Abstract
Tumor contains small population of cancer stem cells (CSC) that are responsible for its maintenance and relapse. Analysis of these CSCs may lead to effective prognostic and therapeutic strategies for the treatment of cancer patients. We report here the identification of CSCs from human lung cancer cells using Aldefluor assay followed by fluorescence-activated cell sorting analysis. Isolated cancer cells with relatively high aldehyde dehydrogenase 1 (ALDH1) activity display in vitro features of CSCs, including capacities for proliferation, self-renewal, and differentiation, resistance to chemotherapy, and expressing CSC surface marker CD133. In vivo experiments show that the ALDH1-positive cells could generate tumors that recapitulate the heterogeneity of the parental cancer cells. Immunohistochemical analysis of 303 clinical specimens from three independent cohorts of lung cancer patients and controls show that expression of ALDH1 is positively correlated with the stage and grade of lung tumors and related to a poor prognosis for the patients with early-stage lung cancer. ALDH1 is therefore a lung tumor stem cell-associated marker. These findings offer an important new tool for the study of lung CSCs and provide a potential prognostic factor and therapeutic target for treatment of the patients with lung cancer.
Introduction
Non-small cell lung cancer (NSCLC) is the most lethal of all cancers in the United States and worldwide mainly because of the lack of effective systemic treatment and rapid development of resistance to chemotherapy (1). Development of effective therapeutics is urgently needed (2). Also needed is the ability to precisely identify early-stage lung cancer patients at high risk for recurrence, who would then benefit most from adjuvant therapies (2).
Accumulating evidence has proposed a model in which tumorigenesis is driven by cancer stem cells (CSC) that are derived from mutated adult stem cells (3). CSCs undergo self-renewal, recapitulate the phenotype of the tumor from which they were derived, develop into phenotypically diverse cancer cell populations, proliferate extensively, and drive both continued expansion of malignant cells and resistance to chemotherapy (3, 4). As is true in most human carcinogenesis, lung tumor growth and metastasis might be promoted by CSCs that are responsible for the aggressiveness of lung cancer (5–8). The existence of lung CSCs could explain why current treatments for lung cancer cannot consistently eradicate tumor cells and “have reached a therapeutic plateau,” because these therapies target the bulk of cancer and are unlikely to eliminate CSCs (3, 4). Furthermore, residual lung CSCs may regenerate a cancer cell population, leading to tumor relapse after therapy. As a result, although conventional chemotherapeutic drugs can shrink lung tumors, their effects are usually transient, and they often do not appreciably extend the life of patients (1–8). Therefore, analysis of molecular genetic aberrations that characterize lung CSCs would deepen our understanding of the tumor biology and development of lung tumorigenesis. Most importantly, the molecular genetic changes could be developed as a new diagnostic system for monitoring the patients’ prognosis and predicting the treatment response of lung cancer, offering the best opportunity to prevent its recurrence. Additionally, the CSC-related molecular genetic changes may enable the development of specific agents for eradicating tumor-maintaining CSCs and thus yield efficient therapeutic approaches to ultimately cure lung cancer.
The aldehyde dehydrogenase (ALDH) family is cytosolic isoenzyme responsible for oxidizing intracellular aldehydes, thus contributing to the oxidation of retinol to retinoic acid in early stem cell differentiation (9). Murine and human hematopoietic and neural stem cells have high ALDH activity (10–15). Class 1 of the ALDH family (ALDH1) is the isoform of ALDH that predominates in mammals (12, 13). Increased ALDH1 activity has been found in stem cell populations in human multiple myeloma, acute myeloid leukemia, and brain and breast cancers (10–17). Therefore, ALDH1 activity might be usable as a common marker for both normal and malignant stem cell populations (17). ALDH1 expression has been reported in some lung cancer cell lines (18), and the increased expression of ALDH1 could result from cigarette smoking and contribute to malignant transformation of lung cells (19). However, the stem cell-related function and clinical significance of the ALDH1 has not yet been investigated in lung cancer. If this is the case in human lung tumorigenesis, it may lead to new targets for developing effective prognostic and therapeutic strategies for treatment of this challenging malignancy.
In this current study, we first used the Aldefluor assay and fluorescence-activated cell sorting (FACS) analysis to isolate ALDH1-positive cells from human lung cancer cell lines. We then evaluated whether ALDH1-positive cells had stem cell-like characteristics. The ALDH1-positive cancer cells exhibited the important CSC properties: in vitro self-renewal, differentiation, and multidrug resistance capacities, expression of stem cell marker, in vivo tumor initiation, and occurrence of a heterogeneous population of cancer cells. Furthermore, by analyzing the expression of ALDH1 in 303 lung tissues from three different populations of patients with lung cancer, we found that relatively high ALDH1 protein levels were positively associated with stage and grade of the tumors and inversely related to the patients’ survival. Our data suggested that ALDH1 might be a lung tumor stem cell marker and a potential prognostic factor and therapeutic target for efficient treatment of lung cancer.
Results
NSCLC Cell Lines Display a Distinctive Fraction of ALDH1-Positive Cells
We used the Aldefluor assay followed by FACS analysis to assess the presence and size of the cell population with ALDH1 enzymatic activity in six human lung cancer cell lines. As shown in Fig. 1A, the NSCLC cell lines H460, H125, H322, and H358 had an average of 2% (2.16 ± 1.28; n = 4) ALDH1-positive cells ranging from 0.6% (H322) to 2.9% (H125) of gated cells. The cell lines H82 and H1299 had no ALDH enzymatic activity. Interestingly, these two cell lines are small cell lung cancer cell lines. We therefore focused on the four NSCLC cell lines, H460, H125, H322, and H358, in the following experiments.
