Abstract
Background
In clinical settings, bacterial infections are usually diagnosed by isolation of colonies after laboratory cultivation followed by species identification with biochemical tests. However, biochemical tests result in misidentification due to similar phenotypes of closely related species. In such cases, 16S rDNA sequence analysis is useful. Herein, we report the first case of an Achromobacter-associated buckle infection that was diagnosed by 16S rDNA sequence analysis. This report highlights the significance of Achromobacter spp. in device-related ophthalmic infections.
Case presentation
A 56-year-old woman, who had received buckling surgery using a silicone solid tire for retinal detachment eighteen years prior to this study, presented purulent eye discharge and conjunctival hyperemia in her right eye. Buckle infection was suspected and the buckle material was removed. Isolates from cultures of preoperative discharge and from deposits on the operatively removed buckle material were initially identified as Alcaligenes and Corynebacterium species. However, sequence analysis of a 16S rDNA clone library using the DNA extracted from the deposits on the buckle material demonstrated that all of the 16S rDNA sequences most closely matched those of Achromobacter spp. We concluded that the initial misdiagnosis of this case as an Alcaligenes buckle infection was due to the unreliability of the biochemical test in discriminating Achromobacter and Alcaligenes species due to their close taxonomic positions and similar phenotypes. Corynebacterium species were found to be contaminants from the ocular surface.
Conclusions
Achromobacter spp. should be recognized as causative agents for device-related ophthalmic infections. Molecular species identification by 16S rDNA sequence analysis should be combined with conventional cultivation techniques to investigate the significance of Achromobacter spp. in ophthalmic infections.
Background
A 16S ribosomal DNA (rDNA) clone library analysis was performed for microbiological diagnosis in a clinical case of buckle infection. This type of analysis has previously been applied to a number of environmental samples to examine the microbial diversity within an ecological niche [1–6]. In clinical settings, it can be used to determine the microbial compositions of specimens, which would be beneficial to human health and would further our understanding of the pathological manifestations due to chronic infections [7–9]. In addition, in acute infections, causative bacteria are expected to be readily identified from the predominant sequences in specimens when a 16S rDNA clone library analysis is employed.
Buckle infection is a rare postoperative complication of retinal detachment. It generally occurs in the late stages of postoperative course. Although resident bacteria on the ocular surface, such as Staphylococcus aureus and Staphylococcus epidermidis, have been reported as the causative pathogens [10–12], environmental bacteria such as Pseudomonas aeruginosa or Stenotrophomonas maltophilia can also cause infections [12–15]. Some of the previous articles describing device-related ophthalmic infections reported isolation of a single pathogen. Considering that we currently know relatively very little about the diversity of microorganisms in nature [16], culture-independent molecular approaches to detect the causative agents may be useful for diagnosis of buckle infections. More than one pathogenic strains and unreported environmental strains could be detected if the molecular genetic approach were applied to those cases. Herein, we report the first case of an Achromobacter species-associated buckle infection diagnosed by use of a 16S rDNA clone library analysis.
Case presentation
A 56-year-old woman complained of purulent discharge and conjunctival hyperemia in her right eye. These symptoms began several months prior to the first visit to our hospital. Eighteen years prior, she had received an uneventful scleral buckling surgery using a solid silicone tire in her right eye for rhegmatogenous retinal detachment. Thirteen years after the surgery, she was administered oral cephem antibiotics once on suspicion of a buckle infection. Although the symptoms temporarily improved, chronic inflammation persisted for several years. Because subsequent topical quinolone and topical steroid treatments were ineffective, she visited our hospital for rigorous diagnosis and radical treatment. On the first visit, the best-corrected visual acuity was 20/200 in the right eye. Observation by a slit lamp microscope revealed conjunctival hyperemia, purulent discharge, and episcleritis. A conjunctival fistula was also observed in the upper quadrants, and large yellowish conjunctival follicles around the exposed buckle material were present (Figure 1). After examination, we removed the buckle material based on the diagnosis of recurrent buckle infection.
Figure 1.
