Abstract
The highly invasive agricultural insect pest Ceratitis capitata (Diptera: Tephritidae) is the most thoroughly studied tephritid fruit fly at the genetic and molecular levels. It has become a model for the analysis of fruit fly invasions and for the development of area-wide integrated pest management (AW-IPM) programmes based on the environmentally-friendly Sterile Insect Technique (SIT). Extensive transcriptome resources and the recently released genome sequence are making it possible to unravel several aspects of the medfly reproductive biology and behaviour, opening new opportunities for comparative genomics and barcoding for species identification. New genes, promotors and regulatory sequences are becoming available for the development/improvement of highly competitive sexing strains, for the monitoring of sterile males released in the field and for determining the mating status of wild females. The tools developed in this species have been transferred to other tephritids that are also the subject of SIT programmes.
Keywords: Medfly, Sterile Insect Technique (SIT), functional genomics, transcriptomics, tephritids
Background
The Mediterranean fruit fly (medfly), Ceratitis capitata Wiedemann, is one of the world's most destructive agricultural insect pests [1-3]. Due to its global distribution and history of rapid and devastating outbreaks [4-6], the medfly is the most thoroughly studied "true" fruit fly (Diptera: Tephritidae) [7] at the genetic and molecular levels. It has thus become a model species for the analysis of fruit fly invasions [8] and for the development of control strategies [9]. Medfly outbreaks have been successfully controlled through area-wide integrated pest management (AW-IPM) programmes based on the environmentally-friendly Sterile Insect Technique (SIT) [10]. In the SIT, the reduction of pest population size is achieved through mass release of reproductively sterile male insects into a wild-type population [11]. Males rendered sterile through ionizing radiation compete with wild-type males for matings and deplete female reproductive success. Preventative sterile male releases have been and are currently applied in areas where the climatic conditions and the availability of suitable hosts for oviposition are particularly favourable for medfly establishment, such as California, Southern Australia and Florida [12-16]. To be most successful, this approach requires i) knowledge of the genetic background of the released males and the genetic structure of the target population, ii) a sexing strain for male-only production, iii) a sterilization system that inflicts the least possible fitness load, and iv) effective procedures to monitor the efficiency of the programmes.
In the last 20 years, enormous progress has been made in understanding medfly biology, with the goal of developing and optimizing a wide range of molecular tools for the implementation of population control strategies (Figure 1). Population genetics provided useful approaches for reconstructing the routes of medfly invasion, highlighting the complexity of the process [4,5,17-25]. Ceratitis capitata was the first non-drosophilid species in which the germ-line was transformed [26], enabling studies on its biology in ways that were previously impossible [27-34].
Figure 1.
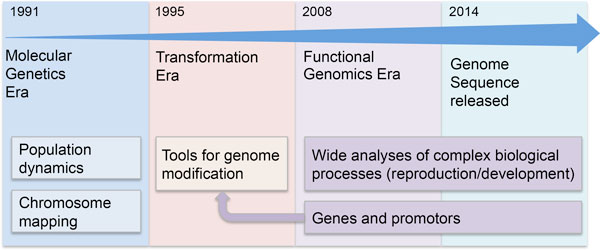
Molecular timeline of medfly research.
The application of functional genomics tools, together with the recent release of the medfly genome sequence (http://arthropodgenomes.org/wiki/i5K;https://www.hgsc.bcm.edu/arthropods/medfly-genome-annotation-groups), allows a more detailed analysis of the complex biological traits that underpin the adaptive potential of this fly at all developmental stages (Figure 2)[8,35]. Indeed, functional genomics provides powerful evolutionary tools to interpret how medfly (either wild or transgenic) develop and respond to the environment. Different aspects of development, behaviour, sexual maturation, and reproduction can now be examined both in terms of gene expression profiles and protein analyses [36-43]. New genes, promotors and regulatory sequences are consequently becoming available for i) the development/improvement of competitive sexing strains, ii) the monitoring of released males in the field, and iii) for determining the mating status of wild females.
Figure 2.
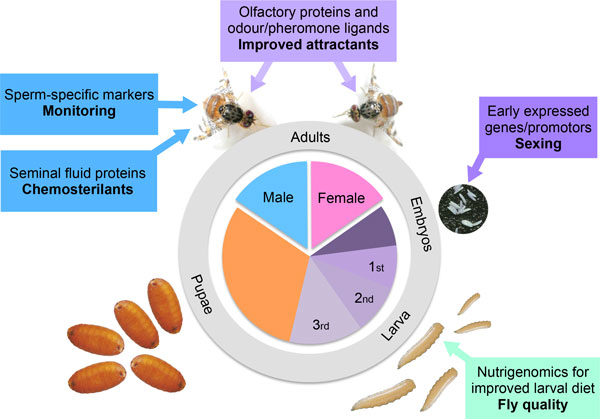
Medfly functional genomics resources and their impact on the improvement of the SIT.
