Abstract
Species belonging to Penicillium section Aspergilloides have a world-wide distribution with P. glabrum, P. spinulosum and P. thomii the most well-known species of this section. These species occur commonly and can be isolated from many substrates including soil, food, bark and indoor environments. The taxonomy of these species has been investigated several times using various techniques, but species delimitation remains difficult. In the present study, 349 strains belonging to section Aspergilloides were subjected to multilocus molecular phylogenetic analyses using partial β-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) sequences. Section Aspergilloides is subdivided into 12 clades and 51 species. Twenty-five species are described here as new and P. yezoense, a species originally described without a Latin diagnosis, is validated. Species belonging to section Aspergilloides are phenotypically similar and most have monoverticillate conidiophores and grow moderately or quickly on agar media. The most important characters to distinguish these species were colony sizes on agar media, growth at 30 °C, ornamentation and shape of conidia, sclerotium production and stipe roughness.
Key words: Eurotiales, Soil fungi, Multigene phylogeny, ITS barcoding
Introduction
In the classification of Raper & Thom (1949), Penicillia that produce monoverticillate conidiophores were placed in the Monoverticillata group. They divided this group into nine series based on colony texture, production of sclerotia and/or cleistothecia and length of conidiophores. A series of species with irregularly branched conidiophores was also included in this classification, namely series Ramigena fide Raper & Thom. Pitt (1980) later excluded this series from his circumscription of subgenus Aspergilloides. In Pitt's classification, subgenus Aspergilloides only included species in which the majority of conidiophore stipes are well defined and terminate in monoverticillate penicilli (Pitt 1980). Additionally, he introduced sections Aspergilloides and Exilicaulis in subgenus Aspergilloides based on the presence or absence of a swelling at the stipe apex. Peterson (2000) was among the first to study the infrageneric relations in Penicillium using DNA sequence data. Based on a phylogeny of nrDNA sequences, the genus was divided into six groups with group 2 containing species mainly belonging to Pitt's section Aspergilloides (P. glabrum, P. purpurescens, P. spinulosum, P. fuscum (syn. E. pinetorum), P. thomii, P. lividum, P. lapidosum (syn. E. lapidosum) and P. asperosporum). Houbraken & Samson (2011) studied the phylogeny of Penicillium in more detail using a combined data set of four genes. Based on the inferred phylogenetic relationships among the Penicillia, they proposed a sectional classification and subdivided Penicillium into two subgenera and 25 sections, with section Aspergilloides being one of them. With exception of P. lapidosum, all species assigned to Peterson's group 2 were included in the re-circumscribed section Aspergilloides and an additional twelve species were included. The majority of species belonging to section Aspergilloides are predominantly monoverticillate and most grow quickly on agar media (Pitt 1980, Houbraken & Samson 2011).
The most well-known species in section Aspergilloides are P. glabrum and P. spinulosum. Phenotype-based identification of these species is problematic and the taxonomy has been studied several times. Raper & Thom (1949) distinguished P. spinulosum from P. glabrum based on colony texture: the colony surface of P. spinulosum was stated to be “loose textured” while P. glabrum was “strictly velvety”. This distinction was also adopted by Ramírez (1982). Pitt (1980) primarily separated P. glabrum from P. spinulosum by conidial wall texture, which was stated to be “smooth or at most finely roughened” for P. glabrum and “rugose or spinose” for P. spinulosum. However, P. spinulosum isolates with smooth to finely roughened conidia were also accepted in his definition of the species, and then the floccose texture of the colony was the key character to separate P. spinulosum from P. glabrum. In 1990, the Subcommission on Penicillium and Aspergillus Systematics (SPAS, currently known as the International Commission on Penicillium and Aspergillus, ICPA) investigated the taxonomy of P. glabrum, P. spinulosum and the related species P. purpurescens and P. montanense (Pitt et al. 1990). Colony diameters on Czapek yeast extract agar (CYA) and 25 % glycerol nitrate agar (G25N), conidial wall texture and width of the phialides proved to be valuable characters for the identification of these species. However, four of the 15 strains could not be identified, indicating the difficulty of a phenotype-based identification of these species and/or the presence of cryptic species. Although P. spinulosum, P. glabrum, P. purpurescens and P. montanense are difficult to distinguish phenotypically, Peterson (2000) could identify them based on nrDNA sequences. These species were closely related in their nrDNA (ITS barcode, including partial LSU) sequences, but displayed 4–8 nucleotide substitutions between each of the pairings. The taxonomy of the “Penicillium glabrum complex” was also studied using partial β-tubulin and calmodulin sequences (Barreto et al. 2011). Based on this data, P. glabrum and P. spinulosum could be clearly distinguished.
In this study, we delimitate Penicillium section Aspergilloides using a phylogenetic analysis of a combined data set of partial β-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) gene sequences. Subsequently, the phylogenetic relationships among species of section Aspergilloides were investigated and species limits were proposed based on the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) concept (Taylor et al. 2000), supplemented with physiological and macro- and microscopic characters. We included 349 strains assigned to section Aspergilloides in our analyses, including type and freshly isolated strains. ITS barcodes were generated and investigated for their suitability for species identification.
Material and methods
Fungal strains
Isolates were obtained from different culture collections (CBS, CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands; IBT, culture collection of the DTU Systems Biology, Lyngby, Denmark; and FRR, culture collection of CSIRO Animal, Food and Health Sciences, North Ryde, Australia). Fresh isolates deposited in the working collection of the Department of Applied and Industrial Mycology (DTO), housed at CBS, were also included in this study and a selection of those strains were accessioned to the CBS culture collection. An overview of strains is listed in Table 1.
Table 1.
Strains used in this study.
| Species name | Collection no. | Substrate, location | GenBank accession nr. |
|||
|---|---|---|---|---|---|---|
| ITS | BenA | RPB2 | CaM | |||
| P. ardesiacum | DTO 093-C1 = CBS 497.73 = ATCC 24719 = FRR 1479 = IFO 30540 = IMI 174719 = VKM F-1749 | Stems of Vitis vinifera during drying; Alma-Ata Region, Kazachstan; ex-neotype of P. ardesiacum | KM189565 | KM088805 | KM089577 | KM089190 |
| P. armarii | DTO 235-F1 = CBS 138171 | House Dust; Hobart, Australia; ex-type of P. armarii | KM189758 | KM089007 | KM089781 | KM089394 |
| DTO 235-F3 | House Dust; Hobart, Australia | KM189759 | KM089008 | KM089782 | KM089395 | |
| DTO 236-D3 | House Dust; Hobart, Australia | KM189760 | KM089009 | KM089783 | KM089396 | |
| P. athertonense | DTO 030-C2 = CBS 138161 | Forest soil (wet); Atherton Tableland, Queensland, Australia | KM189462 | KM088690 | KM089462 | KM089075 |
| P. aurantioviolaceum | DTO 225-E4 = CBS 137777 = NRRL 762 | Unrecorded source; Puerto Rico; ex-neotype of P. aurantioviolaceum | KM189756 | KM089005 | KM089779 | KM089392 |
| DTO 091-A1 = CBS 347.59 = FAT 340 = IFO 6031 = IMI 068221 | Soil; Japan; ex-type of P. thomii var. flavescens nom. inval. | KM189552 | KM088791 | KM089563 | KM089176 | |
| DTO 085-A7 | Soil, 2 mtr. from road; Ranoma fana, Madagascar | KM189542 | KM088780 | KM089552 | KM089165 | |
| DTO 253-H3 = CBS 137779 | Leaves; Zambia | KM189763 | KM089012 | KM089786 | KM089399 | |
| P. austroafricanum | DTO 133-G5 = CBS 137773 | Leaf of Phaenocoma prolifera; Harold Porter Botanical Garden Western Cape, South Africa; ex-type of P. austroafricanum | KM189610 | KM088854 | KM089628 | KM089241 |
| DTO 132-D6 | Leaf of Phaenocoma prolifera; Harold Porter Botanical Garden Western Cape, South Africa | KM189609 | KM088853 | KM089627 | KM089240 | |
| DTO 180-E3 = CV 2851 = KAS 3946 | Fynbos soil; Riverlands, South Africa | KM189637 | KM088881 | KM089655 | KM089268 | |
| DTO 182-B3 = CBS 137756 = CV 851 = KAS 4183 = DAOM 241138 | Air sample from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa | KM189656 | KM088900 | KM089674 | KM089287 | |
| DTO 182-C7 = CBS 137757 = CV 905 = KAS 4197 = DAOM 241141 | Soil from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa | KM189660 | KM088904 | KM089678 | KM089291 | |
| DTO 182-H2 = CBS 137758 = CV 1145 = KAS 3974 = DAOM 241140 | Mite inside Protea repens infructescens; Riverlands, Malmesbury, Western Cape, South Africa | KM189674 | KM088919 | KM089693 | KM089306 | |
| DTO 182-H3 = CBS 137759 = CV 1148 = KAS 3975 = DAOM 241139 | Mite inside Protea repens infructescens; Riverlands, Malmesbury, Western Cape, South Africa | KM189675 | KM088920 | KM089694 | KM089307 | |
| P. brunneoconidiatum | DTO 182-E4 = CBS 137732 = CV 949 = KAS 4214 = DAOM 241359 | Soil; Fynbos, Riverlands, Malmesbury, Western Cape, South Africa; ex-type of P. brunneoconidiatum | KM189666 | KM088911 | KM089685 | KM089298 |
| DTO 182-B7 = CV 875 = KAS 4187 | Fynbos soil; Riverlands, South Africa | KM189657 | KM088901 | KM089675 | KM089288 | |
| DTO 182-C6 = CV 901 = KAS 4196 | Fynbos soil; Riverlands, South Africa | KM189659 | KM088903 | KM089677 | KM089290 | |
| DTO 182-D8 = CV 935 = KAS 4209 | Fynbos soil; Riverlands, South Africa | KM189663 | KM088908 | KM089682 | KM089295 | |
| DTO 182-E2 = CV 946 = KAS 4212 | Fynbos soil; Riverlands, South Africa | KM189665 | KM088910 | KM089684 | KM089297 | |
| DTO 182-F2 = CV 970 = KAS 4222 | Fynbos soil; Riverlands, South Africa | KM189670 | KM088915 | KM089689 | KM089302 | |
| DTO 185-F4 = CV 915 | Fynbos soil; Riverlands, South Africa | KM189691 | KM088937 | KM089711 | KM089324 | |
| DTO 185-F6 = CV 921 | Fynbos soil; Riverlands, South Africa | KM189692 | KM088938 | KM089712 | KM089325 | |
| P. bussumense | DTO 018-B2 = CBS 138160 | Soil; Spanderswoud, Bussum, the Netherlands; ex-type of P. bussumense | KM189458 | KM088685 | KM089457 | KM089070 |
| P. cartierense | DTO 092-H9 = CBS 137956 | Heathland soil; Cartier Heide, Eersel, the Netherlands; ex-type of P. cartierense | KM189564 | KM088804 | KM089576 | KM089189 |
| DTO 091-A6 = CBS 863.71 | Agricultural soil; Wageningen, the Netherlands | KM189557 | KM088796 | KM089568 | KM089181 | |
| P. clavistipitatum | DTO 182-E5 = CBS 138650 = CV 951 = KAS 4216 = DAOM 241125 | Soil; Fynbos, Riverlands, South Africa; ex-type of P. clavistipitatum | KM189667 | KM088912 | KM089686 | KM089299 |
| DTO 182-E8 = CV 960 = KAS 4219 = DAOM 241128 | Fynbos soil; Riverlands, South Africa | KM189668 | KM088913 | KM089687 | KM089300 | |
| DTO 182-E9 = CV 962 = KAS 4220 = DAOM 241126 | Fynbos soil; Riverlands, South Africa | KM189669 | KM088914 | KM089688 | KM089301 | |
| P. contaminatum | DTO 091-A3 = CBS 345.52 = IMI 049057 | Contaminant; Surrey, Kew, UK; ex-type of P. contaminatum | KM189554 | KM088793 | KM089565 | KM089178 |
| DTO 296-G9 = CBS 346.59 | Acidic soil; Unknown location | KM189782 | KM089032 | KM089806 | KM089419 | |
| P. crocicola | DTO 104-E2 = CBS 745.70 = ATCC 18313 = QM 7778 | Corm of Crocus sativus; Japan; ex-isotype of P. crocicola | KM189581 | KM088824 | KM089597 | KM089210 |
| DTO 082-A9 = CBS 137772 | Archive; Den Bosch, the Netherlands | KM189532 | KM088770 | KM089542 | KM089155 | |
| DTO 086-C2 | Swab sample taken in archive; Den Bosch, the Netherlands | KM189545 | KM088783 | KM089555 | KM089168 | |
| DTO 090-F5 | Swab sample in archive; Asperen, the Netherlands | KM189548 | KM088786 | KM089558 | KM089171 | |
| DTO 181-G2 = CBS 137754 = CV 461 = KAS 4133 = DAOM 241137 | Protea repens infructescens; Stellenbosch mountain, Western Cape, South Africa | KM189651 | KM088895 | KM089669 | KM089282 | |
| DTO 210-F5 = CBS 132168 = WSF 2215 | Soil; A1 horizon Soil; S. Wisconsin maple-elm-ash forests, deciduous forest; Wisconsin; USA | KM189750 | KM088999 | KM089773 | KM089386 | |
| DTO 265-H7 | Grapevine; Ajabshir, Iran | KM189768 | KM089018 | KM089792 | KM089405 | |
| DTO 266-A4 = CBS 137780 | Grapevine; Malekan, Iran | KM189769 | KM089019 | KM089793 | KM089406 | |
| P. flavisclerotiatum | DTO 180-I1 = CBS 137749 = CV 77 = KAS 4173 = DAOM 241158 | Soil from Fynbos; Stellenbosch mountain, Western Cape, South Africa; ex-type of P. flavisclerotiatum | KM189643 | KM088887 | KM089661 | KM089274 |
| DTO 180-I8 = CBS 137750 = CV 100 = KAS 3958 = DAOM 241157 | Soil from Fynbos; Stellenbosch mountain, Western Cape, South Africa | KM189644 | KM088888 | KM089662 | KM089275 | |
| DTO 181-H7 = CBS 137748 = CV 537 = KAS 4149 = DAOM 241156 | Soil from Fynbos; Stellenbosch mountain, Western Cape, South Africa | KM189653 | KM088897 | KM089671 | KM089284 | |
| DTO 182-B2 = CV 839 = KAS 4181 = DAOM 241155 | Air sample; Riverlands, South Africa | KM189655 | KM088899 | KM089673 | KM089286 | |
| DTO 182-D3 = CV 924 = KAS 4203 = DAOM 241154 | Fynbos soil; Riverlands, South Africa | KM189661 | KM088906 | KM089680 | KM089293 | |
| DTO 182-D4 = CBS 137751 = CV 925 = KAS 4204 = DAOM 241153 | Soil from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa | KM189662 | KM088907 | KM089681 | KM089294 | |
| DTO 182-D9 = CBS 137752 = CV 938 = KAS 4210 = DAOM 241152 | Soil from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa | KM189664 | KM088909 | KM089683 | KM089296 | |
| DTO 182-F3 = CBS 137753 = CV 971 = KAS 4223 = DAOM 241151 | Soil from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa | KM189671 | KM088916 | KM089690 | KM089303 | |
| DTO 184-D8 = CV 65 | Fynbos soil; Stellenbosch, South Africa | KM189686 | KM088932 | KM089706 | KM089319 | |
| DTO 184-D9 = CV 76 | Fynbos soil; Stellenbosch, South Africa | KM189687 | KM088933 | KM089707 | KM089320 | |
| DTO 184-E1 = CV 80 | Fynbos soil; Stellenbosch, South Africa | KM189688 | KM088934 | KM089708 | KM089321 | |
| DTO 185-A5 = CV 545 | Fynbos soil; Stellenbosch, South Africa | KM189689 | KM088935 | KM089709 | KM089322 | |
| DTO 185-B1 = CV 553 | Fynbos soil; Stellenbosch, South Africa | KM189690 | KM088936 | KM089710 | KM089323 | |
| P. frequentans | DTO 070-E4 = CBS 105.11 | Unrecorded source; ex-type of P. frequentans | KM189525 | KM088762 | KM089534 | KM089147 |
| DTO 070-E2 = CBS 229.28 = FRR 751 = IMI 092231 = MUCL 29111 = NRRL 751 | Soil under conifer; Poland; ex-type of P. paczowskii | KM189524 | KM088761 | KM089533 | KM089146 | |
| DTO 053-F2 = IBT 5635 | Citrus extract; Denmark | KM189485 | KM088722 | KM089494 | KM089107 | |
| DTO 053-F3 = IBT 6178 | Unknown source; Denmark | KM189486 | KM088723 | KM089495 | KM089108 | |
| DTO 053-F4 = IBT 6422 | Indoor air; Denmark | KM189487 | KM088724 | KM089496 | KM089109 | |
| DTO 053-F5 = IBT 6552 = NRRLA-23305 | Barley; Denmark | KM189488 | KM088725 | KM089497 | KM089110 | |
| DTO 053-F6 = IBT 18381 = CCRC 32565 | Melon seed; Hsinchu City, Taiwan | KM189489 | KM088726 | KM089498 | KM089111 | |
| DTO 053-F8 = IBT 23011 | Air of margarin factory; Vejle, Denmark | KM189491 | KM088728 | KM089500 | KM089113 | |
| DTO 053-G1 = IBT 23188 | Saltern; Secovlje salt pans, Slovenia | KM189492 | KM088729 | KM089501 | KM089114 | |
| DTO 053-G2 = IBT 23304 | Artic soil; Svalbard, Norway | KM189493 | KM088730 | KM089502 | KM089115 | |
| DTO 053-G3 = IBT 24700 | Air of factory; Sweden | KM189494 | KM088731 | KM089503 | KM089116 | |
| DTO 053-G4 = IBT 24773 | Saltern; Secovlje salt pans, Slovenia | KM189495 | KM088732 | KM089504 | KM089117 | |
| DTO 053-G5 = IBT 24777 | Saltern; Secovlje salt pans, Slovenia | KM189496 | KM088733 | KM089505 | KM089118 | |
| DTO 053-G6 = IBT 26406 | Ice; Pakitsoq, Greenland | KM189497 | KM088734 | KM089506 | KM089119 | |
| DTO 053-G7 = IBT 26412 | Ice; Pakitsoq, Greenland | KM189498 | KM088735 | KM089507 | KM089120 | |
| DTO 055-B9 | Indoor enviroment; Munchen, Germany | KM189499 | KM088736 | KM089508 | KM089121 | |
| P. frequentans | DTO 174-A2 = CBS 138169 | Leaf of Eucalyptus species; Lavers hill, Tasmania, Australia | KM189620 | KM088864 | KM089638 | KM089251 |
| DTO 249-D1 | Artichoke; Finland | KM189762 | KM089011 | KM089785 | KM089398 | |
| P. fuscum | DTO 111-B7 = CBS 127833 = HDAUPII-06-9026 | Soil; Sichuan Prov., Kangding County; ex-type of Eladia inflata | KM189586 | KM088830 | KM089603 | KM089216 |
| DTO 078-F6 = CBS 203.87 = IBT 16267 | Sandy soil collected on the shore of the Beagle Channel; National Park of ‘Lapataya’ (Tierra del Fuego), Argentina; ex-type of P. lapatayae | KM189531 | KM088768 | KM089540 | KM089153 | |
| DTO 094-D7 = CBS 309.63 = ATCC 18322 | Forest soil; Macedonia; ex-type of P. macedonense | KM189566 | KM088806 | KM089578 | KM089191 | |
| DTO 088-I6 = CBS 295.62 = ATCC 14770 = CCRC 31517 = DSM 2438 = IFO 7743 = IMI 094209 = MUCL 31196 = NRRL 3008 = WSF 15c | Pine-birch forest soil; Vilas County, Wisconsin, USA; ex-type of P. pinetorum and E. pinetorum and ex-neotype of Citromyces fuscus | KM189547 | KM088785 | KM089557 | KM089170 | |
| DTO 097-F1 = CBS 235.60 = ATCC 18483 = QM 8040 | Forest soil; Russia; ex-type of P. silvaticum | KM189568 | KM088811 | KM089583 | KM089196 | |
| DTO 006-I4 = CBS 139.72 | Soil; Alaska, USA | KM189452 | KM088676 | KM089448 | KM089061 | |
| DTO 096-I5 = CBS 311.63 | Forest soil; Netherlands | KM189567 | KM088809 | KM089581 | KM089194 | |
| DTO 181-H5 = CV 531 = KAS 4147 = DAOM 241356 | Fynbos soil; Stellenbosch, South Africa | KM189652 | KM088896 | KM089670 | KM089283 | |
| DTO 202-C9 = CBS 129393 = WSF 15-C | Soil; A1 horizon Soil; Wisconsin conifer-hardwood forests, mixed forest; Wisconsin; USA | KM189724 | KM088973 | KM089747 | KM089360 | |
| DTO 205-H9 = CBS 129541 = RMF 8868 | Soil; A1 horizon Soil; 40 yr old eastern white pine plantation, conifer forest, plantation; Coweeta Long-term Ecological Research (LTER) site; near Otto; North Carolina; USA | KM189730 | KM088979 | KM089753 | KM089366 | |
| DTO 208-D6 = CBS 129806 = RMF 7991,GW 4-4 | Soil; lodgepole pine forest, conifer forest; Yellowstone National Park; Wyoming; USA | KM189737 | KM088986 | KM089760 | KM089373 | |
| DTO 209-A9 = CBS 130039 = RMF 7778 | Soil; A1 horizon Soil; lodgepole pine forest, conifer forest; adjacent to Cinnabar Park; Medicine Bow National Forest; Wyoming; USA | KM189743 | KM088992 | KM089766 | KM089379 | |
| DTO 209-F6 = CBS 130199 = RMF 201 | Soil; A1 horizon Soil; lodgepole pine forest, conifer forest; T16N R81W S28; west slope of Snowy Range; Wyoming; USA | KM189748 | KM088997 | KM089771 | KM089384 | |
| DTO 290-I7 = CBS 138.72 | Soil; Alaska, USA | KM189778 | KM089028 | KM089802 | KM089415 | |
| P. fusisporum | DTO 228-I3 = CBS 137778 | Protea roupelliae var. roupelliae; Buffelskloof, South Africa | KM189757 | KM089006 | KM089780 | KM089393 |
| P. glabrum | DTO 279-F2 = CBS 138433 = NRRL 766 | Unrecorded source; Unknown; ex-neotype of P. aurantiobrunneum | KM189775 | KM089025 | KM089799 | KM089412 |
| DTO 076-G8 = CBS 125543 = IMI 91944 = IBT 22658 = DAOM 227653 | Unrecorded source; ex-neotype of P. glabrum | KM189530 | KM088767 | KM089539 | KM089152 | |
| DTO 265-A9 = CBS 171.81 = IJFM 5072 = IMI 253790 = VKM F-2186 | Culture contaminant of CBS 171.81; Utrecht, the Netherlands; ex-type of P. aragonense | KM189767 | KM089017 | KM089791 | KM089404 | |
| DTO 301-I3 = CBS 260.29 = IMI 092242 = LSHB P79 = MUCL 28650 = MUCL 29119 = NRRL 774 | Unrecorded source; Belgium; ex-type of P. flavidorsum | KM189798 | KM089048 | KM089822 | KM089435 | |
| DTO 301-H8 = CBS 213.28 = FRR 770 = IMI 092265 = IMI 092265ii = LSHB P89 = MUCL 29118 = NRRL 770 | Soil under conifer ; Tatry Mountains, Poland; ex-type of P. oledzkii | KM189795 | KM089045 | KM089819 | KM089432 | |
| DTO 301-I9 = CBS 344.59 = ATCC 18486 = IFO 5359 = IMI 068617 = NRRL 3460 = QM 8152 | Butter; Japan; ex-type of P. spinuloramigenum | KM189803 | KM089053 | KM089827 | KM089440 | |
| DTO 301-I1 = CBS 228.28 = ATHUM 2896 = FRR 752 = IMI 092232 = LSHB P63 = MUCL 29114 = NRRL 752 | Soil under conifer ; Poznan area, Poland; ex-type of P. terlikowskii | KM189797 | KM089047 | KM089821 | KM089434 | |
| DTO 005-G6 | Cork; Portugal | KM189447 | KM088671 | KM089443 | KM089056 | |
| DTO 012-D5 | Wood; Unknown | KM189453 | KM088678 | KM089450 | KM089063 | |
| P. glabrum | DTO 015-I6 | Soil; Los Alerces National Park, Chubut, Argentina | KM189454 | KM088681 | KM089453 | KM089066 |
| DTO 015-I7 | Soil; Los Alerces National Park, Chubut, Argentina | KM189455 | KM088682 | KM089454 | KM089067 | |
| DTO 015-I9 | Soil; Los Alerces National Park, Chubut, Argentina | KM189456 | KM088683 | KM089455 | KM089068 | |
| DTO 016-A5 | Soil; Puerto Piramides, Valdez peninsula, Chubet, Argentina | KM189457 | KM088684 | KM089456 | KM089069 | |
| DTO 036-B5 = CBS 171.81 = IJFM 5072 = IMI 253790 = VKM F-2186 | Air; Madrid, Spain | KM189468 | KM088700 | KM089472 | KM089085 | |
| DTO 039-F6 = CBS 115810 | Indoor environment; Germany | KM189477 | KM088712 | KM089484 | KM089097 | |
| DTO 056-H9 = FRR 6092 | Living leaf of Leptospermum polygofolium; Collaroy, NSW, Australia | KM189500 | KM088737 | KM089509 | KM089122 | |
| DTO 056-I1 = FRR 6093 | Bark of Banksia ericifolia; Lane Cove National Park, NSW, Australia | KM189501 | KM088738 | KM089510 | KM089123 | |
| DTO 056-I2 = FRR 6094 | Living leaf of Acacia suaveolans; Collaroy, NSW, Australia | KM189502 | KM088739 | KM089511 | KM089124 | |
| DTO 057-A4 = FRR 6095 | Litter under Banksia integrifolia; Collaroy, NSW, Australia | KM189513 | KM088750 | KM089522 | KM089135 | |
| DTO 057-A5 = FRR 6096 | Soil; Roadside near Lockhart, NSW, Australia | KM189514 | KM088751 | KM089523 | KM089136 | |
| DTO 057-A7 = FRR 6097 | Tree root; Katandra Nature Reserve, NSW, Australia | KM189515 | KM088752 | KM089524 | KM089137 | |
| DTO 057-B1 = FRR 6098 | Soil, Eucalyptus forest; near Hamilton, Tas, Australia | KM189516 | KM088753 | KM089525 | KM089138 | |
| DTO 057-B3 = FRR 6099 | Living leaf of Leptospermum polygofolium; Collaroy, NSW, Australia | KM189517 | KM088754 | KM089526 | KM089139 | |
| DTO 057-B4 = FRR 6100 | From highly disturbed soil under Banksia integrifolia; School grounds, Terrigal, NSW, Australia | KM189518 | KM088755 | KM089527 | KM089140 | |
| DTO 067-E8 | Cork; Portugal | KM189520 | KM088757 | KM089529 | KM089142 | |
| DTO 067-F2 | Cork; Portugal | KM189521 | KM088758 | KM089530 | KM089143 | |
| DTO 067-F4 | Cork; Portugal | KM189522 | KM088759 | KM089531 | KM089144 | |
| DTO 084-F6 = CBS 127704 | Cork; Portugal | KM189533 | KM088771 | KM089543 | KM089156 | |
| DTO 084-F7 = CBS 127703 | Cork; Portugal | KM189534 | KM088772 | KM089544 | KM089157 | |
| DTO 084-G2 = CBS 126333 | Cork; Portugal | KM189536 | KM088774 | KM089546 | KM089159 | |
| DTO 084-G3 = CBS 126336 | Cork; Portugal | KM189537 | KM088775 | KM089547 | KM089160 | |
| DTO 084-G7 = CBS 127700 | Cork; Portugal | KM189540 | KM088778 | KM089550 | KM089163 | |
| DTO 085-B1 = CBS 138164 | Soil, 2 mtr. from road; Ranoma fana, Madagascar | KM189544 | KM088782 | KM089554 | KM089167 | |
| DTO 087-H6 = CBS 138165 | Swab sample taken in warehouse for fruits; the Netherlands | KM189546 | KM088784 | KM089556 | KM089169 | |
| DTO 099-A6 | Soil in oak forest, taken at 0–10 cm depth; Aîn Hamraia, Tunesia | KM189571 | KM088814 | KM089586 | KM089199 | |
| DTO 119-E6 | Soil in oak forest, taken at 10–20 cm depth; Fej Errih, Tunesia | KM189594 | KM088838 | KM089612 | KM089225 | |
| DTO 121-B6 | Soil in oak forest, taken at 0–10 cm depth; Ras Rajel, Tunesia | KM189603 | KM088847 | KM089621 | KM089234 | |
| DTO 121-D9 | Soil in oak forest, taken at 10–20 cm depth; Ras Rajel, Tunesia | KM189604 | KM088848 | KM089622 | KM089235 | |
| DTO 123-G9 | Inside of chestnut; the Netherlands | KM189605 | KM088849 | KM089623 | KM089236 | |
| DTO 134-B4 = CBS 138166 | Stone inside nectarine; the Netherlands | KM189611 | KM088855 | KM089629 | KM089242 | |
| DTO 153-H2 | Cork; Algeria | KM189612 | KM088856 | KM089630 | KM089243 | |
| DTO 153-H4 | Cork; Algeria | KM189613 | KM088857 | KM089631 | KM089244 | |
| DTO 153-H7 | Cork; Algeria | KM189614 | KM088858 | KM089632 | KM089245 | |
| DTO 154-A1 | Cork; Algeria | KM189615 | KM088859 | KM089633 | KM089246 | |
| DTO 154-F2 | Cork; Algeria | KM189616 | KM088860 | KM089634 | KM089247 | |
| DTO 154-H3 | Cork; Algeria | KM189617 | KM088861 | KM089635 | KM089248 | |
| DTO 155-C8 | Cork; Algeria | KM189618 | KM088862 | KM089636 | KM089249 | |
| DTO 174-A1 | Leaf of Eucryphia cordifolia; Tasmania, Australia | KM189619 | KM088863 | KM089637 | KM089250 | |
| P. glabrum | DTO 174-A3 | Leaf of Eucalyptus sp.; Lavers hill, Tasmania, Australia | KM189621 | KM088865 | KM089639 | KM089252 |
| DTO 174-A7 | Leaf of Eucalyptus ovata; Snake Gully, Kangaroo Island, Australia | KM189622 | KM088866 | KM089640 | KM089253 | |
| DTO 174-D7 | Leaf of Eucalyptus viminalis; Kangaroo Island, Australia | KM189624 | KM088868 | KM089642 | KM089255 | |
| DTO 174-D9 | Leaf of Eucalyptus sp.; Kangaroo Island, Australia | KM189626 | KM088870 | KM089644 | KM089257 | |
| DTO 178-I9 = KAS 3838 | House dust; Stellenbosch, South Africa | KM189631 | KM088875 | KM089649 | KM089262 | |
| DTO 180-F8 = CV 4 = KAS 4125 = DAOM 241361 | Air sample; Stellenbosch, South Africa | KM189639 | KM088883 | KM089657 | KM089270 | |
| DTO 181-C4 = CV 188 = KAS 4054 = DAOM 241132 | Protea repens infructescence bract; Stellenbosch, South Africa | KM189647 | KM088891 | KM089665 | KM089278 | |
| DTO 182-H6 = CV 1181 = KAS 3980 = DAOM 241365 | Mite inside Protea repens infructescence; Riverlands, South Africa | KM189677 | KM088922 | KM089696 | KM089309 | |
| DTO 183-B7 = CV 1494 = KAS 4015 = DAOM 241364 | Protea repens infructescence bract; Riverlands, South Africa | KM189683 | KM088929 | KM089703 | KM089316 | |
| DTO 189-H9 | Soil ; Spanderswoud, the Netherlands | KM189694 | KM088941 | KM089715 | KM089328 | |
| DTO 197-F9 | Air sample; Bakery, Tilburg, The Netherlands | KM189718 | KM088966 | KM089740 | KM089353 | |
| DTO 203-I4 | Soil; Aspear Island, Iran | KM189726 | KM088975 | KM089749 | KM089362 | |
| DTO 206-B4 = CBS 129602 = RMF 9521 | Soil; Iraq | KM189732 | KM088981 | KM089755 | KM089368 | |
| DTO 206-B6 = CBS 129606 = RMF 9242 | Soil; A1 horizon Soil; maple woods, deciduous forest; Cedar Creek Long-term Ecological Research (LTER) site; near East Bethel; Minnesota; USA | KM189733 | KM088982 | KM089756 | KM089369 | |
| DTO 208-B4 = CBS 129784 = RMF 8573 = RMF 8026 | Soil; A1 horizon Soil; tallgrass prairie, grassland; Konza Prairie Research Natural Area; Long-term Ecological Research site (LTER); near Manhattan; Kansas; USA | KM189736 | KM088985 | KM089759 | KM089372 | |
| DTO 259-C6 | Pile of moss; Eindhoven, the Netherlands | KM189764 | KM089013 | KM089787 | KM089400 | |
| DTO 262-G8 | Soft drink; the Netherlands | KM189765 | KM089014 | KM089788 | KM089401 | |
| DTO 266-A8 | Grapevine; Maragheh, Iran | KM189770 | KM089020 | KM089794 | KM089407 | |
| DTO 269-E1 | House dust; South Africa | KM189771 | KM089021 | KM089795 | KM089408 | |
| DTO 296-H5 = CBS 131040 = RMF WT 97 | Soil; Near Zurich; Switzerland | KM189785 | KM089035 | KM089809 | KM089422 | |
| DTO 297-D1 = CBS 328.48 = ATCC 10444 = IMI 040234 = LSHB Ad6 = NRRL 1915 = QM 1924 | Unrecorded source | KM189790 | KM089040 | KM089814 | KM089427 | |
| P. grancanariae | DTO 076-F3 = CBS 687.77 = IJFM 3745 = IMI 253783 | Air; Canary Islands, Gran Canaria, Spain; ex-type of P. grancanariae | KM189529 | KM088766 | KM089538 | KM089151 |
| P. grevilleicola | DTO 174-E6 = CBS 137775 | Leaf of Grevillea ilicifolia; Kingscote, Kangaroo Island; Australia; ex-type of P. grevilleicola | KM189630 | KM088874 | KM089648 | KM089261 |
| DTO 174-E4 | Leaf of Grevillea ilicifolia; Kingscote, Kangaroo Island; Australia | KM189629 | KM088873 | KM089647 | KM089260 | |
| P. hoeksii | DTO 192-H4 = CBS 137776 | Soil under Compact Rush (Juncus conglomeratus); De Ronde Put, Postel, Belgium; ex-type of P. hoeksii | KM189707 | KM088954 | KM089728 | KM089341 |
| DTO 068-D9 = CBS 137952 | Air in factory; Goes, the Netherlands | KM189523 | KM088760 | KM089532 | KM089145 | |
| P. infra-aurantiacum | DTO 183-C3 = CBS 137747 = CV 1518 = KAS 4022 = DAOM 241145 | Bracts of Protea repens infructescens; Riverlands, Malmesbury, Western Cape, South Africa; ex-type of P. infra-aurantiacum | KM189684 | KM088930 | KM089704 | KM089317 |
| DTO 181-F1 = CBS 137746 = CV 362 = KAS 4118 = DAOM 241146 | Protea repens infructescens; Stellenbosch mountain, Western Cape, South Africa | KM189650 | KM088894 | KM089668 | KM089281 | |
| P. jejuense | DTO 174-D3 = CBS 137774 | Leaf of Eucalyptus sp.; Lavers hill, Tasmania, Australia | KM189623 | KM088867 | KM089641 | KM089254 |
| DTO 182-H7 = CBS 137755 = CV 1189 = KAS 3981 = DAOM 241142 | Mite inside Protea repens infructescens; Riverlands, Malmesbury, Western Cape, South Africa | KM189678 | KM088923 | KM089697 | KM089310 | |
| P. kananaskense | DTO 296-G7 = CBS 530.93 = ATCC 90282 = DAOM 216105 = IBT 11775 = IMI 356791 | Soil, FH horizon, in a Pinus contorta var. latifolia forest; Kananaskis Valley, Alberta, Canada; ex-type of P. kananaskense | KM189780 | KM089030 | KM089804 | KM089417 |
| DTO 193-A3 | Moses under Myrica gale (Bog Myrtle); De Ronde Put, Postel, Belgium | KM189710 | KM088957 | KM089731 | KM089344 | |
| P. kiamaense | DTO 056-I6 = CBS 137947 = FRR 6087 | Soil; Barren Grounds National Park, NSW, Australia; ex-type of P. kiamaense | KM189506 | KM088743 | KM089515 | KM089128 |
| P. lividum | DTO 105-H6 = CBS 347.48 = ATCC 14941 = FRR 3407 = IFO 7740 = IHEM 4375 = IMI 099468 = MUCL 31326 = NRRL 3407 = DAOM 226266 | Unrecorded source; Scotland; ex-neotype of P. lividum | KM189582 | KM088825 | KM089598 | KM089211 |
| DTO 006-H8 = CBS 287.65 | Litter of Quercus sp.; Lancashire, Grange-over-Sands, Merlewood Res. Station, UK | KM189450 | KM088674 | KM089446 | KM089059 | |
| DTO 198-I2 = CBS 128415 = WSF 3528 | Soil; amorphus peat; above water table; cedar-fir forests, wetland, conifer forest; Wisconsin; USA | KM189720 | KM088968 | KM089742 | KM089355 | |
| DTO 297-D8 = CBS 132166 = WSF 3895 | Soil; Wisconsin; USA | KM189794 | KM089044 | KM089818 | KM089431 | |
| P. longicatenatum | DTO 180-D9 = CBS 137735 = CV 2847 = KAS 3943 = DAOM 241119 | Soil from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa; ex-type of P. longicatenatum | KM189636 | KM088880 | KM089654 | KM089267 |
| DTO 099-C6 | Soil in oak forest, taken at 10–20 cm depth; Aîn Hamraia, Tunesia | KM189573 | KM088816 | KM089588 | KM089201 | |
| DTO 120-H9 | Soil in oak forest, taken at 0–10 cm depth; Ras Rajel, Tunesia | KM189601 | KM088845 | KM089619 | KM089232 | |
| DTO 174-D8 | Leaf of Dodonea sp.; Australia | KM189625 | KM088869 | KM089643 | KM089256 | |
| DTO 174-E2 | Fruiting body on leaf of unknown sp; Kangaroo Island, Australia | KM189627 | KM088871 | KM089645 | KM089258 | |
| DTO 174-E3 | Fruiting body on leaf of unknown sp; Kangaroo Island, Australia | KM189628 | KM088872 | KM089646 | KM089259 | |
| DTO 180-C8 = CBS 137742 = CV 2829 = KAS 3957 = DAOM 241120 | Soil from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa | KM189633 | KM088877 | KM089651 | KM089264 | |
| DTO 180-D2 = CBS 137734 = CV 2840 = KAS 3935 = DAOM 241118 | Soil from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa | KM189634 | KM088878 | KM089652 | KM089265 | |
| DTO 180-D6CV 2843 = KAS 3940 | Fynbos soil; Riverlands, South Africa | KM189635 | KM088879 | KM089653 | KM089266 | |
| DTO 181-C7 = CBS 137737 = CV 209 = KAS 4069 = DAOM 241122 | Mite inside Protea repens infructescens; Stellenbosch mountain, Western Cape, South Africa | KM189648 | KM088892 | KM089666 | KM089279 | |
| DTO 181-C8 = CV 214 = KAS 4070 | Protea repens infructescence bract; Stellenbosch, South Africa | KM189649 | KM088893 | KM089667 | KM089280 | |
