Abstract
The efficacy of chemotherapeutic drugs is often offset by severe side effects attributable to poor selectivity and toxicity to normal cells. Recently, the enzyme dipeptidyl peptidase IV (DPPIV) was considered as a potential target for the delivery of chemotherapeutic drugs. The purpose of this study was to investigate the feasibility of targeting chemotherapeutic drugs to DPPIV as a strategy to enhance their specificity. The expression profile of DPPIV was obtained for seven cancer cell lines using DNA microarray data from the DTP database, and was validated by RT-PCR. A prodrug was then synthesized by linking the cytotoxic drug melphalan to a proline-glycine dipeptide moiety, followed by hydrolysis studies in the seven cell lines with a standard substrate, as well as the glycyl-prolyl-melphalan (GP-Mel). Lastly, cell proliferation studies were carried out to demonstrate enzyme-dependent activation of the candidate prodrug. The relative RT-PCR expression levels of DPPIV in the cancer cell lines exhibited linear correlation with U95Av2 Affymetrix data (r2 = 0.94), and with specific activity of a standard substrate, glycine-proline-p-nitroanilide (r2 = 0.96). The significantly higher antiproliferative activity of GP-Mel in Caco-2 cells (GI50 = 261 μM) compared to that in SK-MEL-5 cells (GI50 = 807 μM) was consistent with the 9-fold higher specific activity of the prodrug in Caco-2 cells (5.14 pmol/min/μg protein) compared to SK-MEL-5 cells (0.68 pmol/min/μg protein) and with DPPIV expression levels in these cells. Our results demonstrate the great potential to exploit DPPIV as a prodrug activating enzyme for efficient chemotherapeutic drug targeting.
Keywords: drug targeting, dipeptidyl peptidase IV (DPPIV), enzyme-targeted delivery, enzyme-dependent prodrug activation, selective cytotoxic action
1. Introduction
Chemotherapeutic drugs, alone or as an adjuvant therapy to surgery and radiation, are a vital part in cancer treatment. However, their effectiveness is often offset by severe side effects caused by poor selectivity and toxicity to normal cells. In recent years, the rapid advance in the fields of bioinformatics and genomics facilitated the identification of numerous target molecules that are uniquely or overly expressed in cancer cells, including certain receptors1,2 and enzymes.3−5 These discoveries were successfully utilized for the specific delivery of chemotherapeutic drugs to tumor cells, with the double goal of increasing their therapeutic efficacy while decreasing their toxicity to normal cells.6−10
The enzyme dipeptidyl peptidase IV (DPPIV) (EC 3.4.14.5), also known as CD26 (T-cell surface activation antigen) or adenosine deaminase (ADA), was first identified in 1966.11 DPPIV is a member of the prolyl oligopeptidase (POP) family of serine proteases12 and specifically hydrolyzes the peptide bond C-terminal to proline or alanine in the penultimate P1 position.13 It is normally present on the apical membrane surface of several cell types, including T cells and B cells,14,15 natural killer (NK) cells,16,17 epithelial cells of kidney, intestine brush border membranes, plasma,18 and endothelial cells of blood vessels,19 and is involved in many diverse physiological processes.20,21 In addition, it was recently discovered that DPPIV is overly expressed in human renal cell carcinoma tissues,22 and it is thought to play a role in the pathogenesis of other various human cancers as well.23−25 Moreover, proline-containing substrates are hydrolyzed exclusively by proline-specific enzymes and are less susceptible to nonspecific peptidases and proteases.11,13,26,27 Thus, a DPPIV-cleavable prodrug containing a proline dipeptide conjugated to a cytotoxic chemotherapeutic agent may limit nonspecific activation.28−31
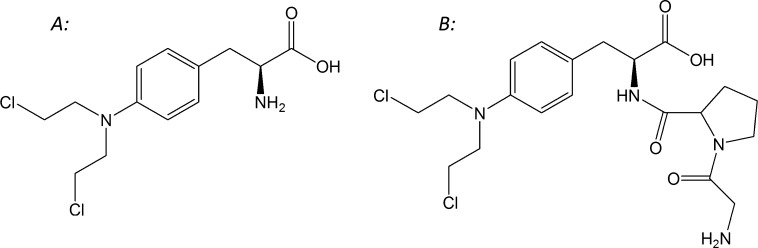
The purpose of this study was to investigate the feasibility of exploiting DPPIV as a prodrug activating enzyme, to allow specific targeting of chemotherapeutic agents to cancer cells. To that end, the expression profiles of DPPIV in 60 cancer cell lines (NCI 60) were obtained, and 7 cell lines, which represent the spectrum of DPPIV expression, were selected. A candidate prodrug was then synthesized by linking the cytotoxic drug melphalan (Figure 1A) to a proline dipeptide, creating glycyl-prolyl-melphalan prodrug (GP-Mel; Figure 1B) with expected DPPIV affinity. Functional activity studies of DPPIV with the prodrug, in the absence vs presence of inhibitor, confirmed that the prodrug is a specific DPPIV substrate. Finally, hydrolysis and cell proliferation studies were performed in high and low DPPIV expression cancer cell lines. Overall, this work indicates that DPPIV may be exploited as a prodrug activating enzyme for efficient chemotherapeutic drug targeting.
Figure 1.
Chemical structures of (A) melphalan and (B) its Gly-Pro dipeptide prodrug, GP-Mel.
2. Materials and Methods
2.1. Materials
Melphalan, porcine kidney dipeptidyl peptidase (porcine DPPIV), glycyl-prolyl-p-nitroanilide (GP-pNA), glycyl-phenylalanyl-p-nitroanilide (GF-pNA), glycyl-arginyl-p-nitroanilide (GR-pNA), p-nitroaniline (pNA), diprotin A (Ile-Pro-Ile), XTT, PMS (N-methyldibenzopyrazine methyl sulfate), and ladder SYBR green were purchased from Sigma Chemical Co. (St. Louis, MO). N,N-Dimethylformamide (DMF), piperidine, triisopropylsilane (TIS), triethanolamine (TEA), and trifluoroacetic acid (TFA) were purchased from Aldrich Chemical Co. (Milwaukee, WI). Boc-l-Gly-l-Pro, 1-hydroxybenzotriazole anhydrous (HOBT), 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), triisopropylsilane (TIS), N-(9-fluorenylmethoxycarbonyloxy)succinimide (Fmoc-OSu), and Wang resin (100–200 mesh) were obtained from Calbiochem-Novabiochem (San Diego, CA). Access RT-PCR kit, DNA blue/orange loading dye, 1 kb ladder and 100 bp ladder were obtained from Promega (Madison, WI). 4–20% TBE gels, TRIzol reagent, and custom-ordered DPPIV sense and antisense primers were from Invitrogen Life Technologies (Carlsbad, CA). The cancer cell lines IGROVI (ovarian), PC-3 (prostate), 786-0 (renal), SK-MEL-5 (melanoma), SK-OV-3 (ovarian), Caco-2 (colon), and HepG2 (liver) were obtained from National Cancer Institute or ATCC. Dulbecco’s modified Eagle’s medium (DMEM), RPMI-1640, fetal bovine serum (FBS), fetal calf serum (FCS), phosphate buffered saline (PBS), and trypsin-EDTA were purchased from GIBCO BRL (Grand Island, NY). Minimal essential medium (MEM) was obtained from ATCC. All HPLC grade solvents (acetonitrile, DMF, ethyl acetate, and piperidine) used for peptide synthesis or HPLC analysis were obtained from EM Sciences (Gibbstown, NJ). Trifluoroacetic acid for HPLC analysis was obtained from Pierce (Rockville, IL). All other chemicals and reagents used were of analytical or HPLC grade.
