Abstract
Though treatment of the ventilated premature infant has experienced many advances over the past decades, determining the best time point for extubation of these infants remains challenging and the incidence of extubation failures largely unchanged. The objective was to provide clinicians with a decision-support tool to determine whether to extubate a mechanically ventilated premature infant by using a set of machine learning algorithms on a dataset assembled from 486 premature infants receiving mechanical ventilation.
Algorithms included artificial neural networks (ANN), support vector machine (SVM), naïve Bayesian classifier (NBC), boosted decision trees (BDT), and multivariable logistic regression (MLR). Results for ANN, MLR, and NBC were satisfactory (area under the curve [AUC]: 0.63–0.76); however, SVM and BDT consistently showed poor performance (AUC ~0.5).
Complex medical data such as the data set used for this study require further preprocessing steps before prediction models can be developed that achieve similar or better performance than clinicians.
I. INTRODUCTION
A heterogeneous set of machine learning algorithms was developed in an effort to provide clinicians with a decision-support tool to predict success or failure for extubation of the ventilated premature infant. Most infants born prematurely, i.e. before their lungs have fully developed, need mechanical ventilatory support soon after birth. To provide this ventilatory support, a tube is placed in the infants’ trachea to allow air fortified with oxygen to be pushed into the infants’ lungs. This support is essential for the survival of the infant; however, it is accompanied by micro trauma to the lung tissue. These and other side effects make it important to extubate the infants as soon as possible; yet, weaning, i.e. reducing the ventilatory support, too early may lead to increased work of breathing, while an extubation undertaken too early may lead to lung collapse (atelectasis) and require re-intubation and increased support [1]–[4]. Despite advances in the treatment of the ventilated premature infant, determination of the best time point to extubate still presents a challenge and the incidence of extubation failure has remained virtually unchanged over the past decade. To provide less experienced clinicians with a decision-support tool our team developed an Artificial Neural Network (ANN) with promising results with 85% accuracy in predicting extubation outcome [5].
This paper reports the extension of the original ANN to a set of algorithms consisting of ANN, support vector machine (SVM), naïve Bayesian classifier (NBC), boosted decision trees (BDT), and multivariable logistic regression (MLR) algorithms.
II. METHODS
A. Machine Learning
A set of heterogeneous algorithms to predict extubation outcome were chosen since different algorithms provide different strengths through their different mathematical approaches and to allow for nonlinear relationships between variables without the need to explicitly specify them [6]. Missing data were imputed based on weight category and variable type using mean or median.
Algorithms used in this study included artificial neural networks (ANNs), multivariable logistic regression (MLR), support vector machines (SVM), naïve Bayesian classifier (NBC), and bagged decision trees (BDT). For the purpose of this study, all algorithms were developed using MATLAB (Version R2009b, Copyright 1984–2009, The MathWorks Inc.). For the development of the ANNs a MATLAB toolbox was used that had been developed previously by one of the authors (JSA) [7]. Application of this toolbox results in ANNs with one hidden layer using conjugate gradient regression to find the weights that minimize the sum of square deviations between the output nodes and the observed values. The toolbox uses a transfer function with sigmoidal form shown in Equation 1 [7].
| (1) |
A nested cross-validation scheme was used for early stopping and variable selection of the ANN regression. The first round of cross validation is used to halt regression when cross validation ceases to decrease. This procedure is nested between another leave-one-out cross validation sampling scheme that is accessing the forward addition of new variables. Instead of sensitivities, this procedure, first described by Mueller et al., uses (cross-validated) prediction error to determine the optimal number of variables included in each model [5], [8]. During the first step of ANN development the sequence of addition of new variables was determined by first obtaining the prediction error for all inputs individually. The variable resulting in the smallest prediction error was then retained. Subsequently variables were added until the prediction error increased if any of the remaining variables was added. In a final step, the prediction error for each resulting ANN configuration was determined using an external validation set.