| DTO 182-B8 = CBS 137738 = CV 885 = KAS 4188 = DAOM 241149 | Soil from Fynbos; Stellenbosch mountain, Western Cape, South Africa | KM189658 | KM088902 | KM089676 | KM089289 | |
| DTO 182-G2 = CV 997 = KAS 4231 | Mite inside Protea repens infructescence; Riverlands, South Africa | KM189672 | KM088917 | KM089691 | KM089304 | |
| DTO 182-G6 = CBS 137739 = CV 1036 = KAS 3964 = DAOM 241148 | Soil from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa | KM189673 | KM088918 | KM089692 | KM089305 | |
| DTO 182-I9 = CBS 137740 = CV 1300 = KAS 3995 = DAOM 241123 | Protea repens infructescens; Riverlands, Malmesbury, Western Cape, South Africa | KM189679 | KM088925 | KM089699 | KM089312 | |
| DTO 183-A1 = CV 1301 = KAS 3996 | Protea repens infructescence bract; Riverlands, South Africa | KM189680 | KM088926 | KM089700 | KM089313 | |
| DTO 183-A3 = CBS 137741 = CV 1335 = KAS 3999 = DAOM 241147 | Soil from Fynbos; Riverlands, Malmesbury, Western Cape, South Africa | KM189681 | KM088927 | KM089701 | KM089314 | |
| DTO 183-C6 = CV 1585 = KAS 4025 | Protea repens infructescence bract; Riverlands, South Africa | KM189685 | KM088931 | KM089705 | KM089318 | |
| DTO 216-B6 | Foliar tissue of Populus angustifolia; Ogden, UT, USA | KM189751 | KM089000 | KM089774 | KM089387 | |
| P. malmesburiense | DTO 182-H5 = CBS 137744 = CV 1180 = KAS 3979 = DAOM 241144 | Mite inside Protea repens infructescens; Riverlands, Malmesbury, Western Cape, South Africa; ex-type of P. malmesburiense | KM189676 | KM088921 | KM089695 | KM089308 |
| P. malmesburiense | DTO 183-A6 = CBS 137745 = CV 1422 = KAS 4003 = DAOM 241143 | Protea repens infructescens; Riverlands, Malmesbury, Western Cape, South Africa | KM189682 | KM088928 | KM089702 | KM089315 |
| P. montanense | DTO 090-I6 = CBS 310.63 = ATCC 14941 = FRR 3407 = IFO 7740 = IHEM 4375 = IMI 099468 = MUCL 31326 = NRRL 3407 = DAOM 226282 | Coniferous forest soil; Ravalli Co., Montana, USA; ex-type of P. montanense | KM189551 | KM088789 | KM089561 | KM089174 |
| DTO 041-D7 | Soil; Poland | KM189482 | KM088718 | KM089490 | KM089103 | |
| DTO 196-B6 = CBS 126808 = WSF 2021 | Soil; amorphus peat; above water table; spruce-larch forests, wetland, conifer forest; Wisconsin; USA | KM189713 | KM088961 | KM089735 | KM089348 | |
| DTO 196-D2 = CBS 126824 = WSF 3733 = WSF 2021 | Soil; amorphus peat; above water table; cedar-fir forests, wetland, conifer forest; Wisconsin; USA | KM189714 | KM088962 | KM089736 | KM089349 | |
| DTO 196-D4 = CBS 126826 = WSF 3952 | Soil; amorphus peat; above water table; open bogs, wetland, shrubland; Wisconsin; USA | KM189715 | KM088963 | KM089737 | KM089350 | |
| DTO 196-E1 = CBS 126832 = WSF 3124 | Soil; amorphus peat; above water table; spruce-larch forests, wetland, conifer forest; Wisconsin; USA | KM189716 | KM088964 | KM089738 | KM089351 | |
| DTO 196-E2 = CBS 126833 | Culture contaminant of WSF 2127 | KM189717 | KM088965 | KM089739 | KM089352 | |
| DTO 198-I4 = CBS 128418 = WSF 3450 | Soil; amorphus peat; above water table; cedar-fir forests, wetland, conifer forest; Wisconsin; USA | KM189721 | KM088969 | KM089743 | KM089356 | |
| DTO 198-I6 = CBS 128426 = WSF 3315 | Soil; amorphus peat; above water table; spruce-larch forests, wetland, conifer forest; Wisconsin; USA | KM189722 | KM088970 | KM089744 | KM089357 | |
| DTO 208-F3 = CBS 129881 = RMF 8750 | Soil; A1 horizon Soil; coniferous forest ecosystem, conifer forest; Andrews Long-term Ecological Research (LTER) site; Willamette National Forest; near Blue River; Oregon; USA | KM189740 | KM088989 | KM089763 | KM089376 | |
| DTO 208-I7 = CBS 130027 = RMF 7785 | Soil; A1 horizon Soil; lodgepole pine forest, conifer forest; adjacent to Cinnabar Park; Medicine Bow National Forest; Wyoming; USA | KM189742 | KM088991 | KM089765 | KM089378 | |
| DTO 209-F4 = CBS 130197 = RMF 199 | Soil; A1 horizon Soil; lodgepole pine forest, conifer forest; T16N R81W S28; west slope of Snowy Range; Wyoming; USA | KM189747 | KM088996 | KM089770 | KM089383 | |
| DTO 209-F9 = CBS 130202 = RMF 204 | Soil; A1 horizon Soil; lodgepole pine forest, conifer forest; T16N R81W S28; west slope of Snowy Range; Wyoming; USA | KM189749 | KM088998 | KM089772 | KM089385 | |
| DTO 263-I9 | Pseudotsuga menziesii var. glauca; White Pass, Cascade Mts. Washington, USA | KM189766 | KM089015 | KM089789 | KM089402 | |
| P. odoratum | DTO 290-I9 = CBS 432.65 = FAT 1138 = IFO 6039 | Soil ; Japan; ex-syntype of P. trzebinskianum | KM189779 | KM089029 | KM089803 | KM089416 |
| DTO 205-B7 = CBS 294.62 = CBS 129423 = WSF 2000 = DAOM 226269 = ATCC 14769 = DSM 2419 = IFO 7741 = IMI 094208ii = NRRL 3007 = DAOM 226269 | Soil; amorphus peat; above water table; spruce-larch forests, wetland, conifer forest; Wisconsin; USA; ex-type of P. odoratum | KM189727 | KM088976 | KM089750 | KM089363 | |
| DTO 296-H8 = CBS 431.65 = FAT 728 = IAM 7193 = IFO 6038 | Soil; Japan; ex-type of P. trzebinskianum | KM189788 | KM089038 | KM089812 | KM089425 | |
| DTO 198-H8 = CBS 128282 = WSF 3201 | Soil; amorphus peat; above water table; spruce-larch forests, wetland, conifer forest; Wisconsin; USA | KM189719 | KM088967 | KM089741 | KM089354 | |
| DTO 201-B2 = CBS 128274 = WSF 3200 | Soil; amorphus peat; above water table; spruce-larch forests, wetland, conifer forest; Wisconsin; USA | KM189723 | KM088972 | KM089746 | KM089359 | |
| DTO 205-C5 = CBS 129440 = WSF 2002 | Soil; amorphus peat; above water table; spruce-larch forests, wetland, conifer forest; Wisconsin; USA | KM189728 | KM088977 | KM089751 | KM089364 | |
| P. odoratum | DTO 206-B7 = CBS 129607 = RMF 9241 | Soil; A1 horizon soil; maple woods, deciduous forest; Cedar Creek Long-term Ecological Research (LTER) site; near East Bethel; Minnesota; USA | KM189734 | KM088983 | KM089757 | KM089370 |
| DTO 208-E5 = CBS 129874 = RMF 8759 | Soil; A1 horizon soil; coniferous forest ecosystem, conifer forest; Andrews Long-term Ecological Research (LTER) site; Willamette National Forest; near Blue River; Oregon; USA | KM189739 | KM088988 | KM089762 | KM089375 | |
| DTO 296-H7 = CBS 129135 = WSF 3894 | Soil; Wisconsin; USA | KM189787 | KM089037 | KM089811 | KM089424 | |
| DTO 301-H9 = CBS 217.30 = NRRL 2062 | Unrecorded source | KM189796 | KM089046 | KM089820 | KM089433 | |
| P. palmense | DTO 076-E2 = CBS 336.79 = ATCC 38669 = IJFM 3840 = VKM F-2181 | Air; Canary Islands, Gran Canaria, Spain; ex-type of P. palmense | KM189528 | KM088765 | KM089537 | KM089150 |
| P. pulvis | DTO 180-B7 = CBS 138432 = KAS 3924 | House dust; South Africa; ex-type of P. pulvis | KM189632 | KM088876 | KM089650 | KM089263 |
| DTO 180-F9 = CV 7 = KAS 4166 = DAOM 241133 | Air sample; Stellenbosch, South Africa | KM189640 | KM088884 | KM089658 | KM089271 | |
| DTO 180-G2 = CV 15 = KAS 4017 = DAOM 241135 | Air sample; Stellenbosch, South Africa | KM189641 | KM088885 | KM089659 | KM089272 | |
| P. purpurescens | DTO 091-D2 = CBS 366.48 = ATCC 10485 = IMI 039745 = NRRL 720 = QM 1959 | Soil; Canada; ex-neotype of P. purpurescens | KM189561 | KM088801 | KM089573 | KM089186 |
| DTO 091-D3 = CBS 126.64 | Soil; Erzurum, Turkey | KM189562 | KM088802 | KM089574 | KM089187 | |
| P. quercetorum | DTO 091-A5 = CBS 417.69 = ATCC 48727 = CCRC 31668 = FRR 516 = IFO 31749 = IMI 140342 = MUCL 31203 = VKM F-1074 | Soil; Es-Euveida, Syria; ex-isotype of P. quercetorum | KM189556 | KM088795 | KM089567 | KM089180 |
| DTO 208-D9 = CBS 129869 = RMF 8789 | Soil; A1 horizon soil; coniferous forest ecosystem, conifer forest; Andrews Long-term Ecological Research (LTER) site; Willamette National Forest; near Blue River; Oregon; USA | KM189738 | KM088987 | KM089761 | KM089374 | |
| P. ranomafanaense | DTO 085-A5 = CBS 137953 | Soil, 2 mtr. from road; Ranoma fana, Madagascar; ex-type of P. ranomafanaense | KM189541 | KM088779 | KM089551 | KM089164 |
| DTO 085-A8 = CBS 137954 | Soil, 2 mtr. from road; Ranoma fana, Madagascar | KM189543 | KM088781 | KM089553 | KM089166 | |
| P. roseomaculatum | DTO 290-I6 = CBS 137254 = IMI 92236 | Unrecorded source; ex-type of P. baiiolum | KM189777 | KM089027 | KM089801 | KM089414 |
| DTO 225-E3 = CBS 137962 = NRRL 728 = FRR 0728 = IMI 189696 = MUCL 29101 | Unrecorded source; ex-type of P. roseomaculatum | KM189755 | KM089004 | KM089778 | KM089391 | |
| DTO 084-F8 = CBS 125096 | Cork; Portugal; ex-type of P. subericola | KM189535 | KM088773 | KM089545 | KM089158 | |
| DTO 035-A1 = CBS 137944 | Soil; New Zealand | KM189465 | KM088697 | KM089469 | KM089082 | |
| DTO 035-A3 | Soil; New Zealand | KM189466 | KM088698 | KM089470 | KM089083 | |
| DTO 053-F7 = CBS 125097 = IBT 23009 | Air; Vejle, Denmark | KM189490 | KM088727 | KM089499 | KM089112 | |
| DTO 057-A2 = CBS 125100 = FRR 4914 | Dried grapes (sultanas) (Vitis vinifera); Mildura, Vic, Australia | KM189511 | KM088748 | KM089520 | KM089133 | |
| DTO 098-E2 = CBS 127706 = KAS 1289 | Lumber; BC, Vancouver, Canada | KM189570 | KM088813 | KM089585 | KM089198 | |
| DTO 100-A7 = CBS 125099 = IBT 20217 | Acified lake; Butte, Montana, USA | KM189579 | KM088822 | KM089595 | KM089208 | |
| DTO 100-A8 = CBS 125098 = IBT 20218 | Acified lake; Butte, Montana, USA | KM189580 | KM088823 | KM089596 | KM089209 | |
| P. roseoviride | DTO 090-I2 = CBS 267.35 = ATCC 10412 = IFO 6089 = IMI 039740ii = NRRL 760 = QM 7485 | Soil in a beech forest; Germany; ex-type of P. roseoviride | KM189549 | KM088787 | KM089559 | KM089172 |
| P. rudallense | DTO 209-C1 = CBS 130049 = RMF 7766 | Soil (beehives); USA; ex-type of P. rudallense | KM189744 | KM088993 | KM089767 | KM089380 |
| DTO 030-G5 | Soil; Barron falls, Queensland, Australia | KM189463 | KM088692 | KM089464 | KM089077 | |
| DTO 056-I4 = CBS 138162 = FRR 6085 | Soil; Rudall River National Park, WA, Australia | KM189504 | KM088741 | KM089513 | KM089126 | |
| DTO 057-B5 = FRR 6091 | Soil; Rudall River National Park, WA, Australia | KM189519 | KM088756 | KM089528 | KM089141 | |
| DTO 180-G4 = CBS 138558 = CV 26 = KAS 4104 = DAOM 241136 | Air sample; Stellenbosch, South Africa | KM189638 | KM088882 | KM089656 | KM089269 | |
| P. saturniforme | DTO 105-I8 = CBS 122276 = AS 3.6886 = HMAS 130355-1-4 | Soil; Little Peony Forest Reserve, Dunhua, Jiling Province, China ; ex-type of E. saturniforme | KM189585 | KM088828 | KM089601 | KM089214 |
| Penicillium sp. | DTO 181-I3 = CBS 137729 = CV 550 = KAS 4156 = DAOM 241129 | Soil from Fynbos; Stellenbosch mountain, Western Cape, South Africa | KM189654 | KM088898 | KM089672 | KM089285 |
| P. spinulosum | DTO 006-H1 = CBS 374.48 = ATCC 10498 = FRR 1750 = IMI 024316 = MUCL 13910 = MUCL 13911 = NCTC 591 = NRRL 1750 = QM 7654 = DAOM 226267 | Culture contaminant; Hannover, Germany; ex-neotype of P. spinulosum | KM189448 | KM088672 | KM089444 | KM089057 |
| DTO 296-G8 = CBS 348.59 = ATCC 22346 = FAT 24 = FRR 3406 = IFO 6239 = IMI 068222 = MUCL 13555 = NRRL 3406 = DAOM 226268 | Soil; Ukaku, Japan; ex-type of P. abeanum and P. trzebinskii var. magnum | KM189781 | KM089031 | KM089805 | KM089418 | |
| DTO 279-F1 = CBS 137964 = NRRL 2051 | Unrecorded source; ex-type of P. flavocinereum | KM189774 | KM089024 | KM089798 | KM089411 | |
| DTO 301-I5 = CBS 269.35 | Forest litter; Germany; ex-type of P. mucosum | KM189800 | KM089050 | KM089824 | KM089437 | |
| DTO 301-I6 = CBS 271.35 | Forest leaf litter; Germany; ex-type of P. tannophilum | KM189801 | KM089051 | KM089825 | KM089438 | |
| DTO 290-I3 = CBS 137257 = IMI 190575 | Unrecorded source; Probably ex-type of P. brunneoviride (Pitt 1980: 180) | KM189776 | KM089026 | KM089800 | KM089413 | |
| DTO 279-E9 = CBS 137963 = NRRL 727 | Unrecorded source; Representative of P. pfefferianum | KM189773 | KM089023 | KM089797 | KM089410 | |
| DTO 056-I8 = CBS 137948 | Bark of Banksia ericifolia; Lane Cove National Park, NSW, Australia | KM189508 | KM088745 | KM089517 | KM089130 | |
| DTO 084-G5 = CBS 127698 | Cork; Portugal | KM189538 | KM088776 | KM089548 | KM089161 | |
| DTO 084-G6 = CBS 127699 | Cork; Portugal | KM189539 | KM088777 | KM089549 | KM089162 | |
| P. sterculiniicola | DTO 031-A4 = CBS 122426 | Spawn run compost; USA; ex-type of P. sterculiniicola | KM189464 | KM088693 | KM089465 | KM089078 |
| DTO 004-B8 = CBS 117778 | Potting soil; the Netherlands | KM189446 | KM088670 | KM089442 | KM089055 | |
| DTO 035-A4 | Soil; New Zealand | KM189467 | KM088699 | KM089471 | KM089084 | |
| DTO 216-I4 = CBS 137960 | Root tissue of Pinus ponderosa; University of Idaho greenhouse, USA | KM189753 | KM089002 | KM089776 | KM089389 | |
| DTO 216-I8 = CBS 137961 | Root tissue of Pinus ponderosa; University of Idaho greenhouse, USA | KM189754 | KM089003 | KM089777 | KM089390 | |
| P. sublectaticum | DTO 076-C5 = CBS 138163 | Unknown marine source; New Zealand; ex-type of P. sublectaticum | KM189527 | KM088764 | KM089536 | KM089149 |
| DTO 244-G2 = CBS 138217 | House dust; New Zealand | KM189761 | KM089010 | KM089784 | KM089397 | |
| P. subspinulosum | DTO 018-C8 | Log of Pinus sp.; Spanderswoud, Bussum, the Netherlands | KM189459 | KM088686 | KM089458 | KM089071 |
| DTO 038-G1 | Forest soil; Rijnsweerd, Utrecht, the Netherlands | KM189476 | KM088711 | KM089483 | KM089096 | |
| DTO 040-E6 | Soil; Poland | KM189478 | KM088713 | KM089485 | KM089098 | |
| DTO 041-F2 = CBS 137946 | Soil; Poland | KM189483 | KM088719 | KM089491 | KM089104 | |
| DTO 042-F7 | Soil; Poland | KM189484 | KM088720 | KM089492 | KM089105 | |
| DTO 056-I9 = CBS 137949 = FRR 4882 | Roots of Wollemi Pine (Wollemia nobilis); Wollemi National Park, NSW, Australia | KM189509 | KM088746 | KM089518 | KM089131 | |
| DTO 057-A1 = CBS 137950 = FRR 4872 | Roots of Wollemi Pine (Wollemia nobilis); Wollemi National Park, NSW, Australia | KM189510 | KM088747 | KM089519 | KM089132 | |
| DTO 057-A3 = CBS 137951 = FRR 6090 | Soil; Barren Grounds National Park, NSW, Australia | KM189512 | KM088749 | KM089521 | KM089134 | |
| DTO 092-G4 = CBS 137955 | Soil under Betula sp.; Cartier Heide, Eersel, the Netherlands | KM189563 | KM088803 | KM089575 | KM089188 | |
| DTO 189-H2 | Soil; Spanderswoud, the Netherlands | KM189693 | KM088940 | KM089714 | KM089327 | |
| DTO 189-I3 | Soil; Spanderswoud, the Netherlands | KM189695 | KM088942 | KM089716 | KM089329 | |
| DTO 190-A1 | Soil; Spanderswoud, the Netherlands | KM189696 | KM088943 | KM089717 | KM089330 | |
| DTO 190-C8 | Soil; Spanderswoud, the Netherlands | KM189698 | KM088945 | KM089719 | KM089332 | |
| DTO 190-D2 = CBS 137957 | Soil; Spanderswoud, the Netherlands | KM189699 | KM088946 | KM089720 | KM089333 | |
| DTO 190-D4 | Soil; Spanderswoud, the Netherlands | KM189700 | KM088947 | KM089721 | KM089334 | |
| P. subspinulosum | DTO 190-D5 = CBS 137958 | Soil; Spanderswoud, the Netherlands | KM189701 | KM088948 | KM089722 | KM089335 |
| DTO 190-D9 | Soil; Spanderswoud, the Netherlands | KM189702 | KM088949 | KM089723 | KM089336 | |
| DTO 206-C4 = CBS 129613 = RMF 9368 = RMF 8940 | Soil; A1 horizon Soil; abandoned agricultural fields; aged 1–10 years, agriculture; Cedar Creek Long-term Ecological Research (LTER) site; near East Bethel; Minnesota; USA | KM189735 | KM088984 | KM089758 | KM089371 | |
| DTO 208-H6 = CBS 129906 = RMF 8736 | Soil; A1 horizon Soil; coniferous forest ecosystem, conifer forest; Andrews Long-term Ecological Research (LTER) site; Willamette National Forest; near Blue River; Oregon; USA | KM189741 | KM088990 | KM089764 | KM089377 | |
| DTO 296-H2 = CBS 345.51 = ATCC 11080 = IMI 046814 = NRRL 2298 = QM 6901 = UPSC 3182 | Soil; England, UK | KM189783 | KM089033 | KM089807 | KM089420 | |
| DTO 297-D5 = CBS 290.53 | Unrecorded source | KM189792 | KM089042 | KM089816 | KM089429 | |
| DTO 297-D6 = CBS 128281 = WSF 3202 | Soil; Wisconsin; USA | KM189793 | KM089043 | KM089817 | KM089430 | |
| P. thiersii | DTO 037-I9 = CBS 117503 = IBT 27050 = NRRL 28162 | Old black stroma of Hypoxylon encrusting the surface of a dead maple ; New Glarus Woods State Park, Wisconsin, USA; ex-type of P. thiersii | KM189474 | KM088709 | KM089481 | KM089094 |
| P. thomii | DTO 091-A9 = CBS 225.81 = IMI 189694 = NRRL 2077 | Pine cone; Spaulding, Georgia, USA; ex-neotype of P. thomii | KM189560 | KM088799 | KM089571 | KM089184 |
| DTO 036-C3 = CBS 257.87 = FRR 2676 | Dried fish, Decapterus sp.; Indonesia; ex-type of P. corynephorum | KM189469 | KM088701 | KM089473 | KM089086 | |
| DTO 105-I6 = CBS 260.87 = FRR 2662 | Dried fish; Rastrelliger kanagurta, Indonesia; ex-type of P. patens | KM189584 | KM088827 | KM089600 | KM089213 | |
| DTO 202-E5 = CBS 129408 = WSF 2003 | Soil; amorphus peat; above water table; spruce-larch forests, wetland, conifer forest; Wisconsin; USA | KM189725 | KM088974 | KM089748 | KM089361 | |
| DTO 205-H3 = CBS 129534 = RMF 8828 | Soil; A1 horizon Soil; native deciduous forest, deciduous forest; Coweeta Long-term Ecological Research (LTER) site; near Otto; North Carolina; USA | KM189729 | KM088978 | KM089752 | KM089365 | |
| P. trzebinskii | DTO 301-I4 = CBS 268.35 | Forest litter; Germany; ex-type of P. mediocre | KM189799 | KM089049 | KM089823 | KM089436 |
| DTO 301-I7 = CBS 289.36 | Tannin solution; Germany; ex-type of P. tannophagum | KM189802 | KM089052 | KM089826 | KM089439 | |
| DTO 006-H2 = CBS 351.51 | Rice; Japan; ex-type of P. toxicarium | KM189449 | KM088673 | KM089445 | KM089058 | |
| DTO 296-H3 = CBS 382.48 = ATCC 10507 = FRR 731 = IFO 6110 = IMI 039749 = MUCL 29102 = NRRL 731 = QM 7678 | Forest soil; Dluga Goslina, Poznan area, Poland; ex-type of P. trzebinskii | KM189784 | KM089034 | KM089808 | KM089421 | |
| DTO 036-E1 | Soil; Poland | KM189470 | KM088703 | KM089475 | KM089088 | |
| DTO 040-F3 | Soil; Poland | KM189479 | KM088714 | KM089486 | KM089099 | |
| DTO 040-H8 = CBS 137945 | Soil; Poland | KM189480 | KM088716 | KM089488 | KM089101 | |
| DTO 040-I8 | Soil; Poland | KM189481 | KM088717 | KM089489 | KM089102 | |
| DTO 190-G2 | Soil; Spanderswoud, the Netherlands | KM189703 | KM088950 | KM089724 | KM089337 | |
| DTO 209-D5 = CBS 130062 = RMF 7822 | Soil; rhizosphere of onion; onion field, agriculture; University of Idaho Experiment Station; Parma; Idaho; USA | KM189745 | KM088994 | KM089768 | KM089381 | |
| DTO 297-D3 = CBS 128424 = WSF 3448 | Soil; Wisconsin; USA | KM189791 | KM089041 | KM089815 | KM089428 | |
| P. tsitsikammaense | DTO 006-I3 = CBS 328.71 = CSIR 1092 | Forest soil; Tsitsikama Forest near Knysna, Cape Province, South-Africa | KM189451 | KM088675 | KM089447 | KM089060 |
| P. turcosoconidiatum | DTO 181-A3 = CBS 138557 = CV 110 = KAS 3970 = DAOM 241130 | Fynbos soil; Stellenbosch, South Africa; ex-type of P. turcosoconidiatum | KM189645 | KM088889 | KM089663 | KM089276 |
| DTO 181-A4 = CBS 137733 = CV 111 = KAS 3971 = DAOM 241131 | Soil from Fynbos; Stellenbosch mountain, Western Cape, South Africa | KM189646 | KM088890 | KM089664 | KM089277 | |
| P. vagum | DTO 180-G3 = CBS 137728 = CV 25 = KAS 4100 = DAOM 241357 | Air sample from Fynbos; Stellenbosch mountain, Western Cape, South Africa; ex-type of P. vagum | KM189642 | KM088886 | KM089660 | KM089273 |
| DTO 038-E7 | Forest soil; Rijnsweerd, Utrecht, the Netherlands | KM189475 | KM088710 | KM089482 | KM089095 | |
| DTO 056-I3 = FRR 4783 | Fresh currants (Vitis vinifera); Mildura, Vic, Australia | KM189503 | KM088740 | KM089512 | KM089125 | |
| P. vagum | DTO 056-I5 = FRR 6086 | Soil; Katandra Nature Reserve, NSW, Australia | KM189505 | KM088742 | KM089514 | KM089127 |
| DTO 056-I7 = FRR 6088 | Soil; Roadside north of Urana, NSW, Australia | KM189507 | KM088744 | KM089516 | KM089129 | |
| DTO 099-A7 | Soil in oak forest, taken at 0–10 cm depth; Aîn Hamraia, Tunesia | KM189572 | KM088815 | KM089587 | KM089200 | |
| DTO 099-D6 | Soil in oak forest, taken at 0–10 cm depth; Aîn Hamraia, Tunesia | KM189574 | KM088817 | KM089589 | KM089202 | |
| DTO 099-F7 | Soil in oak forest, taken at 0–10 cm depth; Aîn Hamraia, Tunesia | KM189576 | KM088819 | KM089591 | KM089204 | |
| DTO 099-G7 | Soil in oak forest, taken at 10–20 cm depth; Aîn Hamraia, Tunesia | KM189577 | KM088820 | KM089592 | KM089205 | |
| DTO 099-I6 | Soil in oak forest, taken at 0–20 cm depth; Aîn Hamraia, Tunesia | KM189578 | KM088821 | KM089594 | KM089207 | |
| DTO 119-A8 | Soil in oak forest, taken at 10–20 cm depth; Fej Errih, Tunesia | KM189588 | KM088832 | KM089606 | KM089219 | |
| DTO 119-C2 | Soil in oak forest, taken at 0–10 cm depth; Fej Errih, Tunesia | KM189589 | KM088833 | KM089607 | KM089220 | |
| DTO 119-C8 | Soil in oak forest, taken at 0–10 cm depth; Fej Errih, Tunesia | KM189590 | KM088834 | KM089608 | KM089221 | |
| DTO 119-D6 | Soil in oak forest, taken at 10–20 cm depth; Fej Errih, Tunesia | KM189591 | KM088835 | KM089609 | KM089222 | |
| DTO 119-D7 | Soil in oak forest, taken at 10–20 cm depth; Fej Errih, Tunesia | KM189592 | KM088836 | KM089610 | KM089223 | |
| DTO 119-E2 | Soil in oak forest, taken at 10–20 cm depth; Fej Errih, Tunesia | KM189593 | KM088837 | KM089611 | KM089224 | |
| DTO 119-G4 | Soil in oak forest, taken at 0–10 cm depth; Fej Errih, Tunesia | KM189595 | KM088839 | KM089613 | KM089226 | |
| DTO 119-H7 | Soil in oak forest, taken at 0–20 cm depth; Ras Rajel, Tunesia | KM189596 | KM088840 | KM089614 | KM089227 | |
| DTO 120-B1 | Soil in oak forest, taken at 0–20 cm depth; Ras Rajel, Tunesia | KM189597 | KM088841 | KM089615 | KM089228 | |
| DTO 120-B4 | Soil in oak forest, taken at 0–20 cm depth; Ras Rajel, Tunesia | KM189598 | KM088842 | KM089616 | KM089229 | |
| DTO 120-C1 | Soil in oak forest, taken at 0–20 cm depth; Ras Rajel, Tunesia | KM189599 | KM088843 | KM089617 | KM089230 | |
| DTO 120-C7 | Soil in oak forest, taken at 0–20 cm depth; Ras Rajel, Tunesia | KM189600 | KM088844 | KM089618 | KM089231 | |
| P. valentinum | DTO 090-I3 = CBS 172.81 = IJFM 5071 | Air; Madrid, Spain; ex-type of P. valentinum | KM189550 | KM088788 | KM089560 | KM089173 |
| DTO 091-A4 = CBS 381.48 = ATCC 10506 = DSM 2214 = IMI 040027 = NRRL 1640 = QM 8002 | Air; Natick, Massachusetts, USA | KM189555 | KM088794 | KM089566 | KM089179 | |
| DTO 205-I6 = CBS 129547 = RMF 9020 | Soil; A1 horizon Soil; oak savanna, savanna; Cedar Creek Long-term Ecological Research (LTER) site; near East Bethel; Minnesota; USA | KM189731 | KM088980 | KM089754 | KM089367 | |
| DTO 296-H6 = CBS 131033 = RMF 2158 | Soil; Grand Teton National Park; T44N R113W S5&6; Wyoming; USA | KM189786 | KM089036 | KM089810 | KM089423 | |
| P. verhagenii | DTO 193-A1 = CBS 137959 | Moses under Myrica gale (Bog Myrtle); De Ronde Put, Postel, Belgium; ex-type of P. verhagenii | KM189708 | KM088955 | KM089729 | KM089342 |
| DTO 023-E1 = CBS 146.83 | Isolated from CBS 145.83 on a synthetic medium; Spain | KM189460 | KM088688 | KM089460 | KM089073 | |
| DTO 028-G1 | Indoor air of house; Eindhoven, the Netherlands | KM189461 | KM088689 | KM089461 | KM089074 | |
| DTO 192-G4 | Soil under Compact Rush (Juncus conglomeratus); De Ronde Put, Postel, Belgium | KM189704 | KM088951 | KM089725 | KM089338 | |
| DTO 192-G7 | Soil under Compact Rush (Juncus conglomeratus); De Ronde Put, Postel, Belgium | KM189705 | KM088952 | KM089726 | KM089339 | |
| DTO 193-A2 | Moses under Myrica gale (Bog Myrtle); De Ronde Put, Postel, Belgium | KM189709 | KM088956 | KM089730 | KM089343 | |
| DTO 193-A5 | Moses under Myrica gale (Bog Myrtle); De Ronde Put, Postel, Belgium | KM189711 | KM088958 | KM089732 | KM089345 | |
| P. yezoense | DTO 091-A2 = CBS 350.59 = ATCC 18333 = FRR 3395 = IFO 5362 = IMI 068615 | Butter; Japan; ex-type of P. yezoense | KM189553 | KM088792 | KM089564 | KM089177 |
| DTO 001-G9 = CBS 117276 | Soil; Alpujarras, Spain | KM189445 | KM088669 | KM089441 | KM089054 | |
| DTO 091-A7 = CBS 140.72 | Soil; Alaska, USA | KM189558 | KM088797 | KM089569 | KM089182 | |
| DTO 091-A8 = CBS 347.78 | Soil under Picea glauca, containing 400 ppm Ni and Cu; 29.3 km SE of Sudbury, Ontario, Canada | KM189559 | KM088798 | KM089570 | KM089183 | |
| DTO 099-E2 | Soil in oak forest, taken at 10–20 cm depth; Aîn Hamraia, Tunesia | KM189575 | KM088818 | KM089590 | KM089203 | |
| DTO 118-E5 | Soil in oak forest, taken at 0–20 cm depth; Fej Errih, Tunesia | KM189587 | KM088831 | KM089604 | KM089217 | |
| DTO 121-A4 | Soil in oak forest, taken at 10–20 cm depth; Ras Rajel, Tunesia | KM189602 | KM088846 | KM089620 | KM089233 | |
| DTO 190-B7 | Soil; Spanderswoud, the Netherlands | KM189697 | KM088944 | KM089718 | KM089331 | |
| DTO 192-G8 | Soil under Compact Rush (Juncus conglomeratus); De Ronde Put, Postel, Belgium | KM189706 | KM088953 | KM089727 | KM089340 | |
| DTO 193-G2 | Moses under Myrica gale (Bog Myrtle); De Ronde Put, Postel, Belgium | KM189712 | KM088959 | KM089733 | KM089346 | |
| DTO 209-F1 = CBS 130194 = RMF 157 | Soil; A1 horizon Soil; narrowleaf cottonwood, deciduous forest; Fort Steele Road; 2 miles south of Interstate 80; 9 miles east of Sinclair; Wyoming; USA | KM189746 | KM088995 | KM089769 | KM089382 | |
| DTO 216-B7 | Foliar tissue of Populus trichocarpa; Nisqually River, WA, USA | KM189752 | KM089001 | KM089775 | KM089388 | |
| DTO 270-H9 | Air in nickelsulfate production facility; Belgium | KM189772 | KM089022 | KM089796 | KM089409 | |
DNA extraction, PCR and sequencing
Strains were grown for 3–14 d on MEA prior to DNA extraction. DNA was extracted using the Ultraclean™ Microbial DNA isolation Kit (MoBio, Solana Beach, U.S.A.) and the extracted DNA was stored at −20 °C. The nuclear ribosomal internal transcribed spacer regions (ITS1-5.8S-ITS2) and parts of the BenA, CaM and RPB2 genes were amplified and sequenced using methods previously described (Houbraken & Samson 2011, Houbraken et al. 2012a, b, Frisvad et al. 2013).
Phylogenetic analysis
Section Aspergilloides was delimitated using a data set combining BenA, CaM and RPB2. The ITS sequences had a low phylogenetic signal and this data was only examined for its applicability in species recognition in the context of DNA barcoding. The phylogeny of individual clades within section Aspergilloides was studied both by comparing single gene phylogenies, to determine whether groups of strains could be recognised as independent evolutionary lineages, and by concatenated analyses of the three genes to resolve relationships among the species. Maximum likelihood (ML) and Maximum Parsimony (MP) analyses were performed using MEGA5 and were applied to most individual BenA, CaM and RPB2 data sets. Exceptions were the P. spinulosum- and P. glabrum-clade data sets, where ML and Bayesian analysis was applied. The robustness of tree topology for each analysis was evaluated by 1 000 bootstrap replicates. All combined data sets were analysed using the RAxML (randomised accelerated maximum likelihood) (Stamatakis et al. 2008) and Bayesian tree inference (BI) analyses using MrBayes v3.1.2 (Ronquist & Huelsenbeck 2003). Prior to analyses, the most suitable substitution model was determined using MrModeltest v. 3.1.2 (Nylander 2004), utilising the Akaike information criterion (AIC). Bayesian analyses were performed with two sets of four chains (one cold and three heated) and the STOPRULE option, stopping analyses at an average standard deviation of split frequencies of 0.01. The sample frequency was set to 100 and the first 25 % of trees were removed as burn-in. The different loci within the combined data sets were analysed as separate partitions. Penicillium expansum ATCC 24692, a member of subgenus Penicillium section Penicillium, was used as outgroup for all analyses. The phylograms were redrawn and annotated using Adobe Illustrator CS5. BI posterior probabilities (pp) values and bootstrap (bs) percentages of the maximum likelihood (ML) analysis are presented at the nodes. Values less than 0.95 pp and less than 70 % bs are not shown. Branches with more than 95 % bs and 1.00 pp are thickened. Newly obtained sequences were deposited in GenBank under accession numbers KM088669–KM089827.
ITS barcoding
The ITS sequence diversity of strains belonging to section Aspergilloides was assessed by determining the number of haplotypes among the ITS sequences. The software programme DnaSP v. 5.10 (Librado & Rozas 2009) was used to find the different haplotypes in the alignment. Gaps and missing data were included in this calculation. ITS sequences were deposited in GenBank under accession numbers KM189445–KM189803.
Phenotypic examination
Macroscopic characters were studied on the agar media Czapek yeast extract agar (CYA), malt extract agar (MEA; Oxoid), creatine sucrose agar (CREA), dichloran 18 % glycerol agar (DG18), yeast extract sucrose agar (YES), oatmeal agar (OA) and CYA supplemented with 5 % NaCl (Samson et al. 2010). Growth of the isolates was also examined on CYA at 15, 30 and 37 °C (CYA15°C, CYA30°C and CYA37°C, respectively). Strains were inoculated at three points onto media in 90 mm Petri dishes and incubated for 7 d in darkness. After incubation, colony diameters on each agar medium were measured. In addition, degree of sporulation, obverse and reverse colony colours, colony shape and texture, and the production of soluble pigments were determined. Acid production on CREA was indicated by a change in the pH sensitive bromocresole purple dye from purple to yellow around growing colonies. Colonies were photographed with a Canon EOS 400D. Species were characterised microscopically by preparing slides from MEA. Lactic acid was used as mounting fluid and a drop of alcohol was added to remove air bubbles and excess conidia. Specimens were examined using a Zeiss AxioSkop2 plus microscope. Strains were also examined for production of alkaloids reacting with Ehrlich reagent using a filter paper method (Lund 1995). The appearance of a violet ring within 10 min was regarded as a positive reaction; all other colours were considered as a negative reaction.
Results and discussion
Overview of section Aspergilloides
The phylogenetic relationships among species belonging to section Aspergilloides were studied using concatenated sequence data of three loci, BenA, CaM and RPB2. In total, 112 mostly ex-type strains were included in the analysis and the total length of the aligned data set was 2 049 characters. The length and the best substitution model for each partition are summarised in Table 2. Members of section Aspergilloides formed a well-supported lineage in the phylogram (100 % ML, 1.00 pp) and section Sclerotiora species form a sister clade to Aspergilloides, although with low statistical support (77 % ML, <0.95 pp). These results largely correspond with those of Houbraken & Samson (2011); however, there are two main differences. In Houbraken & Samson (2011), Penicillium thiersii CBS 117503 occupied a well-supported basal position in section Aspergilloides based on a combined analysis of four genes (Cct8, Tsr1, RPB1 and RPB2). In our phylogeny, this species is basal to section Aspergilloides without statistical support. This species could represent a separate section close to sections Aspergilloides and Sclerotiora, but based on Houbraken & Samson (2011), we opt to provisionally maintain its classification in section Aspergilloides. The other difference is the placement of P. georgiense in section Aspergilloides (Houbraken & Samson 2011). Our data show that this species does not belong to this section and Fig. 1 indicates a relationship with P. ramusculum (CBS 251.56T) in section Ramigena.
Table 2.
Overview details of sequence data sets used in this study.
| Clade | Description data set | No. isolates | Data sets |
|||||
|---|---|---|---|---|---|---|---|---|
| BenA | Substitution model | CaM | Substitution model | RPB2 | Substitution model | |||
| Overview Aspergilloides | 112 | 541 | GTR+G+I | 620 | GTR+G+I | 888 | GTR+G+I | |
| 1 | P. spinulosum-clade | 73 | 438 | K2+G | 520 | GTR+G | 888 | GTR+G+I |
| 2 | P. thomii-clade | 43 | 439 | HKY+G | 527 | GTR+G | 764 | GTR+G |
| 3 | P. glabrum-clade | 104 | 438 | K2P+G | 501 | GTR+G | 887 | GTR+G |
| 4 | P. vagum-clade | 44 | 469 | K2+G | 524 | K2+I | 866 | GTR+G |
| 5 | P. fuscum-clade | 62 | 471 | K2+G | 527 | GTR+G | 755 | GTR+G |
| 6 | P. sublectaticum-clade | 7 | 460 | HKY+I | 524 | GTR+G | 888 | GTR+G |
| 7 | P. verhagenii-clade | 11 | 481 | K2+G | 513 | GTR+G+I | 928 | GTR+G |
| 9 | P. lividum-clade | 20 | 487 | K2+G | 509 | K2+I | 937 | K2+G |
| 10 | P. hoeksii-clade | 10–11 | 473 | GTR+I | 539 | GTR+G | 930 | GTR+G+I |
Fig. 1.
