Abstract
The omnipresent fungal genus Alternaria was recently divided into 24 sections based on molecular and morphological data. Alternaria sect. Porri is the largest section, containing almost all Alternaria species with medium to large conidia and long beaks, some of which are important plant pathogens (e.g. Alternaria porri, A. solani and A. tomatophila). We constructed a multi-gene phylogeny on parts of the ITS, GAPDH, RPB2, TEF1 and Alt a 1 gene regions, which, supplemented with morphological and cultural studies, forms the basis for species recognition in sect. Porri. Our data reveal 63 species, of which 10 are newly described in sect. Porri, and 27 species names are synonymised. The three known Alternaria pathogens causing early blight on tomato all cluster in one clade, and are synonymised under the older name, A. linariae. Alternaria protenta, a species formerly only known as pathogen on Helianthus annuus, is also reported to cause early blight of potato, together with A. solani and A. grandis. Two clades with isolates causing purple blotch of onion are confirmed as A. allii and A. porri, but the two species cannot adequately be distinguished based on the number of beaks and branches as suggested previously. This is also found among the pathogens of Passifloraceae, which are reduced from four to three species. In addition to the known pathogen of sweet potato, A. bataticola, three more species are delineated of which two are newly described. A new Alternaria section is also described, comprising two large-spored Alternaria species with concatenate conidia.
Key words: Alternaria, Early blight of potato, Early blight of tomato, Leaf and stem blight of sweet potato, Multi-gene phylogeny, Purple blotch of onion
Introduction
Alternaria is an important fungal genus with a worldwide distribution. This hyphomycetous ascomycete with phaeodictyospores includes saprophytic, endophytic and pathogenic species, which can be plant pathogens, post-harvest pathogens or human pathogens (Thomma 2003). The genus Alternaria was recently divided into 24 sections (Woudenberg et al. 2013) based on molecular and morphological data, which followed the recent initiative to divide Alternaria into sections (Lawrence et al. 2013). Alternaria sect. Porri is the largest section, containing almost all Alternaria species with medium to large conidia and long beaks. Among them are some important plant pathogens, such as Alternaria bataticola, A. porri, A. solani and A. tomatophila. Alternaria bataticola causes leaf petiole and stem blight of sweet potato in tropical and sub-tropical regions. The disease is most severe in East and Central Africa, with yield losses of over 70 % reported (Osiru et al. 2007). Alternaria porri causes purple blotch of onion, a very destructive disease of onions worldwide. The disease causes a significant reduction in seed and bulb yield, with seed losses of up to 100 % (Abo-Elyousr et al. 2014). Alternaria solani is the causative agent of early blight of potato. This very common disease, which can be found in most potato-growing countries, can cause considerable defoliation. The disease typically reduces yields by ∼20 %, but yield reductions of up to 80 % have been reported (Horsfield et al. 2010). Alternaria tomatophila is known for causing early blight of tomato, attacking the leaves, stems and fruit. This airborne pathogen has spread worldwide, mainly affecting field crops. When left untreated the damage can result in plant defoliation in excess of 60 % (Zitter & Drennan 2005).
The identification of these species has been problematic for many years, with every large-spored Alternaria found on Solanaceae commonly being identified as A. solani. This assumption changed with the treatment of Alternaria species on Solanaceae, in which Simmons (2000) distinguished 22 Alternaria and Nimbya species on solanaceous hosts on the basis of morphology. On potato, Simmons described the large-spored, long-beaked species A. grandis and A. solani, while on tomato he described A. tomatophila, A. cretica and A. subcylindrica. The distinction between potato and tomato pathogens was supported by subsequent molecular studies and chemotaxonomy (Andersen et al. 2008, Rodrigues et al. 2010, Brun et al. 2013, Gannibal et al. 2014).
The taxonomy of Alternaria species on Allium is also confused. Macrosporium porri was first described as pathogen of Allium (Cooke & Ellis 1879), followed by Alternaria allii (Nolla 1927). Both species were later synonymised (Angell 1929) and the name changed to Alternaria porri (Cifferi 1930). The name A. allii was resurrected by Simmons in his identification manual (2007) where he described five large-spored, long-beaked species from Allium, which he could distinguish based on morphology. Large-spored Alternaria from sweet potato were mostly identified as A. bataticola, even if the isolates from some studies (Osiru et al. 2008, Narayanin et al. 2010) showed morphological differences compared with the description of Simmons (2007).
In the present study we aim to use a molecular approach to delineate the medium- to large-spored Alternaria species with long beaks in sect. Porri. A multi-locus analysis based on five partial gene regions, the internal transcribed spacer regions 1 and 2 and intervening 5.8S nrDNA (ITS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), RNA polymerase second largest subunit (RPB2), translation elongation factor 1-alpha (TEF1) and the Alternaria major allergen gene (Alt a 1), was performed. All available ex-type and representative isolates of medium to large-spored, long-beaked species described in Simmons (2007) were included in this study. The present multi-locus analysis supplemented with morphological and cultural data forms the basis for species recognition in sect. Porri.
Materials and methods
Isolates
One hundred eighty-three Alternaria strains including 116 ex-type or representative strains present at the Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands were included in this study (Table 1). With “representative isolate” we refer to the strains used to describe the species based on morphology in Simmons (2007). Freeze-dried strains were revived in 2 mL malt/peptone (50 % / 50 %) and subsequently transferred to oatmeal agar (OA, Crous et al. 2009). Strains stored in the liquid nitrogen collection of the CBS were transferred to OA directly from the −80 °C storage.
Table 1.
Isolates used in this study and their GenBank accession numbers. Bold accession numbers were generated in other studies.
| Name | Old name | Strain number1 | Status2 | Host / Substrate | Locality | GenBank accesion numbers |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | Alt a 1 | TEF1 | RPB2 | ||||||
| Alternaria acalyphicola | CBS 541.94; E.G.S. 38.100; IMI 266969 | T | Acalypha indica | Seychelles | KJ718097 | KJ717952 | KJ718617 | KJ718446 | KJ718271 | |
| Alternaria agerati | CBS 117221; E.G.S. 30.001; QM 9369 | R | Ageratum houstonianum | USA, Illinois | KJ718098 | KJ717953 | KJ718618 | KJ718447 | KJ718272 | |
| Alternaria agripestis | CBS 577.94; E.G.S. 41.034 | T | Euphorbia esula, stem lesion | Canada, Saskatchewan | KJ718099 | JQ646356 | KJ718619 | KJ718448 | KJ718273 | |
| Alternaria allii | Alternaria porri | CBS 107.28; E.G.S. 48.084 | T | Allium cepa, leaf spot | Puerto Rico | KJ718100 | KJ717954 | KJ718620 | KJ718449 | KJ718274 |
| Alternaria porri | CBS 109.41; CBS 114.38 | Allium cepa, seed | Denmark | KJ718101 | KJ717955 | KJ718621 | KJ718450 | KJ718275 | ||
| Alternaria porri | CBS 225.76 | Allium porrum, leaf | Italy | KJ718102 | KJ717956 | KJ718622 | KJ718451 | KJ718276 | ||
| CBS 116701; E.G.S. 33.134 | R | Allium cepa var. viviparum, floral bract | USA, Massachusetts | KJ718103 | KJ717957 | KJ718623 | KJ718452 | KJ718277 | ||
| Alternaria vanuatuensis | CBS 121345; E.G.S. 45.018 | (T) | Allium cepa, leaf | Vanuatu | KJ718104 | KJ717958 | KJ718624 | KJ718453 | KJ718278 | |
| Alternaria alternariacida sp. nov. | Alternaria solani | CBS 105.51; ATCC 11078; IMI 46816; CECT 2997 | T | Solanum lycopersicum, fruit | UK, England | KJ718105 | KJ717959 | KJ718625 | KJ718454 | KJ718279 |
| Alternaria anagallidis | CBS 107.44 | Anagallis arvensis, leaf spot | Denmark, Copenhagen | KJ718106 | JQ646338 | KJ718626 | EU130544 | KJ718280 | ||
| CBS 101004 | Anagallis arvensis, leaf spot | New Zealand, Auckland | KJ718107 | KJ717960 | KJ718627 | KJ718455 | KJ718281 | |||
| CBS 117128; E.G.S. 42.074 | R | Anagallis arvensis, leaf spot | New Zealand, Auckland | KJ718108 | KJ717961 | KJ718628 | KJ718456 | KJ718282 | ||
| CBS 117129; E.G.S. 50.091 | R | Anagallis arvensis, leaf spot | New Zealand, Auckland | KJ718109 | KJ717962 | KJ718629 | KJ718457 | KJ718283 | ||
| Alternaria anodae | PPRI 12376 | Anoda cristata, leaf | South Africa, Gauteng | KJ718110 | KJ717963 | KJ718630 | KJ718458 | KJ718284 | ||
| Alternaria aragakii | CBS 594.93; E.G.S. 29.016; QM 9046 | T | Passiflora edulis | USA, Hawaii | KJ718111 | KJ717964 | KJ718631 | KJ718459 | KJ718285 | |
| Alternaria argyroxiphii | CBS 117222; E.G.S. 35.122 | T | Argyroxiphium sp. | USA, Hawaii | KJ718112 | JQ646350 | KJ718632 | KJ718460 | KJ718286 | |
| PPRI 11848 | Ipomoea batatas, stem lesion | South Africa, Gauteng | KJ718113 | KJ717965 | KJ718633 | KJ718461 | KJ718287 | |||
| PPRI 11971 | Ipomoea batatas, leaf and stem lesion | South Africa, Mpumalanga | KJ718114 | KJ717966 | KJ718634 | KJ718462 | KJ718288 | |||
| Alternaria azadirachtae | CBS 116444; E.G.S. 46.195; BRIP 25386(ss1) | T | Azadirachta indica, leaf spot | Australia, Queensland | KJ718115 | KJ717967 | KJ718635 | KJ718463 | KJ718289 | |
| CBS 116445; E.G.S. 46.196; BRIP 25386(ss2) | R | Azadirachta indica, leaf spot | Australia, Queensland | KJ718116 | KJ717968 | KJ718636 | KJ718464 | KJ718290 | ||
| Alternaria bataticola | CBS 531.63; IFO 6187; MUCL 28916 | T | Ipomoea batatas | Japan | KJ718117 | JQ646349 | JQ646433 | KJ718465 | KJ718291 | |
| CBS 532.63 | Ipomoea batatas | Japan, Tokyo | KJ718118 | KJ717969 | KJ718637 | KJ718466 | KJ718292 | |||
| CBS 117095; E.G.S. 42.157; IMI 350492; BRIP 19470a | R | Ipomoea batatas, leaf spot | Australia, Queensland | KJ718119 | KJ717970 | KJ718638 | KJ718467 | KJ718293 | ||
| CBS 117096; E.G.S. 42.158; BRIP 19470b | R | Ipomoea batatas, leaf spot | Australia, Queensland | KJ718120 | KJ717971 | KJ718639 | KJ718468 | KJ718294 | ||
| PPRI 10502 | Ipomoea batatas, leaf and stem lesion | South Africa, Gauteng | KJ718121 | KJ717972 | KJ718640 | KJ718469 | KJ718295 | |||
| PPRI 11930 | Ipomoea batatas, leaf lesion | South Africa, Kwazulu-Natal | KJ718122 | KJ717973 | KJ718641 | KJ718470 | KJ718296 | |||
| PPRI 11931 | Ipomoea batatas, leaf lesion | South Africa, Kwazulu-Natal | KJ718123 | KJ717974 | KJ718642 | KJ718471 | KJ718297 | |||
| PPRI 11934 | Ipomoea batatas, leaf lesion | South Africa, Gauteng | KJ718124 | KJ717975 | KJ718643 | KJ718472 | KJ718298 | |||
| Alternaria blumeae | Alternaria brasiliensis | CBS 117215; E.G.S. 39.116 | (R) | Phaseolus vulgaris, leaf spot | Brazil, Esperito Santo | KJ718125 | KJ717976 | KJ718644 | KJ718473 | KJ718299 |
| CBS 117364; E.G.S. 40.149; ATCC 201357 | T | Blumea aurita | Thailand, Yala Province | KJ718126 | AY562405 | AY563291 | KJ718474 | KJ718300 | ||
| Alternaria calendulae | CBS 224.76; ATCC 38903; DSM 63161; IMI 205077 | T | Calendula officinalis, leaf spot | Germany | KJ718127 | KJ717977 | KJ718648 | KJ718475 | KJ718301 | |
| CBS 101498 | Calendula officinalis, leaf | New Zealand, Auckland | KJ718128 | KJ717978 | KJ718645 | KJ718476 | KJ718302 | |||
| Alternaria rosifolii | CBS 116439; E.G.S. 42.197 | (T) | Rosa sp., leaf spot | New Zealand, Auckland | KJ718129 | KJ717979 | KJ718646 | KJ718477 | KJ718303 | |
| CBS 116650; E.G.S. 30.142; QM 9561 | R | Calendula officinalis, leaf spot | Japan, Tokyo | KJ718130 | KJ717980 | KJ718647 | KJ718478 | KJ718304 | ||
| Alternaria carthami | CBS 635.80 | Carthamus tinctorius, leaf | Italy, Perugia | KJ718131 | KJ717981 | KJ718649 | KJ718479 | KJ718305 | ||
| Alternaria heliophytonis | CBS 116440; E.G.S. 43.143; IMI 366164 | (T) | Helianthus annuus, leaf | Canada, Saskatchewan | KJ718132 | KJ717982 | KJ718650 | KJ718480 | KJ718306 | |
| CBS 117091; E.G.S. 31.037 | R | Carthamus tinctorius, leaf spot | USA, Montana | KJ718133 | KJ717983 | KJ718651 | KJ718481 | KJ718307 | ||
| Alternaria carthamicola | Alternaria carthami | CBS 117092; E.G.S. 37.057; IMI 276943 | (R)T | Carthamus tinctorius | Iraq | KJ718134 | KJ717984 | KJ718652 | KJ718482 | KJ718308 |
| Alternaria cassiae | CBS 478.81; E.G.S. 33.147 | R | Senna obtusifolia, diseased seedling | USA, Mississippi | KJ718135 | KJ717985 | KJ718653 | KJ718483 | KJ718309 | |
| Alternaria sauropodis | CBS 116119; E.G.S. 47.112; IMI 286317; IMI 392448 | (T) | Sauropus androgynus | Malaysia, Sarawak | KJ718136 | KJ717986 | KJ718654 | KJ718484 | KJ718310 | |
| CBS 117224; E.G.S. 40.121 | R | Senna obtusifolia, leaf spot | Brazil, Federal District | KJ718137 | KJ717987 | KJ718655 | KJ718485 | KJ718311 | ||
| Alternaria hibiscinficiens | CBS 117369; E.G.S. 50.166 | (T) | Hibiscus sabdariffa, leaf | Fiji | KJ718138 | KJ717988 | KJ718656 | KJ718486 | KJ718312 | |
| Alternaria catananches sp. nov. | CBS 137456; PD 013/05703936 | T | Catananche caerulea | Netherlands | KJ718139 | KJ717989 | KJ718657 | KJ718487 | KJ718313 | |
| Alternaria centaureae | CBS 116446; E.G.S. 47.119 | T | Centaurea solstitialis, leaf spot | USA, California | KJ718140 | KJ717990 | KJ718658 | KJ718488 | KJ718314 | |
| Alternaria cichorii | CBS 102.33; E.G.S. 07.017; QM 1760 | T | Cichorium intybus, leaf spot | Cyprus | KJ718141 | KJ717991 | KJ718659 | KJ718489 | KJ718315 | |
| CBS 117218; E.G.S. 52.046; IMI 225641 | R | Cichorium endivia | Greece | KJ718142 | KJ717992 | KJ718660 | KJ718490 | KJ718316 | ||
| Alternaria cirsinoxia | CBS 113261; E.G.S. 41.136 | T | Cirsium arvense, stem lesion | Canada, Saskatchewan | KJ718143 | KJ717993 | KJ718661 | KJ718491 | KJ718317 | |
| Alternaria citrullicola sp. nov. | Alternaria cucumerina | CBS 103.32; VKM F-1881; Nattrass No. 190 | T | Citrullus vulgaris, fruit | Cyprus | KJ718144 | KJ717994 | KJ718662 | KJ718492 | KJ718318 |
| Alternaria conidiophora sp. nov. | CBS 137457 | T | – | Netherlands | KJ718145 | KJ717995 | KJ718663 | KJ718493 | – | |
| Alternaria crassa | CBS 103.18 | Datura sp., leaf spot | USA, Wisconsin | KJ718146 | KJ717996 | KJ718664 | KJ718494 | KJ718319 | ||
| CBS 110.38 | T | Datura stramonium, leaf spot | Cyprus | KJ718147 | KJ717997 | KJ718665 | KJ718495 | KJ718320 | ||
| Alternaria capsici | CBS 109160; E.G.S. 45.075; IMI 262408; IMI 381021 | (T) | Capsicum annuum | Australia | KJ718148 | AY562408 | AY563298 | KJ718496 | KJ718321 | |
| CBS 109162; E.G.S. 46.014 | Nicandra physalodes | USA, Indiana | KJ718149 | GQ180073 | GQ180089 | KJ718497 | KJ718322 | |||
| CBS 116647; E.G.S. 46.013 | R | Datura stramonium, leaf spot | USA, Indiana | KJ718150 | KJ717998 | KJ718666 | KJ718498 | KJ718323 | ||
| CBS 116648; E.G.S. 50.180 | R | Datura stramonium, leaf spot | New Zealand, Auckland | KJ718151 | KJ717999 | KJ718667 | KJ718499 | KJ718324 | ||
| CBS 122590; E.G.S. 44.071 | R | Datura stramonium, leaf spot | USA, Indiana | KJ718152 | GQ180072 | GQ180088 | KJ718500 | KJ718325 | ||
| Alternaria cucumerina | Alternaria loofahae | CBS 116114; E.G.S. 35.123 | (T) | Luffa acutangula | USA, Hawaii | KJ718153 | KJ718000 | KJ718668 | KJ718501 | KJ718326 |
| CBS 117225; E.G.S. 41.127 | R | Cucumis melo, leaf spot | USA, Indiana | KJ718154 | KJ718001 | KJ718669 | KJ718502 | KJ718327 | ||
| CBS 117226; E.G.S. 44.197; BRIP 23060 | R | Cucumis melo, leaf spot | Australia, Queensland | KJ718155 | KJ718002 | KJ718670 | KJ718503 | KJ718328 | ||
| Alternaria cyamopsidis | CBS 364.67; E.G.S. 17.065; QM 8575 | R | Cyamopsis tetragonoloba, leaf spot | USA, Maryland | KJ718156 | KJ718003 | KJ718671 | KJ718504 | KJ718329 | |
| CBS 117219; E.G.S. 13.120; QM 8000 | R | Cyamopsis tetragonoloba, leaf spot | USA, Georgia | KJ718157 | KJ718004 | KJ718672 | KJ718505 | KJ718330 | ||
| Alternaria dauci | CBS 111.38 | T | Daucus carota, seed | Italy | KJ718158 | KJ718005 | KJ718673 | KJ718506 | KJ718331 | |
| CBS 106.48 | Daucus carota, seed | – | KJ718159 | KJ718006 | KJ718674 | KJ718507 | KJ718332 | |||
| CBS 345.79; LEV 14814 | Daucus carota, leaf spot | New Zealand, Ohakune | KJ718160 | KJ718007 | KJ718675 | KJ718508 | KJ718333 | |||
| Alternaria cichorii | CBS 477.83; CBS 721.79; PD 79/954 | Cichorium intybus var. foliosum, leaf spot | Netherlands, Limburg | KJ718161 | KJ718008 | KJ718676 | KJ718509 | KJ718334 | ||
| CBS 101592 | Daucus carota, seed | Netherlands | KJ718162 | KJ718009 | KJ718677 | KJ718510 | KJ718335 | |||
| CBS 117097; E.G.S. 46.006 | R | Daucus carota, commercial seed | USA, California | KC584192 | KC584111 | KJ718678 | KC584651 | KC584392 | ||
| CBS 117098; E.G.S. 46.152 | R | Daucus carota, leaf spot | New Zealand | KJ718163 | KJ718010 | HE796726 | KJ718511 | KJ718336 | ||
| CBS 117099; E.G.S. 47.131 | R | Daucus carota, seed | USA, California | KJ718164 | KJ718011 | KJ718679 | KJ718512 | KJ718337 | ||
| Alternaria poonensis | CBS 117100; E.G.S. 47.138 | (R) | Coriandrum sativum, seedling | Puerto Rico | KJ718165 | JQ646348 | KJ718680 | KJ718513 | KJ718338 | |
| Alternaria deserticola sp. nov. | Alternaria acalyphicola | CBS 110799 | T | desert soil | Namibia | KJ718249 | KJ718077 | KJ718755 | KJ718595 | KJ718424 |
| Alternaria dichondrae | CBS 199.74; E.G.S. 38.007 | T | Dichondra repens, leaf spot | Italy | KJ718166 | JQ646357 | JQ646441 | KJ718514 | KJ718339 | |
| CBS 200.74; E.G.S. 38.008 | T | Dichondra repens, leaf spot | Italy | KJ718167 | KJ718012 | KJ718681 | KJ718515 | KJ718340 | ||
| CBS 346.79 | Dichondra repens, leaf spot | New Zealand | KJ718168 | KJ718013 | KJ718682 | KJ718516 | KJ718341 | |||
| CBS 117127; E.G.S. 40.057 | R | Dichondra sp., leaf | New Zealand, Auckland | KJ718169 | KJ718014 | KJ718683 | KJ718517 | KJ718342 | ||
| Alternaria echinaceae | CBS 116117; E.G.S. 46.081 | T | Echinacea sp., leaf lesion | New Zealand, Gisborne | KJ718170 | KJ718015 | KJ718684 | KJ718518 | KJ718343 | |
| CBS 116118; E.G.S. 46.082 | R | Echinacea sp., leaf lesion | New Zealand, Gisborne | KJ718171 | KJ718016 | KJ718685 | KJ718519 | KJ718344 | ||
| Alternaria grandis | CBS 109158; E.G.S. 44.106 | T | Solanum tuberosum, leaf spot | USA, Pennsylvania | KJ718239 | JQ646341 | JQ646425 | EU130547 | KJ718414 | |
| CBS 116695; E.G.S. 44.108 | R | Solanum tuberosum, leaf spot | USA, Pennsylvania | KJ718241 | KJ718070 | KJ718748 | KJ718587 | KJ718416 | ||
| Alternaria euphorbiicola | CBS 198.86; E.G.S. 38.082 | Euphorbia pulcherrima | USA, Florida | KJ718172 | KJ718017 | KJ718686 | KJ718520 | KJ718345 | ||
| CBS 119410; E.G.S. 41.029 | R | Euphorbia pulcherrima | USA, Hawaii | KJ718173 | KJ718018 | – | KJ718521 | KJ718346 | ||
| CBS 133874; E.G.S. 38.191 | Euphorbia hyssopifolia | USA, Louisiana | KJ718174 | KJ718019 | KJ718687 | KJ718522 | KJ718347 | |||
| Alternaria gypsophilae | CBS 107.41; E.G.S. 07.025; IMI 264349 | T | Gypsophila elegans, seed | Netherlands | KC584199 | KC584118 | KJ718688 | KC584660 | KC584401 | |
| Alternaria ipomoeae sp. nov. | Alternaria cucumerina | CBS 219.79 | T | Ipomoea batatas, stem and petiole | Ethiopia | KJ718175 | KJ718020 | KJ718689 | KJ718523 | KJ718348 |
| PPRI 8988 | Ipomoea batatas, stem | South Africa, Gauteng | KJ718176 | KJ718021 | KJ718690 | KJ718524 | KJ718349 | |||
| Alternaria jesenskae | CBS 133855; CCM 8361 | T | Fumana procumbens, seed | Slovakia | KJ718177 | KJ718022 | KJ718691 | KJ718525 | KJ718350 | |
| Alternaria limicola | CBS 483.90; E.G.S. 39.070 | T | Citrus aurantiifolia, leaf spot | Mexico, Colima | KJ718178 | JQ646329 | JQ646413 | KJ718526 | KJ718351 | |
| CBS 117360; E.G.S. 43.009 | R | Citrus sp. | Mexico, Jalisco | KJ718179 | KJ718023 | – | KJ718527 | KJ718352 | ||
| Alternaria linariae | CBS 105.41; E.G.S. 07.016 | T | Linaria maroccana, seedling | Denmark | KJ718180 | KJ718024 | KJ718692 | KJ718528 | KJ718353 | |
| Alternaria solani | CBS 108.53 | – | – | KJ718181 | KJ718025 | KJ718693 | KJ718529 | KJ718354 | ||
| Alternaria solani | CBS 107.61 | – | Belgium | KJ718182 | KJ718026 | KJ718694 | KJ718530 | KJ718355 | ||
| Alternaria tomatophila | CBS 109156; E.G.S. 42.156 | (T) | Solanum lycopersicum, leaf spot | USA, Indiana | KJ718183 | JQ646347 | GQ180101 | KJ718531 | KJ718356 | |
| Alternaria subcylindrica | CBS 109161; E.G.S. 45.113 | (T) | Solanum lycopersicum var. cerasiforme, leaf spot | USA, Louisiana | KJ718184 | JQ646345 | JQ646429 | KJ718532 | KJ718357 | |
| Alternaria cretica | CBS 109164; E.G.S. 46.188 | (T) | Solanum lycopersicum, leaf spot | Greece, Crete | KJ718185 | JQ646342 | JQ646426 | EU130545 | KJ718358 | |
| Alternaria cucumericola | CBS 116438; E.G.S. 41.057 | (T) | Cucumis sativus, leaf spot | New Zealand | KJ718186 | KJ718027 | KJ718695 | KJ718533 | KJ718359 | |
| Alternaria tabasco | CBS 116441; E.G.S. 45.108 | (T) | Capsicum frutescens, leaf spot | USA, Louisiana | KJ718187 | KJ718028 | KJ718696 | KJ718534 | KJ718360 | |
| Alternaria tomatophila | CBS 116704; E.G.S. 44.074 | (R) | Solanum lycopersicum, leaf spot | USA, Indiana | KJ718188 | KJ718029 | KJ718697 | KJ718535 | KJ718361 | |
| CPC 21620 | Solanum lycopersicum, leaf spot | Thailand, Chiang Mai | KJ718189 | KJ718030 | KJ718698 | KJ718536 | KJ718362 | |||
| Alternaria macrospora | Alternaria porri | CBS 106.29 | Gossypium sp. | Nigeria | KJ718193 | KJ718032 | KJ718701 | KJ718540 | KJ718366 | |
| CBS 117228; E.G.S. 50.190; ATCC 58172 | T | Gossypium barbadense | USA, Arizona | KC584204 | KC584124 | KJ718702 | KC584668 | KC584410 | ||
| Alternaria montanica | CBS 121343; E.G.S. 44.112; IMI 257563 | T | Cirsium arvense | USA, Montana | KJ718194 | KJ718033 | KJ718703 | KJ718541 | KJ718367 | |
| Alternaria multirostrata | CBS 712.68; ATCC 18515; IMI 135454; MUCL 11722; QM 8820; VKM F-2997 | T | Richardia scabra, floral bract | USA, Georgia | KJ718195 | JQ646362 | KJ718704 | EU130546 | KJ718368 | |
| CBS 713.68; ATCC 18517; IMI 135455; MUCL 11715; QM 8821 | R | Richardia scabra, floral bract | USA, Georgia | KJ718196 | KJ718034 | KJ718705 | KJ718542 | KJ718369 | ||
| Alternaria neoipomoeae sp. nov. | PPRI 8990 | Ipomoea batatas | South Africa, North West | KJ718197 | KJ718035 | KJ718706 | KJ718543 | KJ718370 | ||
| PPRI 11845 | T | Ipomoea batatas, stem | South Africa, Gauteng | KJ718198 | KJ718036 | KJ718707 | KJ718544 | KJ718371 | ||
| PPRI 11847 | Ipomoea batatas | South Africa, Mpumalanga | KJ718199 | KJ718037 | KJ718708 | KJ718545 | KJ718372 | |||
| PPRI 13903 | Ipomoea batatas, leaf lesion | South Africa, Gauteng | KJ718200 | KJ718038 | KJ718709 | KJ718546 | KJ718373 | |||
| Alternaria nitrimali | CBS 109163; E.G.S. 46.151 | T | Solanum viarum, leaf spot | Puerto Rico | KJ718201 | JQ646358 | KJ718710 | KJ718547 | KJ718374 | |
| Alternaria novae-guineensis | CBS 116120; E.G.S. 47.198 | T | Citrus sp., dry leaf | Papua New Guinea | KJ718202 | KJ718039 | KJ718711 | KJ718548 | KJ718375 | |
| PPRI 12171 | Galinsoga parviflora, leaf | South Africa, Gauteng | KJ718203 | KJ718040 | KJ718712 | KJ718549 | KJ718376 | |||
| Alternaria obtecta | CBS 117367; E.G.S. 42.063 | R | Euphorbia pulcherrima, leaf | USA, California | KJ718204 | KJ718041 | KJ718713 | KJ718550 | KJ718377 | |
| CBS 134278; E.G.S. 42.064 | Euphorbia pulcherrima | USA, California | KJ718205 | KJ718042 | KJ718714 | KJ718551 | KJ718378 | |||
| Alternaria paralinicola sp. nov. | Alternaria linicola | CBS 116652; E.G.S. 47.157; DAOM 225747 | (R)T | Linum usitatissimum, seed | Canada, Manitoba | KJ718206 | KJ718043 | KJ718715 | KJ718552 | KJ718379 |
| Alternaria passiflorae | CBS 113.38 | Passiflora edulis | Australia, South Queensland | KJ718207 | JQ646353 | JQ646437 | KJ718553 | KJ718380 | ||
| Alternaria solani | CBS 166.77 | Capsicum frutescens, leaf | New Zealand, Waitakere | KJ718208 | KJ718044 | KJ718716 | KJ718554 | KJ718381 | ||
| CBS 629.93; E.G.S. 16.150; QM 8458 | R | Passiflora edulis, fruit | New Zealand | KJ718209 | KJ718045 | KJ718717 | KJ718555 | KJ718382 | ||
| Alternaria hawaiiensis | CBS 630.93; E.G.S. 29.020; QM 9050 | (T) | Passiflora edulis | USA, Hawaii | KJ718210 | JQ646352 | KJ718718 | KJ718556 | KJ718383 | |
| Alternaria gaurae | CBS 116333; E.G.S. 50.121 | (T) | Gaura lindheimeri, leaf | New Zealand, Auckland | KJ718211 | KJ718046 | KJ718719 | KJ718557 | KJ718384 | |
| CBS 117102; E.G.S. 51.165 | R | Passiflora ligularis, fruit spot | New Zealand, Auckland | KJ718212 | KJ718047 | KJ718720 | KJ718558 | KJ718385 | ||
| CBS 117103; E.G.S. 52.032 | R | Passiflora caerulea, leaf spot | New Zealand, Auckland | KJ718213 | KJ718048 | KJ718721 | KJ718559 | KJ718386 | ||
| Alternaria pipionipisi | CBS 116115; E.G.S. 40.096; IMI 340950 | T | Cajanus cajan, seed | India | KJ718214 | KJ718049 | KJ718722 | KJ718560 | KJ718387 | |
| Alternaria obtecta | CBS 117365; E.G.S. 42.048 | (R) | Euphorbia pulcherrima, leaf | USA, California | KJ718215 | KJ718050 | KJ718723 | KJ718561 | KJ718388 | |
| Alternaria obtecta | CBS 134265; E.G.S. 42.047 | Euphorbia pulcherrima | USA, California | KJ718216 | KJ718051 | KJ718724 | KJ718562 | KJ718389 | ||
| Alternaria porri | Alternaria allii | CBS 116649; E.G.S. 17.082; QM 8613 | (R) | Allium cepa, leaf | USA, Nebraska | KJ718217 | KJ718052 | KJ718725 | KJ718563 | KJ718390 |
| CBS 116698; E.G.S. 48.147 | R | Allium cepa, leaf spot | USA, New York | DQ323700 | KC584132 | KJ718726 | KC584679 | KC584421 | ||
| CBS 116699; E.G.S. 48.152 | R,T | Allium cepa, leaf spot | USA, New York | KJ718218 | KJ718053 | KJ718727 | KJ718564 | KJ718391 | ||
| Alternaria protenta | Alternaria solani | CBS 347.79; E.G.S. 44.091; LEV 14726; ATCC 38569 | Solanum lycopersicum, fruit rot | New Zealand, Levin | KJ718219 | KJ718054 | KJ718728 | KJ718565 | KJ718392 | |
| Alternaria hordeiseminis | CBS 116437; E.G.S. 32.076 | (T) | Hordeum vulgare, seed | New Zealand | KJ718220 | KJ718055 | KJ718729 | KJ718566 | KJ718393 | |
| Alternaria solani | CBS 116651; E.G.S. 45.020 | (R) | Solanum tuberosum, tuber | USA, California | KC584217 | KC584139 | GQ180097 | KC584688 | KC584430 | |
| CBS 116696; E.G.S. 45.023; IMI 372955 | R | Helianthus annuus, leaf spot | Israel | KJ718221 | JQ646335 | JQ646419 | KJ718567 | KJ718394 | ||
| CBS 116697; E.G.S. 45.024; IMI 372957 | R | Helianthus annuus, leaf spot | Israel | KJ718222 | KJ718056 | KJ718730 | KJ718568 | KJ718395 | ||
| Alternaria pulcherrimae | CBS 121342; E.G.S. 42.122; IMI 310506 | (R) | Euphorbia pulcherrima | Australia, Queensland | KJ718223 | KJ718057 | KJ718731 | KJ718569 | KJ718396 | |
| Alternaria solani | CBS 135189; E.G.S. 45.053 | (R) | Solanum tuberosum | New Zealand, Hastings | KJ718224 | GQ180082 | GQ180098 | KJ718570 | KJ718397 | |
| Alternaria pseudorostrata | CBS 119411; E.G.S. 42.060 | T | Euphorbia pulcherrima | USA, California | JN383483 | AY562406 | AY563295 | KC584680 | KC584422 | |
| Alternaria ranunculi | CBS 116330; E.G.S. 38.039; IMI 285697 | T | Ranunculus asiaticus, seed | Israel | KJ718225 | KJ718058 | KJ718732 | KJ718571 | KJ718398 | |
| Alternaria ricini | CBS 215.31 | T | Ricinus communis | Japan | KJ718226 | KJ718059 | KJ718733 | KJ718572 | KJ718399 | |
| CBS 353.86 | Ricinus communis | Italy, Sardinia | KJ718227 | JQ646331 | KJ718734 | KJ718573 | KJ718400 | |||
| CBS 117361; E.G.S. 06.181 | R | Ricinus communis | USA, Virginia | KJ718228 | KJ718060 | KJ718735 | KJ718574 | KJ718401 | ||
| Alternaria rostellata | CBS 117366; E.G.S. 42.061 | T | Euphorbia pulcherrima, leaf | USA, California | KJ718229 | JQ646332 | KJ718736 | KJ718575 | KJ718402 | |
| Alternaria scorzonerae | Alternaria linicola | CBS 103.46; Elliot No. 45-190C | Linum usitatissimum | UK, Scotland | KJ718190 | JQ646363 | JQ646447 | KJ718537 | KJ718363 | |
| CBS 478.83; E.G.S. 38.011 | R,T | Scorzonera hispanica, leaf spot | Netherlands, Reusel | KJ718191 | JQ646334 | KJ718699 | KJ718538 | KJ718364 | ||
| Alternaria linicola | CBS 116703; E.G.S. 36.110; IMI 274549 | (R) | Linum usitatissimum, seed | UK, Derbyshire | KJ718192 | KJ718031 | KJ718700 | KJ718539 | KJ718365 | |
| Alternaria sennae sp. nov. | Alternaria cassiae | CBS 477.81; E.G.S. 34.030; IMI 257253 | (R)T | Senna corymbosa, leaf | India, Uttar Pradesh | KJ718230 | JQ646344 | JQ646428 | EU130543 | KJ718403 |
| Alternaria sesami | CBS 240.73 | Sesamum indicum | Egypt | KJ718231 | JQ646343 | KJ718737 | KJ718576 | KJ718404 | ||
| CBS 115264; CBS 117214; E.G.S. 13.027 | R | Sesamum indicum, seedling | India | JF780939 | KJ718061 | KJ718738 | KJ718577 | KJ718405 | ||
| Alternaria sidae | CBS 117730; E.G.S. 12.129 | T | Sida fallax, leaf spot | Kiribati, Phoenix Islands | KJ718232 | KJ718062 | KJ718739 | KJ718578 | KJ718406 | |
| Alternaria silybi | CBS 134092; VKM F-4109 | T | Silybum marianum, leaf | Russia, Vladivistok | KJ718233 | KJ718063 | KJ718740 | KJ718579 | KJ718407 | |
| CBS 134093; VKM F-4117 | Silybum marianum, leaf | Russia, Vladivistok | KJ718234 | KJ718064 | KJ718741 | KJ718580 | KJ718408 | |||
| CBS 134094; VKM F-4118 | Silybum marianum, leaf | Russia, Vladivistok | KJ718235 | KJ718065 | KJ718742 | KJ718581 | KJ718409 | |||
| Alternaria solani | CBS 106.21 | – | – | KJ718236 | KJ718066 | KJ718743 | KJ718582 | KJ718410 | ||
| CBS 111.41 | Solanum aviculare, leaf spot | – | KJ718237 | KJ718067 | KJ718744 | KJ718583 | KJ718411 | |||
| Alternaria danida | CBS 111.44; E.G.S. 07.029; QM 1772 | (T) | Ageratum houstonianum, seed | Italy | Y17070 | KJ718068 | KJ718745 | KJ718584 | KJ718412 | |
| CBS 109157; E.G.S. 44.098 | R | Solanum tuberosum, leaf spot | USA, Washington | KJ718238 | GQ180080 | KJ718746 | KJ718585 | KJ718413 | ||
| Alternaria viciae-fabae | CBS 116442; E.G.S. 46.162; ICMP 10242 | (T) | Vicia faba | New Zealand | KJ718240 | KJ718069 | KJ718747 | KJ718586 | KJ718415 | |
| Alternaria solani-nigri | Alternaria cyphomandrae | CBS 109155; E.G.S. 40.058 | (T) | Cyphomandra betacea, fruit | New Zealand, New Plymouth | KJ718242 | JQ646360 | JQ646444 | KJ718588 | KJ718417 |
| CBS 113403; E.G.S. 51.106; CPC 10620 | R | Solanum nigrum, leaf spot | New Zealand, Waikato | KJ718243 | KJ718071 | KJ718749 | KJ718589 | KJ718418 | ||
| Alternaria herbiculinae | CBS 116332; E.G.S. 49.180 | (T) | Petroselinum crispum, stunted plant | New Zealand, Taranaki | KJ718244 | KJ718072 | KJ718750 | KJ718590 | KJ718419 | |
| Alternaria glyceriae | CBS 116334; E.G.S. 51.107 | (T) | Glyceria maxima, leaf spot | New Zealand, Waikato | KJ718245 | KJ718073 | KJ718751 | KJ718591 | KJ718420 | |
| Alternaria beticola | CBS 116447; E.G.S. 47.196 | (T) | Beta vulgaris, leaf spot | New Zealand, Canterbury | KJ718246 | KJ718074 | KJ718752 | KJ718592 | KJ718421 | |
| CBS 117101; E.G.S. 51.032 | R | Solanum nigrum, leaf spot | New Zealand, Waikato | KJ718247 | KJ718075 | KJ718753 | KJ718593 | KJ718422 | ||
| Alternaria ascaloniae | CBS 121347; E.G.S. 46.052 | (T) | Allium ascalonicum, leaf spot | New Zealand, Hastings | KJ718248 | KJ718076 | KJ718754 | KJ718594 | KJ718423 | |
| Alternaria steviae | CBS 631.88; IFO 31212 | Stevia rebaudiana, leaf spot | Japan, Kagawa | KJ718250 | KJ718078 | KJ718756 | KJ718596 | KJ718425 | ||
| CBS 632.88; IFO 31183 | Stevia rebaudiana, leaf spot | Japan, Kagawa | KJ718251 | JQ646339 | KJ718757 | KJ718597 | KJ718426 | |||
| CBS 117362; E.G.S. 37.019; IFO 31182 | T | Stevia rebaudiana, leaf spot | Japan, Kagawa | KJ718252 | KJ718079 | KJ718758 | KJ718598 | KJ718427 | ||
| Alternaria tagetica | CBS 297.79; GST AM2 | Tagetes sp., seed | UK | KJ718253 | KJ718080 | KJ718759 | KJ718599 | KJ718428 | ||
| CBS 298.79; GST AM3 | Tagetes sp., seed | UK | KJ718254 | KJ718081 | KJ718760 | KJ718600 | KJ718429 | |||
| CBS 479.81; E.G.S. 33.081; GST 556 | R | Tagetes erecta, seed | UK, England | KC584221 | KC584143 | KJ718761 | KC584692 | KC584434 | ||
| CBS 480.81; E.G.S. 33.184 | R | Tagetes sp., seed | USA, South Carolina | KJ718255 | KJ718082 | KJ718762 | KJ718601 | KJ718430 | ||
| CBS 117217; E.G.S. 44.045 | R | Tagetes sp., leaf spot | USA, Ohio | KJ718256 | KJ718083 | KJ718763 | KJ718602 | KJ718431 | ||
| Alternaria thunbergiae | CBS 116331; E.G.S. 41.073; BRIP 14963 | T | Thunbergia alata, leaf spot | Australia, Queensland | KJ718257 | KJ718084 | KJ718764 | KJ718603 | KJ718432 | |
| Alternaria iranica | CBS 120986; E.G.S. 51.075 | (T) | Allium cepa, leaf | Iran, Miandoab | KJ718258 | KJ718085 | KJ718765 | KJ718604 | KJ718433 | |
| CBS 122597 | Thunbergia alata | New Zealand, Auckland | KJ718259 | KJ718086 | KJ718766 | KJ718605 | KJ718434 | |||
| Alternaria tillandsiae | CBS 116116; E.G.S. 43.074 | T | Tillandsia usneoides | New Zealand | KJ718260 | KJ718087 | KJ718767 | KJ718606 | KJ718435 | |
| Alternaria tropica | CBS 631.93; E.G.S. 39.126 | T | Passiflora edulis, fruit | USA, Florida | KJ718261 | KJ718088 | KJ718768 | KJ718607 | KJ718436 | |
| CBS 117216; E.G.S. 39.125 | R | Passiflora edulis, fruit | USA, Florida | KJ718262 | KJ718089 | KJ718769 | KJ718608 | KJ718437 | ||
| Alternaria venezuelensis | CBS 116121; E.G.S. 48.065 | T | Phaseolus vulgaris, leaf spot | Venezuela, Maracay | KJ718263 | KJ718090 | KJ718770 | KJ718609 | KJ718438 | |
| Alternaria zinniae | CBS 118.44 | Callistephus chinensis, seed | Hungary | KJ718264 | JQ646361 | KJ718771 | KJ718610 | KJ718439 | ||
| CBS 107.48 | Zinnia sp., leaf | Netherlands | KJ718265 | KJ718091 | KJ718772 | KJ718611 | KJ718440 | |||
| CBS 117.59 | Zinnia elegans | Italy, Sardinia | KJ718266 | KJ718092 | KJ718773 | KJ718612 | KJ718441 | |||
| CBS 108.61 | Zinnia elegans | – | KJ718267 | KJ718093 | KJ718774 | KJ718613 | KJ718442 | |||
| CBS 299.79 | Zinnia sp., seed | UK | KJ718268 | KJ718094 | KJ718775 | KJ718614 | KJ718443 | |||
| CBS 300.79 | Zinnia sp., seed | UK | KJ718269 | KJ718095 | KJ718776 | KJ718615 | KJ718444 | |||
| CBS 117223; E.G.S. 44.035 | R | Zinnia elegans, leaf spot | New Zealand, Auckland | KJ718270 | KJ718096 | KJ718777 | KJ718616 | KJ718445 | ||
ATCC: American Type Culture Collection, Manassas, VA, USA; BRIP: Queensland Plant Pathology Herbarium, Queensland, Australia; CBS: Culture collection of the Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Utrecht, Netherlands; CCM: Czech Collection of Microorganisms, Brno, Czech Republic; CECT: Spanish Type Culture Collection, Valencia, Spain; CPC: Personal collection of P.W. Crous, Utrecht, Netherlands; DAOM: Canadian Collection of Fungal Cultures, Ottawa, Canada; DSM: German Collection of Microorganisms and Cell Cultures, Leibniz Institute, Braunschweig, Germany; E.G.S.: Personal collection of Dr. E.G. Simmons; Elliott: Personal collection of M.E. Elliott; GST: Personal collection of G.S. Taylor; ICMP: International Collection of Micro-organisms from Plants, Auckland, New Zealand; IFO: Institute for Fermentation Culture Collection, Osaka, Japan; IMI: Culture collection of CABI Europe UK Centre, Egham UK; LEV: Plant Health and Diagnostic Station, Levin, New Zealand; MUCL: (Agro)Industrial Fungi and Yeast Collection of the Belgian Co-ordinated Collections of Micro-organisms (BCCM), Louvain-la Neuve, Belgium; Nattrass: Personal collection of R.M. Nattrass; PD: Plant Protection Service, Wageningen, Netherlands; PPRI: ARC-Plant Protection Research Institute, Roodeplaat, South Africa; QM: Quarter Master Culture Collection, Amherst, MA, USA; VKM: All-Russian Collection of Microorganisms, Moscow, Russia.
T: ex-type strain; R: representative strain; Letters between parentheses refer to synonymised species names; Bold letters are designated in this study.
PCR and sequencing
DNA extraction was performed using the UltraClean Microbial DNA isolation kit (Mobio laboratories, Carlsbad, CA, USA), according to the manufacturer's instructions. The ITS region was amplified with the primers V9G (de Hoog & Gerrits van den Ende 1998) and ITS4 (White et al. 1990), the GAPDH region with gpd1 and gpd2 (Berbee et al. 1999) the RPB2 region with RPB2–5F2 (Sung et al. 2007) and fRPB2–7cR (Liu et al. 1999), the TEF1 gene with the primers EF1-728F and EF1-986R (Carbone & Kohn 1999) or EF2 (O'Donnell et al. 1998) and the Alt a 1 region with the primers Alt-for and Alt-rev (Hong et al. 2005). The ITS, GAPDH, RPB2 and TEF1 PCRs were performed as described in Woudenberg et al. (2013). The reaction mixture for the Alt a 1 PCR consisted of 1 μL genomic DNA, 1 × NH4 reaction buffer (Bioline, Luckenwalde, Germany), 3 mM MgCl2, 20 μM of each dNTP, 0.2 μM of each primer and 0.25 U BIOTAQ DNA polymerase (Bioline). Conditions for PCR amplification consisted of an initial denaturation step of 5 min at 94 °C followed by 40 cycles of 30 s at 94 °C, 30 s at 55 °C and 60 s at 72 °C and a final elongation step of 7 min at 72 °C. The PCR products were sequenced in both directions using the PCR primers and the BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), and analysed with an ABI Prism 3730XL Sequencer (Applied Biosystems) according to the manufacturer's instructions. Consensus sequences were computed from forward and reverse sequences using the BioNumerics v. 4.61 software package (Applied Maths, St-Martens-Latem, Belgium). All newly generated sequences were deposited in GenBank (Table 1).
Phylogenetic analysis
Multiple sequence alignments were generated with MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/index.html), and adjusted by eye where necessary. Bayesian inference and Maximum Likelihood analyses were performed on both the individual sequence datasets as well as the concatenated datasets as described in Woudenberg et al. (2013), with the sample frequency set to 1000 instead of 100 in the Bayesian analysis. For the TEF1 partition an online tool (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html) suggested the K2P model with a gamma-rate variation as nucleotide substitution model, and for the remaining four partitions the TrN model with gamma-distributed rate variation. Sequences from the type species of the phylogenetically closest section, sect. Gypsophilae, A. gypsophilae (Woudenberg et al. 2013), were used as outgroup. The resulting trees were printed with TreeView v. 1.6.6 (Page 1996) and the alignments and trees deposited into TreeBASE (http://www.treebase.org).
Taxonomy
Cultures were incubated on potato carrot agar (PCA, Crous et al. 2009) and synthetic nutrient-poor agar (SNA, Nirenberg 1976) plates at moderate temperatures (∼22 °C) under CoolWhite fluorescent light with an 8 h photoperiod. After 7 d the growth rates were measured and the colony characters noted. Colony colours were rated according to Rayner (1970). Morphological descriptions were made for isolates grown on SNA with a small piece of autoclaved filter paper placed onto the agar surface to enhance sporulation. When sporulation occurred, the sellotape technique was used for making slide preparations (Schubert et al. 2007) with Titan Ultra Clear Tape (Conglom Inc., Toronto, Canada) and Shear's medium as mounting fluid. The 95 % confidence intervals were derived from measurements of 30 structures, with extremes given in parentheses. Photographs of characteristic structures were made with a Nikon Eclipse 80i microscope equipped with a Nikon digital sight DS-Fi1 high definition colour camera, using differential interference contrast (DIC) illumination and the Nikon software NIS-Elements D v. 3.00. Adobe Bridge CS5.1 and Adobe Photoshop CS5 Extended, v. 12.1, were used for the final editing and photographic preparation. Colonies which did not sporulate after 7 d were checked for sporulation up to 3 wk; after this period they were noted as sterile. Nomenclatural data were deposited in MycoBank (Crous et al. 2004).
Results
Phylogeny
Because the amplification/sequencing of the RPB2 region of CBS 137457 and the Alt a 1 region of CBS 119410 and CBS 117360 failed, these genes were included as missing data in the combined analysis of these isolates. The topologies of the trees obtained from the RAxML and Bayesian analyses were overall congruent, resulting in identical species-clades. The phylogenies of the single-gene trees were congruent with one exception, CBS 137456, which swapped between clusters with the different genes used, resulting in a somewhat distorted picture in the combined analysis. The aligned sequences of the ITS (538 characters), GAPDH (581 characters), RPB2 (772 characters), TEF1 (355 characters) and Alt a 1 (476 characters) gene regions of the 183 included Alternaria strains had a total length of 2 722 characters, with respectively 77, 111, 134, 141 and 131 unique site patterns. After discarding the burn-in phase trees, the Bayesian analysis resulted in 7 502 trees from which the 50 % majority rule consensus tree and posterior probabilities were calculated. The multi-gene phylogeny of section Porri (Fig. 1) divided the isolates in 62 species (clades) and one new Alternaria section. The species A. euphorbiicola and A. limicola, previously assigned to sect. Porri (Lawrence et al. 2013, Woudenberg et al. 2013), form a sister-clade to sect. Porri, here described as Alternaria sect. Euphorbiicola sect. nov. A Bayesian phylogeny based on the GAPDH, RPB2 and TEF1 sequences of representative isolates of the closely related sections in Alternaria (sequences obtained from Woudenberg et al. 2013) was constructed for comparison, with A. brassicicola CBS 118699 from sect. Brassisicola, as outgroup (Fig. 2).
Fig. 1.
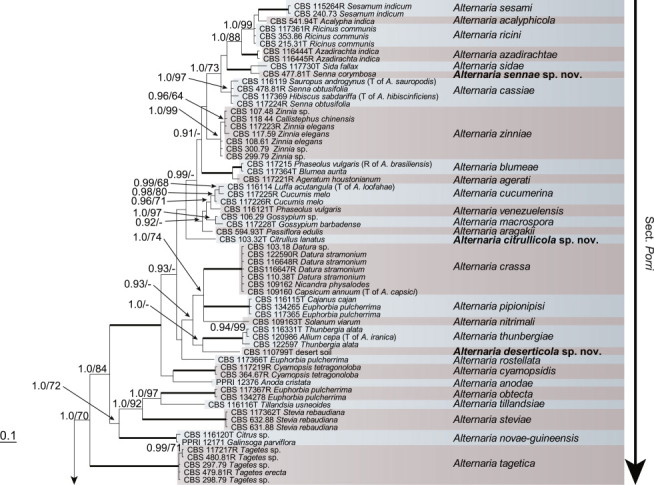
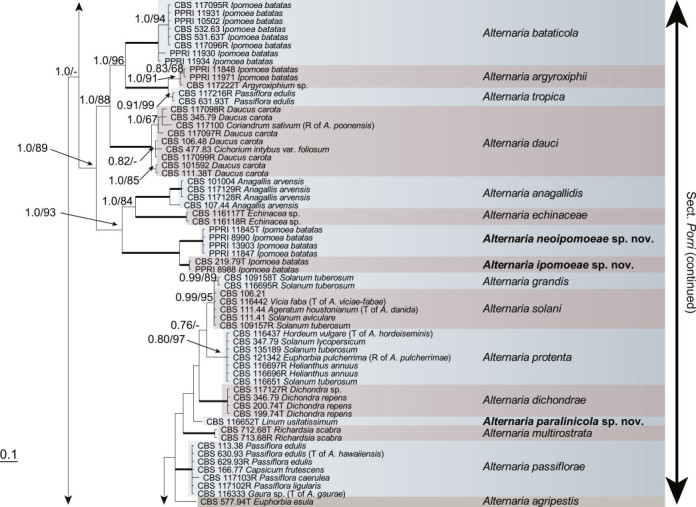
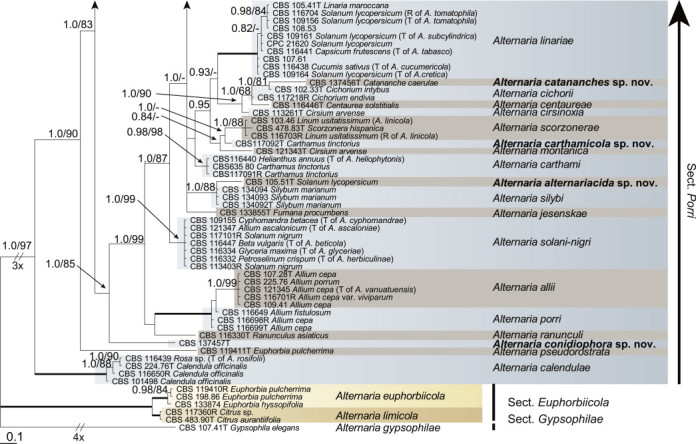
Bayesian 50 % majority rule consensus tree based on the ITS, GAPDH, RPB2, TEF1 and Alt a 1 sequences of 183 Alternaria strains. The Bayesian posterior probabilities > 0.75 (PP) and RAxML bootstrap support values > 65 (ML) are given at the nodes (PP/ML). Thickened lines indicate a PP of 1.0 and ML of 100. Species names between parentheses represent synonymised species names. Ex-type strains are indicated with T and representative strains with R. Novel species names are printed in bold face. The tree was rooted to A. gypsophilae (CBS 107.41).
Fig. 2.
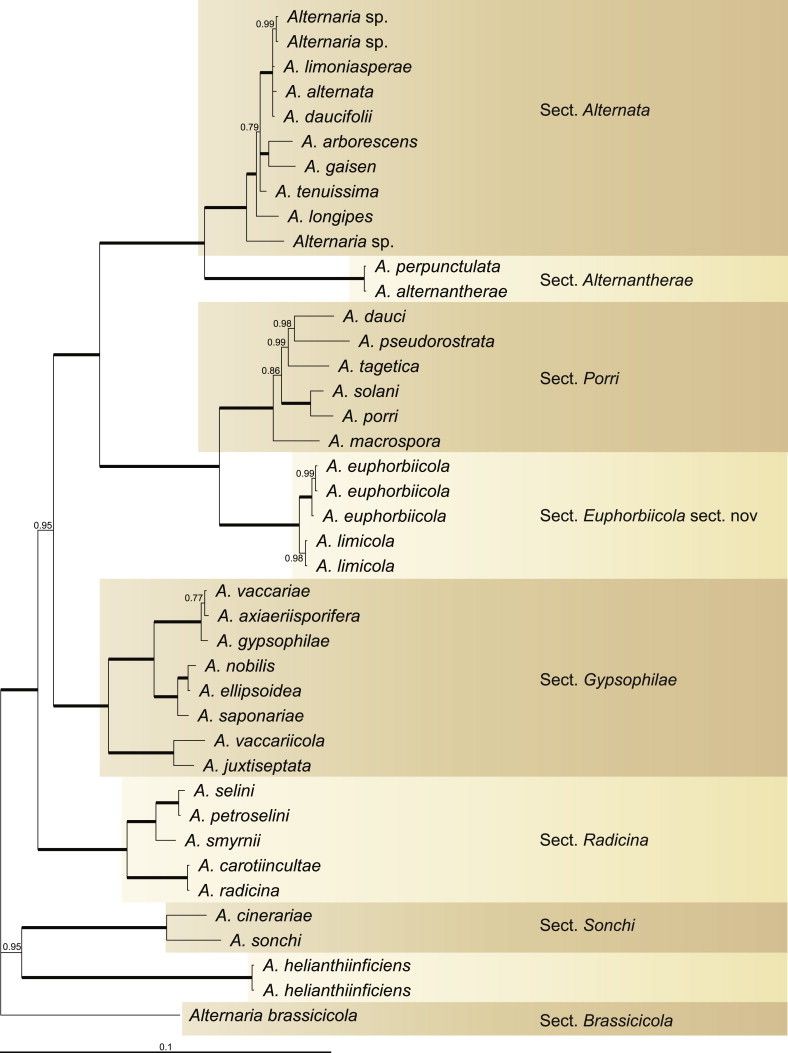
Bayesian 50 % majority rule consensus tree based on the GAPDH, RPB2 and TEF1 sequences of 41 Alternaria strains. The Bayesian posterior probabilities (PP) are given at the nodes. Thickened lines indicate a PP of 1.0. The tree was rooted to A. brassicola (CBS 118699).
Taxonomy
At the onset of this study, Alternaria sect. Porri contained 82 Alternaria species. After extensive phylogenetic analyses and morphological examination we now recognise 63 species in this section (Table 2), of which 10 are newly described. Twenty-seven species names are reduced to synonymy (Table 2). All isolates where taxonomic changes were found based on the multi-gene phylogeny were studied morphologically; photo plates of these species are included. Type details are only listed when typification is proposed.
Table 2.
Current species within Alternaria sect. Porri and their host / substrate.
| Species name | Synonymised names (this study) | Host / Substrate |
|---|---|---|
| Alternaria acalyphicola | Euphorbiaceae (Acalypha indica) | |
| Alternaria agerati | Asteraceae (Ageratum houstonianum) | |
| Alternaria agripestis | Euphorbiaceae (Euphorbia esula) | |
| Alternaria allii | Alternaria vanuatuensis | Amaryllidaceae (Allium cepa, A. porrum) |
| Alternaria alternariacida | Solanaceae (Solanum lycopersicum) | |
| Alternaria anagallidis | Primulaceae (Anagallis arvensis) | |
| Alternaria anodae | Malvaceae (Anoda cristata) | |
| Alternaria aragakii | Passifloraceae (Passiflora edulis) | |
| Alternaria argyroxiphii | Asteraceae (Argyroxiphium sp.), Convolvulaceae (Ipomoea batatas) | |
| Alternaria azadirachtae | Meliaceae (Azadirachta indica) | |
| Alternaria bataticola | Convolvulaceae (Ipomoea batatas) | |
| Alternaria blumeae | Alternaria brasilliensis | Asteraceae (Blumea aurita), Fabaceae (Phaseolus vulgaris) |
| Alternaria calendulae | Alternaria rosifolii | Asteraceae (Calendula officinalis), Rosaceae (Rosa sp.) |
| Alternaria carthami | Alternaria heliophytonis | Asteraceae (Carthamus tinctorius, Helianthus annuus) |
| Alternaria carthamicola | Asteraceae (Carthamus tinctorius) | |
| Alternaria cassiae | Alternaria hibiscinficiens | Fabaceae (Senna obtusifolia), Malvacea (Hibiscus sabdariffa), Phyllanthaceae (Sauropus androgynus) |
| Alternaria sauropodis | ||
| Alternaria catananches | Asteraceae (Catananche caerulea) | |
| Alternaria centaureae | Asteraceae (Centaurea solstitialis) | |
| Alternaria cichorii | Asteraceae (Cichorium endivia, C. intybus) | |
| Alternaria cirsinoxia | Asteraceae (Cirsium arvense) | |
| Alternaria citrullicola | Cucurbitaceae (Citrullus lanatus) | |
| Alternaria conidiophora | Unknown | |
| Alternaria crassa | Alternaria capsici | Solanaceae (Capsicum annuum, Datura stramonium, Nicandra physalodes) |
| Alternaria cucumerina | Alternaria loofahae | Cucurbitaceae (Cucumis melo, Luffa acutangula) |
| Alternaria cyamopsidis | Fabaceae (Cyamopsis tetragonoloba) | |
| Alternaria dauci | Alternaria poonensis | Apiaceae (Daucus carota, Coriandrum sativum), Asteraceae (Cichorium intybus) |
| Alternaria deserticola | Soil | |
| Alternaria dichondrae | Convolvulaceae (Dichondra sp., D. repens) | |
| Alternaria echinaceae | Asteraceae (Echinacea sp.) | |
| Alternaria grandis | Solanaceae (Solanum tuberosum) | |
| Alternaria ipomoeae | Convolvulaceae (Ipomoea batatas) | |
| Alternaria jesenskae | Cistaceae (Fumana procumbens) | |
| Alternaria linariae | Alternaria cretica | Cucurbitaceae (Cucumis sativus), Scrophulariaceae (Linaria maroccana), Solanaceae (Capsicum frutescens, Solanum lycopersicum) |
| Alternaria cucumericola | ||
| Alternaria subcylindrica | ||
| Alternaria tabasco | ||
| Alternaria tomatophila | ||
| Alternaria macrospora | Malvaceae (Gossypium sp., G. barbadense) | |
| Alternaria montanica | Asteraceae (Cirsium arvense) | |
| Alternaria multirostrata | Rubiaceae (Richardia scabra) | |
| Alternaria neoipomoeae | Convolvulaceae (Ipomoea batatas) | |
| Alternaria nitrimali | Solanacaea (Solanum viarum) | |
| Alternaria novae-guineensis | Asteraceae (Galinsoga parviflora), Rutaceae (Citrus sp.) | |
| Alternaria obtecta | Euphorbiaceae (Euphorbia pulcherrima) | |
| Alternaria paralinicola | Linaceae (Linum usitatissimum) | |
| Alternaria passiflorae | Alternaria gaurae | Onagraceae (Gaura lindheimeri), Passifloraceae (Passiflora edulis, P. caerulea, P. ligularis), Solanaceae (Capsicum frutescens) |
| Alternaria hawaiiensis | ||
| Alternaria pipionipisi | Euphorbiaceae (Euphorbia pulcherrima), Fabaceae (Cajanus cajan) | |
| Alternaria porri | Amaryllidaceae (Allium cepa, A. porrum) | |
| Alternaria protenta | Alternaria hordeiseminis | Asteraceae (Helianthus annuus), Euphorbiaceae (Euphorbia pulcherrima), Gramineae (Hordeum vulgare), Solanaceae (Solanum lycopersicum, S. tuberosum) |
| Alternaria pulcherrimae | ||
| Alternaria pseudorostrata | Euphorbiaceae (Euphorbia pulcherrima) | |
| Alternaria ranunculi | Ranunculaceae (Ranunculus asiaticus) | |
| Alternaria ricini | Euphorbiaceae (Ricinus communis) | |
| Alternaria rostellata | Euphorbiaceae (Euphorbia pulcherrima) | |
| Alternaria scorzonerae | Alternaria linicola | Asteraceae (Sorzonerae hispanica), Linaceae (Linum usitatissimum) |
| Alternaria sennae | Fabaceae (Senna corymbosa) | |
| Alternaria sesami | Pedaliaceae (Sesamum indica) | |
| Alternara sidae | Malvaceae (Sida fallax) | |
| Alternaria silybi | Asteraceae (Silybum marianum) | |
| Alternaria solani | Alternaria danida | Asteraceae (Ageratum houstonianum), Fabaceae (Vicia faba), Solanaceae (Solanum aviculare, S. tuberosum) |
| Alternaria viciae-fabae | ||
| Alternaria solani-nigri | Alternaria ascaloniae | Amaryllidaceae (Allium ascalonicum), Apiaceae (Petroselinum crispum), Chenopodiaceae (Beta vulgaris), Gramineae (Glyceria maxima), Solanaceae (Cyphomandra betacea, Solanum nigrum) |
| Alternaria beticola | ||
| Alternaria cyphomandrae | ||
| Alternaria glyceriae | ||
| Alternaria herbiculinae | ||
| Alternaria steviae | Asteraceae (Stevia rebaudiana) | |
| Alternaria tagetica | Asteraceae (Tagetes sp., T. erecta) | |
| Alternaria thunbergiae | Alternaria iranica | Acanthaceae (Thunbergia alata), Amaryllidaceae (Allium cepa) |
| Alternaria tillandsiae | Bromeliaceae (Tillandsia usneoides) | |
| Alternaria tropica | Passifloraceae (Passiflora edulis) | |
| Alternaria venezuelensis | Fabaceae (Phaseolus vulgaris) | |
| Alternaria zinniae | Asteraceae (Callistephus chinensis, Zinnia sp., Z. elegans) |
Section Porri D.P. Lawr., Gannibal, Peever & B.M. Pryor, Mycologia 105: 541. 2013
Type species: Alternaria porri (Ellis) Cif.
Section Porri is characterised by broadly ovoid, obclavate, ellipsoid, subcylindrical or obovoid, medium to large conidia, disto- and euseptate, solitary or in short chains, with a simple or branched, long to filamentous beak. Conidia contain multiple transverse and longitudinal septa and are slightly constricted near some transverse septa. Secondary conidiophores can be formed apically and/or laterally.
Species in sect. Porri
Alternaria acalyphicola E.G. Simmons, Mycotaxon 50: 260. 1994.
Material examined: Seychelles, from Acalypha indica (Euphorbiaceae), before Apr. 1982, C. Kingsland, culture ex-type of A. acalyphicola CBS 541.94 = E.G.S. 38.100 = IMI 266969.
Notes: Alternaria acalyphicola is closely related to A. ricini, with only 1 nt difference in three out of the five genes sequenced; RPB2, TEF1 and GAPDH. Based on this single isolate, the data is inconclusive to support the synonymy of these two species.
Alternaria agerati E.G. Simmons, Mycotaxon 65: 63. 1997.
= Alternaria agerati Sawada, Rep. Dept. Agric. Gov. Res. Inst. Formosa 86: 165. 1943. (nom. inval., Art. 36.1)
Material examined: USA, Illinois, Springfield, from Ageratum houstonianum (Asteraceae) in a commercial greenhouse, Nov. 1968, J.L. Forsberg, representative isolate of A. agerati CBS 117221 = E.G.S. 30.001 = QM 9369.
Alternaria agripestis E.G. Simmons & K. Mort., Mycotaxon 50: 255. 1994.
Material examined: Canada, Saskatchewan, Maxim, from infected stem of Euphorbia esula (Euphorbiaceae), 9 Jul. 1992, P. Harris, culture ex-type of A. agripestis CBS 577.94 = E.G.S. 41.034.
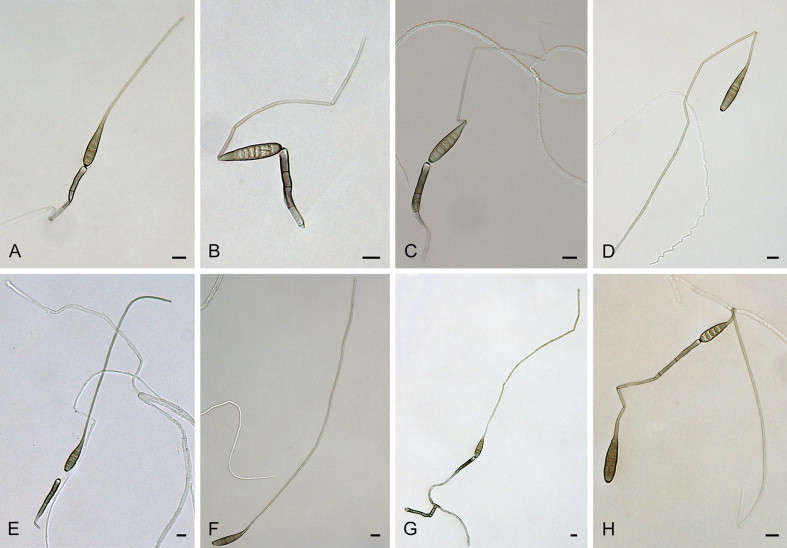
Alternaria allii Nolla, Phytopathology 17: 118. 1927. Fig. 3.
Fig. 3.

