. sublateritium are synonyms, with P. cyaneum (Bainier & Sartory) Biourge, Cellule 33: 102. 1923 representing the oldest name.
Clade 4 includes species classified in section Cinnamopurpurea (Fig. 16). The aligned BenA data set was 390 bp long, with the K2 + G model selected for the ML analysis. One species was identified as P. incoloratum, while a second is described as P. infrapurpureum below.
Clade 5 contains the section Aspergilloides (Fig. 17), which is reviewed in Houbraken et al. (2014b). The alignment was 459 bp long and K2 + G was selected as the most suitable model for ML analysis. Three species were identified, including P. glabrum and two new species, P. sublectatum prov. nom. and P. longicatenatum prov. nom., described in Houbraken et al. (2014b).
Clade 6 contains section Exilicaulis (Fig. 18). The aligned data set was 448 bp long and K2 + G was selected for ML analysis. Species isolated include P. atrosanguineum, P. citreonigrum, P. corylophilum, P. decumbens, P. melinii, P. restrictum and P. rubefaciens. From the phylogeny, it is clear that some species need further study. The P. restrictum complex, including five species, represents one of these. This will be the focus of a future paper. We thus tentatively identify isolates in this complex as P. restrictum, mainly based on their morphological characters.
Clade 7 includes species of section Lanata-Divaricata (Fig. 19). The aligned data set was 472 bp long, with the K2 + G model selected for ML analysis. Isolates were identified as P. oxalicum and a new species is described here as P. singorense.
Clade 8 contains species classified in section Canescentia (Fig. 20). The BenA alignment was 403 bp long, with K2 + G selected for the ML analysis. We describe one new species in this section as P. dunedinense.
Sections Brevicompacta and Ramosa are resolved in clade 9 (Fig. 21). The aligned data set was 394 bp long and K2 + G was selected for the ML analysis. One of the more common species found in dust was P. brevicompactum. Our data suggest that the recently described P. kongii (Wang & Wang 2013) is a synonym of P. brevicompactum. The remaining isolates were identified as P. buchwaldii and P. olsonii. In section Ramosa, we isolated P. swiecickii and one new species closely related to P. soppii, described below as P. lenticrescens.
The species in clade 10 are classified in sections Paradoxa and Turbata (Fig. 22). The aligned data set was 394 bp long and K2 + G was selected for ML analysis. Within section Paradoxa, we describe two new species in the P. atramentosum species complex as P. mexicanum and P. magnielliptisporum. In section Turbata, we identified one of the species as P. madriti.
Clade 11 comprises the recently reviewed section Chrysogena (Houbraken et al. 2012) (Fig. 23). This group of species was well represented in dust samples, especially P. rubens and to a lesser degree P. chrysogenum. The aligned data set was 444 bp long and the K2 + G model selected for the ML analysis. Isolates were identified as P. allii-sativii, P. chrysogenum, P. halotolerans, P. lanosocoeruleum and P. rubens.
Clade 12 mostly includes species classified in sections Penicillium and Fasciculata (Fig. 24). The alignment was 322 bp long and the K2 + G model was selected for ML analysis. Isolates were identified as P. biforme, P. commune, P. coprophilum, P. crustosum, P. cyclopium, P. italicum, P. melanoconidium, P. palitans and P. solitum.
Talaromyces phylogeny
An ITS phylogeny was used to place Talaromyces house dust isolates into their respective sections (Fig. 25), as described by Yilmaz et al. (2014). The aligned ITS data set was 605 bp long and included 125 strains. The ML analysis was done with the GTR + G + I model selected. The phylogeny resolved house dust isolates into four sections, with BenA gene trees subsequently calculated for each section.
Fig. 25.

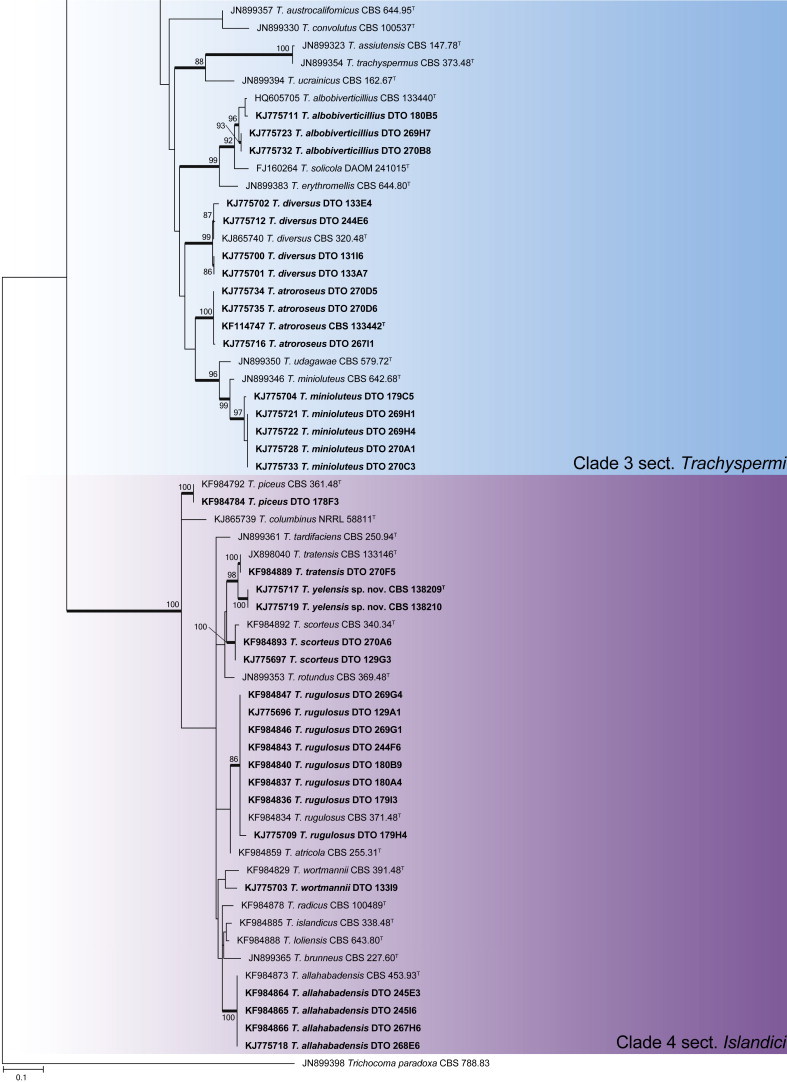
Talaromyces phylogeny of the ITS gene region showing the placement of representative strains isolated from house dust in bold. The coloured blocks indicate the different clades referred to in the text. The tree was rooted to Trichocoma paradoxa.
Clade 1 contains species classified in section Talaromyces (Fig. 26). The aligned BenA data set was 413 bp long, with the K2 + G + I model most suitable for ML analysis. Isolates were identified as the newly described T. cnidii (Sang et al. 2013) and T. amestolkiae (Yilmaz et al. 2012), the previously described T. siamensis and T. verruculosus, and two new species described here as T. sayulitensis and T. oumae-annae.
Fig. 26.

BenA phylogeny of Talaromyces section Talaromyces, showing identities of species isolated from house dust in bold.
Clade 2 contains species that typically produce synnemata after more than one week of growth, which are classified in section Purpurei (Fig. 27). The aligned data set was 389 bp long and K2 + G was selected for ML analysis. Talaromyces ramulosus was isolated from the South African house dust, a species originally described from soil, apples (from the Fynbos biome in South Africa) and moth-damaged grapes (Ontario, Canada) (Visagie et al. 2009).
Fig. 27.

BenA phylogeny of Talaromyces section Purpurei, showing identities of species isolated from house dust in bold.
Clade 3 contains species of section Trachyspermi (Fig. 28). The aligned data set was 373 bp long, with the K2 + G model selected for ML analysis. Isolates were identified as T. albobiverticillius, T. atroroseus, T. diversus and T. minioluteus. Frisvad et al. (2013) recently introduced T. atroroseus, a species that produces large amounts of red pigmentation. In the same paper, T. albobiverticillius was shown to have genetic and phenotypic variation, with either white or green-pigmented conidia produced. Our house dust isolates produced the green phenotype. Phylogenetic data suggest that T. minioluteus represents a species complex. The dust isolates were thus tentatively identified as T. minioluteus, with strains that will be part of a future study on this complex.
Fig. 28.

BenA phylogeny of Talaromyces section Trachyspermi, showing identities of species isolated from house dust in bold.
Clade 4 contains species classified in section Islandici (Fig. 29). The aligned BenA data set was 435 bp long and the K2 + G model was selected for ML analysis. Isolates were identified as T. allahabadensis, T. piceus, T. rugulosus, T. scorteus, T. tratensis, T. wortmanii and the new species described here as T. yelensis.
Fig. 29.
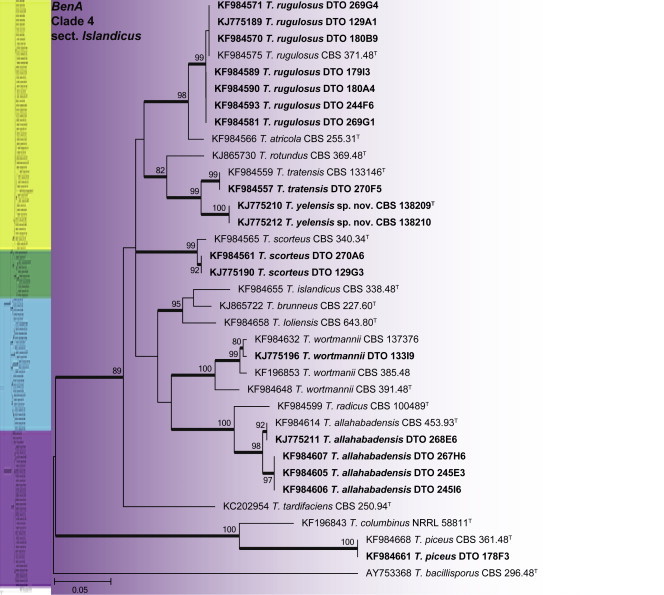
BenA phylogeny of Talaromyces section Islandici, showing identities of species isolated from house dust in bold.
Taxonomy
The genus Aspergillus
Aspergillus section Candidi
Aspergillus subalbidus Visagie, Hirooka & Samson, sp. nov. MycoBank MB809190. Figs 30, 31.
Fig. 30.
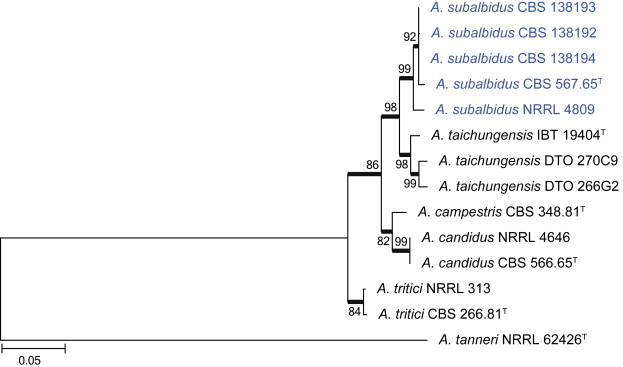
Combined phylogeny for ITS, BenA and CaM of Aspergillus section Candidi. Aspergillus tanneri was used as outgroup. Names in blue are new species described in this study. Model selected: K2 + G, combined alignment 1 529 bp.
Fig. 31.

Aspergillus subalbidus. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, DG18 and obverse CREA. B. Purple to black sclerotia on OA. C–G. Conidiophores on DG18 (E. on MEA). H. Conidia. Scale bars: C, D, F–H = 10 μm; E = 50 μm.
Etymology: Latin, subalbidus, referring to morphological similarity to A. candidus.
Diagnosis: White sporulation dominating colony appearance, purplish black sclerotia produced by some strains, no growth on CYA at 37 °C.
Typus: Brazil, Instituto Biologica, 1939, isolated by Reis (holotype CBS H-21807, culture ex-type CBS 567.65 = ATCC 16871 = IMI 230752 = NRRL 312).
Additional materials examined: Thailand, Songkhla, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138193 = DTO 129F9, CBS 138192 = DTO 129E3. Federated States of Micronesia, Malem on Kosrae Island, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138194 = DTO 266I9.
ITS barcode: KJ866983 (alternative markers: BenA = EU076295; CaM = EF669551)
Colony diam, 7 d (mm): CYA 15–18; CYA 30 °C 17–21; CYA 37 °C no growth; MEA 17–19; YES 25–30; DG18 23–26; CYAS 25–33; OA 14–17; CREA 4–9.
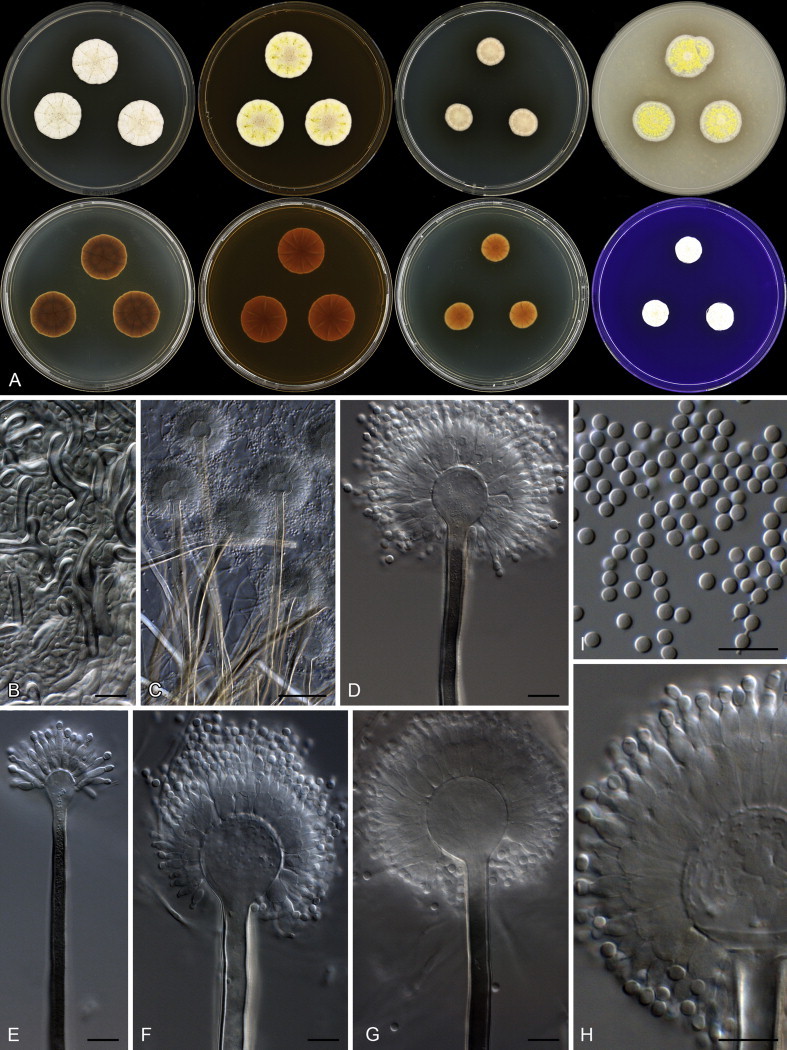
Colony characters: CYA 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas white; soluble pigment absent; exudate absent; reverse olive (3F8) to brownish orange (5C6). CYA 30 °C, 7 d: Colonies similar to CYA at 25 °C, except for purplish black sclerotia present and darker reverse colouration, olive (3F8) to brown (5E8). MEA 25 °C, 7 d: Colony surface floccose; mycelial areas white; sporulation white, centrally brownish grey (5C2); soluble pigment absent; exudate abundant, clear; reverse brown (6E8–8E8). YES 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas white; sclerotia present in some strains, black; soluble pigment absent; exudate absent; reverse centrally light brown (5D6), fading into light yellow (4A5) near margin. DG18 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas white; soluble pigment absent; exudate absent; reverse centrally light to pale yellow (4A5–2A3), elsewhere pale. OA 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas white; sclerotia purplish black; soluble pigment absent; exudate absent; reverse white. CYAS 25 °C, 7 d: Colony surface floccose; white to pale yellow (1A2); soluble pigment absent; exudate absent; reverse beige to light yellow (4C3–B4). CREA 25 °C, 7 d: Colony surface velutinous; mycelial areas white, sporulation white; acid not produced.
Micromorphology: Conidial heads globose; Conidiophores biseriate, sometimes reduced Penicillium-like structures present, on DG18 much larger than on MEA; Stipes hyaline, minor proportion having a brown pigment, smooth, 80–300(–2 000 on DG18) × 3–6 (MEA) or 7–16 (DG18) μm; Vesicles globose to subglobose, on MEA 6–14 μm, on DG18 10–55 μm, covering 100 % of the head; Metulae 6.5–25 × 4–8 μm; Phialides ampulliform, 6–9 × 2.5–3.5 μm; Conidia globose to subglobose, smooth, 3–4 μm (3.5 ± 0.18 × 3.5 ± 1.19, n = 56), average width/length = 0.98, n = 54; Hülle cells absent; Sclerotia purplish to black, especially on OA, 270–620 μm diam.
Notes: Aspergillus subalbidus is morphologically almost identical to A. candidus. The new species also includes strains (CBS 567.65 and CBS 112449) previously identified as A. candidus and most recently, as A. taichungensis (Varga et al. 2007). Morphologically these strains lack the yellow colours observed in A. taichungensis and do not grow on CYA at 37 °C. Phylogenetically, the new species forms a distinct clade closely related to A. campestris, A. candidus, A. taichungensis and A. tritici (Fig. 30). Both A. subalbidus and A. candidus are distinguished from these species by their typical white colonies, smooth conidia and inability to grow at 37 °C. Minor differences were observed when comparing the new species with A. candidus. The new species grew slightly slower on CYA, YES and DG18. The purple to black sclerotia common in A. subalbidus when grown on OA were not observed in A. candidus, as previously reported by Varga et al. (2007). These minor differences make morphological identification very difficult. However, phylogenetically this species is distinct and these minor phenotypic differences warrant describing it as new.
Aspergillus section Flavipedes
Aspergillus templicola Visagie, Hirooka & Samson, sp. nov. MycoBank MB809191. Figs 32, 33.
Fig. 32.
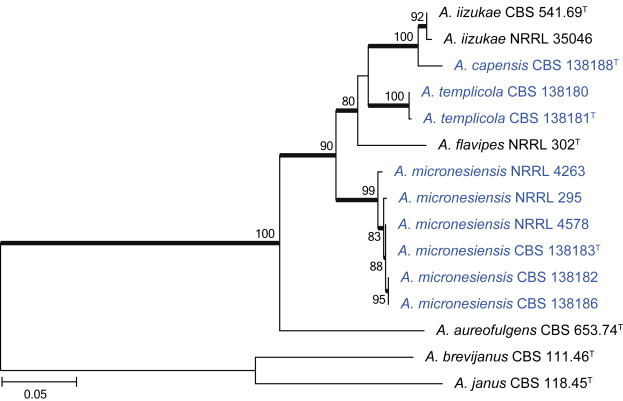
Combined phylogeny for ITS, BenA and CaM of Aspergillus section Flavipedes. Names in blue are new species described in this study. Aspergillus janus and A. brevijanus was used as outgroup. Model selected: Tamura-Nei (TN93) combined alignment 1 695 bp.
Fig. 33.