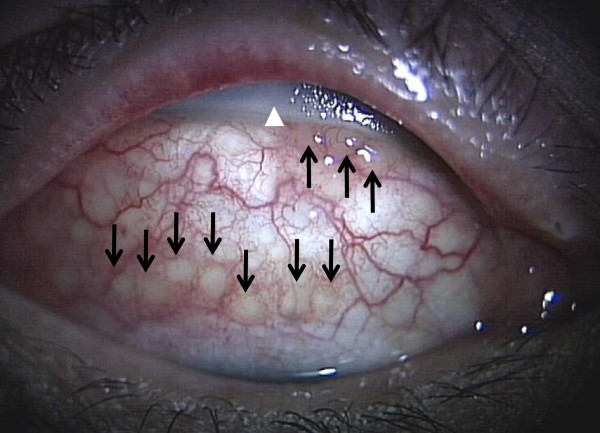
Pre-operative anterior segments photograph. The patient is looking downward. Conjunctival fistula in the upper quadrants and large yellowish conjunctival follicles (black arrows) around the exposed buckle material (white arrowhead) can be observed.
Pre-operatively, Alcaligenes and Corynebacterium species were isolated from the eye discharge. The bacterial identification and drug susceptibility tests were performed automatically using a MicroScan WalkAway 96 SI (Siemens Healthcare Diagnostics, Tokyo, Japan). During the surgery, a 120° solid silicone tire was removed and the scleral bed was irrigated with 0.5% moxifloxacin ophthalmic solution. Post-operatively, 300 mg/day of oral cefdinir was administered for 3 days, and both 0.5% moxifloxacin ophthalmic solution and 0.1% betamethasone sodium phosphate ophthalmic solution were administered 5 times daily for 2 weeks. After removal of the silicone tire, the symptoms improved rapidly. Retinal detachment had not recurred at this point.
Many small yellowish-white deposits were found on the surface of the removed buckle material (Figure 2A). Gram staining of the deposits showed a large number of gram-negative rods. Alcaligenes and Corynebacterium species were also isolated from the buckle material. Species identification and drug susceptibility results were obtained through laboratory procedures identical to those performed preoperatively. The drug susceptibility of the Alcaligenes strain isolated from the buckle was identical to that of the strain preoperatively isolated from the eye discharge (Table 1). In the case of Corynebacterium, there was a definite discrepancy in the drug susceptibilities between the strains obtained pre- and postoperatively; the strain isolated from the eye discharge was resistant to cephalosporin, but the strain isolated from buckle depositions was susceptible to all antibiotics tested (Table 2). Microbiological examination of the removed buckle material indicated that the causative pathogen is a bacterium that belongs to the family Alcaligenaceae. We employed a 16S rDNA clone library analysis to identify the causative bacterium at the species level and to assess the possibility of the involvement of other uncultured species in the buckle infection. Initially, the buckle material was divided into two pieces, and one piece was stained with ruthenium red for examination by scanning electron microscope (SEM) (Figure 2B). The other piece was placed into 15 mL of phosphate-buffered saline (PBS) and sonicated repeatedly using a VialTweeter (Hielscher Ultrasonics Gmbh, Berlin, Germany) at 60 W for 15 min at room temperature. PBS was replaced twice, and the final sonicate was used for DNA extraction.
Figure 2.
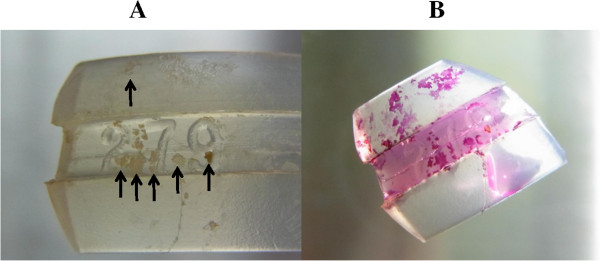
Pictures of the buckle material. (A) Buckle material immediately after the extraction. Many yellowish-white deposits (arrows) on the surface of the buckle material can be observed. (B) Ruthenium red staining. Deposits were stained red by ruthenium red for scanning electron microscopy.
Table 1.
