Abstract
DNA methylation in the genome plays a fundamental role in the regulation of gene expression and is widespread in the genome of eukaryotic species. For example, in higher vertebrates, there is a “global” methylation pattern involving complete methylation of CpG sites genome-wide, except in promoter regions that are typically enriched for CpG dinucleotides, or so called “CpG islands.” Here, we comprehensively examined and compared the distribution of CpG sites within ten model eukaryotic species and linked the observed patterns to the role of DNA methylation in controlling gene transcription. The analysis revealed two distinct but conserved methylation patterns for gene promoters in human and mouse genomes, involving genes with distinct distributions of promoter CpGs and gene expression patterns. Comparative analysis with four other higher vertebrates revealed that the primary regulatory role of the DNA methylation system is highly conserved in higher vertebrates.
Keywords: genome-wide CpG site distribution, CpG sites within promoters, conservation and divergence in DNA methylation, eukaryotes, comparative phylogenetic analysis
Introduction
DNA methylation involves the postreplicative addition of a methyl group to the 5-position of particular cytosines in the DNA sequence and constitutes an important and widely recognized epigenetic mark (Holliday and Pugh 1975; Riggs 1975; Day and Sweatt 2010; Parle-McDermott and Harrison 2011; Zhu and Reinberg 2011). It is highly conserved among eukaryotic species, including protists, fungi, plants and animals, and plays a fundamental role in modulating biological processes, particularly the regulation of transcription (Jaenisch and Bird 2003; Patra et al. 2008; Chen and Riggs 2011; He et al. 2011). Two mechanisms by which DNA methylation regulates gene expression levels have been identified (Attwood et al. 2002; Fahrner et al. 2002; Geiman and Robertson 2002; Li 2002; Herman and Baylin 2003; Goll and Bestor 2005). First, methylated cytosines can physically disrupt the binding of RNA polymerases and transcription factors to the appropriate regions of target genes. Second, methylated DNA may be targeted by multiple proteins, including methyl-CpG-binding domain proteins, histone deacetylases, and chromatin remodeling proteins, to form complex structures, which can inactivate the chromatin and silence gene transcription.
DNA methylation occurs in three sequence contexts. Most frequently it occurs at “CpG” dinucleotides in plants and animals, though it also occurs in both “CpHpG” and “CpHpH” contexts in plants. The level and distribution pattern of DNA methylation can vary dramatically among species. Some eukaryotic organisms including Saccharomyces cerevisiae (budding yeast) and Caenorhabditis elegans (nematode worm) do not encode any DNA methyltransferase family genes and so their DNA is not methylated (Bird 2002; Suzuki and Bird 2008). Other species have a “mosaic” methylation pattern characterized by moderately high methylation levels in many DNA sequence domains, separated by completely unmethylated domains. These species include the fungus Neurospora crassa, plants (e.g., Arabidopsis, corn, rice, and poplar) (Montero 1992; Palmer 2003; Chan et al. 2005; Gehring and Henikoff 2007; Henderson and Jacobsen 2007; Zilberman et al. 2007), and invertebrates (e.g., sea squirt, Drosophila) (Gowher et al. 2000; Salzberg et al. 2004; He et al. 2011). Methylation of these genomes is mainly targeted to gene bodies, or to transposable regions, where it represents a crucial transcriptional silencing mechanism involving small interfering RNAs (Mette et al. 2000; Chan 2004; Suzuki and Bird 2008). In contrast, vertebrate species, particularly mammals, typically exhibit “global” DNA methylation patterns (Robertson 2005; Rollins 2006; Chen and Riggs 2011) where candidate methylation sites across the entire genome are completely methylated, except for those in promoter regions where the methylation level varies highly among different tissues, cells, growth conditions, and developmental stages. The methylation status and local density of CpG dinucleotides within promoter regions is associated with the regulation of gene transcription in vertebrates (Boyes and Bird 1992; Hsieh 1999; Weber 2007), though the same has not been observed in invertebrates. The functional implications of this relationship have not yet been thoroughly explored on a genome-wide basis in vertebrates.
Here we report a comprehensive investigation of the DNA methylation system in eukaryotes through examining the fully sequenced genomes of ten model eukaryotic species, including six higher vertebrates (amniotes): 1) Homo sapiens (human), 2) Mus musculus (mouse), 3) Rattus norvegicus (rat), 4) Bos taurus (cow), 5) Canis familiaris (dog) and 6) Gallus gallus (chicken), one lower vertebrate (Danio rerio, zebrafish), two invertebrates (Drosophila melanogaster [fruitfly] and C. elegans [nematode worm]), and the plant Arabidopsis thaliana (Arabidopsis). We focused on the distribution and roles of CpG dinucleotides and discovered patterns that are highly conserved among the six higher vertebrate species and can be used to accurately assemble the evolutionary relationships among these species. Using extensive data sets of DNA methylation and gene expression from human and mouse tissues, we linked the observed patterns to the regulatory and (most likely) highly conserved role of DNA methylation in modulating gene transcription in higher vertebrate genomes.
Materials and Methods
Whole-Genome Sequences and Genomic Feature Annotation Information
Whole-genome sequence data for each of ten eukaryotic model organisms were downloaded from the University of California–Santa Cruz (UCSC) genome bioinformatics database (http://hgdownload.cse.ucsc.edu/downloads.html, last accessed November 1, 2014) and the Arabidopsis Information Resource (http://www.arabidopsis.org/download/index.jsp, last accessed November 1, 2014), and the corresponding genomic annotation was obtained from the genome annotation database of the UCSC Genome Browser (http://genome-archive.cse.ucsc.edu/downloads, last accessed November 1, 2014), the Exon–Intron Database (http://bpg.utoledo.edu/∼afedorov/lab/eid.html, last accessed November 1, 2014) and the Mammalian Promoter Database (http://mpromdb.wistar.upenn.edu/, last accessed November 1, 2014). The direct links to different types of data sets for each of ten eukaryotic model organisms used in our analysis were listed in supplementary table S1, Supplementary Material online.
Analysis of CpG Dinucleotide Distribution in Promoters
A FORTRAN program was developed to parse the sequence data and identify the locations of CpG dinucleotides and the fraction of GC content for various genomic features. The Poisson distribution was used to test whether the distribution of CpG dinucleotides in promoter regions follows a random pattern. The mean of the distribution, λ = 51, was equal to the average number of CpG sites per 1,000 bases in promoters of the human genome. Hence, we calculated the expected probability of promoters with the number of CpG sites in a 1,000-bp region, k, falling within a number of ranges including “0–25,” “26–40,” “41–50,” “51–60,” “61–75,” and “>75.” Here, we could estimate the expected probability of promoters with 0–25 CpG sites in 1,000 bp length as:
| (1) |
where e was the base of the natural logarithm (e = 2.718), k was the number of occurrences of CpGs in 1,000-bp sequence, and λ was the average number of CpGs in 1,000 bp. In the same way, we also calculated the expected Poisson probabilities of promoters with 26–40, 41–50, 51–60, 61–75, and >75 CpG sites in 1,000 bp length. We then identified the observed proportions of promoters with 0–25, 26–40, 41–50, 51–60, 61–75, and >75 CpG sites in 1,000 bp length. Variation in the length of individual promoters was accounted for by normalizing for a fixed 1,000 bp length. Note that the criterion for grouping the promoters was entirely for convenience of statistical analysis and did not affect the conclusions drawn from the analysis. The Pearson’s chi-square test was implemented to test for the goodness of fit between observed and expected frequency distributions, with degrees of freedom equal to 4:
| (2) |
where Oiis the observed proportion of promoters in the ith category and Ei is the expected proportion of promoters in the ith category.
Identification of High and Low CpG Density Promoters
The ratio of observed/expected (O/E) CpGs in the promoter region of each annotated gene was calculated as follows:
| (3) |
where N is the length of the promoter (Karlin and Mrazek 1997). Two classes of promoter were defined according to Saxonov et al. (2006) and Weber (2007), as follows. First, high CpG density promoters (HCP) with CpG O/E ratio >65% and GC fraction >55%; second, low CpG density promoters (LCP) with CpG O/E ratio <65% and GC fraction <45%; the remaining promoters were classified as intermediate CpG density Promoters (ICP).
