Abstract
The Eastern Afromontane biodiversity hotspot composed of highly fragmented forested highlands (sky islands) harbours exceptional diversity and endemicity, particularly within birds. To explain their elevated diversity within this region, models founded on niche conservatism have been offered, although detailed phylogeographic studies are limited to a few avian lineages. Here, we focus on the recent songbird genus Zosterops, represented by montane and lowland members, to test the roles of niche conservatism versus niche divergence in the diversification and colonization of East Africa's sky islands. The species-rich white-eyes are a typically homogeneous family with an exceptional colonizing ability, but in contrast to their diversity on oceanic islands, continental diversity is considered depauperate and has been largely neglected. Molecular phylogenetic analysis of ∼140 taxa reveals extensive polyphyly among different montane populations of Z. poliogastrus. These larger endemic birds are shown to be more closely related to taxa with divergent habitat types, altitudinal distributions and dispersal abilities than they are to populations of restricted endemics that occur in neighbouring montane forest fragments. This repeated transition between lowland and highland habitats over time demonstrate that diversification of the focal group is explained by niche divergence. Our results also highlight an underestimation of diversity compared to morphological studies that has implications for their taxonomy and conservation. Molecular dating suggests that the spatially extensive African radiation arose exceptionally rapidly (1–2.5 Ma) during the fluctuating Plio-Pleistocene climate, which may have provided the primary driver for lineage diversification.
Keywords: AFLPs, Afromontane, biodiversity hotspot, molecular dating, montane diversification, Plio-Pleistocene
Introduction
Fragmented landscapes such as archipelagos are excellent natural laboratories to assess the influence of geography on genetic and phenotypic divergence (e.g. Roy 1997; Knowles 2000; McCormack et al. 2008; Price 2008; Shepard & Burbrink 2009; Clegg & Phillimore 2010; Lawson 2013). In particular, montane archipelagos harbour some of the highest biological diversity on the planet, making them important regions of interest for understanding the patterns and processes leading to the accumulation of diversity. The hyperdiverse Eastern Afromontane biodiversity hotspot (EABH) (Myers et al. 2000) has received considerable attention from evolutionary biologists investigating how this highly heterogeneous landscape has influenced population differentiation and speciation both temporally and spatially. This region is currently experiencing severe habitat loss (Myers et al. 2000) and has alarming rates of forecasted urban growth (Seto et al. 2012), which places a premium on quantifying the diversity that this key hotspot harbours, as well as understanding the evolutionary processes responsible. Evolutionary insights will furthermore allow a better understanding of organisms' responses to on-going and future habitat fragmentation.
Unlike the continuous mountain ranges of the Himalayas or Andes, the EABH is composed of a chain of ancient isolated massifs (Griffiths 1993) and young volcanoes (<5 Ma e.g. Baker et al. 1971) forming sky islands (Fig.1A). Montane forests typically occur above 800 m on these isolated peaks, so that the climatic conditions and ecosystem are highly differentiated from the surrounding low altitude savannah habitats and thus form ‘ecological islands’. Previously, the montane forests formed a pan-African forest that fragmented in the Early Oligocene due to the onset of aridification (Lovett 1993; Sepulchre et al. 2006) and have therefore had a long period of isolation. The isolation of these habitats potentially allows in situ speciation events to be differentiated from colonization events, which would otherwise be much harder to identify in montane systems exhibiting higher degrees of connectivity (Voelker et al. 2010).
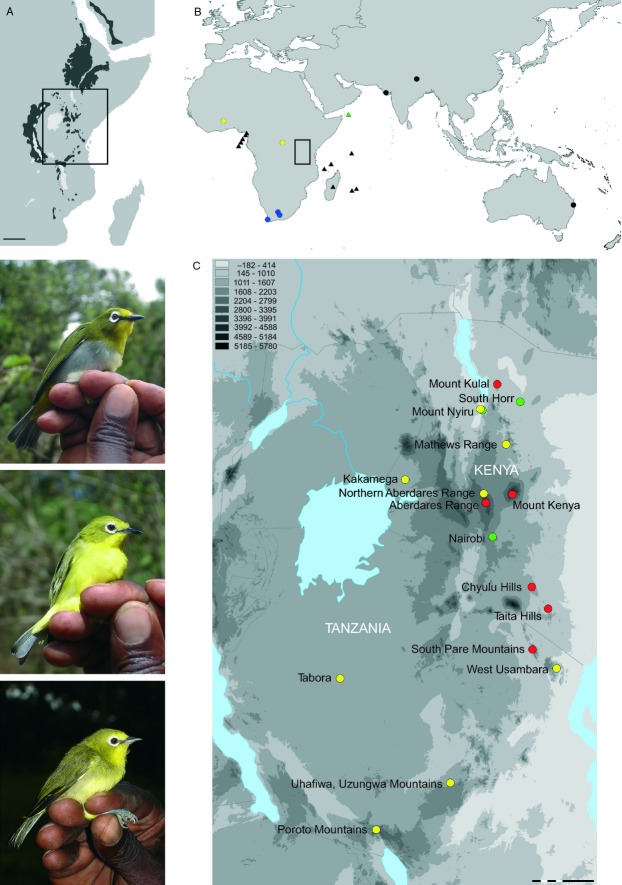
Figure 1.
Distribution of Zosteropidae samples used in this study. (A) The Eastern Afromontane region (highlighted in black), scale = 500 Km; (B) Zosterops samples from outside the focal region; (C) Zosterops samples from the East Afromontane region: Z. poligastrus (red); Z. abyssinicus (green); Z. senegalensis (yellow); Z. pallidus (blue); Zosterops sp. (black); mainland taxa (circles); insular taxa (triangles); altitude in metres; scale=3.2 decimal degrees. Photographs (top to bottom, Cox): Z. poliogastrus kulalensis (K39, Mt Kulal); Z. abyssinicus flavilateralis (T15, foothill of Chyulu Hills); Z. senegalensis jacksoni (T51, Kakamega Forest).
Several competing models have been put forward in an attempt to explain the high levels of endemism observed in tropical montane faunas. Within the EABH, there has been considerable support for the montane speciation model (e.g. Fjeldså & Lovett 1997; Roy 1997; Fjeldså & Bowie 2008). Under this scenario, montane forest habitats (sky islands) separated by intervening lowland areas may have served as historical refugia, where previously widespread populations became geographically isolated as they tracked suitable habitat to higher altitudes in response to climate change during the cool and arid episodes of the Plio-Pleistocene.
An alternative mechanism of climatic zonation, the gradient speciation model (Moritz et al. 2000 and references therein), posits that new species originate as populations adapt to different climatic regimes along an altitudinal gradient, predicting that sister taxa should occupy distinct but adjacent habitats (e.g. Moritz et al. 2000; Ogden & Thorpe 2002; Hall 2005; Kozak & Wiens 2007). In the tropics, the narrowing of climatic profiles between different altitudes produces strong ecological gradients, which in turn selects organisms with narrow ecological tolerances (Moritz et al. 2000; Kozak & Wiens 2007).
The montane and gradient speciation models predict contrasting roles for natural selection, with refuge (i.e. montane speciation) models founded on niche conservatism, in which the inability of populations to adapt to new or changing environmental conditions plays the primary role in geographical isolation, with ecologically similar populations diverging in allopatry (Moritz et al. 2000; Wiens & Donoghue 2004; Kozak & Wiens 2007; Wiens et al. 2010). In contrast, under the gradient model, the ability to adapt to new or changing environmental conditions drives climatic niche divergence (thus population divergence), with differing climatic distributions and/or climatic tolerances limiting gene flow between populations in either allopatry or parapatry (Moritz et al. 2000; Ogden & Thorpe 2002; Hall 2005; Kozak & Wiens 2007). A variation of these two models, the vanishing refuge model (Vanzolini & Williams 1981), proposes that some populations speciate through directional selection towards a tolerance of less favourable habitats as refuges become too small to retain viable populations. Like the gradient model, the vanishing refuge model is based on niche divergence and predicts that sister species occupy distinct habitats; however, the latter model also requires severe population bottlenecks with subsequent range expansion (Moritz et al. 2000). However, the possible contribution of models founded on niche divergence to explain the diversification of lineages from the EABH has been largely ignored.