Aspergillus templicola. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, DG18 and obverse CREA. B–H. Conidiophores. I. Conidia. Scale bars: B = 50 μm; C–I = 10 μm.
Etymology: Latin, templicola, meaning church-dweller, in reference to the ex-type strain, which was isolated from dust collected in a Mexican church.
Diagnosis: Colonies yellowish white to pale yellow, reverse brown to dark brown, Hülle cells absent, conidiophores biseriate with vesicles elongated, diminutive conidiophores present.
Typus: Mexico, Sayulita, dust from church, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21808, culture ex-type CBS 138181 = DTO 270C6).
Additional material examined: Thailand, Bangkok, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138180 = DTO 267H4.
ITS barcode: KJ775545 (alternative markers: BenA = KJ775092; CaM = KJ775394)
Colony diam, 7 d (mm): CYA 25–32; CYA 30 °C 35–36; CYA 37 °C 22–25; MEA 23–26; YES 32–38; DG18 28–34; OA 17–19; CREA 13–21.
Colony characters: Colony surface floccose; sporulation and mycelial areas yellowish white to pale yellow (4A2–3); soluble pigment brown to absent; exudate clear; reverse brown to dark brown (5F8–6F8). CYA 30 °C, 7 d: Colonies similar to CYA at 25 °C, except for yellowish colour in colonies in DTO 267H4. CYA 37 °C, 7 d: Colonies similar to CYA at 25 °C. MEA 25 °C, 7 d: Colony surface floccose; mycelial areas orange white (5A2); sporulation brownish grey (4C2); soluble pigment absent; exudate absent; reverse brown (6E8). YES 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas yellowish white to yellowish grey to greyish yellow (4A2–B2–3); soluble pigment brown; exudate absent; reverse brown (6D8–7E8). DG18 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas pale orange to greyish orange (5A3–B3); soluble pigment absent; exudate absent; reverse light yellow to greyish orange (4A4–5C5). OA 25 °C, 7 d: Colony surface velutinous to somewhat floccose; sporulation and mycelial areas pale orange (5A3); soluble pigment olive, inconspicuous; exudate absent; reverse brownish orange (5C5). CREA 25 °C, 7 d: Colony surface velutinous to somewhat floccose, orange white to pale orange (5A2–3); acid not produced.
Micromorphology: Conidial heads radiating, generally bigger on DG18, often diminutive on MEA and DG18; Conidiophores biseriate; Stipes hyaline to dark brown, smooth walled, some very finely rough walled, 120–1 400 × 5–10 μm; Vesicles, elongate, a minor proportion more subglobose, 9–23 μm wide; Metulae 6–8 × 3–4 μm, covering 75–100 % of head; Phialides ampulliform, 4.5–8.5 × 2.5–3.5 μm; Conidia subglobose, smooth to finely roughened, 2.5–3 × 2–2.5 μm (2.7 ± 0.1 × 2.5 ± 0.1, n = 50), average width/length = 0.93, n = 50; Sclerotia absent.
Notes: Aspergillus templicola is resolved in a clade with A. flavipes, A. iizukae and the two new species described here as A. micronesiensis and A. capensis (Fig. 32). This group of species is morphologically very similar, which makes identification based on phenotypic characters challenging. In their description of A. flavipes, Raper & Fennell (1965) described conidiophore vesicles as subglobose to vertically elongate. We also observed this in strains of A. flavipes. For A. templicola, vesicles were consistently elongated, whilst A. micronesiensis had subglobose vesicles. Compared to A. capensis and A. iizukae, strains of A. templicola had less intense and paler reverses. Hülle cells were observed in A. flavipes and A. micronesiensis, but not in A. iizukae, A. capensis and A. templicola. Morphologically, A. capensis could not be distinguished from A. iizukae using phenotypic characters, although it is phylogenetically distinct. Sequence data is recommended for their identification.
Aspergillus micronesiensis Visagie, Hirooka & Samson, sp. nov. MycoBank MB809192. Figs 32, 34.
Fig. 34.
Aspergillus micronesiensis. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, DG18 and obverse CREA. B. Hülle cells. C–H. Conidiophores. I. Conidia. Scale bars: B, D–I = 10 μm; C = 50 μm.
Etymology: Latin, micronesiensis, in reference to the ex-type strain, which was isolated from dust collected in Micronesia.
Diagnosis: Colonies yellowish white to pale yellow, reverse colour brown to dark brown, Hülle cells present, conidiophores biseriate with vesicles subglobose, diminutive conidiophores present.
Typus: Federated States of Micronesia, Yela of Kosrae Island, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21809, culture ex-type CBS 138183 = DTO 267D5).
Additional materials examined: Haiti, soil, 1960, isolated by J. Rabel, CBS 586.65 = NRRL 4578 = ATCC 16805 = IMI 135423. Mexico, Sayulita, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138182 = DTO 245D7. Thailand, Bangkok, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138186 = DTO 267H5.
ITS barcode: KJ775548 (alternative markers: BenA = KJ775085; CaM = KJ775355)
Colony diam, 7 d (mm): CYA 22–28; CYA 30 °C 30–36; CYA 37 °C 17–25; MEA 20–25; YES 35–44; DG18 15–25; OA 14–24; CREA 12–17.
Colony characters: CYA 25 °C, 7 d: Colony surface floccose; mycelial areas white mycelial areas; sporulation yellowish white to greyish orange (5C3); soluble pigment brown; exudate clear to brown; reverse brown to dark brown (6E8–F8). CYA 30 °C, 7 d: Colonies similar to CYA at 25 °C. CYA 37 °C, 7 d: Colonies similar to CYA at 25 °C. MEA 25 °C, 7 d: Colony surface floccose; mycelial areas yellowish white to pale yellow (3A2–3); sporulation brownish orange (5C4); Hülle cells present, yellow, sexual development not observed; soluble pigment absent; exudate absent or in some strains yellow to brown; reverse brown to dark brown (6D7–7F7). YES 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas yellowish white to pale yellow (3A2–3) to brownish orange (5C4); soluble pigment orange brown; exudate absent; reverse brown (6D7–7E7). DG18 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas yellowish white to light yellow (4A2–4); soluble pigment brown; exudate absent; reverse greyish orange (6B6) to dark brown (7F7). OA 25 °C, 7 d: Colony surface velutinous to floccose; sporulation and mycelial areas orange white (5A3); Hülle cells yellow, sexual development not observed cells; soluble pigment brown; exudate absent; reverse light brown (6D6). CREA 25 °C, 7 d: Colony surface floccose, yellowish white to light yellow to brownish orange (4A2–4–5C4); acid not produced.
Micromorphology: Conidial heads radiating, generally bigger on DG18; Conidiophores biseriate; Stipes hyaline to dark brown, smooth walled, some very finely rough walled, 250–1 900 × 5.5–9.5 μm; Vesicles globose, minor proportion elongated, 13.5–31 μm wide; Metulae 5–13 × 3.5–6.5 μm, covering 75–100 % of head; Phialides ampulliform, 6–8.5 × 2.5–4 μm; Conidia globose to subglobose, smooth to finely roughened, 2.5–3.5 × 2.5–3.5 μm (2.7 ± 0.2 × 2.7 ± 0.2, n = 5), average width/length = 0.98, n = 50; Sclerotia absent.
Notes: See notes for A. templicola above.
Aspergillus capensis Visagie, Hirooka & Samson, sp. nov. MycoBank MB809193. Figs 32, 35.
Fig. 35.
Aspergillus capensis. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, DG18 and obverse CREA. B–F. Conidiophores. G. Conidia. Scale bars: B = 50 μm; C = 20 μm; C–G = 10 μm.
Etymology: Latin, capensis, in reference to the ex-type strain, which was isolated from dust collected in the Cape Town metropolitan area, South Africa.
Diagnosis: Colonies yellowish white to pale yellow, reverse dark brown, Hülle cells absent, conidiophores biseriate with vesicles subglobose, diminutive conidiophores present.
Typus: South Africa, Kuils River in the Cape Town metropolitan area, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21810, culture ex-type CBS 138188 = DTO 179E6).
ITS barcode: KJ775550 (alternative markers: BenA = KJ775072; CaM = KJ775279)
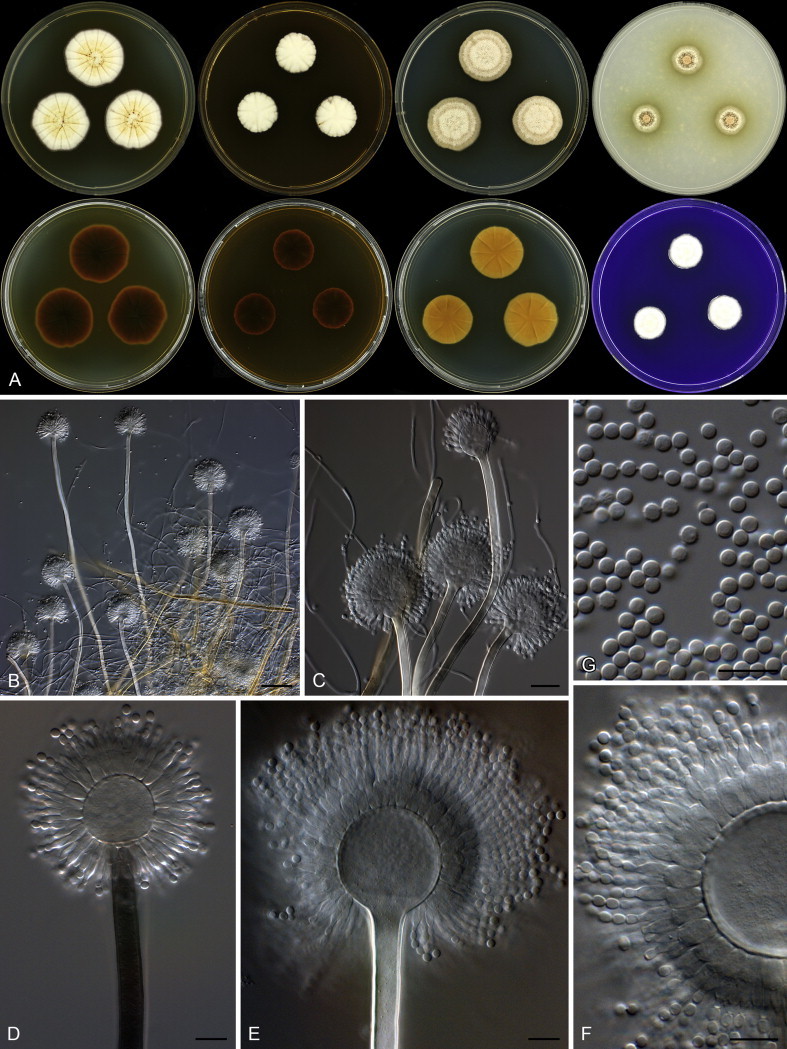
Colony diam, 7 d (mm): CYA 28–29; CYA 30 °C 30–31; CYA 37 °C 18–19; MEA 19–20; YES 39–40; DG18 25–26; OA 12–13; CREA 16–17.
Colony characters: CYA 25 °C, 7 d: Colony surface floccose; mycelial areas white to yellowish white; sporulation yellowish white to white yellow (3A2–5); soluble pigment brown; exudate brown; reverse dark brown (6F5–8). CYA 30 °C, 7 d: Colonies similar to CYA at 25 °C. CYA 37 °C, 7 d: Colony surface floccose, brownish grey (6D2); soluble pigment brown; exudate absent; reverse brownish grey to dark brown (6F3–5). MEA 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas yellowish white (2A2); soluble pigment brown; exudate a few brown droplets; reverse dark brown (6F8). YES 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas dull yellow to greyish yellow (3B3–4B3); soluble pigment brown; exudate absent; reverse brown to dark brown (7E8–F8). DG18 25 °C, 7 d: Colony surface velutinous; sporulation and mycelial areas yellowish white to orange white (4A2–5A2); soluble pigment yellowish brown; exudate absent; reverse greyish yellow to dark yellow (4C6–8). OA 25 °C, 7 d: Colony surface velutinous; sporulation and mycelial areas yellowish white (2A2), olive (3E5) underneath sporulating areas; soluble pigment olive; exudate absent; reverse olive yellow to olive (3D6–F6). CREA 25 °C, 7 d: Colony surface floccose, yellowish white to light yellow (3A3–5); acid not produced.
Micromorphology: Conidial heads radiating; Conidiophores biseriate; Stipes hyaline to dark brown, smooth walled, some very finely rough walled, 235–1 400 × 6.5–11 μm; Vesicles globose to elongated, 18–35 μm wide; Metulae 5.5–10 × 3.5–5.5 μm, metulae cover 100 % of head; Phialides ampulliform, 6–8 × 2.5–4 μm; Conidia globose to subglobose, smooth and finely roughened, 2.5–3.5 × 2.5–3.5 μm (2.9 ± 0.2 × 2.9 ± 0.2, n = 44), average width/length = 0.97, n = 44; Sclerotia absent.
Notes: See notes for A. templicola above.
Aspergillus section Aspergillus
Aspergillus sloanii Visagie, Hirooka & Samson, sp. nov. MycoBank MB809194. Figs 36, 37.
Fig. 36.