Alternaria allii: conidia and conidiophores. A–C. CBS 107.28. D–E. CBS 109.41. F–H. CBS 225.76. I–L. CBS 121345. Scale bars = 10 μm.
= Alternaria vanuatuensis E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 260. 2007.
Materials examined: Denmark, from seed of Allium cepa (Amaryllidaceae), 1937, P. Neergaard, CBS 109.41 = CBS 114.38. Italy, from leaf of Allium porrum (Amaryllidaceae), 1974, H. Nirenberg, CBS 225.76. Puerto Rico, from leaf of Allium cepa, before 1928, J.A.B. Nolla, culture ex-type of A. allii CBS 107.28 = E.G.S. 48.084. USA, Massachusetts, Hadley, from floral bract of Allium cepa var. viviparum, 13 Jul. 1980, E.G. Simmons, representative of A. allii CBS 116701 = E.G.S. 33.134. Vanuatu, from leaves of Allium cepa, 1996, C.F. Hill, culture ex-type of A. vanuatuensis CBS 121345 = E.G.S 45.018.
Notes: Simmons (2007) designated the lectotype of A. allii as Nolla (1927), loc. cit., Pl. III, fig. 11–19, based on the absence of original Nolla specimens. In our study, however, we managed to uncover an original specimen, CBS 107.28, which was deposited in the CBS by J.A.B. Nolla in December 1927 as his “A. allii sp. nov.”, just after he published the new species description. We therefore recognise this isolate as the ex-type strain of A. allii. Isolate CBS 116701 did not sporulate after 3 wk of cultivation on SNA.
Alternaria alternariacida Woudenb. & Crous, sp. nov. MycoBank MB808990. Fig. 4.
Fig. 4.