Birds have long been the subjects of speciation studies on archipelagos due to their success as colonizers (e.g. Mayr 1942; Diamond 1970; Price 2008; Clegg & Phillimore 2010), with studies principally focusing on oceanic islands and to a lesser extent on montane archipelagos (but see e.g. McCormack et al. 2008). Within the EABH, there are approximately 1300 described bird species, of which 110 are endemic, providing substantial comparative systems in which to investigate processes facilitating diversification across the region's sky islands.
To better understand the build-up of biodiversity within the Eastern Afromontane region, we investigated diversification in the songbird genus Zosterops (Passeriformes: Zosteropidae). This species-rich group is composed of small, gregarious, arboreal birds that, aside from some aberrant African taxa traditionally separated in the genus Speirops (e.g. Melo et al. 2011), exhibit remarkable uniformity in their morphological structure, plumage and behaviour (van Balen 2008). As such, the systematics of the Zosteropidae is notoriously problematic, and the affinities of numerous taxa remain enigmatic (Fry et al. 2000). Here, we follow the currently standard taxonomy of Dickinson (2003) and van Balen (2008).
Zosterops are renowned ‘speciators’, displaying spectacular colonizing abilities (Lack 1971), and are suggested to have diversified exceptionally rapidly (∼2 Ma, Moyle et al. 2009). Despite their occurrence across the Old World tropics (including Africa and Asia), research into factors facilitating their diversification has predominately centred on insular taxa (e.g. Warren et al. 2006; Moyle et al. 2009; Clegg & Phillimore 2010). Conversely, continental members have been largely neglected, possibly because less than 10% of their known diversity is attributed to continental landmasses (Moreau 1957).
Where present on East African sky islands (Fig.1), Zosterops (referred to as white-eyes due to their white-eye ring) are often represented by a single endemic subspecies, which is analogous to their distribution on oceanic islands (e.g. Indian Ocean, Gulf of Guinea and Vanuatu archipelagos). Oceanic islands commonly support a single endemic taxon (Dickinson 2003), although two nonsister taxa are present on some islands (e.g. Warren et al. 2006). Just four currently recognized species of Zosterops occur across much of mainland sub-Saharan Africa, of which three are found within the mountain archipelago of East Africa's Rift Valley (Kenyan Highlands and East African Arc); however, subspecies diversity is considerably higher (Dickinson 2003; van Balen 2008). The African montane white-eyes (Z. poliogastrus) of East Africa's sky islands are comparatively larger birds than other mainland species, with rich green backs, yellow or grey bellies and generally broad white-eye rings and bright golden feathers (Fig.1). While some authors have argued that Z. poliogastrus should be split into several species based on vocal differences and ecology (e.g. Collar et al. 1994; Borghesio & Laiolo 2004), plumage variation within this group is subtle. Notably, a recent study based on 15 microsatellite markers indicates that genetic differentiation within Z. poliogaster populations is very high (Habel et al. 2013). All Z. poliogastrus subspecies are endemic to montane forest habitats and are ecologically segregated from parapatric Z. senegalensis or Z. abyssinicus subspecies (Hall & Moreau 1970).
The recent divergence of the Zosteropidae (Moyle et al. 2009) and their montane and lowland distribution within the focal region are attributes that make East African Zosterops a useful system to test current hypotheses of montane diversification and diversity using a phylogenetic approach. An advantage of focusing on recently evolved taxa is that they avoid complicating causal events; thus, their diversity is likely shaped by similar forces happening in a short amount of time, rather than multiple disparate events over a long time. To reconstruct the evolutionary relationships of Zosterops, we generated novel mtDNA (cytochrome b (Cyt b) and NADH dehydrogenase subunit 3 (ND3) genes), and AFLP data based on dense sampling across the Eastern Afromontane region and included additional sampling from outside of the region. Divergence times were estimated using a calibration based on the age of a volcanic island (Grand Comore) and were compared to those generated using the avian molecular clock. Using these data, we address the following questions: First, Have montane forest taxa diversified in situ? Under this scenario, diversification of montane endemics is suggested to be the result of niche conservatism (montane speciation model), in which we would except to find recently evolved montane forms (Plio-Pleistocene time frame) comprising a clade relative to lowland forms (e.g. Roy 1997). In contrast, alternative hypotheses in which diversification is the result of niche divergence (gradient speciation and vanishing refuge models), we would expect sister species to occupy distinct habitats. However, as the latter model requires demographic changes to explain such shifts, we are unable to test this hypothesis here. Second, Did diversification of montane taxa occur within a Plio-Pleistocene time frame, in which montane habitats have been largely buffered from climatic instability? Third, Is montane white-eye diversity underestimated? That a species-rich genus contains so few species in a key hotspot of biological diversity is surprising. Extensive sampling of the region here allows assessment of their diversity within a systematic framework.
Methods
Taxonomic sampling
A total of 148 individuals representing 15 described ingroup taxa (Dickinson 2003) are included in this study based on blood samples (Table S1, Supporting information; Fig.1). To test species monophyly and colonization scenarios, 33 Zosterops samples were obtained from outside East Africa (Fig.1B). Blood sample numbers, collection localities and GenBank Accession nos are listed in Table S1 (accompanying specimen photographs are available from Dryad). Blood samples were taken from mist-netted specimens and stored in ETOH (99%) or Queen's lysis buffer (Seutin et al. 1991).
Molecular sequence data
Total DNA was extracted from blood samples using a DNeasy Blood and tissue kit (Qiagen, UK). Mitochondrial genes (Cyt b, ND3) and a nuclear gene (TGFß2) were selected based on their performance from previous Zosterops studies (Warren et al. 2006; Moyle et al. 2009; Melo et al. 2011). Amplification of ND3 and TGFß2 was performed using published primers (Table S2, Supporting information). To obtain the entire Cyt b gene, the published primer H16065 was used alongside three newly designed primers (Table S2, Supporting information). PCR amplifications and thermal cycling conditions for all three genes are reported in Table S2. Cleaned products were sequenced on an ABI 3730xl DNA analyser (Applied Biosystems, UK).
Amplified Fragment Length Polymorphisms (AFLPs)
Amplified fragment length polymorphism profiles were generated following Vos et al. (1995), with modifications for fluorescent primers implemented in Huang & Sun (1999). Selective PCR was carried out in two stages: a subsample of all restriction fragments was obtained through a preselective amplification, and these were subsequently selectively amplified with more specific dye labelled primers. Thermal cycling conditions and primers for both amplification stages are reported in Appendix S1, Supporting information. This study screened a total of 21 unique primer combinations generated from three selective amplification EcoRI+NNN primers (labelled with different fluorescent dyes) and seven Msel+NNN primers. A subset of eight DNA extracts was chosen to test all 21 AFLP primers combinations, and resulting selective amplification products were electrophoresed on a 3.5% agarose gel against a Hyperladder V (Bioline) size standard to choose the most appropriate primer combinations. Fragment analysis was conducted on a 3730 Applied Biosystems Sanger Sequencer using recommended fluorophores (FAM, NED HEX and LIZ).
AFLP scoring
Peaks were visualized using genemapper version 3.7, and all primer combinations were analysed separately. An initial scoring panel was generated using the automatic panel generation feature of genemapper under default settings. This feature algorithmically generates panels and bins based on the collective peaks present from all samples. The resulting AFLP panels were then checked by eye. As replicates are the only objective measure of quality in AFLP studies (Pompanon et al. 2005), five individuals were repeated from the restriction ligation stage onwards to obtain a relative assessment of the repeatability of AFLP profiles. Additional information is reported in Appendix S1.
Phylogenetic inference
Phylogenetic analyses were performed on a data set of 139 samples comprising 1471 bp of mtDNA sequence data (ND3 348 bp, Cyt b 1123 bp). Sequences were aligned in clustal x 2.0 (Larkin et al. 2007) using default settings, with the resulting alignment checked manually in se-al 2.0 (Rambaut 2002) and translated into amino acids to ensure there were no stop codons. Nuclear DNA (TGFß2) (600 bp) was generated for a subset of the taxa to assess phylogenetic signal, but provided no informative sites and was therefore discounted from subsequent analyses.
partitionfinder 1.01 (Lanfear et al. 2012) was used to select the best-fit partitioning scheme and model of molecular evolution for the mtDNA data using the Bayesian information criterion (BIC) and implementing a heuristic search algorithm (greedy). The resulting partitions and models were implemented in mrbayes 3.1.2 (Huelsenbeck & Ronquist 2001). Starting from a random tree, four metropolis-coupled Markov chain Monte Carlo (MCMC) chains (temp = 0.2) were run simultaneously for 5 000 000 generations three times, sampling every 100 generations with a burn-in of 7500. Convergence of the MCMC runs was assessed graphically using tracer 1.5 (Rambaut & Drummond 2009). Support is assessed by Bayesian posterior probabilities (BPP). Maximum-likelihood (ML) analyses were also performed on the mtDNA data and implemented in garli (Genetic Algorithm for Rapid Likelihood Inference 2.0) (Zwickl 2006). Six search replicates were run to find the best tree, estimating substitution rates, with branch support ascertained by 1000 nonparametric bootstrap (BS) replicates.
