Abstract
In a phylogenetic analysis of vertebrate transferrins (TFs), six major clades (subfamilies) were identified: (1) S, the mammalian serotransferrins; (2) ICA, the mammalian inhibitor of carbonic anhydrase (ICA) homologs; (3) L, the mammalian lactoferrins; (4) O, the ovotransferrins of birds and reptiles; (4) M, the melanotransferrins of bony fishes, amphibians, reptiles, birds, and mammals; and (5) M-like, a newly identified TF subfamily found in bony fishes, amphibians, reptiles, and birds. A phylogenetic tree based on the joint alignment of N-lobes and C-lobes supported the hypothesis that three separate events of internal duplication occurred in vertebrate TFs: (1) in the common ancestor of the M subfamily; (2) in the common ancestor of the M-like subfamily; and (3) in the common ancestor of other vertebrate TFs. The S, ICA, and L subfamilies were found only in placental mammals, and the phylogenetic analysis supported the hypothesis that these three subfamilies arose by gene duplication after the divergence of placental mammals from marsupials. The M-like subfamily was unusual in several respects, including the presence of a uniquely high proportion of clade-specific conserved residues, including distinctive but conserved residues in the sites homologous to those functioning in carbonate binding of human serotransferrin. The M-like family also showed a unusually high proportion of cationic residues in the positively charged region corresponding to human lactoferrampin, suggesting a distinctive role of this region in the M-like subfamily, perhaps in antimicrobial defense.
The transferrins (TFs) constitute a family of single-chain glycoproteins approximately 700 amino acids in length best known for their ability to bind iron and consequent role in iron metabolism (Bowman et al. 1988; Thorstensen and Romslo 1990; Sun et al. 1999; Baker et al. 2002; Farnaud and Evans 2003; Gomme and McCann 2005; Baker and Baker 2009; Jenssen and Hancock 2009; García-Montoya et al. 2012; Giansanti et al. 2012; Gkouvatsos et al. 2012; Harris 2012; Mayle et al. 2012; Rahmanto et al. 2012). The following members of this family are recognized in humans: (1) serotransferrin (or serum transferrin), which is expressed in the liver and secreted into the blood serum, but also has a wider tissue expression and a variety of potential functions (Gkouvatsos et al. 2012); (2) lactoferrin (Figure 1), named for its expression in milk but expressed in most biological fluids (Levay and Viljoen 1995; García-Montoya et al. 2012); and (3) melanotransferrin, identified as a tumor antigen in melanoma but having a wide but much lower level of expression in normal tissues as well (Rahmanto et al. 2012). Well-studied transferrin family members from non-human vertebrates include ovotransferrin, first identified from avian egg white but known to have a wider expression pattern in the domestic chicken (Giansanti et al. 2012); and the inhibitor of carbonic anhydrase (ICA) reported from various non-human mammals (Wang et al. 2007; Eckenroth et al. 2010).
Figure 1.
Primary structure of human lactoferrin (NP_002334). The signal peptide is italicized; the two positively charged regions of the N-lobe are in bold face; the linker between the N-lobe and C-lobe is underlined. Dots above residues indicate iron-binding residues; arrows above residues indicate carbonate-binding residues.
All vertebrate TFs include homologous N-terminal (N-lobe) and C-terminal (C-lobe) lobes (Mizutani et al. 2012), evidently the result of an ancient within-gene duplication event (Lambert et al. 2005; Figure 1). Each of the N-lobes and C-lobes includes four amino acid residues involved in binding iron in the Fe(III) form, along with two amino acid residues that bind a synergistic carbonate anion, which along with the amino acid residues is covalently bound to the iron atom (Kurokawa et al. 1995; Baker et al. 2002; Baker and Baker 2009; Mizutani et al. 2012; Figure 1). Similar three-dimensional structures have been reported for serotransferrins and lactoferrins of a variety of mammalian species and for ovotransferrins of two bird species (Mizutani et al. 2012). Mammalian ICA does not bind iron, but has a three-dimensional structure resembling that seen in other TFs when they are binding iron (Eckenroth et al. 2010). A three-dimensional structure for melanotransferrin has not yet been determined, but melanotransferrin is known to differ from the other vertebrate transferrins in that it attaches to the cell membrane though a glycosyl phostphatidylinositol linkage involving a hydrophobic domain located at the C-terminus of the C-lobe (Alemany et al. 1993). In addition, mammalian melanotransferrin has been found to bind iron only on the N-lobe not the C-lobe (Baker et al. 1992).
Aside from their role in iron metabolism, some members of the TF family are known to have significant roles in immune defense. Both mammalian lactoferrin and avian ovotransferrin have antimicrobial properties because they sequester iron, a needed nutrient for many bacterial pathogens (Jenssen and Hancock 2009; Giansanti et al. 2012). The competition with pathogens for iron has given rise to a co-evolutionary process in which certain bacteria encode transferrin receptors that function to assimilate iron from host TFs (Gray-Owen and Schyvers 1996). In addition, lactoferrin has direct bactericidal effects, apparently mediated by highly positively charged domains in the N-lobe (Nibbering et al. 2001). The latter include a domain located at the N-terminus of the N-lobe (“lactoferricin”) and a second domain toward the C-terminus of the N-lobe (“lactoferrampin”; Sinha et al. 2013). In the three-dimensional structure of lactoferrin, both of these positively charged domains are located on the surface of the protein (Baker et al. 2002). In addition, in human, bovine, and perhaps in many other mammals, a positively charged lactoferricin peptide can be cleaved enzymatically in vivo from the N-terminus of lactoferrin; and has potent bacterical activity (Farnaud and Evans 2003; Hunter et al. 2003; Baker and Baker 2009; Sinha et al. 2013). Both lactoferrin and ovotransferrin have been reported to have anti-fungal and anti-viral activities as well (García-Montoya et al. 2012; Giansanti et al. 2012). Finally lactoferrin serves as a bridge between innate and specific immune systems by activating the NF-κB through the TLR4 receptor pathway (Ando et al. 2010).
Several studies have used phylogenetic methods to reconstruct the pattern of gene duplication that gave rise to the different TF family members in vertebrates (Escrivá et al. 1995; Lambert et al. 2005; Lambert 2012). Lambert et al. (2005) suggested that the internal gene duplication giving rise to separate N-lobe and C-lobe occurred prior to the duplications giving rise to the separate vertebrate genes. In support of this hypothesis, they note that the N-lobe and C-lobe of melanotransferrin are more similar in primary sequence to each other than to either N-lobes or C-lobes of other vertebrate TFs. However, the latter observation seems more consistent with the hypothesis that an independent internal duplication occurred in the melanotransferrin lineage. With regard to the timing of the duplication events giving rise to separate mammalian serotransferrin, lactoferrin, and ICA genes,Lambert et al. (2005) reported evidence that they preceded the radiation of the eutherian (placental) orders of mammals, but were unable to provide more precise timing due to a lack of sequence data.