Combined phylogeny for ITS, BenA and CaM of selected Aspergillus section Aspergillus. Names in blue are new species described in this study. Aspergillus xerophilus was used as outgroup. Model selected: K2 + G, combined alignment 1 695 bp.
Fig. 37.
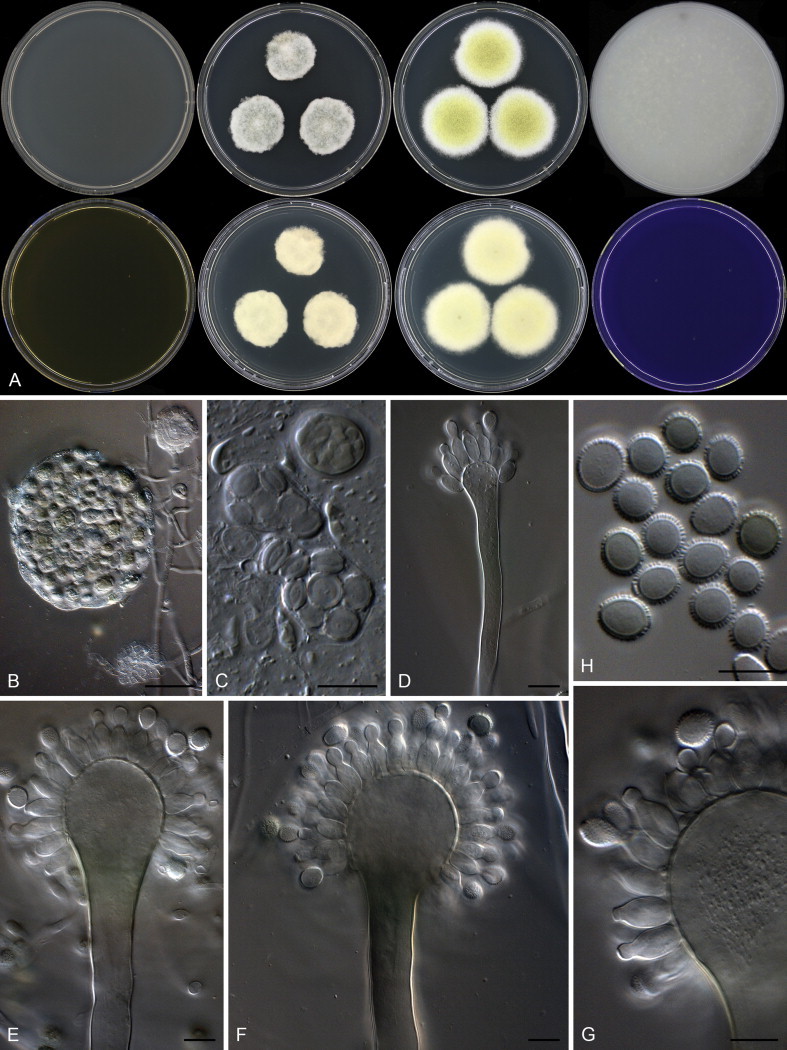
Aspergillus sloanii. A. Colonies: top row left to right, obverse CYA, DG18 of non-sexual strain, DG18 of sexual strain and OA; bottom row left to right, MEA, reverse DG18 of non-sexual strain, reverse DG18 of sexual strain and obverse CREA. B. Ascoma. C. Asci with ascospores. D–G. Conidiophores. H. Conidia. Scale bars: B = 50 μm; C–H = 10 μm.
Etymology: Latin, sloanii, named in honour of Alfred P. Sloan.
Diagnosis: Xerophillic species that does not grow on general media, grows well on DG18 and MEA with 20 % sucrose, eurotium-like sexual state produced with ascospore having lenticular furrows, conidiophores uniseriate with very big rough walled conidia.
Typus: England, Middlesex, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21811, culture ex-type CBS 138177 = DTO 245A1).
Additional materials examined: England, Middlesex, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138176 = DTO 244I8, CBS 138178 = DTO 245A8, CBS 138179 = DTO 245A9, CBS 138231 = DTO 245A6.
ITS barcode: KJ775540 (alternative markers: BenA = KJ775074; CaM = KJ775309)
Colony diam, 7 d (mm): CYA no growth; CYA 30 °C no growth; CYA 37 °C no growth; MEA no growth; YES 3–8; DG18 27–36; OA no growth; CREA no growth.
Colony characters: CYA 25 °C, 7 d: No growth. CYA 30 °C, 7 d: No growth. CYA 37 °C, 7 d: No growth. MEA 25 °C, 7 d: Microcolonies produced after 3 wk. YES 25 °C, 7 d: Microcolony surface floccose, white to greyish white; soluble pigment absent; exudate absent; reverse greyish yellow (4C5). DG18 25 °C, 7 d: Colonies surface floccose; mycelial areas white to greenish yellow (1A6) depending on ascomata produced; sporulation dull green (26D3); ascomata yellow; soluble pigment absent; exudate absent; reverse greenish white to pale green to greyish green (30A2–3–B3). OA 25 °C, 7 d: No growth. CREA 25 °C, 7 d: No growth.
Micromorphology: Conidial heads radiating, produced only on DG18; Conidiophores uniseriate; Stipes hyaline, smooth walled, 160–890 × (6.5–)10–14(–16) μm; Vesicles globose to elongated, sometimes as wide as stipe, (12.5–)25–47(–61) μm wide; Phialides ampulliform, 9–13 × 5–7 μm, covering 100 % of head; Conidia ellipsoidal, minor proportion subglobose, spiny, up to 1.5 μm, (5.5–)7.5–9.5(–11) × 5.5–8.5 μm (8.4 ± 0.85 × 7.36 ± 0.6, n = 45), average width/length = 0.88, n = 42; Ascomata present, Eurotium-like with one layer of mycelia covering ascocarp, 80–170 μm diam; Asci 11–22 μm diam; Ascospores lenticular, furrowed, 4.5–6.5 μm diam.
Notes: Aspergillus sloanii is a monophyletic group within a clade with Aspergillus species that produce a Eurotium-like sexual state (Fig. 36). Its closest relatives, A. glaucus and A. proliferans, grow better on media with high sugar concentrations, but can also grow on the normal CYA, MEA and OA. However, A. sloanii is unable to grow on the latter three media. Other species reported to sometimes not grow on these media include A. penicillioides and A. proliferans. Aspergillus penicillioides, however, produces smaller conidia, 3–5 μm. Aspergillus proliferans produces conidia of similar size to A. sloanii, but has globose to subglobose conidia rather than the predominant ellipsoidal conidia of A. sloanii.
Aspergillus arenarius clade
Aspergillus arenarioides Visagie, Hirooka & Samson, sp. nov. MycoBank MB809195. Figs 38, 39.
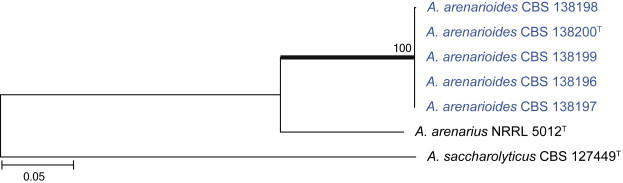
Fig. 38.
Combined phylogeny for ITS, BenA and CaM of Aspergillus arenarius and A. arenarioides. Names in blue are new species described in this study. Aspergillus saccharolyticus was used as outgroup. Model selected: Tamura-3-parameter (T92) +G, combined alignment 1 549 bp.
Fig. 39.
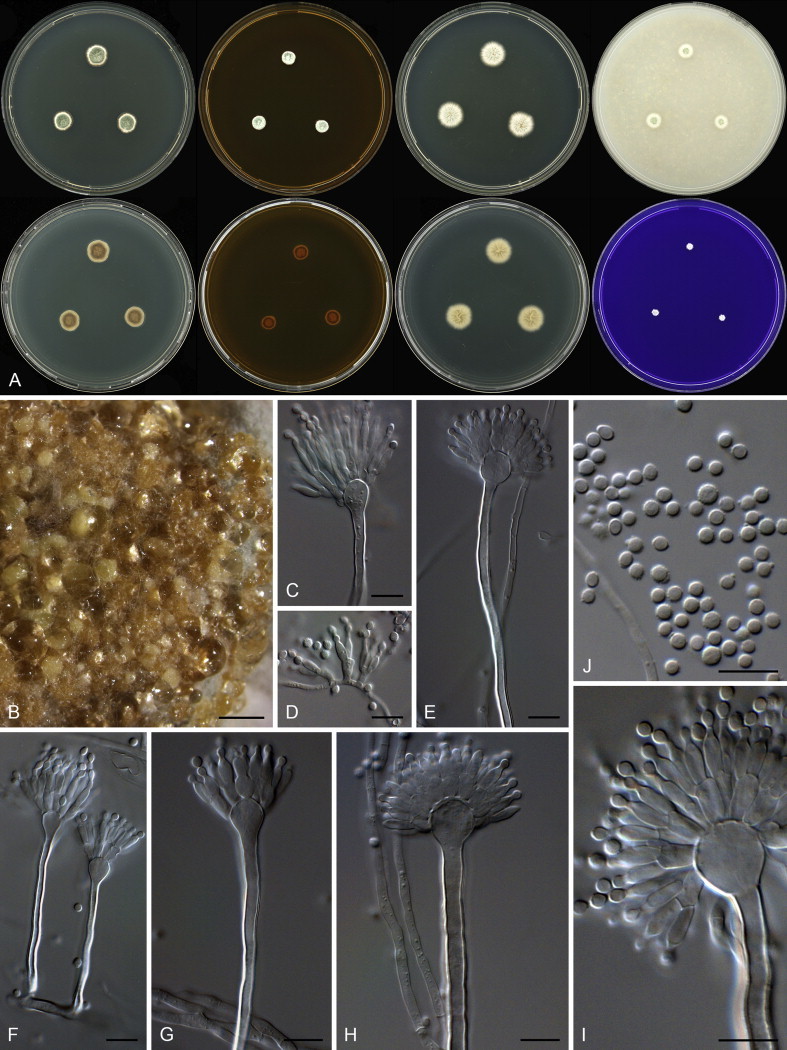
Aspergillus arenarioides. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, DG18 and obverse CREA. B. Sclerotia on CYA. C–I. Conidiophores. J. Conidia. Scale bars: B = 1000 μm; C–J = 10 μm.
Etymology: Latin, arenarioides, referring to the phenotypic similarity of this species to A. arenarius.
Diagnosis: Grows poorly on general media, pale yellow sclerotia produced, conidiophores often Penicillium-like, biseriate, fertile only over 25–50 % of vesicle, conidia globose, rough to echinulate.
Typus: Federated States of Micronesia, Malem of Kosrae Island, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21812, culture ex-type CBS 138200 = DTO 268E3).
Additional materials examined: Federated States of Micronesia, Malem of Kosrae Island, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138198 = DTO 268E1, CBS 138199 = DTO 268E2, CBS 138196 = DTO 267B6, CBS 138197 = DTO 267C7.
ITS barcode: KJ775562 (alternative markers: BenA = KJ775091; CaM = KJ775390)
Colony diam, 7 d (mm): CYA 9–13; CYA 30 °C 13–16; CYA 37 °C no growth; MEA 7–12; YES 10–16; DG18 12–18; CYAS 12–14; OA 7–10; CREA 3–5.
Colony characters: Colony surface velutinous when sporulating, mostly consisting of white sterile mycelia, greyish green (26D5–E5) when sporulating; sclerotia pale yellow in some isolates; soluble pigment mostly absent, some isolates inconspicuously red; exudate mostly absent, clear in some isolates; reverse light yellow to olive brown (4A4–F4), brown (5E6) in isolates with soluble pigment. CYA 30 °C, 7 d: Colonies similar to CYA at 25 °C, except for more abundant sclerotia in some isolates. CYA 37 °C, 7 d: No growth. MEA 25 °C, 7 d: Colony surface velutinous when sporulating, mostly consists of white sterile mycelia, greyish green (2C4) when sporulating; sclerotia pale yellow in some isolates; soluble pigment absent; exudate clear in some isolates; reverse brownish orange to brown (5C5–F8). YES 25 °C, 7 d: Colony surface velutinous in sporulating isolates, mostly consists of white sterile mycelia, greyish green (25D5) when sporulating; soluble pigment absent; exudate absent, reverse light yellow to olive brown (4A4–D4). DG18 25 °C, 7 d: Colony surface floccose, whitish grey to grey to greyish green (30C1–C3); soluble pigment absent; exudate absent, reverse pale yellow to greyish yellow (3A3–C3) to pale orange (5A3). OA 25 °C, 7 d: Colony surface velutinous when sporulating, otherwise floccose, dull green (26D4) when sporulating; sclerotia pale yellow in some isolates; soluble pigment absent; exudate clear; reverse white. CYAS 25 °C, 7 d: Colony surface velutinous when sporulating, mostly consisting of white sterile mycelia, greyish green (26D5–E5) when sporulating; sclerotia pale yellow in some isolates; pigment absent; exudate mostly absent, clear in some isolates; reverse light yellow to olive brown (4A4–F4), brown (5E6) in isolates with soluble pigment. CREA 25 °C, 7 d: Colony surface velutinous, white to greyish green (26C3), acid not produced.
Micromorphology: Conidial heads typically Penicillium-like with some Aspergillus-like conidiphores present, on DG18 the Aspergillus-like head is prominent; Conidiophores biseriate; Stipes mostly hyaline, sometimes brown, smooth walled, 65–200 × 2.5–5.5 μm; Vesicles globose, often elongated on MEA, 5–13 μm; Metulae 5.5–8.5 × 2.5–4.5 μm, covering 25–50 % of head; Phialides ampulliform, 6.5–9.5 × 2.5–4 μm; Conidia globose to subglobose, rough to echinulate, 2.5–3.5(–5.5) μm (2.89 ± 0.2 × 2.87 ± 0.2, n = 44) average width/length = 0.98, n = 43; Hülle cells absent; Sclerotia present, 100–300 μm.
Notes: Aspergillus arenarioides is phylogenetically closely related to A. arenarius (Fig. 38). Both species grow poorly on general media, and produce pale yellow sclerotia and biseriate conidiophores that are often diminutive (Raper & Fennell 1965). Conidia are small and globose, but A. arenarius has smooth walled conidia in contrast to the rough to echinulate conidia of A. arenarioides.
Aspergillus section Usti
Aspergillus porphyreostipitatus Visagie, Hirooka & Samson, sp. nov. MycoBank MB809196. Figs 40, 41.
Fig. 40.

Combined phylogeny for ITS, BenA and CaM of selected Aspergillus section Usti species. Names in blue are new species described in this study. Aspergillus compatibilis was used as outgroup. Model selected: K2 + G, combined alignment 1 345 bp.
Fig. 41.
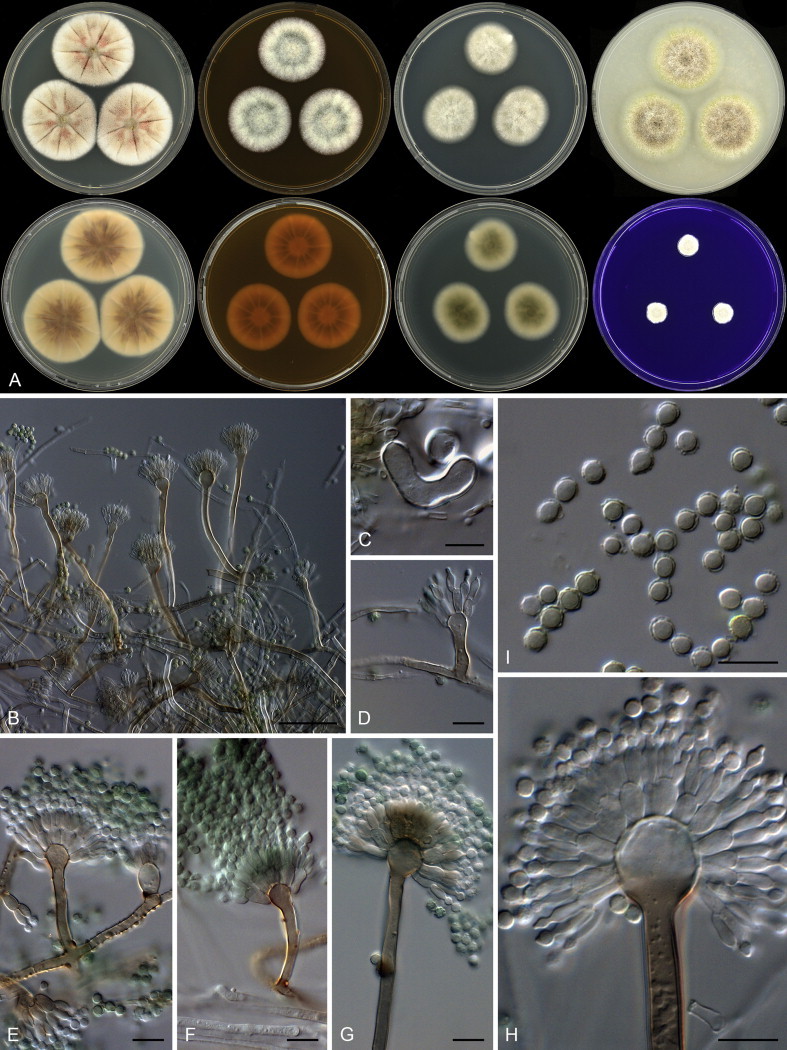
Aspergillus porphyreostipitatus. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, DG18 and obverse CREA. B, D–H. Conidiophores on MEA. C. Hülle cell on OA. I. Conidia. Scale bars: B = 50 μm; C–I = 10 μm.
Etymology: Latin, porphyreostipitatus, meaning red-brown stipe.
Diagnosis: Produces brownish colonies on most media, on MEA and DG18 reverse greyish green, Hülle cells produced on OA, able to grow at 37 °C, conidiophores have reddish brown stipes.
Typus: Mexico, Sayulita, dust from church, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21813, culture ex-type CBS 138203 = DTO 266D9).
Additional material examined: Thailand, Songkhla, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138202 = DTO 132D1.
ITS barcode: KJ775564 (alternative markers: BenA = KJ775080; CaM = KJ775338)
Colony diam, 7 d (mm): CYA 38–41; CYA 30 °C 45–50; CYA 37 °C 5–11; MEA 28–34; YES 40–44; DG18 25–30; CYAS 12–17; OA 30–34; CREA 10–12.
Colony characters: CYA 25 °C, 7 d: Colony surface floccose; sporulation and mycelial areas light brown to brown (5D4–E4); soluble pigment absent; exudate reddish to pink; reverse centrally dark brown to brown (6F7–7E7), elsewhere light yellow (3A5). CYA 30 °C, 7 d: Colonies similar to CYA at 25 °C. CYA 37 °C, 7 d: Colony surface floccose; mycelial areas yellowish white (2A2); soluble pigment yellow; exudate absent; reverse yellowish brown (5D8). MEA 25 °C, 7 d: Colony surface floccose; mycelial areas white; sporulation greyish turquoise to greyish green (24E4–25E4); soluble pigment absent; exudate absent; reverse brown to dark brown (6E8–F8). YES 25 °C, 7 d: Colony surface floccose; mycelial areas white; sporulation brownish grey to light brown (5D2–4); soluble pigment absent; exudate absent; reverse greyish yellow to olive brown (4B5–4D5). DG18 25 °C, 7 d: Colony surface floccose, greyish green (1D3); soluble pigment absent; exudate absent, reverse olive (1FE5–F5). OA 25 °C, 7 d: Colony surface floccose to velutinous; mycelial areas white; sporulation brownish grey (5F2); soluble pigment yellow; exudate minute, clear droplets; reverse greyish yellow (4B4–C4). CYAS 25 °C, 7 d: Colony surface colonies, brownish grey (5D2); soluble pigment absent; exudate absent; reverse olive brown (4F5). CREA 25 °C, 7 d: Colony surface velutinous, greyish brown (5D3) to greyish brown (3B5); acid not produced.
Micromorphology: Conidiophores biseriate, short Penicillium-like conidiophores present, on DG18 less dense (fewer metulae) than on MEA; Stipes reddish brown, hyaline also present, mostly smooth, some areas contain warts, (15–)30–120 × 3.5–6.5 μm; Vesicles globose, sometimes slightly elongated, 8–14 μm; Metulae 5.5–9 × 3–5 μm, covering 75 % of head; Phialides ampulliform, 6–7.5 × 2.5–3.5 μm; Conidia globose to subglobose, often covered by a thick layer (about 0.5 μm), rough, 3–3.5 × 3–3.5 μm (3.3 ± 0.2 × 3.1 ± 0.2, n = 40), average width/length = 0.95, n = 38; Hülle cells produced on OA; Sclerotia absent.
Notes: Aspergillus porphyreostipitatus is resolved within a larger clade with A. baeticus, A. ustus, A. puniceus and A. pseudoustus (Fig. 40). Morphologically these species are similar for producing brownish colours in colonies. The ability of the new species to grow on CYA at 37 °C easily distinguishes it from its morphologically similar relatives. Other species in other clades of section Usti are able to grow at 37 °C (Houbraken et al. 2007, Novakova et al. 2012). However, except for A. compatibilis (≡ Emericella heterothallica), they grow much faster than A. porphyreostipitatus at this temperature. Aspergillus porphyreostipitatus grows slower and sporulates better on MEA compared to A. compatibilis.
Aspergillus section Versicolores
Aspergillus griseoaurantiacus Visagie, Hirooka & Samson, sp. nov. MycoBank MB809197. Figs 42, 43.
Fig. 42.

Combined phylogeny for ITS, BenA and CaM of Aspergillus section Versicolores. Names in blue are new species described in this study. Aspergillus fruticans was used as outgroup. Model selected: T92 + G, combined alignment 1 612 bp.
Fig. 43.