The drug susceptibilities of the strain of Alcaligenes sp. and the strain of Corynebacterium sp.
| Alcaligenes | Corynebacterium | |||
|---|---|---|---|---|
| Antibiotic | Discharge | Buckle | Discharge | Buckle |
| Ampicillin | S | S | - | - |
| Penicillin G | R | R | - | - |
| Cefmenoxime | - | - | R | S |
| Ceftizoxime | R | R | R | S |
| Cefroxime | - | - | R | S |
| Cefepime | - | - | R | S |
| Cefpodoxime pivoxil | - | - | R | S |
| Azithromycin | - | - | R | S |
| Gentamicin | R | R | - | - |
| Tobramycin | R | R | - | - |
| Dibekacin | I | I | - | - |
| Arbekacin | I | I | R | S |
| Levofloxacin | I | I | S | S |
| Ciprofloxacin | S | S | S | S |
| Chloramphenicol | S | S | - | - |
| Imipenem/cilastatin | S | S | S | S |
| Meropenem | S | S | S | S |
Hyphen: not performed. S: susceptible. I: intermediate. R: resistant.
Although the two strains of Alcaligenes sp. show the same profiles, the two strains of Corynebacterium sp. show different profiles.
Table 2.
Summary of 16S rDNA clone library analysis of the infected buckle material
| Sequence type a) | No. of clone b) | Best match c) |
|---|---|---|
| ST1 | 12 | Achromobacter spanius strain LMG 5911 (631/633; 99.7%) |
| ST2 | 1 | Achromobacter spanius strain LMG 5911 (628/631; 99.5%) |
| ST3 | 1 | Achromobacter spanius strain LMG 5911 (628/632; 99.4%) |
| ST4 | 1 | Achromobacter spanius strain LMG 5911 (627/630; 99.5%) |
| ST5 | 1 | Achromobacter spanius strain LMG 5911 (630/632; 99.7%) |
| ST6 | 1 | Achromobacter spanius strain LMG 5911 (623/625; 99.7%) |
| ST7 | 1 | Achromobacter spanius strain LMG 5911 (627/628; 99.8%) |
| ST8 | 1 | Achromobacter spanius strain LMG 5911 (630/632; 99.7%) |
| ST9 | 1 | Achromobacter spanius strain LMG 5911 (627/628; 99.8%) |
| ST10 | 1 | Achromobacter spanius strain LMG 5911 (624/625; 99.8%) |
a)Twenty-one 16S rDNA sequences obtained (631 bp of high quality sequence) are classified basing on SNIPs.
b)Number of sequence belonging to each sequence type is shown.
c)Top hit microorganisims by which Blastn search of each sequence type indicated are listed.
The number in parenthesis shows identical base (bp)/alignment length (bp) to 16S rDNA from indicated species.
Bacterial DNA was extracted from 200 μL of the final PBS sonicate using Extrap Soil Kit Plus ver.2 (Nippon Steel Kankyo Engineering Co., Ltd., Tokyo, Japan). The 16S rDNA gene fragments were amplified with the purified DNA as a template and a universal eubacterial 16S rDNA primer set, 27f (5'-AGAGTTTGATCMTGGCTCAG-3') and Bac1392R (5'-ACGGGCGGTGTGAC-3'). After cloning the amplified products, the sequences were obtained from 24 clones using 27f as the sequencing primer. The low-quality sequences (Phred score <15) were trimmed, and the sequences were analysed for homology to NCBI database sequences using the Blast program. Of the 24 clones, high-quality sequences were obtained from 23 clones, but two of these were from the genomic regions other than 16S rDNAs. All of the partial 16S rDNA sequences obtained from 21 clones showed the best match to those of Achromobacter species (Table 2; identity ranged from 99.4—99.8% over 99% of alignments with query sequences). It is likely that the isolates initially identified as Alcaligenes spp. were in fact Achromobacter spp. This misidentification was probably due to the low discriminatory power of the biochemical test for the species in the family Alcaligenaceae. Single nucleotide polymorphisms (SNPs) were observed among the sequences (12 sequences were identical). These SNPs might indicate that several different Achromobacter strains were present in biofilms on the buckle material, although this was only the sequence diversity among the ribosomal RNA operons in a single Achromobacter chromosome. To further refine the identification of causative bacterial species, the most predominant 16S rDNA sequences obtained were aligned with those from 43 reference species (obtained from Ribosomal Database Project ver. 10) in the family Alcaligenaceae. We aligned the 613-bp regions encompassed within well-conserved regions using the Clustal W program to adjust the positions to be compared. All 21 of the sequences were phylogenetically positioned closely with the sequences from Achromobacter spanius (Figure 3). Based on these results, we conclude that an Achromobacter sp. closely related to A. spanius was the causative agent in this case.SEM of the buckle material showed numerous rod-shaped bacteria surrounded by a biofilm-like material, consistent with our conclusions from the 16S rDNA clone library analysis (Figure 4, A and B).
