Abstract
The Src homology 2 (SH2) domains are participants in metazoan signal transduction, acting as primary mediators for regulated protein-protein interactions with tyrosine-phosphorylated substrates. Here, we describe the origin and evolution of SH2 domain proteins by means of sequence analysis from 21 eukaryotic organisms from the basal unicellular eukaryotes, where SH2 domains first appeared, through the multicellular animals and increasingly complex metazoans. On the basis of our results, SH2 domains and phosphotyrosine signaling emerged in the early Unikonta, and the numbers of SH2 domains expanded in the choanoflagellate and metazoan lineages with the development of tyrosine kinases, leading to rapid elaboration of phosphotyrosine signaling in early multicellular animals. Our results also indicated that SH2 domains coevolved and the number of the domains expanded alongside protein tyrosine kinases and tyrosine phosphatases, thereby coupling phosphotyrosine signaling to downstream signaling networks. Gene duplication combined with domain gain or loss produced novel SH2-containing proteins that function within phosphotyrosine signaling, which likely have contributed to diversity and complexity in metazoans. We found that intra- and intermolecular interactions within and between SH2 domain proteins increased in prevalence along with organismal complexity and may function to generate more highly connected and robust phosphotyrosine signaling networks.
INTRODUCTION
Posttranslational modification by phosphorylation of tyrosine residues is used extensively in metazoan cells as a mechanism to convey signals in response to external and internal cues (1, 2). Phosphotyrosine (pTyr)–mediated signaling plays a central role in many key cellular and developmental processes, including cell proliferation and differentiation. The essential triad of pTyr signaling involves protein tyrosine kinases (PTKs) that phosphorylate substrates, the protein tyrosine phosphatases (PTPs) that dephosphorylate, and the modular protein domains that recognize the phosphorylated ligand and thereby recruit the proteins containing these domains to specify downstream signaling events. Several modular interaction domains are capable of binding to tyrosine-phosphorylated protein ligands. These include most Src homology 2 (SH2) domains (3, 4), a subset of pTyr binding (PTB) domains (5), and at least one C2 domain (6). Among these, SH2 domains are the primary pTyr recognition modules that appear alongside, and coevolve with, PTKs and PTPs (7, 8). Mutations within PTKs, PTPs, and SH2 domain proteins have broad medical relevance, because they are principal players in numerous human malignancies and disorders, including immune deficiencies, diabetes, and cancer (3, 9).
Although PTKs are prevalent in metazoans, they are absent in most unicellular organisms, suggesting that evolution of pTyr signaling is correlated with the development and specialization of multicellularity in the metazoan lineage (10). The emergence of the complete set of pTyr signaling components about 900 million years ago at the premetazoan boundary between single-celled and multicellular organisms suggests that pTyr signaling may have facilitated the evolution of metazoans (11, 12). Additional evolutionary events, such as the apparent global loss of tyrosine residues (13), together with the expansion in the number of genes dedicated to this mode of signaling, suggest that the acquisition of tyrosine phosphorylation and SH2 domain–mediated signaling promoted metazoan development (12, 14, 15). Genes encoding catalytic PTKs, like many gene families involved in cellular communication, evolved from a single or small number of ancestral genes by gene duplication and domain shuffling (16). Concomitantly, SH2 domains expanded, promoting coordinated emergence and increasing sophistication of pTyr signaling during eukaryotic evolution. What sets the components of pTyr signaling apart from other signaling systems are both the timing of their appearance at the unicellular to multicellular transition in metazoa and the rapid expansion in the number of dedicated genes in metazoa. Together, these indicate a central role in metazoan cell signaling and cell type specialization that allowed the rapid evolution of highly complex organisms.
The emergence of cell types with specialized functions that allowed for more elaborate body plans and larger organisms is a hallmark of the increasing complexity in metazoan lineages. It has been postulated that selective intercellular communication is a requirement for such specialization (17). Increasing complexity and robustness in cell communication networks present in multicellular metazoans occurs in lock step with the expansion and diversification of SH2 domain–encoding genes (18, 19). This may explain why 111 SH2 domain–containing proteins are found in humans, whereas the unicellular eukaryotic yeast, Saccharomyces cerevisiae, contains a single protein (3). Domain shuffling of existing genes to generate proteins with novel domain organizations was likely a driving force in the transition from unicellular eukaryotes to differentiated multicellular animals (20). This process placed the modular SH2 domain in the context of other domains that have varied functions, allowing SH2 domains to participate in diverse cellular processes.
A detailed compilation of SH2 proteins is a prerequisite for bioinformatic and systems-level studies, as well as for understanding the evolution of pTyr signaling. This study represents a step toward understanding how SH2 proteins integrated with existing signaling networks to position pTyr signaling as a crucial driver of robust cellular communication networks in metazoans. Several studies have examined SH2 domains within representative organisms (3, 7) or SH2 families across two or three organisms (21-23). To generate a detailed picture of the evolution of SH2 domains and pTyr signaling, we examined the SH2 domain–containing proteins present encoded in the genomes of 21 organisms spanning the Eukaryota, and these included 16 Unikonts and 5 Bikonts. The Unikonta included in this analysis consist of 11 Metazoa (Animalia), 1 Choanozoa, 3 Amoebozoa, and 1 Fungus. Using a combination of sequence comparison and analysis of protein domain architecture and the boundary positions between introns and exons within the SH2 domain or genes encoding these domains, we assigned them into 38 discrete SH2 families and traced these families across the 21 genomes of living organisms analyzed in this study. We identified SH2 domain shuffling events that resulted in the creation of new families of SH2 domain proteins. Our analysis of SH2 domains provides insight into the evolution of modular protein domains and the rapid evolutionary expansion of gene fragments encoding particularly beneficial protein subdomains. Additional information and interactive figures are available at http://www.sh2domain.org.
RESULTS
Expansion of SH2 domains and tyrosine kinases across species
To examine the evolution of SH2 domains, we identified all genes potentially encoding SH2 domains from 21 different organisms from within the two divisions of eukaryotes, the Bikonta (“two flagella”) and the Unikonta (“single flagella”) (24) (Fig. 1A; see table S1 for a complete list of organisms in this study). Using the predictive algorithms of Protein Families (Pfam) (25) and the Simple Modular Architecture Research Tool (SMART) (26), we identified proteins containing SH2 domains for analysis. We found at least one SH2 domain–encoding gene in each of the Eukaryotes sampled (table S2). Included in this analysis are five organisms covering a diverse range of Bikonta: the green plant (Viridaeplantae) thale cress (Arabidopsis thaliana); two Excavates, an amoeba-flagellate (Naegleria gruberi) and a ciliated protozoan Tetrahymena thermophila; and two Alveolata, an omycetes plant pathogen (Phytophthora capsici) and a parasitic protozoan (Trichomonas vaginalis) (Fig. 1B and table S1). Within Unikonta, we examined two major branches, the Amoebozoa and the Opisthokonta. The Amoebozoa examined comprise two social amoeba (slime molds) of the Eumycetozoa (Dictyostelium discoideum and Dictyostelium purpureum), and an Archamoebae (Entamoeba histolytica) (Fig. 1C and table S1). Of the 16 Unikonts, we chose 13 Opisthokonts because they represent important periods in the evolution of metazoan complexity (Fig. 1A; see fig. S1 for an evolutionary time line). These are a yeast (S. cerevisiae), choanoflagellate (Monosiga brevicollis), sea anemone (Nematostella vectensis), roundworm (Caenorhabditis elegans), fruit fly (Drosophila melanogaster), mosquito (Aedes aegypti), sea urchin (Strongylocentrotus purpuratus), sea squirt (Ciona intestinalis), zebrafish (Danio rerio), Western clawed frog (Xenopus tropicalis), opossum (Monodelphis domestica), house mouse (Mus musculus), and humans (Homo sapiens) (Fig. 1C). We included pairs of closely related organisms, such as D. melanogaster and A. aegypti or H. sapiens and M. musculus, to assess variation within closely related lineages. For each organism, all identified genes are listed along with their gene name, gene ID, aliases, chromosomal location, SH2 family, and presence of other domains (table S3). We analyzed seven additional organisms for SH2 domains (section S1); however, because the annotation of these genomes is incomplete, they are described separately (table S4).
Fig. 1.
Coevolution of SH2 domains and tyrosine kinases. (A) A tree of the major divisions within the Eukaryote that are represented by the 21 organisms in this study. The numbers of organisms studied in the different kingdoms, supergroups, and phyla are indicated in yellow circles. (B) SH2 domain proteins and protein kinases were identified in five organisms that fall within the Bikonta, representing three supergroups and five phyla. (C) pTyr-binding SH2 domains appear in the Unikont branch of Eukaryotes. The total complement of kinases, PTKs, and SH2 domain–containing proteins is noted for each species analyzed. An approximate time line for the separation of the major branches is shown in fig. S1. The total number of kinases and PTKs were derived from the literature, as was the number of SH2 domain proteins for human. The total number of SH2 domain–containing proteins from the organisms listed was identified with SMART and Pfam as described in Materials and Methods. (D) The co-expansion of PTKs and SH2 genes is apparent as the numbers of each track one another closely from M. brevicollis to H. sapiens. (E) The percentage of the genome devoted to encoding PTKs and SH2 domains correlates across the different metazoan genomes (see table S1 for the list of two-letter representations of genus species) with a coefficient of determination (R2) of 0.95.
To examine the coevolution of PTKs and SH2 domains, we compared the number of genes encoding SH2 domains to the number encoding kinases and PTKs across all Unikonts (Fig. 1C), including those that lack tyrosine-specific kinases (S. cerevisiae, E. histolytica, D. discoideum, and D. purpureum) (27, 28). For this comparison, we used the comprehensive lists of kinases and PTKs previously described for humans (29), D. discoideum (28), M. brevicollis (7), S. purpuratus (30), and other eukaryotes (31, 32). In general, PTKs expand at a rate similar to that of SH2 domains (Fig. 1D). The correlation between the percentage of PTKs and SH2 domains in their respective genomes is 0.95 (Fig. 1E). Because many SH2 domain–containing proteins also have a tyrosine kinase domain, we also compared their rates of expansion after removing those proteins to determine the rate of expansion of SH2 and PTKs independently of each other. The rate of expansion did not change, with the exception of C. elegans, which among the organisms analyzed contains a uniquely large set of proteins containing both SH2 and PTKs (fig. S2, A and B). In comparison, PDZ domain–containing proteins, which recognize short C-terminal motifs and phosphoinositides, did not expand at the same rate, and their expansion did not correlate across genomes (fig. S2, C to E). Although both SH2 and PDZ domains emerged and proliferated at the metazoan junction in M. brevicollis, their expansion rates differ from M. brevicollis to N. vectensis, N. vectensis to C. elegans, and X. tropicalis to M. domestica (fig. S2C).
Although SH2 domains are present in all five Bikonts in this study, they may not be a universal feature of the Bikonta. Neither the flagellated protozoan parasite Giardia lamblia nor the coccolithophores Emiliania huxleyi have identifiable SH2 domains encoded in the sequences reported to date, although their genome sequences remain incomplete. Relatively few SH2 domains are found in any of the Bikonts, with a maximum of four identified in A. thaliana and N. gruberi (Fig. 1B). These organisms have homologs of the SH2 protein Spt6 (ortholog of Supt6h in humans), which may be a candidate common ancestor to all eukaryotic SH2 domains (table S2).
SH2 domains appear to have proliferated beyond Spt6 homologs across multiple branches of Eukaryota. We found extensive expansion of SH2 domain proteins within Metazoa, as well as the Choanozoa M. brevicollis, and the amoebas D. discoideum, D. purpureum, and E. histolytica (Fig. 1C). However, we observed this expansion only within specific Opisthokont branches and not within others, such as the Fungi. In Amoebozoa, SH2 domains are found either coincident with expansion of tyrosine-like kinases, as in the dictyostelids, which lack conventional PTKs specific for tyrosine (28), or are linked directly to a multifunctional serine/threonine/tyrosine kinase (E. histolytica) (Fig. 1C and table S2). The only SH2 proteins that appear common between living Amoebozoa and Opisthokonts are the transcriptional regulators Spt6 and the signal transducer and activator of transcription (STAT) proteins (table S2). Henceforth, family names of proteins or domains are indicated in all capitalized letters and specific protein names have only first letters capitalized, regardless of the organism of origin. There are no STAT proteins in Fungi, implying either that the STAT proteins in Amoebozoa and Opisthokonts are a product of convergent evolution or that they were lost in Fungi. Given the conserved STAT protein sequence and architecture, it appears likely that these proteins, which predate the split between Opisthokonts and Amoebozoa, were lost in the Fungi lineage (14). This further suggests that the STAT SH2 domains may represent the earliest conserved pTyr-binding SH2 domains. In the Opisthokonts, the emergence and expansion of SH2 domains coincides with the development of kinases and phosphatases specific for tyrosine (8).