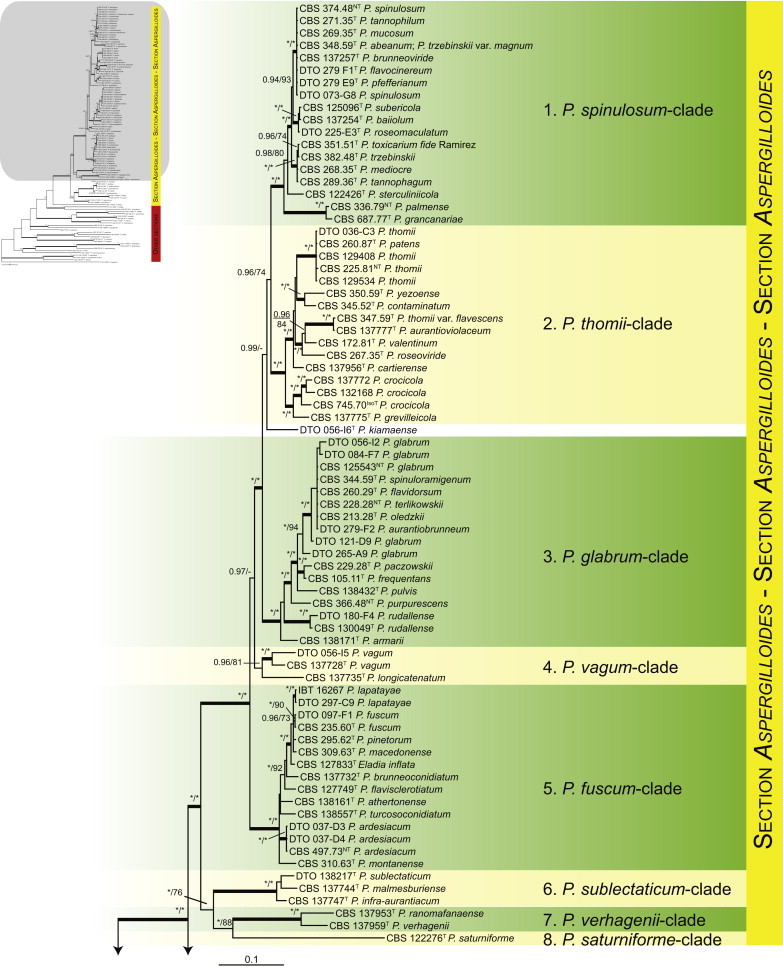
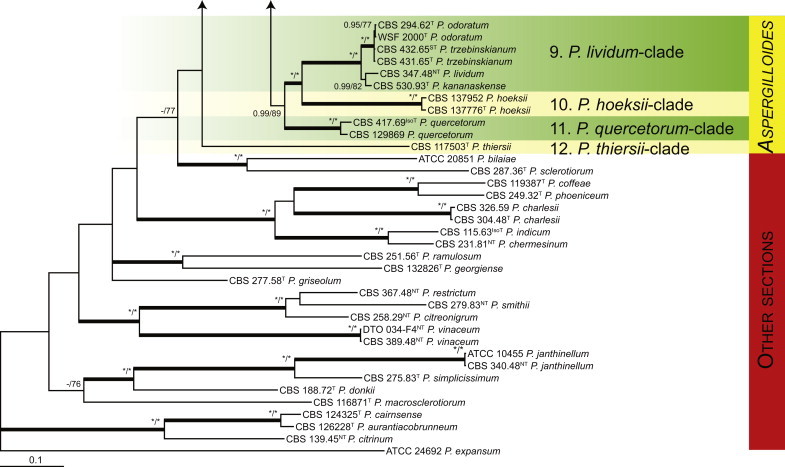
Phylogenetic tree inferred from the concatenated nucleotide matrix (partial BenA, CaM and RPB2 sequences) using Bayesian inference showing the relationship of species accommodated in section Aspergilloides. With exception of P. thiersii, all species of section Aspergilloides for a well-supported lineage. The bar indicates the number of substitutions per site. The phylogram is rooted with Penicillium expansum (ATCC 24692).
Fig. 1 reveals the presence of 12 well-supported lineages in section Aspergilloides. All lineages were fully supported (100 % ML, 1.00 pp) with exception of the P. thiersii (<70 % ML; <0.95 pp) and P. vagum-clades (81 % ML, 0.96 pp). All accepted species, except P. kiamaense, could be assigned to one of the delineated lineages. Bayesian analysis showed that this species is basal to the P. spinulosum- and P. thomii-clade (0.99 pp); however, this was not supported in the ML analysis. Most species of section Aspergilloides share phenotypic characters such as vesiculate, monoverticillate conidiophores, a moderate to fast growth rate on CYA and/or MEA, and a negative Ehrlich reaction. Furthermore, many species of section Aspergilloides produce crusts of conidia on MEA that either shift or fall off in mass, similar to the characteristic colonies of P. crustosum (sect. Penicillium). This feature is most pronounced on DG18. Each clade is treated in detail below. Clades containing multiple species are analysed separately using BenA, CaM and RPB2 sequences, and this data is often supported by phenotypic characters.
Clade 1: Penicillium spinulosum-clade
Species belonging to the P. spinulosum-clade are phenotypically similar to those of the P. glabrum-clade. Both clades contain species that grow rapidly on CYA, YES and MEA. Furthermore, they predominantly produce monoverticillate conidiophores with an inflated apex, and have globose to subglobose conidia that are finely to distinctly roughened, or spirally banded. No consistent phenotypic characters were found to distinguish the two clades. Generally, species that belong to the P. glabrum-clade produce velvety colonies and have darker green conidia on MEA and often an orange-brown reverse on YES, while species of the P. spinulosum-clade are more floccose, produce conidia in shades of pure or dull green and the reverse on YES lacks orange shades. Furthermore, the species of the P. glabrum-clade produce acid on CREA, a feature often absent in species of the P. spinulosum-clade, which grow poorly on CREA.
Eighteen species were placed in synonymy with P. spinulosum by Pitt (1980). Phylogenetically, eleven of these taxa belong to the P. spinulosum-clade: P. abeanum, P. baiiolum, P. brunneoviride, P. flavocinereum, P. mediocre, P. mucosum, P. roseomaculatum, P. trzebinskii, P. trzebinskii var. magnum, P. tannophagum and P. tannophilum. Four of the remaining species belong to other clades of section Aspergilloides: P. paczowskii, P. terlikowskii and P. spinuloramigenum belong to the P. glabrum-clade and P. ardesiacum (CBS 497.73T) is a member of the P. fuscum-clade. Two species are phylogenetically unrelated to section Aspergilloides: P. viridorsum (CBS 269.29T) is close to P. cyclopium and P. citreovirens (CBS 320.59T) is close to P. corylophilum. The type culture of P. janthocitrinum (CBS 268.29T) is dead in the CBS collection and we did not include this strain in our study. We follow Pitt (1980) and treat this species as a synonym of P. spinulosum. Subsequent to Pitt's monograph, P. subericola was described as a new species closely related to P. spinulosum (Barreto et al. 2011).
The type strains of the species that belong to the P. spinulosum-clade, together with freshly isolated strains from various substrates and localities, were subjected to a phylogenetic study. Combined analysis of three genes (BenA, CaM and RPB2) revealed the presence of three well-supported lineages in the P. spinulosum-clade (Fig. 3). One lineage is centred on the type of P. spinulosum (97 % ML, 1.00 pp). Basal to this lineage is a set of strains that is described below as P. sterculiniicola (100 % BS, 1.00 pp). The third lineage comprises P. palmense and P. grancanariae and has a basal position relative to the P. spinulosum and P. sterculiniicola lineages (100 % ML, 1.00 pp).
Fig. 3.
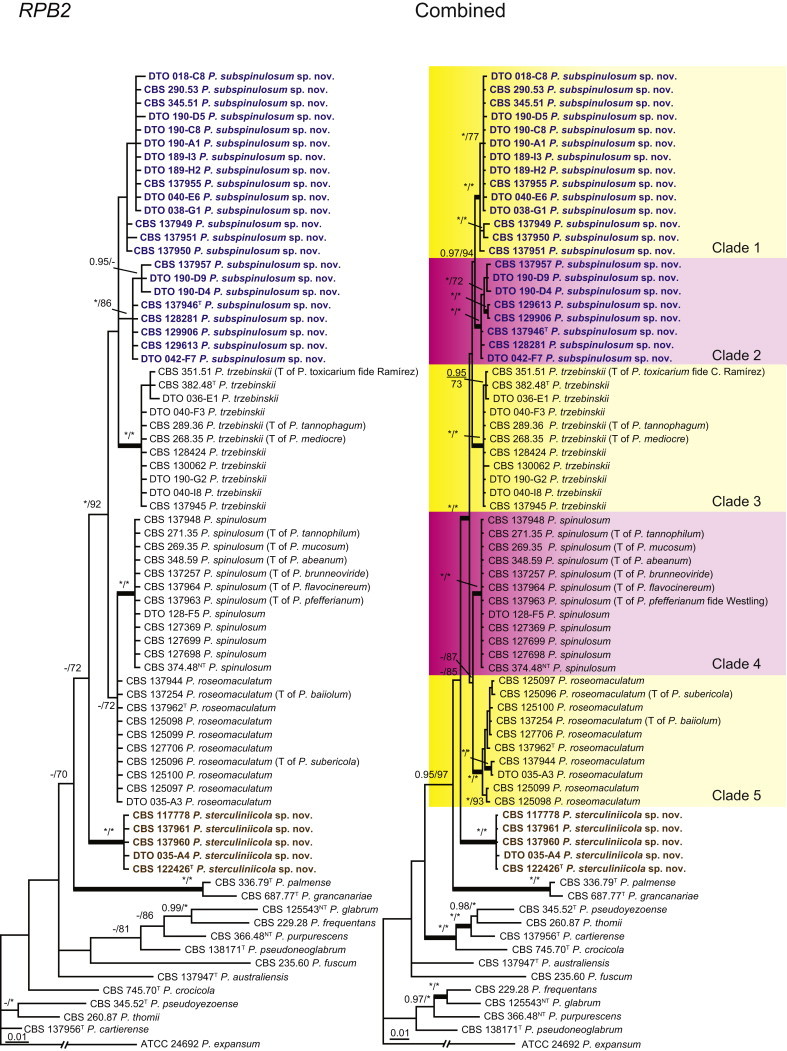
Phylogenetic trees showing the relationship among strains belonging to the P. spinulosum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
The clade containing many other strains previously identified as P. spinulosum, including the ex-neotype of P. spinulosum (CBS 374.48NT) is subdivided into five well-supported lineages (>95 % ML, 1.00 pp) in the combined analysis, which are labelled clades 1–5. The topologies of the single gene phylograms are congruent with the combined phylogram. However, they are generally poorly resolved and only three of the five lineages have support (>70 % and >0.95 pp) (Figs 2, 3). No ex-type strains of any described species occur in clade 1 and 2. The single and combined sequence analysis (Figs 2, 3) shows that the two lineages are related, but distinct. No diagnostic phenotypic differences were observed among strains in these clades. Based on this data, we decided to describe these strains as a single species, P. subspinulosum. The type strains of P. roseomaculatum (CBS 137962), P. baiiolum (CBS 137254) and P. subericola (CBS 125096) belong to clade 5. Barreto et al. (2011) described P. subericola but the types of P. roseomaculatum and P. baiiolum were not included in that study. The latter two species were described by Biourge (1923) and predate P. subericola. As both were described in the same publication, neither has priority. We chose P. roseomaculatum, because the type strain of this species is in better condition than that of P. baiiolum and better resembles the other freshly isolated strains of this species. Clade 4 contains P. spinulosum and the ex-types of P. tannophilum (CBS 271.35T), P. brunneoviride (CBS 137257; probably type, Pitt (1980: 180)), P. mucosum (CBS 269.35T), P. flavocinereum (CBS 137964 = NRRL 2051), P. abeanum and P. trzebinskii var. magnum (CBS 348.59). In addition, NRRL 727 (= CBS 137963), a strain identified as P. pfefferianum by Westling belongs to this clade, confirming the conclusions of Raper & Thom (1949: 184) and Pitt (1980: 177). Pitt (1980) discussed in detail the differences between Citromyces pfefferianus Wehmer and P. pfefferianum (Wehmer) Westling and we follow his conclusions by maintaining C. pfefferianus a synonym of P. glabrum. Penicillium tannophagum (CBS 289.36T), P. mediocre (CBS 268.35T), P. trzebinskii (CBS 382.48T) and P. toxicarium fide Ramírez (CBS 351.51T) belong to the same lineage (clade 3). Penicillium toxicarium Miyake was described without a Latin diagnosis but was validated by Ramírez (1982) based on CBS 351.51. However, Miyake's description of P. toxicarium does not correspond with that of Ramírez, making this validation problematic. Penicillium trzebinskii was described by Zaleski (1927) and predates Penicillium tannophagum and P. mediocre, which were both described in 1935.
Fig. 2.
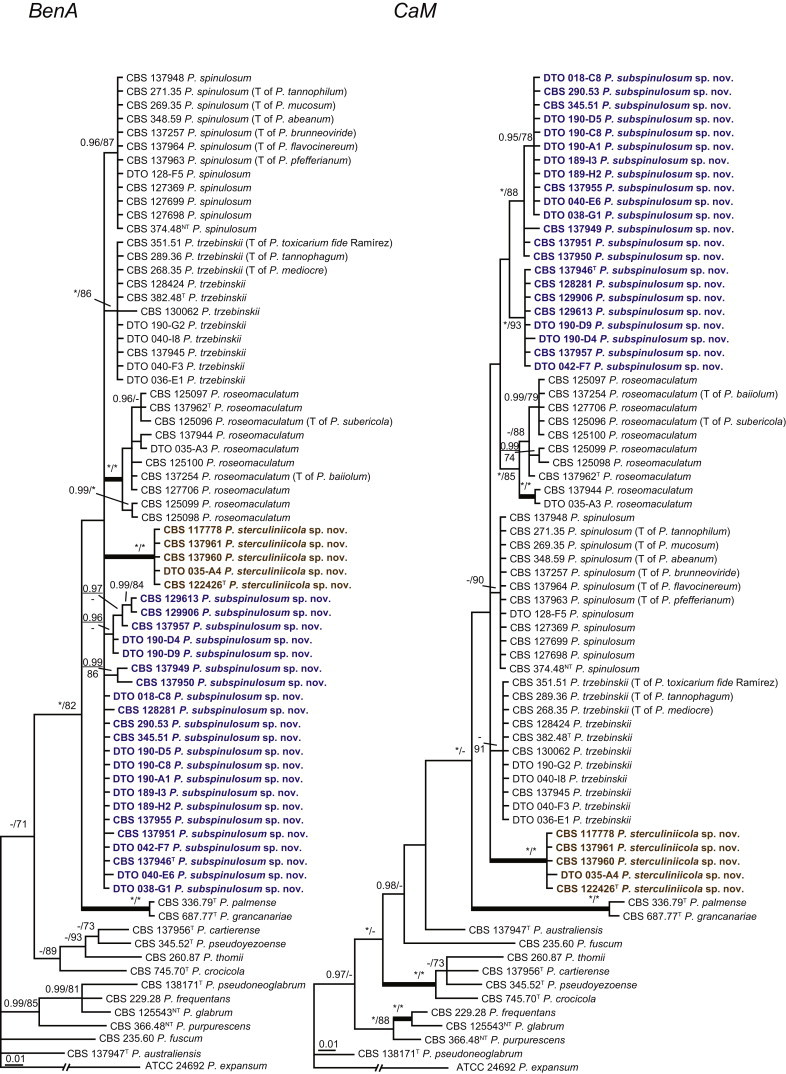
Phylogenetic trees showing the relationship among strains belonging to the P. spinulosum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Thus, seven species are accepted in the P. spinulosum-clade: P. sterculiniicola, P. grancanariae, P. palmense, P. roseomaculatum, P. spinulosum, P. subspinulosum and P. trzebinskii. A summary of their phenotypic characters is given in Table 3. Penicillium spinulosum, P. roseomaculatum, P. trzebinskii and P. subspinulosum are phylogenetically and phenotypically closely related. These species can be differentiated by growth characters on CYA, CREA and MEA. On CYA and MEA, P. spinulosum generally grows faster than the other species; however, there is an overlap in diameters (Table 3). Penicillium spinulosum and P. subspinulosum have similar cream or (pale) brown reverse colours on CYA. In comparison, reverse colours of P. trzebinskii are yellow or light orange-brown in the centre. The reverse of the investigated P. roseomaculatum strains vary and can be cream coloured as in P. spinulosum and P. subspinulosum, but can also be in shades of light brown or yellow. The colony texture on CYA and MEA differentiates P. subspinulosum from the other species. Colonies of P. subspinulosum have a velvety texture on CYA and are velvety with a floccose centre on MEA. The other species have floccose colonies on CYA and MEA. Growth on CREA distinguishes P. subspinulosum and P. trzebinskii from P. spinulosum. The latter grows well on CREA and produces acidic compounds followed by base (delayed reaction, 11 d), while P. subspinulosum and P. trzebinskii grew poorly on CREA and did not produce acidic compounds. Strains belonging to P. roseomaculatum had variable growth characters on CREA. Growth was either poor (n = 2) or good (n = 6). The strains with poor growth on CREA (DTO 035-A1, DTO 057-A2) also did not produce acidic compounds; the others were poor or moderate acid producers. The colony diameter on CYA incubated at 30 °C was informative because P. spinulosum and P. trzebinskii generally have larger colonies than P. subspinulosum. As with its growth on CREA, there was also a large variation among strains of P. roseomaculatum on CYA. Growth at 30 °C was slow (8–12 mm) in some strains, while others grew fast (22–29 mm).
Table 3.
Overview of diagnostic characters of species belonging to the P. spinulosum-clade.
| Species name | Colony diam on CYA (mm) | Sporulation on CYA | Colony diam on MEA (mm) | Colony texture on MEA | Growth on CREA | Acid / base production on CREA | CYA30°C | Conidial ornamentation |
|---|---|---|---|---|---|---|---|---|
| P. grancanariae | 34–38 | Poor | 42–46 | Floccose | Poor | Poor | 16–21 | Finely rough |
| P. palmense | 31–34 | Good | 35–41 | Velvety | Poor | Absent | 24–26 | Finely rough |
| P. spinulosum | (34–)39–52 | Variable: poor to good | (41–)46–52 | Floccose | Good | Poor to moderate | 20–30 | Rough |
| P. sterculiniicola | (27–)40–45 | Absent | (39–)43–53 | Variable: velvety to floccose | Good | Moderate | (29–)39–44 | Distinctly rough |
| P. trzebinskii | (25–)37–41 | Variable: absent to moderate | (26–)30–39(–42) | Floccose | Variable: poor or good | Variable: absent to moderate | Two groups: 8–12 or 22–29 | Finely rough |
| P. subspinulosum | (24–)36–41 | Variable: absent to moderate | (28–)35–42 | Floccose at centre, velvet at edge | Poor | Absent | 4–18(–22) | Finely to distinctly rough |
| P. tannophagum | (25–)42–51 | Variable: poor to moderate | 35–45 | Floccose | Poor | Absent | (10–)17–26(–34) | Finely rough |
Penicillium grancanariae and P. palmense were both isolated from air in Gran Canaria, Spain. These species produce ellipsoidal and finely roughened conidia, while the other P. spinulosum-clade species have globose or subglobose conidia which are (distinctly) spirally banded. Penicillium grancanariae was placed in synonymy with P. thomii, and P. palmense was accepted in the list of accepted Penicillium species (Frisvad et al. 1990, Pitt et al. 2000). Based on CaM and BenA sequences, these species were considered conspecific in the study of Barreto et al. (2011). We treat these species as distinct based on our combined BenA, CaM and RPB2 sequence analysis in combination with phenotypic characteristics (Table 3). Penicillium sterculiniicola is phylogenetically and phenotypically distinct. It differs from other P. spinulosum-clade species by its high growth rate on CYA when incubated at 30 °C ((29–)37–47 mm) and 33 °C (15–24 mm) (Table 3, Fig. 4).
Fig. 4.
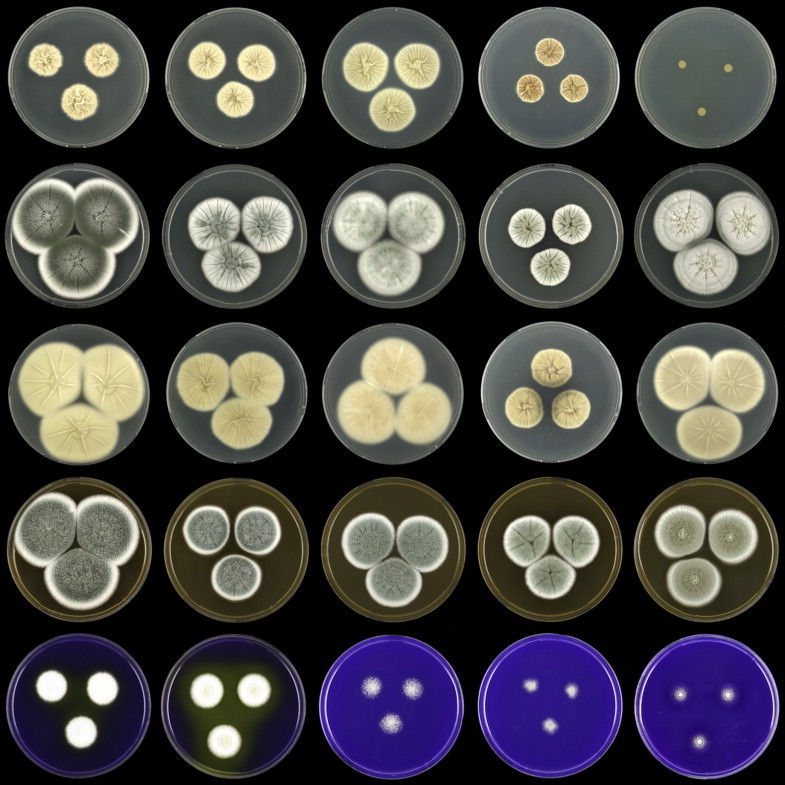
Overview of growth characters of P. spinulosum and related species. Columns, left to right: P. spinulosum, P. roseomaculatum DTO 084-F8, P. roseomaculatum DTO 057-A2, P. trzebinskii, P. subspinulosum. Rows, top to bottom: CYA30°C reverse, CYA obverse, CYA reverse, MEA obverse, CREA obverse.
Clade 2: Penicillium thomii-clade
Species that belong to the P. thomii-clade are phylogenetically related to species of the P. spinulosum-clade (Fig. 1; 74 % ML, 0.96 pp), and basal to these two lineages is the P. glabrum-clade. Species belonging to the P. thomii-clade share characters with species of the P. glabrum- and P. spinulosum-clades such as a fast growth on CYA, MEA and YES, growth on CYA incubated at 30 °C ((5–)15–35(–45) mm) and conidiophores that are predominantly monoverticillate with a vesiculate apex. There are also phenotypic differences among these clades. Species of the P. thomii-clade differ from most other species of section Aspergilloides by the production of hard, gritty sclerotia, which are often in shades of pink on OA (orange-pink, brownish pink). The conidiophores are 200–400 μm long, with roughened walls and conidia are (broadly) ellipsoidal or fusiform. Our description of the P. thomii-clade corresponds with Pitt's description of the morphospecies P. thomii (Pitt 1980).
Based on the presented phylogenies, 12 species are accepted in the P. thomii-clade (Figs 5, 6). Seven species were previously described (P. aurantioviolaceum, P. crocicola, P. fusisporum, P. roseoviride, P. thomii, P. valentinum, P. yezoense), one is in the process of being described elsewhere (P. jejuense, M.S. Park et al. submitted) and four are described here as new (P. austroafricanum, P. cartierense, P. contaminatum, P. grevilleicola). Pitt (1980) synonymised six species and one variety with P. thomii. Penicillium aurantioviolaceum, P. crocicola, P. roseoviride, P. valentinum and P. yezoense were treated as synonyms of P. thomii by Pitt (1980) and are accepted as distinct species in this study. Penicillium yezoense was described without a Latin description and is validated in the taxonomy section below. Penicillium parallelosporum and P. thomii var. flavescens were also regarded as synonyms of P. thomii (Pitt 1980). The type culture of P. parallelosporum (CBS 159.69) is dead in the CBS collection and could not be included in our study. However, this name does not compete with the other names because it was invalidly described, lacking a Latin diagnosis. Penicillium thomii var. flavescens was also described without a Latin diagnosis and the type strain of this species (CBS 347.59T) is placed in synonymy with P. aurantioviolaceum. The accepted species P. valentinum was described in 1980, after the publication of Pitt's monograph. The species was synonymised with P. thomii (Frisvad et al. 1990), but has distinct BenA, CaM and RPB2 sequences. Our phylogenies show that P. patens (CBS 260.87T) is a synonym of P. thomii. It was placed near P. donkii based on its non-vesiculate conidiophores and soft, pale brown sclerotia (Pitt & Hocking 1985), but Frisvad et al. (1990) already noted a close relation of this species with P. thomii.
Fig. 5.
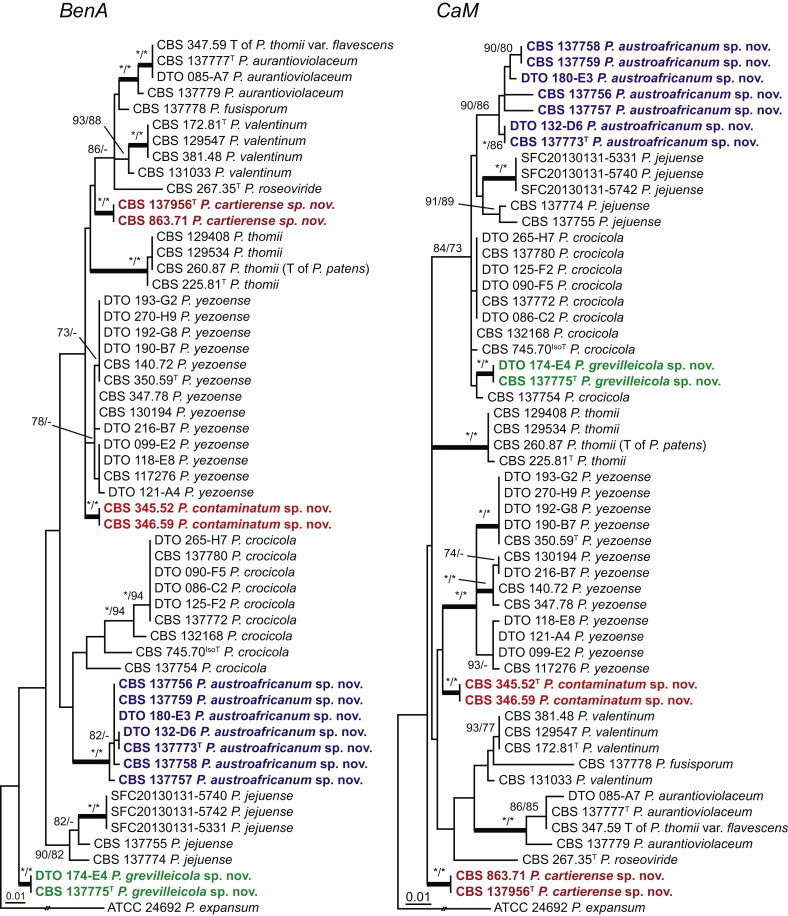
Phylogenetic trees showing the relationship among strains belonging to the P. thomii-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Fig. 6.
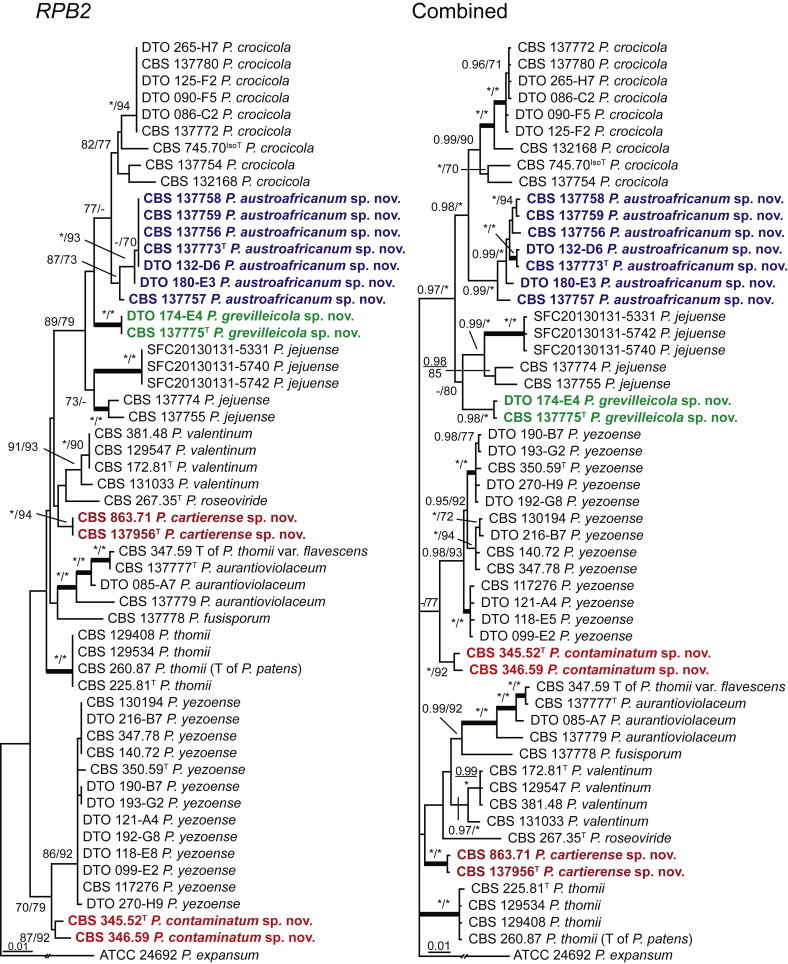
Phylogenetic trees showing the relationship among strains belonging to the P. thomii-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
The result of the combined analysis of BenA, CaM and RPB2 sequences of isolates belonging to the P. thomii-clade is shown in Fig. 6. The deeper nodes of this phylogeny are poorly resolved. Penicillium crocicola, P. austroafricanum, P. jejuense and P. grevilleicola form well-supported lineages on a well-supported branch (0.97 pp, >95 % ML). The other species are resolved in one clade in Fig. 1; however, this is not observed in the combined analysis of the P. thomii-clade (Fig. 6). Penicillium yezoense and P. contaminatum are sister species in Fig. 1, as are P. aurantioviolaceum, P. roseoviride and P. valentinum. No statistical support for this observation is found in Fig. 6. The phylogenies based on the individual data sets are congruent. Sequence variation is present within P. crocicola and P. jejuense. The P. crocicola lineage has no support in the BenA and CaM phylograms and weak support in the RPB2 analysis (82 % ML, 77 % MP). In the individual phylogenies, CBS 132168, CBS 137754 and CBS 745.70 (ex-type strain of P. crocicola) sit outside the main P. crocicola clade. These single strains have slightly different phenotypes than the strains belonging to the main clade. For example, CBS 745.70 grows quickly on CYA30°C (40–45 mm) and does not produce acidic compounds on CREA, and CBS 132168 differs by the formation of light yellow mycelium on YES and a slower lower growth rate on CYA30°C (15–20 mm vs 25–35 mm by the main group of P. crocicola isolates). These isolates might represent new species, but their description is deferred until more strains are collected and examined. Similarly, sequence variation is observed in P. jejuense. In the CaM and RPB2 data sets, CBS 137774 and CBS 137755 cluster together on a well-supported branch, separate from the type of the species. This grouping is not observed in the BenA data set; however, the bootstrap value was low in the MP analysis. The type strain of P. jejuense was unavailable for examination and no phenotypic comparison among the P. jejuense isolates was undertaken. The new species P. austroafricanum, P. cartierense, P. contaminatum and P. grevilleicola are described in the taxonomy part of this paper and phenotypic characters to differentiate between those species and their closest relatives are provided there.
Clade 3: Penicillium glabrum-clade
As mentioned above, species belonging to the P. glabrum-clade are phenotypically closely related to those of the P. spinulosum-clade. A high degree of sequence variation was observed in the BenA, CaM and RPB2 data sets. Based on the phylogenies, combined with phenotypic observations, this study focus on seven lineages (Figs 7, 8). Six of the seven lineages are well-supported in the combined analysis and the lineage containing isolate CBS 138160 is only supported in the ML analysis (82 % ML). The two main lineages in our analysis of the P. glabrum-clade are centred on the ex-types of P. glabrum (CBS 125543NT) and P. frequentans (CBS 105.11T). Another lineage includes the type of P. purpurescens (CBS 366.48T). The remaining lineages are described here as new species, namely P. armarii, P. bussumense, P. pulvis and P. rudallense. Analysis of the individual BenA, CaM and RPB2 partitions resulted in poorly resolved phylograms with many polytomies and generally lacking branch support (Figs 7, 8). In the BenA and CaM phylograms, the P. frequentans and P. glabrum strains do not resolve in two lineages. On the other hand, because of the lack of support of the nodes, this does not result in incongruency between the phylograms, hence the recognition of the two distinct species. In contrast, both species are resolved in the RPB2 phylogram with moderate to good support (80 % ML, 1.00 pp for P. glabrum; 70 % ML, 1.00 pp for P. frequentans). A large sequence variation is present in the currently proposed delineation of P. glabrum. A variation of 4.0 % was observed among the BenA sequences. Less variation is detected in the CaM data set (2.0 %) and the least variation was present in the RPB2 data set (1.4 %). This high degree of sequence variation is not reported in other Penicillium sections. In contrast, some species (e.g. P. commune, P. camemberti and P. caseifulvum) share BenA sequences or can only be differentiated by one base pair difference (Samson et al. 2004, Houbraken et al. 2011a, b). The phylogenetic placement of P. bussumense (CBS 138160T) also needs further attention. Strain CBS 138160T takes a distinct position in the BenA phylogram, and it is among the P. frequentans strains in the CaM analysis. Similarly as in P. glabrum/P. frequentans, both phylograms lack statistical support and this strain is resolved in the RPB2 data set.
Fig. 7.
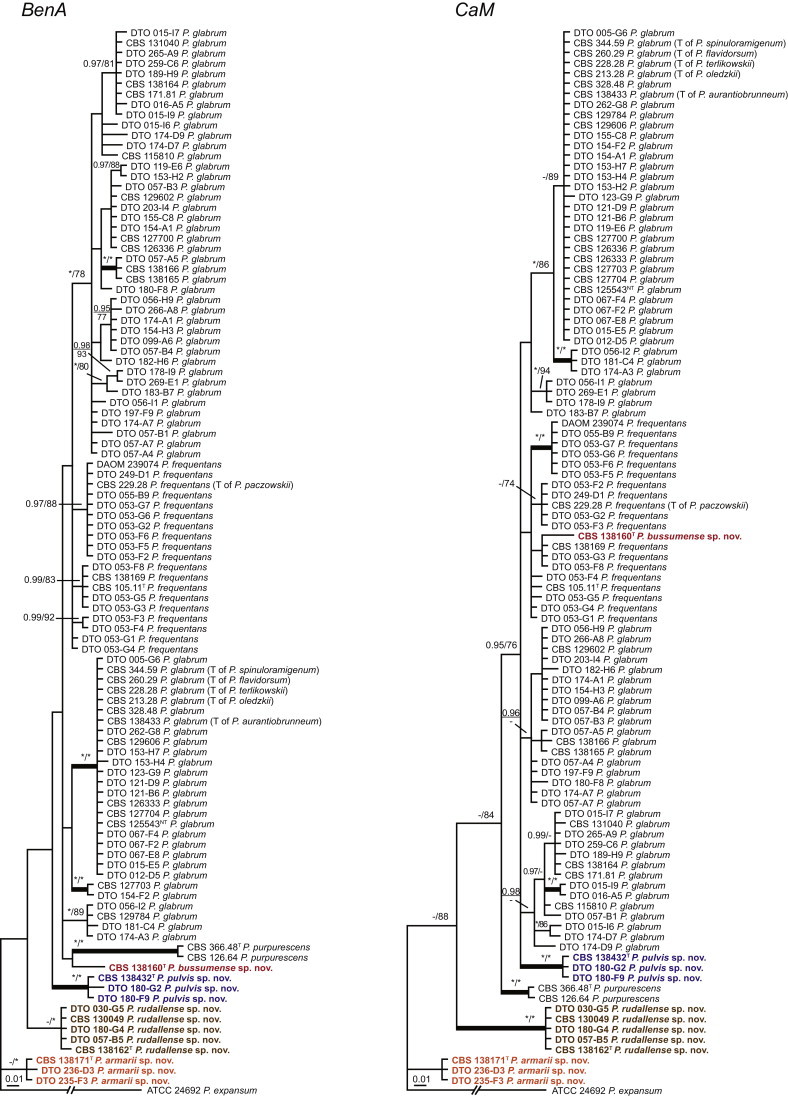
Phylogenetic trees showing the relationship among strains belonging to the P. glabrum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Fig. 8.
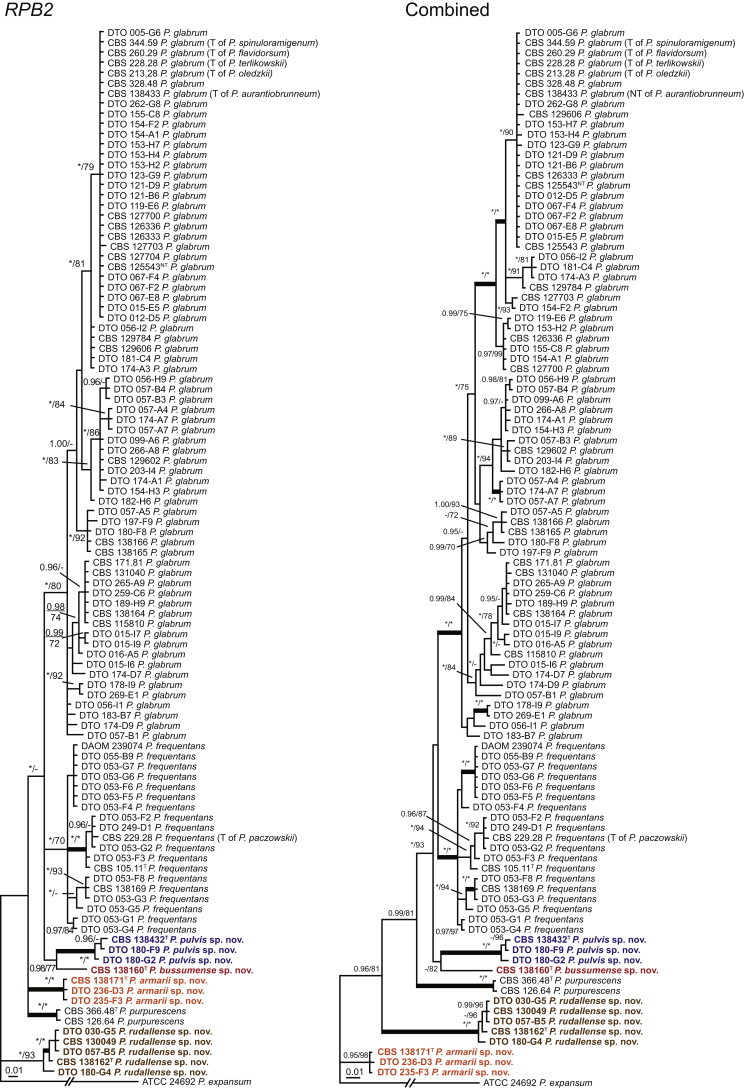
Phylogenetic trees showing the relationship among isolates belonging to the P. glabrum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Two species belonging to the P. glabrum-clade, P. glabrum and P. purpurescens, were accepted in previous studies (Pitt 1980, Pitt et al. 2000, Barreto et al. 2011). Citromyces purpurescens, Citromyces virido-albus, Penicillium virido-album, P. internascens and P. resinae are synonyms of P. purpurescens (see Taxonomy). Houbraken & Samson (2011) incorrectly used CBS 324.83 as a representative of P. asperosporum; this strain is actually the ex-type of P. resinae. Comparison of the RPB2 sequence of this strain deposited in GenBank (JN406574) with sequences generated in this study shows that CBS 324.83 is actually P. purpurescens. Several species were previously placed in synonymy with P. glabrum and one, P. frequentans, is re-introduced here. Based on our sequence data, P. paczowskii (CBS 229.28T) is a synonym of P. frequentans (Figs 7, 8). The name P. frequentans was used by Raper & Thom (1949), but this name is predated by P. glabrum (Subramanian 1971, Pitt 1980). Interestingly, Raper & Thom's concept of P. frequentans was not based on the type strain of P. frequentans, but on NRRL 1915 (= CBS 328.48). This strain is currently identified as P. glabrum and this indicates that Raper & Thom's description of P. frequentans and Pitt's concept of P. glabrum are based on a similar set of isolates that differs from our concept. Recently, a strain identified as P. glabrum (DAOM 239074) had its genome sequenced and this isolate is re-identified here as P. frequentans.
Penicillium aurantiobrunneum (CBS 138433NT), P. flavidorsum (CBS 260.29T), P. oledskii (CBS 213.28T) remain synonyms of P. glabrum. Penicillium terlikowskii (CBS 228.28T) and P. spinuloramigenum (CBS 344.59T), previously listed as synonyms of P. spinulosum, are treated here as synonyms of P. glabrum. Remarkably, combined sequence analysis (Fig. 8) shows that all the ex-type strains of P. glabrum and related synonyms belong to the same lineage, indicating a high genetic similarity. The reason for this bias remains unknown, but could be due to the fact that this sequence-type predominates in nature. Penicillium candidofulvum was listed by Pitt (1980) as a synonym of P. glabrum and a BLAST search with the ITS sequence of the ex-type strain for this species (CBS 254.37T) has 100 % homology with the type strain of P. corylophilum (FRR 802NT; AY373906). No type culture is available of P. fluitans. We follow Raper & Thom (1949) and Pitt (1980) and treat this species as a synonym of P. glabrum. Penicillium trzebinskii was treated as a synonym of P. glabrum by Barreto et al. (2011), but this was based on an incorrectly identified strain (CBS 328.48). Our results shows that the type of P. trzebinskii (CBS 382.48T) is a distinct species belonging to the P. spinulosum-clade.
