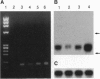
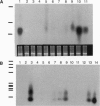
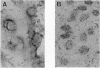
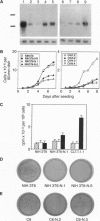
Abstract
Immortalized neural precursor cell lines carrying the polyoma large tumor (T) gene have been shown previously to retain a clear-cut contact inhibition of growth and to differentiate in vitro. In the present study, we have identified and isolated cDNA clones corresponding to RNA expressed preferentially when these cells reach confluence. One of them, NPDC-1, is expressed specifically in the nervous system. The transfection of dividing cells with a NPDC-1 expression vector results in the inhibition of cell proliferation. In addition, the stable introduction of NPDC-1 into transformed cells, even of nonneural origin, leads to the suppression of transformed characteristics.
Full text
PDF

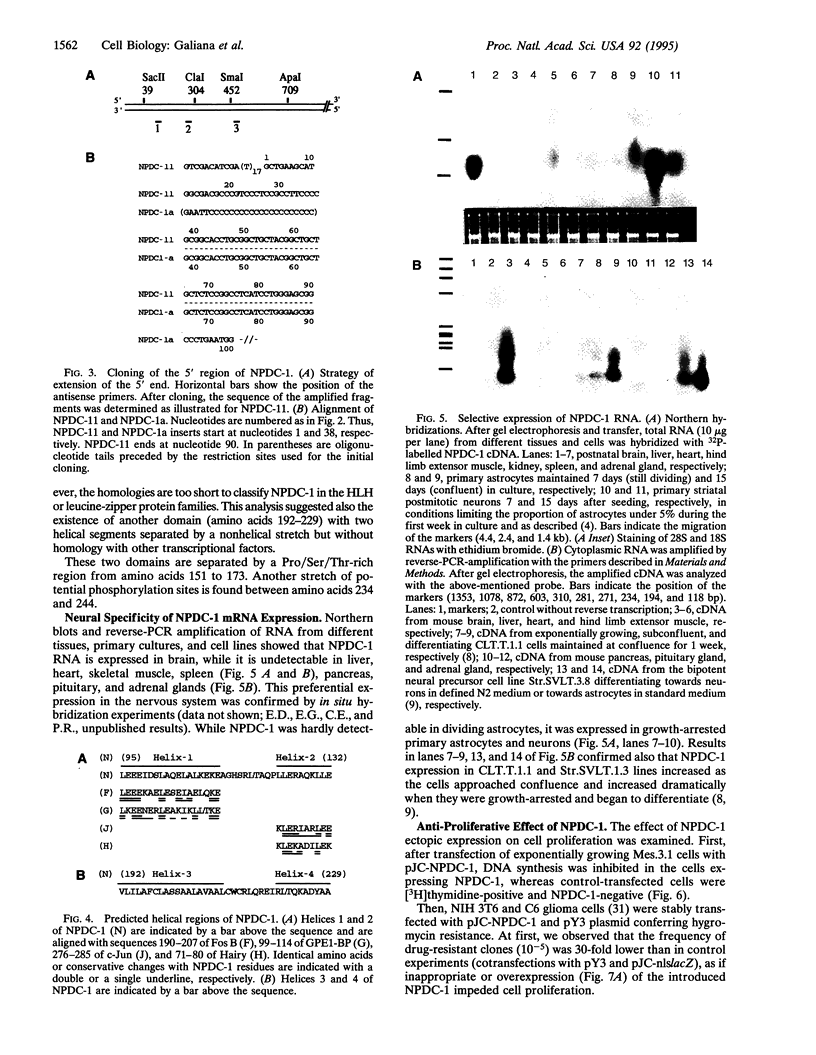
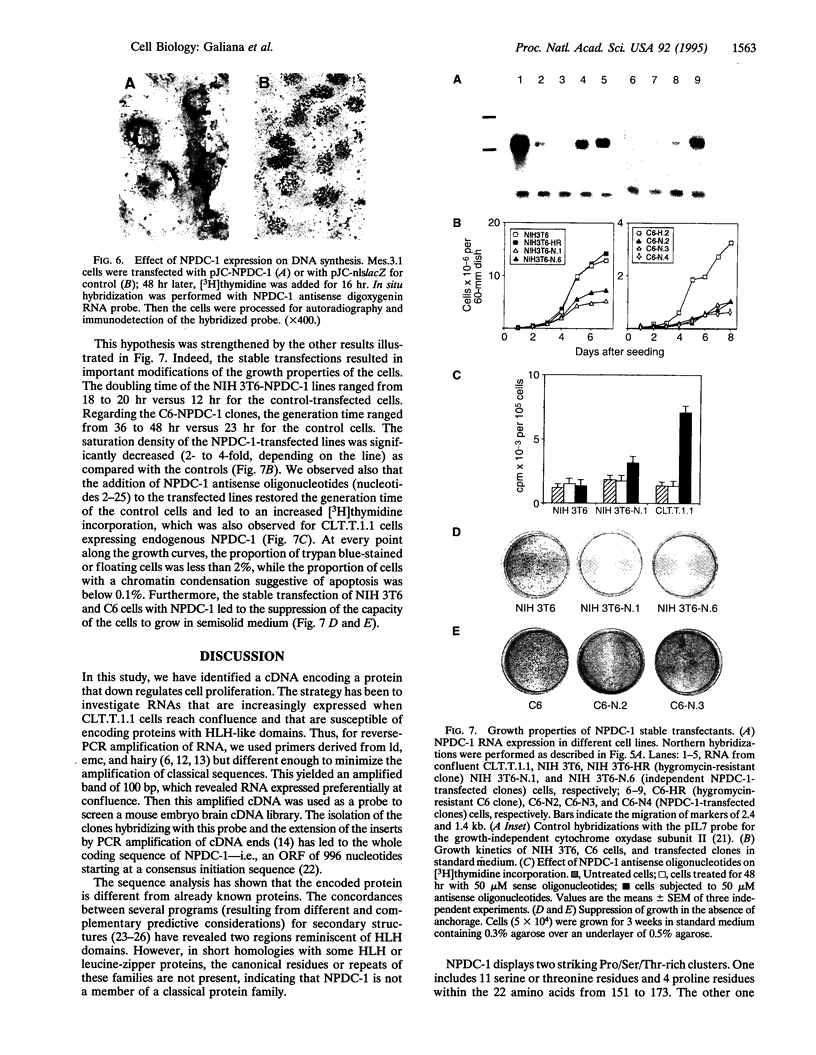

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biou V., Gibrat J. F., Levin J. M., Robson B., Garnier J. Secondary structure prediction: combination of three different methods. Protein Eng. 1988 Sep;2(3):185–191. doi: 10.1093/protein/2.3.185. [DOI] [PubMed] [Google Scholar]
- Blochlinger K., Diggelmann H. Hygromycin B phosphotransferase as a selectable marker for DNA transfer experiments with higher eucaryotic cells. Mol Cell Biol. 1984 Dec;4(12):2929–2931. doi: 10.1128/mcb.4.12.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Romero J. M., Doyle S., Elliott R., Murphy G., Carpenter R. floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell. 1990 Dec 21;63(6):1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Ellis H. M., Spann D. R., Posakony J. W. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990 Apr 6;61(1):27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- Evrard C., Borde I., Marin P., Galiana E., Prémont J., Gros F., Rouget P. Immortalization of bipotential and plastic glio-neuronal precursor cells. Proc Natl Acad Sci U S A. 1990 Apr;87(8):3062–3066. doi: 10.1073/pnas.87.8.