Alternaria alternariacida sp. nov. CBS 105.51: A–H. Conidia and conidiophores. Scale bars = 10 μm.
Etymology: Named after its ability to produce high amounts of alternaric acid.
Alternaria alternariacida differs from the ex-type isolate of its closest phylogenetic neighbour A. silybi (CBS 134092) based on alleles in three loci (positions derived from respective alignments of the separate loci deposited in TreeBASE): ITS position 386 (T), 497 (T), 498 (T); TEF1 position 3 (T), 18 (T); Alt a 1 position 205 (C), 336 (T), 339 (A), 350 (C), 404 (T), 408 (G).
Sporulation is atypical. Primary conidiophores solitary, simple, straight to slightly curved, septate, pale brown with a subhyaline tip, (52–)73–93(–155) × (4–)5–6(–7) μm, bearing a single, darkened, apical conidiogenous locus. Conidia solitary or in unbranched chains of 2(–3) conidia, conidium body pale olive-brown, smooth-walled, narrowly ovoid, solitary, non-catenulate, and secondary conidia (33–)44–49(–56) × (5–)7–8(–9) μm, with (3–)5–6(–8) transverse eusepta and no longitudinal septa; primary conidia in total (85–)99–111(–121) × (6–)7–8(–10) μm. The conidial body can be slightly constricted near the septa. The conidium body gradually tapers into mostly an aseptate, single, unbranched beak, but branched beaks do occur; apical and multiple lateral secondary conidiophores can also occur. Beaks (47−)129−257(−610) μm long, ca. 2 μm wide throughout their length. Sexual morph not observed.
Culture characteristics: After 7 d cultures on SNA flat, fimbriate, white; aerial mycelium sparse, white, colonies reaching 25−30 mm diam; cultures on PCA flat, entire, olivaceous in the centre with three olivaceous concentric circles and a buff to white margin; aerial mycelium fine, felty, white, colonies reaching 50 mm diam; reverse with four olivaceous concentric circles.
Material examined: UK, England, from fruit of Solanum lycopersicum (Solanaceae), 1946, P.W. Brian (holotype CBS H-21734, culture ex-type CBS 105.51 = ATCC 11078 = IMI 46816 = CECT 2997 = IBPG 14 = BRL408).
Note: The atypical sporulation of the single isolate of A. alternariacida, which is over 60 yr old, resulted in our decision to include sequence data in the species description.
Alternaria anagallidis A. Raabe, Hedwigia 78: 87. 1939.
Materials examined: Denmark, Copenhagen, from Anagallis arvensis (Primulaceae), before Mar. 1944, P. Neergaard, CBS 107.44. New Zealand, Auckland, Lynfield, from Anagallis arvensis, 4 May 1998, C.F. Hill, CBS 101004; Auckland, Lynfield, from Anagallis arvensis, 28 Jun. 1995, C.F. Hill, representative isolate of A. anagallidis CBS 117128 = E.G.S. 42.074; Auckland, from leaf spot of Anagallis arvensis, Jan. 2002, C.F. Hill, representative isolate of A. anagallidis CBS 117129 = E.G.S. 50.091.
Notes: Isolate CBS 107.44 differs on 6 nt positions in its RPB2 sequence from the other three A. anagallidis isolates included in this study. Because CBS 107.44 still clusters closest to the other A. anagallidis isolates, and since these isolates, from a single host species, form a distinct clade from all other Alternaria spp., we retained the name A. anagallidis for this isolate.
Alternaria anodae E.G. Simmons, Mycotaxon 88: 198. 2003.
Material examined: South Africa, Gauteng Province, Pretoria, ARC-Roodeplaat VOPI, from leaves of Anoda cristata (Malvaceae), 12 Jan. 2012, A. Thompson, PPRI 12376.
Alternaria aragakii E.G. Simmons, Mycotaxon 46: 181. 1993.
Material examined: USA, Hawaii, from Passiflora edulis (Passifloraceae), before Oct. 1968, M. Aragaki, culture ex-type of A. aragakii CBS 594.93 = E.G.S. 29.016 = QM 9046.
Alternaria argyroxiphii E.G. Simmons & Aragaki, Mycotaxon 65: 40. 1997.
Materials examined: South Africa, Gauteng Province, Pretoria, ARC-Roodeplaat VOPI, from stem lesion of Ipomoea batatas (Convolvulaceae), 20 Apr. 2005, A. Thompson, PPRI 11848; Mpumalanga Province, Marble Hall, from stem and leaf lesion of Ipomoea batatas, 22 Nov. 2011, A. Thompson, PPRI 11971. USA, Hawaii, Maui, Haleakala, from Argyroxiphium sp. (Asteraceae), 1969, M. Aragaki, culture ex-type of A. argyroxiphii CBS 117222 = E.G.S. 35.122.
Note: The host range of A. argyroxiphii is not restricted to Argyroxiphium, but has been broadened with the inclusion of two isolates from Ipomoea batatas (Convolvulaceae).
Alternaria azadirachtae E.G. Simmons & Alcorn, CBS Biodiversity Ser. (Utrecht) 6: 218. 2007.
Materials examined: Australia, Queensland, Tewantin, from Azadirachta indica (Meliaceae), 20 Jul. 1998, A. Bradley, culture ex-type of A. azadirachtae CBS 116444 = E.G.S. 46.195 = BRIP 25386 (ss1); additional strain from the same source, CBS 116445 = E.G.S. 46.196 = BRIP25386 (ss2).
Alternaria bataticola W. Yamam., Trans. Mycol. Soc. Japan 2(5): 89. 1960.
= Macrosporium bataticola Ikata, Agric. Hort. (Tokyo) 22: 241. 1947 (nom. inval., Art. 36.1).
Type: (Lectotype, designated in Simmons 2007) S. Ikata, Agric. & Hort. 22: 241. fig. 1. 1947.
Materials examined: Australia, Queensland, Walkamin, from leaf spot of Ipomoea batatas (Convolvulaceae), 5 Jul. 1991, collector unknown, representative isolate of A. bataticola CBS 117095 = E.G.S. 42.157 = IMI 350492 = BRIP 19470a; additional strain from the same source CBS 117096 = E.G.S. 42.158 = BRIP 19470b. Japan, Tokyo, from Ipomoea batatas, before Nov. 1963, collector unknown, CBS 532.63; from Ipomoea batatas, before Nov. 1963, collector unknown (epitype designated here CBS H-21743, MBT178114, culture ex-epitype CBS 531.63 = IFO 6187 = MUCL 28916). South Africa, Gauteng Province, Pretoria, ARC-Roodeplaat VOPI, from leaf and stem lesion of Ipomoea batatas, 16 Jun. 2010, M. Truter, PPRI 10502; Kwazulu-Natal Province, Empangeni, from leaf lesion of Ipomoea batatas, 4 Jul. 2011, A. Thompson, PPRI 11930; Kwazulu-Natal Province, Empangeni, from leaf lesion of Ipomoea batatas, 4 Jul. 2011, A. Thompson, PPRI 11931; Gauteng Province, Pretoria, ARC-Roodeplaat VOPI, from leaf lesion of Ipomoea batatas, 12 Jan. 2012, A. Thompson, PPRI 11934.
Alternaria blumeae E.G. Simmons & Sontirat, Mycotaxon 65: 81. 1997. Fig. 5.
Fig. 5.