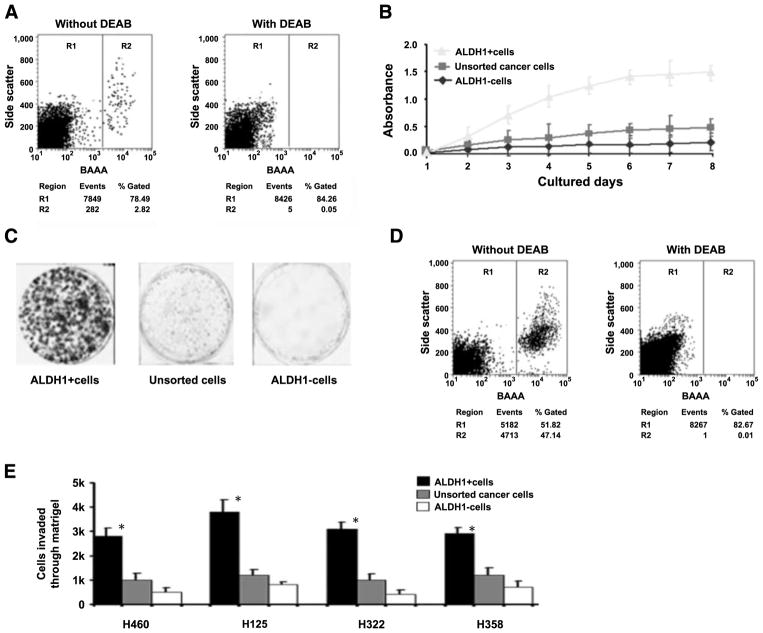
FIGURE 1.
ALDH1-positive lung cancer cells have tumor stem cell properties in vitro. A. FACS analysis of cancer cells using the Aldefluor assay, in which cells were incubated with ALDH substrate, BAAA, which was then converted by intracellular ALDH1 into a negatively charged reaction product BODIPY-aminoacetate. BODIPY-aminoacetate was retained inside cells positively expressing ALDH1, causing the cells to become brightly fluorescent. Brightly fluorescent ALDH1-expressing cells (ALDH1-positive cells) were detected in the green fluorescence channel by using flow cytometry. Cells incubated with BAAA and a specific inhibitor of ALDH, diethylaminobenzaldehyde, were used to establish the baseline fluorescence of these cells (R1) and to define the Aldefluor (ALDH1)-positive region (R2). FACS analysis was done on all cell lines and repeated three times. A. Result from H358 cell line. B. Cell growth curve of parental lung cancer cells and their corresponding ALDH1-positive and ALDH1-negative cancer cells. ALDH1-positive cancer cells grew more rapidly compared with parental and ALDH1-negative cancer cells. Experiments were undertaken on all NSCLC cell lines and repeated three times. B only showed the result from H358 cell line. C. Analysis of cell colony numbers in clonogenicity assays of ALDH1-positive and ALDH1-negative and unsorted cells. ALDH1-positive cell populations formatted larger and more colonies compared with unsorted and ALDH1-negative cells. Experiments were done on all cell lines and repeated three times. C showed the result from H358 cell line. D. Reanalyzing ALDH1-positive and ALDH1-negative cancer cells and measuring their differentiation ability by using the Aldefluor assay and FACS. ALDH1-positive cells (left) gave rise to 52% (52 ± 2.6%) ALDH1-positive population and 48% (48 ± 2.2%) ALDH1-negative cells, whereas ALDH1-negative cells (right) only produced ALDH1-negative cell population, implying that ALDH1-positive cells might undergo asymmetrical division to in vitro self-renew and generate heterogeneous cell populations of both high and low aggressive tumorigenic phenotypes. Experiments were undertaken on all cell lines and repeated three times, whereas D only showed the result from H358 cells. E. Matrigel invasion assay on sorted and unsorted cells. ALDH1-positive and ALDH1-negative lung cancer and unsorted cells (2 × 105) were seeded and incubated for 48 h. Columns, number of cells invaded across the membrane. ALDH1-positive cells were more invasive than ALDH1-negative cells and unsorted parental cancer cells. *, P < 0.05, t test, statistical significance. Each experiment was repeated three times. Points, mean of the independent experiments.
ALDH1-Positive Cells Feature Stem Cell-Like Characteristics In vitro
To examine whether ALDH1-positive cells exhibit increased in vitro growth and clonogenicity, we first cultured the parental NSCLC cells and their corresponding ALDH1-positive and ALDH1-negative cells for 2, 4, 6, and 8 days, respectively. ALDH1-positive cells grew more rapidly than the parental cells and ALDH1-negative cells did (Fig. 1B), indicating increased proliferative capacity of the ALDH1-positive cancer cells. We then examined the clonogenic potential of the different cell populations for their individual proliferative capacity by plating the cells and culturing at clonal density for 14 days. As shown in Fig. 1C, formation of large colonies was substantially greater among ALDH1-positive populations than it was among ALDH1-negative populations, providing further evidence that ALDH1-positive cells display increased proliferative capacity.
Furthermore, after culturing the different cell populations under the same condition, we reanalyzed the cells and measured their differentiation ability by using the Aldefluor assay and FACS analysis. ALDH1-positive cells gave rise to 52% (52 ± 2.6%) ALDH1-positive and 48% (48 ± 2.2%) ALDH1-negative cells (Fig. 1D). In contrast, the ALDH1-negative cells produced only ALDH1-negative cells and could not differentiate into ALDH1-positive cells. These results imply that ALDH1-positive cells might undergo asymmetrical division in vitro to self-renew and generate heterogeneous cell populations of both high and low aggressive tumorigenic phenotypes.
To investigate whether ALDH1-positive cells have greater invasiveness than ALDH1-negative and parental cancer cells do, we conducted the in vitro Matrigel invasion assay on sorted and unsorted NSCLC cells. The experiments were repeated three times. As shown in Fig. 1E, the ALDH1-positive cells were considerably more invasive than the ALDH1-negative cells and the unsorted parental cancer cells (all P < 0.05).
The results of all these investigations suggest that ALDH1-positive lung cancer cells possess CSC-like in vitro properties.
