Abstract
We report on the use of a non-instrumented device for the implementation of a loop-mediated amplification (LAMP) based assay for the select-agent bacterial-wilt pathogen Ralstonia solanacearum race 3 biovar 2. Heat energy is generated within the device by the exothermic hydration of calcium oxide, and the reaction temperature is regulated by storing latent energy at the melting temperature of a renewable lipid-based engineered phase-change material. Endpoint detection of the LAMP reaction is achieved without opening the reaction tube by observing the fluorescence of an innovative FRET-based hybridization probe with a simple custom fluorometer. Non-instrumented devices could maintain reactions near the design temperature of 63°C for at least an hour. Using this approach DNA extracted from the pathogen could be detected at fewer than ten copies within a 25 μL reaction mix, illustrating the potential of these technologies for simple, powerful agricultural diagnostics in the field. Furthermore, the assay was just as reliable when implemented in a tropical environment at 31°C as it was when implemented in an air-conditioned lab maintained at 22°C, illustrating the potential value of the technology for field conditions in the tropics and subtropics.
Keywords: Agricultural diagnostics, Assimilating probe, Bacterial wilt, Biosensor, DNA, LAMP
The bacterial pathogen Ralstonia solanacearum causes wilt in over 200 plant species, including agronomically important crops like potato, tomato, banana, peanut, and ginger (Denny, 2006). The bacterium is widely distributed and causes severe crop losses in the warm and humid tropics, but the host range and other biological characteristics of different subpopulations of the pathogen vary considerably. In recent years, an increasing number of sites in Europe have been affected by cold-adapted populations of the pathogen (Janse et al., 1998; van Elsas et al., 2000). These populations, classified as race 3 biovar 2, primarily affect potato and have been estimated to cause an excess of $950 million in damages each year (Floyd, 2004). While race 3 biovar 2 strains have been repeatedly found on imported geraniums (Swanson et al., 2005), these strains have not established themselves in North America and there is considerable interest in developing technologies to rapidly detect and discriminate them from cold-susceptible strains and other strains already established in subtropical regions of the mainland U.S. Currently, there are no reliable antibodies to allow immunological discrimination of subpopulations of Ralstonia solanacearum, so typing is typically done using metabolic profiling of the cultured isolate, or host typing on susceptible plants (Fegan and Prior, 2005). These methods are cumbersome and slow, and they could introduce catastrophic delays in actions to contain and eradicate an outbreak. Our objective is to develop gene-based diagnostics on an easily applied platform to rapidly discriminate race 3 biovar 2 strains from other populations within the same species, thus allowing accurate and timely management decisions to be made in response to introduction of the disease.
A variety of relatively inexpensive and disposable point-of-care diagnostic systems have been demonstrated that may be transferable to the agricultural setting (Weigl et al., 2008; Yager et al., 2008), including gene-based microfluidic devices in which DNA is extracted and amplified by PCR (Lui et al., 2009; Chen et al., 2010). Initially, our strategy was to adapt a direct electrochemical approach for detection (Fan et al., 2003; Jenkins et al., 2006) of amplicons generated by loop-mediated isothermal amplification (LAMP) (Kubota et al., 2011b) implemented on simple electrodes customized for temperature control (Jenkins et al., 2009) to control DNA replication and hybridization stringency. Difficulties in preventing cross-contamination in the open electrode system and in ensuring reproducibility of the electrochemical interaction resulted in abandoning electrochemical detection in favor of a purely optical probe (“assimilating probes”) (Kubota et al., 2011a) for detecting isothermal replication of DNA in real-time. Isothermal DNA amplification, such as occurs with the LAMP process, allows sensitive detection of pathogen-specific DNA without the tradeoffs in system complexity, power requirements, and speed that is incumbent on PCR-based diagnostics. For a robust and simple field assay, we have focused on using the LAMP reaction (Notomi et al., 2000) because it can be implemented with a single enzyme (strand-displacing polymerase) and does not require preliminary manipulations to construct a nucleic acid fragment that forms the basis for continuous isothermal replication.
Despite the need for gene-based diagnostics to determine the nature of infectious diseases, including host specificity and virulence (Guidot et al., 2009; Kubota et al., 2011b), drug-resistance profiles (Chen et al., 2009; Conceicao et al., 2010), and other biological characteristics (Milling et al., 2009), commercially successful point-of-care diagnostics in medicine and agriculture have thus far been restricted to direct enzyme tests such as glucose test strips and immunochromatographic strips that are used in home pregnancy tests (Bissonnette and Bergeron, 2010; Peeling and Mabey, 2010). Technologies enabling real-time PCR tests for specific pathogens are reaching commercial maturity for the clinical lab (e.g., Gene XPert series tests from Cepheid, and GeneOhm Strep B tests from Beckton Dickinson), but the costs for the instrumentation and disposables are still prohibitive for agricultural applications and medical diagnostics in under-resourced settings.