Alternaria blumeae: conidia and conidiophores. A–D. CBS 117364. E–H. CBS 117215. Scale bars = 10 μm.
= Alternaria brasiliensis F.M. Queiroz, M.F.S. Muniz & M. Menezes, Mycopathologia 150: 63. 2001.
Materials examined: Brazil, Espirito Santo, from leaf spot of Phaseolus vulgaris (Fabaceae), 1989, F.M. Queiroz, representative isolate of A. brasiliensis CBS 117215 = E.G.S. 39.116. Thailand, Yala Province, Amphoe Muang, from Blumea aurita (Asteraceae), 18 Jan. 1992, P. Sontirat, culture ex-type of A. blumeae CBS 117364 = E.G.S. 40.149 = ATCC 201357.
Notes: By synonymising A. brasiliensis with A. blumeae, the host range of this taxon has expanded to include Phaseolus vulgaris. The five sequenced genes are 100 % identical between the two examined specimens.
Alternaria calendulae Ondřej, Čas. Slez. Mus., Ser. A, Hist. Nat. 23: 150. 1974. Fig. 6.
Fig. 6.

Alternaria calendulae: conidia and conidiophores. A–C. CBS 224.76. D–E. CBS 101498. F–H. CBS 116650. I–L. CBS 116439. Scale bars = 10 μm.
= Alternaria calendulae W. Yamam. 1939 (nom. nud.).
= Macrosporium calendulae Nelen, Bull. Centr. Bot. Gard. (Moscow) 35: 90. 1959 (nom. inval., Art. 36.1).
= Macrosporium calendulae Nelen, Bot. Mater. Otd. Sporov. Rast. Bot. Inst. Akad. Nauk S.S.S.R. 15: 144. 1962.
= Alternaria calendulae Nirenberg, Phytopathol. Z. 88: 108. 1977 (nom. illegit., Art. 53.1).
= Alternaria rosifolii E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 192. 2007.
Materials examined: Germany, former West-Germany, from leaf spot of Calendula officinalis (Asteraceae), 1974, H. Nirenberg, culture ex-type of A. calendulae Nirenberg CBS 224.76 = ATCC 38903 = IMI 205077 = DSM 63161. Japan, Tokyo, from leaf spot of Calendula officinalis, before 1964, representative isolate of A. calendulae CBS 116650 = E.G.S. 30.142 = QM 9561. New Zealand, Auckland, Kumeu, from leaf spot of Calendula officinalis, Oct. 1998, C.F. Hill, CBS 101498; Auckland, Mount Albert, from leaf of Rosa sp. (Rosaceae), before Feb. 1995, C.F. Hill, culture ex-type of A. rosifolii CBS 116439 = E.G.S. 42.197.
Note: By synonymising A. rosifolii with A. calendulae, the host range of this taxon has expanded to include Rosa.
Alternaria carthami S. Chowdhury, J. Indian Bot. Soc. 23: 65. 1944. Fig. 7.
Fig. 7.

Alternaria carthami: conidia and conidiophores. A–D. CBS 117091. E–H. CBS 116440. Scale bars = 10 μm.
= Macrosporium anatolicum A. Săvul., Bull. Sect. Sci. Acad. Roumaine 26: 709. 1944.
= Alternaria heliophytonis E.G. Simmons, CBS Biodiversity Ser. (Utrecht) 6: 206. 2007.
Materials examined: Canada, Saskatchewan, Saskatoon, from leaf of Helianthus annuus (Asteraceae), 26 Aug. 1993, C. Jasalavich, culture ex-type of A. heliophytonis CBS 116440 = IMI 366164 = E.G.S. 43.143. Italy, Perugia, from leaf of Carthamus tinctorius (Asteraceae), before Nov. 1980, A. Zazzerini, CBS 635.80. USA, Montana, Sidney, from leaf spot of Carthamus tinctorius, 11 Jul. 1973, E.E. Burns, representative isolate of A. carthami CBS 117091 = E.G.S. 31.037.
Notes: Isolate CBS 635.80 did not sporulate after 3 wk cultivation on SNA. By synonymising A. heliophytonis with A. carthami, the host range of this taxon has expanded to include Helianthus annuus (Asteraceae).
Alternaria carthamicola Woudenb. & Crous, sp. nov. MycoBank MB808991. Fig. 8.
Fig. 8.

Alternaria carthamicola sp. nov. CBS 117092: A–L. Conidia and conidiophores. Scale bars = 10 μm.
Etymology: Named after the host genus from which it was collected, Carthamus.
Primary conidiophores solitary or in small groups, simple, straight to slightly curved, septate, pale to dark brown with a subhyaline tip, (33–)55–71(–108) × 5–6(–7) μm, bearing a single, darkened, apical conidiogenous locus, but may produce geniculate conidiogenous extensions. Conidia solitary, rarely in chains of two conidia, conidium body pale olive-brown, mostly smooth-walled but sometimes ornamented at the base, ovoid, (39–)58–64(–82) × (13–)15–16(–17) μm; with (5–)6–7(–9) transverse and (1–)3(–4) longitudinal septa. Dark coloured eusepta can be formed during development; the conidial body is slightly constricted near the transverse septa. Conidia mostly have a septate, single to double filamentous beak, triple beaks are observed but not common, apical secondary conidiophores can be formed. Beaks (40–)158–186(–219) μm long, ca. 2 μm diam throughout their length and 4 μm at the base. Sexual morph not observed.
Culture characteristics: After 7 d cultures on SNA flat, rhizoid, white to opaque; aerial mycelium sparse, white, floccose, colonies reaching 55–60 mm diam; cultures on PCA flat, entire, olivaceous with three clear concentric circles; aerial mycelium fine, felty, olivaceous to olivaceous-grey, colonies reaching 65–70 mm diam; reverse shows four olivaceous concentric circles with an buff edge.
Material examined: Iraq, from Carthamus tinctorius (Asteraceae), 10 Apr. 1983, M.M. Elsahookie (holotype CBS H-21735, culture ex-type CBS 117092 = IMI 276943 = E.G.S. 37.057).
Notes: The new species A. carthamicola, originally identified as A. carthami, differs only on 9 nt positions in its RPB2 sequence from the other two A. carthami strains studied. Based on its RPB2 sequence it clusters with A. linicola.
Alternaria cassiae Jurair & A. Khan, Pakistan J. Sci. Industr. Res. 3: 72. 1960. Fig. 9.
Fig. 9.

