Abstract
Fragile X syndrome (FXS), the most common genetic form of autism spectrum disorder, is caused by deficiency of the fragile X mental retardation protein (FMRP). Despite extensive research and scientific progress, understanding how FMRP regulates brain development and function remains a major challenge. FMRP is a neuronal RNA-binding protein that binds about a third of messenger RNAs in the brain and controls their translation, stability, and cellular localization. The absence of FMRP results in increased protein synthesis, leading to enhanced signaling in a number of intracellular pathways, including the mTOR, mGLuR5, ERK, Gsk3β, PI3K, and insulin pathways. Until recently, FXS was largely considered a deficit of mature neurons; however, a number of new studies have shown that FMRP may also play important roles in stem cells, among them neural stem cells, germ line stem cells, and pluripotent stem cells. In this review, we will cover these newly discovered functions of FMRP, as well as the other two fragile X-related proteins, in stem cells. We will also discuss the literature on the use of stem cells, particularly neural stem cells and induced pluripotent stem cells, as model systems for studying the functions of FMRP in neuronal development.
2. Introduction
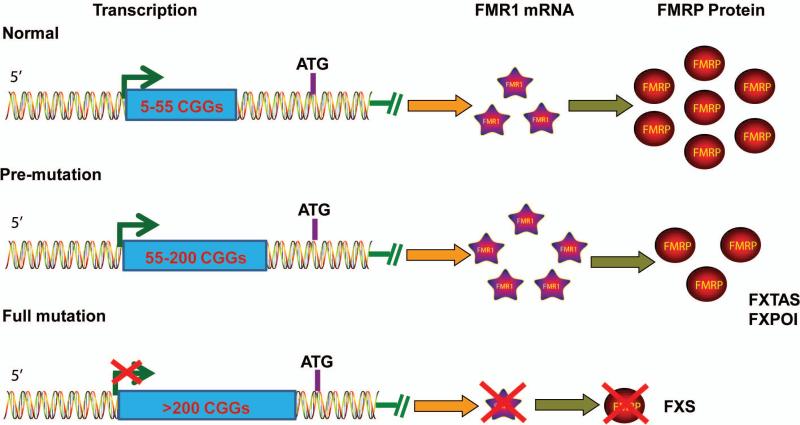
Fragile X syndrome (FXS) affects approximately 1:4000 males and 1:6000 females and is the most common genetic neurodevelopmental disorder, as well as the largest single genetic contributor to autism. The most common cause of FXS is the expansion of CGG trinucleotide repeats at the 5’ untranslated region (5’-UTR) of the FMR1 gene to over 200 repeats, which triggers DNA methylation and aberrant heterochromatinization in the promoter region of the gene, leading to gene silencing and the functional absence of fragile X mental retardation protein (FMRP) (Figure 1) [1,2] Unaffected individuals have fewer than 55 CGG repeats, which are relatively stable across generations. Starting at approximately 55 repeats, the number of CGG repeats becomes unstable and may expand across generations. Individuals with 55 to 200 repeats, termed “premutation” carriers, have increased transcription of the FMR1 gene, with somewhat reduced levels of FMRP. Since FMRP is still present, albeit at reduced levels in most cases, FMR1 premutation carriers do not exhibit the same neurological phenotypes as FXS patients. Nevertheless, premutation carriers may develop two other disorders: fragile X-associated tremor/ataxia syndrome (FXTAS) and fragile X-associated primary ovarian insufficiency (FXPOI), which may result from high levels of FMR1 mRNA containing long CGG repeats [1,3,4]. Hence there is a single FMR1 gene implicated in several different disorders, with FXS being the most severe. Investigating the roles of FMRP in brain development and function is understandably an active area of research, with new functions and pathways of FMRP being continuously discovered [see recent review [1]].
Figure 1. Mutations in the FMR1 gene can lead to several different diseases.
Normal individuals have fewer than 55 CGG repeats. FMR1 premutation carriers can have between 55 and 200 repeats. In these carriers, FMR1 mRNA is expressed at higher levels than in normal individuals, but FMRP protein levels may decrease due to largely unclear mechanisms. These individuals have an increased chance of developing two distinct disorders, FRAXA and FXPOI. When CGG repeat length exceeds 200, the so-called “full mutation,” the FMR1 gene is methylated and silenced, which is the major cause of fragile X syndrome (FXS).
In this review, we will cover both known and potential roles of FMRP in the maintenance and fate determination of several types of stem cells, including germline stem cells (GSCs), neural stem cells in developing brains (eNSCs) and adult brains (aNSCs), embryonic stem cells (ESCs), and inducible pluripotent stem cells (iPSCs). We will discuss the implications of these findings in terms of the roles FMRP plays in development, homeostasis, and regeneration and therapeutic developments for FXS.
3. FMRP
3.1 FMRP Protein Structure Underlying Complex Functions
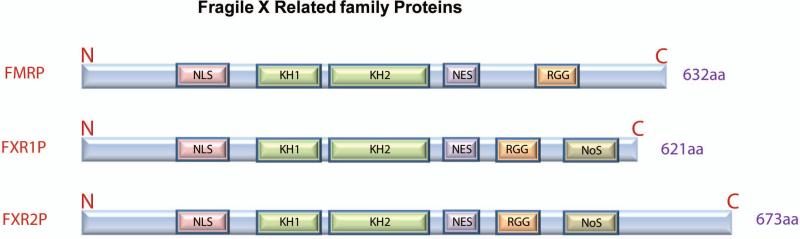
FMRP contains multiple domains that form the basis for its complex functions: two KH RNA-binding domains, an RGG box containing a conserved Arg-Gly-Gly triplet for RNA binding, a nuclear localization signal (NLS), and a nuclear export signal (NES) (Figure 2). FMRP is known to bind mRNA and form a messenger ribonucleoprotein (mRNP) complex that associates with polyribosomes. The second KH (KH2) domain of FMRP seems to be essential for RNA binding because a missense mutation (I304N) in this region abolishes its RNA binding ability [5]. However, other studies imply that the KH2 domain and RGG box can bind RNA independently [6,7]. Moreover, the N-terminal region of FMRP may also play an important role in binding mRNA [8]. How FRMP recognizes its mRNA targets remains an active area of research with a host of unanswered questions. The KH2 domain seems to specifically recognize a specific sequence within a tertiary structure in the target mRNA called the “FMRP kissing complex” [5]. The RGG boxes in FMRP bind mRNAs containing G-quartet motifs, another specific tertiary structure of mRNAs [9]. However, genome-wide sequencing data seem to contradict the existence of such target specificity [10]. A recent study has identified RNA-recognition elements corresponding to different RNA-binding domains of FMRP in a human HEK293 cell line [7]. How these RNA-binding domains independently recognize or coordinately specify the large number of FMRP targets remains unclear. Aside from its complex protein structure, FMRP can also be phosphorylated, and phosphorylated FMRP seems to be associated with stalled ribosomes [11,12]. FMRP phosphorylate may interfere with its interaction with Dicer [13]. Another study demonstrates that the binding of FMRP to the 3’UTR of PSD-95 mRNA depends on FMRP phosphorylation at serine 499 [14]. Therefore, FMRP phosphorylation may be another key regulatory feature.