2.2. Synthesis of l-Glycyl-l-prolyl-melphalan Prodrug
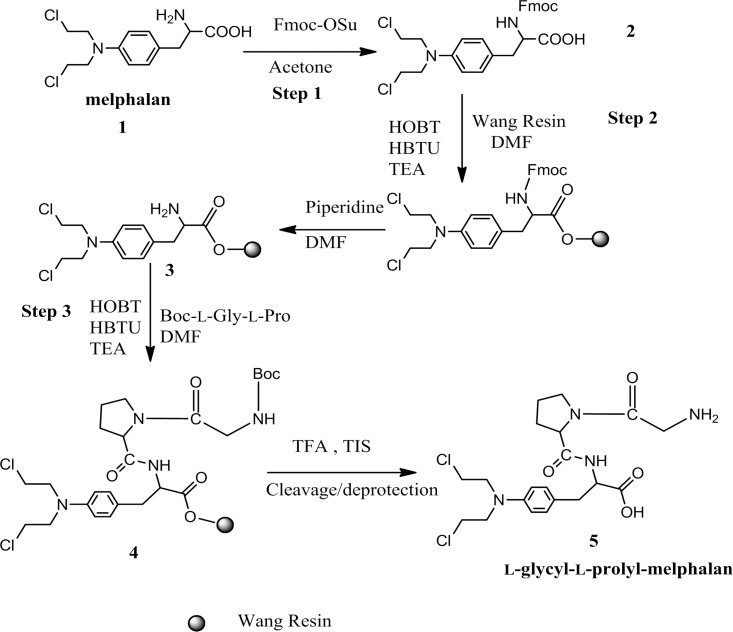
The solid-phase synthesis of the l-glycyl-l-prolyl prodrug of melphalan was carried out in a stepwise fashion with Wang resin as described below and is summarized in the schematic shown in Figure 2.
Figure 2.
Schematic of solid-phase synthesis of l-glycyl-l-prolyl-melphalan prodrug.
-
Step 1.
Fmoc-l-melphalan synthesis: Fmoc-OSu (300 mg, 0.89 mmol) was added to an ice-cold solution of melphalan, 1 (200 mg, 0.82 mmol), in a mixture of acetone (20 mL), distilled water (10 mL), and NaHCO3 (300 mg). The mixture was stirred for 1 h at 0 °C and then reacted at room temperature for 16 h. After the reaction was complete, the mixture was concentrated. 15 mL of ethyl acetate and 15 mL of distilled water were added. The aqueous phase was then extracted with ethyl acetate (3 × 15 mL). The combined organic phases were washed with distilled water and brine and dried over MgSO4. The filtrate was concentrated under vacuum to yield Fmoc-l-melphalan, 2 (330 mg, yield 98%).
-
Step
2.
Fmoc-l-melphalan (330 mg) and Wang resin (0.1 mmol) were added into a 100 mL round reaction bottle with 20 mL of DMF. HOBT (0.3 mmol), HBTU (0.3 mmol), and TEA (0.3 mmol) were then added to the resin mixture and stirred overnight at room temperature. The Wang resin, 3, was washed 3 times with 10 mL of DMF after filtration and then washed 3 times for 10 min each in 10 mL of 20% piperidine in DMF.
-
Step 3.
Boc-l-glycyl-l-proline (0.3 mmol) and Wang resin 3 were added to 20 mL of DMF in a 100 mL round reaction bottle. HOBT (0.3 mmol), HBTU (0.3 mmol), and TEA (0.3 mmol) were added and stirred overnight at room temperature. The resultant Wang resin, 4, was washed 3 times with 10 mL of DMF after filtration and then washed 3 times for 10 min each in 10 mL of 20% piperidine in DMF. Wang resin 4 was then added to 10 mL of cleavage buffer (TFA:TIS:H2O, 95:2.5:2.5) and stirred for 1 h at room temperature. After filtration, the solution was concentrated and cold ether (10 mL) was added to precipitate out the prodrug 5. After removing ether, the residues were reconstituted with 5 mL of distilled water and lyophilized. The TFA salt of the prodrug was obtained as a white powder.
The purity of the melphalan prodrug was determined by HPLC to be 95%. Electrospray ionization mass spectra (ESI-MS) were obtained on a Thermoquest LCQ electrospray ionization mass spectrometer. The observed molecular weight of the prodrug was found to be consistent with that required by its structure. The structural identity of the prodrug was then confirmed using proton nuclear magnetic resonance spectra (1H NMR). 1H NMR spectra were obtained with a 500 mHz Bruker NMR spectrometer.
l-Glycyl-l-prolyl-melphalan: yield 13%; percent purity 95%; 1H NMR (DMSO-d6) δ 1.95–2.20 (m, 4H, CH2 on proline), 3.05–3.35 (m, 12H, CH2), 4.44–4.80 (m, 5H, αCH, αCH2), 6.50–6.90 (m, 4H, phenol H); ESI-MS 459.4 (M + H)+.
2.3. Identification of Target Enzyme in NCI 60 Cancer Cell Lines
The microarray data and the U95Av2 Affymetrix data were downloaded from the DTP database (https://wiki.nci.nih.gov/display/NCIDTPdata/Molecular+Target+Data). The programming language Perl was used to sift through more than 26,000 genes to find all enzymes with names that end with the suffix “ase”. The enzyme text file was then used to separate the enzymes into different classes, such as hydrolases, peptidases, and esterases as described earlier.32 Briefly, the text files were then converted to an Excel sheet. Visual Basic was then used to arrange the data retrieved so that the expression of each enzyme gene in the 60 cancer cell lines could be easily visualized using visual tools such as Cluster and TreeView programs. The arranged data from peptidases was normalized using a pool of 12 cell lines (NCI-H226, COLO 205, MCF-7, HS 578T, OVCAR-3, OVCAR-4, K-562, HL-60 (TB), CAKI-1, LOX IMVI, PC-3, SNB-19) as reference and clustered according to the origin of the cell lines. The sorted expression data for the 60 cancer cell lines was clustered using hierarchical clustering, and a file was created (.cdt) such that the expression of the genes in 60 cancer cell lines could be visualized using TreeView. The potential enzyme target, DPPIV, was selected on the basis of the differential expression in various cell lines or tissues with emphasis on high expression in a particular tissue as compared to other tissues and high substrate specificity of the enzyme.
2.4. Selection of Cancer Cell Lines and Cell Culture
Candidate cancer cell lines were selected from the NCI 60 cell lines based on the expression levels of DPPIV, doubling time, and growth requirements. Thus, the cancer cell lines selected, IGROVI, PC-3, 786-0, SK-OV-3, and SK-MEL-5, represent cells with high, medium, and low expression of DPPIV, reasonably short doubling times, and standard growth requirements. In addition, Caco-2 as well as HepG2 cell lines were also used in the studies.
2.5. Isolation of mRNA and RT-PCR Experiment
Total RNA was lysed and purified from each cancer cell line with TRIzol reagent according to the manufacturer’s protocol. Isolated RNA (0.5 μg) was reverse transcribed to cDNA with AMV reverse transcriptase, Oligo (dT), and Access RT-PCR kit following the standard protocol. Each of the 20 cycles of amplification consisted of denaturation (94 °C for 25 s), annealing (50 °C for 25 s), and extension (68 °C for 40 s). The primer set used for DPPIV amplification was the sense primer (5′-CCTTCTACTCTGATGAGTCACTGC-3′) and the antisense primer (5′-GTGCCACTAAGCAGTTCCATCTTC-3′).33 PCR product was identified by electrophoresis in 4–20% TBE gels followed by SYBR green staining. The gel was then visualized with UV light, and the relative intensities of the RT-PCR product bands were measured using Metamorph software.
2.6. Hydrolysis of Glycyl-prolyl-p-nitroanilide (GP-pNA) by Porcine DPPIV with and without Specific Inhibitor Diprotin A
Porcine kidney DPPIV was used in the studies due to its commercial availability and its close alignment with human DPPIV (88% homology).34 Stock solutions of porcine DPPIV were prepared by reconstituting lyophilized powder (0.87 unit, 23 units/mg) in 1 mL of 0.1 M Tris-HCl (pH 8.0) and stored at −80 °C. Solutions of the various substrates glycyl-prolyl-p-nitroanilide (GP-pNA), glycyl-phenylalaninyl-p-nitroanilide (GF-pNA), or glycyl-argininyl-p-nitroanilide (GR-pNA) at 0.4 mM were mixed with appropriate amounts of diluted DPPIV solution (final DPPIV concentration in mixture, 0.473 μg/mL) and incubated at 37 °C for 15–30 min in flat-bottom 96-well plates.35,36 The hydrolysis of the substrates was monitored by measurement of absorbance at 405 nm (generation of p-nitroanilide) using a precision microplate reader (Emax, Molecular Devices). The competitive inhibition of hydrolysis of the substrates by porcine kidney DPPIV in the presence of the inhibitor diprotin A was determined by including 0.4 mM diprotin A in the diluted DPPIV mixture prior to incubation. Initial hydrolysis rates were determined in all hydrolysis experiments by assaying the amounts of pNA released.