ALDH1-Positive Cells Express a Distinct Stem Cell Marker
To determine whether the ALDH1-positive cells expressed distinct stem cell molecular markers, we tested CD133 antibody, a previously proven lung CSC marker (20), on ALDH1-positive and ALDH1-negative populations. Immunofluorescence analysis revealed that the cytoplasm of ALDH1-positive cells had strong immunoreactivity for CD133 relative to that of ALDH1-negative cells (Supplementary Fig. S1). Sixty-four percent (64 ± 2.9%) of ALDH1-positive cells were positively immunoreactive to CD133, but only 1.2% (1.2 ± 0.6%) of ALDH1-negative cells were positively immunoreactive to CD133 (P < 0.05). This implies that ALDH1-positive cells derived from lung cancer cells are enriched in CSCs.
ALDH1-Positive Cells Are Highly Resistant to Chemo-therapeutic Drugs
Resistance to chemotherapy is another important feature of CSCs. To evaluate whether ALDH1-positive cells showed heightened resistance to the chemotherapeutic agents commonly used in the clinic as first-line therapy for lung cancer, we performed sensitivity assays with those drugs on ALDH1-positive and ALDH1-negative cells. The ALDH1-positive cells displayed greater resistance to the chemotherapeutic drugs than the ALDH1-negative cells did (Supplementary Fig. S2). This finding is in line with the poor therapeutic effect that conventional chemotherapeutic agents have in lung cancer patients, therefore suggesting that ALDH1-positive cells may represent lung CSCs.
Tumor Formation and Self-Renewal of ALDH1-Positive Cancer Cells In vivo
To determine whether ALDH1-positive lung cancer cells were capable of tumor initiation in vivo, we s.c. implanted ALDH1-positive and ALDH1-negative H358 and H126 cells into flanks of the nude mice, respectively. After 4 weeks, the dose of 1 × 103 ALDH1-positive lung cancer cells implanted into five mice yielded tumors with diameters of 10 ± 2.3 mm3 in all mice, whereas the same dose of ALDH1-negative cells did not produce tumors in any mice. Furthermore, the dose of 1 × 105 ALDH1-H125 positive lung cancer cells yielded much larger tumors in all five mice, with diameters of 30 ± 2.9 mm3, whereas the same dose of ALDH1-negative H125 cells generated a small tumor mass (5 mm3) in only one of the five mice (Fig. 2A). Similarly, the dose of 1 × 105 ALDH1-positive lung cancer cells created much larger tumors in all five mice, with diameters of 36 ± 3.2 mm3, whereas the same dose of ALDH1-negative H358 cells generated a small tumor mass (4 mm3) in only one of the five mice.
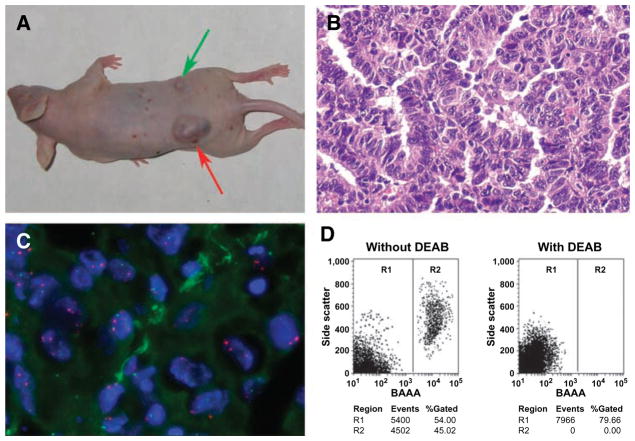
FIGURE 2.
ALDH1-positive lung cancer cells have tumor stem cell properties in vivo. A. Tumor formation ability of ALDH1-positive cells was greater than that of ALDH1-negative cancer cells. ALDH1-positive and ALDH1-negative H125 cells were implanted into flanks of nude mice. After 4 weeks, the dose of 1 × 105 ALDH1-positive lung cancer cells yielded larger tumors (red arrow) in all five mice with diameters of 30 ± 2.9 mm3, whereas the same dose of ALDH1-negative H125 cell only generated a small tumor mass (5 mm3; green arrow) in only one of five mice. Animal experiments were done by using cell lines H358 and H125 and repeated three times. A only showed the result from H125 cell lines. B. Histopathologic examination of the engrafted tumors formed by ALDH1-positive H125 cancer cells revealed a highly cellular mass with characteristics of adenocarcinoma of lung. C. Dual-color FISH analysis of xenograft tumor cells generated from ALDH1-positive H125 lung cancer cells with a probe (green signal) for FHIT gene and a probe (red signal) for chromosome 3 centromeric region. The majority of cells in tumor cells bore deletion of FHIT indicated by fewer green signals than red ones. D. Reanalyzing cells of engrafted tumors generated from ALDH1-positive and ALDH1-negative cancer cells and measuring their differentiation ability by using the Aldefluor assay and FACS. Cells from the tumor produced by ALDH1-positive H125 cancer cells (left) gave rise to 46% (46 ± 3.1%) ALDH1-positive population and 54% (54 ± 3.3%) ALDH1-negative cells, whereas cells from the tumor formed by ALDH1-negative H125 cells (right) only produced ALDH1-negative cell population, implying that ALDH1-positive cells might undergo asymmetrical division to in vivo self-renew and generate heterogeneous cell populations of both high and low aggressive tumorigenic phenotypes. Aldefluor assay and FACS experiments were undertaken on disaggregated cells of tumor engrafts from H358 and H125 cell lines and repeated three times, respectively, whereas D only illustrated the result from H125 cells.
Histopathologic examination revealed a highly cellular mass with characteristics of adenocarcinoma of the lung in the engrafted tumors from the ALDH1-positive H125 cancer cells (Fig. 2B). Further, fluorescence in situ hybridization (FISH) analysis with specific genetic probes exhibited deletion of the FHIF gene (Fig. 2C), one of the most frequently seen genetic characteristics of lung cancer cells, in the tumor masses generated from the ALDH1-positive lung cancer cells.
To elucidate whether ALDH1-positive cancer cell could self-renew and generate lung tumors with heterogeneity in vivo, Aldefluor assay and FACS analysis of the tumor engrafts were conducted after cell disassociation. The disaggregated cells of the tumors generated from the ALDH1-positive cells gave rise to an average of 46% (46 ± 3.1%) ALDH1-positive cells and an average of 54% (54 ± 3.3%) ALDH1-negative cells (Fig. 2D). However, the disassociated cells from the tumor of the ALDH1-negative cancer cells showed only ALDH1-negative populations.