Figure 2. Schematic illustration of fragile X-related family proteins (FXRs).
The three proteins share high homology in amino acid sequences of functional domains. NLS, nuclear localization signal; NES, nuclear export signal; KH domains, RGG, arginine/glycine-rich box; NoS, nucleolar targeting signal.
In addition to FMRP, the family of fragile X-related proteins (FXRs) also contains two autosomal paralogs, FXR1 and FXR2 (also known as FXR1P and FXR2P). The three FXR proteins share similar functional domains, but diverge in the C-termini and in the nucleolar localization signal sequence (NoS) (Figure 2), suggesting they may possess both common and distinct functions [15]. Although studies have shown that FXR proteins can interact with each other, the contribution of FXR1 and FXR2 to FXS is unclear. All three FXRs are highly expressed in the brain. In addition, FXR1 is also expressed in muscle cells and is necessary for the proper development of muscle [16,17,18]. Most genome-wide analyses of brain tissues or cell lines suggest that the three FXRs have largely overlapping RNA targets and therefore functions [7,9,10]. Like FMRP, FXR1 and FXR2 are known to associate predominantly with polyribosome [19]. Both FMRP and FXR2 regulate mammalian circadian behavioral rhythms [20]. Both FMRP and FXR1 can bind the Cdc42 effector PAK1[21]. FXR2 knockout mice display both distinct and similar behavioral phenotypes compared with FMR1 knockout mice [22]. On the other hand, limited studies also suggest that the three FXRs may have differential functions. For example, the RGG box in FMRP, but not the other FXRs, can bind G-quartet structures [9]. FXR2 is present in all neuronal RNA granules containing FXRs (FXGs) and is essential for the formation of these FXGs, whereas FMRP and FXR1 are only found in a subset of FXGs [23,24]. FXR2 and FMRP double mutation mice display exaggerated neurological behavioral deficits, including learning deficits, suggesting that FMRP and FXR2 play a cooperative role in brain development and plasticity [20,25].
3.2 FMRP Regulates Gene Expression through Post-Transcriptional Mechanisms
The construction of neuronal circuits in the developing brain requires the correct assembly of trillions of synaptic connections. Precisely regulated protein synthesis, particularly at neuronal dendrites and synapses, is required in response to developmental cues or synaptic stimuli. The absence of functional FMRP in both FXS patients and animal models leads to overall increased protein synthesis in neurons, suggesting that FMRP acts generally as a repressor for protein expression [1]. FMRP can regulate protein expression in a number of ways, including translational repression, RNA transport, RNA stability, etc. For example, FMRP deficiency leads to increased signaling of several intracellular signaling pathways. Among them, both the mTOR and ERK pathways activate the initiation factor F (eIF) complex required for mRNA Cap-dependent protein translation. Moreover, FMRP is known to interact with CYFIP1, which binds eIF for the initiation of protein translation [26]. FMRP can also inhibit protein translation through ribosome stalling during the elongation phase of translation [10,11]. In addition, FMRP can regulate translation through noncoding small RNAs, particularly microRNAs (miRNAs). Both Drosophila and mammalian FMRP associate with Argonaute 2 and Dicer, two critical components of the RNA-induced silencing complex (RISC)[27,28,29,30]. Overexpression of either miR-125 or miR-132 results in dendritic spine defects in neuronal cells, and knockdown of FMRP in these cells rescues both phenotypes [31]. Another study showed that FMRP and miR-125a together bind the 3’UTR of PSD-95 and inhibit PSD-95 protein translation [14]. These data support an important role for FMRP in the regulation of protein translation.
FMRP is present in both the nucleus and cytoplasm and plays important roles in the activity-dependent transport of specific mRNAs [32]. FMRP transports mRNAs in RNA granules, which is facilitated by other RNA-binding proteins, such as microspherule protein 58 (MSP58) and Caprin-1, and by interacting with motor protein KIF3C [33,34,35]. Dendritic mRNA transport is important for neuronal dendritic spine morphogenesis [36], which may explain the dendritic and spine pathology seen in FXS patients[37,38,39].
The stability of mRNA is highly regulated, which has a major impact on protein expression; however, whether FMRP directly regulates mRNA stability remains unclear. Two recent studies have implicated FMRP as a direct regulator of mRNA stability. FMRP is known to bind PSD-95 mRNA and protect it from decay in the hippocampus. The stabilizing effect of FMRP could also be enhanced upon neuronal activity, such as the stimulation of metabotropic glutamate receptors (mGluRs) [40]. FMRP is also found to reduce the mRNA stability of NXF1, a predominant component of the nuclear export factor family [41]. Nonetheless, a genome-wide analysis failed to detect changes in mRNA stability in FMRP-deficient mouse ovaries [7].
3.3 Regulation of Neuronal Development
Neuronal dendritic spines are critical for neuronal function, and abnormal dendritic spines are often seen in neurodevelopmental disorders [38,39]. Extensive studies of human post-mortem tissues and animal models have revealed immature and sometimes densely packed spines in FMRP-deficient neurons [1], but how FMRP regulates dendritic spine maturation is not fully clear. FMRP interacts with Down's syndrome critical region1 (DSCR1), a protein with a crucial role in intellectual disability, and this interaction is critical for local protein synthesis and dendritic spine morphogenesis [42]. Not surprisingly, new neurons differentiated from FMRP-deficient adult NSCs also exhibit abnormal dendritic spines [43]. The absence of FMRP leads to increased signaling in a number of intracellular pathways, such as PI3K, mGluR5, mTOR, GSK3ß, ERK, and insulin pathways[44,45,46,47,48,49], so antagonists and inhibitors of these signaling pathways are naturally being explored as potential therapeutic treatments for FXS. Among these pathways, the most well established is the mGluR5 pathway. Treatment with CTEP, a selective mGlu5 inhibitor, could correct elevated hippocampal long-term depression, protein synthesis, and audiogenic seizures and rescued cognitive deficits, auditory hypersensitivity, and aberrant dendritic spine density [50,51]. In addition, selective GSK3K inhibitors and lithium could also reverse certain mutant phenotypes of FXS mice [43,52,53,54]. Furthermore, inhibitors for PI3K rescue excess protein synthesis in synaptoneurosomes isolated from a FXS mouse model and FXS patient cells [55]. Notably, most of these signaling pathways also play important albeit sometimes distinct roles in various types of stem cells, therefore alterations in these pathways may contribute to abnormalities in stem cell functions in the absence of FMRP, presenting us with therapeutic targets that can be aimed at and tested in stem cell models.