Currently, diagnostic needs for infectious disease are greatest in under-resourced settings in developing countries where funds, infrastructure, and trained personnel to support and maintain sophisticated instruments are lacking. Because of these challenges, many groups are focused on developing simple platforms to enable reliable, inexpensive, gene-based point-of-care diagnostics for developing countries (Weigl et al., 2008). Agricultural users face many of the same constraints, and may benefit from developments in these resource-appropriate technologies. In this article, we report on adapting a non-instrumented platform (LaBarre et al., 2009; LaBarre et al., 2010a; LaBarre et al., 2010b) to enable LAMP-based detection of an agricultural pathogen with simple portable tools suitable for use in the field and in basic lab settings. While we have developed prototype instruments to implement the LAMP reaction with assimilating probes in real-time directly using a handheld instrument, even the modest power requirements (~1 W) preclude these systems from being deployed as a portable, battery-powered device. Therefore, the non-instrumented approach is especially compelling for field applications where power is not available or is unreliable, as is the case in many developing countries, or in the field in agricultural settings.
Materials and Methods
DNA Isolation
Ralstonia solanacearum race 3 biovar 2 strain UW551 was grown on modified tetrazolium chloride (TZC) agar medium (Norman and Alvarez, 1989) and incubated for 48 h at 28°C. DNA was purified from these cells with the Wizard Genomic DNA Purification Kit (Promega Corp., Madison, Wisc.) according to the manufacturer’s instructions. DNA concentrations were quantified photometrically (absorbance measurements at 260 and 280 nm with an ND-1000 spectrophotometer, NanoDrop Technologies, Inc., Rockland, Del.). The copy number of template genomic DNA was estimated on a mass basis assuming a genome size of approximately 5.93 Mb with 64.5% GC content (Gabriel et al., 2006), resulting in an average base pair mass of 616 Da.
LAMP Reaction and Assimilating Probe
LAMP primers (Kubota et al., 2011b) and probes (Kubota et al., 2011a) (table 1) designed to selectively amplify and detect DNA from race 3 biovar 2 strains of Ralstonia solanacearum were synthesized by Integrated DNA Technologies (Coralville, Iowa). LAMP reactions were performed in 25 μL (total volume) reaction mixtures containing 1.6 μM FIP and BIP, 0.2 μM of the F3 and B3 primers, 0.8 μM of the loop B primer, 0.08 μM of the fluorescent strand of the assimilating probe, 0.16 μM of the quenching strand of the assimilating probe, 400 μM deoxynucleoside triphosphates (dNTPs), 1.0 M betaine (Sigma-Aldrich Corp, St Louis, Mo.), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 6 mM MgSO4, 0.1% Triton X-100, and template DNA extracted as described above. Reaction mixes were kept on ice prior to the addition of 8 U Bst DNA polymerase large fragment (New England Biolabs, Inc., Beverly, Mass.). Reactions were carried out in capped (TCS-0803, Bio-Rad, Hercules, Cal.) 0.2 mL microtubes (TBS-0201, Bio-Rad) using the NINA devices described below for temperature control.
Table 1.
Primer and probe sequences used for specific detection of R. solanacearum race 3 biovar 2.
| Nucleotide Sequence (5′ → 3′) | |
|---|---|
| rk2208.1 Primer Set | |
| F3 | GAGAG ACATG TCCGA TTCCG |
|
| |
| B3 | GCCGA TGTCA TCAAG CTCAA |
|
| |
| FIP | TGTGA CTTCC ACGTC AAGCG TTGCA ATCAC CGACT TCCTC A |
|
| |
| BIP | GCGAG AAGCC CGTGT GCTTG TCACG ATTTT CGGCC AGTT |
|
| |
| Loop B | TGCCG AAGAG CTTTT CGCCA ATCGA CT |
|
| |
| Assimilating Probe | |
| Label strand | FAM[a] - ACGCT GAGGA CCCGG ATGCG AATGC GGATG CGGAT GCCGA TGCCG AAGAG CTTTT CGCCA ATCGA CT |
|
| |
| Quench strand | TCGGC ATCCG CATCC GCATT CGCAT CCGGG TCCTC AGCGT - BHQ[b] |
FAM = 6-carboxyfluorescein.
BHQ = Black Hole Quencher-1 (Biosearch Technologies, Novato, Cal.).