Figure 3.
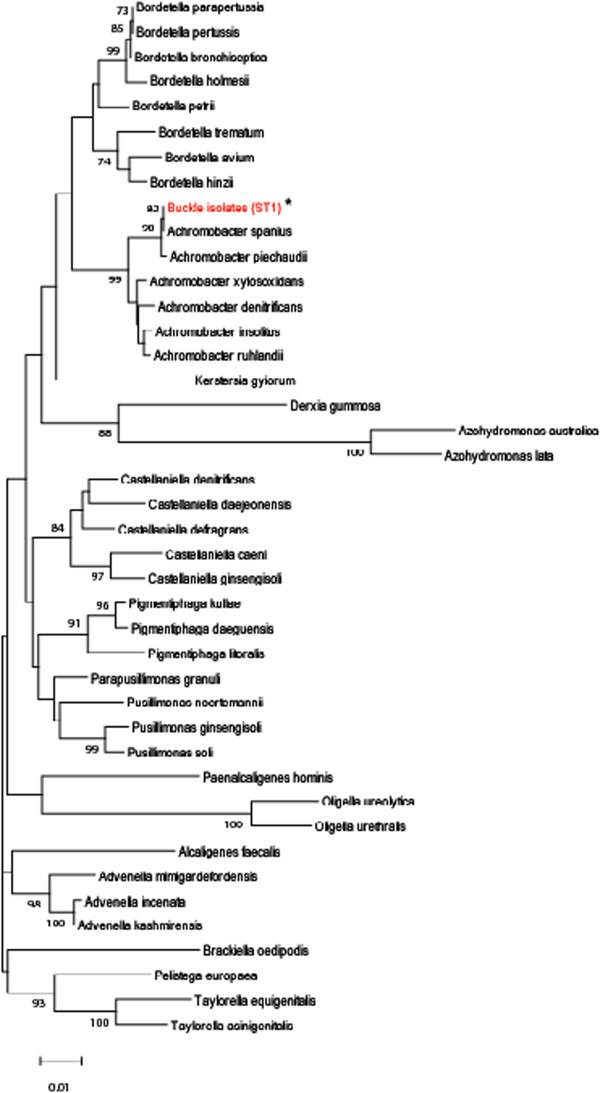
Phylogenetic relationship between the isolate from buckle material and other members of the family Alcaligenaceae . Only the most predominant ST1 sequence (indicated by red and asterisks) was analysed. The tree was constructed using the neighbour-joining algorithm. Numbers at nodes are bootstrap percentages based on 1,000 replications; only values >70% are shown. Bar, 0.01 substitutions per nucleotide position.
Figure 4.
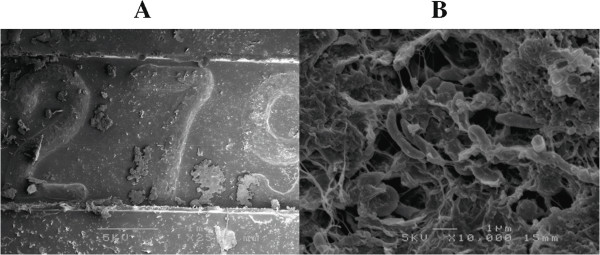
Scanning electron microscopic images of the buckle material. (A) Low magnification. Deposits attached after fixation by glutaraldehyde. (B) High magnification. Numerous rod-shaped bacteria surrounded by biofilm-like material are observed.
Conclusions
In clinical settings, cultivation and phenotypic tests of isolated bacteria employing traditional culture techniques is the first step in diagnosis of infectious diseases. In this case, we aimed to identify the causative pathogens for buckle infection by culturing the eye discharge and buckle material. These cultures resulted in the successful isolation of the two candidates, Alcaligenes and Corynebacterium species. We surmised that Corynebacterium spp. were a contaminant as they are one of the resident bacteria on the ocular surface [17], and Corynebacterium isolates from the discharge and buckle material showed different antimicrobial susceptibilities. Therefore, these different strains of Corynebacterium were most likely from the ocular surface. Correspondingly, the 16S rDNA sequences derived from Corynebacterium spp. were not identified in 16S rDNA clone library analysis. We presume that the Corynebacterium spp. were washed away by irrigation during surgery and sonication because they only attached to the surface of the buckle material and not embedded within biofilm.