A phylogenetic analysis of the 255 character AFLP data based on 92 samples was also performed using mrbayes 3.1.2. Four independent MCMC chains (temp = 0.2) were run for 5 000 000 generations, with a sampling frequency of 1000 and a relative burn-in of 25%. The binary matrix was coded as data-type=restriction and coding=no absence sites, with all other parameters set as default, with support estimated by BPP.
Estimation of divergence times
As there are no suitable fossil calibrations for the Zosteropidae, we employed alternative methods of molecular dating. This included using a geological calibration based on the date of origin of a volcanic island, an approach that has been employed in various other avian studies (e.g. Fleischer et al. 1998; Warren et al. 2006; Moyle et al. 2009; Lerner et al. 2011; Melo et al. 2011). Under this approach, the maximum age of divergence between closely related taxa occupying neighbouring islands is constrained to be the age of the youngest island, representing the earliest possible date for colonization. Following assumptions discussed in previous studies (e.g. Fleischer et al. 1998; Warren et al. 2003), the maximum age estimate for the volcanic origin of Grande Comore at 0.5 Ma (R. Duncan personal communication in Warren et al. 2003) is used to calibrate the node separating the lowland Grande Comore white-eye (Z. maderaspatanus kirki) from other taxa in the ‘maderaspatanus’ clade (Warren et al. 2006). As an avian molecular clock has been used to date many avian studies (e.g. Voelker et al. 2010; Fritz et al. 2011), we also employed the average pairwise substitution rate of 2.1% for Cyt b (Weir & Schluter 2008) used in these studies to investigate whether resulting divergence times are concurrent with those generated from an island calibration.
Cytochrome b and ND3 data were pruned to include one representative from each taxon in order to use a model of speciation with no coalescence and were found to be evolving under a strict molecular clock in beast 1.7.5 (Drummond et al. 2012). In the calibrated approach, divergence time estimates were generated from the concatenated Cyt b and ND3 data, while divergence estimates obtained based on the molecular clock rate (2.1%) were generated from the Cyt b data only. Both approaches used the same starting tree that was generated from the Bayesian analyses of the concatenated data, but retaining only a single sample per taxon. For both analyses, two independent MCMC analyses were run starting from a user specified tree. Chains were run for 10 000 000 generations using a constant rate Yule speciation prior (assumes a constant speciation rate per lineage) implementing the models and partitions generated by partitionfinder, sampling every 1000 generations with a burn-in of 10%. Convergence of the two independent MCMC runs was assessed in tracer 1.5 (Rambaut & Drummond 2009), as were convergence of model parameter values (effective sample size [ESS]) to ensure ESS values >200. The posterior distribution was summarized in the program tree annotator 1.7.5 (Drummond et al. 2012). An empty alignment was also run to investigate the effects of priors on posterior divergence and resulted in no priors needing to be updated.
Biogeographic analysis
To reconstruct whether ancestral areas at nodes for selected clades (A1, A2, B1) in the phylogeny are lowland or highland in origin, we use the event-based method statistical dispersal–vicariance analysis (S-DIVA, Yu et al. 2010) in the program Reconstruct Ancestral States in phylogenies v1.1 (RASP; Yu et al. 2013) using the pruned tree containing one representative from each taxon.
Results
Phylogenetic inference (mtDNA)
Three partitions were identified using PartitionFinder for the concatenated mtDNA with the following models implemented in mrbayes: K80 + I+G (ND3 position 2, Cyt b position 3); HYK+I+G (ND3 position 3, Cyt b position 1); GTR+G (ND3 position 1, Cyt b position 2).
Phylogenetic inference based on Bayesian and ML analyses resulted in well-resolved congruent hypotheses in which support for relationships is generally good (Fig.2). Our results identify an African radiation composed of two major clades (Fig.2A, B) that each contains independent oceanic island clades, with the notable exception of the Ancient Indian Ocean white-eyes (AIO) clade that is not a member of this radiation.
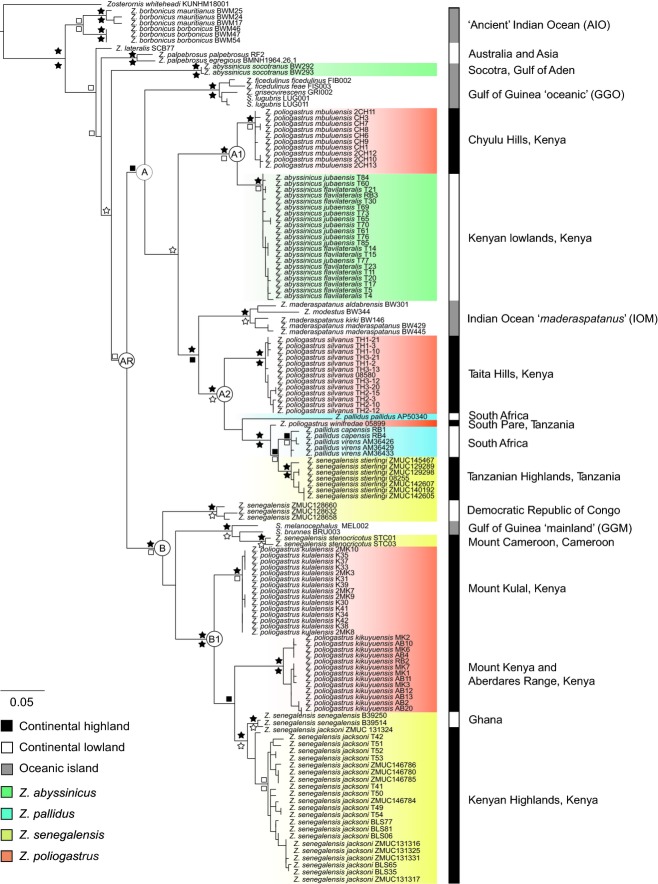
Figure 2.
Phylogenetic tree of African Zosterops generated using Bayesian inference based on ND3 and Cyt b data. Bayesian posterior probabilities (BPP) are indicated above branches, with ML bootstrap (BS) values below branches. Support values are represented by the symbols: black star >95% BPP/BS, white star >90% BPP/BS, black square >80% BPP/BS, white square >50% BPP/BS. Nodes with <50% support are unmarked. Key nodes are labelled AR (African Radiation), A-A2 and B-B1. Taxa are labelled using full trinomial nomenclature following the taxonomy of Dickinson (2003).
A significant and striking finding of this study is the extensive nonmonophyly of mainland African Zosterops taxa, with all continental species (Z. poliogastrus, Z. senegalensis, Z. abyssinicus, Z. pallidus) recovered as nonmonophyletic. In contrast to the nonmonophyly of described species, there is strong support for the monophyly of subspecies, specifically within the regional endemic Z. poliogastrus and the more widely distributed Z. senegalensis.
Each Z. poliogastrus and Z. senegalensis subspecies sampled in this study comprises a well-supported clade, forming independent lineages throughout both major mainland clades (Fig.2, clades A and B). By contrast, two of the three Z. pallidus subspecies (Z. p. capensis and Z. p. virens) comprise a clade (BPP 0.89, BS 70%) with no internal resolution. The position of the single sample of Z. p. pallidus (AP50340) is unclear, although there is no support for its placement as sister to the other Z. pallidus subspecies, supporting the findings of a recent phylogeographic study (Oatley et al. 2012). Likewise, the two mainland parapatric Z. abyssinicus subspecies form a clade (BPP 1, BS 75%) with no support for any division between them. However, our results clearly identify that these mainland Z. abyssinicus taxa are distinct from the insular race Z. a. socotranus from the Island of Socotra. Both Bayesian and ML analyses place Z. a. socotranus as sister to the African radiation, although this finding does not receive high support.