Medfly embryogenesis
A reservoir of early male/female differentially expressed genes and sex regulatory sequences is now available for unravelling the first steps of medfly embryogenesis, i.e. when the maternal-to-zygotic transition (MTZ) occurs and when the sexual fate is established at the molecular level [36,38]. As a practical consequence, promotor and enhancer sequences that are active in early stages of development are becoming available as tools for the future generation and/or improvement of the existing conditional embryonic and female-specific lethality systems developed using conventional techniques. Female-specific lethality systems were developed based on alternative splicing of the Cctransformer gene (Cctra) [31]. Moreover, cellularisation-specific promotors/enhancers allowed the development of a transgenic embryo-specific lethality system [33]. More recently, the combination of the Cctra-based female-specific lethality [31] with the embryonic lethality system [33], yielded a female-specific embryonic lethality (FSEL) system in this species [44].
In this context, the medfly genes with vital functions in early embryonic development, such as those involved in sex determination and cellular blastoderm formation, are of direct use [38]. Their zygotic transcriptional activation follows two waves. The first wave starts within four hours after oviposition and includes the zygotic genes Ccsisterless A (CcsisA), Ccdeadpan (Ccdpn) and Ccslow-as-molasses (Ccslam). The second major burst of expression activation begins five hours after oviposition and includes the maternal-zygotic genes Ccgroucho (Ccgro), CcSex-lethal (CcSxl), Cctransformer (Cctra), Ccfemale-lethal d (Ccfl(2)d), CcRho1 and Ccserendipity-α (Ccsry-α)[38]. During this transition, sexual identity is established at the molecular level, before cellularisation of the embryo occurs. Unlike Drosophila [46,47], Cctra is the key-gene of the sex-determination cascade: it generates mRNAs encoding full-length active proteins only in females and displays an autocatalytic function, that guarantees the female-specific development of cell memory [47]. Cctra, in cooperation with Cctra2, determines the sex-specific splicing of Ccdsx, the transcription factor that is the regulator of the sex-differentiation processes. The mother supplies the embryos with Cctra and Ccdsx female-specific splicing variants. Subsequently, the maternal information for female-specific development is reset in embryos through the reprogramming of Cctra mRNA splicing and the degradation of the maternal Ccdsx mRNAs [38]. The precise timing of sex-specific splicing [38], as well as the proof of evidence that transgenic dsRNA for tra is effective in the conditional production of male-only progeny [48], can be exploited for the development of novel sexing strains.
Metabolic regulation of sexual maturation and mating
The production of highly competitive males is an essential requisite for effective SIT. Transcriptome and microarray-based functional analyses performed on whole flies and specific tissues are providing basic information on the pathways involved in primary metabolism, hormone synthesis, neurological-related processes, gametogenesis, signalling, and sensory perception [36,37,39,42]. The regulation of these specific pathways and biological processes can be affected by long-term artificial rearing, that may translate into reduced quality of individuals released for SIT [42]. In particular, down-regulation of signaling and neurological processes, especially those related to light and chemical stimuli, muscle development, muscle differentiation and locomotion, have been reported as a consequence of mass rearing in artificial conditions [42]. In this context, nutrigenomics can provide valuable information on how nutrition affects gene expression patterns, offering the means to measure male and female medfly responses to changes in the food stream, but also providing information on diet limitations [49]. This is a priority for operational SIT. Transcriptional baseline profiles of key biological pathways involved in sexual maturation of both sexes, and also in response to mating, are available for medflies reared on the standard diet used in mass rearing facilities [37]. Indeed, we know that medfly female maturation requires the activation of fatty acid metabolism as a reflection of the high energy requirements for female reproductive success, such as foraging, nutrient storage and egg development [50]. In addition, Gene Ontology (GO) enrichment analyses revealed that, in mature females, transcript categories related to memory/learning behaviours and visual and olfactory functions are significantly overrepresented [37]. By contrast, male sexual maturation requires the activation of carbohydrate and protein metabolism for energy production and muscle activities, memory formation, smell recognition and pheromone production [37]. All these activities suggest an investment required for lek formation and courtship [51]. Despite extensive post-mating transcriptional changes in the male, changes in the female were surprisingly modest. Indeed, in the male, mating does not down-regulate the transcriptional activities of genes implicated in lek formation/courtship, whereas it increases the activities of genes related to fitness (i.e. double time and Basigin) [37].
Some of these pathways are down-regulated by irradiation [42]. This is the case of processes related to visual and chemical responses, and those associated with muscle development and locomotion. These irradiation-related changes may have an impact on the competitiveness of mass reared flies.
Studies on improved diets or chemical manipulation of the adult environment offer promising options for the improvement of sterile male competitiveness. Approaches aimed at the improvement of the sexual performance of sterile males include i) altering the olfactory environment experienced by freshly eclosed individuals, providing high-quality post-teneral nutrition [52] and ii) inoculating males with probiotic bacteria [53,54].