The most primitive metazoan in this study, the cnidarian N. vectensis, encodes 29 SH2 domains, which is one-fourth of those found in M. brevicollis and less than half the number identified in C. elegans. At the split within deuterostomes (phylum: Chordata and Urochordata), a substantial increase in the number of SH2 domain proteins is observed (Fig. 1C). In chordates, an increase in SH2 domain proteins is accompanied by a concomitant increase in PTKs, yet the SH2 domain proteins exceeded that of the PTKs.
Classification of SH2 domain families
Genes encoding SH2 domains underwent a substantial expansion across the 11 Opisthokont organisms examined. To better understand the evolutionary history of these proteins, we classified them into discrete families. We defined an SH2 domain family as a conserved cluster of SH2 domains that can be traced back through various lineages to a single origin and that likely represents a functionally distinct group, which may be subject to distinct evolutionary pressure. Several approaches have been used to identify relationships between different genes. Examination of protein sequence alignment with ClustalW provides a one-dimensional (1D) method of determining the relationship between different proteins by protein sequence homology. However, this approach alone is insufficient to classify SH2 domains into families (3, 21). Protein domain organization, also referred to as domain architecture, describes the composition and order of domains within a protein (for example, SH3, SH2, SH3) and is useful in detecting evolutionarily distant homologs on the basis of shared domains rather than on the basis of pairwise sequence similarity (33). From our previous analysis of human SH2 domain proteins, many contain identical domain organizations but cannot be classified into the same family because of divergence in sequence homology (3).
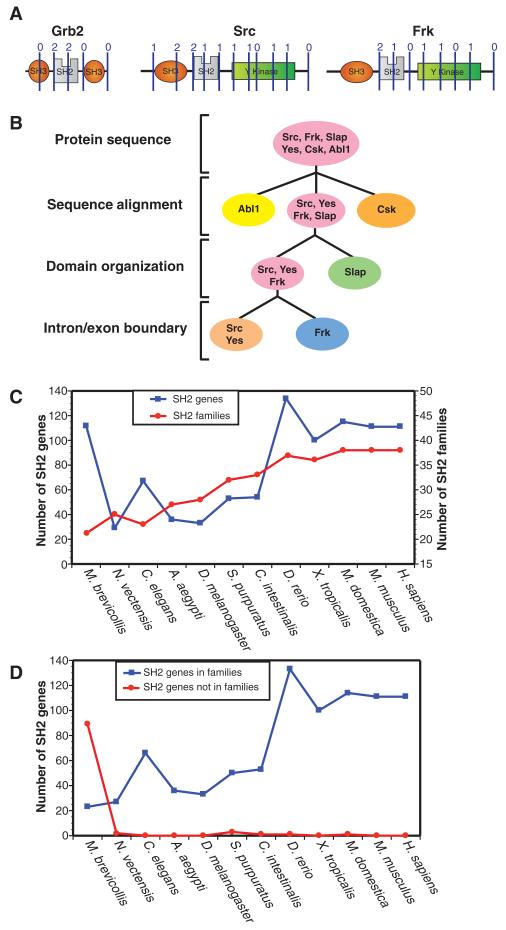
Another analysis of the tyrosine kinase superfamily suggested that homology assignments using an “intron/exon code” is a useful qualitative method in cases when the domain sequence does not provide sufficient phylogenetic information (34). At the genomic level, splicing and exon shuffling represent an evolutionary mechanism of conserving critical sequences and cellular functions (35). Furthermore, a strong correlation exists between domain boundaries and exons across a number of eukaryotic genomes (36). Therefore, we obtained the splicing patterns of all human SH2 domains from the Ensembl database to define the “intron/exon code” of this domain. We complemented this analysis with the domain architecture tool SMART to reveal the position and phase of the intron/exon code (an intron is in phase 0, 1, or 2 if it falls before the first, second, or third base of a codon, respectively). The positions of the identified splice sites (shown as blue lines with the respective phase number listed above) are shown across the domain organization of three different SH2 proteins (Fig. 2A; see section S2 for an analysis of splice patterns). On the basis of available SH2 domain structures, we mapped the intron/exon code for all SH2 domains onto an SH2 domain protein sequence alignment containing secondary-structure elements (fig. S3, A and B) (3). Hierarchical clustering of the intron/exon code aligned to the SH2 domain secondary-structure elements revealed distinct, yet conserved, splice patterns among related SH2 domains (Fig. 3C).
Fig. 2.
The classification of SH2 families. (A) Genomic structure was mapped as intron-exon boundary positions overlaid on the secondary structural motifs of SH2 domains as an additional means of assessing relatedness. Intron and exon splice sites of three example SH2 proteins, indicated with lines and numbers above, were identified with SMART and Ensembl (see Materials and Methods). A ClustalW alignment and hierarchical clustering of human SH2 domains with the splice sites indicated can be found in fig. S3. (B) A hierarchical method for defining SH2 domain families by protein sequence alignment (ClustalW), domain organization (SMART and CCD), and intron/exon splicing patterns (Ensembl and SMART) was developed to define SH2 domain families. The flow chart follows the classification method to distinguish between representative SH2 proteins with similar primary sequences: Abl1, Csk, Slap, Yes, Src, and Frk. (C) The graph represents the number of SH2 proteins and the conserved SH2 families for 11 organisms from H. sapiens and M. brevicollis. (D) The number of proteins that can or cannot be classified into one of the 38 families (table S3).
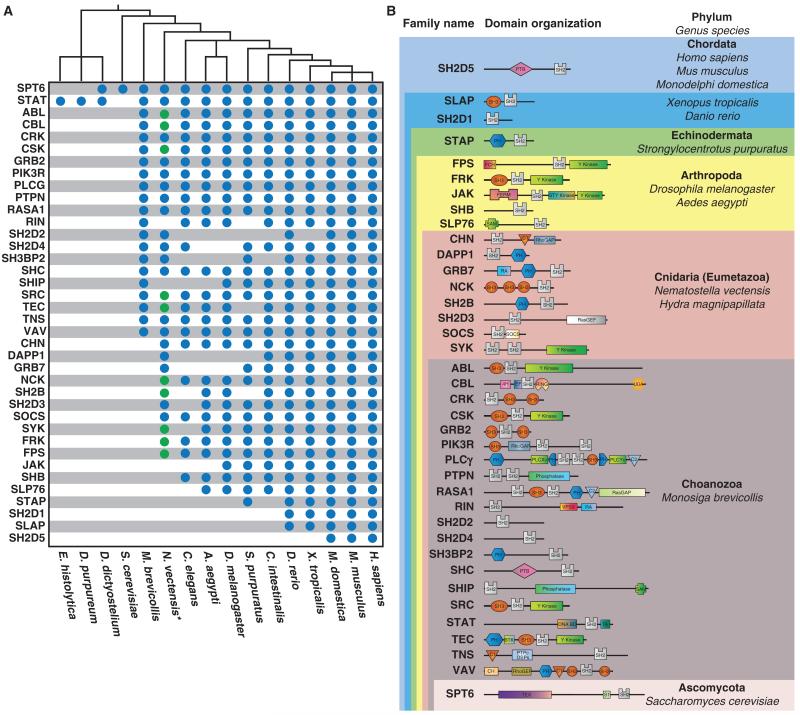
Fig. 3.
The origin of SH2 domain families. (A) The presence or absence of SH2 families across organisms in Unikonta. (*) The cnidarian H. magnipapillata contains SH2 families absent in N. vectensis (green circles) (B) The first appearance of each of the 38 SH2 families is noted with the architecture of each protein represented in domain form (see fig. S4 for the complete key to the domains).
Sorting by the three criteria of sequence alignment, protein domain organization, and splice pattern revealed a total of 38 SH2 domain families within the human genome (see Materials and Methods and Table 1). This analysis confirmed prior functional analysis and provided a robust classification of SH2 proteins into related families. For example, splice site patterns distinguished the FRK family (Brk, Srms, Frk) from the SRC family of kinases (SFKs) composed of Blk, Fgr, Fyn, Hck, Lck, Lyn, Src, and Yes (Fig. 2B). Members of the FRK family have high sequence similarity to SFKs and share the same protein domain organization, yet have different splice junctions and are functionally distinct (21). Examination of intron/exon splicing classified Frk as a member of the FRK family, showing that it is divergent from the SFKs (Fig. 2A). In a related example, Src, Yes, Frk, Slap, Csk, and Abl1 contain both an SH3 and an SH2 domain, and all except Slap contain a tyrosine kinase domain. Sequence alignment clearly distinguished Abl1 and Csk as belonging to distinct families, but failed to distinguish between Src, Yes, Frk, and Slap (Fig. 2B). Slap and Src demonstrate sequence homology and conserved splice patterns, suggesting close evolutionary relatedness, yet are classified as separate families because they differ in domain organization.
Table 1.
The 38 SH2 families. All 111 human SH2 domain proteins were cataloged into 38 distinct SH2 families by means of sequence alignments, domain organization, and intron/exon splicing. For each family we determined whether an individual approach would be sufficient to categorize a family (Yes or No). If a method failed to classify a family, we noted the reason(s) in the comments column.
| Family | Human gene names | Sequence alignment |
Domain organization |
Splice sites | Comments |
|---|---|---|---|---|---|
| ABL | ABL1, ABL2 | Yes | Yes | Yes | |
| CBL | CBL, CBLB, CBLC | Yes | No | Yes | CBLC lacks a UBA domain |
| CHN | CHN1, CHN2 | Yes | Yes | Yes | |
| CRK | CRK, CRKL | Yes | Yes | No | Different splice sites |
| CSK | CSK, MATK | Yes | No | Yes | Same domain organization as SRC and FRK |
| DAPP1 | DAPP1 | Yes | Yes | Yes | |
| FPS | FES, FER | Yes | Yes | Yes | |
| FRK | FRK, BRK, SRMS | No | No | Yes | FRK aligns with SRC, similar domain organization to Src, Csk, Txk |
| GRB | GRB2, GADS, GRAP | Yes | Yes | No | Shares same splice patterns as SHB |
| GRB7 | GRB7, GRB10, GRB14 | Yes | Yes | Yes | |
| JAK | TYK2, JAK1, JAK2, JAK3 | Yes | Yes | Yes | |
| NCK | NCK1, NCK2 | Yes | Yes | Yes | |
| PI3KR | PIK3R1, PIK3R2, PIK3R3 | Yes | No | Yes | PIK3R3 lacks an SH3 and a RhoGAP domain |
| PLCγ | PLCG1, PLCG2 | Yes | Yes | Yes | |
| PTPN | PTPN6, PTPN11 | Yes | Yes | Yes | |
| RASA1 | RASA1 | Yes | Yes | Yes | |
| RIN | RIN1, RIN2, RIN3 | Yes | Yes | Yes | |
| SH2B | APS, LNK, SH2B | Yes | No | Yes | Same domain organization as SH3BP2 and STAP |
| SH2D1 | SH2D1A, SH2D1B | Yes | Yes | No | Shares same splice patterns as SHIP |
| SH2D2 | SH2D2A, HSH2, SH2D7 | Yes | Yes | Yes | |
| SH2D3 | SH2D3A, SH2D3C, BCAR3 | Yes | Yes | Yes | |
| SH2D4 | SH2D4A, SH2D4B | Yes | Yes | Yes | |
| SH2D5 | SH2D5 | Yes | No | Yes | Same domain organization as SHC |
| SH3BP2 | SH3BP2 | Yes | No | Yes | Same domain organization as STAP and SH2B |
| SHB | SHB, SHD, SHE, SHF | Yes | Yes | No | Shares same splice patterns as GRB2 |
| SHC | SHC1, SHC2, SHC3, SHC4 | Yes | No | Yes | Shares the same domain organization as SH2D5 |
| SHIP | SHIP1, SHIP2 | Yes | No | No | SHIP2 contains a SAM domain, shares the same splice pattern as SH2D1 |
| SLAP | SLAP, SLAP2 | No | Yes | No | SLAP and SLAP2 share sequence similarity to SRC and share intron/exon boundaries |
| SLP76 | SLNK, BLNK, SLP76, MIST | Yes | No | Yes | SLP76 contains a SAM domain |
| SOCS |
SOCS1, SOCS2, SOCS3, SOCS4, SOCS5, SOCS6, SOCS7, CISH |
Yes | Yes | No | Different splice patterns |
| SRC |
SRC, FYN, LCK, FGR, YES, LYN, HCK, BLK |
No | No | Yes | Shares sequence similarity and similar domain organization to FRK |
| STAP | BKS, BRDG1 | Yes | Yes | Yes | Same domain organization as SH3BP2 and SH2B |
| STAT |
STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, STAT6 |
Yes | Yes | Yes | |
| SPT6 | SUPT6H | Yes | Yes | Yes | |
| SYK | ZAP70, SYK | Yes | Yes | Yes | |
| TEC | BMX, TEC, BTK, ITK, TXK | Yes | No | Yes | TXK lacks a PH and BTK domain |
| TNS | TNS1, TNS3, TENC1, TNS4 | Yes | No | Yes | TenC1 has a C1 and TNS1 has a PTPc domain |
| VAV | VAV1, VAV2, VAV3 | Yes | Yes | Yes |
Exceptions in family classifications were made in the cases of the group containing Crk and CrkL and the group containing Socs and Cish because of apparent similarity in function, conserved sequence and protein architecture, and lack of clear evidence from genomic structure. Crk and CrkL differ in splice site position, which may have indicated that they should be placed in separate groups; however, this is likely a result of movement of the position of the splice site that accompanied the generation of an extended loop (called the DE loop) within the Crk SH2 domain. The SOCS (suppressor of cytokine signaling) family was grouped together despite Socs1, Socs3, Socs4, Socs5, and Socs6 lacking splice sites in their SH2 domains (fig. S3C). Socs2 shares splice sites with Cish, whereas the splice pattern for Socs7 is unique. Further analysis of evolutionary history and function is necessary to determine whether the SOCS family can be further divided into two or more families. For each family, by detailing the approaches used to define an SH2 domain into a particular family (Table 1), we found that each method has its own limitations; therefore, the combination of the three approaches should be more effective in appropriately defining relationships compared to the assignments from any individual method.
