Abstract
The role of plant growth-promoting rhizobacteria (PGPR) in adaptation of plants in extreme environments is not yet completely understood. For this study native bacteria were isolated from rhizospeheric arid soils and evaluated for both growth-promoting abilities and antagonistic potential against phytopathogenic fungi and nematodes. The phylogentic affiliation of these representative isolates was also characterized. Rhizobacteria associated with 11 wild plant species from the arid soil of Almadinah Almunawarah, Kingdom of Saudi Arabia (KSA) were investigated. From a total of 531 isolates, only 66 bacterial isolates were selected based on their ability to inhibit Fusarium oxysporum, and Sclerotinia sclerotiorum. The selected isolates were screened in vitro for activities related to plant nutrition and plant growth regulation as well as for antifungal and nematicidal traits. Isolated bacteria were found to exhibit capabilities in fix atmospheric nitrogen, produce ammonia, indoleacetic acid (IAA), siderophores, solubilize phosphate and zinc, and showed an antagonistic potential against some phytopathogenic fungi and one nematode species (Meloidogyne incognita) to various extent. Isolates were ranked by their potential ability to function as PGPR. The 66 isolates were genotyped using amplified rDNA restriction analysis (ARDRA) and 16S rRNA gene sequence analysis. The taxonomic composition of the representative genotypes from both rhizosphere and rhizoplane comprised Bacillus, Enterobacter and Pseudomonas. Out of the 10 genotypes, three strains designated as PHP03, CCP05, and TAP02 might be regarded as novel strains based on their low similarity percentages and high bootstrap values. The present study clearly identified specific traits in the isolated rhizobacteria, which make them good candidates as PGPR and might contribute to plant adaption to arid environments. Application of such results in agricultural fields may improve and enhance plant growth in arid soils.
Keywords: arid soil, PGPR, 16SrRNA genes, ARDRA, phylogeny
Introduction
Soil, a dynamic, living matrix is an important resource for agricultural products. Soil is also a storehouse of microbial activity, which is confined to aggregates with accumulated organic matter, the rhizosphere. The rhizosphere both contacts plant roots and supports high populations of active microorganisms and it has attracted much interest (Nautiyal and DasGupta, 2007).
In the rhizospheric plant soil, diversity and community structure of microorganisms are plant species dependent and differ among varieties or cultivars. This may be affected by both specific plant root exudates and soil type (Kremer et al., 1990). Rhizospheric organisms can play a role in governing plant growth and development (Napoli et al., 2008).
Rhizobacteria that exert beneficial effects on plant development are termed “Plant Growth-Promoting Rhizobacteria” (Kloepper and Schroth, 1978). PGPR was found to be mainly involved in enhancing plant nutrition, stress tolerance or health (Vacheron et al., 2013). This is mainly due to their effect associated with enhanced availability of nutrients (Lugtenberg and Kamilova, 2009; Drogue et al., 2012), phytohormones-mediated stimulation of root system (Somers et al., 2004) and induced systemic resistance (Zamioudis and Pieterse, 2012).
PGPR include, for instance, Gram-positive PGPR taxa that include coryneform bacteria, B. subtilis and B. cirulans. Gram-negative PGPR includes fluorescent as well as non-fluorescent pseudomonads (P. gladioli and P. cepacia) and various members of the family Enterobacteriaceae. Also, nitrogen-fixing bacteria such as Azospirillum, Herbaspirillum, Gluconacetobacter, Azotobacter and Azoarcus have been reported as PGPR. These bacteria were mainly found to play a role in increasing nitrogen availability for plant nutrition and induction of minerals uptake (Bashan and de-Bashan, 2005).
Specific studies showed that PGPR either directly or indirectly promote plant growth and yield. The direct growth promoting mechanisms includes (i) N2 fixation; (ii) solubilization of mineral phosphate and zinc; (iii) sequestration of iron by production of siderophores; (iv) production of phytohormones such as auxins, cytokinins and gibberellins; (v) production of the enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase, which hydrolyses ACC, the immediate precursor of ethylene in plants. Lowering of ethylene concentration in seedlings results in stimulating seedlings root length (Bashan and de-Bashan, 2005). PGPR also support plant growth indirectly, by improving growth restricting conditions via (i) production of antibiotics; (ii) depletion of iron from the rhizosphere; (ii) production of fungal cell wall lysing enzymes ß-(1,3)- glucanase and chitinase; (iii) synthesis of antifungal metabolites such as cyanide; (iv) competition for infection sites on roots; (v) induction of systemic resistance (Aeron et al., 2011; Gamalero and Glick, 2011; Jayaprakashvel and Mathivanan, 2011; Saraf et al., 2014).
Arid soils are dominant in the Kingdom of Saudi Arabia, but topographic differences and variations in soil composition suggest a significant number of bacterial species and plant associations may exist. A few literature reports suggest plant species surviving under such extreme conditions may harbor PGPR that have contributed to their fitness. Nonetheless, very little is known about the microbiota that colonizes the roots of desert plants (Jorquera et al., 2012).
The main objective of this study is to isolate rhizobacteria associated with some desert plant species and evaluate their potential contribution to the ability of such plants to survive under extreme conditions. The phylogenetic affiliation of the isolated PGPR and their antagonistic potential against phytopathogenic fungi and nematodes are also described.
Materials and methods
Site description
Almadinah Almunawarahis located at Eastern part of Alhijaz region in the Kingdom of Saudi Arabia at meeting-point Longitude (39°36′6″) and Latitude (24°28′6″). It is located in the north-western part of the kingdom, to the east of the Red Sea. The soils in the arid zone of Almadinah Almunawarah are coarse sandy textured and covered with sand dunes with low available water holding capacity, vulnerability to wind erosion and low fertility along with high salinity, calcareousness and gypsiferous nature. The annual average temperature during the year when the study was conducted was 30.2°C reaching 50°C in the summer months, annual total precipitation was 115 mm and annual average humidity was 17%, harsh conditions for most life forms.
Plant species
A number of wild and native plant species collected during this study from different areas around Almadinah Almunawarah were identified according to the methodologies described by Chaudhary (1999, 2000, 2001). Identified plants included: Zygophyllum simplex L. (family Zygophyllaceae), Peganum harmala L. (family Zygophyllaceae), Notoceras bicorne (Aiton) Amo (family Brassicaceae), Cassia italica (Mill.) (family Caeslpinaceae), Glinus lotoides L. (family Molluginaceae), Calotropis procera (Ait.) Ait. (family Asclepiadaceae), Tamarix amplexicaulis Ehrenb. (family Tamaricaceae), Capparis spinosa L. (family Capparidaceae), Citrullus colocynthis (L.) Schrad. (family Cucurbitaceae), Tribulusterrestris L. (family Zygophyllaceae) and Haloxylon salicornicum (Moq.) (family Chenopodiaceae).
Fungal cultures and root-knot nematode
Cultures of two soil-borne pathogenic fungi namely Fusarium oxysporum (F. oxysporum f. sp. lycopersici), and Sclerotinia sclerotiorum were kindly provided by the Plant Pathology Department, Faculty of Agriculture, Ain Shams University, Egypt. The two fungal pathogens were maintained onto Potato Dextrose Agar (PDA) before use. A population of the root-knot nematode Meloidogyne incognita was routinely maintained on the susceptible tomato cultivar Castle Rock in the glasshouse at 27 ± 5°C in a box filled with sandy loam soil. Nematode eggs were extracted from heavily infested tomato roots using the extraction technique described by Hussey and Barker (1973). To promote the development of eggs and the hatching of second stage juveniles (J2), the nematode eggs/water suspension was kept in darkness at 24°C and aerated with an aquarium pump.