In this study we accept seven species in the P. glabrum-clade: P. glabrum, P. purpurescens, P. frequentans, P. armarii, P. pulvis, P. rudallense and P. bussumense. These species are phenotypically similar and an overview of characters to differentiate them is given in Fig. 9 and Table 4. Penicillium purpurescens and P. armarii differ from the other species by their conidial size (3.2–4.0 μm vs 2.5–3.2(–3.5) μm in other P. glabrum-clade species) and they differ from each other by their colony diameters on CYA and CYA30°C, and the degree of growth on CREA. Penicillium glabrum and P. frequentans are phylogenetically and phenotypically closely related. Penicillium frequentans strains have a (yellow) brown reverse on CYA incubated at 27 °C and 30 °C, while those of P. glabrum are in shades of beige-brown resulting in less warm colours. Furthermore, strains of P. glabrum tend to have a higher growth rate on CYA30°C, but there is an overlap in colony diameters. These observations are supported by extrolite data. The extrolite profiles of these two species differ and P. frequentans strains produce 6-methylisocoumarin and a compound with the same chromophore as pyranonigrin, while P. glabrum isolates are characterised by the production of citromycetin, fulvic acid, asterric acid, bisdechlorgeodin, geodin, sulochrin, and similar polyketides (Hetherington & Raistrick 1931, Mahmoodian & Stickings 1964). Penicillium bussumense can be differentiated from other related species by a CYAS:CYA ratio of 0.95–1.05 and smaller colonies on CYA. Penicillium pulvis has a (dark) brown reverse on CYA and YES, and brown soluble pigments on CYA; a feature not observed in any of the closely related species. Penicillium rudallense has distinctly ornamented dark green conidia which measure 3.0–3.5 μm diam.
Fig. 9.
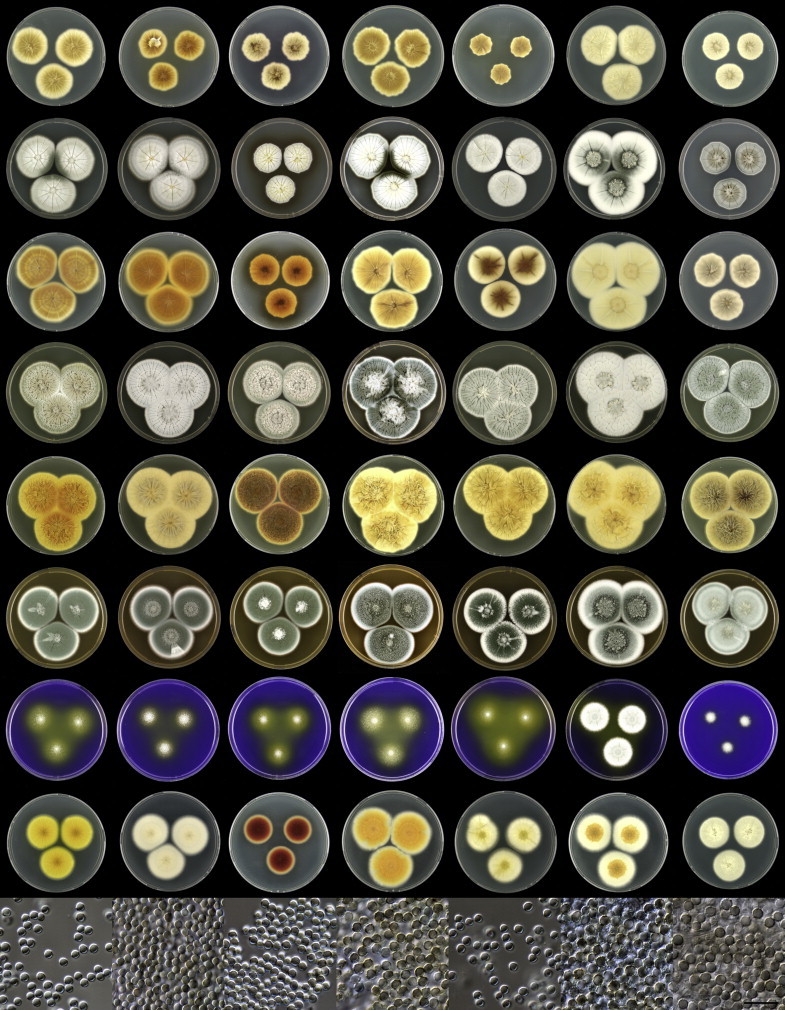
Overview of growth characters of P. glabrum-clade species. Columns, left to right: P. glabrum, P. frequentans, P. pulvis, P. rudallense, P. bussumense, P. armarii, P. purpurescens. Rows, top to bottom: CYA30°C reverse, CYA obverse, CYA reverse, YES obverse, YES reverse, MEA obverse, CREA obverse, DG18 reverse, conidia. Scale bar = 10 μm.
Table 4.
Overview of diagnostic characters of species belonging to the P. glabrum-clade.
| Species name | CYA | MEA | YES | CYA30°C | CYAS:CYA | Growth on CREA | Conidial size (μm) | Conidial ornamentation |
|---|---|---|---|---|---|---|---|---|
| P. armarii | 50–56 | 54–60 | 51–57 | 46–52 | 0.60–0.70 | Good | 3.2–4.0 | Rough |
| P. bussumense | 31–37 | 38–44 | 40–45 | 15–20 | 0.95–1.05 | Weak | 2.7–3.2 | Finely rough |
| P. frequentans | (33–)38–50 | 38–51 | 40–53 | 21–32 | 0.80–0.95 | Weak (occasionally good) | 2.5–3.0 | Finely rough to rough |
| P. glabrum | (30–)35–48 | (30–)38–50 | 40–59 | 25–43 | 0.75–0.85 | Weak | 2.5–3.0 | Finely rough |
| P. pulvis | 26–32 | 37–43 | 39–45 | 24–30 | 0.75–0.90 | Weak | 2.7–3.2 | Finely rough |
| P. purpurescens | 31–37 | 40–50 | 42–50 | 22–28 | 0.92–0.97 | Weak | 3.2–4.0 | Rough |
| P. rudallense | 35–46 | 39–46 | 48–54 | 25–37 | 0.80–0.92 | Weak | 3.0–3.5 | Rough |
Clade 4: Penicillium vagum-clade
The P. vagum-clade forms a well-supported lineage (>95 % ML; 1.00 pp) together with the P. glabrum, P. thomii, P. spinulosum and P. fuscum-clade. The relation between the isolates belonging to the P. vagum-clade is moderately supported (Fig. 1; 81 % ML; 0.96 pp). The isolates that belong to this clade can be divided in two lineages (Figs 10, 11). These two lineages are phenotypically unrelated and share characters such as monoverticillate conidiophores and the production of globose and subglobose conidia; characters that are also observed in many other section Aspergilloides species. The moderate support in the phylogenetic analysis, combined with the phenotypic differences, indicate that these species are only loosely related. Based on the unique phylogenetic placement of these isolates, combined with phenotypic differences, these two lineages are described as new species: P. vagum and P. longicatenatum.
Fig. 10.
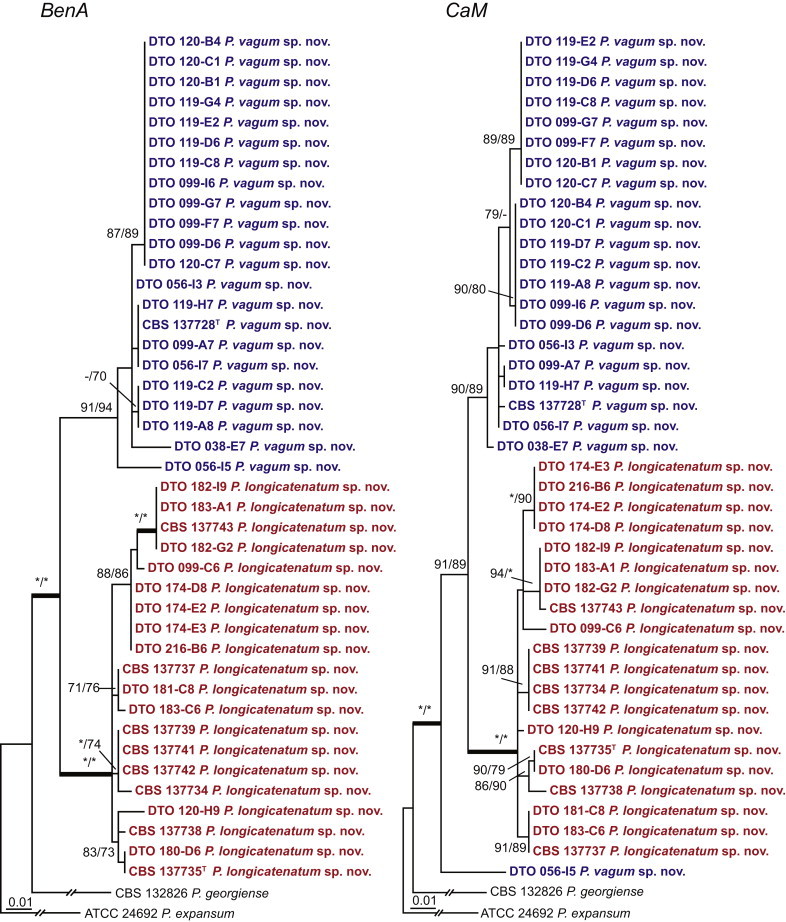
Phylogenetic trees showing the relationship among strains belonging to the P. vagum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Fig. 11.
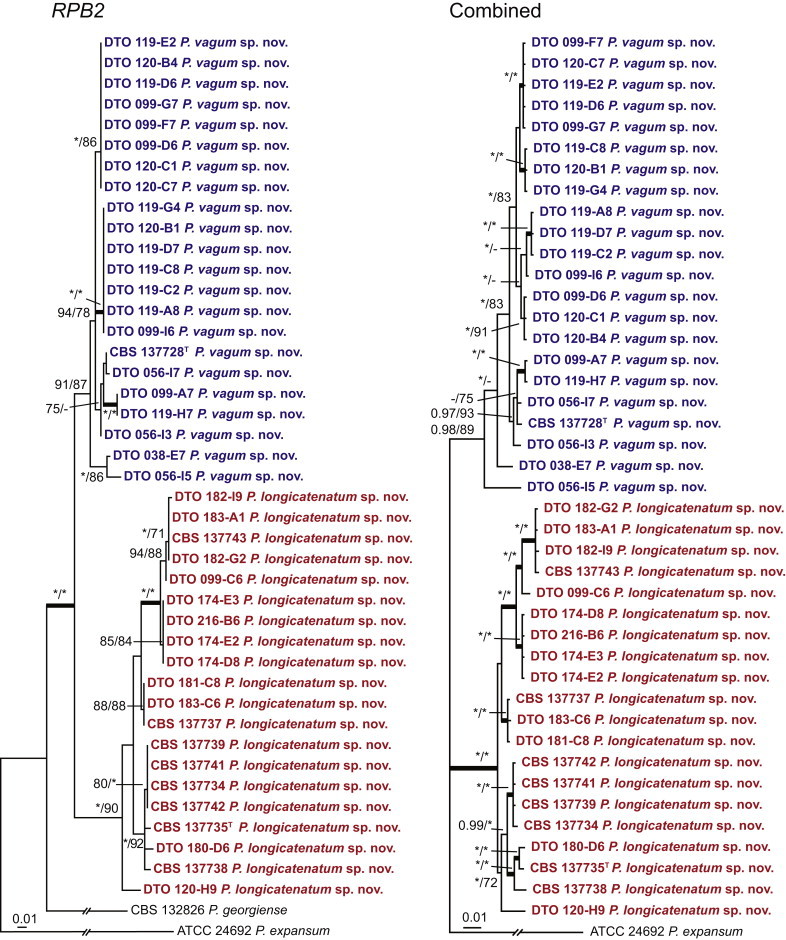
Phylogenetic trees showing the relationship among strains belonging to the P. vagum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Based on the single gene phylogenies, the isolates that belong to the P. vagum-clade can be divided into two supported lineages (Figs 10, 11). The only strongly supported branch contradictory to this arrangement is the position of DTO 056-I5 in the CaM analysis. This isolate is basal to the P. vagum and P. longicatenatum clades in the CaM analysis, but belongs to the P. vagum-clade in the BenA and RPB2 analysis. Penicillium vagum produces thick walled, spiny conidia and has a moderate acid production on CREA. In contrast, conidia of P. longicatenatum are finely roughened and isolates do not produce acidic compounds on CREA. A typical feature of P. longicatenatum is the abundant production of sclerotia on CYA and MEA. A detailed description of both species is given in the taxonomy section below.
Clade 5: Penicillium fuscum-clade
The P. fuscum-clade is phylogenetically related to the P. spinulosum-, P. thomii-, P. glabrum- and P. vagum-clades (>1.00 pp, >95 % ML) (Fig. 1). Species of the P. fuscum-clade differ from other species of sect. Aspergilloides by a slow or moderate growth rate on CYA [(10–)15–25(–30) mm] and CYAS (<15 mm), short stipes [10–80(–150) μm] and distinctly roughened, thick walled, globose to subglobose conidia.
The analysis of the separate BenA and CaM data sets resulted in poorly supported phylograms, while better support was obtained in the RPB2 phylogram (Figs 12, 13). The phylograms are not congruent. In the RPB2 phylogram, the isolates CBS 127833 (ex-type of Eladia inflata), CBS 130199 and CBS 129806 (indicated with ♣ symbol) group together with two other species on a branch with statistical support (86 % ML; 82 % MP) closer to P. brunneoconidiatum and P. flavisclerotiatum. In the BenA and CaM phylogenies, these isolates cluster with P. fuscum (BenA: 84 % ML, 82 % MP; CaM: 74 % ML, 82 % MP). The reason for this inconsistency is unknown, but it might indicate undersampling. The previously mentioned isolates are basal to P. fuscum in the combined analysis of the BenA, CaM and RPB2 data sets (0.99 pp; >95 % ML) and we therefore tentatively identified all of these strains as P. fuscum.
Fig. 12.
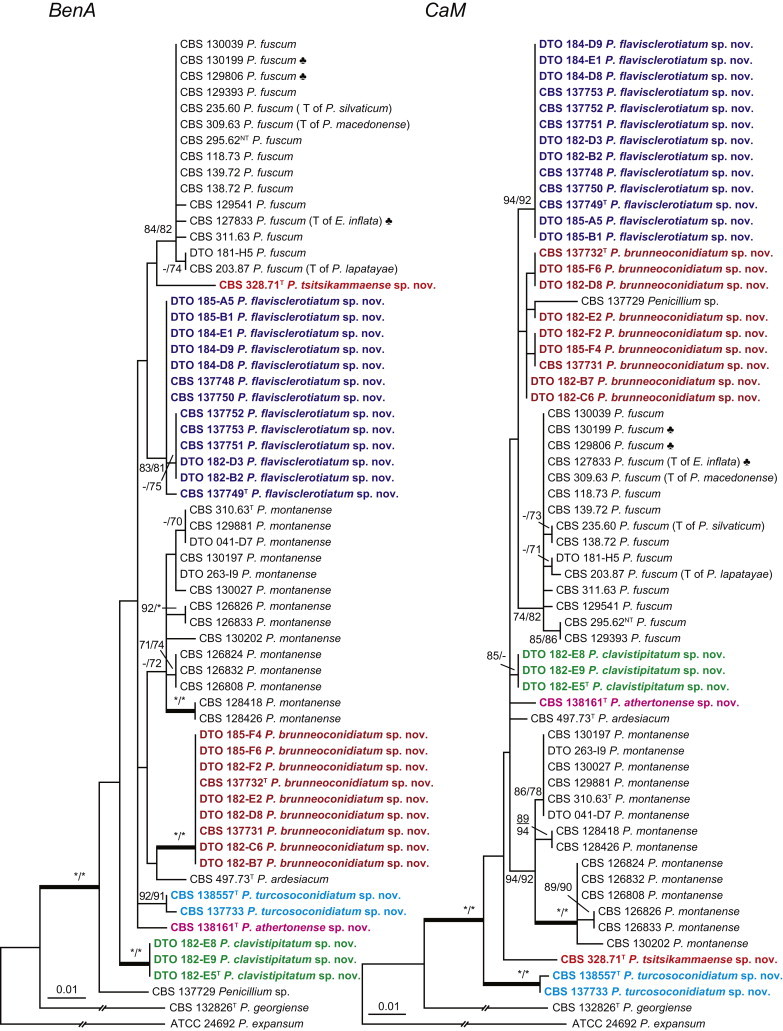
Phylogenetic trees showing the relationship among strains belonging to the P. fuscum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Fig. 13.
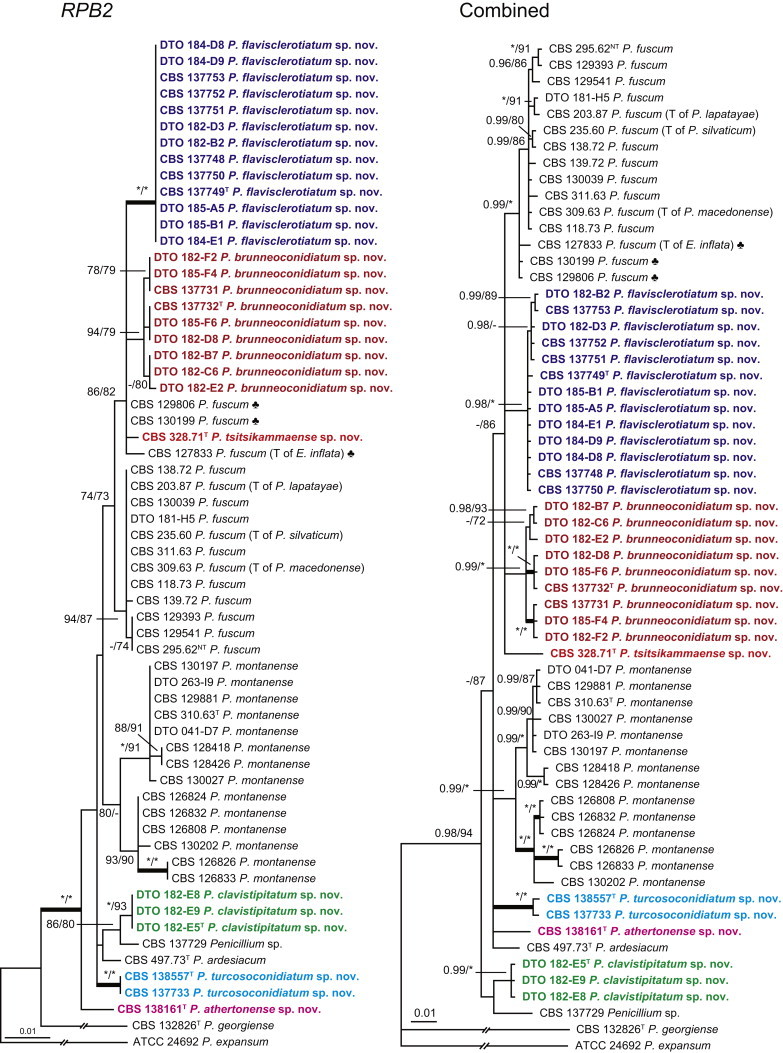
Phylogenetic trees showing the relationship among strains belonging to the P. fuscum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Stolk (1968) found ascospores in an old culture of the type strain of P. pinetorum (CBS 295.62T) and described this strain as Eupenicillium pinetorum. Penicillium fuscum was linked to E. pinetorum based on original descriptions and illustrations of Citromyces fuscus (Stolk & Samson 1983). This name predates P. pinetorum and was therefore accepted by Houbraken & Samson (2011). The species P. silvaticum, P. macedonense (Pitt 1980), P. asperosporum (≡ P. echinosporum G. Sm., non Nehira) and P. montanense were treated as synonyms of P. fuscum (Stolk & Samson 1983). Penicillium macedonense (CBS 309.63T) and P. silvaticum (CBS 235.60T) reside with P. fuscum (CBS 295.62NT) in all phylogenies (Figs 12, 13) confirming their synonymy with P. fuscum. Penicillium montanense was placed in synonymy with P. fuscum by Stolk & Samson (1983), while other studies accepted this species as distinct (Pitt 1980, Pitt et al. 2000). The type strain of P. montanense (CBS 310.63T) is distinct in our phylogenies of the P. fuscum-clade. Penicillium asperosporum was synonymised with P. fuscum by Stolk & Samson (1983), but was considered distinct in other studies (NRLL 3411T = IMI 080450; Peterson 2000). Sequence analysis shows that this species is a synonym of P. montanense (see Taxonomy). The phylogenetic position of ex-type strain of Eladia inflata (CBS 127833T) is uncertain (see above). We tentatively treat this species as a synonym of P. fuscum.
Based on our multigene phylogenies, nine species are accepted in the P. fuscum-clade. Three species were previously described (P. fuscum, P. montanense and P. ardesiacum) and six are newly introduced here: P. athertonense, P. brunneoconidiatum, P. clavistipitatum, P. flavisclerotiatum, P. tsitsikammaense and P. turcosoconidiatum. Conidiophore morphology is diagnostic for distinguishing these species. For example, P. clavistipitatum produces rough-walled stipes and P. brunneoconidiatum and P. turcosoconidiatum produce very short stipes. Penicillium fuscum is the only species in the clade producing ascospores and sclerotia are observed in P. flavisclerotiatum and P. tsitsikammaense. These species can be differentiated by the growth rate on CYA incubated at 25 and 30 °C. An overview of growth characters is given in Table 5.
Table 5.
Overview of diagnostic characters of species belonging to the P. fuscum-clade.
| Species name | CYA | MEA | YES | CYA30 °C | Stipe length (μm) | Stipe ornamentation | Conidial size (μm) | Sclerotia/cleistothecia |
|---|---|---|---|---|---|---|---|---|
| P. ardesiacum | 27–33 | 32–38 | 30–40 | 25–30 | 75–250 | Smooth to finely roughened | 2.5–3.5 | Absent |
| P. athertonense | 27–31 | 27–33 | 27–31 | 20–25 | (30–)60–200 | Smooth | 3.5–4.0 | Absent |
| P. brunneoconidiatum | 18–24 | 10–17 | 20–25 | (0–)10–18 | 7.5–30 | Smooth | 3.5–4.5 | Absent |
| P. clavistipitatum | 17–23 | 20–25 | 18–27 | 0–15 | 20–120 | Rough | 2.5–3.0 | Absent |
| P. flavisclerotiatum | 23–26 | 23–28 | 30–35 | 18–21 | 20–80 | Smooth | 2.0–3.5 | Sclerotia; yellow |
| P. fuscum | 31–37 | 30–36 | 31–37 | 0–10 | 15–120 | Smooth | 2.5–4.0(–5.5) | Cleistothecia; yellow and becoming avellaneous to reddish brown |
| P. montanense | 15–25 | 20–35 | (19–)25–30 | (0–)3–13 | 20–100(–150) | Smooth | Variable: (2.5–)4.0–4.5(–5.5) | Absent |
| P. tsitsikammaense | 10–15 | 19–23 | 20–25 | 5–10 | 25–50 | Smooth | 2.7–3.3(–5.5) | Sclerotia1; white |
| P. turcosoconidiatum | 15–20 | 18–25 | 20–25 | 7–10 | 6–30 | Smooth | 2.0–2.5 | Absent |
Sclerotia on MEA. Stolk & Samson (1983: 127) reported ascospores in the type strain of P. tsitsikammaense (CBS 328.71).
Clade 6: Penicillium sublectaticum-clade
A set of six strains formed a discrete clade within section Aspergilloides, here named the P. sublectaticum-clade. These species are phylogenetically related to the P. verhagenii and P. saturniforme-clade (76 % ML; 1.00 pp) (Fig. 1). The species of the P. sublectaticum-clade are predominantly monoverticillate while those of the latter two clades have a biverticillate branching pattern. Additionally, all isolates of this clade are able to grow on CYA incubated at 30 °C, a feature not observed in the P. verhagenii and P. saturniforme-clades. Phylogenetic analysis of the individual BenA, CaM and RPB2 data sets divided these strains into three well-supported lineages that are described here as new species: P. infra-aurantiacum, P. malmesburiense and P. sublectaticum (Fig. 14). The combined analysis of the BenA, CaM and RPB2 genes indicated that P. sublectaticum and P. malmesburiense are sister species, although with poor statistical support (0.96 pp, <70 % ML) (Fig. 14). Phenotypically, P. sublectaticum differs from P. malmesburiense in having a pale reverse on CYA, CYAS and DG18, and from P. infra-aurantiacum by having irregular margins on CYA and a dark brown reverse on CYA with (yellow) brown margins.
Fig. 14.
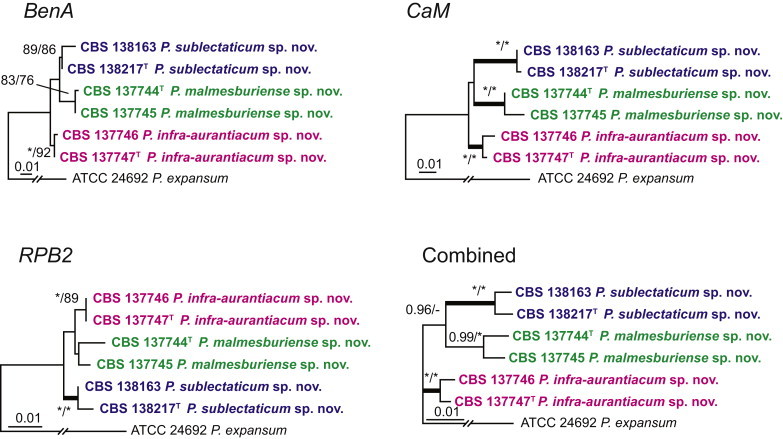
Phylogenetic trees showing the relationship among strains belonging to the P. sublectaticum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Clade 7: Penicillium verhagenii-clade
Isolates belonging to the P. verhagenii-clade are phylogenetically most closely related to P. saturniforme (Fig. 1; 1.00 pp; 88 % ML), and differ from all species in section Aspergilloides by their biverticillate conidiophores. These isolates are characterised by a unique mode of conidiophore development. The conidiophores are initially biverticillate, but can be become divaricate following sympodial branching of the stipe at the apex. Furthermore, the conidia of these species are blue-green or have a blue shade on MEA. Two clades are consistently present in the separate analysis of the BenA, CaM and RPB2 sequences and are treated here as the new species P. verhagenii and P. ranomafanaense (Fig. 15). These two species can be recognised by their reverse colour on CYA, YES and DG18. Penicillium ranomafanaense has orange or reddish reverse colours on CYA and YES and smooth to finely roughened conidiophore stipes, while P. verhagenii isolates have a yellow or yellow-brown reverse and distinctly rough walled stipes.
Fig. 15.
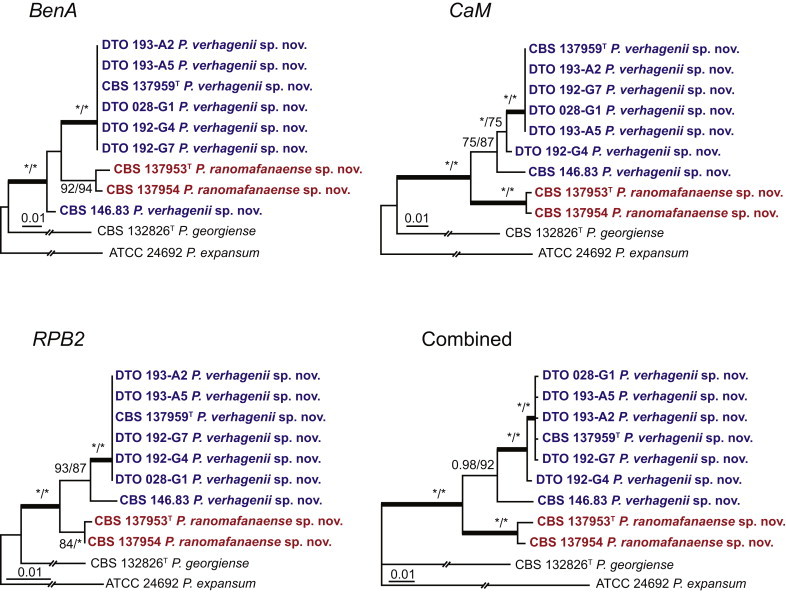
Phylogenetic trees showing the relationship among strains belonging to the P. verhagenii-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Clade 8: Penicillium saturniforme-clade
Penicillium saturniforme (CBS 122276T) occupies a unique and isolated position in the phylogeny of section Aspergilloides (Fig. 1). Phylogenetically, it is most closely related to species of the P. verhagenii-clade (1.00 pp; 88 % ML). Species of the P. verhagenii-clade and P. saturniforme are predominantly biverticillate, a unique feature in section Aspergilloides. Penicillium verhagenii and P. ranomafanaense have divaricate conidiophores following secondary, sympodial growth at the apex of the stipe, a feature not reported in P. saturniforme (Wang & Zhuang, 2009). Furthermore, P. saturniforme produces ascospores and these are not observed in cultures of P. verhagenii and P. ranomafanaense.
Clade 9: Penicillium lividum-clade
The Penicillium lividum-clade is most closely related to the P. hoeksii-clade (1.00 pp; >95 % ML) and basal to these two clades is the P. quercetorum-clade (0.99 pp; 89 % ML). The species of the P. lividum-clade produce (dark) blue-green conidia on MEA, grow moderately fast on CYA (25–35 mm) and very poorly on CREA, without the production of acidic compounds. Furthermore, the conidiophores are densely packed with phialides and the conidia are broadly ellipsoidal or ellipsoidal and distinctly roughened, often striate. Most of these characters are shared with P. hoeksii. The main differences between both the two clades are the absence of growth of species of the P. hoeksii-clade on CYA30°C and the finely roughened conidia produced by P. hoeksii.
Fig. 16 shows the phylogenetic relationships among species of the P. lividum-clade. Three species are accepted: P. lividum, P. odoratum and P. kananaskense. These species are resolved in well-supported lineages, but the relationship among the species is incongruent in the individual BenA, CaM and RPB2 phylogenies. In the CaM data set, P. kananaskense is a sister species of P. odoratum, but with low statistical support (<70 % ML; 73 % MP). In contrast, P. kananaskense and P. lividum are sister species with high bootstrap support (>95 % in ML and MP) in the RPB2 analysis.
Fig. 16.
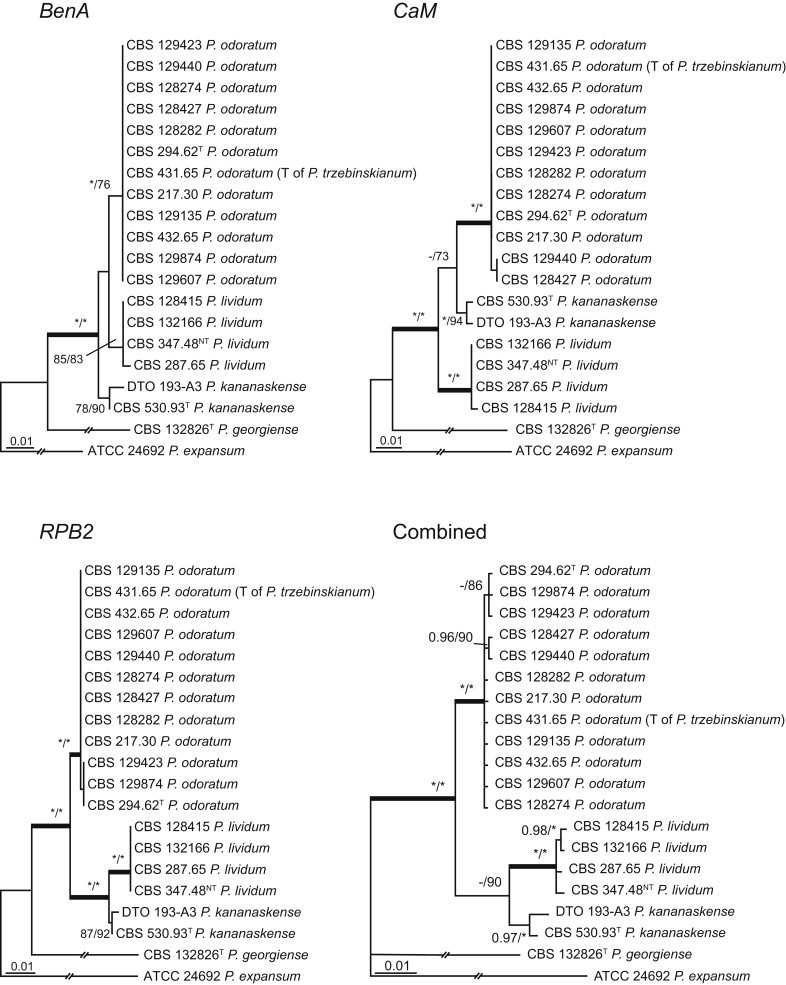
Phylogenetic trees showing the relationship among strains belonging to the P. lividum-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Penicillium lividum was accepted by Pitt (1980) and P. odoratum and P. trzebinskianum were regarded as synonyms. Our phylogenies (Fig. 16) place P. odoratum in a distinct clade separate from P. lividum. Penicillium trzebinskianum (CBS 530.93T) was invalidly described (without a Latin diagnosis) and was subsequently validated by Ramírez (1982). Our data show that this species is a synonym of P. odoratum.
Penicillium kananaskense (CBS 530.93T) was described as a pink-spored Penicillium species (Seifert et al. 1994). An additional strain of this species was examined (DTO 193-A3), which produced dark blue-green conidia on MEA. Although CBS 530.93T and DTO 193-A3 differ in conidial colour, they share some characters that are not observed in other species of the P. lividum-clade. For example, both strains grow slowly on CYA30°C (5–10 mm) and have a vivid yellow or orange reverse on CYA and YES with yellow pigments diffusing into the agar. Penicillium odoratum grows well on CYA30°C (28–35 mm) while P. lividum grows slowly (12–17 mm). The reverse colours on CYA and YES between these two species are similar (pale, beige, pale brown). The colony texture could also be used to distinguish these three species. Penicillium kananaskense produces velvety colonies on MEA, P. odoratum floccose colonies and the colonies of P. lividum have a lanose texture.
Clade 10: Penicillium hoeksii-clade
Seven strains (DTO 006-D8; DTO 068-D9; DTO 192-G9; CBS 137776T, DTO 192-H6; DTO 193-A4; DTO 199-G6) clustered together in our survey of section Aspergilloides isolates. Fig. 1 shows that two representatives of this group included in this analysis (DTO 068-D9, CBS 137776T) form a distinct clade within section Aspergilloides. This unique clade, here named the P. hoeksii-clade, is closely related to the P. lividum- and P. quercetorum-clades (Fig. 1; 0.99 pp; 89 % ML). Species of the P. hoeksii-clade differ from those of the P. lividum-clade by shorter stipes (less than 250 μm), which are smooth-walled. The recently described P. zhuangii belongs to the P. hoeksii-clade (Wang et al. 2014). Our set of isolates is phylogenetically (Fig. 17) and phenotypically distinct from P. zhuangii. We therefore describe this set of seven isolates as a new species, P hoeksii. Phenotypically, P. hoeksii and P. zhuangii share features such as moderate growth on CYA (15–28 mm), absence of growth on CYA incubated at 30 °C and the production of (broadly) ellipsoidal conidia. The species differ mainly in their response to temperatures. Penicillium zhuangii grows faster on CYA incubated at 15 °C than on 25 °C (22–24 vs 15–18 mm), while P. hoeksii isolates grow slower at 15 °C (12–18 mm vs 20–28 mm).
Fig. 17.
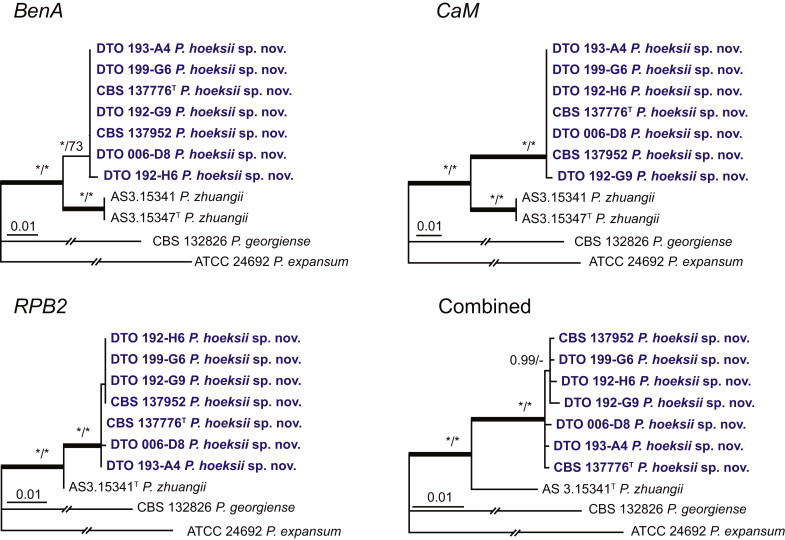
Phylogenetic trees showing the relationship among strains belonging to the P. hoeksii-clade. The bar indicates the number of substitutions per site. The phylogram is rooted with P. expansum (ATCC 24692).
Clade 11: Penicillium quercetorum-clade
Penicillium quercetorum is the only species of the P. quercetorum-clade. Pitt (1980) noted similarities between P. quercetorum and P. thomii, but accepted P. quercetorum based on a slower growth, smooth walled stipes and spheroidal rather than ellipsoidal conidia. However, in other studies, this species was placed in synonymy with P. thomii (Ramírez 1982, Pitt et al. 2000). Our data show that this species is phylogenetically related to the P. hoeksii- and P. lividum-clades (Fig. 1; 0.99 pp; 89 % BS) and more distant from the P. thomii-clade.
Clade 12: Penicillium thiersii-clade
Based on a four gene phylogeny, P. thiersii was convincingly placed in section Aspergilloides by Houbraken & Samson (2011). The phylogenetic placement of this species within section Aspergilloides could not be confirmed in our study because of a lack of statistical support (Fig. 1). Penicillium thiersii is resolved on its own, basal to other species of section Aspergilloides. The long, vesiculate, rough-walled stipes and ellipsoidal conidia suggest a relationship with species of the P. thomii-clade. In contrast, P. thiersii has a slower growth rate than species of the P. thomii-clade (Peterson et al. 2004). No sclerotia or cleistothecia were reported in the original description of P. thiersii. However, macro- and microscopic examination of CBS 117503T showed presence of pale brown cleistothecia on MEA, CYA and DG18. The ascospores of P. thiersii are ellipsoidal, measure 1.7–2.5 × 2.5–3.5 μm, and have an equatorial ridge and smooth to finely roughened valves. P. thiersii produce a series of different extrolites, making it chemically very distinctive: thiersinines, thiersindoles, decaturins and oxalicins (Li et al. 2002, 2003, 2005).
ITS barcoding and identification
The ITS locus is the accepted barcode for fungal identification (Schoch et al. 2012). Sixteen of 51 Aspergilloides species (31.3 %) could be identified using ITS sequences and therefore this locus cannot be used alone for species level identification in this group. This confirms previously published data on the use of the ITS barcodes for species recognition in Penicillium (e.g. Skouboe et al. 1999, Houbraken et al. 2011b, 2012a, b).
ITS sequences were generated from the majority of the strains used in the phylogenies described above. Fifty haplotypes were detected among the 365 investigated ITS sequences. An overview of the various haplotypes is given in Table 6. Fifteen species did not share ITS sequences with other species of sect. Aspergilloides. Among those fifteen species were P. longicatenatum, P. hoeksii, P. quercetorum, P. saturniforme and P. thiersii, species that were also phylogenetically very distinct (Fig. 1). Penicillium ardesiacum and P. tsitsikammaense were represented by only one strain and multiple strains of P. ranomafanaense and P. armarii were included in the analysis; however, these strains were isolated from the same substrate and location. We speculate that these species might also share ITS sequences with other species of sect. Aspergilloides when a more diverse set of isolates can be analysed. Interestingly, the investigated P. subspinulosum isolates had unique ITS sequences, although the species is closely related to P. roseomaculatum, P. trzebinskii and P. spinulosum in our analyses of the BenA, CaM and RPB2 genes. Penicillium trzebinskii could also be distinguished from those species based on its ITS barcode; however, this species also shared sequences with the phenotypically distinct P. grancanariae and P. palmense.
Table 6.
Overview of ITS sequences types of section Aspergilloides species. The GenBank numbers and haplotype numbers in bold font are shared among other species, and those in regular font are species specific. The species in pale yellow can be identified based on ITS sequences only, species in olive green share sequences with other Aspergilloides members.