Identification of Homologous Genes and Interspecies Conservation Analysis
The homologous genes across six higher vertebrate species were downloaded from the National Center for Biotechnology Information (NCBI)-HomoloGene Database (release 65, ftp://ftp.ncbi.nih.gov/pub/HomoloGene/build65/homologene.data), which is built upon both DNA sequence and protein sequence data for homologous gene families, as described at http://www.ncbi.nlm.nih.gov/homologene/build-procedure/ (last accessed November 1, 2014) . Among the genes homologous between two vertebrate species, we inferred a conserved status where the promoters of both genes were classified into the same group (HCP or LCP). By analyzing all annotated protein-coding genes from the NCBI-HomoloGene Database, we could identify the conservation level of the promoter status of homologous genes among six higher vertebrates over evolutionary time. Furthermore, for each pair of homologous genes between two vertebrates with conserved promoter status, we measured their evolutionary conservation at the sequence level. We used two substitution rate statistics to estimate and compare the evolutionary maintenance of homologous genes with either HCP or LCP conserved status: 1) The ratio of nonsynonymous to synonymous substitution rate for sequences in protein-coding regions (Ka/Ks) and 2) the rate of nucleotide substitution for sequences in promoter regions, Kimura80 model (K80) (Kimura 1980). The nonsynonymous (Ka) and synonymous (Ks) substitution rates for each pair of homologous genes were calculated using the “codeml” maximum-likelihood method in PAML4 (Yang 2007). K80 was calculated using the “Kimura80” nucleotide substitution model (Kimura 1980). Only genes with a unique promoter were used in this analysis.
Reconstruction and Comparison of Phylogenetic Relationships among Six Higher Vertebrates
The information of phylogenetic relationships and times of divergence among six higher vertebrates was obtained from published data (Hedges 2002), which used both genome-wide DNA and protein sequences to estimate the phylogenetic tree that minimizes the number of sequence changes. As described in the above section, we inferred the level of conserved status (LCP or HCP) of the promoters of homologous genes between each pair of six higher vertebrates. We directly used these conservation levels as the measurement of divergence distance to build the distance matrix for all six higher vertebrates. Then, we input this distance matrix to Minitab software and used the cluster analysis module “cluster variables” to calculate the similarity (%) among six higher vertebrates and to reconstruct the phylogenetic tree. Default parameter values were used (average linkage method and correlation distance measure). For comparison, a phylogenetic tree was reconstructed in the same way based on the times of divergence among six higher vertebrates calculated according to Hedges (2002).
DNA Methylation and Gene Expression Data Sets
The genome-wide DNA methylation patterns for 28 different human tissues (or cell lines) were assayed using the Infinium HumanMethylation27 DNA analysis BeadChip platform. The raw data from the BeadChip assay were downloaded from the NCBI Gene Expression Omnibus (GEO) database under accession numbers GSE17769, GSE20872, GSE24087, GSE28356, and GSE26133 (http://www.ncbi.nlm.nih.gov/geo, last accessed November 1, 2014). The data consisted of 27,578 probe units representing 27,578 CpG sites across the promoter regions of >14,000 genes. The quantitative estimate of methylation level (β) for each specific CpG site was calculated from the signals of the methylated bead (M) and the unmethylated bead (U) as follows:
| (4) |
This was implemented by the methylation module in the Illumina Genome BeadStudio Software. The methylation levels ranged from 0 (completely unmethylated) to 1 (completely methylated).
Gene expression raw data were obtained from 107 different human tissues (or cell lines) from the NCBI GEO database under accession numbers GSE7127, GSE17768, GSE24089, and GSE11582. The data were generated using the Affymetrix U133 human expression microarray GeneChip, containing over 45,000 probe sets representing approximately 33,000 well-annotated human genes. We analyzed the raw signal intensities of probe sets using the standard Affymetrix strategy MAS5.0 and normalization by the global median scaling method.
The 17 different mouse tissues whose methylation data and expression data were analyzed here came from the C57Bl/6 strain, which has been widely used for genetic research. The whole-genome DNA methylation data for 17 mouse tissues were obtained using whole-genome bisulfite sequencing (bisulfite-seq) using the Illumina Hiseq2000 platform. The raw data were accessed under accession number GSE42836 from the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo, last accessed November 1, 2014). The raw short-read data were preprocessed and mapped with Bowtie to the computationally bisulfite-converted mm9 genome, as described previously (Hon et al. 2013). Methylation level of the cytosine in each CpG site was estimated as the ratio of methylated read-coverage to total read-coverage across the CpG site. Based on bisulfite-seq data, we could identify methylation levels at over 16,000,000 CpG sites across the whole mouse genome. Here, we only focused on the DNA methylation levels in annotated promoter regions of the mouse genome. The genome-wide gene expression of the corresponding 17 mouse tissues was profiled using the Affymetrix mouse genome 430 2.0 GeneChip, consisting of 45,037 probe sets for 21,078 genes. The raw data for mouse gene expression were downloaded from the NCBI GEO database and the accession number for the gene expression data of each mouse tissue was summarized in supplementary table S2, Supplementary Material online. The mouse gene expression data were processed in exactly the same way as described for the human gene expression data.
GO Annotation Data Sets and Overrepresentation Analysis
Gene ontology annotation (GO terms) for each of the six higher vertebrates was downloaded from the Gene Ontology database (http://www.geneontology.org/GO.downloads.annotations.shtml, last accessed November 1, 2014). To identify GO terms overrepresented either in HCP or in LCP groups, the binomial test was employed for each GO term by comparing the number of ORFs in each of the groups associated with a given GO term with the number of genome-wide ORFs associated with the given GO term. For each GO term, a Z statistic was computed as follows:
| (5) |
where Fd is the fraction of HCP (or LCP) promoter genes annotated with the given GO term, FG is the fraction of all annotated genes with the given term, and Nd is the total number of genes with HCP (or LCP). A GO term was determined to be significantly overrepresented in a particular group when Z > 4.75 (P < 10−6 after Bonferroni correction).
The analyses were performed with custom programs/scripts in either Fortran-90 or R languages and are available upon request from the corresponding author.
Results
Genome-Wide Distribution of CpG Sites and GC Content
We first explored the full genome sequences of the ten model eukaryotic species and compared the distribution of the GC content and of CpG dinucleotides across the whole genome and in different genome features (table 1). Among the six higher vertebrate species, the whole genomes and intron regions had the lowest GC content fraction (∼37.95–42.46%), followed by the exon regions (∼48.88–51.57%). The promoter regions had distinctly higher GC content (∼52.21–57.29%), agreeing with the observation that conserved functional sequences have higher GC content compared with the entire genome or intronic regions of the human genome (Pozzoli et al. 2008). The pattern observed for the lower vertebrate (zebrafish), invertebrates, and the plant was clearly different. Although the exon regions had higher GC content compared with the entire genome or introns, the promoter regions did not show enriched GC content, but instead showed a similar GC content to the entire genome or introns.
Table 1.
GC Content and Distribution of CpG Sites in Vertebrate, Invertebrate, and Plant Genome Features
| Higher Vertebrates (Mammals, Birds)a |
Lower Vertebrate, Invertebrates, and Plantb |
|||||
|---|---|---|---|---|---|---|
| GC % | Expected CpG %c | Observed CpG % | GC % | Expected CpG %c | Observed CpG % | |
| Genome-wide | 37.95–42.39 | 3.61–4.49 | 0.95–2.08 | 35.44–41.24 | 3.14–4.25 | 3.48–8.11 |
| Intron | 40.37–42.46 | 4.07–4.50 | 1.75–2.21 | 32.14–39.91 | 2.58–3.98 | 2.65–7.35 |
| Exon | 48.88–51.57 | 5.97–6.65 | 5.35–6.78 | 42.42–50.00 | 4.50–6.25 | 5.89–11.69 |
| Transcripts | 48.47–51.72 | 5.87–6.69 | 4.94–6.82 | 42.59–50.10 | 4.54–6.27 | 5.93–11.71 |
| Promoter | 52.21–57.29 | 6.81–8.21 | 7.66–11.98 | 32.42–41.55 | 2.63–4.32 | 4.19–9.08 |
aRange among six mammalian higher vertebrate species (human, mouse, rat, cow, dog, and chicken).
bRange among four lower vertebrate, invertebrate, or plant species (zebrafish, Drosophila, Arabidopsis, Caenorhabditis elegans).
cExpected CpG percentage calculated based on the observed GC percentage.