2.7. Hydrolysis of GP-pNA in Cancer Cells and Inhibition by Diprotin A
DPPIV activity in the selected cancer cells was determined using the standard substrate GP-pNA. Briefly, cells were grown to 90% confluence in 150 mm culture plates, washed carefully with 0.15 M NaCl, and collected by scraping. The cells were resuspended in 0.15 M NaCl and then centrifuged at 3,000 rpm for 5 min. The cell pellet was resuspended in phosphate buffered saline (PBS, pH 7.4), and sonicated for 5 s two times on ice. The sonicated cell suspension was centrifuged at 18000g for 30 min at 4 °C. The supernatant was then used in hydrolysis studies, and to determine protein content (Bio-Rad DC protein assay). The protein content was adjusted to approximately 1000 μg/mL by appropriate dilutions before use in hydrolysis studies.
Hydrolysis studies were conducted in a 96-well microplate containing the cell homogenate suspensions at 37 °C. The reactions were initiated by the addition of GP-pNA (final concentration 1 mM) to the suspensions. The absorbance was monitored every minute for a total of 30–60 min using a precision microplate reader (Emax, Molecular Devices) at 405 nm. In the competitive inhibition studies, diprotin A at 1 mM was included along with 1 mM GP-pNA, and the mixture was added to the cell homogenate suspensions and absorbance monitored as described above. DPPIV activity was expressed as the amount of pNA (micromoles) released per minute, normalized to the amount of protein.
2.8. Hydrolysis of GP-Mel by Porcine Kidney DPPIV
Hydrolysis of the melphalan prodrug by porcine kidney DPPIV and the effect of the competitive inhibitor diprotin A were assayed using 96-well microplates. Porcine DPPIV enzyme solution (230 μL at a final concentration 4.4 μg/mL) was placed in triplicate wells maintained at 37 °C. The reactions were initiated by the addition of GP-Mel (1 mM final concentration in mixture), and 40 μL aliquots were removed at predetermined time points and added to two volumes of 10% ice-cold TFA to quench the reaction and precipitate protein. In the inhibition studies, diprotin A and GP-Mel were both added (1 mM final concentration) to the enzyme solution, incubated at 37 °C, and sampled as described above. The reactions were monitored for 30–60 min. The quenched precipitated samples were then filtered through a 0.45 μm filter plate and centrifuged at 1800g and 4 °C for 20 min. The recovered filtrate was analyzed by HPLC as described below.
2.9. Hydrolysis of GP-Mel by Caco-2 and SK-MEL-5 Cell Homogenates
The extent of hydrolysis of GP-Mel in Caco-2 and SK-MEL-5 cell homogenates was determined as follows. Caco-2 and SK-MEL-5 cells and cell homogenates were prepared as described earlier. The hydrolysis reactions were carried out in 96-well plates (Corning, Corning, NY). 230 μL of the cell suspensions (1000 μg/mL protein) were placed in triplicate wells and the reactions initiated by the addition of GP-Mel (final concentration 1 mM in mixture) and incubated at 37 °C. At predetermined time points 40 μL aliquots were removed and added to two volumes of 10% ice-cold TFA to quench the reaction and precipitate protein. In the inhibition studies, diprotin A and GP-Mel were both added (1 mM final concentration) to the cell suspensions, incubated at 37 °C, and sampled as described above. The reactions were monitored for 30–60 min. The quenched precipitated samples were then filtered through a 0.45 μm filter plate and centrifuged at 1800g and 4 °C for 20 min. The recovered filtrate was analyzed by HPLC as described below. DPPIV activity was expressed as the amount (picomoles) of melphalan released per minute, normalized to the amount of protein.
2.10. HPLC Analysis
The concentrations of GP-Mel and melphalan were determined on a Waters HPLC system (Waters Inc., Milford, MA). The HPLC system consisted of two Waters pumps (model 515), a Waters autosampler (WISP model 712), and a Waters UV detector (996 photodiode array detector). The system was controlled by Waters Millennium 32 software (Version 3.0.1). Samples were injected onto a Waters XTerra C18 reversed phase column (5 μm, 4.6 × 250 mm) equipped with a guard column. The flow rate was 1 mL/min, and the mobile phase was 70:30 (% v/v) water:acetonitrile (both with 0.1% TFA). The run time was 20 min. Standard curves generated for prodrug and parent drug were utilized for quantitation of integrated area under peaks.
2.11. Cell Proliferation Assays
Cell proliferation assays were conducted to determine the cytotoxic activities of the prodrug GP-Mel and the parent melphalan. The assays were carried out with Caco-2 and SK-MEL-5 cells since the expression of DPPIV was found to be highest and lowest, respectively, in these cells, based on RT-PCR expression results. Caco-2 and SKMEL-5 cells were plated overnight in a 96-well cell culture plate at a density of 5,000 cells/well per 0.1 mL. Stock solutions (1 mM) of GP-Mel and melphalan were prepared in RPMI-1640 phenol red free medium supplemented with FBS. Stock solutions were serially diluted to obtain a total of six drug concentrations, 1 mM, 0.5 mM, 0.25 mM, 0.125 mM, 0.0625 mM, and 0.03125 mM, for cell proliferation studies. After 24 h, the medium in the 96-well plate was aspirated and replaced with drug solutions in the medium. Growth medium alone served as controls. The cells were then incubated at 37 °C and 5% CO2 for 48 h. After 48 h, 50 μL of XTT labeling mixture (5 mL of 1 mg/mL XTT in RPMI-1640 phenol red free medium mixed with 100 μL of 0.383 mg/mL PMS in phosphate buffered saline) was added to each well. The color development, due to formation of formazan dye by metabolically active cells, was monitored for 4 h, after which the plates were read at 490 nm (805 nm as the reference wavelength) with a precision microplate reader (Emax, Molecular Devices). The percent cell viability, at different drug and prodrug concentrations, relative to control was then plotted as a function of drug/prodrug concentration to compute the GI50 values.
3. Results
3.1. Identification of DPPIV and Selection of Cancer Cell Lines
After the genes had been sorted as hydrolases, peptidases, and esterases, they were clustered and their expression in the 60 cancer cell lines was visualized as described previously.32 The expression patterns of proline-specific peptidases such as prolinases/peptidase α, prolyl carboxypeptidase, prolyl oligopeptidase/prolyl endopeptidase, DPPIV/CD26, aminopeptidase P, and prolidase/peptidase D, were selected for further assessment. DPPIV was overexpressed in most renal carcinoma cell lines (UO-31, TK-10, SN12C, RXF 393, CAKI-1, ACHN, A498, and 786-0) compared to cancer cell lines derived from other tissues. The relatively higher DPPIV expression in 786-0 renal carcinoma cell line compared to its expression in SK-OV-3 ovarian carcinoma or SK-MEL-5 melanoma cell lines was consistent with Affymetrix expression data. Thus, in addition to the selection of IGROVI (ovarian cancer cell line), PC-3 (prostate cancer cell line), 786-0 (renal cancer cell line), SK-OV-3 (ovarian cancer cell line), and SK-MEL-5 (melanoma) based on the U95Av2 Affymetrix gene expression of DPPIV (Table 1), HepG2 and Caco-2 cells were also selected for characterization of DPPIV activity with standard substrates and RT-PCR determinations of DPPIV expression.
Table 1. Specific Activity of GP-pNA in the Presence vs Absence of Diprotin A in Various Cancer Cell Lines (pmol/min/μg protein, Mean ± SD; n = 3) and DPPIV Expression Determined with RT-PCR and with U95Av2 Affymetrix GeneChip.
| sp act. (pmol/min/μg) |
|||||
|---|---|---|---|---|---|
| cell line | rel RT-PCR expression | GeneChip expressiona | w/o diprotin A | with diprotin A | % inhibn |
| Caco-2 | 13.49 | 18.40 ± 1.17 | 0.29 ± 0.38 | 98.4 | |
| HepG2 | 12.71 | 20.16 ± 0.88 | 0.23 ± 0.49 | 98.9 | |
| IGROV1 | 7.92 | 171 | 11.42 ± 0.12 | 0.25 ± 0.12 | 97.8 |
| PC-3 | 5.51 | 138 | 4.18 ± 0.11 | 0.30 ± 0.05 | 92.7 |
| 786-0 | 2.81 | 88 | 1.32 ± 0.10 | 0.08 ± 0.11 | 94.0 |
| SK-OV-3 | 1.88 | 33 | 1.50 ± 0.20 | ndb | 100.0 |
| SK-MEL-5 | 1.00 | 6 | 1.25 ± 0.02 | 0.19 ± 0.02 | 84.8 |
Novartis U95Av2 Affymetrix data on the DTP database.
Not detected.