Alternaria cassiae: conidia and conidiophores. A–D. CBS 116119. E–H. CBS 117224. I–L. CBS 117369. Scale bars = 10 μm.
= Alternaria hibiscinficiens E.G. Simmons & C.F. Hill, Mycotaxon 88: 205. 2003.
= Alternaria sauropodis E.G. Simmons, CBS Biodiversity Ser. (Utrecht) 6: 340. 2007.
Materials examined: Brazil, Federal District, from leaf spot of Senna obtusifolia (Fabaceae), May 1990, G. Fiqueiredo, representative isolate of A. cassiae CBS 117224 = E.G.S. 40.121. Fiji, from leaf of Hibiscus sabdariffa (Malvaceae), Jun. 2002, C.F. Hill, culture ex-type of A. hibiscinficiens CBS 177369 = E.G.S. 50.166. Malaysia, Sarawak, Kuching, from Sauropus androgynus (Phyllanthaceae), 25 Apr. 1984, T.K. Kieh, culture ex-type of A. sauropodis CBS 116119 = IMI 286317 = IMI 392448 = E.G.S. 47.112. USA, Mississippi, Stoneville, from diseased seedling of Senna obtusifolia, before Oct. 1980, H.L. Walker, representative isolate of A. cassiae CBS 478.81 = E.G.S. 33.147.
Notes: Isolate CBS 478.81 did not sporulate after 3 wk incubation on SNA. By synonymising A. hibiscinficiens and A. sauropodis with A. cassiae, the host range of this taxon has expanded to include Sauropus androgynus (Euphorbiaceae) and Hibiscus sabdariffa (Malvaceae).
Alternaria catananches Woudenb. & Crous, sp. nov. MycoBank MB808992. Fig. 10.
Fig. 10.

Alternaria catananches sp. nov. A–B. Disease symptoms on Catananche caerulea (photo's K.-H. Nugteren, Florensis B.V., Netherlands). C–L. CBS 137456: conidia and conidiophores. Scale bars = 10 μm.
Etymology: Named after its host genus from which it was isolated, Catananche.
Primary conidiophores solitary, simple, straight to curved, septate, pale brown, (31–)54–67(–94) × (5–)6(–7) μm, bearing a single, darkened, apical conidiogenous locus, but may produce geniculate conidiogenous extensions. Conidia solitary, conidium body pale olive-brown, ornamented in lower half of the conidium, narrowly ovoid, (26–)37–43(–57) × (7–)8–9(–11) μm, with (2–)4(–6) transverse septa and no longitudinal septa. Some darker coloured eusepta can be formed during development. The conidium body gradually tapers into a single, septate, unbranched beak; basal lateral secondary conidiophores can be formed. Beaks (77–)126–160(–260) μm long, ca. 2 μm diam throughout their length. Sexual morph not observed.
Culture characteristics: After 7 d cultures on SNA flat, entire/fimbriate, olivaceous around agar plug, white; aerial mycelium felty, white to olivaceous, colonies reaching 10–15 mm diam; cultures on PCA flat, erose, grey-olivaceous; aerial mycelium fine felty, olivaceous-grey; colonies reaching 25 mm diam; reverse identical.
Material examined: Netherlands, from Catananche caerulea (Asteraceae), 11 Dec. 2013, N. Troost-Riksen (holotype CBS H-21736, culture ex-type CBS 137456 = PD 013/05703936).
Notes: Alternaria catananches seems closely related to the A. cichorii isolates in the multi-gene phylogeny, but this is probably caused by long-branch attraction and incongruency between the different gene trees. Based on the ITS sequence it is identical to A. jesenskae, with RPB2 it is identical to A. cirsinoxia, with TEF1 it clusters with A. cichorii/A. cirsinoxia/A. carthami and with Alt a 1 it is identical to A. cichorii CBS 102.33, A. alternariacida and A. scorzonerae. Only its GAPDH sequences make it distinct from all other Alternaria species. Although the multi-gene tree does not provide strong support for separating it from the A. cichorii isolates, based on the individual gene sequences it is described here as a new Alternaria species.
Alternaria centaureae E.G. Simmons, CBS Biodiversity Ser. (Utrecht) 6: 236. 2007.
Specimen examined: USA, California, Sacramento, from Centaurea solstitialis (Asteraceae), Feb. 1999, D. Fogle, culture ex-type of A. centaureae CBS 116446 = E.G.S. 47.119.
Alternaria cichorii Nattrass, First List of Cyprus Fungi: 29. 1937.
≡ Alternaria porri f. sp. cichorii (Nattrass) T. Schmidt, Pflanzenschutzberichte 32: 181. 1965.
≡ Macrosporium cichorii (Nattrass) Gordenko, Mikol. Fitopatol. 9: 241. 1975.
Materials examined: Cyprus, from leaf spot of Cichorium intybus (Asteraceae), 1933, R.M. Nattrass (holotype IMI 1007, culture ex-type CBS 102.33 = E.G.S. 07.017 = QM 1760). Greece, Attica, from Cichorium endivia (Asteraceae), 24 Feb. 1978, S.D. Demetriades, representative isolate of A. cichorii CBS 117218 = E.G.S. 52.046 = IMI 225641.
Notes: Strain CBS 102.33 was deposited in Aug. 1933 in the CBS by R.M. Nattrass as A. cichorii sp. nov., with the remark that the description of the new species was in preparation. The holotype was subsequently deposited in IMI (IMI 1007) which consists of a dried herbarium specimen. In the present study we link CBS 102.33 as ex-type of A. cichorii to IMI 1007. The two isolates used in this study, CBS 102.33 and CBS 117218, differ only on 7 nt positions in their Alt a 1 sequence. Unfortunately CBS 102.33 is sterile, which does not provide additional information to support them as being two different species. Furthermore, the time difference of 45 yr between isolation of the two strains led to the decision to retain them as one species for now, pending fresh collections.
Alternaria cirsinoxia E.G. Simmons & K. Mort., Mycotaxon 65: 72. 1997.
Material examined: Canada, Saskatchewan, Watrous, from stem lesion and top dieback of Cirsium arvense (Asteraceae), 5 Aug. 1993, K. Mortensen, culture ex-type of A. cirsinoxia CBS 113261 = E.G.S. 41.136.
Alternaria citrullicola Woudenb. & Crous, sp. nov. MycoBank MB808993. Fig. 11.
Fig. 11.
Alternaria citrullicola sp. nov. CBS 103.32: A–H. Conidia and conidiophores. Scale bars = 10 μm.
Etymology: Named after the host genus from which it was collected, Citrullus.
Primary conidiophores solitary, simple, straight or sometimes curved, septate, pale brown with a subhyaline tip, (28–)35–52(–73) × (3–)4(–5) μm, bearing a single, darkened, apical conidiogenous locus. Conidia mostly solitary but chains of two conidia can occur, conidium body pale olive-brown, smooth-walled, narrowly ovoid, (28–)35–41(–56) × (6–)8(–10) μm; with (3–)5–6(–9) transverse distosepta and 0–1(–2) longitudinal septa. Conidia have a single, aseptate, unbranched filamentous beak; apical secondary conidiophores can be formed. Beaks (72–)178–232(–324) μm long, ca. 2 μm diam throughout their length. Sexual morph not observed.
Culture characteristics: After 7 d cultures on SNA flat, fimbriate, white to opaque with primrose sections near the edge; aerial mycelium sparse, fine felty, colonies reaching 45–50 mm diam; cultures on PCA flat, entire, olivaceous with three unclear concentric circles; aerial mycelium is sparse, pale olivaceous-grey, colonies reaching 50–55 mm diam; reverse shows olivaceous-buff to olivaceous rings.
Material examined: Cyprus, from fruit of Citrullus lanatus (Cucurbitaceae), before Jul. 1932, R.M. Nattrass (holotype CBS H-21742, culture ex-type CBS 103.32 = VKM F-1881).
Alternaria conidiophora Woudenb. & Crous, sp. nov. MycoBank MB808995. Fig. 12.
Fig. 12.

Alternaria conidiophora sp. nov. CBS 137457: A–H. Conidia and conidiophores. Scale bars = 10 μm.
Etymology: Named after its characteristically long, thick, conidiophores.
Primary conidiophores solitary, simple, mostly straight but sometimes curved, septate, dark brown with a subhyaline tip, (46–)89–105(–152) × (6–)7(–8) μm, bearing a single to multiple, darkened, long geniculate conidiogenous loci. Conidia solitary, conidium body olive-brown, smooth-walled, narrowly ovoid, (30–)45–52(–66) × (10–)12–13(–18) μm, with (2–)6–7(–9) transverse septa and (0–)1–2(–4) longitudinal septa. Darker coloured eusepta are formed during development. The conidial body is slightly constricted near the transverse septa. Conidia have a single, septate, unbranched, filamentous beak; basal, lateral secondary conidiophores can be formed. Beaks (49–)117–138(–186) μm long; ca. 2 μm diam throughout their length. Sexual morph not observed.
Culture characteristics: After 7 d cultures on SNA flat, fimbriate to rhizoid, white to opaque; aerial mycelium felty, white, colonies reaching 55–60 mm diam; cultures on PCA flat, entire, grey-olivaceous with two concentric circles; aerial mycelium wooly, pale olivaceous-grey, colonies reaching 55–60 mm diam; reverse identical.
Material examined: Netherlands, from unidentified host, Jul. 2011, U. Damm (holotype CBS H-21737, culture ex-type CBS 137457).
Alternaria crassa (Sacc.) Rands, Phytopathology 7: 337. 1917. Fig. 13.
Fig. 13.

Alternaria crassa: conidia and conidiophores. A–D. CBS 109162. E–H. CBS 116648. I–L. CBS 119160. Scale bars = 10 μm.
Basionym: Cercospora crassa Sacc., Michelia 1(no. 1): 88. 1877.
= Macrosporium solani Cooke, Grevillea 12: 32. 1883. (non M. solani Ellis & Martin, 1882)
= Cercospora daturae Peck, Rep. New York State Mus. Nat. Hist. 35: 140. 1884.
= Macrosporium cookei Sacc., Syll. Fungorum 4: 530. 1886. (nom. nov. in Saccardo for M. solani Cooke, 1883, non M. solani Ellis & Martin, 1882)
≡ Alternaria cookei (Sacc.) Bremer, Iʂmen, Karel, Özkan & M. Özkan, Istanbul Üniv. Fak. Mecm., B. 13: 42. 1948.
= Macrosporium daturae Fautrey, Rev. Mycol. (Toulouse) 16: 76. 1894.
≡ Alternaria daturae (Fautrey) Bubák & Ranoj., Fungi Imperf. Exsicc. Fasc. 14: 694. 1911.
= Alternaria capsici E.G. Simmons, Mycotaxon 75: 84. 2000.
Type: (Lectotype, designated in Simmons 2000) PAD, Cercospora crassa, Datura stramonium, S. [elva] ′76. 10.
Materials examined: Australia, from Capsicum annuum (Solanaceae), May 1981, D. Trimboli, culture ex-type of A. capsici CBS 109160 = IMI 262408 = IMI 381021 = E.G.S 45.075. Cyprus, Famagusta, from leaves of Datura stramonium (Solanaceae), Jan. 1936, R.M. Nattrass (epitype designated here CBS H-21744, MBT178115, culture ex-epitype CBS 110.38). New Zealand, Auckland, from leaf spot of Datura stramonium, 2002, C.F. Hill, representative isolate of A. crassa CBS 116448 = E.G.S. 50.180. USA, Indiana, Montgomery County, Nicandra physalodes (Solanaceae), 5 Sep. 1997, E.G. Simmons, CBS 109162 = E.G.S. 46.014; Indiana, from leaf spot of Datura stramonium, 5 Sep. 1997, E.G. Simmons, representative isolate of A. crassa CBS 116447 = E.G.S. 46.013; Indiana, Montgomery County, from leaf spot of Datura stramonium, 1 Aug. 1996, E.G. Simmons, representative isolate of A. crassa CBS 122590 = E.G.S. 44.071; Wisconsin, Madison, from leaf spot of Datura sp., before Apr. 1918, R.D. Rands, CBS 103.18.
Notes: Isolates CBS 110.38 and CBS 116647 did not sporulate after 3 wk incubation on SNA. By synonymising A. capsici with A. crassa, the host range of this taxon expanded to include Capsicum annuum, which also belongs to the Solanaceae.
Alternaria cucumerina (Ellis & Everh.) J.A. Elliott, Amer. J. Bot. 4: 472. 1917. Fig. 14.
Fig. 14.

Alternaria cucumerina: conidia and conidiophores. A–D. CBS 117225. E–H. CBS 117226. I–L. CBS 116114. Scale bars = 10 μm.
Basionym: Macrosporium cucumerinum Ellis & Everh., Proc. Acad. Nat. Sci. Philadelphia 47: 440. 1895.
= Alternaria loofahae E.G. Simmons & Aragaki, CBS Biodiversity Ser. (Utrecht) 6: 316. 2007.
Materials examined: Australia, Queensland, from leaf spot of Cucumis melo (Cucurbitaceae), Oct. 1996, R. O’Brien, representative isolate of A. cucumerina CBS 117226 = E.G.S. 44.197 = BRIP 23060. USA, Hawaii, Oahu, Waialua, from Luffa acutangula (Cucurbitaceae), 1971, M. Aragaki, culture ex-type of A. loofahae CBS 116114 = E.G.S. 35.123; Indiana, Knox County, from leaf spot of Cucumis melo, 1993, R.X. Latin, representative isolate of A. cucumerina CBS 117225 = E.G.S. 41.127.
Notes: The species clade for A. cucumerina does not have a clear support in the multi-gene phylogeny. CBS 117225 and CBS 117226 differ only on 2 nt in their RPB2 sequence, while the ex-type of A. loofahae (CBS 116114) differs on 1 nt from both A. cucumerina isolates in RPB2 and on 1 nt in Alt a 1. This internal variation in the two A. cucumerina isolates and the identical host family, Cucurbitaceae, with A. loofahae, supported the synonymy of A. loofahae. By synonymising A. loofahae with A. cucumerina, the host range of this taxon expanded to include Luffa acutangula.
Alternaria cyamopsidis Rangaswami & A.V. Rao, Indian Phytopathol. 10: 23. 1957.
≡ Alternaria cucumerina var. cyamopsidis (Rangaswami & A.V. Rao) E.G. Simmons, Mycopathol. Mycol. Appl. 29: 131. 1966.
Materials examined: USA, Georgia, from leaf spot of Cyamopsis tetragonoloba (Fabaceae), Jul. 1961, G. Sowell, representative isolate of A. cyamopsidis CBS 117219 = E.G.S. 13.120 = QM 8000; Maryland, Beltsville, from leaf spot of Cyamopsis tetragonoloba, 1964, R.G. Orellana, representative isolate of A. cyamopsidis CBS 364.67 = E.G.S. 17.065 = QM 8575.
Alternaria dauci (J.G. Kühn) J.W. Groves & Skolko, Canad. J. Res., Sect. C, Bot. Sci. 22: 222. 1944. Fig. 15.
Fig. 15.

Alternaria dauci. A. Disease symptoms on Daucus carota. B–L. Conidia and conidiophores. B–C. CBS 117097. D–F. CBS 117098. G–I. CBS 117099. J–L. CBS 117100. Scale bars = 10 μm.
Basionym: Sporidesmium exitiosum var. dauci J.G. Kühn, Hedwigia 1: 91. 1855.
≡ Polydesmus exitiosus var. dauci (J.G. Kühn) J.G. Kühn, Die Krankheiten der Kulturgewächse, ihre Ursachen und ihre Verhütung: 165. 1858.
≡ Macrosporium dauci (J.G. Kühn) Rostr., Tidsskr. Landoekon. ser. 5, 7: 385. 1888.
≡ Alternaria brassicae var. dauci (J.G. Kühn) Lindau, Rabenhorst‘s Kryptog.-Fl., Edn 2 (Leipzig) 1(9): 260. 1908.
≡ Alternaria porri f. sp. dauci (J.G. Kühn) Neerg, Danish species of Alternaria & Stemphylium: 252. 1945.
= Macrosporium carotae Ellis & Langl., J. Mycol. 6: 36. 1890.
≡ Alternaria carotae (Ellis & Langl.) J.A. Stev. & Wellman, J. Wash. Acad. Sci. 34: 263. 1944.
= Alternaria poonensis Ragunath, Mycopathol. Mycol. Appl. 21: 315. 1963.
Type: (Lectotype, designated in Simmons 1995) B, ms. spec. Sporidesmium exitiosum var. dauci Kühn, Leg. Gross Krausche p. Bunzlau, Jul. Kühn.
Materials examined: Italy, from seed of Daucus carota (Apiaceae), Sept. 1937, P. Neergaard (neotype designated here CBS H-21745, MBT178116, culture ex-neotype CBS 111.38). Netherlands, Limburg, Horst, from leaf spot in Cichorium intybus var. foliosum (Asteraceae), 1979, W.M. Loerakker, CBS 477.83 = CBS 721.79 = PD 79/954; from seed of Daucus carota, 1993, S&G Seeds, CBS 101592. New Zealand, from leaf spot of Daucus carota, Mar. 1998, C.F. Hill, representative isolate of A. dauci CBS 117098 = E.G.S. 46.152; Ohakune, from leaf spot of Daucus carota, before Jul. 1979, G.F. Laundon, CBS 345.79 = LEV 14814. Puerto Rico, from seedling of Coriandrum sativum (Apiaceae), 1999, W. Almodovar, representative isolate of A. poonensis CBS 117100 = E.G.S. 47.138. Unknown, from seed of Daucus carota, Jan. 1948, J.W. Groves, CBS 106.48. USA, California, from commercial seed of Daucus carota, Nov. 1994, B.M. Pryor, representative isolate of A. dauci CBS 117097 = E.G.S. 46.006; California, Kern County, from seed of Daucus carota, 1999, D. Fogle, representative isolate of A. dauci CBS 117099 = E.G.S. 47.131.
Notes: The indicated lectotype cannot be traced in B, and appears to be lost. We therefore designate CBS 111.38 as neotype. The isolates CBS 111.38, CBS 345.79 and CBS 101592 did not sporulate after 3 wk incubation on SNA.
Alternaria deserticola Woudenb. & Crous, sp. nov. MycoBank MB808996.
Etymology: Named after the substrate from which it was isolated, namely desert soil.
Culture sterile
Alternaria deserticola differs from the ex-type strain of its closest phylogenetic neighbour A. thunbergiae (CBS 116331) based on alleles in all five loci (positions derived from respective alignments of the separate loci deposited in TreeBASE): ITS position 165 (−), 373 (T), 381 (C), 383 (C), 488 (A); GAPDH position 484 (T); RPB2 position 76 (C), 88 (T), 91 (T), 139 (C), 211 (T), 316 (T), 490 (C), 496 (A), 646 (T), 670 (C), 671 (T), 673 (A), 760 (G); TEF1 position 37 (C), 49 (G), 197 (A), 223 (A), 274 (T), 277(–), 311(T); Alt a 1 position 10 (C), 209 (A), 210 (T), 220 (G), 322 (T), 452 (G).
Culture characteristics: After 7 d cultures on SNA flat, rhizoid, olivaceous-buff; aerial mycelium absent, colonies reaching 55 mm diam; cultures on PCA flat, entire, five grey-olivaceous concentric circles; aerial mycelium sparse, colonies reaching 75–80 mm diam; reverse shows five olivaceous-grey rings.
Material examined: Namibia, from desert soil, 2001, M. Christensen (holotype CBS H-21738, culture ex-type CBS 110799).
Note: The clear phylogenetic distinction of the sterile culture of A. deserticola from all other strains included in this study, resulted in our decision to describe this species based on sequence data only.
Alternaria dichondrae Gambogi, Vannacci & Triolo, Trans. Brit. Mycol. Soc. 65(2): 323. 1975.
Materials examined: Italy, Pisa, from leaf spot of Dichondra repens (Convolvulaceae), Mar. 1974, P. Gambogi, ex-isotype of A. dichondrae CBS 199.74 = E.G.S. 38.007; Pisa, from leaf spot of Dichondra repens, Mar. 1974, P. Gambogi, living lectotype of A. dichondrae CBS 200.74 = E.G.S. 38.008. New Zealand, from leaf spot of Dichondra repens, before 1979, G.F. Laundon, CBS 346.79; Auckland, Lynfield, from leaf of Dichondra sp., Apr. 1991, C.F. Hill, representative isolate of A. dichondrae CBS 117127 = E.G.S. 40.057.
Note: Simmons (2007) designated a lectotype with ex-lectotype strain (CBS 200.74), as he found the ex-isotype strain (CBS 199.74) to be sterile.
Alternaria echinaceae E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 318. 2007.
Materials examined: New Zealand, Gisborne, Makaraka, from leaf of Echinacea sp. (Asteraceae), Jan. 1998, C.F. Hill, culture ex-type of A. echinaceae CBS 116117 = E.G.S. 46.081; Gisborne, Makaraka, from leaf of Echinacea sp., Jan. 1998, C.F. Hill, representative isolate of A. echinaceae CBS 116118 = E.G.S. 46.082.
Alternaria grandis E.G. Simmons, Mycotaxon 75: 96. 2000. Fig. 16.
Fig. 16.
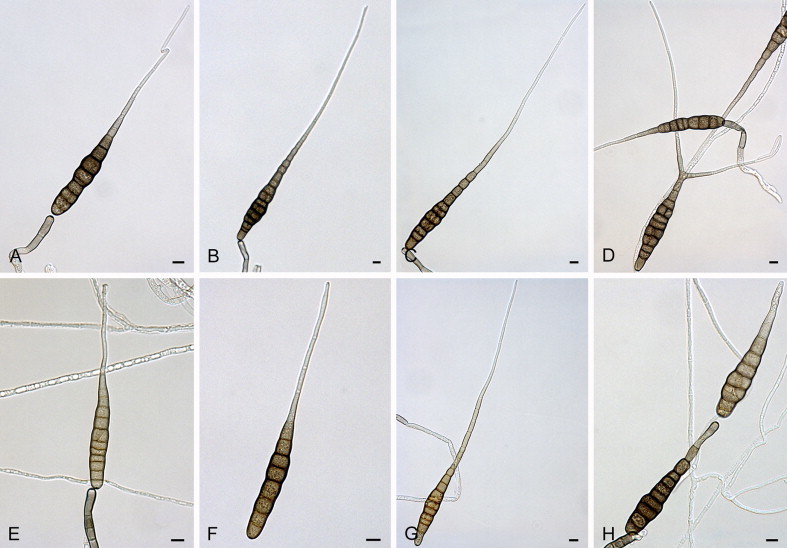
Alternaria grandis: conidia and conidiophores. A–D. CBS 109158. E–H. CBS 116695. Scale bars = 10 μm.
Materials examined: USA, Pennsylvania, Centre County, from leaf lesion of Solanum tuberosum (Solanaceae), Sep. 1966, B.J. Christ, culture ex-type of A. grandis CBS 109158 = E.G.S. 44.106; Pennsylvania, Clarion County, from leaf spot of Solanum tuberosum, Sep. 1966, B.J. Christ, representative isolate of A. grandis CBS 116695 = E.G.S 44.108.
Notes: Although A. grandis differs by only 1 nt in its GAPDH sequence from A. solani, we retain it as a distinct species. Conidia of A. grandis are substantially larger than those of A. solani, and a recently published study could separate A. solani (CBS 109157) and A. grandis (CBS 109158) based on partial calmodulin gene sequence data (Gannibal et al. 2014).
Alternaria ipomoeae M. Truter, Woudenb. & Crous, sp. nov. MycoBank MB808997. Fig. 17.
Fig. 17.