By compiling a list of the SH2 domain proteins of all the Opisthokonta organisms into the appropriate family (table S3), we observed that SH2 domain families increase in number as organismal complexity increases (Fig. 2C). The emergence of new SH2 families rises steadily from M. brevicollis through to the emergence of vertebrates, at which point it plateaus. Only one new family appeared in mammals that was not present in teleosts. Many proteins found within the choanoflagellate M. brevicollis were not classified into any of the 38 conserved SH2 families (Fig. 2D). Choanoflagellates encode a wide variety of SH2 domain proteins, a large fraction of which have no known orthologs outside of choanoflagellates (7, 8), which suggests that many of these unique proteins evolved within the choanoflagellate lineage rather than in an ancestor shared with metazoans. We also noted a small number of proteins across multiple metazoan organisms that could not be classified into one of these families (Fig. 2D and table S3), suggesting that these proteins either were lost or represent forms specific to these lineages and not present in common ancestors.
The origin of SH2 domain families
Both the timing and the rapid expansion in the number of SH2 domain–encoding genes provide a glimpse into the mechanisms for promoting coordinated emergence and increasing sophistication of pTyr signaling during eukaryotic evolution (11, 12). Using the catalog of discrete SH2 families, we traced the origins and diversification of these families (see Materials and Methods and section S3 for descriptions of ortholog predictions). The most common recognizable SH2 domain across all Eukaryota is the conserved gene encoding the transcriptional regulator Spt6, which is present in almost all eukaryotes in our study with the exception of E. histolytica and D. purpureum (Fig. 3A and table S2). We found SH2 diversification beyond Spt6 in various Unikonts and Bikonts, but not Fungi. In general, the SH2 domain proteins in Amoebozoa and Bikonts were distinct from those seen in Metazoa (table S3). Of the 15 SH2 domain proteins in D. discoideum, 11 have no obvious orthologs in metazoans (table S3). The only exceptions are the Spt6 and STAT proteins, which have identifiable orthologs in metazoans and choanoflagellates. In addition to the SH2 domain, a core set of associated domains including, but not limited to, SH3, pleckstrin homology (PH), and tyrosine kinase (TyrK) were present from choanoflagellates to metazoans. However, many unique domain combinations including epidermal growth factor (EGF), tumor necrosis factor (TNF), and Jumonji C (JmJc) domains are present in M. brevicollis (table S3), suggesting widespread experimentation of pTyr signaling components in early choanoflagellates (7, 37). The domain shuffling present in M. brevicollis implies a common ancestor for 20 of the 38 SH2 domain families in humans, which are found in kinases, phosphatases, adaptors, G protein (heterotrimeric guanosine triphosphate–binding protein) signaling, and others (Fig. 3B; see fig. S4 for a key to the domains). Thus, a common ancestor of metazoans and choanoflagellates may have evolved diversification of pTyr signaling through events of domain shuffling and duplication.
Within Metazoa, additional SH2 families arise to create new signaling events to coordinate complex biological systems. Early in eumetazoans, before the radiata-bilateria split, we identified eight new SH2 families that are present in the cnidarians N. vectensis and Hydra magnipapillata, but not in M. brevicollis. These include the adaptor families GRB7, NCK, and SH2B (Fig. 3B). The appearance of adaptors coincides with a broad expansion of receptor tyrosine kinases (RTKs) (34). Before the protostome-deuterostome split, additional families and signaling networks emerge marked by SH2 families, such as SHB, SLP76, FRK, JAK, and FPS (Fig. 3B). Within deuterostomes, only four new SH2 families appear, indicating that the basic domain organization of most SH2 families existed before the protostome-deuterostome split. The sole new SH2 family to appear in Echinodermata is the STAP family (Bks and Brdg1 in humans; Stap2a and Stap2b in zebrafish D. rerio). The other three novel SH2 families that emerge in chordates before the teleost-tetrapod split are SH2D5, SH2D1, and SLAP. Sh2d5 is a scaffold protein containing a PTB and a divergent SH2 (3). Sh2d1a and Sh2d1b (also referred to as Sap and Eat-2, respectively) are regulators of immune signaling (38). The proteins Slap and Slap2 appear to be truncations of an SFK containing the N-terminal SH3 and SH2 domains but lacking the TyrK domain (39), which likely represent the emergence of a new family of SH2 proteins through duplication of an SFK and loss of the kinase domain.
Patterns of SH2 gene duplication and loss
In certain organisms, gene duplication has specifically expanded certain SH2 domain families. For example, C. elegans exhibits a marked spike in total SH2 proteins but not an equivalent increase in SH2 families (Fig. 2C). This increase in SH2 proteins resulted from gene duplication producing 34 copies of the orthologs Fes and Fer, which are in the FPS family (fig. S5). Before the split between Echinodermata and Chordata, 34 of 38 conserved families are present (Fig. 3B). There is, however, a substantial increase in the total number of SH2 proteins within chordates, between sea squirt C. intestinalis (urochordate) and frog X. tropicalis (craniate) (Fig. 2C). Such amplification is not the result of developing new architectures but of gene duplication (Fig. 3, A and B).
Early in chordate evolution, two rounds of whole genome duplication have been proposed to explain the presence of duplicate gene copies (40), and this expansion may underlie the development of novel body forms, such as the origin of the vertebrate skeleton (41). In sea squirt and sea urchin that follow the first such duplication, most SH2 families remain present as single copies, whereas select families (SOCS, GRB2, and SRC) contain multiple copies (fig. S5). A second round of duplication occurs before the emergence of craniata (vertebrates). The highest frequency of duplication for most SH2 families is in the lineage preceding vertebrates represented in this study by D. rerio (fig. S5). Hence, within early chordate evolution, gene duplication to amplify the number of SH2 domain proteins may have been advantageous, but there appears little pressure to develop entirely new architectures. This may indicate that a basic set of SH2 families was sufficient for the essential signaling in chordates and that increasing complexity was accommodated largely by gene duplication and tissue-specific expression.
Although gene duplication may explain the increase in the number of SH2 domain proteins in chordates, we also noted events of gene loss (Fig. 3A). Our classification showed that instances where SH2 domain proteins absent in more complex organisms were not common in the development of pTyr signaling, but did appear to occur in specific lineages. The gene encoding Syk is absent from C. elegans, but is present in both the cnidarian Hydra vulgaris (22) and the various arthropods examined. Thus, the SYK family is not present in C. elegans but is present in the other organisms. STAP is apparently absent from sea squirt but is present in both sponges (simpler) and chordates (more complex). Similarly, RIN is absent from S. purpuratus but is found in the mosquito A. gambiae. This is indicative of different evolutionary pressures driving the retention of specific SH2-mediated signaling pathways while also driving different degrees of expansion during metazoan evolution.
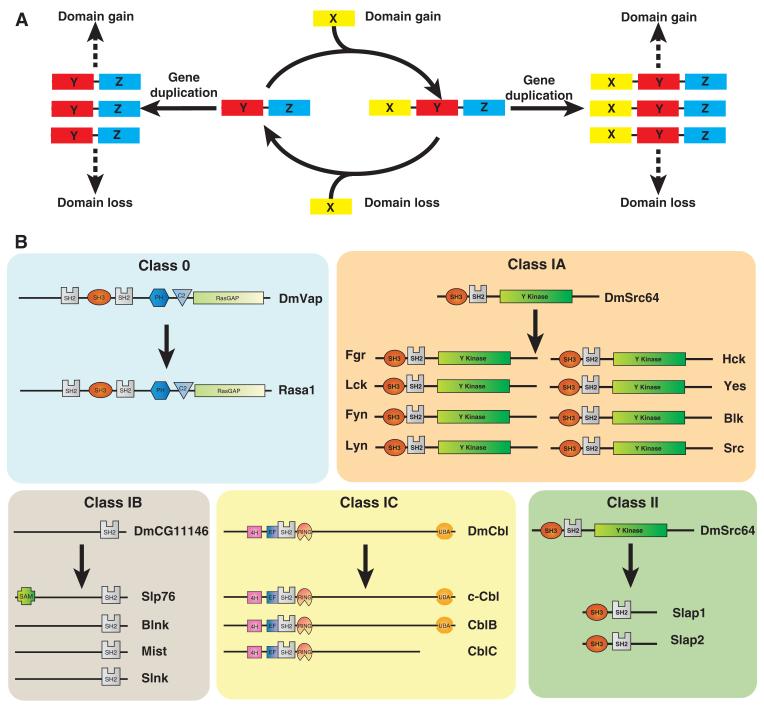
Classifying evolutionary events for SH2 families
Broadly speaking, three major events appear to drive the diversification of SH2-encoding genes: whole gene duplication and domain loss and domain gain (Fig. 4A). To better understand the evolutionary mechanisms, we defined five classes of events that captured the essence of the changes that produced diversification of the modular SH2 domain (Fig. 4B). Class IA, B, and C represent diversification through gene duplication, without (class IA) or with domain gain (class IB) or with domain loss (class IC). The most common event for SH2 families was class IA, which occurred for 25 of 38 SH2 families (table S5). This is particularly prevalent within the chordate lineage. For example, a single copy of the gene encoding Src in fruit fly, DmSrc64, becomes multiplied to create eight SFK members in vertebrates (table S5 and Fig. 4B, class IA).
Fig. 4.
Genetic events that lead to diversification of SH2 families. (A) Diagram representing the events of domain gain or loss either before or after events of gene duplication. (B) Classification of SH2 families on the basis of genome duplication events and domain loss or gain. Class 0, represented by the RASA1 family, does not undergo gene duplication. Class IA, represented by the SRC family, undergoes events of complete gene duplication without domain loss or gain. Class 1B, represented by the SLP76 family, diversifies by acquiring new domains and undergoing gene duplication. Class IC, represented by the CBL family, undergoes gene duplication with domain loss. Class II, represented by the SLAP family, undergoes domain loss followed by gene duplication. For additional information on gene duplications within individual families, see fig. S5. See table S5 for a complete list of SH2 families within their distinct class.
Class IB represents the evolution of new domains through domain acquisition during events of gene duplication (table S5 and Fig. 4B). An example is Slp76, which acquires a SAM domain that is lacking in its relatives Blnk, Slnk, and Mist. This occurs concomitantly with the expansion of this family in the chordate lineage. Additional mechanisms for diversifying related family members include events such as intron or exon shuffling, splice site slippage, or splice site loss or gain, all of which may contribute to evolution through class 1B and 1C. For example, an exon insertion in Gads creates this unique region of sequence within the protein, thus producing a new member of the GRB2 family that is distinct from both the fruit fly ortholog DmDrk and the mammalian family members Grap and Grb2 (fig. S6A).