4. FMRP and Stem Cells
Stem cells are defined as cells with the abilities to self-renew and to differentiate, which are important for development, tissue homeostasis, and injury repair [56]. Stem cells have several characteristics: (1) long-term potential for self-renewal; (2) capability of multi-lineage differentiation; (3) they generate one stem cell and one progenitor that undergoes differentiation through asymmetric cell divisions; (4) reside in a specialized micro-environment that affects their symmetric an asymmetric division; (5) they divide more infrequently than their immediate progenies; (6) they are very rare and constant in number during adulthood. Both extrinsic regulators and intrinsic factors regulate stem cell fate: self-renewal, differentiation, or cell death. Stem cell behaviors are strongly influenced by their specific anatomical locations and surrounding cell types in a tissue, known as the “stem cell niche.” The niche provides physical support to anchor stem cells in a specific area and generate diffusible factors to maintain and regulate them [56]. Stem cells are also regulated by intrinsic factors, some of which are common among all stem cells, and others that are unique to specific types of stem cells. There are many signaling cascades and transcriptional mechanisms important for stem cell proliferation and differentiation, including TGF-β, BMP, Smad, Wnt, Notch, fibroblast growth factor, as well as crosstalk among them[56,57]. Therefore, stem cells are regulated by complex mechanisms and in both temporal- and context-specific manners to maintain their unique characteristics. Understanding stem cell regulation is critical to unlocking their therapeutic potential. On the other hand, stem cells also give us the opportunity to explore mechanisms of development, as well as developmental disorders, such as fragile X syndrome.
Until recently, FXS was largely considered a deficit of mature neurons, so FMRP functions have been investigated largely in post-mitotic neurons and synapses. However, a number of newer studies have shown that FMRP may also play important roles in several types of stem cells [58]. Understanding FMRP regulation of stem cells is significant for two reasons. Post-transcriptional regulation by FMRP likely plays a critical role in these stem cells, providing essential information for the therapeutic application of stem cells. On the other hand, stem cells also give us an opportunity to explore novel mechanisms and test potential treatments for FXS (Figure 3) (Table 1).
Figure 3. FMRP plays important regulatory roles in several types of stem cells.
In germline stem cells (GSCs), FMRP maintains GSCs and inhibits differentiation by interacting with Ago1 and bantam miRNA and by repressing Cbl mRNA expression. In follicle stem cells (FSCs), FMRP maintains FSCs through interacting with Caprin. In eNSCs, FMRP maintains the stem cell population by inhibiting the transition from stem cells to intermediate progenitors (IPCs). However, such a function has not been found in adult NSCs (aNSCs). In aNSCs, FMRP represses translation of cyclin D1 and CDK4, which controls cell proliferation. FMRP also inhibits the translation of Gsk3b, an inhibitor of Wnt signaling that promotes neuronal differentiation, but inhibits glial differentiation. In ESCs and iPSCs, FMRP may also promote neuronal differentiation and inhibit glial differentiation.
Table 1.
The Role of FMRP in Regulating Stem Cell Behaviors
| Type of Stem cells | Species | Effects | Molecular targets/Mechanisms | References |
|---|---|---|---|---|
| GSCs | Drosophila | Maintenance and differentiation inhibition | AGO1 | [62] |
| GSCs | Drosophila | Maintenance and differentiation inhibition | Bantam | [63] |
| GSCs | Drosophila | Proliferation | Cbl | [61] |
| FSCs | Drosophila | Maintenance and differentiation inhibition | Caprin | [65] |
| eNSCs and early postnatal progenitors | Mouse/Human | Neuronal differentiation | N/A | [69] |
| eNSCs and early postnatal progenitors | Mouse/Human | Astroglial differentiation | N/A | [98] |
| eNSC | Human | Proliferation and Differentiation | N/A | [74] |
| eNSCs | Mouse | Differentiation | N/A | [71] |
| eNSCs | Mouse | Maintenance and Depletion | F-actin | [73] |
| aNSCs | Drosophila | Proliferation and Differentiation | N/A | [75] |
| aNSCs | Mouse | Proliferation and Differentiation | CDK4, CyclinD1 and GSK3b/Wnt | [78] |
| aNSCs | Mouse | Neuronal differentiation | GSK3b inhibitor/Wnt | [43] |
| ESCs | Human | Differentiation | Histone modification and DNA methylation | [86] |
| ESCs | Human | Neuronal differentiation | Sox1, Notch1 and Pax6 | [87] |
| iPSCs | Human | Proliferation and Differentiation | Histone modification and DNA methylation | [90] |
| iPSCs | Human | Neuronal maturation | PSD95 | [91] |
4.1 FMRP and Germline Stem Cells (GSCs)
Both male and female gonads contain primordial germ cells (PGCs) that give rise to GSCs for maintaining egg and sperm production throughout adult life. GSCs have the unique ability to transmit genetic information from one generation to the next. Thanks to their relatively simple morphology and ease of genetic manipulation, the Drosophila ovary and testis have become ideal model for the study of GSC biology [59]. Drosophila FMRP (dFMRP) is known to play critical roles in oogenesis by antagonizing the expression of Orb, an RNA-binding protein involved in translational regulation [60]. A later study [61] reported that dFMRP regulates Drosophila oogenesis by controlling the cell cycle progression of germ cells. They found that dFMRP binds and controls the expression of Cbl mRNA that codes for Cbl, an E3 ubiquitin ligase involved in oogenesis (Figure 3). Although the Cbl protein levels are unchanged in dFmrp mutant flies, a genetic reduction in Cbl partially rescues the dFmrp mutant [61]. Yang et al performed studies that focus on the role of dFMRP in GSCs. They found that dFMRP is expressed in both GSCs and somatic cells in Drosophila ovary, and it inhibits PGC and GSC differentiation and maintains the GSC population via interaction with Argonaute protein 1 (AGO1), a key factor in the miRNA pathway [62]. They, subsequently performed dFMRP RNA immunoprecipitation and identified a small group of miRNAs that were bound by dFMRP. They further showed that one of them, bantam miRNA, exhibits a similar function as dFMRP in inhibiting PGC and GSC differentiation and maintaining GSCs in a non-cell-autonomous manner [63] (Figure 3). Another study demonstrated that Ago2, a key component of the endogenous small interfering RNA (siRNA) pathway, represses dFMR1 expression [64]. These studies have uncovered not only a role for FMRP in GSCs, but also noncoding RNAs as functional effectors in this process; however, the precise mechanism underlying dFMRP regulation of GSCs through noncoding RNAs remains unclear. Interestingly, these studies revealed that dFMRP regulation of GSCs is via an extrinsic mechanism [61,62,63].