Divergence estimates
The application of different dating methods to the mitochondrial data (volcanic island calibration versus a 2.1% clock rate) results in divergent time frames for the radiation of African Zosterops. Implementation of the island calibration (Fig.3) estimates a Late Pleistocene divergence of 1.55 Ma (95% highest posterior density (HPD): 0.97–2.5 Ma). This is considerably younger than the time frame estimated by implementing the 2.1% clock (5.79 Ma 95% HPD: 4.8–6.86 Ma, Fig. S1, Supporting information) pushing the divergence of this clade back to the beginning of the Early Pliocene/Late Miocene. Previous estimates of the molecular rate of evolution in Zosteropidae have used island calibrations (Warren et al. 2006; Moyle et al. 2009; Melo et al. 2011) that have resulted in significantly faster rates of evolution than the 2.1% rate, suggesting that the avian clock may be a severe underestimation of the true rate of evolution within the Zosteropidae. Under the avian clock timescale, the divergence of the clade containing Grande Comore taxa Z. maderaspatanus kirki and Z. maderaspatanus maderaspatanus is dated at 1.68 Ma (95% HPD: 0.61–2.85 Ma), with a lower confidence interval marginally outside of the island age calibration of 0.5 Ma for the volcanic origin of Grande Comore.
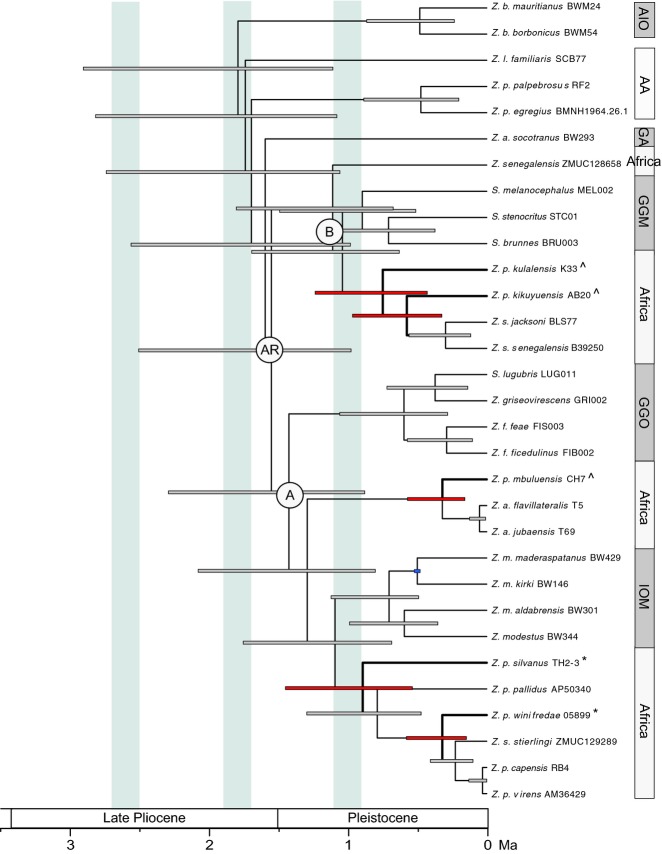
Figure 3.
Divergence times of Zosterops estimated using beast based on the mitochondrial data and calibrated using a geologically determined island date fixing the node indicated by the blue 95% HPD bar at 0.5 Ma following Warren et al. (2006). Thickened branches indicate Z. poliogastrus taxa, and red HPD bars highlight divergence estimates of the focal taxa. Pale blue bars indicate the warmer and wetter periods of the Plio-Plistocene climate following dates from Trauth et al. (2005). AIO, Ancient Indian Ocean; AA, Australia and Asia; GA, Gulf of Aden; GGM, Gulf of Guinea Mainland; GGO, Gulf of Guinea Oceanic; IOM, Indian Ocean ‘maderaspatanus’.
Despite divergence estimates based on median ages for the calibrated tree of the African Zosterops radiation falling within a drier climatic phase (Fig.3), we cannot place much confidence in this finding as 95% HPDs are broad at nodes along the backbone of the tree. However, more confidence can be given to the diversification of montane white-eyes during the cooler and drier East African climate (Fig.3) as our timetree indicates two pulses of diversification of these taxa with Z. poliogastrus silvanus 0.89 Ma (95% HPD: 0.53–1.45 Ma), Z. p. kulalensis 0.75 Ma (95% HPD: 0.43–1.23 Ma) and Z. p kikuyuensis 0.57 Ma (95% HPD: 0.32–0.96 Ma) diverging earlier than the contemporaneous divergence of Z. p. winifredae and Z. p mbuluensis at 0.32 Ma (0.15–0.58 Ma 95% HPD). Diversification of these montane taxa appears to have occurred sometime after the last major wet phase at 1.1–0.9 Ma (Trauth et al. 2007), although 95% HPDs for Z. p. kulalensis and Z. p. silvanus extend across this last wet phase and into the earlier arid phase. Irrespective of dating method, there appears to be little evidence that diversification events correspond to volcanic formation, as these pulses of diversification include members inhabiting both massif (old) and volcanic (young) mountains (Fig.3).
Biogeographic history
While our phylogeny does not include complete sampling, biogeographic analysis indicates at least one instance in which montane forest habitats are ancestral to lowland habitats (clade B1 100%; clade A1 50% + 50% equal probability that highland+lowland are ancestral), whereas ancestral reconstructions are ambiguous for clade A1 (lowland+highland habitats equally probable). However, at a broader scale, lowland habitats are reconstructed as being ancestral for the African radiation.
AFLP profiles
In total, 116 samples were screened to determine template DNA quality and quantity, with 27 dropping out due to poor quality extracts and high levels of noise affecting efficient scoring. The number of bins (alleles) for each of the 15 primer combinations identified by the initial scoring panel ranged from 211 to 563. In general, NED-labelled primer combinations gave the fewest number of fragments, while FAM-labelled primer combinations gave the highest. Average peak amplitude was relatively uniform across primer combinations (∼800 RFU), although the range of peak amplitude varied significantly between bins (100–5000). Shoulder stuttering was present in 11 of the 15 primer combinations used and was most frequently observed for FAM-labelled primers. The signal-to-noise ratio was lowest in FAM-labelled primers and was notably higher in HEX- and NED-labelled primers respectively.
Manual examination of concatenated AFLP profiles identified a large variation in peak amplitude between samples, which subsequently led to a large proportion of the bin being discounted (∼90%). Codominant alleles were evident across all primer combinations examined. However, peak amplitude variability between samples hindered assessments of frequency. Average estimates of genotyping error, measured following Bonin et al. (2004), were 0.8%. The number of fragments scored per sample ranged from 45 to 84, with the mean number of fragment scored being 66.9. Of the 255 AFLP loci examined, 31% (79 alleles) corresponded to private alleles, for which scoring was limited to individuals from the same sampling locality.
AFLP phylogenetic inference
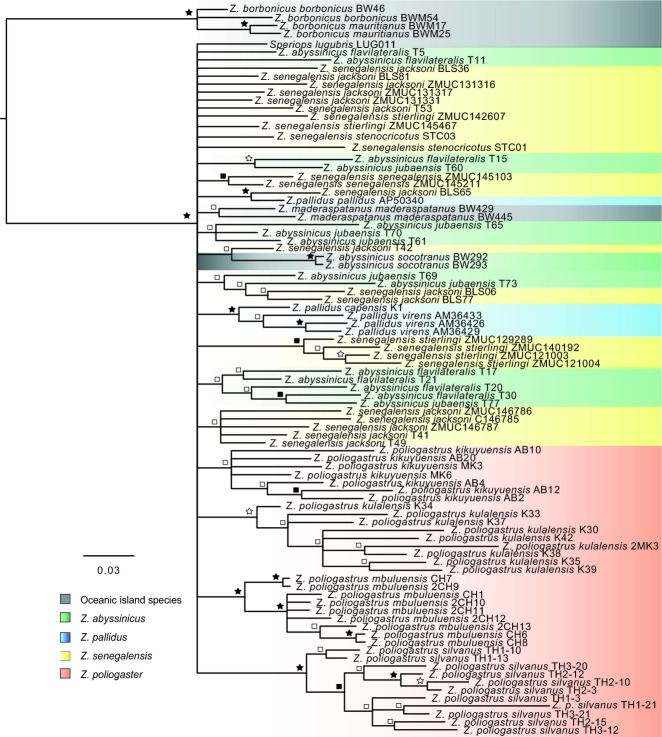
Bayesian inference of the AFLP data identified the two Z. borbonicus subspecies (Ancient Indian Ocean, AIO) as sister to all Africa taxa (BPP = 1.00, Fig.4), which is concordant with the mtDNA phylogeny. However, there is no support for the broader clades (A and B) recovered in the mtDNA phylogeny. Despite the lack of power in the AFLP data to resolve interrelationships, there is good support for the monophyly of range-restricted taxa of continental montane forests (i.e. Z. poliogastrus subspecies). Conversely, there is very little support for the monophyly of subspecies and/or populations of more widely distributed taxa such as Z. abyssinicus and Z. senegalensis. Leaf stability (Thorley & Wilkinson 1999), implemented in the program Phyutility (Smith & Dunn 2008), was used to assess if the lack of resolution in the initial data (92 taxa) was caused by unstable or rogue taxa. On the basis of leaf stability scores, 14 taxa were discarded, and while the resulting analysis of the reduced matrix resulted in a slightly more resolved hypothesis indicating the nonmonophyly of Z. poliogastrus, this result was only weakly supported. Taxon reduction made little impact in resolving deeper level relationships or subspecies monophyly of wide ranging taxa.