Sample collection and isolation of rhizobacteria
Samples of rhizospheric soil and root system from 11 healthy wild plants were collected from different sites at Almadinah Almunawarah during the winter of 2011–2012. To estimate the number of root-associated rhizobacteria, 50 g of roots- adhered soils were used. From the selected plants, roots of three young and healthy plants were collected, shaken vigorously to remove loose soil, placed in sterile paper bags, and maintained in an ice-box. Six roots belonging to one plant species were pooled for an average sample. Sufficient portion of roots were aseptically placed in sterilized 250 ml conical flasks, and a solution of Tween phosphate buffered saline was added to give 100 ml final volume. The flasks were shaken, and tenfold serial dilutions were made in sterile 0.1 M MgSO4 (pH 7.0), and 0.1 ml aliquots from appropriate dilutions were spread- plated onto Tryptic Soy Agar (TSA) medium for total count. Each value presented is an average of three individual counts. The same procedure was followed to enumerate soil rhizospheric bacteria using soil surrounding roots. To estimate spore-forming bacteria from root segment and soil, 9 ml of each serial dilution was placed in a water bath at 80°C for 10 min to kill non-spore forming mesophilic bacteria. The heat-treated samples were serially diluted, inoculated onto TSA medium plates in triplicate and incubated aerobically at 30°C for 48 h (Bai et al., 2002). Developed colonies were counted and those representing different morphological types were selected, and further purified on TSA medium plates.
In vitro screening for antagonism
Bacterial isolates were screened in vitro for growth inhibition of the worldwide distributed soil-borne phytopathogenic fungi F. oxysporum, and S. sclerotiorum. Briefly, 5 μl drops of each bacterial culture (108CFUml−1) were streaked equidistantly on the margins of PDA plates adjusted to pH 7. Mycelial agar plug of 5 mm diameter from a 7-day-old culture of F. oxysporum or S. sclerotiorum grown on PDA plate was placed in the center of the plate between the two parallel streaks of the test bacterium. Control plates not inoculated with bacteria were also prepared. Two independent experiments with each bacterial isolate were performed. Plates were incubated at 25°C for 5 day. Antagonistic activity was assessed by relating mycelia diameter on plates inoculated with bacteria to mycelia diameter on control plates and computing percentage of Growth Inhibition (GI%).
To study the effect of bacterial filtrates on the egg hatching and mortality of M. incognita, three egg masses containing 300 ± 50 eggs per egg mass were mixed with 1 ml of the bacterial suspension in 1.5 ml Eppendorf tube and incubated at 28°C for 5 days. Equal numbers of egg masses in sterile TSA medium were kept as a control. The numbers of dead second-stage juveniles (J2) were recorded after 3 and 5 days using a light microscope and Hawksley counting slide.
Screening isolates with PGPR traits on plant nutrition and growth stimulation
N2-fixation was tested by inoculating bacterial isolates and E. coli (as control bacteria) on plates of N-free agar medium (Haahtela et al., 1983) for 48 h at 28°C. The isolates that grow after being sequentially transferred 10 times to the same medium were considered as presumptive positive for N2-fixation. All isolates were tested for the production of ammonia as described by Cappuccino and Sherman (1992). Phosphate and zinc solubilization was tested by the dissolution of precipitated tricalcium phosphate [Ca3 (PO4)2] and zinc oxide (ZnO) respectively in agar medium (Saravanan et al., 2003; Rodriguez et al., 2004). The solubilization Index (SI) was calculated as the ratio of the total diameter (colony, halo zone) to the colony diameter (Edi-Premona et al., 1996). IAA production was tested according to the procedure described by Loper and Schroth (1986) using TSA medium supplemented with L-tryptophane.
Antifungal and nematicidal traits
Siderophores production was detected as described by Pallai (2005). The assay used the ternary complex chrome azurol-S/Fe(III)/hexadecyl-trimethylammonium bromide as an indicator. Screening of bacterial isolates for hydrogen cyanide (HCN) production was done using cultures grown on TSA supplemented with glycine and alkaline picric acid as indicator (Castric, 1975). Chitinase production was assessed qualitatively by a microbiological method based on spotting of isolates on chitinase medium amended with colloidal chitin (Frändberg and Schnürer, 1998). Cellulase production was visualized by flooding the cellulose decomposition medium plates previously inoculated and incubated at 30°C for 8 days with 0.1% (w/v) Congo red for 15–30 min followed by bleaching the plates with 1M NaCl (Andro et al., 1984). Protease production was indicated by casein degradation after at least 4 days of incubation at 28°C (Abo-Aba et al., 2006). Salicylic acid (SA) production was assessed by the method of Leeman et al. (1996) using succinate medium.
PCR amplification of 16S rRNA genes
A loopful from a bacterial colony was suspended in 50 μl of TE buffer (100 mM Tris HCl, 10 mM EDTA, pH 8.0), heated in a boiling water bath for 5 min, followed by cooling prior to PCR. One μl of cell extract was used as a PCR template to amplify the full length 16S rRNA gene (Arturo et al., 1995). An approximately 1500-bp fragment of the 16S rRNA gene corresponding to positions 8 and 1509 of the E. coli was amplified using the universal primer pair (27F: 5′-AGA GTT TGA TC[A/C] TGG CTC AG-3′, 1492R: 5′-G[C/T]T ACC TTG TTA CGA CTT-3′) (Lane, 1991). Amplifications were performed in a 50 μl reaction volume containing: 5 μl of 10× Taq buffer (100 mM Tris- HCl, pH 8), 1.25 mM MgCl2, 200 μM dNTPs (Invitrogen, USA), 1.2 μM forward primer and reverse primer set (Invitrogen, USA), 1U Taq DNA polymerase (Invitrogen, USA), and about 5 ng of template DNA.
PCR was performed in Thermal Cycler (Applied Biosystem 2720, USA). The PCR conditions were adjusted to 5 min for initial denaturation at 94°C and then 30 cycles of 1 min at 94°C, 1 min at 54°C, and 1 min at 72°C, and finally 10 min at 72°C. The amplified genes were subjected to electrophoresis using 1% agarose gel with the size markers (1 kb DNA ladder, Invitrogen, USA).
Amplified ribosomal DNA restriction analysis (ARDRA)
Twenty microliters of each 16S rRNA gene amplicon was digested for 3 h at 37°C with 2.5 U HaeIII (Fermentas, Vilnius, Lithuania). The DNA restriction fragments were subjected to electrophoresis using 3% agarose gel containing ethidium bromide (50 ng/ml). The gels were made visible by UV transillumination and digitized with the gel documentation system (Gel Doc XR System, Biorad, USA). The images for electrophoretic pattern were analyzed with GelCompar II software (Applied Maths, Kortjik, Belgium). The patterns were used to construct a dendrogram using the unweighted pair group method of arithmetic average (UPGMA) clustering algorithm and Dice similarity coefficient index. Bacterial isolates showing same pattern were grouped into the same group.
Sequencing and analysis of 16S rRNA genes
The nucleotide sequence analysis of the selected isolates based on ARDRA profiles were determined by automated florescent dye terminator sequencing method (Sanger et al., 1977) using DYEynamic ET Terminator Cycle Sequencing Kit, Amersham Pharmacia Biotech with a model ABI 310 genetic sequence analyzer (Applied Biosystems, CA, USA) according to the user manual. One representative isolate of each genotypic profile was chosen for 16S rDNA partial sequencing. Dye terminator-based sequencing was performed using PCR-amplified segments of about 517 bases covering up V3 region of 16S rRNA genes using primer set of EUB8F/EUB517R. Amplified DNA was purified by ethanol precipitation to remove unincorporated dye-labeled terminators. The pellet was then dissolved in 20 μl formamide loading dye and heat shocked at 95°C for 2 min before injection to DNA sequence analyzer.
The sequences were analyzed by Genetyx-Win MFC application software version 4.0. Related sequences were identified using BLAST search program, National Center for Biotechnology Information (NCBI), National Library of Medicine, USA (http://www.ncbi.nlm.nih.gov/) (Altschul et al., 1997). Sequence alignments were performed by Clustal W1.83 XP (Thompson et al., 1997) and phylogenetic trees were constructed using neighbor-joining method (Saitou and Nei, 1987) using MEGA6 software (Kumar et al., 2004).