Here we make use of newly available data from ongoing vertebrate genome sequencing projects in order to provide more precise answers regarding the timing of major gene duplication events in the evolutionary history of vertebrate TFs. Our phylogenetic analysis reveals the existence of an additional ancient subfamily of vertebrate TFs that has not (to our knowledge) been previously reported. Since evolutionary conservation provides evidence of functional importance, we predict that unique functions of a given protein subfamily can be explained by conserved subfamily-specific amino acid replacements (Hughes 2014). Therefore, to understand the functional divergence of the subfamilies of vertebrate TFs, we use statistical methods to reconstruct amino acid sequence evolution across the phylogeny and identify unique conserved replacements in each subfamily. In addition, we examine patterns of amino acid usage and conservation in functionally important residues involved in iron and carbonate binding and in the positively charged regions believed to be important for antimicrobial activity.
Methods
Amino acid sequences for use in phylogenetic analyses were obtained by BLASTP and TBLASTN homology search of the NCBI database using known human and chicken TF family members as query sequences. The data set used in phylogenetic analyses reported here included 90 sequences from 40 vertebrate species (Table 1). Mammalian species were chosen to represent a variety of different orders, with a preference for species for which representatives of multiple different TF subfamilies were available (Table 1). Other vertebrate species were chosen to provide as broad a sampling as possible of the vertebrate classes, thereby enabling us to date gene duplication events relative to the major events of cladogenesis in vertebrate history. Preliminary analyses including additional species yielded similar results (not shown) to those reported here.
Table 1.
Species used in sequence analyses.
| Class | Order | Species |
|---|---|---|
| Actinopterygii | Cichliformes | Maylandia zebra (zebra mbuna) |
| Oreochromis niloticus (Nile tilapia) | ||
| Beloniiformes | Oryzias latipes (Japanese medaka) | |
| Cyprinodontiformes | Xiphophorus maculatus (southern platyfish) | |
| Tetraodontiformes | Takifugu rubripes (torafugu) | |
| Sarcopterygii | Coelocanthiformes | Latimeria chalumnae (coelacanth) |
| Amphibia | Anura | Xenopus laevis (African clawed frog) |
| Xenopus tropicalis (western clawed frog) | ||
| Reptilia | Testudines | Chelonia mydas (green sea turtle) |
| Chrysemys picta belli (western painted turtle) | ||
| Squamata | Chamaeleo chamaeleon (common chameleon) | |
| Lacerta agilis (sand lizard) | ||
| Anolis carolinensis (green anole) | ||
| Natrix natrix (European grass snake) | ||
| Boaedon fuliginosus (brown house snake) | ||
| Python bivittatus (Burmese python) | ||
| Crocodylia | Alligator sinensis (Chinese alligator) | |
| Crocodylus niloticus (Nile crocodile) | ||
| Aves | Struthioniformes | Struthio camelus (ostrich) |
| Casuariformes | Dromaius novaehollandiae (emu) | |
| Galliformes | Gallus gallus (chicken) | |
| Meleagris gallopavo (turkey) | ||
| Psittaciformes | Melopsittacus undulatus | |
| Passeriformes | Ficedula albicollis (collared flycatcher) | |
| Zonotrichia albicollis (white-throated sparrow) | ||
| Taenopygia guttata (zebra finch) | ||
| Mammalia | Monotremata | Ornithorhynchus anatinus (platypus) |
| Marsupialia | Monodelphis domestica (gray short-tailed opossum) | |
| Sarcophilus harrisii (Tasmanian devil) | ||
| Insectivora | Sorex araneus (European shrew) | |
| Primates | Otolemur garnettii (small-eared ga;ago) | |
| Nomascus leucogenys (northern white-cheeked gibbon) | ||
| Homo sapiens (human) | ||
| Rodentia | Cavia porcellus (domestic guinea pig) | |
| Mus musculus (house mouse) | ||
| Carnivora | Canis lupus familiaris (domestic dog) | |
| Perissodactyla | Ceratotherium simum simum (southern white rhinoceros) | |
| Equus caballus (horse) | ||
| Certartiodactyla | Capra hircus (goat) | |
| Bos taurus (bovine) |
Amino acid sequences were aligned by the CLUSTAL algorithm in MEGA 6 (Tamura et al. 2011); the N-lobe sequences and C-lobe sequences were aligned separately, and an additional joint alignment of N-lobe and C-lobe sequences was also produced (Supplementary Figure S1). In preliminary analyses CLUSTAL-OMEGA was also used to align the sequences; the results were similar to those produced by CLUSTAL, the only differences being in regions with numerous indels. One study has suggested that CLUSTAL-OMEGA outperforms CLUSTAL particularly when the sequences to be aligned differ substantially in length (Pais et al. 2014; but see also Torda 2014). In any event, in the present case all sequences were similar in length, and all conserved regions were aligned essentially identically by the two algorithms.
In phylogenetic analyses of a set of sequences, any site at which the alignment postulated a gap in any sequence was excluded from the analyses. The maximum likelihood (ML) analysis was based on the JTT+G+I model, which was chosen using the Bayes Information Criterion in MEGA 6. The reliability of the clustering patterns in ML trees was tested by bootstrapping; 1000 bootstrap pseudo-samples were used. Because an unquestioned outgroup was not available, the phylogenetic trees were unrooted. However, each clade within a given phylogenetic tree was rooted by other clades within the tree.
For a given clade in the phylogeny, a clade-specific conserved amino acid residues was defined as one which was 100% conserved in all members of that clade used in the analysis and which was not conserved in any other clade in the data set. Clade-specific conserved amino acid residues are candidates for playing a role in clade-specific functions; however, not all such replacements are likely to be functionally significant, since some may result from selectively neutral substitutions that are conserved by chance. The number of residues that are conserved within a given clade will be in part a function of the number of clade members (i.e., the number of evolutionary lineages) available for analysis, since more variants at functionally unimportant sites are likely to be seen in a larger sample of lineages than in a smaller sample. For this reason, the percentage of conserved sites which were clade-specific was used as a measure of the extent of amino acid sequence specialization of each clade, since this percentage is expected to be independent of the number of lineages available for analysis (Hughes 2014).