Although Alcaligenes spp. were initially considered to be a causative agents, we had doubts about the microbiological identification based on the following observations. First, the isolate in this case showed resistance to aminoglycosides while the majority of Alcaligenes species have been reported to be susceptible to gentamicin [18]. Second, the taxonomy of the family Alcaligenaceae is continually revised and updated and the biochemical test is unreliable in discriminating Alcaligenes and Achromobacter due to their close phylogenetic relationship [19]. Device-related biofilm infections are often caused by opportunistic environmental pathogens and are often polymicrobial. The frequent discrepancy between direct microscopic counts and the number of culturable bacteria from environmental samples is one of several indications that we currently know very little about the diversity of microorganisms in nature [16]. In addition, precise species identification is typically problematic in environmental isolates. Therefore, we employed a 16S rDNA clone library analysis to precisely classify the isolate at the species level and to test the possibility that the biofilm in this case was polymicrobial and contained uncultivable environmental bacteria. Although 16S rDNA clone library analysis using 24 clones is insufficient for excluding the presence of other pathogenic strains, our results show that this case was buckle infection caused by an Achromobacter species alone that is closely related to A. spanius. To our knowledge, this is the first case report of buckle infection by Achromobacter sp. Reliable epidemiological data on bacterial isolates are important for empirical antimicrobial therapy; therefore, precise identification of bacterial species is essential.
Advances in surgery are expected to increase the opportunities for embedding medical devices within the body with a concomitant increase in the risk for device-related infections by opportunistic environmental pathogens. In fact, there are some reports describing Achromobacter-related infections from artificial devices such as prosthetic knee joints and contact lenses [20, 21]. Clinicians should take into account the inherent limitations of traditional microbiological assays and combine various approaches to obtain precise diagnoses when necessary. These efforts will likely increase the reliability of epidemiological data in the field of infectious diseases.
The taxonomy of the genus Alcaligenes is closely intertwined with that of the genus Achromobacter and is frequently revised [19]. Alcaligenes has also been isolated from clinical specimens, including ophthalmic samples [12, 22–29]. Coenye et al. reported that several isolates identified phenotypically as Alcaligenes species belonged to the genus Achromobacter based on genetic analysis, and they proposed two novel Achromobacter species from these isolates [30]. It is important clinically to discriminate Alcaligenes and Achromobacter because epidemiological data demonstrate that 72.7% of clinical Achromobacter isolates showed multi-drug resistance while all of the Alcaligenes isolates tested were susceptible to imipenem, gentamicin, and ciprofloxacin [18]. With regard to the current clinical case, drugs to which Achromobacter spp. are potentially susceptible were initially administered, followed by the administration of drugs to which Achromobacter spp. are known to be susceptible. However, inflammation around the buckle material continued for several years. SEM observations were indicative of the long clinical course, recurrent symptoms, and Achromobacter’s resistance to antibiotic treatment. Therefore, the Achromobacter-associated buckle infection case reported here is valuable for considering the epidemiology and antimicrobial therapy of ophthalmic infections. The emergence of device-related infections caused by Achromobacter may be intractable, even when efficacious antibiotics are administered.
In conclusion, Achromobacter spp. should be recognized as causative agents for device-related ophthalmic infections. Molecular species identification by 16S rDNA sequence analysis should be combined with conventional cultivation techniques to investigate the significance of Achromobacter spp. in ophthalmic infections.
Consent
Written informed consent was obtained from the patient for publication of this case and the accompanying images.
Acknowledgment
We would like to thank Nippon Steel & Sumikin Eco-Tech Corporation for technical support of the 16S rDNA clone library analysis.
Footnotes
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
FK, KK, and TN treated the patient. HE performed molecular genetic investigations, made the final diagnosis, and wrote the manuscript. YM and TK reviewed the manuscript. All authors read and approved the final manuscript.
Contributor Information
Fumika Hotta, Email: uri0428@yahoo.co.jp.
Hiroshi Eguchi, Email: hiroegu0113@gmail.com.
Takeshi Naito, Email: naito.takeshi@tokushima-u.ac.jp.
Yoshinori Mitamura, Email: ymitaymitaymita@yahoo.co.jp.
Kohei Kusujima, Email: kusujima@abeam.ocn.ne.jp.