Non-Instrumented Nucleic Acid (NINA) Amplification
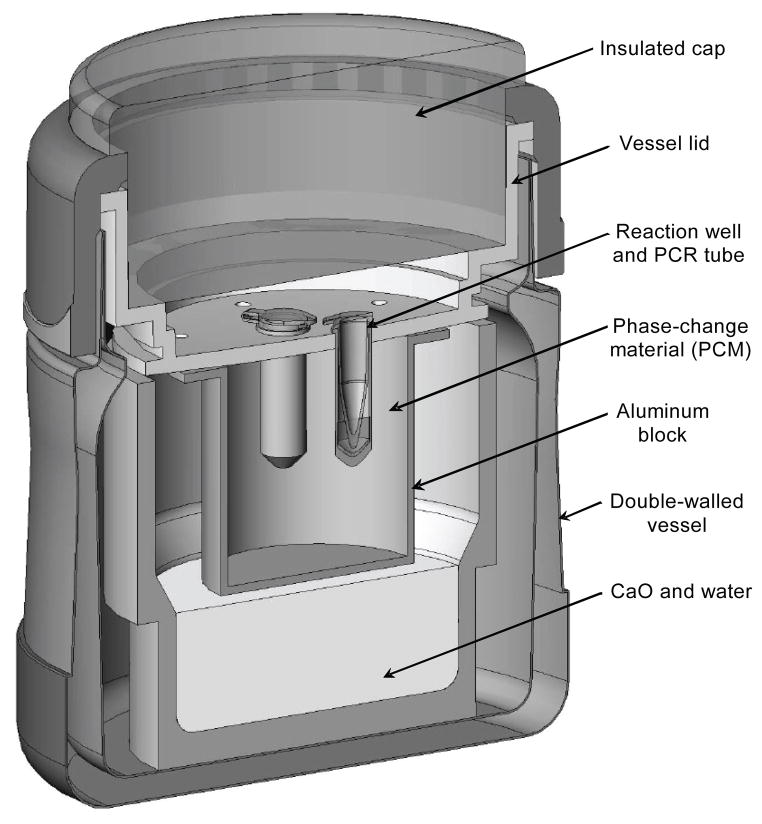
Non-instrumented nucleic acid (NINA) amplification devices to enable the LAMP reaction (fig. 1) were made by modifying ~300 mL (10 oz) double-walled stainless steel vessels with plastic screwtop lids (Part No. B3000BL2, Thermos LLC, Rolling Meadows, Ill.). The plastic lids were modified to accommodate an aluminum block filled with a renewable lipid-based engineered phase-change material (PCM) with a melting point of 65°C (PureTemp 65, Entropy Solutions, Inc., Plymouth, Minn.). To allow for positive and negative controls to be run with each test, each block contained three reaction wells to accommodate PCR tubes in which LAMP reactions could be run. To provide further insulation, a plastic cap filled with a low-density PVC foam insulation (Part No. 9318K77, McMaster-Carr, Robbinsville, N.J.) was designed to slide into the recess above the plastic lid of the reactor vessel. Small boreholes through the plastic portions of the lid and the cap allowed pressure and steam generated from the exothermic reaction to escape from the vessel.
Figure 1.
Isometric cross-section of NINA device to enable non-instrumented temperature control for LAMP reaction. Heat is generated by hydration of calcium oxide, and temperature regulated by the latent energy of the phase-change material (PCM).
Prior to the hydration reaction, 200 μL of distilled water was dispensed into each LAMP reaction well to facilitate thermal equilibration between the PCR tubes and the aluminum block. To run a set of LAMP reactions, 21 g of calcium oxide (CaO) powder (reagent grade, 98%, Catalog No. 248568, Sigma-Aldrich, St. Louis, Mo.) was weighed out into the stainless steel vessel. The plastic lid and cap were then placed over the vessel after adding 7 mL of distilled water uniformly over the CaO powder. After an approximately 10 min pre-heating period to allow the temperature in the block to reach 61°C, the caps were removed from the lids, and freshly prepared PCR tubes with LAMP reagents were loaded into the reaction wells. Tubes were maintained within the NINA amplification devices for 60 min and then removed for fluorometric analysis as described below.
To test the molecular performance of assays run within the NINA devices, reactions were run with standards prepared from serial dilutions of R. solanacearum race 3 biovar 2 DNA isolated as described above. Standards ranged in DNA content from 500 ag (nominally equivalent to about 0.08 genome copies) to 5 μg (equivalent to approximately 8 × 105 genome copies) per reaction, and negative control reactions were run with no template DNA. To generate standard curves, reactions were run for each standard in triplicate, including the non-template controls (NTC). A total of three NINA amplification devices were tested, allowing a full standard curve to be generated with three runs of each device. To test the performance at different ambient conditions, triplicate standards were run once in a lab with no air-conditioning at 31°C ambient temperature and again in an air-conditioned lab maintained at a 22°C ambient temperature. For each complete test, each of the triplicate standards was run in separate NINA amplification devices to control for inter-device variability.