We analyzed amino acid diversity at sites homologous to the iron-binding and carbonate-binding residues of the human serotransferrin sequence (Mizutani et al. 2012). We estimated amino acid diversity (p) within a set of sequences as the number of pairwise differences divided by the number of pairwise comparisons. The standard errors of p were estimated by the bootstrap method (Tamura et al. 2011). We analyzed amino acid composition in two regions homologous to two positively charged domains of human lactoferrin (Figure 1): (1) positively charged region 1: the N-terminal region corresponding to the human lactoferricin antimicrobial peptide (residues 1–48 of the mature protein; Hunter et al. 2005); and (2) positively charged region 2: the lactoferrampin region corresponding to residues 268–284 of mature human lactoferrin (Sinha et al. 2013).
Results
TF Subfamiles
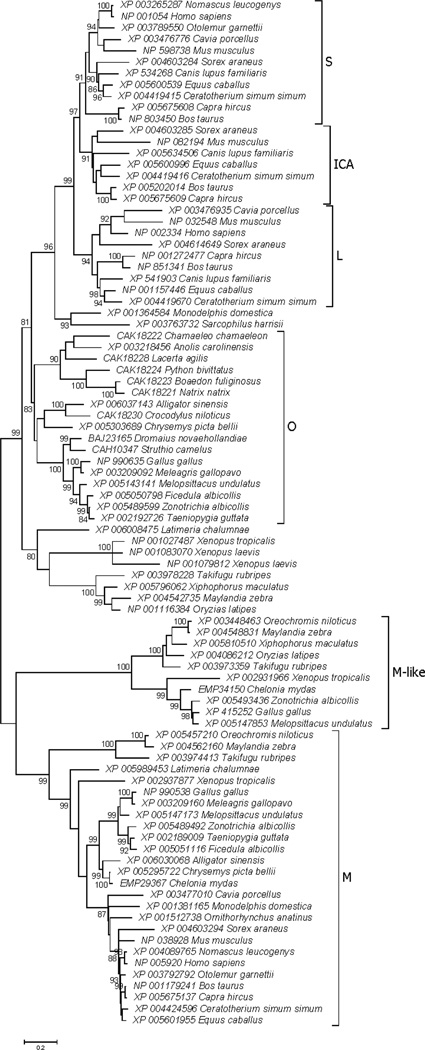
In the unrooted phylogenetic tree of TF N-lobe sequences (Figure 2), six major clades (subfamilies) were identified: (1) S, the mammalian serotransferrins; (2) ICA, the mammalian inhibitor of carbonic anhydrase (ICA) homologs; (3) L, the mammalian lactoferrins; (4) O, the ovotransferrins of birds and reptiles; (4) M, the melanotransferrins of bony fishes, amphibians, reptiles, birds, and mammals; and (5) M-like, a newly identified TF subfamily found in bony fishes, amphibians, reptiles, and birds. Three highly significant internal branches (with at least 99% bootstrap support) supported the separation of three major clusters within the tree: (1) the M subfamily; (2) the M-like subfamily; and (3) all other vertebrate TFs (Figure 2). Since each of these included sequences both from bony fishes and from tetrapods, the phylogenetic tree supported the hypothesis that the gene duplications giving rise to these three clusters occurred prior to the most recent common ancestor (MRCA) of bony fishes and tetrapods.
Figure 2.
ML tree of vertebrate TF N-lobes, based on the JTT+G+I model at 257 aligned amino acid sites. Subfamilies designated in the text are indicated. Numbers on branches correspond to the percentage of 1000 bootstrap pseudo-samples supporting the branch; only values ≥ 80% are shown.
The S, ICA, and L subfamilies were found only in placental mammals, and these three subfamilies clustered together in the phylogenetic tree (Figure 2). Two related sequences from marsupials (from Monodelphis domestica and Sarcophilus harrisii) clustered together but outside S, ICA, and L; and the position of the marsupial sequences outside S, ICA, and L was supported by a highly significant internal branch (99% bootstrap support; Figure 2). This topology supported the hypothesis that the duplications giving rise to separate S, ICA, and L genes occurred after the MRCA of placental and marsupial mammals, and that the S, ICA, and L subfamilies are placental mammal-specific. The O subfamily from birds and reptiles formed a sister group to the cluster including both marsupial TFs and placental S, ICA, and L, although the latter pattern received only moderate (81%) bootstrap support (Figure 2).
Within placental mammals, the L subfamily clustered outside the S and ICA subfamilies, and this pattern was supported by a significant (97% bootstrap support) internal branch (Figure 2). This topology supported the hypothesis that the L subfamily arose first of these three placental subfamilies, with a subsequent gene duplication giving rise to separate S and ICA subfamilies. Each of the S, ICA, and L clusters included sequences from the superorder Laurasiatheria (including the orders Insectivora, Carnivora, Perissodactyla, and Cetartiodactyla) and from the superorder Euarchontoglires (including the orders Primates and Rodentia; Prasad et al. 2008). Therefore, the tree supported the hypothesis that these placental-specific gene duplications occurred prior to the divergence of Laurasiatheria from Euarchontoglires.