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard C., Galiana E., Rouget P. Establishment of 'normal' nervous cell lines after transfer of polyoma virus and adenovirus early genes into murine brain cells. EMBO J. 1986 Dec 1;5(12):3157–3162. doi: 10.1002/j.1460-2075.1986.tb04623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana E., Borde I., Marin P., Rassoulzadegan M., Cuzin F., Gros F., Rouget P., Evrard C. Establishment of permanent astroglial cell lines, able to differentiate in vitro, from transgenic mice carrying the polyoma virus large T gene: an alternative approach to brain cell immortalization. J Neurosci Res. 1990 Jul;26(3):269–277. doi: 10.1002/jnr.490260302. [DOI] [PubMed] [Google Scholar]
- Glaichenhaus N., Léopold P., Cuzin F. Increased levels of mitochondrial gene expression in rat fibroblast cells immortalized or transformed by viral and cellular oncogenes. EMBO J. 1986 Jun;5(6):1261–1265. doi: 10.1002/j.1460-2075.1986.tb04355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F. A., Raden D. L., Davis R. J. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem. 1991 Nov 25;266(33):22159–22163. [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- He X., Rosenfeld M. G. Mechanisms of complex transcriptional regulation: implications for brain development. Neuron. 1991 Aug;7(2):183–196. doi: 10.1016/0896-6273(91)90257-z. [DOI] [PubMed] [Google Scholar]
- Hirai S., Bourachot B., Yaniv M. Both Jun and Fos contribute to transcription activation by the heterodimer. Oncogene. 1990 Jan;5(1):39–46. [PubMed] [Google Scholar]
- Jan Y. N., Jan L. Y. Genes required for specifying cell fates in Drosophila embryonic sensory nervous system. Trends Neurosci. 1990 Dec;13(12):493–498. doi: 10.1016/0166-2236(90)90083-m. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemesle-Varloot L., Henrissat B., Gaboriaud C., Bissery V., Morgat A., Mornon J. P. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2-D-representation of protein sequences. Biochimie. 1990 Aug;72(8):555–574. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- Loeber G., Dörries K. DNA rearrangements in organ-specific variants of polyomavirus JC strain GS. J Virol. 1988 May;62(5):1730–1735. doi: 10.1128/jvi.62.5.1730-1735.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Wakabayashi-Ito N., Nagata S. Molecular cloning of cDNA and a chromosomal gene encoding GPE1-BP, a nuclear protein which binds to granulocyte colony-stimulating factor promoter element 1. FEBS Lett. 1991 Apr 22;282(1):95–97. doi: 10.1016/0014-5793(91)80452-9. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Kim P. S. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992 Feb 21;68(4):699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- Pelech S. L., Sanghera J. S. Mitogen-activated protein kinases: versatile transducers for cell signaling. Trends Biochem Sci. 1992 Jun;17(6):233–238. doi: 10.1016/s0968-0004(00)80005-5. [DOI] [PubMed] [Google Scholar]
- Rushlow C. A., Hogan A., Pinchin S. M., Howe K. M., Lardelli M., Ish-Horowicz D. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 1989 Oct;8(10):3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler H. R. Octamania: the POU factors in murine development. Trends Genet. 1991 Oct;7(10):323–329. doi: 10.1016/0168-9525(91)90422-m. [DOI] [PubMed] [Google Scholar]
- Unger R., Harel D., Wherland S., Sussman J. L. A 3D building blocks approach to analyzing and predicting structure of proteins. Proteins. 1989;5(4):355–373. doi: 10.1002/prot.340050410. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Dwarki V. J., Verma I., Davis R., Hollenberg S., Snider L., Lassar A., Tapscott S. J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991 Aug;5(8):1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- Zerial M., Toschi L., Ryseck R. P., Schuermann M., Müller R., Bravo R. The product of a novel growth factor activated gene, fos B, interacts with JUN proteins enhancing their DNA binding activity. EMBO J. 1989 Mar;8(3):805–813. doi: 10.1002/j.1460-2075.1989.tb03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]