To assess the thermal performance of each NINA device, thermocouples prepared from 40 gauge type-K wire (Part No. TT-K-40-100, Omega Engineering, Inc., Stamford, Conn.) were mounted into the aluminum blocks of each device to monitor the temperature at the interface with the inside of the reaction vessel and within the phase-change material at the middle of the three LAMP reaction wells. Temperature data were logged every second with a commercial data logger (DAQPRO 5300, Omega Engineering, Inc.) throughout the entire operation, starting with the hydration of CaO and ending with the removal of PCR tubes for fluorometric analysis. Collected temperature data from each run were analyzed to determine: time for the phase-change material to reach 61°C; median temperature observed in the phase-change material for 60 min starting 1 min after introducing the PCR tubes; and minimum, maximum, average, and standard deviation about the average temperature values throughout the same 60 min reaction period.
Endpoint Fluorescence Detection
Fluorescence values of completed reactions were measured with a simple customized fluorometer. Sample illumination occurred from the side using a high-intensity blue (470 nm) LED (HLMP CB28 STD00, Agilent Technologies, Santa Clara, Cal.). Fluorescence emission was measured with an adjacent photodiode (T-5 Series, Intor, Inc., Socorro, N.M.) placed at a 90° angle to the light source and interfaced to a high-gain (~5 × 1011 V/A) photoamplifier circuit. Excitation and emission spectra were tuned using narrow band-pass interference filters centered at 470 and 532 nm, respectively (Intor). The first stage of the photoamplifier circuit was built on an electrometer-grade high-impedance diFET operational amplifier (OPA128, Burr-Brown, Tucson, Ariz.), and the second stage included a modest gain first-order low-pass filter with a cutoff frequency near 1 Hz. Voltage from the photoamplifier circuit was digitized by the analog-to-digital converter (ADC) on a commercially available microcontroller (Arduino Duemilenove, Arduino, Italy). In order to improve the resolution and noise immunity of the fluorometer, voltage was sampled by the 10-bit ADC of the microcontroller 64 times at approximately 120 Hz. These 64 samples were summed and stored as a 16-bit number representative of the “relative fluorescence value.” To allow the system to reach optical and electrical equilibrium, fluorescence values were recorded 30 s after introduction of the sample into the fluorometer. While handheld fluorometers are commercially available, they are currently priced at about $2000 and above. In contrast, our custom fluorometer can be assembled with components costing less than $200, including the components required to include temperature control functionality to make a real-time handheld instrument.
For comparison to our custom device, fluorescence values were also recorded using a commercial real-time PCR instrument (iQ5, Bio-Rad, Hercules, Cal.), using the filter settings for SYBR Green.
Statistical Analysis
Samples were classified as positive or negative for LAMP reaction based on comparison of observed fluorescence values to a threshold value. Threshold values were set as the average of a set of six reactions with non-template DNA plus three standard deviations of the mean of these values. All fluorescence data were used to determine a best-fit logistic model of the form:
| (1) |
where F is the relative fluorescence value corrected for the average fluorescence value of non-template controls, x is the quantity (fg) of DNA in the reaction, and a, b, and x0 are empirical coefficients. Regression was implemented numerically using commercially available software (SigmaPlot 10.0, Systat Software, Chicago, Ill.). Detection limits were estimated from the regression results as the DNA quantity equivalent to a relative fluorescence value of three times the root mean squared error of the regression (Kubota et al., 2008).