Alternaria ipomoeae sp. nov. CBS 219.79: A–L. Conidia and conidiophores. Scale bars = 10 μm.
Etymology: Named after the host genus on which it occurs, Ipomoea.
Primary conidiophores simple to branched, straight to slightly curved, septate, pale brown, (10–)51–73(–145) × (4–)5 μm, bearing a single to multiple, darkened, geniculate conidiogenous loci. Conidia mostly solitary but chains of two conidia can occur, conidium body olive-brown, smooth-walled with ornamented base, long ellipsoid to obclavate, (53–)60–65(–76) × (9–)12(–15) μm, with (6–)8–9(–12) transverse septa and (0–)2(–3) longitudinal septa. Up to four dark coloured eusepta can be formed during development; the conidial body is constricted near these eusepta. Conidia have a septate, single to double, filamentous beak; apical and lateral secondary conidiophores can be formed. Beaks (47–)136–162(–221) μm long, single beaks generally longer than multiple beaks, ca. 2 μm diam throughout their length, and approx. 3 μm diam at the base. Sexual morph not observed.
Culture characteristics: After 7 d cultures on SNA are flat, fimbriate, white; aerial mycelium sparse, felty, white, colonies reaching 50 mm diam; cultures on PCA flat, entire, grey-olivaceous with some darker sections; aerial mycelium fine felty, pale olivaceous-grey, colonies reaching 65–70 mm diam; reverse identical.
Materials examined: Ethiopia, from black lesions of Ipomoea batatas (Convolvulaceae), Jun. 1978, A.H.C. van Bruggen (holotype CBS H-21739, culture ex-type CBS 219.79). South Africa, Gauteng Province, Pretoria, ARC-Roodeplaat VOPI, from stem lesions of Ipomoea batatas, 16 Nov. 2006, C.D. Narayanin (paratype PREM 60979, culture ex-paratype PPRI 8988).
Alternaria jesenskae Labuda, P. Eliáš & Sterfl., Microbiol. Res. 163: 209. 2008.
Material examined: Slovakia, district of the village Muzla, Podunajská nizina lowland, from seeds of Fumana procumbens (Cistaceae), Aug. 1999, P. Eliáš jr., culture ex-type of A. jesenskae CBS 133855 = CCM 8361.
Alternaria linariae (Neerg.) E.G. Simmons, CBS Biodiversity Ser. (Utrecht) 6: 677. 2007. Fig. 18.
Fig. 18.

Alternaria linariae. A. Disease symptoms on Solanum lycopersicum. B–P. Conidia and conidiophores. B–C. CBS 105.41. D–F. CBS 109161. G–H. CBS 107.61. I–J. CBS 109156. K–L. CBS 109164. M–N. CBS 116438. O–P. CBS 116441. Scale bars = 10 μm.
Basionym: Alternaria anagallidis var. linariae Neerg., Danish species of Alternaria & Stemphylium: 297. 1945.
= Alternaria cretica E.G. Simmons & Vakal., Mycotaxon 75: 64. 2000.
= Alternaria subcylindrica E.G. Simmons & R.G. Roberts, Mycotaxon 75: 62. 2000.
= Alternaria tomatophila E.G. Simmons, Mycotaxon 75: 53. 2000.
= Alternaria cucumericola E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 210. 2007.
= Alternaria tabasco E.G. Simmons & R.G. Roberts, CBS Biodiversity Ser. (Utrecht) 6: 158. 2007.
Materials examined: Belgium, host unknown, before Mar. 1961, R. Sys, CBS 107.61. Denmark, from seedling of Linaria maroccana (Scrophulariaceae), 13 Nov. 1940, P. Neergaard, culture ex-type of A. linariae CBS 105.41 = E.G.S. 07.016. Greece, Crete, Heraklio, from leaf spot of Solanum lycopersicum (Solanaceae), 1997, D.J. Vakalounakis, culture ex-type of A. cretica, CBS 109164 = E.G.S. 46.188. New Zealand, Northland, Kerikeri, from leaf spot of Cucumis sativus (Cucurbitaceae), Mar. 1993, C.F. Hill, culture ex-type of A. cucumericola CBS 116438 = E.G.S. 41.057. Thailand, Chiang Mai, Royal project, from leaf spot of Solanum lycopersicum, 5 Nov. 2012, P.W. Crous, CPC 21620. Unknown, host unknown, before Apr. 1953, P.W. Brian, CBS 108.53 = No. 408P. USA, Indiana, Montgomery County, from leaf spot of Solanum lycopersicum, 23 Aug. 1995, E.G. Simmons, culture ex-type of A. tomatophila CBS 109156 = E.G.S. 42.156; Indiana, from leaf lesion of Solanum lycopersicum, Aug. 1996, E.G. Simmons, representative isolate of A. tomatophila CBS 116704 = E.G.S. 44.074; Louisiana, Baton Rouge, Louisiana State University Burden Research Plantation, from leaf lesion of Solanum lycopersicum var. cerasiforme, 2 Jul. 1997, R.G. Roberts, culture ex-type of A. subcylindrica CBS 109161 = E.G.S. 45.113; Louisiana, Avery Island, from leaf spot of Capsicum frutescens (Solanaceae), 1 Jul. 1997, R.G. Roberts, culture ex-type of A. tabasco CBS 116441 = E.G.S 45.108 = R.G.R. 97-52.
Notes: By synonymising A. cretica, A. cucumericola, A. subcylindrica, A. tabasco and A. tomatophila with A. linariae, the broad host range of this taxon now consists of Solanaceae, Cucurbitaceae and Scrophulariaceae species. The isolates CBS 108.53 and CBS 116704 did not sporulate on SNA after 3 wk of incubation.
Alternaria macrospora Zimm., Ber. Land-Forstw. Deutsch-Ostafrika 2: 24. 1904.
≡ Macrosporium macrosporum (Zimm.) Nishikado & Oshima, Agric. Res. (Kurashiki) 36: 391. 1944.
= Sporidesmium longipedicellatum Reichert, Bot. Jahrb. Syst. 56: 723. 1921.
≡ Alternaria longipedicellata (Reichert) Snowden, Rep. Dept. Agric. Uganda: 31. 1927 [1926].
Materials examined: Nigeria, from Gossypium sp. (Malvaceae), May 1929, Jones, CBS 106.29. USA, Arizona, from Gossypium barbadense (Malvaceae), before 1984, P.J. Cotty, culture epitype of A. macrospora CBS 117228 = E.G.S. 50.190 = ATCC 58172.
Notes: Isolate CBS 106.29 was preserved in the CBS collection as A. porri, but did not sporulate since 1978. Based on our molecular data this isolate belongs to A. macrospora, which, based on the same host, seems plausible.
Alternaria montanica E.G. Simmons & Robeson, CBS Biodiversity Ser. (Utrecht) 6: 178. 2007.
Material examined: USA, Montana, from Cirsium arvense (Asteraceae), before Apr. 1981, D.J. Robeson, culture ex-type of A. montanica CBS 121343 = E.G.S. 44.112 = IMI 257563.
Alternaria multirostrata E.G. Simmons & C.R. Jacks., Phytopathology 58: 1139. 1968.
Materials examined: USA, Georgia, Tifton, from floral bract of Richardia scabra (Rubiaceae), 1967, C.R. Jackson, culture ex-type of A. multirostrata CBS 712.68 = ATCC 18515 = IMI 135454 = MUCL 11722 = QM 8820 = VKM-F2997; Georgia, Tifton, from floral bract of Richardia scabra, 1967, C.R. Jackson, representative isolate of A. multirostrata CBS 713.68 = ATCC 18517 = IMI 135455 = MUCL 11715 = QM 8821.
Alternaria neoipomoeae M. Truter, Woudenb. & Crous, sp. nov. MycoBank MB808998. Fig. 19.
Fig. 19.

Alternaria neoipomoeae sp. nov. A. Disease symptoms on Ipomoeae batatas (Photo A.H. Thompson, ARC, South Africa). B–L. PPRI 11845: conidia and conidiophores. Scale bars = 10 μm.
Etymology: Named after its close phylogenetic relationship to A. ipomoeae.
Primary conidiophores solitary, simple, straight to slightly curved, septate, pale brown, (10–)23–59(–111) × (4–)5 μm, bearing a single, darkened, apical conidiogenous locus, which may produce 1–2 geniculate conidiogenous extensions. Conidia are mostly solitary but chains of two conidia can occur, conidium body olive-brown, smooth-walled with ornamented base, long ellipsoid to obclavate, (52–)66–77(–93) × (12–)14–16(–18) μm, with (7–)9(–12) transverse and (2–)3–4(–5) longitudinal septa. Up to four dark coloured eusepta can be formed during development; the conidial body is constricted near these eusepta. Conidia mostly have a septate, single to double, filamentous beak, triple beaks are observed but not common; apical and lateral secondary conidiophores can be formed. Beaks (54–)104–136(–200) μm long, ca. 2 μm diam throughout their length, and approx. 3 μm diam at the base. Sexual morph not observed.
Culture characteristics: After 7 d cultures on SNA flat, fimbriate, white to opaque; aerial mycelium sparse, fine felty, white, colonies reaching 60−65 mm diam; cultures on PCA flat, entire, grey-olivaceous with 2 dark and one lighter concentric circles and a pale olivaceous edge; aerial mycelium fine felty, pale olivaceous-grey, colonies reaching 55–60 mm diam; reverse four olivaceous-grey rings.
Materials examined: South Africa, Gauteng Province, Pretoria, ARC-Roodeplaat VOPI, from stem lesion of Ipomoea batatas (Convolvulaceae), 8 Jun. 2011, A. Thompson (holotype PREM 60981, culture ex-type PPRI 11845); North-West Province, Brits, from Ipomoea batatas, 25 Oct. 2007, C.D. Narayanin (paratype PREM 60982, culture ex-paratype PPRI 8990); Mpumalanga Province, Kwamahlanga, from Ipomoea batatas, between 2006 and 2008, C.D. Narayanin (paratype PREM 60983, culture ex-paratype PPRI 11847); Gauteng Province, Pretoria, ARC-Roodeplaat VOPI, from leaf lesion of Ipomoea batatas, Oct. 2013, A. Thompson (paratype PREM 60984, culture ex-paratype PPRI 13903).
Alternaria nitrimali E.G. Simmons & M.E. Palm, Mycotaxon 75: 93. 2000.
Material examined: Puerto Rico, Luquillo, from leaf spot of Solanum viarum (Solanaceae), 26 Feb. 1998, USDA-APHIS, culture ex-type of A. nitrimali CBS 109163 = E.G.S 46.151.
Alternaria novae-guineensis E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 350. 2007.
Materials examined: Papua New Guinea, from dried leaf of Citrus sp. (Rutaceae) imported to New Zealand, 1999, C.F. Hill, culture ex-type of A. novae-guineensis CBS 116120 = E.G.S. 47.198. South Africa, Gauteng, Pretoria, ARC-Roodeplaat VOPI, from leaves of Galinsoga parviflora (Asteraceae), 12 Jan. 2012, A. Thompson, PPRI 12171.
Alternaria obtecta E.G. Simmons, Mycotaxon 50: 250. 1994.
Materials examined: USA, California, Encinitas, from leaf of Euphorbia pulcherrima (Euphorbiaceae), Nov. 1994, C.F. Hill, representative isolate of A. obtecta CBS 117367 = E.G.S. 42.063; California, Encinitas, from Euphorbia pulcherrima (Euphorbiaceae), Nov. 1994, C.F. Hill, CBS 134278 = E.G.S. 42.064.
Alternaria paralinicola Woudenb. & Crous, sp. nov. MycoBank MB808999. Fig. 20.
Fig. 20.

Alternaria paralinicola sp. nov. CBS 116652: A–L. Conidia and conidiophores. Scale bars = 10 μm.
Etymology: Named after its close phylogenetic relationship to A. linicola.
Primary conidiophores solitary, simple, straight to slightly curved, septate, pale brown, (39–)64–82(–133) × (4–)5–6 μm, bearing a single, darkened, apical conidiogenous locus, but may produce geniculate conidiogenous extensions. Conidia are mostly solitary but chains of two conidia can occur, conidium body pale olive-brown, smooth-walled, narrowly ovoid, (31–)39–44(–58) × (8–)10–11(–15) μm, with (3–)5–6(–8) transverse septa and 0–1(–2) longitudinal septa. Dark coloured eusepta are formed during maturation. The conidial body is slightly constricted near the transverse septa. Some transverse blocks of cells can have a conspicuously different width in comparison with neighbouring segments, resulting in specific shape of the conidium body. Conidia mostly have a single, aseptate, unbranched, filamentous beak; double beaks are observed but not common; apical or lateral secondary conidiophores can be formed. Beaks (61–)114–135(–169) μm long, ca. 2 μm diam throughout their length. Sexual morph not observed.
Culture characteristics: After 7 d cultures on SNA flat, fimbriate, white to opaque; aerial mycelium sparse, white, colonies reaching 70–75 mm diam; cultures on PCA flat, entire, grey-olivaceous with four olivaceous clear concentric circles; aerial mycelium is fine felty, olivaceous, colonies reaching 70 mm diam; reverse shows five grey-olivaceous concentric circles.
Material examined: Canada, Manitoba, from seeds of cultivated Linum usitatissimum (Linaceae), 1996, M.E. Corlett (holotype CBS H-21740, culture ex-type CBS 116652 = E.G.S. 47.157 = DAOM 225747).
Note: Alternaria paralinicola, which was originally identified as A. linicola, differs on 16 nt positions in its RPB2 sequence from the other two A. linicola strains studied. Based on its RPB2 sequence it clusters with A. passiflorae.
Alternaria passiflorae J.H. Simmonds, Proc. Roy. Soc. Queensland. 49: 151. 1938. Fig. 21.
Fig. 21.

Alternaria passiflorae: conidia and conidiophores. A–B. CBS 117102. C–D. CBS 117103. E–F. CBS 116333. G–H. CBS 166.77. I–J. CBS 630.93. K–L. CBS 629.93. Scale bars = 10 μm.
= Alternaria hawaiiensis E.G. Simmons, Mycotaxon 46: 184. 1993.
= Alternaria gaurae E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 188. 2007.
Materials examined: New Zealand, from fruit of Passiflora edulis (Passifloraceae), 6 Feb. 1963, F.J. Mortin, representative isolate of A. passiflorae CBS 629.93 = E.G.S. 16.150 = QM 8458; Auckland, from fruit spot of Passiflora ligularis (Passifloraceae), Apr. 2004, C.F. Hill, representative isolate of A. passiflorae CBS 117102 = E.G.S. 51.165; Auckland, from leaf spot of Passiflora caerulea (Passifloraceae), Jul. 2004, C.F. Hill, representative isolate of A. passiflorae CBS 117103 = E.G.S. 52.032; Auckland, from leaf spot of Gaura lindheimeri (Onagraceae), May 2002, C.F. Hill, culture ex-type of A. gaurae CBS 116333 = E.G.S. 50.121; Waitakere, from leaf of Capsicum frutescens (Solanaceae), May 1975, CBS 166.77. USA, Hawaii, from Passiflora edulis, before Oct. 1968, M. Aragaki, culture ex-type of A. hawaiiensis CBS 630.93 = E.G.S. 29.020 = QM 9050.
Notes: By synonymising A. gaurae with A. passiflorae, and including CBS 166.77, formerly identified as A. solani, the host range of A. passiflorae has broadened to include Gaura sp. (Onagraceae) and Capsicum frutescens (Solanaceae).
Alternaria pipionipisi E.G. Simmons, CBS Biodiversity Ser. (Utrecht) 6: 302. 2007.
Materials examined: India, Andhra Pradesh, Hyderabad, from seed of Cajanus cajan (Fabaceae), before Feb. 1990, K.M. & Ch. Reddy, culture ex-type of A. pipionipisi CBS 116115 = E.G.S. 40.096 = IMI 340950. USA, California, Encinitas, from Euphorbia pulcherrima (Euphorbiaceae), Sep. 1994, C.F. Hill, CBS 134265 = E.G.S. 42.047; California, Encinitas, from Euphorbia pulcherrima, Sep. 1994, C.F. Hill, representative isolate of A. obtecta CBS 117365 = E.G.S. 42.048.
Alternaria porri (Ellis) Cif., J. Dept. Agric. Porto Rico 14: 30. 1930 [1929]. Fig. 22.
Fig. 22.

Alternaria porri: conidia and conidiophores. A–D. CBS 116698. E–H. CBS 116699. I–L. CBS 116649. Scale bars = 10 μm.
Basionym: Macrosporium porri Ellis, Grevillea 8 (no. 45): 12. 1879.
≡ Alternaria porri (Ellis) Sawada, Rep. Dept. Agric. Gov. Res. Inst. Formosa, 61: 92. 1930.
Type: (Lectotype, designated in Simmons 2007) NY, Ellis Collection: on leaves of Allium porrum, Newfield, N.J. Sept. 78.
Materials examined: USA, Nebraska, Lincoln, from leaf of Allium cepa (Amaryllidaceae), 1965, D.S. Meredith, representative isolate of A. allii CBS 116649 = E.G.S. 17.082 = QM 8613; New York, Ithaca, from leaf of Allium cepa, 1996, M.J. Yáñes Morales, representative isolate of A. porri CBS 116698 = E.G.S. 48.147; New York, Orange County, from leaf of Allium cepa, 1996, M.J. Yáñes Morales (epitype designated here CBS H-21746, MBT178117, culture ex-epitype CBS 116699 = E.G.S. 48.152).
Alternaria protenta E.G. Simmons, Mycotaxon 25: 207. 1986. Fig. 23.
Fig. 23.