Male reproduction
A better understanding of the reproductive biology of the medfly should permit the development of novel or improved approaches to impact male reproductive success and/or regulate female mating behaviour and fertility. In this respect, transcriptomics and proteomics of reproductive tissues will help to identify genes and promotors. Testes and male accessory glands (MAGs) participate in the maintenance of complementary reproductive functions. In the testes, the key regulatory genes of spermatogenesis tend to be conserved to guarantee the male-specific processes required for sperm production [55,56]. By contrast, the accessory gland secretions act as key factors in male insect reproductive success, and the genes expressed in the MAGs are subject to rapid evolution as a result of sexual conflict and competition [57]. A transcriptome-based analysis performed on medfly testes and male accessory gland tissues resulted in a database of 3344 unique sequences [39]. Transcripts related to spermatogenesis, fertility, sperm-egg binding, as well as those involved in the production of seminal fluid proteins (SFPs), were identified. Some of the SFP transcripts displayed a mating-responsive profile [39]. These will be ideal targets for the development of novel and more specific environmentally-friendly chemosterilants [58,59] that mimic the behaviour-modulating effects of MAG proteins, i.e. by impeding correct sperm storage, or interfering with female remating.
Over a third of the transcripts from these two tissues shared no significant similarities to known genes from other organisms. Considering that they may represent novel and/or fast-evolving sequences, they represent ideal targets for the development of species-specific diagnostic markers.
Improved SIT monitoring strategies
A major issue in the monitoring activities for evaluating SIT effectiveness is the difficulty in assessing the capacity of released sterile males to inseminate wild-type females [60-62]. The availability of the testes- and sperm-specific Ccβ2-tubulin gene has allowed the use of its promotor for fluorescent protein marking of the spermatozoa, and hence to detect females that have mated with released males [32]. Using this marking system, strains have been generated and evaluated for their ability to transfer green or red fluorescent sperm to the female spermathecae. It has been proven that these sperm remain viable and fluorescent for a long time within the spermathecae, also after female death [32] (Figure 3). The transgene previously inserted in one of these lines, namely 1260_F-3_m-1, was then efficiently modified by the use of the site-specific integration system from phage phiC31 [34]. Post-integrational excision of one of the piggyBac inverted terminal repeats resulted in stably integrated transgene insertions that, being inert to the piggyBac transposase, could not be remobilized. This allowed the development of an optimized strain for pest control that minimizes environmental concerns (stab_1260_F-3_m-1)[34]. Once integrated into the medfly GSS Vienna-8 strain, this sperm marking system may offer valuable alternatives to the currently used fluorescent powders [63] that are detected in trapped flies using UV light. Moreover, this sperm marking system can also be integrated into strains carrying diverse transgenes in tandem, for example with conditional embryonic lethality [33] and sexing systems [31].
Figure 3.
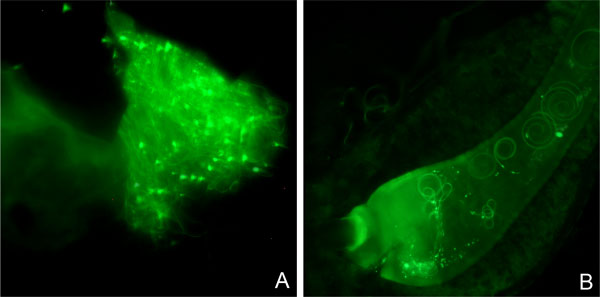
Transgenic sperm can be easily traced in the reproductive tract of laboratory wild-type females. Mechanically opened spermatheca isolated from a laboratory wild-type female mated with a transgenic male with green fluorescent sperm [32], three days after death (A). Spermathecal duct dissected from a laboratory wild-type female mated to a transgenic male with green fluorescent sperm [32] 24 hours after mating (B). Images were captured using an epifluorescence Zeiss Axioplan microscope at 400x magnification with the Zeiss filters set 13.
For monitoring activities, one of the priorities is the development of powerful species- and sex-specific attractants. In this context, it is essential to identify the components of the molecular machinery that recognizes and binds external ligands (odours and pheromone components) and translates this interaction into electrical signals to the central nervous system. Three main groups of molecules are involved: odorant-binding proteins (OBPs), chemosensory proteins (CSPs), and the chemoreceptor superfamily formed by the olfactory (OR), gustatory (GR) and ionotropic (IR) receptor families [64,65]. The chemosensory gene repertoire of the medfly is currently being characterized at the functional genomics and structural level [40,41]. So far, one antennal-enriched OBP appears to be particularly promising for practical applications. Indeed, it displayed highest binding specificity for (E,E)-α-farnesene, a major component of male pheromone blend, and also for Trimedlure, a strong synthetic medfly attractant [41]. The resolution of the three-dimensional structure of this medfly OBP will be the premise for the design of synthetic molecules able to act as antagonists of the natural ligand/s. Such optimized molecules need to be further evaluated and tested for side-effects before they can be used in AW-IPM approaches.
Conclusions
The extensive transcriptome resources now available for the medfly (Table 1) will greatly improve the on-going annotation of the genome. They will also facilitate the generation of genomic data from other tephritid species of agricultural importance [66-71], opening new ways for comparative genomics and barcoding for species identification. In addition, the structural and functional genomics (transcriptomics, proteomics, RNA interference etc) tools that are being developed in the medfly can be extended to other tephritids that are also the subject of SIT programmes, such as Anastrepha and Bactrocera species (A. ludens, A. suspensa, A. obliqua, A. fraterculus, B. cucurbitae, B. tryoni, B. dorsalis, B. correcta)[10].