Figure 4.
Phylogenetic reconstruction of African Zosterops generated by Bayesian inference based on nuclear AFLP fragments. Bayesian posterior probabilities support indicated by the symbols: black star >95%, white star >90%, black square >80% and white square >50%.
Discussion
Evidence for niche divergence
We show for the first time using phylogenetic inference that montane white-eyes from East African sky islands are nonmonophyletic. In contrast to other regional studies that have focused on groups containing highland and lowland members at a similar timescale (e.g. Roy 1997; Voje et al. 2009), white-eyes have independently colonized but have not subsequently diversified within montane forest habitats. The independent colonization and lack of in situ diversification in an insular setting are generally analogous to white-eyes' history on oceanic islands. The repeated transition that has occurred over evolutionary time between lowlands and highlands strongly indicates a lack of niche conservatism. Additionally, irrespective of absolute dates, our timetree (Fig.3) indicates several instances in which endemic forest taxa are not younger than lowland taxa, which is in contrast to previous findings (e.g. Roy 1997; but see Marks 2010), and that for at least one clade, colonization of East African lowlands is suggested to have occurred by montane species. Thus, although the diversification of the focal group is suggested to have occurred rapidly during the Plio-Pleistocene, the phylogenetic hypothesis does not fit a montane speciation (refuge) model as proposed for other regional lineages (e.g. Roy 1997; Voje et al. 2009). Conversely, our results show that the larger and heavier endemic montane Z. poliogaster populations are more closely related to taxa with divergent habitat types, altitudinal distributions and dispersal abilities than they are to populations of restricted endemics that occur in neighbouring montane forest fragments. This is exemplified by the endemic montane Z. p. mubulensis (Chyulu Hills, Kenya) as sister to a clade comprising Z. a. flavilateralis and Z. a. jubaensis (Fig.2, clade A1) that have a wide distribution throughout the dry and arid lowlands of Kenya and Ethiopia.
Divergent selection is potentially also further indicated in the phylogeny (clades A2, B1, Fig.2), although relationships are more complex regarding the ecology of sister species. Within each clade, two endemic Z. poliogastrus subspecies occur in neighbouring montane forest fragments, with at least one of these subspecies sister to a widely dispersed lowland taxon. Despite the proximity of the forest fragments inhabited by the Z. poliogastrus subspecies in clade A2 (<50 km between Taita Hills and S. Pare Mts, and <100 km between Mt Kulal and N. Aberdares), the divergence between these taxa supports the idea that lowland savannah habitat provides a barrier to gene flow causing divergence between isolated forms in neighbouring montane forest fragments (e.g. Fjeldså & Lovett 1997; Roy 1997; Fjeldså & Bowie 2008). Additional highland taxa, Z. senegalensis jacksoni and Z. senegalensis stierlingi, also occupy montane forest habitats throughout Kenya and Tanzania; however, their presence in multiple nonconnected forest fragments indicates that unlike Z. poliogaster, highland Z. senegalensis populations were not restricted by low dispersal abilities.
Our results provide strong support for mechanisms founded on niche divergence. Both the gradient speciation model and the vanishing refuge model have previously been used to explain the occurrence of sister taxa in adjacent but distinct habitats (e.g. Vanzolini & Williams 1981; Moritz et al. 2000; Ogden & Thorpe 2002; Hall 2005; Kozak & Wiens 2007), but in the absence of data on the historical rate of gene flow, it is difficult to distinguish between these two alternative hypotheses (Moritz et al. 2000). While relationships within clade A1 seemingly support a gradient speciation model, those of clades A2 and B1 are more complicated. For example, the broad lowland distributions of Z. pallidus and Z. s. senegalensis are not parapatric with respect to their range-restricted sister taxa, Z. poliogastrus winifredae and Z. p. kikuyensis, and thus, strong directional selection between habitat types along an altitudinal gradient is not supported (Moritz et al. 2000; Kozak & Wiens 2007). Additionally, the presence of highland Z. senegalensis forms also conflicts with the main predictions of the gradient speciation model, in that taxa should occur in distinct habitats that have altitudinal nonoverlapping geographical distributions (Moritz et al. 2000). These sets of relationships, along with several instances of montane forest taxa ancestral to lowland taxa, instead lend some support to the vanishing refuge model (Vanzolini & Williams 1981; Moritz et al. 2000). With their exceptional dispersal abilities and rapid diversification, support for an ecological model of speciation in white-eyes may be a consequence of intrinsic factors as opposed to any common biogeographic pattern. Certainly, it is increasingly apparent from comparative regional studies that no single model or geological period can realistically explain the origin and diversification of the exceptional Afromontane biota.
Recent colonization of Africa and rapid diversification
The phylogenetic hypothesis (supported by mtDNA and AFLPs) identifies a single colonization event of the African continent by the Zosteropidae that, based on an island calibration, is suggested to have occurred as little as 1.55 Ma (95% HPD: 0.97–2.5 Ma) during the latest Pliocene to Early Pleistocene. The divergence into two principal African clades occurred soon after the arrival of Zosterops to the African continent, with subsequent rapid diversification suggested to have occurred in the Lower Pleistocene. Independent lineages diversified into differing habitats across sub-Saharan Africa, as well as colonizing the Gulf of Guinea Islands and making a second colonization of Indian Ocean islands (see Warren et al. 2006 and Melo et al. 2011).
If correct, this time frame, and also that estimated using the conservative avian molecular clock, indicates that Zosterops colonized Africa well after the fragmentation of montane forests during the Early Oligocene due to the onset of aridification (Lovett 1993; Sepulchre et al. 2006). Despite the long-term drying trend of the Plio-Pleistocene climate that reduced forest cover, there have been short alternating periods of extreme humidity and aridity (deMenocal 1995; Trauth et al. 2007). These warmer and wetter periods occurred from approximately 2.7–2.5, 1.9–1.7, 1.1–0.9 Ma (Trauth et al. 2005) and are assumed to have enabled the relic montane forests to expand to lower altitudes, possibly enabling isolated forest patches to have become connected and presenting opportunities for previously allopatric populations to mix; by contrast, cooler and drier periods are postulated to have led to ecological fragmentation with subsequent genetic isolation of montane forest restricted species (deMenocal 1995). Climatic stability of the highland refugia, in conjunction with repeated climate fluctuations affecting lowland areas, may have played an integral role in the diversification of Montane white-eyes, as suggested for other Eastern Afromontane biota (e.g. Roy 1997; deMenocal 2004; Lawson 2010; Measey & Tolley 2011).
Divergence time estimates for our data based on the use of a volcanic island calibration are approximately 4 times younger than those obtained when applying the 2.1% clock rate highlighting the disparity between these two approaches. Our time frame based on an island calibration supports previous estimates of the molecular rate of evolution in Zosteropidae that have documented significantly faster rates of evolution than the avian molecular clock rate (Warren et al. 2006; Moyle et al. 2009; Melo et al. 2011). Support for use of the age of Grande Comore to date divergences within African Zosterops comes from previous studies using geological calibrations that demonstrate consistency in divergence estimates when using different taxon sets, genetic markers and independent calibration points (e.g. New Georgia Group, Soloman Island), as well as different analytical methods (Moyle et al. 2009; Melo et al. 2011). For example, Moyle et al. (2009) dated the divergence of the Gulf of Guinea ‘mainland’ (GGM) clade between 0.89 and 1.35 Ma, which is extremely similar to the estimate produced by the island-calibrated approach used in this study (0.51–1.49 Ma).