It turns out that in addition to GSCs, Drosophila ovary contains a second type of stem cell, follicle stem cells (FSCs), which give rise to all somatic cells in the egg chamber and therefore play critical roles in oogenesis. A recent study shows that dFMRP is also expressed in FSCs and, together with Caprin, regulates the cell cycle, lineage specification, and subsequently the production of functional eggs [65]. dFMRP therefore plays critical roles in GSC maintenance and fate specification, mainly by regulating the niche cells that support GSCs.
The function of FMRP in mammalian GSCs has gone largely unexplored. Both FXS male patients and a FXS male mouse model exhibit macroorchidism, which may be due to the increased proliferation of Sertoli cells, somatic cells in the gonad that are critical for spermatogenesis [66]. Furthermore, FMR1 premutation females have a significantly increased chance of premature ovarian failure due to impaired follicular recruitment and reduced ovarian reserves [67]; however, the mechanism remains unclear.
4.2 FMRP and Embryonic Neural Stem Cells (eNSCs)
The central nervous system (CNS) is generated from a small number of stem cells, and the process, termed “neurogenesis,” has been studied extensively in the last decade [68]. A great deal of experimental evidence demonstrates that radial glia, the eNSCs during mammalian CNS development, first give rise to intermediate progenitors (IPCs), and then differentiate into neurons and later glia. Investigating the regulatory mechanisms governing the self-renewal and fate specification of NSCs and IPCs is critical for understanding how mammalian brains develop and whether these endogenous NSCs and progenitors could be activated for neural repairs in diseases and injuries.
Castren et al first investigated the functions of FMRP on embryonic and early postnatal neurogenesis [69]. They isolated neuroprogenitors from embryonic day 13 (E13) mouse embryos and postnatal day 6 (P6) mouse brains and found that these FMR1 mutant progenitors generated 3- to 4.5-fold more Tuj1+ immature neurons, but ~15% fewer GFAP+ astrocytes in culture compared to wild-type cells. They also found that neuroprogenitors isolated from 18-week-old FXS human fetus had similar phenotypes. Although there was no detectable difference in proliferation in FMR1 mutant neuroprogenitors as assessed by 3H-thymidine incorporation, there was increased BrdU incorporation in embryonic FMR1 mutant mouse SVZ. In addition, these FMRP-deficient immature neurons showed fewer and shorter neurites and a smaller cell body volume, which might be due to altered BDNF/TrkB signaling found by the same group in a later study [70]. Castren et al also showed that differentiating FMR1 mutant mouse progenitors exhibited increased Ca-induced oscillation, which is surprising since these in vitro differentiating neurons are quite immature in nature.
In a subsequent study by the same group [71], FMRP function in embryonic cortical neurogenesis was further assessed by in utero electroporation of a mutant I304N FMRP, leading to a loss-of-function phenotype [72]. Tervonen et al reported that I304N FMRP transfected embryos exhibited an abnormal accumulation of progenitor cells in the SVZ and VZ, with fewer cells migrated to the cortical plate where newly differentiated neurons reside. Consistent with this finding, FMR1 mutant embryos exhibit increased Tbr2+ proliferating (BrdU+) neuroblasts in the SVZ compared to wild-type mouse brains. However, this cannot explain why there are more layer V neurons in FMR1 mutant embryos since cell migration seems impaired in I304N FMRP-transfected brains [71]. A later study by Saffary et al provided a potential answer to this puzzle. During cortical neurogenesis, the transition from NSCs to intermediate progenitor cells (IPCs) is a critical step in maintaining the eNSC population and determining neuronal production. Saffary et al revealed that acute deletion of FMRP using shRNA led to increased production of IPCs at the expense of eNSCs, causing eNSC depletion. The eNSC depletion was a result of altered F-actin organization and could be rescued by overexpressing profilin 1, which promotes F-actin organization [73] (Figure 3). Whether profilin 1 is a direct target for FMRP was not shown. Nevertheless, these results demonstrated that FMRP plays an important role in eNSC maintenance and fate.
Studies of human eNSCs have yielded seemingly contradictory results. While Castren el al showed that eNSCs isolated from a human 18-week-old FXS fetus exhibit increased neuronal production but reduced glial differentiation [69], this result has not been replicated by others. Another study reported that human neural progenitor cells (hNPCs) isolated from a 14-week-old fetal cortex carrying the FMR1 mutation exhibit no significant differences in either self-renewal or multi-lineage differentiation compared to normal NPCs. However, microarray analysis revealed changes in the expression of signal transduction genes in FXS NPCs compared with normal NPCs [74]. Both of these studies are based on cells isolated from a single FXS fetus. It is unclear whether these differences are due to experimental procedures, fetal ages, or both.
Callan et al explored the role of FMRP in regulating NSC proliferation in Drosophila. They found that loss of dFMRP altered cell cycle progression, leading to the overproliferation of NSCs, more neuroblasts exiting from the quiescent state, and increased numbers of newborn neurons persisting in adult brains [75]. These results are similar to those found in mouse and human studies; however, since Drosophila has only one FXR protein, the deficits discovered in dFMRP mutant flies might be the result of triple deletion of all FXRs in mammals, which has not been studied to date, largely due to the perinatal lethality of FXR1 mutant mice.
4.3 FMRP and Adult Neural Stem Cells (aNSCs)
Cognitive impairment and behavioral abnormalities are the common symptoms in FXS patients [76]. Adult neurogenesis in mammalian brains is known to be important for lifelong learning and cognitive ability [2]. Stem cells (aNSCs) exist in adult germinal zones, including the dentate gyrus of the hippocampus and the SVZ of the lateral ventricles, including in human brains. How FMRP contributes to adult neurogenesis, and thus learning and cognition, remains a fascinating question to pursue.
An earlier study found no significant difference in overall BrdU incorporation and differentiation, except for a small increase in neuronal production in the ventral hippocampus of adult FMR1 mutant mice compared to WT mice [77]. Subsequently, Luo et al [78] isolated adult NSCs (aNSCs) from the DG of FMR1 mutant mice and discovered that the loss of FMRP led to an increased proliferative capacity of NSCs, about a 60% reduction in neuronal differentiation, accompanied by a 75% increase in astrocytes compared with wild-type aNSCs. These in vitro results were further confirmed by analysis of in vivo neurogenesis in the DG FMR1 mutant mice. In a follow-up study, Guo et al used an inducible conditional mutant mouse line to delete FMRP and track the cell fate of aNSCs. They confirmed that FMRP deficiency in aNSCs indeed leads to increased production of stem and progenitor cells, but reduced neuronal production and increased glia production [79]. Since adult neurogenesis has been associated with learning and memory, Guo et al further explored how FMRP deficiency in adult-born new neurons contributes to the cognitive deficits in FXS. They showed that the ablation of FMRP specifically in adult-born new neurons leads to impairments in hippocampus-dependent learning in mice. Conversely, restoration of FMRP expression specifically in aNSCs rescues these learning deficits in FMRP-deficient mice [79], demonstrating that defective adult neurogenesis may contribute to the learning impairment seen in FXS.