Sequence accession numbers
The 16S rRNA gene sequences obtained in this study have been deposited in GenBank under accession numbers AB793788–AB793797.
Statistical analysis
Statistical Analysis was performed using the non-parametric Wilcoxon Rank-Sum test to compare traits shown as ranks in different isolates. Each trait was tested in triplicates. The statistical package used was Minitab version 17.
Results
Enumeration and isolation of rhizobacteria
Generally, the population density in this study was considerably higher in rhizoplane than in the rhizosphere for the 11 plant species. The population density of the total bacteria inhabiting the rhizoplane and the rhizosphere ranged from 1.1 × 103 to 7.3 × 105 CFUg−1 roots respectively representing a population of 30.11 and 69.89% respectively. The population density of spore formers inhabiting rhizoplane and rhizosphere ranged from 2 × 101 to 1.3 × 104 CFUg−1 roots respectively.
In vitro evaluation of rhizospheric bacteria as plant growth promoters
Although many studies have been conducted to identify specific traits by which PGPR promote plant growth, usually they were limited to studying just one or two of these traits. Of the 531 total bacterial isolates of the present study only 66 were selected based on their ability to inhibit the world wide distributed soil-borne phytopathogenic fungi F. oxysporum, and S. sclerotiorum. All 66 selected isolates were further screened in vitro for a wide array of PGP traits. Tested PGP traits included N2-fixation, mineral phosphate and zinc solubilization, and IAA production.
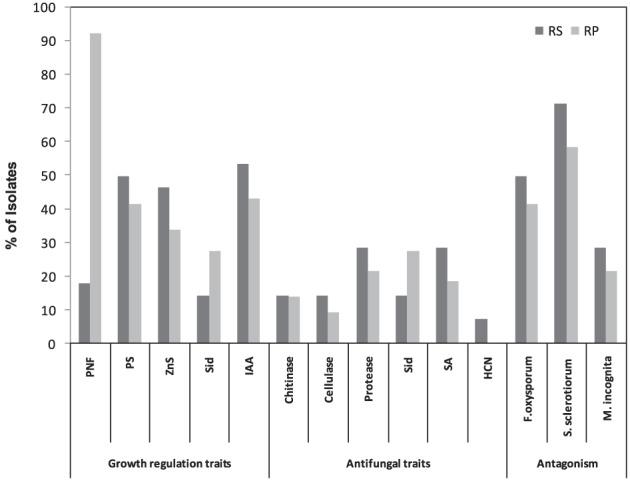
Among all isolates, 69.90% were found to be presumptive nitrogen fixers (PNFs). The percentages of PNFs among rhizosphere and rhizoplane isolates were 17.86%, and 92.3%, respectively (Figure 1). We tested the ability of the screened isolates to solubilize mineral phosphate (P) and zinc (Zn). As shown in Figure 1, 44.09 and 37.63% of the isolates were able to solubilize mineral P and Zn, respectively. These isolates varied in their solubilization abilities as being indicated by differences in solubilization index. On the other hand, proportions of rhizosphere and rhizoplane isolates that were able to solubilize P and Zn were 46.43 and 33.85%, respectively. In the present work, isolates varied greatly in their ability to produce IAA, with rhizosphere IAA producers being higher (53.57%) compared to rhizoplane ones (43.08%) (Figure 1).
Figure 1.
Numerical data of the in vitro screening of bacterial isolates for plant growth promoting traits from rhizosphere (RS) and rhizoplane (RP) of different wild plant species.
Antagonism toward plant pathogenic fungi and nematodes
PGPR have attracted the attention of many researchers because of the potential for developing these bacteria as inocula for plant disease control. Results presented in Figure 1 show that 44.09% of tested isolates had a wide range of antagonistic activity against F. oxysporum, while about 62.37% exhibited an antagonistic activity against S. sclerotiorum. Rhizosphere isolates were found to be highly efficient against fungal pathogens compared to rhizoplane ones. Of the rhizosphere isolates 50% were active against F. oxysporum and 71.43% against S. sclerotiorum, compared to 41.54% and 58.46% of the rhizoplane isolates respectively.
Cell free culture filtrates of isolated strains were also tested in vitro for their nematicidal activity on M. incognita. In general, juvenile mortality increased with increased exposure period to PGPR culture filtrates. A maximum nematode mortality of between 81% and 100% was observed. For reference, nematicidal Pseudomonads showed effects ranging from 84–96% to those of PGPR isolates. About 23.66% of the isolates showed nematicidal activity (Figure 1), with one isolate from rhizosphere and 4 isolates from rhizoplane exhibiting a significant reduction in the number of eggs hatching and a significant increase in M. incognita J2 mortality (97–100%). Interestingly, microscopic examination of the unhatched eggs indicated that a large proportion of them were severely damaged. However, this phenomenon was not observed in unhatched eggs treated with the reference strains.
Screening of rhizospheric bacteria for antifungal and nematicidal traits
Concerning siderophores (Sid), 23.66% of the isolates were able to produce Sid. The percentage of Sid -producers among rhizoplane and rhizosphere isolates was 27.69% and 14.29% respectively (Figure 1). Results in Figure 1 also indicated the widespread ability of these isolates to produce SA (21.51%) (Figure 1). The results revealed that the proportion of HCN-producers varied among the isolates of different plant species and rhizosphere microhabitats. Only 6.45% of isolates produced HCN in rhizosphere habitats (Figure 1). Regarding the lytic enzymes, as shown in Figure 1, the number of protease- producers was the highest (23.66%) followed by chitinase- producers (13.89%), whereas cellulase-producers recorded the lowest number (10.75%) (Figure 1).
Assessment of the in vitro PGPR traits
In an attempt to better select bacterial isolates with high plant growth promotion potential, a bonitur scale similar to that described by Krechel et al. (2002) was generated and used for the assessment of PGPR traits. In this scale, points are given for each in vitro bacterial trait examined. For traits examined here, the maximum possible bonitur score is 29 points (Figure 2). Results of the assessment revealed that out of the 66 isolates screened, 10 isolates according to Σ assessment values varied between 17 points and 23 points. Among those 10 isolates, 3 were isolated from rhizosphere, and 7 from rhizoplane of wild plants (Table 1). Isolate CSP03 from the wild plant C. spinosa showed the highest Σ assessment value of 23 points.
Figure 2.
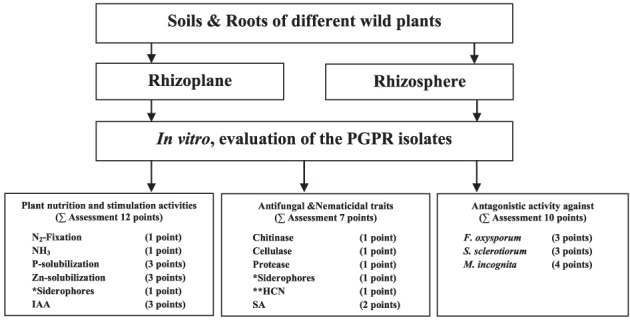
A bonitur scale (of 29 points) used for the assessment of the isolates based on their in vitro PGP traits screening. Two points were given to siderophores production, one as antifungal traits and one for facilitating iron uptake by plants. Points given to HCN production, if positive, were excluded from Σ assessment because it is considered ambiguous with the antifungal and nematicidal traits of HCN offset by deleterious effects on plant growth.
Table 1.
Top 10 rhizosphere and rhizoplane isolates and their plant nutrition and growth promotion, antifungal and nematicidal traits, in addition to their antagonistic activity and general assessment and ranking for their ability to function as PGPR.