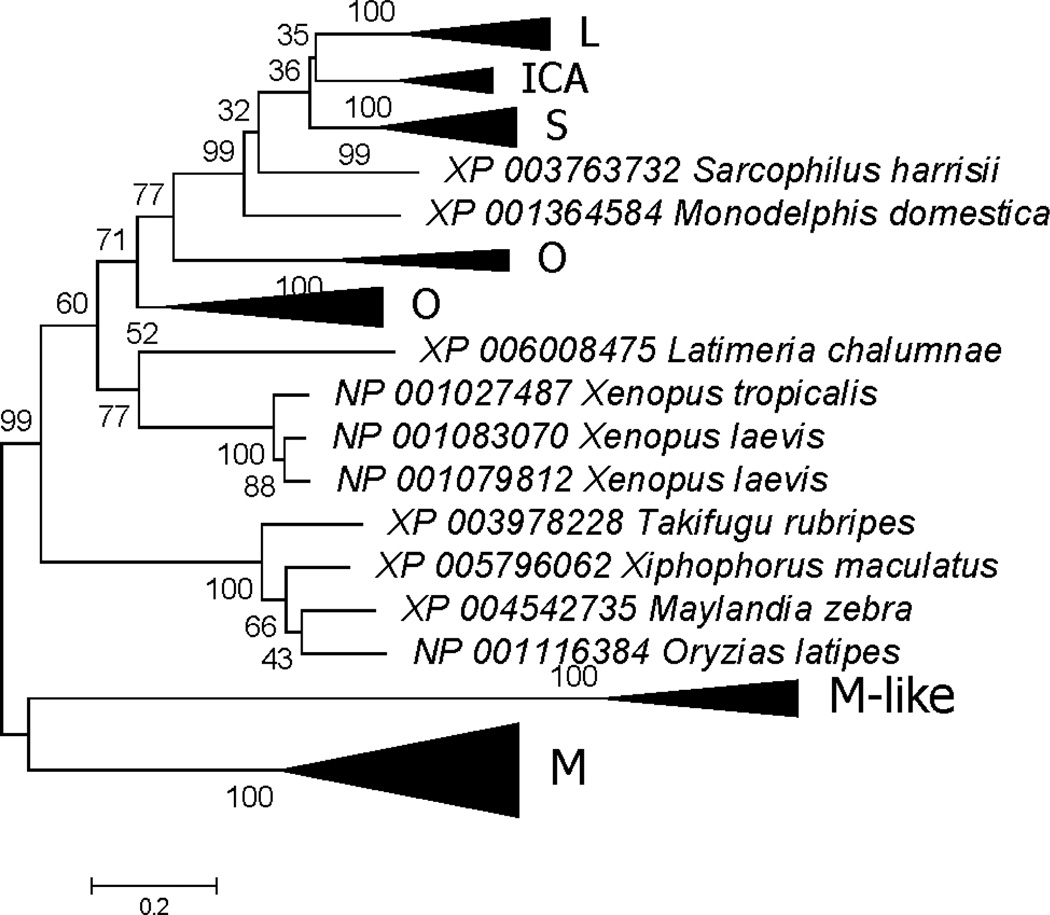
The phylogenetic tree based on the C-lobes shared most of the major clustering patterns seen in that based on the N-lobes (shown in schematic form in Figure 3 and in detail in Supplementary Figure S2). As with the N-lobes, C-lobes of M and M-like subfamilies clustered apart from the other vertebrate C-lobes; and both M and M-like subfamilies corresponded to clusters that received highly significant (100%) bootstrap support (Figure 3 and Supplementary Figure S2). As with N-lobes, the C-lobes of placental mammal S, ICA, and L formed separate clusters, and C-lobes of marsupial homologs fell outside the cluster of placental sequences (Figure 3 and Supplementary Figure S2). However, unlike the N-lobe tree (Figure 2), L rather than S clustered with ICA, although bootstrap support for this pattern was very weak (35%). There was highly significant support (99%) for grouping the marsupial sequences with placental L, ICA, and S; but there was only weak (32%) support for the internal branch supporting the position of the two marsupial C-lobes outside the placental C-lobes (Figure 3 and Supplementary Figure S2). In addition, the two marsupial C-lobe sequences did not cluster together (Figure 3 and Supplementary Figure S2), whereas the two marsupial Nlobe sequences did cluster together (with 93% bootstrap support; Figure 2). Likewise, Clobes for the O subfamily of birds and reptiles did not form a monophyletic group (Figure 3 and Supplementary Figure S2), whereas N-lobe sequences from the O subfamily did form a monophyletic group (Figure 2).
Figure 3.
ML tree of vertebrate TF C-lobes, shown in condensed form (for full tree see Supplementary Figure S2). The phylogenetic analysis was based on the JTT+G+I model at 269 aligned amino acid sites. Subfamilies designated in the text are indicated. Numbers on branches correspond to the percentage of 1000 bootstrap pseudo-samples supporting the branch; only values ≥ 60% are shown.
Internal Gene Duplication
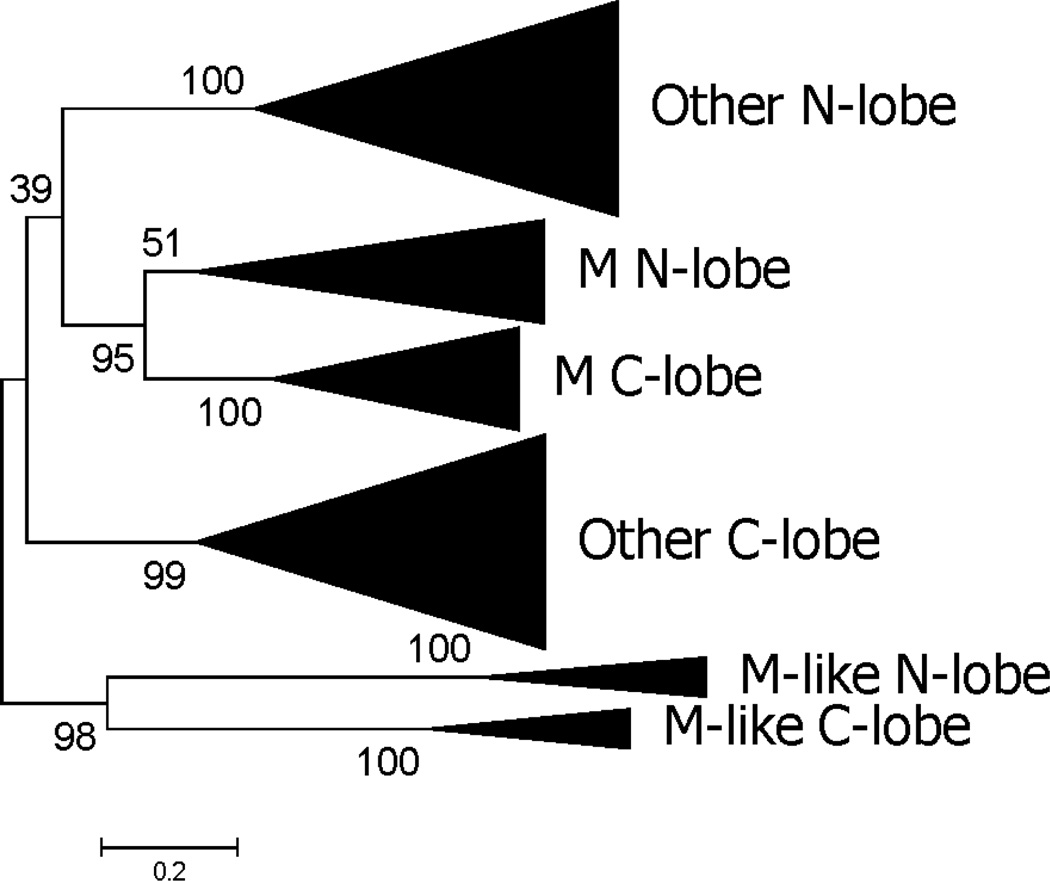
A phylogenetic analysis of the joint alignment of N-lobes and C-lobes was used to provide information regarding the internal gene duplication that gave rise to separate lobes (shown in schematic form in Figure 4 and in detail in Supplementary Figure S3). The N-lobes of M-like subfamily members clustered with the C-lobes of M-like family members, and this pattern was supported by a significant internal branch (96% bootstrap support; Figure 4). Likewise, the N-lobes of M family members clustered with the C-lobes of M family members, and this pattern was supported by a significant internal branch (97% bootstrap support; Figure 4). In the case of all other vertebrate TFs besides the M and M-like subfamilies, the N-lobes all clustered together and the C-lobes clustered together (Figure 4). The relationship between the latter two clusters and the M-like cluster was not well resolved; the N-lobes of all other subfamilies beside M and M-like clustered with M N-lobes and C-lobes, while the C-lobes of all other subfamilies beside M and M-like clustered with M-like N-lobes and C-lobes (Figure 4). However, the latter pattern received very low (39%) bootstrap support (Figure 4). In spite of this unresolved aspect of its topology, the phylogenetic tree supported the hypothesis that there have been at least three separate internal gene duplication events in the TF family of vertebrates: in the MRCA of the M subfamily, in the MRCA of the M-like subfamily, and in the MRCA of all other vertebrate TFs.