Alternaria protenta: conidia and conidiophores. A–B. CBS 116696. C–D. CBS 116697. E–G. CBS 116643. H–J. CBS 116651. K–M. CBS 121342. N–P. CBS 347.79. Scale bars = 10 μm.
= Alternaria pulcherrimae T.Y. Zhang & J.C. David, Mycosystema 8-9: 110. 1996.
= Alternaria hordeiseminis E.G. Simmons & G.F. Laundon, CBS Biodiversity Ser. (Utrecht) 6: 150. 2007.
Materials examined: Australia, Queensland, Brisbane, Chapel Hill, from Euphorbia pulcherrimae (Euphorbiaceae), 25 Aug. 1986, J.L. Alcorn, representative isolate of A. pulcherrimae CBS 121342 = E.G.S. 42.122 = IMI 310506. Israel, from Helianthus annuus (Asteraceae), 1996, collector unknown, representative isolate of A. protenta CBS 116697 = E.G.S. 45.024 = IMI 372957; from Helianthus annuus, 1996, collector unknown, representative isolate of A. protenta CBS 116696 = E.G.S. 45.023 = IMI 372955. New Zealand, Hastings, from Solanum tuberosum (Solanaceae), Mar. 1997, C.F. Hill, representative isolate of A. solani CBS 135189 = E.G.S. 45.053; Levin, from fruit rot of Solanum lycopersicum (Solanaceae), before Jul. 1979, G.F. Laundon, CBS 347.79 = E.G.S. 44.091 = ATCC 38569 = LEV 14726; Palmerston North, from seed of Hordeum vulgare (Gramineae), Jul. 1977, G.F. Laundon, culture ex-type of A. hordeiseminis CBS 116437 = E.G.S. 32.076 = CBS 116443 = E.G.S. 46.163. USA, California, Siskiyou, from Solanum tuberosum, 1996, D. Fogle, representative isolate of A. solani CBS 116651 = E.G.S. 45.020.
Notes: By synonymising A. pulcherrimae and A. hordeiseminis with A. protenta and including three isolates formerly identified as A. solani (CBS 347.79, 116651 and 135189), the host range of A. protenta has expanded extensively. It now comprises plants from the Asteraceae, Euphorbiaceae, Gramineae and Solanaceae. Based on molecular (and morphological) data, A. protenta is closely related to A. solani, and these two species can only be distinguished based on 9 nt differences in their RPB2 sequences (see RPB2 alignment in TreeBASE).
Alternaria pseudorostrata E.G. Simmons, Mycotaxon 57: 398. 1996.
Material examined: USA, California, Encinitas, from Euphorbia pulcherrimae (Euphorbiaceae), Dec. 1994, C.F. Hill, culture ex-type of A. pseudorostrata CBS 119411 = E.G.S. 42.060.
Alternaria ranunculi E.G. Simmons, CBS Biodiversity Ser. (Utrecht) 6: 212. 2007.
Material examined: Israel, Palestine, from seed of Ranunculus asiaticus (Ranunculaceae), 10 Apr. 1984, collector unknown, culture ex-type of A. ranunculi CBS 116330 = E.G.S. 38.039 = IMI 285697.
Alternaria ricini (Yoshii) Hansf., Proc. Linn. Soc. Lond. : 53. 1943.
Basionym: Macrosporium ricini Yoshii, Bult. Sci. Fak. Terk. Kjusu Imp. Univ. 3(4): 327. 1929.
Type: (Lectotype, designated in Simmons 1994) BPI 445446, Macrosporium ricini, Japan, Fukuoka, Ricinus communis, July 1928.
Materials examined: Italy, Sardinia, Sasseri, from Ricinus communis (Euphorbiaceae), before Aug. 1986, J.A. von Arx, CBS 353.86. Japan, Ricinus communis, deposited Feb. 1931 by K. Nakata (epitype designated here CBS H-21747, MBT178118, culture ex-epitype CBS 215.31). USA, Virginia, Holland, from leaf of Ricinus communis, 9 Aug. 1954, C.A. Thomas, representative isolate of A. ricini CBS 117361 = E.G.S. 06.181.
Alternaria rostellata E.G. Simmons, Mycotaxon 57: 401. 1996.
Material examined: USA, California, Encinitas, from leaf of Euphorbia pulcherrimae (Euphorbiaceae), Jan. 1995, C.F. Hill, culture ex-type of A. rostellata CBS 117366 = E.G.S. 42.061.
Alternaria scorzonerae (Aderh.) Loer., Netherlands J. Pl. Pathol. 90(1): 37. 1984.
Basionym: Sporidesmium scorzonerae Aderh., Arbeiten Kaiserl. Biol. Anst. Land-Forstw. 3: 439. 1903.
= Alternaria linicola J.W. Groves & Skolko, Canad. J. Res., Sect. C, Bot. Sci. 22: 223. 1944.
= Alternaria linicola Neerg, Danish species of Alternaria & Stemphylium: 302. 1945. (nom. illegit., Art. 53.1)
Type: (Lectotype, designated in Simmons 1997) Aderhold, Arbeiten Kaiserl. Biol. Anst. Land-Forstw. 3: 440. fig. w/o number. 1903.
Materials examined: Netherlands, Reusel, from leaf spot of Scorzonera hispanica (Asteraceae), 1982, W.M. Loerakker (epitype designated here CBS H-21748, MBT178119, culture ex-epitype CBS 478.83 = E.G.S. 38.011). UK, Scotland, from Linum usitatissimum (Linaceae), 22 Nov. 1945, J.W. Groves, CBS 103.46; Derbyshire, from seed of Linum usitatissimum, 1983, C. Nicholls, representative isolate of A. linicola CBS 116703 = E.G.S. 36.110 = IMI 274549.
Notes: None of the three isolates sporulated on SNA or PCA after 3 wk of incubation, also not after scarification. Corlett & Corlett (1999) already stated that, after sub-cultivation, A. linicola sporulates poorly, or not at all. By synonymising A. linicola with A. scorzonerae, the host range of A. scorzonerae is expanded to include Linum usitatissimum (Linaceae).
Alternaria sennae Woudenb. & Crous, sp. nov. MycoBank MB809000. Fig. 24.
Fig. 24.

Alternaria sennae sp. nov. CBS 477.81: A–L. Conidia and conidiophores. Scale bars = 10 μm.
Etymology: Named after the host genus on which it occurs, Senna.
Primary conidiophores solitary, simple, straight to slightly curved, septate, dark brown with a hyaline tip, (43–)67–81(–108) × (5–)6(–7) μm, bearing a single, darkened, apical conidiogenous locus, but may produce geniculate conidiogenous extensions. Conidia solitary, conidium body pale olive-brown, smooth-walled, narrowly ovoid, (46–)55–62(–69) × (8–)10–12(–14) μm, with (7–)7–8(–10) transverse distosepta and (1–)2–3(–4) longitudinal septa. The conidial body can be slightly constricted near some transverse septa. Conidia have a single, aseptate, filamentous beak, which occasionally branches once; basal lateral secondary conidiophores can be formed. Beaks (38–)99–163(–314) μm long, ca. 2 μm diam. Sexual morph not observed.
Culture characteristics: After 7 d cultures on SNA flat, fimbriate, white to opaque with two olivaceous concentric circles; aerial mycelium sparse, white, floccose, colonies reaching 35−40 mm diam; cultures on PCA flat, undulate, white with grey-olivaceous zones; aerial mycelium felty, pale olivaceous-grey, colonies reaching 50–55 mm diam; reverse with pale olivaceous-grey zones.
Material examined: India, Uttar Pradesh, Gorakhpur, from leaf of Senna corymbosa (Fabaceae), 10 Apr. 1981, R.P. Verma (holotype CBS H-21741, culture ex-type CBS 477.81 = E.G.S. 34.030 = IMI 257253).
Alternaria sesami (E. Kawam.) Mohanty & Behera, Curr. Sci. 27: 493. 1958.
Basionym: Macrosporium sesami E. Kawam., Fungi 1: 27. 1931.
Materials examined: Egypt, from Sesamum indicum (Pedaliaceae), 1972, S.B. Mathur, CBS 240.73. India, from seedlings of Sesamum indicum, Dec. 1959, E.E. Leppik, representative isolate CBS 115264 = CBS 117214 = E.G.S. 13.027.
Alternaria sidae E.G. Simmons, Mycotaxon 88: 202. 2003.
Material examined: Kiribati, Phoenix islands, Canton Island, from leaf spot of Sida fallax (Malvaceae), 11 Feb. 1958, O. & I. Degener, culture ex-type of A. sidae CBS 117730 = E.G.S. 12.129.
Alternaria silybi Gannibal, Mycotaxon 114: 110. 2011.
Materials examined: Russia, Vladivostok, Trudovoe, from leaf lesion of Silybum marianum (Asteraceae), 1 Sep. 2006, Ph. B. Gannibal, culture ex-type of A. silybi CBS 134092 = VKM F-4109; Vladivostok, Trudovoe, from leaf lesion of Silybum marianum, 1 Sep. 2006, Ph. B. Gannibal, CBS 134094 = VKM F-4118; Vladivostok, Botanical Garden-Institute, from leaf lesion of Silybum marianum, 6 Sep. 2006, Ph. B. Gannibal, CBS 134093 = VKM F-4117.
Alternaria solani Sorauer, Z. Pflanzenkrankh. Pflanzenschutz 6: 6. 1896. Fig. 25.
Fig. 25.

Alternaria solani. A. Disease symptoms on Solanum tuberosum (Photo J.E. van der Waals, University of Pretoria, South Africa). B–H. Conidia and conidiophores. B–D. CBS 109157. E–H. CBS 116442. Scale bars = 10 μm.
= Macrosporium solani Ellis & G. Martin, Amer. Naturalist 16(12): 1003. 1882 (non M. solani Cooke, 1883)
≡ Alternaria solani (Ellis & G. Martin) L.R. Jones & Grout, Vermont Agric. Exp. Sta. Annual Rep. 9: 86. 1899. (nom. illegit., Art. 53.1)
≡ Alternaria americana Sawada, Rep. Dept. Agric. Gov. Res. Inst. Formosa 51:117. 1931. (nom. nov. for A. solani (Ellis & G. Martin) L.R. Jones & Grout (1899), non A. solani Sorauer (1896))
≡ Alternaria porri f. sp. solani (Ellis & G. Martin) Neerg, Danish species of Alternaria & Stemphylium: 260. 1945.
= Sporidesmium solani-varians Vañha, Naturwiss. Z. Forst- Landw. 2: 117. 1904.
= Alternaria danida E.G. Simmons, Mycotaxon 65: 78. 1997.
= Alternaria viciae-fabae E.G. Simmons & G.F. Laundon, CBS Biodiversity Ser. (Utrecht) 6: 234. 2007.
Materials examined: Italy, from seed of Ageratum houstonianum (Asteraceae), 27 Aug. 1941, P. Neergaard, culture ex-type of A. danida CBS 111.44 = E.G.S. 07.029 = QM 1772. New Zealand, from Vicia faba (Fabaceae), Jun. 1979, G.F. Laundon, culture ex-type of A. viciae-fabae CBS 116442 = E.G.S. 46.162 = ICMP 10242. Unknown, from leaf spot of Solanum aviculare (Solanaceae), before May 1941, P. Neergaard, CBS 111.41; unknown host, before Nov. 1921, isolated by Künkel, CBS 106.21. USA, Washington, Douglas County, from leaf spot of Solanum tuberosum (Solanaceae), 25 Aug. 1996, E.G. Simmons, representative isolate of A. solani CBS 109157 = E.G.S. 44.098.
Notes: By synonymising A. danida and A. viciae-fabae with A. solani, the host range of this pathogen has expanded to include Asteraceae and Fabaceae host plants. The isolates CBS 106.21 and CBS 111.44 did not sporulate after 3 wk of incubation on SNA (both were already labelled as sterile in the CBS collection database). Isolate CBS 111.41 did sporulate, but the spore formation was atypical.
Alternaria solani-nigri R. Dubey, S.K. Singh & Kamal [as “solani-nigrii”], Microbiol. Res. 154: 120. 1999. Fig. 26.
Fig. 26.
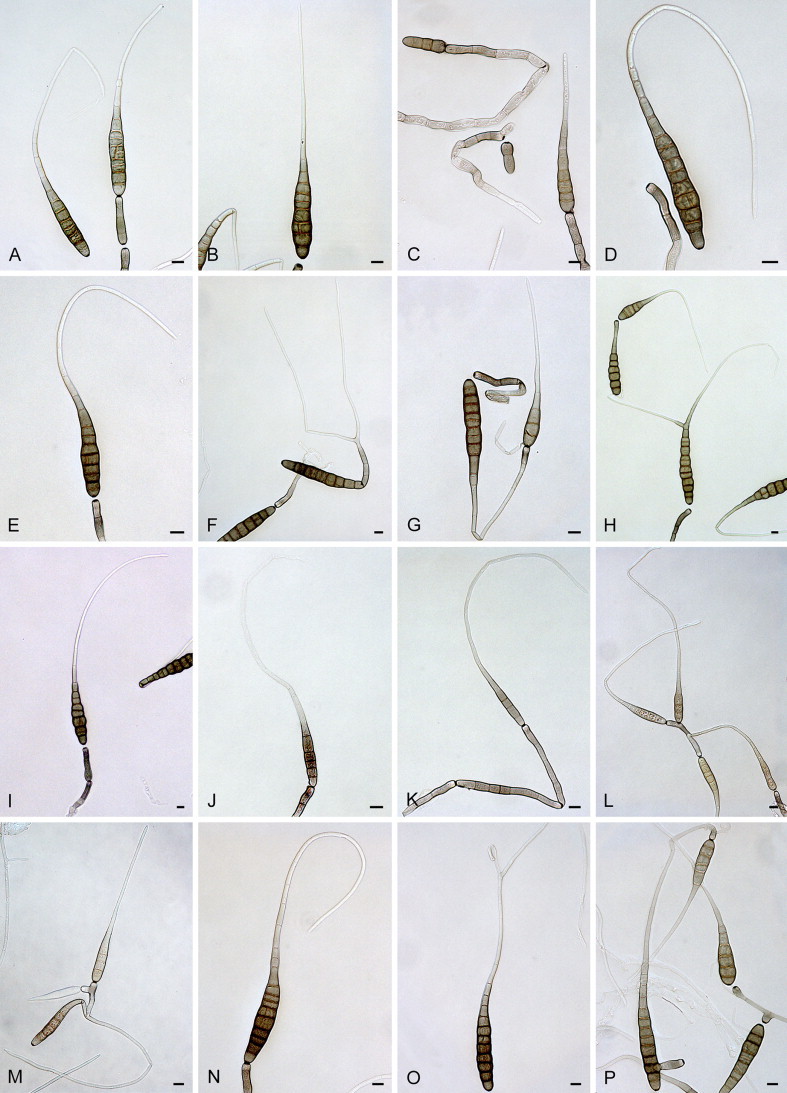
Alternaria solani-nigri: conidia and conidiophores. A–B. CBS 113403. C–D. CBS 116447. E–G. CBS 109155. H–I. CBS 116334. J–K. CBS 121347. L–M. CBS 116332. N–P. CBS 117101. Scale bars = 10 μm.
= Alternaria cyphomandrae E.G. Simmons, Mycotaxon 75: 86. 2000.
= Alternaria ascaloniae E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 168. 2007.
= Alternaria beticola E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 170. 2007.
= Alternaria glyceriae E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 148. 2007.
= Alternaria herbiculinae E.G. Simmons, CBS Biodiversity Ser. (Utrecht) 6: 166. 2007.
Materials examined: New Zealand, Canterbury, Ashburton, from leaf lesion of Beta vulgaris (Chenopodiaceae), Jul. 1999, B. Alexander, culture ex-type of A. beticola CBS 116447 = E.G.S. 47.196; Hastings, from leaf spot of Allium ascalonicum (Amaryllidaceae), Oct. 1997, C.F. Hill, culture ex-type of A. ascaloniae CBS 121347 = E.G.S 46.052; New Plymouth, from fruit of Cyphomandra betacea (Solanaceae), May 1991, C.F. Hill, culture ex-type of A. cyphomandrae CBS 109155 = E.G.S. 40.058; Taranaki, Otaki, from stunted Petroselinum crispum (Apiaceae), 14 Jun. 2001, J.B. Wong, culture ex-type of A. herbiculinae CBS 116332 = E.G.S. 49.180; Waikato, Kopuku, from leaf spot of Glyceria maxima (Gramineae), Apr. 2003, C.F. Hill, culture ex-type of A. glyceriae CBS 116334 = E.G.S. 51.107; Waikato, Whangamarino swamp, from leaf spot of Solanum nigrum (Solanaceae), 21 Jun. 2003, C.F. Hill, representative isolate of A. solani-nigri CBS 113403 = E.G.S. 51.106 = CPC 10620; Waikato, Whangamarino swamp, from leaf spot of Solanum nigrum, 6 Feb. 2003, C.F. Hill, representative isolate of A. solani-nigri CBS 117101 = E.G.S. 51.032.
Notes: By synonymising these five Alternaria species with A. solani-nigri, this becomes a species with a broad host range found on Amaryllidaceae, Apiaceae, Chenopodiaceae, Gramineae and Solanaceae. All studied specimens originate from New Zealand, but the holotype of A. solani-nigri was described from India. The five sequenced genes are 100 % identical between all the specimens studied.
Alternaria steviae Ishiba, T. Yokoy. & Tani, Ann. Phytopathol. Soc. Japan 48(1): 46. 1982.
Materials examined: Japan, Kagawa, Kida-gun, Miki-cho, Ikenobe, from leaf spot of Stevia rebaudiana (Asteraceae), CBS 631.88 = IFO 31212; Kagawa, Kida-gun, Miki-cho, Ikenobe, from leaf spot of Stevia rebaudiana, Jun. 1980, CBS 632.88 = IFO 31183; Kagawa, Zentsuji, Harada-cho, from leaf spot of Stevia rebaudiana, Aug. 1978, C. Ishiba, culture ex-type of A. steviae CBS 117362 = IFO 31182 = E.G.S. 37.019.
Alternaria tagetica S.K. Shome & Mustafee, Curr. Sci. 35: 370. 1966.
Materials examined: UK, from seed of Tagetes sp. (Asteraceae), before May 1979, G.S. Taylor, CBS 297.79; from seed of Tagetes sp., before May 1979, G.S. Taylor, CBS 298.79; England, Manchester, from seed of Tagetes erecta (Asteraceae), before Apr. 1980, G.S. Taylor, representative isolate of A. tagetica CBS 479.81 = E.G.S. 33.081. USA, Ohio, Butler County, Oxford, from leaf of cultivated Tagetes sp., 14 Jun. 1996, M.A. Vincent, representative isolate of A. tagetica CBS 117217 = E.G.S 44.045; South Carolina, Clemson, from seed of Tagetes sp., before Mar. 1981, E. Smallwood Hotchkiss, representative isolate of A. tagetica CBS 480.81 = E.G.S. 33.184.
Alternaria thunbergiae E.G. Simmons & Alcorn, CBS Biodiversity Ser. (Utrecht) 6: 136. 2007. Fig. 27.
Fig. 27.

Alternaria thunbergiae: conidia and conidiophores. A–C. CBS 116331. D–E. CBS 122597. F–H. CBS 120986. Scale bars = 10 μm.
= Alternaria iranica E.G. Simmons & Ghosta, CBS Biodiversity Ser. (Utrecht) 6: 122. 2007.
Materials examined: Australia, Queensland, Brisbane, Chapel Hill, from leaf spot of Thunbergia alata (Acanthaceae), 6 Feb. 1986, J.L. Alcorn, culture ex-type of A. thunbergiae CBS 116331 = E.G.S. 41.073 = BRIP 14963. Iran, Miandoab, from leaf of Allium cepa (Amaryllidaceae), 13 Sep. 2001, Y. Ghosta, culture ex-type of A. iranica CBS 120986 = E.G.S. 51.075. New Zealand, Auckland, Mangere, Tidal Road, from Thunbergia alata, 4 Jun. 2001, C.F. Hill, CBS 122597.
Notes: By synonymising A. iranica with A. thunbergiae, the host range of this taxon has expanded to include Allium cepa. The five sequenced genes are 100 % identical between the ex-type strains of A. thunbergiae and A. iranica. As both species were originally described in the same publication, there is no case for nomenclatural priority. Therefore we chose to synonymise A. iranica under A. thunbergiae because A. thunbergiae is more commonly used in literature (Leahy 1992, Melo et al. 2009).
Alternaria tillandsiae E.G. Simmons & C.F. Hill, CBS Biodiversity Ser. (Utrecht) 6: 314. 2007.
Material examined: USA, from Tillandsia usneoides (Bromeliaceae), Dec. 1995, B. Milnes, culture ex-type of A. tillandsiae CBS 116116 = E.G.S. 43.074.
Alternaria tropica E.G. Simmons, Mycotaxon 46: 187. 1993.
Materials examined: USA, Florida, Homestead, from fruit of Passiflora edulis (Passifloraceae), May 1990, R.T. McMillan Jr., culture ex-type of A. tropica CBS 631.93 = E.G.S. 39.126; Florida, Homestead, from fruit of Passiflora edulis, May 1990, R.T. McMillan Jr., representative isolate of A. tropica CBS 117216 = E.G.S. 39.125.
Alternaria venezuelensis E.G. Simmons & Rumbos, CBS Biodiversity Ser. (Utrecht) 6: 128. 2007.
Material examined: Venezuela, Maracay, from leaf spot of Phaseolus vulgaris (Fabaceae), before Oct. 1999, R. Rumbos, culture ex-type of A. venezuelensis CBS 116121 = E.G.S. 48.065.
Alternaria zinniae M.B. Ellis, Mycol. Pap. 131: 22. 1972.
= Alternaria zinniae H. Pape, Angew. Bot. 24: 61. 1942. (nom. inval., Art. 36.1)
Materials examined: Hungary, from seed of Callistephus chinensis (Asteraceae), 12 Aug. 1942, P. Neergaard, CBS 118.44. Italy, Sardinia, Sasseri, from Zinnia elegans (Asteraceae), 18 Oct. 1958, U. Prota, CBS 117.59. Netherlands, Huizum, from leaf of Zinnia sp., 27 Jul. 1948, A. Jaarsveld, CBS 107.48. New Zealand, Auckland, Royal Oak, from leaf spot of Zinnia elegans, May 1996, C.F. Hill, representative isolate of A. zinniae CBS 117223 = E.G.S. 44.035. UK, from seed of Zinnia sp., 1979, G.S. Taylor, CBS 299.79; from seed of Zinnia sp., 1979, G.S. Taylor, CBS 300.79. Unknown, from Zinnia elegans, summer 1961, Smith, CBS 108.61.
Section Euphorbiicola Woudenb. & Crous, sect. nov. MycoBank MB809001. Fig. 28
Fig. 28.