3.2. Expression of DPPIV in Human Cancer Cell Lines: RT-PCR
RT-PCR of extracellular mRNA in the seven selected human cancer cell lines was performed with primers specific for DPPIV. The expression profiles of DPPIV in the seven cancer cell lines evidenced by the band at 315 bp are shown in Figure 3. The results of semiquantitative RT-PCR analysis (Table 1) indicated that DPPIV was expressed at high levels in Caco-2 and HepG2 cells, and at very low levels in SK-MEL-5 cells. Thus, the relative RT-PCR expression (expression in SK-MEL-5 set at unity) in the seven cell lines listed in Table 1 were Caco-2 (13.49), HepG2 (12.71), IGROVI (7.92), PC-3 (5.51), 786-0 (2.81), SK-OV-3 (1.89), and SK-MEL-5 cells (1.0). The relative RT-PCR expression in the cancer cell lines exhibited an excellent linear correlation (r2 = 0.94) with U95Av2 Affymetrix microarray data that was available for five of the seven cell lines (Figure 4).
Figure 3.
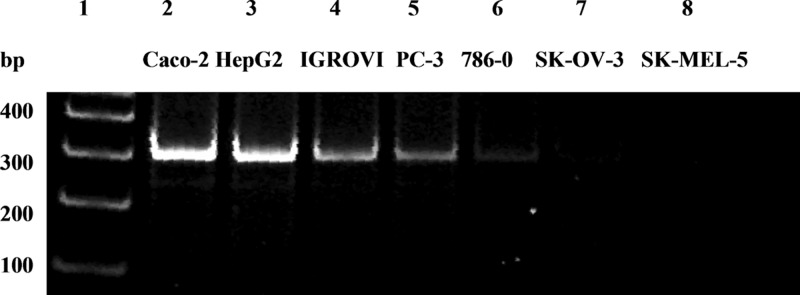
Expression profiles of DPPIV (315 bp product) in 7 cancer cell lines determined by semiquantitative RT-PCR: 100 base pair DNA ladder (lane 1), Caco-2 (lane 2), HepG2 (lane 3), IGROVI (lane 4), PC-3 (lane 5), 786-0 (lane 6), SK-OV-3 (lane 7), and SK-MEL-5 (lane 8).
Figure 4.
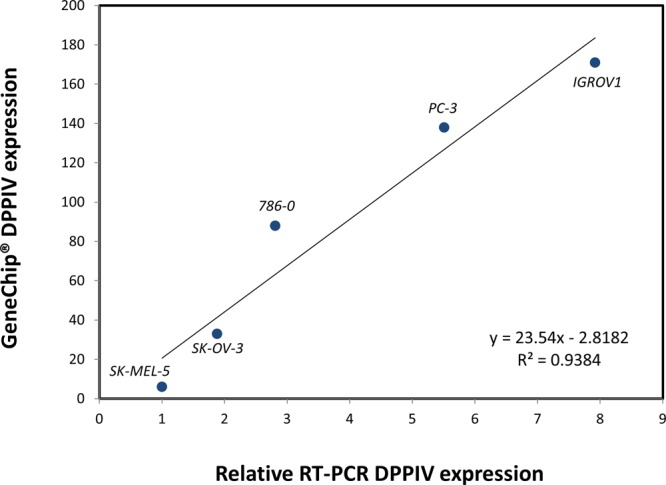
Linear correlation of Affymetrix GeneChip expression of DPPIV with DPPIV expression determined using RT-PCR in 5 cancer cell lines.
3.3. Hydrolysis of Standard Substrates by Porcine DPPIV
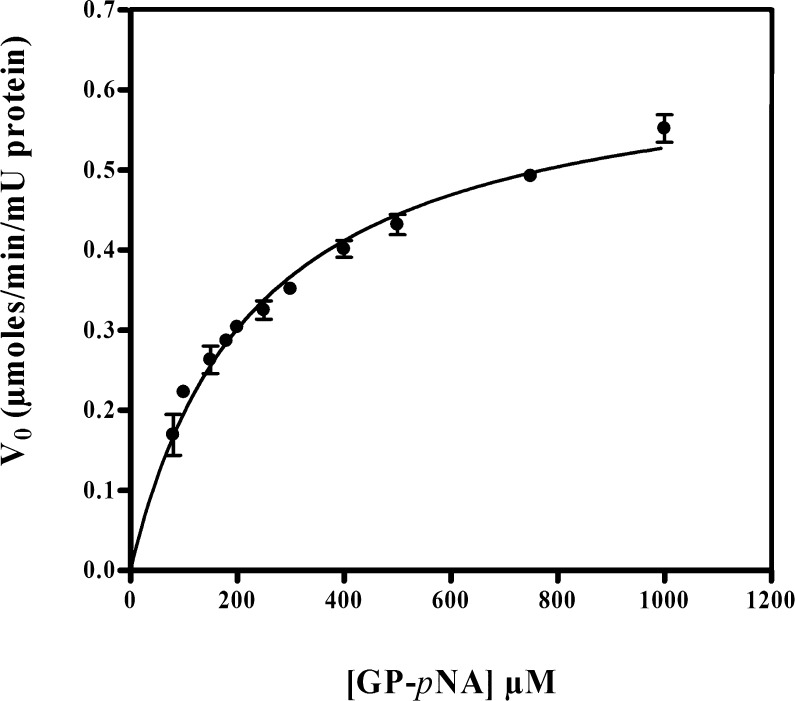
The specific activity of a standard substrate GP-pNA for porcine DPPIV was determined by the release of p-nitroanilide and the resultant effect on the clarity of the solution. The baseline activity was 14.02 ± 0.90 nmol/min/μg DPPIV. In the presence of the specific inhibitor diprotin A, it decreased substantially to 0.15 nmol/min/μg DPPIV. The kinetic parameters Km and Vmax obtained from a plot of V0 versus [S] (Figure 5) were 231 ± 33 μM and 0.650 ± 0.037 μmol/min/mU DPPIV, respectively (fit estimate ± SD; n = 3). The kcat value determined from Vmax and DPPIV concentration (E0) was 36 s–1. In contrast, GF-pNA or GR-pNA produced no detectable release of p-nitroanilide (negligible absorbance at 405 nm), indicating that these two molecules were resistant to cleavage by porcine DPPIV.
Figure 5.
GP-pNA hydrolysis kinetic profile with porcine DPPIV (mean ± SD, n = 3).
3.4. Hydrolysis of GP-pNA in Cancer Cell Homogenates and Inhibition by Diprotin A
The activity of DPPIV in various cancer cell homogenates was assessed using the standard substrate GP-pNA. The results shown in Figure 6 indicate that DPPIV activity was in the order HepG2 ≈ Caco-2 ≫ IGROVI ≫ PC-3 ≫ 786-0 ≥ SK-OV-3 ≥ SK-MEL-5 cells. Caco-2 and HepG2 cells exhibited high DPPIV activity (around 20 pmol/min/μg protein) whereas SK-MEL-5 cells showed the lowest activity (1.3 pmol/min/μg protein) among the cells tested (Figure 6). These results are in excellent correlation (r2 = 0.96) with the relative RT-PCR expression of DPPIV in the cell lines (Figure 7). The hydrolysis of GP-pNA in the presence of diprotin A was significantly inhibited in all cancer cell homogenates examined (Figure 6), with an average inhibition of 95% (range: 85–100%).
Figure 6.
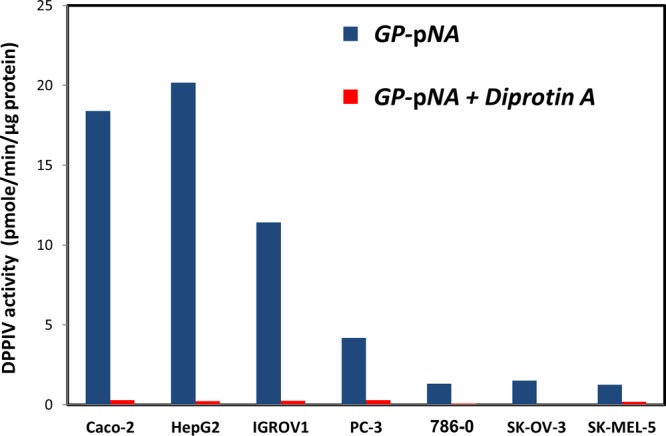
Specific activity of DPPIV against GP-pNA (pmol/min/μg protein) in 7 cell homogenates, in the absence vs presence of diprotin A (mean ± SD, n = 3).
Figure 7.