Luo et al investigated the mechanism underlying FMRP regulation of aNSCs using RNA immunoprecipitation followed by microarray and real-time PCR; they identified a number of direct mRNA targets of FMRP in aNSCs, including two cell cycle regulators, cyclin-dependent kinase 4 (CDK4) and CyclinD1, and a Wnt signaling inhibitor, GSK3β (Figure 3). They further showed that increased GSK3β in the absence of FMRP led to reduced Wnt signaling and subsequent reduced expression of the neuronal master regulator Neurogenin 1, which is responsible for reduced neuronal production but increased glial differentiation. Based on this discovery, Guo et al used a GSK3β inhibitor, SB216763, to restore Wnt signaling, which rescued neurogenesis and improved hippocampus-dependent learning in a mouse model of FXS [43]. This result together with another recent finding [54] points to the possibility that GSK3b inhibition could be a potential treatment for the learning deficits seen in FXS.
The results obtained from mouse analyses differ from those from Drosophila neuroblasts [75,80]. The likely reason is species differences. Drosophila has only one FXR protein, dFMRP, which probably carries out the functions of all three mammalian FXRs. In another study, Guo et al found that deletion of FXR2 leads to increased neuronal production [81], which is the opposite of what they found in FMRP-deficient mice, but similar to results found in FMRP-deficient eNSCs and dFMRP-deficient neuroblasts [58]. FMRP and FXR2 apparently play different roles in the regulation of aNSCs during adult neurogenesis. It is also likely that both common and unique functions of these FXRs vary depending on cell type and developmental stage. Future investigations on pure populations of cells or even single cells may reveal a much clearer picture of the specific roles of FXR1 and FXR2. It remains to be seen whether FMRP and FXR2 play distinct regulatory roles in human neurogenesis.
4.4 FMRP and Embryonic Stem Cells (ESCs)/Inducible Pluripotent Stem Cells (iPSCs)
ESCs are pluripotent cells derived from the inner-cell mass of blastocyst stage embryos. In 1981, Evans and Kaufman isolated the first mouse embryonic stem cells (mESCs)[82]. About 17 years later, the first human ESC lines were generated [83]. Human ESCs are invaluable in the study of early embryonic development, since they can recapitulate embryogenesis by expressing developmentally regulated genes and by activating the same molecular pathways as those that occur in vivo[84]. Thus, they can be used to assess the impact of specific gene mutations on certain developmental events, allowing us to identify critical factors that play a role in the processes of cell commitment, differentiation, and adult cell reprogramming [85].
Dalit Ben-Yosef's group published the first analysis of a human ESC line derived from a preimplantation FXS embryo, which allowed them to assess the early events of FMR1 gene inactivation. They discovered that, in undifferentiated FXS-ESCs with full expansion of CGG repeats, the FMR1 gene is expressed and its promoter is enriched with acetylated histone H3, an active chromatin mark. In differentiated cells, the FMR1 gene is methylated and its promoter is enriched with methylated H3 at lysine 9, a repressive chromatin mark. This is the first evidence to show that FMR1 inactivation is triggered by differentiation [86]. Subsequently, Telias et al [87] showed that FMR1 expression exhibits steady upregulation during human ESC neural differentiation, whereas FXS-ESCs failed to upregulate FMR1 gene expression during differentiation and exhibited aberrant expression of several critical genes for neurogenesis, including SOX1, NOTCH1, and PAX6. FXS-ESCs also differentiated into fewer neurons but more astrocytes, reminiscent of what was seen in adult mouse NSCs[78,79]. Furthermore, even though FXS-ESCs can differentiate into functional neurons, these neurons have reduced synaptic connections [87]. The FXS-ESCs have proved to be an important model for exploring the disease mechanism of FXS in humans. For example, a recent study using two male FXS ESC lines showed that DNA replication is altered at the CGG repeat in FXS-hESCs due to both failed usage of a replication origin upstream of the FMR1 gene and replication fork stalling at CGG repeats. However, the DNA replication phenotype seems to be specific to ESCs, not differentiated cells, which is puzzling[88].
The development of inducible pluripotent cell (iPSC) technology has allowed us to reprogram somatic cells into pluripotent stem cells, revolutionizing our ability to study human development and diseases. Despite intense interest, very few FXS iPSC studies have been published to date. Urbach et al derived iPSCs from fibroblasts of three FXS individuals [89]. Surprisingly, they found that the FMR1 gene remains transcriptionally silent and the promoter region of FMR1 retained DNA methylation and repressive chromatin marks in all FXS-iPSC lines. These data not only highlight critical differences between hESCs and hiPSCs for modeling FXS, but also reveal that the reprogramming process has little effect on the silenced FMR1 gene [89]. Later, Sheridan et al confirmed the lack of FMR1 gene reactivation in iPSCs from three different FXS patients. They further analyzed their differentiation and showed that FXS-iPSC differentiated neurons exhibit shorter neurites and fewer neurons, but more glia [90]. Although these data are consistent with mouse data [78,79], more iPSC lines and more detailed quantification are needed to draw meaningful conclusions. Fragile X premutation iPSCs have also been generated from both premutation carriers [91] and from FXS individuals carrying premutation alleles [90]. The FMR1 gene is actively expressed in premutation iPSCs, but the FMRP protein levels vary. Liu et al showed that, even with nearly normal levels of FMRP expression, premutation iPSC differentiated neurons exhibit reduced post-synaptic protein PSD-95 puncta, reduced neurite complexity, and sustained Ca elevation in response to glutamate stimulation [91].
These studies demonstrate how pluripotent human stem cells have given us exciting opportunities to study FXS and related disorders in human systems and to investigate questions that cannot be be answered using rodent models, such as epigenetic silencing of FMR1 due to CGG expansion. The lack of activation of the FMR1 gene during standard iPSC derivation is both puzzling and intriguing, and it presents a great challenge for the study of FXS in human models. Recently, Ganfi et al [92] developed a chemically defined cell culture condition that allows for derivation of iPSCs with a ground-state naïve nature. Using this condition, they showed that iPSCs derived from two different FXS fibroblast lines have active FMR1 gene expression. One of the fibroblast lines is the same line used by Sherian et al, but it did not show FMR1 gene expression upon reprogramming using standard iPSC methods [90].