Some haplotypes were predominantly present. Haplotype 11 contained only species of the P. lividum-clade and none of P. lividum-clade species belonged to another haplotype. With the exception of P. crocicola and P. austroafricanum, all P. thomii-clade species are represented by haplotype 18 and 23. However, these haplotypes do not exclusively accommodate P. thomii-clade species and can therefore not be used for “clade” level identification. Other P. thomii-clade species have haplotypes 25 (P. austroafricanum) and 26 (P. austroafricanum and P. crocicola). Eighty-one isolates have haplotype 30. All P. frequentans and the majority of P. glabrum share this haplotype. The only other species with this haplotype is P. pulvis (two of the three examined sequences).
BenA is recommended as a general purpose secondary barcode for precise species identification in Penicillium (Samson et al. 2010, Visagie et al. 2014). Either partial CaM, BenA and RPB2 sequences can be used for identification purposes in section Aspergilloides and all sequences obtained were species specific. The use of BenA might be hampered by a high intra-specific variation in P. glabrum (4.0 %) and P. frequentans (3.0 %). It is unclear whether this variation is only present in these two species. Perhaps it occurs in other species of sect. Aspergilloides, but we may not have observed it because of our limited sample size. Alternatively, it is feasible that there are additional phylogenetic species, or incipient speciation, in P. glabrum and P. frequentans that are only hinted at by our data. The difficulties of correct identification linked to a high intra-specific variation will be solved by continued sampling of BenA sequences to assay intra-species diversity, leading to higher similarity matches.
Phenotypic and physiologic characters can be used for species identification. Although there are differences in phenotype, identification based on these features remains difficult for non-specialists and a sequence-based approach is recommended, followed by assessment of confirmatory phenotypic characters. Various factors can hinder a phenotype-based identification, including lab-to-lab or batch-to-batch differences in agar media, degeneration of strains and subjective interpretation of structures and colours. If only a phenotype-based identification is possible, then it is of major importance to follow the recommendations listed in Visagie et al. 2014.
Taxonomy
Our phylogenetic analysis revealed that section Aspergilloides currently contains 51 species. The 26 new species that belong to this section are described below.
Penicillium armarii Houbraken, Visagie, Samson & Seifert, sp. nov. MycoBank MB809955. Fig. 18.
Fig. 18.
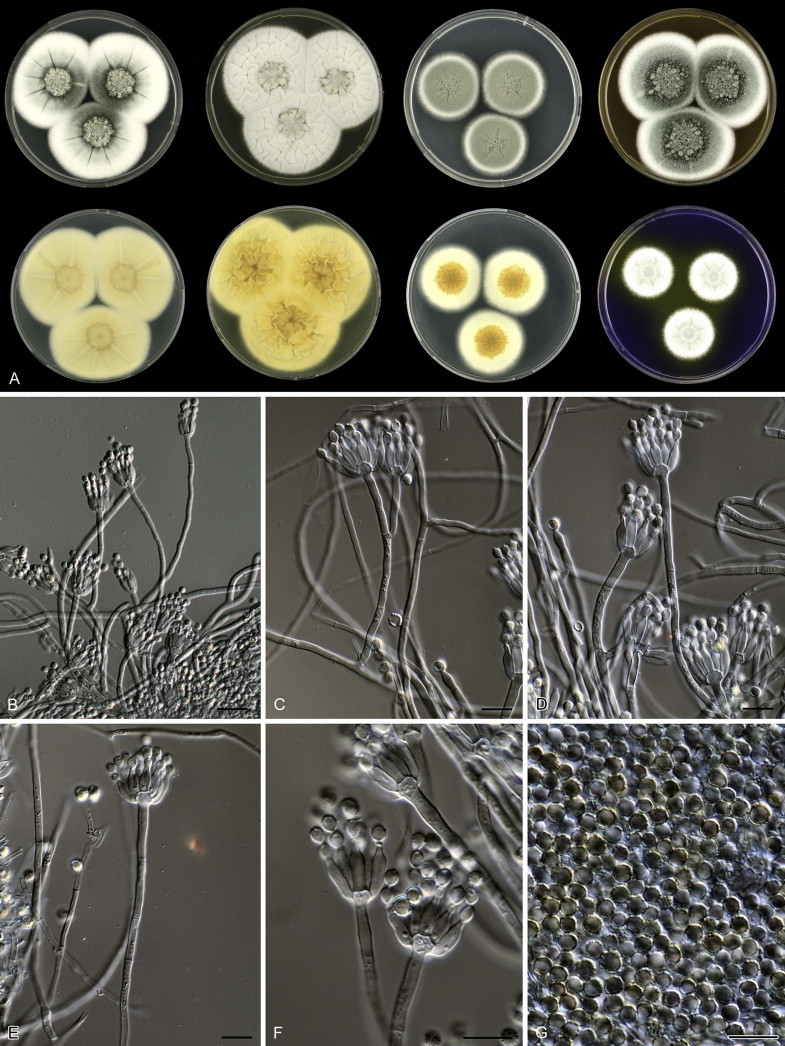
Penicillium armarii, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Named after the collection site, in a closet.
Diagnosis: This species belongs to the P. glabrum-clade and can be differentiated by its fast growth rate on CYA (25 and 30 °C) and MEA, and good growth on CREA.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium glabrum-clade.
Typus: Australia, Tasmania, Hobart, house dust, collected by G. Gates, 2009, isolated by E. Whitfield and K. Mwange (holotype CBS H-21870, culture ex-type CBS 138171 = DTO 235-F1).
Barcode and molecular based ID: ITS barcode: KM189758 (alternative markers: BenA = KM089007; CaM = KM089394; RPB2 = KM089781).
Description: Colony diam, 7 d, in mm: CYA 50–56 CYA15°C 25–31; CYA30°C 46–52; CYA37°C no growth; MEA 54–60; YES 51–57; DG18 34–40; CYAS 31–37; ratio CYAS:CYA 0.60–0.70; CREA 25–31, good growth, moderate acid production, followed by base production.
Sporulation on CYA moderate to good, colony texture weakly floccose, light radially sulcate; conidia dark dull green; mycelium inconspicuous; exudate present in the centre as large clear exudate droplets; soluble pigments absent; margin entire; reverse pale yellow. Sporulation on YES absent or very poor, mycelium white; soluble pigments absent; reverse yellow with orange centre. Good sporulation on DG18, colony texture crustose; conidia dull green; reverse pale with orange-brown centre. Good sporulation on MEA, colony texture floccose; conidia dark green in the centre, dull green toward the edge; exudate present as large clear droplets; reverse colour not affecting medium. Ehrlich reaction negative.
Sclerotia absent. Conidiophores 50–300 μm long, apices vesiculate, especially in older parts of the colony, up to 5.5 μm, smooth walled, predominantly monoverticillate, sometimes with divergent additional branch up to 15 μm long, stipe 2.0–3.5 μm wide. Phialides ampulliform with distinct neck, 6–14 per stipe, 8.5–10.5 × 2.5–3.0 μm. Conidia in long distorted chains, globose, distinctly ornamented 3.2–4.0 μm diam.
Penicillium athertonense Houbraken, sp. nov. MycoBank MB809956. Fig. 19.
Fig. 19.
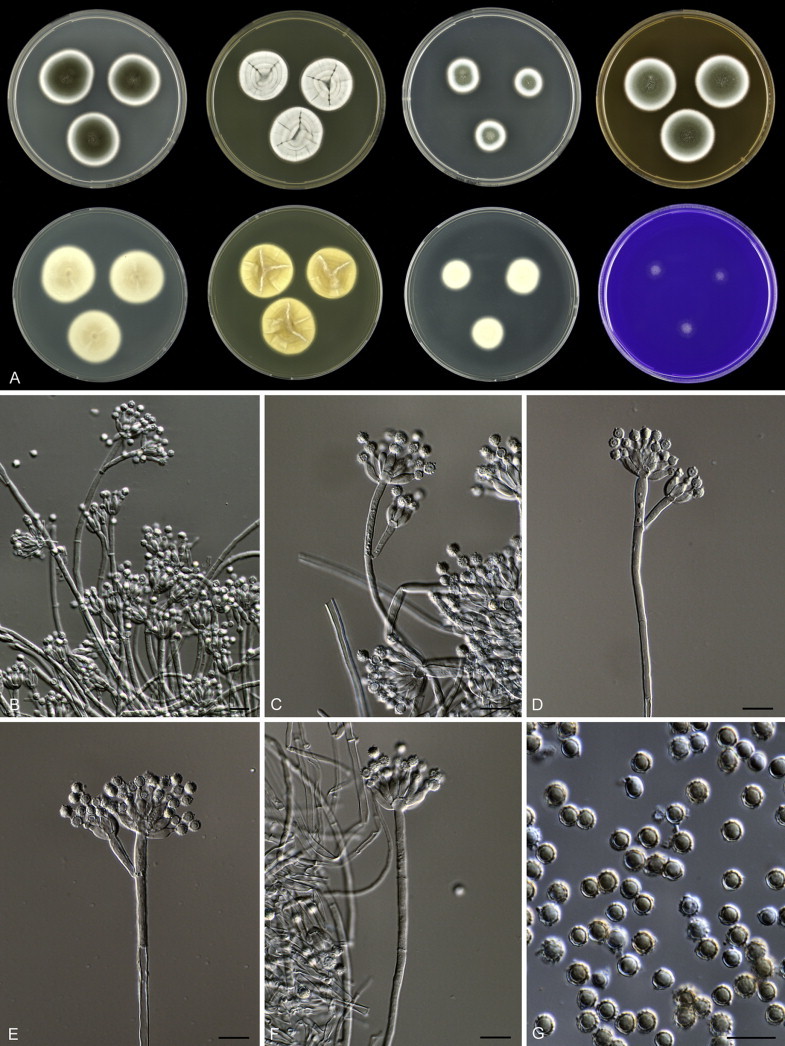
Penicillium athertonense, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Referring to Atherton Tablelands, the location of the type strain.
Diagnosis: The species is phylogenetically distinct from other species of the P. fuscum-clade. Phenotypically, it can be distinguished by a fast growth rate on CYA at 30 °C (20–25 mm).
In: subgenus Aspergilloides, section Aspergilloides, Penicillium fuscum-clade.
Typus: Australia, Queensland, Atherton Tableland, forest soil, isolated by J. Houbraken & R. van Leeuwen (holotype CBS H-21874, culture ex-type CBS 138161 = DTO 030-C2).
Barcode and molecular based ID: ITS barcode: KM189462 (alternative markers: BenA = KM088690; CaM = KM089075; RPB2 = KM089462).
Description: Colony diam, 7 d, in mm: CYA 27–31; CYA15°C 9–13; CYA30°C 20–25; CYA37°C no growth; MEA 27–33; YES 27–31; DG18 17–21; CYAS 15–20; ratio CYAS:CYA 0.6–0.65; CREA 8–12, weak growth and no acid production.
Sporulation on CYA strong; colony texture velvety, radially sulcate, low; conidia dark green; mycelium white; exudate absent; soluble pigment not produced; margin entire; reverse pale yellow to yellow in colony centre, pale beige at the margin. Sporulation on YES poor, conidia pale green; mycelium white; soluble pigment not produced; reverse pale yellow. Moderate to good sporulation on DG18, colony texture velvety; conidia dark green in the centre, dull blue-green on the edge of colony; mycelium white; reverse pale. Sporulation on MEA moderate to good; colony texture velvety; conidia pure (dark) green; exudate present as small, pale droplets; reverse centre brown, reverse of not affecting; Ehrlich reaction negative.
Sclerotia absent. Conidiophores, (30–)60–200 μm long, smooth walled; slightly vesiculate up to 5 μm wide; frequently with an additional branch, 13–19 μm long. Phialides divergently arranged, widest above the middle and with a short neck, 4–14 per stipe, 8.0–9.0 × 2.5–3.0 μm. Conidia in long distorted chains, globose, distinctly roughened, spinose, 3.5–4.0 μm diam.
Penicillium austroafricanum Houbraken & Visagie, sp. nov. MycoBank MB809957. Fig. 20.
Fig. 20.
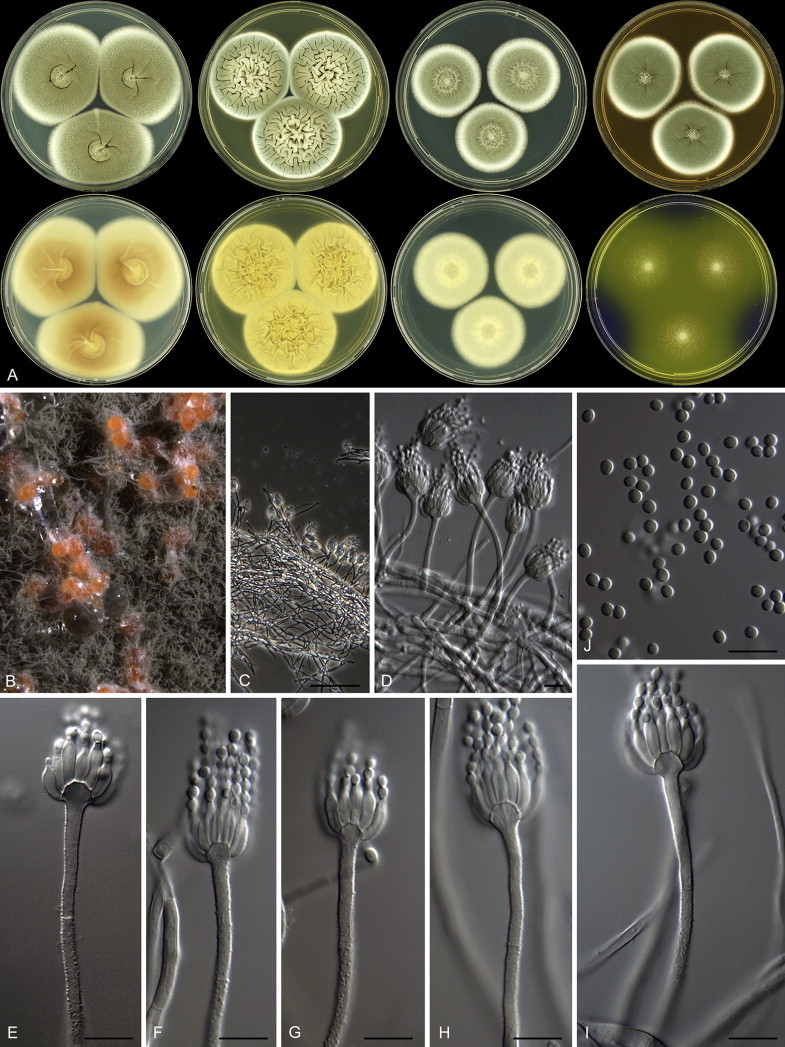
Penicillium austroafricanum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B, C. Sclerotia. D–I. Conidiophores. J. Conidia. Scale bars = 10 μm.
Etymology: Named after South Africa, the origin of the type strain.
Diagnosis: This species is phylogenetically related to P. crocicola and both species have a strong acid production on CREA. Penicillium austroafricanum has a pale yellow or pale brown reverse colour on DG18, while P. crocicola has a pale or a pinkish-brown reverse colour.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium thomii-clade.
Typus: South Africa, Western Cape, Harold Porter Botanical Garden, leaf of Phaenocoma prolifera, isolated by J. Houbraken & P. Crous (holotype CBS H-21864, culture ex-type CBS 137773 = DTO 133-G5).
Barcode and molecular based ID: ITS barcode: KM189610 (alternative markers: BenA = KM088854; CaM = KM089241; RPB2 = KM089628).
Description: Colony diam, 7 d, in mm: CYA 51–62; CYA15°C 23–35; CYA30 °C 30–50; CYA37°C no growth; MEA 40–55; YES 45–55; DG18 27–45; CYAS 35–42; ratio CYAS:CYA 0.67–0.77; CREA 30–35, poor growth, strong acid and no base production.
Sporulation on CYA moderately dense to dense; conidia dull green to greyish green, colony texture velutinous and floccose, low, sulcate, creamish sclerotia produced; mycelium white; exudate clear, sometimes absent; soluble pigment not produced; margin low, wide, entire; reverse greenish white to light yellow to light brown, more yellowish in some isolates. Sporulation on YES moderately dense; conidia dull green, colony texture velutinous, moderately deep, sulcate, greyish colour in non-sporulating areas; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse light yellow to dull yellow greyish yellow. Sporulation on DG18 strong; conidia dull green to greyish green, colony texture velvety or floccose, low; mycelium white; exudate absent; soluble pigment not produced; reverse pale yellow, pale brown, greenish white. Sporulation on MEA moderately dense to dense; conidia dull green to greyish green, colony texture velutinous, low, plane, creamish sclerotia produced; mycelium white; exudate clear; soluble pigment not produced; margin low, wide, entire; reverse brownish yellow to yellowish brown. Ehrlich reaction negative.
Sclerotia on MEA, hard, consisting of polygonal cells, 200–400 × 150–250 μm. Conidiophores monoverticillate; stipes distinctly roughened, 45–220 × 2–3.5 μm, vesicles 4–7.5 μm diam. Phialides 13–24 per stipe, ampulliform, 8–10.5 × 2.5–3.5 μm. Conidia smooth to finely rough walled, broadly ellipsoidal or ellipsoidal, 2.5–4 × 2.5–3 μm.
Penicillium brunneoconidiatum Visagie, Houbraken & K. Jacobs, sp. nov. MycoBank MB809958. Fig. 21.
Fig. 21.
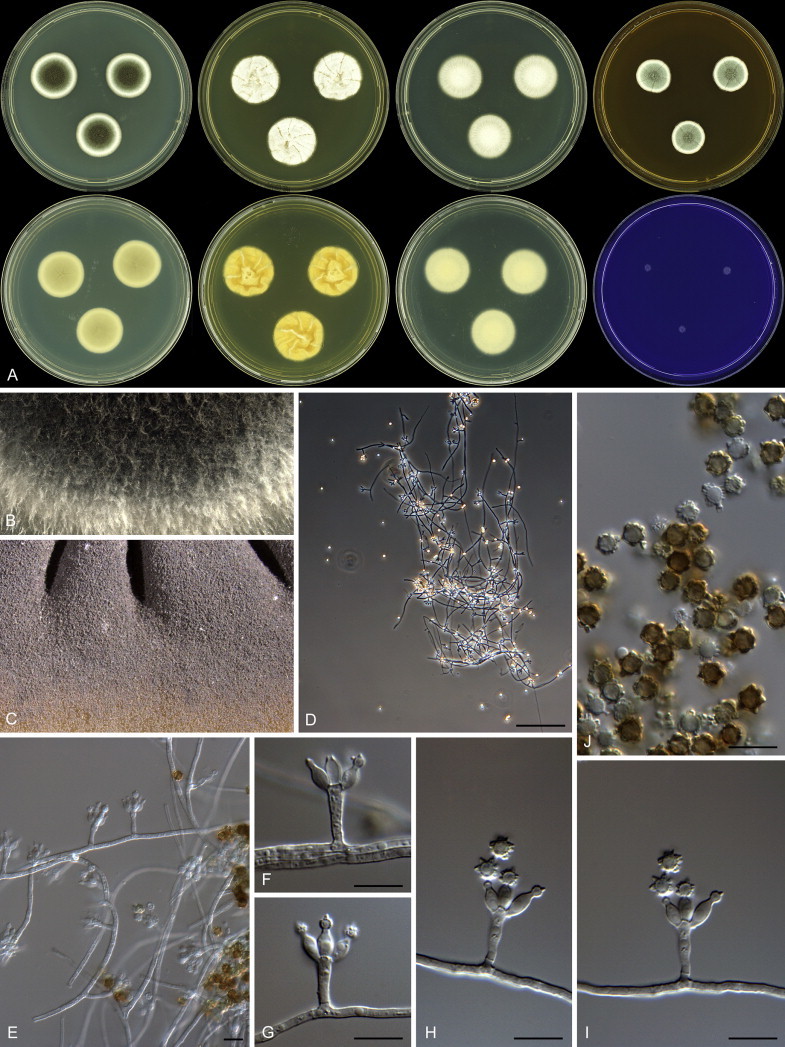
Penicillium brunneoconidiatum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B, C. Colony texture. D–I. Conidiophores. J. Conidia. Scale bars = 10 μm.
Etymology: Referring to the brown conidia of the species.
Diagnosis: Penicillium brunneoconidiatum differs from the other species belonging to the P. fuscum-clade by its restricted growth on CYA, YES and MEA, short stipes, large conidia (3.5–4.5 μm) and dark green to olive green conidia that become dark brown with age (Table 5).
In: subgenus Aspergilloides, section Aspergilloides, Penicillium fuscum-clade.
Typus: South Africa, Western Cape, Malmesbury, Riverlands, Fynbos, soil, isolated by C.M. Visagie (holotype CBS H-21873, culture ex-type CBS 137732 = DTO 182-E4 = CV 949 = DAOM 241359).
Barcode and molecular based ID: ITS barcode: KM189666 (alternative markers: BenA = KM088911; CaM = KM089298; RPB2 = KM089685).
Description: Colony diam, 7 d, in mm: CYA 18–24; CYA15°C 7–15; CYA30°C (0–)10–18; CYA37°C no growth; MEA 10–17; YES 20–25; DG18 21–24; CYAS 6–10; ratio CYAS:CYA 0.3–0.4; CREA 3–6, weak growth, no acid production.
Sporulation on CYA moderately dense to dense; conidia dull green to dark green, becoming brown with age, colony texture velutinous, low to moderately deep, plane to slightly sulcate in some strains; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to greyish green to dull green. Sporulation on YES absent to sparse; conidia greenish white, colony texture floccose, deep, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse pale yellow to light yellow to olive. Sporulation on DG18 absent to sparse; conidia greenish white, colony texture velutinous, low, slightly sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to greyish yellow. Sporulation on MEA dense; conidia dull green, becoming brown with age, colony texture velutinous, moderately deep, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish brown. Ehrlich reaction negative.
Conidiophores monoverticillate; stipes smooth walled, 7.5–30 × 1.5–2.5 μm, with a minor proportion up to 120 μm long, vesicles 2.5–5 μm diam. Phialides ampulliform, 3–6 per stipe, 5–7.5 × 2.5–3.5 μm. Conidia thick and rough walled, globose, 3.5–4.5 μm.
Penicillium bussumense Houbraken, sp. nov. MycoBank MB809959. Fig. 22.
Fig. 22.
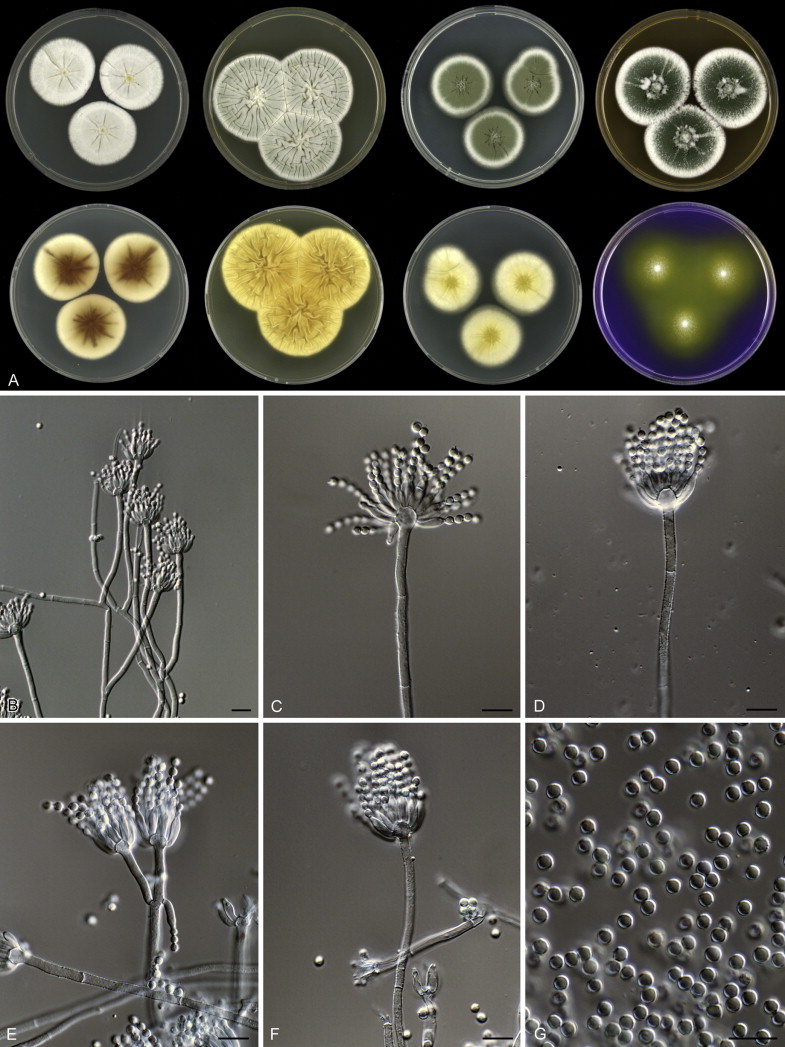
Penicillium bussumense, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Referring to Bussum, the location of the type strain.
Diagnosis: This species belongs to the P. glabrum-clade and can be differentiated by its CYAS:CYA ratio between 0.99–1.02 and the production of dark green conidia, floccose colonies and with the broad non-sporulating margin on MEA.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium glabrum-clade.
Typus: the Netherlands, Bussum, Spanderswoud, soil, isolated by J. Houbraken (holotype CBS H-21869, culture ex-type CBS 138160 = DTO 018-B2).
Barcode and molecular based ID: ITS barcode: KM189458 (alternative markers: BenA = KM088685; CaM = KM089070; RPB2 = KM089457).
Description: Colony diam, 7 d, in mm: CYA 31–37; CYA15°C 22–26; CYA30°C 15–20; CYA37°C no growth; MEA 38–44; YES 40–45; DG18 30–35; CYAS 34–39; ratio CYAS:CYA 0.95–1.05; CREA 18–23, poor growth, weak acid production.
Sporulation on CYA absent; radially sulcate, deep; mycelium white; exudate sparsely produced as yellow droplets; soluble pigments present, weak, yellow-brown; margin entire; reverse pale beige at the margin, dark brown in the centre. Sporulation on YES poor; conidia pale grey-green; mycelium white; soluble pigments absent; reverse yellow-brown. Good sporulation on DG18, colony texture velvety; conidia dark green; reverse pale in the centre, transparent at the margin. Good sporulation on MEA, colony texture floccose; conidia dark green; exudate present as small clear droplets; reverse colour not affecting medium. Ehrlich reaction negative.
Sclerotia absent. Conidiophores short, 50–250 μm long, stipes vesiculate up to 5.5 μm diam, finely to distinctly rough-walled, predominantly monoverticillate, sometimes with divergent additional branch up to 20 μm long, stipe 2.5–3.5 μm wide. Phialides ampulliform, densely packed, 6–20 per stipe, 8.0–10.0 × 2.5–3.0 μm. Conidia in long distorted chains, globose, finely ornamented, 2.7–3.2 μm diam.
Penicillium cartierense Houbraken, sp. nov. MycoBank MB809960. Fig. 23.
Fig. 23.
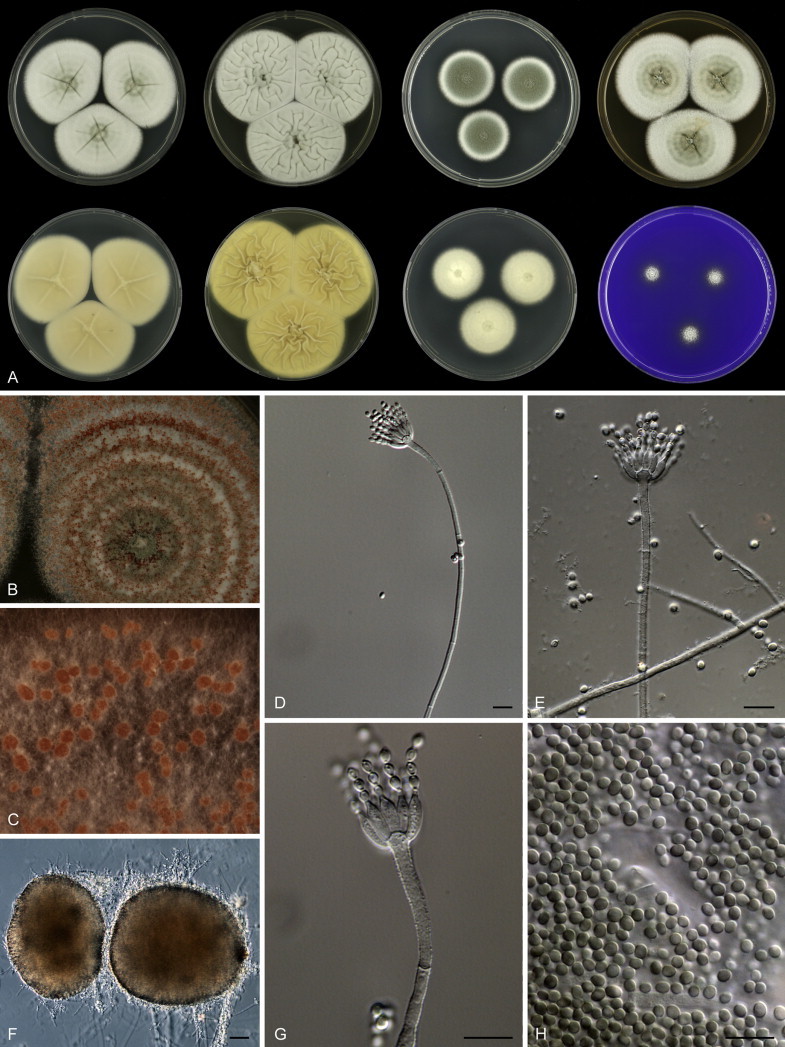
Penicillium cartierense, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–C, F. Sclerotia. D–E, G. Conidiophores. H. Conidia. Scale bars = 10 μm.
Etymology: Named after Cartierheide, the Netherlands, origin of the type strain.
Diagnosis: The species is phylogenetically unique and similar to other species of the P. thomii-clade, differing by the production of reddish brown sclerotia.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium thomii-clade.
Typus: the Netherlands, Eersel, Cartierhiede, soil, isolated by J. Houbraken (holotype CBS H-21861, culture ex-type CBS 137956 = DTO 092-H9).
Barcode and molecular based ID: ITS barcode: KM189564 (alternative markers: BenA = KM088804; CaM = KM089189; RPB2 = KM089576).
Description: Colony diam, 7 d, in mm: CYA 42–49; CYA15°C 23–29; CYA30°C 17–32; CYA37°C no growth; MEA 46–50; YES 51–55; DG18 28–34; CYAS 30–37; ratio CYAS:CYA 0.66–0.81; CREA 13–19, weak growth and no or very poor acid production.
Weak sporulation on CYA and only in the centre; colony texture floccose, radial sulcate; conidia pale dull green or dull green; mycelium white; exudate absent or present as small pale yellow droplets; soluble pigments absent; margin entire; reverse pale brown with dark brown centre becoming reddish brown in time (CBS 863.71). Sporulation on YES absent or poor, conidia dull grey green; mycelium white or very pale crème; soluble pigments absent; reverse yellow-brown. Strong sporulation on DG18, conidia dull green; reverse pale to pale brown or yellow-brown. Moderate to good sporulation on MEA, colony texture floccose; conidia dull green; sclerotia abundantly present, pale brown when young becoming red-brown at age; exudate present as large, clear to light brown droplets; colony reverse yellow in centre, colour medium under colony margins not affected. Ehrlich reaction negative.
Sclerotia pale brown when young, becoming reddish brown after three weeks of incubation at RT on MEA, 250–500(–650) μm; hard; consisting of polygonal cells; no asci or ascospores observed. Conidiophores 150–250 μm long, vesicles 4–7 μm diam, (finely) roughened with ornamentation up to apex, monoverticillate; stipe 3.0–4.0 μm wide. Phialides ampulliform, 10–14 per vesicle, 9–11 × 2.5–3.5 μm. Conidia in long irregular columns, broadly ellipsoidal, smooth to finely roughened, 3.0–4.0 μm.
Penicillium clavistipitatum Visagie, Houbraken & K. Jacobs, sp. nov. MycoBank MB809961. Fig. 24.
Fig. 24.
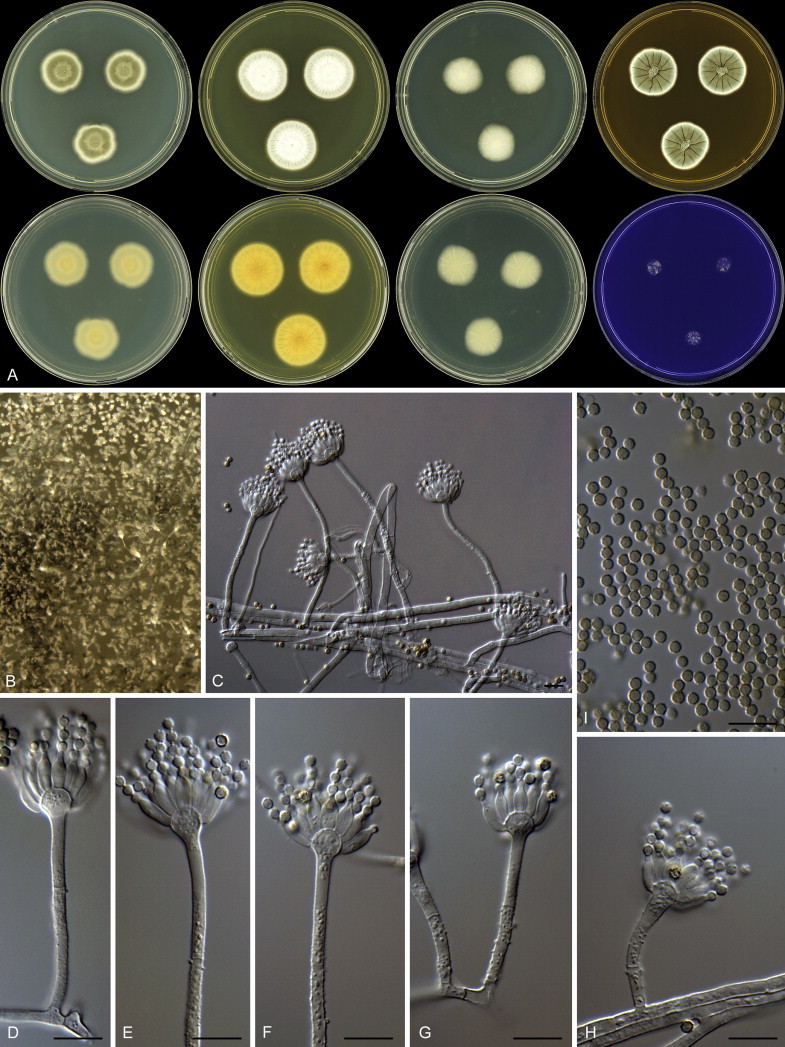
Penicillium clavistipitatum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B. Colony texture. C–H. Conidiophores. I. Conidia. Scale bars = 10 μm.
Etymology: Referring to the vesiculate stipe apices of the species.
Diagnosis: Penicillium clavistipitatum grows restrictedly on CYA and MEA and has rough walled stipes that end in vesicles up to 10 μm diam.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium fuscum-clade.
Typus: South Africa, Malmesbury, Riverlands, Fynbos, soil, isolated by C.M. Visagie (holotype CBS H-21882, culture ex-type CBS 138650 = DTO 182-E5 = CV 336 = KAS 4112 = DAOM 241092).
Barcode and molecular based ID: ITS barcode: KM189667 (alternative markers: BenA = KM088912; CaM = KM089299; RPB2 = KM089686).
Description: Colony diam, 7 d, in mm: CYA 17–23; CYA15°C 11–13; CYA30°C no growth to 15 mm; CYA37°C no growth; MEA 20–25; YES 18–27; DG18 18–20; CYAS 6–9; ratio CYAS:CYA 0.35–0.4; CREA 7–10, weak growth, no acid production.
Sporulation on CYA dense; conidia greenish grey to dark green, colony texture velutinous, low, plane; mycelium white; exudate absent; soluble pigment not produced; margin subsurface, narrow, entire; reverse yellowish white to greyish green to olive grey. Sporulation on YES moderately dense; conidia dull green, colony texture velutinous, low, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish grey to light yellow to olive brown. Sporulation on DG18 absent to sparse; conidia greyish green, colony texture velutinous, low, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to olive. Sporulation on MEA dense; conidia greyish green to dark green, colony texture velutinous, low, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse light yellow to yellowish brown. Ehrlich reaction negative.
Sclerotia absent. Conidiophores monoverticillate; stipes rough walled, 20–120 × 2.5–3.5 μm, vesicles 5.5–10 μm diam. Phialides ampulliform, 22–35 per stipe, 7–9 × 2.5–3.5 μm. Conidia globose, distinctly rough walled, 2.5–3 × 2.5–3 μm.
Penicillium contaminatum Houbraken, sp. nov. MycoBank MB809962. Fig. 25.
Fig. 25.
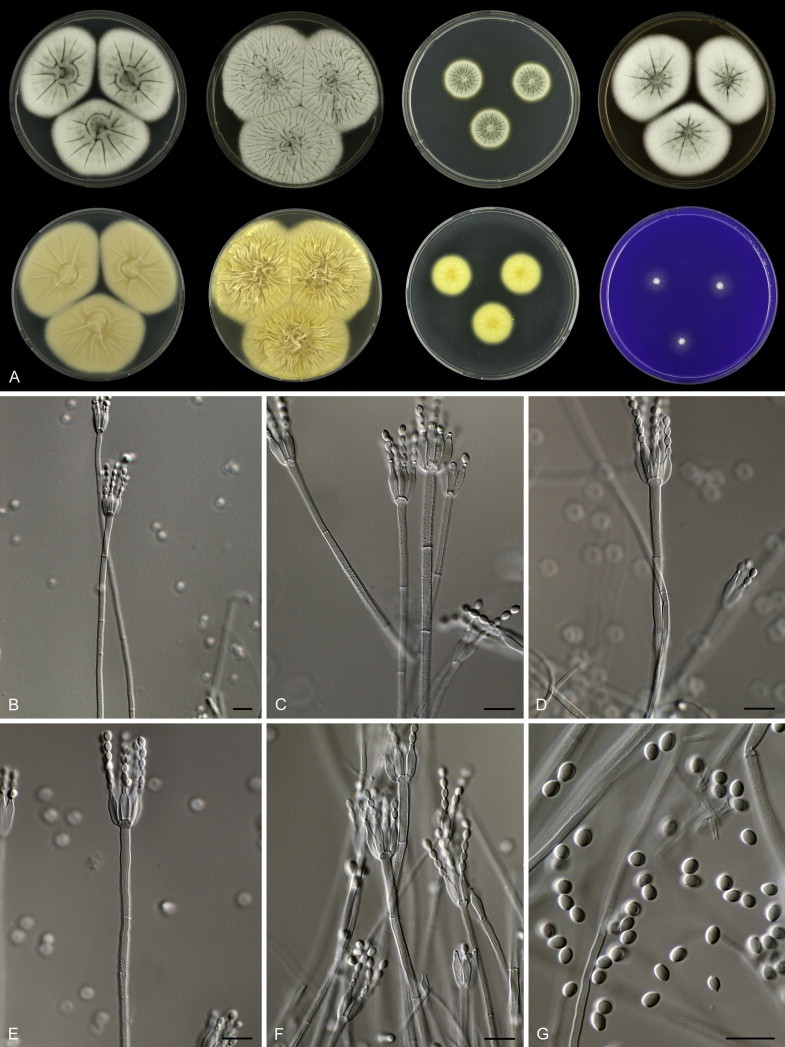
Penicillium contaminatum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Named after the origin of the type strain, a culture contaminant.
Diagnosis: The species is phylogenetically closely related to P. yezoense, but differs by more restricted growth on DG18 (21–25 vs 30–39 mm) and CYAS (30–35 vs 37–47 mm), less sporulation on YES and CYAS and conidia that are broadly ellipsoidal compared to the ellipsoidal conidia of P. yezoense. The reverse of P. contaminatum on YES is in shades of yellow, while that of P. yezoense is often in brown shades.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium thomii-clade.
Typus: United Kingdom, Kew, Surrey, culture contaminant (holotype CBS H-21866, culture ex-type CBS 345.52 = DTO 091-A3 = IMI 049057).
Barcode and molecular based ID: ITS barcode: KM189554 (alternative markers: BenA = KM088793; CaM = KM089178; RPB2 = KM089565).
Description: Colony diam, 7 d, in mm: CYA 42–55; CYA15°C 16–22; CYA30°C 28–38; CYA37°C no growth; MEA 46–50; YES 51–57; DG18 21–25; CYAS 30–35; ratio CYAS:CYA 0.61–0.73; CREA 11–17, weak growth and no acid production.