Results and Discussion
Thermal Performance of Non-Instrumented Devices
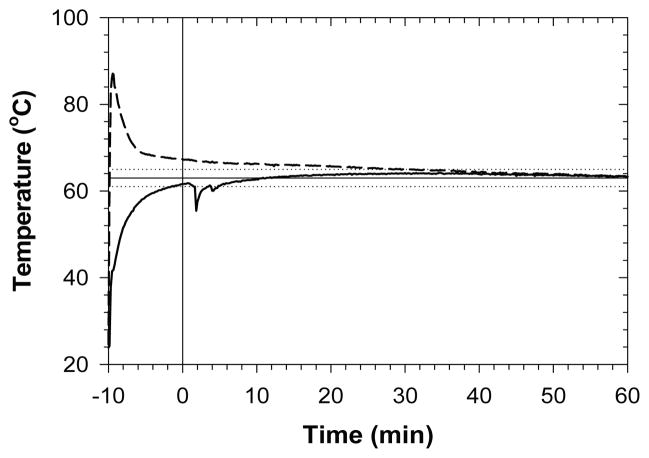
The NINA amplification devices generally operated close to the defined specifications of heating the PCM up to 61°C within 10 min of the start of the CaO hydration reaction, and maintaining the PCM at 63°C ±2°C over the course of a 1 h LAMP reaction (table 2). The most common deviation was from the minimum temperature specification during the LAMP reaction, with 14 out of 18 trials dipping below the minimum temperature at some point during the reaction (data not shown). These deviations typically occurred only briefly at the very start of reactions (e.g., fig. 2) as temperatures dropped after removing the caps to place the LAMP reaction tubes into the wells of the thermostated block. It was also relatively common (10 out of 18 trials) for blocks to take more than 10 min to warm up to 61°C, but in all cases the blocks heated up to specification within about 16 min. All design specifications were predicated on an ambient temperature of 22°C, so deviations occurred more frequently when devices were used at an ambient temperature of 31°C. The maximum temperature specification was exceeded five out of nine times that devices were used at 31°C ambient temperature. In general, variability in performance was significantly higher for the NINA devices when run at the higher ambient temperature (table 2). Even so, average and median temperatures within the devices were close to specification under all conditions (table 2; fig. 2).
Table 2.
Summary statistics for thermal performance of NINA amplification device.
| Statistic | 22°C Ambient Temperature
|
31°C Ambient Temperature
|
||
|---|---|---|---|---|
| Mean[a] | SD[b] | Mean[a] | SD[b] | |
| Time to 61°C[c] | 10.91 | 1.76 | 11.42 | 3.46 |
| Average[d] | 63.4 | 0.6 | 64.1 | 1.2 |
| SD[d] | 0.7 | 0.3 | 1.3 | 0.8 |
| Median[d] | 63.5 | 0.7 | 64.2 | 1.2 |
| Minimum[d] | 60.8 | 0.3 | 60.5 | 0.7 |
| Maximum[d] | 64.3 | 1.0 | 65.9 | 2.3 |
Mean of observed parameter over all trials of devices (n = 9 at each ambient temperature).
Sample standard deviation of observed parameter over all trials of devices (n = 9 at each temperature).
Time for regulated chamber to reach 61°C after adding water to CaO (min).
Temperature parameter observed at 1 h intervals after introducing sample (°C).
Figure 2.
Representative temperature profiles within a NINA amplification device. The dashed curve represents the CaO hydration chamber, and the solid curve represents the phase-change regulated chamber, where the LAMP reaction occurs. Water is added to CaO at −10 min, and samples are inserted into the regulated chamber at or shortly after time 0. The horizontal lines show the design temperature (solid line) with upper and lower bounds (dotted lines) within the regulated chamber.
Validation of Custom Fluorometer against Commercial Instrument
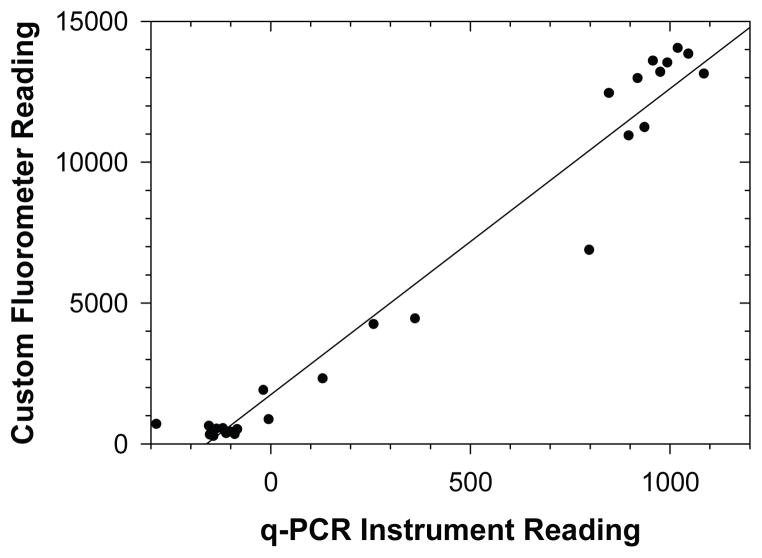
The fluorescence values measured with the custom fluorometer correlated reasonably well with those measured using the commercial real-time PCR instrument (fig. 3). In addition, classification of samples according to fluorescence values read by the real-time PCR instrument did not differ from the classifications made by the custom fluorometer (data not shown). These findings suggest that the simple handheld instrument was sufficient in range, resolution, and reproducibility to detect fluorescence changes due to the LAMP reaction and probe. In previous research, LAMP reactions were stopped by thermally denaturing the polymerase at 80°C, a step that was not possible in the NINA amplification devices. In this work, fluorescence values of completed reactions stored at 4°C were stable over a period of at least 48 h (data not shown), even when the reactions were not stopped. This indicated that the assimilating probes were stable within the amplicon, and that non-specific amplifications or other processes occurring post-reaction did not interact with the sequence-specific probe. This demonstrates a further advantage of the assimilating probe technology compared to non-sequence-specific detection technologies for LAMP, especially when using non-instrumented heaters or simple water baths to implement LAMP. In these situations, there is no need for an additional reaction stop treatment, and completed reactions can still be reliably read for days after the reaction.