Figure 4.
ML based on joint alignment of vertebrate TF N-lobes and C-lobes, shown in condensed form (for full tree see Supplementary Figure S3). The phylogenetic analysis was based on the JTT+G+I model at 230 aligned amino acid sites. Numbers on branches correspond to the percentage of 1000 bootstrap pseudo-samples supporting the branch; only values ≥ 60% are shown.
Amino Acid Diversity and Composition
Amino acid sequence diversity within clades was estimated separately for the two positively charged domains, the remainder of the N-lobe, and the C-lobe (Table 2). There was significantly greater amino acid sequence diversity in positively charged region 1 of S, L, O, and M subfamilies than in the remainder of the N-lobe (Table 2). However, positively charged region 2 did not differ significantly from the remainder of the N-lobe with respect to amino acid sequence diversity in any subfamily except (Table 2). The Clobe differed significantly in amino acid sequence diversity from the remainder of the Nlobe in the ICA and O subfamilies (Table 2).
Table 2.
Amino acid diversity (expressed as a percentage ± S.E.) in domains of vertebrate transferrin clades1.
| Clade | Protein Region | ||||
|---|---|---|---|---|---|
| Positively Charged Region 1 | Positively Charged Region 2 | Remainder N-lobe | C-lobe | Entire Protein | |
| S | 34.5 ± 4.5 %*2 | 23.5 ± 5.8% | 24.4 ± 1.6% | 27.1 ± 1.7% | 26.1 ± 1.2% |
| ICA | 34.9 ± 4.0 % | 28.0 ± 6.5% | 26.6 ± 1.7% | 20.1 ± 1.5%** | 23.5 ± 1.0% |
| L | 47.6 ± 4.9 %*** | 32.7 ± 7.2% | 29.7 ± 1.7% | 27.0 ± 1.6% | 29.6 ± 1.1% |
| O | 48.6 ± 5.1 %** | 54.0 ± 6.5%** | 34.0 ± 1.7% | 40.3 ± 1.7%** | 39.1 ± 1.2% |
| M | 42.6 ± 4.8 %* | 40.6 ± 7.4% | 30.8 ± 1.6% | 32.2 ± 1.4% | 32.4 ± 1.0% |
| M-like | 30.0 ± 4.0 % | 40.8 ± 8.2% | 32.6 ± 1.8% | 32.3 ± 1.7% | 32.4 ± 1.2% |
Clades are as in Figure 2.
Z tests (2-tailed) of the hypothesis that the amino acid diversity equals that of the remainder of the N-lobe:
P < 0.05;
P < 0.01;
P < 0.001.
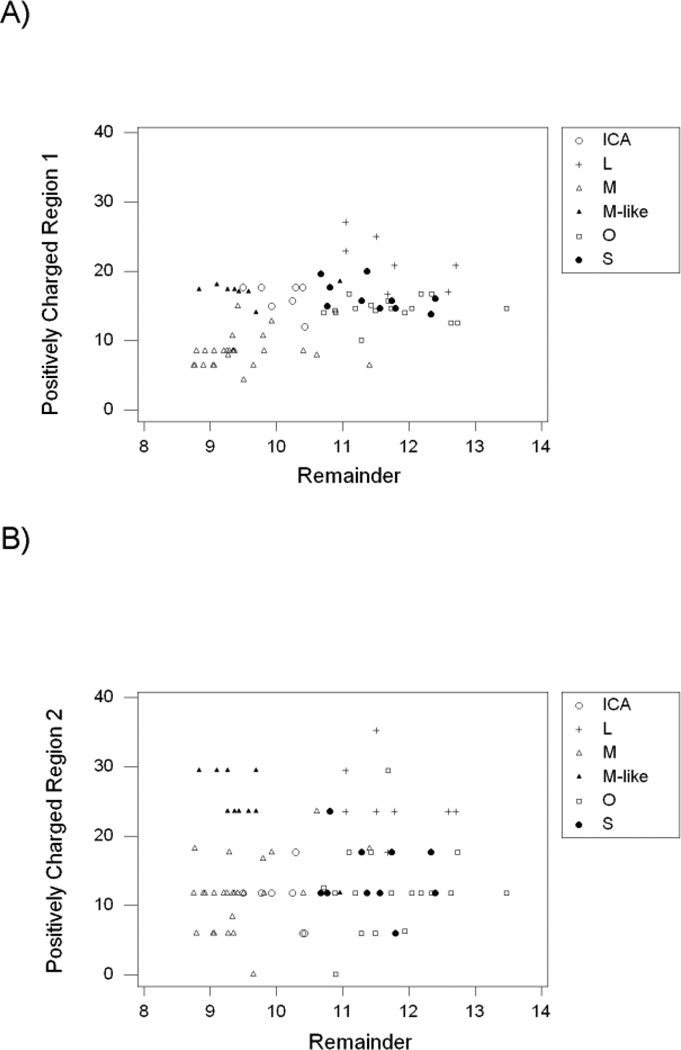
We examined the percentage of the two strongly positively charged amino acid residues, Arginine and Lysine (%K+R) in the two positively charged regions of the N-lobe and in the remainder of the protein (excluding the two positively charged regions). Across the six subfamilies, %K+R in positively charged region 1 was positively correlated with that in the remainder of the protein (r = 0.528; P < 0.001; Figure 5A). The highest %K+R values in positively charged region 1 were seen in the L subfamily, and the lowest in the M subfamily (Figure 5A). On the other hand, %K+R in positively charged region 2 was not significantly correlated with that in the remainder of the protein (r = 0.067; n.s.; Figure 5B). In positively charged region 2, mean %K+R was highest in the L subfamily (mean = 24.0 ± 1.6 %) and in the M-like subfamily (mean = 24.7 ± 1.0%; Figure 5B).
Figure 5.
Percent arginine and lysine (%K+R) in (A) positively charged region 1 and (B) positively charged region 2, plotted against that in the remainder of the TF sequence. In positively charged region 1, r = 0.528 (P < 0.001); in positively charged region 2, r = 0.067 (n.s.).