Weak sporulation on CYA; colony texture velvety, radially sulcate, deep; conidia grey green; mycelium white; exudate absent or present in the centre as small clear droplets; soluble absent or brown (CBS 346.59); margin entire to slightly irregular; reverse cream or brown. Sporulation on YES absent or poor, conidia dull grey green; mycelium white; soluble pigments absent; reverse yellow. Moderate sporulation on DG18, conidia dull green; reverse (bright) yellow. Weak sporulation on MEA, colony texture velvety to slightly floccose; conidia dull green; exudate absent; reverse with yellow centre, colour medium under margins unaffected. Ehrlich reaction yellow.
Sclerotia absent. Conidiophores 150–250 μm long, apices vesiculate 4–7 μm diam, roughened, monoverticillate; stipe 2.5–3.5 μm wide. Phialides ampulliform with short narrow neck, 10–14(–16) per stipe, 8.5–11.5 × 2.5–3.5 μm. Conidia in long irregular columns, broadly ellipsoidal, smooth when young, finely roughened in older parts of the colony.
Penicillium flavisclerotiatum Visagie, Houbraken & K. Jacobs, sp. nov. MycoBank MB809963. Fig. 26.
Fig. 26.
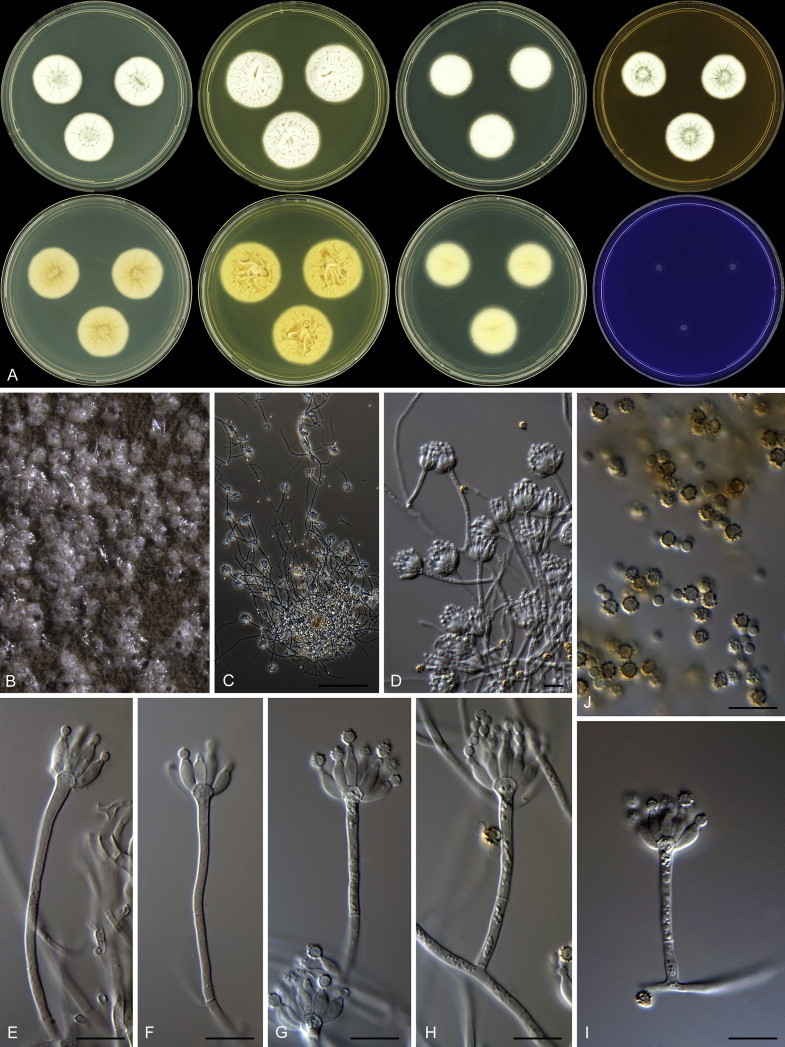
Penicillium flavisclerotiatum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B. Colony texture showing sclerotia. C–I. Conidiophores. J. Conidia. Scale bars = 10 μm.
Etymology: Referring to the yellow sclerotia produced by this species.
Diagnosis: This species belongs to the P. fuscum-clade and is characterised by a slow growth rate on CYA, YES and MEA and the production of (pale) yellow sclerotia.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium fuscum-clade.
Typus: South Africa, Western Cape, Stellenbosch mountain, Fynbos soil, isolated by C.M. Visagie (holotype CBS H-21879, culture ex-type CBS 137750 = DTO 180-I8 = CV 100 = DAOM 241157).
Barcode and molecular based ID: ITS barcode: KM189644 (alternative markers: BenA = KM088888; CaM = KM089275; RPB2 = KM089662).
Description: Colony diam, 7 d, in mm: CYA 23–26; CYA15°C 10–18; CYA30°C 18–21; CYA37°C no growth; MEA 23–28; YES 30–35; DG18 25–27; CYAS 9–10; ratio CYAS:CYA 0.4; CREA 7–8, weak growth, acid not produced.
Sporulation on CYA moderately dense; conidia greyish green to dull green, colony texture velutinous and floccose, low, sulcate, cream to light yellow sclerotia produced; mycelium white; exudate clear; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to greyish orange. Sporulation on YES absent; conidia , colony texture floccose, moderately deep, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to light yellow. Sporulation on DG18 very sparse; conidia white to greenish white, colony texture floccose, low, lightly sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to light yellow. Sporulation on MEA moderately dense; conidia greyish green to dull green, colony texture floccose, low, sulcate, yellow sclerotia sometimes present; mycelium white; exudate absent to clear; soluble pigment not produced; margin low, narrow, entire; reverse brownish yellow to yellowish brown. Ehrlich reaction negative.
Sclerotia produced on CYA and MEA, 80–160 × 70–150 μm. Conidiophores monoverticillate; stipes smooth walled, 23–80 × 2–3.5 μm, vesicles 4–5.5 μm diam. Phialides ampulliform, 8–20 per stipe, 6–9 × 2.5–3.5 μm. Conidia heavy rough to spiny walls, some strains only finely rough walled, globose to somewhat subglobose, 2–3.5 × 2–3.5 μm.
Penicillium grevilleicola Houbraken & Quaedvlieg, sp. nov. MycoBank MB809964. Fig. 27.
Fig. 27.
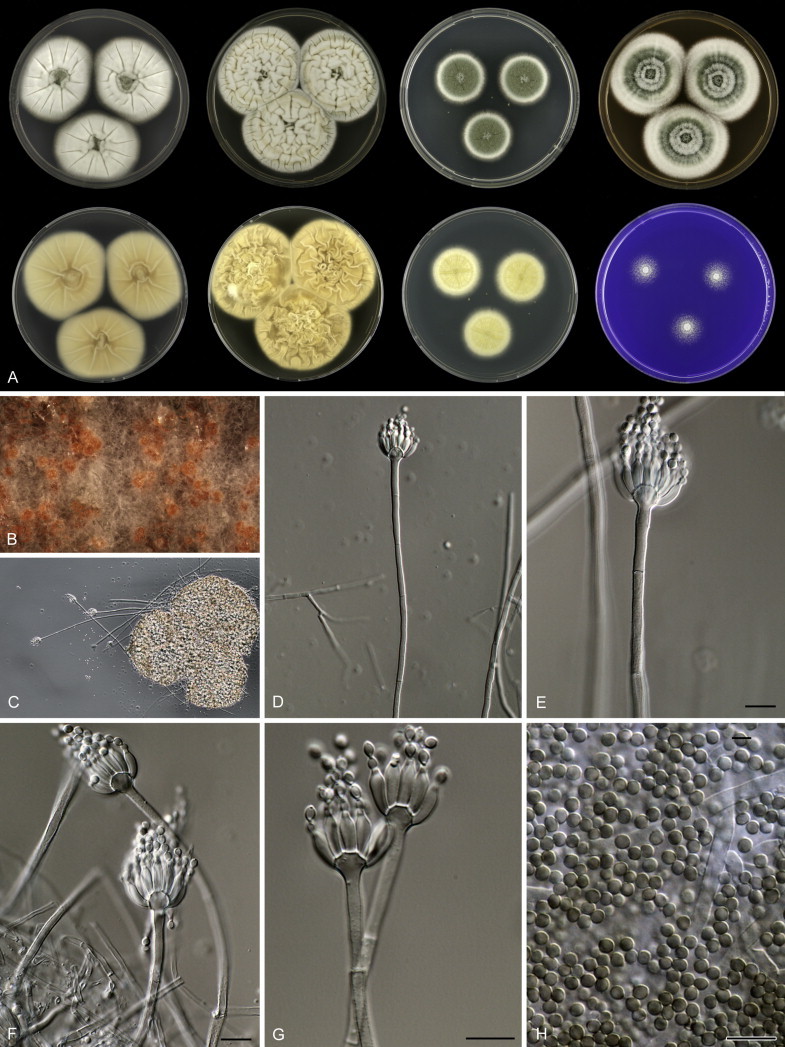
Penicillium grevilleicola, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B, C. Sclerotia. D–G. Conidiophores. H. Conidia. Scale bars = 10 μm.
Etymology: Referring to the host from which the type strain was isolated, Grevillea ilicifolia.
Diagnosis: The species has dark pure green conidia on MEA, whereas the phylogenetically related species P. crocicola, P. austroafricanum, P. jejuense have dull or dull grey-green conidia. Furthermore, P. grevilleicola has strongly floccose colonies and broadly ellipsoidal conidia.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium thomii-clade.
Typus: Australia, Kangaroo Island, Kingscote, leaf of Grevillea ilicifolia, isolated by J. Houbraken & W. Quaedvlieg (holotype CBS H-21871, culture ex-type CBS 137775 = DTO 174-E6).
Barcode and molecular based ID: ITS barcode: KM189630 (alternative markers: BenA = KM088874; CaM = KM089261; RPB2 = KM089648).
Description: Colony diam, 7 d, in mm: CYA 44–48; CYA15°C 26–30; CYA30°C 18–22; CYA37°C no growth; MEA 43–47; YES 48–52; DG18 26–30; CYAS 33–37; ratio CYAS:CYA 0.74–0.78; CREA 18–22, weak growth and no acid production.
Weak sporulation on CYA and only in the centre; colony texture floccose, radially sulcate, deep; mycelium white; exudate present in the centre as small clear droplets; soluble pigments absent; margin entire; reverse pale crème brown with. Sporulation on YES absent or very poor, mycelium white; soluble pigments absent; reverse orange-yellow. Strong sporulation on DG18, conidia dark green; reverse yellow. Moderate to good sporulation on MEA, colony texture floccose; conidia pure green; few exudate droplets, small, pale yellow; reverse with yellow centre, colour medium under margins unaffected; sclerotia present under mycelium and becoming visible after 14 d incubation. Ehrlich reaction negative.
Sclerotia present, orange-brown, 200–350 μm; hard; consisting of polygonal cells; no asci or ascospores observed. Conidiophores 200–400 μm long, stipes strongly vesiculate, 5–8 μm diam, roughened, monoverticillate; stipe 3.0–4.0 μm wide. Phialides ampulliform with short neck, densely packed, up to 18 per stipe, 10–12 × 3.0–3.5 μm. Conidia in long irregular columns, broadly ellipsoidal, smooth to finely roughened.
Penicillium hoeksii Houbraken, sp. nov. MycoBank MB809965. Fig. 28.
Fig. 28.
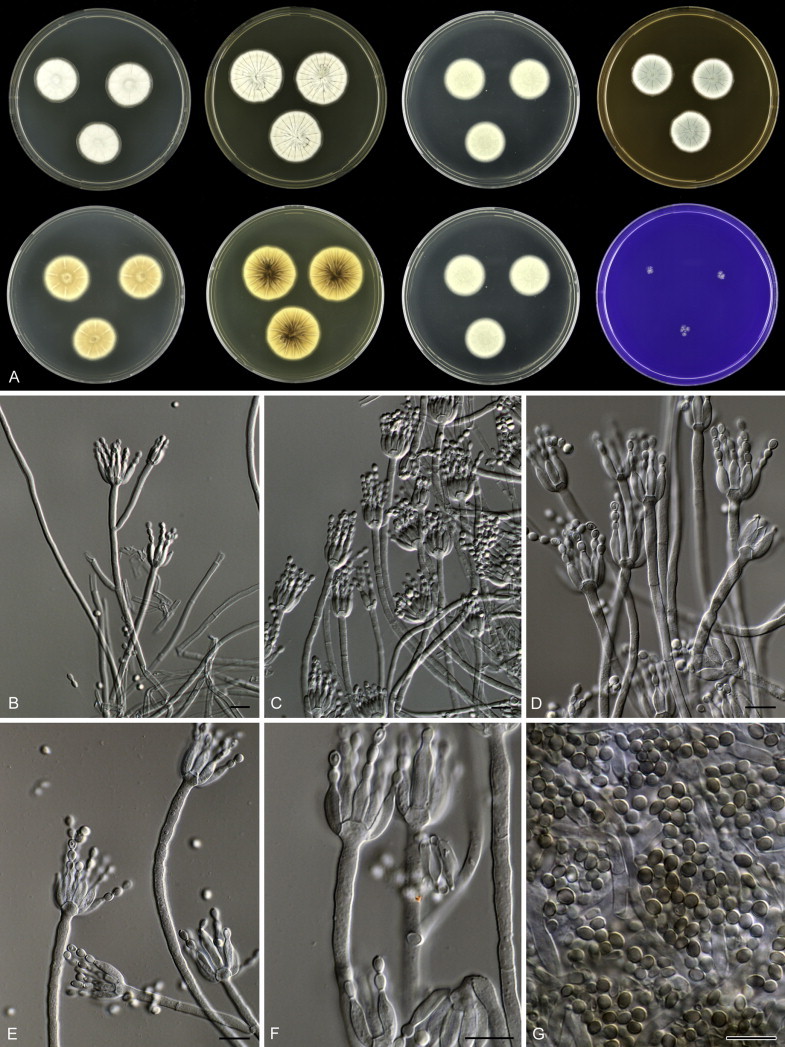
Penicillium hoeksii, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Named after Toon Hoeks, who assisted during the collection of the soil sample from which the type strain was isolated.
Diagnosis: The species is characterised by the production of brown soluble pigments on CYA, OA and/or CYAS, smooth walled stipes, finely roughened (broadly) ellipsoidal conidia and no growth on CYA at 30 °C. This species grows better on CYA at 25 °C than at 15 °C, in contrast to P. zhuangii.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium hoeksii-clade.
Typus: Belgium, Postel, soil under Compact Rush (Juncus conglomeratus), isolated by J. Houbraken (holotype CBS H-21860, culture ex-type CBS 137776 = DTO 192-H4).
Barcode and molecular based ID: ITS barcode: KM189707 (alternative markers: BenA = KM088954; CaM = KM089341; RPB2 = KM089728).
Description: Colony diam, 7 d, in mm: CYA 20–28; CYA15°C 12–18; CYA30°C no growth; CYA37°C no growth; MEA 20–28; YES 26–33; DG18 18–26; CYAS 13–20; ratio CYAS:CYA 0.60–0.67(–0.85); CREA 2–7, weak growth and no acid production.
Sporulation on CYA absent or weak; colony texture velvety, radially sulcate, deep; conidia grey green; mycelium white; exudate absent or present in the centre as small clear or yellow droplets; soluble pigment production strong, brown; margin entire; reverse in shades of brown, sometimes with dark brown centre. Sporulation on YES absent or poor, conidia grey green; mycelium white or pale crème; soluble pigment production weak, brown; reverse brown with dark brown centre. Good sporulation on DG18, colony texture floccose to slightly funiculose; conidia dull green; reverse transparent, pale brown or brown. Moderate sporulation on MEA, colony texture velvety, sometimes with slightly floccose centre; conidia greyish blue green; exudate absent; reverse in shades of brown. Ehrlich reaction negative.
Sclerotia absent. Conidiophores 50–250 μm long, stipes slightly vesiculate up to 5.0 μm diam, smooth walled, monoverticillate, in older parts divaricate with metulae in intercalary positions, intergrading with monoverticillate conidiophores; stipe 3.0–3.5 μm wide. Phialides ampulliform, (1–)2–5(–8) per stipe, 9.0–11.5 × 2.5–3.5 μm. Conidia in short distorted chains, (broadly) ellipsoidal, finely roughened, 3.0–3.7 × 2.5–3.0 μm.
Penicillium infra-aurantiacum Visagie, Houbraken & K. Jacobs, sp. nov. MycoBank MB809966. Fig. 29.
Fig. 29.
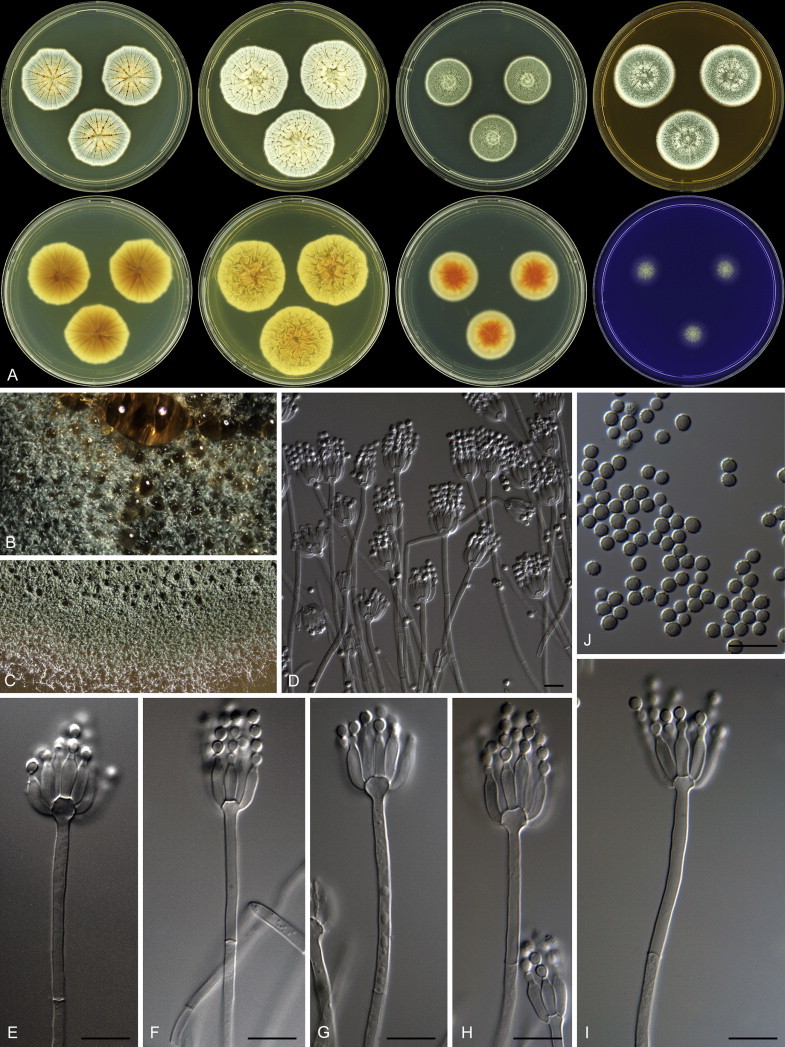
Penicillium infra-aurantiacum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B, C. Sclerotia. D–I. Conidiophores. J. Conidia. Scale bars = 10 μm.
Etymology: Referring to the orange reverse pigmentation that is diagnostic for the species.
Diagnosis: This species is phylogenetically related to P. sublectaticum and P. malmesburiense. Penicillium infra-aurantiacum differs from P. sublectaticum having irregularly shaped colonies on CYA, while those of P. malmesburiense are polygonal in outline.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium sublectaticum-clade.
Typus: South Africa, Western Cape, Malmesbury, Riverlands, bracts of Protea repens infructescence, isolated by C.M. Visagie (holotype CBS H-21880, culture ex-type CBS 137747 = DTO 183-C3 = CV 1518 = DAOM 241145).
Barcode and molecular based ID: ITS barcode: KM189684 (alternative markers: BenA = KM088930; CaM = KM089317; RPB2 = KM089704).
Description: Colony diam, 7 d, in mm: CYA 30–34; CYA15°C 21–23; CYA30°C 12–15; CYA37°C no growth; MEA 30–33; YES 33–35; DG18 25–30; CYAS 18–21; ratio CYAS:CYA 0.6; CREA 15–16, weak growth, no acid production.
Sporulation on CYA moderately dense; conidia greyish turquoise, colony texture velutinous and floccose, low, sulcate; margin slightly polygonal; mycelium white; exudate clear to orange; soluble pigment yellow, inconspicuous; margin low, narrow, polygonal in face view; reverse yellowish white to brownish orange to yellowish brown to brown. Sporulation on YES moderately dense; conidia greyish green, colony texture velutinous and floccose, moderately deep, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to yellowish grey to olive to greyish orange. Sporulation on DG18 dense; conidia dark green, colony texture velutinous and floccose, low, slightly sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to greyish green to light yellow to orange. Sporulation on MEA moderately dense; conidia greyish green, colony texture velutinous and floccose, low, sulcate; mycelium white; exudate clear; soluble pigment not produced; margin low, narrow, entire; reverse light yellow to yellowish brown to brown. Ehrlich reaction negative.
Conidiophores monoverticillate; stipes smooth walled, 100–230 × 2–3 μm, vesicles 4.5–6 μm diam. Phialides ampulliform, 10–18 per stipe, 8.5–11 × 2.5–3 μm. Conidia rough walled, globose, 2.5–3.5 μm.
Penicillium kiamaense Houbraken & Pitt, sp. nov. MycoBank MB809967. Fig. 30.
Fig. 30.
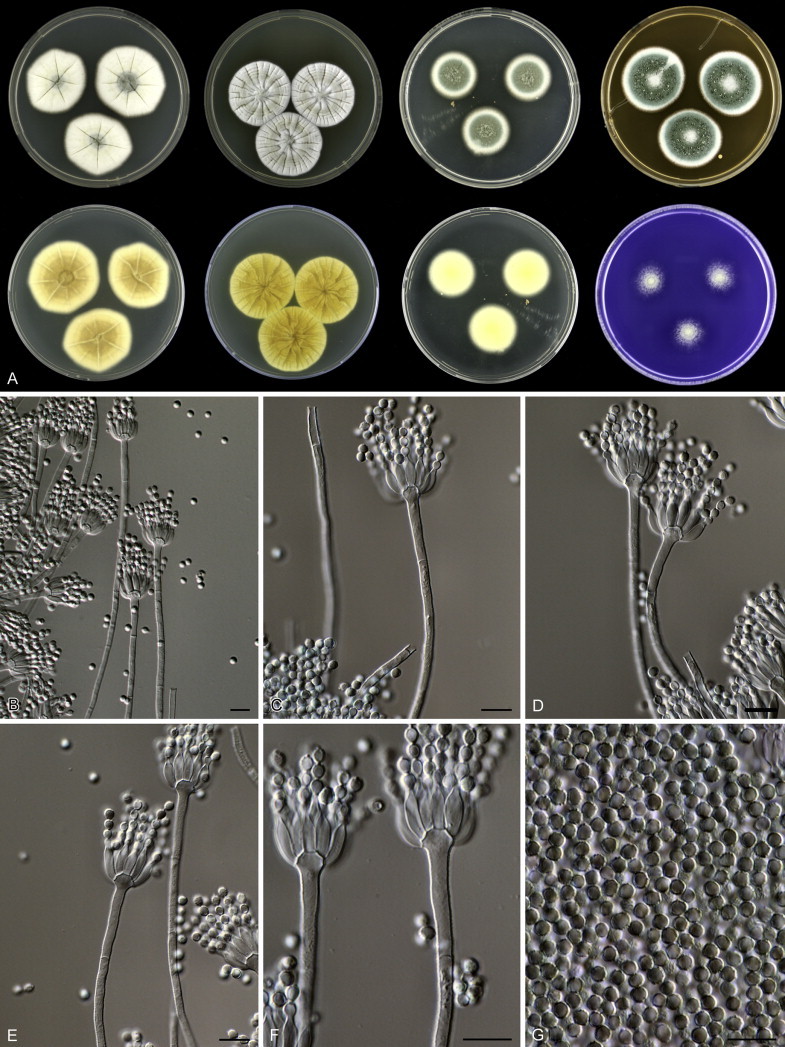
Penicillium kiamaense, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Named after Kiama, the location of the type strain of this species.
Diagnosis: This species is phylogenetically unique and most closely related to species of the P. glabrum- and P. thomii-clades. Phenotypically is most closely related to P. subspinulosum, but can be differentiated by the production of dark (dull) green conidia on OA and MEA and a reverse on YES in shades of orange.
In: subgenus Aspergilloides, section Aspergilloides, undefined clade (basal to P. glabrum and P. thomii-clade).
Typus: Australia, NSW, Barren Grounds Nature Reserve, near Kiama, soil, isolated by J.I. Pitt (holotype CBS H-21857, culture ex-type CBS 137947 = FRR 6087 = DTO 056-I6).
Barcode and molecular based ID: ITS barcode: KM189506 (alternative markers: BenA = KM088743; CaM = KM089128; RPB2 = KM089515).
Description: Colony diam, 7 d, in mm: CYA 33–37; CYA15°C 22–26; CYA30°C 12–16; CYA37°C no growth; MEA 33–37; YES 37–41; DG18 23–27; CYAS 24–28; ratio CYAS:CYA 0.73–0.75; CREA 18–22, weak growth and no acid production.
Sporulation on CYA weak, only in the centre; colony texture velvety, radially sulcate, deep; conidia dull grey-green; mycelium white; exudate present in the centre as small pale yellow droplets; soluble pigment production weak, yellow-brown; colony in face view polygonal; reverse yellowish brown in the centre, brown at the margins. Sporulation on YES poor; mycelium white to pale cream; soluble pigment production strong, orange-brown; reverse orange with an orange-brown centre. Good sporulation on DG18, colony texture crustose; conidia dull green; reverse yellow. Good sporulation on MEA, colony texture velvety; conidia dark green; exudate present, pale; reverse with yellow centre, colour medium under margins unaffected. Ehrlich reaction negative.
Sclerotia absent. Conidiophores 50–250 μm long, stipes vesiculate up to 6.0 μm diam, finely rough walled, predominantly monoverticillate, sometimes with a short branch up to 14 μm long, stipe 3.0–4.0 μm wide. Phialides ampulliform, 5–12 per stipe, 9.0–11.0 × 2.5–3.5 μm. Conidia in moderately long chains, globose to subglobose, distinctly ornamented with striations, 3.0–3.5 μm.
Penicillium longicatenatum Visagie, Busby, Houbraken & K. Jacobs, sp. nov. MycoBank MB809968. Fig. 31.
Fig. 31.
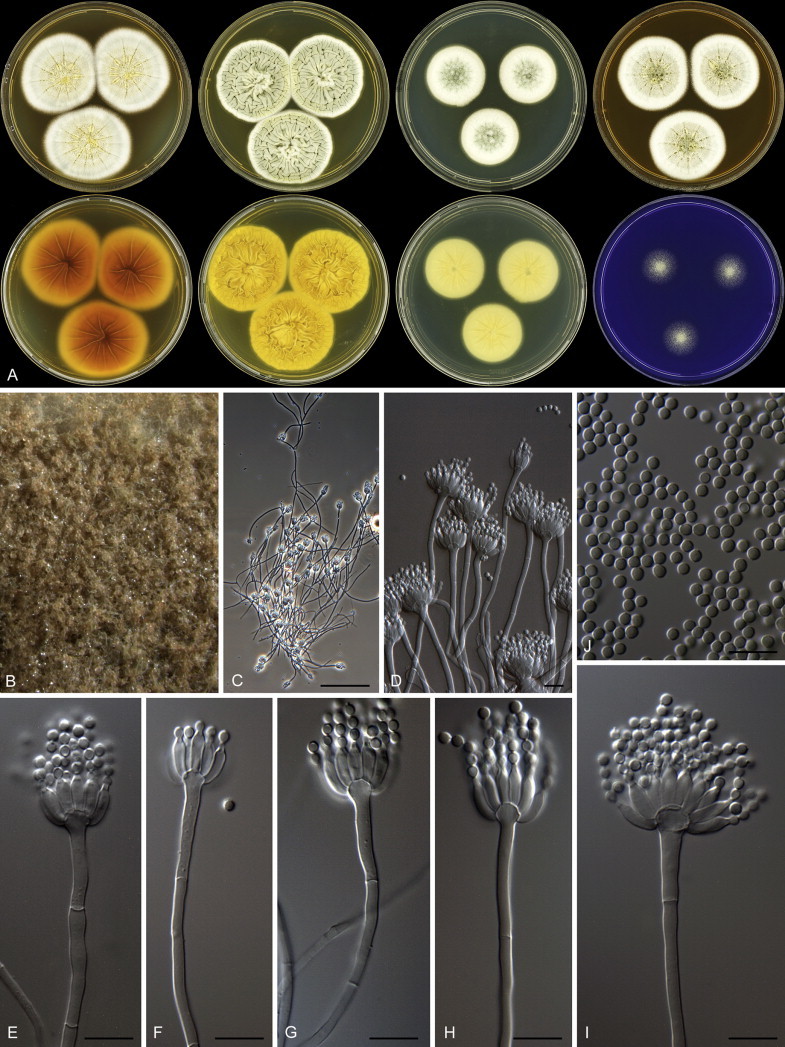
Penicillium longicatenatum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B. Colony texture. C–I. Conidiophores. J. Conidia. Scale bars = 10 μm.
Etymology: Referring to the long chains of conidia produced by this species in culture.
Diagnosis: This species is phylogenetically unique and in common with other species of the P. thomii-clade, it produces sclerotia on CYA and MEA and grows well on CYA at 30 °C. Conidia are subglobose.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium vagum-clade.
Typus: South Africa, Western Cape, Malmesbury, Riverlands, Fynbos, soil, isolated by C.M. Visagie (holotype CBS H-21875, culture ex-type CBS 137735 = DTO 180-D9 = CV 2847 = DAOM 241119).
Barcode and molecular based ID: ITS barcode: KM189636 (alternative markers: BenA = KM088880; CaM = KM089267; RPB2 = KM089654).
Description: Colony diam, 7 d, in mm: CYA 40–45; CYA15°C 23–27; CYA30°C 25–35; CYA37°C no growth; MEA 35–45; YES 45–50; DG18 30–35; CYAS 35–45; ratio CYAS:CYA 0.9–1; CREA 15–20, weak growth, acid not produced.
Sporulation on CYA moderately dense; conidia greyish green to dull green to greyish green, colony texture velutinous and floccose, low, sulcate, cream to light brown sclerotia produced; mycelium white; exudate clear to yellow; soluble pigment yellow to yellowish brown; margin low, narrow, entire; reverse greyish yellow near margin, elsewhere brown. Sporulation on YES moderately dense; conidia greyish green to dull green, colony texture velutinous, low to moderately deep, sulcate; mycelium white; exudate absent; soluble pigment yellow; margin low, narrow, entire; reverse light yellow to greyish yellow. Sporulation on DG18 moderately dense to dense; conidia greyish green to dull green to greyish green, colony texture velutinous, low, slightly sulcate; mycelium white; exudate absent; soluble pigment yellow; margin low, narrow, entire; reverse light yellow to greyish green. Sporulation on MEA moderately dense; conidia greyish green to dull green to greyish green, colony texture velutinous and floccose, low to moderately deep, sulcate, cream to greyish brown sclerotia present; mycelium white; exudate clear to yellow; soluble pigments not produced; margin low, narrow to wide, entire; reverse greyish orange to brownish yellow to yellowish brown. Ehrlich reaction negative.
Sclerotia produced on CYA and MEA, 50–250 × 40–250 μm. Conidiophores monoverticillate; stipes smooth walled, 60–330 × 2.5–3.5 μm, vesicles 4–9 μm diam. Phialides ampulliform, 12–25 per stipe, 7–11 × 2.5–4 μm. Conidia finely rough walled, subglobose, 2.5–3.5 × 2–3 μm.
Penicillium malmesburiense Visagie, Houbraken & K. Jacobs, sp. nov. MycoBank MB809969. Fig. 32.
Fig. 32.
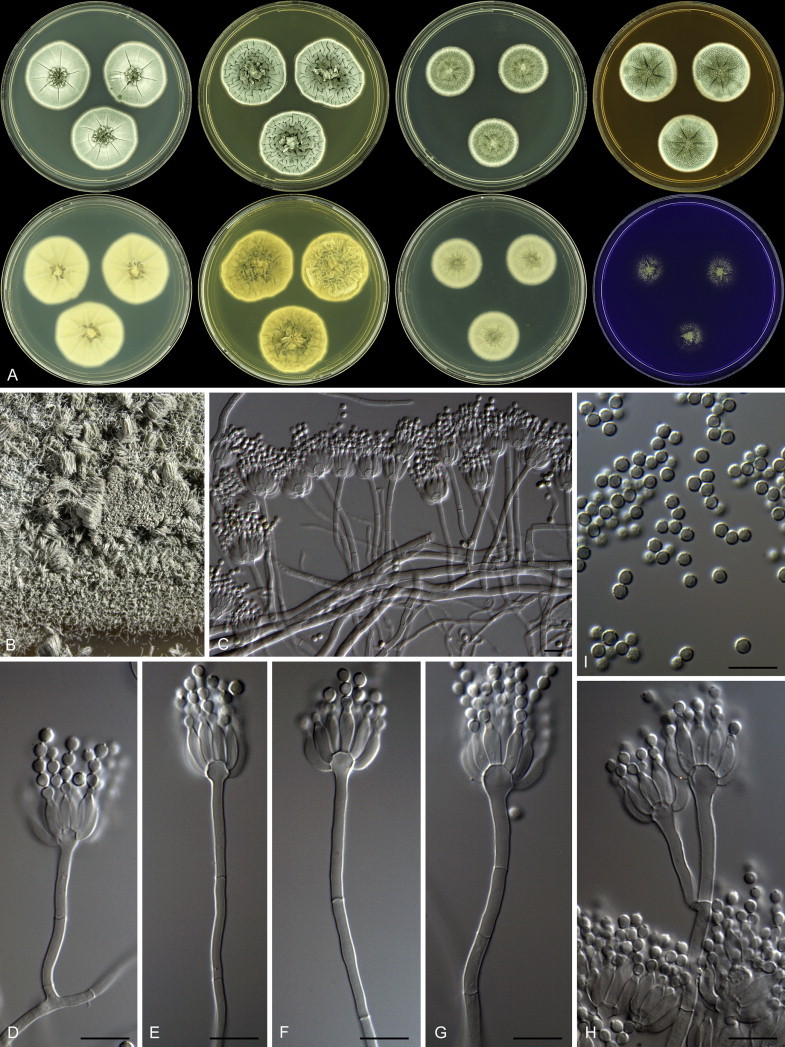
Penicillium malmesburiense, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B. Colony texture. C–H. Conidiophores. I. Conidia. Scale bars = 10 μm.
Etymology: Referring to the collection site of the type strain, Malmesbury.
Diagnosis: The species is phylogenetically closely related to P. infra-aurantiacum and P. sublectaticum, but differs by its pale reverse on CYA, CYAS and DG18.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium sublectaticum-clade.
Typus: South Africa, Western Cape, Malmesbury, Riverlands, mite from Protea repens infructescence, isolated by C.M. Visagie (holotype CBS H-21872, culture ex-type CBS 137744 = DTO 182-H5 = CV 1180 = DAOM 241144).
Barcode and molecular based ID: ITS barcode: KM189676 (alternative markers: BenA = KM088921; CaM = KM089308; RPB2 = KM089695).
Description: Colony diam, 7 d, in mm: CYA 34–35; CYA15°C 20–23; CYA30°C 20–22; CYA37°C no growth; MEA 30–33; YES 34–35; DG18 24–26; CYAS 24–28; ratio CYAS:CYA 0.7–0.8; CREA 15–20, weak growth, acid not produced.
Sporulation on CYA moderately dense; conidia dull green to greyish green, colony texture velutinous, low, sulcate; mycelium white; exudate clear; soluble pigment not produced; margin low, narrow, entire; reverse greenish grey to greyish yellow, with dark olive spots at centre. Sporulation on YES moderately dense; conidia dull green to dark green, colony texture velutinous, moderately deep, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to olive. Sporulation on DG18 moderately dense; conidia dull green to dark green, colony texture velutinous with some floccose areas, low, slightly sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse greenish grey to greyish yellow, with dark olive spots at centre. Sporulation on MEA moderately dense to dense; conidia greyish green to dark green, colony texture velutinous, low, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish brown to brown. Ehrlich reaction negative.
Conidiophores monoverticillate; stipes smooth walled, 35–115 × 2–3 μm, vesicles 4–6.5 μm diam. Phialides ampulliform, 10–20 per stipe, 8.5–10 × 3–3.5 μm. Conidia finely rough walled, subglobose, 2.5–3.5 × 2.5–3 μm.
Penicillium pulvis Houbraken, Visagie, Samson & Seifert, sp. nov. MycoBank MB809970. Fig. 33.
Fig. 33.
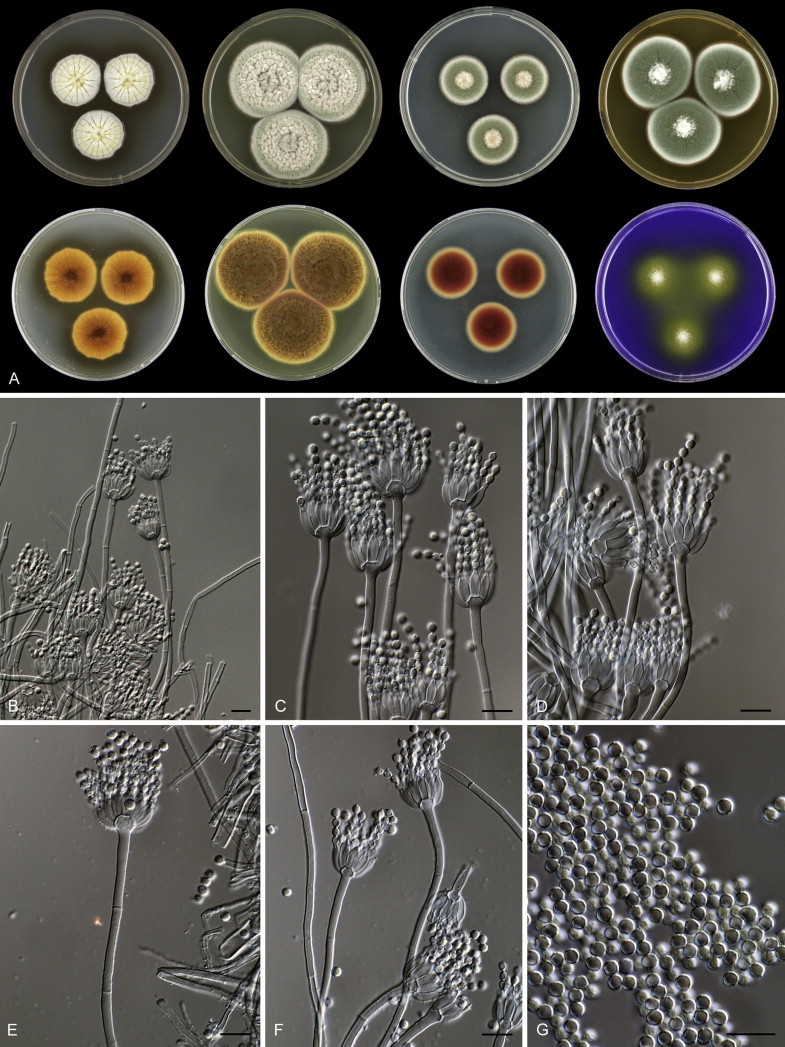
Penicillium pulvis, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Referring to dust, the substrate from which the type strain was isolated.
Diagnosis: This species grows more restrictedly on CYA than other species of the P. glabrum-clade. It has a (dark) brown reverse on CYA, YES and DG18 and produces brown soluble pigment on CYA.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium glabrum-clade.
Typus: South Africa, Kuils River, house dust, collected by K. Jacobs, isolated by E. Whitfield & K. Mwange (holotype CBS H-21878, culture ex-type CBS 138432 = DTO 180-B7).
Barcode and molecular based ID: ITS barcode: KM189632 (alternative markers: BenA = KM088876; CaM = KM089263; RPB2 = KM089650).
Description: Colony diam, 7 d, in mm: CYA 26–32; CYA15°C 22–28; CYA30°C 24–30; CYA37°C no growth; MEA 37–43; YES 39–45; DG18 23–29; CYAS 21–27; ratio CYAS:CYA 0.75–0.90; CREA 19–25, weak growth and moderate acid production.
Sporulation on CYA weak; radially sulcate, deep; mycelium pale beige; exudate present as small orange-yellow droplets; soluble pigment production strong, brown; margin irregular; reverse brown, with dark brown centre. Sporulation on YES absent weak, mycelium white; conidia grey green; soluble pigment not produced; reverse dark brown. Moderate to good sporulation on DG18; colony texture velvety to slightly floccose; conidia dull green; reverse dark brown. Good sporulation on MEA, colony texture velvety; conidia dull green; exudate absent; reverse dark brown in the centre, reverse colour not affecting medium. Ehrlich reaction negative.