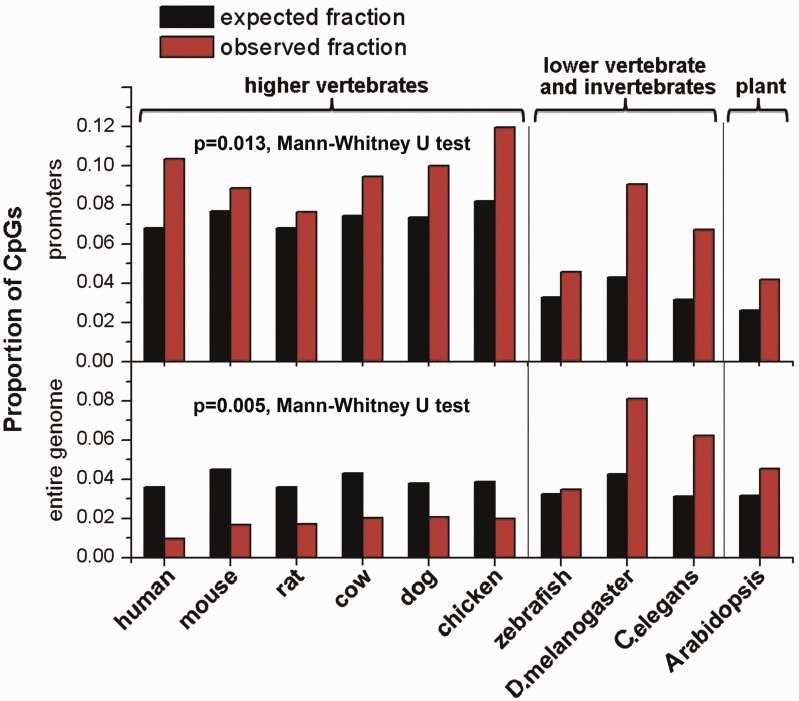
Next, we calculated the expected proportions of CpG sites based on the random union of C and G nucleotides and compared the expectations with the observed proportions. We found that CpG dinucleotides were consistently and significantly enriched (observed > expected) in promoter regions in the six higher vertebrate genomes (P = 0.013, Mann–Whitney U test) (fig. 1 and table 1), consistent with the higher level of promoter GC content in these species. Meanwhile, the genome-wide CpG content was significantly lower than that expected in the higher vertebrate genomes (P < 0.005, Mann–Whitney U test). However, the lower vertebrate, invertebrate, and plant species showed a different pattern, in which the observed proportion of CpGs consistently but not significantly exceeded the expected in both the promoter regions and the entire genome (P = 0.061, Mann–Whitney U test) (fig. 1).
Fig. 1.—
Observed and expected proportions of CpGs across the entire genome or in gene promoters of ten model species.
Distributional Divergence of CpG Sites in Promoter Regions across Species
To test whether the CpG sites were randomly distributed in the promoter regions, we examined the distribution of the number of CpGs occurring per 1,000 bp of promoter sequence and grouped the promoters into six categories according to the number of CpG sites (0–25, 26–40, 41–50, 51–60, 61–75, and >75). We modeled the occurrence of CpGs as independent random events that follow a Poisson distribution with parameter λ (i.e., the mean of the distribution) in a fixed promoter length (here, 1,000 bp). Pearson’s chi-square test was employed to test for significance of concordance between the observed CpGs and the expected CpGs under the Poisson model. For example, the average number of CpG sites per 1,000 bases in promoters of the human genome was equal to 51. Thus, we calculated the observed proportion of promoters and expected Poisson probability of promoters in each of six categories for the human genome (table 2). Overall, the analysis showed that P values of the test were less than 10−15 across all six higher vertebrates. The highly significant deviation of CpG sites in the promoters from the Poisson expectation strongly supports their nonrandom distribution. In sharp contrast, CpG sites in the promoters of the four lower vertebrate, invertebrate, and plant species perfectly follow the Poisson distribution and scatter randomly over the promoters (P > 0.05).
Table 2.
Observed Proportion and Expected Poisson Probability of Promoters Classified into Each of the Six CpG Density Categories in the Human Genome
| Number of CpG Sites per 1,000 Bases of Promoter Sequence |
Total | ||||||
|---|---|---|---|---|---|---|---|
| 0–25 | 26–40 | 41–50 | 51–60 | 61–75 | >75 | ||
| Observed number of promoters | 10,028 | 3,306 | 2,205 | 2,674 | 4,888 | 11,056 | 34,157 |
| Observed proportion of promoters (%)a | 29.36 | 9.68 | 6.46 | 7.83 | 14.41 | 32.37 | 100 |
| Expected number of promoters | 5 | 2,272 | 14,155 | 14,493 | 3,187 | 38 | 34,157 |
| Expected proportion of promoters (%) | 0.01 | 6.65 | 41.44 | 42.43 | 9.33 | 0.11 | 100 |
| Pearson’s chi-square statistic | 861.42 | 0.01 | 0.30 | 0.28 | 0.03 | 94.61 | 956.65 |
aExpected proportion is calculated based on a Poisson distribution with mean parameter equal to 51.
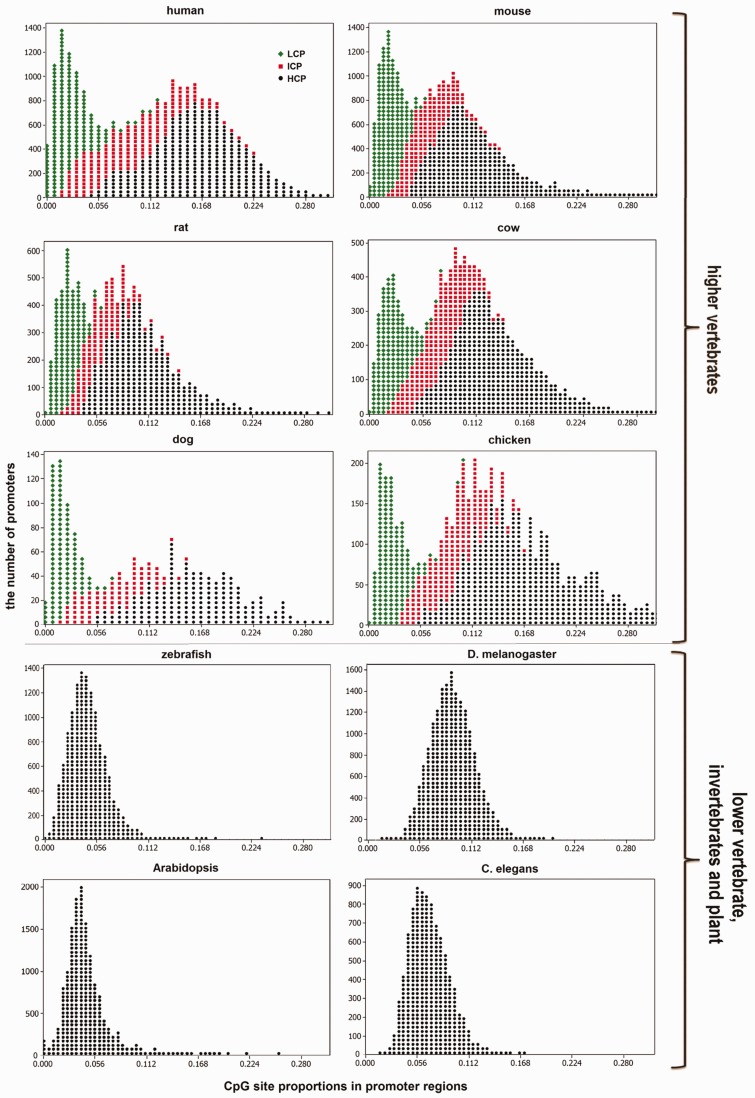
To further characterize the nonrandom distribution of CpGs in higher vertebrate promoters, we looked at the occurrence of “CpG islands,” which are recognized as small dispersed regions of DNA sequence that contain highly dense clusters of CpG dinucleotides relative to the whole genome. The widely accepted definition of a CpG island is a genomic region at least 200 bp in length, with GC content fraction >50% and an observed/expected CpG percentage ratio of >60% (Gardiner-Gardner and Frommer 1987). Among the 34,257 annotated promoters of the human genome, we found 21,890 (63.9%) promoters containing CpG islands, whereas the other 12,367 (36.1%) have only few CpG dinucleotides. For the other five higher vertebrate species, CpG islands were detected in over half of their annotated promoters. The density of CpG sites in the promoters of all six higher vertebrates showed a bimodal distribution (fig. 2), which was reported previously only in the human genome (Saxonov et al. 2006; Glass 2007). In contrast, no CpG islands were found in the four lower vertebrate, invertebrate, or plant genomes, and a unimodal distribution of CpG sites was observed (fig. 2). This difference cannot be attributed to the difference in the GC content distribution between the two groups because the distribution does not clearly differ between the two groups (supplementary fig. S1, Supplementary Material online). We proceeded to explore the functional roles of DNA methylation in regulating gene expression and attempted to explain the bimodal distribution pattern of CpGs in the promoters of higher vertebrates.
Fig. 2.—
Proportion of CpG sites in gene promoters across ten model species. The horizontal axis represents the proportion of CpG sites in gene promoters, whereas the vertical axis represents the number of promoters for each model species. Color is used to distinguish HCP (black), LCP (green), and ICP (red).