Table 1.
Transcriptome and microarray resources available for the medfly.
| Database | Tissue/status | Strain | Type | Accession numbers | Ref. |
|---|---|---|---|---|---|
| NCBI dbEST database | Embryo (from 30 min to 36 hr after oviposition) | Ispra | Expressed sequence tags | FG068301-FG078567 | [36] |
| NCBI dbEST database | Adult male and female heads (from 30 min to 8 days after emergence). | Ispra | Expressed sequence tags | FG078568-FG089553 | [36] |
| NCBI dbEST database | Adult testes and male accessory glands. | Ispra | Expressed sequence tags | JK832450-JK838363 | [39] |
| NCBI dbEST database | Adult male accessory glands. | Guatemala mass-rearing strain (Moscamed) | Expressed sequence tags | DQ406805-DQ406817 | [72] |
| NCBI GEO Dataset | Adult female head. Immature versus mature. | Ispra | Microarray | GSE19571 | [37] |
| NCBI GEO Dataset | Adult male head. Immature versus mature. | Ispra | Microarray | GSE19572 | [37] |
| NCBI GEO Dataset | Adult female head. Mated versus Virgin. | Ispra | Microarray | GSE19573 | [37] |
| NCBI GEO Dataset | Adult Male head. Mated versus Virgin. | Ispra | Microarray | GSE19608 | [37] |
| NCBI Sequence Read Archive | Adult, whole body, Irradiated | Vienna 7 | Illumina Hiseq 2000 sequencing | SRX312172-SRX312174 | [42] |
| NCBI Sequence Read Archive | Adult, whole body, Non-irradiated | Vienna 7 | Illumina Hiseq 2000 sequencing | SRX312183-SRX312185 | [42] |
| NCBI Sequence Read Archive | Pupae, Irradiated | Vienna 7 | Illumina Hiseq 2000 sequencing | SRX312176-SRX312180 | [42] |
| NCBI Sequence Read Archive | Pupae, Non-irradiated | Vienna 7 | Illumina Hiseq 2000 sequencing | SRX312186-SRX312188 | [42] |
| NCBI Sequence Read Archive | Adult, whole body, Non-irradiated | Wild Hawaii | Illumina Hiseq 2000 sequencing | SRX312189-SRX312191 | [42] |
| NCBI Sequence Read Archive | Pupae, Non-irradiated | Wild Hawaii | Illumina Hiseq 2000 sequencing | SRX312192-SRX312194 | [42] |
The increased knowledge of the biology of the medfly acquired through genomic approaches will also facilitate the further development of regulations for the transfer and potential field release of genetically modified fruit flies.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Francesca Scolari, Email: francesca.scolari@unipv.it.
Ludvik M Gomulski, Email: gomulski@unipv.it.
Paolo Gabrieli, Email: paolo.gab@gmail.com.
Mosè Manni, Email: mose.manni01@universitadipavia.it.
Grazia Savini, Email: grazia.savini01@universitadipavia.it.
Giuliano Gasperi, Email: gasperi@unipv.it.
Anna R Malacrida, Email: malacrid@unipv.it.
Acknowledgements
The work was carried out within the FAO/IAEA research CRP programme ''Development and evaluation of improved strains of insect pests for SIT'' (GG) and the IAEA Technical Contract No:16966 (GG). The work was also partially funded by a PRIN grant from the Italian Ministry of Education, University and Research (MIUR) (20077 RCHRW) (LMG).
The authors would like to thank the two anonymous referees for their valuable comments which helped to improve the manuscript.
This article has been published as part of BMC Genetics Volume 15 Supplement 2, 2014: Development and evaluation of improved strains of insect pests for SIT. The full contents of the supplement are available online at http://www.biomedcentral.com/bmcgenet/supplements/15/S2.
Publication of this supplement was funded by the International Atomic Energy Agency. The peer review process for articles published in this supplement was overseen by the Supplement Editors in accordance with BioMed Central's peer review guidelines for supplements. The Supplement Editors declare that they have no competing interests.