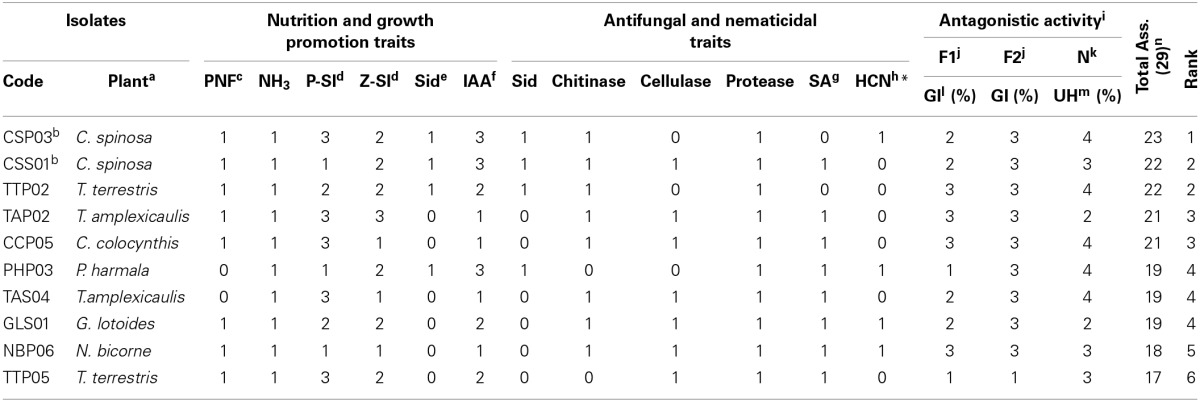
aWild Plant.
bS, Rhizosphere - P, Rhizoplane.
cPNF, Putative N2-fixation.
dSI, Phosphate solubilization index (1 = 2.99; 2 = 3 -4.99; 3 = 5), Zinc solubilization index (1 = 1.99; 2 = 2–3; 3 = 3).
eSid, Siderophores production.
fIAA, Indoleacetic acid production (1 = 1.99; 2 = 2 −2.99; 3 = 3 μg ml−1).
gSA, Salicylic acid production (1 = 50–100; 2 = 100–200 μg ml−1).
hHCN, Hydrogen cyanide.
iAntagonistic activity against fungi & nematode.
jF1, Fusarium oxysporum, F2, Sclerotinia sclerotiorum.
kN, Meloidogyne incognita.
lGI (%), Growth inhibition percentage, (1 = 30–49.9%; 2 = 50–69.9%; 3 = 70%).
mUH (%), Unhatched % (1 = 30–49.9%; 2 = 50–69.9%; 3 = 70–89.9%; 4 = 90%).
nTotal assessment points.
*The point given for HCN production was excluded.
Molecular characterization of potential PGPR isolates in restriction analysis of the amplified 16S rRNA genes
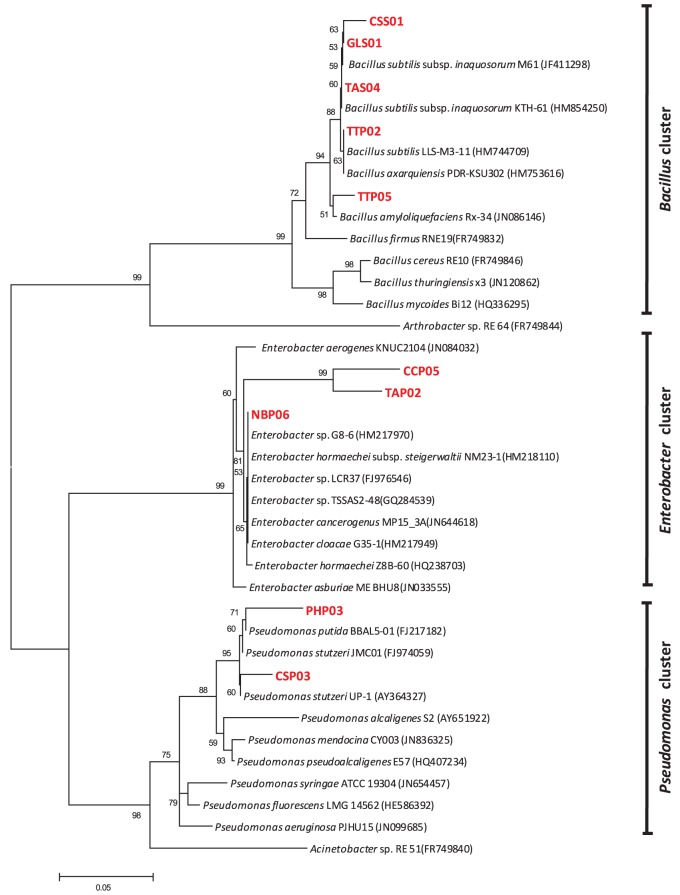
All of the 66 isolates that have been tested for PGPR traits were subjected to ARDRA and therefore were sorted into distinguished groups. Different profiles were generated by restriction digestion with the enzymes HaeIII indicating the presence of different genotypes. In accordance with the dendrogram of genetic similarity using Dice similarity coefficient index, all 66 isolates were grouped into 23 different groups (Figure 3). Selected genotypes with highest PGP rank were therefore categorized and subjected to molecular identification using 16S rRNA gene partial sequence analysis.
Figure 3.
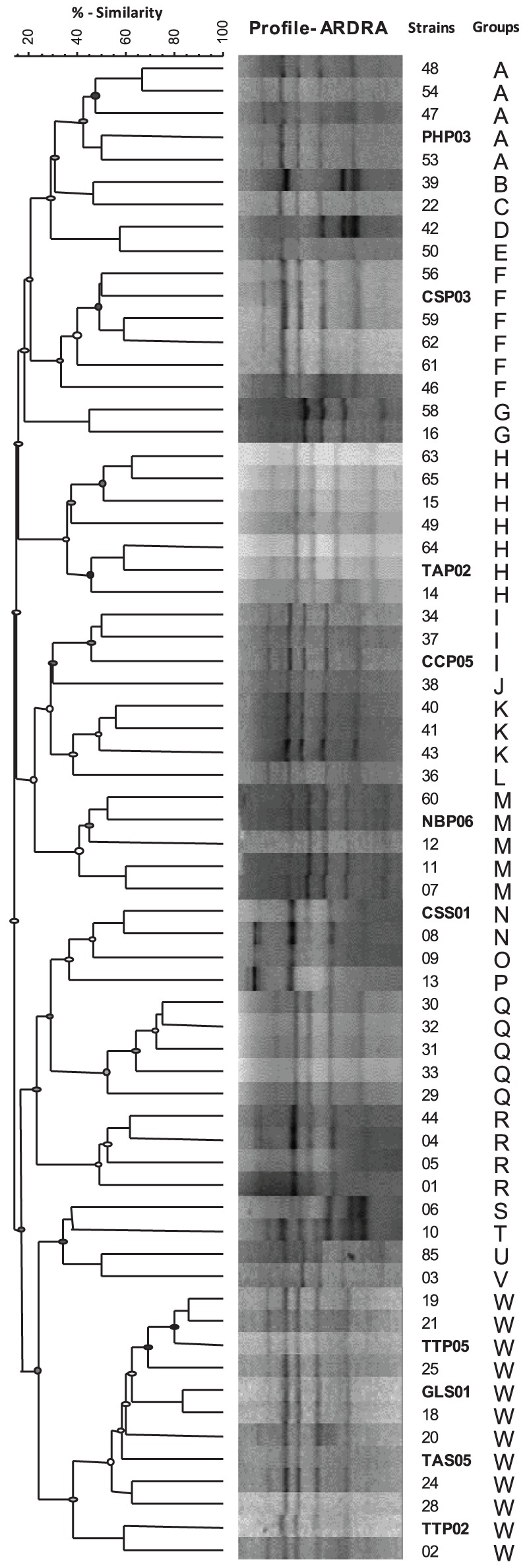
Dendrogram of genetic similarity using Dice similarity coefficient index for bacterial isolates from rhizosphere and rhizoplane of different wild plant species. ARDRA banding patterns were obtained after restriction digestion of the amplified 16S rRNA with HaeIII. Banding patterns were analyzed with GelCompar software and dendrogram was constructed after grouping using UPGMA.