We classified gene promoters of the higher vertebrate species into two main groups as previously defined for all human genes according to the GC fraction and observed to expected ratio of CpG sites (O/E), as in Weber (2007). First is the HCP with GC fraction ≥55% and CpG O/E ≥65%; second is the LCP with GC fraction <45% and CpG O/E <65%. The remaining genes were not assigned into either HCP or LCP group and were grouped as the ICP, as in previous work (Saxonov et al. 2006; Weber 2007). For each of the six higher vertebrates, promoters were classified as HCP (∼50% of promoters), LCP (∼25%), or ICP (∼25%). We investigated the extent to which the annotated CpG islands overlapped with each of the three types of promoter in the six higher vertebrate genomes using the CpG islands data set downloaded from the UCSC annotation database. A promoter was recognized to contain a CpG island if the CpG island covered more than 25% of the promoter region. Between 50% and 63% of all promoters in the higher vertebrate genomes contain CpG islands as shown in supplementary table S3, Supplementary Material online. CpG islands are significantly overrepresented in the HCP (P < 0.005, Mann–Whitney U test), over 80% of which contained CpG islands. In contrast, CpG islands are significantly underrepresented in the LCP (P < 0.004, Mann–Whitney U test) and there are only a few (<6%) LCP containing CpG islands. Additionally, the distribution of CpG islands in ICP does not differ from the distribution of CpG islands in all promoters (P = 0.471, Mann–Whitney U test). In the following analyses, we focused on the two most divergent classes (HCP and LCP).
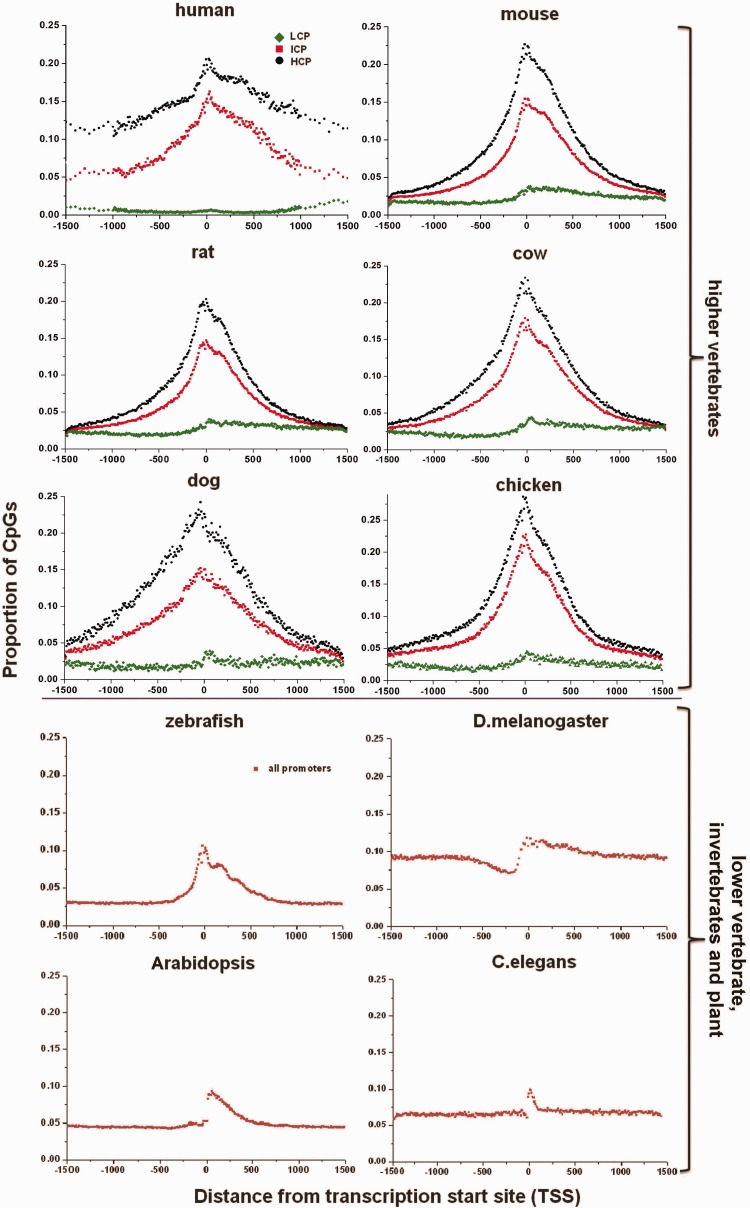
A striking difference was apparent between HCP and LCP, both for the GC content fraction and the occurrence of CpG sites in relation to the transcription start site (TSS) (fig. 3 and supplementary fig. S3, Supplementary Material online). For the HCP in the higher vertebrates, both the proportion of CpG sites and the GC content fraction peaked consistently in the vicinity of the TSS and declined with increasing distance from the TSS. On the other hand, the proportions of CpG sites in LCP were approximately zero, despite a mild peak for the GC content fraction occurring immediately downstream of the TSS. These results indicate a high level of conservation of CpG site distribution among higher vertebrate species, suggestive of an important biological function. For zebrafish, the patterns of GC content fraction and CpG site density at all promoters were similar to those of the HCP of the higher vertebrates. The pattern was noticeably different for the invertebrate species, with the GC content fraction and CpG density exhibiting a sharp peak immediately downstream of the TSS, but either a flat curve (Arabidopsis and C. elegans), or surprisingly a valley (D. melanogaster), upstream of the TSS (fig. 3).
Fig. 3.—
Distribution of CpGs with respect to the TSS. The horizontal axis represents the distance from the TSS, whereas the vertical axis represents the CpG fraction.
Evolutionary Conservation of Promoter HCP or LCP Status in Higher Vertebrates
To further explore the level of conservation of promoter status in the six higher vertebrates, we grouped homologous genes between each pair of the six higher vertebrate species into a conserved pair if the genes were classified into the same category of either HCP or LCP (table 3). The number of genes in each category for each of the six higher vertebrate species is given on the diagonal, whereas the off-diagonal elements show the proportion of conserved homologous genes between the corresponding species pair. For example, 93.7% of genes with HCP in human also had HCP in mouse, whereas 97.6% of genes with HCP in mouse also had HCP in human. It can be seen from table 3 that the HCP or LCP status of promoters is highly conserved among homologous genes in mammals. Between 77.9% and 97.6% of homologous genes among the five mammals are conserved in either HCP or LCP categories. A similar level of conservation is observed between the five mammals and the bird (chicken) for genes with HCP. Although the level of conservation is reduced to a range of 33–56% between the mammals and chicken for genes with LCP, it is still significantly higher than the conservation of LCP status in the promoters of nonhomologous (randomly paired) genes (P = 0.022, Mann–Whitney U test).
Table 3.
Conservation of Two Classes of Promoter in Higher Vertebrates
| Proportion of Conserved HCP (%) |
Proportion of Conserved LCP (%) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Rat | Cow | Dog | Chicken | Human | Mouse | Rat | Cow | Dog | Chicken | |
| Human | 7,139 | 97.6 | 97.4 | 96.9 | 88.8 | 85.7 | 2,895 | 86.7 | 87.3 | 79.5 | 89.8 | 42.1 |
| Mouse | 93.7 | 8,097 | 96.7 | 93.7 | 85.8 | 84.7 | 85.1 | 4,365 | 89.9 | 83.2 | 87.4 | 48.2 |
| Rat | 91.9 | 92.6 | 5,634 | 90.4 | 83.4 | 89.7 | 85.0 | 94.6 | 2,596 | 86.9 | 77.9 | 56.3 |
| Cow | 93.6 | 94.1 | 95.2 | 1,536 | 84.5 | 84.3 | 90.6 | 96.0 | 79.3 | 577 | 88.9 | 54.8 |
| Dog | 82.2 | 80.5 | 84.9 | 86.3 | 435 | 80.6 | 81.1 | 90.6 | 87.2 | 97.1 | 187 | 40.0 |
| Chicken | 89.6 | 87.0 | 89.7 | 84.4 | 82.5 | 913 | 47.3 | 53.6 | 51.6 | 53.7 | 33.3 | 251 |
Note.—The diagonal cells show the number of genes with HCP or LCP in each species. The upper and lower triangles show the percentage of genes in the column species also given the same classification for the row species.
To compare the conservation level of HCP or LCP status of the promoter among homologous genes between any pair of the six higher vertebrates, we explored the difference between the corresponding rates of conservation. We found that HCP were considerably more conserved between species compared with LCP (P = 0.006, Mann–Whitney U test) (table 3). Thus, HCP have been more conserved over evolutionary time than LCP among six higher vertebrates. We next investigated whether the differential conservation of HCP versus LCP status was associated with differential conservation at the sequence level. Table 4 summarized two substitution rate statistics to compare the sequence level evolution of homologous genes with either HCP or LCP conserved status. K80 is the rate of nucleotide substitution in promoter regions under the Kimura 80 of promoter sequence evolution, whereas Ka/Ks is the ratio of nonsynonymous and synonymous substitution rate, which measures protein evolution and is a possible indicator of selection pressure. The results clearly showed that both K80 and Ka/Ks values varied significantly between HCP and LCP conserved homologous gene groups (P < 0.001 in all cases, student’s t-test). Both of these measurements suggested that homologous genes with conserved HCP status were more highly conserved at the sequence level than those with LCP status.