The following built-in MATLAB functions were used for the remaining four algorithms: 1) MLR: (“glmfit”) generalized linear regression models of the form
| (2) |
as described in [9, 10] were developed using binomial distributions with the logit link function,
| (3) |
[11], the default for the binomial distribution. Full models as well as parsimonious models using stepwise regression were developed. 2) SVM: (“svmtrain”) Models were developed using the Gaussian Radial Basis Function kernel and the Sequential Minimal Optimization method. 3) NBC: (“NaiveBayes”) The NB classifier was built using a Kernel smoothing density estimate obtained from equation
| (4) |
with K being the kernel and h a smoothing function as described in [12], as distribution for all features to model the data. Prior probabilities were estimated from the relative frequencies of the variables in the dataset (default). 4) BDT: (“TreeBagger”) The TreeBagger function was used to build ensembles of 100 decision trees using the square root of the total number of features for each decision split with the minimal leaf size set to 1 (default).
Each of the methods was initially provided the full data set consisting of 54 input variables. Each algorithm was used with 100 data sets that were created through resampling. For this process we repeatedly (100 times) randomly split the total sample into 2/3 vs. 1/3 for training/cross-validation and testing data and applied each of the algorithms to each dataset. The median performer for each algorithm among the 100 applications was determined using the area under the curve (AUC) obtained from Receiver Operating Characteristic (ROC) curves. Similarly, performance of all algorithms was compared using AUCs from training and validation data.
In addition to the main data set, several different sub data sets were used following the procedure described above. These subsets were created based on: a) birth weight (<1000g vs. ≥1000g and 500g–999g vs. 1000g–1499g vs. 1500g–1999g); b) birth year (2006–2007 vs. 2008–2009); c) use of weaning protocol (yes vs. no); d) correlation - variables that were highly correlated were excluded (for example birth weight vs. current weight, lag time from last blood gas to extubation vs. lag time between last two blood gases, HCO3 vs. BE); e) Principal Components Analysis - variables were excluded if factor loadings were below 0.4, 0.35 and 0.3; and f) tests for statistical significance (t-tests/chi-square tests) – variables were excluded if p was found to be above 0.1.
The goal of this study was to identify those algorithms with high predictive capabilities (>90% accuracy) to be implemented as a web-based tool accessible to clinicians for use in clinical routine.
B. Data Collection
After receiving approval from the local IRB, data were collected on 486 eligible premature infants born at the Medical University of South Carolina (MUSC) between January 2005 and September 2009. Variables included demographic characteristics of the infant, clinical measures such as birth weight and APGAR scores determined at birth, medication given to the mother (prior to delivery) or infant, ventilator information including FiO2 content and air pressures, whether the extubation was successful or failed, and type of ventilatory support infants received after extubation within 48 and 72 hours (Table 1). Fifty-four input variables containing information prior to the extubation time point are available routinely as part of the infants’ medical chart and used by clinicians for clinical decision-making at the bedside, and were used for model development. Of the 486 infants included in the data set, 59 (12.1%) infants failed extubation, making the outcome variable severely unbalanced.
Table 1.
Input Variables listed by type
| Input Variable Type | Input Variable |
|---|---|
| Demographic characteristics | Age in days |
| Gender | |
| Race/ethnicity | |
| Gestational age | |
| Birth weight | |
| Weight at extubation | |
| Clinical characteristics | APGAR scores at 1 and 5 minutes |
| Heart rate | |
| Respiratory rate | |
| Blood pressure | |
| Medication | Mother: Betamethasone |
| Infant: | |
| Surfactant | |
| Saline | |
| Methylxanthines | |
| Ventilator information | Time to intubation from birth |
| Time from last blood gas until extubation | |
| Type of ventilator | |
| Ventilator settings at extubation and at the last time point prior to extubation | |
| Blood gas values | pH, CO2, CaO2, HCO3, BE |
| prior to, at and after extubation |
Figure 1 presents normalized data for all 54 input variables used in the development of the prediction models sorted by the outcome variable (success). Quantiles are presented with 0 representing the minimum value, and 1 representing the maximum value for each variable.
Fig. 1.
Quantiles for values between the minimum (set as 0) and maximum (set as 1) values observed for all input variables used in the development of models.