Altogether, our in vivo data strongly suggest that, in contrast to the ALDH1-negative tumor cells, the ALDH1-positive lung cancer cells show unique features of cancer stem-like cells, including initiation of tumorigenesis and active tumor cell proliferation with multipotent differentiation potential.
ALDH1 Overexpression Is Associated with Aggressive Biological Behavior of NSCLC
To assess whether ALDH1 up-regulation is involved in the aggressive biological behavior of NSCLC, we first analyzed the expression of ALDH1 by using immunohistochemical analysis of freshly collected transbronchial needle aspiration (TBNA) specimens. There were 18 of 60 lung cancer samples showing positive staining for ALDH1 (Fig. 3A; Table 1). This ALDH1 overexpression was statistically significantly related to clinical stage of the patients’ disease (P = 0.02). We then examined ALDH1 expression in lung tissue microarrays obtained from a cohort of 59 patients with different clinical stages and histologic types and grades of NSCLC. Consistent with the results of our evaluations of the TBNA specimens, ALDH1 protein expression was found in cytoplasm and confined mainly to the tumor cells (Fig. 3B). Overexpression of ALDH1 occurred in 17 of 59 (29%), lung tumors occurred in 8 of 30 (27%) adenocarcinomas, and 8 of 25 (32%) CSCs, and 1 of 4 (25%) large cell carcinomas of the lung. The results from these two independent sets of clinical samples suggested that the high ALDH1 expression significantly correlated with histologic grade and clinical stage of those patients with the NSCLCs (all P < 0.05).
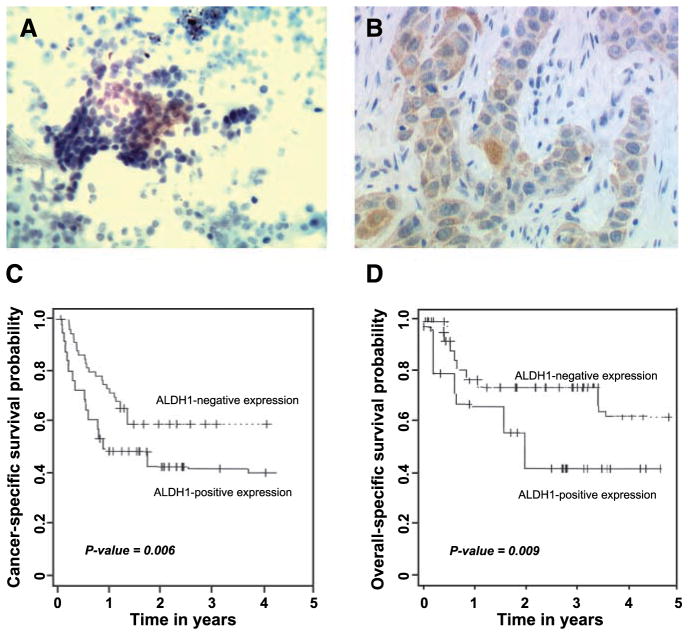
FIGURE 3.
ALDH1 overexpression is associated with aggressive biological behavior of NSCLC and a poor prognosis for patients with NSCLC. A. Analysis of TBNA specimen obtained from a lung tumor mass from a stage IV NSCLC patient by using immunohistochemistry showed positive ALDH1 expression. B. immunohistochemical analysis of a paraffin-embedded lung tumor tissue of a patient diagnosed with stage IV lung adenocarcinoma showed that overexpression of ALDH1 occurred predominately in cytoplasm of cancer cells. C. Probability of cancer-specific survival by levels of ALDH1 expression in stage I NSCLC. D. Probability of overall survival by levels of ALDH1expression in stage I NSCLC. Kaplan-Meier method was used to determine the survival probability, and log-rank test was used to compare the survival curves between groups.
Table 1.
Immunohistochemical Staining of ALDH1 on TBNA Samples According to Clinicopathologic Characteristics
| Characteristics | No. patients | ALDH1-positive expression | P* |
|---|---|---|---|
| All cases | 96 | ||
| Age (y) | 0.90 | ||
| ≤55 | 37 | 7 | |
| >55 | 59 | 11 | |
| Gender | 0.80 | ||
| Female | 40 | 8 | |
| Male | 56 | 10 | |
| Diagnosis | |||
| Normal | 36 | 0 | |
| NSCLC | 60 | 18 | 0.02 |
| Stage I | 9 | 2 | |
| Stage II | 18 | 5 | |
| Stage III | 16 | 5 | |
| Stage IV | 17 | 6 | |
| Differentiation of NSCLC | 0.01 | ||
| Well | 18 | 3 | |
| Moderate | 22 | 6 | |
| Poor | 20 | 9 | |
NOTE: Number (%) of patients unless otherwise indicated.
All P values were determined by two-sided tests. P values ≤ 0.05 were considered statistically significant.
Furthermore, staining with CD133 antibody for the immunohistochemical analysis of the tissue microarrays showed that a subset of ALDH1-positive cancers cells expressed CD133. Overall, 10 of the 17 (60%) cancer specimens that showed ALDH1-positive expression also showed positive expression for CD133, whereas none of the samples that had ALDH1-negative expression showed CD133 expression. In addition, all of the CD133-positve samples were also positive for ALDH1 staining. These data confirm our observation in our in vitro cell culture experiments: ALDH1-positive cells might be rich with lung CSCs.