Table 4.
Means and Standard Errors for Two Substitution Rate Statistics of Homologous Genes with Conserved Promoter Status
| Homologous Genes with Conserved HCP |
Homologous Genes with Conserved LCP |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Rat | Cow | Dog | Chicken | Human | Mouse | Rat | Cow | Dog | Chicken | |
| Human | 0.63 ± 0.005 | 0.62 ± 0.006 | 0.63 ± 0.011 | 0.54 ± 0.019 | 0.76 ± 0.008 | 0.74 ± 0.012 | 0.71 ± 0.016 | 0.72 ± 0.029 | 0.63 ± 0.063 | 0.89 ± 0.028 | ||
| Mouse | 0.10 ± 0.001 | 0.21 ± 0.004 | 0.71 ± 0.008 | 0.63 ± 0.040 | 0.91 ± 0.010 | 0.22 ± 0.005 | 0.34 ± 0.007 | 0.77 ± 0.013 | 0.77 ± 0.040 | 1.05 ± 0.013 | ||
| Rat | 0.10 ± 0.001 | 0.09 ± 0.001 | 0.70 ± 0.009 | 0.67 ± 0.080 | 0.90 ± 0.010 | 0.20 ± 0.007 | 0.24 ± 0.005 | 0.79 ± 0.009 | 0.83 ± 0.041 | 1.03 ± 0.018 | ||
| Cow | 0.13 ± 0.004 | 0.10 ± 0.003 | 0.11 ± 0.003 | 0.45 ± 0.046 | 0.83 ± 0.025 | 0.29 ± 0.015 | 0.22 ± 0.007 | 0.19 ± 0.008 | 0.62 ± 0.015 | 1.01 ± 0.033 | ||
| Dog | 0.08 ± 0.007 | 0.05 ± 0.004 | 0.06 ± 0.005 | 0.07 ± 0.013 | 0.78 ± 0.049 | 0.16 ± 0.031 | 0.15 ± 0.018 | 0.16 ± 0.019 | 0.20 ± 0.044 | 0.99 ± 0.025 | ||
| Chicken | 0.09 ± 0.003 | 0.09 ± 0.003 | 0.08 ± 0.003 | 0.11 ± 0.009 | 0.07 ± 0.009 | 0.18 ± 0.020 | 0.15 ± 0.011 | 0.15 ± 0.011 | 0.19 ± 0.023 | 0.13 ± 0.040 | ||
Note.—The upper triangles show the rates of nucleotide substitution under the K80 in promoter regions for paired homologous genes with conserved promoter status (mean ± standard error). The lower triangles show the ratio of nonsynonymous and synonymous substitution rates (Ka/Ks) in protein-coding regions for paired homologous genes with conserved promoter status (mean ± standard error).
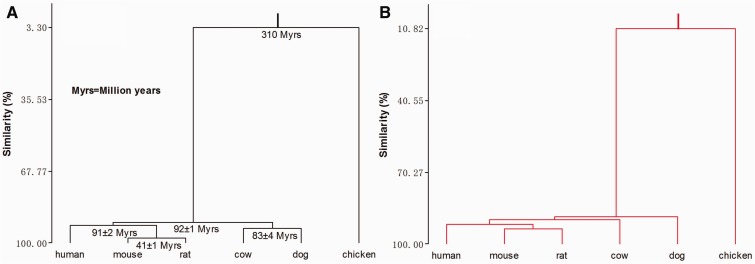
Divergence in the conservation level of promoter status reflected evolutionary divergence between the species. We therefore reconstructed the phylogeny among these species using the conservation level of promoter status and compared it with the phylogeny constructed from DNA/protein sequence data of each species (Hedges 2002). Figure 4 shows that the two phylogenetic trees are remarkably similar. The main discrepancy between the two trees occurs at the point where the dog links into the phylogenies. In our tree based on promoter status conservation level, the dog species diverged prior to all of the other mammals (fig. 4B), whereas in the tree based on DNA and protein sequence data (fig. 4A), the dog and cow diverged from the other three mammals around 92 Ma, before the two separated around 83 Ma (Hedges 2002). This discrepancy can most likely be attributed to the poor quality of sequence annotation for the dog genome. In fact, promoters have been identified for only 11% (1,481/13,410) of all dog genes.
Fig. 4.—
Phylogenies of six higher vertebrate species reconstructed either from DNA and protein sequence data (A) or from conservation level of HCP or LCP status in gene promoters (B).
Distinct Methylation Patterns between HCP and LCP
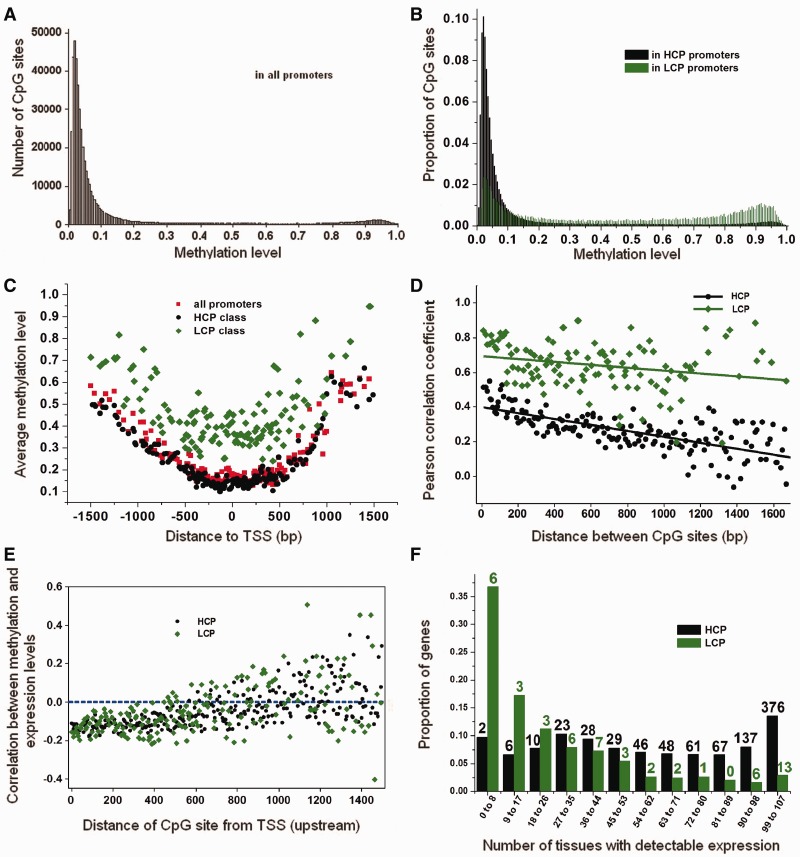
We analyzed genome-wide DNA methylation profiles from 28 different human tissues (or cell lines), which were assayed by the Illumina Human Methylation27 BeadChip platform (Bonazzi et al. 2011; Chari et al. 2011; Loudin et al. 2011). This BeadChip assessed 27,578 CpG sites located within the promoter regions of 14,475 genes. Multiple sites (on average, two CpGs) were interrogated per promoter region. We confirmed that CpG sites have much lower methylation levels in promoter regions when compared with the genome-wide average, as previously shown (Lister et al. 2009). Figure 5A shows a slightly bimodal distribution: The majority (72.7%) of CpG sites in all promoter regions across 28 tissues were unmethylated (methylation level ≤ 0.1), whereas 18.5% were semimethylated (methylation level between 0.1 and 0.7) and 8.8% were considered methylated (methylation level ≥ 0.7), according to the criteria established in Bell et al. (2011). The distribution of methylation levels showed two distinct patterns for HCP compared with LCP (fig. 5B). The HCP showed a unimodal distribution, with 77.1% unmethylated, 16.6% semimethylated and 6.3% methylated CpG sites, whereas the CpG sites in LCP tended to be more highly methylated, with corresponding proportions of 25.8%, 37.9% and 36.3%. Both HCP and LCP showed a similar distribution of methylation levels with respect to distance from the TSS (fig. 5C). The lowest methylation levels are found in the vicinity of the TSS, whereas the methylation level increases with increasing distance from the TSS. However, CpG sites in the LCP showed consistently higher methylation levels than those in the HCP throughout the promoter region. Within an individual promoter, the methylation levels of adjacent CpG pairs were positively correlated, and the correlation tends to be weakened when the CpG pairs are distantly separated (fig. 5D). Moreover, the CpG pairs within LCP exhibited a higher correlation in methylation levels compared with HCP across all distances.