Class IC is defined by a gene duplication event followed by loss of a domain or region. The CBL family contains an example in which mammalian Cbl-C lacks a C-terminal region containing a ubiquitin-associated (UBA) domain that is present in its ortholog encoded by DmCbl in fruit fly and human paralogs c-Cbl and Cbl-B (Fig. 4B).
We defined class II events as the creation of SH2 families from domain loss, which then appears to be followed by gene duplication (table S5 and Fig. 4B). This is exemplified by the members of the SLAP, SH2D1, and SYK families. The proteins in the SLAP family appear to originate from a truncation of an SFK, whereas Sh2d1a and Sh2d1b appear to have arisen from a fragment of the phosphatase Ship. In both cases, the truncated forms (Slap1 and 2, Sh2d1a and b) lack the catalytic region of their ancestral form and appear to act as regulators of signaling pathways that also involve their parent proteins (42-44). The ancestral origins of these genes can be supported by sequence alignment and conservation of intron and exon splice sites (Fig. 2A). The loss of domains within SH2-encoding genes, particularly by truncation, is a feature of higher metazoans because both SLAP and SH2D1 families arise in chordates (Fig. 3B). Another example of a class II event is represented by the PTKs of the SYK family in chordates, which lack the ankyrin repeats that are present in all lower organisms from sea squirt C. intestinalis to the arthropods A. aegypti and D. melanogaster (22) (table S3). Such genetic events allow evolutionary divergence to occur in a stepwise process.
Although most SH2 families exhibit gene duplication events, this is not a universal feature. Several genes encoding SH2 proteins, notably Spt6, Rasa1, Sh3bp2, and Dapp1, appear not to have undergone productive duplication events to produce multiple family members in the species studied and thus are classified as class 0 (Fig. 4B and table S5). This may reflect that any duplicates of these genes either conferred no selective advantage and were not retained, or were actively selected against as deleterious (fig. S5).
Evolving new protein interactions between SH2 family members
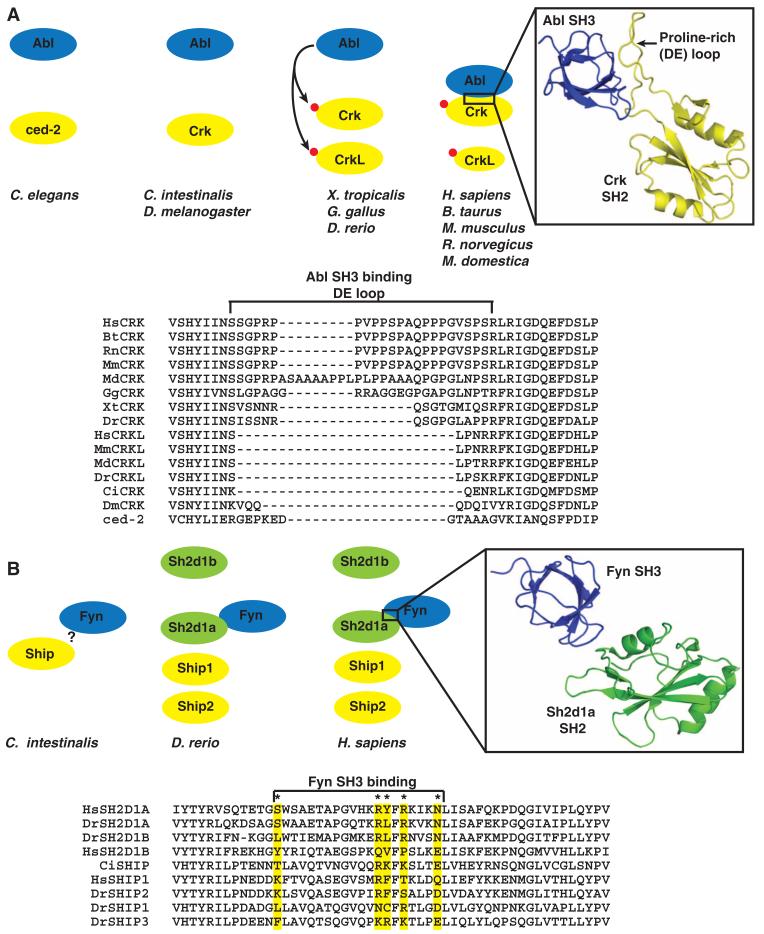
The appearance of duplicate genes may have multiple outcomes (18). One possibility is that duplication results in a duplicate gene free from the selective pressure constraining the parental gene. Whereas an essential function is maintained by one copy, the second copy may tolerate mutations that would otherwise be detrimental to fitness. This may sanction the creation of novel proteins through domain gain or loss that in turn promote increased complexity and diversity in signaling. A duplicate version, freed from the requirement to maintain binding partners, may evolve to discriminate pTyr motifs different from the parental SH2 domain (45). From both sequence alignments and protein interactions, it is apparent that in some cases, duplicated SH2 domain proteins evolved distinct and novel interactions. A case in point is the CRK family, composed of Crk and its paralog CrkL in mammals and their common orthologs DmCrk in D. melanogaster and ced-2 in C. elegans. Crk and CrkL likely result from gene duplication before vertebrates (Fig. 5A). Clustal alignment of the CRK family SH2 domains from multiple chordates, such as human, cow, opossum, chicken, frog, zebrafish, and the urochordate sea squirt, to invertebrates, such as fruit fly and worm, revealed that the human Crk has an extended proline-rich region of sequence in the DE loop (Fig. 5A). This loop interacts with the SH3 domain of Abl (46, 47), effectively creating a unique interaction face within an existing SH2 domain (Fig. 5A). This connection links two existing signaling networks, potentially serving as a foundation for achieving complexity. This proline-rich insertion is of recent origin and appears only in the mammalian orthologs of Crk, whereas only a short insertion in the DE loop is present in other nonmammalian vertebrates, such as zebrafish, frog, or chicken. No extended proline-rich sequence and no visible insertion are present either in CrkL or in the D. melanogaster or C. elegans orthologs. Although Crk and Abl orthologs are both present in D. melanogaster and C. elegans, only in mammals is this loop, which is needed for the interaction between Crk and Abl, present. Furthermore, using intron and exon splice site analysis, we found a splice site within the unique proline-rich sequence (DE loop) of the SH2 domain of Crk (fig. S6B), which suggests that an analysis of splice sites may be a mechanism to determine evolutionary divergence between related SH2 domain proteins.
Fig. 5.
Evolving new SH2 interactions between duplicate genes. (A) In vertebrates, both Crk and CrkL contain a tyrosine residue that is phosphorylated by Abl or other tyrosine kinases (red circle). Phosphorylation on this conserved site promotes the intramolecular interaction with the respective SH2 domain. In mammals, but not teleosts, the Crk SH2 domain has evolved a loop between β strands D and E (DE loop) that creates a binding site for the SH3 domain of Abl to promote this interaction and phosphorylation of Crk. A ribbon diagram shows the human Crk (yellow) SH2 domain with the extended proline-rich loop interacting with the SH3 domain of Abl (blue). The structure was adapted from PDB 1JU5. The sequences of the SH2 domain from the CRK family in C. elegans (ced-2), D. melanogaster (DmCrk), C. intestinalis (CiCrk), D. rerio (DrCrk, DrCrkL), M. domestica (MdCrk, MdCrkL), Bos taurus (BtCrk, BtCrkL), M. musculus, (MmCrk, MmCrkL), and H. sapiens (HsCrk, HsCrkL) were aligned with ClustalX. For a detailed sequence analysis, see also fig. S6B. (B) The Sh2d1a and Sh2d1b adaptors arose through gene duplication and domain loss from the Ship-encoding gene in C. intestinalis, resulting in four related SH2 domains in vertebrates. The SH3 domain of Fyn makes contact with residues within the SH2 domain of Sh2d1a, but not with Sh2d1b and Ship. These contact residues are conserved in only vertebrate Sh2d1a. The structure was adapted from PDB1M27. The * indicates key contact residues previously reported (48).
The adaptor molecule Sh2d1a, also known as Sap, has evolved to diversify its biological function from its apparent parental protein Ship and paralog Sh2d1b (Fig. 5B). Sh2d1a and Sh2d1b belong in the SH2D1 family and are classified as class II because they arose from the phosphatase Ship through duplication and loss of the catalytic domain. Lacking the enzymatic phosphatase domain, Sh2d1a evolved as a modulator of signaling and an adaptor to recruit the SH3 domain of Fyn (48, 49). The paralog Sh2d1b does not interact with Fyn, but appears to facilitate signaling in antigen-presenting cells by acting as a scaffold through the multiple tyrosine residues in its short C-terminal tail, which propagate downstream signaling (50). Members of the SH2D1 family may also modulate signaling by competing for Ship1 for binding to tyrosine-based motifs in certain receptors (51). All members of this family are detected almost exclusively in cells of the immune system (42, 52) and act chiefly in immune cell signaling in mammals. These examples demonstrate the evolution of SH2 domains to acquire additional complexity not only through duplication events with potential domain loss or gain, but also through the ability to acquire interactions that are independent of pTyr within the SH2 domain itself.
Emergence of pTyr-binding sites in SH2 domain proteins
Numerous SH2 domain proteins function as scaffolds and have the capacity to recruit other protein interaction domains to build robust signaling networks. Phosphorylation of SH2 domain proteins to recruit other SH2 domain proteins or mediate an intramolecular interaction may function as a more general paradigm in the evolution of complex signaling networks. We analyzed several human SH2 domain proteins that are phosphorylated on Tyr residues, which recruit other SH2 domains, and determined whether these sites are present in early eukaryotes (53). We found numerous examples of SH2-mediated interactions with other SH2 domain proteins that appear to have evolved late in metazoans and are present in vertebrates, and we categorized these interactions into three major groups (Fig. 6).
Fig. 6.
Tyrosine pTyr sites evolved late in SH2 domain proteins. New sites of tyrosine phosphorylation create inter- and intramolecular binding sites for SH2 domains, as illustrated by the proteins Crk, Shc, and Plcγ. (A) The adaptor protein Crk contains an intramolecular tyrosine phosphorylation site that is present in the H. sapiens and D. rerio orthologs but absent in the fruit fly D. melanogaster (DmCrk). Tyrosine phosphorylation at this site by the kinase Abl creates an intramolecular binding site for the Crk SH2 domain. (B) The scaffold Shc contains several Y-x-N (x, any amino acid) motifs that recruit the adaptor protein Grb2. This motif is present in D. melanogaster and all vertebrate paralogs (Shc1-4) but not in the Shc protein from the choanoflagellate M. brevicollis or the nematode C. elegans. (C) SH2 domain proteins within a family can differ in the presence of pTyr sites. Plcγ1 and Plcγ2 contain identical domain organizations, yet only Plcγ1 contains a C terminus pTyr site capable of recruiting SH2 domains. This binding site is present among vertebrates but absent in the choanoflagellate M. brevicollis and fruit fly D. melanogaster. For more examples, see fig. S8.
The first group of interactions is represented by intramolecular regulation through tyrosine phosphorylation, which is exemplified by the CRK family (Fig. 6A). Within the CRK family, an intramolecular reorganization between the SH2 domain and the site phosphorylated by the tyrosine kinase Abl on Crk (Tyr221) and CrkL (Tyr207) plays a role in CRK function (54). Examination by sequence alignment revealed that this tyrosine phosphorylation site is present in vertebrates including zebrafish and mammals, but is absent in the orthologs in D. melanogaster and C. elegans (fig. S6B). The insertion of the intramolecular phosphorylation site on both Crk and CrkL evolved after these two paralogs were produced from a gene duplication event (fig. S7) and, therefore, may have evolved for regulatory control between the duplicate copies. The protein-binding activity of Crk is therefore likely regulated by a mechanism similar to that of the SFKs in which the pTyr site within the C-terminal tail creates an intramolecular SH2 binding site, leading to autoinhibition of the kinase (55, 56). Consistent with a regulatory role for this phosphorylation site, aviral version of Crk (v-Crk) is truncated before Crk Tyr221 and forms constitutive complexes with Abl and other proteins, which plays a role in v-Crk transformation (57).
Most SH2 domain proteins present across different families are tyrosine phosphorylated within their mammalian orthologs. Tyrosine phosphorylation can act to recruit other SH2 domains, which we categorize as a separate group (Fig. 6B). For the scaffold protein Shc, tyrosine phosphorylation of the pY-x-N (x represents any amino acid) motif recruits the adaptor protein Grb2 to the complex (Fig. 6B) (58). This motif is present at two sites in all four paralogs (Shc1, Shc2, Shc3, Shc4) in mammals and only a single site in D. melanogaster. Although Grb2 is present in the nematode C. elegans, these phosphorylation sites are absent in Shc in this organism. This can be observed among numerous SH2 domain proteins including Vav, Slp76, Cbl, Syk, and possibly many other SH2 domain proteins (fig. S8).