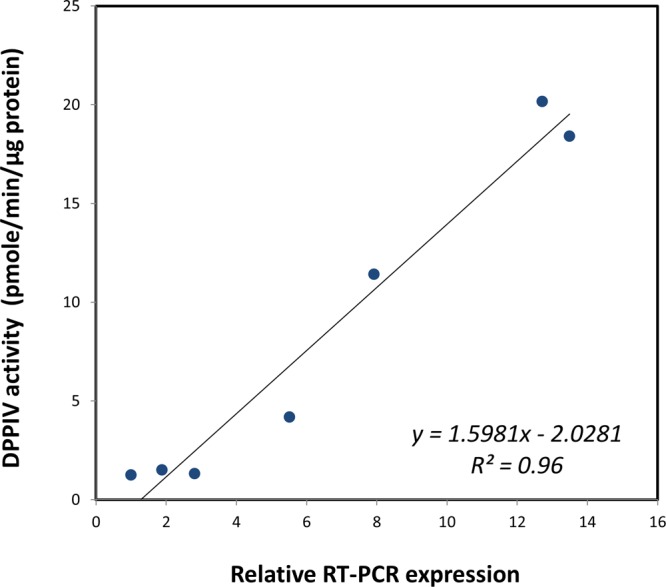
Linear correlation of specific activity of DPPIV against GP-pNA (pmol/min/μg protein) in 7 cancer cell homogenates with DPPIV expression determined using RT-PCR.
3.5. GP-Mel Model Prodrug Activation by Porcine Kidney DPPIV and Cell Homogenates
The specific activity of pure porcine enzyme against GP-Mel was 1.2 ± 0.1 nmol/min/μg protein. The activity was substantially lowered to 100 pmol/min/μg protein, a 93% inhibition, in the presence of diprotin A. DPPIV activity against the prodrug determined in Caco-2 and SK-MEL-5 homogenates was 5.14 ± 0.01 and 0.68 ± 0.03 pmol/min/μg protein, respectively, which was significantly inhibited by an average of 85% in the presence of diprotin A (Figure 8).
Figure 8.
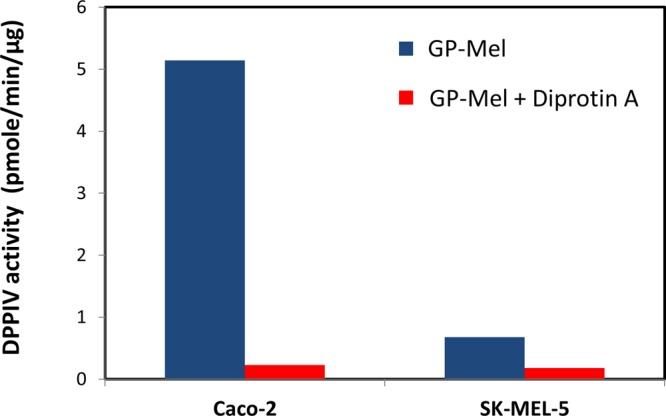
Specific activity of DPPIV against GP-Mel (pmol/min/μg protein) in Caco-2 and SK-MEL-5 cell homogenates, in the absence vs presence of diprotin A (mean ± SD, n = 3).
3.6. Cell Proliferation Studies
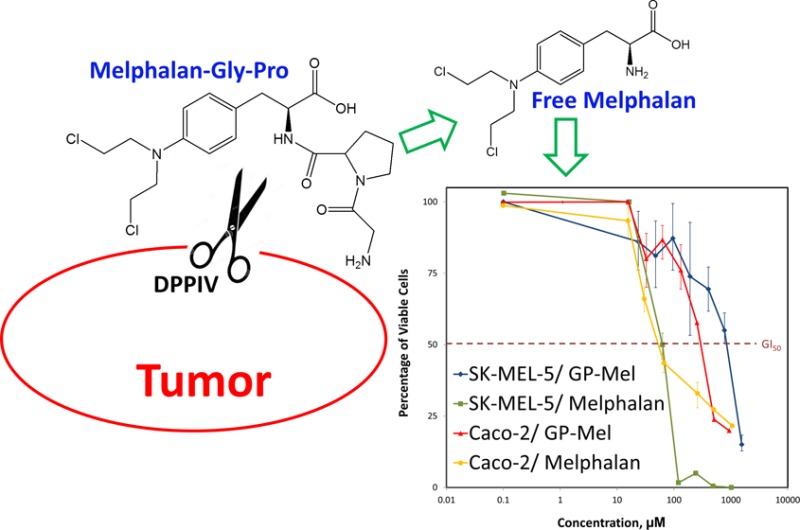
The antiproliferative activity of GP-Mel was determined in Caco-2 and SK-MEL-5 cancer cells and compared with that obtained with the parent drug melphalan. The percent cell viability profiles as a function of drug or prodrug concentration are shown in Figure 9. The profiles indicate that the antiproliferative action of melphalan was similar in Caco-2 and SK-MEL-5 cells with GI50 values of 34.9 μM and 56.6 μM, respectively. The GI50 value for GP-Mel in Caco-2 cells (261.3 μM), however, was significantly lower than that obtained with SK-MEL-5 cells (806.7 μM). The cytotoxic activity of GP-Mel in Caco-2 and SK-MEL-5 cells was consistent with the expression levels of DPPIV in these cells.
Figure 9.
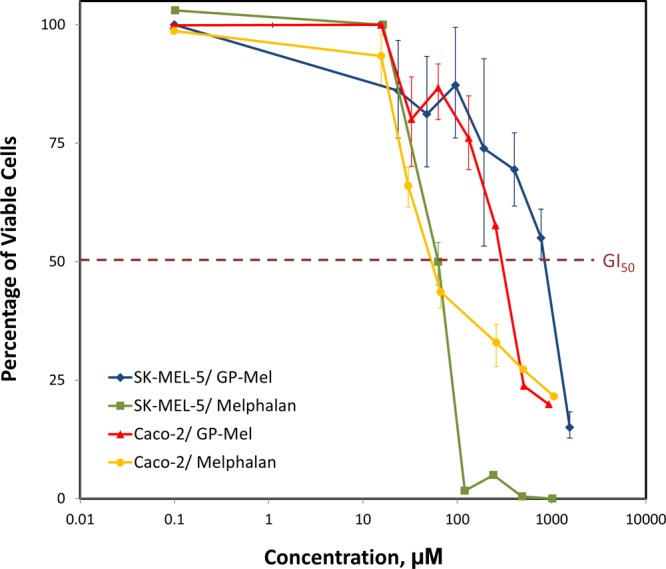
Cell proliferation assay of melphalan and GP-Mel after 48 h incubation with Caco-2 and SK-MEL-5 cells. Data are expressed as mean ± SD; n = 3 for each treatment at each concentration.
4. Discussion
In the past three decades, the science of molecular biology was revolutionized by the rapid advancement of the complementing fields of genomics and bioinformatics. This revolution had a profound effect on cancer research.37 Indeed, new molecular characteristics of cancer cells are discovered almost daily.100 Recently, a number of enzymes from the peptidase/protease class were found to be highly expressed in several types of tumors, and to play an important role in the pathophysiology of tumor cells.38,39 DPPIV has been previously associated with the onset and progression of several cancer types; Inamoto et al. demonstrated that the blockage of DDPIV reduced several cancer-related processes in the human renal carcinoma cell line Caki-2. It also reduced the tumor size and increased the survival of mice in a xenograft model.22 Furthermore, in patients with clear cell renal cell carcinoma (CCRCC), higher DPPIV activity was linked with a significant decrease in patients’ 5 year survival rates.40 DPPIV was also shown to be overexpressed in several human colon cancer tissues and in human colon cancer cell lines,33,41,42 and its inhibition reduced carcinogenesis in a rat model.43 In the prostate, DPPIV activity in cancerous versus benign prostatic hyperplasia was increased 2-fold.44,45 An elevation of DPPIV activity was also found in the prostatic secretions and the peripheral zone of the prostate, where most prostate cancers arise.44 Higher DPPIV levels in cancerous versus normal prostate tissue was correlated with PSA level, tumor residue, cancer stage, and tumor size in humans.46 Lastly, DPPIV activity was suggested as a marker for thyroid carcinomas.46,47
In addition to its expression and involvement in cancer, DPPIV is one of the few proline-specific proteases that is able to cleave proline-associated peptide bonds, as the unique cyclic structure of proline serves as a structure regulation element which limits the susceptibility for nonspecific enzymatic degradation.48−52 Thus, a high level of expression in cancer cells, combined with high substrate specificity, indicated that DPPIV may be a potential target molecule for the delivery of chemotherapeutic drugs, and sparked our interest in developing a DPPIV-cleavable anticancer prodrug.