Aspergillus griseoaurantiacus. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, DG18 and obverse CREA. B, C, E–H. Conidiophores. D. Hülle cells. I. Conidia. Scale bars: B, C, E–I = 10 μm; D = 20 μm; E = 100 μm.
Etymology: Latin, griseoaurantiacus, meaning greyish orange, referring to the colour of colonies on CYA and MEA.
Diagnosis: Colonies have a white to light orange to greyish orange colour on CYA and MEA, producing globose Hülle cells, growth on CYA at 37 °C, conidiophores biseriate, vesicles spathulate or elongated, conidia finely roughened and ellipsoidal.
Typus: Federated States of Micronesia, Yela of Kosrae Island, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21814, culture ex-type CBS 138191 = DTO 267D8).
Additional materials examined: Thailand, Songkhla, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138190 = DTO 267D2. Mexico, Sayulita, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138189 = DTO 245F5.
ITS barcode: KJ775553 (alternative markers: BenA = KJ775086; CaM = KJ775357)
Colony diam, 7 d (mm): CYA 25–28; CYA 30 °C 25–26; CYA 37 °C 3–5; MEA 18–21; YES 32–34; DG18 14–16; OA 15–20; CREA 18–20.
Colony characters: Colony surface floccose; mycelial areas white to light orange (5A5) to greyish orange (5B3); sporulation dull green (28D3–E3); soluble pigment brownish red; exudate minute red droplets; reverse reddish brown to dark brown (8F7–8), margin light orange (5A4). CYA 30 °C, 7 d: Colonies similar to CYA at 25 °C. CYA 37 °C, 7 d: Colony surface floccose, white; soluble pigment absent; exudate absent; reverse olive (2E5). MEA 25 °C, 7 d: Colony surface floccose; mycelial areas white to brownish orange (5C3); sporulation dull green (30D3), sometimes (26D4); soluble pigment absent; exudate minute red droplets; reverse light brown (6D6–8) centrally, sometimes greyish orange (5B6). YES 25 °C, 7 d: Colony surface floccose; mycelial areas white to light orange (5A5) to greyish orange (5B3); sporulation sparse, dull green (28D3–E3); soluble pigment absent; exudate absent; reverse deep orange to orange (5A8–B8), margin yellow to light yellow (3A6–4A5). DG18 25 °C, 7 d: Colony surface floccose; mycelial areas white to light orange (5A5) to greyish orange (5B3); sporulation dull green (28D3–E3); soluble pigment reddish brown; exudate absent; reverse deep orange to orange (6A8–B8) to brown (6D8) to olive brown (4E6). OA 25 °C, 7 d: Colony surface floccose; mycelial areas white to greyish; sporulation dull green (27E3 to 29E3) to greyish green (28C3); soluble pigment absent; exudate clear to brownish; reverse greyish yellow (3B5–C5). CREA 25 °C, 7 d: Colony surface floccose, mycelial areas white to light yellow (3A4) to greyish orange to brown (5B5–E5); acid not produced.
Micromorphology: Conidial heads radiating, diminutive Penicillium-like conidiophores typically present in aerial hyphae; Conidiophores biseriate, sometimes greenish; Stipes hyaline to brown, smooth walled, 100–500 × 3.5–8 μm; Vesicles spathulate or elongated, (3.5–)9–18(–26.5) μm wide; Metulae 4–10 × 3–5.5 μm, covering 75 % of head; Phialides ampulliform, 5.5–7 × 2.5–3.5 μm; Conidia ellipsoidal, finely roughened, 2.5–4 × 2–3 μm (3 ± 0.3 × 2.5 ± 0.2, n = 53), average width/length = 0.84, n = 53; Sclerotia absent.
Notes: Aspergillus griseoaurantiacus forms a coherent species within a clade closely related to A. tabacinus, A. versicolor, A. fructus, A. amoenus, A. austrocalifornicus and A. protuberus (Fig. 42). Four of these, A. griseoaurantiacus, A. amoenus, A. fructus and A. versicolor, are able to grow on CYA at 37 °C (Jurjević et al. 2012). Aspergillus griseoaurantiacus produces smooth walled, globose to subglobose conidia, with a small proporation ellipsoidal, whereas A. amoenus produces finely roughened ellipsoidal conidia. Aspergillus fructus and A. versicolor both have finely roughened conidia, but all other characters are very similar to the new species. Jurjević et al. (2012) considered phenotypic characters too similar for A. versicolor and A. fructus, and recommended the use of sequences for identification. This makes identification of our new species based on morphology similarly challenging. However, sequences easily distinguish the species.
The genus Penicillium
Penicillium alfredii Visagie, Seifert & Samson, sp. nov. MycoBank MB809180. Figs 44, 45.
Fig. 44.

RPB2 phylogeny of the genus Penicillium, showing the unique position of P. alfredii. Names in blue are new species described in this study. Talaromyces wortmanii was used as outgroup. Model selected: K2 + G, combined alignment 953 bp.
Fig. 45.
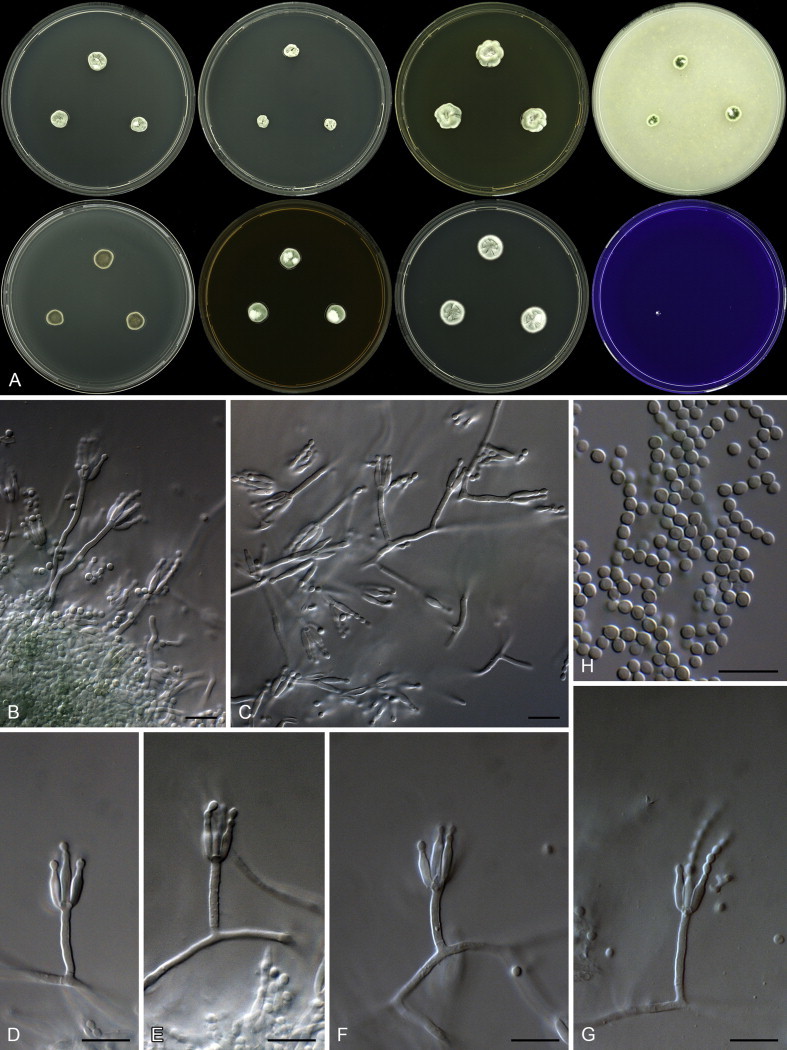
Penicillium alfredii. A. Colonies: top row left to right, obverse CYA, CYA 30 °C, YES and OA; bottom row left to right, reverse CYA, obverse MEA, DG18 and CREA. B–G. Conidiophores. H. Conidia. Scale bars: B–H = 10 μm.
Etymology: Latin, alfredii, named in honour of Alfred P. Sloan.
Diagnosis: Growth poor on all media, colonies dense, producing monoverticillate conidiophores with short stipes, short phialides and smooth, globose conidia.
Typus: Federated States of Micronesia, Lelu of Kosrae Island, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21800, culture ex-type CBS 138224 = DTO 269A4).
ITS barcode: KJ775684 (alternative markers: BenA = KJ775177; CaM = KJ775411; RPB2 = KJ834520)
Colony diam, 7 d (mm): CYA 8–10; CYA 30 °C 5–6; CYA 37 °C no growth; MEA 9–10; YES 13–14; DG18 13–15; CYAS 5–8; OA 6–7; CREA no growth to microcolonies.
Colony characters: Colonies moderately deep, sunken at centre, plane; margins moderately deep, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish green (26C4); soluble pigments absent; exudates absent; reverse dull green (27E4). MEA 25 °C, 7 d: Colonies moderately deep, sunken at centre, plane; margins low, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish green (26C4); soluble pigments absent; exudates absent; reverse brownish orange (5C6) with some brown (5F4) areas. YES 25 °C, 7 d: Colonies moderately deep, sunken at centre, sulcate; margins low, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish green (26C4); soluble pigments absent; exudates absent; reverse dull green (27E4). DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse dull green (26D4); soluble pigments absent; exudates absent; reverse greyish green (30B3). OA 25 °C, 7 d: Colonies low, plane; margins low, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse dark green (27F8); soluble pigments absent; exudates clear. CREA 25 °C, 7 d: Acid not produced.
Micromorphology: Conidiophores monoverticillate; Stipes smooth walled, 8–45 × 1.5–2.5 μm; Vesicles 2.5–3 μm; Phialides ampulliform, sometimes more slender and elongated, 6–8 × 1.5–3 μm (7.1 ± 0.6 × 2.4 ± 0.2); Conidia smooth, globose to subglobose, 2–2.5 × 2–2.5 μm (2.3 ± 0.1 × 2.2 ± 0.1), average width/length = 0.94, n = 44.
Notes: This species is distinct from all Penicillium species and phylogenetically cannot be classified in any of the 25 sections proposed in Houbraken & Samson (2011). ITS sequences (Fig. 12) place the species closest to section Ramigena, whilst RPB2 resolves it on a long branch related to sections Torulomyces and Fracta (Fig. 44). Phenotypically, Penicillium alfredii grows poorly on all media, and colonies resemble those of species in section Torulomyces. However, the latter section includes species that were generally classified in the genus Torulomyces and produce monophialidic conidiophores. This contrasts to the monoverticillate conidiophores of P. alfredii. As such, P. alfredii probably represents a new section. However, the phylogenetic data presented is inconclusive for introducing a new section. This is mainly due to the unresolved position of P. cryptum and P. lassenii (Fig. 44) currently classified in section Torulomyces, which will be addressed in a future study.
Penicillium section Cinnamopurpurea
Penicillium infrapurpureum Visagie, Seifert & Samson, sp. nov. MycoBank MB809181. Figs 46, 47.
Fig. 46.
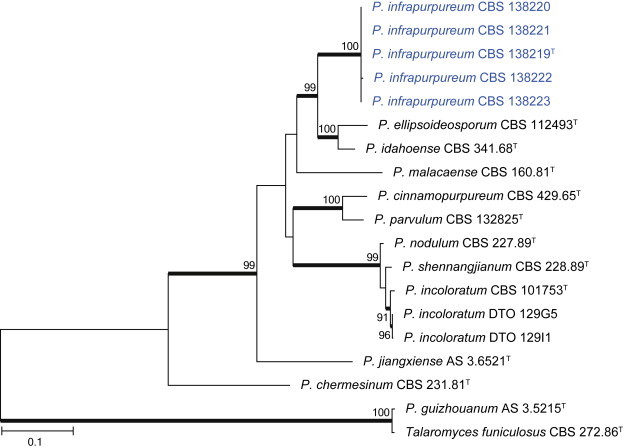
Combined phylogeny for ITS, BenA and CaM of Penicillium section Cinnamopurpurea. Names in blue are new species described in this study. Talaromyces funiculosus and P. guizhouanum was used as outgroup. Model selected: TN93 + G, combined alignment 1 449 bp.
Fig. 47.

Penicillium infrapurpureum. A. Colonies: top row left to right, obverse CYA, CYA 30 °C, YES and OA; bottom row left to right, reverse CYA, obverse MEA, DG18 and CREA. B–G. Conidiophores. H. Conidia. Scale bars: B–H = 10 μm.
Etymology: Latin, infrapurpureum, meaning purple reverse, referring to the purple reverse on CYA.
Diagnosis: Dense, slow growing colonies, purplish to bluish grey reverse on CYA, no growth on CYA at 30 °C, monoverticillate conidiophores producing smooth broadly ellipsoidal conidia.
Typus: Australia, Hobart, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21801, culture ex-type CBS 138219 = DTO 235F6).
Additional materials examined: Australia, Hobart, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138220 = DTO 235G2, CBS 138221 = DTO 235G5, CBS 138222 = DTO 235G6, CBS 138223 = DTO 235H5.
ITS barcode: KJ775679 (alternative markers: BenA = KJ775172; CaM = KJ775406)
Colony diam, 7 d (mm): CYA 14–17; CYA 30 °C no growth; CYA 37 °C no growth; MEA 14–17; YES 17–22; DG18 16–18; CYAS 16–18; OA 8–10; CREA 4–5.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, dense, sunken at centre, sulcate; margins low, narrow, irregular; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish green to dark green (25E5–F5); soluble pigments absent; exudates absent; reverse purplish to bluish grey (20F3), some isolates less intensely coloured. MEA 25 °C, 7 d: Colonies moderately dense, dense, sunken at centre, sulcate; margins low, narrow, irregular; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish green to dark green (25E5–F5); soluble pigments absent; exudates absent; reverse violet brown (10F8) to light brown (7D5). YES 25 °C, 7 d: Colonies deep, sulcate; margins low, narrow, irregular; mycelia white; texture velutinous; sporulation moderately dense to dense, conidia en masse greyish green to dark green (25E5–F5); soluble pigments absent; exudates absent; reverse olive brown to brown (4F5–5F5). DG18 25 °C, 7 d: Colonies low, sulcate; margins low, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense to dense, conidia en masse dull to greyish green (26E3–4); soluble pigments absent; exudates absent; reverse greyish red (8B6–9B6) at centre, yellowish grey (3B2) near margin. OA 25 °C, 7 d: Colonies low, plane; margins low, narrow, irregular; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse dark green (25F6); soluble pigments absent; exudates absent. CREA 25 °C, 7 d: Acid not produced.
Micromorphology: Conidiophores monoverticillate, sub-terminal branching sometimes observed; Stipes smooth walled, 20–70 × 2.5–3.5 μm; Vesicles 3.5–5.5 μm; Phialides ampulliform, sometimes more slender and elongated, 8.5–13.5 × 2.5–3.5 μm (10.8 ± 1.4 × 3.1 ± 0.3); Conidia smooth, broadly ellipsoid, 2.5–3.5 (–5.5) × 2.5–3.5 μm (3.1 ± 0.3 × 2.8 ± 0.3), average width/length = 0.9, n = 46.
Notes: Penicillium infrapurpureum is classified in section Cinnamopurpurea with other species that grow slowly on CYA and MEA (Fig. 46). The new species produces a striking purple to bluish reverse on CYA. A similar colouration was reported for P. cinnamopurpureum (Pitt 1979). Phylogenetically, P. infrapurpureum is resolved in a clade with P. idahoense and P. ellipsoideosporum. Penicillium idahoense also produces a purple reverse and smooth walled conidia (Paden 1971), but a cleistothecial morph is commonly observed, whereas P. ellipsoideosporum produces similar conidia, but lacks the colourful reverse (Wang & Kong 2000). Penicillium ellipsoideosporum also produces shorter phialides, 6.5–8.5 μm, than P. infrapurpureum and P. idahoense. Penicillium infrapurpureum does not grow on CYA at 30 °C or above, with P. idahoense sometimes growing at 37 °C. Two species described from China, P. guizhouanum and P. jiangxiense, were thought to be close relatives of P. cinnamopurpureum (Kong 2000, Kong & Liang 2003). Sequences from the ex-type strains show that this is not the case with P. guizhouanum, which has almost identical sequences to Talaromyces funiculosus. Penicillium jianxiense is tentatively placed in section Cinnamopurpurea. Based on its ITS barcode, it does not belong in the section, but BenA and CaM is most similar to other species in the section (Fig. 46).
Penicillium section Lanata-Divaricata
Penicillium singorense Visagie, Seifert & Samson, sp. nov. MycoBank MB809182. Figs 48, 49.
Fig. 48.

Combined phylogeny for ITS, BenA and CaM of Penicillium section Lanata-Divaricata. Names in blue are new species described in this study. Penicillium oxalicum was used as outgroup. Model selected: K2 + G, combined alignment 1 535 bp.
Fig. 49.