Molecular typing and phylogenetic affiliation of potential PGPR isolates
Representative isolates belonging to different ARDRA groups, showing high growth promoting potential, were selected for the identification purposes. Isolates were identified by partial sequencing of their 16S rRNA genes, which led to the classification of the isolates into three specific genera, namely; Bacillus, Enterobacter, and Pseudomonas (Table 2).
Table 2.
Genotypic analysis and assignment of 16S rRNA gene sequence for selected isolates with in vitro PGPR traits.
| ARDRA profile | No. of Strainsa | Codeb | Blast match | |||
|---|---|---|---|---|---|---|
| Identity | Accession noc | Similarity (%) | Accession nod | |||
| F | 6 | CSP03 | Pseudomonas stutzeri GAPP4 | GU396288 | 98.11 | AB793795 |
| N | 2 | CSS01 | Bacillus sp. ZB2 | EU236757 | 99.58 | AB793788 |
| W | 12 | TTP02 | Bacillus subtilis LLS-M3-11 | HM744709 | 99.40 | AB793794 |
| H | 7 | TAP02 | Enterobacter sp. LCR37 | FJ976546 | 94.00 | AB793792 |
| I | 3 | CCP05 | Enterobacter cloacae 1245 | JF322972 | 95.53 | AB793791 |
| A | 5 | PHP03 | Pseudomonas putida BBAL5-01 | FJ217182 | 96.82 | AB793789 |
| W | 12 | TAS04 | Bacillus subtilis subsp. inaquosorum M61 | JF411298 | 99.20 | AB793793 |
| W | 12 | GLS01 | Bacillus subtilis subsp. inaquosorum KTH-61 | HM854250 | 99.60 | AB793790 |
| M | 5 | NBP06 | Enterobacter sp. G8-6 | HM217970 | 99.19 | AB793797 |
| W | 12 | TTP05 | Bacillus subtilis CD-6 | EU090295 | 97.95 | AB793796 |
Numbers of strains in the corresponding ARDRA profile.
Code for the selected strains with best PGP traits.
GeneBank sequence accession numbers of most closely related sequences.
GeneBank sequence accession numbers of selected strains.
Phylogenetic studies confirmed the affiliation of the selected genotypes into Bacillus, Enterobacter, and Pseudomonas clusters revealed by clustering of each genus to its corresponding group. Figure 4 represents the phylogenetic tree based on 16S rRNA sequences analysis and showing the relationship between selected isolates and representative species along with other related genera. Although most of the genotypes were closely related to reference strains, three strains designated as PHP03, CCP05, and TAP02 were regarded as novel species based on their low similarity and high bootstrap values.
Figure 4.
Neighbor-joining tree showing the phylogenetic relationship between selected potential PGPR isolates and reference strains from GenBank database. The bar represents 0.05 substitutions per site, bootstrap values (n = 1000) are displayed.
According to Blast similarity matches, the Bacillus group was found to include five different strains, CSS01 (99.58% similarity to Bacillus sp. strain ZB2), GLS01 (99.6% similarity to Bacillus subtilis strain KTH-61), TAS04 (99.2% similarity to Bacillus subtilis strain M61), TTP02 (99.4% similarity to Bacillus subtilis strain LLS- M3-11), and TTP05 (97.95% similarity to Bacillus subtilis strain CD-6). The five representative Bacillus strains showed a high growth promoting potentials. In this study we have demonstrated the dominance of endospore-forming bacilli in rhizospheric arid soil and confirmed their potential as PGPR. The tested antifungal traits also confirmed the high ranking of all Bacillus strains as a PGPR isolates. The distribution of Bacillus strains in rhizosphere was found to be much higher than in rhizoplane perhaps due to their higher tolerance to arid conditions and therefore independent survival from plant roots.
Enterobacter group on the other hand was found to include three different strains: CCP05 (95.53% similarity to Enterobacter cloacae strain 1245), TAP02 (94% similarity to Enterobacter asburiae strain E53), and NBP06 (99.1% similarity to Enterobacter cloacae strain G35-1). The three tested strains also possessed plant growth promoting and anti-phytopathogenic fungi traits. The third group included the Pseudomonades which had two different strains: PHP03 (96.82% similarity to Pseudomonas stutzeri strain p4) and CSP03 (98.11% similarity to P. stutzeri strain GAPP4). Pseudomonas strains PHP03 and CSP03 showed a high assessment rank for nutrition and growth regulation traits. The two strains were characterized for their higher nematicidal and fungicidal effects.
Discussion
A considerable worldwide research has focused on the exploration of varied agro- ecological niches for the existence of native beneficial micro-organisms. (Lugtenberg and Kamilova, 2009; Dastager et al., 2010). Wild plants are likely to harbor unique rhizobacterial communities that differ from those found in cultivated plants, which are extensively bred and subjected to intensive applications of various agrochemicals (Gamal-Eldin et al., 2008).
In the current study, rhizobacteria with in vitro PGPR traits were isolated from arid rhizospheric soil of 11 different wild plant species. These potential PGPR isolates were enumerated and screened in vitro for a broad spectrum of plant growth-promoting abilities as well as for antagonistic potential against phytopathogenic fungi and nematodes. All isolates were grouped according to their ARDRA banding pattern. Selected strains scoring high rank as PGPR were identified and their phylogenetic affiliations were determined.
In general, the population density of both bacteria and spore formers isolated from the 11 plant species in the present study was higher in the rhizoplane than in the rhizosphere. Differences in numbers and composition of rhizosphere microbes for different plant species and even different varieties within species have been reported (Kremer et al., 1990; Bisseling et al., 2009). The higher numbers of bacteria in the rhizoplane of the arid soil may be attributed to the presence of the root exudates that favor their existence. Rhizosphere on the other hand was found to harbor higher spore formers bacteria compared to rhizoplane. This may be best explained on the ground of lacking root exudates that support microbial activity.
Isolation of bacteria from rhizosphere and rhizoplane usually results in a large number of strains. It is almost impossible to test and evaluate every bacterial strain isolated for all PGP traits. In this study, a strategy has been established to sort out bacterial strains and restrict them to those highly suspected to have PGP traits. Antagonistic potential against phytopathogenic fungi was selected as criteria to have a preliminary judgment for the isolates whether they possess PGP traits or not. Amongst 531 bacteria isolated from the rhizospheric soil, only 66 isolates were able to inhibit F. oxysporum and S. sclerotiorum and were therefore recruited as PGPR. Isolates having antagonist potentials were considered as good candidates for being PGPR because of their indirect effect for promoting plant growth via inhibition of various plant pathogens. Selected isolates were further screened for PGPR traits like N2-fixation, mineral phosphate and zinc solubilization, and IAA production. 69.90% of the isolates were found to be nitrogen fixers with 92.3% isolated from the rhizoplane. Biological N2- fixation (BNF) by soil microorganisms is considered one of the major mechanisms by which plants benefit from the association of micro-partners. Several PGPR have been reported to fix N2 (Andrade et al., 1997). Lugtenberg and Kamilova (2009) attributed a similar distribution of nitrogen fixers to chemo-attraction of the microorganisms moving toward carbon exudates, allowing them to colonize and multiply in both the rhizosphere and the rhizoplane. Malik et al. (1997) investigated the nitrogen fixing ability of Azospirillum strain N-4 in rice and found a significant contribution of nitrogen fixed by the PGPR.
In soil, both macro and micronutrients undergo a complex dynamic equilibrium between soluble and insoluble forms. The equilibrium is strongly influenced by the soil pH which can be shifted by the microbiota ultimately affecting their accessibility to plant roots for absorption. Soluble phosphorus and zinc are deficient in most natural soils, However, insoluble forms of phosphorus and zinc can be made available to plants by the aids of rhizospheric microorganisms (Saravanan et al., 2003).