Gene duplication can give rise to multiple copies from a single copy of a SH2 domain protein, which may produce two proteins with identical domain organizations, but with diverse functions, depending on the context of the signaling network (18). We grouped examples of the evolution of sites of tyrosine phosphorylation in individual family members into a distinct set (Fig. 6C). In the case of the phospholipase C–γ (PLCγ) domain family, tyrosine phosphorylation may not play a role in protein function in early eukaryotes, such as M. brevicollis or D. melanogaster. However, after gene duplication, human PLCγ1 gains a Tyr that is phosphorylated and capable of recruiting the SH2 domains of Abl, Crk, Nck, Rasa1, or Vav (59), whereas Plcγ2 lacks this site (Fig. 6C). This further highlights additional levels of complexity by which a duplicated member of a single family can adapt within a signaling context.
Evolution of pTyr networks
The evolution of phosphorylation networks can prove useful for exploring and understanding the conserved or fast-evolving signaling events implicated in complex human diseases (13). SH2 domain proteins can functionally diverge through alterations in regions within the SH2 domain itself, which can lead to novel interactions, or through the process of the evolution of phosphorylation sites within the full-length protein. To better understand the network around pTyr signaling, we analyzed specific interaction networks from fruit flies to humans. Because experimentally confirmed pTyr interactions are limited in lower organisms, such as C. elegans and D. melanogaster, it remains a challenge to map conserved networks with experimental data. To circumvent this issue, we used sequence alignments (from D. melanogaster to H. sapiens) and algorithmic predictions of SH2 domain families with conserved SH2 binding specificities to identify interactions likely to be conserved across multiple organisms and thus identify conserved pTyr networks. First, to determine whether the core specificity pocket is conserved, we examined the conservation of the pTyr peptide binding pocket by mapping the SH2 domain sequence alignments onto known crystal structures from the Protein Data Bank (PDB; http://www.rcsb.org). In this analysis, we selected structures of SH2 domains bound to peptide ligands from the human SH3- and SH2-containing adaptors Nck, Crk, and Grb2. The pTyr peptide binding core of these three SH2 domains exhibited >80% conservation in sequence identity (Fig. 7A). This suggests that for these select adaptor proteins, the amino acid sequences within the SH2 domain that determine specificity and pTyr peptide binding is likely constrained or fixed in lower organisms. When we mapped the sequence conservation, using alignments of SH2 domain sequences from D. melanogaster to H. sapiens, onto other human SH2 domain structures, we observed conservation of the specificity pocket for several, but not all, SH2 domain families (fig. S9). The high level of conservation in the binding motif for SH2 domains may suggest that the specificity of this domain is constrained and that the mechanism of network rewiring occurs through the evolution of changes within the pTyr motif.
Fig. 7.
Evolution of pTyr networks. (A) The structures indicate the degree of sequence conservation of the SH2 domains of Grb2 (PDB 1BMB), Crk (PDB 1JU5), and Nck (PDB 2CI9) mapped onto the tertiary structure. The surface representations are colored according to the amount of conservation and the bound pTyr peptide is indicated (gray). The region of the PTB pocket is highlighted with a black box. (B) A regional pTyr interaction network for Grb2, Crk, and Nck (Dock in fruit fly) is shown for the fruit fly D. melanogaster. SH2 interactions for these domains were predicted with mammalian SH2 specificity data. Solid black lines indicate SH2-mediated interactions; dashed lines represent PTB-mediated interactions. A blue line indicates an SH3-dependent interaction. Components of the pTyr network shown as colored circles are SH2 adaptor proteins (yellow), scaffolds (blue), tyrosine kinases (green), or signal regulators (orange). (C) The equivalent mammalian pTyr network for these SH2 adaptor proteins from experimentally validated and tested interactions reveals that additional connections are present in mammals.
Analysis of conservation in the pTyr network from lower organisms to humans for the adaptors Nck, Crk, and Grb2 may provide insight into how these networks may have evolved. The sequences of proteins in protein interaction networks in the fruit fly D. melanogaster (Fig. 7B) and well-characterized human SH2 interaction networks were analyzed (Fig. 7C) for conserved pTyr motifs. For Grb2, one network that is highly conserved from fruit fly to human is the interaction with the scaffold Shc and the RTK epidermal growth factor receptor (EGFR) (60, 61). The SH2 domain of Grb2 recognizes pY-x-N motifs in both the scaffold Shc and EGFR. Two sets of Grb2-binding motifs are present in the human ortholog of Shc, whereas only one is present in D. melanogaster (fig. S8). For the Crk network, both Crk and Abl interact through their SH2 domains with p130Cas in flies and humans (Fig. 7) (62). The network becomes more complex in mammals because the integration of phosphorylation sites into Cbl links Abl and Crk together (63, 64). The Nck adaptor network reveals both conserved and novel interactions between D. melanogaster and H. sapiens. The scaffold Dok in flies lacks pTyr sites for the Dock (the Nck ortholog) SH2, but the pTyr sites evolve in mammalian Dok1 and Dok2, enabling SH2-mediated interactions between Nck and Abl (65, 66). Such examples highlight how complexity in pTyr signaling networks can be achieved through the evolution of phosphorylation motifs to mediate SH2 domain interactions.
Tissue specialization of SH2 domain proteins
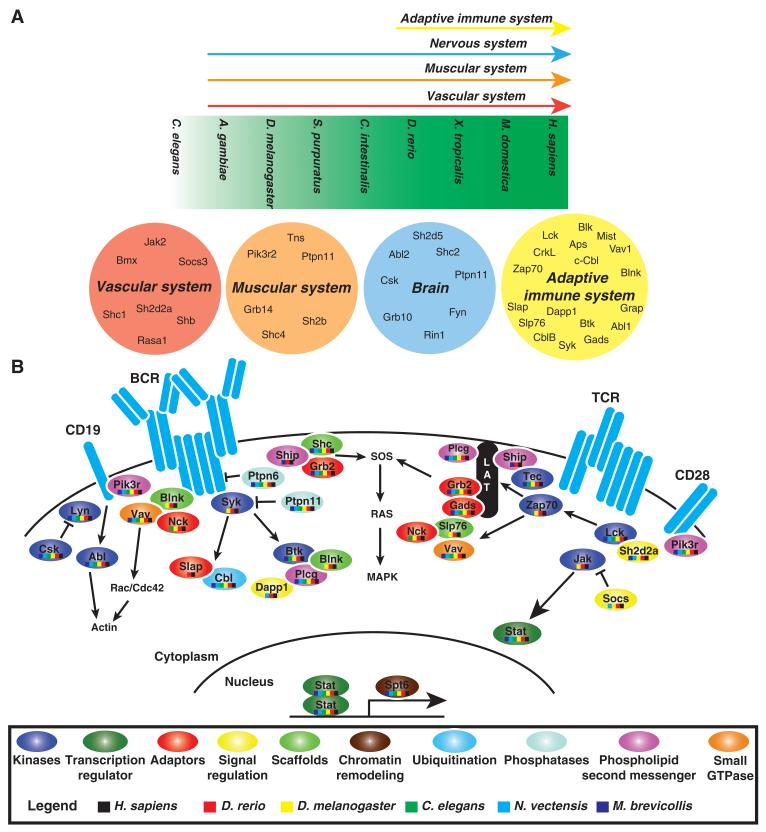
pTyr signaling occurs early in premetazoans and is further used in more complex organisms to evolve and develop complex biological systems, such as the nervous system, vascular system, and immune system (Fig. 8A). At specific points in evolution, complex biological systems emerge concomitantly with particular SH2 families. The number of SH2 domain families, arising through gene duplication, correlates with the number of different cell types (19). To understand whether duplicated SH2-encoding genes within families develop specialized tissue expression patterns, we collected gene expression data from UniGene (fig. S10A) and analyzed tissues affected by gene disruption in mice (3). Numerous SH2 domain proteins regulate the signaling events in the adaptive immune system, particularly downstream of the T cell receptor (TCR) and B cell receptor (BCR) (Fig. 8B). Many proteins including Syk, Zap70, Blnk, Slp76, Gads, and several SFKs (fig. S10B) play an important role in signaling mediated by TCRs and BCRs, and their respective paralogs have specialized functions in other tissue types. The utilization of SH2 domain proteins in specific tissues is reflected in the phenotypes observed upon loss of function. The genes encoding 81 SH2-containing proteins have been disrupted in mice (3). Whereas several SH2 domain proteins are critical for early to mid embryogenesis, a number are required in specific tissues postnatally. These include the SH2-containing proteins, such as Lck, Zap70, Slp76, and Gads, that are involved in TCR signaling and whose corresponding genes are required for thymic development and functional signaling in thymocytes (67-70). In humans, mutations in the genes encoding 23 distinct SH2 domain proteins are linked to multiple clinical disorders, including cancers and leukemias, developmental disorders, diabetes, and immunodeficiencies (3). The flexibility of SH2 proteins to promote the development of novel signaling cascades such as these highlights the ability of gene duplication to accommodate specialization and tissue-specific signaling.
Fig. 8.
Evolution of complex biological systems. (A) The components of the vascular, muscular, nervous, and immune systems evolve in the respective organisms indicated by the colored arrows. SH2-encoding genes disrupted by mouse knockout that results in defects in specialized tissues (3) or genes found exclusively in specific tissues with gene expression data are included within the circles (fig. S10). (B) The evolution of pTyr signaling within the adaptive immune system suggests that such complex systems make extensive use of existing SH2 components to develop new and robust signaling networks. The TCR and BCR signaling networks are indicated along with SH2 domain-containing proteins and the general signaling circuitry that they connect. Specific SH2 domains are represented as ovals colored on the basis of their functional categories (3). The evolutionary origin of each SH2 domain protein is indicated with a colored bar code displaying whether that gene and its ortholog were present in the indicated organisms studied.
DISCUSSION
Transition of SH2 domains to the recognition of pTyr
We examined SH2 domain–containing proteins encoded within the genomes of 28 different eukaryotic organisms to understand the mechanisms that have facilitated the development of pTyr signaling (Fig. 1A). SH2 domains are present in Bikonts, including the unicellular organisms N. gruberi and P. capsici (Fig. 1B and table S2), and thus predate the emergence of PTKs (71). A common SH2 domain protein Spt6 is found in Bikonts and is universally present in metazoans. Spt6 was first described in the budding yeast S. cerevisiae (72), and structural studies showed that Spt6 has a tandem SH2 fold (73, 74). Only the first SH2 domain of Spt6 is a canonical SH2 domain; the second SH2 fold is highly variant and not detected with standard SH2 domain models, such as SMART or Pfam. Both SH2 domains are necessary for this region to recognize phosphorylated serine residues on the C-terminal domain of RNA polymerase (75). The structure of the yeast Spt6 SH2 domain contained features that clearly resemble pTyr-binding SH2 domains in mammals (73, 76, 77). It thus appears likely that Spt6 SH2 represents an ancestral pSer/pThr-binding SH2 domain fold.
Most branches of the Unikonta, including Amoebozoa, Choanozoa, and Animalia, contain diverse SH2 domains, suggesting that pTyr-binding SH2 domains developed early in the Unikont lineage, perhaps even predating the Unikont-Bikont divergence. Amoebozoa, Choanozoa, and Animalia all encode SH2 domain–containing STAT proteins. This raises an intriguing question as to why Fungi, which together with Metazoa are in the Opisthokont branch of Unikonta, lack any apparent pTyr-binding SH2 domains. The simplest explanation is that the Fungi lost SH2 domains as a result of deleterious outcomes from the occurrence of pTyr and failed to make use of pTyr signaling in any important manner. Alternatively, it is conceivable that SH2 domains in Amoebozoa represent examples of horizontal gene transfer from the choanoflagellate line, or coincident convergent evolution. Whatever the origin of pTyr-binding SH2 domains in Amoebozoa, it is evident that members of the Amoebozoa lineage, such as the Entamoeba and Mycetozoa, expanded SH2 domains into a largely independent set of proteins with relatively fewobvious homologs in Metazoa (table S2). At the junction between unicellular and multicellular Unikonts, SH2 domains apparently evolved the ability to recognize pTyr-based motifs, allowing rapid evolution of pTyr signaling by shuffling SH2 domains into various proteins (12).