ALDH1 Overexpression Is Associated with a Poor Prognosis for Patients with Early-Stage NSCLC
To assess the potential use of ALDH1 as a prognostic marker in early-stage lung tumors, we analyzed the relationship between ALDH1 expression and clinical outcomes of patients with early NSCLC by using the lung tissue microarrays consisting of stage I NSCLC tissues. Although ALDH1-positive cells were found infrequently (19%) in the specimens of the early-stage lung cancer specimens, the probabilities of overall survival at 5 years after surgery were 0.32 (95% confidence interval, 0.26–0.49) for patients whose tumors had positive ALDH1 expression compared with 0.72 (95% confidence interval, 0.66–0.82) for those whose tumors had no ALDH1 expression. Also, the probabilities of cancer-specific survival were 0.62 (95% confidence interval, 0.46–0.83) and 0.96 (95% confidence interval, 0.88–0.98) for patients whose tumors showed positive and negative ALDH1 expression, respectively. These data suggest that the cancer and overall-specific survival probabilities were significantly different between the two groups (P = 0.006 and 0.009, respectively, log-rank test; Fig. 3C and D).
Moreover, by performing multivariate analysis (age, sex, histologic type of lung cancer, and patient’s smoking status) using the Cox model, we found that overexpression of ALDH1 was an independent predictor of disease-free and cancer-specific survival (P = 0.019 and 0.016, respectively, log-rank test) from among the clinical and histopathologic variables tested.
Collectively, the observation that relatively high ALDH1 expression was statistically significantly associated with a more advanced pathologic grade and stage of disease and with a poor clinical outcome of lung cancers suggests that ALDH1 plays an important role in the progression of lung cancer. Detection of ALDH1 protein expression could be usable as a prognostic biomarker for the disease.
Discussion
In this study, by using Aldefluor assay for detecting ALDH1 expression followed by standard FACS analysis, we successfully isolated ALDH1-positive cells from lung cancer cells. We also showed that ALDH1-positive cells exhibited properties consistent with those of tumor stem cells, including high capacities for proliferation, self-renewal, resistance to chemotherapy in vitro, and expression of the CSC surface marker CD133. Furthermore, our in vivo experiments showed that ALDH1-positive cells could generate tumors that recapitulated the heterogeneity of lung cancer cells. More importantly, ALDH1 overexpression was positively associated with aggressive biological behavior of NSCLC and a poor prognosis for patients with early-stage NSCLC.
CSCs and normal stem cells express similar surface markers, by which they may be identified and characterized (3). However, the rigorous identification and isolation of tissue-specific CSCs has thus far been accomplished in only a few human organ systems (21). Furthermore, although some potential CSC markers have been developed in animal (e.g., Sca-1, a cell surface marker for bronchioalveolar stem cells in mice), there is no functional human homologue for them (8). CD133 could be used with exclusion of CD24, CD31, CD34, CD44, CD45, CD90, and CD113 for isolating human lung CSCs (17, 21). However, several other cell populations have been identified that share the same marker phenotype, including circulating fibrocytes (17, 21), thus limiting its application in enriching lung CSCs (6). A Hoechst 33342 efflux assay in combination with flow cytometry has been used to identify and isolate “side populations,” which might be enriched in cancer stem-like cells (17). Although the assay shows high efficiency in isolating side populations, the toxicity associated with the Hoechst 33342 dye create bias by selectively injuring non-side population cells that might affect subsequent functional stem cell assays in vitro and in vivo (22). Furthermore, several recent reports even suggested that side populations in some tumors might not represent CSCs (23–25).
Our findings showed that using an Aldefluor-FACS assay could successfully separate ALDH1-positive cells. Because ALDH1-positive cells are viable and selectable based on their fluorescence, they are readily available for stem cell studies both in vitro and in vivo, thus avoiding the toxicity associated with Hoechst 33342 on the isolated cells. Furthermore, the Aldefluor assay can detect ALDH1 activity rather than cell surface phenotype and hence overcomes the necessity to identify specific cell surface markers that discriminate stem cells from their differentiated progeny, the expression of which may also be dynamic and influenced by the local microenvironment (17). Therefore, unlike the previously described CSC phenotype, which might require the use of a combination of several surface antigens (26), the Aldefluor-FACS would be a simple and effective approach for defining and isolating an enriched lung CSC population from cancer cell lines and could be potentially amenable to clinical applications. We are currently developing a protocol using the Aldefluor-FACS assay that could be applied to surgically obtained human tissues for isolating lung CSCs.
There are following pieces of evidence from this study supporting that the ALDH1-positive cancer cells might be enriched in lung tumor-stem-like cells. First, the tumor stem cells should have properties of self-renewal, proliferation, and recapitulating the phenotype of the tumor from which they were derived. Our in vitro experiments revealed that ALDH1-positive cancer cells grew faster and had higher clone formation efficiency than did ALDH1-negative cancer cells. Furthermore, in vivo experiments using mice showed that only 1 × 103 ALDH1-positive cancer cells were required to form tumors, whereas at least 1 × 105 ALDH1-negative cancer cells were necessary, indicating that the tumor formation ability of ALDH1-positive cells was dramatically greater than that in the latter. In addition, the H&E staining and FISH analysis of engrafted tumors illustrated histopathologic and genetic patterns similar to those of the primary lung cancer cells, implying that the ability of the ALDH1-positive population in vivo could recapitulate the characteristics of the tumor subtype. Second, asymmetrical cell division is an important characteristic of CSCs because it requires one division for self-renewal and differentiation and further generates additional CSCs and phenotypically diverse cancer cells (27). Our in vitro tests showed that the ALDH1-positive cancer cells rather than ALDH1-negative cancer cells could undergo asymmetrical cell division and differentiate into both ALDH1-positive and ALDH1-negative cells, indicating their multipotent differentiation potential. The asymmetrical cell division of ALDH1-positive cells was also confirmed in mice: the dissociated cells of the engraftments generated from ALDH1-positive cancer cells displayed an average of 46% ALDH1-positive cells and an average of 54% ALDH1-negative cells. The data further showed that ALDH1-positive cells could give rise to a heterogeneous property of lung tumor. Third, according to the tumor stem cell theory, cancer can survive chemotherapy and relapse because of the resident CSCs that are drug resistant and can repopulate the tumor even when the bulk of nontumorigenic cells are killed (28–30). In the chemosensitivity assay, we found that ALDH1-positive cancer cells were more tolerant to common chemotherapeutic agent than ALDH1-negative populations, indicating that ALDH1-positive cancer cells were widely resistant to chemotherapy. Finally, we showed that the ALDH1-positive cells shared the stem cell marker, CD133, to a great degree in vitro and in vivo.