The L subfamily thus showed high % K +R in both positively charged regions, as is not unexpected since these regions were defined based on human lactoferrin (Sinha et al. 2013). The L subfamily also showed relatively high %K+R in the remainder of the protein (Figure 5). By contrast, the M-like subfamily showed high %K+R in positively charged region 2 but not in the remainder of the protein (Figure 5B). The unique pattern of charged residues in the M-like subfamily was a major reason why %K+R in positively charged region 2 was not significantly correlated with that in the remainder of the protein (Figure 5B).
Clade-Specific Conserved Residues
Identification of clade-specific conserved amino acid residues showed far more such residues in both the N-lobes and C-lobes of the M-like subfamily than in those of any other subfamily (Table 3; Supplementary Figure S1). It might be hypothesized that limited number of taxa for which M-like sequences were available may have contributed to the relatively high number of such sites in this subfamily. However, the overall amino acid sequence diversity in the M-like subfamily was very similar to and slightly higher than that in the M subfamily (Table 2). Thus, the limited number of taxa with M-like subfamily sequences did not appear to affect the overall amino acid sequence diversity in this subfamily. Moreover, our results also showed that clade-specific residues constituted a significantly greater proportion of all conserved residues in both N-lobes (21.3%) and C-lobes (30.1%) of the M-like subfamily than in those of any other subfamily (Table 3). The next highest percentages of clade-specific conserved residues were 11.7% in the C-lobes of the M subfamily and 11.3% in the C-lobes of the ICA subfamily (Table 3). Thus, the M-like subfamily appeared to be unique among vertebrate TF subfamilies with respect to the level of subfamily-specific conserved amino acid residues.
Table 3.
Conserved residues in major clades of vertebrate transferrins.
| Clade1 | N-lobe | C-lobe | ||
|---|---|---|---|---|
| Clade-Specific (%) | Other (%)2 | Clade-Specific ((%) | Other (%)2 | |
| S | 0 (0.0%) | 120 (100.0%)*** | 2 (1.4%) | 140 (98.6%)*** |
| ICA | 0 (0.0%) | 119 (100.0%)*** | 17 (11.3%) | 151 (88.7%)*** |
| L | 1 (1.0%) | 103 (99.0%)*** | 7 (5.0%) | 132 (95.0%)*** |
| O | 0 (0.0%) | 77 (100.0%)*** | 0(0.0%) | 78 (100.0%)*** |
| M-like | 23 (21.3%) | 85 (78.7%) | 37 (30.1%) | 85 (69.9%) |
| M | 3 (5.4%) | 53 (94.6%)* | 9 (11.7%) | 68 (88.3%)** |
Clades are as in Figure 2.
Fisher’s exact tests (2-tailed) of the hypothesis that the proportion equals that of the M-like clade in the same lobe:
P < 0.05;
P < 0.01;
P < 0.001.
Iron- and Carbonate-Binding Residues
All 10 iron- and carbonate-binding residues observed in human serotransferrin were completely conserved in other S subfamily members analyzed and partially conserved in other subfamilies (Table 4). The amino acid diversity at the 10 sites was low in the S, L, and O subfamilies compared to the ICA, M, and M-like subfamilies (Table 4). Only the O subfamily did not show significantly greater amino acid diversity at these sites than did the S subfamily (Table 4).
Table 4.
Residues at positions homologous to active site residues of human serotransferrin.
| Site1 | Clade2 | ||||||
|---|---|---|---|---|---|---|---|
| Iron-binding | S | L | ICA | O | M | M-like | |
| N-lobe | 63 | D | D,G | D,G | D,G | D,F,N,S | D |
| 95 | Y | Y | Y,H | Y | Y | I,T,V | |
| 188 | Y | Y | Y,S | Y | Y,G | N,S | |
| 249 | H | H,N | H | H | H,R | G,K,R | |
| C-lobe | 392 | D | D,G | D | D | D,G,K,R,S | D |
| 426 | Y | Y | Y,H | Y | Y,H | A,L,V | |
| 517 | Y | Y | F,P,S | Y | Y,D | N | |
| 585 | H | H | H,R | H | H,Q,R | N,S | |
| Carbonate-binding | |||||||
| N-lobe | 124 | R | R,W | R,C,W | R,K | R,Q | W3 |
| C-lobe | 456 | R | R | T3 | R | R,M,S | S3 |
| Mean amino acid diversity (%)4,5 | 0.0% | 5.0% * | 29.3% ** | 2.4% | 30.1% ** | 28.4% * | |
| No. conserved residues shared with S | 10 | 6 | 2 | 8 | 1 | 2 | |
Eight of the 10 residues were conserved in all O subfamily members analyzed, and six of 10 were conserved in all L member families analyzed (Table 4). By contrast, only one of these 10 residues was conserved in the M subfamily, and only two in each of the ICA and M-like subfamilies (Table 4). The presence of numerous different residues at one of these 10 positions suggests the absence of strong functional constraint (Table 4). On the other hand, there were certain cases where clade-specific conserved residues were found at these positions. The M-like subfamily showed clade-specific conserved residues at the positions where carbonate-binding arginine residues are found in both N-lobe and C-lobe of human serotransferrin; M-like subfamily members showed tryptophan instead of arginine at the N-lobe site, and serine instead of arginine at the C-lobe site (Table 4). The ICA subfamily also showed a clade-specific conserved residue (threonine) at the site corresponding to the C-lobe carbonate-binding site of serotransferrin (Table 4).
Discussion
A phylogenetic analysis of vertebrate TF sequences supported the hypothesis that three major groups of vertebrate TFs arose by gene duplication prior to the MRCA of bony fishes and tetrapods: (1) the M subfamily; (2) the M-like subfamily; and (3) the remaining vertebrate TFs. Each of these three major groups included paralogs from bony fishes and tetrapods. Thus the phylogenetic analysis supported the hypothesis that the gene duplication giving rise to the each of these groups occurred prior to the MRCA of bony fishes and tetrapods, which is estimated to have occurred about 455 Mya (Hedges 2009).
Aside from mammalian members of the M subfamily, TFs from placental mammals formed three distinct subfamilies, designated S, ICA, and L. Our phylogenetic analysis supported the hypothesis that, aside from M, the other three placental mammal subfamilies arose by gene duplication that occurred after the MRCA of the placental (eutherian) mammals, which is estimated to have occurred about 176 Mya (Madsen 2009). The two successive gene duplications giving rise to separate S, ICA, and L subfamilies were estimated to have occurred before the MRCA of the superorders Euarchontoglires and Laurasiatheria, which occurred about 97 Mya (Murphy and Eizirik 2009). Although the phylogeny based on C-lobe sequences did not resolve the relationship among the three placental mammal-specific subfamilies, the phylogeny based on N-lobe sequences supported the hypothesis that the L subfamily was the first of the three to arise. Avian transferrins were found in the M and M-like subfamilies and in a third subfamily designated O. The tree based on N-lobe sequences suggested that the O subfamily of birds and reptiles form a sister group to a cluster including both marsupial TFs and placental mammal S, ICA, and L.