Fig. 5.—
Methylation and gene expression patterns across 28 human tissues. Methylation levels of CpG sites in all promoters (A), and in HCP and LCP (B), across 28 different human tissues. The average methylation levels with respect to the TSS, with each point representing the average methylation level in an interval of 10 bp (C). The correlation of methylation levels between all pairwise CpGs sites in the same promoter, with each point showing the average correlation in 10-bp intervals according to the distance between CpG sites (D). The correlation coefficient between methylation and gene expression level with increasing distance from the TSS (E). Distribution of the number of tissues in which HCP and LCP genes are expressed. Each bar is labeled with the corresponding number of expressed housekeeping genes as identified in Zhu et al. (2008) (F).
The corresponding results based on the mouse promoter methylation information were obtained from bisulfite-seq data and are summarized in supplementary figure S3A–D, Supplementary Material online. Bisulfite-seq can more precisely detect the methylation level of CpG sites than the Methylation BeadChip platform based on hybridization technology. Moreover, the bisulfite-seq data could simultaneously measure the methylation level for over 1 million CpG sites in mouse promoter regions, whereas the Human Methylation BeadChip can detect the methylation level for only approximately 27,000 CpG sites in human promoter regions. Despite these differences, the results from mouse bisulfite-seq data were similar to those obtained from the human Methylation BeadChip data. The overall methylation pattern of CpG sites in promoters also showed a bimodal distribution (supplementary fig. S3A, Supplementary Material online). Furthermore, the CpG sites in LCP exhibited higher methylation levels than the CpG sites in HCP (supplementary fig. S3B, Supplementary Material online). The methylation pattern with respect to distance from the TSS (supplementary fig. S3C, Supplementary Material online) and distance between adjacent CpG pairs (supplementary fig. S3D, Supplementary Material online) in mouse HCP and LCP was consistent with the pattern identified in human promoters (fig. 5). Overall, the distinct methylation patterns between HCP and LCP genes are consistent between the two species, indicating a remarkable level of conservation between HCP and LCP over evolutionary time.
Distinct Expression Patterns between HCP and LCP across 107 Human Tissues and 17 Mouse Tissues
We next investigated the relationship between promoter DNA methylation and gene expression levels in human and mouse tissues. The human promoter methylation data were collected from the 28 tissues as analyzed above, and gene expression was measured across 107 human tissues (including the same 28 tissues) using Affymetrix U133 human expression microarrays (Johansson et al. 2007; Bell et al. 2011; Chari et al. 2011). The mouse promoter methylation and gene expression data were measured across a set of 17 tissues of C57Bl/6 mice, using whole-genome bisulfite-seq on the Illumina Hiseq2000 platform, and the Affymetrix mouse genome 430 2.0 GeneChip, respectively. First, we observed a clear negative correlation (from −0.05 to −0.18) between the gene expression level and methylation level of each profiled CpG site across the 28 human tissues (fig. 5E). This correlation was confined to CpGs located in the core and proximal promoter regions (0–250 bp upstream of the TSS), with the average correlation coefficient equal to −0.10 and −0.12 in HCP and LCP, respectively. For the CpG sites located further upstream (>250 bp) from the TSS, the strength of correlation decreased and no obvious relationship with gene expression level was apparent. No differences in the correlation were observed between LCP and HCP. The corresponding results based on the mouse data were shown in supplementary figure S3E, Supplementary Material online. There is a similar negative correlation pattern between gene expression level and methylation level of CpG sites located in the core and proximal promoter regions, with the average correlation coefficient equal to −0.04 and −0.16 in HCP and LCP, respectively. Therefore, methylation of CpG sites in the core and proximal promoter regions must play a crucial role in regulating gene expression levels in both human and mouse.
Next, we compared the number of tissues from which each gene was detectably expressed, from a total of 107 human tissues (fig. 5F). The difference between LCP and HCP genes was striking. Genes with LCP tended to be expressed in only a small number of tissues compared with genes with HCP. More than 35% of genes with LCP were expressed in no more than eight tissues, whereas only less than 5% were expressed in 99–107 tissues. On the other hand, genes with HCP showed a reasonably uniform distribution (from 0 to 107) for the number of tissues in which they were expressed, and approximately 15% of genes were expressed in 99–107 tissues. The corresponding gene expression results from 17 mouse tissues were shown in supplementary figure S3F, Supplementary Material online. A similar expression pattern was found in mouse data that the genes with LCP tended to be expressed in fewer tissues compared with genes with HCP. More than 32% of genes with LCP were expressed in no more than one tissue, whereas only 6% were expressed in all 17 tissues. The genes with LCP were therefore more likely to be “tissue-specific,” a finding consistent with our earlier observation of increased divergence between mammals and chicken for genes in the LCP group (table 3). Meanwhile, genes with HCP were more likely to be “housekeeping” genes, expressed in many different tissues or all tissues to maintain cellular functions. In fact, among 885 housekeeping genes identified in the human genome (Zhu et al. 2008) that were also present in the gene expression data set, only 5.9% had LCP, whereas 94.1% had HCP. We further investigated the expression patterns of these 885 annotated housekeeping genes in 107 different human tissues. In figure 5F, the number labeled above each bar represented the corresponding number of expressed housekeeping genes in different expression categories. It indicated that the annotated housekeeping genes tend to be expressed in a broad range of human tissues. For instance, 376 (42.4%) of these annotated housekeeping genes have HCP and were expressed in almost all (99–107) human tissues.
Troukhan et al. (2009) reported that the expression of genes with TATA-boxes tends to be tissue specific, whereas genes without TATA-boxes tend to be expressed more broadly. We investigated the distribution of TATA-boxes in different classes (categories) of promoters for each of the six higher vertebrate species, and the analysis was summarized in supplementary table S4, Supplementary Material online. It shows that only a small proportion of promoters contained the canonical TATA-box in higher vertebrates. For instance, only about 13.8% of promoters contained a TATA-box in the human genome, consistent with a previous report showing a minority of mammalian promoters having the TATA-box architecture and about 10% of promoters having TATA-boxes in the human genome (Yang et al. 2007). Furthermore, we observed a marked difference in the TATA-box structure between HCP and LCP. In the genomes of the six higher vertebrates, the TATA-box was significantly enriched in LCP in comparison to HCP (P < 0.001, one-tailed paired student’s t-test).
Distinct and Conserved Functions of Genes with HCP or LCP
Functional annotation of the human genome into GO terms led to the discovery that promoters with CpG islands are more likely to be associated with genes performing basic cellular functions, whereas promoters without CpG islands are associated with genes delivering tissue-specific functions (Larsen et al. 1992; Ponger et al. 2001; Saxonov et al. 2006). We carried out a binomial test to identify overrepresentation of GO classes for genes with HCP versus LCP. From the six higher vertebrates, we found approximately 100 GO terms significantly overrepresented in either HCP or LCP groups. Genes with LCP were particularly enriched for functions (GO terms) characteristic of differentiated or highly regulated cells, for example immunological functions, whereas those with HCP were enriched for more basic cellular processes, such as regulation of transcription and cell cycle activity. Comparison of significant GO terms among the six higher vertebrate species allowed us to identify GO terms shared by at least four species as “conserved” terms. Accordingly, 16 and 12 GO terms were identified as conserved in HCP and LCP gene groups, respectively (table 5). As expected, these conserved GO terms were enriched in tissue-specific functions for the LCP group and enriched in housekeeping functions for the HCP genes.
Table 5.
Conserved and Overrepresented GO Terms for Genes with HCP and LCP in Six Higher Vertebrates
| GO ID | Conservationa | Subontology | GO Term Description |
|---|---|---|---|
| Overrepresented among genes with HCP | |||
| 0000122 | 4 | BP | Regulation of transcription from RNA polymerase promoter |
| 0003676 | 4 | MF | Nucleic acid binding |
| 0003677 | 4 | MF | DNA binding |
| 0003723 | 4 | MF | RNA binding |
| 0004672 | 4 | MF | Protein kinase activity |
| 0004930 | 4 | MF | G-protein coupled receptor activity |
| 0005634 | 4 | CC | Nucleus |
| 0005730 | 4 | CC | Nucleolus |
| 0006915 | 4 | BP | Apoptotic process |
| 0016021 | 4 | CC | Integral to membrane |
| 0016301 | 4 | MF | Kinase activity |
| 0043234 | 4 | CC | Protein complex |
| 0043565 | 5 | MF | Sequence-specific DNA binding |
| 0044212 | 4 | MF | Transcription regulatory region DNA binding |
| 0045892 | 4 | BP | Negative regulation of transcription, DNA-dependent |
| 0045893 | 4 | BP | Positive regulation of transcription, DNA-dependent |
| Overrepresented among genes with LCP | |||
| 0004869 | 4 | MF | Cysteine-type endopeptidase inhibitor activity |
| 0004984 | 4 | MF | Olfactory receptor activity |
| 0006955 | 4 | BP | Immune response |
| 0006958 | 5 | BP | Complement activation, classical pathway |
| 0006974 | 4 | BP | Response to DNA damage stimulus |
| 0007596 | 4 | BP | Blood coagulation |
| 0007601 | 4 | BP | Visual perception |
| 0008009 | 4 | MF | Chemokine activity |
| 0008270 | 4 | MF | Zinc ion binding |
| 0009897 | 4 | CC | External side of plasma membrane |
| 0015711 | 4 | BP | Organic anion transport |
| 0032729 | 4 | BP | Positive regulation of interferon-gamma production |
Note.—CC, cellular component; BP, biological process; MF, molecular function.
aThe number of higher vertebrate species for which the corresponding GO term is overrepresented.