The functional activity of pure porcine DPPIV was assessed against three Gly-X dipeptide chromogenic compounds, GP-pNA, a well-known standard DPPIV substrate,53 as well as GF-pNA and GR-pNA, two dipeptide analogues that do not contain proline in the P1 position. The kinetic parameters obtained in this study were consistent with those previously reported for GP-pNA with porcine DPPIV.54,55 The negligible activity observed with GF-pNA and GR-pNA is consistent with DPPIV substrate requirements, and eliminates the possibility of nonspecific degradation. Thus, substrate-specific and enzyme-specific responses were established. Similarly, in the seven investigated cancer cell lines, GP-pNA exhibited an excellent linear correlation (r2 = 0.96) with RT-PCR DPPIV expression (Figure 7), and DPPIV activity was significantly inhibited in the presence of the specific inhibitor diprotin A (average inhibition ∼95%). The similarity of the extent of inhibition in cell homogenates with that observed with porcine DPPIV underscores the similarity of human and porcine DPPIV.
Melphalan, phenylalanine mustard (L-PAM), is an established anticancer agent that was originally approved for the treatment of multiple myeloma. Recently it has been suggested as a therapeutic agent in the treatment of a variety of cancers such as ovarian cancer,56 breast cancer,57 colorectal cancer,58 and melanoma.59 However, its use has been limited due to severe side effects such as bone marrow suppression, leukopenia, and thrombocytopenia,60−62 largely due to lack of selectivity for cancer cells, and so it was selected as the model drug for investigating our approach. A proline dipeptide prodrug of melphalan was designed to follow DPPIV substrate specificity requirements. These include the positioning of proline exclusively at the P1 position. While alanine can also be used, it results in lower cleavage rates.63 Additionally, the peptide bond between P1 and P1′ requires a trans-configuration.27,64
The release of melphalan from the GP-Mel prodrug by pure porcine DPPIV was extensive, and was 93% reduced in the presence of diprotin A, indicating that the activation of GP-Mel was DPPIV-dependent. This was further corroborated in Caco-2 and SK-MEL-5 homogenates when the prodrug activation was significantly inhibited by diprotin A (Figure 8). The 8-fold greater activation of the prodrug in Caco-2 compared to SK-MEL-5 homogenates (Figure 8) was consistent with the 14-fold higher RT-PCR DPPIV expression levels, and the 15-fold higher DPPIV activity against GP-pNA in Caco-2 cells compared to SK-MEL-5 cells (Figure 7).
As a preliminary assessment of GP-Mel potential to serve as an anticancer prodrug, we have investigated the antiproliferative activity of GP-Mel in Caco-2 and SK-MEL-5 cell lines. The antiproliferative activity of the prodrug was shown to be dependent on DPPIV expression level in the cells; the cytotoxicity (represented by GI50) of GP-Mel in Caco-2 cells was 3-fold higher (260 μM) than that in SK-MEL-5 cells (800 μM) (Figure 9). In contrast, for the free parent drug melphalan, similar GI50 values were obtained in Caco-2 and in SK-MEL-5 cells (35 and 44 μM, respectively), demonstrating the nonselective cytotoxic action of melphalan (Figure 9). The cytotoxic effect of GP-Mel on both cell lines was significantly lower in comparison to melphalan; the prodrug failed to show significant cytotoxic effect in concentrations equimolar to those required for maximal growth inhibition by melphalan. This result may indicate that the GP-Mel prodrug is not likely to be cytotoxic by itself, and further validates the key role of DPPIV in the activation of the GP-Mel prodrug. On the other hand, it may indicate insufficient activation, resulting in too low free drug levels. Overall, our results demonstrate the potential to exploit DPPIV as a prodrug activating enzyme for efficient chemotherapeutic drug targeting.
5. Conclusions
In conclusion, DPPIV was identified as a potential prodrug target due to its differential expression levels in tumor and normal tissues and relatively strict substrate specificity. A Gly-Pro dipeptide prodrug of melphalan, GP-Mel, was designed and synthesized, based on the highly specific substrate requirements of DPPIV. The finding that the activation and antiproliferative activity of GP-Mel in cells were highly dependent on DPPIV expression levels confirmed our hypothesis that DPPIV is a feasible functional prodrug target for effective and selective chemotherapeutic action.
Acknowledgments
This work was supported by NIH Grant R01-GM37188 and by the College of Pharmacy, University of Michigan. We thank Dr. Gustavo Rosania for providing the IGROV1, PC3, and 786-0 cell lines. We thank Dr. Victor Yang for use of the 96-well plate reader in colorimetric assays.
Glossary
Abbreviations Used
- HOBT
1-hydroxybenzotriazole
- HBTU
2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- TIS
triisopropylsilane
- Fmoc-OSu
N-(9-fluorenylmethoxycarbonyloxy)succinimide
- DPPIV
dipeptidyl peptidase
- GP-pNA
glycyl-prolyl-p-nitroanilide
- GF-pNA
glycyl-phenylalanyl-p-nitroanilide
- GR-pNA
glycyl-arginyl-p-nitroanilide
- diprotin A
isoleucyl-prolyl-isoleucine-Pro-Ile
- GP-Mel
glycyl-prolyl-melphalan
- SK-MEL-5
melanoma
- Caco-2
colon adenocarcinoma
- HepG2
hepatocellular carcinoma
- IGROVI
ovarian cancer cell line
- PC-3
prostate cancer cell line
- 786-0
renal cancer cell line
- SK-OV-3
ovarian cancer cell line
- XTT
sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzenesulfonic acid hydrate
- PMS
N-methyldibenzopyrazine methyl sulfate
Author Present Address
§ Merck Research Laboratories (MRL), West Point, PA, United States.
Author Present Address
∥ Institute of Biochemistry and Molecular Medicine, University of Bern, Switzerland.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Nielsen T. O.; Friis-Hansen L.; Poulsen S. S.; Federspiel B.; Sorensen B. S. Expression of the EGF family in gastric cancer: downregulation of HER4 and its activating ligand NRG4. PLoS One 2014, 94e94606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jethon A.; Pula B.; Piotrowska A.; Wojnar A.; Rys J.; Dziegiel P.; Podhorska-Okolow M. Angiotensin II type 1 receptor (AT-1R) expression correlates with VEGF-A and VEGF-D expression in invasive ductal breast cancer. Pathol. Oncol. Res. 2012, 184867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C. S.; Cheung A. H.; Wong S. K.; Wan T. M.; Ng L.; Chow A. K.; Cheng N. S.; Pak R. C.; Li H. S.; Man J. H.; Yau T. C.; Lo O. S.; Poon J. T.; Pang R. W.; Law W. L. Prognostic significance of CD26 in patients with colorectal cancer. PLoS One 2014, 95e98582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.; Fan X. M. Role of cyclooxygenase-2 in gastric cancer development and progression. World J. Gastroenterol. 2013, 19427361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R.; Jain M.; Madhusudhan N.; Sheppard N. G.; Strittmatter L.; Kampf C.; Huang J.; Asplund A.; Mootha V. K. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014, 5, 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroux C. J.; Rankin G. M.; Moeker J.; Bornaghi L. F.; Katneni K.; Morizzi J.; Charman S. A.; Vullo D.; Supuran C. T.; Poulsen S. A. A prodrug approach toward cancer-related carbonic anhydrase inhibition. J. Med. Chem. 2013, 56239623–34. [DOI] [PubMed] [Google Scholar]