III. RESULTS
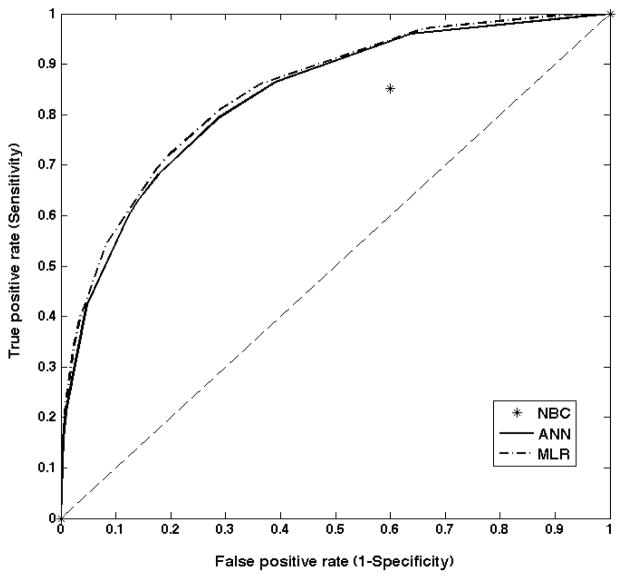
Table 2 reports the performance of the different prediction methods for training/cross-validation and testing sets as measured by the area under the curve (AUC) determined from the Receiver Operating Characteristic (ROC) curves for the full data set. ROC curves for the test set for ANN and MLR methods are displayed in figure 2; NBC is displayed as a single prediction point (except 0 and 1). Though several algorithms resulted in highly accurate predictions for the training data, using the test data two of the methods did not perform much better than chance – equivalent to an AUC of 0.5 with a 50/50 chance to predict the correct outcome. Only ANN and MLR showed satisfactory performance with MLR having slightly higher AUC than ANN but none of the methods performed above 0.8, which would be considered minimally acceptable performance for this population. The median performing ANN used 19 input variables, one hidden layer with 11 nodes and 1 output node (details are provided in Appendix 1). The high results for the training sets indicate overfitting of the algorithms to the available data resulting in a diminished capability to predict outcomes for unseen data, reflected in the poor performance in the test data.
Table 2.
Performance of algorithms as measured by area under the curve (median AUC)
| Full data set | Data set containing only statistically significant input variables | |||
|---|---|---|---|---|
| Algorithm | Training | Validation | Training | Validation |
| ANN | 0.930 | 0.753 | 0.921 | 0.682 |
| BDT | 1.000 | 0.513 | 1.000 | 0.507 |
| MLR | 0.880 | 0.762 | 0.853 | 0.776 |
| NBC | 0.610 | 0.626 | 0.769 | 0.607 |
| SVM | - | 0.493 | - | 0.519 |
Fig. 2.
ROC curves for test data for ANN, MLR and NBC (full set).
Performance was consistently low across all algorithms regardless of sub-setting of data to more homogenous groups such as birth weight, year or use of weaning protocol, removal of highly correlated input variables, removal of input variables identified through Principal Components Analysis and combinations of these techniques (results not shown). Only the subset including only input variables that exhibited statistical significant differences between infants extubated successfully and infants who failed extubation showed somewhat similar performance as indicated by slightly increased AUC for MLR and similar AUCs for ANN and NBC (table 1). Input variables included in the subset comprised of birth weight, APGAR score at 5 minutes, maternal betamethasone, lag time, rate ratio, FiO2, PIP, inspiratory time, tidal volume, pH, PaCO2, PAO2, SaO2, pulse, blood pressure, minute volume, surfactant, caffeine. Due to the uniformly low performance of all algorithms as indicated by the AUCs, no decision-support tool using the most predictive approaches to determine the final prediction was developed.