The M-like subfamily was not found in mammals, and indeed it has not been identified as a distinct subfamily in previous reviews of the TF family. We found M-like subfamily genes in several of the bird species included in our analyses, including the chicken. In addition to birds, sequences belonging to the M-like family were found in bony fishes, amphibians, and reptiles. The M-like family sequences showed no evidence of the hydrophobic domain at the C-terminus of the C-lobe that is involved in the glycosyl phostphatidylinositol linkage found in M subfamily members (Alemany et al. 1993).
Contrary to the hypothesis of a single internal gene duplication event (Lambert et al. 2005), our phylogenetic analysis supported the hypothesis that at least three separate events of internal duplication occurred in vertebrate TFs: (1) in the common ancestor of the M subfamily; (2) in the common ancestor of the M-like subfamily; and (3) in the common ancestor of other vertebrate TFs. In the phylogenetic tree based on the joint alignment of N-lobes and C-lobes, N-lobes of the M subfamily clustered with C-lobes of the M subfamily; and N-lobes of the M-like subfamily clustered with C-lobes of the M-like subfamily (Figure 4). Thus, the phylogenetic tree provided strong support for separate internal duplications in the latter two subfamilies. As regards the vertebrate TFs other than the M and M-like subfamilies, the phylogenetic tree did not clearly resolve the relationship between their N-lobes and C-lobes. In fact the N-lobes of TFs other than M and M-like clustered with both lobes of the M subfamily, suggesting an ancient interlocus recombination event; however, the latter topology did not receive significant support (Figure 4). In spite of this unresolved branch, the N-lobes and C-lobes of other vertebrate TFs formed separate clusters distinct from those of the M and M-like subfamily, supporting the hypothesis of a third internal duplication.
It has been reported that the C-lobes of TFs are generally more conserved than the N-lobes (Nakamusu et al. 1999), but the present results showe that the non-conserved portion of the N-lobes is confined to the N-terminal region that in human lactotransferrin is cleaved to form the positively charged lactoferricin peptide (positively charged region 1). The subfamilies that showed most evidence of rapid amino acid evolution in the N-terminal regions included L and O, in which this region showed unusually high proportions of the positively charged amino acid residues arginine and lysine (Table 2 and Figure 5). On the other hand, there was also rapid evolution in this region of the M subfamily, which showed relatively low proportions of arginine and lysine (Table 2 and Figure 5). In the only subfamilies (ICA and O) that showed a significant difference in amino acid diversity between the C-lobes and the N-lobes excluding the positively charged regions, diversity was actually higher in the C-lobes.
The M-like subfamily showed an unusually high proportion of arginine and lysine in positively charged region 2 (corresponding to human lactoferrampin), comparable to that in the L subfamily (Figure 5). This observation suggests that positively charged region 2 may play a uniquely important role in the M-like subfamily, perhaps in antimicrobial defense.
Among all vertebrate TF subfamilies, the M-like subfamily was found to be unique with respect both to the number of conserved clade-specific residues and the proportion of conserved residues that are clade-specific. Interestingly, two of the conserved clade-specific residues in the M-like family were located at sites involved in carbonate binding by other TFs, suggesting that these sites may have assumed a divergent but conserved function in the M-like subfamily. The prevalence of conserved clade-specific residues suggests that members of the M-like subfamily have evolved distinctive functions not shared by other vertebrate TFs.
Aside from the M-like family, the C-lobes of the ICA subfamily and the M subfamily showed the highest proportions of conserved residues that were clade-specific (Table 3). Both lobes of ICA and the C-lobe of the M subfamily have lost iron-binding capacity (Baker et al. 1992; Wang et al. 2007). The presence of numerous conserved clade-specific residues in the C-lobes of both of these subfamilies suggests that they have adapted to novel functions.
The vertebrate TFs are multifunctional proteins whose diverse physiological roles are only beginning to be understood. By identifying well-suported subfamilies of vertebrate TFs, the present analysis provides a framework for future studies. In addition, by examining patterns of amino acid sequence conservation and change in functionally important regions of TFs, we provide insights into potential functional diversification. The latter analyses pinpoint the unique sequence features of the newly identified M-like subfamily, which although absent from mammals and the domestic chicken, may play important roles in the biology of bony fishes, reptiles and amphibians, and certain avian species.
Supplementary Material
References
- Alemany R, Vilá MR, Franci C, Egea G, Real FX. Glycosyl phosphatidylinositol membrane anchoring of melanotransferrin (p97): apical compartmentalization in intestinal epithelial cells. J Cell Sci. 1993;104:1155–1162. doi: 10.1242/jcs.104.4.1155. [DOI] [PubMed] [Google Scholar]
- Ando K, Hasegawa K, Shindo K, Furusawa T, Fujino T, Kikugawa K, Nakano H, Takeuchi O, Akira S, Akiyama T, Gohda J, Inoue J, Hayakawa M. Human lactoferrin activates NF-κB through the Toll-like receptor 4 pathway while it interferes with the lipopolysaccharide-stimulated TLR4 signaling. FEBS J. 2010;277:2051–2068. doi: 10.1111/j.1742-4658.2010.07620.x. [DOI] [PubMed] [Google Scholar]
- Baker EN, Baker HM, Smith CA, Stebbins MR, Kahn M, Hellström KE, Hellström I. Human melanotransferrin (p97) has only one functional iron-binding site. FEBS Lett. 1992;298:215–218. doi: 10.1016/0014-5793(92)80060-t. [DOI] [PubMed] [Google Scholar]
- Baker EN, Baker HM, Kidd RD. Lactoferrin and transferrin: functional variations on a common structural framework. Biochem Cell Biol. 2002;80:27–34. doi: 10.1139/o01-153. [DOI] [PubMed] [Google Scholar]
- Baker EN, Baker HM. A structural framework for understanding the multifunctional character of lactoferrin. Biochimie. 2009;91:3–10. doi: 10.1016/j.biochi.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Bowman BH, Yang F, Adrian GS. Transferrin: evolution and genetic regulation and expression. Adv Genet. 1988;25:1–38. doi: 10.1016/s0065-2660(08)60457-5. [DOI] [PubMed] [Google Scholar]