Tomomi Kuwahara, Email: tomomi@med.kagawa-u.ac.jp.
References
- 1.Nishiyama M, Yamamoto S, Kurosawa N. Microbial community analysis of a coastal hot spring in Kagoshima, Japan, using molecular- and culture-based approaches. J Microbiol. 2013;51:413–422. doi: 10.1007/s12275-013-2419-z. [DOI] [PubMed] [Google Scholar]
- 2.Someya N, Ohdaira Kobayashi Y, Tsuda S, Ikeda S. Microbes Environ. 2013. Molecular characterization of the bacterial community in a potato phytosphere. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chouari R, Le Paslier D, Daegelen P, Dauga C, Weissenbach J, Sghir A. Molecular analyses of the microbial community composition of an anoxic basin of a municipal wastewater treatment plan reveal a novel lineage of proteobacteria. Microb Ecol. 2010;60:272–281. doi: 10.1007/s00248-009-9632-7. [DOI] [PubMed] [Google Scholar]
- 4.Britschgi TB, Giovannoni SJ. Phylogenetic analysis of a natural marine bacterioplankton by rRNA gene cloning and sequencing. Appl Environ Microbiol. 1991;57:1707–1713. doi: 10.1128/aem.57.6.1707-1713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt TM, DeLong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caton IR, Schneegurt MA. Culture-independent analysis of the soil bacterial assemblage at the Great Salt Plains of Oklahoma. J Basic Microbiol. 2012;52:16–26. doi: 10.1002/jobm.201100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrera-Rubio R, Garcia-Nunez M, Seto L, Anto JM, Moya A, Monso E, Mira A. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2012;50:3562–3568. doi: 10.1128/JCM.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M, Wei H, Chen Y, Lu H, Zuo J, Su M, Qiu Y, Jia W, Xiao C, Smith LM, Yang S, Holmes E, Tang H, Zhao G, Nicholson JK, Li L, Zhao L. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simón-Soro A, Pignatelli M, Mira A. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland SP, Pulido JS, Miller D, Ellis D, Alfonso E, Scott M, Costerton JW. Biofilm and scleral buckle-associated infections. A mechanism for persistence. Ophthalmology. 1991;98:933–938. doi: 10.1016/S0161-6420(91)32199-7. [DOI] [PubMed] [Google Scholar]
- 11.Oshima Y, Ohji M, Inoue Y, Harada J, Motokura M, Saito Y, Emi K, Tano Y. Methicillin-resistant Staphylococcus aureus infections after scleral buckling procedures for retinal detachments associated with atopic dermatitis. Ophthalmology. 1999;106:142–147. doi: 10.1016/S0161-6420(99)90025-8. [DOI] [PubMed] [Google Scholar]
- 12.Wirostko WJ, Covert DJ, Han DP, Connor TB, Jr, Kim JE, Hammersley J, Lindgren K. Microbiological spectrum of organisms isolated from explanted scleral buckles. Ophthalmic Surg Lasers Imaging. 2009;40:201–202. doi: 10.3928/15428877-20090301-22. [DOI] [PubMed] [Google Scholar]
- 13.Liu DT, Lee VY, Chi-Lai L, Lam DS. Stenotrphomonas maltophilia and Mycobacterium chelonae coinfection of the extraocular scleral buckle explant. Ocul Immunol Inflamm. 2007;15:441–442. doi: 10.1080/09273940701732263. [DOI] [PubMed] [Google Scholar]
- 14.Nishikiori N, Ohguro H. An intractable case of Pseudomonas aeruginosa infection after scleral buckling for rhegmatogenous retinal detachment. Clin Ophthalmol. 2008;2:223–226. doi: 10.2147/OPTH.S2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhry NA, Tabandeh H, Rosenfeld PJ, Miller D, Davis J. Scleral buckle infection with ciprofloxacin-resistant Pseudomonas aeruginosa. Arch Ophthalmol. 1998;116:1251. [PubMed] [Google Scholar]