IV. DISCUSSION
Diverse approaches could be used in committee formations to obtain better generalization and performance of the combined results compared to the individual results. Improvement of performance, however, can only be expected if the individual committee members are accurate, i.e. better than guessing, and diverse, producing different errors, both false negative and false positive, for a given variable set. In our results none of the algorithms was sufficiently accurate to be included individually or in combination in a decision-support tool. Highest achieved median performance was 0.78 for MLR in the data set including only input variables with a statistically significant relationship with extubation identified in prior descriptive analyses, some methods resulted in AUC as low as 0.5, i.e. no better than chance.
These results suggest that any method that relies on variable selection will not do well for these clinical data. We hypothesize that the difficulties found in scaling up predictive modeling to larger numbers of subjects reflect batch effects associated with aggregating datasets, which cause validation by resampling to fail: for each data point, it is likely that a similar one exists in the test set from the same batch, which will cause variables selected in response to be irrelevant for other batches. This hypothesis is strengthened by the better results obtained with MLR compared to ANN, of which they are special cases. In both the full set and the last subset examined (based on statistically significant differences between the groups) ANN, MLR and NBC performed better as evidenced by AUCs (from 0.63–0.76). The two methods that performed poorly in all sets were SVM and BDT, which over-fitted the data in each dataset used resulting in close to perfect predictions for the training data but very poor predictions for new, unseen data.
A pre-processing step, then, seems to be needed to reduce the dimensionality in the dataset, i.e., the number of variables that are considered by each of the algorithms for determination of the prediction. This step may aid in improving the performance of the individual algorithms sufficiently to develop a decision-support tool using, for example, a committee of algorithms. This use would allow taking advantage of the heterogeneity in algorithms within the committee by utilizing predictions methods with different strengths, therefore improving generalization and performance in the combined results compared to the individual predictions. This variable selection procedure would have to be designed specifically to deal with batch effects. Such a variable selection procedure could, for example, be configured to rank variables as to how much batch effects they exhibit. As discussed in Leek et al. [13], this is an active area of research, with current solution largely depending on multivariate exploratory analysis rather than on a reliable preprocessing step in the discriminant analysis procedure.
Limitations
Though we assembled a large dataset to carry out the development of our machine learning algorithms, data from a period of several years were collected retrospectively. During this time, procedures in the NICU may have changed, along with personnel. For example, starting in summer 2006 a weaning protocol was gradually implemented that may not have been fully accounted for by inclusion of a variable whether or not the protocol had been followed for a given infant. Additionally, variables that may have been of interest but were not collected in the medical record could not be used in this study. Further, extubation failures comprised only 12% of the available data, resulting in an extremely unbalanced dataset, thus limiting the ability of the prediction methods to learn from these data. Moreover, though the data could be obtained from an electronic database, at the time of data collection this task involved reading of pages from handwritten medical records that had been electronically scanned into the clinical database. The information obtained from this database was then entered into the electronic study database. This process has inherent data quality issues typical for “real world” data sets such as missing forms, missing data points, illegible handwriting, and data entry errors and requires extensive data cleaning as well as imputation of missing values.
V. CONCLUSION
Though artificial intelligence methodologies are making strides toward taking over routine tasks that have been carried out by humans, the medical field remains a challenge for the implementation of automated tasks. Since any of the methods employed rely on a transformation of the data they are given, these methods may be inherently disadvantaged where decisions are made based on “gut feelings” and intuition that reflect implicit awareness of covariates gained in the course of a long exposure to the problem (“experience”). Currently, there is no method available to capture the kind of contextual data needed to feed statistical classifiers with the same information that is being processed in the brains of the clinical experts. However, because such subjective decisions are all based on having spent years at the bedside and are not readily available to inexperienced clinicians, we feel that any tool providing reasonable decision-support to improve the decision-making of these clinicians would be valuable for clinical practice. Yet even that modest goal still has not been accomplished, especially when dealing with premature infants where even “pretty good” is just not good enough. The results obtained suggest a critical missing component in computational statistics workflows to handle aggregates of datasets, where batch effects are very likely to occur. This conclusion is aligned with a growing drive towards relatively undirected data-intensive approaches (“data science”) that rely on the “unreasonable effectiveness of data” [14]. In a nutshell, the results reported here suggest that maximizing data capture describing the context of complex biomedical processes may not be enough by itself. It is also necessary that the data include enough batch/cohorts such that the predictive model can capture the co-variation associated with those data.