We also explored the association of genes with either HCP or LCP with annotated tumor suppressor genes. So far there are 861 annotated tumor suppressor genes for over 54 different human tumors in the most up-to-date TSGene database (Tumor Suppressor Gene database, http://bioinfo.mc.vanderbilt.edu/TSGene/, last accessed November 1, 2014). Of the 861 suppressors, 365 can be mapped uniquely to one class in our annotated promoter data set (supplementary table S5, Supplementary Material online). Among tumor suppressor genes, 91.2% contained CpG islands, with 70%, 23% and 7% of the 365 suppressors in the HCP, ICP or LCP groups, respectively, showing a significant association of tumor suppressor genes with HCP (P < 0.001, Pearson’s chi-square test). Additionally, we investigated the methylation level in promoters of the 365 tumor suppressor genes across 28 human tissues or cell lines, of which 18 were tumor tissues or tumor cell lines. We found that the pattern of methylation level in the promoter regions of the tumor suppressors was comparable to that of other nonsuppressor genes. In addition, the LCP had a much higher level of methylation than the HCP in the tumor suppressor genes, particularly in the region surrounding the TSS. The CpG sites in tumor suppressor gene HCP trend to have markedly higher methylation levels in the tumor samples compared with nontumor samples (supplementary fig. S4, Supplementary Material online), which is consistent with widely observed methylation of tumor suppressor gene promoters occurring in human cancers (Esteller 2002).
Discussion
DNA methylation has an essential role in the modulation of gene transcription in eukaryotic species, particularly vertebrates (Antequera and Bird 1993; Bennetzen et al. 1994; Attwood et al. 2002; Zilberman and Henikoff 2007; Suzuki and Bird 2008). Although several studies have explored the relationship between regulation of gene transcription by DNA methylation and the CpG content of gene promoters (Boyes and Bird 1992; Hsieh 1999; Robinson et al. 2004; Robertson 2005; Weber 2007), studies in the current literature have been either based on limited data sets or focused only on analysis of the human genome. Our study presents the first comprehensive and comparative investigation of the DNA methylation system and its impact on gene transcription between ten model eukaryotic species, including higher vertebrates, a lower vertebrate, invertebrates, and a plant.
Our analysis revealed that the genome-wide distribution patterns of GC content and CpG dinucleotides vary dramatically for higher vertebrates compared with lower vertebrate, invertebrate, and plant species. In higher vertebrates, both the GC content and CpG dinucleotides were consistently enriched in functional regions of the genome, particularly in promoter regions, compared with putative “nonfunctional” regions, including introns and intergenic sequences. This pattern may be explained by the following two observations. First, methylated cytosines have a higher probability than unmethylated cytosines to be converted to thymine over evolutionary time (Ehrlich et al. 1982; Kerry Lee 2001). Second, nearly all CpG sites from nonfunctional sequences are completely methylated in higher vertebrate species. Functional constraints within genic regions would limit the frequency of such mutations. However, we did not detect this pattern for lower vertebrate, invertebrate, or plant species; instead, CpG dinucleotides were enriched across all regions of the genome, meaning that we could not detect evidence of higher levels of functional constraint in putative “functional” compared with putative nonfunctional regions of these genomes, though the reasons for this are unclear.
Focusing next on gene promoters, we discovered that far from being randomly distributed within the promoter sequence, CpG dinucleotides consistently showed a bimodal distribution pattern in each of the six higher vertebrate species (human, mouse, rat, cow, dog, and chicken). The previously defined “CpG rich” promoters (HCP) and “CpG poor” promoters (LCP) could be observed in all six higher vertebrate species, but not in the lower vertebrate, invertebrate, or plant species. For both groups of genes, CpGs were concentrated in the core and proximal promoter regions. Furthermore, the classification of genes into HCP or LCP groups was highly conserved among the homologous genes of the six higher vertebrate species. Indeed, the level of conservation of promoter sequences between species could be used to accurately reconstruct the evolutionary relationships between these species. Remarkably, we found that genes with HCP have significantly higher levels of conservation among vertebrates, in both promoter and protein-coding sequences, compared with genes with LCP. This indicates that among vertebrates, genes with HCP are likely to be under stronger purifying selection pressure than genes with LCP. All of these observations led us to conclude that the DNA methylation system is highly conserved among higher vertebrate species and to further explore a functional role for the distribution of the CpG dinucleotides within promoter sequences.
DNA methylation of CpGs within both HCP and LCP of the human and mouse genome is nonrandom; the level of methylation across the length of the promoter shows a u-shaped distribution, with the lowest levels corresponding with the core promoter regions. This distribution is likely to facilitate transcription initiation, whereas the increased methylation level in the proximal and distal promoter regions could modulate transcription by modulating the binding of transcription factors. Methylation, specifically in the core and proximal promoter regions, negatively regulated the gene expression level across multiple human and mouse tissues and human cell lines. This could be explained by the physical distribution of protein-binding sites in promoter regions; the binding sites for RNA polymerase and most essential transcription factors are located in the core and proximal promoter regions (Koudritsky and Domany 2008), whereas only few additional transcription factor-binding sites are located in the distal promoter region (>250 bp upstream of the TSS).
Moreover, we discovered distinct characteristics of HCP and LCP that ultimately relate to their underlying biological functions. The level of CpG methylation was consistently higher within LCP compared with HCP. Methylation levels of CpGs within the same promoter were highly correlated among different cell types or tissues, particularly for two CpGs located in close proximity. These differences in the pattern of DNA methylation between the two classes of promoter were reflected in different patterns of gene expression. Genes with HCP were expressed in a broader range of tissues, and were associated using GO analysis with housekeeping functions, whereas genes in the LCP group were enriched in tissue-specific functions. We further discovered that 94% of annotated housekeeping genes contained HCP, confirming previous reports of HCP being more frequently associated with housekeeping genes expressed in a large number of tissues, whereas LCP are associated with tissue-specific genes (Larsen et al. 1992; Ponger et al. 2001; Saxonov et al. 2006; Weber 2007). Moreover, we observed a higher level of conservation in both coding and promoter sequences in the HCP genes than in the LCP genes among six higher vertebrates. This agrees with the observation that housekeeping genes in mice and human evolve more slowly than tissue-specific genes (Zhang and Li 2004), which can also be associated with the increased breadth of expression of such genes compared with tissue-specific genes (Park and Choi 2010). In addition, tissue-specific genes tend to locate in late replicating regions of the human genome (Cui et al. 2012), which may also contribute to a higher mutation rate compared with housekeeping genes. In conclusion, for genes with HCP, the DNA methylation system regulates the expression level in a wide spectrum of tissues, whereas for genes with LCP, the DNA methylation system provides a functional “on–off” switch to determine whether the gene is expressed or not. Most importantly, we have shown here that this relationship is conserved among all six model higher vertebrate species.
VanderKraats et al. (2013) performed a comprehensive analysis of the relationship between methylation around the TSS region and gene expression using high-resolution RNA sequencing and DNA methylation sequencing data in several human tumor and normal tissues. They observed that hyper- or hypomethylation spanning the TSS may negatively correlate with gene expression changes in tumor and normal tissues. This study confirms this observation on a larger scale covering six vertebrate genomes, and further reveals evolutionary conservation of the methylation pattern surrounding the TSS between the two distinct promoter groups. VanderKraats et al. observed that gene expression could be negatively regulated by methylation downstream of the TSS (mainly within 3 kb of the TSS), though this methylation pattern was only seen in a small group of genes (37 genes). In this study, we did not observe this phenomenon. This long distance negative regulation of gene expression may not be directly due to methylation but to a repressive chromatin environment in the promoters of these genes (Hon et al. 2012).