In the case of P and Zn solubilization, 44.09 and 37.63% of the isolates from the present study had such traits respectively. Proposed mechanisms for solubilization abilities have been reported, among which production of organic acids was the major mechanism of action by which insoluble compounds were converted to more soluble forms (Pietr et al., 1990).
IAA is also one of the most physiologically active growth regulator (auxins), and a common product of L-tryptophane that is metabolized by several microorganisms including PGPR (Arshad and Frankenberger, 1991). The present study showed that the percentage of rhizosphere IAA producers was higher compared to that of the rhizoplane. Several studies proved a good correlation between induction of root elongation and phytohormone production (Asghar et al., 2002). It is hypothesized that the rhizospheric bacteria influenced the growth of wild plants by producing auxins in the plant rhizotic zone upon the release of tryptophan in the root exudates (Asghar et al., 2002).
Besides stimulating plant growth by direct mechanisms, PGPR can also indirectly induce plant growth by protecting plants against soil-borne pathogens (Bloemberg and Lugtenberg, 2001; Mercado-Blanco and Bakker, 2007). Rhizosphere isolates showed higher antagonistic activity against F. oxysporum and S. sclerotiorum compared to those isolated from the rhizoplane. It has been reported previously that the plant species or cultivars and especially the composition of root exudates plays a key role in the diversity of rhizobacterial populations and can influence the frequency of antagonistic bacteria (Kremer et al., 1990; Siciliano et al., 1998).
Nematicidal activity against M. incognita was also observed especially by the rhizosphere isolates. Similar results were reported by Bin et al. (2005) who found that whole cultures and culture filtrates of some rhizobacteria showed nematicidal effects on J2 of M. javanica ranging from 62–64 and 62–70% respectively. This finding provides evidence that such isolates may produce nematicidal compounds to control M. incognita. The damage shown under the microscope by unhatched eggs was presumably due to lytic enzymes secreted by the tested isolates. In this context, Khan et al. (2004) found that M. javanica eggs treated with chitinase or protease in a liquid culture of Paecilomyces lilacinus displayed large vacuoles in the chitin layer. This phenomenon may also be the case in the top 5 isolates in our studies as they were positive for chitinase and protease production.
PGPR may enhance plant growth via suppression of phytopathogens by a variety of indirect mechanisms. This may include, the ability to produce siderophores to chelate iron, antifungal metabolites, lytic enzymes (e.g., chitinase, protease, cellulase and β-1, 3- glucanase), and hydrogen cyanide (Bloemberg and Lugtenberg, 2001). Among the total PGPR isolates, 23.66% were Sid-producers with higher percentage being rhizoplane isolates. It has been reported that the ability of PGPR to antagonize pathogenic fungi was related to the production of extracellular siderophores which deprive phytopathogenic microflora of iron, thus limiting their growth (Kloepper et al., 1980).
Many studies indicated that SA also plays an important role in plant defense response against pathogen attack and is essential for the development of both systemic acquired resistance (SAR) and induced systemic resistance (ISR) in plants (Zhang et al., 2002; Elbadry et al., 2006). A percentage of 21.51% of our isolates showed ability to produce SA.
Cyanide production is an ambiguous trait and is sometimes associated with deleterious as well as beneficial rhizobacteria (Bakker and Schippers, 1987; Alström and Burns, 1989). Only 6.45% of our rhizosphere isolates were HCN-producers.
Lytic enzymes is another trait associated with PGPR enabling them to limit fungal pathogens growth, as in vitro studies showed that the exposure of selected plant pathogenic fungi to lytic enzymes such as chitinase, protease, gluconase or cellulase can result in degradation of the structural matrix of fungal cell wall (Dunne et al., 1998). In our study, we observed that 23.66% were protease-producers, 13.89% were chitinase- producers and only 10.75% were cellulose-producers.
During the molecular study, 66 isolates were genotyped using amplified rDNA restriction analysis (ARDRA and 16S rRNA gene sequence analysis. The taxonomic composition of the representative genotypes from both rhizosphere and rhizoplane comprised Bacillus, Enterobacter, and Pseudomonas.
Bacillus spp. is known for their wide distribution in many soil types and was suggested for their possible role in the adaptation of desert plants to support their growth (Jorquera et al., 2012). Among all Bacillus spp. that have been isolated and characterized as PGPR, spore-forming bacilli have received much attention for commercial purposes due to their stability in the environment (Brannen and Backman, 1993).
Mirza et al. (2001) reported the isolation of strains of Enterobacter spp. from rhizosphere of sugarcane and illustrated their ability to function as PGPR. Enterobacter spp. were also found in diversity of PGPR isolated from sugarcane cultivated in South of Brazil (Beneduzi et al., 2013). In an effort to study the diversity of PGPR associated with rhizopheric soil and roots of canola, Farina et al. (2012) found that Pseudomonas and Enterobacter were among the most abundant rhizospheric bacteria showing several PGPR traits.
It has also been reported that Pseudomonas spp. is one of the most important bacteria inhabiting the rhizosphere of a diverse group of plants (Costa et al., 2006).
It has been shown that, in comparison to other plant microenvironments, the rhizosphere is one of the main reservoirs of antagonistic bacteria (Berg et al., 2005) with Pseudomonads as being most dominant bacteria showing antagonistic properties and therefore potential PGPR (Berg et al., 2006). However, in contrast to these reports, our results suggest that rhizoplane of wild plants in arid soil is a potential source for Pseudomonades with PGP properties rather than rhizosphere. Pseudomonas aeruginosa was found to play important role in oxidative stress tolerance in wheat (Islam et al., 2014). Moreover, Jorquera et al. (2012) found more diverse flora of rhizobacterial composition of the ancient clones of L. tridentata, with the hypothesis that those might play an important role in the adaptation of this plant and others to arid environments.
Conclusion
Rhizobacteria isolated from native wild desert plants was found to harbor antagonistic activity against F. oxysporum and S. sclerotiorum as well as unique PGP traits e.g., N2-fixation, indoleacetic acid and siderophores production, mineral phosphate and zinc solubilization, and antagonistic potentials. These antagonistic strains with their broad spectrum of in vitro PGP abilities and antagonistic potentials in such harsh environment make them good candidates as growth supporting agents for plants thriving at desert environment. As, in vitro studies should be considered prior to any green house and field studies, the present study provides a compelling evidence that rhizobacteria associated with plants growing at harsh environments do possess the traits which may contribute to plant adaptation to arid environments. Characterizing strains with in vitro PGPR traits from native plants adapted to harsh environment like that of Saudi Arabia would help scientists understand the behavior of PGPR in extreme environments and also funding strains that can be used in such environment to improve the growth of agricultural plants.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors acknowledge the Deanship of the Scientific Research, Taibah University, KSA, for the financial support to the Project No. 429/285. We are grateful to Dr. Thomas R. Glass, Senior Scientist, Sapidyne Instruments Inc., USA for his help in English editing of the manuscript.