Coevolution of pTyr signaling
Within the Metazoan lineage, diversification of SH2 domains and PTKs is tightly linked, suggesting that they coevolved to create diverse functional signaling systems (Fig. 1). Within the Unikont lineages, SH2 domain expansion may be partially driven by the kinases that generate the binding partners for SH2 domains through phosphorylation. True PTKs first appeared in the choanoflagellate M. brevicollis (11), but are absent in amoeba and slime molds (28). However, some Ser-Thr kinases, such as mitogen-activated protein kinases (MAPKs), are dual-function enzymes capable of phosphorylating tyrosine residues (78), which may explain the expansion of SH2 domain proteins in D. discoideum. Indeed, it is in Amoebozoa that we first encounter SH2 domains linked to a dual-specificity kinase, the Ser/Thr/Tyr kinase Shk (table S2). Tyrosine phosphorylation is used in D. discoideum and can be observed in the phosphorylation of STATc, at a site near its C terminus (79, 80). When STATc is tyrosine phosphorylated, it homodimerizes through reciprocal SH2 domain–pTyr interactions and accumulates in the nucleus, functioning in a manner analogous to that of the mammalian STAT orthologs. Because D. discoideum diverged from the lineage that gave rise to Metazoa before Fungi but after the Bikonta-Unikonta split, it remains unclear why STATs were apparently lost in Fungi (27). In addition to the STATs, 24 kinase subfamilies shared between D. discoideum and metazoan kinomes are absent in yeast, suggesting that yeast developed a specialized biological program not requiring many of the extant kinases or SH2 domains (28). Thus, functional use of pTyr and SH2 domains predates dedicated PTKs (81).
The choanoflagellate M. brevicollis is the most primitive organism in our analysis that encodes a core set of PTKs, PTPs, and SH2 domains (8) and thus exhibits a functional pTyr signaling network reminiscent of that of metazoans. A total of 20 SH2 families originate in M. brevicollis, several of which have distinct roles in this lineage (Fig. 3). The cytoplasmic tyrosine kinase MbCsk is coexpressed and can phosphorylate MbSrc1 in a manner similar to that of the mammalian Src and Csk pair. However, the negative regulation of Src through phosphorylation is absent from the M. brevicollis pair, suggesting that this allosteric regulation of Src developed more recently in the metazoan lineage (82, 83). The presence of an extensive and varied collection of signaling molecules suggests that M. brevicollis may have depended on pTyr to mediate many of its cellular functions, which we have yet to understand. It will be fascinating to see what roles the genes unique to specific lineages, such as those found in M. brevicollis, serve in coordinating organism-specific pTyr signaling.
Throughout the different metazoan lineages, new SH2 families arise, including seven in cnidarians and five in arthropods (Fig. 3B). The appearance of these families may indicate the evolution of specific signaling networks that underpin the evolution of phenotypic complexity. Extensive use of cell polarity is a hallmark of cnidarians, and this includes the first appearance of the canonical Wnt and β-catenin signaling pathways (84, 85) along with several SH2 domain families, including NCK and SOCS (Fig. 3). Both Nck and Socs7 play an important role in regulating cell polarity through Septins (86), which suggests that they may have coevolved with the proteins controlling cell adhesion and polarity that emerge in cnidarians (87). In arthropods, complex tissues appear, such as the heart and the vascular system (88). Several proteins from these specific families that appear in arthropods may have coevolved to create protein interaction networks that play a role in the vascular system, including Shb (89) and Jak2 (90). In addition, protein interaction studies have revealed that the adaptor protein Shb can interact with Jak (91) and other SH2 proteins that appear in the same evolutionary jump, such as Frk (92) and Slp76 (93). This supports the notion that these proteins have undergone coevolution as a set of connected nodes within a signaling network in a manner that may have promoted the development of tissue-specialized signaling.
Whereas some protein interactions coevolve and remain conserved through evolution, other pTyr signaling networks may evolve rapidly by integrating pTyr motifs recognized by SH2 domains into scaffolds and other SH2 domain proteins (Figs. 6 and 7). The identification of conserved and fast-evolving phosphorylation sites is important for understanding human diseases (13). An understanding of the evolutionary mechanisms for creating new pTyr networks by modification of SH2 domain specificity or integration with different signaling pathways may lead to new approaches for cellular rewiring and creating customized circuits (94).
SH2 domain expansion underlies increased complexity in biological systems
The rate of co-expansion of SH2 domains and PTKs in animals is the singular indication that the components of pTyr signaling are of broad utility and are under evolutionary pressure to not only be retained, but also to expand into additional areas of cellular control. One possible mechanism driving duplication is tissue-specific expression, because gene duplication enables tissue or developmental specialization to evolve (17, 95). Many factors may drive expression divergence between gene duplicates, including biotic and abiotic stresses in the environment (96). The expansion of SH2 domains has a strong correlation (R = 0.93) with the number of different cell types (19), suggesting the utility of pTyr signaling for the evolution of organisms with higher complexity. Both tissue expression patterns and knockout phenotypes of SH2 domain proteins in mice (3) reveal specialized roles in specific cell types and tissues for certain SH2 proteins (Fig. 8A and fig. S10). In more complex organisms, more specialized biological systems develop, such as the vascular system, muscular system, central and peripheral nervous systems, and adaptive immune system (Fig. 8A). In SH2 families, such as SLP76, SYK, and GRB2, duplication events produce multiple gene copies with specialized functions in specific tissues. In the case of the GRB2 family, Grb2 itself is broadly expressed, and a targeted gene disruption in mice results in embryonic lethality (97). By contrast, GRB2 family members Gads and Grap are present only in certain lymphoid lineages and perform specialized functions in signaling relevant to the adaptive immune response (98-102) (Fig. 8A). In other cases, knockouts of multiple related SH2-containing proteins cause embryonic lethality, whereas knockout of a single family member has only subtle effects that manifest phenotypically in specific tissues. This suggests that duplication of genes can retain many redundant functions but also evolve to acquire specialized functions within specific tissues.
Indeed, many examples of duplicated genes are found within the adaptive immune response. These also include the Jak-to-STAT signaling network (103), even though both Jak and STAT proteins predate the adaptive immune response and arose at different points in evolution (Fig. 8B). An evolutionary viewpoint of SH2 domains in the adaptive immune system reveals numerous tissue-specific connections between family members, including Slp76 with Gads and Lck with Zap70 (Fig. 8B).
Duplication events may also promote diversity in SH2 domain binding specificity in some cases, as well as allowing development of allosteric secondary interactions. In this way, the product of a gene duplication allows for the emergence of a new gene that preserves certain functions of the parent while acquiring specialized functions to coordinate novel interactions (104). As an example, the Socs proteins have evolved to fine-tune their primary binding interface to recognize specific sets of targets. Thus, Socs2 binds specifically to the growth hormone receptor, whereas Socs4 recognizes phosphorylated EGFR (105). In contrast, SH2 specificities within families, such as GRB2 (consisting of Grb2, Gads, and Grap), NCK (Nck1 and Nck2), and CRK (Crk and CrkL), are highly conserved (Fig. 7) and share a common core set of binding peptides (59). Both Crk and Sh2d1a each evolved from existing family members and acquired new interactions through recruitment of SH3 domains through a non–pTyr-dependent, secondary binding surface within the SH2 domain (Fig. 5). Evolving new binding sites not only is a mechanism for diversifying duplicate genes but also enables the development of new signaling networks within specific tissues (Figs. 6 and 7). Collectively, these observations suggest that gene duplication may have allowed SH2 domain proteins to coevolve and diverge within tissue types to create specialized cellular signaling networks such as those involved in signaling from the TCR and BCR in lymphocytes (Fig. 8B).
Concluding remarks
The study of SH2 domains provides a view into the evolution of pTyr signaling and the development of multicellularity in the metazoan lineage. More generally, it provides a glimpse at how modular protein interaction domains diversify to produce highly connected interaction networks that drive signaling that is both selective and complex. Yet, the current analysis of SH2 domains in eukaryotes also draws attention to the fact that we still have much to understand in terms of the evolution of new protein architectures and the signal transduction networks that they enable. Indeed, we are far from understanding the significance of pTyr signaling within early eukaryotes that drove the rapid expansion of SH2 domains and PTKs. It is our hope that the more comprehensive and codified view outlined here will improve our understanding of not only SH2 domains, but also the evolutionary process by which protein domains are combined in novel ways to create new connections in robust signaling networks.
MATERIALS AND METHODS
SH2 domain identification in eukaryotic genomes
We used the Pfam hidden Markov models (HMMs) (http://www.sanger.ac.uk/Software/Pfam/) (25) and SMART HMM (http://smart.embl-heidelberg.de/) (26, 106) domain descriptions to search the protein sequence data for H. sapiens and M. musculus as previously described (3). The genomes of A. aegypti, C. elegans, E. histolytica, M. domestica, S. cerevisiae, and S. purpuratus were assembled from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). The genomes of C. intestinalis, D. purpureum, M. brevicollis, N. gruberi, N. vectensis, P. capsici, T. vaginalis, Trichoplax adhaerens, and X. tropicalis were obtained from the Department of Energy Joint Genome Institute (http://genome.jgi-psf.org/). SH2 domain proteins were identified from the D. discoideum and D. purpureum genomes with NCBI and UniProt. Chromosomal locations for D. discoideum were derived from dictyBase (http://dictybase.org/). The FlyBase version FB2006_01 was used to determine genomic locations for D. melanogaster. The T. thermophila genome sequences were obtained from the Tetrahymena Genome Database (http://www.ciliate.org/). The A. thaliana genomic sequences were obtained from both NCBI and the Arabidopsis Information Resource (http://www.arabidopsis.org/). The complete set containing sequence duplication and redundancy was filtered for proteins identified as having identical genetic loci. Where possible, the organisms were cross-referenced between JGI, NCBI, Ensembl (http://www.ensembl.org/index.html), and UniProt (http://www.pir.uniprot.org). Because of the draft nature of several genomes, some SH2-encoding genes may have not been detected or identified in our survey. A complete list of organisms in this study containing all the common names, genus species, and taxonomy ID can be found in table S1. An evolutionary time line for the organisms listed in this study can be found in fig. S1.
Splice pattern analysis of human SH2 domains
Splice sites from human SH2 domains were identified with the nucleotide and protein sequence information from the Ensembl database in accompaniment with the domain architecture tool SMART. The splice site junctions within the SH2 domain were visualized at the protein sequence level within the Ensembl database. To verify these splice sites, we performed transcript alignment with genomic sequences using MegAlign (DNASTAR). The Ensembl sequence identifier for each SH2 domain protein was entered into SMART, because this generated the domain organization with splice site positions and phase indication (0, 1, 2) at the splice junction. Splice sites with phase 0 are located between codons, phase 1 introns are located between the first and the second nucleotides of a codon, and phase 2 introns are located between the second and the third nucleotides. The data gathered by SMART were then overlaid with the Ensembl splice site position to generate a comprehensive set of splice patterns for the 121 human SH2 domains. Using our previous ClustalW alignment of all human SH2 domains (3), we mapped the splice sites onto the SH2 domain protein secondary structure with published 3D structures. We identified 225 structures covering 61 SH2 domains in 55 proteins (3) (see http://www.sh2domain.org for a complete list), many of which were solved with peptide or synthetic ligands. When two SH2 domains contain a splice site with the same phase that falls at identical splice junctions (within a window of one to two positions N- or C-terminal with respect to the splice site) in the sequence alignment, we consider this splice site position to be conserved. Standard hierarchical clustering was performed on the compiled splice sites with the sequence alignment as a background template. The phases (0, 1, 2) and the positions within the secondary structure were used for weighting the conservation of the splice sites.
Classification of human SH2 domain families
The collection of human SH2 domain protein sequences was previously described (3). A phylogenetic tree containing all 121 human SH2 domains was generated by means of the neighbor-joining method with bootstrap replicates implemented in MEGA2 (107). Multiple substitutions were corrected for with the Poisson correction. The branch points were used as a primary method for distinguishing SH2 domains into unique families. Secondary disambiguation of SH2 families was assisted by analysis of protein domain organization. The HMM definitions from SMART, Pfam, and CDD were used to identify the domain descriptions within the 111 human protein sequences. Tertiary disambiguation of SH2 families used splice patterns within the SH2 domain to assign SH2 proteins into families with identical splice-junction patterns when protein domain organization and near-neighbor sequence phylogeny were not sufficient to make the distinction.