Taken together, these observations indicate that ALDH1-positive cells could fulfill the criteria that are sufficient to indicate the persistence of a malignant stem cell pattern; therefore, the ALDH1-positive lung cancer cells might be potential CSCs of lung tumorigenesis.
In addition to its important applications for better understanding the molecular mechanisms of carcinogenesis, the CSC hypothesis has fundamental clinical implications for cancer risk assessment, prognostication, and development of potential therapeutics for human malignancies. We here showed that ALDH1 overexpression occurred in the TBNA samples from lung cancer patients and was significantly associated with the clinical stage of the disease, although the ALDH1-positive cells represented a relatively small population. This observation was consistent with the notion that CSCs constituted a minority of the tumor population. Furthermore, when tested in two large independent cohorts of archived lung tissue specimens, ALDH1 overexpression was positively correlated with the stage and grade of NSCLCs. In addition, we found that ALDH1 expression inversely correlated with survival of the patients with stage I NSCLC. Moreover, ALDH1 could function as a prognostic factor for predicting outcome in patients with early-stage lung cancer. Therefore, our data imply that elevated expression of ALDH1 could contribute to lung cancer development and progression and that detection of the ALDH1 aberrations might be a useful biomarker for identifying a poor prognosis in patients with NSCLC. ALDH1 may also provide a therapeutic target for developing specific agents for eradiating lung CSCs and thus could potentially yield efficient therapeutic approaches to cure human NSCLCs.
In summary, using in vitro and in vivo experimental systems, we showed that Aldefluor assay followed by FACS analysis might provide a useful approach for isolating lung cancer cells with increased ALDH1. These ALDH1-positive cancer cells are endowed with extensive proliferation and self-renewal potential, being able to generate tumors that recapitulate the heterogeneity of the parental lung tumor cells, and therefore have stem cell properties. This assay for ALDH1 is readily available and thus could be added to the currently available panel of diagnostic markers used to improve the accuracy of clinical outcome predictions and choice of appropriate therapy. ALDH1 would also facilitate the development of an effective therapeutic target for NSCLC cancer. Nevertheless, further molecular biologic analysis of ALDH1 and a longitudinal clinical study in large population to validate its prognostic value to develop a novel strategy for improving treatment efficiencies of lung cancer will be needed.
Materials and Methods
Cell Lines and Cultures
Six human lung cancer cell lines H460, H125, H322, H358, H82, and H889 were obtained from the American Type Culture Collection. The cells were maintained in the culture medium recommended by the American Type Culture Collection and harvested by using treatment with 0.25% trypsin (Invitrogen) when they were in the logarithmic phase of growth for use in the following experiments.
Aldefluor Assay and Separation of the ALDH1-Positive Cell Population by FACS Analysis
To isolate an Aldefluor-stained cell population with ALDH1 enzymatic activity, we used the Aldefluor kit (Stem Cell Technologies), which is designed for optimal identification and isolation of stem cells through specific interaction with human ALDH1 (15). The experiments were undertaken according to the manufacturer’s instruction. Briefly, cells were suspended in Aldefluor assay buffer containing uncharged ALDH1-substrate, BODIPY-aminoacetaldehyde (BAAA), and incubated for 40 min at 37°C. BAAA was taken up by living cells through passive diffusion and then converted by intracellular ALDH into a negatively charged reaction product BODIPY-aminoacetate, which was retained inside cells expressing high levels of ALDH1, causing the cells to become brightly fluorescent. The brightly fluorescent ALDH1-expressing cells (ALDH1-positive cells) were detected in the green fluorescence channel (520–540 nm) of a FACScan instrument (BD Biosciences). A set of cells was stained using the identical conditions with the specific ALDH inhibitor, diethylaminobenzaldehyde (Sigma), to serve as a negative control for each experiment. Propidium iodide (Sigma) fluorescence was detected using the orange fluorescence channel. Because only cells with an intact cellular membrane could retain the Aldefluor reaction product, only viable ALDH1-positive cells were identified. Cells incubated with BAAA and diethylaminobenzaldehyde were used to establish the baseline fluorescence of these cells (R1) and to define the (ALDH1)-positive region (R2). Incubation of cells with the substrate in the absence of diethylaminobenzaldehyde induced a shift in BAAA fluorescence defining the Aldefluor-positive population. Data were analyzed by using Cell Quest software (BD Biosciences). Each experiment was repeated three times.
Cell Proliferation Assay
Parental cells and sorted ALDH1-positive and ALDH1-negative cells were plated in six-well plates (1 × 104 per well) in growth medium supplemented with fibroblast growth factor (Sigma) and epidermal growth factor (Sigma). One well every week was used for cell counting by using trypan blue exclusion. To determine their in vitro self-renewal ability, a single cell per well was seeded in 96-well plates. After seeding, the single cell-containing wells were identified and analyzed for the ability of the cells to generate spheres. Each experiment was repeated three times.
Cell Invasion Assay
Cellular potential for invasiveness was determined using Matrigel invasion chambers (BD Biosciences Discovery Labware) according to the manufacturer’s instruction. Briefly, cells were seeded into the upper chambers at 2 × 105 per chamber in serum-free DMEM. The outer chambers were filled with the same medium but containing fetal bovine serum as the chemoattractant. Cells were incubated for 48 h, and the noninvading cells were removed by swabbing the top layer of Matrigel. The membranes containing the invasive cells were stained with hematoxylin and mounted on slides, and the entire membrane with invading cells was counted on light microscopy. Each experiment was repeated three times.
Immunofluorescence Assay
Sorted ALDH1-positive and ALDH1-negative cells were grown on glass coverslips for 48 h and fixed in 4% paraformaldehyde (Sigma). Immunofluorescence staining was done as described previously (31) with a primary antibody against CD133 (Miltenyi Biotec). Finally, cells were examined and imaged under a Leica microscope (Leica Microsystems) equipped with Hamamatsu 9792-95 digital camera (Hamamatsu).