Penicillium singorense. A. Colonies: top row left to right, obverse CYA, CYA 30 °C, YES and OA; bottom row left to right, reverse CYA, obverse MEA, DG18 and CREA. B–G. Conidiophores. H. Conidia. Scale bars: B–H = 10 μm.
Etymology: Latin, singorense, in reference to the ex-type strain, which was isolated from house dust collected in the city Singora/Songkhla, Thailand.
Diagnosis: Fast growing colonies on all media, strong growth on CYA at 30 and 37 °C, conidiophores irregular, mono- to biverticilliate, producing roughened subglobose to ellipsoidal conidia.
Typus: Thailand, Songkhla, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21802, culture ex-type CBS 138214 = DTO 133C6).
Additional materials examined: Thailand, Songkhla, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138211 = DTO 129H7, CBS 138212 = DTO 129H8, DTO 131H8, CBS 138213 = DTO 131I8, DTO 132C8.
ITS barcode: KJ775674 (alternative markers: BenA = KJ775167; CaM = KJ775403)
Colony diam, 7 d (mm): CYA (35–)40–45; CYA 30 °C (35–)40–50; CYA 37 °C 40–43; MEA (35–)45–48; YES (37–)42–45; DG18 21–26; CYAS 19–25; OA 40–45; CREA 20–25.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, wide, entire; mycelia white; texture floccose; sporulation sparse, conidia en masse dull green (26D3–4); soluble pigments absent; exudates absent, sometime clear; reverse greyish yellow to olive brown (4C5–D7). MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, wide, entire; mycelia white; texture floccose; sporulation sparse, conidia en masse greenish grey (26C2); soluble pigments absent; exudates absent, sometimes clear; reverse light brown to brown (5D7–6D7). YES 25 °C, 7 d: Colonies low to moderately deep, sulcate; margins low, wide, entire; mycelia white; texture floccose; sporulation sparse, conidia en masse greenish grey (26B2); soluble pigments absent; exudates absent; reverse reddish to greyish yellow (4A6–B6). DG18 25 °C, 7 d: Colonies low to moderately deep, sulcate; margins low, narrow, entire; mycelia white; texture floccose; sporulation sparse, conidia en masse greenish grey (25B2); soluble pigments absent; exudates absent; reverse yellow (2A6) at centre, greyish green (30B5) elsewhere. OA 25 °C, 7 d: Colonies low, plane; margins low, wide, entire; mycelia white; texture floccose; sporulation sparse to moderately dense, conidia en masse greyish green (28C3); soluble pigments absent; exudates absent. CREA 25 °C, 7 d: Acid not produced.
Micromorphology: Conidiophores irregular, mono- to biverticillate; stipes smooth to finely rough walled, 50–1 000 × 1.5–2.5 μm; Vesicles 2–3 μm; Metulae/branches divergent, when present only two, 10–33 × 1.5–2.5 μm (20.8 ± 6.7 × 2.1 ± 0.3); Phialides ampulliform, 7–10 × 2.5–3 μm (8.4 ± 0.8 × 2.7 ± 0.2); Average length metula/phialide 2.5; Conidia finely rough to rough, subglobose to ellipsoidal, 2.5–3 × 2–3 μm (2.7 ± 0.1 × 2.4 ± 0.1), average width/length = 0.88, n = 39.
Notes: Penicillium singorense is a close relative to P. penajorense and P. vanderhammenii (Fig. 48). The latter species do not grow on CYA at 37 °C, in contrast to the fast growing colonies of P. singorense. In addition, yellow cleistothecia were reported for P. vanderhammenii (Houbraken et al. 2010). This was not observed in P. singorense.
Penicillium section Canescentia
Penicillium dunedinense Visagie, Seifert & Samson, sp. nov. MycoBank MB809183. Figs 50, 51.
Fig. 50.
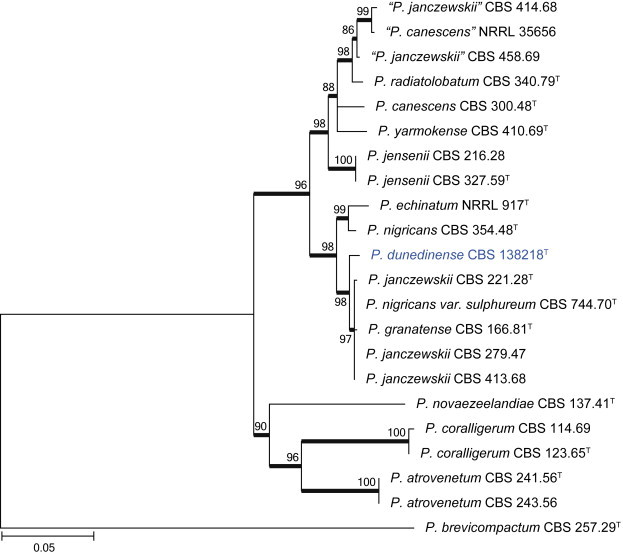
Combined phylogeny for BenA and CaM of Penicillium section Canescentia species. Names in blue are new species described in this study. Penicillium brevicompactum was used as outgroup. Model selected: K2 + G, combined alignment 789 bp.
Fig. 51.
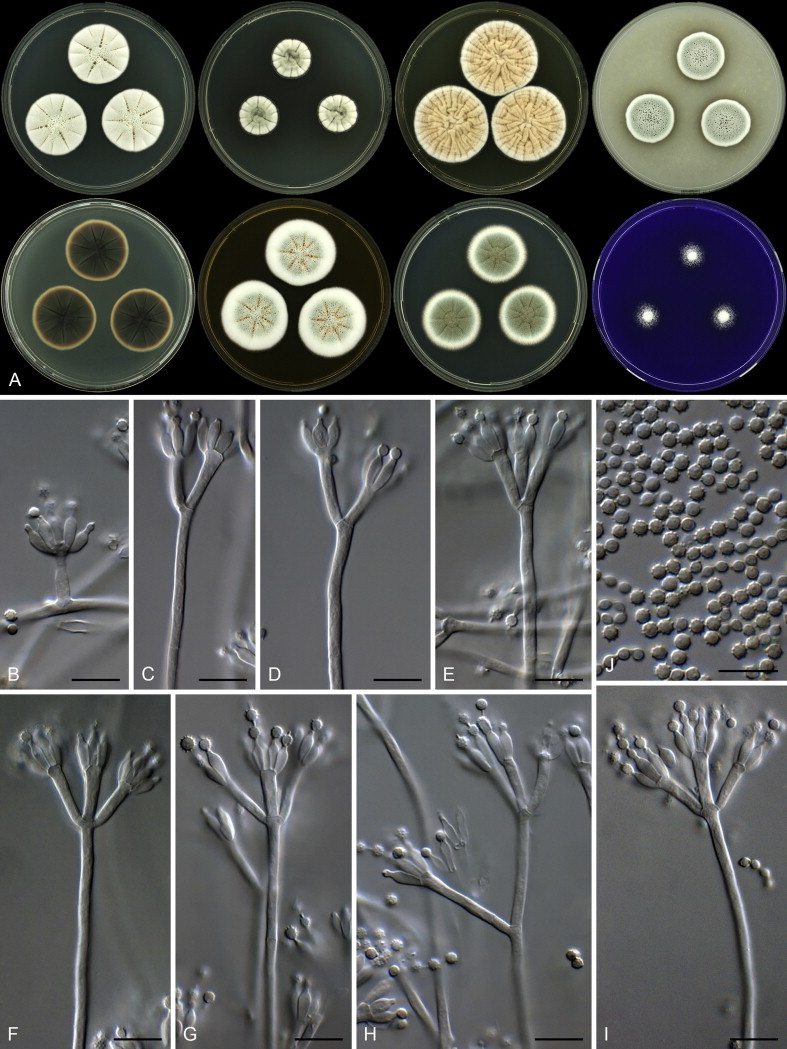
Penicillium dunedinense. A. Colonies: top row left to right, obverse CYA, CYA 30 °C, YES and OA; bottom row left to right, reverse CYA, obverse MEA, DG18 and CREA. B–I. Conidiophores. J. Conidia. Scale bars: B–J = 10 μm.
Etymology: Latin, dunedinense, in reference to the ex-type strain, which was isolated from dust collected in Dunedin, New Zealand.
Diagnosis: Fast growing colonies on MEA, brownish grey reverse on CYA, greyish orange colonies on YES, conidiophores with smooth walled stipes and rough walled conidia.
Typus: New Zealand, Dunedin, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21803, culture ex-type CBS 138218 = DTO 244G1).
ITS barcode: KJ775678 (alternative markers: BenA = KJ775171; CaM = KJ775405)
Colony diam, 7 d (mm): CYA 29–31; CYA 30 °C 19–20; CYA 37 °C no growth; MEA 35–36; YES 38–40; DG18 31–32; CYAS 23–25; OA 21–22; CREA 14–15.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate, having an inconspicuous orange colour in non-sporulating areas; margins low, narrow, entire; mycelia white; texture floccose; sporulation sparse, conidia en masse greyish green to greenish grey (25C3–26C2); soluble pigments absent; exudates abundant, clear; reverse brownish grey (7F2). MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, entire; mycelia white; texture floccose; sporulation moderately dense, conidia en masse greyish green (25C4); soluble pigments absent; exudates orange to clear; reverse brown to dark brown (7E7–F7). YES 25 °C, 7 d: Colonies moderately deep, sulcate, having a greyish orange (6B3) colour; margins low, narrow, entire; mycelia white to greyish orange (6B3); texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse brown to dark brown (7E7–F7). DG18 25 °C, 7 d: Colonies low to moderately deep, sulcate; margins low, wide, entire; mycelia white; texture floccose; sporulation moderately dense, conidia en masse dull green (29D3) near centre, greyish green (25D5) elsewhere; soluble pigments absent; exudates absent; reverse greyish orange to brownish orange (5B4–C4). OA 25 °C, 7 d: Colonies moderately deep, plane; margins moderately deep, narrow, entire; mycelia white; texture floccose; sporulation sparse to moderately dense, conidia en masse greyish green (25C3–4); soluble pigments inconspicuously brown; exudates clear. CREA 25 °C, 7 d: Acid not produced.
Micromorphology: Conidiophores biverticillate, subterminal branching sometimes present; Stipes smooth walled, 120–350 × 2.5–3 μm; rami/branches 13–25 × 2.5–3 μm; metulae divergent, 9.5–16 × 2.5–3 μm (12.4 ± 1.5 × 2.7 ± 0.2); Phialides ampulliform, 6–8 × 2.5–3 μm (7.2 ± 0.45 × 2.7 ± 0.2); Average length metula/phialide 1.73; Conidia echinulate, globose, 2–3 μm (2.5 ± 0.1), average width/length = 0.99, n = 32.
Notes: Penicillium dunedinense is a distinct clade sister to P. janczewskii, P. nigricans and P. echinatum (Fig. 50). This group of species has conidiophores with smooth walled stipes and rough walled conidia. This section is currently being revised by Visagie et al. (in prep.), with P. nigricans and P. echinatum, considered synonyms of P. janczewskii by Pitt (1979) both considered distinct. Penicillium dunedinense can be distinguished from all the strains examined by its faster growth on MEA, greyish orange colonies on YES and the very dark brownish grey reverse on CYA. Penicillium janczewskii sensu stricto never grows more than 30 mm, more commonly close to 20 mm on MEA.
Penicillium section Ramosa
Penicillium lenticrescens Visagie, Seifert & Samson, sp. nov. MycoBank MB809184. Figs 52, 53.
Fig. 52.
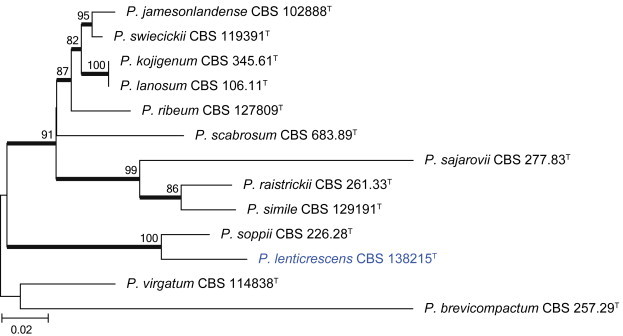
Combined phylogeny for ITS, BenA and CaM of Penicillium section Ramosa species. Names in blue are new species described in this study. Penicillium brevicompactum was used as outgroup. Model selected: GTR + G, combined alignment 1 360 bp.
Fig. 53.
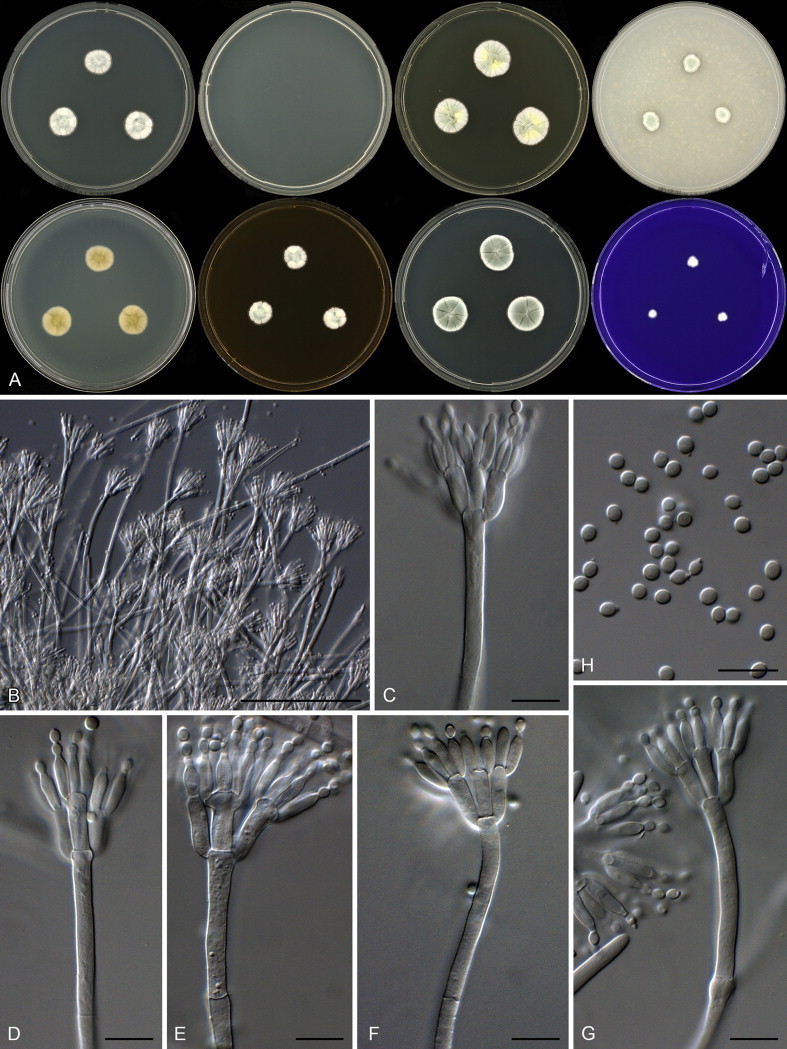
Penicillium lenticrescens. A. Colonies: top row left to right, obverse CYA, CYA 30 °C, YES and OA; bottom row left to right, reverse CYA, obverse MEA, DG18 and CREA. B–G. Conidiophores. H. Conidia. Scale bars: B = 100 μm; C–H = 10 μm.
Etymology: Latin, lenticrescens, meaning slow growing, referring to the restricted growth of the species on all media.
Diagnosis: Slow growth on general media, no growth at 30 °C, conidiophores biverticillate, producing smooth walled stipes and smooth walled subglobose conidia.
Typus: New Zealand, Dunedin, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21804, culture ex-type CBS 138215 = DTO 129A8).
ITS barcode: KJ775675 (alternative markers: BenA = KJ775168; CaM = KJ775404).
Colony diam, 7 d (mm): CYA 12–14; CYA 30 °C no growth; CYA 37 °C no growth; MEA 10–11; YES 17–18; DG18 17–18; CYAS 19–20; OA 6–8; CREA 4–5.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, entire; mycelia white; texture floccose; sporulation moderate, conidia en masse greyish green (25B4–C4); soluble pigments absent; exudates minute droplets, clear; reverse greyish green (30C4) centrally, fading to pale yellow (2A2) near margin. MEA 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, entire; mycelia white and inconspicuously yellow; texture floccose; sporulation moderately dense, conidia en masse greyish green (25B4–C4); soluble pigments absent; exudates minute, clear droplets; reverse brownish orange (5C6). YES 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, entire; mycelia white and yellow; texture floccose; sporulation sparse to moderate, conidia en masse greyish green (26B3); soluble pigments absent; exudates absent; reverse pale to light yellow (4A3–5). DG18 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, entire; mycelia white; texture floccose; sporulation moderately dense, conidia en masse dull green (25D4); soluble pigments absent; exudates absent; reverse pale green (30A3). OA 25 °C, 7 d: Colonies moderately deep, plane; margins low, narrow, entire; mycelia white; texture floccose; sporulation moderately dense, conidia en masse greyish green (25B4–C4); soluble pigments absent; exudates absent. CREA 25 °C, 7 d: Acid not produced.
Micromorphology: Conidiophores biverticillate; Stipes smooth walled, 150–415 × 3–4 μm; Metulae divergent, swollen at apex up to 7.5 μm, 9.5–15 × 3–4.5 μm (11.9 ± 1.4 × 3.89 ± 0.4); Phialides ampulliform, 7.5–10.5 × 2.5–3.5 μm (8.8 ± 0.6 × 2.9 ± 0.2); Average length metula/phialide 1.35; Conidia smooth, subglobose, with a minor proportion ellipsoidal, 2.5–3.5 × 2.5–3 μm (2.9 ± 0.15 × 2.6 ± 0.2), average width/length = 0.90, n = 37.
Notes: Penicillium lenticrescens forms a monophyletic clade in section Ramosa closely related to P. soppii (Fig. 52). Both species sporulate rather sparsely after 7 d of growth. However, generally P. soppii grows faster and produces abundant sclerotia, features not observed in P. lenticrescens. Conidiophores of the two species are similar.
Penicillium section Paradoxa
Penicillium mexicanum Visagie, Seifert & Samson, sp. nov. MycoBank MB809185. Figs 54, 55.
Fig. 54.
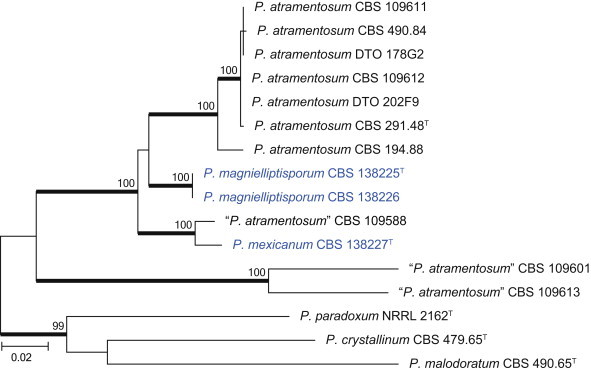
Combined phylogeny for BenA and CaM of Penicillium section Paradoxa species. Names in blue are new species described in this study. The tree was rooted to P. paradoxum, P. crystallinum and P. malodoratum. Model selected: K2 + G, combined alignment 853 bp.
Fig. 55.
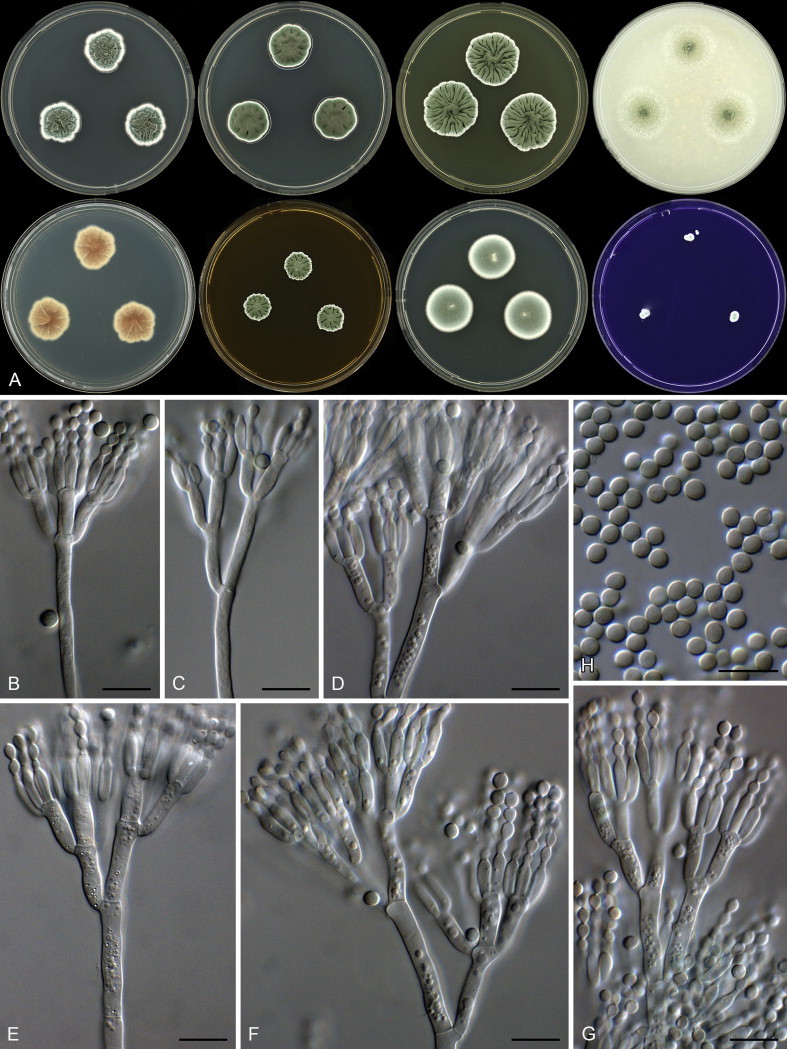
Penicillium mexicanum. A. Colonies: top row left to right, obverse CYA, CYA 30 °C, YES and OA; bottom row left to right, reverse CYA, obverse MEA, DG18 and CREA. B–G. Conidiophores. H. Conidia. Scale bars: B–H = 10 μm.
Etymology: Latin, mexicanum, in reference to the ex-type strain, which was isolated from Mexico.
Diagnosis: Slow growth on general media, on CYA at 30 °C colonies 19–21 mm, conidiophores with smooth walled stipes, smooth walled, broadly ellipsoidal to ellipsoidal conidia (3–4 × 3–3.5 μm).
Typus: Mexico, Sayulita, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21805, culture ex-type CBS 138227 = DTO 270F1).
ITS barcode: KJ775685 (alternative markers: BenA = KJ775178; CaM = KJ775412)
Colony diam, 7 d (mm): CYA 20–22; CYA 15C 18–20; CYA 30 °C 19–21; CYA 37 °C no growth; MEA 12–14; YES 25–28; DG18 23–26; CYAS 21–24; OA 21–25; CREA 5–8.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, in some isolates irregular; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish green (26E6); soluble pigments absent; exudates abundant, clear to purplish; reverse centrally brown (6D5–6), elsewhere orange white (5A2). MEA 25 °C, 7 d: Colonies low, radially sulcate, raised at centre; margins low, narrow, irregular; mycelia white; texture velutinous; sporulation dense, conidia en masse greyish green (26E5); soluble pigments absent; exudates absent; reverse yellowish brown (5E8) at centre, margin brown (5E8) at margin YES 25 °C, 7 d: Colonies moderately deep, randomly sulcate, raised at centre; margins low, narrow, irregular; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse dull green (26D4–E4); soluble pigments absent; exudates absent; reverse dull yellow (3B3), olive (3D5–E5). DG18 25 °C, 7 d: Colonies low, very lightly radially sulcate; margins low, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish green (25C5–E5); soluble pigments absent; exudates absent; reverse greenish grey (29B2–C2). OA 25 °C, 7 d: Colonies low, plane, sporulating in rings; margins low, wide, entire; mycelia white; texture velutinous; sporulation dense, conidia en masse greyish green (25F8–26F8); soluble pigments absent; exudates abundant clear. CREA 25 °C, 7 d: Acid not produced.
Micromorphology: Conidiophores terverticillate, sometimes quarterverticillate; Stipes smooth walled, 65–370 × 3–4.5 μm; Branches/rami 1–4 per stipe, 11–20 × 3–4.5 μm; Metulae appressed, 8.5–13.5 × 2.5–4.5 μm (11.3 ± 1.1 × 3.6 ± 0.4); Phialides ampulliform, 7–10 × 2.5–3.5 μm (8.2 ± 0.8 × 2.9 ± 0.2); Average length metula/phialide 1.37; Conidia smooth, broadly ellipsoidal to ellipsoidal, 3–4 × 3–3.5 μm (3.6 ± 0.2 × 3.2 ± 0.1), average width/length = 0.89, n = 45.
Notes: Penicillium mexicanum is closely related to P. atramentosum and P. magnielliptisporum (described below) in section Paradoxa (Fig. 54). Frisvad & Samson (2004) considered the smooth walled globose conidia, good growth on CREA and absence of growth at 30 °C diagnostic for P. atramentosum. Phylogenetically, the strains identified as P. atramentosum in Frisvad & Samson (2004) represent a species complex. In our studies, 0–6 mm growth was observed on CYA at 30 °C for strains previously assigned to P. atramentosum. Slightly faster growth, 12–14 mm, was seen in CBS 109588, the closest relative of P. mexicanum. Penicillium mexicanum and P. magnielliptisporum grew 19–21 mm and 9–10 mm respectively at 30 °C. In addition, conidia of the two new species were consistently larger than those of P. atramentosum (< 3 μm). Except for the faster growth of P. mexicanum at 30 °C, its growth is more restricted on most media than P. magnielliptisporum. In addition, P. magnielliptisporum produces much bigger conidia than P. mexicanum.
Penicillium magnielliptisporum Visagie, Seifert & Samson, sp. nov. MycoBank MB809186. Figs 54, 56.
Fig. 56.
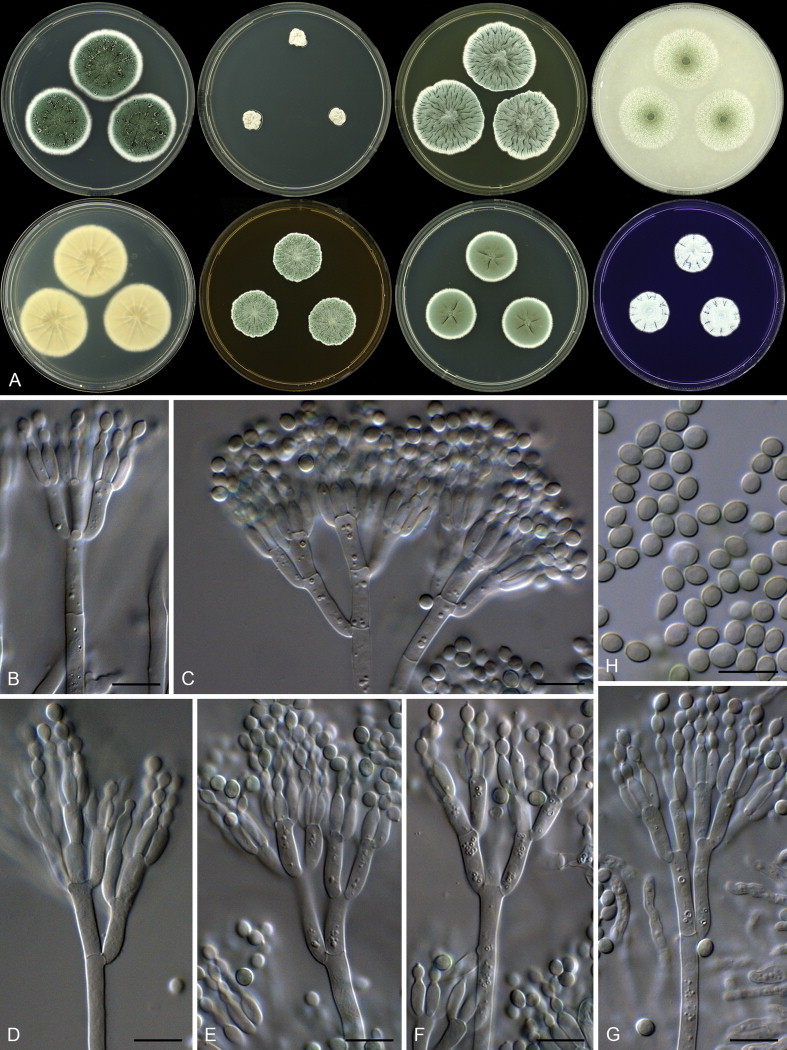
Penicillium magnielliptisporum. A. Colonies: top row left to right, obverse CYA, CYA 30 °C, YES and OA; bottom row left to right, reverse CYA, obverse MEA, DG18 and CREA. B–G. Conidiophores. H. Conidia. Scale bars: B–H = 10 μm.
Etymology: Latin, magnielliptisporum, meaning large ellipsoidal conidia, in reference to the conidia of this species, which are larger than those of its closest relatives.
Diagnosis: Good growth on general media, also on CREA, restricted growth on CYA at 30 °C, conidiophores with smooth walled stipes, large, smooth broadly ellipsoidal to ellipsoidal conidia, 3.5–5 × 3–4 μm.
Typus: New Zealand, Dunedin, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21806, culture ex-type CBS 138225 = DTO 128H8).
Additional material examined: New Zealand, Dunedin, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138226 = DTO 128I1.
ITS barcode: KJ775686 (alternative markers: BenA = KJ775179; CaM = KJ775413)
Colony diam, 7 d (mm): CYA 35–38; CYA 15C 26–29; CYA 30 °C 9–10; CYA 37 °C no growth; MEA 21–23; YES 31–35; DG18 22–25; CYAS 19–20; OA 31–34; CREA 8–15.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep, sulcate; margins low, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish green (26E6); soluble pigments absent; exudates abundant, clear; reverse yellowish grey (2B3–C3). MEA 25 °C, 7 d: Colonies low, radially sulcate, slightly raised at centre; margins low, narrow, irregular; mycelia white; texture velutinous; sporulation dense, conidia en masse greyish green (26D5–E5); soluble pigments absent; exudates minute, clear droplets; reverse yellowish brown (5E8) at centre, margin brown (5E8). YES 25 °C, 7 d: Colonies moderately deep, randomly sulcate, raised at centre; margins low, narrow, irregular; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish to dull green (25D4–5); soluble pigments absent; exudates absent; reverse dull yellow (3B3), olive (3D5–E5). DG18 25 °C, 7 d: Colonies low, very lightly radially sulcate; margins low, narrow, entire; mycelia white; texture velutinous; sporulation moderately dense, conidia en masse greyish green (25C5–E5); soluble pigments absent; exudates absent; reverse greenish grey (29B2–C2). OA 25 °C, 7 d: Colonies low, plane; margins low, wide, entire; mycelia white; texture velutinous; sporulation dense, conidia en masse greyish green (25F8–26F8); soluble pigments absent; exudates abundant clear. CREA 25 °C, 7 d: Acid not produced.
Micromorphology: Conidiophores terverticillate, sometimes quarterverticillate; Stipes smooth walled, 80–280 × 3–4.5 μm; Branches/rami 1–4 per stipe, 12–20 × 3–4.5 μm; Metulae appressed, 9–14 × 3–4.5 μm (11.8 ± 1.3 × 3.5 ± 0.3); Phialides ampulliform, 7.5–10 × 2.5–3.5 μm (8.7 ± 0.6 × 2.7 ± 0.2); Average length metula/phialide 1.36; Conidia smooth, broadly ellipsoidal to ellipsoidal, 3.5–5 × 3–4 μm (4.3 ± 0.3 × 3.4 ± 0.2), average width/length = 0.79, n = 33.
Notes: See notes for P. mexicanum above.
The genus Talaromyces
Talaromyces section Talaromyces
Talaromyces oumae-annae Visagie, Yilmaz, Seifert & Samson, sp. nov. MycoBank MB809187. Figs 57, 58.
Fig. 57.
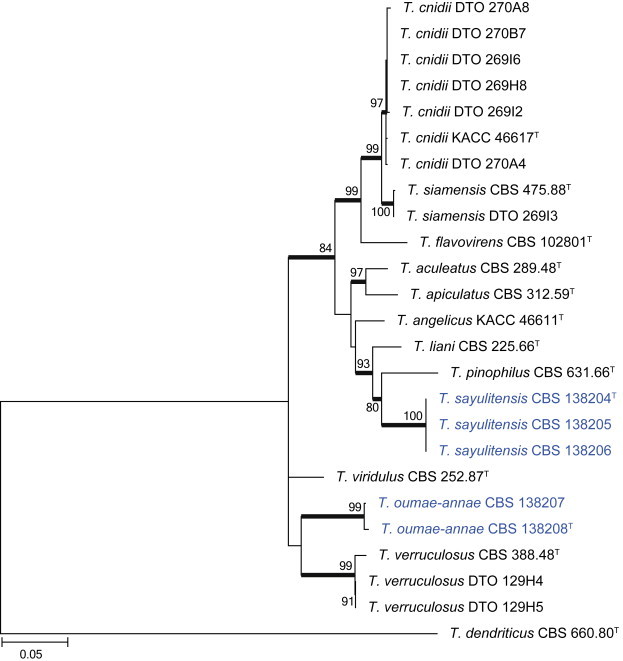
Combined phylogeny for ITS, BenA and CaM of Talaromyces section. Talaromyces species closely related to the new species from house dust. The tree was rooted to T. dendriticus. Model selected: K2 + G, combined alignment 1 452 bp.
Fig. 58.