References
- Bateman MA. The ecology of fruit flies. Annu Rev Entomol. 1972;17:493–518. doi: 10.1146/annurev.en.17.010172.002425. [DOI] [Google Scholar]
- Aluja M, Mangan RL. Fruit fly (Diptera: Tephritidae) host status determination: critical conceptual, methodological, and regulatory considerations. Annu Rev Entomol. 2008;53:473–502. doi: 10.1146/annurev.ento.53.103106.093350. [DOI] [PubMed] [Google Scholar]
- Papadopoulos NT, Plant RE, Carey JR. From trickle to flood: the large-scale, cryptic invasion of California by tropical fruit flies. Proc Biol Sci. 2013;280(1768):20131466. doi: 10.1098/rspb.2013.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperi G, Bonizzoni M, Gomulski LM, Murelli V, Torti C, Malacrida AR, Guglielmino CR. Genetic differentiation, gene flow and the origin of infestations of the medfly, Ceratitis capitata. Genetica. 2002;116(1):125–135. doi: 10.1023/A:1020971911612. [DOI] [PubMed] [Google Scholar]
- Malacrida AR, Gomulski LM, Bonizzoni M, Bertin S, Gasperi G, Guglielmino CR. Globalization and fruitfly invasion and expansion: the medfly paradigm. Genetica. 2007;131(1):1–9. doi: 10.1007/s10709-006-9117-2. [DOI] [PubMed] [Google Scholar]
- Carey JR. The Mediterranean fruit fly (Ceratitis capitata) invasion of California deepens: response to an alternative explanation for recurring outbreaks. Am Entomol. 2010;56:158–163. [Google Scholar]
- White IM, Elson-Harris MM. Fruit flies of economic significance: Their identification and bionomics. Wallingford, Oxon, UK: CAB International; 1992. [Google Scholar]
- Diamantidis AD, Carey JR, Nakas CT, Papadopoulos NT. Population-specific demography and invasion potential in medfly. Ecol Evol. 2011;1(4):479–488. doi: 10.1002/ece3.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrichs J, Robinson AS, Cayol JP, Enkerlin W. Medfly Area Wide Sterile Insect Technique Programmes for Prevention, Suppression or Eradication: the importance of Mating Behavior Studies. Fla Entomol. 2002;85(1):1–13. doi: 10.1653/0015-4040(2002)085[0001:MASITP]2.0.CO;2. [DOI] [Google Scholar]
- Dyck VA, Hendrichs J, Robinson AS. Sterile Insect Technique: Principles and practice in Area-wide Integrated Pest Management. Dordrecht The Netherlands: Springer; 2005. [Google Scholar]
- Knipling EF. Possibilities of insect control or eradication through the use of sexually sterile males. J Econ Entomol. 1955;48:459–462. [Google Scholar]
- Smallridge CJ, Hopkins DC. In: Proceedings of the 6th International Symposium on Fruit Flies of Economic Importance. Brian BN, editor. Johannesburg: Ultra Litho (Pty) Ltd; 2004. Preventative sterile fly release for the management of a Mediterranean fruit fly outbreak in South Australia; pp. 213–215. [Google Scholar]
- Dowell RV, Siddiquw IA, Meyer F, Spaugy EL. In: International conference on area-wide control of insect pests and the fifth international symposium on fruit flies of economic importance; Penang, Malaysia. Tan K-H, editor. Penerbit Universiti Sains Malaysia, Penang, Malaysia; 2000. Mediterranean fruit fly preventative release programme in southern California; pp. 369–377. [Google Scholar]
- Hood-Nowotny R, Mayr L, Islam A, Robinson A, Caceres C. Routine isotope marking for the Mediterranean fruit fly (Diptera: Tephritidae) J Econ Entomol. 2009;102(3):941–947. doi: 10.1603/029.102.0312. [DOI] [PubMed] [Google Scholar]
- CDFA. Mediterranean fruit fly preventive release program. March 2002. CDFA, Sacramento, CA, USA; 2002. [Google Scholar]
- Andress E, Walters I, del Toro M, Shelly T. Release-Recapture of Sterile Male Mediterranean Fruit Flies (Diptera: Tephritidae) in Southern California. Proceedings of the Hawaiian Entomological Society. 2013;45:11–29. [Google Scholar]
- Baruffi L, Damiani G, Guglielmino CR, Bandi C, Malacrida AR, Gasperi G. Polymorphism within and between populations of Ceratitis capitata: comparison between RAPD and multilocus enzyme electrophoresis data. Heredity (Edinb) 1995;74(Pt 4):425–437. doi: 10.1038/hdy.1995.60. [DOI] [PubMed] [Google Scholar]
- Gasparich GE, Silva JG, Han HY, McPheron BA, Steck GJ, Sheppard WS. Population genetic structure of Mediterranean fruit fly (Diptera: Tephritidae) and implications for worldwide colonization patterns. Annals of the Entomological Society of America. 1997;90:790–797. [Google Scholar]
- Gomulski LM, Bourtzis K, Brogna S, Morandi PA, Bonvicini C, Sebastiani F, Torti C, Guglielmino CR, Savakis C, Gasperi G. et al. Intron size polymorphism of the Adh1 gene parallels the worldwide colonization history of the Mediterranean fruit fly, Ceratitis capitata. Molecular Ecology. 1998;7:1729–1741. doi: 10.1046/j.1365-294x.1998.00509.x. [DOI] [Google Scholar]