Identifying orthologs of SH2 families
The protein domain organization (also referred to as domain architecture) provided another parameter for identifying orthologs of SH2 families. The approaches described above were used to trace orthologs to identify precursors of genes and domains involved. Human orthologs were identified through sequence alignment with NCBI’s BLASTP and by matching conserved domain organization. The top alignment hit from BLASTP or the lowest E-value was used to best identify orthologs and paralogs. A comparative analysis of ortholog and paralog predictions using alternative approaches, such as Ensembl and InParanoid (http://inparanoid.sbc.su.se), was examined (see section S3 and tables S6 to S8). This analysis found BLAST to be the most reliable ortholog prediction method compared to Ensembl and InParanoid alone. However, we used the Ensembl ortholog prediction to confirm our BLAST results. In certain cases where the sequence has diverged to a point where BLAST recognition was difficult to determine, we examined domain organization to categorize proteins into families. When neither BLAST nor domain organization was capable of determining a family, we excluded these proteins from our 38 families and considered these proteins as unique to a specific lineage or organism.
Sequence and structural alignments
SH2 domain sequences were obtained from NCBI and run through the domain tool SMART. SH2 domain sequences from SMART were extended 10 to 15 amino acids on both ends to capture the complete boundaries of the domain. Sequence alignments were compiled with ClustalX. SH2 domain structures noted were downloaded from the Research Collaboratory for Structural Bioinformatics PDB (http://www.rcsb.org). Sequence conservation mapped onto the SH2 domain structure was achieved with the program UCSF Chimera (http://www.cgl.ucsf.edu/chimera). Alignments from ClustalX and the structure files were manually uploaded into the Chimera software. Colors for sequence conservations were manually adjusted with the percentage conservation tool.
Supplementary Material
Acknowledgments
We thank G. Manning (Salk Institute) and T. Pawson (Samuel Lunenfeld Research Institute) for helpful discussions and A. Pasculescu, S. Guettler, R. Nalluru, D. Djordjevic and C. Lee for technical assistance.
Funding: Supported by funds provided by NSF (MCB-0819125), the University of Chicago Cancer Research Center, a pilot project grant from the University of Chicago Diabetes Research Training Center, and the Cancer Research Foundation to P.D.N. B.A.L. was a recipient of an Abbott Graduate Fellowship. B.E. was funded in part by NIH grant TG-GM07183.
Footnotes
www.sciencesignaling.org/cgi/content/full/4/202/ra83/DC1
Section S1. SH2 domain proteins in organisms with incomplete genomes.
Section S2. Analysis of the intron/exon code of human SH2 domains.
Section S3. Comparative analysis of ortholog and paralog predictions.
Fig. S1. An evolutionary time line for the organisms represented in this study.
Fig. S2. Evolutionary expansion of SH2 domains, tyrosine kinases, and PDZ domains.
Fig. S3. Splice site positions within the protein sequence alignment of human SH2 domains.
Fig. S4. The bead on a string representation for protein domains.
Fig. S5. Gene duplication and loss within SH2 domain families.
Fig. S6. Clustal alignments of the GRB2 and CRK families.
Fig. S7. Insertion of an intramolecular phosphorylation site for autoinhibition of CRK.
Fig. S8. Evolving new SH2 interactions through novel pTyr sites.
Fig. S9. Conservation of the pTyr ligand-binding pocket in SH2 domains.
Fig. S10. Tissue expression of human SH2 domain proteins.
Table S1. A complete list of organisms in this study.
Table S2. SH2 domains in Bikonta and Amoebozoa.
Table S3. Comprehensive list of SH2 domain–containing proteins in Eukaryotes (Excel file).
Table S4. Comprehensive list of SH2 domain–containing proteins in organisms with incomplete genome annotations (Excel file).
Table S5. Classification of SH2 domain family divergence.
Table S6. Defining SH2 families with Ensembl paralog predictions versus family organization.
Table S7. Ortholog predictions of human SH2 proteins to proteins in lower organisms.
Table S8. Ortholog predictions from C. elegans to H. sapiens.
References
Author contributions: B.A.L., E.S., K.J., B.E., and A.S. analyzed the data. K.J., B.A.L., and E.S. designed the Web site. B.A.L. and P.D.N. designed the research and wrote the paper.
Data availability: http://www.sh2domain.org
REFERENCES AND NOTES
- 1.Hunter T. Tyrosine phosphorylation: Thirty years and counting. Curr. Opin. Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 3.Liu BA, Jablonowski K, Raina M, Arcé M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins—Establishing the boundaries of phosphotyrosine signaling. Mol. Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 5.Smith MJ, Hardy WR, Murphy JM, Jones N, Pawson T. Screening for PTB domain binding partners and ligand specificity using proteome-derived NPXY peptide arrays. Mol. Cell. Biol. 2006;26:8461–8474. doi: 10.1128/MCB.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCδ is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter T. The role of tyrosine phosphorylation in cell growth and disease. Harvey Lect. 1998-1999;94:81–119. [PubMed] [Google Scholar]
- 10.Hunter T, Cooper JA. Protein-tyrosine kinases. Annu. Rev. Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 11.King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: Molecular insights into early animal evolution. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15032–15037. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim WA, Pawson T. Phosphotyrosine signaling: Evolving a new cellular communication system. Cell. 2010;142:661–667. doi: 10.1016/j.cell.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan CS, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner MO, Jørgensen C, Bader GD, Aebersold R, Pawson T, Linding R. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal. 2009;2:ra39. doi: 10.1126/scisignal.2000316. [DOI] [PubMed] [Google Scholar]
- 14.Kawata T, Shevchenko A, Fukuzawa M, Jermyn KA, Totty NF, Zhukovskaya NV, Sterling AE, Mann M, Williams JG. SH2 signaling in a lower eukaryote: A STAT protein that regulates stalk cell differentiation in Dictyostelium. Cell. 1997;89:909–916. doi: 10.1016/s0092-8674(00)80276-7. [DOI] [PubMed] [Google Scholar]
- 15.Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD. Multivesicular body sorting: Ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol. Biol. Cell. 2004;15:468–480. doi: 10.1091/mbc.E03-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiu SH, Li WH. Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol. Biol. Evol. 2004;21:828–840. doi: 10.1093/molbev/msh077. [DOI] [PubMed] [Google Scholar]
- 17.Li WH, Yang J, Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–607. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Chothia C, Gough J. Genomic and structural aspects of protein evolution. Biochem. J. 2009;419:15–28. doi: 10.1042/BJ20090122. [DOI] [PubMed] [Google Scholar]
- 19.Vogel C, Chothia C. Protein family expansions and biological complexity. PLoS Comput. Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin J, Xie X, Chen C, Park JG, Stark C, James DA, Olhovsky M, Linding R, Mao Y, Pawson T. Eukaryotic protein domains as functional units of cellular evolution. Sci. Signal. 2009;2:ra76. doi: 10.1126/scisignal.2000546. [DOI] [PubMed] [Google Scholar]
- 21.Serfas MS, Tyner AL. Brk, Srm, Frk, and Src42A form a distinct family of intracellular Src-like tyrosine kinases. Oncol. Res. 2003;13:409–419. doi: 10.3727/096504003108748438. [DOI] [PubMed] [Google Scholar]
- 22.Steele RE, Stover NA, Sakaguchi M. Appearance and disappearance of Syk family protein-tyrosine kinase genes during metazoan evolution. Gene. 1999;239:91–97. doi: 10.1016/s0378-1119(99)00373-x. [DOI] [PubMed] [Google Scholar]
- 23.Colicelli J. ABL tyrosine kinases: Evolution of function, regulation, and specificity. Sci. Signal. 2010;3:re6. doi: 10.1126/scisignal.3139re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 25.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg JM, Manning G, Liu A, Fey P, Pilcher KE, Xu Y, Smith JL. The Dictyostelium kinome—Analysis of the protein kinases from a simple model organism. PLoS Genet. 2006;2:e38. doi: 10.1371/journal.pgen.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 30.Bradham CA, Foltz KR, Beane WS, Arnone MI, Rizzo F, Coffman JA, Mushegian A, Goel M, Morales J, Geneviere AM, Lapraz F, Robertson AJ, Kelkar H, Loza-Coll M, Townley IK, Raisch M, Roux MM, Lepage T, Gache C, McClay DR, Manning G. The sea urchin kinome: A first look. Dev. Biol. 2006;300:180–193. doi: 10.1016/j.ydbio.2006.08.074. [DOI] [PubMed] [Google Scholar]
- 31.Miranda-Saavedra D, Barton GJ. Classification and functional annotation of eukaryotic protein kinases. Proteins. 2007;68:893–914. doi: 10.1002/prot.21444. [DOI] [PubMed] [Google Scholar]
- 32.Martin DM, Miranda-Saavedra D, Barton GJ. Kinomer v. 1.0: A database of systematically classified eukaryotic protein kinases. Nucleic Acids Res. 2009;37:D244–D250. doi: 10.1093/nar/gkn834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fong JH, Geer LY, Panchenko AR, Bryant SH. Modeling the evolution of protein domain architectures using maximum parsimony. J. Mol. Biol. 2007;366:307–315. doi: 10.1016/j.jmb.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Aniello S, Irimia M, Maeso I, Pascual-Anaya J, Jiménez-Delgado S, Bertrand S, Garcia-Fernàndez J. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol. Biol. Evol. 2008;25:1841–1854. doi: 10.1093/molbev/msn132. [DOI] [PubMed] [Google Scholar]
- 35.Moore AD, Björklund AK, Ekman D, Bornberg-Bauer E, Elofsson A. Arrangements in the modular evolution of proteins. Trends Biochem. Sci. 2008;33:444–451. doi: 10.1016/j.tibs.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Liu M, Grigoriev A. Protein domains correlate strongly with exons in multiple eukaryotic genomes—Evidence of exon shuffling? Trends Genet. 2004;20:399–403. doi: 10.1016/j.tig.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 37.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat. Rev. Immunol. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- 39.Pandey A, Duan H, Dixit VM. Characterization of a novel Src-like adapter protein that associates with the Eck receptor tyrosine kinase. J. Biol. Chem. 1995;270:19201–19204. doi: 10.1074/jbc.270.33.19201. [DOI] [PubMed] [Google Scholar]
- 40.Prince VE, Pickett FB. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 41.Zhang G, Cohn MJ. Genome duplication and the origin of the vertebrate skeleton. Curr. Opin. Genet. Dev. 2008;18:387–393. doi: 10.1016/j.gde.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Latour S, Veillette A. The SAP family of adaptors in immune regulation. Semin. Immunol. 2004;16:409–419. doi: 10.1016/j.smim.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 43.Sosinowski T, Pandey A, Dixit VM, Weiss A. Src-like adaptor protein (SLAP) is a negative regulator of T cell receptor signaling. J. Exp. Med. 2000;191:463–474. doi: 10.1084/jem.191.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dragone LL, Shaw LA, Myers MD, Weiss A. SLAP, a regulator of immunoreceptor ubiquitination, signaling, and trafficking. Immunol. Rev. 2009;232:218–228. doi: 10.1111/j.1600-065X.2009.00827.x. [DOI] [PubMed] [Google Scholar]
- 45.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, Neel BG, Birge RB, Fajardo JE, Chou MM, Hanafusa H, Schaffhausen B, Cantley LC. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 46.Anafi M, Rosen MK, Gish GD, Kay LE, Pawson T. A potential SH3 domain-binding site in the Crk SH2 domain. J. Biol. Chem. 1996;271:21365–21374. doi: 10.1074/jbc.271.35.21365. [DOI] [PubMed] [Google Scholar]
- 47.Donaldson LW, Gish G, Pawson T, Kay LE, Forman-Kay JD. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14053–14058. doi: 10.1073/pnas.212518799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ. SAP couples Fyn to SLAM immune receptors. Nat. Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- 49.Latour S, Roncagalli R, Chen R, Bakinowski M, Shi X, Schwartzberg PL, Davidson D, Veillette A. Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation. Nat. Cell Biol. 2003;5:149–154. doi: 10.1038/ncb919. [DOI] [PubMed] [Google Scholar]