Talaromyces oumae-annae. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, obverse YES and CREA. B–H. Conidiophores. I. Conidia. Scale bars: B–I = 10 μm.
Etymology: Latin, oumae-annae, named in honour of “Ouma Anna”, grandmother of Visagie, this species was isolated from dust collected in her house in Kuils River, Cape Town.
Diagnosis: Growing restrictedly on CYA and DG18, grows well on other media, conidiophores biverticillate with some subterminal branches formed, stipes smooth walled, conidia rough walled and ellipsoidal.
Typus: South Africa, Kuils River in the Cape Town metropolitan area, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21797, culture ex-type CBS 138208 = DTO 269E8).
Additional materials examined: South Africa, Kuils River, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138207 = DTO 180B4.
ITS barcode: KJ775720 (alternative markers: BenA = KJ775213; CaM = KJ775425)
Colony diam, 7 d (mm): CYA 16–18; CYA 30 °C 16–17; CYA 37 °C 10–11; MEA 29–30; YES 20–23; DG18 14–17; CYAS No growth; OA 30–35; CREA 5–6.
Colony characters: CYA 25 °C, 7 d: Colonies low, slightly raised at centre, plane; margins low, narrow, entire; mycelia white; texture floccose and velutinous; sporulation moderately dense to dense, conidia en masse dull green (25D4–E4); soluble pigments yellow; exudates absent; reverse greyish green (29B6–C6). MEA 25 °C, 7 d: Colonies low, plane; margins low, narrow, entire; mycelia white and pastel yellow; texture velutinous, centrally floccose with sterile aerial mycelia; sporulation dense, conidia en masse greyish green (27D5–E5); soluble pigments absent; exudates absent; reverse light brown (7D6) in the centre fading into brownish orange (6C6). YES 25 °C, 7 d: Colonies low, raised at centre, lightly sulcate; margins low, narrow, entire; mycelia white to yellow; texture velutinous to floccose; sporulation dense, conidia en masse dull green (25D4–E4); soluble pigments yellow; exudates absent; reverse centre light yellow to greyish yellow (2A5–B5), at margins greyish green (27E5). DG18 25 °C, 7 d: Colonies moderately deep, lightly sulcate; margins low, narrow, entire; mycelia white to yellow; texture floccose; sporulation dense, conidia en masse greyish green (26E5–27E5); soluble pigments absent; exudates absent; reverse light orange (6A4–5). OA 25 °C, 7 d: Colonies low, plane; margins low, narrow, entire; mycelia white; texture velutinous; sporulation dense, conidia en masse greyish green to dark green (26E5–F5); soluble pigments absent; exudates absent. CREA 25 °C, 7 d: Acid not produced.
Micromorphology: Conidiophores biverticillate, subterminal branches sometimes present; Stipes smooth walled, 85–240 × 2.5–3.5 μm; Branches up to 30 μm long; Metulae appressed, 8–11 (–12.5) × 2.5–3.5 μm (10.6 ± 1.0 × 3 ± 0.2); Phialides acerose, 9–11.5 × 2–3 μm (10.6 ± 0.8 × 2.7 ± 0.3); Average length metula/phialide 1.01; Conidia rough, ellipsoidal, 3–3.5 × 2.5–3 μm (3.2 ± 0.2 × 2.6 ± 0.2), average width/length = 0.83, n = 38.
Notes: Talaromyces oumae-annae is phylogenetically closely related to T. verruculosus and T. viridulus (Fig. 57). However, T. oumae-annae produces ellipsoidal conidia compared to the globose conidia of T. verruculosus. The latter species also grows much faster on CYA at all temperatures (CYA 32–35; CYA 30 °C 37–38; CYA 37 °C 25–26). Talaromyces viridulus, originally described as Geosmithia viridis, produces rod-shaped conidia, in contrast to the ellipsoidal conidia of T. oumae-annae.
Talaromyces sayulitensis Visagie, Yilmaz, Seifert & Samson, sp. nov. MycoBank MB809188. Figs 57, 59.
Fig. 59.
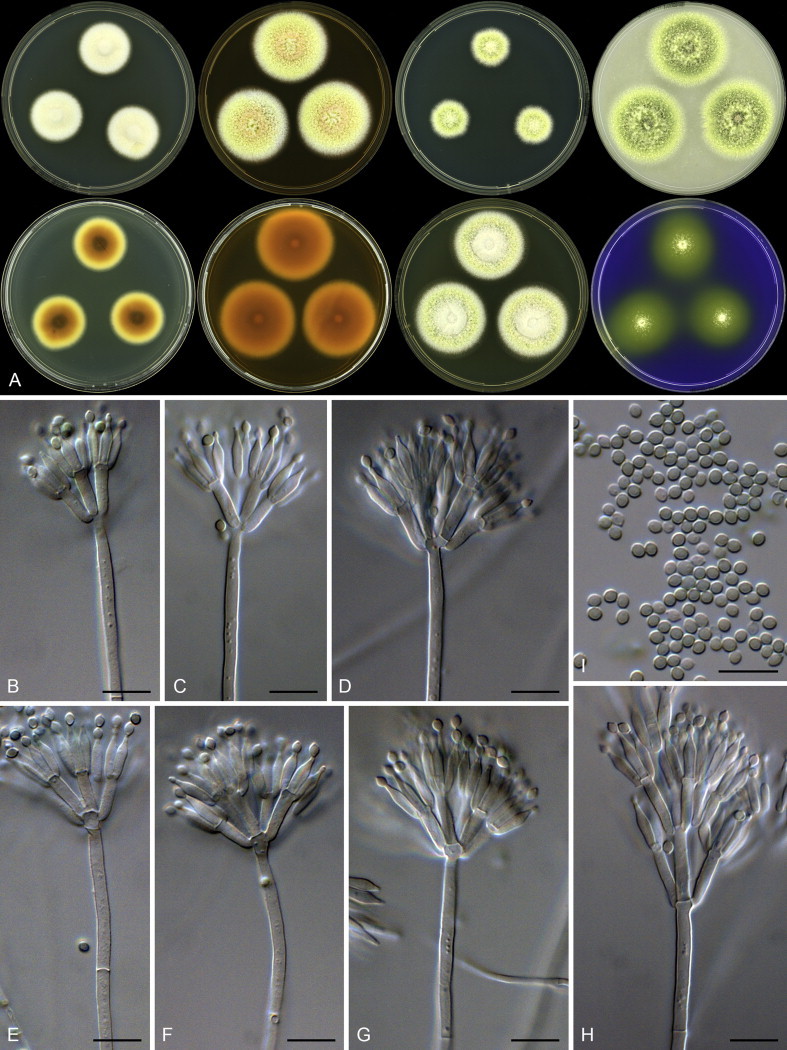
Talaromyces sayulitensis. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, obverse YES and CREA. B–H. Conidiophores. I. Conidia. Scale bars: B–I = 10 μm.
Etymology: Latin, sayulitensis, in reference to the ex-type strain, which was isolated from dust collected in Sayulita.
Diagnosis: Yellow mycelia dominate colony appearance, good growth on CYA at 37 °C, acid produced on CREA, conidiophores biverticillate, stipes smooth walled, conidia smooth and subglobose to broadly ellipsoidal.
Typus: Mexico, Sayulita, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21798, culture ex-type CBS 138204 = DTO 245H1).
Additional materials examined: Mexico, Sayulita, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138205 = DTO 245H2, CBS 138206 = DTO 245H3.
ITS barcode: KJ775713 (alternative markers: BenA = KJ775206; CaM = KJ775422)
Colony diam, 7 d (mm): CYA 24–29; CYA 30 °C 35–43; CYA 37 °C 32–40; MEA 37–40; YES 37–40; DG18 18–22; CYAS 5–8; OA 40–42; CREA 15–18.
Colony characters: CYA 25 °C, 7 d: Colonies low, raised at centre, slightly sulcate; margins low, narrow, entire; mycelia white to yellow to red; texture floccose; sporulation absent; soluble pigments absent; exudates absent to clear in some isolates; reverse brown (6E6) centrally, fading into brownish orange (6C7) and light yellow (4A5). MEA 25 °C, 7 d: Colonies low, slightly raised at centre, plane; margins low, narrow, entire; mycelia white, pastel yellow and pastel red; texture loosely funiculose to floccose; sporulation sparse, conidia en masse greyish green (27D5–E5); soluble pigments absent; exudates absent; reverse brownish orange (6C6–7). YES 25 °C, 7 d: Colonies low, raised at centre, sulcate; margins low, narrow, entire; mycelia white to yellow; texture loosely funiculose to floccose; sporulation sparse to moderately dense, conidia en masse greyish green (27D5–E5); soluble pigments absent; exudates absent; reverse brownish orange (6C6–7). DG18 25 °C, 7 d: Colonies low, plane; margins low, narrow, entire; mycelia white to yellow; texture floccose; sporulation moderately dense, conidia en masse greyish green (26D5–E5); soluble pigments absent; exudates absent; reverse light yellow (3A5–4A5). OA 25 °C, 7 d: Colonies low, slightly raised at centre, plane; margins low, wide, entire; mycelia white to yellow; texture loosely funiculose and floccose, especially in the centre sterile aerial hyphae; sporulation dense, conidia en masse greyish green (27C5–D5); soluble pigments absent; exudates absent. CREA 25 °C, 7 d: Acid strongly produced.
Micromorphology: Conidiophores biverticillate, subterminal branches sometimes present; Stipes smooth walled, (40–)85–300 × 2–3.5 μm; Branches up to 40 μm long; Metulae appressed, 8–11.5(–14) × 2.5–3 μm (10.2 ± 1.3 × 2.8 ± 0.2); Phialides acerose, 8–11 × 2.5–3 μm (9.4 ± 0.6 × 2.6 ± 0.2); Average length metula/phialide 1.09; Conidia smooth, subglobose to broadly ellipsoidal, 2.5–3 × 2–2.5 μm (2.6 ± 0.1 × 2.2 ± 0.1), average width/length = 0.87, n = 37.
Notes: Phylogenetically, T. sayulitensis forms a coherent clade closely related to T. pinophilus and T. liani (≡ P. liani). Talaromyces liani lacks the acid production characteristic of T. sayulitensis, produces larger conidia 2.5–4 μm, and typically produces a sexual state. Talaromyces pinophilus also produces acid on CREA and also lacks a sexual state and other colony characters are very similar to T. sayulitensis, although some minor differences are observed in colony growth rates. This does not allow unequivocal morphological identification of the new species. Phylogenetically it is distinct and this justifies introducing it as a new species.
Talaromyces section Islandici
Talaromyces yelensis Visagie, Yilmaz, Seifert & Samson, sp. nov. MycoBank MB809189. Figs 60, 61.
Fig. 60.