- Malacrida AR, Marinoni F, Torti C, Gomulski LM, Sebastiani F, Bonvicini C, Gasperi G, Guglielmino CR. Genetic aspects of the worldwide colonization process of Ceratitis capitata. J Hered. 1998;89(6):501–507. doi: 10.1093/jhered/89.6.501. [DOI] [PubMed] [Google Scholar]
- Kourti A. Estimates of heterozygosity and patterns of geographic differentiation in natural populations of the medfly (Ceratitis capitata) Hereditas. 2002;137:173–179. doi: 10.1034/j.1601-5223.2002.01653.x. [DOI] [Google Scholar]
- Kourti A. Estimates of gene flow from rare alleles in natural populations of medfly Ceratitis capitata (Diptera: Tephritidae) Bulletin of Entomological Research. 2004;94:449–456. doi: 10.1079/ber2004324. [DOI] [PubMed] [Google Scholar]
- Kourti A. Patterns of variation within and between Greek populations of Ceratitis capitata suggest extensive gene flow and latitudinal clines. Journal of Economic Entomology. 2004;97:1186–1190. doi: 10.1603/0022-0493(2004)097[1186:POVWAB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bonizzoni M, Guglielmino CR, Smallridge CJ, Gomulski M, Malacrida AR, Gasperi G. On the origins of medfly invasion and expansion in Australia. Mol Ecol. 2004;13(12):3845–3855. doi: 10.1111/j.1365-294X.2004.02371.x. [DOI] [PubMed] [Google Scholar]
- Barr NB. Pathway analysis of Ceratitis capitata (Diptera: Tephritidae) using mitochondrial DNA. J Econ Entomol. 2009;102(1):401–411. doi: 10.1603/029.102.0153. [DOI] [PubMed] [Google Scholar]
- Loukeris TG, Livadaras I, Arcà B, Zabalou S, Savakis C. Gene transfer into the medfly, Ceratitis capitata, with a Drosophila hydei transposable element. Science. 1995;270(5244):2002–2005. doi: 10.1126/science.270.5244.2002. [DOI] [PubMed] [Google Scholar]
- Handler AM, McCombs SD, Fraser MJ, Saul SH. The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7520–7525. doi: 10.1073/pnas.95.13.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophides GK, Livadaras I, Savakis C, Komitopoulou K. Two medfly promoters that have originated by recent gene duplication drive distinct sex, tissue and temporal expression patterns. Genetics. 2000;156(1):173–182. doi: 10.1093/genetics/156.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel K, Stamenova A, Pinkerton AC, Franz G, Robinson AS, Gariou-Papalexiou A, Zacharopoulou A, O'Brochta DA, Atkinson PW. Hermes-mediated germ-line transformation of the Mediterranean fruit fly Ceratitis capitata. Insect Mol Biol. 2001;10(2):155–162. doi: 10.1046/j.1365-2583.2001.00250.x. [DOI] [PubMed] [Google Scholar]
- Gong P, Epton MJ, Fu G, Scaife S, Hiscox A, Condon KC, Condon GC, Morrison NI, Kelly DW, Dafa'alla T. et al. A dominant lethal genetic system for autocidal control of the Mediterranean fruitfly. Nat Biotechnol. 2005;23(4):453–456. doi: 10.1038/nbt1071. [DOI] [PubMed] [Google Scholar]
- Fu G, Condon KC, Epton MJ, Gong P, Jin L, Condon GC, Morrison NI, Dafa'alla TH, Alphey L. Female-specific insect lethality engineered using alternative splicing. Nat Biotechnol. 2007;25(3):353–357. doi: 10.1038/nbt1283. [DOI] [PubMed] [Google Scholar]
- Scolari F, Schetelig MF, Bertin S, Malacrida AR, Gasperi G, Wimmer EA. Fluorescent sperm marking to improve the fight against the pest insect Ceratitis capitata (Wiedemann; Diptera: Tephritidae) N Biotechnol. 2008;25(1):76–84. doi: 10.1016/j.nbt.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Schetelig MF, Caceres C, Zacharopoulou A, Franz G, Wimmer EA. Conditional embryonic lethality to improve the sterile insect technique in Ceratitis capitata (Diptera: Tephritidae) BMC Biol. 2009;7:4. doi: 10.1186/1741-7007-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetelig MF, Scolari F, Handler AM, Kittelmann S, Gasperi G, Wimmer EA. Site-specific recombination for the modification of transgenic strains of the Mediterranean fruit fly Ceratitis capitata. Proc Natl Acad Sci USA. 2009;106(43):18171–18176. doi: 10.1073/pnas.0907264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuval B, Hendrichs J. In: Fruit flies (Tephritidae): Phylogeny and Evolution of Behavior. Aluja M, Norrbom A, editor. Boca Raton, Florida: CRP Press; 2000. Behavior of flies in the genus Ceratitis (Dacinae: Ceratitidini) pp. 429–457. [Google Scholar]