Drug Sensitivity Assay
The lung cancer cells were seeded in 96-well plates at 100 μL/well. After 24 h recovery, six chemotherapeutic drugs, cisplatin, gemcitabine, doxorubicin, daunorubicin, vinorelbine, and docetaxel, were added at their 50% inhibitory concentrations to both sorted and unsorted cells. The cells were then incubated for another 24 h. Sensitivity was determined using a colorimetric 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (Sigma) and inner salt cell proliferation assay (Promega) according to the manufacturer’s protocol. Absorbance was measured and drug resistance was calculated as described previously (32).
Tumor Cell Implantation Experiments
Five mice per group of athymic Swiss nu/nu/Ncr nu (nu/nu) mice were s.c. inoculated with ALDH1-positive and ALDH1-negative H358 and H125 cells at doses of 1 × 103 and 1 × 105, respectively. The mice were observed for 4 weeks to allow tumor growth and then euthanized under deep anesthesia with pentobarbital (Sigma). The tumors were surgically removed and their volume was calculated by using formula: 0.52 × length × width2. A portion of the tumor tissues was fixed in 10% formaldehyde and embedded in paraffin. The embedded tissues were stained with H&E for histopathologic examination and FISH for genetic characterization. Disassociated cells were obtained from the rest of the tumor grafts (33) and then were reanalyzed by using Aldefluor assay and FACS analysis.
FISH Analysis
A specific probe for FHIT, a tumor suppressor gene that is commonly deleted in lung cancer (34), was labeled with green fluorescent dye and tested by using FISH on the tissue sections prepared from the tumor xenografts as described previously (34). Briefly, the centromeric probe for chromosome 3 (Vysis) was labeled with red fluorescent dye and used as the internal control probe. The centromeric probe for chromosome 3 probe was mixed with the FHIT probe in hybridization buffer (Vysis). After hybridization for overnight, the slides were then examined using a Leica microscope (Leica Microsystems).
Clinical Specimens and Immunohistochemical Analysis
To determine correlation between ALDH1 expression and clinicopathologic features in NSCLCs, we first prospectively collected TBNA samples through bronchoscopy from 96 patients including 36 cancer-free and 60 lung cancer patients (Table 1). Cytocentrifuge slides were made from each TBNA sample and then fixed in 4% paraformaldehyde as described in our previous work (35). Second, we obtained tissue microarrays (Express Biotech) constructed from surgically resected lung tissues of 59 patients with different stages and histologic types of NSCLC (32). Furthermore, to evaluate the associations between ALDH1 expression and the outcomes of NSCLC patients, we used lung tissue microarrays consisting of tumor specimens obtained from 148 patients with stage I NSCLC, for whom we had complete medical records and follow-up data (36).
Immunohistochemical staining was done on the biopsy samples and tissue microarrays by using rabbit polyclonal antibody against ALDH1 (Santa Cruz Biotechnology) and 3,3′-diaminobenzidine (Santa Cruz Biotechnology) as chromogens. We also performed immunohistochemical analysis with antibody against CD133 (Miltenyi Biotec) on the commercial tissue microarray slides. Each batch of slides contained a positive control and a negative control. The immunoreactive score of the samples for ALDH1 was determined based on previously published criteria (15, 19). Briefly, scoring of the ALDH1 staining was done independently by two experienced investigators who examined the samples by light microscopy and in a blinded manner with respect to the clinical information of the subjects. Staining intensity was rated according to the following scale: 0 = no visible staining, 1 = faint staining, 2 = moderate staining, and 3 = strong staining. Percentage of cells with positive ALDH1 was graded as 0%, <10%, 10% to 25%, 25% to 50%, and 50% to 75% or higher. Considering 0.6 mm diameter tissue core biopsy specimens on tissue microarrays, we reviewed entire area of each tissue spot under high power (×400). To compare all of the available data, an overall score was assigned to each case by multiplying the intensity score by the mean percentage of cells staining. Cutoff points were chosen to categorize samples as ALDH1 positive or negative in term of the overall score. The sample had ≤10% of cells for ALDH1 expression with faint staining (the overall score was ≤10%) was regarded as a negative ALDH1 specimen. The sample had >10% of cells for ALDH1 expression with faint or higher staining (the overall score was >10%) was regarded as a positive ALDH1 one. The immunoreactive score of the samples for CD133 was determined according to previously published criteria (37).
Statistical Analysis
The statistical software SPSS12.0 (SPSS) was used to analyze the differences between ALDH1-positive and ALDH1-negative cells with unpaired or paired t tests for statistical significance. P values < 0.05 were considered significant. To evaluate the association between ALDH1 expression and histopathologic characteristics of lung tumors and clinical stages and outcomes of the patients, survival curves were calculated by using the product-limit estimate (Kaplan-Meier method) and the curves were examined by using the log-rank test. Univariate and multivariate analyses were also done using Cox’s proportional hazards model to determine which factors might have significant influence on the patients’ survival.
Supplementary Material
Acknowledgments
Grant support: National Cancer Institute grants CA-126710, CA-CA137742, and CA-133956, Wendy Will Case Cancer Award, The University of Maryland Statewide Health Network award, seed money from the American Cancer Society Institutional Research Grant, Associate Member Award from National Cancer Institute-The Early Detection Research Network, Flight Attendant Medical Research Institute clinical innovator award, and Department of Pathology of University of Maryland School of Medicine intramural award (F. Jiang).