Combined phylogeny for ITS and BenA of Talaromyces section Islandici. Names in blue are new species described in this study. The tree was rooted to T. piceus. Model selected: K2 + G, combined alignment 738 bp.
Fig. 61.
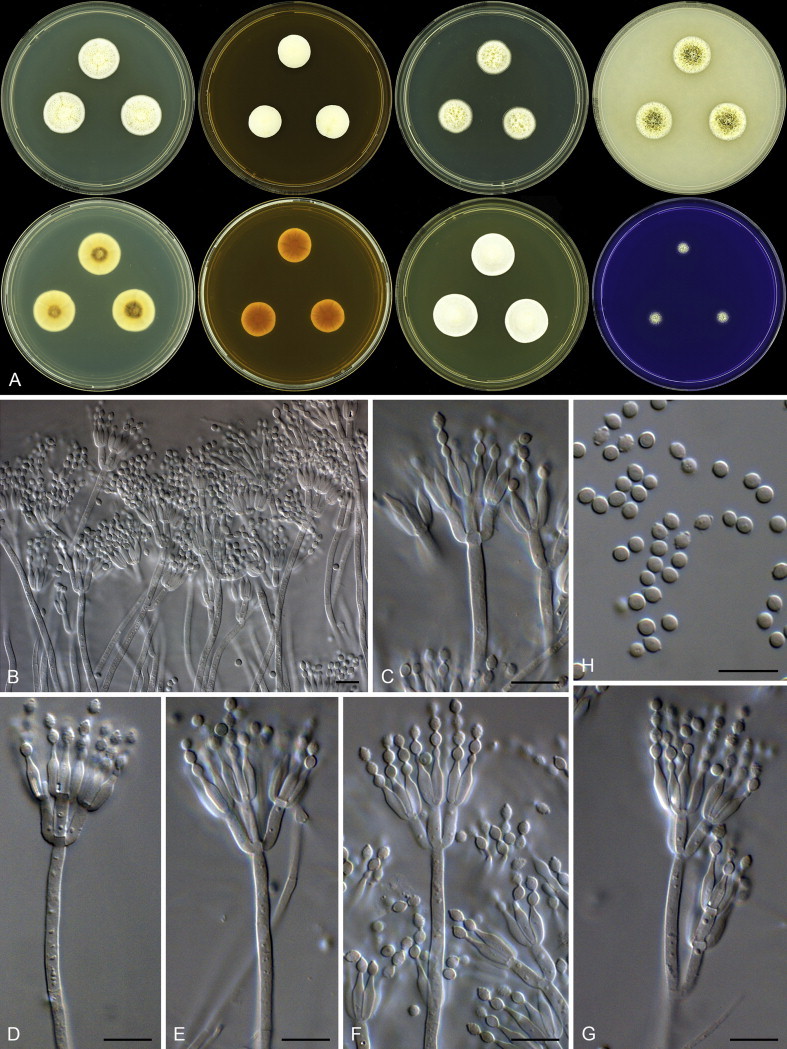
Talaromyces yelensis. A. Colonies: top row left to right, obverse CYA, MEA, DG18 and OA; bottom row left to right, reverse CYA, MEA, obverse YES and CREA. B–G. Conidiophores. H. Conidia. Scale bars: B–H = 10 μm.
Etymology: Latin, yelensis, in reference to the ex-type strain, which was isolated from dust collected in Yela, Micronesia.
Diagnosis: Very dense, deep and yellow colonies produced on general media, conidiophores biverticillate, ampulliform phialides end in fine apical pores, roughened subglobose to broadly ellipsoidal conidia.
Typus: Federated States of Micronesia, Yela of Kosrae Island, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange (holotype CBS H-21799, culture ex-type: CBS 138209 = DTO 268E5).
Additional material examined: Federated States of Micronesia, Yela of Kosrae Island, house dust, 2010, isolated by Ed Whitfield & Kalima Mwange, CBS 138210 = DTO 268E7.
ITS barcode: KJ775717 (alternative markers: BenA = KJ775210)
Colony diam, 7 d (mm): CYA 20–22; CYA 30 °C 25–26; CYA 37 °C 14–16; MEA 15–16; YES 20–21; DG18 16–17; CYAS 13–14; OA 18–20; CREA 9–10.
Colony characters: CYA 25 °C, 7 d: Colonies moderately deep; margins low, narrow, entire; mycelia white to yellowish to orange; texture floccose; sporulation absent; soluble pigments absent; exudates clear and sticky; reverse yellowish white (2A2) to light yellow (3A5) to brown (5F6). MEA 25 °C, 7 d: Colonies very deep, plane; margins deep, narrow, entire; mycelia white to yellow to orange; texture floccose; sporulation absent; soluble pigments absent; exudates yellow; reverse brownish yellow to yellowish brown to brown (5C8–E8). YES 25 °C, 7 d: Colonies very deep, plane; margins low, narrow, entire; mycelia white to yellow to orange; texture floccose; sporulation absent; soluble pigments absent; exudates absent; reverse yellowish white (4A2) to greyish orange (5B5). DG18 25 °C, 7 d: Colonies deep, plane; margins low, narrow, entire; mycelia white to yellow; texture floccose; sporulation sparse, conidia en masse greyish green (26C3); soluble pigments absent; exudates yellow and sticky; reverse yellowish white to yellow (3A2–6). OA 25 °C, 7 d: Colonies moderately deep, plane; margins low, narrow, entire; mycelia white to yellow; texture floccose; sporulation moderately dense, conidia en masse dark green (26F6); soluble pigments absent; exudates clear and sticky. CREA 25 °C, 7 d: Acid not produced.
Micromorphology: Conidiophores biverticillate, subterminal branches sometimes present; stipes smooth walled, 60–190 × 2.5–3.5 μm; branches up to 30 μm long; metulae appressed, 8–11 × 2.5–3.5 μm (9.7 ± 0.7 × 2.9 ± 0.3); phialides ampulliform, ending in a fine apical pore, 8–10 × 2.5–3 μm (9.1 ± 0.6 × 2.7 ± 0.1); average length metula/phialide 1.06; conidia rough, subglobose to broadly ellipsoidal, 2.5–3.5 × 2.5–3 μm (2.96 ± 0.1 × 2.64 ± 0.2), average width/length = 0.89, n = 43.
Notes: Talaromyces yelensis is closely related to T. tratensis in section Islandici (Fig. 60). The latter species typically produces a sexual state with roughened ascospores and ellipsoidal smooth walled conidia. Talaromyces yelensis produces subglobose to broadly ellipsoidal conidia that have rough walls and lacks a sexual state.
Discussion
Phylogenetic species recognition
ITS is the most commonly sequenced gene for fungi and was recently accepted as the official DNA barcode (Schoch et al. 2012). Curated reference data sets are currently limited, with several publications addressing the issue (Kõljalg et al. 2005, Santamaria et al. 2012, Kõljalg et al. 2013, Schoch et al. 2014). With regards to Aspergillus, Penicillium and Talaromyces, the accepted species list endorsed by ICPA provides accession numbers of ITS barcodes to all ex-type strains. Although ITS does not distinguish among all species, with some species sharing identical sequences (Skouboe et al. 1999, Peterson 2000a,b, Samson et al. 2011), it does provide valuable information on sectional classification and often provides enough information for making a species identification. In order to compensate for the lack of variability in ITS, the ICPA list also include accession numbers for BenA and CaM sequences, meant to serve as secondary identification markers.
Our data shows that BenA works well for Penicillium and Talaromyces identifications, while CaM performs well in Aspergillus. However, some problems were experienced. Aspergillus steynii and A. elegans share identical CaM sequences (Fig. 2), something very uncommon in Aspergillus. In this case, these two species have unique ITS and BenA sequences.
In Penicillium, BenA has limitations in the P. chrysogenum (Fig. 23) and P. camembertii (Fig. 24) species complexes. Houbraken et al. (2012) reviewed section Chrysogena and distinguished several phylogenetically closely related species, and showed that different genes suggest different phylogenies. For example, although P. chrysogenum has unique BenA sequences, variation among strains makes distinguishing it from P. allii-sativii complicated. On the other hand, CaM does not distinguish well between P. chrysogenum and P. rubens (Houbraken et al. 2012), but BenA easily distinguishes the two. As such, a combination of the two genes is often required for identifying isolates within the clade. Another difficult clade is the P. camemberti complex. The ex-type cultures for P. commune, P. camemberti and P. caseifulvum have identical ITS, BenA and CaM sequences, as also reported by Giraud et al. (2009) for elongation factor-1α. The importance and different roles of these species in the cheese industry makes it unsatisfactory to synonymise them, and as a result the white sporulating P. camembertii are considered a domesticated form of P. commune or P. caseifulvum (green sporulating). A similar situation exists in Aspergillus, where A. oryzae is considered a domestic form of A. flavus (Varga et al. 2011). None of these problems were experienced for Talaromyces, where BenA worked very well for identifications.
In some cases, an ex-type sequence alone is insufficient reference data for making a conclusive identification, a reflection of intraspecific variation, for example in P. italicum (Fig. 24), P. sumatraense (Fig. 13) (Houbraken et al. 2011b) and in Talaromyces section Trachyspermi (Fig. 28) (Frisvad et al. 2013). As such, a verified reference data set that includes non-ex-type strains representing the sequence diversity within phylogenetically delineated species is the next crucial step for sequence-based identifications in these genera.
Fungi in house dust
Samson et al. (2010) and Flannigan et al. (2011) listed 100 fungal species common in indoor environments. From this list, we also found A. fumigatus, A. sydowii, P. brevicompactum and P. citrinum to be common in the collected house dust. Of significance is the effect of taxonomic revisions on this type of information. For example, A. versicolor used to be considered very common in indoor environments. However, it was recently shown to represent a species complex, with nine new species introduced (Jurjević et al. 2012). From our data, A. versicolor was still isolated from four different countries and A. creber was isolated in higher numbers from three countries. From unpublished data, we are also noting that most of the “A. versicolor” strains collected from indoor environments over many years in the DTO collection housed at CBS should now be identified as A. creber. Another example is Aspergillus section Circumdati. The ochratoxin producer A. westerdijkiae is reported to have a wide distribution indoors. From dust, we could only recover this species from Mexico and South Africa, whereas A. subramanianii was found in high numbers from four countries. Penicillium chrysogenum is also considered to have a worldwide distribution from indoor environments. However, after Houbraken et al. (2011a) reintroduced P. rubens as the name for the commercial penicillin producing strain closely related to P. chrysogenum, we are finding P. rubens to be very common indoors and not P. chrysogenum.
The origin of common indoor species is difficult to determine. Aspergillus sydowii is a good example. We found A. sydowii to be one of the most common species in collected dust samples and the species is generally considered as widespread. The species is often isolated from soil (Domsch et al. 1980), is very common on mouldy gypsum wallboard, dust, paint and various foods (Gorbushina et al. 2007, Samson et al. 2010, Flannigan et al. 2011) and is commonly found in marine environments where it acts as an opportunistic pathogen of sea corals (Roth et al. 1964, Smith et al. 1996, Geiser et al. 1998, Toledo-Hernández et al. 2008, Rypien et al. 2008, Rypien 2008, Kirkwood et al. 2009). The source or origin of this species is still unknown, even though most studies suggest it being a terrestrial soil-borne fungus. The suggestion thus is that A. sydowii, along with a number of other soil-borne fungi, gets carried into indoor environments. Its ability to grow in such a wide range of niches is intriguing and needs further studies.
Recent studies suggested that the indoor fungal communities as observed with metagenomic analyses exploiting next generation sequencing are mostly determined by the outdoor fungal communities (Adams et al. 2013a,b). In our study, the highest diversity was observed in countries that are also listed as biodiversity hotspots of the world (Myers et al. 2000). This might suggest that at least a considerable proportion of these species isolated from house dust originated from outdoors. However, the prevalence of specific species commonly isolated from indoor surveys suggests that the indoor environments do select for the growth of specific species. In addition, much of the metagenomics diversity may come from transient, dormant or dead spores.
From various indoor culture-independent surveys, it is apparent that the ITS database is not yet sufficient for identification of Aspergillus, Penicillium and Talaromyces (Amend et al. 2010, Adams et al. 2013a,b). These studies often cite Aspergillus sp. and Penicillium sp. as the most abundant. It would be valuable, even if species identification were not feasible, to identify to which clade or taxonomic section or series the sequences belong. The Last Common Ancestor (LCA) analysis commonly employed for identifying OTU's in metagenomic studies employ the GenBank taxonomic hierarchy to assign query sequences to taxonomic nodes. This hierarchy generally lacks ranks between genus and species, which means that the analysis suffers from a regrettable lack of precision for large genera, such as those studied here. In order to at least partly alleviate this kind of issue, our ITS barcodes of ex-type sequences and reference barcodes created from dust isolates will be uploaded into the UNITE database as part of a planned curated set on indoor moulds. As part of a future study, these reference sequences will be used for comparing d2e and 454-pyrosequencing data (Amend et al. 2010) in order to better understand the communities of Aspergillus, Penicillium and Talaromyces in indoor environments.
Acknowledgements
This research was supported by grants from the Alfred P. Sloan Foundation Program on the Microbiology of the Built Environment. We thank Toni Atkinson, Martin Bidartondo, Bryony Horton, Karin Jacobs, Wayne Law, Endang Rehayu, Health Canada, and many others who assisted with dust collection. Our thanks go out to Long Wang of Institute of Microbiology, Chinese Academy of Sciences, who sent us the ex-type strains of P. jianxiense and P. guizhouanum. We would like to acknowledge Uwe Braun who provided nomenclatural assistance for this manuscript and Neriman Yilmaz who assisted with descriptions of the new Talaromyces species.
Footnotes
Peer review under responsibility of CBS-KNAW Fungal Biodiversity Centre.
Contributor Information
K.A. Seifert, Email: keith.seifert@agr.gc.ca.
R.A. Samson, Email: r.samson@cbs.knaw.nl.
References
- Adams R.I., Miletto M., Taylor J.W. Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. The ISME Journal. 2013;7:1262–1273. doi: 10.1038/ismej.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R.I., Miletto M., Taylor J.W. The diversity and distribution of fungi on residential surfaces. PLoS One. 2013;8:e78866. doi: 10.1371/journal.pone.0078866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimanianda V., Bayry J., Bozza S. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460:1117–1121. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- Amend A.S., Seifert K.A., Samson R.A. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. PNAS. 2010;107:13748–13753. doi: 10.1073/pnas.1000454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen B., Frisvad J.C., Søndergaard I. Associations between fungal species and water-damaged building materials. Applied and Environmental Microbiology. 2011;77:4180–4188. doi: 10.1128/AEM.02513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian P.W., Curtis P.J., Hemming H.G. Wortmannin, an antibiotic produced by Penicillium wortmanni. Transactions of the British Mycological Society. 1957;40:365–368. [Google Scholar]
- Chunduri J. Indoor fungal populations inhabiting cement structures-remedial measures. IOSR Journal of Environmental Science, Toxicology and Food Technology. 2014;8:19–24. [Google Scholar]
- Codner R.C., Sargeant K., Yeo R. Production of aflatoxin by the culture of strains of Aspergillus flavus-oryzae on sterilized peanuts. Biotechnology and Bioengineering. 1963;5:185–192. [Google Scholar]
- Collado J., Platas G., Paulus B. High-throughput culturing of fungi from plant litter by a dilution-to-extinction technique. FEMS Microbiology Ecology. 2007;60:521–533. doi: 10.1111/j.1574-6941.2007.00294.x. [DOI] [PubMed] [Google Scholar]
- Devi S.S., Sreenivasulu Y., Rao K.V.B. Talaromyces verruculosus, a novel marine fungi as a potent polyhydroxybutyrate degrader. Research Journal of Pharmacy and Technology. 2014;7:433–438. [Google Scholar]
- Domsch K.H., Gams W., Anderson T.-H. IHW-Verlag; Eching: 1980. Compendium of Soil Fungi. [Google Scholar]
- Dufossé L., Fouillaud M., Caro Y. Filamentous fungi are large-scale producers of pigments and colorants for the food industry. Current Opinion in Biotechnology. 2014;26:56–61. doi: 10.1016/j.copbio.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Ezekiel C.N., Udom I.E., Frisvad J.C. Assessment of aflatoxigenic Aspergillus and other fungi in millet and sesame from Plateau State, Nigeria. Mycology. 2014;5:16–22. doi: 10.1080/21501203.2014.889769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannigan B., Samson R.A., Miller J.D. CRC Press; Boca Raton: 2011. Microorganisms in Home and Indoor Work Environments: Diversity, Health Impacts, Investigation and Control. [Google Scholar]
- Frisvad J.C., Frank J.M., Houbraken J. New ochratoxin A producing species of Aspergillus section Circumdati. Studies in Mycology. 2004;50:23–43. [Google Scholar]
- Frisvad J., Samson R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology. 2004;49:1–174. [Google Scholar]
- Frisvad J.C., Smedsgaard J., Larsen T.O. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Studies in Mycology. 2004;49:201–241. [Google Scholar]
- Frisvad J.C., Yilmaz N., Thrane U. Talaromyces atroroseus, a new species efficiently producing industrially relevant red pigments (SE Baker, Ed.) PLoS One. 2013;8:e84102. doi: 10.1371/journal.pone.0084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Hoshino T., Inoue H. Taxonomic revision of the cellulose-degrading fungus Acremonium cellulolyticus nomen nudum to Talaromyces based on phylogenetic analysis. FEMS Microbiology Letters. 2013;351:32–41. doi: 10.1111/1574-6968.12352. [DOI] [PubMed] [Google Scholar]
- Garber G. An overview of fungal infections. Drugs. 2001;61(Suppl. 1):1–12. doi: 10.2165/00003495-200161001-00001. [DOI] [PubMed] [Google Scholar]
- Geiser D.M., Taylor J.W., Ritchie K.B. Cause of sea fan death in the West Indies. Nature. 1998;394:137–138. [Google Scholar]