- Gomulski LM, Dimopoulos G, Xi Z, Soares MB, Bonaldo MF, Malacrida AR, Gasperi G. Gene discovery in an invasive tephritid model pest species, the Mediterranean fruit fly, Ceratitis capitata. BMC Genomics. 2008;9:243. doi: 10.1186/1471-2164-9-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomulski LM, Dimopoulos G, Xi Z, Scolari F, Gabrieli P, Siciliano P, Clarke AR, Malacrida AR, Gasperi G. Transcriptome profiling of sexual maturation and mating in the Mediterranean fruit fly, Ceratitis capitata. PLoS One. 2012;7(1):e30857. doi: 10.1371/journal.pone.0030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrieli P, Falaguerra A, Siciliano P, Gomulski LM, Scolari F, Zacharopoulou A, Franz G, Malacrida AR, Gasperi G. Sex and the single embryo: early development in the Mediterranean fruit fly, Ceratitis capitata. BMC Dev Biol. 2010;10:12. doi: 10.1186/1471-213X-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolari F, Gomulski LM, Ribeiro JM, Siciliano P, Meraldi A, Falchetto M, Bonomi A, Manni M, Gabrieli P, Malovini A. et al. Transcriptional profiles of mating-responsive genes from testes and male accessory glands of the Mediterranean fruit fly, Ceratitis capitata. PLoS One. 2012;7(10):e46812. doi: 10.1371/journal.pone.0046812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano P, Scolari F, Gomulski LM, Falchetto M, Manni M, Gabrieli P, Field LM, Zhou JJ, Gasperi G, Malacrida AR. Sniffing out chemosensory genes from the Mediterranean fruit fly, Ceratitis capitata. PLoS One. 2014;9(1):e85523. doi: 10.1371/journal.pone.0085523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano P, He XL, Woodcock C, Pickett JA, Field LM, Birkett MA, Kalinova B, Gomulski LM, Scolari F, Gasperi G. et al. Identification of pheromone components and their binding affinity to the odorant binding protein CcapOBP83a-2 of the Mediterranean fruit fly, Ceratitis capitata. Insect Biochem Mol Biol. 2014;48:51–62. doi: 10.1016/j.ibmb.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calla B, Hall B, Hou S, Geib SM. A genomic perspective to assessing quality of mass-reared SIT flies used in Mediterranean fruit fly (Ceratitis capitata) eradication in California. BMC Genomics. 2014;15:98. doi: 10.1186/1471-2164-15-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Andrés V, Castañera P, Sabater-Muñoz B. Transcriptome analysis in Ceratitis capitata to unveil genes involved in ageing-maturation process. Spanish Journal of Agricultural Research. 2013;11(3):842–854. doi: 10.5424/sjar/2013113-3987. [DOI] [Google Scholar]
- Ogaugwu CE, Schetelig MF, Wimmer EA. Transgenic sexing system for Ceratitis capitata (Diptera: Tephritidae) based on female-specific embryonic lethality. Insect Biochem Mol Biol. 2013;43(1):1–8. doi: 10.1016/j.ibmb.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Salz HK, Erickson JW. Sex determination in Drosophila: The view from the top. Fly (Austin) 2010;4(1):60–70. doi: 10.4161/fly.4.1.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham P, Penn JK, Schedl P. Masters change, slaves remain. Bioessays. 2003;25(1):1–4. doi: 10.1002/bies.10207. [DOI] [PubMed] [Google Scholar]
- Pane A, Salvemini M, Delli Bovi P, Polito C, Saccone G. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development. 2002;129(15):3715–3725. doi: 10.1242/dev.129.15.3715. [DOI] [PubMed] [Google Scholar]
- Saccone G, Pane A, de Simone A, Salvemini M, Milano A, Annunziata L, Mauro U, Polito L. In: New sexing strains for Mediterranean fruit fly Ceratitis capitata: transforming females into males. Edited by Vreysen MJB, Robinson AS, Hendrichs J, editor. Dordrecht, the Netherlands: Springer; 2007. pp. 95–102. [Google Scholar]
- Yocum GD, Coudron TA, Brandt SL. Differential gene expression in Perillus bioculatus nymphs fed a suboptimal artificial diet. J Insect Physiol. 2006;52(6):586–592. doi: 10.1016/j.jinsphys.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Drew RAI, Yuval B. In: Fruit flies (Tephritidae): phylogeny and evolution of behavior. Aluja M, Norrbom AL, editor. Boca Raton, Florida: CRC Press; 2000. The evolution of fruit fly feeding behaviour; pp. 731–749. [Google Scholar]
- Eberhard W. In: Fruit Flies (Tephritidae): Phylogeny and Evolution of Behavior. Aluja M, Norrbom A, editor. Boca Raton, Florida: CRC Press; 2000. Sexual behavior and sexual selection in the Mediterranean fruit fly, Ceratitis capitata (Dacinae: Ceratitidini) pp. 457–489. [Google Scholar]
- Yuval B, Maor M, Levy K, Kaspi R, Taylor P, Shelly T. Breakfast of champions or kiss of death? Survival and sexual performance of protein-fed, sterile Mediterranean fruit flies (Diptera: Tephritidae) Florida Entomologist. 2007;90:115–122. doi: 10.1653/0015-4040(2007)90[115:BOCOKO]2.0.CO;2. [DOI] [Google Scholar]
- Pereira R, Yuval B, Liedo P, Teal PEA, Shelly TE, McInnis DO, Hendrichs J. Improving sterile male performance in support of programmes integrating the sterile insect technique against fruit flies. Journal of Applied Entomology. 2013;137(S1):178–190. [Google Scholar]
- Ben Ami E, Yuval B, Jurkevitch E. Manipulation of the microbiota of mass-reared Mediterranean fruit flies Ceratitis capitata (Diptera: Tephritidae) improves sterile male sexual performance. ISME J. 2010;4(1):28–37. doi: 10.1038/ismej.2009.82. [DOI] [PubMed] [Google Scholar]