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.American Cancer Society. Cancer facts and figures 2007–2008. [Google Scholar]
- 2.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49– 52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 3.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;1:105– 11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 4.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;12:1253– 61. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 5.Berns A. Stem cells for lung cancer? Cell. 2005;121:811– 3. doi: 10.1016/j.cell.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Kim CF. Paving the road for lung stem cell biology: bronchioalveolar stem cells and other putative distal lung stem cells. Am J Physiol Lung Cell Mol Physiol. 2007;293:1092– 8. doi: 10.1152/ajplung.00015.2007. [DOI] [PubMed] [Google Scholar]
- 7.Giangreco A, Groot KR, Janes SM. Lung cancer and lung stem cells: strange bedfellows? Am J Respir Crit Care Med. 2007;175:547– 53. doi: 10.1164/rccm.200607-984PP. [DOI] [PubMed] [Google Scholar]
- 8.Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823– 35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida A, Hsu LC, Davé V. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme. 1992;46:239– 44. doi: 10.1159/000468794. [DOI] [PubMed] [Google Scholar]
- 10.Armstrong L, Stojkovic M, Dimmick I, et al. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22:1142– 51. doi: 10.1634/stemcells.2004-0170. [DOI] [PubMed] [Google Scholar]
- 11.Chute JP, Muramoto GG, Whitesides J, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707– 12. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hess DA, Craft TP, Wirthlin L, et al. Widespread non-hematopoietic tissue distribution by transplanted human progenitor cells with high aldehyde dehydrogenase activity. Stem Cells. 2006;26:611– 20. doi: 10.1634/stemcells.2007-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hess DA, Wirthlin L, Craft TP, et al. Selection based on CD133 and high aldehyde dehydrogenase activity isolates long-term reconstituting human hematopoietic stem cells. Blood. 2006;107:2162– 9. doi: 10.1182/blood-2005-06-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearce DJ, Taussig D, Simpson C, et al. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;6:752– 60. doi: 10.1634/stemcells.2004-0292. [DOI] [PubMed] [Google Scholar]
- 15.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;15:555– 67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldmann G, Dhara S, Fendrich V, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187– 96. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balicki D. Moving forward in human mammary stem cell biology and breast cancer prognostication using ALDH1. Cell Stem Cell. 2007;15:485– 7. doi: 10.1016/j.stem.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Sreerama L, Sladek NE. Class 1 and class 3 aldehyde dehydrogenase levels in the human tumor cell lines currently used by the National Cancer Institute to screen for potentially useful antitumor agents. Adv Exp Med Biol. 1997;414:81– 94. doi: 10.1007/978-1-4615-5871-2_11. [DOI] [PubMed] [Google Scholar]
- 19.Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340– 9. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 20.Eramo A, Lotti F, Sette G, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504– 14. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 21.Suetsugu A, Nagaki M, Aoki H, et al. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun. 2006;29:820– 4. doi: 10.1016/j.bbrc.2006.10.128. [DOI] [PubMed] [Google Scholar]
- 22.Wu A, Oh S, Wiesner SM, et al. Persistence of CD133+ cells in human and mouse glioma cell lines: detailed characterization of GL261 glioma cells with cancer stem cell-like properties. Stem Cells Dev. 2008;17:173– 84. doi: 10.1089/scd.2007.0133. [DOI] [PubMed] [Google Scholar]
- 23.Burkert J, Otto WR, Wright NA. Side populations of gastrointestinal cancers are not enriched in stem cells. J Pathol. 2008;5:564– 73. doi: 10.1002/path.2307. [DOI] [PubMed] [Google Scholar]
- 24.Pearce DJ, Ridler CM, Simpson C, Bonnet D. Multiparameter analysis of murine bone marrow side population cells. Blood. 2004;103:2541– 6. doi: 10.1182/blood-2003-09-3281. [DOI] [PubMed] [Google Scholar]
- 25.Morita Y, Ema H, Yamazaki S, Nakauchi H. Non-side-population hematopoietic stem cells in mouse bone marrow. Blood. 2006;108:2850– 6. doi: 10.1182/blood-2006-03-010207. [DOI] [PubMed] [Google Scholar]
- 26.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983– 8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Guo LP, Chen LZ, et al. Identification of cancer stem cell-like side population cells in human nasopharyngeal carcinoma cell line. Cancer Res. 2007;8:3716– 24. doi: 10.1158/0008-5472.CAN-06-4343. [DOI] [PubMed] [Google Scholar]
- 28.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895– 902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 29.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275– 84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 30.Brabletz T, Jung A, Spaderna S, et al. Opinion: migrating cancer stem cells—an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744– 9. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 31.Jiang F, Caraway NP, Li R, Katz RL. RNA silencing of S-phase kinase-interacting protein 2 inhibits proliferation and centrosome amplification in lung cancer cells. Oncogene. 2005;21:3409– 18. doi: 10.1038/sj.onc.1208459. [DOI] [PubMed] [Google Scholar]
- 32.Fan T, Qiu Q, Li R, et al. Up-regulation of 14-3-3Z in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67:7901– 6. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 33.Jiang F, Caraway NP, Katz RL. Genomic copy number aberration of surfactant protein a (SFTPA) gene in patients with stage I non-small cell lung cancer. Clin Cancer Res. 2005;11:2005. doi: 10.1158/1078-0432.CCR-04-2087. [DOI] [PubMed] [Google Scholar]
- 34.Li R, Todd NW, Qiu Q, et al. Genetic deletions in sputum as diagnostic markers for early detection of stage I non-small cell lung cancer. Clin Cancer Res. 2007;13:482– 7. doi: 10.1158/1078-0432.CCR-06-1593. [DOI] [PubMed] [Google Scholar]
- 35.Barkan GA, Caraway NP, Jiang F, et al. Comparison of molecular abnormalities in bronchial brushings and tumor touch preparations. Cancer. 2004;105:35– 43. doi: 10.1002/cncr.20800. [DOI] [PubMed] [Google Scholar]
- 36.Wang HJ, Caraway NP, Katz RL, Jiang F. Overexpression of potential oncogene S100A2 protein as a prognostic marker for patients with stage I non-small cell lung cancer. Int J Cancer. 2005;116:666– 9. doi: 10.1002/ijc.21035. [DOI] [PubMed] [Google Scholar]
- 37.Hilbe W, Dirnhofer S, Oberwasserlechner F, et al. CD133 positive endothelial progenitor cells contribute to the tumour vasculature in non-small cell lung cancer. J Clin Pathol. 2004;9:965– 9. doi: 10.1136/jcp.2004.016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.