Alternaria section Euphorbiicola: conidia and conidiophores. A–G. Alternaria limicola. H–P. Alternaria euphorbiicola. A–D. CBS 117360. E–G. CBS 483.90. H–J. CBS 198.86. K–M. CBS 119410. N–P. CBS 133874. Scale bars = 10 μm.
Type species: Alternaria euphorbiicola E.G. Simmons & Engelhard.
Section Euphorbiicola is characterised by ovoid, obclavate, medium to large conidia that are disto- and euseptate, in short to moderately long chains, with no or a simple long beak in the terminal conidia. Conidia contain multiple transverse and some longitudinal septa and are slightly constricted near some transverse septa. Short to long, broad, apical, and sometimes lateral, secondary conidiophores are formed.
Note: The new Alternaria sect. Euphorbiicola can be easily distinguished from sect. Porri based on the formation of conidia in chains in sect. Euphorbiicola.
Alternaria euphorbiicola E.G. Simmons & Engelhard, Mycotaxon 25: 196. 1986.
≡ Macrosporium euphorbiae Reichert, Bot. Jahrb. Syst. 56: 723. 1921. Non Macrosporium euphorbiae Bartholomew 1908. (nom. illegit., Art 53.1).
Materials examined: USA, Florida, from Euphorbia pulcherrima (Euphorbiaceae), 1985, A.W. Engelhard, CBS 198.86 = E.G.S. 38.082; Hawaii, Oahu, from Euphorbia pulcherrima, Mar. 1984, M. Aragaki, representative isolate CBS 119410 = E.G.S. 41.029; Louisiana, from Euphorbia hyssopifolia (Euphorbiaceae), 1986, L. Walker, CBS 133874 = E.G.S 38.191.
Alternaria limicola E.G. Simmons & M.E. Palm, Mycotaxon 37: 82. 1990.
Materials examined: Mexico, Colima, from leaf of Citrus aurantiifolia (Rutaceae), May 1989, M. Palm, culture ex-type of A. limicola CBS 483.90 = E.G.S. 39.070; Jalisco, from Citrus sp., Sep. 1995, M. Palm, representative isolate CBS 117360 = E.G.S. 43.009.
Discussion
In the present phylogenetic study aiming to delimit Alternaria species in sect. Porri, we reduced the 82 known morphospecies in this section to 63 based on our polyphasic approach. Some important plant pathogens have now been assigned to specific clades in the phylogenetic tree and correlated with their distinct morphology, which will aid plant pathologists to identify their newly collected isolates.
The 10 isolates named A. solani at the onset of this study cluster within five different species-clades, and only three of them retain the name A. solani. This is not surprising, as almost all large-spored, narrow-beaked Alternaria strains hitherto isolated from Solanaceae were called A. solani, following the concept of M.B. Ellis (1971). Simmons (2000) already noted that early blight of tomato is actually caused by A. tomatophila rather than A. solani, and also described two additional species on tomato, A. cretica and A. subcylindrica. These tomato pathogens all cluster in one clade based on our phylogenetic analysis, which also includes the ex-type strain of A. linariae. The basionym of A. linariae, A. anagallidis var. linariae, is the oldest name in this cluster, which therefore applies to this clade mainly represented by tomato pathogens. When Neergaard (1945) described this species he found the fungus on seeds and seedlings with damping-off symptoms from Linaria marroccana (Scrophulariaceae), Antirrhinum majus (Scrophulariaceae) and on a healthy seedling of Papaver rhoeas (Papaveraceae). His pathogenicity tests (Neergaard 1945) showed that A. linariae could also attack Brassica oleracea (Brassicaceae), Solanum lycopersicum (Solanaceae), Lactuca sativa (Asteraceae), Godetia hybrida (Onagraceae), Nicotiana affinis (Solanaceae) and Papaver paeoniflorum (Papaveraceae), indicating a very broad host range. The isolates included in this study also show that, besides its broad host range, A. linariae is also widespread, found in Europe, USA, New Zealand and Asia. Three other isolates formerly identified as A. solani, including a former representative isolate used by Simmons (2007), cluster with A. protenta, an Alternaria species originally described from Helianthus annuus (Asteraceae). CBS 116651 is mentioned as a representative strain of A. solani by Simmons (2007), but he later expressed doubt as to the identity of this isolate (Simmons pers. comm.). The host range of A. protenta has expanded extensively, now comprising plants from the Asteraceae, Euphorbiaceae, Gramineae and Solanaceae. A pathogenicity test performed on A. protenta isolated from sunflower seed (Wu & Wu 2003) concluded that sunflower was the only susceptible host among the 10 host plants tested. One of the host plants tested was Solanum lycopersicum, which we include as host of A. protenta. However, the authors did not clearly state how the A. protenta isolates, which they only found on seed of one out of seven cultivars of sunflower seeds tested, were identified. The manuscript also lacks molecular data, which could affirm their identification of A. protenta. To our knowledge, no pathogenicity tests have thus far been performed with the species synonymised under A. protenta, A. hordeiseminis or A. pulcherrimae. Based on molecular and morphological data, A. protenta is closely related to A. solani, and these two species can only be distinguished by the 9 nt differences in their RPB2 sequences. To confirm the potato pathogen clade, called A. solani, we sequenced the RPB2 region of multiple isolates collected from Solanum tuberosum, which are present in E.G. Simmons collection, now deposited at the CBS. Almost all (22/24 strains) cluster within the now recognised A. solani species clade (data not shown). The ex-type strain of A. danida (CBS 111.44), now a synonym of A. solani, was originally deposited in the CBS collection by P. Neergaard as A. porri f. sp. solani. Pathogenicity tests performed on this strain (Neergaard 1945) showed that it could attack hosts from several plant families [e.g. Allium cepa (Amaryllidaceae), Brassica oleracea (Brassicaceae), Solanum lycopersicum (Solanaceae) and Lactuca sativa (Asteraceae)], indicating a very broad host range. Our sequences of A. danida differ from those deposited in GenBank by Lawrence et al. (2013), and therefore we repeated the cultivation and DNA extraction to confirm our results and the resulting synonymy with A. solani. Although the other large-spored, long-beaked Alternaria species described from potato, A. grandis (Simmons 2000), differs only by 1 nt in its GAPDH sequence (position 99, T instead of C, see locus alignment in TreeBASE) within the 2 722 positions used in the phylogeny, we did not synonymise A. grandis under A. solani. The two isolates included, CBS 109158 and CBS 116695, have substantially larger conidia than the other A. solani isolates, and a recently published study revealed that A. solani (CBS 109157) and A. grandis (CBS 109158) differ on 8 out of 770 nt in their calmodulin sequence (Gannibal et al. 2014).
The oldest large-spored onion pathogens, A. porri and A. allii, form two closely related but distinct clades, which only differ based on 8 nt in their RPB2 sequences (see locus alignment in TreeBASE). The three newer species described from Allium, A. ascaloniae, A. iranica and A. vanuatuensis (Simmons 2007), are all synonymised with other species. Alternaria ascaloniae is synonymised under A. solani-nigri, a species with a broad host range, mainly found in New Zealand. To our knowledge, no pathogenicity tests have been performed with the species now placed in synonomy with A. solani-nigri, which could affirm the broad host range for this species. Alternaria iranica is synonymised under A. thunbergiae, known as the causative agent of Alternaria leaf spot on Thunbergia (Leahy 1992), reported from Australia, USA and Brazil. Alternaria vanuatuensis clusters in the Allium clade, comprising A. allii and A. porri. Based on the sequence data generated here, it is synonymised under A. allii. According to Simmons (2007), the conidia of A. allii are distinguishable from those of A. porri and other large-spored species known on Allium, based on their multiple beaks and branches. However, the representative isolates of A. allii used by Simmons (2007) do not cluster in a single clade; CBS 116649 clusters with the two A. porri representative isolates. On the other hand, A. vanuatuensis is described as a single-beaked species, but clusters with the A. allii isolate deposited in the CBS collection by J.A.B. Nolla on 27 December 1927 as A. allii sp. nov. (CBS 107.28, recognised as the ex-type strain here). Simmons obtained this isolate from the CBS in February 2000 (E.G.S. 48.084), but was unable to induce sporulation. We observed few conidia, but these were only single-beaked. Unfortunately we could not induce CBS 116701 to sporulate, which leaves us at odds with Simmons's notes, with only single- to double-beaked conidia in the A. allii clade, and double- to triple-beaked conidia in the A. porri clade. The number of beaks and branches from the Allium isolates therefore is not suitable to make a distinction between the two major Allium species. The species can be easily differentiated on the basis of sequence data of the RPB2 gene region generated in this study.
Based on morphology, four large-spored Alternaria species with long beaks were described as Passifloraceae pathogens. Our phylogeny only supports three of these: A. tropica, A. aragakii and the more common A. passiflorae. The fourth species, A. hawaiiensis, is synonymised under A. passiflorae based on sequence data. Simmons (2007) described A. hawaiiensis as a new species lacking multiple beaks, which is a characteristic of A. passiflorae. Our sequence data led us to conclude that this characteristic is not suitable for species delimitation, which we also concluded from the data of the onion pathogens, A. allii, A. vanuatuensis and A. porri. The clustering of two isolates within our A. passiflorae clade, which originate from different host families (Onagraceae and Solanaceae), renders A. passiflorae as unspecific to Passifloraceae.
An ongoing study in South Africa on sweet potato pathogens reveals multiple Alternaria species on this host associated with blight symptoms on leaves, petioles, and stems. In addition to the known pathogen of sweet potato, A. bataticola, three other pathogenic species are delineated of which two are newly described as A. ipomoeae and A. neoipomoea. A new unknown Alternaria pathogen, causing sweet potato stem blight in Ethiopia, was reported by van Bruggen in 1984. This isolate (CBS 219.79) was sent to the CBS for identification, but the author did not agree with the morphological identification made at that time as A. cucumerina, a name under which it was still stored in the CBS collection. Our data indicate that this pathogen, which also is found in stem lesions of Ipomoea batatas in South Africa, should be recognised as a new species, now named A. ipomoeae. Most isolates from South Africa however cluster in a clade close to A. ipomoeae, now named A. neoipomoea, which can clearly be distinguished from A. ipomoeae morphologically and by sequence data. Two more isolates from sweet potato in South Africa are identified as A. argyroxiphii, an Alternaria species originally described from Argyroxiphium sp. This finding is a new host report for A. argyroxiphii, and a first report of the fungus from South Africa.
Based on the sequence data generated in this study, A. euphorbiicola and A. limicola clearly separate from the other species in sect. Porri (Fig. 1). This separation is supported by morphological differences, and we therefore propose the new section, sect. Euphorbiicola. However, when we examined the phylogeny displaying the neighbouring sections of sect. Porri (Fig. 2), questions arose concerning sect. Gypsophilae and sect. Radicina. These two sections display almost similar branch length differences within the respective sections, comparable to what sect. Porri displays with sect. Euphorbiicola. An additional character of sect. Gypsophilae and sect. Radicina is that the species within these sections share the same host family, respectively Caryophyllaceae and Apiaceae. We therefore choose to retain these sections at present, but additional molecular and morphological studies could eventually lead to the recognition of additional sections.
The present polyphasic approach displays the current species delimitation in Alternaria sect. Porri. We recognise 63 Alternaria species in this section with medium to large conidia and a long (filamentous) beak, which can be distinguished based on molecular data. Not all species distinctions are 100 % clear based on DNA data only; nevertheless, we tried to be consistent in synonymising or not synonymising species: the number of genes with nt differences and the number of nt differences are taken into account, together with the morphology, host, country and time of isolation. All Alternaria isolates currently stored in the CBS collection, which cluster within sect. Porri based on their gene sequences, were included in our study. Some species, however, are under-sampled, which results in some uncertainty in keeping isolates as separate species or reducing them to synonymy. Although we attempted to use the available data as best as possible, with the inclusion of additional isolates some uncertain species boundaries are bound to be better resolved.
The finding of the third species on potato (A. protenta) is a good example of the importance of fungal systematics. Multiple manuscripts report on the high level of genetic variability observed among A. solani isolates (van der Waals et al. 2004; Lourenco et al. 2011, Leiminger et al. 2013) and based on secondary metabolite profiling A. solani isolates cluster in two distinct groups (Andersen et al. 2008). Furthermore, two genotypes are described based on the cytochrome b gene structure of A. solani isolates (Leiminger et al. 2014), which is an important gene in fungicide resistance. However, our study indicates that previous reports could actually be dealing with three (or more) different species. Without knowing the correct identity of your pathogen, many incorrect conclusions can be drawn about diversity, evolutionary mechanisms, host range, and options for disease control.
Acknowledgements
This research was supported by the Dutch Ministry of Education, Culture and Science through an endowment of the FES programme “Making the tree of life work”.
Footnotes
Peer review under responsibility of CBS-KNAW Fungal Biodiversity Centre.
References
- Abo-Elyousr K.A.M., Abdel-Hafez S.I.I., Abdel-Rahim I.R. Isolation of Trichoderma and evaluation of their antogonistic potential against Alternaria porri. Journal of Phytopathology. 2014;162:567–574. [Google Scholar]
- Andersen B., Dongo A., Pryor B.M. Secondary metabolite profiling of Alternaria dauci, A. porri, A. solani, and A. tomatophila. Mycological Research. 2008;112:241–250. doi: 10.1016/j.mycres.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Angell H.R. Purple blotch of onion (Macrosporium porri Ell.) Journal of Agricultural Research. 1929;38(9):467–487. [Google Scholar]
- Berbee M.L., Pirseyedi M., Hubbard S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 1999;91:964–977. [Google Scholar]
- Bruggen A.H.C. van. Sweet potato stem blight caused by Alternaria sp.: a new disease in Ethiopia. Netherlands Journal of Plant Pathology. 1984;90:155–164. [Google Scholar]
- Brun S., Madrid H., Gerrits van den Ende A.H.G. Multilocus phylogeny and MALDI-TOF analysis of the plant pathogenic species Alternaria dauci and relatives. Fungal Biology. 2013;117:32–40. doi: 10.1016/j.funbio.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- Cifferi R. Phytopathological survey of Santo Domingo, 1925–1929. Journal of the Department of Agriculture of Porto Rico. 1930;14:5–44. [Google Scholar]
- Cooke M.C., Ellis J.B. New Jersey fungi. Grevillea. 1879;8:11–16. [Google Scholar]
- Corlett M., Corlett M.E. Fungi Canadenses. No. 341. Alternaria linicola. Canadian Journal of Plant Pathology. 1999;21(1):55–57. [Google Scholar]
- Crous P.W., Gams W., Stalpers J.A. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology. 2004;50:19–22. [Google Scholar]
- Crous P.W., Verkley G.J.M., Groenewald J.Z., editors. Fungal Biodiversity. CBS-KNAW Fungal Biodiversity Centre; Utrecht, Netherlands: 2009. (CBS laboratory Manual Series 1). [Google Scholar]
- Ellis M.B. Commonwealth Mycological Institute; Kew, UK: 1971. Dematiaceous hyphomycetes. [Google Scholar]
- Gannibal P.B., Orina A.S., Mironenko N.V. Differentiation of the closely related species, Alternaria solani and A. tomatophila, by molecular and morphological features and aggressiveness. European Journal of Plant Pathology. 2014;139:609–623. [Google Scholar]
- Hong S.G., Cramer R.A., Lawrence C.B. Alt a 1 allergen homologs from Alternaria and related taxa: analysis of phylogenetic content and secondary structure. Fungal Genetics and Biology. 2005;42:119–129. doi: 10.1016/j.fgb.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Hoog G.S. de, Gerrits van den Ende A.H.G. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Horsfield A., Wicks T., Davies K. Effect of fungicides use strategies on the control of early blight (Alternaria solani) and potato yield. Australasian Plant Pathology. 2010;39:368–375. [Google Scholar]
- Lawrence D.P., Gannibal P.B., Peever T.L. The sections of Alternaria: formalizing species-groups concepts. Mycologia. 2013;105:530–546. doi: 10.3852/12-249. [DOI] [PubMed] [Google Scholar]
- Leahy R.M. Florida Department of Agriculture and Consumer Services, Division of Plant Industry; 1992. Alternaria leaf spot of Thunbergia. Plant pathology circular No. 352. [Google Scholar]
- Leiminger J.H., Auinger H.-J., Wenig M. Genetic variability among Alternaria solani isolates from potatoes in Southern Germany based on RAPD-profiles. Journal of Plant Diseases and Protection. 2013;120:164–172. [Google Scholar]
- Leiminger J.H., Adolf B., Hausladen H. Occurence of the F129L mutation in Alternaria solani populations in Germany in response to QoI application, and its effect on sensitivity. Plant Pathology. 2014;63:640–650. [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Lourenço V., Jr., Rodrigues T.T.M.S., Campos A.M.D. Genetic structure of the population of Alternaria solani in Brazil. Journal of Phytopathology. 2011;159:233–240. [Google Scholar]
- Melo M.P., Soares D.J., Araújo J.S.P. Alternaria leaf spot, caused by Alternaria thunbergiae, recorded for the first time on Thunbergia alata from Brazil. Australasian Plant Disease Notes. 2009;4:23–25. [Google Scholar]
- Narayanin C.D., Thompson A.H., Slabbert M.M. First report of Alternaria blight of sweet potato caused by Alternaria bataticola in South Africa. African Plant Protection. 2010;16:7–9. [Google Scholar]
- Neergaard P. Oxford University Press; London: 1945. Danish species of Alternaria and Stemphylium. [Google Scholar]
- Nirenberg H.I. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Section Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem. 1976;169:1–117. [Google Scholar]
- Nolla J.A.B. A new Alternaria disease of onions (Allium cepa L.) Phytopathology. 1927;17(2):115–132. [Google Scholar]
- O'Donnell K., Kistler H.C., Cigelnik E. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osiru M., Adipala E., Olanya O.M. Occurrence and distribution of Alternaria leaf petiole and stem blight on sweetpotato in Uganda. Plant Pathology Journal. 2007;6(2):112–119. [Google Scholar]
- Osiru M.O., Adipala E., Olanya O.M. Leaf petiol and stem blight of sweet potato caused by Alternaria bataticola in Uganda. Plant Pathology Journal. 2008;7(1):118–119. [Google Scholar]
- Page R.D.M. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Rayner R.W. Commonwealth Mycological Institute; Kew, UK: 1970. A Mycological Colour Chart. [Google Scholar]
- Rodrigues T.T.M.S., Berbee M.L., Simmons E.G. First report of Alternaria tomatophila and A. grandis causing early blight on tomato and potato in Brazil. New Disease Reports. 2010;22:28. [Google Scholar]
- Schubert K., Groenewald J.Z., Braun U. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales) with standardisation of methods for Cladosporium taxonomy and diagnostics. Studies in Mycology. 2007;58:105–156. doi: 10.3114/sim.2007.58.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons E.G. Alternaria themes and variations (74–105) Mycotaxon. 1994;50:219–270. [Google Scholar]
- Simmons E.G. Alternaria themes and variations (112–144) Mycotaxon. 1995;55:55–163. [Google Scholar]
- Simmons E.G. Alternaria themes and variations (151–223) Mycotaxon. 1997;65:1–91. [Google Scholar]
- Simmons E.G. Alternaria themes and variations (244–286). Species on Solanaceae. Mycotaxon. 2000;75:1–115. [Google Scholar]
- Simmons E.G. CBS Fungal Biodiversity Centre; Utrecht, Netherlands: 2007. Alternaria: an Identification Manual. CBS Biodiversity Series 6. [Google Scholar]
- Sung G.-H., Sung J.-M., Hywel-Jones N.L. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Thomma B.P.H.J. Alternaria spp.: from general saprophyte to specific parasite. Molecular Plant Pathology. 2003;4:225–236. doi: 10.1046/j.1364-3703.2003.00173.x. [DOI] [PubMed] [Google Scholar]
- Waals J.E. van der, Korsten L., Slippers B. Genetic diversity among Alternaria solani isolates from potatoes in South Africa. Plant Disease. 2004;88:959–964. doi: 10.1094/PDIS.2004.88.9.959. [DOI] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: a Guide to Methods and Applications. Academic Press; San Diego, California, USA: 1990. pp. 315–322. [Google Scholar]
- Woudenberg J.H.C., Groenewald J.Z., Binder M. Alternaria redefined. Studies in Mycology. 2013;75:171–212. doi: 10.3114/sim0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.C., Wu W.S. Sporulation, pathogenicity and chemical control of Alternaria protenta a new seedborne pathogen on sunflower. Australasian Plant Pathology. 2003;32:309–312. [Google Scholar]
- Zitter T.A., Drennan J.L. Proceedings in the 20th Annual Tomato Disease Workshop. Ohio State University; Ohio: 2005. Shift in performance of fungicides for the control of tomato early blight; pp. 28–30. [Google Scholar]