This study has focused on methylation in promoter regions and its impact on repression of gene expression. It has been reported that gene body methylation is not associated with repression of gene expression in vertebrates (Jones 2012). In support of this, our analysis showed that the average methylation level downstream of the TSS in human and mouse genomes could become very high (even in genes with HCP), but it did not in turn repress gene transcription (fig. 5C and supplementary fig. S3C, Supplementary Material online). Recently, many studies have reported a positive relationship between gene-body methylation and gene expression levels in nonvertebrate species including Arabidopsis (Cokus et al. 2008), silkworm (Xiang et al. 2010), honeybee (Foret et al. 2012), and several eukaryotic species (Zemach et al. 2010). For vertebrates, a positive relationship between alternative splicing and gene body methylation has been reported in both human (Anastasiadou et al. 2011; Shukla et al. 2011) and mouse (Wan et al. 2013) genomes. Therefore gene body methylation may have a significant role in the repression of gene expression in nonvertebrate genomes, and in the regulation of alternative splicing in vertebrate genomes. We also investigated whether there is a relationship between genic GC3 and promoter methylation in the species under study here. For each gene with an annotated promoter, the GC3 content in the coding region was calculated as GC3 = (C3 + G3)/(L/3), where C3 and G3 were counts of cytosine and guanine in the third position of codons and L was the length of the coding region (Tatarinova et al. 2013). Across the six higher vertebrates, the Pearson’s correlation coefficient between genic GC3 content and the promoter CpG density was only weak and varied from −0.05 to 0.10. In contrast, it is reported that GC3-rich genes are usually tissue specific, whereas GC3-poor genes are usually housekeeping in rice, bee, and Arabidopsis genomes (Tatarinova et al. 2013).
Supplementary Material
Supplementary figures S1–S4 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to two anonymous reviewers for their constructive criticisms which have helped improve both context and presentation of an earlier version of the article. This work was supported by research grants from the National Basic Research Program of China (grant number 2012CB316505), the National Natural Science Foundation of China (grant numbers 81172006, 91231114, and 31401126), China Postdoctoral Science Foundation (grant number 2014M561406), and the Leverhulme Trust and BBSRC, United Kingdom to Z.W.L.
Literature Cited
- Anastasiadou C, Malousi A, Maglaveras N, Kouidou S. Human epigenome data reveal increased CpG methylation in alternativelyspliced sites and putative exonic splicing enhancers. DNA Cell Biol. 2011;30(5):267–275. doi: 10.1089/dna.2010.1094. [DOI] [PubMed] [Google Scholar]
- Antequera A, Bird A. DNA methylation: molecular biology and biological significance. Basel (Birkhauser): Nature Publishing Group. 1993 [Google Scholar]
- Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene transcription. Cell Mol Life Sci. 2002;59:241–257. doi: 10.1007/s00018-002-8420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Schrick K, Springer PS, Brown WE, SanMiguel P. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bonazzi VF, et al. Cross-platform array screening identifies COL1A2, THBS1, TNFRSF10D and UCHL1 as genes frequently silenced by methylation in melanoma. PLoS One. 2011;6:e26121. doi: 10.1371/journal.pone.0026121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Chari R, Coe B, Vucic E, Lockwood W, Lam W. An integrative multi-dimensional genetic and epigenetic strategy to identify aberrant genes and pathways in cancer. BMC Syst Biol. 2011;4:67. doi: 10.1186/1752-0509-4-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z-X, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286:18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P, et al. Distinct contributions of replication and transcription to mutation rate variation of human genomes. Genomics Proteomics Bioinformatics. 2012;10(1):4–10. doi: 10.1016/S1672-0229(11)60028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucl Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- Foret S, et al. DNA methylation dynamics, metabolic fluxes, genesplicing, and alternative phenotypes in honey bees. Proc Natl Acad Sci U S A. 2012;109:4968–4973. doi: 10.1073/pnas.1202392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Gardner M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gehring M, Henikoff S. DNA methylation dynamics in plant genomes. Biochim Biophys Acta. 2007;1769:276–286. doi: 10.1016/j.bbaexp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation—how does it all fit together? J Cell Biochem. 2002;87:117–125. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- Glass JL. CG dinucleotide clustering is a species-specific property of the genome. Nucl Acids Res. 2007;35:6798–6807. doi: 10.1093/nar/gkm489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gowher H, Leismann O, Jeltsch A. DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J. 2000;19:6918–6923. doi: 10.1093/emboj/19.24.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X-J, Chen T, Zhu J-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Mechanisms of disease: gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Hon GC, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45(10):1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL. Evidence that protein binding specifies sites of DNA demethylation. Mol Cell Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Johansson P, Pavey S, Hayward N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 2007;20:216–221. doi: 10.1111/j.1600-0749.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrazek J. Compositional differences within and between eukaryotic genomes. Proc Natl Acad Sci U S A. 1997;94:10227–10232. doi: 10.1073/pnas.94.19.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerry Lee T. Methylated cytosine and the brain: a new base for neuroscience. Neuron. 2001;30:649–652. doi: 10.1016/s0896-6273(01)00325-7. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Koudritsky M, Domany E. Positional distribution of human transcription factor binding sites. Nucl Acids Res. 2008;36:6795–6805. doi: 10.1093/nar/gkn752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudin MG, et al. Genomic profiling in Down syndrome acute lymphoblastic leukemia identifies histone gene deletions associated with altered methylation profiles. Leukemia. 2011;25:1555–1563. doi: 10.1038/leu.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero LM. The distribution of 5-methylcytosine in the nuclear genome of plants. Nucl Acids Res. 1992;20:3207–3210. doi: 10.1093/nar/20.12.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LE. Maize genome sequencing by methylation filtration. Science. 2003;302:2115–2117. doi: 10.1126/science.1091265. [DOI] [PubMed] [Google Scholar]
- Park SG, Choi SS. Expression breadth and expression abundance behave differently in correlations with evolutionary rates. BMC Evol Biol. 2010;10:241. doi: 10.1186/1471-2148-10-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parle-McDermott A, Harrison A. DNA methylation: a timeline of methods and applications. Front Genet. 2011 doi: 10.3389/fgene.2011.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra S, Patra A, Rizzi F, Ghosh T, Bettuzzi S. Demethylation of (Cytosine-5-C-methyl) DNA and regulation of transcription in the epigenetic pathways of cancer development. Cancer Metastasis Rev. 2008;27:315–334. doi: 10.1007/s10555-008-9118-y. [DOI] [PubMed] [Google Scholar]
- Ponger L, Duret L, Mouchiroud D. Determinants of CpG Islands: expression in early embryo and isochore structure. Genome Res. 2001;11:1854–1860. doi: 10.1101/gr.174501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzoli U, et al. Both selective and neutral processes drive GC content evolution in the human genome. BMC Evol Biol. 2008;8:99. doi: 10.1186/1471-2148-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD. X-inactivation, differentiation and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Robinson PN, Bohme U, Lopez R, Mundlos S, Nurnberg P. Gene-Ontology analysis reveals association of tissue-specific 5′CpG-island genes with development and embryogenesis. Hum Mol Genet. 2004;13:1969–1978. doi: 10.1093/hmg/ddh207. [DOI] [PubMed] [Google Scholar]
- Rollins RA. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg A, Fisher O, Siman-Tov R, Ankri S. Identification of methylated sequences in genomic DNA of adult Drosophila melanogaster. Biochem Biophys Res Commun. 2004;322:465–469. doi: 10.1016/j.bbrc.2004.07.134. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SE, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Tatarinova T, Elhaik E, Pellegrini M. Cross-species analysis of genic GC3 content and DNA methylation patterns. Genome Biol Evol. 2013;5:1443–1456. doi: 10.1093/gbe/evt103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troukhan M, Tatarinova T, Bouck J, Flavell RB, Alexandrov NN. Genome-wide discovery of cis-elements in promoter sequences using gene expression. OMICS. 2009;13:139–151. doi: 10.1089/omi.2008.0034. [DOI] [PubMed] [Google Scholar]
- VanderKraats ND, Hiken JF, Decker KF, Edwards JR. Discovering high-resolution patterns of differential DNA methylation that correlate with gene expression changes. Nucleic Acids Res. 2013;41(14):6816–6827. doi: 10.1093/nar/gkt482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, et al. Integrative analysis of tissue-specific methylation and alternative splicing identifies conserved transcription factor binding motifs. Nucleic Acids Res. 2013;41(18):8503–8514. doi: 10.1093/nar/gkt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Xiang H, et al. Single base-resolution methylome of the silkwormreveals a sparse epigenomic map. Nat Biotechnol. 2010;28:516–520. doi: 10.1038/nbt.1626. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;4:551–556. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389(1):52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel I, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li W-S. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21(2):236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- Zhu B, Reinberg D. Epigenetic inheritance: Uncontested? Cell Res. 2011;21:435–441. doi: 10.1038/cr.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, He F, Song S, Wang J, Yu J. How many human genes can be defined as housekeeping with current expression data? BMC Genomics. 2008;9:172. doi: 10.1186/1471-2164-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.