- 16.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue Y, Usui M, Ohashi Y, Shiota H, Yamazaki T, Preoperative Disinfection Study Group Preoperative disinfection of the conjunctival sac with antibiotics and iodine compounds: a prospective randomized multicenter study. Jpn J Ophthalmol. 2008;52:151–161. doi: 10.1007/s10384-008-0517-y. [DOI] [PubMed] [Google Scholar]
- 18.Misawa S, Oguri T, Nakamura A, Tabe Y, Kondo S, Miyake K, Miyake N, Igari J, Ohsaka A. Prevalence and antimicrobial susceptibility of metallo-β -lactamase-producing gram-negative bacilli from clinical specimens (Japanese) Jpn J of Infect Chemother. 2007;55:211–219. [Google Scholar]
- 19.Yabuuchi E, Kawamura Y, Kosako Y, Ezaki T. Emendation of genus Achromobacter and Achromobacter xylosoxidans (Yabuuchi and Yano) and proposal of Achromobacter ruhlandii (Packer and Vishniac) comb. Nov., Achromobacter piechaudii (Kiredjian et al.) comb. Nov., and Achromobacter xylosoxidans subsp. Denitrificans (Rüger and Tan) comb. Nov. Microbiol Immunol. 1998;42:429–438. doi: 10.1111/j.1348-0421.1998.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 20.Taylor P, Fischbein L. Prosthetic knee infection due to Achromobacter xylosoxidans. J Rheumatol. 1992;19:992–993. [PubMed] [Google Scholar]
- 21.Wiley L, Bridge DR, Wiley LA, Odom JV, Elliott T, Olson JC. Bacterial biofilm diversity in contact lens-related disease: emerging role of Achromobacter, Stenotrophomonas, and Delftia. Invest Ophthalmol Vis Sci. 2012;53:3896–3905. doi: 10.1167/iovs.11-8762. [DOI] [PubMed] [Google Scholar]
- 22.Krause ML, Sohail MR, Patel R, Wittich CM. Achromobacter piechaudii bloodstream infection in an immunocompetent host. Am J Case Rep. 2012;13:265–267. doi: 10.12659/AJCR.883527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambiase A, Catania MR, Del Pezzo M, Rossano F, Terlizzi V, Sepe A, Raia V. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 2011;30:973–980. doi: 10.1007/s10096-011-1182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teng SO, Ou TY, Hsieh YC, Lee WC, Lin YC, Lee WS. Complicated intra-abdominal infection caused by extended drug-resistant Achromobacter xylosoxidans. J Microbiol Immunol Infect. 2009;42:176–180. [PubMed] [Google Scholar]
- 25.Tsay RW, Lin LC, Chiou CS, Liao JC, Chen CH, Liu CE, Young TG. Alcaligenes xylosoxidans bacteremia: clinical features and microbiological characteristics of isolates. J Microbiol Immunol Infect. 2005;38:194–199. [PubMed] [Google Scholar]
- 26.Aisengerg G, Rolston KV, Safdar A. Bacteremia caused by Achromobacter and Alcaligenes species in 46 patients with cancer (1989–2003) Cancer. 2004;101:2134–2140. doi: 10.1002/cncr.20604. [DOI] [PubMed] [Google Scholar]
- 27.Huang ZL, Chen YF, Chang SW, Lin KK, Hsiao CH. Recurrent Alcaligenes xylosoxidans keratitis. Cornea. 2005;24:489–490. doi: 10.1097/01.ico.0000151562.17366.77. [DOI] [PubMed] [Google Scholar]
- 28.Kiernan DF, Chin EK, Sclafani LA, Saidel MA. Multiple drug-resistant Alcaligenes xylosoxidans keratitis in a sanitation worker. Eye Contact Lens. 2009;35:212–214. doi: 10.1097/ICL.0b013e3181aac4fd. [DOI] [PubMed] [Google Scholar]
- 29.Newman PE, Hider P, Waring GO, 3rd, Hill EO, Wilson LA, Harbin TS. Corneal ulcer due to Achromobacter xylosoxidans. Br J Ophthalmol. 1984;68:472–474. doi: 10.1136/bjo.68.7.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coenye T, Vancanneyt M, Cnockaert MC, Falsen E, Swings J, Vandamme P. Kerstersia gyiorum gen. nov., sp. nov., a novel Alcaligenes faecalis-like organism isolated from human clinical samples, and reclassification of Alcaligenes denitrificans Rüger and Tan 1983 as Achromobacter denitrificans cpmb. Nov. Int J Syt Bacteriol. 2003;53:1825–1831. doi: 10.1099/ijs.0.02609-0. [DOI] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2415/14/142/prepub