- Dahan A.; Khamis M.; Agbaria R.; Karaman R. Targeted prodrugs in oral drug delivery: the modern molecular biopharmaceutical approach. Expert Opinion on Drug Delivery 2012, 981001–13. [DOI] [PubMed] [Google Scholar]
- Krais J. J.; De Crescenzo O.; Harrison R. G. Purine nucleoside phosphorylase targeted by annexin v to breast cancer vasculature for enzyme prodrug therapy. PLoS One 2013, 810e76403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick C. A.; Jiang Y. L.; Paromov V.; Brannon M.; Krishnan K.; Stone W. L. Identification of oxidized protein hydrolase as a potential prodrug target in prostate cancer. BMC Cancer 2014, 14, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satsangi A.; Roy S. S.; Satsangi R. K.; Vadlamudi R. K.; Ong J. L. Design of a paclitaxel prodrug conjugate for active targeting of an enzyme upregulated in breast cancer cells. Mol. Pharmaceutics 2014, 1161906–18. [DOI] [PubMed] [Google Scholar]
- Hopsu-Havu V. K.; Glenner G. G. A new dipeptide naphthylamidase hydrolyzing glycyl-prolyl-beta-naphthylamide. Histochemie 1966, 73197–201. [DOI] [PubMed] [Google Scholar]
- Polgar L. The prolyl oligopeptidase family. Cell. Mol. Life Sci. 2002, 592349–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G.; Heins J.; Barth A. The conformation around the peptide bond between the P1- and P2-positions is important for catalytic activity of some proline-specific proteases. Biochim. Biophys. Acta 1983, 7423452–62. [DOI] [PubMed] [Google Scholar]
- Buhling F.; Junker U.; Reinhold D.; Neubert K.; Jager L.; Ansorge S. Functional role of CD26 on human B lymphocytes. Immunol. Lett. 1995, 451–247–51. [DOI] [PubMed] [Google Scholar]
- Ohnuma K.; Dang N. H.; Morimoto C. Revisiting an old acquaintance: CD26 and its molecular mechanisms in T cell function. Trends Immunol. 2008, 296295–301. [DOI] [PubMed] [Google Scholar]
- Dinjens W. N.; ten Kate J.; van der Linden E. P.; Wijnen J. T.; Khan P. M.; Bosman F. T. Distribution of adenosine deaminase complexing protein (ADCP) in human tissues. J. Histochem. Cytochem. 1989, 37121869–75. [DOI] [PubMed] [Google Scholar]
- Shingu K.; Helfritz A.; Zielinska-Skowronek M.; Meyer-Olson D.; Jacobs R.; Schmidt R. E.; Mentlein R.; Pabst R.; von Horsten S. CD26 expression determines lung metastasis in mutant F344 rats: involvement of NK cell function and soluble CD26. Cancer Immunol. Immunother. 2003, 529546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A.; Goebel-Stengel M.; Teuffel P.; Hofmann T.; Buße P.; Kobelt P.; Rose M.; Klapp B. F. Obese patients have higher circulating protein levels of dipeptidyl peptidase IV. Peptides 2014, 61075–82. [DOI] [PubMed] [Google Scholar]
- McCaughan G. W.; Wickson J. E.; Creswick P. F.; Gorrell M. D. Identification of the bile canalicular cell surface molecule GP110 as the ectopeptidase dipeptidyl peptidase IV: an analysis by tissue distribution, purification and N-terminal amino acid sequence. Hepatology 1990, 114534–44. [DOI] [PubMed] [Google Scholar]
- Boonacker E.; Van Noorden C. J. The multifunctional or moonlighting protein CD26/DPPIV. Eur. J. Cell Biol. 2003, 82253–73. [DOI] [PubMed] [Google Scholar]
- Cortes A.; Gracia E.; Moreno E.; Mallol J.; Lluis C.; Canela E. I.; Casado V. Moonlighting Adenosine Deaminase: A Target Protein for Drug Development. Med. Res. Rev. 2014, 10.1002/med.21324. [DOI] [PubMed] [Google Scholar]
- Inamoto T.; Yamochi T.; Ohnuma K.; Iwata S.; Kina S.; Inamoto S.; Tachibana M.; Katsuoka Y.; Dang N. H.; Morimoto C. Anti-CD26 monoclonal antibody-mediated G1-S arrest of human renal clear cell carcinoma Caki-2 is associated with retinoblastoma substrate dephosphorylation, cyclin-dependent kinase 2 reduction, p27(kip1) enhancement, and disruption of binding to the extracellular matrix. Clin. Cancer Res. 2006, 1211 Part 13470–7. [DOI] [PubMed] [Google Scholar]
- Hama T.; Okada M.; Kojima K.; Kato T.; Matsuyama M.; Nagatsu T. Purification of dipeptidyl-aminopeptidase IV from human kidney by anti dipeptidyl-aminopeptidase IV affinity chromatography. Mol. Cell. Biochem. 1982, 43135–42. [DOI] [PubMed] [Google Scholar]
- Pala L.; Rotella C. M. The role of DPP4 activity in cardiovascular districts: in vivo and in vitro evidence. J. Diabetes Res. 2013, 2013, 590456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Kate J.; Dinjens W. N.; Meera Khan P.; Bosman F. T. Adenosine deaminase complexing protein in cancer studies. Anticancer Res. 1986, 65983–8. [PubMed] [Google Scholar]
- Cunningham D. F.; O’Connor B. Proline specific peptidases. Biochim. Biophys. Acta 1997, 13432160–86. [DOI] [PubMed] [Google Scholar]
- Devasthale P.; Wang Y.; Wang W.; Fevig J.; Feng J.; Wang A.; Harrity T.; Egan D.; Morgan N.; Cap M.; Fura A.; Klei H. E.; Kish K.; Weigelt C.; Sun L.; Levesque P.; Moulin F.; Li Y. X.; Zahler R.; Kirby M. S.; Hamann L. G. Optimization of activity, selectivity, and liability profiles in 5-oxopyrrolopyridine DPP4 inhibitors leading to clinical candidate (Sa)-2-(3-(aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5H-pyrrolo[3,4-b]py ridin-6(7H)-yl)-N,N-dimethylacetamide (BMS-767778). J. Med. Chem. 2013, 56187343–57. [DOI] [PubMed] [Google Scholar]
- Diez-Torrubia A.; Cabrera S.; de Castro S.; García-Aparicio C.; Mulder G.; De Meester I.; Camarasa M.-J.; Balzarini J.; Velázquez S. Novel water-soluble prodrugs of acyclovir cleavable by the dipeptidyl-peptidase IV (DPP IV/CD26) enzyme. Eur. J. Med. Chem. 2013, 700456–68. [DOI] [PubMed] [Google Scholar]
- Diez-Torrubia A.; García-Aparicio C.; Cabrera S.; De Meester I.; Balzarini J.; Camarasa M. a.-J.; Velázquez S. Application of the dipeptidyl peptidase IV (DPPIV/CD26) based prodrug approach to different amine-containing drugs. J. Med. Chem. 2010, 532559–72. [DOI] [PubMed] [Google Scholar]
- García-Aparicio C.; Bonache M.-C.; De Meester I.; San-Félix A.; Balzarini J.; Camarasa M.-J.; Velázquez S. Design and discovery of a novel dipeptidyl-peptidase IV (CD26)-based prodrug approach. J. Med. Chem. 2006, 49175339–51. [DOI] [PubMed] [Google Scholar]
- García-Aparicio C.; Diez-Torrubia A.; Balzarini J.; Lambeir A.-M.; Velázquez S.; Camarasa M.-J. Efficient conversion of tetrapeptide-based TSAO prodrugs to the parent drug by dipeptidyl-peptidase IV (DPPIV/CD26). Antiviral Res. 2007, 762130–9. [DOI] [PubMed] [Google Scholar]
- Mittal S.; Song X.; Vig B. S.; Landowski C. P.; Kim I.; Hilfinger J. M.; Amidon G. L. Prolidase, a potential enzyme target for melanoma: design of proline-containing dipeptide-like prodrugs. Mol. Pharmaceutics 2005, 2137–46. [DOI] [PubMed] [Google Scholar]
- Darmoul D.; Voisin T.; Couvineau A.; Rouyer-Fessard C.; Salomon R.; Wang Y.; Swallow D. M.; Laburthe M. Regional expression of epithelial dipeptidyl peptidase IV in the human intestines. Biochem. Biophys. Res. Commun. 1994, 20321224–9. [DOI] [PubMed] [Google Scholar]
- Bar J.; Weber A.; Hoffmann T.; Stork J.; Wermann M.; Wagner L.; Aust S.; Gerhartz B.; Demuth H. U. Characterisation of human dipeptidyl peptidase IV expressed in Pichia pastoris. A structural and mechanistic comparison between the recombinant human and the purified porcine enzyme. Biol. Chem. 2003, 384121553–63. [DOI] [PubMed] [Google Scholar]