Figure 3.
Relative fluorescence values read by custom device compared to real-time PCR instrument readings immediately following reaction; R2 = 0.964; y = 10.87x + 1745).
LAMP Amplification and Endpoint Detection
Despite brief deviations from thermal performance specifications, LAMP assays implemented within the NINA amplification devices were robust and reliable for detecting DNA from R. solanacearum race 3 biovar 2 at all ambient conditions tested (fig. 4). Standard dilutions resulting in less than a single genome copy equivalent did not result in positive amplification, consistent with low likelihoods of sampling the intact gene at these high dilution rates. Standards nominally containing about eight genome copy equivalents were classified as positive for the LAMP reaction 50% of the time, indicating that the assay implemented in the NINA amplification approach is capable of detecting an infectious dose of fewer than ten bacteria. Statistically, the detection limit for reliable detection was equivalent to about 137 genome copies, significantly lower than LAMP detection based on turbidimetric analysis (~105 CFU) (Kubota et al., 2008). Despite higher incidence of deviation from thermal specifications within the NINA devices, the molecular assay worked at least as well at 31°C ambient temperature as at 22°C ambient temperature, demonstrating the robustness of the LAMP assay and the value of the NINA amplification technology even at prevailing conditions in typical tropical environments.
Figure 4.
Endpoint fluorescence values for DNA standards subjected to non-instrumented nucleic acid amplification with assimilating probe. Standards classified as positive (black circles) and negative (white circles) for amplification describe a best-fit logistic curve (a = 11380; b = −0.6413; x0 = 1009; R2 = 0.841, RMSE = 2130) resulting in a detection limit of 861 fg DNA (vertical solid line, equivalent to 137 copies of the R. solanacearum genome). A single genome copy equivalent is represented by the vertical dashed line.
Conclusions and Future Work
Innovative new technologies were successfully demonstrated for detection of pathogen-specific DNA with a minimal amount of specialized instrumentation. Molecular amplification was robust and reliable when implemented in a completely non-instrumented device, and detection of the reaction could be readily achieved using a customized, battery-powered handheld fluorometer without any further processing or manipulation of the amplification product. Reagent and disposable costs were approximately $0.60 per test. Retail costs of components were approximately $25 for a single NINA amplification device and approximately $100 for the optical components and controller of the custom fluorometer. Hardware was simple and could be used with a minimal amount of training by following simple instructions, and the entire system was shown to be reliable even in conditions representative of tropical and subtropical field conditions.
For practical adaptation by agricultural users in the field, disposables need to be available in a ready-to-use form. Stabilized ready-to-use reaction tubes can be prepared by freeze-drying the LAMP reagents (Pack and Deng, 2008). While nucleic acid based analyses can be implemented directly in agricultural samples (e.g., Kubota et al., 2008), agricultural samples such as soil slurries, irrigation water, and plant tissues contain numerous inhibitors that can impair the sensitivity and detection limit of these methods. In addition, to improve detection limit and reliability, it is often desirable to concentrate DNA from highly dilute, dispersed pathogens in large volumes of soil, irrigation water, or plant tissues. To address these issues, we are currently investigating a number of commercially available systems for rapid DNA extraction, as well as our own approaches to rapidly process large volumes of these materials and extract purified DNA from them using simple handheld devices.
To improve the functionality of the NINA devices, we are also developing and testing new prototypes that can accommodate greater numbers of tests simultaneously, that can use alternative geometries for improved heat transfer, and that can use a small quantity (~100 mL) of boiling water instead of hydrated CaO as an energy source.
Acknowledgments
We gratefully acknowledge support from USDA-NRI projects 2005-55605-16683 and 2007-55605-17843 for the development of molecular tools for detection of agricultural pathogens. The work on the NINA device at PATH is supported by grants under the Health Innovation Portfolio, which is supported by a partnership of the U.S. Agency for International Development, private foundations, and individual donors.
Contributor Information
Ryo Kubota, Laboratory Manager, Department of Molecular Biosciences and Bioengineering, University of Hawaii, Honolulu, Hawaii.