References
- Abo-Aba S. E. M., Soliman E. A. M., Nivien A. A. (2006). Enhanced production of extra cellular alkaline protease in Bacillus ciculance through plasmid transfer. Res. J. Agric. Biol. Sci. 16, 526–530. [Google Scholar]
- Aeron A., Pandey P., Kumar S., Maheshwari D. K. (2011). Emerging role of plant growth promoting rhizobacteria, in Bacteria in Agrobiology: Cropecosystem, ed Maheshwari D. K. (Berlin/Heidelberg: Springer Verlag; ), 1–26 10.1007/978-3-642-18357-7_1 [DOI] [Google Scholar]
- Alström S., Burns R. G. (1989). Cyanide production by rhizobacteria as a possible mechanism of plant growth Inhibition. Biol. Fert. Soils. 7, 232–238 10.1007/BF00709654 [DOI] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade G., Esteban E., Velasco L., Lortie M. J., Bedmar E. J. (1997). Isolation and identification of N2-fixing microorganisms from the rhizoplane of Capparis spinosa (L.). Plant Soil 197, 19–23 10.1023/A:1004211909641 [DOI] [Google Scholar]
- Andro T., Chambost J. P., Kotoujansky A., Cattaneo J., Bertheau Y., Barras F., et al. (1984). Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J. Bacteriol. 160, 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad M., Frankenberger W. T. (1991). Microbial production of plant hormones. Plant Soil. 133, 1–8 10.1007/BF00011893 [DOI] [Google Scholar]
- Arturo A. M., Oldelson D. A., Hichey R. F., Tiedje J. M. (1995). Bacterial community fingerprinting of amplified 16-23S ribosomal DNA gene and restriction endonuclease analysis, in Molecular Microbial Ecology Manual, eds Akkermans A. D. L., van Elsas J. D., Bruijn F. J. (Dordrecht: Kluwer Academic Publications; ), 1–8. [Google Scholar]
- Asghar H. N., Zahir Z. A., Arshad M., Khaliq A. (2002). Relationship between in vitro production of auxins by rhizobacteria and their growth promoting activities in Brassica juncea L. Biol. Fert. Soils 35, 231–237 10.1007/s00374-002-0462-8 [DOI] [Google Scholar]
- Bai Y., D'Aoust F., Smith D. L., Driscoll B. T. (2002). Isolation of plant growth promoting Bacillus strains from soybean root nodules. Can. J. Microbiol. 48, 230–238. 10.1139/w02-014 [DOI] [PubMed] [Google Scholar]
- Bakker A. W., Schippers B. (1987). Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp. mediated plant growth- stimulation. Soil Biol. Biochem. 19, 451–457 10.1016/0038-0717(87)90037-X [DOI] [Google Scholar]
- Bashan Y., de-Bashan L. E. (2005). Bacteria/plant growth-promoting, in Encyclopedia of Soils in the Environment, Vol. 1, ed Hillel D. (London; Oxford: Elsevier; ), 103–115. [Google Scholar]
- Beneduzi A., Moreira F., Costa P. B., Vargas L. K., Lisboa B. B., Favreto R., et al. (2013). Diversity and plant growth promoting evaluation abilities of bacteria isolated from sugarcane cultivated in the south of Brazil. App. Soil Ecol. 63, 94–104 10.1016/j.apsoil.2012.08.010 [DOI] [Google Scholar]
- Berg G., Krechel A., Ditz M., Sikora R. A., Ulrich A., Hallmann J. (2005). Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microb. Ecol. 51, 215–229. 10.1016/j.femsec.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Berg G., Opelt K., Zachow C., Lottmann J., Götz M., Costa R., et al. (2006). The rhizosphere effect on bacteria antagonistic toward the pathogenic fungus Verticillium differs depending on plant species and site. FEMS Microb. Ecol. 56, 250–261. 10.1111/j.1574-6941.2005.00025.x [DOI] [PubMed] [Google Scholar]
- Bin L., Xie G., Soad A., Coosemans J. (2005). Suppression of Meloidogyne javanica by antagonistic and plant growth-promoting rhizobacteria. J. Zhejiang Univ. Sci. 6, 496–501. 10.1613/jzus.2005.B0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisseling T., Dangl J. L., Schulze-Lefert P. (2009). Next-generation communication. Science 324, 691. 10.1126/science.1174404 [DOI] [PubMed] [Google Scholar]
- Bloemberg G. V., Lugtenberg B. J. (2001). Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4, 343–350. 10.1016/S1369-5266(00)00183-7 [DOI] [PubMed] [Google Scholar]
- Brannen P. M., Backman P. A. (1993). Cotton colonization by Bacillus subtilis inoculants to augment seedling disease control and promote season-long root health, in Proceedings of the Beltwide Cotton Production Research Conference (New Orleans, LA: ), 194–196. [Google Scholar]
- Cappuccino J. C., Sherman N. (1992). Microbiology: A Laboratory Manual, 3rd Edn. New York, NY: Benjamin/cummings Pub Co. [Google Scholar]
- Castric P. A. (1975). Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can. J. Microbiol. 21, 613–618. 10.1139/m75-088 [DOI] [PubMed] [Google Scholar]
- Chaudhary S. A. (1999). Flora of the Kingdom of Saudi Arabia. Vol. 1. Riyadh: Ministry of Agriculture and Water. [Google Scholar]
- Chaudhary S. A. (2000). Flora of the Kingdom of Saudi Arabia. Vol. II. Riyadh: Ministry of Agriculture and Water. [Google Scholar]
- Chaudhary S. A. (2001). Flora of the Kingdom of Saudi Arabia. Vol. III. Riyadh: Ministry of Agriculture and Water. [Google Scholar]
- Costa R., Gomes N. C. M., Peixoto R. S., Rumjanek N., Berg G., Mendonca-Hagler L. C. S., et al. (2006). Diversity and antagonistic potential of Pseudomonas spp. associated to the rhizosphere of maize grown in a subtropical organic farm. Soil Biol. Biochem. 38, 2434–2447 10.1016/j.soilbio.2006.03.003 [DOI] [Google Scholar]
- Dastager S. G., Deepa C. K., Pandey A. (2010). Potential plant growth promoting activity of Serratia nematophila NII- 0.928 on black pepper (Piper nigrum L.). World J. Microbiol. Biotechnol. 27, 259–265 10.1007/s11274-010-0454-z [DOI] [Google Scholar]
- Drogue B., Doré H., Borland S., Wisniewski-Dyé F., Prigent-Combaret C. (2012). Which specificity in cooperation between phytostimulating rhizobacteria and plants? Res. Microbiol. 163, 500–510. 10.1016/j.resmic.2012.08.006 [DOI] [PubMed] [Google Scholar]
- Dunne C., Moënne-Loccoz Y., McCarthy J., Higgins P., Powell J., Dowling D. N., et al. (1998). Combining proteolytic and phloroglucinol-producing bacteria for improved biocontrol of Pythium-mediated damping-off of sugar beet. Plant Pathol. 47, 299–307. [Google Scholar]
- Edi-Premona M., Moawad M. A., Vlek P. L. G. (1996). Effect of phosphate- solubilizing Pseudomonas putida on growth of maize and its survival in the rhizosphere. Indones. J. Crop Sci. 11, 13–23. [Google Scholar]
- Elbadry M., Taha R. M., Eldougdoug K. A., Gamal-Eldin H. (2006). Induction of systemic resistance in faba bean (Vicia feba, L.) to bean yellow mosaic potyvirus (BYMV) via seed bacterization with plant growth promoting rhizobacteria. J. Plant Dis. Protect. 113, 247–251. [Google Scholar]
- Farina R., Beneduzi A., Ambrosini A., de Campos S. B., Lisboa B. B., Wendisch V., et al. (2012). Diversity of plant growth promoting rhizobacteria communities associated with the stages of canola growth. App. Soil Ecol. 55, 44–52 10.1016/j.apsoil.2011.12.011 [DOI] [Google Scholar]
- Frändberg E., Schnürer J. (1998). Antifungal activity of chitinolytic bacteria isolated from airtight stored cereal grain. Can. J. Microbiol. 44, 121–127 10.1139/w97-141 [DOI] [Google Scholar]
- Gamal-Eldin H., Elbadry M., Mahfouz S., Abdelaziz S. (2008). Screening of fluorescent pseudomonad bacteria isolated from rhizospheres of cultivated and wild plants in vitro for plant growth promoting traits. J. Agric. Sci. 33, 3365–3383. [Google Scholar]