5. Conclusions and Perspectives
Although there has been great progress towards an understanding of the role FMRP plays in neuronal synaptic plasticity, exploring its functions in stem cells and development is still in its infancy Stem cells not only make excellent systems for understanding the function of FMRP, they also give us a unique opportunity to develop novel therapeutics and treatments for FXS.
Although there are now a number of chemicals, such as GABA agonists, p110 beta selective inhibitors, PI3K inhibitors, minocycline, and lithium, which can rescue certain behavioral deficits in FXS patients or mouse models[54,55,93,94,95,96,97] , few effective drugs are actually available to patients. It is critical that we identify novel drug targets and develop new and effective therapies. Various types of stem cells in different species, with their ability to model developmental processes in vitro and in vivo, serve as great models for achieving these goals. As evidenced by the literature, these cells have helped unveil novel pathways and mechanisms. In addition, pluripotent human ESCs and iPSCs allow us to study the functions of FMRP in human development and answer the questions that cannot be answered using animal models, such as when, how, and why the FMR1 gene is silenced during cellular differentiation. Furthermore, creating a platform that allows for high-throughput drug screening is a challenge. Here stem cells, especially iPS cells derived from FXS patients, could serve as an invaluable source of cells. Therefore, advances in stem cell biology will certainly give us new insight into the function of FMRP, providing hope for a better quality of life for FXS patients.
Acknowledgements
We thank Cheryl T. Strauss for editing. This work was supported by grants from the NIH (R01MH080434 and R01MH078972 to X.Z.), FRAXA (to X.Z.), and a Center Grant from the NIH to the Waisman Center (P30HD03352).
Footnotes
Author Contributions: Yue Li: Conception and design, Collection and/or assembly of data, Manuscript writing, Final approval of manuscript
Xinyu Zhao: Conception and design, Financial support , Collection and/or assembly of data, Manuscript writing, Final approval of manuscript
The authors declare no conflict of interest.
References
- 1.Wang T, Bray SM, Warren ST. New perspectives on the biology of fragile X syndrome. Curr Opin Genet Dev. 2012;22:256–263. doi: 10.1016/j.gde.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng W, Aimone JB, Gage FH. Adult hippocampal neurogenesis, synaptic plasticity and memory: facts and hypotheses. Nat Rev Neurosci. 2010;11:339–350. doi: 10.1038/nrn2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagerman PJ, Hagerman RJ. The fragile-X premutation: A maturing perspective. Am J Hum Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tassone F, Hagerman PJ. Expression of the FMR1 gene. Cytogenet Genome Res. 2003;100:124–128. doi: 10.1159/000072846. [DOI] [PubMed] [Google Scholar]
- 5.Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, et al. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes & Development. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung YJ, Conti J, Currie JR, Brown WT, Denman RB. RNAs that interact with the Fragile X syndrome RNA binding protein FMRP. Biochem Biophys Res Commun. 2000;275:973–980. doi: 10.1006/bbrc.2000.3405. [DOI] [PubMed] [Google Scholar]
- 7.Ascano M, Jr., Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reeve SP, Lin XD, Sahin HB, Jiang FF, Yao A, et al. Mutational analysis establishes a critical role for the N terminus of fragile X mental retardation protein FMRP. Journal of Neuroscience. 2008;28:3221–3226. doi: 10.1523/JNEUROSCI.5528-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 10.Darnell JC, Van Driesche SJ, Zhang CL, Hung KYS, Mele A, et al. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, et al. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 12.Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, et al. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. Journal of Neuroscience. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheever A, Ceman S. Phosphorylation of FMRP inhibits association with Dicer. RNA. 2009;15:362–366. doi: 10.1261/rna.1500809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Molecular Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum Mol Genet. 2009;18:3164–3177. doi: 10.1093/hmg/ddp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siomi MC, Siomi H, Sauer WH, Srinivasan S, Nussbaum RL, et al. FXR1, an autosomal homolog of the fragile X mental retardation gene. Embo Journal. 1995;14:2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mientjes EJ, Willemsen R, Kirkpatrick LL, Nieuwenhuizen IM, Hoogeveen-Westerveld M, et al. Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum Mol Genet. 2004;13:1291–1302. doi: 10.1093/hmg/ddh150. [DOI] [PubMed] [Google Scholar]
- 18.Engels B, van 't Padje S, Blonden L, Severijnen LA, Oostra BA, et al. Characterization of Fxr1 in Danio rerio; a simple vertebrate model to study costamere development. J Exp Biol. 2004;207:3329–3338. doi: 10.1242/jeb.01146. [DOI] [PubMed] [Google Scholar]
- 19.Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Molecular and Cellular Biology. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, et al. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am J Hum Genet. 2008;83:43–52. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Say E, Tay HG, Zhao ZS, Baskaran Y, Li R, et al. A functional requirement for PAK1 binding to the KH(2) domain of the fragile X protein-related FXR1. Molecular Cell. 2010;38:236–249. doi: 10.1016/j.molcel.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Bontekoe CJ, McIlwain KL, Nieuwenhuizen IM, Yuva-Paylor LA, Nellis A, et al. Knockout mouse model for Fxr2: a model for mental retardation. Hum Mol Genet. 2002;11:487–498. doi: 10.1093/hmg/11.5.487. [DOI] [PubMed] [Google Scholar]
- 23.Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: A Presynaptic Fragile X Granule Expressed in a Subset of Developing Brain Circuits. Journal of Neuroscience. 2009;29:1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akins MR, Leblanc HF, Stackpole EE, Chyung E, Fallon JR. Systematic mapping of fragile X granules in the mouse brain reveals a potential role for presynaptic FMRP in sensorimotor functions. J Comp Neurol. 2012;520:3687–3706. doi: 10.1002/cne.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Hou L, Klann E, Nelson DL. Altered hippocampal synaptic plasticity in the FMR1 gene family knockout mouse models. J Neurophysiol. 2009;101:2572–2580. doi: 10.1152/jn.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 27.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Development. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Development. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, et al. Dicer-derived MicroRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. Journal of Biomedicine and Biotechnology. 2006:64347–64358. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nature neuroscience. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 31.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCaffery P, Deutsch CK. Macrocephaly and the control of brain growth in autistic disorders. Prog Neurobiol. 2005;77:38–56. doi: 10.1016/j.pneurobio.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Davidovic L, Bechara E, Gravel M, Jaglin XH, Tremblay S, et al. The nuclear MicroSpherule protein 58 is a novel RNA-binding protein that interacts with fragile X mental retardation protein in polyribosomal mRNPs from neurons. Hum Mol Genet. 2006;15:1525–1538. doi: 10.1093/hmg/ddl074. [DOI] [PubMed] [Google Scholar]