Paul LaBarre, Technical Officer/Portfolio Leader, Center for POC for Global Health, Program for Appropriate Technologies in Health (PATH), Seattle, Washington.
Jered Singleton, Product Development Technician, Center for POC for Global Health, Program for Appropriate Technologies in Health (PATH), Seattle, Washington.
Andy Beddoe, Product Development Officer, Center for POC for Global Health, Program for Appropriate Technologies in Health (PATH), Seattle, Washington.
Bernhard H. Weigl, Director, Center for POC for Global Health, Program for Appropriate Technologies in Health (PATH), Seattle, Washington
Anne M. Alvarez, Professor, Department of Plant and Environmental Protection Sciences
Daniel M. Jenkins, ASABE Member, Associate Professor, Department of Molecular Biosciences and Bioengineering, University of Hawaii, Honolulu, Hawaii.
References
- Bissonnette L, Bergeron MG. Diagnosing infections: Current and anticipated technologies for point-of-care diagnostics and home-based testing. Clin Microbiol and Infection. 2010;16(8):1044–1053. doi: 10.1111/j.1469-0691.2010.03282.x. [DOI] [PubMed] [Google Scholar]
- Chen DF, Mauk M, Qiu XB, Liu CC, Kim JT, Ramprasad S, Ongagna S, Abrams WR, Malamud D, Corstjens P, Bau HH. An integrated, self-contained microfluidic cassette for isolation, amplification, and detection of nucleic acids. Biomed Microdevices. 2010;12(4):705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FJ, Hiramatsu K, Huang IW, Wang CH, Lauderdale TLY. Panton-Valentine leukocidin (PVL)-positive methicillin-susceptible and resistant Staphylococcus aureus in Taiwan: Identification of oxacillin-susceptible mecA-positive methicillin-resistant S. aureus. Diagnostic Microbiol and Infectious Dis. 2009;65(4):351–357. doi: 10.1016/j.diagmicrobio.2009.07.024. [DOI] [PubMed] [Google Scholar]
- Conceicao T, Tavares A, Miragaia M, Hyde K, Aires-de-Sousa M, de Lencastre H. Prevalence and clonality of methicillin-resistant Staphylococcus aureus (MRSA) in the Atlantic Azores islands: Predominance of SCCmec types IV, V, and VI. European J Clinical Microbiol and Infectious Dis. 2010;29(5):543–550. doi: 10.1007/s10096-010-0892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny TP. Plant pathogenic Ralstonia species. In: Gnanamanickam SS, editor. Plant-Associated Bacteria. Dordrecht, The Netherlands: Springer; 2006. pp. 573–644. [Google Scholar]
- Fan CH, Plaxco KW, Heeger AJ. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc Natl Acad Sci. 2003;100(16):9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fegan M, Prior P. How complex is “the Ralstonia solanacearum species complex”? In: Allen C, Prior P, Hayward AC, editors. Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex. St. Paul, Minn: American Phytopathology Society; 2005. pp. 449–461. [Google Scholar]
- Floyd J. New pest response guidelines: Ralstonia solanacearum race 3 biovar 2 southern wilt of geranium. Riverdale, Md: USDA-APHIS Plant Protection and Quarantine; 2004. [Google Scholar]
- Gabriel DW, Allen C, Schell M, Denny TP, Greenberg JT, Duan YP, Flores-Cruz Z, Huang Q, Clifford JM, Presting G, Gonzalez ET, Reddy J, Elphinstone J, Swanson J, Yao J, Mulholland V, Liu L, Farmerie W, Patnaikuni M, Balogh B, Norman D, Alvarez A, Castillo JA, Jones J, Saddler G, Walunas T, Zhukov A, Mikhailova N. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Molecular Plant-Microbe Interactions. 2006;19(1):69–79. doi: 10.1094/MPMI-19-0069. [DOI] [PubMed] [Google Scholar]
- Guidot A, Elbaz M, Carrere S, Siri MI, Pianzzola MJ, Prior P, Boucher C. Specific genes from the potato brown rot strains of Ralstonia solanacearum and their potential use for strain detection. Phytopathology. 2009;99(9):1105–1112. doi: 10.1094/PHYTO-99-9-1105. [DOI] [PubMed] [Google Scholar]
- Janse JD, Araluppan FAX, Schans J, Wenneker M, Westerhuis W. Experiences with bacterial brown rot Ralstonia solanacearum biovar 2, race 3 in The Netherlands. In: Prior P, Allen C, Elphinstone JG, editors. Bacterial Wilt Disease: Molecular and Ecological Aspects. Berlin, Germany: Springer-Verlag; 1998. pp. 146–154. [Google Scholar]