Acknowledgments
The authors would like to thank Kathy Ray for her dedication to this work and her many suggestions during data collection.
This work was supported by NIH/NHLBI (grant #5R21HL090598).
APPENDIX
Table 3.
Details of median performing ANN architecture with 19 inputs, 11 hidden nodes, 1 output.
| Inputs | Weights | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| Birth weight | 0.708 | 0.204 | 0.123 | −0.188 | −0.048 | −0.495 | 0.844 | 3.060 | 0.708 | 0.204 | 0.123 |
| APGAR score at 5 minutes | 2.162 | 0.540 | −1.505 | 0.250 | −0.146 | −1.040 | −0.411 | −0.206 | 2.162 | 0.540 | −1.505 |
| Maternal Betamethasone | −0.747 | 1.280 | 0.190 | 0.729 | 0.468 | −1.547 | −0.141 | 0.591 | −0.747 | 1.280 | 0.190 |
| Ratio: spontaneous breathing/ventilatory rate | −1.391 | −4.480 | 2.480 | −0.455 | −0.381 | −0.604 | −1.164 | −0.157 | −1.391 | −4.480 | 2.480 |
| Lag time between last blood gas result and extubation | 1.001 | 2.885 | −1.587 | −0.582 | −0.357 | 1.844 | 1.505 | 2.945 | 1.001 | 2.885 | −1.587 |
| Current weight | 0.439 | 1.697 | 0.298 | −0.304 | −0.389 | 0.658 | 0.493 | 2.244 | 0.439 | 1.697 | 0.298 |
| FiO2 | 1.426 | −2.196 | −1.548 | 0.234 | 0.423 | 3.593 | −2.749 | −1.665 | 1.426 | −2.196 | −1.548 |
| Peak inspiratory pressure | 2.613 | −0.204 | 2.018 | 0.145 | 0.370 | −1.000 | 0.567 | −1.100 | 2.613 | −0.204 | 2.018 |
| Inspiratory time | −0.008 | −0.584 | −1.414 | −0.380 | −0.157 | 1.988 | −1.442 | 0.502 | −0.008 | −0.584 | −1.414 |
| Tidal volume | 0.536 | −1.266 | −1.072 | 0.063 | 0.193 | −2.214 | 0.227 | 2.261 | 0.536 | −1.266 | −1.072 |
| pH | 5.182 | −0.878 | −0.190 | 0.478 | −0.141 | 0.328 | 0.603 | 0.105 | 5.182 | −0.878 | −0.190 |
| PaCO2 | 2.716 | 0.396 | −0.242 | −0.384 | −0.214 | −0.368 | −1.143 | −1.489 | 2.716 | 0.396 | −0.242 |
| PaO2 | 0.145 | −1.127 | 0.775 | 0.096 | −0.185 | 0.823 | 1.100 | 2.455 | 0.145 | −1.127 | 0.775 |
| SaO2 | −2.228 | −0.098 | −0.444 | 0.181 | −0.157 | −2.968 | 0.206 | 1.305 | −2.228 | −0.098 | −0.444 |
| Heart rate | −2.945 | −0.728 | −1.347 | −1.113 | −0.495 | 4.543 | 1.318 | 2.965 | −2.945 | −0.728 | −1.347 |
| Blood pressure | 0.304 | 1.693 | −1.700 | −0.199 | 0.068 | −1.690 | −0.745 | 0.087 | 0.304 | 1.693 | −1.700 |
| Minute volume | −1.502 | −0.669 | 1.476 | 0.859 | 0.555 | −1.859 | 2.171 | 3.284 | −1.502 | −0.669 | 1.476 |
| Surfactant | −2.287 | −0.188 | 0.944 | 0.536 | 0.094 | −3.164 | −0.149 | −2.163 | −2.287 | −0.188 | 0.944 |
| Caffeine | −0.089 | −1.865 | −0.337 | 0.003 | 0.277 | 1.968 | −0.818 | 0.778 | −0.089 | −1.865 | −0.337 |
| Bias 1 | −1.121 | 0.960 | −0.315 | −0.195 | −0.166 | 2.066 | −0.093 | −2.701 | −1.121 | 0.960 | −0.315 |
| Weights | 5.974 | −4.067 | −3.756 | −1.596 | −1.259 | −8.055 | −1.894 | 7.848 | 5.974 | −4.067 | −3.756 |
| Bias 2 | −1.210 | ||||||||||
Contributor Information
Martina Mueller, Medical University of South Carolina, Charleston, SC 29425 USA.