Sclerotia absent. Conidiophores, 100–300 μm long, apices vesiculate up to 6.5 μm wide, smooth to finely walled, strictly monoverticillate, stipe 2.5–3.5 μm wide. Phialides ampulliform, 4–16 per stipe, 9.0–10.0 × 2.5–3.0 μm. Conidia in long well-defined chains, globose to subglobose, finely ornamented, 2.7–3.2 μm.
Penicillium ranomafanaense Houbraken & Hagen, sp. nov. MycoBank MB809971. Fig. 34.
Fig. 34.
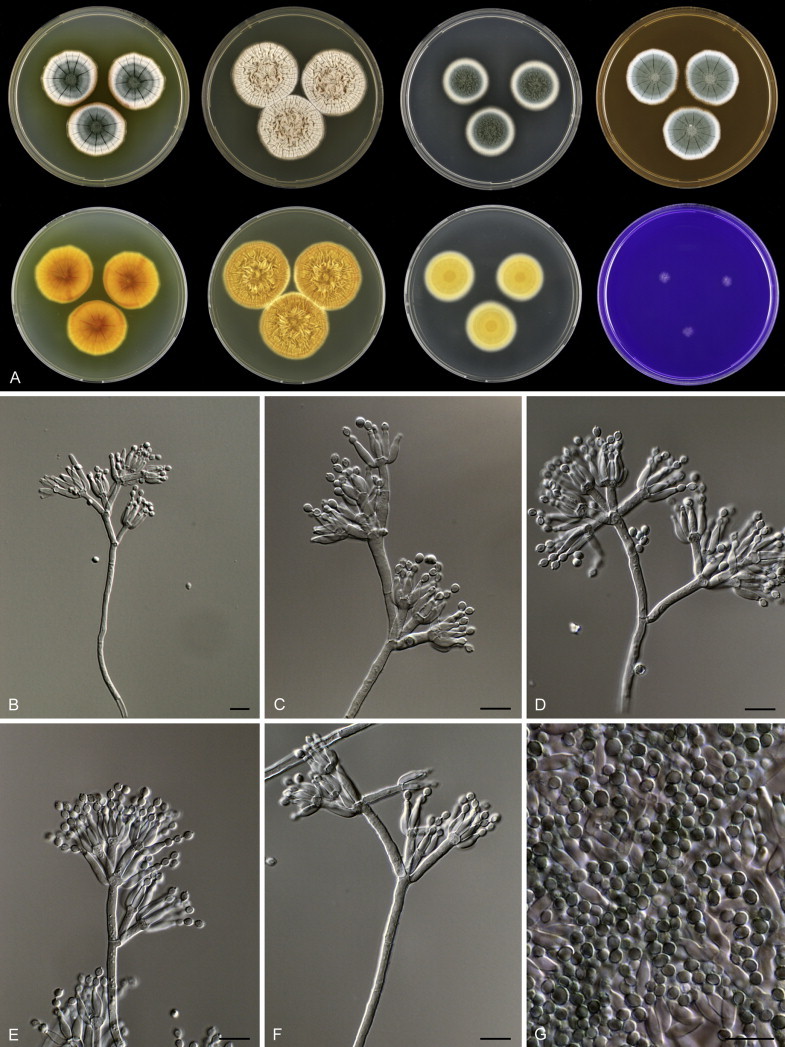
Penicillium ranomafanaense, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Named after Ranoma fana, the location of the type specimen.
Diagnosis: The species is phylogenetically related to P. verhagenii, but differs in having an orange reverse on DG18 and smooth to finely roughened stipes.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium verhagenii-clade.
Typus: Madagascar, Ranoma fana, soil, isolated by F. Hagen & J. Houbraken (holotype CBS H-21862, culture ex-type CBS 137953 = DTO 085-A5).
Barcode and molecular based ID: ITS barcode: KM189541 (alternative markers: BenA = KM088779; CaM = KM089164; RPB2 = KM089551).
Description: Colony diam, 7 d, in mm: CYA 26–33; CYA15°C 20–26; CYA30°C no growth; CYA37°C no growth; MEA 25–33; YES 33–43; DG18 22–30; CYAS 12–27; ratio CYAS:CYA 0.50–0.85; CREA 5–10, weak growth and no acid production.
Sporulation on CYA weak to moderate; colony texture velvety, radially sulcate, deep; conidia dull green becoming dark green in the centre; mycelium white; exudate absent; soluble pigment production strong, yellow to orange; margin entire or slightly polygonal in face view; reverse yellow-orange with orange-red centre. Sporulation on YES absent; mycelium pale crème; soluble pigment not produced; reverse orange. Good sporulation on DG18, colony texture slightly floccose; conidia dull to dark green; mycelium white, reverse orange. Good sporulation on MEA, colony texture velvety; conidia dark green in the centre, towards dull green at the edge, always with a blue element; exudate absent; reverse not affecting the medium. Ehrlich reaction negative.
Sclerotia absent. Conidiophores 100–250 μm long, with smooth or finely roughened stipes; irregularly branched, predominantly symmetrically biverticillate, older conidiophores becoming divaricate due to having secondary growth of the stipe by proliferation at the apex, stipe 2.5–3.5 μm wide. Metulae in terminal whorl of 3–6, of unequal length, (9.0–)11.0–16.5(–22) × 2.5–3.5 μm. Phialides, two types present, predominantly ampulliform, short, 8.5–9.5 × 2.5–3.5 μm, also larger phialides present, cylindrical, (9–)10.5 × 14(–16) μm, (2–)4–10(–14) per metulae. Conidia in short distorted chains, roughened, occasionally with striations, variable in shape: subglobose to broadly ellipsoidal in DTO 085-A5, 2.5–3.0 μm in size, (broadly) ellipsoidal in DTO 085-A8 2.5–3.0 × 2.0–2.7 μm.
Penicillium rudallense Houbraken, Visagie & Pitt, sp. nov. MycoBank MB809972. Fig. 35.
Fig. 35.
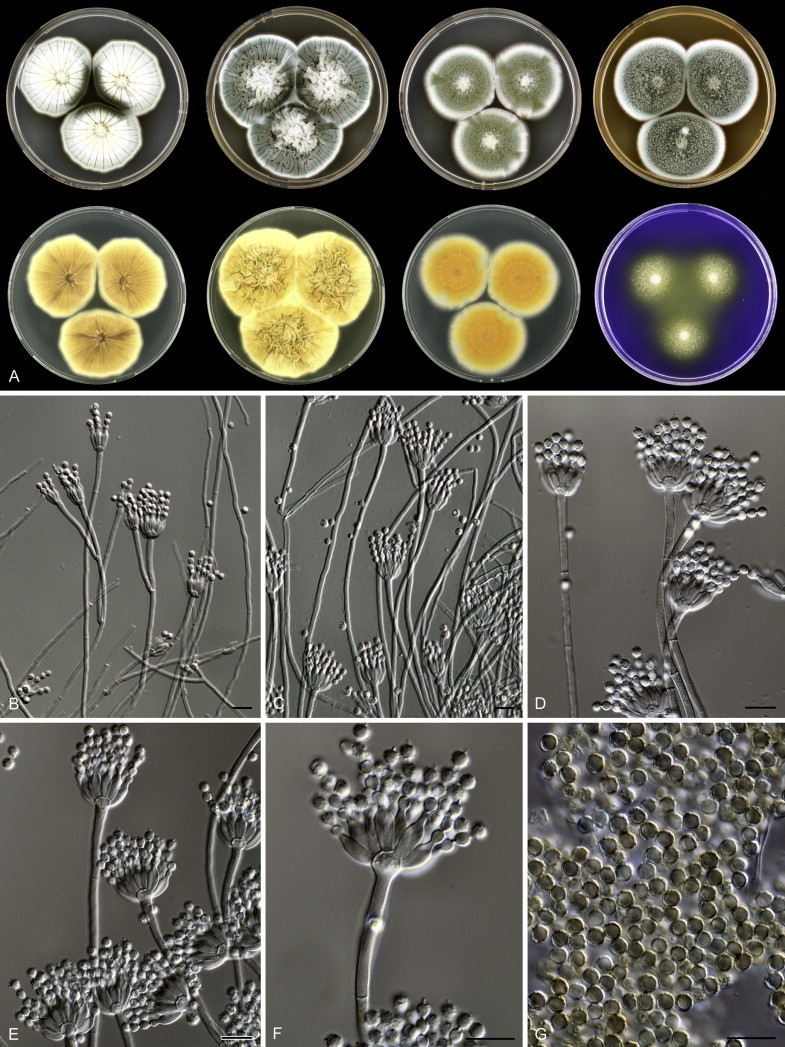
Penicillium rudallense, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Referring to the location of the type strain, Rudall River National Park.
Diagnosis: This species belongs phylogenetically to the P. glabrum-clade and can be differentiated by the production of distinctly ornamented, dark green conidia on CYA and MEA, and good growth on CYA.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium glabrum-clade.
Typus: Australia, WA, Rudall River National Park, soil, isolated by A.D. Hocking (holotype CBS H-21867, culture ex-type CBS 138162 = FRR 6085 = DTO 056-I4).
Barcode and molecular based ID: ITS barcode: KM088741 (alternative markers: BenA = KM089126; CaM = KM189504; RPB2 = KM089513).
Description: Colony diam, 7 d, in mm: CYA 35–46; CYA15°C 22–28; CYA30°C 25–37; CYA37°C 0–7; MEA 39–46; YES 48–54; DG18 35–43; CYAS 35–40; ratio CYAS:CYA 0.80–0.92; CREA 18–32, poor growth and moderate acid production.
Sporulation on CYA good; radially sulcate, deep; colony texture velvety; conidia dark green; mycelium white; exudate absent or present as yellow droplets; soluble pigment absent or light brown; margin slightly polygonal; reverse (pale) orange-brown or brown with dark brown centre. Sporulation on YES strong; conidia dark dull green, mycelium beige; soluble pigments absent; reverse yellow, orange-yellow or crème with brown centre. Good sporulation on DG18, colony texture granular or floccose; conidia dull green; reverse pale or orange. Good sporulation on MEA, colony texture velvety, sometimes slightly floccose; conidia dark green; exudate absent. Ehrlich reaction negative.
Sclerotia absent. Conidiophores 150–400 μm long, stipes slightly vesiculate in young parts of the colony, inflated in older parts, up to 6.0 μm, smooth or finely rough walled, predominantly monoverticillate, sometimes symmetrically biverticillate or with additional branch, stipe 2.0–3.0 μm wide. Phialides ampulliform with short neck, 6–14 per stipe, 8.0–10.0 × 2.5–3.5 μm. Conidia in long well-defined chains, globose, distinctly ornamented, slightly striated and inner and outer cell wall visible, 3.0–3.5 μm.
Penicillium sterculiniicola Houbraken, sp. nov. MycoBank MB809973. Fig. 36.
Fig. 36.
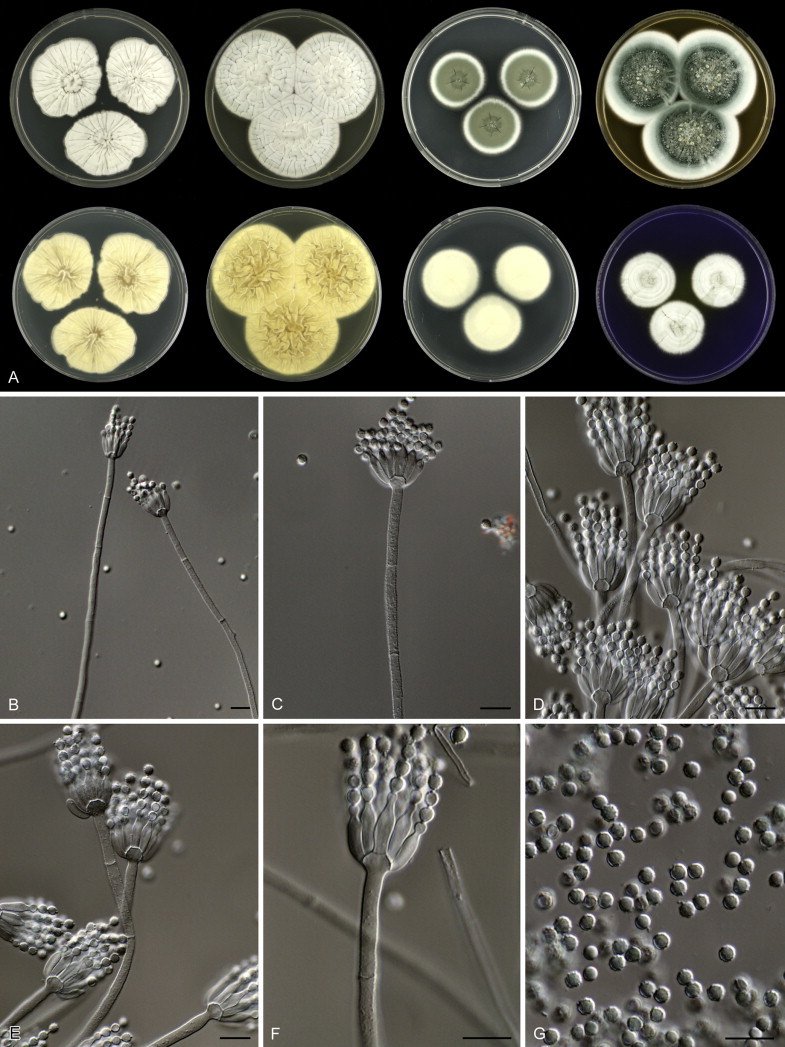
Penicillium sterculiniicola, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Named after compost (sterculinium), the substrate from which the type was isolated.
Diagnosis: This species grows well on CREA, colonies at CYA at 30 °C after 7 d (29–) are 37–47 mm diam, and no sporulation occurs on CYA and YES. Stipes are rough walled and the conidia 2.7–3.7 μm diam.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium spinulosum-clade.
Typus: USA, spawn run compost (holotype CBS H-21877, culture ex-type CBS 122426 = DTO 031-A4).
Barcode and molecular based ID: ITS barcode: KM189464 (alternative markers: BenA = KM088693; CaM = KM089078; RPB2 = KM089465).
Description: Colony diam, 7 d, in mm: CYA (25–)39–45; CYA15°C 17–22; CYA30°C (29–)37–47; CYA37°C no growth; MEA 37–55; YES 35–45; DG18 22–35; CYAS 20–28; ratio CYAS:CYA 0.55–0.63(–0.84); CREA 24–34, good growth and weak to moderate acid production, followed by a delayed base production.
Sporulation on CYA absent or very weak; radially sulcate, deep; mycelium white; exudate absent; soluble pigment not produced; margin entire, irregular in DTO 216-I4; reverse in shades of brown, pale brown, crème or brown. Sporulation on YES absent, mycelium white; soluble pigment not produced; reverse yellow-brown. Good sporulation on DG18, colony texture crustose; conidia dull green; reverse pale or transparent. Good sporulation on MEA, colony texture floccose, occasionally velvety to floccose; conidia dark green or dark dull green; exudate present as large droplets, clear or pale brown. Ehrlich reaction negative.
Sclerotia absent. Conidiophores 50–250 μm long, apices vesiculate in older parts of the colony, less pronounced in younger parts, up to 6.5 μm diam, stipe finely roughened to distinct rough walled, predominantly monoverticillate, sometimes with divergent additional branch up to 15 μm long, stipe 2.5–3.5 μm wide. Phialides ampulliform, 6–14 per stipe, 9.0–11.0 × 2.5–3.5 μm. Conidia in long distorted chains, globose, distinctly ornamented with striations, 2.5–4.0 μm.
Penicillium sublectaticum Houbraken, Frisvad, Samson & Seifert, sp. nov. MycoBank MB809974. Fig. 37.
Fig. 37.
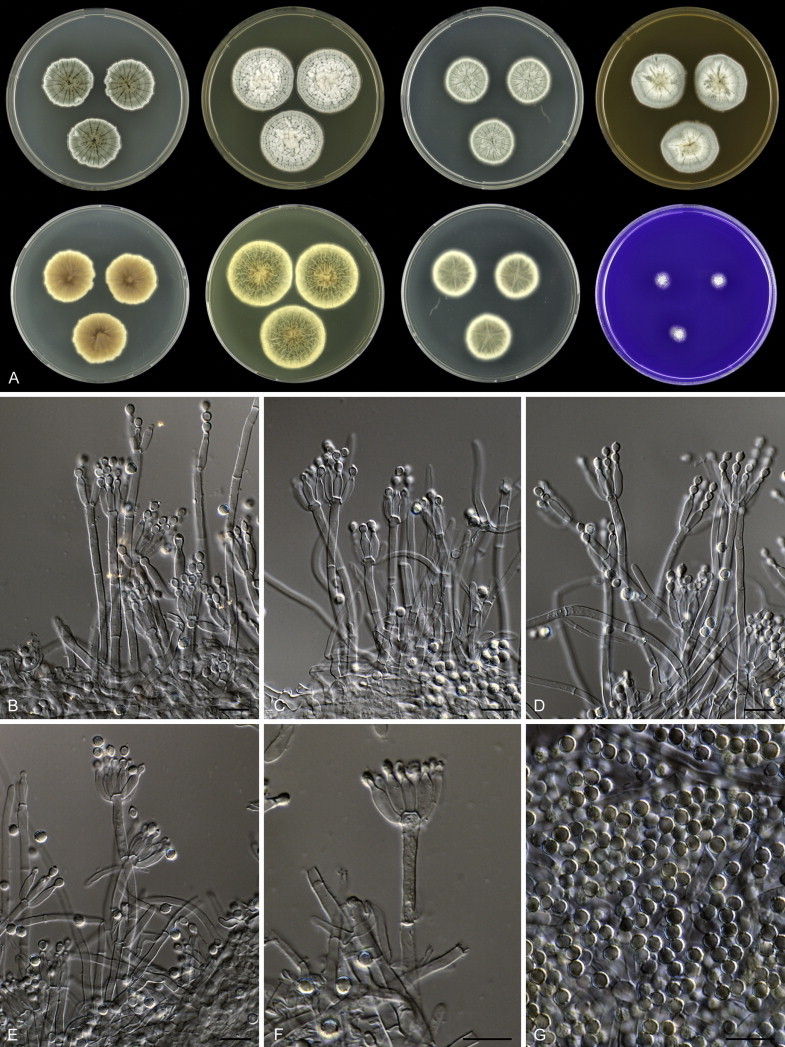
Penicillium sublectaticum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Referring to “under the bed”, the location from where the type strain was isolated from.
Diagnosis: The species is phylogenetically closely related to P. infra-aurantiacum and P. malmesburiense. Phenotypically, it is most similar to P. infra-aurantiacum and differs by irregular margins on CYA and a dark brown reverse on CYA with (yellow) brown margins.
In: subgenus Aspergilloides, section Aspergilloides, P. sublectaticum-clade.
Typus: New Zealand, Dunedin, house dust, collected by T. Atkinson, 2009, isolated by E. Whitfield and K. Mwange (holotype CBS H-21955, culture ex-type: CBS 138217 = DTO 244-G2).
Barcode and molecular based ID: ITS barcode: KM189761 (alternative markers: BenA = KM089010; CaM = KM089397; RPB2 = KM089784).
Description: Colony diam, 7 d, in mm: CYA 25–35; CYA15°C 17–25; CYA30°C 5–15; CYA37°C no growth; MEA 27–35; YES 30–37; DG18 23–30; CYAS 18–25; ratio CYAS:CYA 0.67–0.73; CREA 8–17, weak growth and no acid production.
Moderate sporulation on CYA and only in the centre; colony texture velvety; conidia grey green; radially sulcate, deep; mycelium white; exudate present as red-brown droplets; soluble pigments present, poor, red-brown; margin irregular; reverse brown or dark brown centre with (yellow-)brown edge. Sporulation on YES absent, mycelium white; soluble pigments absent; reverse reddish brown. Good sporulation on DG18; colony texture floccose; conidia dull green; reverse red-brown. Moderate sporulation on MEA, colony texture floccose; conidia dark dull green; few exudate droplets, small, pale yellow or clear; reverse brown. Ehrlich reaction negative.
Sclerotia absent. Conidiophores 40–250 μm long, stipes predominantly non-vesiculate, occasionally slightly vesiculate in older parts of the colony, up to 4.0 μm, smooth walled, predominantly monoverticillate, occasionally with additional monoverticillate branch; stipe 2.5–3.5 μm wide. Phialides ampulliform to cylindrical, (2–)4–12 per stipe, 8–10 × 2.0–3.0 μm. Conidia in moderate to long well-defined columns, globose to subglobose, distinctly ornamented with striations, 3.0–3.5 μm.
Penicillium subspinulosum Houbraken, sp. nov. MycoBank MB809975. Fig. 38.
Fig. 38.
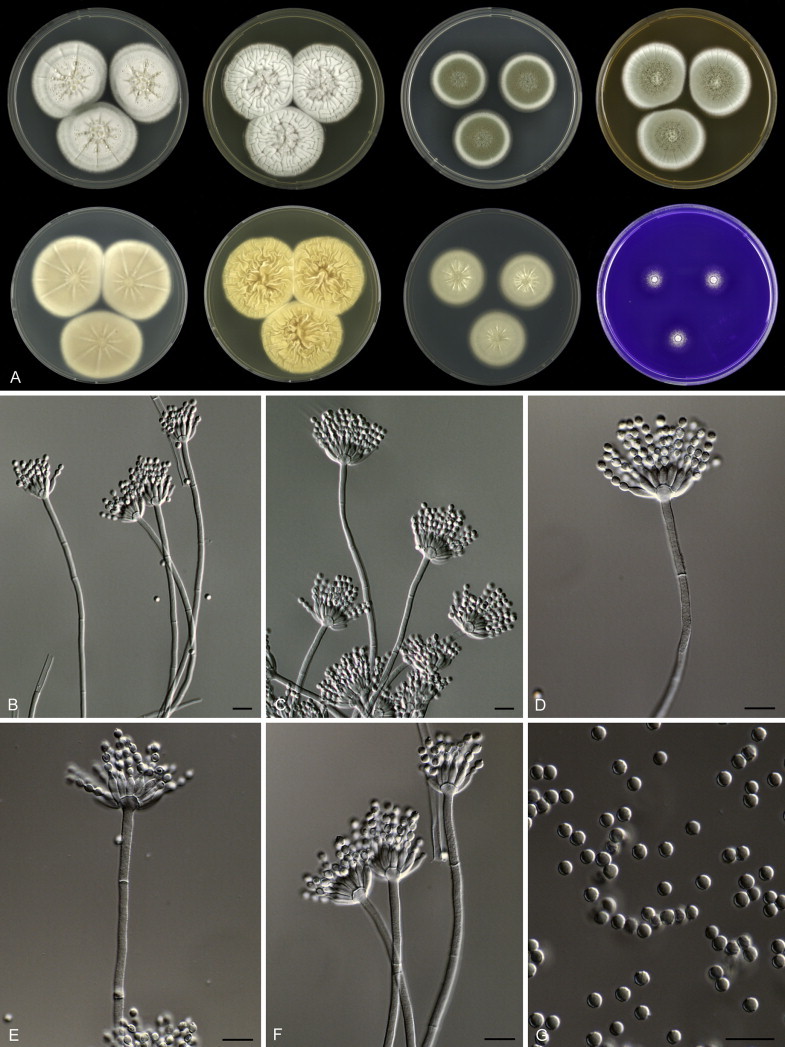
Penicillium subspinulosum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Referring to the close relationship with P. spinulosum.
Diagnosis: The species is phylogenetically close to P. spinulosum and related species, but differs by its poor on CREA, velvety colonies on CYA and restricted growth on CYA30°C (4–18(–22) mm).
In: subgenus Aspergilloides, section Aspergilloides, Penicillium spinulosum-clade.
Typus: Poland, soil, isolated by J. Houbraken & B. Byskal (holotype CBS H-21856, culture ex-type CBS 137946 = DTO 041-F2).
Barcode and molecular based ID: ITS barcode: KM189483 (alternative markers: BenA = KM088719; CaM = KM089104; RPB2 = KM089491).
Description: Colony diam, 7 d, in mm: CYA (24–)36–41; CYA15°C 18–30; CYA30°C 4–18(–22); CYA37°C no growth; MEA (28–)35–42; YES 35–45; DG18 22–35; CYAS 25–35; ratio CYAS:CYA 0.65–0.90; CREA 10–18, poor growth and no acid production.
Sporulation on CYA poor; colony texture velvety, radially sulcate, deep; conidia pure green or dull green; mycelium white; exudate present, clear or pale yellow; soluble pigment absent except in DTO 297-D5 and then pale yellow; margin entire, occasionally lobate; reverse generally in shades of pale brown or cream (pale yellow). Sporulation on YES absent or poor, conidia pale dull green; mycelium white; soluble pigment not produced; reverse cream or pale brown. Good sporulation on DG18, colony texture velvety; conidia dull green; mycelium inconspicuous. Good sporulation on MEA, colony texture floccose in the centre, velvety at the edge; conidia pure green or pure to dull green; exudate if present as clear or pale yellow droplets; reverse brown or not affecting the medium. Ehrlich reaction negative.
Sclerotia absent. Conidiophores 200–400 μm long, with finely roughened stipes, apices vesiculate up to 7 μm diam, predominantly monoverticillate, occasionally with additional branch up to 20 μm long, stipe 2.5–3.5 μm wide. Phialides ampulliform, densely packed, 6–16 per stipe, 7.5–9.5 × 2.0–3.0 μm. Conidia in moderately long, distorted chains, roughened, sometimes with striation or bars, globose, 2.5–3.3 μm.
Penicillium tsitsikammaense Houbraken, sp. nov. MycoBank MB809976. Fig. 39.
Fig. 39.
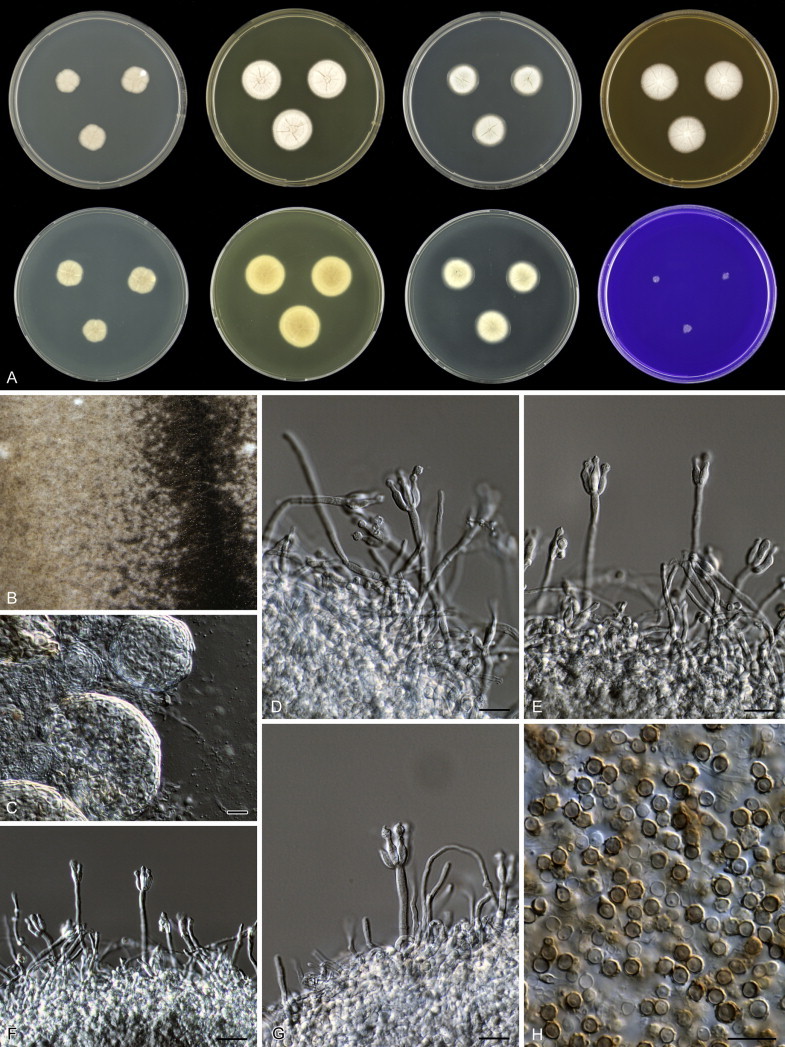
Penicillium tsitsikammaense, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–C. Sclerotia. D–G. Conidiophores. H. Conidia. Scale bars = 10 μm.
Etymology: Named after the collection site of the type strain, Tsitsikamma forest, South Africa.
Diagnosis: The species belongs to the P. fuscum-clade and can be distinguished from the other species by the absence of or poor growth on CYAS (0–2 mm), slow growth rate on CYA and YES, with sporulation absent or poor on all media.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium fuscum-clade.
Typus: South Africa, Cape Province, Tsitsikamma Forest near Knysna, forest soil, isolated by D.B. Scott (holotype CBS H-21881, culture ex-type CBS 328.71 = DTO 006-I3 = CSIR 1092).
Barcode and molecular based ID: ITS barcode: KM189451 (alternative markers: BenA = KM088675; CaM = KM089060; RPB2 = KM089447).
Description: Colony diam, 7 d, in mm: CYA 10–15; CYA15°C 8–12; CYA30°C 5–10; CYA37°C no growth; MEA 19–23; YES 20–25; DG18 17–21; CYAS 0–2; ratio CYAS:CYA 0.15–0.2; CREA 3–5, weak growth and no acid production.
Sporulation on CYA absent; mycelium pale pink or pinkish-beige; exudate absent; soluble pigment not produced; margin lobate (irregular); reverse pale yellow in colony centre, pale beige at the margin. Sporulation on YES absent, mycelium white; soluble pigment not produced; reverse pale yellow. Poor sporulation on DG18; colony texture velvety to floccose; conidia pale green; mycelium white; reverse pale green in the centre, pale at the margins. Sporulation on MEA absent; mycelium white; conidia produced after prolonged incubation, brown-green; sclerotia visible on the edge of the colony periphery, white; exudate absent; reverse centre pale orange and pale yellow at edge; Ehrlich reaction negative.
Sclerotia present on MEA, white, soft, consisting of polygonal cells, (30–)50–120 μm. Conidiophores monoverticillate, short, 25–50 μm long, smooth walled; non-vesiculate. Phialides ampulliform, often with a conspicuous neck, 2–4 per stipe, 8.5–10.0(–13) × 2.5–3.5 μm. Conidia in short distorted chains, globose to subglobose, thick walled, distinctly roughened, a proportion smooth walled, 2.5–3.5 μm diam, a minor proportion larger, up to 5.5 μm.
Notes: Stolk & Samson (1983: 127) reported ascospores in CBS 328.71; however, we did not observe any ascospores on OA, MEA, CYA or YES after ten weeks incubation at room temperature. Cleistothecia of P. fuscum, a closely related species, mature slowly and a longer incubation time also might be needed for P. tsitsikammaense (Scott 1968). Another possibility might be that the strain is degenerated after prolonged maintenance.
Penicillium turcosoconidiatum Visagie, Houbraken & K. Jacobs, sp. nov. MycoBank MB809977. Fig. 40.
Fig. 40.
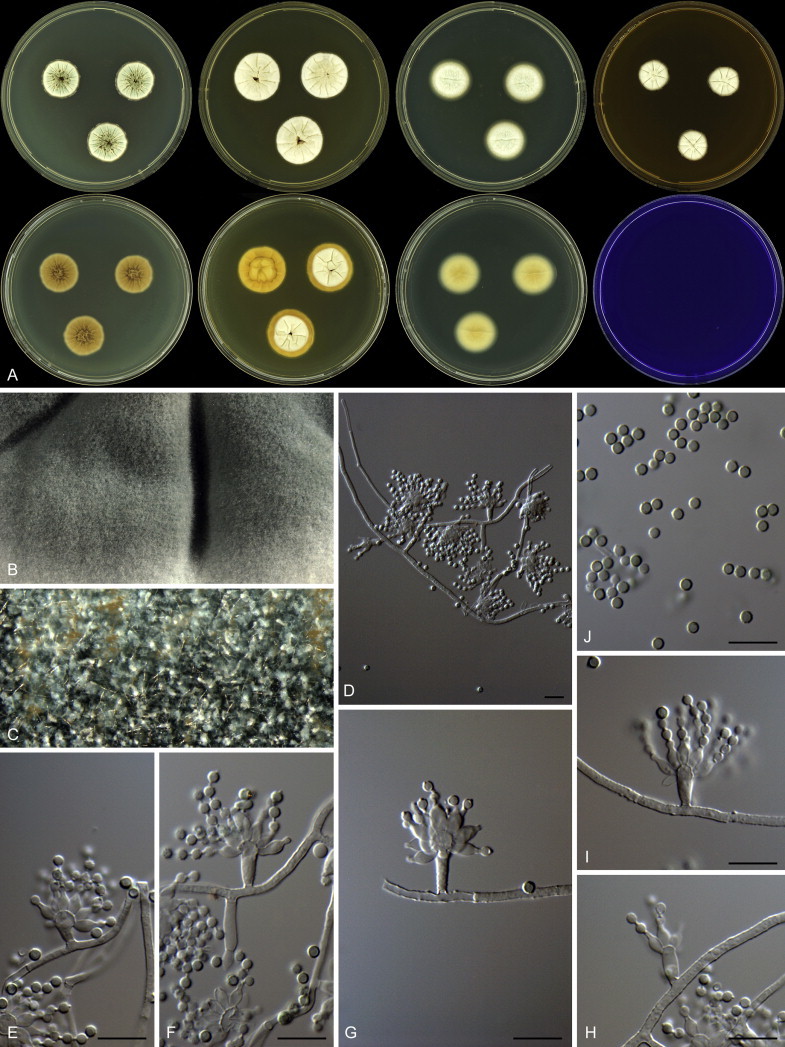
Penicillium turcosoconidiatum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B, C. Colony texture. D–I. Conidiophores. J. Conidia. Scale bars = 10 μm.
Etymology: Named after the light blue turquoise conidia on MEA.
Diagnosis: Penicillium turcosoconidiatum differs from the other species of the P. fuscum-clade by its restricted growth on CYA, YES and MEA, short stipes, size of conidia (2–2.5 μm) and turquoise conidial colour (Table 5).
In: subgenus Aspergilloides, section Aspergilloides, Penicillium fuscum-clade.
Typus: South Africa, Stellenbosch, soil, isolated by C.M. Visagie (holotype CBS H-21876, culture ex-type CBS 138557 = DTO 181-A3 = CV 110 = DAOM 241130).
Barcode and molecular based ID: ITS barcode: KM189645 (alternative markers: BenA = KM088889; CaM = KM089276; RPB2 = KM089663).
Description: Colony diam, 7 d, in mm: CYA 15–20; CYA15°C 7–8; CYA30°C 7–10; CYA37°C no growth; MEA 18–25; YES 22–23; DG18 18–19; CYAS 6–8; ratio CYAS:CYA 0.4; CREA no growth, no acid production.
Sporulation on CYA sparse; conidia greyish green, colony texture velutinous, moderately deep, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to brownish orange. Sporulation on YES absent; colony texture floccose, moderately deep, sulcate, orange white colour; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to greyish yellow. Sporulation on DG18 very sparse; conidia greenish white, colony texture floccose, low, plane; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse pale to pale yellow. Sporulation on MEA absent after 7 d; conidia greyish turquoise after prolonged incubation, colony texture velutinous, low, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse yellowish white to brownish orange. Ehrlich reaction negative.
Sclerotia absent. Conidiophores monoverticillate, mycelia on which conidiophores are borne often rough walled; stipes smooth walled, 6–30 × 1.5–3 μm, vesicles 3–6 μm diam; phialides ampulliform, 12–18 per stipe, 5–6.5 × 2.5–3.5 μm; conidia rough walled, globose, 2–2.5 μm.
Penicillium vagum Houbraken, Pitt, Visagie & K. Jacobs, sp. nov. MycoBank MB809978. Fig. 41.
Fig. 41.
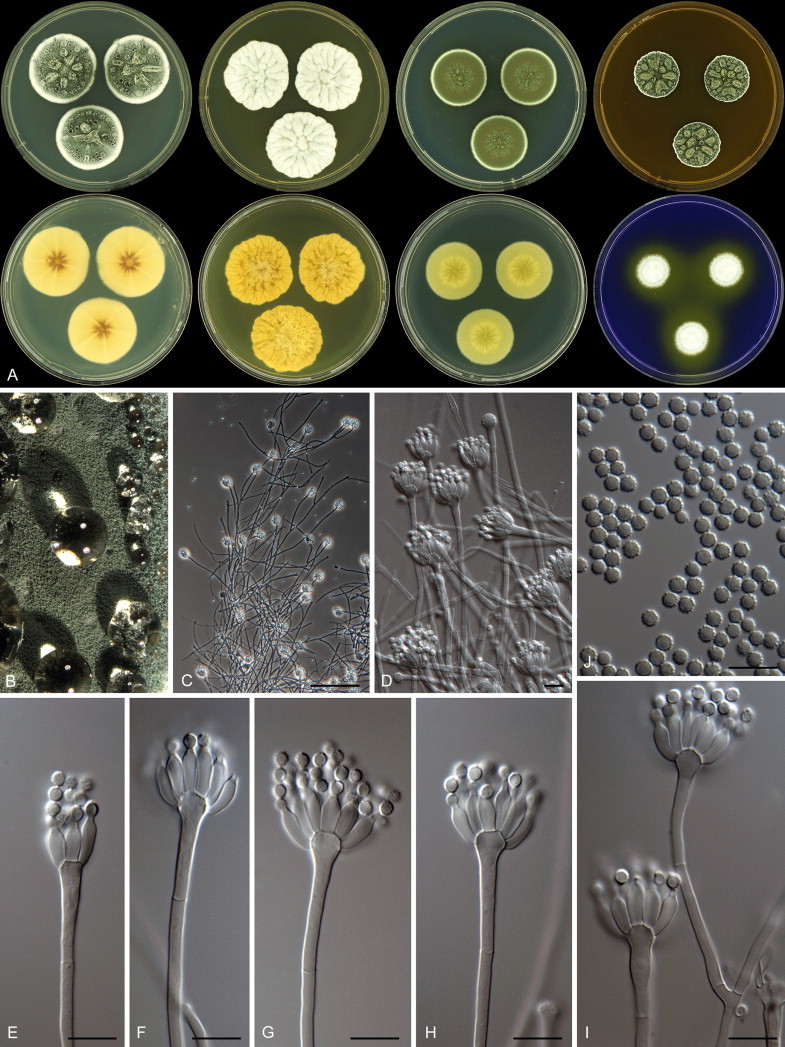
Penicillium vagum, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B. Colony texture. C–I. Conidiophores. J. Conidia. Scale bars = 10 μm.
Etymology: Latin, vagum = meaning wanderer; named in reference to the phylogenetic relationships of this species, which change according to the different genes analysed.
Diagnosis: This species is phylogenetically distinct. It is characterised by floccose colony texture on CYA, dark green conidia on MEA and distinctly roughened globose conidia.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium vagum-clade.
Typus: South Africa, Western Cape, Stellenbosch mountain, air sample, isolated by C.M. Visagie (holotypus CBS H-21926, cultures ex-type: CBS 137728 = DTO 180-G3 = CV 25 = DAOM 241357).
Barcode and molecular based ID: ITS barcode: KM189642 (alternative markers: BenA = KM088886; CaM = KM089273; RPB2 = KM089660).
Description: Colony diam, 7 d, in mm: CYA 30–43; CYA15°C 14–22; CYA30°C 30–31; CYA37°C no growth, sometimes 4 mm; MEA 20–44; YES 34–54; DG18 27–30; CYAS 26–32; ratio CYAS:CYA 0.75–0.85; CREA 20–22, weak to good growth, moderate acid production, no base production.
Sporulation on CYA moderately dense; conidia dull or pure green, colony texture floccose, moderately deep, sulcate; mycelium white; exudate abundant, clear; soluble pigment absent or present, yellow-brown; margin low, narrow, entire; reverse yellowish white to greyish yellow, (yellow-) brown. Sporulation on YES poor to strong; conidia variable, greenish white, dull green or dark green, colony texture floccose, moderately deep, sulcate; mycelium white or yellow; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse light yellow to greyish yellow. Sporulation on DG18 dense; conidia dark green, colony texture velutinous, low, sulcate; mycelium white; exudate absent; soluble pigment not produced; margin low, narrow, entire; reverse transparent, greyish green, pale yellow or pale brown in the centre. Sporulation on MEA dense; conidia dark green (26F5–F8), colony texture velutinous, moderately deep, sulcate; mycelium white; exudate abundant clear; soluble pigment not produced; margin low, narrow, entire; reverse brownish yellow to yellowish brown (5C8–D8). Ehrlich reaction negative.