- Jenkins DM, Chami B, Kreuzer M, Presting G, Alvarez AM, Liaw BY. Hybridization probe for femtomolar quantification of selected nucleic acid sequences on a disposable electrode. Analytical Chem. 2006;78(7):2314–2318. doi: 10.1021/ac051619s. [DOI] [PubMed] [Google Scholar]
- Jenkins DM, Song CY, Fares S, Cheng H, Barrettino D. Disposable thermostated electrode system for temperature-dependent electrochemical measurements. Sensors and Actuators B. 2009;137(1):222–229. [Google Scholar]
- Kubota R, Vine BG, Alvarez AM, Jenkins DM. Detection of Ralstonia solanacearum by loop-mediated isothermal amplification. Phytopathology. 2008;98(9):1045–1051. doi: 10.1094/PHYTO-98-9-1045. [DOI] [PubMed] [Google Scholar]
- Kubota R, Jenkins DM, Alvarez AM, Su WW. FRET-based assimilating probe for sequence-specific real-time monitoring of loop-mediated isothermal amplification (LAMP) Biological Eng Trans. 2011a;4(2):81–100. [Google Scholar]
- Kubota R, Schell MA, Peckham GD, Rue J, Alvarez AM, Allen C, Jenkins DM. In silico genomic subtraction guides development of highly accurate, DNA-based diagnostics for Ralstonia solanacearum race 3 biovar 2 and blood disease bacterium. J General Plant Pathology. 2011b;77(3):182–193. [Google Scholar]
- LaBarre P, Wilmoth J, Beddoe A, Singleton J, Gerlach J, Weigl B. Proc 41st Annual AACC Oak Ridge Conf. Washington, D.C: American Association for Clinical Chemistry; 2009. LAMP without electric heat: A chemically heated, non-instrumented nucleic acid amplification assay platform for point-of-care use. [Google Scholar]
- LaBarre P, Gerlach J, Wilmoth J, Beddoe A, Singleton J, Weigl B. Proc 32nd Annual Intl Conf IEEE EMBS. Piscataway, N.J: IEEE; 2010a. Non-instrumented nucleic acid amplification (NINA): Instrument-free molecular malaria diagnostics for low-resource settings. [DOI] [PubMed] [Google Scholar]
- LaBarre P, Gerlach J, Wilmoth J, Beddoe A, Singleton JL, Hawkins K, Weigl B. Proc 42nd Annual AACC Oak Ridge Conf. Washington, D.C: American Association for Clinical Chemistry; 2010b. Non-instrumented nucleic acid amplification (NINA): Instrument-free molecular diagnostics for low-resource settings. [DOI] [PubMed] [Google Scholar]
- Lui C, Cady NC, Batt CA. Nucleic acid-based detection of bacterial pathogens using integrated microfluidic platform systems. Sensors. 2009;9(5):3713–3744. doi: 10.3390/s90503713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milling A, Meng FH, Denny TP, Allen C. Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2. Phytopathology. 2009;99(10):1127–1134. doi: 10.1094/PHYTO-99-10-1127. [DOI] [PubMed] [Google Scholar]
- Norman D, Alvarez A. A rapid method for presumptive identification of Xanthomonas campestris pv. dieffenbachiae and other xanthomonads. Plant Disease. 1989;73(8):654–658. [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack TD, Deng X. Stable reagents and kits useful in loop-mediated amplification (LAMP) 20080182312. Washington, D.C: U.S. Patent and Trademark Office; Application. 2008
- Peeling RW, Mabey D. Point-of-care tests for diagnosing infections in the developing world. Clin Microbiol and Infection. 2010;16(8):1062–1069. doi: 10.1111/j.1469-0691.2010.03279.x. [DOI] [PubMed] [Google Scholar]
- Swanson JK, Yao J, Tans-Kersten J, Allen C. Behavior of Ralstonia solanacearum race 3 biovar 2 during latent and active infection of geranium. Phytopathology. 2005;95(2):136–143. doi: 10.1094/PHYTO-95-0136. [DOI] [PubMed] [Google Scholar]
- van Elsas JD, Kastelein P, van Bekkum P, van der Wolf JM, de Vries PM, van Overbeek LS. Survival of Ralstonia solanacearum biovar 2, the causative agent of potato brown rot, in field and microcosm soils in temperate climates. Phytopathology. 2000;90(12):1358–1366. doi: 10.1094/PHYTO.2000.90.12.1358. [DOI] [PubMed] [Google Scholar]
- Weigl B, Domingo G, LaBarre P, Gerlach J. Towards non- and minimally instrumented, microfluidics-based diagnostic devices. Lab on a Chip. 2008;8(12):1999–2014. doi: 10.1039/b811314a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annual Review Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]