- Dahan A.; Duvdevani R.; Dvir E.; Elmann A.; Hoffman A. A novel mechanism for oral controlled release of drugs by continuous degradation of a phospholipid prodrug along the intestine: In-vivo and in-vitro evaluation of an indomethacin–lecithin conjugate. J. Controlled Release 2007, 119186–93. [DOI] [PubMed] [Google Scholar]
- Dahan A.; Duvdevani R.; Shapiro I.; Elmann A.; Finkelstein E.; Hoffman A. The oral absorption of phospholipid prodrugs: In vivo and in vitro mechanistic investigation of trafficking of a lecithin-valproic acid conjugate following oral administration. J. Controlled Release 2008, 12611–9. [DOI] [PubMed] [Google Scholar]
- Lockhart D. J.; Winzeler E. A. Genomics, gene expression and DNA arrays. Nature 2000, 4056788827–36. [DOI] [PubMed] [Google Scholar]
- Dahan A.; Zimmermann E. M.; Ben-Shabat S. Modern prodrug design for targeted oral drug delivery. Molecules 2014, 191016489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauvois B. Transmembrane proteases in cell growth and invasion: new contributors to angiogenesis?. Oncogene 2004, 232317–29. [DOI] [PubMed] [Google Scholar]
- Chang C.; Werb Z. The many faces of metalloproteases: cell growth, invasion, angiogenesis and metastasis. Trends Cell Biol. 2001, 1111S37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrinaga G.; Blanco L.; Sanz B.; Perez I.; Gil J.; Unda M.; Andres L.; Casis L.; Lopez J. I. The impact of peptidase activity on clear cell renal cell carcinoma survival. Am. J. Physiol. 2012, 30312F1584–91. [DOI] [PubMed] [Google Scholar]
- Abe M.; Havre P. A.; Urasaki Y.; Ohnuma K.; Morimoto C.; Dang L. H.; Dang N. H. Mechanisms of confluence-dependent expression of CD26 in colon cancer cell lines. BMC Cancer 2011, 11, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweibaum A.; Hauri H. P.; Sterchi E.; Chantret I.; Haffen K.; Bamat J.; Sordat B. Immunohistological evidence, obtained with monoclonal antibodies, of small intestinal brush border hydrolases in human colon cancers and foetal colons. Int. J. Cancer 1984, 345591–8. [DOI] [PubMed] [Google Scholar]
- Femia A. P.; Raimondi L.; Maglieri G.; Lodovici M.; Mannucci E.; Caderni G. Long-term treatment with Sitagliptin, a dipeptidyl peptidase-4 inhibitor, reduces colon carcinogenesis and reactive oxygen species in 1,2-dimethylhydrazine-induced rats. Int. J. Cancer 2013, 133102498–503. [DOI] [PubMed] [Google Scholar]
- Wilson M. J.; Haller R.; Li S. Y.; Slaton J. W.; Sinha A. A.; Wasserman N. F. Elevation of dipeptidylpeptidase IV activities in the prostate peripheral zone and prostatic secretions of men with prostate cancer: possible prostate cancer disease marker. J. Urol. 2005, 17431124–8. [DOI] [PubMed] [Google Scholar]
- Wilson M. J.; Ruhland A. R.; Quast B. J.; Reddy P. K.; Ewing S. L.; Sinha A. A. Dipeptidylpeptidase IV activities are elevated in prostate cancers and adjacent benign hyperplastic glands. J. Androl. 2000, 212220–6. [PubMed] [Google Scholar]
- Lu Z.; Qi L.; Bo X. J.; Liu G. D.; Wang J. M.; Li G. Expression of CD26 and CXCR4 in prostate carcinoma and its relationship with clinical parameters. J. Res. Med. Sci. 2013, 188647–52. [PMC free article] [PubMed] [Google Scholar]
- Aratake Y.; Kotani T.; Tamura K.; Araki Y.; Kuribayashi T.; Konoe K.; Ohtaki S. Dipeptidyl aminopeptidase IV staining of cytologic preparations to distinguish benign from malignant thyroid diseases. Am. J. Clin. Pathol. 1991, 963306–10. [DOI] [PubMed] [Google Scholar]
- Barlow D. J.; Thornton J. M. Helix geometry in proteins. J. Mol. Biol. 1988, 2013601–19. [DOI] [PubMed] [Google Scholar]
- Bradbury A. F.; Finnie M. D.; Smyth D. G. Mechanism of C-terminal amide formation by pituitary enzymes. Nature 1982, 2985875686–8. [DOI] [PubMed] [Google Scholar]
- Mentlein R. Dipeptidyl-peptidase IV (CD26)--role in the inactivation of regulatory peptides. Regul. Pept. 1999, 8519–24. [DOI] [PubMed] [Google Scholar]
- Persson B.; Flinta C.; von Heijne G.; Jornvall H. Structures of N-terminally acetylated proteins. Eur. J. Biochem. 1985, 1523523–7. [DOI] [PubMed] [Google Scholar]
- Vanhoof G.; Goossens F.; De Meester I.; Hendriks D.; Scharpe S. Proline motifs in peptides and their biological processing. FASEB J. 1995, 99736–44. [PubMed] [Google Scholar]
- Shibuya-Saruta H.; Sugiyama M.; Kasahara Y. Colorimetrical rate assay for urinary dipeptidyl peptidase IV (DPPIV) activity using a new substrate. J. Clin. Lab. Anal. 1995, 92113–8. [DOI] [PubMed] [Google Scholar]
- Pascual I.; Gomez H.; Pons T.; Chappe M.; Vargas M. A.; Valdes G.; Lopez A.; Saroyan A.; Charli J. L.; de los Angeles Chavez M. Effect of divalent cations on the porcine kidney cortex membrane-bound form of dipeptidyl peptidase IV. Int. J. Biochem. Cell Biol. 2011, 433363–71. [DOI] [PubMed] [Google Scholar]
- Svensson B.; Danielsen M.; Staun M.; Jeppesen L.; Noren O.; Sjostrom H. An amphiphilic form of dipeptidyl peptidase IV from pig small-intestinal brush-border membrane. Purification by immunoadsorbent chromatography and some properties. Eur. J. Biochem. 1978, 903489–98. [DOI] [PubMed] [Google Scholar]
- Mougenot P.; Fabbro M.; Bressolle F.; Pouessel D.; Culine S.; Pinguet F. Phase II study of melphalan as a single-agent infused over a 24-h period with individual adapted dosing in patients with recurrent epithelial ovarian cancer. Oncol. Rep. 2006, 151237–41. [PubMed] [Google Scholar]
- Vahdat L. T.; Balmaceda C.; Papadopoulos K.; Frederick D.; Donovan D.; Sharpe E.; Kaufman E.; Savage D.; Tiersten A.; Nichols G.; Haythe J.; Troxel A.; Antman K.; Hesdorffer C. S. Phase II trial of sequential high-dose chemotherapy with paclitaxel, melphalan and cyclophosphamide, thiotepa and carboplatin with peripheral blood progenitor support in women with responding metastatic breast cancer. Bone Marrow Transplant. 2002, 303149–55. [DOI] [PubMed] [Google Scholar]
- Marinelli A.; Vahrmeijer A. L.; van de Velde C. J. Phase I/II studies of isolated hepatic perfusion with mitomycin C or melphalan in patients with colorectal cancer hepatic metastases. Recent Results Cancer Res. 1998, 147, 83–94. [DOI] [PubMed] [Google Scholar]
- Nieweg O. E.; Kroon B. B. Isolated limb perfusion with melphalan for melanoma. J. Surg. Oncol. 2014, 1094332–7. [DOI] [PubMed] [Google Scholar]
- Davis-Perry S.; Hernandez E.; Houck K. L.; Shank R. Melphalan for the treatment of patients with recurrent epithelial ovarian cancer. Am. J. Clin. Oncol. 2003, 264429–33. [DOI] [PubMed] [Google Scholar]
- Sarosy G.; Leyland-Jones B.; Soochan P.; Cheson B. D. The systemic administration of intravenous melphalan. J. Clin. Oncol. 1988, 6111768–82. [DOI] [PubMed] [Google Scholar]
- Vigneau C.; Ardiet C.; Bret M.; Laville M.; Fiere D.; Tranchand B.; Fouque D. Intermediate-dose (25mg/m2) IV melphalan for multiple myeloma with renal failure. J. Nephrol. 2002, 156684–9. [PubMed] [Google Scholar]
- Bongers J.; Lambros T.; Ahmad M.; Heimer E. P. Kinetics of dipeptidyl peptidase IV proteolysis of growth hormone-releasing factor and analogs. Biochim. Biophys. Acta 1992, 11222147–53. [DOI] [PubMed] [Google Scholar]
- Rahfeld J.; Schierhorn M.; Hartrodt B.; Neubert K.; Heins J. Are diprotin A (Ile-Pro-Ile) and diprotin B (Val-Pro-Leu) inhibitors or substrates of dipeptidyl peptidase IV?. Biochim. Biophys. Acta 1991, 10762314–6. [DOI] [PubMed] [Google Scholar]