- 50.Roncagalli R, Taylor JE, Zhang S, Shi X, Chen R, Cruz-Munoz ME, Yin L, Latour S, Veillette A. Negative regulation of natural killer cell function by EAT-2, a SAP-related adaptor. Nat. Immunol. 2005;6:1002–1010. doi: 10.1038/ni1242. [DOI] [PubMed] [Google Scholar]
- 51.Li C, Iosef C, Jia CY, Han VK, Li SS. Dual functional roles for the X-linked lymphoproliferative syndrome gene product SAP/SH2D1A in signaling through the signaling lymphocyte activation molecule (SLAM) family of immune receptors. J. Biol. Chem. 2003;278:3852–3859. doi: 10.1074/jbc.M206649200. [DOI] [PubMed] [Google Scholar]
- 52.March ME, Ravichandran K. Regulation of the immune response by SHIP. Semin. Immunol. 2002;14:37–47. doi: 10.1006/smim.2001.0340. [DOI] [PubMed] [Google Scholar]
- 53.Miller ML, Jensen LJ, Diella F, Jørgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobashigawa Y, Sakai M, Naito M, Yokochi M, Kumeta H, Makino Y, Ogura K, Tanaka S, Inagaki F. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat. Struct. Mol. Biol. 2007;14:503–510. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- 55.Martin GS. The hunting of the Src. Nat. Rev. Mol. Cell Biol. 2001;2:467–475. doi: 10.1038/35073094. [DOI] [PubMed] [Google Scholar]
- 56.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 57.Feller SM, Knudsen B, Hanafusa H. c-Abl kinase regulates the protein binding activity of c-Crk. EMBO J. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salcini AE, McGlade J, Pelicci G, Nicoletti I, Pawson T, Pelicci PG. Formation of Shc-Grb2 complexes is necessary to induce neoplastic transformation by overexpression of Shc proteins. Oncogene. 1994;9:2827–2836. [PubMed] [Google Scholar]
- 59.Liu BA, Jablonowski K, Shah EE, Engelmann BW, Jones RB, Nash PD. SH2 domains recognize contextual peptide sequence information to determine selectivity. Mol. Cell. Proteomics. 2010;9:2391–2404. doi: 10.1074/mcp.M110.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olivier JP, Raabe T, Henkemeyer M, Dickson B, Mbamalu G, Margolis B, Schlessinger J, Hafen E, Pawson T. A Drosophila SH2-SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos. Cell. 1993;73:179–191. doi: 10.1016/0092-8674(93)90170-u. [DOI] [PubMed] [Google Scholar]
- 61.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 62.Mayer BJ, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr. Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- 63.Miyoshi-Akiyama T, Aleman LM, Smith JM, Adler CE, Mayer BJ. Regulation of Cbl phosphorylation by the Abl tyrosine kinase and the Nck SH2/SH3 adaptor. Oncogene. 2001;20:4058–4069. doi: 10.1038/sj.onc.1204528. [DOI] [PubMed] [Google Scholar]
- 64.Sattler M, Salgia R, Okuda K, Uemura N, Durstin MA, Pisick E, Xu G, Li JL, Prasad KV, Griffin JD. The proto-oncogene product p120CBL and the adaptor proteins CRKL and c-CRK link c-ABL, p190BCR/ABL and p210BCR/ABL to the phosphatidylinositol-3′ kinase pathway. Oncogene. 1996;12:839–846. [PubMed] [Google Scholar]
- 65.Noguchi T, Matozaki T, Inagaki K, Tsuda M, Fukunaga K, Kitamura Y, Kitamura T, Shii K, Yamanashi Y, Kasuga M. Tyrosine phosphorylation of p62Dok induced by cell adhesion and insulin: Possible role in cell migration. EMBO J. 1999;18:1748–1760. doi: 10.1093/emboj/18.7.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamanashi Y, Baltimore D. Identification of the Abl- and rasGAP-associated 62 kDa protein as a docking protein, Dok. Cell. 1997;88:205–211. doi: 10.1016/s0092-8674(00)81841-3. [DOI] [PubMed] [Google Scholar]
- 67.Pivniouk V, Tsitsikov E, Swinton P, Rathbun G, Alt FW, Geha RS. Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell. 1998;94:229–238. doi: 10.1016/s0092-8674(00)81422-1. [DOI] [PubMed] [Google Scholar]
- 68.Yoder J, Pham C, Iizuka YM, Kanagawa O, Liu SK, McGlade J, Cheng AM. Requirement for the SLP-76 adaptor GADS in T cell development. Science. 2001;291:1987–1991. doi: 10.1126/science.1057176. [DOI] [PubMed] [Google Scholar]
- 69.Arpaia E, Shahar M, Dadi H, Cohen A, Roifman CM. Defective T cell receptor signaling and CD8+ thymic selection in humans lacking Zap-70 kinase. Cell. 1994;76:947–958. doi: 10.1016/0092-8674(94)90368-9. [DOI] [PubMed] [Google Scholar]
- 70.Molina TJ, Kishihara K, Siderovski DP, van Ewijk W, Narendran A, Timms E, Wakeham A, Paige CJ, Hartmann KU, Veillette A, Davidson D, Mak TW. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357:161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- 71.Williams JG, Zvelebil M. SH2 domains in plants imply new signalling scenarios. Trends Plant Sci. 2004;9:161–163. doi: 10.1016/j.tplants.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 72.Maclennan AJ, Shaw G. A yeast SH2 domain. Trends Biochem. Sci. 1993;18:464–465. doi: 10.1016/0968-0004(93)90006-9. [DOI] [PubMed] [Google Scholar]
- 73.Diebold ML, Loeliger E, Koch M, Winston F, Cavarelli J, Romier C. Non-canonical tandem SH2 enables interaction of elongation factor Spt6 with RNA polymerase II. J. Biol. Chem. 2010;285:38389–38398. doi: 10.1074/jbc.M110.146696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun M, Larivière L, Dengl S, Mayer A, Cramer P. A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD) J. Biol. Chem. 2010;285:41597–41603. doi: 10.1074/jbc.M110.144568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dengl S, Mayer A, Sun M, Cramer P. Structure and in vivo requirement of the yeast Spt6 SH2 domain. J. Mol. Biol. 2009;389:211–225. doi: 10.1016/j.jmb.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 77.Gao Q, Hua J, Kimura R, Headd JJ, Fu XY, Chin YE. Identification of the linker-SH2 domain of STAT as the origin of the SH2 domain using two-dimensional structural alignment. Mol. Cell. Proteomics. 2004;3:704–714. doi: 10.1074/mcp.M300131-MCP200. [DOI] [PubMed] [Google Scholar]
- 78.Schieven G, Thorner J, Martin GS. Protein-tyrosine kinase activity in Saccharomyces cerevisiae. Science. 1986;231:390–393. doi: 10.1126/science.2417318. [DOI] [PubMed] [Google Scholar]
- 79.Fukuzawa M, Araki T, Adrian I, Williams JG. Tyrosine phosphorylation-independent nuclear translocation of a Dictyostelium STAT in response to DIF signaling. Mol. Cell. 2001;7:779–788. doi: 10.1016/s1097-2765(01)00222-2. [DOI] [PubMed] [Google Scholar]
- 80.Langenick J, Araki T, Yamada Y, Williams JG. A Dictyostelium homologue of the metazoan Cbl proteins regulates STAT signalling. J. Cell Sci. 2008;121:3524–3530. doi: 10.1242/jcs.036798. [DOI] [PubMed] [Google Scholar]
- 81.Williams JG, Noegel AA, Eichinger L. Manifestations of multicellularity: Dictyostelium reports in. Trends Genet. 2005;21:392–398. doi: 10.1016/j.tig.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 82.Li W, Young SL, King N, Miller WT. Signaling properties of a non-metazoan Src kinase and the evolutionary history of Src negative regulation. J. Biol. Chem. 2008;283:15491–15501. doi: 10.1074/jbc.M800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Segawa Y, Suga H, Iwabe N, Oneyama C, Akagi T, Miyata T, Okada M. Functional development of Src tyrosine kinases during evolution from a unicellular ancestor to multicellular animals. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12021–12026. doi: 10.1073/pnas.0600021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee PN, Pang K, Matus DQ, Martindale MQ. A WNT of things to come: Evolution of Wnt signaling and polarity in cnidarians. Semin. Cell Dev. Biol. 2006;17:157–167. doi: 10.1016/j.semcdb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 85.Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, Xu R, Martindale MQ. An ancient role for nuclear β-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–450. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- 86.Kremer BE, Adang LA, Macara IG. Septins regulate actin organization and cell-cycle arrest through nuclear accumulation of NCK mediated by SOCS7. Cell. 2007;130:837–850. doi: 10.1016/j.cell.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Magie CR, Martindale MQ. Cell-cell adhesion in the cnidaria: Insights into the evolution of tissue morphogenesis. Biol. Bull. 2008;214:218–232. doi: 10.2307/25470665. [DOI] [PubMed] [Google Scholar]
- 88.Hartenstein V, Mandal L. The blood/vascular system in a phylogenetic perspective. Bioessays. 2006;28:1203–1210. doi: 10.1002/bies.20497. [DOI] [PubMed] [Google Scholar]
- 89.Funa NS, Kriz V, Zang G, Calounova G, Akerblom B, Mares J, Larsson E, Sun Y, Betsholtz C, Welsh M. Dysfunctional microvasculature as a consequence of Shb gene inactivation causes impaired tumor growth. Cancer Res. 2009;69:2141–2148. doi: 10.1158/0008-5472.CAN-08-3797. [DOI] [PubMed] [Google Scholar]
- 90.Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 91.Lindholm CK. IL-2 receptor signaling through the Shb adapter protein in T and NK cells. Biochem. Biophys. Res. Commun. 2002;296:929–936. doi: 10.1016/s0006-291x(02)02016-8. [DOI] [PubMed] [Google Scholar]
- 92.Annerén C, Lindholm CK, Kriz V, Welsh M. The FRK/RAK-SHB signaling cascade: A versatile signal-transduction pathway that regulates cell survival, differentiation and proliferation. Curr. Mol. Med. 2003;3:313–324. doi: 10.2174/1566524033479744. [DOI] [PubMed] [Google Scholar]
- 93.Lindholm CK, Henriksson ML, Hallberg B, Welsh M. Shb links SLP-76 and Vav with the CD3 complex in Jurkat T cells. Eur. J. Biochem. 2002;269:3279–3288. doi: 10.1046/j.1432-1033.2002.03008.x. [DOI] [PubMed] [Google Scholar]
- 94.Lim WA. Designing customized cell signalling circuits. Nat. Rev. Mol. Cell Biol. 2010;11:393–403. doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wolfe KH, Li WH. Molecular evolution meets the genomics revolution. Nat. Genet. 2003;33(suppl.):255–265. doi: 10.1038/ng1088. [DOI] [PubMed] [Google Scholar]
- 96.Ha M, Li WH, Chen ZJ. External factors accelerate expression divergence between duplicate genes. Trends Genet. 2007;23:162–166. doi: 10.1016/j.tig.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng AM, Saxton TM, Sakai R, Kulkarni S, Mbamalu G, Vogel W, Tortorice CG, Cardiff RD, Cross JC, Muller WJ, Pawson T. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell. 1998;95:793–803. doi: 10.1016/s0092-8674(00)81702-x. [DOI] [PubMed] [Google Scholar]
- 98.Asada H, Ishii N, Sasaki Y, Endo K, Kasai H, Tanaka N, Takeshita T, Tsuchiya S, Konno T, Sugamura K. Grf40, a novel Grb2 family member, is involved in T cell signaling through interaction with SLP-76 and LAT. J. Exp. Med. 1999;189:1383–1390. doi: 10.1084/jem.189.9.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trüb T, Frantz JD, Miyazaki M, Band H, Shoelson SE. The role of a lymphoid-restricted, Grb2-like SH3-SH2-SH3 protein in T cell receptor signaling. J. Biol. Chem. 1997;272:894–902. doi: 10.1074/jbc.272.2.894. [DOI] [PubMed] [Google Scholar]
- 100.Bourette RP, Arnaud S, Myles GM, Blanchet JP, Rohrschneider LR, Mouchiroud G. Mona, a novel hematopoietic-specific adaptor interacting with the macrophage colony-stimulating factor receptor, is implicated in monocyte/macrophage development. EMBO J. 1998;17:7273–7281. doi: 10.1093/emboj/17.24.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qiu M, Hua S, Agrawal M, Li G, Cai J, Chan E, Zhou H, Luo Y, Liu M. Molecular cloning and expression of human Grap-2, a novel leukocyte-specific SH2- and SH3-containing adaptor-like protein that binds to Gab-1. Biochem. Biophys. Res. Commun. 1998;253:443–447. doi: 10.1006/bbrc.1998.9795. [DOI] [PubMed] [Google Scholar]
- 102.Liu SK, McGlade CJ. Gads is a novel SH2 and SH3 domain-containing adaptor protein that binds to tyrosine-phosphorylated Shc. Oncogene. 1998;17:3073–3082. doi: 10.1038/sj.onc.1202337. [DOI] [PubMed] [Google Scholar]
- 103.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 104.Hughes AL. Adaptive evolution after gene duplication. Trends Genet. 2002;18:433–434. doi: 10.1016/s0168-9525(02)02755-5. [DOI] [PubMed] [Google Scholar]
- 105.Bullock AN, Rodriguez MC, Debreczeni JE, Songyang Z, Knapp S. Structure of the SOCS4-ElonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation. Structure. 2007;15:1493–1504. doi: 10.1016/j.str.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.