- Gamalero E., Glick B. R. (2011). Mechanisms used by plant growth-promoting bacteria, in Bacteria in Agrobiology: Plant Nutrient Management, ed Maheshwari D. K. (Berlin; Heidelberg: Springer-Verlag; ), 17–46. [Google Scholar]
- Haahtela K., Helander I., Nurmiaha-Lassila E. L., Sundman V. (1983). Morphological and physiological characteristics and lipopolysaccharide composition of N2-fixing (C2H2-reducing) root-associated Pseudomonas sp. Can. J. Microbiol. 29, 874–880 [DOI] [PubMed] [Google Scholar]
- Hussey R. S., Barker K. R. (1973). A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Dis. Rep. 57, 1025–1028. [Google Scholar]
- Islam F., Yasmeen T., Ali S., Arif M. S., Hussain S., Rizvi H. (2014). Influence of Pseudomonas aeruginosa as PGPR on oxidative stress tolerance in wheat under Zn stress. Ecotoxicol. Environ. Saf. 104, 285–293. 10.1016/j.ecoenv.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Jayaprakashvel M., Mathivanan N. (2011). Management of plant diseases by microbial metabolites, in Bacteria in Agrobiology: Plant Nutrient Management, ed Maheshwari D. K. (Berlin/Heidelberg: Springer-Verlag; ), 237–265. [Google Scholar]
- Jorquera M. A., Shaharoona B., Nadeem S. M., de la Luz-Mora M., Crowley D. E. (2012). Plant growth-promoting rhizobacteria associated with ancient clones of creosote bush (Larreatridentata). Microb. Ecol. 64, 1008–1017. 10.1007/s00248-012-0071-5 [DOI] [PubMed] [Google Scholar]
- Khan A., Williams K. L., Nevalainen H. K. M. (2004). Effects of Paecilomyces lilacinus protease and chitinase on the eggshell structures and hatching of Meloidogyne javanica juveniles. Biol. Control 31, 346–352 10.1016/j.biocontrol.2004.07.011 [DOI] [Google Scholar]
- Kloepper J. W., Leong J., Teintze M., Schroth M. N. (1980). Pseudomonas siderophores: a mechanism explaining disease-suppressive soils. Curr. Microbiol. 4, 317–320 10.1007/BF02602840 [DOI] [Google Scholar]
- Kloepper J. W., Schroth M. N. (1978). Plant growth promoting rhizobacteria on radishes, in Proceedings of the IVth International Conference on Plant Pathogenic Bacteria, Station de PathologieVégétableet et Phytobactériologie INRA (Angers: ), 879–882. [Google Scholar]
- Krechel A., Faupel A., Hallmann J., Ulrich A., Berg G. (2002). Potato-associated bacteria and their antagonistic potential towards plant-pathogenic fungi and the plant-parasitic nematode Meloidogyne incognita (Kofoid and White) Chitwood. Can. J. Microbiol. 48, 772–786. 10.1139/w02-071 [DOI] [PubMed] [Google Scholar]
- Kremer R. J., Begonia M. F. T., Stanley L., Lanham E. T. (1990). Characterization of rhizobacteria associated with weed seedlings. Appl. Environ. Microbiol. 56, 1649–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: an integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5, 150–163. 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Lane D. J. (1991). 16S/23S rRNA sequencing, in Nucleic acid Techniques in Bacterial Systematics, eds Stackebrandt E., Goodfellow M. (New York, NY: John Wiley and Sons; ), 115–175. [Google Scholar]
- Leeman M., DenOuden F. M., Van Pelt J. A., Cornelissen C., Matamala-Garros A., Bakker P. A. H. M., et al. (1996). Suppression of Fusarium wilt of radish by co-inoculation of fluorescent Pseudomonas spp. and root-colonizing fungi. Eur. J. Plant Pathol. 102, 21–31 10.1007/BF01877112 [DOI] [Google Scholar]
- Loper J. E., Schroth M. N. (1986). Influence of bacterial sources of indole-3-acetic acid on root elongation of sugar beet. Phytopathology 76, 386–389 10.1094/Phyto-76-386 [DOI] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Malik K. A., Bilal R., Mehnaz S., Rasul G., Mirza M. S., Ali S. (1997). Association of nitrogen-fixing plant-growth promoting rhizobacteria (PGPR) with kallar grass and rice. Plant Soil 194, 37–44 10.1023/A:1004295714181 [DOI] [Google Scholar]
- Mercado-Blanco J., Bakker P. A. (2007). Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie Van Leeuwenhoek 92, 367–389. 10.1007/s10482-007-9167-1 [DOI] [PubMed] [Google Scholar]
- Mirza M. S., Ahmed W., Latif F., Haurat J., Bally R., Normand P., et al. (2001). Isolation, partial characterization, and the effect of plant growth- promoting bacteria (PGPB) on micro-propagated sugarcane in vitro. Plant Soil 237, 47–54 10.1023/A:1013388619231 [DOI] [Google Scholar]
- Napoli C., Mello A., Bonfante P. (2008). Dissecting the rhizosphere complexity: the truffle-ground study case. Rend. Lincei. 19, 241–259 10.1007/s12210-008-0017-5 [DOI] [Google Scholar]
- Nautiyal C. S., DasGupta S. M. (2007). Screening of plant growth-promoting rhizobacteria, in Advanced Techniques in Soil Microbiology, eds Varma A., Oelmüller R. (New York, NY: Springer; ), 363–375. [Google Scholar]
- Pallai R. (2005). Effect of Plant Growth-Promoting Rhizobacteria on Canola (Brassica Napus. L) and Lentil (Lens Culinaris. Medik) Plants. M.Sc. thesis, Department of Applied Microbiology and Food Science, University of Saskatchewan, Saskatoon. [Google Scholar]
- Pietr S. J., Koran B., Stankiewcz M. (1990). Influence of rock phosphate-dissolving rhizobacteria on the growth and the P-uptake by oat-preliminary results, in 2nd International Workshop on Plant Growth Promoting Rhizobacteri (Interlaken: ), 14–19. [Google Scholar]
- Rodriguez H., Gonzalez T., Goire I., Bashan Y. (2004). Gluconic acid production and phosphate solubilization by the plant growth promoting bacterium Azospirillum spp. Naturwissenschaften 91, 552–555. 10.1007/s00114-004-0566-0 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. (1977). DNA sequencing with chain- termination inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467. 10.1073/pnas.74.12.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraf M., Pandya U., Thakkar A. (2014). Role of allelochemicals in plant growth promoting rhizobacteria for biocontrol of phytopathogens. Microbiol. Res. 169, 18–29. 10.1016/j.micres.2013.08.009 [DOI] [PubMed] [Google Scholar]
- Saravanan V. S., Subramoniam S. R., Raj S. A. (2003). Assessing in vitro solubilization potential of different zinc solubilizing bacterial (ZSB) isolates. Braz. J. Microbiol. 34, 121–125 10.1590/S1517-83822004000100020 [DOI] [Google Scholar]
- Siciliano S. D., Theoret C. M., de Freitas J. R., Hucl P. J., Germida J. J. (1998). Differences in the microbial communities associated with the roots of different cultivars of canola and wheat. Can. J. Microbiol. 44, 844–851. [Google Scholar]
- Somers E., Vanderleyden J., Srinivasan M. (2004). Rhizosphere bacterial signalling: a love parade beneath our feet. Crit. Rev. Microbiol. 30, 205–240. 10.1080/10408410490468786 [DOI] [PubMed] [Google Scholar]
- Thompson D., Gibson J., Plewinak F., Jeanmougin F., Higgins G. (1997). The Clastal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuc. Acid Res. 25, 4867–4887. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron J., Desbrosses G., Marie-Lara B., Touraine B., Moënne-Loccoz Y., Muller D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4:356. 10.3389/fpls.2013.00356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C., Pieterse C. M. J. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant-Microbe Interact. 25, 139–150. 10.1094/MPMI-06-11-0179 [DOI] [PubMed] [Google Scholar]
- Zhang S., Moyne A. L., Reddy M. S., Kloepper J. W. (2002). The role of salicylic acid in induced systemic resistance elicited by plant growth-promoting rhizobacteria against blue mold of tobacco. Biol. Control. 25, 288–296 10.1016/S1049-9644(02)00108-1 [DOI] [Google Scholar]