Previous phylogenetic avian studies of this region have relied on the avian molecular clock (∼2%) to estimate sequence divergence times (e.g. Roy 1997; Roy et al. 2001; Fjeldså & Bowie 2008), although only Voelker et al.'s (2010) study has applied this rate using quantitative methods (e.g. nonparametric rate smoothing) making comparisons of divergence estimates between studies problematic. Unsurprisingly, by applying the conservative 2.1% clock rate to our data (Fig. S1), we estimate a similar time frame to Voelker et al.'s (2010) study on African forest robins who suggest Pliocene forest retraction c. 5–3 Ma as facilitating diversification in Africa's avifauna. The reliance on a molecular clock for the majority of avian studies is unfortunately due to the paucity of suitable fossil or geological calibrations. However, with broader taxonomic mitogenomic studies (e.g. Pacheco et al. 2011) identifying great variation of mutation rates within different bird groups, there is increasing support questioning a universal mitochondrial avian molecular clock (e.g. García-Moreno 2004; Lovette 2004). Nevertheless, time frames based on island ages are not without uncertainty (see Heads 2011) as these studies assume that the island endemic evolved in situ. This scenario does not account for endemic taxa being older than the island they inhabit having survived on nearby islands or mainland and later going extinct there (Heads 2011), and thus, our results based on a single island calibration should be viewed cautiously.
Underestimation of sky island diversity
Moreau's (1957) assessment that 10% of Zosterops species-level diversity occurs on continental landmasses would appear an underestimation, as our results reveal significant nonmonophyly of mainland Africa taxa, specifically endemic montane Z. poliogastrus and the widespread Z. senegalensis. Based on external morphological data, notably plumage, the montane populations of Z. poliogastrus have been classified as subspecies of a wider species complex (Dickinson 2003; van Balen 2008). However, the extensive sampling in this study of five of the eight Z. poliogastrus subspecies demonstrates this species to be polyphyletic. Strong support based on mitochondrial and AFLP data of the subspecific montane populations in these polyphyletic species indicates that populations should be elevated to species rank as independent taxonomic units rather than remain intraspecific taxa. As well as being scientifically interesting, this finding is important to considerations of species vulnerability, as conservation organizations (e.g. Birdlife International 2014) follow classifications of these polytypic species simply as ‘Z. poligastrus’ (currently considered to be a species of least concern) and thus incorrectly assume range size. The conservation status of these species should therefore be reassessed, particularly as the EABH is currently experiencing severe habitat loss (Myers et al. 2000).
Mitochondrial data also identify the polyphyly of the widespread African species Z. senegalensis and the northern East Africa species Z. abyssinicus, although denser sampling of subspecies is needed to determine a more complete picture of intraspecific relationships. Overall, our results question the utility in Zosterops of the traditional phenotypical characters used to delineate bird species. This highlights the need for a complete molecular systematic review of African Zosterops, applying species delimitation methods to quantitatively infer taxonomic boundaries (cf Fujita et al. 2013).
The use of AFLPs for avian phylogenetic studies
Given the lack of phylogenetic resolution of the ncDNA marker TGFß2, AFLPs were selected here as potentially suitable nuclear markers due to their utility in resolving rapidly evolving vertebrate clades (e.g. Sullivan et al. 2004; Joyce et al. 2011) as previous studies estimated very recent divergence dates for Zosterops (e.g. Moyle et al. 2009). However, the AFLP data proved largely disappointing for resolving internal relationships within the African radiation compared to the mtDNA data. While AFLPs have been increasingly used to resolve recently diverged fish radiations (e.g. Sullivan et al. 2004; Joyce et al. 2011), the use of AFLPs in avian phylogenetic studies is scarce (but see Humphries & Winker 2010), albeit they have been used successfully at the population level (e.g. Parchman et al. 2006; Mila et al. 2010). Although our AFLP matrix may have contained limited signal with only 255 AFLP loci, another study (Dasmahapatra et al. 2009) using a similar sized data matrix (310 AFLP loci) generated a well-resolved tree. Inspection of our data reveals that a large proportion of characters corresponded to alleles that were specific to a single population (private alleles). The larger number of private alleles in the data is likely to have resulted in the strong support for the monophyly of independent populations, specifically range-restricted taxa, with limited phylogenetic resolution between populations. As such, our data compared to those of a similar size (e.g. Dasmahapatra et al. 2009) are likely to have underperformed due to a much lower information content as opposed to insufficient data. However, a recent study focusing on auklet relationships (Humphries & Winker 2010) using AFLPs was also unable to generate a well-supported tree despite their data containing a greater number of polymorphic sites than did ours. It is possible that both of these avian studies selected taxa that are too divergent or that the stochastic process of incomplete lineage sorting has masked any phylogenetic signal (Humphries & Winker 2010). More avian studies using AFLPs are needed to determine the utility of these markers for phylogenetic inference.
Conclusion
Understanding how highly fragmented landscapes have influenced population differentiation and speciation is important not only regarding evolutionary processes responsible for generating elevated diversity, but in quantifying the biodiversity present for future conservation planning. This is particularly important in the Eastern Afromontane region that contains several thousand endemic species, but which is threatened by habitat loss and climate change. Our genetic study focusing on endemic Eastern Afromontane white-eyes indicates that contrary to other studies on unrelated taxonomic groups, niche divergence was likely as a major driver of speciation in this very recent clade. However, although our data support a speciation model in which reproductive isolation accumulates in allopatry with a significant contribution from ecologically mediated divergent natural selection, we are unable to test between different ecological models of speciation (i.e. gradient and vanishing speciation hypotheses). In addition to our findings highlighting how a highly fragmented landscape impacts speciation processes, we suggest that under a revised taxonomy, a number of East African sky islands would gain a new endemic Zosterops species. Based on our findings, montane Zosterops should be re-evaluated regarding their conservation status, particularly given the vulnerability of this biodiversity hotspot. These results further highlight the need for additional phylogeographic studies of taxa from this region to assess its unique diversity.
Acknowledgments
This work was supported by NERC funding NE/G523612/1. SCC acknowledges UCL's Graduate School and the Department of Zoology, The Natural History Museum for fieldwork funds. We are grateful to Phillista Malaki and Nicodemous Nalianya (National Museums of Kenya) for facilitating and providing field assistance, Nigel Cleere (NHM, Tring) for his invaluable field knowledge and mist-netting skills, and Mari-Wyn Burley (UCL) for sequencing. We thank Ben Warren, Martim Melo, Jon Fjeldså, Luca Borghesio and Tim Crowe for providing important blood and DNA samples. Ben Warren, Dany Garant and anonymous referees provided helpful comments that greatly improved this manuscript.
Glossary
- J.J.D., S.C.C. and R.P.P-J.
designed the study.
- J.J.D. and S.C.C.
wrote the manuscript with input from R.P.P-J.
- S.C.C., J.C.H. and B.A.
conducted the fieldwork.
- S.C.C.
performed the laboratory work. J.J.D.
- S.C.C.
analysed the data.
Data accessibility
DNA sequences: GenBank accession nos LK056696–LK056918.
Phylogenetic data matrices (mtDNA and AFLP) and bird specimen photographs have been deposited in the Dryad data repository (doi:10.5061/dryad.st56 h).
Supporting Information
Additional supporting information may be found in the online version of this article.
Details of AFLP amplification and scoring.
Divergence estimates for Zosterops generated using BEAST based on the 2.1% avian molecular clock for the Cyt b dataset. 95% highest posterior density (HPD) bars in grey, while those in red are superimposed from Fig. 3 (i.e. based on the volcanic island-calibrated timetree).
Taxa, sampling locations, altitude and Genbank accession numbers used in this study.
Primers and thermal cycling conditions used in amplification and sequencing of genes.
References
- Baker BH, Williams LAJ, Miller JA, Fitch FJ. Sequence and geochronology of the Kenya rift volcanics. Tectonophysics. 1971;11:191–215. [Google Scholar]
- van Balen S. Family Zosteropidae (White-eyes) In: del Hoyo J, Elliot A, Christie DA, editors. Handbook of the Birds of the World. Vol. 13. Penduline-Tits to Shrikes. Barcelona: Lynx Edicions; 2008. pp. 402–485. [Google Scholar]
- Birdlife International. Species factsheet: Zosterops poliogastrus. 2014. URL: http://www.birdlife.org.
- Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Molecular Ecology Notes. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Borghesio L, Laiolo P. Habitat use and feeding ecology of Kulal white-eye Zosterops kulalensis. Bird Conservation International. 2004;14:11–24. [Google Scholar]
- Clegg SM, Phillimore AB. The influence of gene flow and drift on genetic and phenotypic divergence in two species of Zosterops in Vanuatu. Philosophical Transactions of the Royal Society of London B Biological Sciences. 2010;365:1077–1092. doi: 10.1098/rstb.2009.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collar NJ, Crosby M, Stattersfield AJ. Birds to Watch 2: The World List of the Threatened Birds. Cambridge, UK: BirdLife International; 1994. (BirdLife Conservation Series 4) [Google Scholar]
- Dasmahapatra KK, Hoffman JI, Amos W. Pinniped phylogenetic relationships inferred using AFLP markers. Heredity. 2009;103:168–177. doi: 10.1038/hdy.2009.25. [DOI] [PubMed] [Google Scholar]
- Diamond JM. Ecological consequences of island colonization by Southwest Pacific birds, I. types of niche shifts. Proceedings of the National Academy of Sciences, USA. 1970;67:529–536. doi: 10.1073/pnas.67.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson EC. The Howard and Moore Complete Checklist of the Birds of the World. 3rd edn. London: Christopher Helm; 2003. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the beast 1.7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjeldså J, Bowie RCK. New perspectives on the origin and diversification of Africa's forest avifauna. African Journal of Ecology. 2008;46:235–247. [Google Scholar]
- Fjeldså J, Lovett JC. Geographical patterns of old and young species in African forest biota: the significance of specific montane areas as evolutionary centres. Biodiversity and Conservation. 1997;6:325–346. [Google Scholar]
- Fleischer RC, McIntosh CE, Tarr CL. Evolution on a volcanic conveyor belt: using phylogeographic reconstructions and K-Ar-based ages of the Hawaiian Islands to estimate molecular evolutionary rates. Molecular Ecology. 1998;7:533–545. doi: 10.1046/j.1365-294x.1998.00364.x. [DOI] [PubMed] [Google Scholar]
- Fritz SA, Jønsson KA, Fjeldså J, Rahbek C. Diversification and biogeographic patterns in four island radiations of passerine birds. Evolution. 2011;66:179–190. doi: 10.1111/j.1558-5646.2011.01430.x. [DOI] [PubMed] [Google Scholar]
- Fry CH, Keith S, Urban EK. The Birds of Africa. London: Academic Press; 2000. [Google Scholar]
- Fujita MK, Leache AD, Burbrink FT, McGuire JZ, Moritz C. Coalescent-based species delimitation in an integrative taxonomy. Trends in Ecology and Evolution. 2013;27:480–488. doi: 10.1016/j.tree.2012.04.012. [DOI] [PubMed] [Google Scholar]
- García-Moreno J. Is there a universal mtDNA clock for birds? Journal of Avian Biology. 2004;35:465–468. [Google Scholar]
- Griffiths CJ. The geological evolution of East Africa. In: Lovett JC, Wasser SK, editors. Biogeography and Ecology of the Rain Forests of Eastern Africa. Cambridge: Cambridge University Press; 1993. pp. 9–21. [Google Scholar]
- Habel JC, Cox S, Gassert F, Mulwa RK, Meyer J, Lens L. Population genetics of the East African White-eye species complex. Conservation Genetics. 2013;14:1019–1028. [Google Scholar]
- Hall JPW. Montane speciation patterns in Ithomiola butterflies (Lepidoptera: Riodinidae): are they consistently moving up in the world? Proceedings of the Royal Society B. 2005;272:2457–2466. doi: 10.1098/rspb.2005.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BP, Moreau RE. An Atlas of Speciation in African Passerine Birds. London: British Museum (Natural History); 1970. [Google Scholar]
- Heads M. Old taxa on young islands: a critique of the use of island age to date island-endemic clades and calibrate phylogenies. Systematic Biology. 2011;60:204–218. doi: 10.1093/sysbio/syq075. [DOI] [PubMed] [Google Scholar]
- Huang J, Sun M. A modified AFLP with fluorescence-labelled primers and automated DNA sequencer detection for efficient fingerprinting analysis in plants. Biotechnology Techniques. 1999;13:277–278. [Google Scholar]
- Huelsenbeck JP, Ronquist F. mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics Applications Note. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Humphries EM, Winker K. Working through polytomies: Auklets revisited. Molecular Phylogenetics and Evolution. 2010;54:88–96. doi: 10.1016/j.ympev.2009.07.023. [DOI] [PubMed] [Google Scholar]
- Joyce DA, Lunt DH, Genner MJ, Turner G, Bills R, Seehausen O. Repeated colonization and hybridization in Lake Malawi cichlids. Current Biology. 2011;21:108–109. doi: 10.1016/j.cub.2010.11.029. [DOI] [PubMed] [Google Scholar]
- Knowles LL. Tests of Pleistocene speciation in montane grasshoppers (genus Melanoplus) from the sky islands of western North America. Evolution. 2000;54:1337–1348. doi: 10.1111/j.0014-3820.2000.tb00566.x. [DOI] [PubMed] [Google Scholar]
- Kozak KH, Wiens JJ. Climatic zonation drives latitudinal variation in speciation mechanisms. Proceedings of the Royal Society B. 2007;274:2995–3003. doi: 10.1098/rspb.2007.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack D. Ecological Isolation in Birds. Cambridge, Massachusetts: Harvard University Press; 1971. [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP. clustal w and clustal x version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lawson LP. The discordance of diversification: evolution in the tropical-montane frogs of the Eastern Arc Mountains of Tanzania. Molecular Ecology. 2010;19:4046–4060. doi: 10.1111/j.1365-294X.2010.04788.x. [DOI] [PubMed] [Google Scholar]
- Lawson LP. Diversification in a biodiversity hot spot: landscape correlates of phylogeographic patterns in the African spotted reed frog. Molecular Ecology. 2013;22:1947–1960. doi: 10.1111/mec.12229. [DOI] [PubMed] [Google Scholar]
- Lerner HRL, Meyer M, James HF, Hofreiter M, Fleischer RC. Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian Honeycreepers. Current Biology. 2011;21:1838–1844. doi: 10.1016/j.cub.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Lovett JC. Climatic history and forest distribution in eastern Africa. In: Lovett JC, Wasser SK, editors. Biogeography and Ecology of the Rain Forests of Eastern Africa. Cambridge: Cambridge University Press; 1993. pp. 23–29. [Google Scholar]
- Lovette IJ. Mitochondrial dating and mixed support for the “2% rule” in birds. The Auk. 2004;121:1–6. [Google Scholar]
- Marks BD. Are lowland rainforests really evolutionary museums? Phylogeography of the green hylia (Hylia prasina) in the Afrotropics. Molecular Phylogenetics and Evolution. 2010;55:178–184. doi: 10.1016/j.ympev.2009.10.027. [DOI] [PubMed] [Google Scholar]
- Mayr E. Systematics and the Origin of Species. New York: Columbia University Press; 1942. [Google Scholar]
- McCormack JE, Bowen BS, Smith TB. Integrating paleoecology and genetics of bird populations in two sky island archipelagos. BMC Biology. 2008;6:28. doi: 10.1186/1741-7007-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measey GJ, Tolley KA. Sequential fragmentation of Pleistocene forests in an East Africa biodiversity hotspot: chameleons as a model to track forest history. PLoS ONE. 2011;6:e26606. doi: 10.1371/journal.pone.0026606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo M, Warren BH, Jones PJ. Rapid parallel evolution of aberrant traits in the diversification of the Gulf of Guinea white-eyes (Aves, Zosteropidae) Molecular Ecology. 2011;20:4953–4967. doi: 10.1111/j.1365-294X.2011.05099.x. [DOI] [PubMed] [Google Scholar]
- deMenocal PB. Plio-Pleistocene African Climate. Science. 1995;270:53–59. doi: 10.1126/science.270.5233.53. [DOI] [PubMed] [Google Scholar]
- deMenocal PB. African climate change and faunal evolution during the Pliocene-Pleistocene. Earth and Planetary Science Letters. 2004;220:3–24. [Google Scholar]