- 34.Davidovic L, Jaglin XH, Lepagnol-Bestel AM, Tremblay S, Simonneau M, et al. The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum Mol Genet. 2007;16:3047–3058. doi: 10.1093/hmg/ddm263. [DOI] [PubMed] [Google Scholar]
- 35.El Fatimy R, Tremblay S, Dury AY, Solomon S, De Koninck P, et al. Fragile X mental retardation protein interacts with the RNA-binding protein Caprin1 in neuronal RiboNucleoProtein complexes [corrected]. PLoS One. 2012;7:e39338. doi: 10.1371/journal.pone.0039338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dictenberg JB, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. International Journal of Developmental Neuroscience. 2008;26:831–831. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antar LN, Li CX, Zhang HL, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Molecular and Cellular Neuroscience. 2006;32:37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Bear M. The mGluR theory of fragile X mental retardation. Neuropsychopharmacology. 2005;30:S59–S59. doi: 10.1111/j.1601-183X.2005.00135.x. [DOI] [PubMed] [Google Scholar]
- 39.Smrt RD, Zhao X. Epigenetic regulation of neuronal dendrite and dendritic spine development. Frontiers of biology. 2010;5:304–323. doi: 10.1007/s11515-010-0650-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang MQ, Wang QQ, Huang YQ. Fragile X mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize Nxf1 mRNA in neuronal cells. Proc Natl Acad Sci U S A. 2007;104:10057–10062. doi: 10.1073/pnas.0700169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Zhu JZ, Chang KT, Min KT. DSCR1 interacts with FMRP and is required for spine morphogenesis and local protein synthesis. Embo Journal. 2012;31:3655–3666. doi: 10.1038/emboj.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo W, Murthy AC, Zhang L, Johnson EB, Schaller EG, et al. Inhibition of GSK3beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21:681–691. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min WW, Yuskaitis CJ, Yan QJ, Sikorski C, Chen SQ, et al. Elevated glycogen synthase kinase-3 activity in Fragile X mice: Key metabolic regulator with evidence for treatment potential. Neuropharmacology. 2009;56:463–472. doi: 10.1016/j.neuropharm.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gross C, Nakamoto M, Yao XD, Chan CB, Yim SY, et al. Excess Phosphoinositide 3-Kinase Subunit Synthesis and Activity as a Novel Therapeutic Target in Fragile X Syndrome. Journal of Neuroscience. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fatemi SH, Folsom TD, Kneeland RE, Yousefi MK, Liesch SB, et al. Impairment of fragile X mental retardation protein-metabotropic glutamate receptor 5 signaling and its downstream cognates ras-related C3 botulinum toxin substrate 1, amyloid beta A4 precursor protein, striatal-enriched protein tyrosine phosphatase, and homer 1, in autism: a postmortem study in cerebellar vermis and superior frontal cortex. Molecular Autism. 2013;4:21–39. doi: 10.1186/2040-2392-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lohith TG, Osterweil EK, Fujita M, Jenko KJ, Bear MF, et al. Is metabotropic glutamate receptor 5 upregulated in prefrontal cortex in fragile X syndrome? Molecular Autism. 2013;4:15–22. doi: 10.1186/2040-2392-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng N, Weiler IJ, Sumis A, Berry-Kravis E, Greenough WT. Early-Phase ERK Activation as a Biomarker for Metabolic Status in Fragile X Syndrome. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2008;147B:1253–1257. doi: 10.1002/ajmg.b.30765. [DOI] [PubMed] [Google Scholar]
- 49.Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, et al. Dysregulation of mTOR Signaling in Fragile X Syndrome. Journal of Neuroscience. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, et al. Chronic Pharmacological mGlu5 Inhibition Corrects Fragile X in Adult Mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huber KM, Roder JC, Bear MF. Chemical induction of mGluR5- and protein synthesis--dependent long-term depression in hippocampal area CA1. J Neurophysiol. 2001;86:321–325. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- 52.Min WW, Yuskaitis CJ, Yan Q, Sikorski C, Chen S, et al. Elevated glycogen synthase kinase-3 activity in Fragile X mice: key metabolic regulator with evidence for treatment potential. Neuropharmacology. 2009;56:463–472. doi: 10.1016/j.neuropharm.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuskaitis CJ, Mines MA, King MK, Sweatt JD, Miller CA, et al. Lithium ameliorates altered glycogen synthase kinase-3 and behavior in a mouse model of Fragile X syndrome. Biochem Pharmacol. 2010;79:632–646. doi: 10.1016/j.bcp.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.King MK, Jope RS. Lithium treatment alleviates impaired cognition in a mouse model of fragile X syndrome. Genes Brain and Behavior. 2013;12:723–731. doi: 10.1111/gbb.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gross C, Bassell GJ. Excess Protein Synthesis in FXS Patient Lymphoblastoid Cells Can Be Rescued with a p110 beta-Selective Inhibitor. Molecular Medicine. 2012;18:336–345. doi: 10.2119/molmed.2011.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X, Zhao X. Epigenetic regulation of mammalian stem cells. Stem Cells Dev. 2008;17:1043–1052. doi: 10.1089/scd.2008.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jobe EM, McQuate AL, Zhao X. Crosstalk among Epigenetic Pathways Regulates Neurogenesis. Frontiers in neuroscience. 2012;6:59–73. doi: 10.3389/fnins.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callan MA, Zarnescu DC. Heads-Up: New Roles for the Fragile X Mental Retardation Protein in Neural Stem and Progenitor Cells. Genesis. 2011;49:424–440. doi: 10.1002/dvg.20745. [DOI] [PubMed] [Google Scholar]
- 59.Spradling A, Fuller MT, Braun RE, Yoshida S. Germline stem cells. Cold Spring Harb Perspect Biol. 2011;3:a002642. doi: 10.1101/cshperspect.a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costa A, Wang Y, Dockendorff TC, Erdjument-Bromage H, Tempst P, et al. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Developmental cell. 2005;8:331–342. doi: 10.1016/j.devcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 61.Epstein AM, Bauer CR, Ho A, Bosco G, Zarnescu DC. Drosophila Fragile X Protein controls cellular proliferation by regulating cbl levels in the ovary. Dev Biol. 2009;330:83–92. doi: 10.1016/j.ydbio.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 62.Yang LL, Duan RH, Chen DS, Wang J, Chen DH, et al. Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila. Hum Mol Genet. 2007;16:1814–1820. doi: 10.1093/hmg/ddm129. [DOI] [PubMed] [Google Scholar]