- Giraud F., Giraud T., Aguileta G. Microsatellite loci to recognize species for the cheese starter and contaminating strains associated with cheese manufacturing. International Journal of Food Microbiology. 2009;137:204–213. doi: 10.1016/j.ijfoodmicro.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Glass N.L., Donaldson G.C. Development of premier sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbushina A.A., Kort R., Schulte A. Life in Darwin's dust: intercontinental transport and survival of microbes in the nineteenth century. Environmental Microbiology. 2007;9:2911–2922. doi: 10.1111/j.1462-2920.2007.01461.x. [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Green B.J., Tovey E.R., Beezhold D.H. Surveillance of fungal allergic sensitization using the fluorescent halogen immunoassay. Journal de mycologie medicale. 2009;19:253–261. doi: 10.1016/j.mycmed.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean P., Weber R. Fungus balls of the paranasal sinuses: a review. European Archives of Oto-Rhino-Laryngology. 2007;264:461–470. doi: 10.1007/s00405-007-0281-5. [DOI] [PubMed] [Google Scholar]
- Höppe P., Martinac I. Indoor climate and air quality. Review of current and future topics in the field of ISB study group 10. International Journal of Biometeorology. 1998;42:1–7. doi: 10.1007/s004840050075. [DOI] [PubMed] [Google Scholar]
- Hong S.-B., Cho H.-S., Shin H.-D. Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology. 2006;56:477–486. doi: 10.1099/ijs.0.63980-0. [DOI] [PubMed] [Google Scholar]
- Horner W.E., Helbling A., Salvaggio J.E. Fungal allergens. Clinical Microbiology Reviews. 1995;8:161–179. doi: 10.1128/cmr.8.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoog G.S. de. Centraalbureau voor Schimmelcultures; Utrecht, Netherlands: 2000. Atlas of Clinical Fungi. [Google Scholar]
- Houbraken J., Due M., Varga J. Polyphasic taxonomy of Aspergillus section Usti. Studies in Mycology. 2007;59:107–128. doi: 10.3114/sim.2007.59.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Frisvad J.C., Samson R.A. Fleming's penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus. 2011;2:87–95. doi: 10.5598/imafungus.2011.02.01.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Frisvad J.C., Samson R.A. Taxonomy of Penicillium section Citrina. Studies in Mycology. 2011;70:53–138. doi: 10.3114/sim.2011.70.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Frisvad J.C., Seifert K.A. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia. 2012;29:78–100. doi: 10.3767/003158512X660571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Lopez-Quintero C.A., Frisvad J.C. Five new Penicillium species, P. araracuarense, P. elleniae, P. penarojense, P. vanderhammenii and P. wotroi, from Colombian leaf litter. International Journal of Systematic and Evolutionary Microbiology. 2010;61:1462–1475. doi: 10.1099/ijs.0.025098-0. [DOI] [PubMed] [Google Scholar]
- Houbraken J., Samson R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Visagie C.M., Seifert K.A. Review of Penicillium section Aspergilloides. Studies in Mycology. 2014;78:373–451. doi: 10.1016/j.simyco.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Vries R.P. de, Samson R.A. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Advances in Applied Microbiology. 2014;86:199–249. doi: 10.1016/B978-0-12-800262-9.00004-4. [DOI] [PubMed] [Google Scholar]
- Hubka V., Lyskova P., Frisvad J.C. Aspergillus pragensis sp. nov. discovered during molecular reidentification of clinical isolates belonging to Aspergillus section Candidi. Medical Mycology. 2014;52:565–576. doi: 10.1093/mmy/myu022. [DOI] [PubMed] [Google Scholar]
- Jaakkola J.J.K., Hwang B.-F., Jaakkola M.S. Home dampness and molds as determinants of allergic rhinitis in childhood: a 6-year, population-based cohort study. American Journal of Epidemiology. 2010;172:451–459. doi: 10.1093/aje/kwq110. [DOI] [PubMed] [Google Scholar]
- Jurjević Ž., Peterson S.W., Horn B.W. Aspergillus section Versicolores: nine new species and multilocus DNA sequence based phylogeny. IMA Fungus. 2012;3:59–79. doi: 10.5598/imafungus.2012.03.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanse O.S., Whitelaw-Weckert M., Kadam T.A. Phosphate solubilization by stress-tolerant soil fungus Talaromyces funiculosus SLS8 isolated from the Neem rhizosphere. Annals of Microbiology. 2014 http://dx.doi.org/10.1007/s13213-014-0839-6 In press. [Google Scholar]
- Karvala K., Toskala E., Luukkonen R. Prolonged exposure to damp and moldy workplaces and new-onset asthma. International Archives of Occupational and Environmental Health. 2011;84:713–721. doi: 10.1007/s00420-011-0677-9. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauserud H., Svegården I.B., Saetre G.-P. Asian origin and rapid global spread of the destructive dry rot fungus Serpula lacrymans. Molecular Ecology. 2007;16:3350–3360. doi: 10.1111/j.1365-294X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- Kirkwood M., Todd J.D., Rypien K.L. The opportunistic coral pathogen Aspergillus sydowii contains dddP and makes dimethyl sulfide from dimethylsulfoniopropionate. The ISME Journal. 2009;4:147–150. doi: 10.1038/ismej.2009.102. [DOI] [PubMed] [Google Scholar]
- Kõljalg U., Larsson K.-H., Abarenkov K. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytologist. 2005;166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- Kõljalg U., Nilsson R.H., Abarenkov K. Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Kong H.Z. A new Penicillium species isolated from Guizhou. Mycosystema. 2000;19:1–2. [Google Scholar]
- Kong H.Z., Liang Z.Q. A new Penicillium species isolated from Jiangxi, China. Mycosystema. 2003;22:4–5. [Google Scholar]
- Kornerup A., Wanscher J.H. Methuen & Co Ltd; Copenhagen: 1967. Methuen Handbook of Colour. [Google Scholar]
- Latgé J.-P. Aspergillus fumigatus and Aspergillosis. Clinical Microbiology Reviews. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.M., Horie Y., Wang Y. Three new Aspergillus species isolated from clinical sources as a causal agent of human aspergillosis. Mycoscience. 1998;39:299–305. [Google Scholar]
- Lin S.J., Schranz J., Teutsch S.M. Aspergillosis case-fatality rate: systematic review of the literature. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2001;32:358–366. doi: 10.1086/318483. [DOI] [PubMed] [Google Scholar]
- Lyratzopoulos G., Ellis M., Nerringer R. Invasive infection due to Penicillium species other than P. marneffei. Journal of Infection. 2002;45:184–195. doi: 10.1053/jinf.2002.1056. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Gershwin M.E. Sick building syndrome. III. Stachybotrys chartarum. The Journal of Asthma: Official Journal of the Association for the Care of Asthma. 2000;37:191–198. doi: 10.3109/02770900009055442. [DOI] [PubMed] [Google Scholar]
- Manoch L., Dethoup T., Yilmaz N. Two new Talaromyces species from soil in Thailand. Mycoscience. 2013;54:335–342. [Google Scholar]
- 59.McNeill J., Barrie F.F., Buck W.R., editors. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code) Koeltz Scientific Books; Königstein: 2012. [Regnum vegetabile no. 154.] [Google Scholar]
- McNeill J., Turland N.J. Major changes to the Code of Nomenclature-Melbourne, July 2011. Taxon. 2011;60:1495–1497. [Google Scholar]
- Myers N., Mittermeier R.A., Mittermeier C.G. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Novakova A., Hubka V., Saiz-Jimenez C. Aspergillus baeticus sp. nov. and Aspergillus thesauricus sp. nov., two species in section Usti from Spanish caves. International Journal of Systematic and Evolutionary Microbiology. 2012;62:2778–2785. doi: 10.1099/ijs.0.041004-0. [DOI] [PubMed] [Google Scholar]
- Okuda T., Klich M.A., Seifert K.A. Media and incubation effect on morphological characteristics of Penicillium and Aspergillus. In: Samson R.A., Pitt J.I., editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 83–99. [Google Scholar]
- Paden J.W. Three new species of Eupenicillium from soil. Mycopathologia et Mycologia applicata. 1971;43:259–268. doi: 10.1007/BF02051745. [DOI] [PubMed] [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences. In: Samson R.A., Pitt J.I., editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 163–178. [Google Scholar]
- Peterson S.W. Phylogenetic relationships in Aspergillus based on rDNA sequence analysis. In: Samson R.A., Pitt J.I., editors. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 323–355. [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.3852/mycologia.100.2.205. [DOI] [PubMed] [Google Scholar]
- Peterson S.W. Aspergillus and Penicillium identification using DNA sequences: barcode or MLST? Applied Microbiology and Biotechnology. 2012;95:339–344. doi: 10.1007/s00253-012-4165-2. [DOI] [PubMed] [Google Scholar]
- Peterson S.W., Jurjevic Z. Talaromyces columbinus sp. nov., and genealogical concordance analysis in Talaromyces Clade 2a. PLoS One. 2013;8:e78084. doi: 10.1371/journal.pone.0078084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pildain M.B., Frisvad J.C., Vaamonde G. Two novel aflatoxin-producing Aspergillus species from Argentinean peanuts. International Journal of Systematic and Evolutionary Microbiology. 2008;58:725–735. doi: 10.1099/ijs.0.65123-0. [DOI] [PubMed] [Google Scholar]
- Pitt J.I. Academic Press Inc; London: 1979. The Genus Penicillium and its Teleomorphic States Eupenicillium and Talaromyces. [Google Scholar]
- Pitt J.I., Hocking A.D. Springer; New York: 2009. Fungi and Food Spoilage. [Google Scholar]
- Raper K.B., Fennell D.I. Williams & Wilkins; Baltimore: 1965. The Genus Aspergillus. [Google Scholar]
- Rivera K.G., Díaz J., Chavarría-Díaz F. Penicillium mallochii and P. guanacastense, two new species isolated from Costa Rican caterpillars. Mycotaxon. 2012;119:315–328. [Google Scholar]
- Roth F.J., Jr., Orpurt P.A., Ahearn D.G. Occurrence and distribution of fungi in a subtropical marine environment. Canadian Journal of Botany. 1964;42:375–383. [Google Scholar]
- Rypien K.L. African dust is an unlikely source of Aspergillus sydowii, the causative agent of sea fan disease. Marine Ecology Progress Series. 2008;367:125–131. [Google Scholar]
- Rypien K.L., Andras J.P., Harvell C.D. Globally panmictic population structure in the opportunistic fungal pathogen Aspergillus sydowii. Molecular Ecology. 2008;17:4068–4078. doi: 10.1111/j.1365-294X.2008.03894.x. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Houbraken J., Thrane U. 2010. Food and Indoor Fungi. CBS Laboratory manual. [Google Scholar]
- Samson R.A., Visagie C.M., Houbraken J. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Yilmaz N., Houbraken J. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Studies in Mycology. 2011;70:159–183. doi: 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang H., An T., Kim C.S. Two novel Talaromyces species isolated from medicinal crops in Korea. Journal of Microbiology. 2013;51:704–708. doi: 10.1007/s12275-013-3361-9. [DOI] [PubMed] [Google Scholar]
- Santamaria M., Fosso B., Consiglio A. Reference databases for taxonomic assignment in metagenomics. Briefings in Bioinformatics. 2012;13:682–695. doi: 10.1093/bib/bbs036. [DOI] [PubMed] [Google Scholar]
- Schmidt O. Indoor wood-decay basidiomycetes: damage, causal fungi, physiology, identification and characterization, prevention and control. Mycological Progress. 2007;6:261–279. [Google Scholar]
- Schoch C.L., Robbertse B., Robert V. Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi. Database. 2014;2014 doi: 10.1093/database/bau061. http://dx.doi.org/10.1093/database/bau061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Seifert K.A., Huhndorf S. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. PNAS. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H.W., Boller R.A. Aflatoxin production of species and strains of the Aspergillus flavus group isolated from field crops. Applied Microbiology. 1973;25:885–889. doi: 10.1128/am.25.6.885-889.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouboe P., Frisvad J.C., Taylor J.W. Phylogenetic analysis of nucleotide sequences from the ITS region of terverticillate Penicillium species. Mycological Research. 1999;103:873–881. [Google Scholar]
- Smith G.W., Ives L.D., Nagelkerken I.A. Caribbean sea-fan mortalities. Nature. 1996;383:487. [Google Scholar]
- Straus D.C. Molds, mycotoxins, and sick building syndrome. Toxicology and Industrial Health. 2009;25:617–635. doi: 10.1177/0748233709348287. [DOI] [PubMed] [Google Scholar]
- Takahata Y., Hiruma M., Sugita T. A case of onychomycosis due to Aspergillus sydowii diagnosed using DNA sequence analysis. Mycoses. 2008;51:170–173. doi: 10.1111/j.1439-0507.2007.01458.x. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanney J.B., Seifert K.A. Rasamsonia pulvericola sp. nov., isolated from house dust. IMA Fungus. 2013;4:205–212. doi: 10.5598/imafungus.2013.04.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terr A.I. Sick building syndrome: is mould the cause? Medical Mycology. 2009;47(Suppl 1):S217–S222. doi: 10.1080/13693780802510216. [DOI] [PubMed] [Google Scholar]
- Toledo-Hernández C., Zuluaga-Montero A., Bones-González A. Fungi in healthy and diseased sea fans (Gorgonia ventalina): is Aspergillus sydowii always the pathogen? Coral Reefs. 2008;27:707–714. [Google Scholar]
- Varga J., Frisvad J.C., Samson R.A. Polyphasic taxonomy of Aspergillus section Candidi based on molecular, morphological and physiological data. Studies in Mycology. 2007;59:75–88. doi: 10.3114/sim.2007.59.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Frisvad J.C., Samson R.A. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Studies in Mycology. 2011;69:57–80. doi: 10.3114/sim.2011.69.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartivarian S.E., Anaissie E.J., Bodey G.P. Emerging fungal pathogens in immunocompromised patients: classification, diagnosis, and management. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 1993;17(Suppl 2):S487–S491. doi: 10.1093/clinids/17.supplement_2.s487. [DOI] [PubMed] [Google Scholar]
- Visagie C.M., Jacobs K. Three new additions to the genus Talaromyces isolated from Atlantis sandveld fynbos soils. Persoonia. 2012;28:14–24. doi: 10.3767/003158512X632455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Houbraken J., Frisvad J.C. Identification and nomenclature of the genus Penicillium. Studies in Mycology. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Houbraken J., Rodriques C. Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia. 2013;31:42–62. doi: 10.3767/003158513X667410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Llimona X., Vila J. Phylogenetic relationships and the newly discovered sexual state of Talaromyces flavovirens, comb. nov. Mycotaxon. 2012;122:399–411. [Google Scholar]
- Visagie C.M., Roets F., Jacobs K. A new species of Penicillium, P. ramulosum sp. nov., from the natural environment. Mycologia. 2009;101:888–895. doi: 10.3852/08-149. [DOI] [PubMed] [Google Scholar]
- Visagie C.M., Varga J., Houbraken J. Ochratoxin production and taxonomy of the yellow aspergilli (Aspergillus section Circumdati) Studies in Mycology. 2014;78:1–61. doi: 10.1016/j.simyco.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Wang L. Penicillium kongii, a new terverticillate species isolated from plant leaves in China. Mycologia. 2013;105:1547–1554. doi: 10.3852/13-022. [DOI] [PubMed] [Google Scholar]
- Wang L., Kong H.Z. Penicillium ellipsoideosporum, a new species isolated from China. Mycosystema. 2000;19:463–465. [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Shinsky T.J., editors. PCR Protocols: a Guide to Methods and Applications. Academic Press Inc; New York: 1990. pp. 315–322. [Google Scholar]
- Wilson S., Straus D. The presence of fungi associated with sick building syndrome in North American Zoological Institutions. Journal of Zoo and Wildlife Medicine. 2002;33:322–327. doi: 10.1638/1042-7260(2002)033[0322:TPOFAW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Yilmaz N., Houbraken J., Hoekstra E.S. Delimitation and characterisation of Talaromyces purpurogenus and related species. Persoonia. 2012;29:39–54. doi: 10.3767/003158512X659500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz N., Visagie C.M., Houbraken J. The polyphasic taxonomy of the genus Talaromyces. Studies in Mycology. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