- Mila B, Warren BH, Heeb P, Thebaud C. The geographic scale of diversification on islands: genetic and morphological divergence at a very small spatial scale in the Mascarene grey white-eye (Aves: Zosterops borbonicus) BMC Evolutionary Biology. 2010;10:158. doi: 10.1186/1471-2148-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau RE. Variation in the western Zosteropidae (Aves) Bulletin of the British Museum. 1957;4:318–433. [Google Scholar]
- Moritz C, Patton JL, Schneider CJ, Smith TB. Diversification of rainforest faunas: an integrated molecular approach. Annual Review of Ecology and Systematics. 2000;31:533–563. [Google Scholar]
- Moyle RG, Filardi CE, Smith CE, Diamond J. Explosive Pleistocene diversification and hemispheric expansion of a “great speciator”. Proceedings of the National Academy of Sciences, USA. 2009;106:1863–1868. doi: 10.1073/pnas.0809861105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Oatley G, Voelker G, Crowe TM, Bowie RCK. A multi-locus phylogeny reveals a complex pattern of diversification related to climate and habitat heterogeneity in southern African white-eyes. Molecular Phylogenetics and Evolution. 2012;64:633–644. doi: 10.1016/j.ympev.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Ogden R, Thorpe RS. Molecular evidence for ecological speciation in tropical habitats. Proceeding of the National Academy of Science. 2002;99:13612–13615. doi: 10.1073/pnas.212248499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Battistuzzi FU, Lentino M, Aguilar RF, Kumar S, Escalante AA. Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Molecular Biology and Evolution. 2011;28:1927–1942. doi: 10.1093/molbev/msr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchman TL, Benkman CW, Britch SC. Patterns of genetic variation in the adaptive radiation of New World crossbills (Aves: Loxia) Molecular Ecology. 2006;15:1873–1887. doi: 10.1111/j.1365-294X.2006.02895.x. [DOI] [PubMed] [Google Scholar]
- Pompanon F, Bonin A, Bellemain E, Taberlet P. Genotyping errors: causes, consequences and solutions. Nature Reviews Genetics. 2005;6:847–859. doi: 10.1038/nrg1707. [DOI] [PubMed] [Google Scholar]
- Price T. Speciation in Birds. Boulder, Colorado: Roberts and Co; 2008. [Google Scholar]
- Rambaut A. Sequence Alignment Editor. 2002. Se-Al Carbon v2.0a11. Available from: URL http://tree.bio.ed.ac.uk/software/seal/
- Rambaut A, Drummond AJ. 2009. Tracer v1.5. Available from: URL http://beast.bio.ed.ac.uk/Tracer.
- Roy MS. Recent diversification in African greenbuls (Pycnonotidae: Andropadus) supports a montane speciation model. Proceedings of the Royal Society B. 1997;264:1337–1344. [Google Scholar]
- Roy MS, Sponer R, Fjeldså J. Molecular systematics and evolutionary history of Akalats (Genus Sheppardia); A pre-Pleistocene radiation in a group of African forest birds. Molecular Phylogenetics and Evolution. 2001;18:74–83. doi: 10.1006/mpev.2000.0862. [DOI] [PubMed] [Google Scholar]
- Sepulchre P, Ramstein G, Fluteau F, Schuster M, Tiercelin JJ, Brunet M. Tectonic uplift and eastern Africa aridification. Science. 2006;313:1419–1423. doi: 10.1126/science.1129158. [DOI] [PubMed] [Google Scholar]
- Seto KC, Güneralp B, Hutyra LR. Global forecasts of urban expansion to 2030 and direct impacts on biodiversity and carbon pools. Proceedings of the National Academy of Sciences, USA. 2012;109:16083–16088. doi: 10.1073/pnas.1211658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seutin G, White BN, Boag PT. Preservation of avian blood and tissue samples for DNA analyses. Canadian Journal of Zoology. 1991;69:82–90. [Google Scholar]
- Shepard DB, Burbrink FT. Phylogeographic and demographic effects of Pleistocene climatic fluctuations in a montane salamander, Plethodon fourchensis. Molecular Ecology. 2009;18:2243–2262. doi: 10.1111/j.1365-294X.2009.04164.x. [DOI] [PubMed] [Google Scholar]
- Smith SA, Dunn C. Phyutility: a phyloinformatics utility for trees, alignments, and molecular data. Bioinformatics. 2008;24:715–716. doi: 10.1093/bioinformatics/btm619. [DOI] [PubMed] [Google Scholar]
- Sullivan JP, La voue ' S, Arnegard ME, Hopkins CD. AFLPs resolve phylogeny and reveal mitochondrial introgression within a species flock of African electric fish (Mormyroidea: Teleostei) Evolution. 2004;58:825–841. doi: 10.1111/j.0014-3820.2004.tb00415.x. [DOI] [PubMed] [Google Scholar]
- Thorley JL, Wilkinson M. Testing the phylogenetic stability of early tetrapods. Journal of Theoretical Biology. 1999;200:343–344. doi: 10.1006/jtbi.1999.0999. [DOI] [PubMed] [Google Scholar]
- Trauth MH, Maslin MA, Deino A, Strecker MR. Late Cenozoic moisture history of East Africa. Science. 2005;309:2051–2053. doi: 10.1126/science.1112964. [DOI] [PubMed] [Google Scholar]
- Trauth MH, Maslin MA, Deino AL, Strecker MR, Bergner AGN, Dühnforth M. High- and Low- latitude forcing of Plio-Plistocene East African climate and human evolution. Journal of Human Evolution. 2007;53:475–486. doi: 10.1016/j.jhevol.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Vanzolini PE, Williams EE. The vanishing refuge: a mechanism for ecogeographic speciation. Papéis Avulsos de Zoologia. 1981;34:251–255. [Google Scholar]
- Voelker G, Outlaw RK, Bowie RCK. Pliocene forest dynamics as a primary driver of African bird speciation. Global Ecology and Biogeography. 2010;19:111–121. [Google Scholar]
- Voje KL, Hemp C, Flagstad Ø, Sætre G-P, Stenseth NC. Climate change as an engine for speciation in flightless Orthoptera species inhabiting African mountains. Molecular Ecology. 2009;18:93–108. doi: 10.1111/j.1365-294X.2008.04002.x. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren BH, Bermingham E, Bowie RCK, Prys-Jones RP, Thébaud C. Molecular phylogeography reveals island colonization history and diversification of western Indian Ocean sunbirds (Nectarinia: Nectariniidae) Molecular Phylogenetics and Evolution. 2003;29:67–85. doi: 10.1016/s1055-7903(03)00063-0. [DOI] [PubMed] [Google Scholar]
- Warren B, Bermingham E, Prys-Jones RP, Thebauds C. Immigration, species radiation and extinction in a highly diverse songbird lineage: white-eyes on the Indian Ocean islands. Molecular Ecology. 2006;15:3769–3786. doi: 10.1111/j.1365-294X.2006.03058.x. [DOI] [PubMed] [Google Scholar]
- Weir JT, Schluter D. Calibrating the avian molecular clock. Molecular Ecology. 2008;17:2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Donoghue MJ. Historical biogeography, ecology and species richness. Trends in Ecology and Evolution. 2004;19:639–644. doi: 10.1016/j.tree.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Ackerly DD, Allen AP, et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecology Letters. 2010;13:1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- Yu Y, Harris AJ, He X. S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Molecular Phylogenetics and Evolution. 2010;56:848–850. doi: 10.1016/j.ympev.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Yu Y, Harris AJ, He X-J. RASP (Reconstruct Ancestral State in Phylogenies) 2.1 beta. 2013. Available at http://mnh.scu.edu.cn/soft/blog/RASP. [DOI] [PubMed]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin, Texas: The University of Texas; 2006. PhD Dissertation, [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of AFLP amplification and scoring.
Divergence estimates for Zosterops generated using BEAST based on the 2.1% avian molecular clock for the Cyt b dataset. 95% highest posterior density (HPD) bars in grey, while those in red are superimposed from Fig. 3 (i.e. based on the volcanic island-calibrated timetree).
Taxa, sampling locations, altitude and Genbank accession numbers used in this study.
Primers and thermal cycling conditions used in amplification and sequencing of genes.
Data Availability Statement
DNA sequences: GenBank accession nos LK056696–LK056918.
Phylogenetic data matrices (mtDNA and AFLP) and bird specimen photographs have been deposited in the Dryad data repository (doi:10.5061/dryad.st56 h).