- Eckenroth BE, Mason AB, McDevitt ME, Lambert LA, Everse SJ. The structure and evolution of the murine inhibitor of carbonic anhydrase: a member of the transferrin superfamily. Protein Sci. 2010;19:1616–1626. doi: 10.1002/pro.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escrivá H, Pierce A, Coddeville B, González F, Benaissa M, Léger D, Wieruszeski J-M, Spik G, Pamblanco M. Rat mammary-gland transferrin: nucleotide sequence, phylogenetic analysis and glycan structure. Biochem J. 1995;367:47–55. doi: 10.1042/bj3070047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnaud S, Evans RW. Lactoferrin – a multifunctional protein with antimicrobial properties. Mol Immunol. 2003;40:395–400. doi: 10.1016/s0161-5890(03)00152-4. [DOI] [PubMed] [Google Scholar]
- García-Montoya IA, Cendón TS, Arévalo-Gallegos S, Rascón-Cruz Q. Lactoferrin a multiple bioactive protein: an overview. Biochim Biophy Acta. 2012;1820:226–236. doi: 10.1016/j.bbagen.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti F, Leboffe L, Pitari G, Ippoliti R, Antonini G. Physiological roles of ovotransferrin. Biochim Biophys Acata. 2012;1820:218–225. doi: 10.1016/j.bbagen.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Gkouvatsos K, Papanikolaou G, Pantopoulos K. Regulation of iron transport and the role of transferrin. Biochim Biophys Acta. 2012;1820:188–202. doi: 10.1016/j.bbagen.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Gomme PT, McCann KB. Transferrin: structure, function and potential therapeutic actions. Drug Discov Today. 2005;10:267–273. doi: 10.1016/S1359-6446(04)03333-1. [DOI] [PubMed] [Google Scholar]
- Gray-Owen SD, Schyvers AB. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 1996;4:185–191. doi: 10.1016/0966-842x(96)10025-1. [DOI] [PubMed] [Google Scholar]
- Harris WR. Anion binding properties of the transferrins. Implications for function. Biochim Biophys Acta. 2012;1820:348–361. doi: 10.1016/j.bbagen.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Hedges SB. Vetrebrates (Vertebrata) In: Hedges SB, Kumar S, editors. The Timetree of Life. Oxford: Oxford University Press; 2009. pp. 309–314. [Google Scholar]
- Hughes AL. Evolutionary diversification of aminopeptidase N in Lepidoptera by conserved derived amino acid residues. Mol Phyl Evol. 2014;76:127–133. doi: 10.1016/j.ympev.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter HN, Demcoe AR, Jenssen H, Gutteberg TJ, Vogel HJ. Human lactoferricin is partially folded in aqueous solution and is better stabilized in a membrane mimetic solvent. Antimicrob Agents Chemother. 2005;49:3387–3395. doi: 10.1128/AAC.49.8.3387-3395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennssen H, Hancock RE. Antimicrobial properties of lactoferrin. Biochimie. 2009;91:19–29. doi: 10.1016/j.biochi.2008.05.015. [DOI] [PubMed] [Google Scholar]
- Kurokawa H, Mikami B, Hirose M. Crystal structure of diferric hen ovotratransferrin at 2.4 Å resolution. J Mol Biol. 1995;254:196–207. doi: 10.1006/jmbi.1995.0611. [DOI] [PubMed] [Google Scholar]
- Lambert LA. Molecular evolution of the transferrin family and associated receptors. Biochim Biophy Acta. 2012;1820:244–255. doi: 10.1016/j.bbagen.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Lambert LA, Perri H, Meehan TJ. Evolution of duplications in the transferrin family of proteins. Comp Biochem Physiol B. 2005;140:11–25. doi: 10.1016/j.cbpc.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Levay PF, Viljoen M. Lactoferrin: a general review. Haematologica. 1995;80:252–267. [PubMed] [Google Scholar]
- Mayle KM, Le AM, Kamei DT. The intracellular trafficking pathway of transferrin. Biochim Biophys Acta. 2012;1820:264–281. doi: 10.1016/j.bbagen.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K, Toyoda M, Mikami B. X-ray structures of transferrins and related proteins. Biochim Biophys Acta. 2012;1820:203–211. doi: 10.1016/j.bbagen.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Madsen . Mammals (Mammalia) In: Hedges SB, Kumar S, editors. The Timetree of Life. Oxford: Oxford University Press; 2009. pp. 459–461. [Google Scholar]
- Murphy WJ, Eizirik E. Placental mammals (Eutheria) In: Hedges SB, Kumar S, editors. The Timetree of Life. Oxford: Oxford University Press; 2009. pp. 471–474. [Google Scholar]
- Nakamusu K, Kawamoto T, Shen M, Gotoh O, Teramoto M, Noshiro M, Kato Y. Membrane-bound transferrin-like protein (MTf): structure, evolution and selective expression during chondrogenic differentiation of mouse embryonic cells. Biochim Biophys Acta. 1999;1447:258–264. doi: 10.1016/s0167-4781(99)00173-6. [DOI] [PubMed] [Google Scholar]
- Nibbering PH, Ravensbergen E, Welling MM, van Berkel LA, van Berkel PH, Pauwels EK, Nuijens JH. Human lactoferrin and peptides derived from its N terminus are highly effective against interactions with antibiotic-resistant bacteria. Infect Immun. 2001;69:1469–1476. doi: 10.1128/IAI.69.3.1469-1476.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais FS-M, de Cássia Ruy P, Oliveira G, Coimbra RS. Assessing the efficiency of multiple sequence alignment programs. Algorithms Mol Biol. 2014;2014:9, 4. doi: 10.1186/1748-7188-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad AB, Allard MW NISC Comparative Sequencing Program, Green ED. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmanto YS, Bal S, Loh KH, Yu Y, Richardson DR. Melanotransferrin: search for a function. Biochim Biophys Acta. 2012;1820:237–243. doi: 10.1016/j.bbagen.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Sinha M, Kaushik S, Kaur P, Sharma S, Singh TP. Antimicrobial lactoferrin peptides: the hidden players in the protective function of a multifunctional protein. Int J Peptides. 2013:390230. doi: 10.1155/2013/390230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Li H, Sadler PJ. Transferrin as a metal ion mediator. Chem Rev. 1999;99:2817–2842. doi: 10.1021/cr980430w. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstensen K, Romslo I. The role of transferrin in the mechanism of cellular iron uptake. Biochem J. 1990;271:1–10. doi: 10.1042/bj2710001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torda AE. Not assessing the efficiency of multiple sequence alignment programs. Algorithms Mol Biol. 2014;2014:9–18. doi: 10.1186/1748-7188-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Lothrop AP, James NG, Griffiths TA, Lambert LA, Leverence R, Kaltashov IA, Andrews NC, MacGillivray RT, Mason AB. A novel murine protein with no effect on iron homeostasis is homologous with transferrin and is the putative inhibitor of carbonic anhydrase. Biochem J. 2007;406:85–95. doi: 10.1042/BJ20070384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.