- Bonilla E, Xu EY. Identification and characterization of novel mammalian spermatogenic genes conserved from fly to human. Mol Hum Reprod. 2008;14(3):137–142. doi: 10.1093/molehr/gan002. [DOI] [PubMed] [Google Scholar]
- White-Cooper H, Doggett K, Ellis R. In: Sperm biology: an evolutionary perspective. Birkhead TR, Hosken DJ, Pitnick SS, editor. New York: Academic Press; 2009. The evolution of spermatogenesis; pp. 151–183. [Google Scholar]
- Ravi Ram K, Wolfner MF. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr Comp Biol. 2007;47(3):427–445. doi: 10.1093/icb/icm046. [DOI] [PubMed] [Google Scholar]
- Baldini F, Gabrieli P, Rogers DW, Catteruccia F. Function and composition of male accessory gland secretions in Anopheles gambiae: a comparison with other insect vectors of infectious diseases. Pathog Glob Health. 2012;106(2):82–93. doi: 10.1179/2047773212Y.0000000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BV, Klöck C, Schatz A, Nguyen JB, Kakani EG, Catteruccia F, Khosla C, Baxter RH. Dihydroisoxazole inhibitors of Anopheles gambiae seminal transglutaminase AgTG3. Malar J. 2014;13(1):210. doi: 10.1186/1475-2875-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnis DO. Size differences between normal and irradiated sperm heads in mated female Mediterranean fruit flies (Diptera: Tephritidae) Annals of the Entomological Society of America. 1993;86:305–308. [Google Scholar]
- McInnis DO, Tam S, Grace C, Miyashita D. Population suppression and sterility rates induced by variable sex ratio, sterile insect releases of Ceratitis capitata (Diptera: Tephritidae) in Hawaii. Annals of the Entomological Society of America. 1994;87:231–240. [Google Scholar]
- San Andrés V, Urbaneja A, Sabater-Muñoz B, Castañera P. A novel molecular approach to assess mating success of sterile Ceratitis capitata (Diptera: Tephritidae) males in sterile insect technique programs. J Econ Entomol. 2007;100(4):1444–1449. doi: 10.1603/0022-0493(2007)100[1444:ANMATA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- FAO/IAEA/USDA - Food and Agriculture Organisation IAEA; United States Department of Agriculture. Manual for product quality control and shipping procedures for sterile mass-reared tephritid fruit flies. Version 5.0. Vienna, Austria: International Atomic Energy Agency; 2003. [Google Scholar]
- Pelosi P, Mastrogiacomo R, Iovinella I, Tuccori E, Persaud KC. Structure and biotechnological applications of odorant-binding proteins. Appl Microbiol Biotechnol. 2014;98(1):61–70. doi: 10.1007/s00253-013-5383-y. [DOI] [PubMed] [Google Scholar]
- Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- Nirmala X, Schetelig MF, Yu F, Handler AM. An EST database of the Caribbean fruit fly, Anastrepha suspensa (Diptera: Tephritidae) Gene. 2013;517(2):212–217. doi: 10.1016/j.gene.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Pavlidi N, Dermauw W, Rombauts S, Chrisargiris A, Van Leeuwen T, Vontas J. Analysis of the Olive fruit fly Bactrocera oleae transcriptome and phylogenetic classification of the major detoxification gene families. PLoS One. 2013;8(6):e66533. doi: 10.1371/journal.pone.0066533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, Robertson HM, Feder JL, Varala K, Hudson ME, Ragland GJ, Hahn DA, Berlocher SH. Sympatric ecological speciation meets pyrosequencing: sampling the transcriptome of the apple maggot Rhagoletis pomonella. BMC Genomics. 2009;10:633. doi: 10.1186/1471-2164-10-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Peng T, He W, Zhang H. High-throughput sequencing to reveal genes involved in reproduction and development in Bactrocera dorsalis (Diptera: Tephritidae) PLoS One. 2012;7(5):e36463. doi: 10.1371/journal.pone.0036463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JC, Chien TY, Hu CC, Chen MJ, Wu WJ, Feng HT, Haymer DS, Chen CY. Discovery of genes related to insecticide resistance in Bactrocera dorsalis by functional genomic analysis of a de novo assembled transcriptome. PLoS One. 2012;7(8):e40950. doi: 10.1371/journal.pone.0040950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdell KM, Lyons-Sobaski SA, Robertson HM, Walden KK, Feder JL, Wanner K, Berlocher SH. Expressed sequence tags from cephalic chemosensory organs of the northern walnut husk fly, Rhagoletis suavis, including a putative canonical odorant receptor. J Insect Sci. 2010;10:51. doi: 10.1673/031.010.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SJ, Chapman T. Identification of genes expressed in the accessory glands of male Mediterranean Fruit Flies (Ceratitis capitata) Insect Biochem Mol Biol. 2006;36(11):846–856. doi: 10.1016/j.ibmb.2006.08.009. [DOI] [PubMed] [Google Scholar]