- 63.Yang YY, Xu SL, Xia LX, Wang J, Wen SM, et al. The Bantam microRNA Is Associated with Drosophila Fragile X Mental Retardation Protein and Regulates the Fate of Germline Stem Cells. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pepper AS, Beerman RW, Bhogal B, Jongens TA. Argonaute2 suppresses Drosophila fragile X expression preventing neurogenesis and oogenesis defects. PloS one. 2009;4:e7618. doi: 10.1371/journal.pone.0007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reich J, Papoulas O. Caprin controls follicle stem cell fate in the Drosophila ovary. PLoS One. 2012;7:e35365. doi: 10.1371/journal.pone.0035365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Slegtenhorst-Eegdeman KE, de Rooij DG, Verhoef-Post M, van de Kant HJ, Bakker CE, et al. Macroorchidism in FMR1 knockout mice is caused by increased Sertoli cell proliferation during testicular development. Endocrinology. 1998;139:156–162. doi: 10.1210/endo.139.1.5706. [DOI] [PubMed] [Google Scholar]
- 67.Oatley MJ, Racicot KE, Oatley JM. Sertoli cells dictate spermatogonial stem cell niches in the mouse testis. Biology of reproduction. 2011;84:639–645. doi: 10.1095/biolreprod.110.087320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annual review of neuroscience. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castren M, Tervonen T, Karkkainen V, Heinonen S, Castren E, et al. Altered differentiation of neural stem cells in fragile X syndrome. Proc Natl Acad Sci U S A. 2005;102:17834–17839. doi: 10.1073/pnas.0508995102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Louhivuori V, Vicario A, Uutela M, Rantamaki T, Louhivuori LM, et al. BDNF and TrkB in neuronal differentiation of Fmr1-knockout mouse. Neurobiol Dis. 2011;41:469–480. doi: 10.1016/j.nbd.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 71.Tervonen TA, Louhivuori V, Sun X, Hokkanen ME, Kratochwil CF, et al. Aberrant differentiation of glutamatergic cells in neocortex of mouse model for fragile X syndrome. Neurobiol Dis. 2009;33:250–259. doi: 10.1016/j.nbd.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 72.Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, et al. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 2009;5:e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saffary R, Xie ZG. FMRP Regulates the Transition from Radial Glial Cells to Intermediate Progenitor Cells during Neocortical Development. Journal of Neuroscience. 2011;31:1427–1439. doi: 10.1523/JNEUROSCI.4854-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhattacharyya A, McMillan E, Wallace K, Tubon TC, Jr., Capowski EE, et al. Normal Neurogenesis but Abnormal Gene Expression in Human Fragile X Cortical Progenitor Cells. Stem Cells Dev. 2008;17:107–117. doi: 10.1089/scd.2007.0073. [DOI] [PubMed] [Google Scholar]
- 75.Callan MA, Cabernard C, Heck J, Luois S, Doe CQ, et al. Fragile X protein controls neural stem cell proliferation in the Drosophila brain. Hum Mol Genet. 2010;19:3068–3079. doi: 10.1093/hmg/ddq213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bagni C, Tassone F, Neri G, Hagerman R. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. Journal of Clinical Investigation. 2012;122:4314–4322. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eadie BD, Zhang WN, Boehme F, Gil-Mohapel J, Kainer L, et al. Fmr1 knockout mice show reduced anxiety and alterations in neurogenesis that are specific to the ventral dentate gyrus. Neurobiology of disease. 2009;36:361–373. doi: 10.1016/j.nbd.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 78.Luo Y, Shan G, Guo W, Smrt RD, Johnson EB, et al. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo WX, Allan AM, Zong RT, Zhang L, Johnson EB, et al. Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nat Med. 2011;17:559–U575. doi: 10.1038/nm.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Callan MA, Clements N, Ahrendt N, Zarnescu DC. Fragile X Protein is required for inhibition of insulin signaling and regulates glial-dependent neuroblast reactivation in the developing brain. Brain Res. 2012;1462:151–161. doi: 10.1016/j.brainres.2012.03.042. [DOI] [PubMed] [Google Scholar]
- 81.Guo W, Zhang L, Christopher DM, Teng ZQ, Fausett SR, et al. RNA-binding protein FXR2 regulates adult hippocampal neurogenesis by reducing Noggin expression. Neuron. 2011;70:924–938. doi: 10.1016/j.neuron.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 83.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 84.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20–36. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dvash T, Ben-Yosef D, Eiges R. Human embryonic stem cells as a powerful tool for studying human embryogenesis. Pediatr Res. 2006;60:111–117. doi: 10.1203/01.pdr.0000228349.24676.17. [DOI] [PubMed] [Google Scholar]
- 86.Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 87.Telias M, Segal M, Ben-Yosef D. Neural differentiation of Fragile X human Embryonic Stem Cells reveals abnormal patterns of development despite successful neurogenesis. Dev Biol. 2013;374:32–45. doi: 10.1016/j.ydbio.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 88.Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, et al. The DNA Replication Program Is Altered at the FMR1 Locus in Fragile X Embryonic Stem Cells. Molecular cell. 2014;53:19–31. doi: 10.1016/j.molcel.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential Modeling of Fragile X Syndrome by Human Embryonic Stem Cells and Induced Pluripotent Stem Cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu J, Koscielska KA, Cao Z, Hulsizer S, Grace N, et al. Signaling defects in iPSC-derived fragile X premutation neurons. Hum Mol Genet. 2012;21:3795–3805. doi: 10.1093/hmg/dds207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 93.Berry-Kravis E, Potanos K. Psychopharmacology in fragile X syndrome--present and future. Ment Retard Dev Disabil Res Rev. 2004;10:42–48. doi: 10.1002/mrdd.20007. [DOI] [PubMed] [Google Scholar]
- 94.Paribello C, Tao L, Folino A, Berry-Kravis E, Tranfaglia M, et al. Open-label add-on treatment trial of minocycline in fragile X syndrome. Bmc Neurology. 2010;10:91–99. doi: 10.1186/1471-2377-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Olmos-Serrano JL, Corbin JG, Burns MP. The GABA(A) Receptor Agonist THIP Ameliorates Specific Behavioral Deficits in the Mouse Model of Fragile X Syndrome. Dev Neurosci. 2011;33:395–403. doi: 10.1159/000332884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu ZH, Yang Q, Feng B, Liu SB, Zhang N, et al. Group I mGluR antagonist rescues the deficit of D1-induced LTP in a mouse model of fragile X syndrome. Molecular Neurodegeneration. 2012;7:24–37. doi: 10.1186/1750-1326-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu ZH, Huang TJ, Smith CB. Lithium reverses increased rates of cerebral protein synthesis in a mouse model of fragile X syndrome. Neurobiol Dis. 2012;45:1145–1152. doi: 10.1016/j.nbd.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]