Carol C. Wagner, Email: wagnercl@musc.edu, Medical University of South Carolina, Charleston, SC, USA
Romesh Stanislaus, Email: Romesh.Stanislaus@sanofipasteur.com, Sanofi Pasteur, Cambridge, MA, USA.
Jonas S. Almeida, Email: jalmeida@uab.edu, University of Alabama, Birmingham, AB, USA
References
- 1.Halliday HL. What interventions facilitate weaning from the ventilator? A review of the evidence from systematic reviews. Paediatr Respir Rev. 2004;5(Suppl A):S347–S352. doi: 10.1016/s1526-0542(04)90060-7. [DOI] [PubMed] [Google Scholar]
- 2.Greenough A, Prendergast M. Difficult extubation in low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 2008;93:F242–F245. doi: 10.1136/adc.2007.121335. [DOI] [PubMed] [Google Scholar]
- 3.Barrington KJ. Extubation failure in the very preterm infant. J Pediatr (Rio J) 2009;85(5):375–377. doi: 10.2223/JPED.1937. [DOI] [PubMed] [Google Scholar]
- 4.Gupta S, Sinha SK, Tin W, Donn SM. A Randomized Controlled Trial of Post-extubation Bubble Continuous Positive Airway Pressure Versus Infant Flow Driver Continuous Positive Airway Pressure in Preterm Infants with Respiratory Distress Syndrome. J Pediatr. 2009;154:645–50. doi: 10.1016/j.jpeds.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 5.Mueller M, Wagner CL, Annibale DJ, Hulsey TC, Knapp RG, Almeida JS. Predicting extubation outcome in preterm newborns: a comparison of neural networks with clinical expertise and statistical modeling. Pediatr Res. 2004;56(1):11–8. doi: 10.1203/01.PDR.0000129658.55746.3C. [DOI] [PubMed] [Google Scholar]
- 6.Aksela M. Comparison of Classifier Selection Methods for Improving Committee Performance. In: Windeatt T, Roli F, editors. MCS 2003. Springer-Verlag; Berlin Heidelberg: 2003. pp. 84–93. LNCS 2709. [Google Scholar]
- 7.Almeida JS. Predictive non-linear modeling of complex data by artificial neural networks. Curr Opin Biotech. 2002;13(1):72–76. doi: 10.1016/s0958-1669(02)00288-4. [DOI] [PubMed] [Google Scholar]
- 8.Mueller M, Wagner CL, Annibale DJ, Hulsey TC, Knapp RG, Almeida JS. Parameter Selection for and Implementation of a Web-based Decision-Support Tool to Predict Extubation Outcome in Premature Infants. BMC Medical Informatics and Decision Making. 2006;6:11. doi: 10.1186/1472-6947-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen J, Cohen P, West SG, Aiken LS. Applied Multiple Regression / Correlation Analysis for the Behavioral Sciences. Lawrence Erlbaum Associates, Publishers; London: 2003. [Google Scholar]
- 10.Logistic regression. http://en.wikipedia.org/wiki_Logistic_regression.
- 11.Matlab glmfit. http://www.mathworks.com/help/stats/glmfit.html.
- 12.Gaussian Parzen Kernel. http://en.wikipedia.org/wiki/Kernel_density_estimation.
- 13.Leek JT, Scharpf R, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halevy A, Norvig P, Pereira F. The Unreasonable Effectiveness of Data. IEEE Intell Syst. 2009;24( 2):8–12. [Google Scholar]