Sclerotia absent. Conidiophores monoverticillate. Stipes smooth walled, 36–310 × 2.5–3.5 μm, vesicles 4.5–7 μm diam. Phialides ampulliform, 15–20 per stipe, 8.5–11 × 3–4 μm. Conidia conspicuously spiny walled, globose, 2.5–3.5 μm.
Penicillium verhagenii Houbraken, sp. nov. MycoBank MB809979. Fig. 42.
Fig. 42.
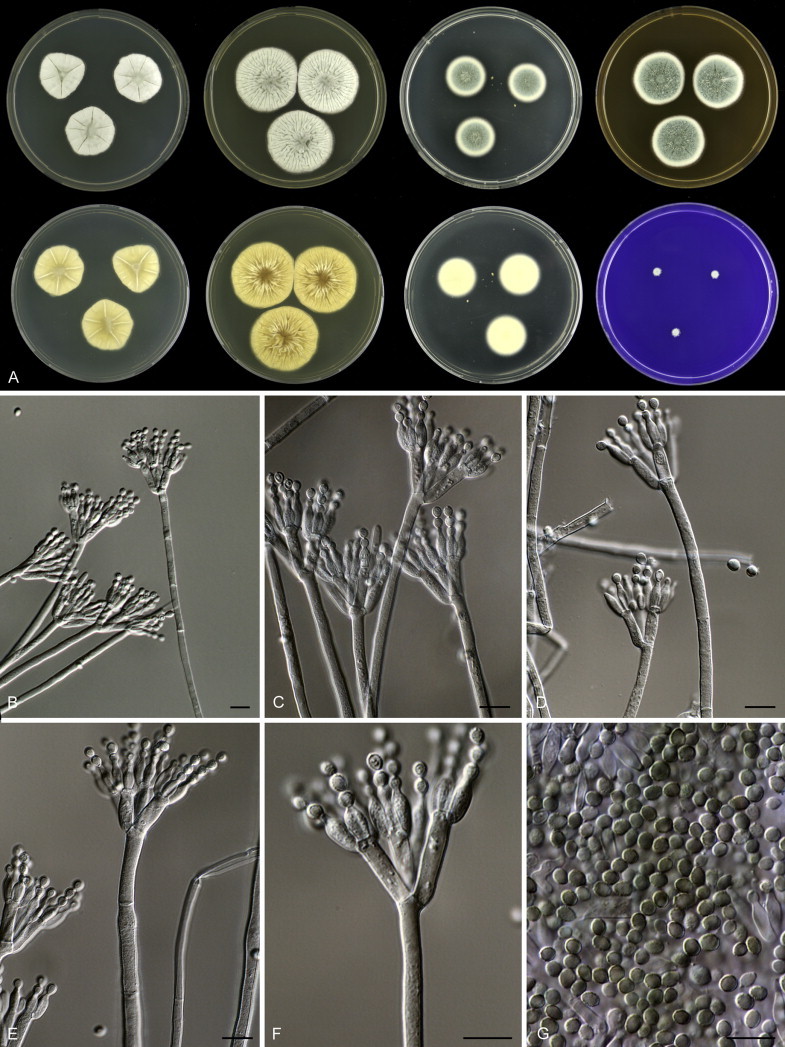
Penicillium verhagenii, A. 7 d old cultures, 25 °C, left to right; first row, all obverse, CYA, YES, DG18, MEA; second row, CYA reverse, YES reverse, DG18 reverse, CREA obverse. B–F. Conidiophores. G. Conidia. Scale bars = 10 μm.
Etymology: Named after Cor Verhagen, who assisted with the collection of the soil sample from which the type strain was isolated.
Diagnosis: The species is characterised by the production of long stipes, up to 400 μm, biverticillate conidiophores, restricted growth on CYA at 27 °C and CYAS and bluish green conidia on MEA.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium verhagenii-clade.
Typus: Belgium, Postel, mosses under Myrica gale (Bog Myrtle), isolated by J. Houbraken (holotype CBS H-21865, culture ex-type CBS 137959 = DTO 193-A1).
Barcode and molecular based ID: ITS barcode: KM189708 (alternative markers: BenA = KM088955; CaM = KM089342; RPB2 = KM089729).
Description: Colony diam, 7 d, in mm: CYA 20–27; CYA15°C 17–25; CYA30°C no growth; CYA37°C no growth; MEA 21–30; YES 28–35; DG18 18–25; CYAS 3–9; ratio CYAS:CYA 0.15–0.30(–0.40); CREA 2–7, weak growth and no acid production.
Sporulation on CYA absent or weakly present in the centre of colony; colony texture velvety to floccose, radially sulcate, deep; conidia pale grey green; mycelium white; exudate absent; soluble pigment absent or poor, pale yellow; margin entire or slightly polygonal in face view; reverse yellow or yellow-brown. Sporulation on YES absent or poor, conidia pale grey green; mycelium white or pale crème; soluble pigment not produced; reverse yellowish brown. Good sporulation on DG18, colony texture velvety; conidia dull green; mycelium inconspicuous in the centre, in some isolates pale yellow at the edge, reverse pale or pale yellow. Good sporulation on MEA, colony texture floccose; conidia blue-green or dull to blue-green; exudate present as pale or yellow droplets; colony reverse not affecting the medium colour, sometimes with yellow centre. Ehrlich reaction negative.
Sclerotia absent. Conidiophores 150–400 μm long, finely rough walled in young parts of the colony, distinctly rough walled in older parts; symmetrically biverticillate, some conidiophores becoming divaricate following having secondary growth of the stipe by sympodial branching at the apex, stipe 3.0–4.5 μm wide. Metulae in terminal whorl of 3–6, of unequal length, 13.0–16.5 × 3.0–4.0 μm. Phialides ampulliform, 2–8 per stipe, 9.0–10 × 3.0–3.5 μm. Conidia in short distorted chains, (broadly) ellipsoidal, roughened, slightly striate, 3.3–4.0 × 2.5–3.3 μm.
Notes: The type strain of P. rubefaciens (CBS 145.83T) and two substrains (CBS 146.83 and CBS 147.83) were deposited in the CBS collection by J.A. Quintanilla in 1983. Quintanilla (1982) noted that this species produced floccose sectors and one of the cultures derived from these sectors (CBS 146.83) is identified here as P. verhagenii.
Penicillium yezoense Hanzawa ex Houbraken, sp. nov. MycoBank MB809980.
= Penicillium yezoense Hanzawa, J. Agric. Chem. Soc. Japan: 774. 1943. MB335775 (nom. inval., Art. 39.1.).
Diagnosis: The species is phenotypically and phylogenetically related to P. contaminatum, but differs by a faster growth rate on DG18 (21–25 vs 30–39 mm) and CYAS (30–35 vs 37–47 mm) and a better sporulation on YES and CYAS. The conidia of P. yezoense are ellipsoidal while those of P. contaminatum are broadly ellipsoidal.
Typus: Japan, butter, isolated by Y. Sasaki (holotype CBS H-21863, culture ex-type CBS 350.59 = ATCC 18333 = FRR 3395 = IFO 5362 = IMI 068615).
In: subgenus Aspergilloides, section Aspergilloides, Penicillium thomii-clade.
Barcode and molecular based ID: ITS barcode: KM189553 (alternative markers: BenA = KM088792; CaM = KM089177; RPB2 = KM089564).
Notes: Penicillium yezoense was described without a Latin diagnosis. To validate the species an English diagnosis is given above, with the name of the original author maintained. This species was considered a synonym of P. thomii (Pitt 1980, Ramírez 1982, Pitt et al. 2000); however, this study shows that P. yezoense is phylogenetically unique (Figs 5, 6).
List of currently accepted species and their synonyms in Penicillium section Aspergilloides
The following list includes accepted species in sect. Aspergilloides and their synonyms.
Penicillium ardesiacum Novobr., Novosti Sist. Nizsh. Rast. 11: 228. 1974. MB319257.
= Penicillium thomii var. flavescens S. Abe, J. Gen. Appl. Microbiol. Tokyo 2: 50. 1956. MB347377 (nom. inval., Art. 39.1.).
In: Penicillium fuscum-clade
Typus: Kazachstan, Alma-Ata Region, Vitis vinifera, T.I. Novobranova. Culture ex-type: IMI 174719 = CBS 497.73 = ATCC 24719 = FRR 1479 = IFO 30540 = IMI 174719 = VKM F-1749.
Penicillium armarii Houbraken, Visagie, Samson & Seifert, this study. MycoBank MB809955.
In: Penicillium glabrum-clade
Typus: Australia, Tasmania, Hobart, house dust, collected by G. Gates, 2009, isolated by E. Whitfield and K. Mwange. Culture ex-type: CBS 138171 = DTO 235-F1.
Penicillium athertonense Houbraken, this study. MycoBank MB809956.
In: Penicillium fuscum-clade
Typus: Australia, Queensland, Atherton Tableland, forest soil, J. Houbraken & R. van Leeuwen. Culture ex-type: CBS 138161 = DTO 030-C2.
Penicillium aurantioviolaceum Biourge, Cellule 33: 282. 1923. MycoBank MB257885.
In: Penicillium thomii-clade
Neotypus: Puerto Rico, unrecorded source, R.A. Toro. Neotype, designated here: CBS H-21954; culture ex-neotype: CBS 137777 = NRRL 762.
Penicillium austroafricanum Houbraken & Visagie, this study. MycoBank MB809957.
In: Penicillium thomii-clade
Type: South Africa, Western Cape, Harold Porter Botanical Garden, leaf of Phaenocoma prolifera, J. Houbraken & P. Crous. Culture ex-types: CBS 137773 = DTO 133-G5.
Penicillium brunneoconidiatum Visagie, Houbraken & K. Jacobs, this study. MycoBank MB809958.
In: Penicillium fuscum-clade
Typus: South Africa, Western Cape, Malmesbury, Riverlands, Fynbos, soil, C.M. Visagie. Culture ex-type: CBS 137732 = DTO 182-E4 = CV 949 = DAOM 241359.
Penicillium bussumense Houbraken, this study. MycoBank MB809959.
In: Penicillium glabrum-clade
Typus: the Netherlands, Bussum, Spanderswoud, soil, J. Houbraken. Culture ex-type: CBS 138160 = DTO 018-B2.
Penicillium cartierense Houbraken, this study. MycoBank MB809960.
In: Penicillium thomii-clade
Typus: the Netherlands, Eersel, Cartierhiede, soil, J. Houbraken. Culture ex-type: CBS 137956 = DTO 092-H9.
Penicillium clavistipitatum Visagie, Houbraken & K. Jacobs, this study. MycoBank MB809961.
In: Penicillium fuscum-clade
Typus: South Africa, Malmesbury, Riverlands, Fynbos, soil, C.M. Visagie. Culture ex-type: CBS 138650 = DTO 182-E5 = CV 336 = KAS 4112 = DAOM 241092.
Penicillium contaminatum Houbraken, this study. MycoBank MB809962.
In: Penicillium thomii-clade
Typus: United Kingdom, Kew, Surrey, culture contaminant. Culture ex-type CBS 345.52 = DTO 091-A3 = IMI 049057.
Penicillium crocicola W. Yamam., Sci. Rep. Hyogo Univ. Agric. 2: 28. 1956. MycoBank MB302391.
In: Penicillium thomii-clade
Type: Japan, corm of Crocus sativus, W. Yamamoto. Culture ex-type: CBS H-7528 = CBS 745.70 = ATCC 18313 = QM 7778.
Penicillium flavisclerotiatum Visagie, Houbraken & K. Jacobs, this study. MycoBank MB809963.
In: Penicillium fuscum-clade
Typus: South Africa, Western Cape, Stellenbosch mountain, Fynbos, soil, isolated by C.M. Visagie. Culture ex-type: CBS 137750 = DTO 180-I8 = CV 100 = DAOM 241157.
Penicillium frequentans Westling, Ark. Bot. 11: 133. 1911. MycoBank MB152118.
= Penicillium paczoskii K.M. Zaleski, Bull. Int. Acad. Polon. Sci., Sér. B 1927: 505. 1927. MB273253.
In: Penicillium glabrum-clade
Typus: Unknown source, K. Westling. CBS 105.11.
Penicillium fuscum (Sopp) Biourge, Cellule 33: 103. 1923. MycoBank MB289082.
≡ Citromyces fuscus Sopp, Skr. Vidensk.-Selsk. Christiana Math.-Nat. Kl. 11: 120. 1912. MB178643.
= Penicillium silvaticum Suprun, Byull. Mosk. Obshch. Ispyrt. Prir.: 90. 1956. MB492648.
= Penicillium pinetorum M. Chr. & Backus, Mycologia 53: 457. 1961. MB335758.
= Penicillium macedoniense Verona & Mick., Mycopathol. Mycol. Appl. 18: 289. 1962. MB335746.
= Eupenicillium pinetorum Stolk, Antonie van Leeuwenhoek 34: 37. 1968. MB330740.
= Penicillium lapatayae C. Ramírez, Mycopathologia 91: 96. 1985. MB105610.
= Eladia inflata Y.L. Jiang & T.Y. Zhang, Mycotaxon 108: 128. 2009. MB512859.
In: Penicillium fuscum-clade
Neotypus: USA, Wisconsin, pine-birch forest soil, Vilas County, M. Christensen. Culture ex-neotype: WSF 15-C = CBS 295.62 = ATCC 14770 = CCRC 31517 = DSM 2438 = IFO 7743 = IMI 094209 = MUCL 31196 = NRRL 3008.
Notes: The type culture of P. lapatayae maintained in the CBS collection (CBS 203.87) is dead. The IBT culture collection contains two ex-type isolates, one directly sent by C. Ramírez to the IBT collection (IJFM 19012 = IBT 10870 = DTO 297-C8) and another received the ATCC (ATCC 60197 = IBT 16267 = DTO 297-C9). IBT 16267 is identified here as P. fuscum (Figs 12, 13) and IBT 10870 as P. adametzioides. Neither strain formed the pink soft sclerotia described in the original description. However, IBT 16267 resembles the original description best because this isolate produces conspicuously ornamented subglobose conidia and pure to dark green conidia on MEA. In contrast, IBT 10870 produces broadly ellipsoidal, smooth to finely roughened conidia, and grey-green colonies on MEA. Based on this data, we consider P. lapatayae a synonym of P. fuscum.
Penicillium fusisporum L. Wang, PLoS ONE 9:e101454-P2. 2014. MB806119.
In: Penicillium thomii-clade
Typus: China, Shaanxi, Nangongshan Forest Park, leaves of Rhododendron sp., P-J Han. Culture ex-type: HMAS 244961 = CBS 137463 = AS3.15338 = NRRL 62805.
Penicillium glabrum (Wehmer) Westling, Ark. Bot. 11: 131. 1911. MB120545.
≡ Citromyces glaber Wehmer, Beitr. Kenntn. Einh. Pilze 1: 24. 1893. MB178959.
= Citromyces pfefferianus Wehmer, Ber. Deutsch. Bot. Ges. 11: 333. 1893. MB157685.
= Penicillium aurantiobrunneum Dierckx, Ann. Soc. Sci. Bruxelles 25: 86. 1901. MB237393.
?= Penicillium fluitans Tiegs, Ber. Deutsch. Bot. Ges. 37: 500. 1919. MB151731; fide Raper & Thom (1949) and Pitt (1980). No culture examined.
= Penicillium flavidorsum Biourge, Cellule 33: 290. 1923. MB265032.
= Penicillium oledzkii K.M. Zaleski, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 499. 1927. MB272809.
= Penicillium terlikowskii K.M. Zaleski, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 501. 1927. MB280026.
= Penicillium spinuloramigenum Y. Sasaki, J. Appl. Mycol., Japan: 58. 1946. MB302426 (nom. inval., Art. 39.1.).
= Penicillium spinuloramigenum Y. Sasaki ex C. Ramírez, Manual and Atlas of the Penicillia: 162. 1982. MB115801.
In: Penicillium glabrum-clade
Neotypus: unrecorded source, K. Westling. Culture ex-neotype: IMI 91944 = IMI 91944 = CBS 125543.
Penicillium grancanariae C. Ramírez, A.T. Martínez & Ferrer, Mycopathologia 66: 79. 1978. MycoBank MB319273.
In: Penicillium spinulosum-clade
Typus: Spain, Canary Islands, Gran Canaria, air. Culture ex-type: IJFM 3745 = CBS 687.77 = IJFM 3745 = IMI 253783.
Penicillium grevilleicola Houbraken & Quaedvlieg, this study. MycoBank MB809964.
In: Penicillium thomii-clade
Typus: Australia, Kangaroo Island, Kingscote, leaf of Grevillea ilicifolia, J. Houbraken & W. Quaedvlieg. Culture ex-type: CBS 137775 = DTO 174-E6.
Penicillium hoeksii Houbraken, this study. MycoBank MB809965.
In: Penicillium hoeksii-clade
Typus: Belgium, Postel, soil under Compact Rush (Juncus conglomeratus), J. Houbraken. Culture ex-type: CBS 137776 = DTO 192-H4.
Penicillium infra-aurantiacum Visagie, Houbraken & K. Jacobs, this study. MycoBank MB809966.
In: Penicillium sublectaticum-clade
Typus: South Africa, Western Cape, Malmesbury, Riverlands, bracts of Protea repens infructescence, C.M. Visagie. Culture ex-type: CBS 137747 = DTO 183-C3 = CV 1518 = DAOM 241145.
Penicillium jejuense M.S. Park & Y.W. Lim, submitted. MycoBank MB808392.
In: Penicillium thomii-clade
Typus: Republic of Korea, Jeju Island, Pollicipes mitella. Culture ex-type: SFC20140101-M756T.
Penicillium kananaskense Seifert, Frisvad & McLean, Can. J. Bot. 72: 20. 1994. MB362160.
In: Penicillium lividum-clade
Typus: Canada, Alberta, Kananaskis Valley, soil, FH horizon, in a Pinus contorta var. latifolia forest, M.A. McLean. Culture ex-type: CBS 530.93 = ATCC 90282 = DAOM 216105 = IBT 11775 = IMI 356791.
Penicillium kiamaense Houbraken & Pitt, this study. MycoBank MB809967.
In: undefined clade (basal to P. glabrum and P. thomii-clade).
Typus: Australia, NSW, Barren Grounds Nature Reserve, near Kiama, soil, J.I. Pitt. Culture ex-type: CBS 137947 = FRR 6087 = DTO 056-I6.
Penicillium lividum Westling, Ark. Bot. 11: 134. 1911. MycoBank MB178817.
In: Penicillium lividum-clade
Neotypus: Unrecorded source, Scotland. Culture ex-neotype: IMI 39736 = CBS 347.48 = ATCC 10102 = CCRC 31286 = DSM 1180 = IFO 6102 = NRRL 754 = QM 1930 = VKM F-303.
Note: Some sub-cultures of the ex-neotype culture of P. lividum received from a few collections were contaminated with P. spinulosum. Through the courtesy of Dr. John David, former curator of IMI, we re-examined the neotype specimen IMI 39736 and confirmed that it conforms to the concept of this species proposed by Pitt (1980), and the more restricted concept adopted in this paper.
Penicillium longicatenatum Visagie, Busby, Houbraken & K. Jacobs, this study. MycoBank MB809968.
In: Penicillium vagum-clade
Typus: South Africa, Western Cape, Malmesbury, Riverlands, Fynbos, soil, C.M. Visagie. Culture ex-type: CBS 137735 = DTO 180-D9 = CV 2847 = DAOM 241119.
Penicillium malmesburiense Visagie, Houbraken & K. Jacobs, this study. MycoBank MB809969.
In: Penicillium sublectaticum-clade
Typus: South Africa, Western Cape, Malmesbury, Riverlands, mite from Protea repens infructescence, C.M. Visagie. Culture ex-type: CBS 137744 = DTO 182-H5 = CV 1180 = DAOM 241144.
Penicillium montanense M. Chr. & Backus, Mycologia 54: 574. 1962. MycoBank MB335752.
= Penicillium echinosporum G. Sm., Trans. Brit. Mycol. Soc. 45: 387. 1962. MB335724 (nom. illeg., Art. 53).
= Penicillium asperosporum G. Sm., Trans. Brit. Mycol. Soc. 48: 275. 1965. MB335714.
In: Penicillium fuscum-clade
Typus: USA, Montana, Ravalli County Lodgepole, pine-Douglas fir soil. Culture ex-type: CBS 310.63 = ATCC 14941 = FRR 3407 = IFO 7740 = IHEM 4375 = IMI 099468 = MUCL 31326 = NRRL 3407.
Notes: There is some taxonomic confusion around P. echinosporum and P. asperosporum. Penicillium echinosporum Nehira was described in 1933 without a Latin diagnosis. The name was incorrectly validated by Ramírez (1982) but Latin descriptions were compulsory only after 1935. Samson et al. (2011) showed that the type of this species (CBS 344.51T) belongs to Talaromyces and the combination Talaromyces echinosporus was proposed. Currently, this species is placed in synonymy with Talaromyces rugulosus (Yilmaz et al. 2014). In 1962, unaware of the existence P. echinosporum Nehira, Smith described P. echinosporum G. Sm. and typified it with IMI 080450T. Three years later he corrected this error and renamed his species P. asperosporum. Sequence analysis confirm that NRRL 3411 (= IMI 080450T) is conspecific with P. montanense.
Penicillium odoratum M. Chr. & Backus, Mycologia 53: 459. 1961. MycoBank MB335755.
= Penicillium trzebinskianum S. Abe, J. Gen. Appl. Microbiol., Tokyo 2: 63. 1956. MB302427.
= Penicillium trzebinskianum S. Abe ex C. Ramírez, Manual and Atlas of the Penicillia: 79. 1982. MB115803.
In: Penicillium lividum-clade
Typus: USA, Wisconsin, soil, spruce-larch bog, M. Christensen. Culture ex-type: WSF 2000 = DTO 205-B7 = CBS 294.62 = CBS 129423 = DAOM 226269 = ATCC 14769 = DSM 2419 = IFO 7741 = IMI 094208ii = NRRL 3007 = DAOM 226269.
Penicillium palmense C. Ramírez & A.T. Martínez, Mycopathologia 66: 80. 1978. MycoBank MB319289.
In: Penicillium spinulosum-clade
Typus: Air, Canary Islands, Gran Canaria, Spain. Culture ex-type: IJFM 3840 = CBS 336.79 = ATCC 38669 = VKM F-2181.
Penicillium pulvis Houbraken, Visagie, Samson & Seifert, this study. MycoBank MB809970.
In: Penicillium glabrum-clade
Typus: South Africa, South Africa, Kuils River, house dust, K. Jacobs. Culture ex-type: CBS 138432 = DTO 180-B7.
Penicillium purpurescens (Sopp) Raper & Thom, A manual of the Penicillia: 177. 1949. MycoBank MB335761.
≡ Citromyces purpurescens Sopp, Skr. Vidensk.-Selsk. Christiana, Math.-Naturvidensk. Kl. 11: 117. 1912. MB157120.
?= Citromyces virido-albus Sopp, Skr. Vidensk.-Selsk. Christiana, Math.-Naturvidensk. Kl. 11: 131. 1912. MB454083; fide Raper & Thom (1949); no culture available.
?= Penicillium virido-album (Sopp) Biourge, Cellule 33: 106. 1923. MB492659; fide Raper & Thom (1949).
= Penicillium internascens Szilvinyi, Zentralbl. Bakteriol. Parasitenk., Abt. 2 103: 148. 1941. MB289091.
= Penicillium resinae Z.T. Qi & H.Z. Kong, Acta Mycol. Sin.: 103. 1982. MB110236.
In: Penicillium glabrum-clade
Neotypus: Canada, soil, G.R. Bisby. Culture ex-neotype: IMI 39745 = CBS 366.48 = ATCC 10485 = NRRL 720 = QM 1959.
Penicillium quercetorum Baghd., Novosti Sist. Nizsh. Rast. 5: 110. 1968. MB335762.
In: Penicillium quercetorum-clade
Typus: Syria, soil near Es-Euveida, V.C. Baghdadi. Culture ex-type: CBS H-7527 = CBS 417.69 = ATCC 48727 = CCRC 31668 = FRR 516 = IFO 31749 = IMI 140342 = MUCL 31203 = VKM F-1074.
Penicillium ranomafanaense Houbraken & Hagen, this study. MycoBank MB809971.
In: Penicillium verhagenii-clade
Typus: Madagascar, Ranoma fana, soil, F. Hagen & J. Houbraken. Culture ex-type: CBS 137953 = DTO 085-A5.
Penicillium roseomaculatum Biourge, Cellule 33: 301. 1923. MycoBank MB276785.
= Penicillium baiicola Biourge, Cellule 33: 305. 1923. MB258101.
= Penicillium subericola Barreto, Frisvad & Samson, Fungal Diver. 49: 32. 2011. MB517383
In: Penicillium spinulosum-clade
Typus: Unrecorded source, P. Biourge. Culture ex-type: CBS 137962 = NRRL 728 = FRR 0728 = IMI 189696 = MUCL 29101.
Penicillium roseoviride Stapp & Bortels, Zentralbl. Bakteriol. Parasitenk., Abt. 2 93: 51. 1935. MycoBank MB492646.
In: Penicillium thomii-clade
Typus: Germany, soil in a beech forest. Culture ex-type: CBS 267.35 = ATCC 10412 = IFO 6089 = IMI 039740ii = NRRL 760 = QM 7485).
Penicillium rudallense Houbraken, Visagie & Pitt, this study. MycoBank MB809972.
In: Penicillium glabrum-clade
Typus: Australia, WA, Rudall River National Park, soil, A.D. Hocking. Culture ex-type: CBS 138162 = FRR 6085 = DTO 056-I4.
Penicillium saturniforme (L. Wang & W.Y. Zhuang) Houbraken & Samson, Stud. Mycol. 70: 48. 2011. MycoBank MB561958.
≡ Eupenicillium saturniforme L. Wang & W.Y. Zhuang, Mycopathologia 167: 300. 2009. MB541663.
In: Penicillium saturniforme-clade
Typus: China, Jiling Province, Dunhua Little Peony Forest Reserve, soil. Culture ex-type: AS 3.6886 = CBS 122276 = HMAS 130355-1-4.
Penicillium spinulosum Thom, U.S.D.A. Bur. Animal Industr. Bull. 118: 76. 1910. MycoBank MB215401.
= Penicillium pfefferianum (Wehmer) Westling, Ark. Bot. 11: 132. 1911. MB492636.
= Penicillium flavocinereum Biourge, Cellule 33: 293. 1923. MB265060.
?= Penicillium janthocitrinum Biourge, Cellule 33: 311. 1923. MB119135; fide Pitt (1980).
= Penicillium mucosum Stapp & Bortels, Zentralbl. Bakteriol. Parasitenk., Abt. 2 93: 51. 1935. MB492626.
= Penicillium tannophilum Stapp & Bortels, Zentralbl. Bakteriol. Parasitenk., Abt. 2 93: 52. 1935. MB492654.
= Penicillium brunneoviride Szilvinyi, Zentralbl. Bakteriol. Parasitenk., Abt. 2 103: 144. 1941. MB289078.
= Penicillium trzebinskii var. magnum Sakag. & S. Abe, J. Gen. Appl. Microbiol., Tokyo 2: 62. 1956. MB352367.
= Penicillium abeanum G. Sm., Trans. Brit. Mycol. Soc. 46: 333. 1963. MB335704.
In: Penicillium spinulosum-clade
Neotypus: Germany, Hannover, culture contaminant, C. Wehmer. Culture ex-neotype: IMI 24316i = CBS 374.48 = ATCC 10498 = FRR 1750 = MUCL 13910 = MUCL 13911 = NCTC 591 = NRRL 1750 = QM 7654.
Penicillium sterculiniicola Houbraken, this study. MycoBank MB809973.
In: Penicillium spinulosum-clade
Typus: USA, spawn run compost. Culture ex-type: CBS 122426 = DTO 031-A4.
Penicillium sublectaticum Houbraken, Frisvad, Samson & Seifert, this study. MycoBank MB809974.
In: subgenus Aspergilloides, section Aspergilloides, P. sublectaticum-clade.
Typus: New Zealand, Dunedin, house dust, collected by T. Atkinson, 2009, isolated by E. Whitfield and K. Mwange. Culture ex-type: CBS 138217 = DTO 244-G2.
Penicillium subspinulosum Houbraken, this study. MycoBank MB809975.
In: subgenus Aspergilloides, section Aspergilloides, Penicillium spinulosum-clade.
Typus: Poland, soil, J. Houbraken & B. Byskal. Culture ex-type CBS 137946 = DTO 041-F2.
Penicillium thiersii S.W. Peterson, E.M. Bayer & Wicklow, Mycologia 96: 1283. 2005. MycoBank MB487738.
In: Penicillium thiersii-clade
Typus: USA, Wisconsin, New Glarus Woods State Park, old black stroma of Hypoxylon encrusting the surface of a dead maple log, H.D. Thiers. Culture ex-type: BPI 842269 = CBS 117503 = IBT 27050 = NRRL 28162.
Penicillium thomii Maire, Bull. Soc. Hist. Nat. Afrique N. 8: 189. 1917. MycoBank MB202819.
?= Penicillium parallelosporum Y. Sasaki, J. Fac. Agric. Hokkaido Imp. Univ. 49: 147. 1950. MB302414 (nom. inval., Art. 39.1.).
= Penicillium patens Pitt & A.D. Hocking, Mycotaxon 22: 197. 1985. MB105611.
In: Penicillium thomii-clade
Neotypus: USA, Spaulding, pine cone. Culture ex-neotype: IMI 189694 = CBS 225.81 = NRRL 2077.
Penicillium trzebinskii K.M. Zaleski, Bull. Int. Acad. Polon. Sci., Sér. B., Sci. Nat. 1927: 498. 1927. MycoBank MB280795.
= Penicillium tannophagum Stapp & Bortels, Zentralbl. Bakteriol. Parasitenk., Abt. 2 93: 52. 1935. MB492653.
= Penicillium mediocre Stapp & Bortels, Zentralbl. Bakteriol. Parasitenk., Abt. 2 93: 50. 1935. MB492624.
= Penicillium toxicarium I. Miyake ex C. Ramírez, Manual and Atlas of the Penicillia: 125. 1982. MB115802.
In: Penicillium spinulosum-clade
Typus: Poland, Poznan area, Dluga Goslina Forest, soil, K. Zaleski. Culture ex-type: CBS 382.48 = ATCC 10507 = FRR 731 = IFO 6110 = IMI 039749 = MUCL 29102 = NRRL 731 = QM 7678.
Penicillium tsitsikammaense Houbraken, this study. MycoBank MB809976.
In: Penicillium fuscum-clade
Typus: South Africa, Cape Province, Tsitsikamma Forest near Knysna, forest soil, D.B. Scott. Culture ex-type: CBS 328.71 = DTO 006-I3 = CSIR 1092.
Penicillium turcosoconidiatum Visagie, Houbraken & K. Jacobs, this study. MycoBank MB809977.
In: Penicillium fuscum-clade
Typus: South Africa, Stellenbosch, soil, C.M. Visagie. Culture ex-type: CBS 138557 = DTO 181-A3 = CV 110 = DAOM 241130.
Penicillium vagum Houbraken, Pitt, Visagie & K. Jacobs, this study. MycoBank MB809978.
In: Penicillium vagum-clade
Typus: South Africa, Western Cape, Stellenbosch mountain, air sample, C.M. Visagie. Culture ex-type: CBS 137728 = DTO 180-G3 = CV 25 = DAOM 241357.
Penicillium valentinum C. Ramírez & A.T. Martínez, Mycopathologia 72: 183. 1980. MycoBank MB113027.
In: Penicillium thomii-clade
Typus: Spain, Madrid, air. C. Ramírez. Culture ex-type: IJFM 5071 = CBS 172.81.
Penicillium verhagenii Houbraken, this study. MycoBank MB809979.
In: Penicillium verhagenii-clade
Typus: Belgium, Postel, mosses under Myrica gale (Bog Myrtle), J. Houbraken. Culture ex-type CBS 137959 = DTO 193-A1.
Penicillium yezoense Hanzawa ex Houbraken, this study. MycoBank MB809980.
= Penicillium yezoense Hanzawa, J. Agric. Chem. Soc. Japan: 774. 1943. MB335775 (nom. inval., Art. 39.1.).
In: Penicillium thomii-clade
Typus: Japan, butter, Y. Sasak. Culture ex-type: CBS 350.59 = ATCC 18333 = FRR 3395 = IFO 5362 = IMI 068615.
Penicillium zhuangii L. Wang, PLoS ONE 9 e101454-P4. 2014. MycoBank MB805945.
In: Penicillium hoeksii-clade
Typus: China, Shaanxi, Nangongshan Forest Park, leaves of Rhododendron sp., 1 500 m, P-J Han. Culture ex-type: HMAS 244961 = CBS 137464 = AS3.15338 = NRRL 62806.
Acknowledgements
This research was in part supported by grants from the Alfred P. Sloan Foundation Program on the Microbiology of the Built Environment. The research of Karin Jacobs was funded by a grant from the South African Biosystematics Initiative (SABI) of the National Research Foundation (NRF), South Africa. We are grateful to the many collectors of soil samples and house dust samples that yielded many of the new species described here. Uwe Braun is acknowledged for his advice on the new species names and the nomenclatural issues. We are grateful to John David, former curator of herb. IMI, for sending us a microscopic preparation from the neotype specimen of P. lividum.
Footnotes
Peer review under responsibility of CBS-KNAW Fungal Biodiversity Centre.
Contributor Information
J. Houbraken, Email: j.houbraken@cbs.knaw.nl.
K. Jacobs, Email: kj@sun.ac.za.
References
- Barreto M.C., Houbraken J., Samson R.A. Taxonomic studies of the Penicillium glabrum complex and the description of a new species P. subericola. Fungal Diversity. 2011;49:23–33. [Google Scholar]
- Biourge P. Les moisissures du groupe Penicillium Link. Cellule. 1923;33:7–331. [Google Scholar]
- Frisvad J.C., Houbraken J., Popma S. Two new Penicillium species P. buchwaldii and P. spathulatum, producing the anticancer compound asperphenamate. FEMS Microbiology Letters. 2013;339:77–92. doi: 10.1111/1574-6968.12054. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Samson R.A., Stolk A.C. Disposition of recently described species in Penicillium. Persoonia. 1990;14:209–232. [Google Scholar]
- Hetherington A.C., Raistrick H. Studies in the biochemistry of microorganisms XI. On citromycetin, a new yellow colouring matter produced from glucose by a species of Citromyces. Philosophical Transactions of the Royal Society, London, Series B. 1931;220:209–244. [Google Scholar]
- Houbraken J., Frisvad J.C., Samson R.A. Fleming's penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus. 2011;2:87–95. doi: 10.5598/imafungus.2011.02.01.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Frisvad J.C., Samson R.A. Taxonomy of Penicillium section Citrina. Studies in Mycology. 2011;70:53–138. doi: 10.3114/sim.2011.70.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Frisvad J.C., Seifert K.A. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia. 2012;29:78–100. doi: 10.3767/003158512X660571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Samson R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Spierenburg H., Frisvad J.C. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie van Leeuwenhoek. 2012;101:403–421. doi: 10.1007/s10482-011-9647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Gloer J.B., Wicklow D.T. Thiersindoiloes A-C: new indole diterpenoids from Penicillium thiersii. Journal of Natural Products. 2003;66:1232–1235. doi: 10.1021/np030192m. [DOI] [PubMed] [Google Scholar]
- Li C., Gloer J.B., Wicklow D.T. Thiersinines A and B: novel antiinsectan indole diterpenoids from a new fungicolous Penicillium species (NRRL 28147) Organic Letters. 2002;4:3095–3098. doi: 10.1021/ol026424a. [DOI] [PubMed] [Google Scholar]
- Li C., Gloer J.B., Wicklow D.T. Antiinsectan decaturin and oxalicine analogues from Penicillium thiersii. Journal of Natural Products. 2005;68:319–322. doi: 10.1021/np0496486. [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lund F. Differentiating Penicillium species by detection of indole metabolites using a filter paper method. Letters in Applied Microbiology. 1995;20:228–231. [Google Scholar]
- Mahmoodian A., Stickings C.E. 15 metabolites of Penicillium frequentans Westling, isolation of sulochrin, asterric acid, (+)-bisdecchlorogeodin + 2 new substituted anthraquinones questin + questinol. Biochemical Journal. 1964;92:369–378. doi: 10.1042/bj0920369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J.A.A. Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest v2. Program distributed by the author. [Google Scholar]
- Park MS, Fong JJ, Oh SY, et al. Penicillium jejuense sp. nov., isolated from the marine environments of Jeju Island, Korea. Submitted. [DOI] [PubMed]
- Peterson S.W. Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 163–178. [Google Scholar]
- Peterson S.W., Bayer E.M., Wicklow D.T. Penicillium thiersii, Penicillium angulare and Penicillium decaturense, new species isolated from wood-decay fungi in North America and their phylogenetic placement from multilocus DNA sequence analysis. Mycologia. 2004;96:1280–1293. [PubMed] [Google Scholar]
- Pitt J.I. Academic Press Inc; London: 1980 [“1979”]. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. [Google Scholar]
- Pitt J.I., Hocking A.D. New species of fungi from Indonesian dried fish. Mycotaxon. 1985;22:197–208. [Google Scholar]
- Pitt J.I., Klich M.A., Shaffer G.P. Differentiation of Penicillium glabrum from Penicillium spinulosum and other closely related species: an integrated taxonomic approach. Systematic and Applied Microbiology. 1990;13:304–309. [Google Scholar]
- Pitt J.I., Samson R.A., Frisvad J.C. List of accepted species and their synonyms in the family Trichocomaceae. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam: 2000. pp. 9–79. [Google Scholar]
- Quintanilla J.A. Three new species of Penicillium isolated from soil. Mycopathologia. 1982;80:73–82. [Google Scholar]
- Ramírez C. Elsevier Biomedical Press; Amsterdam: 1982. Manual and atlas of the Penicillia. [Google Scholar]
- Raper K., Thom C. The Williams & Wilkins Company; Baltimore: 1949. A manual of the penicillia. [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes version 3.0: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Houbraken J., Thrane U. 2010. Food and indoor fungi. (CBS Laboratory manual). [Google Scholar]
- Samson R.A., Seifert K.A., Kuijpers A. Phylogenetic analysis of Penicillium subgenus Penicillium using partial B-tubulin sequences. Studies in Mycology. 2004;49:175–200. [Google Scholar]
- Samson R.A., Yilmaz N., Houbraken J. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Studies in Mycology. 2011;70:159–183. doi: 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Seifert K.A., Huhndorf S. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D.B. The genus Eupenicillium Ludwig. CSIR Research Report. 1968;272:1–150. [Google Scholar]
- Seifert K.A., Frisvad J.C., McLean M.A. Penicillium kananaskense, a new species from Alberta soil. Canadian Journal of Botany. 1994;72:20–24. [Google Scholar]
- Skouboe P., Frisvad J.C., Taylor J.W. Phylogenetic analysis of nucleotide sequences from the ITS region of terverticillate Penicillium species. Mycological Research. 1999;103:873–881. [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML Web-Servers. Systematic Biology. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stolk A.C. Studies on the genus Eupenicillium Ludwig. III. Four new species of Eupenicillium. Antonie van Leeuwenhoek. 1968;34:37–53. doi: 10.1007/BF02046412. [DOI] [PubMed] [Google Scholar]
- Stolk A.C., Samson R.A. The Ascomycete genus Eupenicillium and related Penicillium anamorphs. Studies in Mycology. 1983;23:1–149. [Google Scholar]
- Subramanian C.V. Indian Council of Agricultural Research; New Delhi: 1971. Hyphomycetes: an account of Indian species except Cercosporae. [Google Scholar]
- Taylor J.W., Jacobson D.J., Kroken S. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Visagie C.M., Houbraken J., Frisvad J.C. Identification and nomenclature of the genus Penicillium. Studies in Mycology. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Yu Y., Wang L. Penicillium fusisporum and P. zhuangii, two new monoverticillate species with apical-swelling stipes of section Aspergilloides isolated from plant leaves in China. PLoS ONE. 2014;9:e101454. doi: 10.1371/journal.pone.0101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhuang W.-Y. Eupenicillium saturniforme, a new species discovered from northeast China. Mycopathologia. 2009;167:297–305. doi: 10.1007/s11046-008-9179-z. [DOI] [PubMed] [Google Scholar]
- Yilmaz N., Visagie C.M., Houbraken J. Polyphasic taxonomy of the genus Talaromyces. Studies in Mycology. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaleski K.M. Über die in Polen gefundenen Arten der Gruppe Penicillium Link. I, II and III Teil. Bulletin de l'Académie Polonaise des Sciences et des Lettres, Classe des Sciences Mathématiques et Naturelles – Série B: Sciences Naturelles. 1927:417–563. pls 36–44 (printed in 1928) [Google Scholar]


